Summary:
Myelodysplastic syndrome (MDS) describes a family of blood disorders driven by the clonal expansion of mutated blood cells that can evolve into secondary acute myeloid leukemia (sAML). Two new studies use single-cell and deep sequencing to elucidate the progression of MDS to AML, revealing discrete clonal architectures and the driving role of signaling mutations.
The progression of myelodysplastic syndrome (MDS) into secondary acute myeloid leukemia (sAML) is driven by the expansion of clones that harbor leukemia driver mutations (1). sAML has an exceptionally poor prognosis compared with de novo AML and recent studies have shown that, in AML, clonal heterogeneity and mutation frequency correlate with clinical outcomes (2). However, our understanding of the clonal architecture and mutation acquisition as MDS transforms into sAML is limited.
The clonal evolution of MDS into sAML has traditionally relied on bulk sequencing analyses that described the clonal evolution as a linear process, whereby one premalignant clone gives rise to a second clone by sequential mutation acquisition. These views were challenged by the identification of patients with MDS that exhibited polyclonality that could only arise from a branched evolution (3, 4). Furthermore, taking into account the differentiation state of cells in which mutations occur adds a level of complexity (5–7). A more granular understanding of clonal and cellular trajectories during the transition from MDS to AML may enable earlier, more precise, and more durable therapeutic interventions.
In this issue of Blood Cancer Discovery, Guess and colleagues and Menssen and colleagues use high sensitivity and single-cell sequencing approaches to characterize clonal evolution of MDS to sAML (8, 9). Guess and colleagues implement high-throughput single-cell DNA sequencing, which is a powerful tool to study complex clonal structures in myeloid (pre-) malignancies (10). They sequenced 45 commonly mutated AML-associated genes in 37 paired MDS and sAML samples from 18 patients. Between MDS and sAML samples, the authors found that the number of mutations did not differ, nor did the number of mutations per clone or the clonal heterogeneity (by Shannon diversity index), although the variant allele frequency (VAF) of mutations increased. Mutations in TP53 and DNMT3A were the most common, and additional mutations were classified into epigenetic, transcription factor, signaling, cohesin, TP53, and other types—a rationale for the functional gene categories is provided in the supplements. The single-cell data allowed the authors to determine that 12 patients displayed linear evolution and six displayed branching evolution. Signaling mutations were enriched in the group with branching evolution. Furthermore, TP53 and epigenetic mutations were more likely present in the founding clone, whereas signaling mutations were often subclonal.
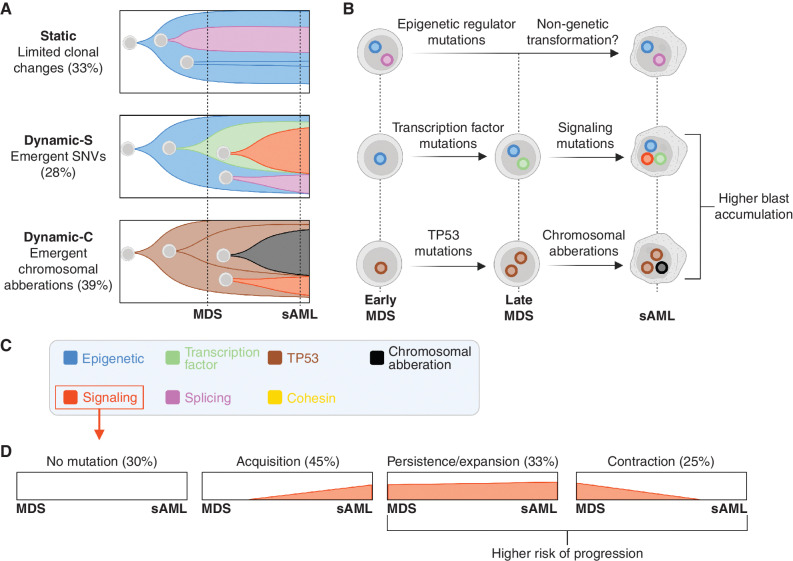
Guess and colleagues further characterized how the mutational landscape drives disease progression by analyzing the changes in clonal phylogenies in each patient from their MDS to sAML stage. They describe three patterns of clonal trajectories based on clonal emergence/stability and mutation types (Fig. 1A–C). The first pattern, Static, described patients who had relatively stable clones, present in both MDS and matched sAML samples. These samples were enriched for epigenetic modifying mutations such as DNMT3A, TET2, and IDH1/2, indicating that—rather than clonal expansion—epigenetic mechanisms may drive disease progression in some cases. The remaining two patterns, Dynamic-C and Dynamic-S, described samples in which a clone that was small or undetectable in the MDS sample emerged as a dominant clone in sAML, indicative of a subclonal sweep. These two patterns differed based on the mutational patterns that drove the clonal expansion. Dynamic-S described emergent clones that were driven by single-nucleotide variations (SNV) and were enriched for both transcription factor and signaling mutations. Dynamic-C described emergent clones with chromosomal aberrations and were enriched for TP53 mutations. The Dynamic patterns were associated with increased blast fold changes, linking genomic evolution to clinical parameters. While independent studies and larger patient numbers are required to confirm the three identified patterns of clonal evolution, they provide a useful framework of potential clonal architectures.
Figure 1.
Schematic detailing the main findings from paired MDS and sAML sequencing. A and B, Three patterns of MDS to sAML clonal evolution are observed by Guess and colleagues, the percentages indicate the proportion of patients following each. Both studies found that progenitor cells acquire mutation types in preferred orders. In the schematic, each cell represents a different clone, and nuclei are color-coded based on driver mutations. C, Both studies classify genetic lesions into functional categories, including epigenetic (e.g., TET2, ASXL1), signaling (e.g., FLT3, NRAS), transcription factor (e.g., GATA2, ETV6), splicing (e.g., U2AF1, SRSF2) and cohesin mutations (e.g., STAG2, RAD21). D, Multiple patterns of clonal evolution of signaling mutations emerged from the work of Menssen and colleagues. The percentages indicate the proportions of patients that follow indicated patterns; the total exceeds 100% because patients can have multiple signaling mutations (which can exhibit all patterns of evolution). Created with BioRender.com.
Guess and colleagues combined single-cell DNA-sequencing with surface marker expression in 2 patients, extending earlier data in clonal hematopoiesis (10). Leukemia-associated mutations were enriched in primitive and myeloid cells, and rare in lymphoid cells, suggesting that most of the detected mutations occur in myeloid-biased stem/progenitor cells, or that the mutations confer a myeloid bias. Finally, the authors performed scRNA-seq in paired MDS and sAML samples from 2 patients. Transcriptional changes associated with disease progression were analyzed separately in primitive and mature cells. In both patients, an increase in primitive cells or primitive cell markers was seen with progression, and some markers of inflammation were upregulated in mature cells. However, most of the transcriptional analyses indicated complex and heterogeneous changes. For example, one patient displayed downregulation of MHC genes in mature cells and downregulation of interferon signatures in both primitive and mature cells, whereas the other patient displayed upregulation of interferon signatures in mature cells. These results illustrate that changes in pathway activity are heterogeneous between patients and between cell types.
In complementary studies, Menssen and colleagues evaluated paired bone marrow samples from 44 patients at MDS and sAML, initially using a targeted sequencing panel. They classified mutations into DNA methylation, chromatin modifiers, transcription, signaling, spliceosome, cohesin, and TP53 categories and found that signaling mutations were less common in the MDS samples compared with sAML. The absence of these mutations at MDS indicated that signaling mutations may have been acquired between MDS diagnosis and sAML transformation. In other patients, signaling and transcription factor mutations were present at diagnosis, and deeper sequencing was required to gain more precise insight into the architecture of clonal evolution.
To characterize mutation types in founding clones and subclones, Menssen and colleagues used enhanced whole-genome sequencing, error-corrected sequencing, digital droplet PCR, and single-cell DNA sequencing. One of the advantages of deeper sequencing is the detection of small subclones, which revealed signaling gene mutations in 43% of patients with MDS and 61% of sAML. From enhanced whole-genome sequencing on 12 patients, the number and genetic composition of subclones was imputed from mutation VAFs. From this, the authors found that TP53 mutations were almost exclusively in founding clones, epigenetic regulator mutations were equally distributed amongst founder and subclones, and transcription factor, signaling, and cohesin mutations were mainly present in subclones. Similar to findings from Guess and colleagues, the authors noted that epigenetic regulator mutations tend to be acquired prior to splicing mutations. Furthermore, the authors found that transcription factor mutations were acquired prior to signaling mutations, in subclones where these mutation types cooccur. Taken together, these analyses provide new insights into the preferred order of mutations in founding clones and in subclones.
Menssen and colleagues found that clones with signaling mutation exhibited diverse patterns of clonal evolution during MDS to sAML transformation. Of the signaling mutations detected in MDS, 62% were still detectable after progression to sAML, while the rest were lost during the transition as confirmed by ddPCR. During the MDS stage, NRAS and KRAS mutations were mutated at similar rates but at sAML there were predominantly NRAS mutations, indicating that NRAS may have a greater potential to drive leukemic expansion. The authors then describe three main patterns of clonal evolution for signaling mutation-bearing clones: (i) emergent signaling mutation clones, (ii) signaling mutation clones that persist or expand during transformation, and (iii) signaling mutations that were present at MDS but lost in subsequent sAML (Fig. 1D). These different patterns of signaling mutation clonal evolution can exist in a patient simultaneously. For example, one MDS patient harbored three signaling gene mutations with one disappearing and two expanding as they transitioned into sAML. Patients with MDS who had a signaling mutation had a higher chance of progressing to AML, particularly in low-intermediate risk groups. With further validation, this finding could provide a new biomarker by which to stratify MDS patients for different treatment modalities.
Although sensitive and single-cell DNA sequencing can reveal biologically and clinically important features of cancer progression, the cell states in which these mutations occur are incompletely integrated. For example, the underlying stem cell pool may contain more complex clonal architectures (5–7). Combined mapping of clonal evolution and differentiation states (by transcriptional, epigenetic, or protein analysis), with computational innovations, will facilitate the incorporation of cell state heterogeneity. Nevertheless, the analyses by Guess and colleagues and Menssen and colleagues expand our knowledge of clonal evolution throughout the transition from MDS to sAML. They have identified patterns of clonal evolution that are predominantly associated with different mutation types and demonstrated that mutations are acquired in preferred orders. Dynamic patterns of clonal evolution are associated with higher blast fold changes and the presence of signaling mutations at MDS may be a poor prognostic indicator, indicating important clinical correlates. These new insights into clonal evolution provide a starting point to identify and track clones that are most likely to transform. This could lead to the design of therapeutic interventions that specifically target these clones, such as small-molecule inhibitors targeting clones with signaling mutations. Our increasing molecular understanding of genetic and nongenetic mechanisms of cancer evolution will result in earlier interventions to prevent transformation and more effective personalized therapies.
Authors’ Disclosures
No disclosures were reported.
References
- 1. Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H, et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2017;49:204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benard BA, Leak LB, Azizi A, Thomas D, Gentles AJ, Majeti R. Clonal architecture predicts clinical outcomes and drug sensitivity in acute myeloid leukemia. Nat Commun 2021;12:7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med 2012;366:1090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia 2013;27:1275–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J, Kao Y-R, Sun D, Todorova TI, Reynolds D, Narayanagari S-R, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med 2019;25:103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwenger E, Steidl U. An evolutionary approach to clonally complex hematologic disorders. Blood Cancer Discov 2021;2:201–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547:104–8. [DOI] [PubMed] [Google Scholar]
- 8. Guess T, Potts CR, Bhat P, Cartailler JA, Brooks A, Holt C, et al. Distinct patterns of clonal evolution drive myelodysplastic syndrome progression to secondary acute myeloid leukemia. Blood Cancer Discov 2022;3:316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Menssen AJ, Khanna A, Miller CA, Srivatsan SN, Chang GS, Shao J, et al. Convergent clonal evolution of signaling gene mutations is a hallmark of myelodysplastic syndrome progression. Blood Cancer Discov 2022;3:330–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]