Abstract
Human ACE2 Human angiotensin converting enzyme 2 (hACE2) is the key cell attachment and entry receptor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with the original SARS-CoV-2 isolates unable to use mouse ACE2 (mACE2). Herein we describe the emergence of a SARS-CoV-2 strain capable of ACE2-independent infection and the evolution of mouse-adapted (MA) SARS-CoV-2 by in vitro serial passaging of virus in co-cultures of cell lines expressing hACE2 and mACE2. MA viruses evolved with up to five amino acid changes in the spike protein, all of which have been seen in human isolates. MA viruses replicated to high titers in C57BL/6J mouse lungs and nasal turbinates and caused characteristic lung histopathology. One MA virus also evolved to replicate efficiently in several ACE2-negative cell lines across several species, including clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) ACE2 knockout cells. An E484D substitution is likely involved in ACE2-independent entry and has appeared in only ≈0.003 per cent of human isolates globally, suggesting that it provided no significant selection advantage in humans. ACE2-independent entry reveals a SARS-CoV-2 infection mechanism that has potential implications for disease pathogenesis, evolution, tropism, and perhaps also intervention development.
Keywords: SARS-CoV-2, COVID-19, ACE2-independent infection, mouse adaptation
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in 2019, causing a global pandemic of coronavirus disease 2019 (COVID-19) (Wu et al. 2020). The human angiotensin converting enzyme 2 (ACE2) (hACE2) receptor is generally held to be the key cell attachment and entry receptor for SARS-CoV-2, with the receptor-binding domain (RBD) of the spike protein binding to hACE2 (Rawle et al. 2021). ACE2 binding has deep ancestral origins within the sarbecovirus lineage of coronaviruses (Starr et al. 2022), with SARS-CoV, SARS-CoV-2, and SARS-related bat coronaviruses all using ACE2 as their entry receptor (Hu et al. 2017). Genetically diverse spike proteins are able to bind ACE2 (Hu et al. 2017; Starr et al. 2022), with all the SARS-CoV-2 variants of concern (Alpha, Beta, Gamma, Delta, and Omicron) reported to require ACE2 binding for efficient infection (Shuai et al. 2021; Cele et al. 2022).
The original isolates of SARS-CoV-2 (Wuhan strain) are unable to bind with mouse ACE2 (mACE2) for infection (Rawle et al. 2021); therefore, to study these viruses, a series of genetically modified mouse models were developed that express hACE2 (Muñoz-Fontela et al. 2020; Amarilla et al. 2021; Rawle et al. 2021; Shou et al. 2021; Bishop et al. 2022). Some subsequent SARS-CoV-2 variants of concern emerged to be able to use mACE2, including Alpha, Beta, Gamma, and Omicron, but not Delta (Shuai et al. 2021; Halfmann et al. 2022). In addition, mouse-adapted (MA) SARS-CoV-2 have been generated by serial passage of original SARS-CoV-2 isolates in mouse lungs (Gu et al. 2020; Leist et al. 2020; Wang et al. 2020a; Huang et al. 2021; Zhang et al. 2021) or by reverse genetics (Dinnon et al. 2020) to introduce amino acid changes that allow binding to mACE2.
As infection of wild animals by SARS-CoV-2 is increasingly being reported (Chandler et al. 2021; Diaz, Walker, and Webster 2021; Griffin et al. 2021; Langereis et al. 2021; Palermo et al. 2021; Welkers et al. 2021; Hale et al. 2022), with speculation that Omicron may have potentially arisen from rodents (Wei et al. 2021a; Mallapaty 2022), we further sought to characterize the process of mouse adaptation and virus evolution in vitro. We have previously reported the use of HEK293T cells that express hACE2 (HEK293T-hACE2) or mACE2 (HEK293T-mACE2) by virtue of lentiviral transduction, with the former, but not the latter, able to support the efficient replication of an original SARS-CoV-2 isolate, hCoV-19/Australia/QLD02/2020 (SARS-CoV-2QLD02) (Rawle et al. 2021). We also showed that transduction with mACE2 containing the N31K and H353K mouse-to-human amino acid changes (to generate HEK293T-mACE2N31K/H353K cells) represented the minimum changes required to support SARS-CoV-2QLD02 replication (Rawle et al. 2021). Herein we serially passaged SARS-CoV-2QLD02 in HEK293T-hACE2 or HEK293T-mACE2N31K/H353K cells co-cultured with HEK293T-mACE2 cells, followed by passaging in HEK293T-mACE2 cells to generate five different MA viruses. These MA viruses were able to replicate in both HEK293T-hACE2 and HEK293T-mACE2 cells. These viruses were also able to replicate in C57BL/6J mice, leading to characteristic COVID-19 histopathological lesions and inflammatory responses. Remarkably, one of these viruses (MA1) was also able to replicate in a number of cell lines that do not express significant levels of ACE2 and do not support SARS-CoV-2QLD02 infection. Furthermore, MA1 showed the efficient and productive infection of clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) ACE2-knockout cells, the first report of SARS-CoV-2 infection of cells that unequivocally express no ACE2. ACE2-independent infection has potential implications for SARS-CoV-2 biology, disease, and perhaps also intervention development.
Results
SARS-CoV-2QLD02 co-culture passaging to select for MA viruses
To investigate the process of evolution to mACE2 utilization, SARS-CoV-2QLD02 was passaged in co-cultures of HEK293T-hACE2 and HEK293T-mACE2 or HEK293T-mACE2N31K/H353K and HEK293T-mACE2 cells, followed by passage in HEK293T-mACE2 cells (Fig. 1A). After four co-culture passages, no viruses emerged that were able to replicate efficiently in HEK293T-mACE2 cells (Fig. 1B, Passage 4). However, after nine co-culture passages, five viruses (MA1-5) evolved a clear ability to replicate in HEK293T-mACE2 cells (Fig. 1B, Passage 9). As expected, SARS-CoV-2QLD02 did not replicate in HEK293T-mACE2 cells (Fig. 1B). All five MA viruses showed overt cytopathic effects (CPEs) in HEK293T-mACE2 cells within 2 days postinfection (Fig. 1C). Importantly, these viruses were also able to replicate in lungs of wild-type C57BL/6J mice after intrapulmonary inoculation (via the intranasal route) with 1.5 × 104 CCID50 per mouse (Fig. 1D). MA1 and MA2 showed the highest lung titers, replicating to >106 CCID50/g (Fig. 1D). As expected, SARS-CoV-2QLD02 was unable to replicate in C57BL/6J mice (Fig. 1D). The co-culture and in vitro passaging of SARS-CoV-2QLD02 therefore generated five viruses able to replicate in wild-type mice.
Figure 1.
In vitro evolution of SARS-CoV-2QLD02 to mACE2 utilization. (A) Schematic of SARS-CoV-2QLD02 passaging in HEK293T-hACE2 or HEK293T-mACE2N31K/H353K cells, co-cultured with HEK293T-mACE2 cells (nine passages), followed by passaging in HEK293T-mACE2 cells (three passages) and mouse infections. Viruses used to infect mice and viruses derived from mice lungs (2 dpi) were sequenced. Stock virus was prepared for MA1 and MA2 from mouse lungs (2 dpi) for use in subsequent experiments. (B) Supernatants from passages 4 and 9 (from the co-cultures—blue arrows) were used to infect HEK93T-mACE2 cells and viral growth over 72 h determined by CCID50 assays. Dotted line—limit of detection. MA1, MA3, and MA6 (underlined) were derived from HEK293T-hACE2/HEK293T-mACE2 co-cultures, MA2, MA4, and MA5 from HEK293T-mACE2N31K/H353K/HEK293T-mACE2 co-cultures. (C) Inverted light microscopy images of CPE in HEK293T-mACE2 cells infected with passage 9 supernatants at 72 h postinfection. Images are representative of at least three replicates. (D) MA viruses obtained after three passages in HEK293T-mACE2 cells were used to infect C57BL/6J mice. Lung tissue titers are shown for 2 dpi for MA1-5 (n = 1 for each) and for SARS-CoV-2QLD02 (n = 6).
Amino acid changes that evolved in MA viruses have previously been reported in human isolates
MA1-5 viruses were sequenced before and after infection of C57BL/6J mice (Fig. 1A, Sequencing) by RNA sequencing (RNA-Seq). Sequences for MA viruses before and after infection of C57BL/6J mice were almost identical (Supplementary Dataset 1), illustrating that the single passage in mice did not evolve further significant changes. There were thirteen different amino acid changes and two deletions in the spike protein across the five MA viruses (Fig. 2A, B). All these changes have previously been identified in human isolates (Fig. 2C). The Q498H and Q493R substitutions (Fig. 2A) have been identified previously for MA viruses and increase the affinity for mACE2 (Dinnon et al. 2020; Gu et al. 2020; Huang et al. 2021; Zhang et al. 2021). The E484D substitution has previously been associated with ACE2-independent infection (Puray-Chavez et al. 2021; Ramirez et al. 2021; Hoffmann et al. 2022). The deletion of ‘QTQTN’ at Positions 675–679 (flanking the polybasic furin cleavage site) commonly arises after in vitro passage of SARS-CoV-2 (Liu et al. 2020). This deletion impairs S1/S2 processing in a manner similar to the deletion of the polybasic furin cleavage site, with the QTQTN deletion likely reducing the accessibility of furin to the cleavage site (Lemmin et al. 2020; Peacock et al. 2021; Vu et al. 2021). MA3 contained a deletion of Amino Acids 69–72, accompanied by an I68R substitution (Fig. 2A, B), changes similar to the 69–70 deletion present in Alpha and Omicron BA.1 variants that may increase spike incorporation into virions (Meng et al. 2021). The functional implications of the other spike amino acid changes identified in MA viruses (P812L, E96A, R408G, D215G, N501T, N74T, W64R, and D769N) are not well understood; however, some additional information is provided in Supplementary Fig. S1. To be able to tease apart the functional implications of these individual amino acid changes, new viruses would need to be constructed via reverse genetics (Amarilla et al. 2021).
Figure 2.
Sequencing of MA viruses. (A) Amino acid changes and deletions (∆) in MA1, MA2, MA3, MA4, and MA5 (full dataset in Supplementary Dataset 1). Amino acid changes that only appear in one MA virus are in red, with other colors used to show amino acid changes common between at least two MA viruses. (B) Amino acids changes and deletions in any of the MA viruses as shown on the structure of SARS-CoV-2QLD02 spike bound to hACE2 (PDB: 7DF4). Underlining indicates the non-conservative amino changes. (C) Spike changes in the MA viruses. *—amino acid changes located within the RBD. Underlining indicates the non-conservative amino acid changes. GISAID n—number of GISAID submissions that contain this change; GISAID %—percentage of all GISAID submissions with this change. Alpha, Beta, Gamma, Delta, and Omicron: black text—hallmark changes in variants of concern; gray text—not a hallmark change in variants of concern. Match of amino acid changes in MA viruses with hallmark changes in variants of concern (green—exact match, blue—conservative, and brown—non-conservative change).
The non-spike changes comprise nine different amino acid substitutions in Orf1ab (Fig. 2A), with all these changes also identified in human isolates (Supplementary Fig. S2). Apart from one study that computationally predicted that the C51S amino acid change in nsp1 reduced protein stability (Mou et al. 2021), to the best of our knowledge the other amino acid changes in Orf1ab have not been described in the available literature, and thus functional implications are unknown. The deletion of Amino Acids 20–31 in Orf3a of MA1, MA3, and MA4 resides in the extracellular N-terminal domain; unfortunately, protein structure is not available for Amino Acids 1–39 (Kern et al. 2021). SARS-CoV-2 Orf3a is a major contributor to viral pathogenesis (Liu et al. 2022b). SARS-CoV-2 Orf3a forms ion channels in cell membranes and is thus known as a viroporin. Viroporins have various roles in virus replication including viral entry, replication, release, and virion morphogenesis (Zhang et al. 2022). The deletion of the N-terminal domain of SARS-CoV-2 Orf3a increases localization to the plasma membrane (Kern et al. 2021). In general, the non-spike amino acid changes likely have no impact on mACE2 binding but instead likely represent adaptation to replication in HEK293T cells.
Modeling the spike RBD changes indicate the Q493R/K and Q498H increase interactions with mACE2
The amino acid changes in the MA viruses RBD (R408G, E484D, Q493R/K, Q498H, and N501T) were modeled using PyMOL to visualize their potential impact on mACE2 binding (Fig. 3). Interactions between SARS-CoV-2QLD02 RBD and mACE2 are predicted between Q493 and N31/Q34, N501 and H353, and Q498 and Y41 (Fig. 3A). These interactions do not support SARS-CoV-2QLD02 replication and are largely retained for MA viruses. However, MA viruses have additional predicted interactions between H498 and Y41 (Fig. 3B) or K/R493 and N31/Q34 (Fig. 3C–F). Our results thus support the previously reported contention that Q493K/R and Q498H represent key changes for mouse adaptation of SARS-CoV-2 (Leist et al. 2020; Huang et al. 2021).
Figure 3.
Modeling RBD changes. RBD substitutions for MA viruses were modeled using PyMOL (PDB: 7DF4) to visualize their potential effects on mACE2 binding. (A) Interactions between SARS-CoV-2QLD02 RBD and mACE2 are predicted between Q493 and N31/Q34, N501, and H353, and Q498 and Y41 (dashed lines/pale turquoise ovals). These interactions are insufficient to support replication and are largely retained for MA viruses and mACE2. (B) MA1 has additional predicted interactions between H498 and Y41 (magenta oval). (C–F) MA2-5 have additional predicted interactions between K/R493 and N31/Q34 (C–F, magenta ovals). Green = ACE2. Yellow = hACE2 residues changed to mACE2. Blue = SARS-CoV-2 spike RBD. Black-dotted interactions represent hydrogen bonds, and yellow-dotted lines represent any interactions within 3.5 Å.
N501T was selected in all three HEK293T-mACE2 + HEK293T-mACE2N31K/H353K co-cultures, but none of the HEK293T-mACE2 + HEK293T-hACE2 co-cultures. N501T was also reported to be selected for mink (Welkers et al. 2021) and ferrets (Richard et al. 2020; Zhou et al. 2022). N501T is a conservative change, and modeling suggested that the N501T change would not significantly affect interactions of MA viruses with H353 of mACE2 (Fig. 3).
MA1 and MA2, the viruses with the highest capacity to infect the lungs of wild-type mice (Fig. 1D), had two other amino acid changes that modeling predicted are not directly involved in interactions with mACE2: R408G for MA2 and E484D for MA1. The R408G change in MA2 and the R408S change in most Omicron BA.2 sequences remove a side chain that is not involved in the RBD-hACE2 interface (Supplementary Fig. S3).
MA1 replicates efficiently in several ACE2-negative cell lines
In vitro growth kinetics of MA1 and MA2 were studied in more detail using stock virus derived from infected mouse lungs and prepared in HEK293T-mACE2 cells (Fig. 1A). As expected, MA1 and MA2, as well as SARS-CoV-2QLD02, and Alpha and Beta strain viruses, all replicated in HEK293T-hACE2 cells (Fig. 4A). MA1, MA2, and Alpha, Beta, and Omicron BA.1 strain viruses, but not SARS-CoV-2QLD02, replicated in HEK293T-mACE2 cells (Fig. 4B), consistent with Fig. 1B–D and the reported ability of Alpha, Beta, and Omicron BA.1 variants to replicate in mice (Shuai et al. 2021). Perhaps of interest, given the postulated rodent origins of Omicron (Wei et al. 2021a; Mallapaty 2022), an Omicron BA.1 isolate (SARS-CoV-2QIMR01) replicated more efficiently in HEK293T-mACE2 than in HEK293T-hACE2 cells (Fig. 4A, B). Other studies also show that Omicron viruses have reduced replication capacity in Vero E6, Caco2, and Calu3 (all monkey or human ACE2 positive) cells compared to all previous variants of concern (Mautner et al. 2022; Shuai et al. 2022).
Figure 4.
In vitro growth kinetics of mACE2-adapted viruses reveal an ACE2-independent entry mechanism. Growth kinetics of the indicated viruses in HEK293T-hACE2 cells (A), HEK293T-mACE2 cells (B), untransduced HEK293T cells (C), 3T3 cells (D), AE17 cells (E), BHK-21 cells (F), A549 cells (G), HeLa cells (H), and LLC-PK1 cells (I) after infection at MOI ≈ 0.1. (J) Caco2 and (K) Caco2-ACE2 knockout cells were infected at MOI ≈ 1. n = 3–6 replicates per virus strain per cell line. Dotted line—limit of detection. *P < 0.05; statistics by t-test or the Kolmogorov–Smirnov test for 3 or 4 dpi versus 0 dpi.
Remarkably, when replication was examined in control untransduced HEK293T cells, both MA1 and MA2 showed significant replication, with MA1 replicating to ≈8 log10CCID50/ml in 2 days (Fig. 4C). None of the other viruses were able to replicate efficiently in untransduced HEK293T cells (Fig. 4C), consistent with previous reports (Cantuti-Castelvetri et al. 2020; Rawle et al. 2021; Wang et al. 2021a). Replication was also tested in a series of cell lines (Fig. 4D-I) in which (1) publically available RNA-Seq data showed negligible levels of ACE2 messenger RNA (mRNA) expression (Supplementary Fig. S4A) or (2) surface expression of ACE2 is below detection (LLC-PK1) (Mori et al. 2022). MA1 was able to replicate in all the aforementioned cell lines (Fig. 4D–I). In contrast, MA2, SARS-CoV-2QLD02, and Omicron BA.1 were unable to replicate in any of these cell lines (Fig. 4D–I); the SARS-CoV-2QLD02 results are consistent with previous reports (Chu et al. 2020; Hoffmann et al. 2020; Rawle et al. 2021; Trimarco et al. 2021; Mori et al. 2022). Overall, the data suggest that ACE2-independent infection by MA1 is robust across multiple cell lines and is functional, at least in vitro, across four different species (human, mouse, hamster, and pig). None of these cell lines expressed TMPRSS2 mRNA (Supplementary Fig. S4B), with virus fusion instead likely to rely on cathepsin L (Laporte et al. 2021), with cathepsin L mRNA abundantly expressed in all the cell lines (except BHK-21 which does not have an annotated orthologue) (Supplementary Fig. S4C).
While ACE2 mRNA levels were negligible in HEK293T, 3T3, HeLa, and A549 cells, they are not zero (Supplementary Fig. S4A). To evaluate infection by MA1 in the unambiguous absence of ACE2, a human CRISPR/Cas9 ACE2 knockout Caco-2 cell line was tested. This cell line has been validated by the manufacturer using immunocytochemistry, western blot, and Sanger sequencing to show complete ACE2 knockout (data available online at the distributer website, see Abcam ab273731). Wild-type Caco2 cells express ACE2, TMPRSS2, and cathepsin L (Supplementary Fig. S4A–C) and support replication of SARS-CoV-2QLD02, MA1, and MA2 (Fig. 4J). There was no detectable replication of SARS-CoV-2QLD02 or MA2 in the Caco2-ACE2 knockout cell line (Fig. 4K), indicating ACE2 dependence. Importantly, MA1 replicated in the Caco2-ACE2 knockout cell line (Fig. 4K), indicating unambiguous ACE2-independent infection.
MA1 and MA2 infection of C57BL/6J mice
To characterize the in vivo behavior of SARS-CoV-2 that evolved mACE2-binding capacity through in vitro passage, C57BL/6J mice were infected by intrapulmonary inoculation (via the intranasal route) with 105 CCID50 MA1 or MA2 per mouse. Virus titers in lungs reached 6–8 log10 CCID50/g on Day 2, with no significant differences seen between MA1 and MA2 (Fig. 5A). These results are broadly similar to those reported previously for MA viruses (Dinnon et al. 2020; Leist et al. 2020). Nasal turbinate titers on Day 2 were significantly higher for MA1 (≈7 log10 CCID50/g) when compared with MA2 (Fig. 5B). MA1 and MA2 virus titers were below the level of detection for all other mice and tissues tested (similar to other reports (Huang et al. 2021)), with the exception of the heart where MA1 infection resulted in detectable heart infection on Day 2 in 2/4 mice (Fig. 5C). (Infectious virus can also be detected in the heart of K18-hACE mice—Supplementary Fig. S5A). There was no significant weight loss observed for MA1- or MA2-infected mice (Supplementary Fig. S5B), consistent with the absence of fulminant brain infection (Carossino et al. 2022).
Figure 5.
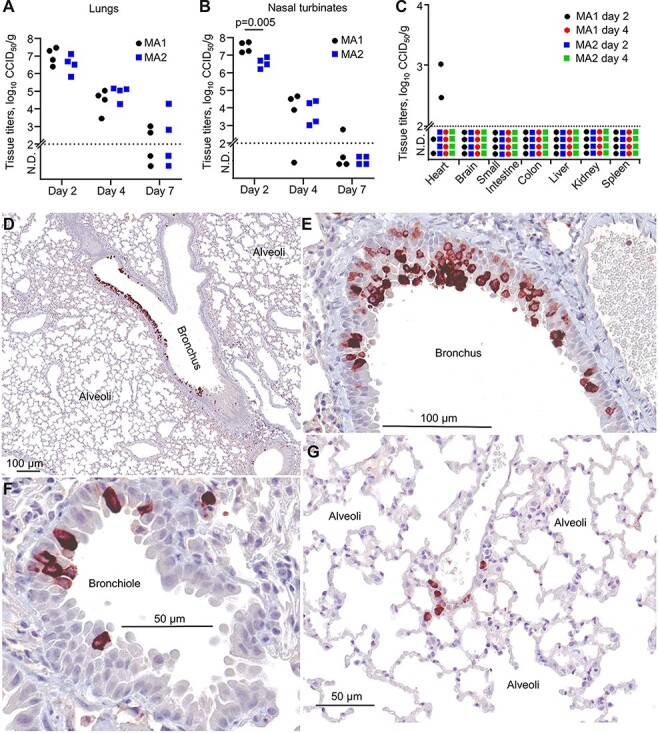
MA1 and MA2 infection in C57BL/6J mice. (A) C57BL/6J mice were infected with MA1 or MA2, lungs were collected at Days 2, 4, or 7 postinfection, and tissue titers were determined by CCID50 assays. (B) As for (A) for nasal turbinates. Statistics by t-test. (C) As for (A) for the indicated tissues. (D–G) IHC using an anti-SARS-CoV-2 spike monoclonal antibody and lungs taken on Day 2 after infection of C57BL/6J mice with MA1. Images are representative of lung sections from four mice. Dark brown staining (Nova Red) indicates infected cells, often with the expected, clearly discernable, cytoplasmic staining pattern. The large unstained areas in (D) and (G) are included to illustrate the specificity of the IHC staining.
Immunohistochemistry (IHC) staining with anti-SARS-CoV-2 spike monoclonal antibody showed that MA1 virus infection was localized primarily to the epithelium of bronchi (Fig. 5D-E) and bronchioles (Fig. 5F) in C57BL/6J mouse lungs, with occasional staining of cells in the alveoli (Fig. 5G). A similar picture emerged for MA2 (Supplementary Fig. S6). There was also occasionally prodigious staining of bronchial epithelium in MA1-infected lungs (Supplementary Fig. S7A). Other studies using MA SARS-CoV-2 viruses also showed viral staining in bronchial epithelium (Dinnon et al. 2020; DiPiazza et al. 2021; Huang et al. 2021), and such staining is also seen after infection with non-MA viruses of mice expressing hACE2 from the mACE2 promoter (Winkler Emma et al. 2021). Staining of alveoli was not a prominent feature in MA1- (Fig. 5D, G) or MA2- (Supplementary Fig. S7) infected C57BL/6J mice, with similar results reported for SARS-CoV-2 infection of mice expressing hACE2 from the mACE2 promoter (Winkler Emma et al. 2021). This contrasts markedly with SARS-CoV-2 infection of K18-hACE2 mice, which show widespread infection of alveoli (Winkler Emma et al. 2021). Occasional staining of MA1 was also observed in tracheal columnar epithelial cells (Supplementary Fig. S7B) and nasal epithelium (Supplementary Fig. S8), indicating upper respiratory tract infection. Omicron viruses similarly show a propensity to replicate in the upper respiratory tract, rather than in lung parenchyma (Hui et al. 2022; McMahan et al. 2022; Peacock et al. 2022).
Mouse-to-mouse (C57BL/6J to K18-hACE2) transmission was not seen for either MA1 or MA2 (Supplementary Fig. S9A, B). Such transmission has been reported for K18-hACE2 to K18-hACE2 mice (Bao et al. 2020) and for deer mice (Griffin et al. 2021). The loss of the furin cleavage site flanking region (QTQTN) in MA1 and MA2 (Supplementary Fig. S9C) has been shown to ablate furin cleavage (Lemmin et al. 2020; Peacock et al. 2021; Vu et al. 2021), with a functional furin cleavage site required for transmission in ferrets (Peacock et al. 2021). Furin cleavage site deletion was also identified as the most promising attenuation strategy (Sasaki et al. 2021) for the development of a live attenuated vaccine (Wang et al. 2021b; Goławski et al. 2022), with such deletions reducing pathogenicity in mice (Johnson et al. 2021; Hossain et al. 2022). Loss of this site commonly arises after in vitro passage of SARS-CoV-2 (Liu et al. 2020) and provides a built-in safety feature for our MA viruses.
Lung pathology after MA1 and MA2 infections of C57BL/6J mice
C57BL/6J mice were infected with MA1 and MA2 and lungs were harvested on Days 0, 2, 4, and 7 postinfection and analyzed by hematoxylin and eosin (H&E) staining. Examples of low magnification images of whole lung sections illustrate the loss of alveolar airspace (Fig. 6A), which reached significance by image analysis on Day 7 for MA1-infected mice (Fig. 6B). The ratio of dark blue (nuclear) to total red (cytoplasmic) pixels in H&E-stained sections is a measure of cellular infiltration (Prow et al. 2019), with lungs from MA1- or MA2-infected mice showing significantly higher cellular infiltrates compared to uninfected mice when all days postinfection are taken together (Fig. 6C).
Figure 6.
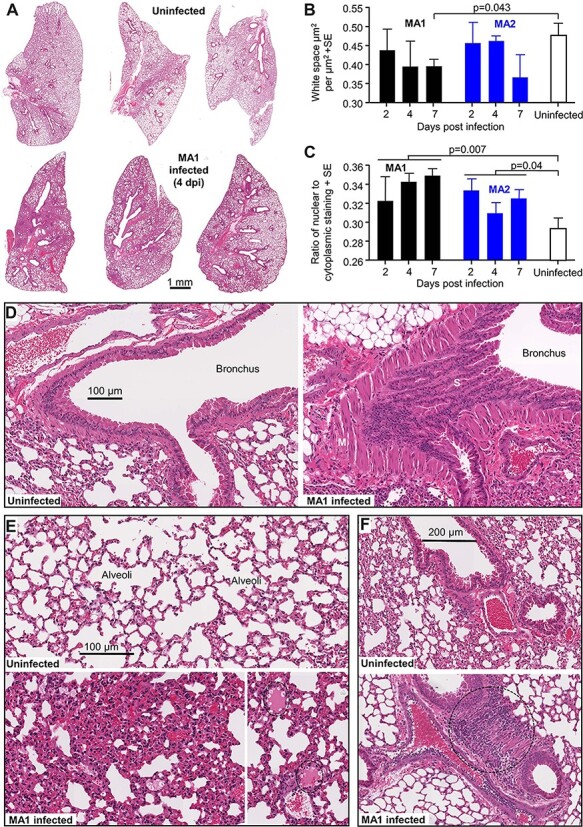
H&E staining of lungs after infection of C57BL/6J mice with MA1 and MA2. (A) Representative low magnification images of H&E-stained lung sections for uninfected and MA1-infected C57BL/6J mice taken 4 days postinfection. (B) Image analysis to measure lung consolidation quantitated as an area of white space (unstained air spaces within the lung) per µm2 (n = 4 mice per group, with three sections scanned per lung and values averaged to produce one value for each lung). Statistics by t-test. (C) Image analysis to quantitate leukocyte infiltration. The ratios of nuclear (blue/dark purple) to cytoplasmic (red) staining of H&E-stained lung (n = 4 mice per group, with three sections scanned per lung and values averaged to produce one value for each lung). Statistics by t-tests, with data from the three time points for infected mice combined. (D) High magnification image showing bronchus from uninfected mice (left) and MA1-infected mice (right) on Day 4 postinfection. The latter shows sloughing of the bronchial epithelium (S) and smooth muscle hyperplasia (M). (E) High magnification image showing uninfected lungs (top) and MA1-infected lungs (4 dpi) (bottom). The latter shows lung consolidation and loss of alveolar air spaces, as well as pulmonary edema (dotted ovals, bottom right). (F) High magnification images of uninfected lungs (top) and in MA1-infected lungs on 4 dpi (bottom). The latter shows dense cellular infiltrate (dotted circle).
H&E staining revealed clear evidence of bronchiolar sloughing and smooth muscle hyperplasia in MA1-infected mice (Fig. 6D). The collapse of alveolar spaces, with some pulmonary edema, is also evident (Fig. 6E) in MA1-infected lungs, consistent with Fig. 6B. Areas with dense cellular infiltrates were occasionally pronounced in MA1-infected lungs (Fig. 6E, dotted circle), consistent with Fig. 6C. Overall, similar lung pathology was seen in MA2-infected mice (Supplementary Fig. S10). MA infection of C57BL/6J mice results in robust lung pathology, with no overtly unique features when compared, for instance, with the infection of K18-hACE2 mice with SARS-CoV-2QLD02 (Amarilla et al. 2021).
Inflammatory responses in MA1-infected C57BL/6J mouse lungs are similar to, but less severe than, SARS-CoV-2QLD02 infection of K18-hACE2 mouse lungs
RNA-Seq was used to determine the inflammatory responses in C57BL/6J mouse lungs on Day 4 post MA1 infection (full gene list in Supplementary Dataset 2a). Differentially expressed genes (DEGs) (n = 1,027, Supplementary Dataset 2b) were analyzed for ‘UpStream Regulators’ (USRs) (Supplementary Dataset 2c) and ‘Diseases and Functions’ (Supplementary Dataset 2d) tools of the Ingenuity Pathway Analysis (IPA) software. The same analyses were performed for Day 4 lungs from K18-hACE2 mice infected with SARS-CoV-2QLD02 (Mills et al. 2021) (n = 1,349 DEGs; Supplementary Dataset 2e–h, respectively). A highly significant correlation emerged for cytokine USRs (Fig. 7A; Supplementary Dataset 2i). A number of USR annotations (Fig. 7A, red) indicated that inflammation in K18-hACE2 is more severe (1) with higher z-scores for the pro-inflammatory cytokine tumor necrosis factor (TNF) and osteopontin (SPP1), a proposed severity marker of COVID-19 (MacDonald et al. 2021), and (2) lower z-scores for anti-inflammatory cytokines IL10 and IL37 and Secretoglobin family 1a member 1 (SCGB1A1), a pulmonary surfactant protein that blunts alveolar macrophage responses and mitigates against cytokine surges (Xu et al. 2020).
Figure 7.
RNA-Seq comparison of MA1-infected C57BL/6J versus SARS-CoV-2QLD02-infected K18-hACE2 mouse lungs. RNA-Seq was performed on mouse lungs on 4 dpi for MA1 infection of C57BL/6J mice and SARS-CoV-2QLD02 infection of K18-hACE2 mice. DEGs (n = 1,027 for C57BL/6J, n = 1,349 for K18-hACE2) were analyzed by IPA, and z-scores for significant USRs (A) or Diseases and Functions (B) were plotted with C57BL/6J + MA1 on the x-axis and K18-hACE2 + QLD02 on the y-axis. Statistics by Pearson’s correlations. Dotted black lines—95 per cent confidence limits. Solid black line—line of best fit. Annotations outside the 95 per cent confidence limits are colored in red. Selected annotations are labeled (red text). Full data sets are provided in Supplementary Dataset 2.
The disease and functions annotation z-scores also showed a highly significant correlation for SARS-CoV-2QLD02 infection of K18hACE2 mice and MA1 infection of C57BL/6J mice, although the former again indicated a more severe inflammatory response, with increased cell movement, cell death, and weight loss annotations (Fig. 7B; Supplementary Dataset 2j), consistent with histology (Amarilla et al. 2021) (Fig. 6) and body weight measurements (Mills et al. 2021) (Supplementary Fig. S5B). Thus, although MA1 can infect cells lacking ACE2, in addition to cells expressing ACE2, the inflammatory responses in C57BL/6J mice were similar (even less severe) to those seen in the K18-hACE2/SARS-CoV-2QLD02 model, which is dependent on hACE2.
Discussion
Herein we describe the rapid in vitro adaptation of an original isolate of SARS-CoV-2 to evolve into viruses capable of infecting mice, with one virus (MA1) also capable of ACE2-independent infection. The in vitro co-culture adaptation method for generating MA SARS-CoV-2 is easier, cheaper, and more ethical than the serial passage in vivo and simpler than reverse genetics cloning. ACE2-independent infection by MA1 allows studies of SARS-CoV-2 infection in non-ACE2 expressing cells in vitro and in vivo, which other MA strains do not provide (Dinnon et al. 2020; Gu et al. 2020; Leist et al. 2020; Wang et al. 2020a; Huang et al. 2021; Zhang et al. 2021). ACE2-independent infection was robust across multiple cell lines (including ACE2 knockout cells) and was functional in at least four different species (human, mouse, hamster, and pig). Our study again highlights the ability of SARS-CoV-2 to evolve rapidly in the face of selection pressure (Ramirez et al. 2021), a feature that has also been documented in individual patients (Jary et al. 2022). Our study also highlights the relatively small number of changes (that include Q498H, Q493R/K, and N501T) that allow SARS-CoV-2 to jump species (Dinnon et al. 2020; Leist et al. 2020; Wang et al. 2020a; Huang et al. 2021; Rawle et al. 2021; Zhang et al. 2021).
The ability to infect cells in an ACE2-independent manner would appear to require the E484D substitution, identified herein and previously (Puray-Chavez et al. 2021; Ramirez et al. 2021; Hoffmann et al. 2022). Despite being identified in relatively few human sequences, this substitution has been identified across all variants of concern and in a large range of geographical locations around the world (Supplementary Fig. S11), as well as in deer (Hale et al. 2022). Omicron strains have an E484A substitution in this position, with Omicron BA.1 requiring ACE2 expression for infection (Fig. 4) (Hui et al. 2022). Modeling suggests that the E484D substitution retracts the carboxyl group located between Y489 and F490 of the RBD (Supplementary Fig. S12), perhaps thereby providing a novel binding site. A series of alternative or additional receptors have been reported for the SARS-CoV-2 Wuhan strain and include Dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN)/lymph node-specific ICAM-3 grabbing non-integrin (L-SIGN) (Amraei et al. 2021), Kidney Injury Molecule-1/T cell immunoglobulin mucin domain 1 (Mori et al. 2022), AXL Receptor Tyrosine Kinase (Wang et al. 2021a), neuropilin 1 (Cantuti-Castelvetri et al. 2020), and CD147 (Wang et al. 2020b). The analysis of publically available RNA-Seq data indicated that only CD147 was expressed at high levels in all cell lines tested herein (Supplementary Fig. S13). However, Wang et al. reported replication of an original isolate of SARS-CoV-2 in BHK-21 cells only when human CD147 was over-expressed (Wang et al. 2020b). Many have speculated that integrins, which are highly expressed in lungs, could be a potential SARS-CoV-2 entry receptor (Sigrist, Bridge, and Le Mercier 2020; Dakal 2021; Makowski, Olson-Sidford, and Weisel 2021; Nader et al. 2021; Park et al. 2021; Simons et al. 2021; Bugatti et al. 2022; Liu et al. 2022a). SARS-CoV-2 contains an ‘RGD’ amino acid motif in the spike receptor binding domain (RBD) (Supplementary Fig. S14A), and this motif commonly binds cell surface integrins (Makowski, Olson-Sidford, and Weisel 2021), although one molecular dynamics microscale simulation study disputes the involvement of the spike RGD in integrin binding (Othman et al. 2022). The ACE2-negative cell lines that supported MA1 replication express a range of integrins (Supplementary Fig. S14B). It is possible that the E484D and/or Q498H amino acid changes in MA1 increase the accessibility of the RGD motif for integrin binding; however, this is difficult to determine using current PyMOL modeling capabilities (Supplementary Fig. S14A).
The MA viruses are likely to enter via the endosomal route, given the loss of the furin cleavage site (Bestle et al. 2020; Peacock et al. 2022). This loss improves cleavage by cathepsin L, which substitutes for TMPRSS2 by cleaving S2ʹ in endosomes and releasing viral RNA into the cytoplasm (Laporte et al. 2021). Omicron, for different unknown reasons, shows reduced reliance on TMPRSS2 and relies more on endosomal entry (Hui et al. 2022; Meng et al. 2022; Shuai et al. 2022). The D614G change, which is present in all SARS-CoV-2 variants (except the original Wuhan strain), also increases entry via the cathepsin L route (Laporte et al. 2021). Cathepsin L is widely expressed in human nasal and lung epithelial cells (Laporte et al. 2021) and in the cell lines tested herein (Supplementary Fig. S4C).
The concept that Omicron might have arisen via rodents (Wei et al. 2021b; Mallapaty 2022) is perhaps consistent with the observation that the Omicron BA.1 virus replicated more efficiently in mACE2-expressing cells when compared to hACE2-expressing cells (Fig. 4A, B). Omicron binding to mACE2 is significantly higher than Alpha and Beta variants, which are also able to use mACE2 as a receptor (Cameroni et al. 2022). Increased mACE2-binding by Omicron viruses may involve the Q493R, Q498R, and N501Y substitutions (Fig. 2C, Omicron BA.1), given that these changes are selected in MA viruses (Dinnon et al. 2020; Gu et al. 2020; Leist et al. 2020; Huang et al. 2021; Zhang et al. 2021). Our MA viruses have Q493R, Q498H, and N501T substitutions (Fig. 2C; Supplementary Fig. S1), with modeling suggesting Q493R and Q498H increase mACE2 binding (Fig. 3). The newer Omicron sub-variants BA.4 and BA.5 have a Q493 reversion, but whether these viruses have lower mACE2-binding affinity and/or higher hACE2-binding affinity remains to be established. The BA.4 and BA.5 variants are more transmissible in humans than previous Omicron sub-variants (Tegally et al. 2022), and the Q493 reversion may contribute by slightly increased hACE2 affinity (Starr et al. 2020). Finally, like Omicron viruses (Hui et al. 2022), MA1, MA2, and other MA viruses (Dinnon et al. 2020; Huang et al. 2021) show a preference for replication in the upper respiratory tract.
The pathological consequences of ACE2-independent infection remain to be characterized, and one might speculate that ACE2-independent infection may play a role in long COVID (Lopez-Leon et al. 2021; Mehandru and Merad 2022). Our studies of acute MA1 infections in C57BL/6J mice argue that when ACE2-dependent infection remains possible, the ability also to infect via an ACE2-independent mechanism does not result in overtly unique pathology or immunopathology. The development of mouse models of long COVID, evaluation of infection and disease in ACE2−/− mice and the identification of the alternate receptor used by MA1 are the focus of our future studies.
Materials and methods
Ethics statement and regulatory compliance
All mouse work was conducted in accordance with the ‘Australian code for the care and use of animals for scientific purposes’ as defined by the National Health and Medical Research Council of Australia. Mouse work was approved by the QIMR Berghofer Medical Research Institute (MRI) Animal Ethics Committee (P3600, A2003-607). For intrapulmonary inoculations, mice were anesthetized using isoflurane. Mice were euthanized using CO2 or cervical dislocation. Obtaining samples from consenting COVID-19 patients to obtain human SARS-CoV-2 isolates was approved by the QIMR Berghofer MRI Human Research Ethics Committee (P3600).
Breeding and use of GM mice were approved under a Notifiable Low Risk Dealing (NLRD) Identifier: NLRD_Suhrbier_Oct2020: NLRD 1.1(a). Cloning and use of lentiviral vectors for transduction of ACE2 into cell lines were approved under an NLRD (OGTR identifier: NLRD_Suhrbier_Feb2021: NLRD 2.1(l), NLRD 2.1(m)).
All infectious SARS-CoV-2 work was conducted in a dedicated suite in a biosafety level-3 (PC3) facility at the QIMR Berghofer MRI (Australian Department of Agriculture, Water and the Environment certification Q2326 and Office of the Gene Technology Regulator certification 3445). All work was approved by the QIMR Berghofer MRI Safety Committee (P3600).
Cell lines and SARS-CoV-2 culture
Vero E6 (C1008, ECACC, Wiltshire, England; obtained via Sigma Aldrich, St. Louis, MO, USA), Lenti-X 293T (Takara Bio), Caco2 (a gift from Dr Yan Lu, QIMR Berghofer MRI), Caco2-ACE2 knockout (Abcam, ab273731), AE17 (a gift from Dr Delia Nelson, Faculty of Health Sciences, Curtin Medical School), NIH-3T3 (American Type Culture Collection, ATCC, CRL-1658), LLC-PK1 (a gift from Prof. Roy Hall, University of Queensland), A549 (ATCC CCL-185), BHK-21 (ATCC# CCL-10), and HeLa (ATCC-CLL 2) cells were cultured in medium comprising DMEM for Lenti-X 293T and Caco2 (WT and ACE2 KO) and A549 cells, M199 for LLC-PK1 cells, or RPMI1640 for all the others (Gibco), supplemented with (endotoxin free (Johnson et al. 2005)) 10 per cent fetal bovine serum (FBS), penicillin (100 IU/ml)/streptomycin (100 μg/ml) (Gibco/Life Technologies), and L-glutamine (2 mM) (Life Technologies). Cells were cultured at 37°C and 5 per cent CO2. Cells were routinely checked for mycoplasma (MycoAlert Mycoplasma Detection Kit MycoAlert, Lonza), and FBS was assayed for endotoxin contamination before purchase (Johnson et al. 2005).
The SARS-CoV-2 original, Alpha, and Beta isolates were kindly provided by Dr Alyssa Pyke (Queensland Health Forensic & Scientific Services, Queensland Department of Health, Brisbane, Australia). The viruses (hCoV-19/Australia/QLD02/2020, Alpha variant hCoV-19/Australia/QLD1517/2021, and Beta variant hCoV-19/Australia/QLD1520/2020) were isolated from patients, and sequences deposited at GISAID (https://www.gisaid.org/). Virus stocks were generated by infection of Vero E6 cells at a multiplicity of infection (MOI) ≈ 0.01, with supernatant collected after 2–3 days, cell debris removed by centrifugation at 3,000× g for 15 min at 4°C, and virus aliquoted and stored at −80°C. Virus titers were determined using standard CCID50 assays (as mentioned later). The virus was determined to be mycoplasma-free using co-culture with a non-permissive cell line (i.e. HeLa) and Hoechst staining as described (La Linn et al. 1995).
An Omicron isolate (SARS-CoV-2QIMR01) was isolated from a consenting COVID-19 patient in Brisbane, Australia. A nasal swab was self-inserted into the nose by the patient, then submerged and agitated in 1 ml RPMI + 10 per cent FBS + penicillin (100 IU/ml)/streptomycin (100 μg/ml) (Gibco/Life Technologies), and was transported to the laboratory on ice. After centrifugation at 20,000× g for 10 min, the supernatant was cultured on 2 × 106 Vero E6-TMPRSS2 cells (Amarilla et al. 2021) in a T25 flask for 3 days at 37°C and 5 per cent CO2; 1 ml was passaged on to a confluent T75 flask of Vero E6-TMPRSS2 cells, and supernatant harvested and aliquoted after 3 days. Virus titers were determined using standard CCID50 assays (as mentioned later). For sequencing, viral RNA was isolated from infected Vero E6-TMPRSS2 cells by TRIzol extraction as per manufacturer’s instructions. Reverse transcription was performed using ProtoScript® II First Strand complementary DNA (cDNA) Synthesis Kit (New England Biolabs) as per manufacturer’s instructions. Polymerase chain reaction (PCR) of a spike gene fragment was performed using Q5 High-Fidelity 2X Master Mix (New England Biolabs) and the following primers: Forward 5ʹ-TTGAACTTCTACATGCACCAGC-3ʹ and Reverse 5ʹ-CCAGAAGTGATTGTACCCGC-3ʹ. The fragment was gel purified using Monarch DNA Gel Extraction Kit (New England Biolabs), and Sanger sequencing was performed using BigDye Terminator v3.1 and either the forward or the reverse primer. The virus was confirmed as the Omicron variant due to containing the following amino acid changes compared to SARS-CoV-2QLD02: N547K, D614G, H655Y, N679K, P681H, N764K, D796Y, and N856K. The whole genome of the virus was sequenced by next-generation RNA-Seq (as mentioned later), and the genome sequence can be found on GenBank (Accession number ON819429) and GISAID (EPI_ISL_13414183). Analyses using GISAID indicated that the Omicron virus, SARS-CoV-2QIMR01 (B.1.1.529), belongs to the BA.1.17 lineage.
CCID50 assays
Vero E6 cells were plated into ninety-six well flat bottom plates at 2 × 104 cells per well in 100 µl of the medium. For tissue titer, tissue was homogenized in tubes each containing four ceramic beads twice at 6,000× g for 15 s, followed by centrifugation twice at 21,000× g for 5 min before 5-fold serial dilutions in 100 µl RPMI1640 supplemented with 2 per cent FBS. For cell culture supernatant, 10-fold serial dilutions were performed in 100 µl RPMI1640 supplemented with 2 per cent FBS. 100 µl of serially diluted samples were added to Vero E6 cells, and the plates were cultured for 5 days at 37°C and 5 per cent CO2. The virus titer was determined by the method of Spearman and Karber.
SARS-CoV-2 passaging in hACE2 and mACE2 co-cultures
Lentivirus encoding hACE2, mACE2, or mACE2N31K/H353K were produced in HEK293T cells by plasmid transfection and were used to transduce HEK293T cells, as described previously (Rawle et al. 2021). HEK293T-hACE2 or HEK293T-mACE2N31K/H353K were mixed with equal cell numbers of HEK293T-mACE2, and 500,000 cells (i.e. 250,000 HEK293T-hACE2 or mACE2N31K/H353K + 250,000 HEK293T-mACE2) were seeded in six well plates overnight. Co-cultures were infected with SARS-CoV-2 at MOI ≈ 0.1. Every 3–4 days, supernatant was collected and centrifuged at 2000× g at 4°C for 5 min, and 200 µl was added to fresh co-cultures and the remaining was stored at −80°C. This was performed a total of nine times before 1 ml was passaged on to HEK293T-mACE2 cells. After one passage in HEK293T-mACE2 cells, supernatant was used to infect new HEK293T-mACE2 cells for 2 h, then inoculum was removed and cells were washed three times with phosphate-buffered saline (PBS), and media replaced. Supernatant was harvested at 0 and 72 h postinfection for virus titration by CCID50 to confirm virus replication in mACE2 expressing cells. Virus was then passaged one additional time in HEK293T-mACE2 cells, and supernatant was stored at −80°C for use in sequencing and mouse infections.
For growth kinetics experiments, HEK293T, HEK293T-hACE2 and HEK293T-mACE2, NIH-3T3, AE17, BHK-21, A549, HeLa, or LLC-PK1 cells were infected with SARS-CoV-2 (SARS-CoV-2QLD02, MA1, MA2, Alpha, Beta, or Omicron) at MOI ≈ 0.1 for 1 h at 37°C, cells were washed with PBS, and media replaced. Caco2 and Caco2-ACE2 knockout cells were infected with SARS-CoV-2QLD02, MA1, or MA2 at MOI ≈ 1 for 1 h at 37°C, cells were washed twice with PBS, and media replaced. Culture supernatant was harvested at the indicated time points and titered by CCID50 assay as described earlier.
Mouse intrapulmonary SARS-CoV-2 infection
Female C57BL/6J mice (∼6 months old at the time of infection) were purchased from Animal Resources Centre (Canning Vale, WA, Australia). The conditions the mice were kept are as follows: light = 12:12 h dark/light cycle, 7:45 a.m. sunrise and 7:45 p.m. sunset, 15 min light dark and dark light ramping time; enclosures: M.I.C.E cage (Animal Care Systems, Colorado, USA); ventilation: 100 per cent fresh air, eight complete air exchange/h/rooms; in-house enrichment: paper cups (Impact-Australia), tissue paper, cardboard rolls; bedding: PuraChips (Able scientific) (aspen fine); food: double bagged norco rat and mouse pellet (AIRR, Darra, QLD); and water: deionized water acidified with HCl (pH = 3.2). Mice were anesthetized using isoflurane and given an intrapulmonary inoculation ≈5 × 104 CCID50 SARS-CoV-2 delivered via the intranasal route in 50 µl. Mice were sacrificed by cervical dislocation at Days 2, 4, or 7, and lungs, nasal turbinates, brain, small intestine, colon, liver, kidney, and spleen were collected. The right lung and all other organs were immediately homogenized in tubes each containing four beads twice at 6,000× g for 15 s and used in tissue titration as described earlier. Left lungs were fixed in 10 per cent formalin for histology.
K18-hACE2 mice (strain B6.Cg-Tg(K18-ACE2)2Prlmn/J, JAX Stock No.: 034860) (McCray et al. 2007) were purchased from The Jackson Laboratory, USA, and bred and maintained in-house at QIMRB Berghofer Medical Research Institute as heterozygotes by crossing with C57BL/6J mice (Bishop et al. 2022). Mice were genotyped using Extract-N-Amp Tissue PCR Kit (Sigma Aldrich) according to manufacturers’ instructions with the following primers: Forward 5ʹ-CTTGGTGATATGTGGGGTAGA-3ʹ and Reverse 5ʹ-CGCTTCATCTCCCACCACTT-3ʹ (recommended by NIOBIOHN, Osaka, Japan). Thermocycling conditions were as follows: 94°C 3 min, 35 cycles of 94°C 30 s, 55.8 °C 30 s, 72°C 1 min, and final extension of 72°C 10 min.
RNA sequencing
For viral RNA purification, cell culture supernatants were processed using the NucleoSpin RNA Virus kit (Machery-Nagel) as per manufacturers’ instructions. For mouse lung RNA isolation, lungs were transferred from RNAlater to TRIzol (Life Technologies) and were homogenized twice at 6,000× g for 15 s. Homogenates were centrifuged at 14,000× g for 10 min, and RNA was isolated as per manufacturers’ instructions.
RNA concentration and quality were measured using TapeStation D1K TapeScreen assay (Agilent). cDNA libraries were prepared using the Illumina TruSeq Stranded mRNA library prep kit, and the sequencing was performed on the Illumina Nextseq 550 platform generating 75-bp paired-end reads.
For virus sequences, per base sequence quality for >90 per cent bases was above Q30 for all samples. The quality of raw sequencing reads was assessed using FastQC (Andrews 2010) (v0.11.80) and trimmed using Cutadapt (Martin 2011) (v2.3) to remove adapter sequences and low-quality bases. Trimmed reads were aligned using STAR (Dobin et al. 2013) (v2.7.1a) to a SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2; 29,903 bp). Aligned reads were viewed using Integrative Genome Viewer (Robinson et al. 2011), and any position with >20 per cent change compared to the reference genome was manually curated. SAMtools mpileup was used to produce a consensus sequence from mapped reads (Li et al. 2009).
For mouse lungs, per base sequence quality for >90 per cent bases was above Q30 for all samples. The quality of raw sequencing reads was assessed using FastQC (Andrews 2010) (v0.11.80) and trimmed using Cutadapt (Martin 2011) (v2.3) to remove adapter sequences and low-quality bases. Trimmed reads were aligned using STAR (Dobin et al. 2013) (v2.7.1a) to a combined reference that included the mouse GRCm39 primary assembly and the GENCODE M27 gene model (Harrow et al. 2012), SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2; 29,903 bp). Mouse gene expression was estimated using RSEM (Li and Dewey 2011) (v1.3.0). Reads aligned to SARS-CoV-2 were counted using SAMtools (Li et al. 2009) (v1.9). Differential gene expression in the mouse was analyzed using EdgeR (3.22.3) and modeled using the quasi-likelihood F-test, glmQLFTest().
Pathway analysis
USRs and Diseases and Functions enriched in DEGs in direct and indirect interactions were investigated using IPA (QIAGEN).
SARS-CoV-2 amino acid change analyses and modeling
The GISAID (https://www.gisaid.org/phylodynamics/global/nextstrain/) (Elbe and Buckland-Merrett 2017) EpiCoV ‘search’ function (https://www.epicov.org/epi3/frontend#577a0f) was used to identify the number of GISAID SARS-CoV-2 sequence submissions containing each amino acid change in the MA viruses. Data were filtered for submissions that selected ‘Human’ as the host. Data were accessed on 21 February 2021 when the number of total submissions was 8,601,773, and this was used to calculate the proportion of each amino acid change in the MA viruses among total GISAID sequence submissions.
PyMOL v4.60 (Schrodinger) was used for mutagenesis of the crystal structure of SARS-CoV-2 spike bound with ACE2 from the protein data bank (7DF4) (Xu et al. 2021).
Histopathology and IHC
Lungs, trachea (via pluck necropsy, i.e. removal of tongue, larynx, trachea, lungs, heart, and part of the esophagus in one piece), and whole mouse head were fixed in 10 per cent formalin, embedded in paraffin, and sections stained with H&E (Sigma Aldrich). Mouse heads were decalcified in ethylenediaminetetraacetic Acid (EDTA) for 6 weeks before paraffin embedding. Slides were scanned using Aperio AT Turbo (Aperio, Vista, CA, USA) and analyzed using Aperio ImageScope software (LeicaBiosystems, Mt Waverley, Australia) (v10) and the Positive Pixel Count v9 algorithm. Automatic quantitation of white space was undertaken using QuPath v0.2.3 (Bankhead et al. 2017). IHC for SARS-CoV-2 antigen was undertaken using mouse anti-SARS-CoV-2 spike monoclonal antibody 1E8 (Hobson-Peters et al. in preparation) as described previously (Rawle et al. 2021).
Statistics
Statistical analyses of experimental data were performed using IBM SPSS Statistics for Windows, Version 19.0 (IBM Corp., Armonk, NY, USA). The t-test was used when the difference in variances was <4, skewness was greater than −2, and kurtosis was <2. Otherwise, the nonparametric Kolmogorov–Smirnov test was used.
Supplementary Material
Acknowledgements
From QIMR Berghofer MRI, we thank Dr I Anraku for managing the PC3 (BSL3) facility, animal house staff for mouse breeding and agistment, Paul Collins for library preparation and RNA-Seq, and Dr Clay Winterford, Sang-Hee Park, and Crystal Chang for the histology and IHC. We thank Dr Alyssa Pyke and Mr Fredrick Moore (Queensland Health, Brisbane) for providing the SARS-CoV-2 isolates. We thank the Monash Genome Modification Platform for providing the plasmid containing the mouse-codon optimized hACE2 gene.
Contributor Information
Kexin Yan, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia.
Troy Dumenil, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia.
Bing Tang, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia.
Thuy T Le, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia.
Cameron R Bishop, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia.
Andreas Suhrbier, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia; Australian Infectious Disease Research Centre, GVN Center of Excellence, Brisbane, 300 Herston Road, Herston, 4029 and The University of Queensland, St Lucia, 4072, Australia.
Daniel J Rawle, Infection and Inflammation Department, QIMR Berghofer Medical Research Institute, 300 Herston Road, Herston, 4029, Queensland, Australia.
Supplementary data
Supplementary data are available at VEVOLU online.
Funding
We thank the Brazil Family Foundation and Clive Berghofer (and many others) for their generous philanthropic donations to support SARS-CoV-2 PC3 research at QIMR Berghofer MRI. A.S. holds an Investigator grant from the National Health and Medical Research Council of Australia (APP1173880). The project was also partly funded by an intramural QIMR Berghofer MRI seed grant awarded to D.J.R.
Conflict of interest:
The authors declare no competing interests.
Data availability
All raw sequencing data (fastq files) are available from the Sequence Read Archive, BioProject accession: PRJNA804321. Our Omicron BA.1.17 sequence (SARS-CoV-2_QIMR01) is available online at GenBank (Accession number ON819429) and GISAID (EPI_ISL_13414183). All other data are available within the paper and supporting information files.
Author contributions
D.J.R. participated in the conceptualization; D.J.R. and A.S. participated in the methodology; D.J.R., A.S., T.D., and C.B. participated in the formal analysis; K.Y., D.J.R., T.T.L., and B.T. participated in the investigation; A.S. participated in the resources; D.J.R., A.S., and T.D. participated in the data curation; D.J.R. and A.S. participated in the writing; D.J.R., A.S., and T.D. participated in the visualization; D.J.R. and A.S. participated in the supervision; D.J.R. and A.S. participated in the project administration; and A.S. and D.J.R. participated in the funding acquisition.
References
- Amarilla A. A. et al. (2021) ‘A Versatile Reverse Genetics Platform for SARS-CoV-2 and Other Positive-Strand RNA Viruses’, Nature Communications, 12: 3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amraei R. et al. (2021) ‘CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2’, ACS Central Science, 7: 1156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010) ‘A Quality Control Tool for High Throughput Sequence Data’. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed 21 July 2022.
- Bankhead P. et al. (2017) ‘QuPath: Open Source Software for Digital Pathology Image Analysis’, Scientific Reports, 7: 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L. et al. (2020) ‘Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 via Close Contact and Respiratory Droplets among Human Angiotensin-Converting Enzyme 2 Mice’, The Journal of Infectious Diseases, 222: 551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D. et al. (2020) ‘TMPRSS2 and Furin are Both Essential for Proteolytic Activation of SARS-CoV-2 in Human Airway Cells’, Life Science Alliance, 3: e202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C. R. et al. (2022) ‘Mouse Models of COVID-19 Recapitulate Inflammatory Pathways Rather than Gene Expression’, bioRxiv, 2022.02.24.481866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugatti A. et al. (2022) ‘SARS-CoV-2 Infects Human ACE2-Negative Endothelial Cells Through an α(v)β(3) Integrin-Mediated Endocytosis Even in the Presence of Vaccine-Elicited Neutralizing Antibodies’, Viruses, 14: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E. et al. (2022) ‘Broadly Neutralizing Antibodies Overcome SARS-CoV-2 Omicron Antigenic Shift’, Nature, 602: 664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L. et al. (2020) ‘Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity’, Science, 370: 856–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carossino M. et al. (2022) ‘Fatal Neurodissemination and SARS-CoV-2 Tropism in K18-hACE2 Mice Is Only Partially Dependent on hACE2 Expression’, Viruses, 14: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S. et al. (2022) ‘SARS-CoV-2 Omicron Has Extensive But Incomplete Escape of Pfizer BNT162b2 Elicited Neutralization and Requires ACE2 for Infection’, Nature, 602: 654–6.35016196 [Google Scholar]
- Chandler J. C. et al. (2021) ‘SARS-CoV-2 Exposure in Wild White-Tailed Deer’, Proceedings of the National Academy of Sciences, 118: e2114828118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H. et al. (2020) ‘Comparative Tropism, Replication Kinetics, and Cell Damage Profiling of SARS-CoV-2 and SARS-CoV with Implications for Clinical Manifestations, Transmissibility, and Laboratory Studies of COVID-19: An Observational Study’, The Lancet Microbe, 1: e14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakal T. C. (2021) ‘SARS-CoV-2 Attachment to Host Cells Is Possibly Mediated via RGD-integrin Interaction in a Calcium-Dependent Manner and Suggests Pulmonary EDTA Chelation Therapy as a Novel Treatment for COVID 19’, Immunobiology, 226: 152021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A. V., Walker M., and Webster J. P. (2021) ‘Surveillance and Control of SARS-CoV-2 in Mustelids: An Evolutionary Perspective’, Evolutionary Applications, 14: 2715–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K. H. et al. (2020) ‘A Mouse-Adapted Model of SARS-CoV-2 to Test COVID-19 Countermeasures’, Nature, 586: 560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPiazza A. T. et al. (2021) ‘COVID-19 Vaccine mRNA-1273 Elicits a Protective Immune Profile in Mice That Is Not Associated with Vaccine-Enhanced Disease upon SARS-CoV-2 Challenge’, Immunity, 54: 1869–82.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. (2013) ‘STAR: Ultrafast Universal RNA-seq Aligner’, Bioinformatics, 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., and Buckland-Merrett G. (2017) ‘Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health’, Global Challenges (Hoboken, NJ), 1: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goławski M. et al. (2022) ‘The Reassessed Potential of SARS-CoV-2 Attenuation for COVID-19 Vaccine Development-A Systematic Review’, Viruses, 14: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. D. et al. (2021) ‘SARS-CoV-2 Infection and Transmission in the North American Deer Mouse’, Nature Communications, 12: 3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H. et al. (2020) ‘Adaptation of SARS-CoV-2 in BALB/c Mice for Testing Vaccine Efficacy’, Science, 369: 1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale V. L. et al. (2022) ‘SARS-CoV-2 Infection in Free-ranging White-Tailed Deer’, Nature, 602: 481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P. J. et al. (2022) ‘SARS-CoV-2 Omicron Virus Causes Attenuated Disease in Mice and Hamsters’, Nature, 603: 687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J. et al. (2012) ‘GENCODE: The Reference Human Genome Annotation for the ENCODE Project’, Genome Research, 22: 1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M. et al. (2020) ‘SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor’, Cell, 181: 271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— et al. (2022) ‘Evidence for an ACE2-Independent Entry Pathway That Can Protect from Neutralization by an Antibody Used for COVID-19 Therapy’, mBio, 13: e00364–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. G. et al. (2022) ‘Roles of the Polybasic Furin Cleavage Site of Spike Protein in SARS-CoV-2 Replication, Pathogenesis, and Host Immune Responses and Vaccination’, Journal of Medical Virology, 94: 1815–20. [DOI] [PubMed] [Google Scholar]
- Hu B. et al. (2017) ‘Discovery of a Rich Gene Pool of Bat SARS-related Coronaviruses Provides New Insights into the Origin of SARS Coronavirus’, PLOS Pathogens, 13: e1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. et al. (2021) ‘Q493K and Q498H Substitutions in Spike Promote Adaptation of SARS-CoV-2 in Mice’, EBioMedicine, 67: 103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K. P. Y. et al. (2022) ‘SARS-CoV-2 Omicron Variant Replication in Human Bronchus and Lung Ex Vivo’, Nature, 603: 715–20.doi: 10.1038/s41586-022-04479-6 [DOI] [PubMed] [Google Scholar]
- Jary A. et al. (2022) ‘Spike Gene Evolution and Immune Escape Mutations in Patients with Mild or Moderate Forms of COVID-19 and Treated with Monoclonal Antibodies Therapies’, Viruses, 14: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. J. et al. (2005) ‘Heat Shock Protein 10 Inhibits Lipopolysaccharide-Induced Inflammatory Mediator Production’, Journal of Biological Chemistry, 280: 4037–47. [DOI] [PubMed] [Google Scholar]
- Johnson B. A. et al. (2021) ‘Loss of Furin Cleavage Site Attenuates SARS-CoV-2 Pathogenesis’, Nature, 591: 293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D. M. et al. (2021) ‘Cryo-EM Structure of SARS-CoV-2 ORF3a in Lipid Nanodiscs’, Nature Structural & Molecular Biology, 28: 573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Linn M. et al. (1995) ‘Complete Removal of Mycoplasma from Viral Preparations Using Solvent Extraction’, Journal of Virological Methods, 52: 51–4. [DOI] [PubMed] [Google Scholar]
- Langereis M. A. et al. (2021) ‘An Alphavirus Replicon-Based Vaccine Expressing a Stabilized Spike Antigen Induces Protective Immunity and Prevents Transmission of SARS-CoV-2 between cats’, npj Vaccines, 6: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte M. et al. (2021) ‘The SARS-CoV-2 and Other Human Coronavirus Spike Proteins Are Fine-Tuned Towards Temperature and Proteases of the Human Airways’, PLOS Pathogens, 17: e1009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S. R. et al. (2020) ‘A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice’, Cell, 183: 1070–85.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmin T. et al. (2020) ‘Structures and Dynamics of the Novel S1/S2 Protease Cleavage Site Loop of the SARS-CoV-2 Spike Glycoprotein’, Journal of Structural Biology: X, 4: 100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C. N. (2011) ‘RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome’, BMC Bioinformatics, 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2009) ‘The Sequence Alignment/Map Format and SAMtools’, Bioinformatics, 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. (2022a) ‘Integrin Mediates Cell Entry of the SARS-CoV-2 Virus Independent of Cellular Receptor ACE2’, Journal of Biological Chemistry, 298: 101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. (2022b) ‘A Live-Attenuated SARS-CoV-2 Vaccine Candidate with Accessory Protein Deletions’, bioRxiv, 2022.02.14.480460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. (2020) ‘Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2’, Journal of Virology, 94: e00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Leon S. et al. (2021) ‘More than 50 Long-term Effects of COVID-19: A Systematic Review and Meta-Analysis’, Scientific Reports, 11: 16144–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald L. et al. (2021) ‘COVID-19 and RA Share an SPP1 Myeloid Pathway That Drives PD-L1+ Neutrophils and CD14+ Monocytes’, JCI Insight, 6: e147413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Olson-Sidford W., and Weisel J. W. (2021) ‘Biological and Clinical Consequences of Integrin Binding via a Rogue RGD Motif in the SARS CoV-2 Spike Protein’, Viruses, 13: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. (2022) ‘Where Did Omicron Come from? Three Key Theories. The Hunt for the Origins of Omicron’, Nature, 602: 26–8. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011) ‘Cutadapt Removes Adapter Sequences from High-throughput Sequencing Reads’, EMBnet.journal, 17: 10–12.doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mautner L. et al. (2022) ‘Replication Kinetics and Infectivity of SARS-CoV-2 Variants of Concern in Common Cell Culture Models’, Virology Journal, 19: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P. B. Jr. et al. (2007) ‘Lethal Infection of K18- hACE2 Mice Infected with Severe Acute Respiratory Syndrome Coronavirus’, Journal of Virology, 81: 813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K. et al. (2022) ‘Reduced Pathogenicity of the SARS-CoV-2 Omicron Variant in Hamsters’, Med (New York, N.Y.), 3: 262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S., and Merad M. (2022) ‘Pathological Sequelae of Long-haul COVID’, Nature Immunology, 23: 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B. et al. (2021) ‘Recurrent Emergence of SARS-CoV-2 Spike Deletion H69/V70 and Its Role in the Alpha Variant B.1.1.7’, Cell Reports, 35: 109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— et al. (2022) ‘Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Tropism and Fusogenicity’, Nature, 603: 706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. J. et al. (2021) ‘BET Inhibition Blocks Inflammation-Induced Cardiac Dysfunction and SARS-CoV-2 Infection’, Cell, 184: 2167–82.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y. et al. (2022) ‘KIM-1/TIM-1 Is a Receptor for SARS-CoV-2 in Lung and Kidney’, medRxiv, 2020: 16.20190694. [Google Scholar]
- Mou K. et al. (2021) ‘Emerging Mutations in Nsp1 of SARS-CoV-2 and Their Effect on the Structural Stability’, Pathogens, 10: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C. et al. (2020) ‘Animal Models for COVID-19’, Nature, 586: 509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader D. et al. (2021) ‘SARS-CoV-2 Uses Major Endothelial Integrin αvβ3 to Cause Vascular Dysregulation In-vitro during COVID-19’, PLoS One, 16: e0253347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman H. et al. (2022) ‘SARS-CoV-2 Spike Protein Unlikely to Bind to Integrins via the Arg-Gly-Asp (RGD) Motif of the Receptor Binding Domain: Evidence from Structural Analysis and Microscale Accelerated Molecular Dynamics’, Frontiers in Molecular Biosciences, 9: e834857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo P. M. et al. (2021) ‘SARS-CoV-2 Neutralizing Antibodies in White-Tailed Deer from Texas’, Vector Borne and Zoonotic Diseases (Larchmont, N.Y.), 22: 62–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. J. et al. (2021) ‘The Spike Glycoprotein of SARS-CoV-2 Binds to β1 Integrins Expressed on the Surface of Lung Epithelial Cells’, Viruses, 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T. P. et al. (2021) ‘The Furin Cleavage Site in the SARS-CoV-2 Spike Protein Is Required for Transmission in Ferrets’, Nature Microbiology, 6: 899–909. [DOI] [PubMed] [Google Scholar]
- ——— et al. (2022) ‘The SARS-CoV-2 Variant, Omicron, Shows Rapid Replication in Human Primary Nasal Epithelial Cultures and Efficiently Uses the Endosomal Route of Entry’, bioRxiv, 2021.12.31.474653. [Google Scholar]
- Prow N. A. et al. (2019) ‘Exacerbation of Chikungunya Virus Rheumatic Immunopathology by a High Fiber Diet and Butyrate’, Frontiers in Immunology, 10: 2736–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puray-Chavez M. et al. (2021) ‘Systematic Analysis of SARS-CoV-2 Infection of an ACE2-negative Human Airway Cell’, Cell Reports, 36: 109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S. et al. (2021) ‘Overcoming Culture Restriction for SARS-CoV-2 in Human Cells Facilitates the Screening of Compounds Inhibiting Viral Replication’, Antimicrobial Agents and Chemotherapy, 65: e00097–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawle D. J. et al. (2021) ‘ACE2-Lentiviral Transduction Enables Mouse SARS-CoV-2 Infection and Mapping of Receptor Interactions’, PLOS Pathogens, 17: e1009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M. et al. (2020) ‘SARS-CoV-2 Is Transmitted via Contact and via the Air Between Ferrets’, Nature Communications, 11: 3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T. et al. (2011) ‘Integrative Genomics Viewer’, Nature Biotechnology, 29: 24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M. et al. (2021) ‘SARS-CoV-2 Bearing a Mutation at the S1/S2 Cleavage Site Exhibits Attenuated Virulence and Confers Protective Immunity’, mBio, 12: e01415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou S. et al. (2021) ‘Animal Models for COVID-19: Hamsters, Mouse, Ferret, Mink, Tree Shrew, and Non-human Primates’, Frontiers in Microbiology, 12: 626553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai H. et al. (2022) ‘Attenuated Replication and Pathogenicity of SARS-CoV-2 B.1.1.529 Omicron’, Nature, 603: 693–9.doi: 10.1038/s41586-022-04442-5 [DOI] [PubMed] [Google Scholar]
- ——— et al. (2021) ‘Emerging SARS-CoV-2 Variants Expand Species Tropism to Murines’, EBioMedicine, 73: 103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J., Bridge A., and Le Mercier P. (2020) ‘A Potential Role for Integrins in Host Cell Entry by SARS-CoV-2’, Antiviral Research, 177: 104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons P. et al. (2021) ‘Integrin Activation Is an Essential Component of SARS-CoV-2 Infection’, Scientific Reports, 11: 20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T. N. et al. (2020) ‘Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding’, Cell, 182: 1295–310.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— et al. (2022) ‘ACE2 Binding Is an Ancestral and Evolvable Trait of Sarbecoviruses’, Nature, 603: 913–8.doi: 10.1038/s41586-022-04464-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H. et al. (2022) ‘Continued Emergence and Evolution of Omicron in South Africa: New BA.4 And BA.5 Lineages’, medRxiv. 2022.05.01.22274406. [Google Scholar]
- Trimarco J. D. et al. (2021) ‘TMEM41B Is a Host Factor Required for the Replication of Diverse Coronaviruses Including SARS-CoV-2’, PLOS Pathogens, 17: e1009599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu M. N. et al. (2021) ‘QTQTN Motif Upstream of the Furin-cleavage Site Plays Key Role in SARS-CoV-2 Infection and Pathogenesis’, bioRxiv. 2021.12.15.472450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. (2020a) ‘Mouse-Adapted SARS-CoV-2 Replicates Efficiently in the Upper and Lower Respiratory Tract of BALB/c and C57BL/6J Mice’, Protein & Cell, 11: 776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. (2020b) ‘CD147-Spike Protein Is a Novel Route for SARS-CoV-2 Infection to Host Cells’, Signal Transduction and Targeted Therapy, 5: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. (2021a) ‘AXL Is a Candidate Receptor for SARS-CoV-2 That Promotes Infection of Pulmonary and Bronchial Epithelial Cells’, Cell Research, 31: 126–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2021b) ‘Scalable Live-attenuated SARS-CoV-2 Vaccine Candidate Demonstrates Preclinical Safety and Efficacy’, Proceedings of the National Academy of Sciences, 118: e2102775118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. et al. (2021a) ‘Evidence for a Mouse Origin of the SARS-CoV-2 Omicron Variant’, Journal of Genetics and Genomics, 48: 1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— et al. (2021b) ‘Evidence for a Mouse Origin of the SARS-CoV-2 Omicron Variant’, Journal of Genetics and Genomics, 48: 1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkers M. R. A. et al. (2021) ‘Possible Host-Adaptation of SARS-CoV-2 Due to Improved ACE2 Receptor Binding in Mink’, Virus Evolution, 7: veaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler Emma S. et al. (2021) ‘SARS-CoV-2 Causes Lung Infection Without Severe Disease in Human ACE2 Knock-in Mice’, Journal of Virology, 96: e0151121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. et al. (2020) ‘A New Coronavirus Associated with Human Respiratory Disease in China’, Nature, 579: 265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. et al. (2021) ‘Conformational Dynamics of SARS-CoV-2 Trimeric Spike Glycoprotein in Complex with Receptor ACE2 Revealed by cryo-EM’, Science Advances, 7: eabe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. et al. (2020) ‘Lung Secretoglobin Scgb1a1 Influences Alveolar Macrophage-Mediated Inflammation and Immunity’, Frontiers in Immunology, 11: 584310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. (2022) ‘Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19’, Frontiers in Microbiology, 13: e854567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2021) ‘SARS-CoV-2 Rapidly Adapts in Aged BALB/c Mice and Induces Typical Pneumonia’, Journal of Virology, 95: e02477-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. et al. (2022) ‘Mutations That Adapt SARS-CoV-2 to Mink or Ferret Do Not Increase Fitness in the Human Airway’, Cell Reports, 38: 110344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data (fastq files) are available from the Sequence Read Archive, BioProject accession: PRJNA804321. Our Omicron BA.1.17 sequence (SARS-CoV-2_QIMR01) is available online at GenBank (Accession number ON819429) and GISAID (EPI_ISL_13414183). All other data are available within the paper and supporting information files.


