Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), is a new coronavirus strain that was first reported in December 2019 in Wuhan, China. A specific treatment for COVID-19 has yet to be identified. Potential therapeutic targets include SARS-CoV-2 main protease (Mpro) and the SARS-CoV-2 spike-ACE2 interaction. Molecular docking, molecular dynamics (MD), solvent screening for the extraction of the specified compounds, and prediction of the drug properties of certain molecules were the methods used in this study to investigate compounds from the medicinal plant Myristica fragrans, which is one of twelve herbs in Prasachandaeng remedy (PSD). ArgusLab, AutoDock Vina, and AutoDock were used to perform docking tasks. The examined ligands were compared with panduratin A as a standard (Kanjanasirirat et al., 2020), which is a promising medicinal plant molecule for the treatment of COVID-19. Molecular docking revealed that malabaricones B and C and licarins A, B and C bound to SARS-CoV-2/ACE2 and SARS-CoV-2 Mpro with low binding energies compared to that of the standard ligand. Furthermore, appropriate solvent usage is important. Acetone was selected by COSMOquick software for compound extraction in this investigation because it can extract large amounts of all five of the abovementioned M. fragrans compounds. Furthermore, the drug-like properties of these compounds were studied utilizing the Lipinski, Veber, and Ghose criteria. The results revealed that these M. fragrans compounds have potential as effective medicines to combat the COVID-19 pandemic. However, to assess the therapeutic potential of these ligands, additional research is needed, which will use our findings as a foundation.
Keywords: Myristica fragrans (houtt.), SAR-CoV-2 main protease, SARs-CoV-2 spike-ACE-2, Molecular docking, Molecular Dynamics, Pharmacokinetics
1. Introduction
The coronavirus (CoV) family includes severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which was first discovered in Wuhan, China. Coronavirus disease 2019 (COVID-19) is a highly contagious human virus that has spread rapidly and caused an unprecedented pandemic, resulting in a large number of deaths and economic crises around the world (Prajapat et al., 2020, Sohrabi et al., 2020). As of December 14, 2021, the World Health Organization (WHO) reported that over 269.5 million cases of COVID-19 had been confirmed, with 5,304,248 deaths (World Health Organization, 2021). People in developing countries have been the most affected in terms of public health because they lack access to modern health care systems and treatments (Dandara et al., 2021). However, phytotherapy, which uses natural compounds to treat viral infections, could be a viable option (Antonio et al., 2021). Traditional medicines, primarily medicinal herbs, are used by approximately 80 % of the population in underdeveloped nations, according to WHO estimates (Series, 2018). Random selection of natural sources, chemotaxonomy (screening of comparable compounds in organisms belonging to the same family or genus), and ethnomedicine are the three major approaches used to choose natural products for the study of their biological qualities (Sheng-Ji, P.J.P.b., 2001, Singh et al., 2020). The most effective strategy has been determined to be the study of natural products that have a long history of use in diverse communities for the treatment of certain diseases and are part of the phytotherapeutic arsenal of common knowledge (Wu and Tan, 2019).
Prasachandaeng remedy (PSD), which is on Thailand's National List of Essential Treatment, has been used as an antipyretic medicine for children and adults in Thailand's hospitals and TTM clinics (Prommee et al., 2021). PSD is made up of twelve herbs, i.e., Citrus aurantiifolia (Christm.) Swingle (Ma-Nao), Dracaena cochinchinensis (Lour.) S.C. Chen (Chan-Daeng), Bouea macrophylla Griff. (Ma-Prang), Ligusticum chuanxiong Hort. (Khod-Hua-Bua), Caesalpinia sappan L. (Faang), Kaempferia galanga L. (Por-Hom), Mesua ferrea L. (Boon-Nak), Mammea siamensis T. Anderson. (Sa-ra-Pee), Jasminum sambac (L.) Aiton (Ma-li), Heliciopsis terminalis (Kurz) Sleumer (Mhuad-Kon), Nelumbo nucifera Gaertn (Bua-Luang), and Myristica fragrans Houtt. (Chan-Thet). Although PSD contains twelve herbs, the M. fragrans extract was determined to be the most efficient against COVID-19; however, there is no scientific evidence that it inhibits COVID-19. As a result, the current study focused on phytochemicals from M. fragrans Houtt., which are indicated in Table 1 as COVID-19 inhibitors.
Table 1.
Molecular docking results and predicted IC50 values of SARS-CoV-2/ACE-2 and SARS-CoV-2 main protease inhibitory activity of 13 Myristica fragrans (houtt.) extracts and Panduratin A (positive control).
| Compounds |
Arguslab |
Autodock |
Autodock Vina |
Inhibitory activity IC50 |
||||
|---|---|---|---|---|---|---|---|---|
| ACE2 | Mpro | ACE2 | Mpro | ACE2 | Mpro | ACE2 | Mpro | |
| Binding Energy | Binding Energy | Binding Energy | Binding Energy | Binding Energy | Binding Energy | |||
| panduratin A | −13.8811 | −10.668 | −7.21 | −6.98 | −9.3 | −6.6 | 5.19 uM | 7.59 uM |
| isoelemicin | −9.38539 | −8.48386 | −5.32 | −5.0 | −6.0 | −5.1 | 125.47 uM | 217.58 uM |
| myristicin | −9.3564 | −7.88654 | −6.02 | −4.91 | −6.4 | −5.2 | 38.93 uM | 251.26 uM |
| surinamensin | −9.87815 | −8.8307 | −6.09 | −5.52 | −7.8 | −6.4 | 34.42 uM | 90.57 uM |
| malabaricone C | −12.692 | −11.2303 | −6.04 | −4.77 | −8.1 | −5.9 | 37.6 uM | 319.17 uM |
| elemicin | −8.38233 | −8.06298 | −4.99 | −4.68 | −5.8 | −4.8 | 220.34 uM | 371.89 uM |
| licarin A | −12.1667 | −9.6228 | −7.43 | −8.42 | −8.7 | −8.2 | 3.59 uM | 668.63 nM |
| malabaricone B | −12.0989 | −10.7849 | −7.8 | −5.92 | −7.2 | −5.8 | 1.93 uM | 45.4 uM |
| licarin C | −11.2124 | −9.15462 | −6.86 | −7.25 | −8.2 | −7.0 | 9.42 uM | 4.84 uM |
| licarin B | −11.6741 | −9.21127 | −8.69 | −8.53 | −9.7 | −7.9 | 430.11 nM | 558.99 nM |
| sabinene | −12.6372 | −8.408 | −5.96 | −5.08 | −5.7 | −4.8 | 42.93 uM | 189.95 uM |
| β-Pinene | −12.9771 | −9.13638 | −5.43 | −5.61 | −5.5 | −5.1 | 104.34 uM | 77.65 uM |
| α-Pinene | −12.9521 | −8.38199 | −5.57 | −5.55 | −5.5 | −5.0 | 82.61 uM | 84.92 uM |
| terpinen-4-ol | −9.82619 | −7.94893 | −6.51 | −5.1 | −6.0 | −5.1 | 16.85 uM | 183.1 uM |
M. fragrans (Houtt.) is an aromatic evergreen tree with spreading branches and golden fleshy fruits that resemble apricots or peaches that grows to a height of 30–39 feet (1 ft = 30.48 cm). Widely known as nutmeg, it is one of twelve herbs in PSD. There are several additional species of this plant, but M. fragrans is the most common. Both nutmeg and mace are used to flavour a variety of meals, including breakfast cereals, snacks, soups, sweet sauces, fruit juices, and gravies (Hallström and Thuvander, 1997). Because of its essential oil, M. fragrans has been used to treat rheumatoid arthritis and gastrointestinal ailments in China, Malaysia, and Indonesia. Additionally, the administration of M. fragrans extract to hyperlipidaemic rabbits was shown to lower blood lipoprotein lipid levels (Ram et al., 1996).
Many studies have shown that active substances with antiviral effects can be extracted from M. fragrans, such as two neolignan compounds and the well-recognized as resorcinols malabaricone B and malabaricone C, which also possess known antiviral activities (Simpson and Amos, 2017, Rosmalena et al., 2019). Moreover, licarins were reported to have antioxidant (Lin et al., 2015), antiviral (Sawasdee et al., 2013), and neuroprotective activities (Ma et al., 2005).
Our study relies on an in silico approach similar to that used in the early stages of today's state-of-the-art drug discovery pipelines, which includes (1) protein–ligand docking of all M. fragrans bioactive compounds with the binding interfaces of the SARS–CoV-2 main protease Mpro and the receptor binding domain (RBD)–ACE2 complex; (2) ligand pathway simulations of the best predicted compounds from step 1 to evaluate convenient mechanisms by which the compounds can enter the binding site; (3) molecular dynamics (MD) simulations to determine the stability of the best fit protein–ligand complexes from step 1; (4) solvent evaluation to determine the maximum yields of the selected compounds; and (5) calculation of the pharmacokinetic parameters for the most qualified compounds based on the docking protocol's preceding sections. This study shows that M. fragrans-based compounds have antiviral activity and can be used as quick phytotherapeutic alternatives for treating COVID-19.
2. Materials and methods
2.1. Materials
In this investigation, the following software programs were used: i) AutoDock 1.5.6, ii) Python 3.8.2, iii) MGLTools 1.5.4, iv) Discovery Studio 2017, v) Ligplot, vi) ArgusLab 4.0.1, vii) ChemSketch, viii) Avogadro, ix) OpenBabel, and x) COSMOquick for screening the appropriate extraction solvents; xi) SwissADME (a free pharmacokinetics evaluation web tool), and xii) PlayMolecule (a free MD online tool). CavityPlus is a web server that can detect protein cavities. The current investigation was conducted with a system with the following properties: processor, Intel Xeon-E5-2678v3 12C/24 T CPU @ 2.50 GHz −3.10 GHz; system memory, 32 GB RAM DDR4-2133 RECC; graphics processor, VGA GTX 1070 TI 8G; operating system type, 64-bit; and operating system, Windows 10. These prerequisites were specified in the documentation of the abovementioned software.
2.2. Protein structure and setup
The calculated binding affinities of the main constituents of M. fragrans (Table 1) and their capacity to destroy the SARS-CoV-2/ACE2 complex and inhibit SARS-CoV-2 Mpro, a simple therapeutic strategy for anti-coronavirus therapy, were investigated. The crystal structures of the SARS-CoV-2/ACE2 complex and SARS-CoV-2 Mpro were retrieved from the Protein Data Bank (PDB entry codes 7DMU and 7KVL, respectively) (Higuchi et al., 2021, Noske et al., 2021), and Discovery Studio 2017 was used to manually remove all bound ligands, ions, and solvent molecules. AutoDock Tools was used to parameterize the structures of the chosen proteins for docking investigations (Eawsakul et al., 2021, Nasongkla et al., 2021). Polar hydrogens were added to aid in the creation of hydrogen bonds.
2.3. Molecular docking
ArgusLab was used to dock 13 compounds derived from M. fragrans into the target sites of SARS-CoV-2/ACE2 and SARS-CoV-2 Mpro. The binding values were verified using AutoDock Vina, and the binding energies were calculated using the following parameters: the grid box with xyz points was set to a size of 86 × 66 × 80 with grid positions of 39.905, 3.092, and 22.477 and an exhaustiveness number of 20. The xyz points of the SARS-CoV-2 major protease binding pocket were set to 52 × 70 × 64 with grid positions of −26.283, 12.599, and 58.966 and an exhaustiveness number of 20. Furthermore, AutoDock was utilized to confirm the binding affinities using the same box parameters as AutoDock Vina. The Lamarckian genetic algorithm (LGA) was used to find the probable docking conformations of the ligands in the targeted proteins, with the number of GA iterations set to 50. Default values were used for all other parameters (Eawsakul et al., 2021, Nasongkla et al., 2021). The results are expressed in terms of binding energy (kcal/mol), and Eq. (1) was used to predict the 50 % inhibition concentration (IC50). Docking effects were considered when the binding energy values were less than those of panduratin A to the target proteins in at least 1 program.
| (1) |
where Ki is the inhibition constant, which was calculated from Ki = exp(ΔG/R × T); Kd is the dissociation constant; and L is the ligand concentration.
Discovery Studio and Ligplot were used to display the chemical interactions (hydrogen bonds and hydrophobic interactions) between the best-binding ligand and the site on the target protein.
2.4. Analysis of the active pocket binding site
CavityPlus software (Xu et al., 2018) was used to assess the binding location of the ligand–protein combination (pkumdl.cn). Druggability was quantitatively calculated using the binding sites of the entire protein. The druggability value indicates whether a cavity is a good target for binding M. fragrans compounds.
2.5. MD simulations
Starting with the output model from the docking experiments, the ligand–protein complex simulations were carried out using PlayMolecule software (Martínez-Rosell et al., 2017) (playmolecule.com). A ligand was created using the GAFF2 force field (Gianoncelli et al., 2020) and the parametrization function. Using pH = 7.4, the AMBER force field, and the default experimental parameters, the complexes were built for simulation using the ProteinPrepare and SystemBuilder modules (Coghi et al., 2021). SimpleRun was used to perform a 25 ns simulation using the default settings (Gianoncelli et al., 2020, Coghi et al., 2021). The outputs included the root mean square deviation (RMSD) for each frame against time and the root mean square fluctuation (RMSF) of the protein.
2.6. Solvent considerations for selected compounds
Following our previous study (Eawsakul et al., 2021), COSMOquick was used to determine which solvents were suitable for extraction to improve the extraction yields of the selected compounds (Achinivu et al., 2021). Five compounds were evaluated in nine different solvents, including water, ethanol, methanol, n-butanol, dichloromethane (DCM), chloroform, acetone, n-hexane, and diethyl ether. Physical and chemical criteria, such as solubility, dielectric constant, and donor and acceptor properties, were used to classify the solvents. Four factors were evaluated in this investigation for the selected compounds: heat of fusion (Hfus), temperature (T; set to room temperature), and the melting temperature (Tm) of the compounds. Thus, using the COSMOquick mathematical software technique and Eq. (2), the solubilities of the selected compounds in various solvents were computationally predicted.
| (2) |
2.7. Drug-likeness and absorption, distribution, metabolism, excretion, and toxicity (ADMET) prediction for M. Fragrans compounds
Computer-based ADME evaluations have recently gained popularity in drug discovery and development (Ntie-Kang et al., 2013). The SwissADME server (https://www.swissadme.ch/index.php) was used to estimate the pharmacokinetic properties and drug-likeness of the drug candidate molecules (Ranjith et al., 2019). Additionally, the Lipinski, Ghose, and Veber bioavailability criteria and scores were also applied for toxicological predictions (Kadri and Aouadi 2020).
3. Results
3.1. Molecular docking
Plant chemicals have long attracted scientific interest in the development of new drugs. Despite the use of various antiviral medications to treat COVID-19, experimental and clinical therapeutic development investigations still take a long time. In this study, molecular docking revealed robust interactions between certain compounds and SARS-CoV-2/ACE2 and SARS-CoV-2 Mpro. The scoring function based on the AScore and LGA was used to determine the binding strength. Electrostatic, hydrogen bonding, and van der Waals interactions may all contribute to the binding free energy (Mihajlovic and Mitrasinovic 2009). The protein–ligand complex with the lowest binding energy reflects the greatest stability. Table 1 shows the binding energy results, which were determined by ArgusLab, AutoDock, and AutoDock Vina. Discovery Studio 2017 (Biovia 2017) and Ligplot were used to visualize all of the docked structures (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, Fig. 12). Panduratin A, a positive control, binds with high affinity to SARS-CoV-2/ACE2 and SARS-CoV-2 Mpro (-13.9 and −10.7 kcal/mol, respectively, as determined by ArgusLab software; −7.2 and −7.0, respectively, as determined by AutoDock software; and −9.3 and −6.6, respectively, as determined by AutoDock Vina software).
Fig. 1.
Binding position of licarin A (purple), licarin B (yellow), licarin C (red), and positive control as panduratin A (green) on ACE2.
Fig. 2.
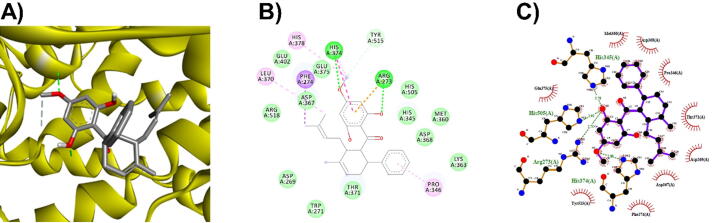
3D (A), 2D (B) interaction visualization of panduratinA binding ACE2 with discovery studio and 2D (C) visualization with Ligplot.
Fig. 3.
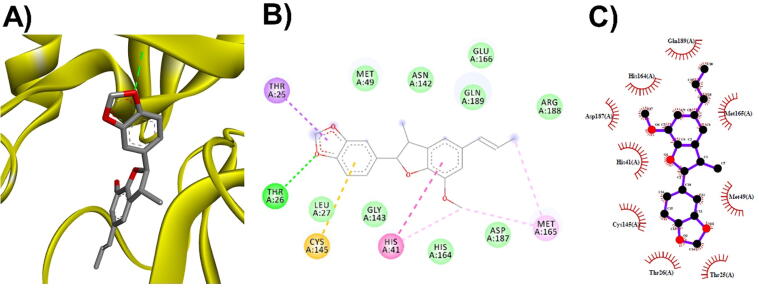
3D (A), 2D (B) interaction visualization of licarin A binding ACE2 with discovery studio and 2D (C) visualization with Ligplot.
Fig. 4.
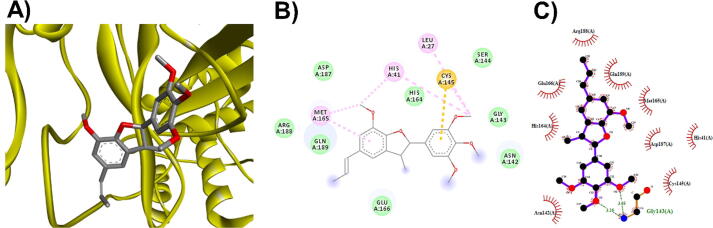
3D (A), 2D (B) interaction visualization of licarin B binding ACE2 with discovery studio and 2D (C) visualization with Ligplot.
Fig. 5.
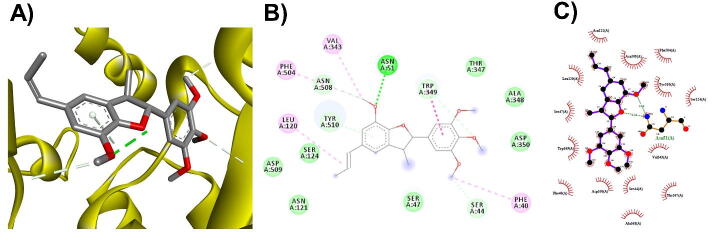
3D (A), 2D (B) interaction visualization of licarin C binding ACE2 with discovery studio and 2D (C) visualization with Ligplot.
Fig. 6.
Binding position of licarin A (purple), licarin B (yellow), licarin C (red), malabaricone B (orange), malabaricone C (sky blue) and positive control as panduratin A (green) on Mpro.
Fig. 7.
3D (A), 2D (B) interaction visualization of panduratin A binding Mpro with discovery studio and 2D (C) visualization with Ligplot.
Fig. 8.
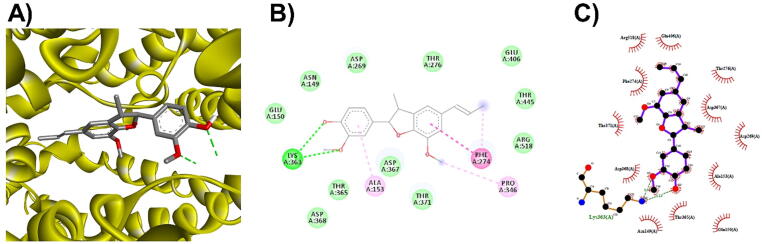
3D (A), 2D (B) interaction visualization of licarin A binding Mpro with discovery studio and 2D (C) visualization with Ligplot.
Fig. 9.
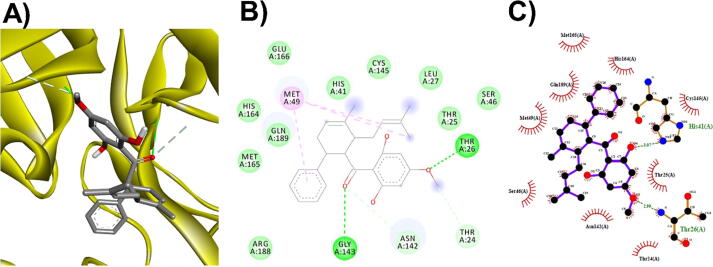
3D (A), 2D (B) interaction visualization of licarin B binding Mpro with discovery studio and 2D (C) visualization with Ligplot.
Fig. 10.
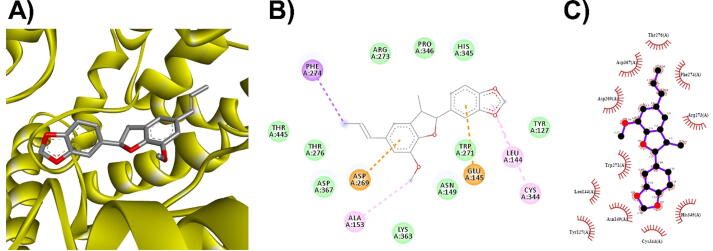
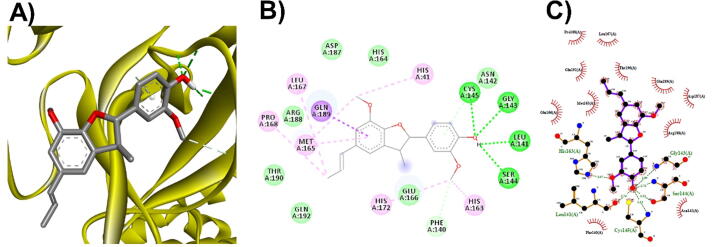
3D (A), 2D (B) interaction visualization of licarin C binding Mpro with discovery studio and 2D (C) visualization with Ligplot.
Fig. 11.
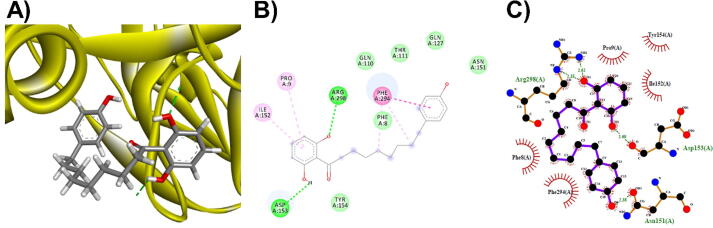
3D (A), 2D (B) interaction visualization of malabaricone B binding Mpro with discovery studio and 2D (C) visualization with Lgplot.
Fig. 12.
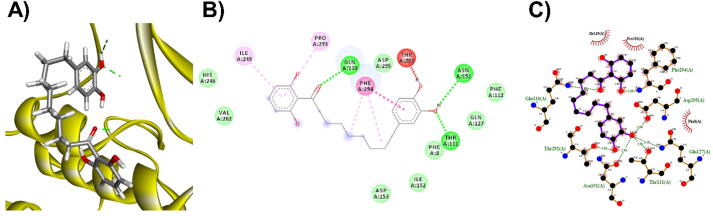
3D (A), 2D (B) interaction visualization of malabaricone C binding Mpro with discovery studio and 2D (C) visualization with Ligplot.
3.2. MD simulations
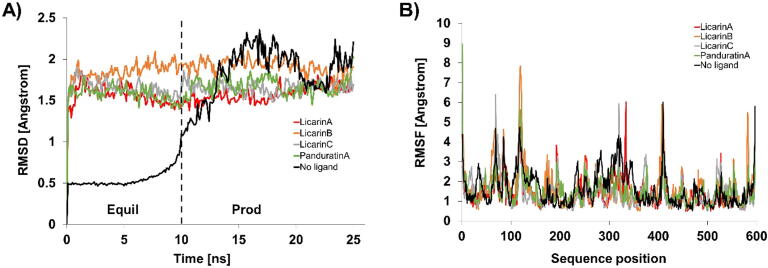
Since molecular docking studies of compound candidates binding to target proteins may not be sufficient, the use of MD is very important. MD is performed to investigate the stability of ligand–protein complexes under a system designed to approximate the conditions in the human body. In this study, stability and flexibility were determined by calculating the RMSD and RMSF values, respectively. During simulations, high deviations and protein fluctuation can indicate a lack of stability.
According to the MD results, the RMSD values of ACE2 with licarins A, B, and C seem to be stable compared to those of ACE2 without a ligand. The initial RMSDs after the first 10 ns and the final RMSDs for SARS-CoV-2/ACE2 in complex with panduratin A as a positive control were 1.537 and 2.026 nm, respectively, over the entire simulation period (range: 0.489 nm). Thus, good chemical candidates should have an RMSD range of less than 0.489 nm. During the simulation period, the initial RMSD after the first 10 ns and the final RMSDs for the ACE2 protein-licarin A complex were 1.445 and 1.681 nm, respectively (range: 0.236 nm). The corresponding values for the ACE2 protein-licarin B complexes were 1.956 and 1.907 nm, respectively (range: 0.049 nm), and those of the ACE2 protein-licarin C complex were 1.718 and 1.694 nm, respectively (range: 0.024 nm). Thus, the RMSD values of licarins A, B, and C were smaller than that of the positive control.
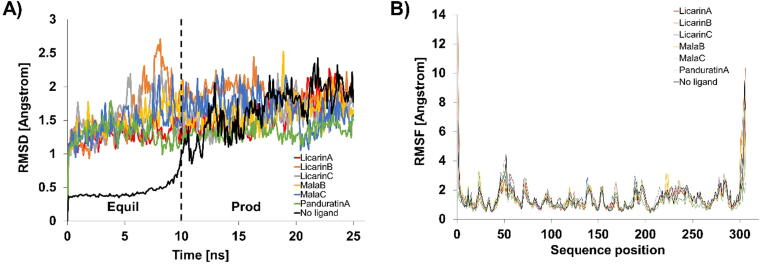
The residual atom fluctuations of the ACE2-panduratin A complex (positive control) were found to be in the range of 0.56–8.95 nm (fluctuation: 8.39 nm), as illustrated in Fig. 13B. On the other hand, licarins A, B, and C had ranges of 0.49–6.02 nm (fluctuation: 5.53 nm), 0.51–7.85 (fluctuation: 7.34 nm), and 0.50–6.36 nm (fluctuation: 5.85 nm), respectively. The residual atom fluctuations of Mpro with panduratin A were found to lie in the range of 0.47–9.52 nm (fluctuation: 9.05 nm), as illustrated in Fig. 14B. In contrast, licarin A, licarin C, malabaricone B and malabaricone C in complex with Mpro were found to have residual atom fluctuation ranges of 0.45–7.71 nm (fluctuation: 7.26 nm), 0.38–7.43 nm (fluctuation: 7.05 nm), 0.40–7.52 nm (fluctuation: 7.12 nm), and 0.46–7.87 nm (fluctuation: 7.41 nm), respectively.
Fig. 13.
Analysis of RMSD and RMSF of the best licarin A (red), licarin B (orange), licarin C (gray), panduratin A as positive control (green) on ACE-2 (black). A) RMSD B) RMSF.
Fig. 14.
Analysis of RMSD and RMSF of the best licarin A (red), licarin B (orange), licarin C (gray), MalabaricineB (yellow), malabariconeC (blue), panduratin A as positive control (green) on Mpro (black). A) RMSD B) RMSF.
In addition to inhibiting COVID-19 through its interaction with ACE2, the inhibition mechanism of Mpro is also important for effectively controlling COVID-19. Therefore, stability studies of the binding between the compound candidates and Mpro need to be investigated. The RMSD values of Mpro with licarins A, B, and C and malabaricones B and C seemed to be stable compared to that of Mpro with no ligand. The initial RMSDs after the first 10 ns and the final RMSDs for Mpro complexed with panduratin A as a positive control were 1.22 and 1.50 nm, respectively, over the entire simulation period (range: 0.28 nm). As a result, good chemical candidates should have an RMSD range of less than 0.28 nm. During the simulation period, the initial RMSDs after the first 10 ns and the final RMSDs for the Mpro-licarin B complex were 1.91 and 1.82 nm, respectively (range: 0.09 nm). The Mpro-licarin C complex had values of 1.52 and 1.70 nm (range: 0.18 nm), and the Mpro-malabaricone C complex had values of 1.44 and 1.58 nm (0.14 nm), respectively. Thus, the RMSD values of licarin B, licarin C, and malabaricone C were smaller than that of the positive control.
3.3. Solvent considerations for extraction of the selected compounds
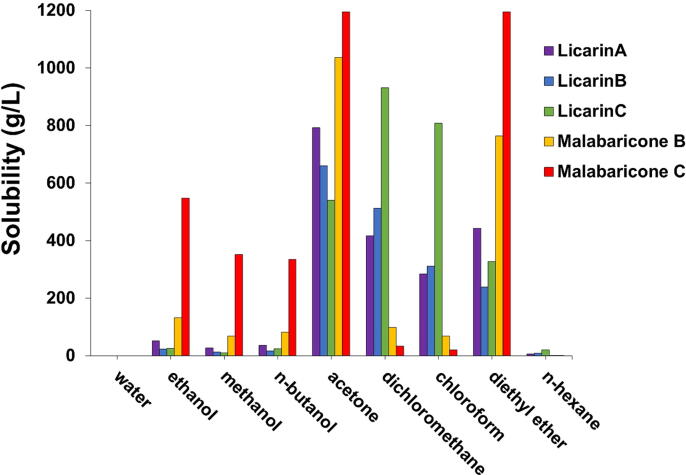
Eq. (2) was used to find the optimum solubilities of the selected compounds. The highest extraction yields of licarin A, licarin B, malabaricone B, and malabaricone C occurred in acetone, as shown in Fig. 15. DCM extraction yielded the largest amount of licarin C. Although licarin C has lower solubility in acetone than DCM, its solubility in acetone (540 g/L) is nevertheless good. With decreasing iLOGP, solubility showed an increasing trend, as expected. Table 2 shows that the iLOGP values of malabaricone B and malabaricone C indicate that they are the most soluble in acetone, with values ranging from 1,036 to 1,194 g/L, and the iLOGP values of licarins A, B, and C also indicate this, with values ranging from 540 to 792 g/L.
Fig. 15.
Correlations between extraction with various solvents and the corresponding yield.
Table 2.
Lipophilicity of the Phytoconstituents of Myristica fragrans (houtt.).
| SI. No. | Small molecule | iLOGP | XLOGP3 | WLOGP | MLOGP | SILICOS-IT | Consensus Log Po/w |
|---|---|---|---|---|---|---|---|
| 1 | licarin A | 3.70 | 4.38 | 4.25 | 2.73 | 4.36 | 3.88 |
| 2 | licarin B | 3.89 | 4.58 | 4.26 | 3.13 | 4.67 | 4.11 |
| 3 | licarin C | 4.28 | 4.68 | 4.56 | 2.61 | 4.98 | 4.22 |
| 4 | malabaricone B | 2.70 | 6.05 | 4.96 | 3.08 | 5.01 | 4.36 |
| 5 | malabaricone C | 2.25 | 5.69 | 4.67 | 2.51 | 4.54 | 3.93 |
3.4. Solubility of the compounds derived from M. Fragrans
Lipophilicity and hydrophilicity can be used to predict a compound's solubility because they reflect the single most informative and successful physicochemical characteristic in medicinal chemistry (Malinowska et al., 2019). Therefore, lipophilicity is a critical metric in drug discovery and design (Freeman-Cook et al., 2013). Lipophilicity is expressed experimentally as the partition coefficient (log P), which is the partition equilibrium of the unionized solute in the presence of water and an immiscible organic solvent. Greater lipophilicity is associated with a higher log P value (Arnott and Planey, 2012). This work used six available models to examine the lipophilicity characteristics of compounds, namely, iLOGP, XLOGP3, WLOGP, MLOGP, SILICOS-IT, and Log Po/w. Four of the six models yielded the same results, which are sorted in descending order by their ability to dissolve in lipids: malabaricone B, malabaricone C, licarin C, licarin B, and licarin A. The solubility results are given in Table 3. The ESOL model showed results consistent with the lipophilicity: licarin A, licarin B, licarin C, malabaricone C, and malabaricone B, ranked from highest to lowest solubility in water.
Table 3.
Water solubility of the Phytoconstituents of Myristica fragrans (houtt.).
| Small molecule |
ESOL |
Ali |
SILICOS-IT |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log S (ESOL) |
Solubility |
Log S (ESOL) |
Solubility |
Log S (ESOL) |
Solubility |
|||||||
| mg/mL | mol/L | Class | mg/mL | mol/L | class | mg/mL | mol/L | class | ||||
| Licarin A | −4.73 | 6.09e-03 | 1.87e-05 | Moderately soluble | −5.1 | 2.58e-03 | 7.90e-06 | Moderately soluble | −5.26 | 1.77e-03 | 5.43e-06 | Moderately soluble |
| Licarin B | −4.91 | 4.00e-03 | 1.23e-05 | Moderately soluble | −5.08 | 2.70e-03 | 8.33e-06 | Moderately soluble | −5.47 | 1.11e-03 | 3.42e-06 | Moderately soluble |
| Licarin C | −5.02 | 3.55e-03 | 9.59e-06 | Moderately soluble | −5.38 | 1.56e-03 | 4.20e-06 | Moderately soluble | −6.07 | 3.18e-04 | 8.59e-07 | Poorly soluble |
| Malabaricone B | −5.47 | 1.16e-03 | 3.39e-06 | Moderately soluble | −7.46 | 1.18e-05 | 3.45e-08 | Poorly soluble | −6.34 | 1.56e-04 | 4.55e-07 | Poorly soluble |
| Malabaricone C | −5.33 | 1.68e-03 | 4.69e-06 | Moderately soluble | −7.51 | 1.10e-05 | 3.07e-08 | Poorly soluble | −5.76 | 6.29e-04 | 1.75e-06 | Moderately soluble |
3.5. Pharmacokinetics of the compounds from M. Fragrans
The pharmacokinetics of the aforementioned drugs are predictably shown in Table 4. Notably, CYP enzymes could be associated with reduced medication clearance from the body. As a result, drug-associated CYP inhibition may play a role in lowering drug metabolism (Almazroo et al., 2017). In Table 4, inhibition of CYP2C19, CYP2C9, and CYP2D6 was shown to be related to increases in the licarin A, licarin B, licarin C, and malabaricone B contents, helping to keep those compounds in the bloodstream longer by reducing drug metabolism in the liver. Licarin B and malabaricone B can both inhibit CYP3A4, whereas malabaricone B and malabaricone C can also inhibit CYP1A2.
Table 4.
Pharmacokinetic Parameters of the Phytoconstituents of Myristica fragrans (houtt.).
| Small molecule | GI absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Log Kp (cm/s) |
|---|---|---|---|---|---|---|---|---|---|
| Licarin A | High | Yes | No | No | Yes | Yes | Yes | No | −5.18 |
| Licarin B | High | Yes | No | No | Yes | Yes | Yes | Yes | −5.03 |
| Licarin C | High | Yes | No | No | Yes | Yes | Yes | No | −5.24 |
| Malabaricone B | High | No | No | Yes | Yes | Yes | Yes | Yes | −4.09 |
| Malabaricone C | High | No | No | Yes | No | Yes | Yes | No | −4.45 |
3.6. Drug-likeness of the phytoconstituents of M. Fragrans
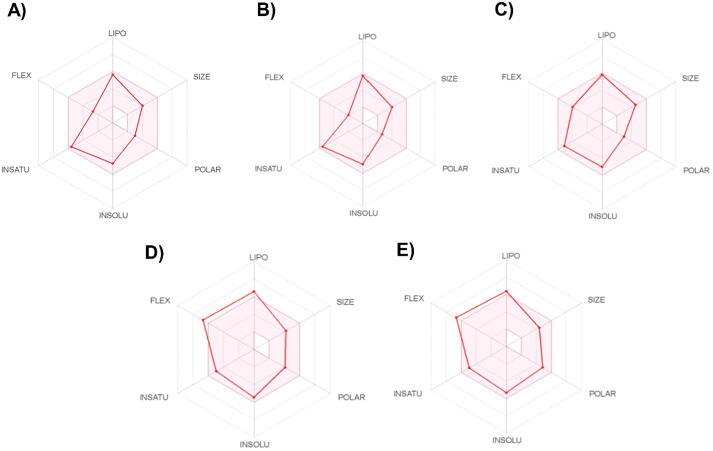
Fig. 16 depicts the oral bioavailability predictions for the compounds based on six physicochemical characteristics. The red lines depict the properties of licarin A, licarin B, licarin C, malabaricone B, and malabaricone C, while the pink area represents the ideal range of these features (Lovering et al., 2009, Ritchie et al., 2011). The red lines of three compounds (licarins A, B, and C) in Fig. 16A-16C lie within the pink area. As a result, we can deduce that these compounds are orally bioavailable.
Fig. 16.
the bioavailability radar of the compounds A) licarinA, B) licarinB, C) licarin C, D) malabaricone B, and E) malabaricone C for six physicochemical characteristics.
4. Discussion
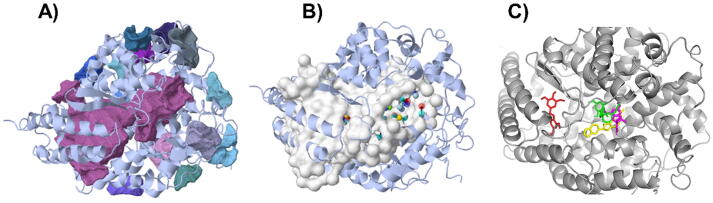
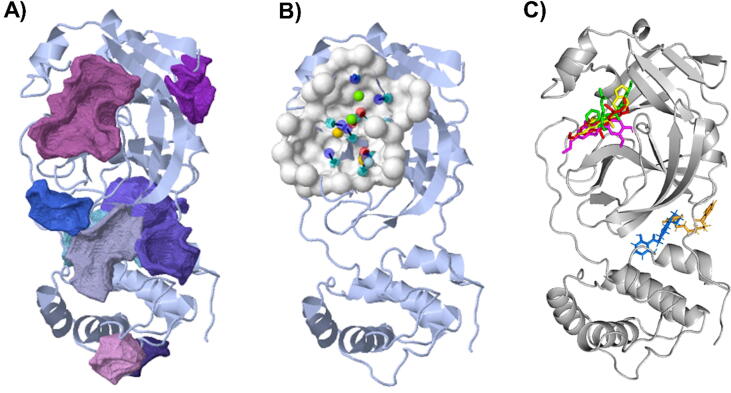
The M. fragrans phytochemicals under consideration here must have a lower binding energy value than that of the positive control after analysis by each software utilized. The results showed that a total of five active substances from M. fragrans had lower binding energies than the positive control, as shown in Table 1. Binding pocket analysis of ACE2 assessed by CavityPlus software revealed that ACE2 had a total of 15 pocket binding sites, as shown in Fig. 1A. However, only one of these sites had strong druggability, as shown in Fig. 1B, and the other 14 were found to have low druggability. Three compounds, licarins A, B and C, were able to bind to the highly druggable site of ACE2, as shown in Fig. 1C. In addition to binding to the proper position of ACE2, all three compounds were able to bind to the target protein strongly based on the chemical interactions. Licarin A can form 2 hydrogen bonds (LYS363) and 4 hydrophobic interactions (ALA153, PRO346, and PHE274) with ACE2, as shown in Fig. 3. Licarin B can form 6 hydrophobic interactions (PHE274, ASP269, ALA153, GLU145, LEU144, and CYS344) with ACE2, as shown in Fig. 4. Licarin C can form 5 hydrogen bonds (ASN51, ASN508, TYR510, SER44, and TRP349) and 5 hydrophobic interactions (VAL343, PHE504, LEU120, PHE40, and TRP349) with ACE2, as shown in Fig. 5. In addition to the inhibition of COVID-19 via ACE2, there is a main protease (Mpro) inhibition mechanism that plays an important role in suppressing COVID-19. In this study, binding pocket analysis displayed a total of 9 binding sites on Mpro, as shown in Fig. 6A. However, only one of the Mpro sites had medium druggability, as shown in Fig. 6B, whereas the other 8 had low druggability. Three compounds, licarins A, B and C, were able to bind to the medium druggable site of Mpro, as shown in Fig. 6C. Two compounds, malabaricones B and C, were able to bind to sites with low druggability. The abilities of these compounds to suppress COVID-19 through Mpro binding were mediated by several interactions with the target protein. Licarin A can form 7 hydrogen bonds (CYS145, GLY143, LEU141, SER144, HIS163, and PHE140) and 8 hydrophobic interactions (LEU167, GLN189, PRO168, MET165, HIS41, HIS163, and HIS172) with Mpro, as shown in Fig. 8. Licarin B can form 1 hydrogen bond (THR26) and 6 hydrophobic interactions (THR25, CYS145, HIS41, and MET165) with Mpro, as shown in Fig. 9. Licarin C can form 1 hydrogen bond (GLY143) and 7 hydrophobic interactions (MET165, HIS41, LEU27, CYS145) with Mpro, as shown in Fig. 10. Malabaricone B can form 3 hydrogen bonds (ASP153, ARG298, and ASN151) and 5 hydrophobic interactions (ILE152, PRO9, and PHE294) with Mpro, as shown in Fig. 11. Malabaricone C can form 7 hydrogen bonds (GLN110, GLN127, ASN151, ASP295, PHE294, THR111, and THR292) and 5 hydrophobic interactions (ILE249, PRO293, and PHE294) with Mpro, as shown in Fig. 12. Although the binding sites of malabaricones B and C with Mpro are sites with low druggability, the binding of both compounds with Mpro involves abundant hydrogen bonds. This reflects the ability of both compounds to strongly bind to Mpro. Thus, it was concluded that licarins A, B, and C and malabaricones A and B have the potential to inhibit Mpro.
Although molecular docking studies of the binding between compound candidates and the target proteins may not be sufficient, the use of MD is very important. The findings here demonstrate that these compound candidates bind stably to ACE2, suggesting that they could act as inhibitors. Furthermore, changes in the protein residues upon binding to the compound candidates were identified, indicating that they were not very flexible. To infer good binding affinities, stable RMSD and RMSF values are needed. The residual atom fluctuations of ACE2-panduratin A (positive control) were found to be in the range of 0.56–8.95 nm (fluctuation: 8.39 nm), as illustrated in Fig. 13B. On the other hand, licarins A, B, and C had ranges of 0.49–6.02 nm (fluctuation: 5.53 nm), 0.51–7.85 nm (fluctuation: 7.34 nm), and 0.50–6.36 nm (fluctuation: 5.85 nm), respectively. During the simulation period, the ACE2 fluctuations with licarins A, B, and C were minimal compared to that of the positive control. This result might allow the deduction of their high binding affinity during the simulation period, demonstrating that these compound candidates bind stably to Mpro and suggesting that they could be beneficial as inhibitors. Furthermore, changes in the protein residues were identified upon binding to the compound candidates, indicating that these residues were not very flexible. To infer good binding affinities, stable RMSD and RMSF values are needed. The residual atom fluctuations of Mpro bound to panduratin A were found to lie in the range of 0.47–9.52 nm (fluctuation: 9.05 nm), as illustrated in Fig. 14B. In contrast, licarin A, licarin C, malabaricone B and malabaricone C were found to have ranges of 0.45–7.71 nm (fluctuation: 7.26 nm), 0.38–7.43 nm (fluctuation: 7.05 nm), 0.40–7.52 nm (fluctuation: 7.12 nm), and 0.46–7.87 nm (fluctuation: 7.41 nm), respectively. During the simulation period, the Mpro fluctuations after binding licarin A, licarin C, malabaricone B and malabaricone C were minimal, in the range between those for no ligand and with panduratin A. Thus, these results indicated high binding affinities for the abovementioned compounds during the simulation period.
Our results further demonstrated that acetone was a good choice for M. fragrans extraction to obtain large quantities of the five active compounds. These compounds had moderate solubility in water, and all are suitable for oral delivery due to their high GI absorption and CYP inhibition.
To select oral drug candidates, drug-likeness was determined based on the physicochemical properties of the compounds (Daina et al., 2017). Rule-based filters were divided into the following four categories.
1) The following parameters are included in Lipinski’s filter: number of hydrogen bond acceptors of less than or equal to 10, number of hydrogen bond donors of less than or equal to 5, MLOGP (lipophilicity) of less than 4.15, and molecular weight of less than or equal to 500 (Lipinski et al., 1997).
2) The following parameters are included in Veber’s filter: total polar surface area of less than or equal to 140 and a number of rotatable bonds of less than or equal to 10 are included in Veber's filter (Veber et al., 2002).
3) The following parameters are included in Egan’s filter: total polar surface area of less than or equal to 131.6 and WLOGP (lipophilicity) of less than or equal to 5.88 are included in Egan's filter (Egan et al., 2000).
4) The following parameters are included in Muegge's filter: number of hydrogen bond donors of less than or equal to 5, number of hydrogen bond acceptors of less than or equal to 10, number of rotatable bonds of less than or equal to 15, number of heteroatoms of more than 1, number of carbon atoms of more than 4, number of rings of less than or equal to 7, total polar area of less than or equal to 150, XLOGP3 (lipophilicity) o between −2 and 5, and molecular weight between 200 and 600 (Muegge et al., 2001).
Table 5 shows the results of the drug-likeness evaluation of licarin A, licarin B, licarin C, malabaricone B, and malabaricone C, and we can deduce the following points.
Table 5.
Drug likeness of the Phytoconstituents of Myristica fragrans (houtt.).
| Small molecule | Lipinski | Veber | Egan | Muegge |
Bioavailability score |
|---|---|---|---|---|---|
| Licarin A | Yes; 0 violation | Yes | Yes | Yes | 0.55 |
| Licarin B | Yes; 0 violation | Yes | Yes | Yes | 0.55 |
| Licarin C | Yes; 0 violation | Yes | Yes | Yes | 0.55 |
| Malabaricone B | Yes; 0 violation | Yes | Yes | No: 1 violation XLOGP3 greater than 5 | 0.55 |
| Malabaricone C | Yes; 0 violation | Yes | Yes | No: 1 violation XLOGP3 greater than 5 | 0.55 |
1) All of the compounds (licarin A, licarin B, licarin C, malabaricone B, and malabaricone C) were in accordance with Lipinski's rule (Chen et al., 2020).
2) All of the compounds were in accordance with Veber's and Egan's rules (Oyinloye et al., 2021).
3) All of the compounds, with the exception of malabaricones B and C, which have lipophilicity (XLOGP3) values greater than 5, satisfy Muegge's criteria (Muegge, 2003).
The general study of the antiviral activities of alternative medicines has been used in the past for treatment. Most of the herbs used to treat viruses have a bitter taste or can be studied from a similar genus. The herb used in this research is one from traditional Thai medicine that has been used to treat fever for a long time. Another well-known herb that has been reported to treat fever, Tinospora crispa (Rakib et al., 2020), is also effective against COVID-19. However, the results showed that no active substance from T. crispa was able to treat this coronavirus better than lopinavir. In addition, the highest number of hydrogen bonds formed with T. crispa extract was only 3. However, most substances extracted from M. fragrans can form more than three hydrogen bonds.
5. Conclusion
This study performed molecular docking of compounds extracted from M. fragrans, one of twelve herbs in PSD, to the targets SARS-CoV-2 Mpro and the SARS-CoV-2/ACE2 interaction. Licarins A, B, and C and malabaricones B and C had higher binding affinities (less than −7.0 kcal/mol) than other compounds as determined by each program utilized. Furthermore, the binding affinities of these five active compounds were higher than that of panduratin A, the natural compound and positive control. Both ACE2 and Mpro were predicted to be inhibited by licarin A and licarin B. Mpro was predicted to be inhibited by licarin C, malabaricone B, and malabaricone C. Licarin A and licarin B bind to the same ACE2 location as panduratin A, according to the active site study. Although licarin C binds to ACE2 in a different position, it has a low binding affinity.
As a result of their frequent involvement in ligand–protein binding, ALA153, PRO346, PHE274, and PRO346 are among the most important amino acid residues in ACE2. Licarins A, B, and C bound at the same Mpro site as panduratin A. Malabaricones B and C bound to Mpro at sites different from that of panduratin A, resulting in low binding affinities. At the active binding site, THR26, GLY143, and MET165 are among the important amino acids of Mpro. Furthermore, during the MD simulations period, licarins A, B, and C stably bound to ACE2 with low RMSD and RMSF values. Additionally, licarins A, B and C and malabaricones B and C showed strong binding affinity to Mpro with low RMSDs and RMSFs during the simulation time.
Most of the compounds (licarin A, licarin B, licarin C, malabaricone B, and malabaricone C) met the criteria of Lipinski, Veber, and Egan, and all but malabaricones B and C met the Muegge recommendations. According to the oral bioavailability predictions, these early data imply that, with the exception of malabaricone C, these compounds might be utilized to treat COVID-19 orally.
Acetone was used to extract M. fragrans, which yielded high levels of licarins A, B and C and malabaricones B and C. As a result, the acetone extract of M. fragrans may aid in the prevention of COVID-19. However, more in vivo experimental research is needed to confirm our findings and develop more effective compounds for COVID-19 prevention and treatment.
Funding
This research was financially supported by the new strategic research project (P2P) fiscal year 2022, Walailak University, Thailand. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank RECIHP for providing the facilities and materials that helped to complete this research. This research was financially supported by the new strategic research project (P2P) fiscal year 2022, Walailak University, Thailand.
Footnotes
Peer review under responsibility of King Saud University.
References
- Achinivu, E.C., Mohan, M., Choudhary, H., Das, L., Huang, K., Magurudeniya, H.D., Pidatala, V.R., George, A., Simmons, B.A., Gladden, J.M.J.G.C., 2021. A predictive toolset for the identification of effective lignocellulosic pretreatment solvents: A case study of solvents tailored for lignin extraction.
- Almazroo, O.A., Miah, M.K., Venkataramanan, R.J.C.i.l.d., 2017. Drug metabolism in the liver. [DOI] [PubMed]
- Antonio, A., Wiedemann, L., Galante, E., Guimarães, A., Matharu, A., Veiga-Junior, V.J.H., 2021. Efficacy and sustainability of natural products in covid-19 treatment development: Opportunities and challenges in using agro-industrial waste from citrus and apple. [DOI] [PMC free article] [PubMed]
- Arnott, J.A., Planey, S.L.J.E.o.o.d.d., 2012. The influence of lipophilicity in drug discovery and design. [DOI] [PubMed]
- Biovia D.S. Discovery studio visualizer. San Diego, CA, USA. 2017 [Google Scholar]
- Chen, X., Li, H., Tian, L., Li, Q., Luo, J., Zhang, Y.J.J.o.c.b., 2020. Analysis of the physicochemical properties of acaricides based on lipinski's rule of five. [DOI] [PubMed]
- Coghi, P., Yang, L.J., Ng, J.P., Haynes, R.K., Memo, M., Gianoncelli, A., Wong, V.K.W., Ribaudo, G.J.P., 2021. A drug repurposing approach for antimalarials interfering with sars-cov-2 spike protein receptor binding domain (rbd) and human angiotensin-converting enzyme 2 (ace2). [DOI] [PMC free article] [PubMed]
- Coronavirus, W. H. O. (2021). Dashboard (https://covid19.who.int/). (Accessed 14 December 2021).
- Daina, A., Michielin, O., Zoete, V.J.S.r., 2017. Swissadme: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. [DOI] [PMC free article] [PubMed]
- Dandara, C., Dzobo, K., Chirikure, S.J.O.A.J.o.I.B., 2021. Covid-19 pandemic and africa: From the situation in zimbabwe to a case for precision herbal medicine. [DOI] [PubMed]
- Eawsakul, K., Panichayupakaranant, P., Ongtanasup, T., Warinhomhoun, S., Noonong, K., Bunluepuech, K.J.H., 2021. Computational study and in vitro alpha-glucosidase inhibitory effects of medicinal plants from a thai folk remedy. [DOI] [PMC free article] [PubMed]
- Egan, W.J., Merz, K.M., Baldwin, J.J.J.J.o.m.c., 2000. Prediction of drug absorption using multivariate statistics. [DOI] [PubMed]
- Freeman-Cook, K.D., Hoffman, R.L., Johnson, T.W.J.F.m.c., 2013. Lipophilic efficiency: The most important efficiency metric in medicinal chemistry. [DOI] [PubMed]
- Gianoncelli, A., Ongaro, A., Zagotto, G., Memo, M., Ribaudo, G.J.M., 2020. 2-(3, 4-dihydroxyphenyl)-4-(2-(4-nitrophenyl) hydrazono)-4h-chromene-3, 5, 7-triol.
- Hallström, H., Thuvander, A.J.N.t., 1997. Toxicological evaluation of myristicin. [DOI] [PubMed]
- Higuchi, Y., Suzuki, T., Arimori, T., Ikemura, N., Mihara, E., Kirita, Y., Ohgitani, E., Mazda, O., Motooka, D., Nakamura, S.J.N.C., 2021. Engineered ace2 receptor therapy overcomes mutational escape of sars-cov-2. [DOI] [PMC free article] [PubMed]
- Kadri, A., Aouadi, K.J.J.A.P.S., 2020. In vitro antimicrobial and α-glucosidase inhibitory potential of enantiopure cycloalkylglycine derivatives: Insights into their in silico pharmacokinetic, druglikeness, and medicinal chemistry properties.
- Kanjanasirirat, P., Suksatu, A., Manopwisedjaroen, S., Munyoo, B., Tuchinda, P., Jearawuttanakul, K., Seemakhan, S., Charoensutthivarakul, S., Wongtrakoongate, P., Rangkasenee, N.J.S.r., 2020. High-content screening of thai medicinal plants reveals boesenbergia rotunda extract and its component panduratin a as anti-sars-cov-2 agents. [DOI] [PMC free article] [PubMed]
- Lin, S.Y., Ko, H.H., Lee, S.J., Chang, H.S., Lin, C.H., Chen, I.S.J.C., biodiversity, 2015. Biological evaluation of secondary metabolites from the root of machilus obovatifolia. [DOI] [PubMed]
- Lipinski, C.A., Lombardo, F., Dominy, B.W., Feeney, P.J.J.A.d.d.r., 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. [DOI] [PubMed]
- Lovering, F., Bikker, J., Humblet, C.J.J.o.m.c., 2009. Escape from flatland: Increasing saturation as an approach to improving clinical success. [DOI] [PubMed]
- Ma, C.J., Kim, S.R., Kim, J., Kim, Y.C.J.B.j.o.p., 2005. Meso-dihydroguaiaretic acid and licarin a of machilus thunbergii protect against glutamate-induced toxicity in primary cultures of a rat cortical cells. [DOI] [PMC free article] [PubMed]
- Malinowska, M., Miroslaw, B., Sikora, E., Ogonowski, J., Wojtkiewicz, A.M., Szaleniec, M., Pasikowska-Piwko, M., Eris, I.J.P.o., 2019. New lupeol esters as active substances in the treatment of skin damage. [DOI] [PMC free article] [PubMed]
- Martínez-Rosell, G., Giorgino, T., De Fabritiis, G.J.J.o.c.i., modeling, 2017. Playmolecule proteinprepare: A web application for protein preparation for molecular dynamics simulations. [DOI] [PubMed]
- Mihajlovic, M.L., Mitrasinovic, P.M.J.M.S., 2009. Applications of the arguslab4/ascore protocol in the structure-based binding affinity prediction of various inhibitors of group-1 and group-2 influenza virus neuraminidases (nas).
- Muegge, I., Heald, S.L., Brittelli, D.J.J.o.m.c., 2001. Simple selection criteria for drug-like chemical matter. [DOI] [PubMed]
- Muegge, I.J.M.r.r., 2003. Selection criteria for drug‐like compounds. [DOI] [PubMed]
- Nasongkla, N., Tuchinda, P., Munyoo, B., Eawsakul, K.J.E.-B.C., Medicine, A., 2021. Preparation and characterization of muc-30-loaded polymeric micelles against mcf-7 cell lines using molecular docking methods and in vitro study. [DOI] [PMC free article] [PubMed]
- Noske, G., Nakamura, A., Gawriljuk, V., Fernandes, R., Lima, G., Rosa, H., Pereira, H., Zeri, A., Nascimento, A., Freire, M.J.b., 2021. A crystallographic snapshot of sars-cov-2 main protease maturation process. [DOI] [PMC free article] [PubMed]
- Ntie-Kang, F., Lifongo, L.L., Mbah, J.A., Owono, L.C.O., Megnassan, E., Mbaze, L.M.a., Judson, P.N., Sippl, W., Efange, S.M.J.I.s.p., 2013. In silico drug metabolism and pharmacokinetic profiles of natural products from medicinal plants in the congo basin. [DOI] [PMC free article] [PubMed]
- Oyinloye, B.E., Iwaloye, O., Ajiboye, B.O.J.S.A., 2021. Polypharmacology of gongronema latifolium leaf secondary metabolites against protein kinases implicated in parkinson's disease and alzheimer's disease.
- Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., Kumar, S., Bhattacharyya, A., Kumar, H., Bansal, S.J.I.j.o.p., 2020. Drug targets for corona virus: A systematic review. [DOI] [PMC free article] [PubMed]
- Prommee, N., Itharat, A., Panthong, S., Makchuchit, S., Ooraikul, B.J.J.o.E., 2021. Ethnopharmacological analysis from thai traditional medicine called prasachandaeng remedy as a potential antipyretic drug. [DOI] [PubMed]
- Rakib, A., Paul, A., Chy, M., Uddin, N., Sami, S.A., Baral, S.K., Majumder, M., Tareq, A.M., Amin, M.N., Shahriar, A.J.M., 2020. Biochemical and computational approach of selected phytocompounds from tinospora crispa in the management of covid-19. [DOI] [PMC free article] [PubMed]
- Ram, A., Lauria, P., Gupta, R., Sharma, V.J.J.o.e., 1996. Hypolipidaemic effect of myristica fragrans fruit extract in rabbits. [DOI] [PubMed]
- Ranjith, D., Ravikumar, C.J.J.o.P., Phytochemistry, 2019. Swissadme predictions of pharmacokinetics and drug-likeness properties of small molecules present in ipomoea mauritiana jacq.
- Ritchie, T.J., Ertl, P., Lewis, R.J.D.D.T., 2011. The graphical representation of adme-related molecule properties for medicinal chemists. [DOI] [PubMed]
- Rosmalena, R., Elya, B., Dewi, B.E., Fithriyah, F., Desti, H., Angelina, M., Hanafi, M., Lotulung, P.D., Prasasty, V.D., Seto, D.J.P., 2019. The antiviral effect of indonesian medicinal plant extracts against dengue virus in vitro and in silico. [DOI] [PMC free article] [PubMed]
- Sawasdee, K., Chaowasku, T., Lipipun, V., Dufat, T.-H., Michel, S., Likhitwitayawuid, K.J.T.L., 2013. New neolignans and a lignan from miliusa fragrans, and their anti-herpetic and cytotoxic activities.
- Series, W.H.O.J.W.T.R., 2018. Who guidelines on good herbal processing practices for herbal medicines.
- Sheng-Ji, P.J.P.b., 2001. Ethnobotanical approaches of traditional medicine studies: Some experiences from asia. [DOI] [PubMed]
- Simpson D., Amos S. Elsevier; Pharmacognosy: 2017. Other plant metabolites; pp. 267–280. [Google Scholar]
- Singh, Y.D., Panda, M.K., Satapathy, K.B.J.A.i.p.b., Springer, Singapore, 2020. Ethnomedicine for drug discovery.
- Sohrabi, C., Alsafi, Z., O'neill, N., Khan, M., Kerwan, A., Al-Jabir, A., Iosifidis, C., Agha, R.J.I.j.o.s., 2020. World health organization declares global emergency: A review of the 2019 novel coronavirus (covid-19). [DOI] [PMC free article] [PubMed]
- Veber, D.F., Johnson, S.R., Cheng, H.-Y., Smith, B.R., Ward, K.W., Kopple, K.D.J.J.o.m.c., 2002. Molecular properties that influence the oral bioavailability of drug candidates. [DOI] [PubMed]
- Wu, X.M., Tan, R.X.J.N.p.r., 2019. Interaction between gut microbiota and ethnomedicine constituents. [DOI] [PubMed]
- Xu, Y., Wang, S., Hu, Q., Gao, S., Ma, X., Zhang, W., Shen, Y., Chen, F., Lai, L., Pei, J.J.N.a.r., 2018. Cavityplus: A web server for protein cavity detection with pharmacophore modelling, allosteric site identification and covalent ligand binding ability prediction. [DOI] [PMC free article] [PubMed]