Loss of Ca2+ binding causes ER stress and creates a unique dependency on the IRE1α/XBP1 pathway in type I and not type II mutant CALR-driven MPNs
Abstract
Approximately 20% of patients with myeloproliferative neoplasms (MPN) harbor mutations in the gene calreticulin (CALR), with 80% of those mutations classified as either type I or type II. While type II CALR-mutant proteins retain many of the Ca2+ binding sites present in the wild-type protein, type I CALR-mutant proteins lose these residues. The functional consequences of this differential loss of Ca2+ binding sites remain unexplored. Here, we show that the loss of Ca2+ binding residues in the type I mutant CALR protein directly impairs its Ca2+ binding ability, which in turn leads to depleted endoplasmic reticulum (ER) Ca2+ and subsequent activation of the IRE1α/XBP1 pathway of the unfolded protein response. Genetic or pharmacologic inhibition of IRE1α/XBP1 signaling induces cell death in type I mutant but not type II mutant or wild-type CALR-expressing cells, and abrogates type I mutant CALR-driven MPN disease progression in vivo.
Significance:
Current targeted therapies for CALR-mutated MPNs are not curative and fail to differentiate between type I- versus type II-driven disease. To improve treatment strategies, it is critical to identify CALR mutation type–specific vulnerabilities. Here we show that IRE1α/XBP1 represents a unique, targetable dependency specific to type I CALR-mutated MPNs.
This article is highlighted in the In This Issue feature, p. 265
INTRODUCTION
Myeloproliferative neoplasms (MPN) are clonal hematopoietic disorders that arise in the stem cell compartment and result in the excess production of mature myeloid cells. They include polycythemia vera (PV; increased red cell production, hemoglobin, and hematocrit), essential thrombocythemia (ET; increased megakaryocyte and platelet production), and primary myelofibrosis [PMF; characterized by the overproliferation of megakaryocytes and granulocytes with abnormal collagen deposition in the bone marrow (BM) stroma; refs. 1–3]. Approximately 40% of patients with ET and PMF harbor somatic, heterozygous mutations in the gene calreticulin (CALR), which encodes a calcium (Ca2+)-binding chaperone protein that primarily resides in the endoplasmic reticulum (ER; refs. 4, 5). CALR contains three distinct domains: the globular N-domain and proline-rich P-domain are responsible for the chaperone function of the protein, although the P domain also contains one high-affinity Ca2+ binding site; the highly disordered C-terminal domain binds Ca2+ with a series of acidic amino acids, and terminates at an ER retention signal (KDEL; ref. 6; Fig. 1A, top left). All CALR mutations occur as +1 base pair frameshift-induced indels in exon 9 of the gene, which encodes the C-terminal Ca2+ binding domain, and produce an identical 36 amino acid mutant C-terminal tail. The mutant C-terminus is characterized by the replacement of the acidic Ca2+ binding residues with positively charged arginine and lysine residues, and loss of the KDEL sequence (Fig. 1A, bottom left). This neomorphic sequence is required for the oncogenic activity of mutant CALR, wherein the mutant protein aberrantly binds to the thrombopoietin receptor MPL to constitutively activate pathogenic JAK/STAT signaling and drive the disease (7–9).
Figure 1.
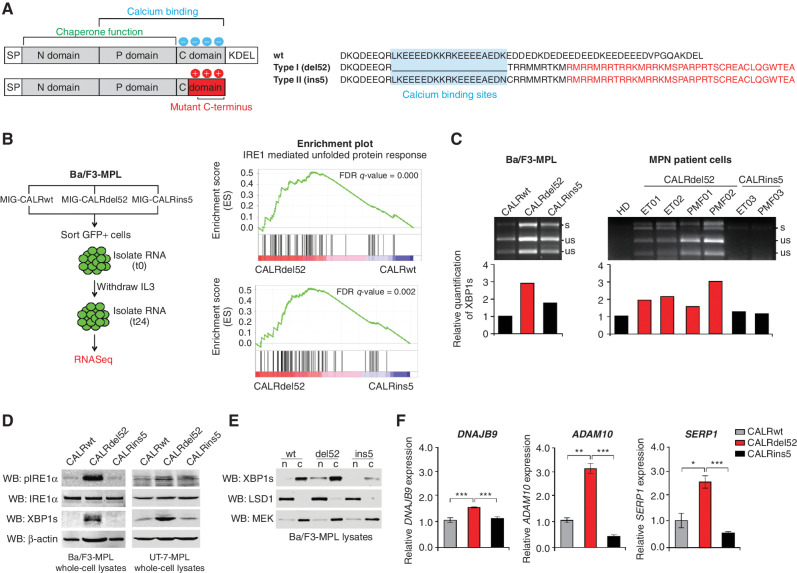
Type I CALRdel52 mutations differentially activate the IRE1α/XBP1 pathway of the unfolded protein response (UPR). A, Left, structural comparison of wild-type (top) and mutant (bottom) calreticulin (CALR). CALR contains three distinct domains: the N-domain and P-domain are responsible for the chaperone function of the protein, although the P domain also contains one high-affinity Ca2+ binding site; the C-terminal domain binds Ca2+ with a series of acidic amino acids (denoted by blue circles), and terminates at an ER retention signal (KDEL). SP, ER signal peptide. CALR mutations occur in the C-terminal Ca2+ binding domain, and produce an identical 36 amino acid mutant C-terminal tail (red shaded region). The mutant C-terminus is characterized by the replacement of the acidic Ca2+ binding residues with positively charged residues (denoted by red circles), and loss of the KDEL sequence. SP, ER signal peptide. Right, schematic depicting C-terminal amino acid sequence of CALRwt versus type I CALRdel52 and type II CALRins5. CALRins5 retains many of the Ca2+ binding residues present in the wild-type protein, which are lost in the del52-mutant protein (highlighted in blue). 36 amino acid mutant C-terminal tail shared between all CALR-mutant proteins is depicted in red. B, Left, schematic depicting workflow for RNA-sequencing (RNA-seq) experiment in Ba/F3-MPL cells expressing CALR variants in MSCV-IRES-GFP (MIG) backbone. Right, GSEA plots for IRE1α-mediated UPR in Ba/F3-MPL-CALRdel52 cells versus Ba/F3-MPL-CALRwt cells (top) and Ba/F3-MPL-CALRins5 cells (bottom). C, Top, XBP1 splicing assay performed in Ba/F3-MPL cells and MPN patient cells. Top band shows the spliced form of XBP1 (s), bottom bands show the Pst1-digested unspliced form of XBP1 (us). Bottom, quantification of spliced XBP1 band. D, Western blot analysis for phospho-IRE1α, total IRE1α, and spliced XBP1 (XBP1s) in Ba/F3-MPL and UT-7-MPL cells expressing CALR variants. β-Actin was used as a loading control. E, Western blot analysis for XBP1s in CALR-expressing Ba/F3-MPL nuclear and cytosolic extracts. LSD1 was used as a nuclear marker and MEK was used as a cytosolic marker. F, qPCR for XBP1 targets DNAJB9, ADAM10, and SERP1 in UT-7-MPL cells. Each bar represents the average of three replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Eighty percent of CALR mutations are classified as either type I (52 bp deletion; CALRdel52) or type II (5 bp insertion; CALRins5). Despite their shared mutant C-termini and mutual ability to bind and activate MPL, patients with type I and type II CALR mutations display significant clinical and prognostic differences. Type I mutations are more common in PMF, and are associated with a higher risk of fibrotic transformation from ET, while type II mutations are primarily associated with ET (10, 11). The mechanisms underlying these divergent clinical phenotypes remain unknown, but suggest that the molecular pathogenesis is not limited to activation of the MPL/JAK/STAT signaling axis. To date, however, there has been no identification of differentially activated pathways or dependencies in type I versus type II mutant CALR-driven disease. Although patients with CALR+ MPNs can be treated with JAK2 inhibitors, these drugs are not curative and primarily alleviate symptom burden without altering the natural course of the disease due to an inability to eradicate the malignant clone (12–19). Thus, to improve treatment strategies for patients with CALR+ MPNs, it is critical to identify specific dependencies unique to CALR mutation type that can be exploited for therapeutic gain.
Type I and type II CALR mutations are classified on the basis of the extent of amino acid homology with the wild-type protein. While type II CALR-mutant proteins retain many of the Ca2+ binding sites present in the wild-type protein, type I CALR-mutant proteins lose these residues (Fig. 1A, right). Importantly, it has not yet been shown that type I mutations directly result in a loss of Ca2+ binding ability. It has, however, been demonstrated that patients with type I CALR mutations, but not those with type II, exhibit abnormal intracellular Ca2+ signals, characterized by significantly increased store-operated Ca2+ entry (SOCE) activity, a regulator of Ca2+ flux into the cell (11). A subsequent study reported that this abnormal SOCE activity is a result of diminished interaction between mutant CALR and stromal interaction molecule 1 (STIM1; ref. 20), a protein of SOCE machinery that functions as a Ca2+ sensor in the ER (21). However, this study did not identify any significant differences in SOCE activation between type I and type II mutant CALR megakaryocytes. This suggests that there may be other regulatory mechanisms at play that mediate intracellular Ca2+ signaling in type I versus type II mutant CALR-expressing cells outside of SOCE machinery. Moreover, in addition to understanding how Ca2+ signaling differs in type I versus type II mutant CALR cellular contexts, and whether differential signaling is a direct result of the loss of Ca2+ binding sites on the type I mutant protein, it is critical to understand the functional consequences of these differences. Ca2+ affects myriad cellular pathways, but those which are differentially regulated by abnormal Ca2+ signaling in type I versus type II mutant CALR have yet to be explored.
Perturbations in ER Ca2+ concentration can cause accumulation of misfolded proteins in the ER. In response, activation of stress sensors IRE1α, PERK, and ATF6 triggers the unfolded protein response (UPR), which activates gene programs aimed at restoring ER function and adapting to microenvironmental changes. If ER stress becomes irremediable, the UPR commits the cell to apoptosis (22). Sustained ER stress has been documented in many cancer types, and some tumor cells exploit the UPR to withstand stress and promote survival (23, 24). Although CALR, as an ER chaperone, is an important UPR effector, the role of the UPR in mutant CALR-driven MPNs remains unclear, with previous studies demonstrating both impairment (25) and upregulation (26, 27) of the UPR in mutant CALR cells.
Here, we sought to first functionally validate whether the UPR is indeed activated by CALR mutations. Furthermore, given the differences in both amino acid composition and clinical phenotype between type I and type II CALR mutants, we examined whether the UPR is differentially activated between mutation types, and in doing so provide the first evidence for unique molecular dependencies in type I versus type II mutant CALR-driven MPNs. In addition, although traditionally considered gain-of-function mutations, we sought to directly answer the enduring question of whether type I CALR mutations serve as loss-of-function mutations in the context of Ca2+ binding ability. Finally, we show that the functional consequence of the differential loss of Ca2+ binding sites induced by type I versus type II CALR mutations is the activation of and dependency on the IRE1α/XBP1 pathway of the UPR, and that this pathway can be targeted for therapeutic intervention in type I mutant CALR-driven disease.
RESULTS
Type I CALRdel52 Mutations Differentially Activate the IRE1α/XBP1 Pathway of the UPR
To identify whether the UPR is differentially regulated in mutant versus wild-type CALR (CALRwt)-expressing cells, and in type I CALRdel52 versus type II CALRins5-expressing cells, we performed whole-transcriptome RNA sequencing (Fig. 1B, left) using the Ba/F3 cell line model system in which we previously dissected the molecular mechanisms underlying mutant CALR-mediated hematopoietic transformation (9, 28). IRE1α−regulated UPR transcripts were enriched in CALRdel52 cells as compared with both CALRwt and CALRins5-expressing cells (Fig. 1B, right; Supplementary Table S1).
To validate these findings, we employed an XBP1 splicing assay (29) in CALRwt, CALRdel52, and CALRins5 cells. Consistent with our RNA-sequencing (RNA-seq) results, we found that CALRdel52-expressing Ba/F3-MPL (Fig. 1C, left), human megakaryocytic UT-7-MPL (Supplementary Fig. S1), and primary peripheral blood mononuclear cells (PBMC) from patients with MPNs (Supplementary Table S2) demonstrate increased levels of spliced XBP1 (XBP1s) compared with CALRwt- and CALRins5-expressing cells (Fig. 1C, right). We further validated activation of this pathway at the protein level by Western blot analysis in Ba/F3-MPL and UT-7-MPL cells (Fig. 1D).
To assess XBP1s nuclear localization as a metric for its activity as a transcription factor, we performed nuclear/cytosolic fractionation of Ba/F3-MPL cells expressing CALRwt, CALRdel52, or CALRins5. Increased nuclear XBP1s was observed in CALRdel52-expressing cells compared with CALRwt or CALRins5 (Fig. 1E). Finally, we performed quantitative PCR (qPCR) of canonical XBP1 target genes in UT-7-MPL cells. In accordance with our previous results, CALRdel52-expressing cells displayed significantly increased expression of XBP1 target genes DNAJB9 (Fig. 1F, left), ADAM10 (Fig. 1F, middle), and SERP1 (Fig. 1F, right), compared with CALRwt and CALRins5-expressing cells. Together, these results confirm that the IRE1α/XBP1 pathway of the UPR is differentially activated in CALRdel52 compared with CALRwt and CALRins5 cells.
Type I CALRdel52 Mutations Lead to Loss of Ca2+ Binding Function, Resulting in ER Ca2+ Depletion
Given the differential activation of IRE1α/XBP1 by CALRdel52 compared with CALRins5, we hypothesized that the molecular mechanism underlying this activation may be related to the loss of Ca2+ binding residues unique to the CALRdel52 protein (Fig. 1A, right). We first analyzed our RNA-seq data for global transcriptomic changes in Ca2+ signaling pathways between CALRdel52 versus CALRwt or CALRins5-expressing cells. Ca2+ signaling pathway genes were significantly enriched in CALRdel52- as compared with CALRwt- (Fig. 2A, top) and CALRins5-expressing cells (Fig. 2A, bottom; Supplementary Table S1). This suggests that there are indeed differences in Ca2+ regulation unique to CALRdel52 cells.
Figure 2.
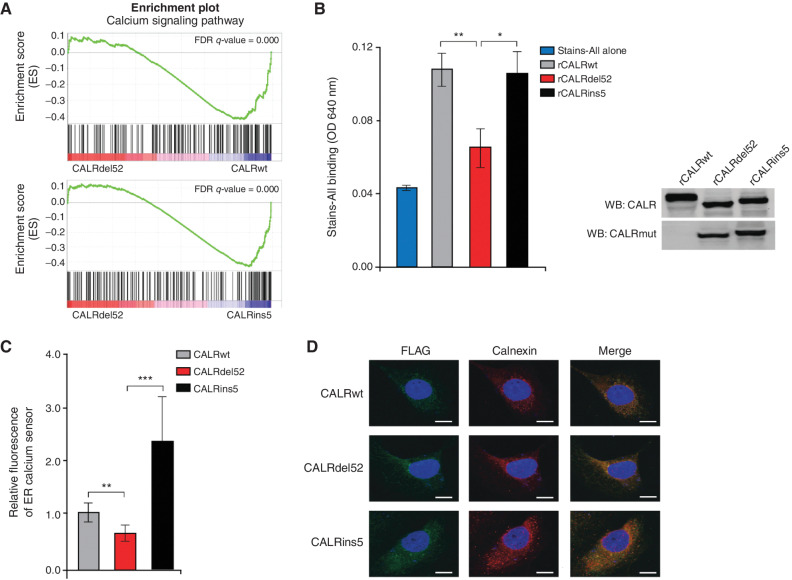
Type I CALRdel52 mutations lead to loss of Ca2+ binding function, resulting in ER Ca2+ depletion. A, GSEA plots for Ca2+ signaling pathway in Ba/F3-MPL-CALRdel52 cells versus Ba/F3-MPL-CALRwt cells (top) and Ba/F3-MPL-CALRins5 cells (bottom). B, Left, absorbance (640 nm) of 20 μg of recombinant CALRwt, CALRdel52, CALRins5 incubated with 0.025% Stains-All solution to indirectly measure the Ca2+ binding ability of each rCALR protein. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; **, P < 0.01). Right, Western blot analysis for calreticulin (CALR; which detects both wild-type and mutant CALR) and mutant CALR (which only detects the mutant C-terminus) for rCALR proteins used in Stains-All assay. C, Quantification of relative fluorescence of Ca2+ sensor in U2OS cells expressing iV2-CALRwt, CALRdel52, and CALRins5. Each bar represents the average of five individual cells. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; **, P < 0.01). D, Immunofluorescence of FLAG-tagged CALR variants (green) and ER marker calnexin (red) in U2OS cells. Nuclear staining with DAPI is shown in blue. Merge column depicts the overlay of FLAG-CALR in green with the ER in red to demonstrate colocalization (yellow). Scale bar, 10 μm.
Next, we asked whether the CALRdel52 protein does in fact lose its ability to bind Ca2+ as a result of the loss of Ca2+ binding residues in the C-terminus. To examine this, we performed a Stains-All assay as a metric of Ca2+ binding ability (30). Incubation of purified, recombinant CALR proteins (rCALR) with Stains-All led to similar increases in absorbance in CALRwt and CALRins5 protein–dye complexes compared with Stains-All alone, suggesting similar Ca2+ binding capabilities between the two proteins. However, CALRdel52 protein–dye complexes resulted in a significantly decreased absorbance compared with CALRwt and CALRins5, and no significant increase in absorbance compared to Stains-All alone (Fig. 2B). Together, this suggests that CALRdel52 mutations, but not CALRins5 mutations, lead to loss of Ca2+ binding function.
To assess the functional effects of the loss of CALRdel52 Ca2+ binding ability, we performed Ca2+ imaging studies using a genetically encoded ER-targeted red fluorescent Ca2+ indicator, R-CEPIA1er (31). This sensor is designed to bind free intraluminal ER Ca2+ that is not bound by other proteins, and thus serves to qualitatively measure ER Ca2+ concentration. Here, we used the human bone osteosarcoma cell line U2OS, an adherent cell line with abundant ER that has been shown to be optimal for live imaging studies. Cells were transiently cotransfected with plasmids encoding CALRwt, CALRdel52, and CALRins5 in a backbone expressing Venus fluorescent protein (iV2; ref. 32) and the plasmid encoding the R-CEPIA1er Ca2+ sensor 48 hours prior to imaging. Cell imaging studies revealed that cells transfected with CALRdel52 plasmid demonstrated significantly decreased fluorescence of the ER Ca2+ sensor, indicating decreased ER Ca2+ concentration, compared with cells expressing CALRwt or CALRins5 (Fig. 2C; Supplementary Fig. S2A and S2B). Notably, this was not due to an inability of CALRdel52 to localize to the ER, as immunofluorescence studies demonstrated similar ER localization of FLAG-tagged CALRwt, CALRdel52, and CALRins5, evidenced by the overlap of staining for ER marker calnexin (red) with anti-FLAG (green; Fig. 2D). Together, these data suggest that loss of Ca2+ binding sites on the CALRdel52 protein leads to a loss of Ca2+ binding function, resulting in diminished ER Ca2+ storage capacity and depleted ER Ca2+ levels.
Type I Mutant CALRdel52-driven ER Ca2+ Depletion Activates the IRE1α/XBP1 Pathway
We next tested whether differential activation of IRE1α/XBP1 by CALRdel52 is due to CALRdel52-induced ER Ca2+ depletion. For this, we generated a rescue construct designed to restore CALR-mediated Ca2+-binding capacity to the ER. The construct encodes the ER signal peptide at the N-terminus, followed by the P- and C-domains of CALRwt, which govern all of its Ca2+ binding function and contain the KDEL ER retention signal (Fig. 3A). Notably, the P- and C-domains alone, when expressed as a recombinant protein in bacteria, have been shown to bind Ca2+ at concentrations equal to the full-length protein (33).
Figure 3.
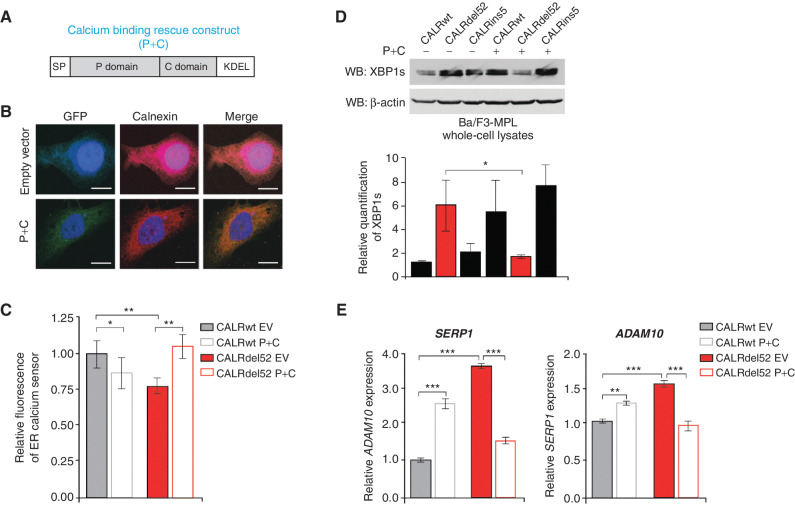
Type I mutant CALRdel52-driven ER Ca2+ depletion activates the IRE1α/XBP1 pathway. A, Schematic of the P+C rescue construct. The ER signal peptide was cloned just prior to the P domain to ensure proper localization of the protein. B, Immunofluorescence of empty GFP-expressing vector or P+C rescue construct in GFP-expressing backbone (green) and ER marker calnexin (red) in U2OS cells. Nuclear staining with DAPI is shown in blue. Merge column depicts the overlay of P+C in green with the ER in red to demonstrate colocalization (yellow). Scale bar, 10 μm. C, Quantification of relative fluorescence of Ca2+ sensor in U2OS cells expressing CALRwt + empty vector (control), CALRwt + P+C, CALRdel52 + empty vector, or CALRdel52 + P+C. Each bar represents the average of five independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; **, P < 0.01). D, Top, Western blot analysis for spliced XBP1 (XBP1s) in Ba/F3-MPL cells expressing calreticulin (CALR) variants and either empty vector (−) or P+C (+) rescue construct. β-Actin was used as a loading control. Bottom, quantification of XBP1s band relative to β-actin control from two independent Western blots (shown above and in Supplementary Fig. S3B). Analysis was performed using Thermo Fisher Scientific iBright Analysis Software. Each bar represents the average of two independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05). E, qPCR for XBP1 targets ADAM10, and SERP1 in Ba/F3-MPL cells expressing CALRwt and CALRdel52 with or without the P+C rescue construct. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01; ***, P < 0.001).
First, we performed immunofluorescence to confirm that the protein encoded by the P+C rescue construct does indeed localize to the ER. Here, U2OS cells were transiently transfected with either empty vector or the P+C rescue construct in a GFP-expressing backbone, and stained with a calnexin antibody to detect the ER (red). We observed overlap between staining for calnexin and GFP in the presence of the P+C construct, suggesting that the rescue construct does indeed localize to the ER (Fig. 3B). Finally, to confirm that expression of the P+C rescue construct is sufficient to restore Ca2+ binding ability to the ER, we performed imaging studies using the Ca2+ sensor described in Fig. 2. U2OS cells stably expressing either an empty vector or the P+C rescue construct were cotransfected with iV2-CALRdel52 plasmids and the R-CEP1A1er sensor 48 hours prior to imaging. U2OS cells stably expressing an empty vector or the P+C rescue construct and cotransfected with iV2-CALRwt plasmids and the R-CEP1A1er sensor were included as controls. ER Ca2+ levels in CALRdel52-expressing cells were completely rescued upon introduction of the P+C rescue construct compared with introduction of the empty vector alone. (Fig. 3C; Supplementary Fig. S3A).
With the function of our Ca2+ binding rescue construct established, we next investigated its effects on the IRE1α/XBP1 pathway. We stably expressed empty vector or the P+C rescue construct in Ba/F3-MPL-CALRwt, CALRdel52, and CALRins5 cells, and performed a Western blot analysis for XBP1s. XBP1s levels were markedly decreased in CALRdel52 cells expressing the P+C rescue construct compared with CALRdel52 cells expressing the empty vector (Fig. 3D, top; Supplementary Fig. S3B, quantified below). Notably, we found that the Ca2+ binding domain of CALRins5 was also capable of rescuing CALRdel52 XBP1s levels (Supplementary Fig. S3C). In addition, mRNA expression of XBP1 targets ADAM10 and SERP1 were also significantly decreased upon introduction of the CALRwt P+C rescue construct (Fig. 3E). Together, these data suggest that depletion of ER Ca2+ by CALRdel52 mediates activation of the IRE1α/XBP1 pathway, and that this activation can be rescued by introduction of a functional CALR Ca2+ binding construct.
Type I Mutant CALRdel52-Expressing Cells Are Dependent on Depleted ER Ca2+ to Activate IRE1α/XBP1, Which Promotes Cell Survival Via Upregulation of BCL-2
Having discerned the mechanism of ER stress underlying activation of the IRE1α/XBP1 pathway in CALRdel52 cells, we next asked whether this pathway represents a unique molecular dependency that promotes CALRdel52 cell survival.
Because we previously observed that the P+C rescue construct led to downregulation of XBP1s expression levels in CALRdel52 cells, we first asked how cell viability is affected in response to this rescue phenotype. We performed a cell viability assay in Ba/F3-MPL cells expressing CALRwt, CALRdel52, and CALRins5 with empty vector or stable expression of the P+C rescue construct. Because Ba/F3-MPL cells expressing CALRwt die upon withdrawal of IL3, we included Ba/F3-MPL-CALRwt cells in the presence of IL3 to determine whether the P+C rescue construct has any detrimental effects on cell viability in the absence of CALR mutations. All other cells were subjected to IL3 withdrawal to induce mutant CALR-driven IL3 independent cell growth. As expected, CALRwt-expressing cells in the absence of IL3 died, while CALRwt cells in the presence of IL3 were unaffected by introduction of the P+C rescue construct. Likewise, CALRins5 cells displayed no growth defect in the presence of the P+C rescue construct. CALRdel52 cells, however, exhibited significantly decreased cell viability in the presence of the P+C rescue construct compared with CALRdel52 cells expressing an empty vector (Fig. 4A). This suggests that CALRdel52 cells are dependent on depleted ER Ca2+ levels to activate IRE1α/XBP1 signaling, which promotes cell survival.
Figure 4.
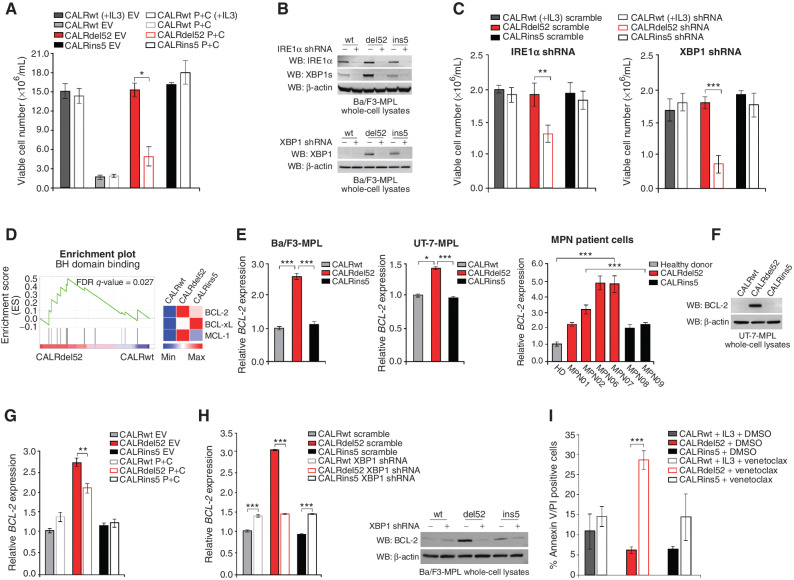
Type I mutant CALRdel52-expressing cells are dependent on depleted ER Ca2+ to activate IRE1α/XBP1, which promotes cell survival via upregulation of BCL-2. A, Total viable cell number at 48 hours post IL3 withdrawal in Ba/F3-MPL cells expressing calreticulin (CALR) variants and either empty vector or P+C rescue construct. Ba/F3-MPL-CALRwt cells grown in the presence of IL3 was included as a control. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05). B, Top, Western blot analysis for IRE1α and XBP1s in Ba/F3-MPL cells expressing CALR variants and either a scrambled shRNA or shRNA against IRE1α. Bottom, Western blot analysis for XBP1 in Ba/F3-MPL cells expressing CALR variants and either a scrambled shRNA or shRNA against XBP1. C, Total viable cell number at 48 hours post IL3 withdrawal in Ba/F3-MPL cells expressing CALR variants and either a scrambled shRNA or shRNA against IRE1α (left) or XBP1 (right). Ba/F3-MPL-CALRwt cells grown in the presence of IL3 were included as a control. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01; ***, P < 0.001). D, Left, GSEA plots for BH domain binding in Ba/F3-MPL-CALRdel52 cells versus Ba/F3-MPL-CALRwt cells. Right, Heat map displaying relative expression levels of BCL-2 family genes BCL-2, BCL-xL, and MCL-1 in Ba/F3-MPL cells expressing CALR variants. E, qPCR for BCL-2 expression in Ba/F3-MPL cells (left) and UT-7-MPL cells (middle) expressing CALRwt, CALRdel52, and CALRins5, and in peripheral blood mononuclear cells (PBMC) from a healthy donor (HD) or patients with myeloproliferative neoplasms (MPN; patient number depicted below each bar) with CALRdel52 or CALRins5 mutations (right). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; ***, P < 0.001). F, Western blot analysis for BCL-2 in UT-7-MPL cells expressing CALR variants. β-Actin was used as a loading control. G, qPCR for BCL-2 expression in Ba/F3-MPL cells expressing CALR variants and either empty vector or P+C rescue construct. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01). H, Left, qPCR for BCL-2 expression in Ba/F3-MPL cells expressing CALR variants and a scramble shRNA or shRNA against XBP1. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (***, P < 0.001). Right, Western blot analysis for BCL-2 in Ba/F3-MPL cells expressing CALR variants and a scramble shRNA (−) or shRNA against XBP1 (+). I, Quantification of flow cytometric analysis for Annexin V/PI double positivity in Ba/F3-MPL cells expressing CALR variants and treated with or without venetoclax (1 μmol/L for 24 hours). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (***, P < 0.001).
To further interrogate the specific requirement of IRE1α and XBP1s for CALRdel52 cell survival, we generated shRNA knockdowns against IRE1α and XBP1 in Ba/F3-MPL cells. Knockdown efficiency of the top scoring shRNAs against IRE1α (Fig. 4B, top) and XBP1 (Fig. 4B, bottom) were validated by Western blot analysis. Trypan blue exclusion cell counting assays demonstrated that cell viability was significantly decreased only in CALRdel52 cells upon knockdown of IRE1α (Fig. 4C, left) and XBP1 (Fig. 4C, right), suggesting that this pathway is required by CALRdel52 cells, but not CALRwt or CALRins5 cells for survival.
To understand how the IRE1α/XBP1 pathway promotes CALRdel52 cell survival, we focused on the antiapoptotic protein BCL-2. Although its function is best characterized in mitochondria, BCL-2 has also been shown to localize to the ER (34), where it plays a role in the control of Ca2+ homeostasis. Interestingly, BCL-2 overexpression has been shown to inhibit apoptosis in the face of depleted ER Ca2+ (35–37). Given the CALRdel52-driven phenotype of ER Ca2+ depletion and consequential IRE1α/XBP1 activation we observed, we asked whether BCL-2 may act downstream of IRE1α/XBP1 to promote continued cell survival. Notably, BCL-2 has been shown to regulate the IRE1α/XBP1 pathway, where BCL-2 enhances IRE1α endonuclease activity to promote sustained production of XBP1s (38). However, whether BCL-2 can also act downstream of IRE1α/XBP1 to promote cell survival remains unclear.
To address this question, we first analyzed our RNA-seq data (Fig. 1A) for BH (BCL-2 homology) domain binding signatures in CALRwt-, CALRdel52-, and CALRins5-expressing cells. BH domain binding genes were significantly enriched in CALRdel52 cells compared with CALRwt (Fig. 4D, left; Supplementary Table S1), with BCL-2 expression increased in CALRdel52 compared with both CALRwt and CALRins5 (Fig. 4D, right).
To validate BCL-2 upregulation in CALRdel52 cells, we performed qPCR for BCL-2 in Ba/F3-MPL cells (Fig. 4E, left), UT-7-MPL cells (Fig. 4E, middle), and primary MPN patient PBMCs (Fig. 4E, right; Supplementary Table S2). Significant upregulation of BCL-2 mRNA expression was observed in each cell type expressing CALRdel52 compared with those expressing CALRwt or CALRins5. Similarly, all MPN patient samples harboring a CALRdel52 mutation demonstrated significantly increased BCL-2 mRNA expression compared with healthy donor (HD). Likewise, all but one CALRdel52 sample demonstrated significantly increased BCL-2 mRNA expression compared with both samples harboring a CALRins5 mutation. We confirmed this finding at the protein level by Western blot analysis in UT-7-MPL cells (Fig. 4F). To determine whether the observed BCL-2 upregulation is mediated by the IRE1α/XBP1 pathway, we performed qPCR in both the stable P+C rescue cell line (Fig. 4G) and qPCR (Fig. 4H, left) and Western blot analysis (Fig. 4H, right) for BCL-2 in XBP1 knockdown cells, which both display downregulation of XBP1 (Fig. 3E; Fig. 4C). In all cases, we saw significantly decreased BCL-2 expression in CALRdel52 cells when XBP1 levels were downregulated. Having validated that BCL-2 is downstream of XBP1 in CALRdel52 cells, we next confirmed that XBP1 upregulates BCL-2 to promote cell survival in CALRdel52 cells. To do this, we treated cells with BCL-2 inhibitor venetoclax and performed Annexin V/PI staining to assay for apoptosis (Fig. 4I). We found that while neither CALRwt nor CALRins5 cells exhibited a significant increase in apoptosis upon venetoclax treatment, CALRdel52 cells underwent significant apoptotic cell death upon treatment. We validated this finding in primary MPN patient cells (Supplementary Fig. S4). This suggests that the IRE1α/XBP1 pathway mediates upregulation of BCL-2 specifically in CALRdel52 cells to promote cell survival in the face of CALRdel52-driven ER Ca2+ depletion.
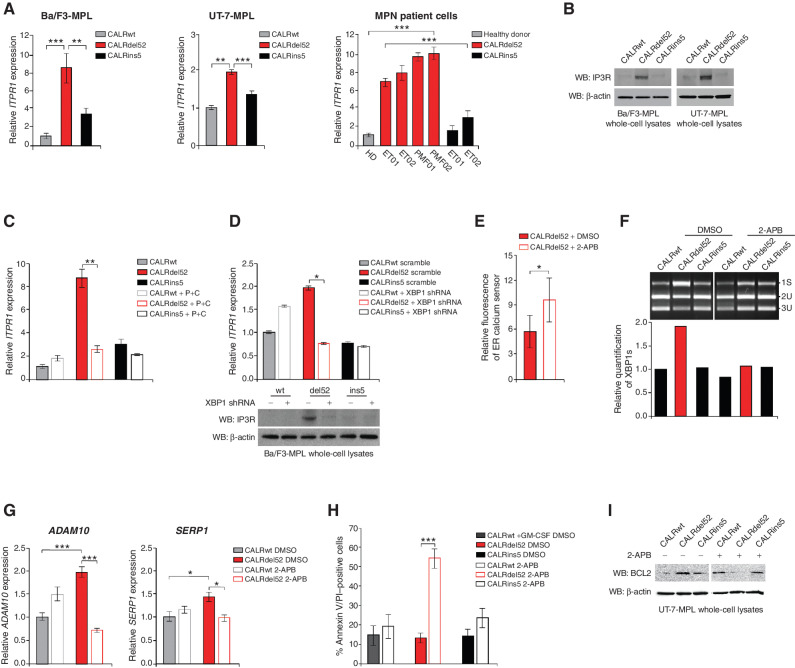
XBP1 Upregulates the IP3 Receptor to Induce a Positive Feedback Loop of Sustained Depleted ER Ca2+ and IRE1a/XBP1 Pathway Activation in Type I CALRdel52-Expressing Cells
We next turned our focus to how CALRdel52 cells sustain depleted ER Ca2+ in order to promote continued activation of IRE1α/XBP1 survival signaling. Inositol 1,4,5-trisphosphate (IP3) receptors (IP3R), the main Ca2+-release channels in the ER, and play a key role in the control of Ca2+ signaling (39). Notably, IP3Rs initiate SOCE, which has been shown to be differentially regulated in mutant versus wild-type CALR expressing cells (11, 20). Interestingly, BCL-2 has been shown to bind IP3R to inhibit Ca2+ release from the ER, which in turn prevents apoptosis (40). Here, we hypothesized that because CALRdel52 mutations engender dependency on depletion of ER Ca2+, this mechanism is rewired to promote IP3R-regulated Ca2+ release from the ER and sustain IRE1α/XBP1 activation, which upregulates BCL-2 to promote cell survival independent of IP3R inhibition.
To first assess whether IP3R is upregulated in CALRdel52-expressing cells, we performed qPCR for the IP3R gene ITPR1 in Ba/F3-MPL cells (Fig. 5A, left), UT-7-MPL cells (Fig. 5A, middle), and primary MPN patient PBMCs (Fig. 5A, right). Significant upregulation of ITPR1 mRNA expression was observed in each cell type expressing CALRdel52 compared with those expressing CALRwt and CALRins5. Likewise, all MPN patient samples harboring a CALRdel52 mutation demonstrated significantly increased ITPR1 mRNA expression compared with healthy donor (HD) and to both samples harboring a CALRins5 mutation. We confirmed this finding at the protein level by Western blot in Ba/F3-MPL and UT-7-MPL cells (Fig. 5B). To determine whether this upregulation is mediated by the IRE1α/XBP1 pathway, we performed qPCR in the stable P+C rescue cell line (Fig. 5C) and qPCR (Fig. 5D, top) and Western blot analysis (Fig. 5D, bottom) for ITPR1 in XBP1 knockdown cells. ITPR1 mRNA and IP3R protein expression were both decreased only in CALRdel52 cells upon XBP1 downregulation. This suggests that CALRdel52 cells mediate upregulation of IP3R at least in part via the IRE1α/XBP1 pathway.
Figure 5.
XBP1 upregulates the IP3 receptor to induce a positive feedback loop of sustained depleted ER Ca2+ and IRE1α/XBP1 pathway activation in type I CALRdel52-expressing cells. A, qPCR for ITPR1 in Ba/F3-MPL cells (left) and UT-7-MPL cells (middle) expressing CALRwt, CALRdel52, and CALRins5, and in peripheral blood mononuclear cells (PBMC) from a healthy donor (HD) or patients with myeloproliferative neoplasms (MPN; patient number depicted below each bar) with CALRdel52 or CALRins5 mutations (right). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01; ***, P < 0.001). B, Western blot analysis for IP3R in Ba/F3-MPL and UT-7-MPL cells expressing calreticulin (CALR) variants. β-Actin was used as a loading control. C, qPCR for ITPR1 expression in Ba/F3-MPL cells expressing CALR variants and either empty vector or P+C rescue construct. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01). D, Top, qPCR for ITPR1 expression in Ba/F3-MPL cells expressing CALR variants and a scramble shRNA or shRNA against XBP1. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05). Bottom, Western blot analysis for IP3R in Ba/F3-MPL cells expressing CALR variants and a scramble shRNA (−) or shRNA against XBP1 (+). E, Quantification of relative fluorescence of Ca2+ sensor in U2OS cells expressing iV2-CALRdel52 treated with or without 2-APB (100 μmol/L for 1.5 minutes). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05). F, Top, XBP1 splicing assay performed in Ba/F3-MPL cells expressing CALR variants treated with or without 2-APB (100 μmol/L for 24 hours). Top band shows the spliced form of XBP1 (s), bottom bands show the Pst1-digested unspliced form of XBP1 (us). Bottom, quantification of spliced XBP1 band. Analysis was performed using Thermo Fisher Scientific iBright Analysis Software. G, qPCR for ADAM10 and SERP1 expression in UT-7-MPL cells expressing CALR variants treated with or without 2-APB (100 μmol/L for 24 hours). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01). H, Quantification of flow cytometric analysis for Annexin V/PI double positivity in Ba/F3-MPL cells expressing CALR variants and treated with or without 2-ABP (100 μmol/L for 72 hours). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (***, P < 0.01). I, Western blot analysis for BCL-2 in Ba/F3-MPL cells expressing CALR variants and treated with or without 2-ABP (100 μmol/L for 24 hours). β-Actin was used as a loading control.
Next, we tested whether IP3R is the channel by which CALRdel52-expressing cells mediate efflux of ER Ca2+ to sustain ER Ca2+ depletion. To do so, we utilized the allosteric IP3R inhibitor, 2-aminoethoxydiphenyl borate (2-APB), which inhibits IP3R Ca2+ release (41). U2OS cells transiently cotransfected with CALR variants and the CEPIA1er Ca2+ sensor were treated with vehicle or 2-ABP for 90 seconds and imaged as previously described in Figs. 2 and 3. We found that CALRdel52-expressing cells treated with 2-APB demonstrated significantly increased ER Ca2+ levels compared with CALRdel52 cells treated with vehicle, as measured by fluorescence of the ER Ca2+ sensor (Fig. 5E; Supplementary Fig. S5A). This suggests that IP3R is at least partially responsible for the depleted ER Ca2+ stores in CALRdel52 cells.
Finally, we tested the hypothesis that XBP1 upregulates IP3R to produce a positive feedback loop whereby IP3R sustains ER Ca2+ depletion to promote continued activation of IRE1α/XBP1 in CALRdel52 cells. To do this, we treated Ba/F3-MPL cells with 2-APB and measured XBP1 splicing as previously described in Fig. 1. Strikingly, we found that inhibition of IP3R with 2-APB led to decreased levels of XBP1s mRNA only in CALRdel52 cells, suggesting that not only does XBP1 up-regulate IP3R, but IP3R activity promotes IRE1α/XBP1 signaling in a positive feedback loop (Fig. 5F). To interrogate the effects of IP3R inhibition on XBP1 transcriptional activity, we performed qPCR for XBP1 targets ADAM10 and SERP1, and found that 2-APB treatment led to significantly decreased expression of both genes in CALRdel52 cells (Fig. 5G). Finally, to determine whether IP3R activity is important for the survival of CALRdel52 cells, we treated UT-7-MPL cells with 2-APB and performed Annexin V/PI staining to measure apoptosis. We found that while 2-APB had no significant proapoptotic effect on CALRwt or CALRins5-expressing cells, CALRdel52 cells displayed significantly increased Annexin V/PI positivity in response to 2-APB treatment (Fig. 5H). To determine whether this increased cell death is mediated through downregulation of BCL-2, we performed a Western blot analysis for BCL-2 in 2-ABP treated UT-7-MPL cells. Indeed, we found that BCL-2 expression is decreased in CALRdel52 cells upon 2-ABP treatment (Fig. 5I, quantified in Supplementary Fig. S5B). Together, these results indicated that CALRdel52 cells promote ER Ca2+ depletion via XBP1-mediated up-regulation of IP3R, which in turn leads to continued activation of IRE1α/XBP1 to promote cell survival through BCL-2.
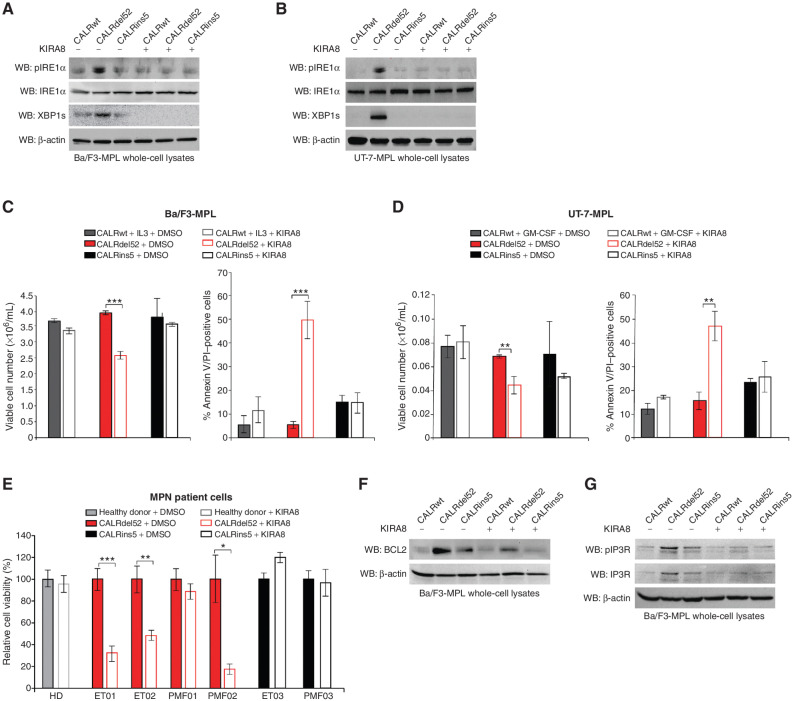
The IRE1α/XBP1 Pathway Represents a Potential Target for Therapy in CALRdel52-Driven MPNs
Given the dependence of CALRdel52 cells on the IRE1α/XBP1 pathway, we reasoned that IRE1α may represent a potential target for therapy in CALRdel52-driven MPNs. To test this, we employed the highly selective IRE1α inhibitor, KIRA8, that binds to the kinase domain of IRE1α and allosterically inhibits its RNase activity (42).
To first determine the lowest dose at which KIRA8 inhibits IRE1α/XBP1 signaling, we performed a dose–response assay for XBP1 splicing (Supplementary Fig. S6A), as well as Western blot analysis for phospho-IRE1α (Fig. 6A and B). A dose of 5 μmol/L was sufficient to inhibit IRE1α phosphorylation and XBP1 splicing. KIRA8 specifically inhibited viability of and induced apoptosis in CALRdel52-expressing Ba/F3-MPL cells (Fig. 6C), UT-7-MPL cells (Fig. 6D), and primary MPN patient PBMCs (Fig. 6E; Supplementary Table S2), without affecting cells expressing CALRwt or CALRins5. This suggests that inhibiting IRE1α may have minimal off-target effects on cells not expressing CALRdel52. Finally, to assess the effects of KIRA8 treatment on XBP1 targets BCL-2 and IP3R, we performed Western blot analysis in KIRA8-treated Ba/F3-MPL-CALR cells. We found that KIRA8 treatment led to downregulation of BCL-2 (Fig. 6F, quantified in Supplementary Fig. S6B), phospho-IP3R, and total IP3R expression levels (Fig. 6G, quantified in Supplementary Fig. S6C) in CALRdel52 cells, confirming that KIRA8 treatment is able to mitigate the expression and activity of these critical targets that drive CALRdel52 cell survival.
Figure 6.
The IRE1α/XBP1 pathway represents a potential target for therapy in CALRdel52-driven MPNs. A, Western blot analysis for phospho-IRE1α, total IRE1α and XBP1s in Ba/F3-MPL cells expressing calreticulin (CALR) variants treated with or without KIRA8 (5 μmol/L for 4 hours). β-Actin was used as a loading control. B, Western blot analysis for phospho-IRE1α, total IRE1α, and XBP1s in UT-7-MPL cells expressing CALR variants treated with or without KIRA8 (5 μmol/L for 4 hours). β-Actin was used as a loading control. C, Left, total viable cell number at 72 hours post IL3 withdrawal in Ba/F3-MPL cells expressing CALR variants treated with or without KIRA8 (5 μmol/L). Right, quantification of flow cytometric analysis for Annexin V/PI double positivity in Ba/F3-MPL cells expressing CALR variants and treated with or without KIRA8 (5 μmol/L for 48 hours). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (***, P < 0.001). D, Left, total viable cell number at 72 hours post GM-CSF withdrawal in UT-7-MPL cells expressing CALR variants treated with or without KIRA8 (5 μmol/L). Right, quantification of flow cytometric analysis for Annexin V/PI double positivity in UT-7-MPL cells expressing CALR variants and treated with or without KIRA8 (5 μmol/L for 48 hours). Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01). E, Relative cell viability in peripheral blood mononuclear cells from a healthy donor (HD) or patients with myeloproliferative neoplasms (MPN; patient number depicted below each bar) with CALRdel52 or CALRins5 mutations, treated with or without KIRA8 (500 nmol/L for 48 hours). Viability was determined by CellTiter-Glo assay. Each bar represents the average of three independent replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). F, Western blot analysis for BCL-2 in Ba/F3-MPL cells expressing CALR variants treated with or without KIRA8 (5 μmol/L for 24 hours). β-Actin was used as a loading control. G, Western blot analysis for IP3R in Ba/F3-MPL cells expressing CALR variants treated with or without KIRA8 (5 μmol/L for 24 hours). β-Actin was used as a loading control.
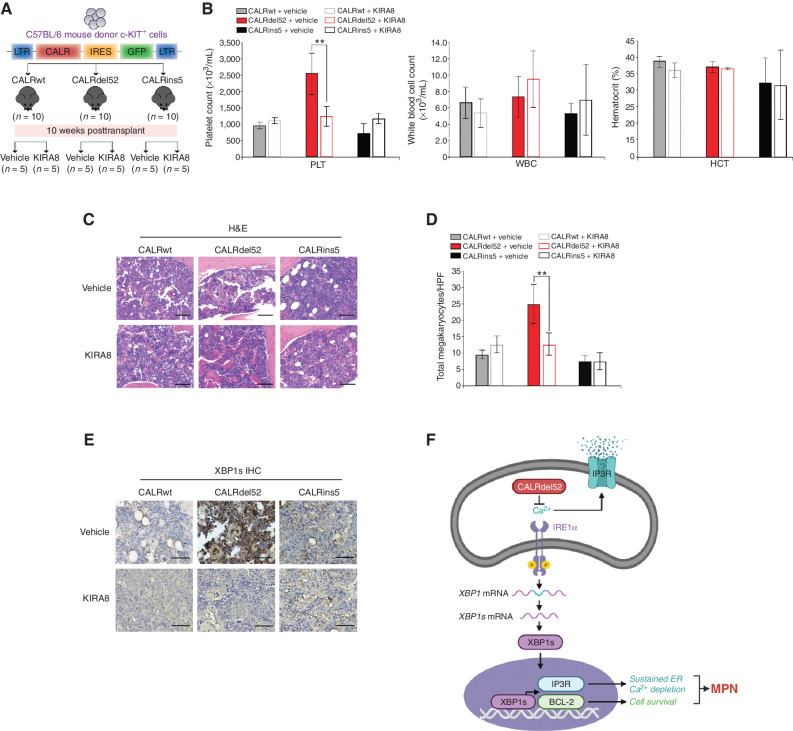
Inhibition of IRE1α Signaling Abrogates MPN Disease Progression In Vivo
Finally, to study the effects of IRE1α inhibition in vivo, we used a BM transplantation (BMT) model of mutant CALR-driven ET as previously described (9, 43). Ten weeks post-transplantation, mice were bled retro-orbitally to confirm onset of thrombocytosis in CALRdel52 mice, then randomly divided into two groups per genotype (n = 5 per group). Mice from each group were treated with intraperitoneal injection of either vehicle or 50 mg/kg KIRA8 on a 7 day on/7 day off dose schedule for a total of 6 weeks (Fig. 7A). At 16 weeks posttransplantation, mice were sacrificed for end point analysis, which included complete blood counts (CBC) and histopathological analysis. CBC analysis revealed that, as expected, mice receiving CALRdel52-expressing cells demonstrated significantly increased platelet (PLT) counts at 16 weeks, compared with mice receiving CALRwt and CALRins5-expressing cells. Notably, although CALRins5 mutations are associated with ET in human patients, this oncogene has been shown to be a weaker driver of ET in mice, with no detectable thrombocytosis at 16 weeks in a BMT model (43). Likewise, we saw no evidence of thrombocytosis in CALRins5 mice at this time point. Remarkably, we found that just 7 days of KIRA8 treatment in CALRdel52 mice was sufficient to induce a significant decrease in platelet counts (Fig. 7B, left). Hematocrit (HCT; Fig. 7B, middle) and white blood cell (WBC) counts (Fig. 7B, right) were generally consistent across all genotypes, as previously reported (15), and unaffected by KIRA8 treatment.
Figure 7.
Inhibition of IRE1α signaling abrogates myeloproliferative neoplasm (MPN) disease progression in vivo. A, Schematic of retroviral bone marrow transplantation assay (BMT). B, Platelet counts (PLT; left), white blood cell counts (WBC; middle), and hematocrit (HCT; right) at 16 weeks posttransplantation in the peripheral blood of recipient mice receiving CALRwt, CALRdel52, or CALRins5-expressing c-KIT+ BM cells treated as indicated (n = 10 in each group). Each bar represents the average of 5 mice. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01). C, Histopathologic hematoxylin and eosin (H&E) sections of BM from representative CALRwt, CALRdel52, and CALRins5 mice treated with vehicle or 50 mg/kg/day KIRA8 (20× magnification, black scale bars = 100 μm). D, Megakaryocyte counts per high-power field (HPF) in BM of CALRwt, CALRdel52, or CALRins5 mice treated with vehicle or 50 mg/kg/day KIRA8. Each bar represents the average of 5 mice. Error bars, SD. Significance was determined by two-tailed Student t test (**, P < 0.01). E, IHC analysis for XBP1 in BM from representative CALRwt, CALRdel52, and CALRins5 mice treated with vehicle or 50 mg/kg/day KIRA8 (40× magnification, black scale bars = 25 μm). F, Working model: left, type I CALRdel52 proteins exhibit loss of Ca2+ binding function. This leads to depleted ER Ca2+, which in turn activates the IRE1α/XBP1 pathway. XBP1 mediates upregulation of BCL-2, which promotes cell survival, and IP3R, which facilitates continued ER Ca2+ efflux to sustain ER Ca2+ depletion and activation of IRE1α/XBP1, ultimately driving MPN. Inhibiting this pathway leads to rapid cell death in vitro and abrogation of disease progression in vivo. Right, in contrast, CALRwt and CALRins5 cells retain calreticulin (CALR)-mediated ER Ca2+ binding function, and thus do not hijack IRE1α/XBP1 to promote survival. Instead, canonical IRE1α/XBP1 signaling in these cells may be activated upon ER stress, wherein this pathway would initially promote adaptation and survival to restore proteostasis, but commit cells to apoptosis if stress cannot be resolved (figure created with BioRender).
Histopathologic analysis of mouse BM sections further revealed that CALRdel52 mice displayed hallmark characteristics of ET, which were abrogated following 7 days of KIRA8 treatment. BM from vehicle-treated CALRdel52 mice displayed hypercellularity with enlarged, hyperlobulated megakaryocytes that demonstrated a tendency towards clustering and signs of emperipolesis. In contrast, BM from KIRA8-treated CALRdel52 mice show mostly normocellularity with megakaryocytes that are decreased in size, frequency, and clustering (Fig. 7C and D). Finally, to validate that the effects of KIRA8 on CALRdel52-driven disease attenuation were indeed mediated through inhibition of IRE1α/XBP1 signaling, we performed IHC for total XBP1 on mouse BM sections (Fig. 7E). In vehicle-treated CALRdel52 mouse BM, we saw prominent positive staining for XBP1, including nuclear expression in megakaryocytes. Importantly, KIRA8 treatment led markedly decreased expression of XBP1 in CALRdel52 mouse BM.
Finally, given the upregulation of BCL-2 in CALRdel52-expressing cells, and the ability of BCL-2 inhibitor venetoclax to induce apoptosis and reduce cell viability specifically in CALRdel52 cell lines and primary MPN patient samples, respectively (Fig. 4; Supplementary Fig. S4; Supplementary Table S2), we tested venetoclax in our BMT model to determine its efficacy in abrogating disease compared with KIRA8. The BMT was performed as above, and 10 weeks posttransplantation, mice were bled retro-orbitally to confirm onset of thrombocytosis in CALRdel52 mice, then randomly divided into two groups per genotype (n = 10 per group). Mice from each group were treated for 5 days with vehicle, 50 mg/kg KIRA8 as described above, or 25 mg/kg venetoclax administered by oral gavage (Supplementary Fig. S7). At 11 weeks posttransplantation, CBC analysis revealed that, as expected, mice receiving CALRdel52-expressing cells demonstrated significantly increased platelet (PLT) counts compared with mice receiving CALRwt and CALRins5-expressing cells. KIRA8 and venetoclax treatment both led to significant PLT reduction in CALRdel52 mice, with KIRA8 showing slightly more efficacy than venetoclax.
Together, these results demonstrate that pharmacologic inhibition IRE1α signaling, where targets include IRE1α itself as well as BCL-2, leads to substantial abrogation of CALRdel52-driven MPN disease progression in vivo. Further studies are warranted to more thoroughly test BCL-2 inhibition in this model (including treating mice until at least the 16-week end point). In addition, we conducted preliminary studies to test the efficacy of combined IRE1α and BCL-2 inhibition on CALRdel52-driven disease, but found that the combination at the doses used (50 mg/kg/day KIRA8 + 25 mg/kg/day venetoclax for 7 days) was lethal for all animals tested. Thus, additional studies are warranted to identify a therapeutic window for this combination that may result in synergistic inhibition of CALRdel52-driven disease.
DISCUSSION
The identification of the distinct phenotypic and prognostic outcomes of type I versus type II CALR mutations has provided important insight into how these mutations differ clinically, despite their shared mutant C-terminus (10, 11). However, since this finding, there has been little in the way of understanding how these mutations differ in cellular consequence, and what their differential molecular dependencies may be. Here, we show the first evidence of a unique molecular dependency in type I versus type II CALR mutations, and demonstrate that this dependency is a functional consequence of the molecular difference between type I and type II CALR mutations.
Because CALR mutations are classified based on extent of homology with the C-terminal Ca2+ binding domain of wild-type protein, we sought to investigate pathways that would be most influenced by the loss of Ca2+ binding sites in the type I versus type II protein. This, together with the fact that CALR is an ER chaperone that is heavily implicated in the regulation of ER stress, led us to focus on the UPR as a potential differential dependency between type I and type II mutant CALR-expressing cells. In doing so, we found that type I, but not type II CALR mutations lead to activation of the most ancient and conserved arm of the UPR, the IRE1α/XBP1 pathway. We hypothesized that type I CALR mutations may lead to depleted ER Ca2+ due to the inability of the type I mutant protein to bind and store Ca2+ in the ER. Indeed, we found that type I CALR-mutant proteins demonstrate significantly impaired Ca2+ binding ability, and that this results in chronically depleted ER Ca2+ levels. Furthermore, we found that this ER Ca2+ depletion directly activates the IRE1α/XBP1 pathway of the UPR, and that the ER Ca2+ efflux receptor IP3R is upregulated downstream of XBP1 to sustain a state of depleted ER Ca2+, which in turn creates a positive feedback loop to maintain IRE1α/XBP1 activation. We further show that this pathway represents a unique molecular dependency for type I- compared to type II-mutant CALR-expressing cells or wild-type cells, in part through upregulation of BCL-2, and can be pharmacologically targeted both in vitro and in vivo to specifically induce death of type I mutant CALR-expressing cells and abrogate type I mutant CALR-driven disease (Fig. 7F).
Intriguingly, we found that the P+C rescue plasmid has the opposite effect in CALRwt and CALRins5 cells compared with CALRdel52 cells, where IRE1α/XBP1 activity is substantially increased (Fig. 3D). This is paradoxically accompanied by decreased rather than increased ER Ca2+ in CALRwt cells (Fig. 3C), with no significant effect on cell viability (Fig. 4A) nor ITRP1 expression (Fig. 5C). While detailed analysis of this is outside the scope of the current work, we speculate that introduction of the P+C rescue plasmid in cells with intact Ca2+ signaling and homeostasis itself causes ER stress, leading to increased IRE1α/XBP1 activation. We expect this pathway to behave more canonically in these cells, as it is not being hijacked to promote continued cell survival here as it is in CALRdel52 cells. Thus, we anticipate that CALRwt and CALRins5 cells may activate this pathway to rapidly restore Ca2+ homeostasis. Because these cells stably express the P+C rescue plasmid, it is likely that there is an acute increase in ER Ca2+ and ITPR1 expression, and perhaps even cell death, to reestablish ER homeostasis. However, over the course of selection for stably expressing cells, we may also be selecting for cells that have already gone through this acute rebalancing via their increased IRE1α/XBP1 activation.
Additional outstanding questions relate to the mechanisms underlying BCL-2 and IP3R in this model. BCL-2 has been shown to bind to the IP3 receptor to inhibit Ca2+ release from the ER, which in turn prevents apoptosis. In contrast, we show that although BCL-2 is upregulated by XBP1, IP3R is also upregulated and activated by this pathway, and in fact serves to sustain IRE1α/XBP1 survival signaling. Further investigation into this paradoxical rewiring of BCL-2-mediated regulation of IP3R and its underlying mechanisms in the context of type I CALR mutations is certainly warranted.
Taken together, this work is the first to demonstrate that type I and type II mutant CALR-expressing cells display differential molecular dependencies, and that those differential dependencies can be targeted for therapeutic gain in type I versus type II-driven MPNs. Moreover, this study answers an enduring question regarding the functional consequence of the loss of Ca2+ binding sites on the type I mutant CALR protein, and demonstrates how type I CALR mutant–expressing cells rewire the UPR and downstream Ca2+ signaling and apoptotic pathways to drive MPNs.
METHODS
Cell Lines and Cell Culture
293T and U2OS cells were obtained from ATCC and their identity was authenticated by short tandem repeat (STR) profiling. Ba/F3 cells and UT-7 cells were purchased from German Collection of Microorganisms and Cell Cultures (DSMZ) and were not further authenticated. All cell lines were intermittently tested for Mycoplasma. 293T and U2OS cells were cultured in DMEM (Corning) medium with 10% FBS (Corning) and penicillin/streptomycin (Corning). Ba/F3 and UT-7 cells were cultured in RPMI1640 medium with 10% FBS and penicillin/streptomycin. Ba/F3 cells were supplemented with 2 ng/mL murine IL3 (R&D Systems); UT-7 cells were supplemented with 10 ng/mL human GM-CSF (R&D Systems). All cell lines were grown at 37°C, 5% CO2, 95% humidity. Cells are tested for Mycoplasma bi-monthly.
Stable Overexpression Cell Line Generation
Ba/F3 and UT-7 cell lines stably expressing the thrombopoietin receptor (MPL) were generated by retroviral transduction. In brief, retroviral supernatants were generated by cotransfection of pMSCV-hygro-hMPL with packaging plasmids in 293T cells. Viral supernatants were collected at 24 and 48 hours posttransfection. Ba/F3 or UT-7 cells were subjected to spin infection with viral supernatants, followed by 7 days of hygromycin selection at 1 mg/mL. Stable expression of CALR variants was achieved by cotransfecting pMSCV-neo-FLAG–tagged-CALR variants (pMSCV-neo-signal peptide-FLAG-CALRwt, -CALRdel52, -CALRins5), pMSCV-IRES-GFP-CALR variants, or pMSCV-puro-P+C with packaging plasmids in 293T cells. Viral supernatants were collected at 24 and 48 hours posttransfection. Ba/F3, UT-7, or U2OS cells were subjected to spin infection with viral supernatants, followed by 7 days of antibiotic selection (neomycin 1 mg/mL; puromycin 2 μg/mL), or cell sorting for GFP positivity.
RNA-Seq
The sequencing procedure was carried out using the NovaSeq 6000 sequencing (Illumina) at the University of Chicago Genomics Facility (Chicago, IL). RNA extracted from Ba/F3-MPL cells was prepared following the standard protocols recommended by the RNeasy Mini Kit (Qiagen) and sequenced in two runs to generate paired-end 100 bp reads. For each sample, the raw FASTQ files from two flow cells were combined before downstream processing. RNA-seq data was processed using a local Galaxy 20.05 instance for the following steps: quality and adapter trimming were performed on the raw sequencing reads using Trim Galore! 0.6.3. The reads were mapped to the mouse genome (GRCm38.p4 with GENCODE annotation) using RNA STAR 2.7.5b. The resulting mapped reads from each sample were counted by featureCounts 1.6.4 for per gene read counts.
Statistical Analysis for RNA-Seq
The raw counts were analyzed for differential expression between experimental conditions using DESeq2 1.22.1, which also generated a normalized gene expression matrix. Gene expression data normalized by DESeq2, where the transcripts had previously been mapped to the genome and relevant genes as described in RNA-seq method, were used for gene-set enrichment analyses and heat mapping using Morpheus from the Broad Institute; gene sets utilized for gene-set enrichment analysis were those predetermined from the GO and KEGG databases referenced in Supplementary Table S1. For calculation of percent spliced XBP1, mapped reads for XBP1 gene were used from RNA-seq replicates 1 and 3 to calculate the extent of alternative splicing for each genotype. Specifically, the percentage of spliced reads for a sample was calculated by diving the number of alternatively spliced reads over the total number of XBP1 reads for that sample. Splicing data was analyzed and plotted using Prism.
Accession Number
Gene expression data are available in the GEO database with the accession number GSE173805.
XBP1 Splicing Assay
Total RNA was isolated from cells following the standard protocols recommended by the RNeasy Mini Kit (Qiagen) or the Absolutely RNA Nanoprep Kit (Agilent) depending on cell yield. cDNA was synthesized from RNA templates using Reverse Transcriptase and Reverse Transcriptase Reaction Mix (Bio-Rad). Real-time RT-PCR was performed in the Applied Biosystems ProFlex PCR System using GoTaq Green Master Mix 2X (Promega) and XBP1 forward primer (5′-AGG AAA CTG AAA AAC AGA GTA GCA GC-3′) and reverse primer (5′-TCC TTC TGG GTA GAC CTC TGG-3′). Samples were subject to 1 hour of incubation at 37°C with PST1-HF restriction enzyme diluted in CutSmart Buffer (NEB). Samples were run on a 3% agarose gel for 1 hour at 120V.
Cell Fractionation
Cells were harvested and prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) according to the manufacturer's instructions. LSD1 was used to confirm purity of nuclear extract, and MEK was used to confirm purity of cytoplasmic extracts.
Western Blotting
Cells were harvested in ice-cold PBS and lysed in Nonidet P-40 lysis buffer supplemented with phosphatase and protease inhibitors. Cell lysates were clarified by centrifugation, and supernatants were normalized via Bradford assay using BSA as a standard. Normalized lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes utilizing the iBlot 2 Gel Transfer Device. Membranes were probed using the designated antibodies and visualized with either SuperSignal Wester Dura Extended Duration Substrate (Thermo Fisher Scientific) or SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific).
Quantitative PCR
Total RNA was extracted from cells using the PureLink RNA Mini Kit (Invitrogen). cDNA was synthesized from 1 μg of RNA template using the iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using 2X SYBR Green qPCR Master Mix (Applied Biosystems). Primers used for qPCR are listed below. β-Actin was used for normalization. See Supplementary Table S3 for qPCR primer sequences.
Protein Purification
CALR variants were cloned into the pBAD-DEST49 expression vector, then transformed into One Shot BL21 (DE3) chemically competent E. coli cells. Transformed cells were grown in LB under ampicillin selection followed by the addition of 0.02% l-arabinose for induction of CALR expression. After 4 hours, cells were harvested and lysed via sonication. Proteins were purified using ProBond resin (Invitrogen). Purification was performed as directed by the ProBond purification system (Invitrogen). The eluted proteins were validated via Western blot analysis.
Stains-All Assay
Purified recombinant proteins were incubated in 0.025% Stains-All solution (Sigma) at room temperature in the dark for 30 minutes. Samples were then read for spectral absorbance in a plate reader. Absorbance from wavelengths ranging from 610 to 650 nm were analyzed for each of the samples.
Immunofluorescence
U2OS cells were grown onto 13 mmol/L Deckgläser Cover Glasses. After transient transfection using FLAG-tagged CALR variants or P+C constructs, cells were washed twice with PBS and fixed with 4% paraformaldehyde for 10 minutes at room temperature. Cells were permeabilized with 0.1% triton X-100 and blocked in normal donkey serum for 1 hour at room temperature. Cells were then incubated with the corresponding primary antibodies overnight in 4°C. Cells were washed twice and incubated with secondary antibody for 1 hour at room temperature. The cells were then washed twice and mounted onto slides using ProLong Gold Antifade Reagent with DAPI.
Intracellular Ca2+ Imaging
U2OS cells were seeded at 0.5 × 106 cells per 1 mL of DMEM. After 24 hours, cells were transiently cotransfected with venus fluorescent iV2-CALR variant plasmids and red fluorescent pCMV-R-CEPIAer, which was a gift from Masamitsu Iino (Addgene plasmid # 58216; http://n2t.net/addgene:58216; RRID:Addgene_58216). Cells were imaged on the Leica Stellaris 8 Laser Scanning Confocal the following day.
Ca2+ Imaging Analysis
Venus-expressing cells were segmented using the ROI manager on ImageJ. The intensity of the red fluorescent expression for each cell (5 cells per condition) was calculated and then averaged.
Generation of shRNA Stable Knockdown Cell Lines
Stable knockdown of endogenous IRE1α or XBP1 was achieved using pLKO.1-based lentiviral vector shRNA constructs targeting mouse IRE1α (sh-1 5′ CCG GGC TCG TGA ATT GAT AGA GAA ACT CGA GTT TCT CTA TCA ATT CAC GAG CTT TTT 3′, sh-2 CCG GCC CAC TTC TCT TTC TTT CTA ACT CGA GTT AGA AAG AAA GAG AAG TGG GTT TTT 3′) or mouse XBP1 (sh-1 5′ CCG GAG ATA GAA AGA AAG CCC GGA TCT CGA GAT CCG GGC TTT CTT TCT ATC TTT TTT 3′, sh-2 5′ CCG GCC AGG AGT TAA GAA CAC GCT TCT CGA GAA GCG TGT TCT TAA CTC CTG GTT TTT 3′). A scrambled shRNA was used as a control. Lentivirus was produced by cotransfecting the shRNA construct with packaging plasmids VSVG and psPAX2 into 293T cells using TransIT LT-1 Reagent (Mirus Bio). Twenty-four hours posttransfection, medium was replaced by fresh complete medium. Lentivirus from the supernatant was then collected at 24 and 48 hours. Ba/F3-MPL-CALR cells growing in IL3 were transduced with lentiviral supernatant in the presence of 8 μg/mL polybrene (Sigma). Cell selection began 48 hours posttransduction with 2 μg/mL puromycin (Gibco). Selected cells were then expanded and validated by qPCR and Western blot analyses.
Cell Proliferation Assay
Cultured Ba/F3-MPL or UT-7-MPL cells were washed four times with PBS to starve of cytokine, then grown in RPMI1640 supplemented with 10% FBS (Ba/F3) or 20% FBS (UT-7) and 5% penicillin/streptomycin. Cells were then seeded in triplicate with the indicated treatments at 1.25 × 105 cells/mL in the presence or absence of cytokine for the indicated time points. Living cells were counted at each time point using a Vi-CELL automated cell counter (Beckman Coulter).
Mice
All animal use and experiments performed were approved by Institutional Animal Care and Use Committee (ACUP#72596) at the University of Chicago. Six- to 8-week-old C57BL/6 female mice were purchased from The Jackson Laboratory. For BMT experiments, retroviral supernatants were generated by transient cotransfection of 293T cells with MSCV-IRES-GFP-CALRwt, -CALRdel52 or -CALRins5 and EcoPak constructs using TransIT LT-1 Reagent (Mirus Bio). Viral supernatant was collected 24 and 48 hours after transfection. Two days before the transplant, BM cells were collected from the femurs and tibia of donor mice. Cells were incubated with CD117 (c-KIT) MicroBeads (Miltenyi Biotec) and subjected to positive selection using an autoMACS Pro Separator (Miltenyi Biotec). c-KIT–enriched cells were then cultured overnight in SFEM medium supplemented with 50 ng/mL recombinant murine TPO, 50 ng/mL recombinant murine SCF, 10 ng/mL recombinant murine IL3, and 10 ng/mL recombinant murine IL6. Eighteen hours later, cells were infected with retroviral supernatant by spin infection on RetroNectin-coated plates (Takara Bio) and cultured overnight in SFEM media containing 50 ng/mL recombinant murine TPO, 50 ng/mL recombinant murine SCF, 10 ng/mL recombinant murine IL3, and 10 ng/mL recombinant murine IL6. The following day, cells (1 × 106 per mouse) were resuspended in Hank Balanced Salt Solution and injected retro-orbitally into lethally irradiated (900 cGy) C57 BL/6 recipient mice. Peripheral blood was collected retro-orbitally at indicated time points to measure CBC.
Human Material
Peripheral blood cells from 7 patients with MPNs were kindly provided by Steffen Koschmieder (RWTH Aachen University, Aachen, Germany). Peripheral blood cells from 6 additional patients with MPNs were kindly provided by and Elisa Rumi and Daniela Pietra, University of Pavia and Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy. Inclusion criteria for sample collection were diagnosis of an MPN and presence of a CALR mutation. Peripheral blood samples from patients with MPNs were collected on the dates indicated in Supplementary Table S2, and stored at the central biomaterial bank of the Faculty of Medicine at RWTH Aachen University (Aachen, Germany), or the Hematology Oncology Division Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo (San Matteo, Italy). Samples were obtained after written informed consent and approval of the local ethics committee according to the Declaration of Helsinki.
CellTiter-Glo Assay
PBMCs were cultured in StemSpan serum-free expansion medium (SFEM; StemCell Technologies) supplemented with 1% penicillin/streptomycin (Corning), SCF (50 ng/mL), Flt3-L (50 ng/mL), IL3 (10 ng/mL), and IL6 (10 ng/mL) for 24 hours prior to treating with KIRA8 (500 nmol/L) and seeding in a 96-well white-walled plate (10,000 cells per well). CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used to determine cell viability via ATP luminescence. One-hundred microliter of CellTiter-Glo Reagent was added to PBMCs in 100 μL of media. Cells were lysed via orbital shaking and luminescence was recorded.
Annexin V/Propidium Iodide Stain Flow Cytometry
Ba/F3-MPL or UT7-MPL cells were harvested, washed three times with PBS and stained with FITC-Annexin V (BD) and propidium iodide, and incubated in the dark at room temperature for 20 minutes. Cells were then re-suspended in 1× Annexin V Binding Buffer (BD) and the percentage of apoptotic and dead cells was measured on the Beckman CytoFLEX S flow cytometer.
Drug Treatments
Venetoclax (ABT-199) was purchased from Abcam. 2-Aminoethoxydiphenyl borate (2-APB) was purchased from Santa Cruz Biotechnology. KIRA8 was a gift from Scott Oakes.
IHC
Spleens and BM were resected, fixed in formalin, and embedded in paraffin before being mounted onto Vectabond-coated Superfrost Plus Slides. Slides were baked at 60°C overnight, deparaffinized by 5-minute washes three times in xylene, hydrated in a graded series of ethanol washes, and rinsed with distilled water. Epitopes were retrieved by heating samples in 10 mmol/L Tris-EDTA buffer, pH 9 on high until boiling followed by 10 minutes on low. For XPB1 staining, sections were incubated in an XBP1 antibody (Abcam), diluted 1:500 using RTU horse serum overnight at 4°C. After incubation in the primary antibody, sections were washed and incubated in ImmPRESS secondary antibody for 1 hour at room temperature. Sections were then washed three times and nuclear stained with hematoxylin before two graded series of ethanol washes and one xylene wash for 30 minutes prior to mounting using Permount. Images were captured using the Zeiss Axioskop upright histology microscope at 40× magnification and analyzed using ImageJ software and 3D hisTECH.
Histopathology
Mouse tissues (BM and spleen) were fixed in 10% neutral buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Images of histologic slides were obtained on the CRi Pannoramic MIDI scanner (Cambridge Research and Instrumentation). BM megakaryocytes were analyzed and quantified by two independent pathologists who were blinded to mouse genotype, and represent an average megakaryocyte count from 10 high-power fields assessed.
CBC Analysis
Mice were bled retro-orbitally every 4 weeks post BM transplant. Seventy-five microliters of blood was collected in heparinized capillary tubes and transferred to EDTA tubes for analysis. Complete blood counts were obtained by running the blood on the Hemavet 950 FS Auto Blood Analyzer (Drew Scientific).
Statistical Analysis
All comparisons represent two-tailed unpaired Student t test analysis unless otherwise specified (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Data Sharing Statement
Gene expression data are available in the GEO database with the accession number GSE173805.
Supplementary Material
Acknowledgments
We would like to thank Vytas Bindokas and Christine Labnoe in the Integrated Light Microscopy Core at the University of Chicago for invaluable microscopy training and guidance in all of our cell imaging studies, Pieter Faber in the Genomics Facility at the University of Chicago for executing our RNA-seq experiments during the COVID-19 shutdown, and Akash Patnaik, Alex Muir, Kay Macleod, Lucy Godley and Marsha Rosner for providing feedback and suggestions that greatly improved our work. We also thank Dongbo (Peter) Yang for his gracious assistance with our RNA-seq analysis. We would also like to thank Jing Chen and Alex Rosencrance for careful and critical review of our manuscript. This work was supported by the Goldblatt Endowment (to J. Ibarra), the Multi-disciplinary Training Program in Cancer Research (NIH T32 CA009594; to J. Ibarra), the Italian Association for Cancer Research (to M. Ciboddo), the Deutsche Forschungsgemeinschaft [German Research Foundation - KO2155/6-1 and KO2155/7-1 (Clinical Research Unit CRU344) to S. Koschmieder], the NIH [R00HL136924, to S.E. Elf; R01CA219815, to S.A. Oakes; R01EY027810, to S.A. Oakes; U01DK127786, to S.A. Oakes], the American Society of Hematology Scholar Award (to S.E. Elf), the Leukemia and Lymphoma Society Career Development Program (CDP) Special Fellow Award (to S.E. Elf), the Leukemia and Lymphoma Society Career Achievement Award (to S.E. Elf), Gabrielle's Angel Foundation in partnership with the Mark Foundation for Cancer Research (to S.E. Elf), and the Ludwig Fund for Cancer Research (to S.E. Elf).
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
J. Ibarra reports grants from NIH T32 MTRC, Cancer Research Foundation Goldblatt Fellowship, Robert C and Mary Jane Gallo Scholarship, and grants from American Society of Hematology Abstract Achievement Award during the conduct of the study. S. Koschmieder reports grants, personal fees, and non-financial support from Novartis, Bristol-Myers Squibb, Janssen/Geron, AOP Pharma; personal fees and non-financial support from Pfizer, Incyte, Ariad, Celgene, CTI, Roche, Baxalta, Sanofi, Abbvie, Alexion, Karthos; and grants, personal fees, and other support from RWTH Aachen University outside the submitted work; in addition, S. Koschmieder reports funding from Novartis, Bristol-Myers Squibb, Janssen/Geron; advisory board honoraria from Pfizer, Incyte, Ariad, Novartis, AOP Pharma, BMS, Celgene, Geron, Janssen, CTI, Roche, Baxalta, Sanofi; patent for BET inhibitor at RWTH Aachen University; honoraria from Novartis, BMS, Celgene, Geron, Janssen, Pfizer, Incyte, Ariad, Shire, Roche, Abbvie, and AOP Pharma; and other financial support (e.g., travel support) from Alexion, Novartis, BMS, Incyte, Ariad, AOP Pharma, Baxalta, CTI, Pfizer, Sanofi, Celgene, Shire, Janssen, Geron, Abbvie, and Karthos. S. Gurbuxani reports honorarium from AbbVie for consulting. S.A. Oakes reports personal fees from OptiKira, LLC outside the submitted work; in addition, S.A. Oakes has a patent for Modulation of IRE1 (publication number: 20160024094 issued, licensed, and with royalties paid, a patent for inventors: Bradley J. Backes, Dustin J. Maly, Scott A. Oakes, Feroz R. Papa, Gayani Perera, Likun Wang) and Substituted Imidazo[1,5-a]pyrazines for Modulation of Ire1 (patent number: 10131668 issued and licensed, a patent for inventors: Dustin J. Maly, Bradley J. Backes, Scott A. Oakes, Feroz R. Papa, Rajarshi Ghosh, Likun Wang), Combined Modulation of IRE1 (publication number: 20170165259; abstract: described herein, inter alia, are combined compositions of an Ire1 kinase modulating compound and an Ire1 ribonuclease modulating compound and methods of using the same; type: application; filed: December 21, 2016; publication date: June 15, 2017; issued, a patent for inventors: Richard M. Keenan, Bradley J. Backes, Dustin J. Maly, Charles Reynolds, Ben Whittaker, Jamie Knight, Jon Sutton, George Hynd, Feroz R. Papa, Scott A. Oakes), Compounds and Compositions for Ire1 Inhibition (issued, and a patent for inventors: Raymond E. Moellering, Zeyu Qiao, Scott A. Oakes, Shannon E. Elf, Marsha Rosner, Chi Long Nguyen), and a patent for Compositions and Therapeutic Methods for DNA Binding and Transcriptional Regulation (application number: 63/282,647 pending). No disclosures were reported by the other authors.
Authors’ Contributions
J. Ibarra: Formal analysis, validation, investigation, visualization, methodology, writing–original draft. Y.A. Elbanna: Formal analysis, validation, investigation, visualization, methodology, writing–original draft. K. Kurylowicz: Formal analysis, validation, investigation, visualization, methodology, writing–original draft. M. Ciboddo: Data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft. H.S. Greenbaum: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. N.S. Arellano: Conceptualization, resources, data curation, formal analysis, validation, investigation, writing–original draft, writing–review and editing. D. Rodriguez: Formal analysis, supervision, investigation, writing–review and editing. M. Evers: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. A. Bock-Hughes: Software, formal analysis, validation, investigation, visualization, methodology. C. Liu: Software, formal analysis, validation, investigation, visualization, methodology. Q. Smith: Formal analysis, validation, investigation, visualization, methodology. J. Lutze: Formal analysis, supervision, validation, investigation, visualization, methodology. J. Baumeister: Resources, data curation, formal analysis, supervision. M. Kalmer: Resources, data curation. K. Olschok: Resources, data curation. B. Nicholson: Resources, data curation, methodology. D. Silva: Resources, data curation, investigation, methodology. L. Maxwell: Investigation. J. Dowgielewicz: Investigation. E. Rumi: Resources, data curation, formal analysis, investigation. D. Pietra: Resources, data curation. I.C. Casetti: Resources, data curation. S. Catricala: Conceptualization, resources, data curation, formal analysis, validation, investigation, writing-original draft, writing–review and editing. S. Koschmieder: Resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft. S. Gurbuxani: Resources, data curation, formal analysis. R.K. Schneider: Data curation, formal analysis, investigation, visualization, methodology. S.A. Oakes: Data curation, formal analysis, supervision, investigation, visualization, methodology, writing–review and editing. S.E. Elf: Conceptualization, resources, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing-original draft, project administration, writing–review and editing.
References
- 1. Campbell PJ, Green AR. The myeloproliferative disorders. N Engl J Med 2006;355:2452–66. [DOI] [PubMed] [Google Scholar]
- 2. Levine RL, Gilliland DG. Myeloproliferative disorders. Blood 2008;112:2190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spivak JL. Myeloproliferative neoplasms. N Engl J Med 2017;376:2168–81. [DOI] [PubMed] [Google Scholar]
- 4. Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013;369:2379–90. [DOI] [PubMed] [Google Scholar]
- 5. Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 2013;369:2391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michalak M, Corbett EF, Mesaeli N, Nakamura K, Opas M. Calreticulin: one protein, one gene, many functions. Biochem J 1999;344:281–92. [PMC free article] [PubMed] [Google Scholar]
- 7. Chachoua I, Pecquet C, El-Khoury M, Nivarthi H, Albu RI, Marty C, et al. Thrombopoietin receptor activation by myeloproliferative neoplasm associated calreticulin mutants. Blood 2016;127:1325–35. [DOI] [PubMed] [Google Scholar]
- 8. Araki M, Yang Y, Masubuchi N, Hironaka Y, Takei H, Morishita S, et al. Activation of the thrombopoietin receptor by mutant calreticulin in CALR-mutant myeloproliferative neoplasms. Blood 2016;127:1307–16. [DOI] [PubMed] [Google Scholar]
- 9. Elf S, Abdelfattah NS, Chen E, Perales-Patôn J, Rosen EA, Ko A, et al. Mutant calreticulin requires both its mutant C-terminus and the thrombopoietin receptor for oncogenic transformation. Cancer Discov 2016;6:368–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tefferi A, Wassie EA, Guglielmelli P, Gangat N, Belachew AA, Lasho TL, et al. Type I versus Type II calreticulin mutations in essential thrombocythemia: a collaborative study of 1027 patients. Am J Hematol 2014;89:E121–4. [DOI] [PubMed] [Google Scholar]
- 11. Pietra D, Rumi E, Ferretti VV, Di Buduo CA, Milanesi C, Cavalloni C, et al. Differential clinical effects of different mutation subtypes in CALR-mutant myeloproliferative neoplasms. Leukemia 2016;30:431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guglielmelli P, Biamonte F, Rotunno G, Artusi V, Artuso L, Bernardis I, et al. Impact of mutational status on outcomes in myelofibrosis patients treated with ruxolitinib in the COMFORT-II study. Blood 2014;123:2157–60. [DOI] [PubMed] [Google Scholar]
- 13. Wang X, Ye F, Tripodi J, Hu CS, Qiu J, Najfeld V, et al. JAK2 inhibitors do not affect stem cells present in the spleens of patients with myelofibrosis. Blood 2014;124:2987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deininger M, Radich J, Burn TC, Huber R, Paranagama D, Verstovsek S. The effect of long-term ruxolitinib treatment on JAK2p.V617F allele burden in patients with myelofibrosis. Blood 2015;126:1551–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vannucchi AM, Kantarjian HM, Kiladjian JJ, Gotlib J, Cervantes F, Mesa RA, et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica 2015;100:1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, et al. Efficacy, safety, and survival with ruxolitinib in patients with myelofibrosis: results of a median 3-year follow-up of COMFORT-I. Haematologica 2015;100:479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison CN, Vannucchi AM, Kiladjian JJ, Al-Ali HK, Gisslinger H, Knoops L, et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia 2016;30:1701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verstovsek S, Mesa RA, Gotlib J, Gupta V, DiPersio JF, Catalano JV, et al. Long-term treatment with ruxolitinib for patients with myelofibrosis: 5-year update from the randomized, double-blind, placebo-controlled, phase 3 COMFORT-I trial. J Hematol Oncol 2017;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vainchenker W, Leroy E, Gilles L, Marty C, Plo I, Constantinescu SN. JAK inhibitors for the treatment of myeloproliferative neoplasms and other disorders. F1000Res. 2018;7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Buduo CA, Abbonante V, Marty C, Moccia F, Rumi E, Pietra D, et al. Defective interaction of mutant calreticulin and SOCE in megakaryocytes from patients with myeloproliferative neoplasms. Blood 2020;135:133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, et al. STIM is a calcium sensor essential for calcium-store-depletion-triggered calcium influx. Curr Biol 2005;15:1235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 2011;334:1081–6. [DOI] [PubMed] [Google Scholar]
- 23. Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer 2014;14:581–97. [DOI] [PubMed] [Google Scholar]
- 24. Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress–activated cell reprogramming in oncogenesis. Cancer Discov 2015;5:586–97. [DOI] [PubMed] [Google Scholar]
- 25. Salati S, Genovese E, Carretta C, Zini R, Bartalucci N, Prudente Z, et al. Calreticulin Ins5 and Del52 mutations impair unfolded protein and oxidative stress responses in K562 cells expressing CALR-mutants. Sci Rep 2019;9,10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nam AS, Kim KT, Chaligne R, Izzo F, Ang C, Taylor J, et al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 2019;571:355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pronier E, Cifani P, Merlinsky TR, Berman KB, Somasundara AVH, Rampal RK, et al. Targeting the CALR interactome in myeloproliferative neoplasms. JCI Insight 2018;3:e122703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elf S, Abdelfattah NS, Baral AJ, Beeson D, Rivera JF, Ko A. Defining the requirements for the pathogenic interaction between mutant calreticulin and MPL in MPN. Blood 2018;131:782–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, et al. Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell 2014;158:534–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campbell KP, MacLennan DH, Jorgensen AO. Staining of the calcium-binding proteins, calsequestrin, calmodulin, troponin C, and S-100, with the cationic carbocyanine dye “Stains-all.” J Biol Chem 1983;258:11267–73. [PubMed] [Google Scholar]
- 31. Suzuki J, Kanemaru K, Ishii K, Ohkura M, Okubo Y, Masamisu I. Imaging intraorganellar calcium at subcellular resolution using CEPIA. Nat Commun 2014;5:4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol Ther 2008;16:698–706. [DOI] [PubMed] [Google Scholar]
- 33. Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its calcium binding domains. J Biol Chem 1991;266:21458–65. [PubMed] [Google Scholar]
- 34. Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J 1996;15:4130–41. [PMC free article] [PubMed] [Google Scholar]
- 35. Distelhorst CW, McCormick TS. Bcl-2 acts subsequent to and independent of calcium fluxes to inhibit apoptosis in thapsigargin- and glucocorticoid-treated mouse lymphoma cells. Cell calcium 1996;19:473–83. [DOI] [PubMed] [Google Scholar]
- 36. Foyouzi-Youssefi R, Arnaudeau S, Borner C, Kelley WL, Tschopp J, Lew DP, et al. Bcl-2 decreases the free calcium concentration within the endoplasmic reticulum. Proc Natl Acad Sci U S A 2000;97:5723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oakes SA, Opferman JT, Pozzan T, Korsmeyer SJ, Scorrano L. Regulation of endoplasmic reticulum calcium dynamics by proapoptotic BCL-2 family members. Biochem Pharmacol 2003;66:1335–40. [DOI] [PubMed] [Google Scholar]
- 38. Chonghaile TN, Gupta S, John M, Szegezdi E, Logue SE, Samali A. BCL-2 modulates the unfolded protein response by enhancing splicing of X-box binding protein-1. Biochem Biophys Res Commun 2015;466:40–5. [DOI] [PubMed] [Google Scholar]
- 39. Mak DO, Foskett JK. Inositol 1,4,5-trisphosphate receptors in the endoplasmic reticulum: a single-channel point of view. Cell calcium 2015;58:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen R, Valencia I, Zhong F, McColl KS, Roderick HL, Bootman MD, et al. Bcl-2 functionally interacts with inositol 1,4,5-trisphosphate receptors to regulate calcium release from the ER in response to inositol 1,4,5-trisphosphate. J Cell Biol 2004;166:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peppiatt CM, Collins TJ, Mackenzie L, Conway SJ, Holmes AB, Bootman MD, et al. 2-Aminoethoxydiphenyl borate (2-APB) antagonises inositol 1,4,5-trisphosphate-induced calcium release, inhibits calcium pumps and has a use-dependent and slowly reversible action on store-operated calcium entry channels. Cell calcium 2003;34:97–108. [DOI] [PubMed] [Google Scholar]
- 42. Thamsen M, Ghosh R, Auyeung VC, Brumwell A, Chapman HA, Backes BJ, et al. Small molecule inhibition of IRE1α kinase/RNase has anti-fibrotic effects in the lung. PLoS One 2019;14:e0209824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marty C, Pecquet C, Nivarthi H, El-Khoury M, Chachoua I, Tulliez M, et al. Calreticulin mutants in mice induce an MPL-dependent thrombocytosis with frequent progression to myelofibrosis. Blood 2016;127:1317–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.