Abstract
Salicinoids are salicyl alcohol-containing phenolic glycosides with strong antiherbivore effects found only in poplars and willows. Their biosynthesis is poorly understood, but recently a UDP-dependent glycosyltransferase, UGT71L1, was shown to be required for salicinoid biosynthesis in poplar tissue cultures. UGT71L1 specifically glycosylates salicyl benzoate, a proposed salicinoid intermediate. Here, we analyzed transgenic CRISPR/Cas9-generated UGT71L1 knockout plants. Metabolomic analyses revealed substantial reductions in the major salicinoids, confirming the central role of the enzyme in salicinoid biosynthesis. Correspondingly, UGT71L1 knockouts were preferred to wild-type by white-marked tussock moth (Orgyia leucostigma) larvae in bioassays. Greenhouse-grown knockout plants showed substantial growth alterations, with decreased internode length and smaller serrated leaves. Reinserting a functional UGT71L1 gene in a transgenic rescue experiment demonstrated that these effects were due only to the loss of UGT71L1. The knockouts contained elevated salicylate (SA) and jasmonate (JA) concentrations, and also had enhanced expression of SA- and JA-related genes. SA is predicted to be released by UGT71L1 disruption, if salicyl salicylate is a pathway intermediate and UGT71L1 substrate. This idea was supported by showing that salicyl salicylate can be glucosylated by recombinant UGT71L1, providing a potential link of salicinoid metabolism to SA and growth impacts. Connecting this pathway with growth could imply that salicinoids are under additional evolutionary constraints beyond selective pressure by herbivores.
Knockout of a recently discovered enzyme of salicinoid biosynthesis in poplar leads to changes in salicylic acid and jasmonic acid metabolism and has unexpected effects on growth.
IN A NUTSHELL.
Background: Poplars and willows make abundant anti-herbivore compounds called salicinoids, the original source of the widely used analgesic aspirin. Salicinoids are derived from a phenylpropanoid compound called cinnamic acid, but how the saliciniods are synthesized is still unknown. Previously, we reported that a glycosyl transferase enzyme called UGT71L1 is needed for salicinoid biosynthesis, but the other enzymes in the pathway are not known.
Question: Since UGT71L1 is necessary for the biosynthesis of the salicinoids, what happens if we disrupt this pathway by CRISPR/Cas9 in transgenic poplar plants? We hypothesized that in plants that lack the UGT71L1 enzyme and have disrupted salicinoid biosynthesis, we might be able to identify potential intermediates and other chemicals that would provide clues to help us elucidate the salicinoid pathway.
Findings: Transgenic poplars with CRISPR/Cas9 disruption of UGT71L1 accumulated only small amounts of salicinoids as predicted. These plants also showed unexpected growth abnormalities such as smaller serrated leaves, reduced internode length, and smaller stature. This was surprising, since salicinoids and other phenylpropanoid products are not known to affect plant growth. We therefore performed a rescue experiment and reinserted a functional UGT71L1 gene back into the knockouts. This corrected both the chemical and physical alterations, thus demonstrating directly that UGT71L1 disruption alone can cause developmental abnormalities. We then carried out metabolomic and transcriptomic analyses of the UGT71L1 knockouts. We discovered that the UGT71L1 knockouts have much higher levels of salicylic acid and jasmonic acid, well-known plant defense hormones. These hormones inhibit growth and could thus explain the physical phenotype. We hypothesized that extra salicylic acid comes from an unstable intermediate, and thus we proposed that salicyl salicylate is a UGT71L1 substrate and pathway intermediate.
Next steps: Next steps are to validate salicyl salicylate glucoside as an intermediate and to determine how salicyl salicylate glucoside is processed to salicortin. We want to identify new enzymes that carry out the subsequent transformations, and ultimately to complete the biosynthetic pathway to the salicinoids.
Introduction
Poplars (Populus spp.) have been widely adopted as the model species for tree genomics, molecular biology, and physiology. A major advantage of Populus as an experimental system is its susceptibility to Agrobacterium-mediated transformation (Brunner et al., 2004), and clustered regularly interspaced short palindromic repeats (CRISPR)/associated protein 9 (Cas9) generated knockouts have become powerful resources for understanding gene function in these trees (Elorriaga et al., 2018). Poplars contain abundant and diverse phenolic metabolites, which make them attractive plants for studies on ecological biochemistry. The most abundant phenolics found in poplar leaves are the proanthocyanidins (condensed tannins), hydroxycinnamate esters, and the phenolic glycosides known as salicinoids. These are unique compounds built from glucosylated salicyl alcohol, typically further esterified with other organic acids including benzoate, p-coumarate, acetate, or a 6-hydroxy-2-cyclohexen-on-oyl (HCH) moiety. Salicinoids are found only in the Salicaceae family, composed of poplars and willows (Salix). Approximately 35 salicinoid structures have been described from various Salix and Populus species (Boeckler et al., 2011; Keefover-Ring et al., 2014). Populus tremula � alba, a hybrid commonly used in research for its ease of genetic transformation (Han et al., 2000), accumulates tremulacin, salicortin, salicin, and tremuloidin as the predominant salicinoids (Figure�1). Due to their bioactivity, salicinoids are important in traditional medicine among many indigenous communities (Desborough and Keeling, 2017).
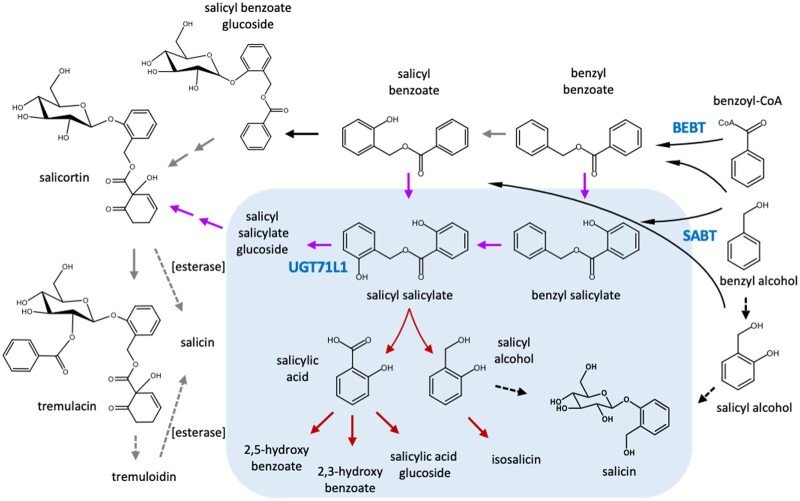
Figure 1.
Proposed biosynthetic pathway for salicinoids based on prior work in various Populus or Salix species and the current experiments. Alternate entry points starting with BEBT and SABT are shown. Solid black arrows and blue font indicate established enzymatic reactions (Chedgy et al., 2015; Fellenberg et al., 2020; Kulasekaran et al., 2021); dashed black arrows are proposed biosynthetic steps based on published in vivo labeling experiments (Zenk, 1967; Babst et al., 2010); dashed gray arrows represent known nonenzymatic or enzymatic decomposition steps (Ruuhola et al., 2003; Julkunen-Tiitto and Sorsa, 2001); gray arrows indicate hypothesized biosynthetic reactions. Compounds within the shaded background and purple arrows indicate proposed biosynthetic steps and products based on the current work. Red arrows in the shaded area represent additional reactions deduced to be enhanced in UGT71L1-KO plants based on metabolomic analysis in this study.
Salicinoids are abundant in vegetative tissues including leaves, where they can reach concentrations of over 15% dry weight (Donaldson et al., 2006). Complex salicinoids such as salicortin and tremulacin that contain the unstable HCH moiety are toxic when ingested and act as antiherbivory compounds (Boeckler et al., 2011; Lindroth and Clair, 2013). High salicinoid concentrations correlate with poor performance of gypsy moth (Lymantria dispar) and forest tent caterpillar larvae (Malacosoma disstria; Hwang and Lindroth, 1997). Salicinoids also have an inhibitory effect on mammalian herbivores (Tahvanainen et al., 1985; Basey et al., 1988). Unlike many other defensive secondary metabolites, herbivory or tissue damage does not appreciably induce the accumulation of salicinoids and they are considered constitutive defense compounds (Boeckler et al., 2011, 2013).
Despite the ecological and potential medical importance of the salicinoids, their biosynthesis remains largely unelucidated. Early radiolabeling experiments by Zenk (1967) in Salix purpurea demonstrated that salicin is derived from cinnamic acid or benzoic acid via salicylaldehyde. Similar experiments to probe the biosynthesis of more complex salicortin were reported by Babst et al. (2010), who determined that benzoic acid and benzaldehyde are readily incorporated into both salicyl alcohol and HCH moieties of salicortin, but benzyl alcohol contributes exclusively to the salicyl alcohol moiety. In contrast, the labeling data indicate that free salicyl alcohol or salicylaldehyde are not intermediates in the biosynthesis of salicortin (Babst et al., 2010). Rather, salicyl alcohol and salicylaldehyde are incorporated into salicin, suggesting that the biosynthetic routes for salicin and salicortin are at least partly distinct. However, salicylic acid (SA) is not a precursor for salicinoids. Babst et al. (2010) also proposed benzyl benzoate as a potential intermediate in salicinoid biosynthesis, and enzymes for the condensation of benzyl alcohol and benzoyl-CoA to benzyl benzoate had been previously identified in Petunia hybrida and Nicotiana benthamiana. Furthermore, the conversion of cinnamate to benzaldehyde, benzyl alcohol, or benzoic acid via cinnamoyl-CoA-dependent and -independent pathways has been demonstrated (Widhalm and Dudareva, 2015). In poplar, benzyl benzoate and other benzenoid leaf volatiles are induced by herbivory and are synthesized from cinnamoyl-CoA by β-oxidative pathway enzymes (Lackus et al., 2021); however, the pathway for these volatiles appears to be separate from the biosynthesis of salicinoids. Details of benzenoid biosynthesis relevant for salicinoid biosynthesis remain unknown but are actively under investigation.
The idea of potential involvement of benzyl benzoate or salicyl benzoate in salicinoid synthesis was strengthened by the identification of poplar benzoyl-CoA: benzyl alcohol O-benzoyltransferase (BEBT) and benzoyl-CoA: salicyl alcohol O-benzoyltransferase (SABT), two acyl transferases that specifically synthesize benzyl benzoate and salicyl benzoate, respectively (Chedgy et al., 2015). The involvement of these potential intermediates was further corroborated when we identified two uridine diphosphate-dependent glucosyl transferases (UGTs), UGT71L1 and UGT78M1, which are co-expressed with SABT and show high specificity for salicyl benzoate as the glucose acceptor (Fellenberg et al., 2020). BEBT, SABT, and the UGTs thus provide the outlines of a pathway to salicortin and other complex salicinoids via benzyl benzoate, salicyl benzoate, and its glucoside (Figure�1). We demonstrated the central importance of salicyl benzoate and UGT71L1 using CRISPR/Cas9 knockout hairy root lines of UGT71L1, which are unable to make salicortin, tremulacin, and tremuloidin. These knockout lines did retain about 50% of salicin content, however, confirming the existence of an alternate biosynthetic route to salicin (Babst et al., 2010). Similar results have been obtained using RNAi suppression of UGT71L1 carried out independently (Kulasekaran et al., 2021). How salicyl benzoate glucoside is further oxidized to give rise to the HCH moiety is not known, but likely begins with hydroxylation of the aromatic ring. To date, no other enzymes or intermediates of the pathway have been identified.
The presence of salicinoids in the Salicaceae is notable because willow bark was the source of the original identification of SA as an active analgesic in the 19th century (Ding and Ding, 2020). The source of SA in willow and poplar extracts is now known to be the salicinoids, which decompose spontaneously or enzymatically (Ruuhola et al., 2003). However, SA itself is not a direct precursor of salicinoids. Although SA is well known as a bioactive plant phenolic for over a century, its function as a stress hormone was not discovered until 1979, when its role in inducing resistance against viruses and as an inducer of plant defense genes was described (Raskin, 1992). How SA acts as a signal for plant defense has been intensively studied since then, leading to detailed current knowledge of NPR1 and NPR3/NPR4 SA receptors and signaling pathways (Ding et al., 2018; Innes, 2018; Liu et al., 2020). Following pathogen detection, SA levels rise and signal a transcriptional response leading to the local expression of defense genes as well as triggering secondary signals that activate a systemic defense response (Ding and Ding, 2020). Treatment of Arabidopsis plants with SA alters the expression of thousands of genes, upregulating defense as well as downregulating growth-related genes (van Butselaar and Van den Ackerveken, 2020). SA is involved in regulating senescence, modulating effects of other phytohormones, and controlling plant growth within a defense-growth trade-off (Zhang et al., 2013; Liu et al., 2016; Ding and Ding, 2020).
Detailed investigations in Arabidopsis and N. benthamiana have determined that SA is synthesized by distinct plastidic and cytosolic pathways. In the plastidic pathway, SA is derived from isochorismate via chorismate in the shikimate and aromatic amino acid pathway. Isochorismate is exported into the cytosol and converted to isochorismate-9-glutamate, which then spontaneously or enzymatically decomposes to release SA (Rekhter et al., 2019). In the cytosolic SA pathway, SA is derived from trans-cinnamic acid produced by phenylalanine ammonia lyase (PAL) of the phenylpropanoid pathway (Ryan et al., 1995; Zhang and Li, 2019). In Arabidopsis, the isochorismate pathway is responsible for the majority of pathogen-induced SA accumulation (Wildermuth et al., 2001). In contrast, in soybean, isochorismate-, and phenylpropanoid-derived SA contribute equally to pathogen-induced SA accumulation (Shine et al., 2016). SA biosynthesis and the relative importance of the isochorismate-derived SA and phenylpropanoid-derived SA in poplar has not been investigated. The importance of SA in defense against biotrophic rust fungi in poplar was recently demonstrated, however (Ullah et al., 2019).
To obtain additional insight into salicinoid biosynthesis, we generated transgenic CRISPR/Cas9 UGT71L1 knockout poplars. While hairy root cultures had provided initial proof of function of UGT71L1, rooted plants grown under greenhouse conditions accumulate higher levels of phenolics than tissue cultures and are thus more likely to show metabolic connections. Our objective was to use nontargeted and targeted metabolomics to identify changes in phenolic profiles caused by loss of UGT71L1 in order to identify potential intermediates in salicinoid biosynthesis. Our experiments confirmed the central in planta role of UGT71L1 in the salicinoid pathway, but also indicate that additional UGT enzymes may be play minor roles. Surprisingly, the loss of UGT71L1 in whole poplar plants manifested novel phenotypic effects including leaf curling and reduced expansion and growth. Additional experiments suggested that the effects on growth could be due to SA release from the disrupted salicinoid pathway, thus connecting a secondary metabolic pathway to growth and development.
Results
CRISPR/Cas9 generates diverse biallelic mutations in the UGT71L1 gene in transgenic P. tremula � alba plants
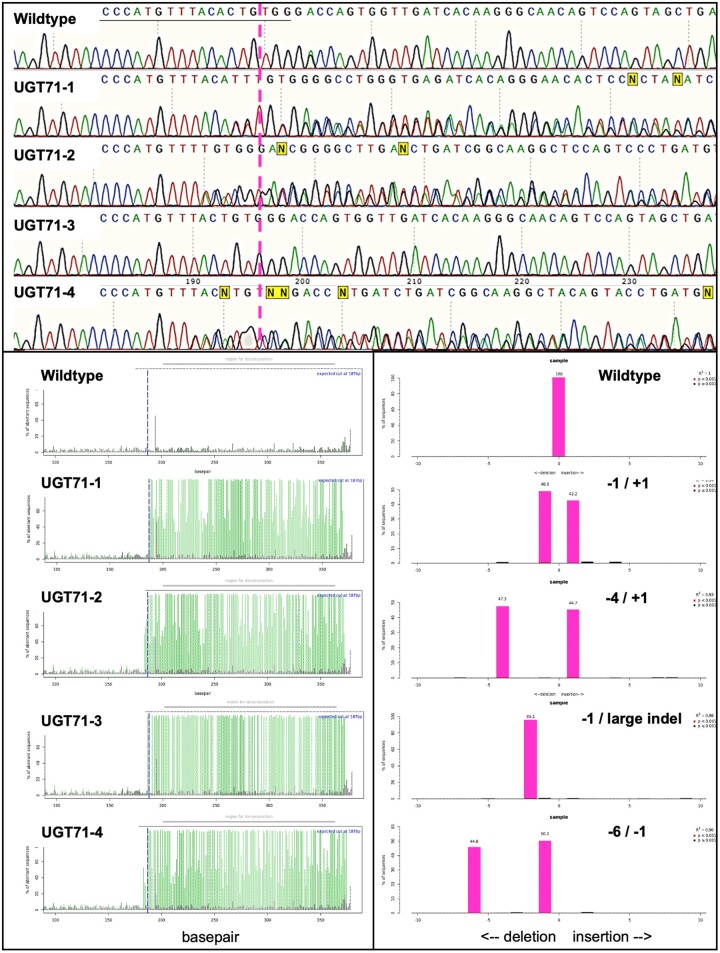
We generated independent P. tremula � alba UGT71L1 knockout lines using CRISPR/Cas9 and the previously utilized guide RNA (gRNA) sequence (Fellenberg et al., 2020). Preliminary analysis of regenerating in vitro UGT71L1 plantlets confirmed very low concentrations of tremulacin, tremuloidin, and salicortin compared to wild-type (WT) plants. Transgenic lines with low salicinoid accumulation were sequenced at the UGT71L1 gRNA site, and four lines carrying mutations in both UGT71L1 alleles were studied further. Mutations in these lines generally consisted of small indels with a mix of single base-pair (bp) insertions, single bp deletions, and slightly larger deletions (Figure�2). We also observed a large deletion in one allele of line UGT71-3. Large deletions are often reported in CRISPR/Cas9 knockouts in plants utilizing nonhomologous end joining (Shen et al., 2017). In addition, one allele in transgenic line UGT71-4 contained an in-frame 6-bp deletion and likely encoded a functional gene product. This transgenic line was considered a partial knockout and could be expected to show an intermediate phenotype. To guard against nontarget gene mutations, we used the polymerase chain reaction (PCR) to amplify and sequence the five genomic regions most similar to the target gRNA. No off-target mutations were detected (Supplemental Table S1).
Figure 2.
Detection of biallelic mutations and indels four independent UGT71L1-knockout lines. Upper panel shows Sanger sequencing traces of the UGT71L1 gRNA target site. The gRNA is underlined, and the magenta dashed line marks the Protospacer Adjacent Motif (PAM) necessary for Cas9 double-strand cleavage. N indicates ambiguous bases in the sequence readout. The lower panels show read-outs of the TIDE (shinyapps.datacurators.nl/tide/) webtool used to detect mutations in the target sequence. The vertical blue dashed line (left panel) indicates the position of the gRNA PAM site, while the vertical green traces indicate variant sequences in chromatograms indicating a mutation in the downstream sequence. Lower right panel shows the number of nucleotides inserted or deleted along the horizontal axis for each mutated line. Lines UGT71-1 to UGT71-3 are biallelic knockouts with frameshifts in both alleles. Line UGT71-4 has mutations in both alleles but only one results in a frameshift. This line is designated as partial-KO.
Metabolomic analysis of UGT71L1 knockout poplars demonstrates a substantial reduction in salicinoid content and a major shift in phenolic metabolites
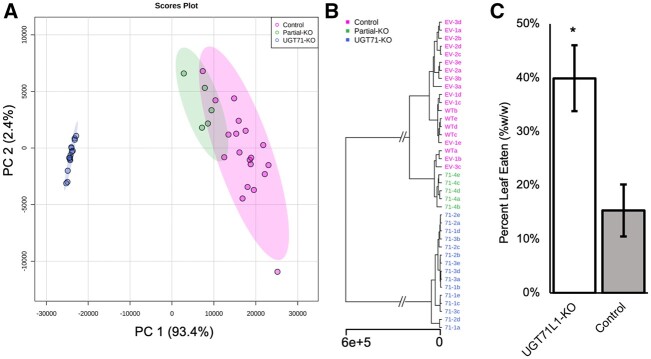
Both nontargeted and targeted metabolomic approaches were used to investigate the impact of eliminating UGT71L1 activity on poplar secondary metabolism. Methanolic leaf extracts from greenhouse-grown saplings were first profiled by ultra-high pressure liquid chromatography–high-resolution mass spectrometry (UPLC–HMRS). The experiment consisted of the replicated UGT71L1 biallelic and monoallelic knockout plants along with three independent empty vector lines and WT controls. Unsupervised multivariate data analyses revealed clear trends within this data set. Principal component analysis of nontargeted metabolomic data clearly separated the UGT71L1-knockout (KO) mutant from the UGT71L1 partial-KO (monoallelic) and control lines (Figure�3) along principal component 1 explaining 93.4% of the variance in the data model. Results from hierarchical cluster analysis indicated no difference between the empty vector and WT lines, and supported combining both into a single control group (Figure�3B). These untargeted analyses demonstrated the strong effect of disrupting UGT71L1; it also showed the quantitative effects of the gene on the metabolome, since the monoallelic mutant line was grouped by principal component analysis as an intermediate phenotype. Overall, the nontargeted analysis identified 3,019 features that differed significantly between UGT71L1-KO and control plants. From these, a list of 68 tentatively identified differentially expressed metabolites in the UGT71L1-KO lines was generated by manual inspection of spectra, annotation, accurate masses, and MS2 fragmentation analysis (Supplemental Table S2).
Figure 3.
Nontargeted metabolomic analysis of UGT71L1-KO plants. A, Principal component score plots of non-targeted metabolomic analysis of three independent UGT71L1-KO lines (five biological replicates each), one partial-KO line (four biological replicates), and controls consisting of three empty vector lines and a WT (four or five biological replicates each). B, Unweighted pair group method with arithmetic mean hierarchical cluster analysis of nontargeted metabolomic samples. Multivariate analysis was conducted using Metaboanalyst 5.0 (Chong et al., 2019). Blue color (left-most cluster in A, lowest branch in B) indicates UGT71L1-KO lines, green (middle cluster in A, central branch in B) indicates partial-KO lines, and magenta (right cluster in A, top branch in B) indicates empty vector or WT control lines. C, Orgyia leucostigma feeding preference (third and fourth instar larvae) expressed as percent leaf mass consumed. Leaf disks from two independent UGT71L1-KO lines were pooled for choice bioassays with disks from two independent empty vector lines. The entire choice test was conducted twice for a total of 25 arena assays (error bars indicate �se). *Indicates significant difference (two-tailed t test, P < 0.005).
We also probed the effect of UGT71L1 disruption in our transgenic plants by targeted metabolomic analysis (Lackus et al., 2020; 2021). This method targeted compounds known to occur in poplar and was validated with authentic standards, providing a positive identification for many of the features and compounds seen in the nontargeted analyses. We again carried out a multivariate analysis of the targeted metabolomic data set, which consisted of 40 identified poplar salicinoids and phenolics (Supplemental Table S3). Principal component analysis and hierarchical clustering again showed a clear separation of the transgenic types into three chemically distinct groups as in the nontargeted metabolomic analysis (Supplemental Figure S1).
Loss of UGT71L1 in whole plant transgenics led to a pronounced reduction in the concentrations of major salicinoids in leaves (Table�1). Tremulacin and salicortin exhibited reductions of more than 90%, whereas salicin concentration was reduced by 80% compared to control lines. Therefore, unlike our previous observations in hairy root cultures where disruption of UGT71L1 leads to the complete loss of salicortin and tremulacin, the transgenic plants were observed to retain a basal level of salicinoid biosynthesis. The biological impact of these phytochemical changes on herbivore preference was tested with a generalist tree-feeding lepidopteran, Orygia leucostigma. Choice assays with third and fourth instar larvae demonstrated that UGT71L1-KO leaf disks were consumed significantly more than control disks (Figure�3C).
Table 1.
Salicinoid content in leaves of transgenic UGT71L1-KO (biallelic), partial-KO (monoallelic), and control P. tremula�alba plants as determined by targeted metabolomic analysis.1
| Compound | UGT71L1-KO | Partial-KO | Control |
|---|---|---|---|
| Salicortin | 178.5 (�83.16)a | 2813.45 (�76.07)c | 3185.16 (�96.53)c |
| Tremulacin | 96.23 (�52.76)a | 1289.85 (�20.87)b | 1647.52 (�43.00)c |
| Salicin | 33.25 (�4.61)a | 149.04 (�4.88)c | 157.91 (�4.41)c |
| Salicin 7-sulfate | 13876.9 (�807.39)ns | 14517.65 (�654.9)ns | 12418.06 (�377.08)ns |
Values represent mean peak area�mg DW-1�10−3 � SE. UGT71L1-KO data are from three independent transgenic lines (four biological replicates/line); partial-KO data are from a single line (five biological replicates); control data are from a pool of three independently generated empty vector lines (four or five biological replicates/line) and a WT line (three biological replicates). Significant differences (P < 0.05) between genotypes for individual compounds are designated by different letters as determined by Tukey’s post hoc honest significant difference test. ns, non-significant.
The metabolomic analysis indicated that salicin-7-sulfate, a recently discovered poplar salicinoid (Noleto-Dias et al., 2018; Lackus et al., 2020) was only slightly affected by the loss of UGT71L1. Our targeted data showed equivalent concentrations across genotypes, whereas the nontargeted data showed a relative increase in UGT71L1-KO lines. The discrepancy between both data sets is likely due to a difference in leaf age, as the nontargeted data were derived from mature fully expanded leaf samples, whereas targeted analysis was conducted on younger expanding leaves. In addition, we observed an increase in the minor salicinoids such as salireposide and trichocarpin in UGT71L1-KO lines (Supplemental Table S3), suggesting parallel pathways and redundant UGTs. Grandidentatin, a related phenolic glycoside, also increased in concentration in UGT71L1 knockouts.
Both targeted and nontargeted metabolomic analyses showed that many nonsalicinoid phenolic compounds are severely impacted in UGT71L1-KO plants (Table�2). Reductions were observed for some flavonoids including kaempferol, dihydrokaempferol, apigenin and naringenin, and their glycosides (Supplemental Tables S2 and S3). Lignin content was also reduced, as determined through acetyl-bromide analysis (Supplemental Figure S2). We observed reduced levels of several caffeoyl quinic and coumaroyl quinic acids, which are abundant constituents of poplar leaves (Ma et al., 2018). In general, most identified phenolic compounds decreased in UGT71L1-KO plants (Table�2). However, some phenolics became more abundant, and we observed significant increases in concentration of small benzoic acid-derived compounds. For example, gentisic acid (2,5-dihydroxy benzoic acid) and 2,3-dihydroxy benzoic acid concentrations increased substantially in UGT71L1-KO plants. Benzoic acid and benzyl alcohol glucosides were elevated in UGT71L1-KO plants, though at overall low concentrations (Supplemental Tables S2 and S3). A compound tentatively identified via MS2 fragmentation analysis as benzoic acid glucoside-2,3-hydroxy-2-ethoxy-4,6-cyclohexene was significantly elevated in concentration in UGT71L1-KO plants. This compound shares structural similarities with benzoic acid glucoside and may be an intermediate molecule in the biosynthesis of the HCH moiety. Altogether, we observed broad impacts of UGT71L1 knockout on phenolic pathways, suggesting direct and indirect metabolic effects of UGT71L1.
Table 2.
Phenolic content in leaves of transgenic UGT71L1-KO (biallelic), partial-KO (monoallelic), and control P. tremula�alba plants as determined by nontargeted metabolomic analysis
| Compound type1 | UGT71L1-KO | Partial-KO | Control |
|---|---|---|---|
| Salicinoids | 5,196.27 (�285.35)a | 43,089.75 (�2,430.75)b | 58,145.9 (�2,127.27)c |
| Flavonols and flavanones | 369.08 (�25.16)a | 10,770.07 (�655.44)b | 12,380.45 (�433.35)c |
| Procyanidins | 583.94 (�36.78)a | 3,922.67 (�208.93)b | 4,658.04 (�174.13)c |
| Chlorogenic acids | 676.73 (�43.95)a | 1,515.57 (�138.87)c | 1,441.4 (�83.29)c |
| Lignin (percentage in PFCW)2 | 7.50% (�0.14)a | 11.10% (�0.23)c | 11.05% (�0.13)c |
Values indicate total combined average peak areas�10−4 (�se) of all identified compounds for each type of phenolic compound. Each biological replicate is an average value of three technical replicates. UGT71L1-KO data are from a pool of five biological replicates for each of three independent lines; partial-KO data represent five biological replicates of a single line; control data represents a pool of three independent empty vector lines each with four to five biological replicates and five biological replicates of a WT line.
Lignin concentrations are expressed as percentage by weight of the Protein-Free Cell Wall (PFCW), calculated as the mean of two technical replicates for each biological replicate. For UGT71L1-KO, three independent lines were used each with two biological replicates. Partial-KO concentrations were determined from a single independent line with three biological replicates, and in controls from a pool of three independent empty vector lines each with three biological replicates and one WT line with three biological replicates. Significant differences (P < 0.05) between genotype for individual compounds are designated by different letters as determined by a Tukey’s post hoc honest significant difference test.
Disruption of UGT71L1 leads to significant growth alterations that can be reversed by retransformation with a functional enzyme
While UGT71L1-KO plantlets in tissue culture showed minor and nonsignificant growth alterations, these became progressively more apparent after plants were acclimated and moved to the greenhouse. This was seen in all independently generated transgenic UGT71L1-KO poplar lines. We first observed that leaves from UGT71L1-KO plants had a serrated or irregular margin, with a narrower leaf shape than WT or empty vector controls and were smaller overall (Figure�4; Supplemental Figure S3). UGT71L1-KO plants also showed reduced internode length and grew more slowly. This was manifested as a reduction in stem diameter, plant height, internodal space, and leaf size for all biallelic knockout lines. In contrast, the monoallelic partial-KO lines were physically indistinguishable from control lines.
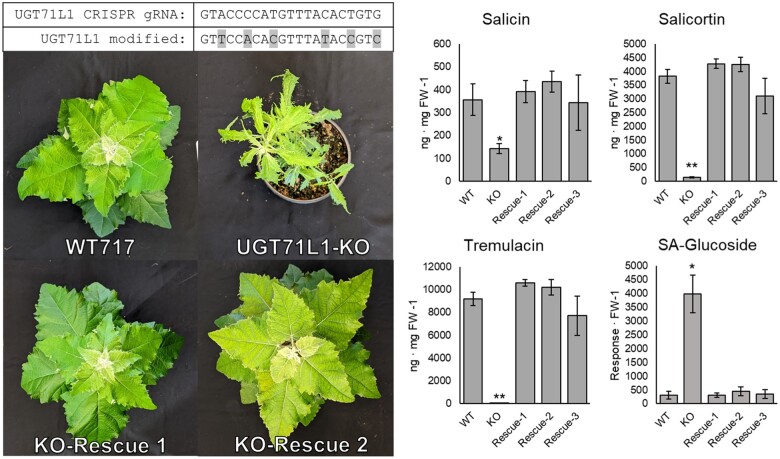
Figure 4.
Phenotypic effects of UGT71L1 disruption in transgenic poplar. A, Fully expanded leaves from 3-month-old greenhouse grown UGT71L1-KO plants (left) and empty vector control plants (right). Scale bar represents 10 cm. B, Abaxial view of fully expanded 3-month-old greenhouse grown UGT71L1-KO leaf. C, Abaxial view of corresponding control leaf. D, Whole plant image of UGT71L1-KO (left) and control (right) plants after 3 months of greenhouse growth. E, Change in plant height for UGT71L1-KO, partial-KO line, and control plants grown in greenhouse for 9 weeks. Data for seven to eight biological replicates each of three independent UGT71L1-KO lines, nine replicates of the partial-KO line, and pooled control samples from three independent empty vector lines and a WT plant (four to five replicates per line) are shown. Data are shown as mean � se.
The observed leaf and growth alterations were surprising, since physiological functions or growth effects for salicinoids have not been previously described. To directly test whether the severe growth phenotypes of UGT71L1-KO were due specifically to UGT71L1 disruption and not off-target or other effects, we carried out a transgenic rescue experiment. One of the biallelic UGT71L1-KO transgenic lines was retransformed with binary vector to insert a synthetic UGT71L1 sequence back into the mutant. The coding sequence of the synthetic gene had been modified to eliminate the gRNA binding site, making it immune to editing by CRISPR/Cas9. Transgenic rescued plants were regenerated, moved into soil and acclimated to the greenhouse in parallel with the original UGT71L1-KO line and the WT control. All transgenic rescue lines developed normally without any visible growth abnormalities, and rescued plantlets were physically indistinguishable from WT plants (Figure�5). UPLC quantification of the major salicinoids confirmed that salicinoid biosynthesis was also restored (Figure�5, right). These data also established that the growth phenotype observed in UGT71L1-KO is caused by the disruption of UGT71L1 and is not due to other effects of the CRISPR/Cas9 construct.
Figure 5.
Rescue of UGT71L1-KO plant phenotype by retransformation with a synthetic UGT71L1 coding sequence. Upper left (top) shows the gRNA target sequence for native UGT71L1 and the corresponding sequence modified to be resistant to the CRISPR/Cas9 construct. Degenerate codons were modified as indicated in gray highlights in the lower nucleotide sequence. Lower left images show two independently transformed 2-month-old rescue plant lines, together with a wild-type and a UGT71L1-KO line. Right show concentrations of major salicinoids and salicylic acid glucoside in expanded leaves of 2-month-old greenhouse-grown poplars. Values shown are mean � se. WT, wild-type with five biological replicates; KO, UGT71L1-KO with four biological replicates. Rescue-1, -2, and -3 represent independently transformed rescue lines, with four to five biological replicates per line. Statistical significance was determined using a t test. *P < 0.05 indicates significant differences from WT plants; **P < 0.01 indicates.
We next considered that plant growth regulators and hormones may be affected and analyzed these using a previously established method (Irmisch et al., 2014). This analysis revealed that both SA and SA-glucoside were 10-fold higher in UGT71L1-KO plants compared to control lines (Table�3), an intriguing observation given the structural similarity of SA to salicinoids. When we measured SA-glucoside content in the transgenic rescue plants described above, we found that it was back to WT levels (Figure�5). This finding suggested that the elevated SA content is directly linked to the loss of UGT71L1. High SA content in the UGT71L1-KO plants also correlated with elevated levels of 2,5-and 2,3-hydroxybenzoic acids, both SA breakdown products. Abscisic acid and indole-3-acetic acid were quantified but were not consistently altered in transgenic lines. However, concentrations of jasmonic acid (JA) and several other jasmonates were strongly enhanced. Importantly, the most active jasmonates, JA, and JA-Ile, were elevated by 27- and 17-fold, respectively. 12-Hydroxy-JA sulfate production exhibited an opposite trend and was detected at very high concentrations in control lines and was less abundant in the UGT71L1-KO lines. The biological role of 12-hydroxy-JA sulfate is unclear, but its presence at such high concentrations suggest it may not be active as a regulatory molecule (Fern�ndez-Milmanda et al., 2020). We also measured soluble amino acid concentrations in leaves, which overall increased approximately three-fold in UGT71L1-KO plants. Most amino acids, apart from threonine, showed an increased concentration in UGT71L1-KO lines. Asp, Glu, and Trp were among the most abundant amino acids and exhibited the greatest changes, with 26-fold, 5-fold, and 2.5-fold concentration increases, respectively (Table�4).
Table 3.
Phytohormone content of young UGT71L1-KO, partial-KO, and control P. tremula�alba leaves1
| Compound | UGT71L1-KO | Partial-KO | Control |
|---|---|---|---|
| SA | 30.27 (�6.04)a | 7.95 (�0.75)c | 3.81 (�1.6)c |
| SA-glucoside | 328.87 (�29.78)a | 95.69 (�35.99)c | 32.97 (�1.83)c |
| JA | 17.27 (�1.95)a | 0.99 (�0.14)c | 0.63 (�0.06)c |
| JA-glucoside | 168.66 (�7.41)a | 640.63 (�23.1)c | 588.65 (�17.44)c |
| 12-OH-JA | 8.92 (�0.55)a | 2.42 (�0.34)c | 3.06 (�0.19)c |
| JA-Ile | 0.16 (�0.02)a | 0.01 (�0.001)c | 0.01 (�0.002)c |
| OH-JA-Ile | 0.22 (�0.01)a | 0.07 (�0.01)c | 0.06 (�0.005)c |
| COOH-JA-Ile | 0.02 (�0.008)a | 0.007 (�0.002)a,c | 0.007 (�0.001)c |
| OPDA | 9.01 (�0.92)a | 7.48 (�0.25)a | 4.46 (�0.18)c |
| Sulfo-JA | 4,873.8 (�194.73)a | 20,903.2 (�648.3)c | 20,957.39 (�680.02)c |
| ABA | 6.86 (�0.82)a | 20.37 (�4.47)b | 12.11 (�1.17)c |
| Indole acetic acid2 | 297.14 (�10.9)a | 193.15 (�11.56)b | 246.43 (�9.03)c |
Content in ng�mg DW−1 (� se). UGT71L1-KO data represent a pool of five biological replicates for each of three independent lines; partial-KO data represent five biological replicates of a single line; control data are from a pool of three independent empty vector lines each with five biological replicates and a WT line with five biological replicates. Significant differences (P < 0.05) between genotype for individual compounds are designated by different letters as determined by Tukey’s post hoc honest significant difference test.
Indole acetic acid values represent response g DW−1 (�se).
OH, hydroxy; OPDA, 12-oxo-phytodienoic acid; Ile, isoleucine; ABA, abscisic acid; Sulfo-JA: 12-hydroxy JA sulfate.
Table 4.
Soluble amino acid content in young leaves of UGT71L1-KO, partial-KO, and control poplar1
| Amino acid | UGT71L1-KO | Partial-KO | Control |
|---|---|---|---|
| Alanine | 8.19 (�0.39)a | 3.63 (�0.29)c | 4.60 (�0.25)c |
| Serine | 17.13 (�1.59)a | 4.04 (�0.28)c | 3.15 (�0.23)c |
| Proline | 0.83 (�0.05)a | 0.36 (�0.02)c | 0.50 (�0.02)c |
| Valine | 0.59 (�0.04)a | 0.20 (�0.01)c | 0.18 (�0.007)c |
| Threonine | 5.14 (�0.34)ns | 4.54 (�0.2)ns | 5.45 (�0.22)ns |
| Isoleucine | 0.56 (�0.04)a | 0.16 (�0.009)c | 0.14 (�0.004)c |
| Aspartate | 9.03 (�0.41)a | 4.97 (�0.35)b | 7.45 (�0.23)c |
| Glutamate | 25.59 (�1.4)a | 15.62 (�1.07)b | 21.22 (�0.89)c |
| Methionine | 0.31 (�0.02)a | 0.12 (�0.008)c | 0.12 (�0.004)c |
| Histidine | 2.72 (�0.31)a | 0.38 (�0.06)c | 0.32 (�0.01)c |
| Phenylalanine | 0.93 (�0.09)a | 0.54 (�0.05)c | 0.29 (�0.01)c |
| Arginine | 4.26 (�0.55)a | 0.11 (�0.02)c | 0.11 (�0.02)c |
| Tyrosine | 0.10 (�0.007)a | 0.05 (�0.002)c | 0.05 (�0.001)c |
| Asparagine | 43.24 (�5.78)a | 2.32 (�0.33)c | 2.71 (�0.23)c |
| Glutamine | 23.73 (�2.27)a | 4.14 (�0.74)c | 4.14 (�0.54)c |
| Lysine | 7.00 (�0.6)a | 1.46 (�0.28)c | 1.45 (�0.16)c |
| Tryptophan | 53.67 (�4.91)a | 25.09 (�0.7)c | 19.23 (�0.69)c |
| Leucine | 0.34 (�0.02)a | 0.07 (�0.005)c | 0.07 (�0.001)c |
| S-methylmethionine | 0.07 (�0.005)a | 0.005 (�0.0004)c | 0.004 (�0.0002)c |
| Totals | 203.53 (�18.89)a | 67.88 (�4.49)c | 71.29 (�3.56)c |
Values represent concentrations in nmol�mg DW−1 (�se). UGT71L1-KO data represent five biological replicates each of three independent lines, partial-KO are five biological replicates of a single line, and controls are a pool of three independent empty vector lines each with five biological replicates and five biological replicates of a WT line. Significant differences (P < 0.05) between genotype for individual compounds are designated by different letters as determined by a Tukey’s post hoc honest significant difference test. ns, not-significant.
Transcriptome analysis indicates a general upregulation of defense metabolism and downregulation of photosynthetic processes in UGT71L1-KO plants
To further investigate the effect of UGT71L1 disruption on poplar growth and metabolism an RNA-seq experiment was conducted. We generated RNA-seq libraries for three biological replicates each of an individual UGT71L1-KO, WT, empty vector control, and partial-KO line, which were subjected to high-throughput Illumina sequencing. Mapping of reads to the P. tremula transcriptome (AspenDB; Xue et al., 2015) identified 16,343 unique transcripts, which were then used to identify differentially expressed genes. Comparison of the biallelic UGT71L1-KO plants with WT identified a set of 5,982 differential genes at P < 0.05. Enrichment analysis using the PoplarGene webtool (Liu et al., 2016) identified striking trends in the UGT71L1-KO transcriptome. First, the most highly enriched GO categories were related to pathogen defense including chitinase activity, cell wall catabolism, phenylpropanoid biosynthesis, and SA glucoside biosynthetic genes (Figure�6). Second, most primary metabolic GO categories including carbon fixation were under-represented in the UGT71L1-KO transcriptome. Both shifts in gene expression suggest the activation of plant defense in UGT71L1-KO plants. Such activation is consistent with the elevated SA levels we observed (Table�3).
Figure 6.
Gene ontology (GO) terms significantly enriched in UGT71L1-KO transcriptome compared to WT analyzed by the PoplarGene webtool. Green bars indicate on left significant enrichment of GO terms in UGT71L1-KO lines (log2FC >2); magenta bars on right show significant enrichment of GO terms in WT plants (log2FC >2).
We next inspected the list of differentially regulated genes from key pathways related to salicinoid, shikimate, and phenylpropanoid biosynthesis and defense (Table�5). Several enzyme-encoding genes of the shikimic acid and aromatic amino acid pathway were downregulated in UGT71L1-KO plants, in particular those encoding 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, shikimate dehydrogenase (SDH), chorismate mutase (CM) as well as prephenate aminotransferase (PAT). In contrast, general phenylpropanoid and flavonoid genes were not significantly affected by UGT71L1 disruption, except for a gene for 4-coumaroyl CoA ligase (4CL) and one for flavonoid-3′,5′-hydroxylase and flavonol synthase. PAL and chalcone synthase are all encoded by multiple genes in poplar, none of which were significantly differentially expressed.
Table 5.
Differentially expressed genes (P ≤0.05) in leaves of 3-month-old UGT71L1-KO plants compared to WT controls1
| Gene | Poplar gene ID2 | Fold-change | P-value | Adj. P-value | |||
|---|---|---|---|---|---|---|---|
| Shikimate pathway | |||||||
| DAHPS | Potri.002G099200 | 0.62 | <0.01 | 0.02 | |||
| Shikimate dehydrogenase (SDH) | Potri.013G029800 | 0.24 | 0.01 | 0.05 | |||
| CM | Potri.010G162300 | 0.49 | 0.02 | 0.07 | |||
| Arogenate dehydratase (ADT/PAT) | Potri.009G148800 | 1.57 | <0.01 | <0.01 | |||
| Salicinoid and phenylpropanoid pathway | |||||||
| UGT71L1 | Potri.016G014500 | 0.24 | <0.01 | <0.01 | |||
| 4-Coumarate CoA ligase (4CL) | Potri.003G188500 | 0.47 | 0.04 | 0.12 | |||
| Flavonoid 3′5′-hydroxylase (F3′5′H) | Potri.009G069100 | 0.23 | <0.01 | <0.01 | |||
| Flavonol synthase (FLS) | Potri.004G139600 | 0.18 | <0.01 | <0.01 | |||
| Potri.004G139700 | 0.32 | <0.01 | <0.01 | ||||
| Salicylic acid glycosyltransferase (SAGT) | Potri.007G140500 | 4.52 | <0.01 | <0.01 | |||
| Jasmonate (JA) metabolism | |||||||
| 13-Lipoxygenase 2 (LOX2) | Potri.001G015600 | 3.86 | <0.01 | <0.01 | |||
| OPC CoA ligase 1 (OPCL1) | Potri.017G033600 | 4.82 | <0.01 | <0.01 | |||
| Potri.015G092300 | 1.65 | 0.02 | 0.07 | ||||
| Potri.012G095000 | 2.97 | <0.01 | <0.01 | ||||
| Jasmonate-amido synthetase (JAR1) | Potri.002G168200 | 2.48 | <0.01 | 0.01 | |||
| JA carboxyl methyltransferase (JMT) | Potri.005G230100 | 1.83 | 0.04 | 0.13 | |||
| CYP94B – JA-Ile 12 hydroxylase | Potri.005G220700 | 4.35 | <0.01 | <0.01 | |||
| Potri.001G277300 | 0.58 | <0.01 | <0.01 | ||||
| CYP94C1 – JA-Ile 12 hydroxylase | Potri.004G185300 | 63.56 | 0.01 | 0.03 | |||
| IAA-alanine hydrolase (IAR3) | Potri.010G035900 | 2.83 | <0.01 | <0.01 | |||
| Potri.003G045200 | 2.03 | <0.01 | <0.01 | ||||
| Jasmonate-ile amino hydrolase (ILL6) | Potri.002G082400 | 7.89 | <0.01 | <0.01 | |||
| Jasmonate ZIM domain (JAZ) (JAZ6) | Potri.003G068900 | 24.59 | <0.01 | <0.01 | |||
| Jasmonate ZIM domain (JAZ3) | Potri.015G035800 | 1.68 | <0.01 | <0.01 | |||
| Coronatine Insensitive 1 (COI1) | Potri.008G064400 | 1.32 | <0.01 | 0.01 | |||
| JA-Sulfotransferase (JA-SOT) | Potri.003G188800 | 4.72 | <0.01 | 0.02 | |||
| Potri.003G193500 | 9.78 | <0.01 | 0.01 | ||||
| Potri.003G005200 | 11.88 | <0.01 | <0.01 | ||||
RNA-seq data from three biological replicates of one UGT71L1-KO line and three biological replicates of the WT control line were analyzed by HiSat2 (Kim et al., 2019) and Cufflinks (Trapnell et al., 2012). Differentially expressed genes were identified using the DeSeq2 (Love et al., 2014) R package.
More than one poplar gene ID may have the same Arabidopsis gene correspondence and annotation.
DAHPS, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase; OPC, 3-oxo-2-(cis-Z-pentenyl)-cyclopentane-1-octanoic acid; JAR1, jasmonate resistant 1; IAA, indole acetic acid; ILE, isoleucine; ZIM, zinc finger expressed in inflorescence meristem.
JA biosynthesis and metabolic genes were significantly upregulated in the UGT71L1-KO plants (Table�5). Using the Plant Metabolic Network POPLARCYC 3.0 metabolic network database we explored genome-wide metabolic pathways and gene clusters in UGT7L1-KO and WT lines (Schl�pfer et al., 2017). JA biosynthetic genes 13-lipoxygenase 2 (LOX2), 3-oxo-2-(2′-[Z]-pentenyl)cyclopentane-1-octanoic acid (OPC–8:0) CoA ligase 1 (OPCL1), jasmonate-amido synthetase (JAR1), and JA carboxyl methyltransferase (JMT) were all significantly upregulated in UGT71L1-KO lines. JA modifying and degradation genes including those encoding putative JA-Ile 12 hydroxylases, JA-Ile amino hydrolase (ILL6), IAA-alanine hydrolase (IAR3), JA-ZIM domain (JAZ), coronatine insensitive 1 (COI1), and JA-sulfotransferases (JA-SOT) were also significantly upregulated alongside the biosynthetic genes (Liechti and Farmer, 2002; Mugford et al., 2009; Huang et al., 2017). It thus appears both JA synthesis and inactivation genes were upregulated in these plants; together with our observed enhancement of JA and JA-Ile levels, this suggests that JA metabolism was stimulated. No cohesive trend in GA-related gene expression was identified; GA biosynthetic genes exhibited both significant up- and downregulation (Supplemental Table S4). Transcripts encoding salicylic acid glycosyltransferase, an enzyme that converts SA to its inactive glucoside (Dean and Delaney, 2008), accumulated more than four-fold higher levels in UGT71L1-KO plants. This gene was likely upregulated in response to the high SA content and contributed to the 10-fold increase in SA-glucoside we observed (Table�3).
The correspondence of higher SA and JA content as well as greater abundance of SA- and JA-related transcripts in the UGT71L1-KO plants motivated us to look closely at the growth impacts of these phytohormones. SA and JA both modulate growth-defense tradeoffs and cause growth inhibition in many plants (Zhang and Turner, 2008; Huang et al., 2017; Guo et al., 2018). The elevated levels of these stress hormones could therefore underlie the unique physical UGT71L1-KO plant phenotype and reduced growth and expansion. As a preliminary test of this hypothesis, we treated plantlets with exogenous SA or the potent and highly stable JA analog coronatine (Feys et al., 1994; Xiao et al., 2004). No effect was observed after SA treatment, perhaps because SA can be readily inactivated by plants (Supplemental Figure S4). However, after 5 weeks of treating plantlets with 25-�M coronatine, reduced growth and smaller leaves relative to water-treated plants was observed (Supplemental Figure S4). These effects confirmed the interaction of JA with growth in poplar.
SA is a potential product of disrupted salicinoid biosynthesis and possible driver of the UGT71L1-KO phenotype
Although SA is not a precursor or otherwise involved in salicinoid biosynthesis, we hypothesized that it could be released from a salicinoid intermediate when UGT71L1 is blocked. Specifically, SA could be generated during salicinoid biosynthesis if the benzoate ring moiety of salicyl benzoate is first hydroxylated to salicyl salicylate prior to the UGT71L1-catalyzed glucosylation step (Figure�1). In the absence of UGT71L1, salicyl salicylate could not continue into salicinoid biosynthesis. As an unstable molecule, it could be readily hydrolyzed into salicyl alcohol and free SA. Postulating that salicyl benzoate is hydroxylated prior to glucosylation is a variation of our previously proposed pathway (Fellenberg et al., 2020). It would be a direct mechanism for the substantial accumulation of SA observed in UGT71L1-KO plants, however (Figure�1). It also represents a novel pathway for the generation of SA in plants (see “Discussion”).
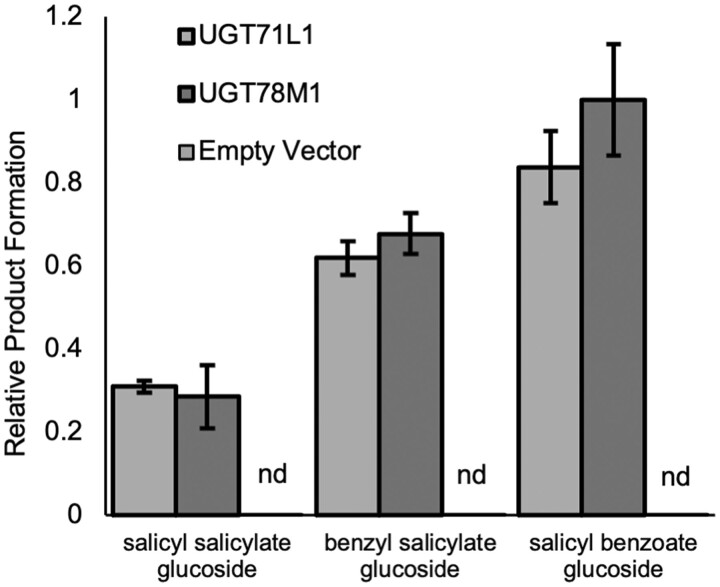
This hypothesis predicts that salicyl salicylate should also be a substrate for UGT71L1. Salicyl salicylate is not commercially available, but it was synthesized for enzyme assays with recombinant UGT71L1, together with salicyl benzoate and benzyl salicylate. The enzyme was able to efficiently convert salicyl salicylate to the corresponding glucoside (Figure�7; Supplemental Figure S5), although we did not determine kinetic parameters due to a limited supply of this compound. We also repeated assays using salicyl salicylate with UGT78M1, a second UGT hypothesized to be involved in biosynthesis of other minor salicinoids, but only a very small or no role in biosynthesis of salicortin or tremulacin (Fellenberg et al., 2020). The profile of activity with all three substrates with UGT78M1 was similar to that with UGT71L1. The acceptance of salicyl salicylate as a glucose acceptor for both enzymes is notable, since both enzymes take a narrow range of substrates (Fellenberg et al. 2020). The ability of these UGTs to glucosylate salicyl salicylate provided indirect experimental evidence for the hypothesized sequence of reactions and potential source of SA suggested above.
Figure 7.
Enzymatic activity of recombinant UGT71L1 and UGT78M1 with potential salicinoid biosynthetic intermediates. Bars show means (n = 3) of relative product formation relative to UGT78M1 glucosylation of salicyl benzoate under standard conditions. Empty vector controls were carried out in duplicate and consisted of extracts from bacterial host strains without the recombinant enzyme. Each assay contained 5-�g protein and 250-�M substrate and was conducted as described in “Materials and methods”. Product formation was quantified by UPLC-UV and absorbance at 280 nm. Data shown are mean � se.
Discussion
The demonstration that UGT71L1 is required for the biosynthesis of salicortin, tremulacin, and tremuloidin in poplar tissue cultures (Fellenberg et al., 2020; Kulasekaran et al., 2021) was a key advance in our understanding of salicinoid biochemistry. The present work with UGT71L1 whole-plant knockouts broadens these findings and suggests the presence of additional enzymes and routes in a biosynthetic network. Metabolomic and transcriptomic data indicated the unexpected impact of disruption of a key step in salicinoid biosynthesis on other phenolic pathways as well as primary metabolism and growth, potentially via feedbacks to the shikimate pathway and highly elevated levels of SA and JA. Together, our experiments suggest that excess SA is released in UGT71L1-KO poplars from salicinoid metabolites, thus driving the chemical and physical phenotype of these plants and revealing a novel mechanism for generating SA in poplar.
UGT71L1 is a central enzyme in the pathway to salicortin and tremulacin, but additional UGTs are likely involved in the biosynthesis of other salicinoids
Our knockout plants confirm that UGT71L1 is a core enzyme in the pathway to salicortin and tremulacin, the major salicinoids in P. tremula � P. alba. Unlike UGT71L1-KO root cultures where tremulacin and salicortin could not be detected, in whole plants UGT71L1 disruption by CRISPR/Cas9 reduced these compounds by 90%–95% but did not eliminate them. This suggests redundancy in the pathway and implies the presence of other UGT enzymes and genes with parallel functions (see below). Salicin content in UGT71L1-KO plants was less affected than salicortin or tremulacin and only reduced to 20% of WT concentrations. Prior in vivo labeling experiments had already demonstrated that salicin biosynthesis proceeds by a route that is distinct from the pathway to larger salicinoids, since salicyl alcohol and salicyl aldehyde are incorporated efficiently into salicin but not salicortin (Zenk, 1967; Babst et al., 2010). For example, salicin may also be derived via o-coumarate via salicylaldehyde as suggested by Babst et al. (2010). Such alternate routes to salicin would require additional UGT enzymes. In addition, salicin may also be derived via hydrolysis of the ester bond in salicortin or salicyl benzoate glucoside (Figure�1), which could explain the partial reduction by the loss of UGT71L1 we observed.
Interestingly, UGT71L1 disruption did not reduce salicin-7-sulfate concentrations, and in older leaves enhanced its accumulation. This compound was recently found to be present in some, but not all, willow and poplar species (Noleto-Dias et al., 2018; Lackus et al., 2020) and is highly abundant in the P. tremula � P. alba hybrid. The function of salicin-7-sulfate is not known, but it does not affect gypsy moth feeding preference and thus appear not to be an important compound for defense against lepidopteran herbivores (Lackus et al., 2020). Our metabolomic data show that while young leaves (targeted analysis) contain equivalent concentrations of salicin-7-sulfate in UGT71L1-KO lines and controls, older UGT71L1-KO leaves (nontargeted analysis) showed elevated concentrations. This pattern correlated with a 12-fold increase in the expression of the corresponding sulfotransferase gene PtSOT1 (Lackus et al., 2020) in UGT71L1-KO plants relative to WT seen in our RNA-Seq data. It suggests that synthesis and accumulation of salicin-7-sulfate is not disrupted and perhaps even favored in UGT71L1 knockouts. How the biosynthesis of salicin-7-sulfate and other sulfated salicinoids is related to that of the nonsulfated versions, and which UGT enzymes are involved, requires further study. Since salicin-7-sulfate is not found in some species of Populus that do synthesize other salicinoids, this compound may not have a central role in salicinoid metabolism.
Likewise, we observed that the minor salicinoids trichocarpin and salireposide were not impacted by UGT71L1 disruption, but rather increased in concentration. Their presence implies that they are synthesized by distinct UGTs. These salicinoids contain benzyl alcohol or 2,5-hydroxybenzyl alcohol, respectively, rather than salicyl alcohol (2-hydroxybenzyl alcohol) at their core, which suggests there may be UGTs with substrate preferences distinct from UGT71L1. These enhanced levels of trichocarpin and salireposide, the persistence of low concentrations of salicortin, tremulacin, and salicin, and the presence of large concentrations of salicin-7-sulfate all indicated that additional UGT71L1-like enzymes are active in UGT71L1-KO plants and likely contribute to overall salicinoid accumulation. For example, we previously proposed that UGT78M1 could be involved in salicinoid biosynthesis based on its substrate preferences for salicyl benzoate (Fellenberg et al., 2020), but direct evidence for its function using knockout plants is still lacking. Surprisingly, Kulasekaran et al. (2021) report that Salix does not have a UGT78M1 homolog. However, additional candidate genes similar to UGT71L1 are present in the poplar genome and are promising targets for future study.
Metabolomic analysis of UGT71L1 knockout plants provides insight into salicinoid biosynthesis
The comparative metabolomic analyses provided a strategy for identifying salicinoid pathway intermediates, since these are expected to accumulate in UGT71L1-KO plants where flux into the salicinoids is blocked. According to our earlier model (Fellenberg et al., 2020), a block at the UGT71L1 step would be expected to result in the accumulation of salicyl benzoate. Alternatively, if as suggested above, the 2-hydroxylation of salicyl benzoate occurs prior the glycosylation step, salicyl salicylate can be expected to accumulate in UGT71L1-KO plants instead (Figure�1). However, we were not able to detect salicyl salicylate or salicyl benzoate in our metabolomic data. Kulasekaran et al. (2021) also noted that in similar experiments using tissue cultured UGT71L1 RNAi plants, neither salicyl salicylate nor salicyl benzoate was detected. These esters may not be stable and could hydrolyze spontaneously or enzymatically. The inability of cells to glucosylate salicyl salicylate or salicyl benzoate due to UGT71L1 disruption would therefore generate a release of benzoic acid or SA, as we observed in UGT71L1-KO leaf extracts. Since SA concentration in tissues is strictly controlled, free SA is also expected to be rapidly conjugated or modified by metabolic reactions involved in SA homeostasis. This explains the increase in gentisic acid (2,5-dihydroxybenzoate) and 2,3-dihydroxybenzoic acid glucosides in UGT71L1-KO lines, compounds known from work in Arabidopsis to constitute a key pathway for inactivating SA (Zhang et al., 2013; Zhang et al., 2017; Zhang and Li, 2019). Therefore, we propose that the increase in these hydroxybenzoates reflects a metabolic response to the buildup and degradation of salicyl salicylate and/or salicyl benzoate and release of SA. Support for this idea also comes from work in Nicotiana benthamiana, where the application of benzyl salicylate to leaves enhances expression of systemic acquired resistance genes and reduces the number and size of tobacco mosaic virus lesions (Kamatham et al., 2017), suggesting that SA is released from this compound. Overall, our metabolomic data are consistent with a proposed salicinoid pathway in which the salicyl benzoate pool is 2-hydroxylated to salicyl salicylate prior to glucosylation by UGT71L1. The SA moiety of salicyl salicylate glucoside could then further oxidized to the HCH moiety by as yet unknown enzymes.
An alternate pathway proposes that the HCH moiety is synthesized from benzyl glucose and subsequently transferred to salicinoid acceptor molecules. This could also include oxidation of benzoyl-CoA to HCH-CoA followed by transesterification to other salicinoids, as suggested by Kulasekaran et al. (2021). There is no direct evidence for these alternate routes to date. However, the presence of HCH-salicortin in P. tremula � alba and many other poplar species (Feistel et al., 2015) suggests a mechanism to transfer HCH via acyltransferase reactions from a donor molecule. Alternatively, HCH may be synthesized from salicyl benzoate glucoside or salicyl salicylate glucoside, but subsequently transferred to other salicinoids. While the ultimate source of HCH is benzoic acid (Babst et al., 2010), how this moiety is derived remains a central question for salicinoid biosynthesis. In vivo-labeling experiments may help to provide an answer.
UGT71L1-KO plants manifest broad perturbations in shikimate and phenylpropanoid metabolism
UGT71L1-KO plants accumulated reduced concentrations of flavonoids, procyanidins, and lignin, all abundant phenylpropanoid-derived compounds. The plants also showed a reduction in caffeoyl quinate (chlorogenic acid) and the corresponding coumaroyl- and shikimoyl esters (Ma et al., 2018). These compounds are directly linked to the shikimate pathway (Clifford et al., 2017). Such broad effects of UGT71L1 disruption were unexpected based on the specific substrate preference of UGT71L1, and they indicate that the phenylpropanoid and/or shikimate pathways were perturbed in UGT71L1-KO plants by additional feedbacks. Consistent with this, our transcriptomic analysis indicated that expression of shikimic acid pathway and aromatic amino acid biosynthesis genes was downregulated: DAHP synthase, SDH, CM, and PAT all showed reduced transcript levels. However, few phenylpropanoid pathway genes were found to be downregulated. Transcriptional mechanisms for regulating the shikimate pathway in plants are poorly understood, but are assumed to involve feedbacks from aromatic amino acids and other downstream products (Maeda and Dudareva, 2012).
In contrast, at the post-transcriptional level complex mechanisms for regulating the shikimate pathway in plants have been described. In particular, feedback regulation of DAHPS, the enzyme controlling pathway influx, appears to play a central role (Maeda and Dudareva, 2012). Negative feedback mechanisms for DAHPS in Arabidopsis were recently characterized in detail (Yokoyama et al., 2021). Whereas the three DAHPS isoforms can be regulated differently by several allosteric regulators, all are inhibited by chorismate and caffeate (Yokoyama et al., 2021). These are both key intermediates in the shikimate and phenylpropanoid pathways, respectively. Interestingly, both caffeate and phenylethanol caffeate levels were enhanced in the UGT71L1-KO plants, as were the aromatic amino acids. We therefore speculate that the broad reduction in flavonoids, lignin, hydroxycinnamate esters, and other phenylpropanoids in UGT71L1-KO plants is caused by allosteric downregulation of the shikimate pathway due to the accumulation of a downstream product or shikimate pathway intermediate in UGT71L1-KO plants. The downregulation of shikimate and phenylpropanoid products is reminiscent of the broad impact of the overexpression of the flavonoid repressors MYB165 and MYB194 (Ma et al., 2018). Although the mechanisms are likely different, in both transgenic experiments, it is apparent that the shikimate pathway in poplar is sensitive to perturbation in phenylpropanoid and phenolic metabolism.
The increase in nonsalicinoid phenylpropanoids, specifically grandidentatin, in UGT71L1-KO plants remains an area requiring further investigation (Supplemental Table S2). Grandidentatin is a glucoside of p-coumarate and dihydroxycylcohexane first isolated in Populus grandidentata (Pearl et al., 1962; Zhang et al., 2006; Kim and Bae, 2009) but its biosynthesis has not been studied. We previously noted a small increase of grandidentatin in RNAi knockdown plants of BEBT and SABT (Chedgy, 2015), and Coleman et al. (2008) found an increase in grandidentatin after the downregulation of the shikimoyl 3-coumarate hydroxylase. Grandidentatin thus appears to be particularly impacted by diverse metabolic disruptions.
Elevated SA and JA levels may connect disrupted salicinoid biosynthesis to plant growth phenotypes
The most striking result from our study is the altered phenotype of UGT71L1-KO plants, which includes reduced leaf and plant size, and which is correlated with strong upregulation in SA-regulated defense genes. The unexpected effect of the UGT71L1 disruption on growth and morphology motivated us to carry out a rescue experiment, retransforming knockouts with a functional UGT71L1 coding sequence. The recovery of normal leaf shape and growth of these plants, together with WT levels of salicinoids and SA-glucoside confirmed that the strong phenotype is due to the loss of UGT71L1 and not caused by off-target effects. The phenotypic abnormalities observed in these poplar lines are particularly striking, since our earlier experiments with MYB repressor-overexpressing poplars showed that such plants also produce very low concentrations of many phenolics and phenylpropanoids, but their growth or form was not affected to an appreciable extent (Yoshida et al., 2015; Ma et al., 2018). This indicates that reduced levels of phenylpropanoids per se are unlikely to cause the physical phenotype observed. However, analysis of phytohormones altered in UGT71L1-KO plants suggested that elevated SA and JA may be potential causes of the phenotype. Hyperaccumulation of either SA or JA is known to reduce growth and lead to dwarfed growth forms. For example, smaller plant size is observed in Arabidopsis mutants that lack the SA 5-hydroxylase gene which contributes to SA inactivation, leading to elevated SA content (Zhang et al., 2017). In contrast, overexpression of this hydroxylase gene leads to larger leaves and rosettes compared to WT plants due to reduced SA levels. Similar effects were observed in Medicago, where transgenic downregulation of monolignols and the phenylpropanoid pathway caused dwarfism; since SA is also derived from the phenylpropanoid pathway, a reversal of the phenotype was achieved by applying exogenous SA (Gallego-Giraldo et al., 2011). These negative effects of SA on growth are manifested as the apparent tradeoff between growth and defense metabolism, which allows the plant to prioritize resources depending on environmental conditions (Zhang et al., 2017; Van Butselaar and Van den Ackerveken, 2020). SA homeostasis is thus critical to normal growth and development in Arabidopsis and other species, and SA metabolism is tightly regulated at both transcriptional and post-transcriptional levels (Ding and Ding, 2020).
How SA metabolism is organized and regulated in poplar is not known. SA content of WT poplar leaves in our study was approximately three-fold greater than what is reported for healthy 3-week-old Arabidopsis leaves, prior to elicitation after which SA increases sharply (Liu et al., 2016). Previous work in poplar had attempted to manipulate SA concentrations by overexpressing a bacterial IrP9 gene, part of the microbial pathway to synthesize SA from chorismate, or by expressing a bacterial nahG salicylate hydroxylase gene in transgenics in order to reduce SA content (Xue et al., 2013). Transgenic poplar expressing IrP9 in plastids showed a two- to three-fold enhanced SA content, while SA-glucoside content increased up to 800-fold, suggesting that the plant is conjugating excess SA. In contrast, overexpressing nahG in transgenic poplar caused no change in free SA, emphasizing that SA accumulation is carefully regulated (Morse et al., 2007; Xue et al., 2013). No growth phenotypes were observed in either the IrP6 or NahG transgenic poplars. This could reflect the much smaller changes in SA concentrations observed in these transgenics, compared to a 10-fold increase in free SA measured for UGT71L1-KO plants; it may also reflect localization of SA biosynthesis. In Arabidopsis, 90% of SA is synthesized via the isochorismate (ICS) pathway from plastidic isochorismate, which is moved across the plastid envelope into the cytosol where it is converted to SA (Rekhter et al., 2019). This may limit the potential of plastidic overexpression of IrP6. SA can also be produced in the cytosol from phenylpropanoid metabolism and cinnamate, though in most species this pathway produces much less SA. In contrast, we expect that in the UGT71L1-KO plants SA derived from salicyl salicylate is generated entirely in the cytosol based on the localization of the enzyme. This source of SA is separate from the standard cytosolic and plastidic SA biosynthetic pathways and may thus be outside the normal SA homeostatic mechanism that regulates SA concentrations in cells. This could explain the high degree of SA overaccumulation, in turn leading to strong effects on growth.
In addition to SA, the UGT71L1-KO plants showed very high JA and JA-Ile content and also overaccumulated several inactivate forms of JA including 12-hydroxy JA and sulfo-JA. The expression of both JA biosynthetic and catabolic genes was enriched in the UGT71L1-KO transcriptome, further suggesting that JA metabolism was stimulated. An increase in both SA and JA/JA-Ile was unexpected, since these signaling molecules often act antagonistically (Rivas-San Vicente and Plasencia, 2011; Wasternack and Hause, 2013). However, an induction of JA due to SA release has also been observed, for example during effector-triggered immunity in Arabidopsis (Liu et al., 2016). We therefore speculate that in our transgenic plants JA overaccumulation could have been stimulated by SA. It is important to note that the antagonism between SA and JA seen in other species appears not to be present in poplar. Both hormones are upregulated in parallel during pathogen stress (Ullah et al., 2019), and treatment with MeJa enhances SA content of poplar leaves in vitro (Park et al., 2017). Conversely, treatment of P. trichocarpa with SA induces multiple JAZ transcripts (Xia et al., 2017). Interestingly, JAZ6 was strongly upregulated in UGT71L1-KO in our data (Potri.003G068900, Table�5), and was also the most strongly SA-induced JAZ identified in Xia et al. (2017). Together with the high JA and JA-Ile content of our UGT71L1-KO plants, these data suggest that JA metabolism is activated by UTG71L1 disruption, and that SA could explain the upregulation of JAZ and JA-related gene expression.
Like SA, JA inhibits growth as part of a defense-growth tradeoff, including the inhibition of root elongation, leaf expansion, and hypocotyl elongation (Huang et al., 2017; Guo et al., 2018). In our experiments, the effect of coronatine in inhibiting growth and partially mimicking the CRISPR/Cas9-generated phenotype confirmed that JA growth inhibition is also operational in poplar. Our transcriptomic data set showed that a gene for GA20 oxidase is significantly downregulated, and some GA2 and GA3 oxidase genes are upregulated in UGT71L1-KO lines. This pattern suggested a reduction in active gibberellins (Supplemental Table S4). However, we did not measure gibberellins as part of our experiments. Growth inhibition by JA may be in part mediated by the downregulation of gibberellin signaling (Yang et al., 2012; Guo et al., 2018; Howe et�al, 2018). Although both JA and SA have different effects on gene expression overall, common expression modules associated with growth inhibition have been identified for poplars treated with SA or JA (Luo et al., 2019).
Alternative explanations for the UGT71L1-KO phenotype include disruptions of primary metabolism. For example, analysis of amino acids indicated significantly enhanced accumulation of several amino acids, specifically Asn, Gln, and Trp. Whether high amino acid content could be a direct cause of growth inhibition is not clear. Recent work in Arabidopsis with the jazD mutant, which is deficient in 10 JAZ genes and constitutively expresses JA-mediated defense and shows extreme growth inhibition, also reported elevated Trp content and increased expression of Trp biosynthesis genes (Major et al., 2020). This suggests that high JA and growth inhibition can lead to significant adjustment of primary metabolism including elevated Trp, although it may not be the primary driver of the phenotype (Guo et al., 2018). As an auxin biosynthetic precursor, Trp could influence auxin function or metabolism and thus effect growth. However, Arabidopsis differs from poplar in that it requires significant biosynthesis of Trp for the indole glucosinolate group of secondary metabolites. In contrast, most of the poplar defense molecules including salicinoids are phenylpropanoids and thus derived from phenylalanine. Overall, based on the multiple lines of evidence, we favor the hypothesis that the combined effect of salicinoid-derived SA and JA on growth and development is most likely the primary cause for the phenotypic effects observed. We summarize this hypothesis in Figure�8.
Figure 8.
Proposed metabolic and hormonal interactions leading to growth and metabolic phenotypes in UGT71L1-KO plants. Flat magenta arrows represent inhibited responses, and pointed green arrows represent stimulated processes. The interruption of salicinoid biosynthesis by CRISPR/Cas9 is shown with a magenta X, suggested to cause the hyperaccumulation of SA in these plants. The dashed arrow represents a hypothesized but not yet demonstrated stimulation of jasmonate by SA in poplar. Stimulation of biotic stress responses and inhibition of growth reflect the GO categories of enhanced and repressed genes observed in RNA-seq data (Figure�5), as well as the direct effects of coronatine on poplar (Figure�4). Feedback inhibition of the shikimate pathway is hypothesized based on RNA-seq and metabolomics data (Table�5;Supplementary Tables S3 and S4), including the accumulation of possible allosteric regulator caffeic acid.
Broader implications of the interaction of salicinoid metabolism with growth and development
The growth phenotype of UGT71L1-KO plants points to previously unknown connections of poplar secondary metabolites to primary processes. Interactions of growth regulators and defensive secondary metabolism have been observed in other species and may be more widespread than previously realized (Erb and Kliebenstein, 2020, Duran-Medina et al., 2021). Several studies in Arabidopsis have demonstrated that glucosinolates and their breakdown products can influence growth. Katz et al. (2020) showed that allyl glucosinolate catabolites interact with components of the auxin signaling pathway, leading an inhibition of root growth and development. Similar effects on auxin signaling or growth were observed with indole and aliphatic glucosinolates (Katz et al., 2015; Malinovsky et al., 2017). In P. trichocarpa, the herbivore-induced volatile, benzyl cyanide, can be converted to the auxin phenyl acetic acid (G�nther et al., 2019). Flavonoids have been observed to affect auxin transport in some experimental systems such as Arabidopsis seedling roots (Peer and Murphy, 2007). In apple, flavonoids and dihydochalcones have been suggested to interact with auxin transport or uptake (Dare et al., 2017). Interestingly, disruption of biosynthesis of the dihydrochalcone glycoside phloridzin by RNAi-inhibition of MdUGT88F1 in apple causes stunted growth and narrow leaves (Dare et al., 2017), reminiscent of the poplar UGT71L1-KO phenotype described here. Although we cannot rule out hormonal interactions by other phenolics, our data are most consistent with the hypothesis that SA accumulation is the ultimate cause of the phenotype in the UGT71L1-KO plants. Regardless of the mechanism, our work implies a potential interaction of the salicinoid pathway with growth regulatory processes and adds to a growing list of secondary metabolic pathways impacting plant development.
The linkage of salicinoid metabolism to growth processes could have important implications for the evolution of this pathway. Secondary chemical profiles are generally considered strongly heritable but under weak evolutionary constraints, compared to morphological and phenological traits (Kessler and Kalske, 2018). In P. tremuloides, leaf salicinoid content is highly heritable and under strong genetic control, and usually varies several-fold among individual genotypes (Hwang and Lindroth, 1997; Cole et al., 2021). Salicinoids are effective defensive chemicals against both insect and mammalian herbivores (Boeckler et al., 2011), and thus expected to be maintained in populations by herbivore pressure. In a common garden experiment using a suite of P. tremuloides genotypes, Cope et al. (2021) used salicinoid content as a proxy for defensive capacity to demonstrate a growth–defense tradeoff. However, based on the apparent persistence of such tradeoffs across many environments, the authors suggest that although salicinoid content may have evolved as a result of herbivore pressure, “covariance with other ecologically important traits renders the evolution of defense traits sensitive to selection pressures other than herbivory” (Cope et al., 2021). Such selective pressures could include additional, metabolic constraints on the control and plasticity of salicinoid biosynthesis, such as the effects of salicinoid-derived SA on growth and development shown here. Tighter metabolic constraints may also explain why salicinoids are generally constitutively expressed and developmentally regulated (Boeckler et al., 2011). This is in contrast to the strong stress-induction of the other major phenolic pathways in poplar, including flavonoids and condensed tannins, hydroxycinnamic acid esters, and benzenoid volatiles (Mellway et al., 2009; Lackus et al., 2021). Investigations of salicinoid regulatory genes could shed further light on potential developmental linkages with salicinoid biosynthesis. Such genes still await identification, however.
Materials and methods
Poplar transformation and growth
CRISPR/Cas9 UGT71L1 knockout constructs were created using the p201N vector (Jacobs et al., 2015) containing the previously described gRNA sequence for UGT71L1 (Fellenberg et al., 2020). P201N:gRNA was moved into Agrobacterium tumefaciens strain GV3101 by electroporation. Leaf explants from in vitro P. tremula � alba (INRA 717-1B4) leaves were excised and used for transformation experiments as previously described (James et al., 2017). Positive transformants were selected on medium containing kanamycin. UGT71L1 mutations were screened using the TIDE Webtool (Brinkman et al., 2014) with PCR amplification products of the gRNA target site. Mutations were confirmed in both alleles by Sanger sequencing of the PCR product. Three independent empty-vector control lines expressing the Cas9 protein with no gRNA were also generated. Confirmed transgenic and mutant lines were propagated as in vitro cultures, and when rooted were transferred to soil in two-inch pots and grown in a mist chamber for 3 weeks. Following 3 weeks in the mist chamber, plants were transferred to one-gallon pots containing peat-moss based soilless mix (Sunshine Mix 4, Sungro, Seba Beach, AB, Canada). Soil was supplemented with slow-release fertilizer as described by Major and Constabel (2006). Plants were grown in a greenhouse under ambient light conditions, with supplemental lighting provided by 600-W high-pressure sodium lights with an average intensity of 300 �mol m−2 s−1 for a 16-h photoperiod. Plants were irrigated as required and rotated within the greenhouse weekly.
For coronatine and SA treatments, in vitro plantlets were removed from tissue culture, potted in rooting media, and moved into a greenhouse mist chamber. Following one week of acclimation, coronatine in water was applied via handheld spray bottles. Coronatine solution (10 �M or 25 �M) was applied as a mist until droplets formed. For SA treatments solutions of SA in water were prepared at 1-mM, 100-�M, and 10-�M concentrations. SA solutions were adjusted to pH 6 with 0.2-M KOH and 0.1% (v/v) Tween-20 was added. Plantlets were maintained in the mist chamber for the duration of the experiment. The mist chamber was under ambient daylight with supplemental lighting from 400-W high-pressure sodium lights with an average intensity of 75 �mol m−2s−1 for a 16-h photoperiod. Plants were rotated periodically and physical measurements were taken weekly throughout the experiment.
Orygia leucostigma feeding assays
Small leaves (<4 cm long) from 5-month-old P. alba � tremula saplings were harvested and used in choice feeding assays with O. leucostigma. Insect eggs and diet were obtained from Insect Production Services of the Great Lakes Forestry Centre (Natural Resources Canada, Sault Ste. Marie, Canada). Single third or fourth instar larvae were placed in small petri dish arenas for 30 min prior to the addition of a pre-weighed leaf from both a UGT71L1-KO plant and an empty vector control plant. Feeding assays were carried out in a darkened room for 20 h. Following feeding, individual leaves were re-weighed to determine the amount of weight eaten per leaf. Control dishes with no caterpillars were used to correct for water loss. Only dishes where caterpillars had fed were used for data collection. Data were recorded as weight of foliage consumed.
Nontargeted metabolomic analysis
For all chemical analysis of greenhouse experiments, biological replication consisted of four or five individual copies (clonal greenhouse plants) for each transgenic or control line. Typically three or more independent transgenic lines were included in the experiment. Leaves of 3-month-old greenhouse-grown poplar were labeled, with leaf #1 designated as the youngest leaf with a lamina breadth of >1 cm. Expanded leaves #10–13 were harvested, the midvein excised, and the leaf blades frozen in liquid nitrogen. Samples were stored at −80�C prior to extraction and analysis. Samples were homogenized under liquid nitrogen using a ceramic mortar and pestle. Homogenized samples were freeze-dried and stored at room temperature. For extraction, 50 mg of freeze-dried tissue was weighed into a 2-mL cryo-vial with 4 � 5 mm steel ball bearings and 750 mL of chilled methanol. Samples were homogenized 2 � 45 s at 5,500 rpm using a Precellys 24 Homogenizer, then sonicated for 2 min in a sonicating water bath. Samples were centrifuged at 13,000g for 2 min and the supernatant was filtered through 0.22-�m PTFE filters into glass vials. Methanol extractions were repeated twice and extracts were pooled. Extracts were dried under vacuum at ambient temperature. Final extraction weight was determined, vials were sealed and stored at −20�C until analysis. Each biological replicate included an extraction blank. For analysis, samples were resuspended to 500 �g�mL−1 in methanol and stored briefly at 4�C prior to injection.
Samples were analyzed in a randomized injection order using a Thermo Ultimate 3000 (UPLC) coupled to Thermo LTQ Orbitrap XL (HRMS) and UltiMate Corona VeoRS charged aerosol detector (Thermo Fisher Scientific Inc, Waltham, MS, USA). Samples were separated using a Phenomenex Kinetex 1.7 �m C18 100 � 50 � 2.1 mm column with a SecurityGuard ULTRA C18 guard cartridge. For positive ionization mode samples were run with a continuous flow rate of 0.35 mL�min−1. Solvent A was liquid chromatography-mass spectrometry (LC/MS) grade water with 5-mM ammonium acetate and 0.1% formic acid; solvent B was acetonitrile with 0.1% formic acid. Separation occurred on a gradient of 2%B to 45%B for 10 min; followed by 45%B to 100%B over 5 min and then 100%B to 2%B over 3 min for a total run time of 18 min. The electrospray ionization (ESI) source had a capillary voltage of 35 V. Negative ionization mode samples were run under the same separation gradient, and the ESI source had a capillary voltage of −35 V. Both modes collected m/z from 100 to 2,000.00. For MS/MS collection alternate scans were fragmented from the previous scan with an m/z range of 300–2,000.00. MS/MS was conducted on representative samples for the UGT71L1-KO and control lines.
Raw MS1 data were processed using MZMine2 (Pluskal et al., 2010). Peaks were filtered for duplicates and then peak area and height data were exported to Microsoft Excel. Mass features were processed, and any mass features present in extraction and solvent blanks were eliminated from the data set. Peaks were also removed if <50% of a sample group (UGT71L1-KO, partial-KO, and control) were below the noise threshold. MS2 data were processed through MZMine2 (Pluskal et al., 2010) as previously described, except that MS1 data were paired with MS2 fragmentation data and exported to SIRIUS 4 utilizing CSI:fingerID (D�hrkop et al., 2015, 2019) was used to query molecular structure databases assign putative structural identities.
Targeted metabolomic, hormone, and amino acid analyses
For targeted metabolomics, phytohormone and amino acid analysis tissue was prepared and freeze-dried as per the nontargeted metabolomic methodology. Pooled tissue harvested from leaves # 4–8 was analyzed. Ten milligram of freeze-dried material was weighed into deep 96-well plates and 1 mL of methanol containing internal standards (40 ng�mL−1 D6-abscisic acid (D6-ABA), 40 ng�mL−1 D6-JA (D6-JA), D4-salicylic acid, 8 ng�mL−1 D6-jasmonic-acid isoleucine (D6-JA-Ile; Santa Cruz Biotechnology, Dallas, TX, USA), 40 ng�mL−1 D5-indole acetic acid (D5-IAA, OlChemIm s.r.o., Olomouc, Czech Republic)) was added. Samples were shaken vigorously for 30 s and then shaken at 200 rpm for 30 min. Samples were centrifuged at 1,400g for 2 min. Fifty microliters of supernatant was aliquoted into 96-well plate for amino acid analysis and 400 �L of supernatant was aliquoted into 96-well plates for targeted metabolomics and phytohormone analysis. All samples were stored at −20�C prior to analysis. For amino acid analysis 450 �L of MilliQ water containing 10-�g amino acids was added to each sample.
Targeted metabolomics
Chromatographic separation was achieved using an Agilent 1260 Infinity II LC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a Zorbax Eclipse XDB-C18 column (50 � 4.6 mm, 1.8 μm, Agilent Technologies), using aqueous formic acid (0.05% (v/v)) and acetonitrile as mobile phases A and B, respectively. The mobile phase flow rate was 1.1 mL�min−1. The elution profile is shown in Supplemental Table S5 as gradient A. The column temperature was maintained at 20�C. The LC system was coupled to a QTRAP 6500 tandem mass spectrometer (Sciex, Darmstadt, Germany) equipped with a turbospray ion source, operated in negative ionization mode. The ion spray voltage was maintained at −4,500 eV and the turbo gas temperature was set at 600�C. Nebulizing gas was set at 60 psi, curtain gas at 45 psi, heating gas at 60 psi, and collision gas at medium level. Multiple reaction monitoring (MRM) was used to monitor analyte parent ion → product ion formation for each analyte as displayed in Supplemental Table S6. Identification of compounds was performed using authentic standards. Data acquisition and processing was performed using Analyst 1.6.3 (Sciex) and MultiQuant 3.0.3 (Sciex) software.
Phytohormone analysis
Analysis was conducted as described previously in Irmisch et al. (2014). In brief, an Agilent 1260 Infinity II LC system (Agilent Technologies) coupled to a QTRAP 6500 tandem mass spectrometer (Sciex) was used for the analysis. Chromatographic separation was achieved using a Zorbax Eclipse XDB-C18 column (50 � 4.6 mm, 1.8 μm, Agilent Technologies), and aqueous formic acid (0.05% (v/v)) and acetonitrile as mobile phases A and B, respectively. The mobile phase flow rate was 1.1 mL�min−1. The elution profile is listed in Supplemental Table S5 as gradient B. The tandem mass spectrometer was equipped with a turbospray ion source, operated in negative ionization mode. The ion spray voltage was maintained at −4,500 eV and the turbo gas temperature was set at 650�C. Nebulizing gas was set at 60 psi, curtain gas at 40 psi, heating gas at 60 psi, and collision gas at medium level. Indoleacetic acid was quantified using the same LC–MS/MS system with the same chromatographic conditions but using positive mode ionization with an ion spray voltage at 5,500 eV. MRM was used to monitor analyte parent ion → product ion formation for each analyte as displayed in Supplemental Table S6. Data acquisition and processing was performed using Analyst 1.6.3 (Sciex) and MultiQuant 3.0.3 (Sciex) software.
Amino acid analysis
Chromatography was performed on an Agilent 1200 HPLC system, equipped with a Zorbax Eclipse XDB-C18 column (50 �3 �4.6 mm, 1.8 �m; Agilent Technologies). Aqueous formic acid (0.05%) and acetonitrile were used as mobile phases A and B, respectively, and the elution profile is listed in Supplemental Table S5 as gradient C. The mobile phase flow rate was 1.1 mL min−1, and the column temperature was maintained at 25�C. The liquid chromatograph was coupled to a QTRAP 6500 tandem mass spectrometer (Sciex, Darmstadt, Germany) equipped with a turbospray ion source, operated in positive ionization mode (ion spray voltage, 5,500 eV; turbo gas temperature, 650�C; nebulizing gas, 70 psi; curtain gas, 40 psi; heating gas, 70 psi; collision gas at medium level). MRM parameters are listed in Supplemental Table S6. Analyst 1.5 software (Applied Biosystems) was used for data acquisition and processing. Data acquisition and processing was performed using Analyst 1.6.3 (Sciex) and MultiQuant 3.0.3 software (Sciex).
Generation and analysis of UGT71L1-KO rescue plants
A modified UGT71L1 without the Cas9 gRNA target sites used to create the original UGT71L1-KO transgenics was designed based on the validated UGT71L1 coding sequence of P. tremula � alba (INRA 717-1B4) genome v2 (http://aspendb.uga.edu/databases/spta-717-genome) and synthesized in a PBK+ vector (BioMatik, ON, Canada). Six nucleotide mismatches were introduced into degenerate codon positions into the UGT71L1 gRNA target site. The modified UGT71L1 sequence (UGT71L1m) was cloned into a pMDC32 vector (Curtis and Grossniklaus, 2003) using added 5′-KpnI and 3′-Spe1 restriction sites. PMDC32:UGT71L1m was moved into A. tumefaciens GV3103 via electroporation. One UGT71L1-KO transgenic line was re-transformed with this construct using previously described poplar transformation methods (James et al., 2017). Successfully transformed calli were sequentially selected on woody plant medium (Caisson) containing hygromycin (10 μg�mL−1). Successful transformants were rooted in vitro and acclimated to greenhouse growth as described above.
For chemical analysis, expanded leaves of 2-month-old rescue plants were flash frozen and homogenized in a mortar and pestle in liquid nitrogen. Approximately 100 mg of frozen powder were extracted into 1 mL of methanol, sonicated in a bath sonicator for 5 min and shaken on a benchtop orbital shaker for 3 min. Extracts were centrifuged for 10 min at 20,000g and the supernatant was collected and stored at −20�C prior to phenolic analysis using a Waters Acquity UPLC system equipped with QDa MS detector (Waters, Mississauga, ON, Canada). Two-microliter sample injections were separated on a Waters Acquity UPLC Ethylene Bridged Hybrid column (C18, 2.1 � 50 mm, pore size 1.7 �m) and a binary solvent gradient comprising ultra-pure water containing 0.1% formate (A) and LC/MS grade acetonitrile with 0.1% formate (B). The flow rate was 0.5 mL�min−1. Separation gradient was 99.9% A, 0–0.5 min; 99.9%–80.0% A, 0.5–6.0 min; 80.0%–50.0% A, 6.0–9.0 min; 50.0%–10.0% A, 9.0–10.0 min; 10% A, 10.0–11.5 min; 10%–99.9% A, 11.5–12.0 min; 99.9% A, 12.0–13.0 min. The QDa detector using electrospray ionization in negative mode was used to detect and quantify salicortin, tremulacin, tremuloidin, and salicin. Salicin formate adduct at m/z = 331.0 with cone voltage (V) of 10 was detected at 2.9 min. Salicortin (-H) at m/z = 423.0 with V = 10 was detected at 5.6 min. Tremulacin (-H) at m/z = 537.0 with V = 10 was detected at 8.4 min. Tremulacin was quantified using linear regression of authentic standard peak areas provided by Dr. Richard Lindroth. Salicortin was quantified using linear regression of an authentic standard (Biosynth, San Diego, USA). Salicin was quantified using commercially available standards from Sigma-Aldrich (Oakville, ON, Canada). Peak integration was conducted using MassLynx 4.2 (Waters, Mississauga, ON, Canada).
RNA-seq analysis
RNA was extracted as per Yoshida et al., (2015) from four clonal plants (biological replicates) for one UGT71L1-KO, the partial KO, and the WT control line. mRNA isolation and enrichment was conducted using a MagJET enrichment kit (ThermoScientific, Waltham, MA, USA). mRNA was fragmented at 94�C for 13 min to achieve an approximate fragment size of 350 bp. cDNA libraries were prepared as per Ma et al. (2018) using commercially available kits for Illumina sequencing. Twelve libraries were sequenced by the University of British Columbia Sequencing + Bioinformatics Consortium (Vancouver, BC, Canada) on one NextSeq High Output Lane. Sequencing results were de-multiplexed, adaptor sequences were trimmed, and raw data were aligned to the P. tremula transcriptome V.2 (Xue et al., 2015) using HiSat2 (Kim, et al., 2019). Cufflinks (Trapnell et al., 2012) was used to analyze differentially expressed genes and DeSeq2 package in R was used to generate the final differentially expressed gene table. Gene annotation was done using the annotated transcriptome in Popgenie (popgenie.org; Sj�din et al., 2009). Enrichment analysis was conducted using highly expressed genes in WT and UGT71L1 samples and the PoplarGene database (Liu et al., 2016). Metabolic pathway expression of identified transcripts was assessed using POPLARCYC 3.0 (Schl�pfer et al., 2017).
Synthesis of salicyl salicylate
2-Hydroxy benzoyl chloride was prepared from salicylic acid (Sigma-Aldrich) according to Zhao et al. (2018). To a mixture of salicyl alcohol (248 mg, 2 mmol, Merck KGaA) and 2-hydroxy benzoyl chloride (368 mg, 2 mmol) in anhydrous CH2Cl2 (10 mL), dry triethylamine (280 �L, 2 mmol, Sigma-Aldrich) was added at 0�C. The mixture was stirred for overnight at room temperature under Ar, then CH2Cl2 and saturated aqueous NH4Cl were added. The mixture was extracted twice with CH2Cl2. The organic phase was washed with brine, dried over Na2SO4, and concentrated in vacuo. The residue was purified twice by silica gel flash column chromatography (n-hexane: EtOAc gradient, 96:4–66:34, v/v) to give salicyl salicylate (2-hydroxybenzyl 2-hydroxybenzoate; 50 mg, 0.2 mmol, 10%). NMR spectra (1H and 13C NMR, 1H-1H COSY, 1H-13C HSQC, and 1H-13C HMBC) of the product were measured on a Bruker Avance III HD 500 NMR spectrometer, operating at 500.13 MHz for 1H and 125.75 MHz for 13C. The spectrometer was equipped with a TCI cryoprobe (5 mm for Avance III HD 500). Spectrometer control and data processing was accomplished using Bruker Topspin ver. 3.6.1. All NMR measurements were recorded at 25�C in CDCl3. Chemical shifts were referenced to the residual solvent signals for measurements in CDCl3 (δH 7.26/δC 77.16). HRMS was carried out on a Bruker Compact ESI-OTOF spectrometer (Bruker Daltonics GmbH, Bremen, Germany) using ESI ionization in the negative mode. For 1H and 13C NMR data, see Supplemental Table S7; HRMS (ESI, negative mode) m/z [M-H]−: calcd. for C14H11O4, 243.0663; found, 243.0668.
UGT enzyme assays
Recombinant UGT71L1 and UGT78M1 was produced and purified as per Fellenberg et al., (2020). Enzyme assays were performed for 3 h at 30�C, in a 200-�L reaction volume in 50-mM sodium phosphate (pH 7.4). Reactions contained UDP-glucose (5 mM), substrates at a final concentration of 250 �M and 5 �g of purified recombinant protein. Reactions were halted with the addition of 20 �L 30% trichloroacetic acid (w/v), and 6 min of centrifuging at 16,100g. Supernatant was collected and stored at −20�C prior to analysis. Analysis and product identification of enzyme reaction products was conducted on a Waters Acquity UPLC LC/MS system. For analysis, an 8-�L sample injection was separated using a Waters Acquity Ethylene Bridged Hybrid (C18, pore size: 1.7 �m, dimensions: 2.1 � 50 mm) column, and glucoside products were identified based on mass and retention time. Product peak areas were determined using UV absorbance at 280 nm. Compounds were separated on a gradient binary consisting of ultra-pure water containing 0.1% formate (solvent A) and LC/MS grade acetonitrile with 0.1% formate (solvent B). The flow rate was 0.5 mL�min−1. Separation gradient was 90.0% A, 0–0.5 min; 90.0%–10.0% A, 0.5–6.0 min; 10.0%–0.5% A, 6.0–7.0 min; 0.5%–10.0% A, 7.0–8.0 min; 10.0%–90.0% A, 8.0–8.5 min. A single quadrupole MS in positive ionization mode (ES+) continually scanned masses 50.0 to 1,000.0 m/z from 0.2 to 8.5 min. Cone voltage was set at 35 V. Sodium adducts of benzyl salicylate glucoside and salicyl benzoate glucoside were identified at 413 m/z. The sodium adduct of salicyl salicylate glucoside was identified at 429 m/z.
Statistical analysis
Multivariate and univariate statistics for metabolomic samples were processed using a combination of Rstudio (RStudio Team, 2018) and the MUMA package (Gaude et al., 2012), as well as MetaboAnalyst (Chong et al., 2019). Statistical processing of growth data, insect feeding assays was conducted using R-Studio (RStudio Team, 2018). Detailed results of statistical analysis are available in Supplemental Data Set S1.
Accession numbers
RNA-seq data for this work has been deposited in the National Centre for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under ID GSE186781 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186781).
[reviewer access token: qnqzyiwgpzazxox]
Metabolomic data are deposited in the NIH database Metabolomics Workbench under ID:ST001962 and temporarily available at: http://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST001962&Access=DpdN6315
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1. Sanger sequencing analysis to detect potential off-target mutations.
Supplemental Table S2. List of tentatively identified differential compounds detected by nontargeted metabolomics.
Supplemental Table S3. Targeted analysis of phenolic secondary metabolites.
Supplemental Table S4. Differentially expressed genes related to gibberellin metabolism.
Supplemental Table S5. HPLC gradients used for analysis of targeted metabolites.
Supplemental Table S6. Parameters used for targeted metabolomic analysis (LC–MS/MS).
Supplemental Table S7. NMR analysis and validation of salicyl salicylate.
Supplemental Figure S1. Multivariate statistical analysis of-targeted metabolomic data set.
Supplemental Figure S2. Acetyl bromide lignin analysis.
Supplemental Figure S3. Whole-plant photographs of 3.5-month-old greenhouse-grown UGT71L1 knockout and WT poplars.
Supplemental Figure S4. Effects of coronatine and SA on the growth of WT poplar.
Supplemental Figure S5. Representative chromatogram of enzyme assay products.
Supplemental Methods. Acetyl bromide lignin extraction and quantification.
Supplemental Data Set S1. Summary of statistical analysis.
Supplementary Material
Acknowledgments
Funding for this work from the Natural Science and Engineering Research Council of Canada, Agriculture and Agri-food Canada, and the Max-Planck Society is gratefully acknowledged. We thank the Centre for Forest Biology at the University of Victoria for travel and greenhouse support. We thank Lynn Yip for her expert help with plant transformation and tissue culture, Megan Loland for help with the rescue experiment, Brad Binges for help managing greenhouse experiments, and Carmen Lea for providing tussock moth larvae.
Conflict of interest statement. None declared.
Contributor Information
Harley Gordon, Department of Biology, Centre for Forest Biology, University of Victoria, Victoria, BC V8P 5C2, Canada.
Christin Fellenberg, Department of Biology, Centre for Forest Biology, University of Victoria, Victoria, BC V8P 5C2, Canada.
Nathalie D Lackus, Department of Biochemistry, Max Planck Institute for Chemical Ecology, Jena 07745, Germany.
Finn Archinuk, Department of Biology, Centre for Forest Biology, University of Victoria, Victoria, BC V8P 5C2, Canada.
Amanda Sproule, Agriculture and Agri-Food Canada, Ottawa, Ontario K1A 0C6, Canada.
Yoko Nakamura, Department of Nuclear Magnetic Resonance, Max Planck Institute for Chemical Ecology, Jena 07745, Germany; Department of Natural Product Biosynthesis, Max Planck Institute for Chemical Ecology, Jena 07745, Germany.
Tobias G K�llner, Department of Biochemistry, Max Planck Institute for Chemical Ecology, Jena 07745, Germany.
Jonathan Gershenzon, Department of Biochemistry, Max Planck Institute for Chemical Ecology, Jena 07745, Germany.
David P Overy, Agriculture and Agri-Food Canada, Ottawa, Ontario K1A 0C6, Canada.
C Peter Constabel, Department of Biology, Centre for Forest Biology, University of Victoria, Victoria, BC V8P 5C2, Canada.
H.G. carried out experiments, analyzed the data, and drafted and edited the manuscript. C.F. created transgenics, N.D.L. performed the phytohormone, amino acids, and targeted metabolite analysis, F.A. genotyped the transgenics and determined mutations, A.S. carried out nontargeted metabolomic analysis, Y.N. synthesized substrates, T.G.K. helped with phytohormone, amino acids, and targeted analyses, D.P.O. planned and supervised nontargeted metabolomics and edited the manuscript. C.P.C. conceived and planned experiments and co-wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: C. Peter Constabel (cpc@uvic.ca)
References
- Babst BA, Harding SA, Tsai CJ (2010) Biosynthesis of phenolic glycosides from phenylpropanoid and benzenoid precursors in Populus. J Chem Ecol 36: 286–297 [DOI] [PubMed] [Google Scholar]
- Basey JM, Jenkins SH, Busher PE (1988) Optimal central-place foraging by beavers: tree-size selection in relation to defensive chemicals of quaking aspen. Oecologia 76: 278–282 [DOI] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, Van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AM, Busov VB, Strauss SH (2004) Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends Plant Sci 9: 49–56 [DOI] [PubMed] [Google Scholar]
- Boeckler G, Gershenzon J, Unsicker S (2011) Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 72: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Boeckler GA, Gershenzon J, Unsicker SB (2013) Gypsy moth caterpillar feeding has only a marginal impact on phenolic compounds in old-growth black poplar. J Chem Ecol 39: 1301–1312 [DOI] [PubMed] [Google Scholar]
- Chedgy RJ (2015) The Role of BAHD Acyltransferases in Poplar. PhD thesis. University of Victoria, Victoria, BC, Canada
- Chedgy RJ, K�llner TG, Constabel CP (2015) Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry 113: 149–159 [DOI] [PubMed] [Google Scholar]
- Chong J, Wishart D, Xia J (2019) Using Metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinfor 68: e86. [DOI] [PubMed] [Google Scholar]
- Clifford MN, Jaganath IB, Ludwig IA, Crozier A (2017) Chlorogenic acids and the acyl-quinic acids: discovery, biosynthesis, bioavailability and bioactivity. Nat Prod Rep 34: 1391–1421 [DOI] [PubMed] [Google Scholar]
- Cole CT, Morrow CJ, Barker HL, Rubert-Nason KF, Riehl JFL, K�llner TG, Lackus ND, Lindroth RL (2021) Growing up aspen: ontogeny and trade-offs shape growth, defence and reproduction in a foundation species. Ann Bot 127:505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope OL, Keefover-Ring K, Kruger EL, Lindroth RL (2021) Growth-defense trade-offs shap population genetic composition in an iconic forest tree species. Proc Natl Acad Sci USA 118: e2103162118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HD, Park JY, Nair R, Chapple C, Mansfield SD (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3’-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci USA 105: 4501–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dare AP, Yauk YK, Tomes S, McGhie TK, Rebstock RS, Cooney JM, Atkinson RG (2017) Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. Plant J 91: 237–250 [DOI] [PubMed] [Google Scholar]
- Dean JV, Delaney SP (2008) Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol Plant 132: 417–425 [DOI] [PubMed] [Google Scholar]
- Desborough M, Keeling D (2017) The aspirin story – from willow to wonder drug. Brit J Haematol 177: 674–683 [DOI] [PubMed] [Google Scholar]
- Ding P, Ding Y (2020) Stories of salicylic acid: a plant defense hormone. Trends Plant Sci 25: 549–656 [DOI] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y (2018) Opposite roles of salicylic acid receptors npr1 and npr3/npr4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467 [DOI] [PubMed] [Google Scholar]
- Donaldson JR, Stevens MT, Barnhill HR, Lindroth RL (2006) Age-related shifts in leaf chemistry of clonal aspen (Populus tremuloides). J Chem Ecol 32: 1415–1429 [DOI] [PubMed] [Google Scholar]
- Duran-Medina Y, Ruiz-Cortes BE, Guerrero-Largo H, Marsch-Martinez N (2021) Specialized metabolism and develoment: An unexpected friendship. Curr Op Plant Biol 64: 102142. [DOI] [PubMed] [Google Scholar]
- D�hrkop K, Fleischauer M, Ludwig M, Aksenov AA, Melnik AV, Meusel M, Dorrestein P, Rousu J, B�cker S (2019) SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods 16: 299–302 [DOI] [PubMed] [Google Scholar]
- D�hrkop K, Shen H, Meusel M, Rousu J, B�cker S (2015) Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA 112: 12580–12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorriaga E, Klocko AL, Ma C, Strauss SH (2018) Variation in mutation spectra among CRISPR/Cas9 mutagenized poplars. Front Plant Sci 9: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Kliebenstein DJ (2020) Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol 184: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feistel F, Paetz C, Lorenz S, Schneider B (2015) The absolute configuration of salicortin, HCH-salicortin and tremulacin from Populus trichocarpa x deltoides Beaupr�. Molecules 20: 5566–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellenberg C, Corea O, Yan LH, Archinuk F, Piirtola EM, Gordon H, Reichelt M, Brandt W, Wulff J, Ehlting J, et al. (2020) Discovery of salicyl benzoate UDP-glycosyltransferase, a central enzyme in poplar salicinoid phenolic glycoside biosynthesis. Plant J 102: 99–115 [DOI] [PubMed] [Google Scholar]
- Fern�ndez-Milmanda GL, Crocco CD, Reichelt M, Mazza CA, K�llner TG, Zhang T, Cargnel MD, Lichy MZ, Fiorucci AS, Fankhauser C, et al. (2020) A light-dependent molecular link between competition cues and defence responses in plants. Nat Plants 6: 223–230 [DOI] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude E, Chignola F, Spiliotopoulos D, Mari S, Spitaleri A, Ghitti M (2012) Muma: Metabolomics Univariate and Multivariate Analysis version 1.4 from CRAN. https://rdrr.io/cran/muma/� (July 11, 2019)
- G�nther J, Lackus ND, Schmidt A, Huber M, Stodtler HJ, Reichelt M, Gershenzon J, K�llner TG (2019) Separate pathways contribute to the herbivore-induced formation of 2-phenylethanol in poplar. Plant Physiol 180: 767–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA (2018) Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44: 72–81 [DOI] [PubMed] [Google Scholar]
- Han R, Meilan MC, Strauss SH (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19: 315–320 [DOI] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ (2018) Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol 69: 387–415 [DOI] [PubMed] [Google Scholar]
- Huang H, Liu B, Liu L, Song S (2017) Jasmonate action in plant growth and development. J Exp Bot 68: 1349–1359 [DOI] [PubMed] [Google Scholar]
- Hwang SY, Lindroth RL (1997) Clonal variation in foliar chemistry of aspen: Effects on gypsy moths and forest tent caterpillars. Oecologia 111: 99–108 [DOI] [PubMed] [Google Scholar]
- Innes R (2018) The positives and negatives of NPR: a unifying model for salicylic acid signaling in plants. Cell 173: 1314–1315 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Jiang Y, Chen F, Gershenzon J, K�llner TG (2014) Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa). BMC Plant Biol 14: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AM, Ma D, Mellway R, Gesell A, Yoshida K, Walker V, Tran L, Stewart D, Reichelt M, Suvanto J, et al. (2017) Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol 174: 154–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkunen-Tiitto R, Sorsa S (2001) Testing the effects of drying methods on willow flavonoids, tannins, and salicylates. J Chem Ecol 27: 779–789 [DOI] [PubMed] [Google Scholar]
- Kamatham S, Pallu R, Pasupulati AK, Singh SS, Gudipalli P (2017) Benzoylsalicylic acid derivatives as defense activators in tobacco and Arabidopsis. Phytochemistry 143: 160–169 [DOI] [PubMed] [Google Scholar]
- Katz E, Bagchi R, Jeschke V, Rasmussen ARM, Hopper A, Burow M, Estelle M, Kliebenstein DJ (2020) Diverse allyl glucosinolate catabolites independently influence root growth and development. Plant Physiol 183:1376–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E, Nisani S, Yadav BS, Woldemariam MG, Shai B, Obolski U, Ehrlich M, Shani E, Jander G, Chamovitz DA (2015) The glucosinolate breakdown product indole-3-carbinol acts as an auxin antagonist in roots of Arabidopsis thaliana. Plant J 82:547–555 [DOI] [PubMed] [Google Scholar]
- Keefover-Ring K, Ahnlund M, Abreu IN, Jansson S, Moritz T, Albrectsen BR (2014) No evidence of geographical structure of salicinoid chemotypes within Populus tremula. PLoS One 9: e107189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Kalske A (2018) Plant secondary metabolite diversity and species interactions. Annu Rev Ecol Evol Syst 49: 115–138 [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Bae YS (2009) Grandidentatin isomer from bark of suwon poplar (Populus alba L.�Populus glandulosa Uyeki). Holzforschung 63: 315–318 [Google Scholar]
- Kulasekaran S, Cerezo-Medina S, Harflett C, Lomax C, de Jong F, Rendour A, Ruvo G, Hanley S, Beale M, Ward JL (2021) A willow UDP-glycosyltransferase involved in salicinoid biosynthesis. J Exp Bot 72: 1634–1648 [DOI] [PubMed] [Google Scholar]
- Lackus ND, M�ller A, Kr�ber TDU, Reichelt M, Schmidt A, Nakamura Y, Paetz C, Luck K, Lindroth R, Constabel CP, et al. (2020) The occurrence of sulfated salicinoids in poplar and their formation by sulfotransferase. Plant Physiol 183: 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackus ND, Schmidt A, Gershenzon J, K�llner TG (2021) A peroxisomal β-oxidative pathway contributes to the formation of C6–C1 aromatic volatiles in poplar. Plant Physiol 186: 891–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti R, Farmer EE (2002) The jasmonate signal pathway. Plant Cell 296: 1649–1650 [DOI] [PubMed] [Google Scholar]
- Lindroth RL, Clair SBS (2013) Adaptations of quaking aspen (Populus tremuloides Michx.) for defense against herbivores. Forest Ecol Manag 299: 14–21 [Google Scholar]
- Liu L, Sonbol FM, Huot B, Gu Y, Withers J, Mwimba M, Yao JHS, Dong X (2016) Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat Commun 7: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ding C, Chu Y, Chen J, Zhang W, Zhang B (2016) PoplarGene: poplar gene network and resource for mining functional information for genes from woody plants. Sci Rep 6: 31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sun T, Sun Y, Zhang Y, Radojičić A, Ding Y, Tian H, Huang X, Lan J, Chen S, et al. (2020) Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell 32: 4002–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Xia W, Cao P, Xiao Z, Zhang Y, Liu M, Zhan C, Wang N (2019) Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Reichelt M, Yoshida K, Gershenzon J, Constabel CP (2018) Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J 96: 949–965 [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Major IT, Constabel CP (2006) Molecular analysis of poplar defense against herbivory: comparison of wound- and insect elicitor-induced gene expression. New Phytol 172: 617–635 [DOI] [PubMed] [Google Scholar]
- Major IT, Guo G, Zhai J, Kapali G, Kramer DM, Howe GA (2020) A phytochrome B-independent pathway restricts growth at high levels of jasmonate defense. Plant Physiol 183: 733–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Thomsen MLF, Nintemann SJ, Jagd LM, Bourgine B, Burow M, Kliebenstein DJ (2017) An evolutionarily young defense metabolite influences the root growth of plants via the ancient TOR signaling pathway. eLife 6: e29353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP (2009) The wound-, pathogen-, and ultraviolet B-Responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse AM, Tschaplinski TJ, Dervinis C, Pijut PM, Schmelz EA, Day W, Davis JM (2007) Salicylate and catechol levels are maintained in nahG transgenic poplar. Phytochemistry 68: 2043–2052 [DOI] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford ST, Nakazato Y, Noji M, Takahashi H, Kramell R, et al. (2009) Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noleto-Dias C, Ward JL, Bellisai A, Lomax C, Beale MH (2018) Salicin-7-sulfate: a new salicinoid from willow and implications for herbal medicine. Fitoterapia 127: 166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SB, Kim J, Han J, Ahn CH, Park EJ, Choi Y (2017) Exploring genes involved in benzoic acid biosynthesis in the Populus davidiana transcriptome and their transcriptional activity upon methyl jasmonate treatment. J Chem Ecol 43:1097–1108 [DOI] [PubMed] [Google Scholar]
- Pearl IA, Darling SF, Justman O (1962) Studies on the leaves of the family Salicaceae. I. populin from the leaves of Populus grandidentata and Populus tremuloides. J Org Chem 27: 2685–2687 [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Pluskal T, Castillo S, Villar-Briones A, Oresic M (2010) MZmine2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin I (1992). Salicylate, a new plant hormone. Plant Physiol 99: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, L�dke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I (2019) Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365: 498–502 [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- RStudio Team (2018) RStudio: Integrated Development for R. RStudio Inc., Boston, MA [Google Scholar]
- Ruuhola T, Julkunen-Tiitto R, Vainiotalo P (2003) In vitro degradation of willow salicylates. J Chem Ecol 29: 1083–1097 [DOI] [PubMed] [Google Scholar]
- Ryan CA, Lamb CJ, Jagendorf AT, Kolattukudy PE, Lee H, Leon J, Raskin I (1995) Biosynthesis and metabolism of salicylic acid. Plant Signal Behav 92: 493–496 [Google Scholar]
- Schl�pfer P, Zhang P, Wang C, Kim T, Banf M, Chae L, Dreher K, Chavali A, Nilo-Poyanco R, Bernard T, et al. (2017) Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol 173: 2041–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Strunks GD, Klemann BJPM, Hooykaas PJJ, de Pater S (2017) CRISPR/Cas9-induced double-strand break repair in Arabidopsis nonhomologous end-joining mutants. G3: Gen Genom Genet 7: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine MB, Yang JW, El-Habbak M, Nagyabhyru P, Fu DQ, Navarre D, Ghabrial S, Kachroo P, Kachroo A (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol 212: 627–636 [DOI] [PubMed] [Google Scholar]
- Sj�din A, Street NR, Sandberg G, Gustafsson P, Jansson S (2009) The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol 182: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Tahvanainen AJ, Helle E, Julkunen-Tiitto R, Lavola A (1985) Phenolic compounds of willow bark as deterrents against feeding by mountain hare. Oecologia 65: 319–323 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg S, Rinn J, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C, Tsai CJ, Unsicker SB, Xue L, Reichelt M, Gershenzon J, Hammerbacher A (2019) Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol 221: 960–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Butselaar T, Van den Ackerveken G (2020) Salicylic acid steers the growth–immunity tradeoff. Trends Plant Sci 25: 566–576 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhalm JR, Dudareva N (2015) A familiar ring to it: biosynthesis of plant benzoic acids. Mol Plant 8: 83–97 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 417: 562–565 [DOI] [PubMed] [Google Scholar]
- Xia W, Yu H, Cao P, Luo J, Wang N (2017) Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Front Plant Sci 8: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Dai L, Liu F, Wang Z, Peng W, Xie D (2004) COS1: an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 16: 1132–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue LJ, Alabady MS, Mohebbi M, Tsai CJ (2015) Exploiting genome variation to improve next-generation sequencing data analysis and genome editing efficiency in Populus tremula � alba 717-1B4. Tree Genet Genom 11: 82 [Google Scholar]
- Xue LJ, Guo W, Yuan Y, Anino EO, Nyamdari B, Wilson MC, Frost C, Chen H, Babst B, Harding S, et al. (2013) Constitutively elevated salicylic acid levels alter photosynthesis and oxidative state but not growth in transgenic Populus. Plant Cell 25: 2714–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao L, Sun T, Li J, Deng X, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Oliveira MVV, Kleven B, Maeda HA (2021) The entry reaction of the plant shikimate pathway is subjected to highly complex metabolite-mediated regulation. Plant Cell 33: 671–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Constabel CP (2015) The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol 167: 693–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenk MH (1967) Pathways of salicyl alcohol and salicin formation in Salix purpurea L. Phytochemistry 6: 245–252 [Google Scholar]
- Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS (2013) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA 110: 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Thuong PT, Min BS, Ngoc TM, Hung TM, Lee IS, Na M, Seong Y, Song K, Bae KH (2006) Phenolic glycosides with antioxidant activity from the stem bark of Populus davidiana. J Nat Prod 69: 1370–1373 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turner JG (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X (2019) Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol 50: 29–36 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao L, Zhao J, Li Y, Wang J, Guo R, Gan S, Liu CJ, Zhanga K (2017) S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol 175: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Chen MY, Poisson T, Pannecoucke X, Bouillon JP, Besset T (2018) Pd-Catalyzed trifluoromethylthiolation of unsaturated compounds: a general approach. Euro J Org Chem 2018: 6167–6175 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.