Abstract
Plants have evolved a lignin-based Casparian strip (CS) in roots that restricts passive diffusion of mineral elements from the soil to the stele. However, the molecular mechanisms underlying CS formation in rice (Oryza sativa), which contains a CS at both the exodermis and endodermis, are poorly understood. Here, we demonstrate that CS formation at the rice endodermis is redundantly regulated by three MYELOBLASTOSIS (MYB) transcription factors, OsMYB36a, OsMYB36b, and OsMYB36c, that are highly expressed in root tips. Knockout of all three genes resulted in a complete absence of CS at the endodermis and retarded plant growth in hydroponic conditions and in soil. Compared with the wild-type, the triple mutants showed higher calcium (Ca) levels and lower Mn, Fe, Zn, Cu, and Cd levels in shoots. High Ca supply further inhibited mutant growth and increased Ca levels in shoots. Transcriptome analysis identified 1,093 downstream genes regulated by OsMYB36a/b/c, including the key CS formation gene OsCASP1 and other genes that function in CS formation at the endodermis. Three OsMYB36s regulate OsCASP1 and OsESB1 expression by directly binding to MYB-binding motifs in their promoters. Our findings thus provide important insights into the mechanism of CS formation at the endodermis and the selective uptake of mineral elements in roots.
Three MYB36 transcription factors regulate multiple genes implicated in Casparian strip formation at the endodermis of roots for the selective uptake of mineral elements in rice.
IN A NUTSHELL.
Background: The Casparian strip (CS) is a ring-like lignin-rich structure in the anticlinal cell wall between endodermal cells in the roots of vascular plants. The CS prevents unfavorable inflow and backflow of nutrients between the soil and stele. In Arabidopsis, AtMYB36 is the master regulator of CS formation at the endodermis. However, little is known about the molecular mechanism of CS formation in rice, which has a CS at both the exodermis and endodermis.
Question: Which homologs of Arabidopsis AtMYB36 are critical regulators of CS formation in rice? How do these genes regulate CS formation? What is the difference in CS role between rice and Arabidopsis?
Finding: Loss of OsMYB36a/b/c interfered only with CS formation in the endodermis, but not the exodermis. Three OsMYB36s redundantly regulate the expression of CS-associated genes. Knocking out all three genes resulted in plants that lacked a CS in the endodermis and reduced growth under both soil and hydroponic conditions. Furthermore, the severe growth inhibition in the triple mutants was mainly caused by over-accumulation of calcium (Ca). The absence of a CS and compensatory lignin deposited at the endodermis in Osmyb36abc disrupts the apoplastic flow of Ca to the stele, which was blocked by the CS at the endodermis in wild-type rice. These changes were not observed in Atmyb36 and other CS mutants in Arabidopsis, indicating that the rice CS defect has distinct effects on the selective uptake of mineral elements in roots.
Next steps: We will functionally characterize the downstream genes regulated by OsMYB36a/b/c, with the aim of elucidating the molecular mechanisms of CS formation at the endodermis and the role of the CS in mineral element uptake in rice.
Introduction
Plants rely on their roots to take up mineral nutrients from the soil to ensure growth and development. The concentrations of these nutrients in the soil fluctuate broadly during plant growth. This fluctuation is especially pronounced in paddy soil, where rice (Oryza sativa) is cultivated. Rice is able to grow under both upland and flooded conditions, where both the concentrations and chemical forms of mineral nutrients are quite different (Fageria et al., 2011). For example, under upland conditions, manganese (Mn) and iron (Fe) are present mainly in insoluble manganic and ferric forms, whereas under flooded conditions, they exist in the more reduced, soluble manganous (Mn2+) and ferrous (Fe2+) forms at significantly higher concentrations (Ponnamperuma, 1972). In response to these environmental differences, plants have evolved diverse strategies that control nutrient uptake for optimal growth. One of these strategies is the formation of a physical barrier called the Casparian strip (CS) at the endodermis and sometimes also at the exodermis, depending on the species in roots. The CS seals the cell wall space between neighboring endodermal cells, thereby playing a key role in preventing the unfavorable inflow and backflow of nutrients between the soil and the root stele (Robbins et al., 2014).
The CS is primarily composed of a hydrophobic lignin polymer (Naseer et al., 2012; Geldner, 2013). Genetic and molecular analyses have identified several key genes involved in CS formation in the Arabidopsis thaliana endodermis. Five CS domain proteins (CASPs) were initially reported to function in CS formation (Roppolo et al., 2011). These CASPs localize specifically at the CS membrane domain (CSD) and form a stabilizing transmembrane scaffold that assembles lignin-polymerizing proteins for CS lignification, including ENHANCED SUBERIN 1 (ESB1), RESPIRATORY BURST OXIDASE HOMOLOG F (RBOHF), and PEROXIDASE 64 (PER64) (Hosmani et al., 2013; Lee et al., 2013). UCLACYANIN 1 (UCC1) was recently shown to be involved in lignification in this central CS nanodomain (Reyt et al., 2020). Genes including ESB1, CASPs, PER64, and UCC1 are positively regulated by the R2R3-MYELOBLASTOSIS (MYB) transcription factor AtMYB36 (Kamiya et al., 2015; Liberman et al., 2015; Reyt et al., 2020). The expression of AtMYB36 is activated by SHORTROOT (SHR) (Li et al., 2018; Barbosa et al., 2019). Furthermore, SHR serves as a master regulator to direct the subcellular CS positioning of these proteins via SCARECROW (SCR) (Li et al., 2018). Both SHR and SCR belong to the GRAS-domain transcription factor family.
In Arabidopsis, CS integrity is under surveillance by the CIF1/2-SGN3-SGN1 signaling pathway (Doblas et al., 2017; Nakayama et al., 2017; Okuda et al., 2020). The leucine-rich repeat receptor-like kinase SCHENGEN3 (SGN3) flanks the CSD (Pfister et al., 2014), whereas the receptor-like kinase SGN1 localizes to the cortex-facing plasma membrane of endodermal cells (Alassimone et al., 2016). When the CS is defective, two small peptides expressed in the stele, CASPARIAN STRIP INTEGRITY FACTOR1 (CIF1) and CIF2, diffuse to the outer face of the endodermis and bind to SGN3, thereby activating the SGN3-SGN1 pathway to trigger compensatory lignification in the cortex and pericycle cell corners (Doblas et al., 2017; Nakayama et al., 2017). Once the CS is sealed, the diffusion of CIF1/2 is inhibited and they cannot interact with the co-receptor complex SGN3-SGN1. Therefore, a network of transcription factors (SHR, SCR, and AtMYB36) acts in parallel with the CIF1/2-SGN3-SGN1 signaling pathway to control CS development in Arabidopsis.
Almost all Arabidopsis mutants with defects in CS formation reported to date exhibit ionomic alterations in shoot tissues. For instance, the mutation of MYB36 led to lower concentrations of Ca, Mn, and Fe and higher concentrations of Na, Mg, and Zn in leaves (Kamiya et al., 2015). The ionomic alteration of the casp1-1 casp3-1 double mutant in leaves was similar to that of myb36-1 (Kamiya et al., 2015). The esb1 mutant showed decreased Ca, Mn, and Zn levels and increased K, S, As, Na, Se, and Mo levels in leaves (Hosmani et al., 2013). However, the concentrations of all ions examined except Mg and K were unaltered in the sgn3 mutant (Pfister et al., 2014). The sgn3 mutant exhibited high Mg and low K levels in leaves. Therefore, the CS plays a critical role in controlling the selective uptake of mineral elements into the stele for transfer to the shoot in Arabidopsis.
Unlike Arabidopsis roots, rice roots contain two CSs, which are located at the exodermis and endodermis (Enstone et al., 2002). OsCASP1 was recently shown to function in CS formation at the rice endodermis, but not at the exodermis (Wang et al., 2019a). The loss of function of OsCASP1 resulted in reduced growth, primarily due to the overaccumulation of calcium (Ca) in shoots. In addition, CS defects at the endodermis in the Oscasp1 mutant reduced the abundance of the silicon (Si) transporter Low Si 1 (Lsi1), leading to reduced Si uptake. These changes were not observed in Atcasp1 and other CS mutants in Arabidopsis, indicating that the CS plays different roles in selective mineral element uptake in rice versus Arabidopsis (Wang et al., 2019a). However, relatively little is known about the molecular mechanisms of rice CS formation and the role of the CS in controlling mineral uptake in rice roots.
In this study, we functionally identified three rice MYB36-like genes, OsMYB36a, OsMYB36b, and OsMYB36c, which are homologs of Arabidopsis AtMYB36. Detailed functional analysis revealed that OsMYB36s are critical transcription factors that coordinately regulate the temporal and spatial expression of genes essential for CS formation at the rice endodermis. Finally, compared with AtMYB36, OsMYB36s, or their encoded proteins showed distinct patterns of expression, cellular localization, effects on the selective uptake of mineral elements in roots, and effects on the regulation of CS-related genes.
Results
Sequence analysis of OsMYB36s
Using the amino acid sequence of AtMYB36 as a query, we identified three rice MYB36-like genes in the rice genome database, which we named OsMYB36a (LOC_Os08g15020), OsMYB36b (LOC_Os02g54520), and OsMYB36c (LOC_Os03g56090) (Supplemental Figure S1A and Supplemental Data Set S1). We amplified the full-length open-reading frames (ORFs) of three OsMYB36 genes from root cDNA. Sequence analysis of the amplified ORFs showed that the sequence of OsMYB36a and OsMYB36c is identical to the predicted transcripts based on the Rice Genome Annotation Project Database (http://rice.uga.edu/index.shtml) while that of OsMYB36b is not. OsMYB36b consists of three exons and two introns and encodes a protein of 316 amino acids (Supplemental Figure S1B). The N-terminal regions of these three OsMYB36s contain a highly conserved R2R3-type MYB domain that resembles that of AtMYB36, while their C-terminal regions are highly divergent (Supplemental Figure S1C). OsMYB36a shares the highest similarity to AtMYB36, with 45.3% identity, while OsMYB36b and OsMYB36c share 41.8% and 34.7% identity with AtMYB36, respectively (Supplemental Data Set S1).
OsMYB36s are highly expressed in the roots and their proteins together localize to the nuclei of root cells in the elongation and differentiation zones
To examine the organ specificity of OsMYB36s expression in rice, we performed reverse-transcription quantitative PCR (RT-qPCR) to evaluate their transcript levels in various organs, including roots, stems, leaf blades, and leaf sheath tissues. We determined that all three OsMYB36 genes were more highly expressed in roots than in other organs (Figure�1A). A more detailed expression analysis of OsMYB36s in different root regions showed that OsMYB36a expression was higher in the root tip (0–1 cm from the root apex) and 3–4 cm from the root apex than in other regions of the root. OsMYB36c transcripts were most abundant in the root tip (0–1 cm from the root apex), while OsMYB36b transcript levels were comparable across different root segments (Figure�1B). Furthermore, OsMYB36c showed higher expression in the roots than OsMYB36a and OsMYB36b (Figure�1B).
Figure 1.
Expression pattern of OsMYB36 and localization of OsMYB36a protein in rice. A, Relative expression of OsMYB36 genes in various tissues of WT rice (cv. Nipponbare). B, The spatial expression of OsMYB36 genes in the roots. RNA was extracted from different segments of the roots (0–1, 1–2, 2–3, and 3–4 cm from root apex) of 5-day-old seedlings. Expression of OsMYB36 genes was analyzed by RT-qPCR. Histone H3 was used as an internal standard. Data are means � sd of three biological replicates. C–N, Immunostaining of the roots of ProOsMYB36a:OsMYB36a cDNA-GFP transgenic plants using an anti-GFP antibody. F, J, and N represent the magnified image of the yellow boxed area in E, I, and M, respectively. K–N, Immunostaining of the roots of the WT using the anti-GFP antibody. Magenta indicates the signal of the anti-GFP antibody; blue indicates autofluorescence of the cell wall and nuclei stained with DAPI. These images were observed in the magenta (C, G, and K), blue (D, H, and L), and their merged (E, I, and M) channels. Root cross sections were made at 10 mm (C–F) or 5 mm (G–N) from the apex. ep, epidermis; ex, exodermis; sc, sclerenchyma; co, cortex; en, endodermis; st, stele. Six roots of each line were tested and showed a similar stain pattern. Scale bars, 50 μm.
To determine the subcellular localization of OsMYB36s in rice cells, we co-expressed the OsMYB36s-GFP fusion protein in rice protoplasts with the nuclear marker fusion protein OsGhd7-RFP (a fusion between Grain number, plant height, and heading date 7 [OsGhd7] and the fluorescent protein RFP) (Fu et al., 2019). OsMYB36a/b/c fluorescent signals strongly overlapped with signals from the nuclear marker (Supplemental Figure S2, A–L), indicating that three OsMYB36 members are nuclear proteins.
To investigate the cellular localization of OsMYB36s in roots, we performed immunostaining of transgenic rice harboring ProOsMYB36s:OsMYB36s cDNA-GFP. Immunostaining signals of an anti-GFP antibody in different root sections showed that OsMYB36a/b/c was expressed in all root cells examined, both at the root tip (where they were most abundant) and within the elongation zone (Figure�1, C–J and Supplemental Figures S3 and S4). To exclude the possibility that OsMYB36a introns can affect its cellular localization in roots, we also performed immunostaining of transgenic rice carrying ProOsMYB36a:genomic OsMYB36a-GFP. Again, we detected OsMYB36a in all root cells examined, suggesting that introns do not participate in regulating the expression pattern of OsMYB36a in roots (Supplemental Figure S3, A–A3 and B–B3). We also stained the nuclei with 4′,6-diamidino-2-phenylindole (DAPI) during immunolocalization with the anti-GFP antibody, which indicated that OsMYB36a and OsMYB36c mainly localized to the nucleus in the elongation and differentiation zones, but not in the meristem zone, while OsMYB36b localized to the nuclei of all root cells examined (Figure�1, C–J and Supplemental Figures S3 and S4). There was no GFP signal observed in the wild-type (WT) roots (Figure�1, K–N).
OsMYB36s redundantly control normal deposition of lignin at the endodermis
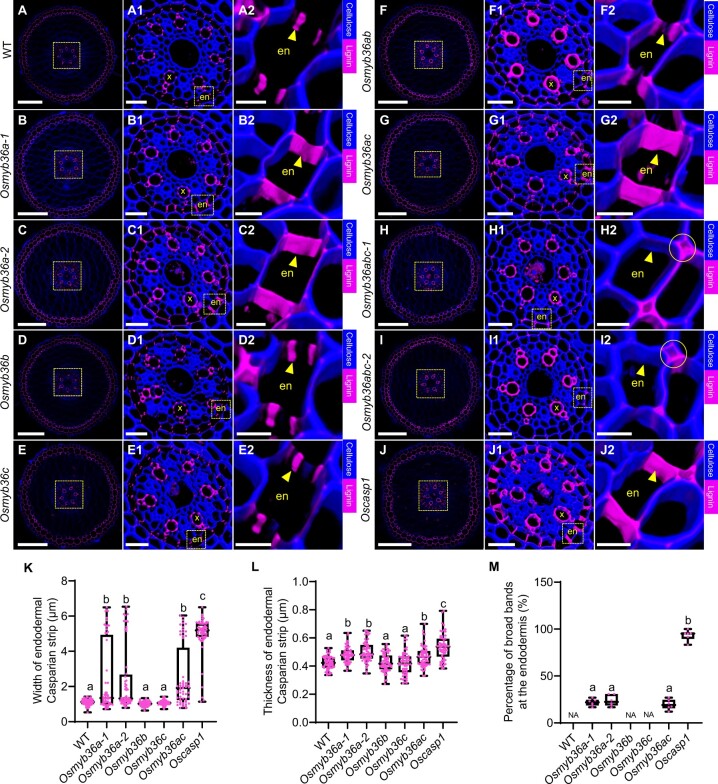
To investigate the role of OsMYB36s in CS formation, we generated single, double, and triple knockout lines of OsMYB36s by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene editing: Osmyb36a-1, Osmyb36a-2, Osmyb36b, Osmyb36c, Osmyb36ab, Osmyb36ac, Osmyb36abc-1, and Osmyb36abc-2 (Supplemental Figure S5, A–C). Among these mutant lines, root growth was slightly affected only in Osmyb36abc triple mutants at the early seedling stage (Supplemental Figure S5, D and E). Lignin staining showed that the CS at the anticlinal side of the exodermal cells formed normally in both the WT and Osmyb36s mutants (Supplemental Figure S6). However, a defect in CS formation at the anticlinal side of endodermal cells was observed in the mutants containing the Osmyb36a mutations. In the root tip region (10 mm from the root apex), lignin at the CS was incorrectly deposited, forming a broad band in some anticlinal walls of endodermal cells in root cross-sections from the Osmyb36a single and Osmyb36ac double mutants and Oscasp1 (Supplemental Figure S7, B–B2, C–C2, G–G2, J–J2, K, and L) rather than a fine band, as observed in the WT and single mutants of Osmyb36b and Osmyb36c (Supplemental Figure S7, A–A2, D–D2, E–E2, K, and L). Strikingly, no lignin deposition at the site of CS formation in the endodermis was detected in the Osmyb36ab double and Osmyb36abc triple mutants (Supplemental Figure S7, F–F2, H–H2, and I–I2). Further from the root apex (15 mm), endodermal CSs of the Osmyb36a single and Osmyb36ac double mutants became wider than that of the WT and the Osmyb36b and Osmyb36c single mutants but narrower than that of the CS mutant Oscasp1 (Figure�2, A–A2, B–B2, C–C2, D–D2, E–E2, G–G2, J–J2, and K–M). Interestingly, we observed ectopic lignin deposition at the corner/sides of the pericycle- and cortex-facing cell walls in the Osmyb36ab double mutant (Figure�2, F–F2). However, ectopic lignin deposition was mainly detected at the cortex-facing cell corner/walls between the endodermis and the cortex in Osmyb36abc triple mutants (Figure�2, H–H2 and I–I2). We observed a similar deposition pattern for lignin in longitudinal root sections from all lines (Supplemental Figure S8). These results indicate that three OsMYB36 members redundantly regulate the correct deposition of lignin at the anticlinal walls of endodermal cells.
Figure 2.
Observation of CS formation in WT, single and multiple knockout lines of OsMYB36s, and Oscasp1 mutant. Lignin staining was performed in the roots (15 mm from the apex) of the WT (A–A2), Osmyb36a-1 (B–B2), Osmyb36a-2 (C–C2), Osmyb36b (D–D2), Osmyb36c (E–E2), Osmyb36ab (F–F2), Osmyb36ac (G–G2), Osmyb36abc-1 (H–H2), Osmyb36abc-2 (I–I2), and Oscasp1-1 (J–J2). (A1–J1) magnified images of the yellow boxed area in (A–J), respectively. (A2–J2) magnified images of the yellow boxed area in (A1–J1), respectively. Magenta and blue show signal of lignin and cellulose, respectively. en, endodermis; x, xylem vessel. Yellow arrows indicate CS. Yellow circles indicate the corners between endodermis and cortex. Scale bars: 100 μm (A–J), 20 μm (A1–J1), 5 μm (A2–J2). K, Width of endodermal CS. L, Thickness of endodermal CS. Data in K and L are means � sd of 60 replicates derived from individual plants (10 replicates per plant and 6 plants per line). M, Percentage of broad bands at the endodermis. Six independent root sections for each line were analyzed. Every measurement of the width and thickness of CS or percentage of broad bands was labeled as a dot on the box plot. The box extends from the 25th to 75th percentiles and plots each individual value as a point superimposed on the graph. The line in the middle of the box marks the median. Significant difference was determined by Tukey’s test and labeled with different lowercase letters (P < 0.05).
We also generated transgenic lines overexpressing OsMYB36a or OsMYB36b under the control of the maize (Zea mays) Ubiquitin 1 promoter. OsMYB36a or OsMYB36b was expressed at high levels in their respective transgenic lines (Supplemental Figure S9, A and B). Overexpression of OsMYB36a or OsMYB36b did not affect the root growth of the early seedlings (Supplemental Figure S9, C and D). Lignin staining showed that CS formation at the endodermis in OsMYB36a or OsMYB36b overexpression lines was similar to that in WT rice (Supplemental Figures S10 and S11). Furthermore, ectopic lignin deposition in nonendodermal cells in the roots was not observed in the OsMYB36a- or OsMYB36b-overexpressing lines (Supplemental Figures S10 and S11). These results indicate that increasing the expression level of OsMYB36a or OsMYB36b individually in roots is not sufficient to induce lignin accumulation in root cells, including at the site of CS formation.
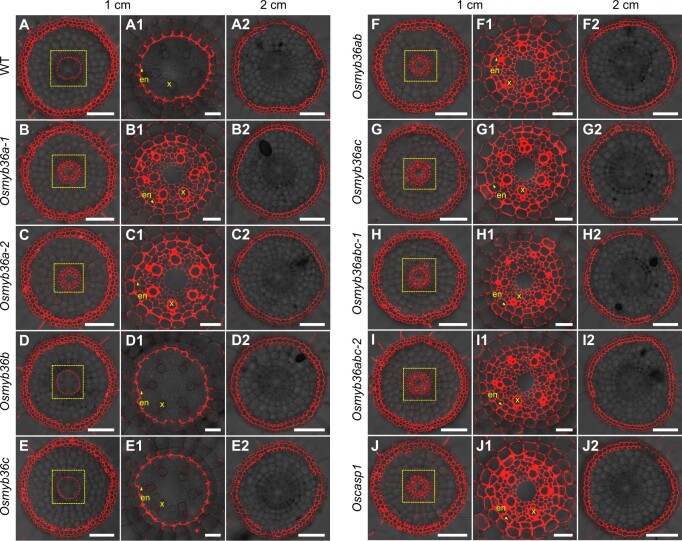
Mutants possessing the Osmyb36a mutations lack a functional apoplastic barrier at the endodermis
To determine whether the function of an apoplastic barrier at the endodermis was damaged in these osmyb36s mutants, we analyzed the permeability of the fluorescent dye propidium iodide (PI), an apoplastic tracer, into the stele of roots following incubation in PI for 40 min. At the root tip zone (1 cm from the root apex), where the CS had not completely formed at the exodermis, PI penetrated only into the cortical side of the WT endodermis and was blocked completely at the site of the CS (Figure�3, A and A1). In contrast, PI was able to penetrate into the stele in all the mutants except the Osmyb36b and Osmyb36c single mutants, a pattern similar to that of Oscasp1 (Figure�3, B–J and B1–J1). However, in the more mature region of the root (2 cm from the apex), where the CS had formed at the exodermis, the penetration of PI into the cortex was blocked at the exodermis in all lines examined (Figure�3, A2–J2). These results indicate that the functional apoplastic barrier at the endodermis was disrupted in the Osmyb36a, Osmyb36ab, Osmyb36ac, and Osmyb36abc mutants and that OsMYB36a played a more predominant role in the formation of a functional apoplastic barrier than OsMYB36b or OsMYB36c.
Figure 3.
Analysis of PI permeability in the roots of the WT, single and multiple knockout lines of OsMYB36s, and the Oscasp1 mutant. Roots of 5-day-old seedlings were incubated in PI solution for 40 min. Cross sections of the WT (A–A2), Osmyb36a-1 (B–B2), Osmyb36a-2 (C–C2), Osmyb36b (D–D2), Osmyb36c (E–E2), Osmyb36ab (F–F2), Osmyb36ac (G–G2), Osmyb36abc-1 (H–H2), Osmyb36abc-2 (I–I2), and Oscasp1-1 (J–J2) were observed at 10 mm (A–J) and 20 mm (A2–J2) from the root apex. (A1–J1) Magnified images of the yellow boxed area in (A–J), respectively. Yellow arrows indicate CS. en, endodermis; x, xylem vessel. Six roots of each line were tested and showed the same phenotype. Scale bars: 100 μm (A–J), 20 μm (A1–J1), and 100 μm (A2–J2).
OsMYB36s regulate suberin deposition on the exodermal and endodermal cells in roots
In Arabidopsis, CS mutants exhibit compensatory ectopic deposition of suberin at the endodermis. To explore whether the monogenic, digenic, and trigenic mutations of OsMYB36s resulted in a similar phenotype, we examined the accumulation of suberin in different root regions via suberin staining. At the region 30 mm from the root apex, suberin accumulation at several endodermal cells was observed in the WT as well as the Osmyb36b and Osmyb36c single mutants (Supplemental Figure S12, A, D, and E), whereas an increased suberization of most endodermal cells in a discontinuous pattern was observed in the Osmyb36a single and Osmyb36ac double mutants (Supplemental Figure S12, B, C, and F). This increase was not as strong as that in Oscasp1 (Supplemental Figure S12, J, U, and W). In contrast, little suberin accumulation at the endodermis was detected in the Osmyb36ab double and Osmyb36abc triple mutants compared with the WT (Supplemental Figure S12, G–I). Further from the root apex (40 mm), the trend of different suberin deposition patterns among them was not altered although endodermal suberization was enhanced in all lines (Supplemental Figure S12, K–T). Interestingly, we observed ectopic suberin deposition at the cortex-facing cell corner/walls between the endodermis and the cortex in both Osmyb36abc triple mutants (Supplemental Figure S12, R and S). We observed a similar pattern of suberin deposition in longitudinal root sections from all lines (Supplemental Figure S13). These results suggest that the single knockout of OsMYB36a, but not OsMYB36b or OsMYB36c, induced earlier and stronger suberin accumulation in the endodermis compared with the WT while the double knockout of OsMYB36ab and triple knockout of OsMYB36abc had the opposite effect.
In addition, suberin accumulation in the exodermis at different root regions was similar among all the lines except the Osmyb36ab and Osmyb36abc mutants (Supplemental Figures S12 and S13). More suberized cells at the exodermis were observed in the Osmyb36ab and Osmyb36abc mutants (Supplemental Figure S12, G, H, I, Q, R, S, Y, and Z). These results indicate that simultaneous knockout of OsMYB36a/b or OsMYB36a/b/c stimulates suberin deposition on the exodermal cells in roots.
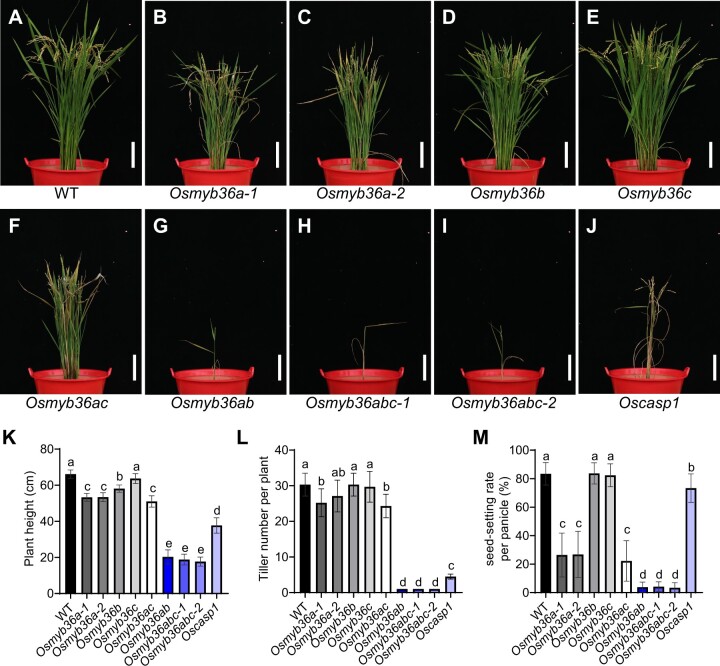
Mutants harboring the Osmyb36a mutations display pleiotropic growth defects
When grown in paddy soil, mutants carrying the Osmyb36a mutations showed necrosis of older leaves and inhibited growth to different extents at the vegetative stage, as observed in the previously described CS mutant Oscasp1 (Supplemental Figure S14). At maturity, the growth and yield of the plants harboring the Osmyb36a mutations were significantly reduced compared with WT plants, including plant height, tiller number, and seed-setting rate in flooded soils (Figure�4, B, C, F–I, and K–M). The Osmyb36ab double and Osmyb36abc triple mutants exhibited stronger inhibitory effects on rice growth than Oscasp1 (Figure�4, G–J). However, knockout of single OsMYB36b or OsMYB36c hardly affected growth. To validate the phenotype of the Osmyb36a mutants, we performed a complementation experiment by introducing a construct harboring the ProOsMYB36a:OsMYB36a into Osmyb36a-2 plants. Transgenic plants carrying the ProOsMYB36a:OsMYB36a construct exhibited phenotypes resembling those of the WT (Supplemental Figure S15), indicating that the mutation in OsMYB36a was responsible for the abnormal phenotype observed in the CRISPR-OsMYB36a lines.
Figure 4.
Phenotypic analysis of single and multiple knockout lines of OsMYB36s. A–J, Growth phenotype of WT rice, single and multiple knockout lines of OsMYB36s, and Oscasp1-1 grown in soil at the booting stage. Scale bars, 10 cm. K–M, Agronomic traits of WT rice, OsMYB36s-knockout lines, and Oscasp1-1. Plant height (K), Tiller number per plant (L), and seed-setting rate per panicle (M). Data are means � sd of 10 (K, L) and 30 (M) biological replicates. Significant difference was determined by Tukey’s test and labeled with different letters (P < 0.05).
Mutants containing the Osmyb36a mutations exhibit ionomic alterations in shoots
To examine whether CS defects in the Osmyb36s mutants had a significant effect on mineral element uptake, we performed ionomic analysis of roots and shoots from the WT and Osmyb36s knockout lines under different growth conditions. When grown in hydroponic solution, Osmyb36a single and Osmyb36ac double mutants showed increased Ca, Sr, and P concentrations and reduced Mg, Fe, Cd, and Rb concentrations in shoots compared with the WT, while the single knockout of OsMYB36b or OsMYB36c hardly affected the ionome (Figure�5). The Osmyb36ab double and Osmyb36abc triple mutants displayed large ionomic changes with a higher accumulation of Ca, Sr, and P and a lower accumulation of K, Mn, Fe, Rb, Ge, Zn, Cu, and Cd, which were similar to but more pronounced than the changes in Oscasp1 (Figure�5). The concentrations of most mineral elements in roots were not altered in the Osmyb36ab and Osmyb36abc mutants (Supplemental Figure S16), except that Cu and Cd concentrations were lower in Osmyb36ab and Osmyb36abc than in the WT (Supplemental Figure S16C). However, under both flooded and upland soil conditions, the Ca concentrations in shoots of the mutants containing the Osmyb36a mutation were the most affected among the mineral elements examined (Supplemental Figure S17). In particular, both triple mutants showed an approximately seven-fold increase in Ca concentrations in shoots compared with the WT (Supplemental Figure S17A). These results indicate that simultaneous mutation of three OsMYB36s led to large changes in the concentrations of multiple mineral elements in shoots, especially Ca.
Figure 5.
Comparison of the shoot ionome of WT rice, OsMYB36s-knockout lines, and Oscasp1-1 mutant. A, Concentration of Ca, Sr, K, and P in the shoots. B, Concentration of Mg and As in the shoots. C, Concentration of Mn, Fe, Cu, Zn, Cd, Ge, and Rb in the shoots. The WT and OsMYB36s-knockout lines were grown in a nutrient solution for 30 days. Before harvest, the plants were transferred to a solution containing 0.2 �M of As and Cd, and 1 �M of Ge, Rb, and Sr, for 1 day. Data are means � sd of four biological replicates. Significant difference was determined by Tukey’s test and labeled with different letters (P < 0.05).
Ca accumulation represses the growth of various Osmyb36a mutants
We previously demonstrated that the growth of the CS-defective mutant Oscasp1 is inhibited, primarily due to the overaccumulation of Ca in shoots (Wang et al., 2019a). Therefore, we compared Ca accumulation and growth in the WT and Osmyb36s mutants as well as Oscasp1 as a control under different Ca concentrations. Similar to Oscasp1 plants, the growth of the mutants containing the Osmyb36a mutation was significantly inhibited by Ca treatment, and older leaves exhibited necrosis with increasing Ca supply (Figure�6A), which phenocopied the mutants grown in soil. In contrast, the growth of WT plants was not affected by any of the Ca concentrations tested (Figure�6, A–C).
Figure 6.
Comparison of growth and Ca accumulation of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant under different Ca concentrations. A, Plant growth of WT plants, OsMYB36s-knockout lines, and Oscasp1 mutant exposed to different Ca concentrations (0.18, 1, and 5 mM) for 12 days. Scale bars, 10 cm. B and C, Dry weight of shoot and root. D, Concentration of Ca in the shoots. E, Concentration of Ca in the xylem sap. Thirty-day-old seedlings of WT and mutants were exposed to 5 mM Ca for 6 h before collecting xylem sap. Data are means � sd of four biological replicates. Significant difference was determined by Tukey’s test and labeled with different letters (P < 0.05).
Mineral analysis showed that the shoot Ca concentration of the mutants containing the Osmyb36a mutation was significantly higher than that of the WT under different Ca concentrations (Figure�6D). Especially, the Osmyb36ab and Osmyb36abc mutants contained approximately 2.5, 2.5, and 3.3 times the Ca concentration of WT plants in shoots in response to 0.18, 1, and 5 mM Ca treatment, respectively (Figure�6D). In contrast, there were no significant differences in Ca concentrations in the roots of all the lines except the Osmyb36ab and Osmyb36abc mutants in response to these treatments (Supplemental Figure S18). Additionally, the Ca concentrations in xylem sap from the mutants containing the Osmyb36a mutation was sharply increased by up to 3–10-fold compared with those in the WT in response to 5 mM Ca treatment (Figure�6E). Furthermore, the Ca concentrations in the shoots and xylem sap of the Osmyb36ab and Osmyb36abc mutants were markedly higher than those of other lines under the same conditions (Figure�6, D and E). These results suggest that the growth retardation of the mutants possessing the Osmyb36a mutation is largely due to high Ca accumulation in shoots and that OsMYB36a/b/c have an important effect on Ca accumulation in rice.
OsMYB36s are transcriptional activators and redundantly regulate multiple CS-associated genes in roots
To examine whether OsMYB36s have transcriptional activity, a yeast assay was employed. We fused the full-length, N terminus, or C terminus of OsMYB36s to the DNA-binding domain of GAL4, producing the BD-OsMYB36s, BD-OsMYB36s-N, and BD-OsMYB36s-C constructs, respectively. Yeast cells transformed with the BD-OsMYB36s or BD-OsMYB36s-C construct grew well and formed blue colonies on selective growth medium, whereas those transformed with BD-OsMYB36s-N or the negative control did not (Supplemental Figure S19). These results suggest that three OsMYB36s possess transcription activation activity in yeast cells and that their activation domains are located at their C termini.
To identify genes that are directly or indirectly regulated by OsMYB36s, we analyzed the root transcriptomes of WT, Osmyb36a-1, Osmyb36ab, and Osmyb36abc-1 plants by RNA-seq. We found that the Osmyb36abc-1 triple mutant had a larger number of upregulated or downregulated genes than either the Osmyb36a-1 single or Osmyb36ab double mutant compared with the WT. In total, we identified 5,587 differentially expressed genes (DEGs) between the WT and Osmyb36abc-1 mutant. Among these, 2,540 and 3,047 DEGs were upregulated and downregulated, respectively, in Osmyb36abc-1 roots, and were therefore defined as candidate genes regulated by OsMYB36a/b/c (Figure�7A and Supplemental Data Set S2). To rule out the genes whose expression in the Osmyb36abc mutant was indirectly affected by a loss of functional CS and ectopic deposition of lignin and suberin at the endodermis, we subtracted genes whose expression was also altered in the Oscasp1 mutant, because Oscasp1 also displayed the dysfunctional CS and ectopic lignin and suberin accumulation at the endodermis, but OsCASP1 is not a transcriptional regulator. Since Oscasp1 is a CRISPR/Cas9 mutant, in which the mRNA accumulation of mutated targeted gene sometimes is suppressed (Wang et al., 2019b), OsCASP1 was not eliminated as a candidate, although its expression level was reduced in the Oscasp1 mutant (Supplemental Data Sets S3–S5). Finally, 1,093 downregulated genes were detected in Osmyb36abc, which were considered to be OsMYB36a/b/c-regulated candidate genes implicated in CS formation at the endodermis (Figure�7A and Supplemental Data Set S3).
Figure 7.
Regulation of CS-associated genes by OsMYB36a/b/c. A, Venn diagrams of OsMYB36a/b/c downregulated genes using the RNA-seq of the roots compared the Osmyb36abc mutant with the WT, Osmyb36a, Osmyb36ab, and Oscasp1. B, Cluster analysis of CS associated genes regulated by OsMYB36a/b/c based on z-score-normalized expression of genes. Red and blue indicate high and low expression level, respectively. Heatmap is constructed by R package. C, Cluster analysis of gene expression levels in various Osmyb36s mutants and Oscasp1 relative to the WT based on RT-qPCR results (Supplemental Data Set S5). The color gradient from low (blue) to high (yellow) represents relative levels of gene expression. Heatmap was generated by MeV software. D, Schematic diagram of the reporter and effector constructs used in the dual LUC assay system. E and F, Activation of the promoters of OsCASP1 (E) and OsESB1 (F) by OsMYB36s in rice protoplasts. The LUC/REN ratio relative to the control (a basal signal without OsMYB36s) is shown. Data are means � sd of three biological replicates. G and H, OsMYB36 proteins binding to the promoters of OsCASP1 (G) and OsESB1 (H) in yeast. The truncated OsMYB36s cDNA (∼360 bp) was fused to the GAL4 activation domain in the prey vector (pGAD-MYB36s-N). The promoter sequences of OsCASP1 (2,082-bp fragment immediately upstream of the start codon) and OsESB1 (2,126-bp fragment immediately upstream of the start codon) were fused to the HIS reporter gene in the pHIS2.1 vector (pHIS-ProCASP1 and pHIS-ProOsESB1), respectively. Yeast cells transformed with different pairs of plasmids were cultured on SD medium containing 0, 10, or 100 mM 3-amino-1,2,4-triazole, a competitor of HIS3 at 30�C for 3 days in the absence of His. I, EMSA showing binding of recombinant His-OsMYB36s-N to the MYB-binding motif of target promoters. The sequence of the MYB-binding motif is indicated by red letters. FAM-labeled fragments of the OsCASP1and OsESB1 promoter (FAM-P1) are shown in lanes 1–4. Unlabeled OsCASP1and OsESB1 promoter fragment competitors (Cold-P1) were used in 50-fold molar excess (lane 3). Unlabeled OsCASP1fragment competitor without the MYB-binding motif (Cold-P2) was used in 50-fold molar excess (lane 4).
Among these, 52 genes are homologous to or belong to the same gene family as known CS-related genes in Arabidopsis (Figure�7B and Supplemental Data Set S4). To validate the genes identified by RNA-seq, we performed RT-qPCR analysis. We selected six genes with no known homologs in Arabidopsis, as well as another 21 genes whose homologs were identified as major players in CS formation in Arabidopsis, such as OsCASPs, OsESBs, and OsSGN1- and OsSGN3-like genes (Figure�7C and Supplemental Data Set S5). The RT-qPCR data for most genes were in close agreement with the transcriptome data, indicating that the RNA-seq data were highly reliable. We also found that in general, the double mutant had a lower expression level of these CS-related genes than the single mutant, with the lowest expression level observed in the Osmyb36abc triple mutant. These data suggest that OsMYB36a, OsMYB36b, and OsMYB36c redundantly regulate the transcription of these genes.
To determine whether OsMYB36s can activate the promoters of known or candidate CS-related genes, we performed a transient transactivation assay in rice protoplasts using the luciferase (LUC) gene as the reporter and CaMV35-driven OsMYB36s as the effector (Figure�7D). LUC driven by the promoter of eight individual genes (ProOsCASP1:LUC, ProOsESB1:LUC, ProOs03g0245500:LUC, ProOs08g0515700:LUC, ProOs09g0493400:LUC, ProOs09g0535400:LUC, ProOs10g0155100:LUC, or ProOs11g0499600:LUC) was introduced alone or co-introduced with Pro35S:OsMYB36s in rice protoplasts. The luminescence intensity in cells carrying both OsMYB36s and the promoter-LUC fusion was significantly stronger than those in cells carrying the promoter-LUC fusion alone (Figure�7, E and F and Supplemental Figure S20), suggesting that OsMYB36a, OsMYB36b, or OsMYB36c does indeed activate the transcriptional activity of the promoters of these eight genes, including the key CS formation gene OsCASP1 and OsESB1.
To further test the notion that OsMYB36s transcriptionally regulates OsCASP1 and OsESB1, we performed a yeast one-hybrid (Y1H) assay. A fragment (∼2.0 kb) upstream of the OsCASP1 and OsESB1 start codon was used as bait for the binding assays. Yeast cells carrying OsMYB36s and either the OsCASP1 or OsESB1 promoter grew better on these selective media than yeast cells carrying the empty vector pGADT7 and the promoter fragments (Figure�7, G and H), indicating that OsMYB36a, OsMYB36b, or OsMYB36c directly interacts with the OsCASP1 and OsESB1 promoters.
We also performed an electrophoretic mobility shift assay (EMSA) to determine whether OsMYB36s directly bind to the MYB-binding motifs within the promoter regions of OsCASP1 and OsESB1. The promoter sequences of OsCASP1 and OsESB1 contain several MYB-binding motifs, as determined by plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) analysis. We designed two probes covering the MYB-binding sites: OsCASP1 (−236 to −217 bp from the start codon) and OsESB1 (−217 to −200 bp). We detected the formation of a MYB36s/FAM-labeled probe complex. The presence of the same probe lacking the FAM label significantly inhibited the formation of this complex, while the unlabeled probe without the MYB-binding motif did not (Figure�7I). These results indicate that OsMYB36a, OsMYB36b, or OsMYB36c physically interacts with the promoter sequences of OsCASP1 and OsESB1 via their MYB-binding motifs.
OsMYB36s affects the establishment of the OsCASP1 microdomain on endodermal cells
To examine whether OsMYB36s are involved in regulating the localization of OsCASP1, we performed an immunostaining assay for GFP to observe the localization of OsCASP1 by introducing ProOsCASP1:OsCASP1-GFP into the WT, Osmyb36a-1, OsMYB36a- and OsMYB36b- overexpressing lines, and Osmyb36abc (Supplemental Figure S21). In the region 1 mm from the root apex, GFP signals were not detected in any line (Figure�8, A–D and Supplemental Figure S22, A–D). However, in the region 3 mm from the root apex, OsCASP1-GFP was localized throughout the plasma membrane and was enriched in the anticlinal sides of endodermal cells in both the WT and Osmyb36a-1 (Figure�8, E, F, andU and Supplemental Figure S22, E and F). In contrast, OsCASP1-GFP in the overexpressing line of OsMYB36a but not OsMYB36b formed a fine band that was specifically localized to the middle of the anticlinal sides of endodermal cells, where the CS would be deposited (Figure�8, G, K, O, S, andU–W and Supplemental Figures S22, G, K, O, S and S23). Finally, a fine band of OsCASP1-GFP signals was detected in the region 5 mm from the root apex in the WT (Figure�8, J, N, R, andV and Supplemental Figure S22, J, N, and R) and in the region 7 mm from the root apex in Osmyb36a (Figure�8, I, M, Q, and W and Supplemental Figure S22, I, M, and Q). However, no OsCASP1-GFP accumulation in all the root sections was observed in the Osmyb36abc triple mutant (Figure�8, H, L, P, and T and Supplemental Figure S22, H, L, P, and T). Additionally, RNA-seq and RT-qPCR analysis showed that OsCASP1 transcript was hardly detected in the triple mutant (Supplemental Data Sets S2 and S5). These results indicate that the mutation of single OsMYB36a delayed the formation of the OsCASP1 microdomain on the anticlinal side of the endodermal cells, whereas the overexpression of OsMYB36a not OsMYB36b promoted this process, but they did not alter the cellular or subcellular localization of OsCASP1 in the root, and that OsMYB36a/b/c redundantly affects the establishment of its microdomain through the transcriptional regulation of OsCASP1.
Figure 8.
Spatial localization of OsCASP1 in WT rice, Osmyb36a, Osmyb36abc, and an OsMYB36a-overexpressing line. A–T, Double staining for GFP (green) and cellulose (blue) was performed to observe OsCASP1 localization at different root positions (1, 3, 5, 7, and 10 mm from the apex) of WT (A, E, I, M, and Q), Osmyb36a-1 (B, F, J, N, and R), OsMYB36a overexpressing line (C, G, K, O, and S) and Osmyb36abc-1 (D, H, L, P, and T). 1 mm (A–D), 3 mm (E–H), 5 mm (I–L), 7 mm (M–P), and 10 mm (Q–T) from the root apex. en, Endodermis. Scale bars, 20 μm. (U–W) Quantification of pixel intensity (0–255) across the OsCASP1 microdomain (distance in micrometer) at 3 mm (U), 5 mm (V), or 7 mm (W) from the root apex using surface views of (E–O). Light curves represent individual replicates (n = 4). Data of each individual plant are the average pixel intensity of six OsCASP1 microdomains. Mean values for ProOsCASP1:OsCASP1-GFP in Osmyb36a-1 (red), WT (black), and ProUbi:OsMYB36a (OX, green).
Knockout of all three OsMYB36s alters the expression of the Si transporter Lsi1 in roots
The disruption of OsCASP1 significantly reduced the protein abundance of Lsi1 in root endodermal cells but did not affect its polar localization (Wang et al., 2019a). To determine whether the knockout of OsMYB36s would affect the expression and/or polarity of Lsi1, we performed double staining for GFP and cellulose in the roots of transgenic plants generated by introducing ProLsi1:Lsi1genomic-GFP into the WT, Osmyb36a-2, and Osmyb36abc backgrounds (Supplemental Figure S21). The protein abundance and polarity of Lsi1 at the distal side of the exodermis and endodermis were similar between WT and Osmyb36a roots (Supplemental Figure S24, A and B). However, Lsi1 was polarly localized at the distal side of the exodermis, and its expression was not detected at the endodermis in the Osmyb36abc roots (Supplemental Figure S24, C and D). Furthermore, its abundance at the exodermis was not changed. These results indicated that simultaneous knockout of OsMYB36a/b/c resulted in undetectable Lsi1 expression at the endodermis, but did not affect its expression and polarity at the exodermis.
Discussion
Genes regulated by OsMYB36a/b/c function in different aspects of CS formation at the endodermis
The rice genome contains three MYB36-like genes (OsMYB36a, OsMYB36b, and OsMYB36c). These genes encode proteins with highly conserved R2R3-type MYB DNA binding domains in their N-terminal regions, whereas their C-terminal transcriptional activation domains are relatively variable. Although all three OsMYB36 members can activate the expression of CS-associated genes such as OsCASP1 and OsESB1 in rice protoplasts, individually they have a weak or no effect on the expression level of these genes in rice roots. However, OsMYB36ab or OsMYB36abc possess a gradually enhanced ability to modulate the expression of these genes. A similar trend was observed for endodermal CS formation, with the strongest effect on disrupting CS formation also found in the Osmyb36abc mutants. These results indicate that three OsMYB36 members redundantly regulate CS formation at the endodermis through activating the expression of CS-associated genes.
Rice roots contain one CS at the endodermis and one at the exodermis (Enstone et al., 2002). Although three OsMYB36s were expressed in all types of root cells, the loss of OsMYB36a/b/c interfered with CS formation at the endodermis, but not at the exodermis (Supplemental Figure S6), suggesting that CS formation at these tissues is regulated by different mechanisms. Transcriptomic analysis revealed candidate downstream genes regulated by OsMYB36a/b/c that are implicated in the positioning, lignification, and integrity of endodermal CS formation (Supplemental Data Sets S3 and S4 and Supplemental Figure S25). For example, four candidate genes, OsCASP1, OsCASP2, OsCASP3, and OsCASP5, belong to the CASP family. OsCASP1 is thought to interact with itself and OsCASP2 at the CSD (Wang et al., 2020b) and to form a scaffold to mediate CS formation at the endodermis (Wang et al., 2019a). Os01g0827600 is a subunit of the Exo70 exocyst complex. In Arabidopsis, the exocyst subunit AtEXO71A1 mediates the localization and positioning of CASP1 at the CSD where the CS is to be deposited (Kalmbach et al., 2017). Eleven candidate genes appear to encode lignin-polymerizing proteins, including peroxidase, laccase (LAC), uclacyanin, and dirigent proteins. Os01g0263300 and Os01g0263000 are homologs of Arabidopsis PER64, which is involved in monolignol oxidation at the CSD (Lee et al., 2013; Barbosa et al., 2019). Endodermis-enriched PERs are required for CS lignification (Rojas-Murcia et al., 2020). Os03g0297900 encodes a LAC. LACs are thought to function as secretory multicopper oxidases that participate in lignin polymerization (Boerjan et al., 2003; Barbosa et al., 2019; Zhuang et al., 2020). Os06g0218600, Os04g0545400, Os07g0101000, Os04g0561900, and Os02g0653200 encode uclacyanin proteins. Recently, this family was reported to be required for the lignification of a central CS nanodomain in Arabidopsis (Reyt et al., 2020). Three genes encode the dirigent protein in rice, namely Os03g0280750, Os01g0155300, and Os03g0143900. These genes are homologous to the Arabidopsis gene encoding the CS-localized protein ESB1, which is involved in CS lignification (Hosmani et al., 2013).
Nine candidate genes encoding receptor-like kinases were also identified as downstream genes of OsMYB36a/b/c such as Os05g0170300, Os09g0513600, Os03g0556600, and Os06g0544100 (Figure�7C and Supplemental Data Set S5). These genes are homologous to Arabidopsis SGN1 or SGN3, encoding a component of the CIF1/2-SGN3-SGN1 signaling pathway (Doblas et al., 2017; Nakayama et al., 2017). This signaling pathway controls CS integrity at the endodermis by activating the compensatory lignification machinery. These nine genes are likely involved in maintaining the integrity of the CS in rice. However, most candidate genes that function downstream of OsMYB36a/b/c are uncharacterized, and their functions remain to be examined.
Interestingly, knockout of a single OsMYB36 gene did not affect exodermal suberization, while simultaneous knockout of OsMYB36a/b or OsMYB36a/b/c resulted in an increased suberization of most exodermal cells. These results suggested that three OsMYB36s might redundantly modulate suberin accumulation at the exodermis. Alternatively, a compensatory suberin accumulation at the exodermis might be indirectly induced by the loss of CS at the endodermis in the Osmyb36ab and Osmyb36abc mutants. Additionally, the broad expression patterns of three OsMYB36 genes imply that they may process other physiological functions in rice. For instance, mutants containing the Osmyb36a mutation exhibited multiple growth defects such as a reduced number of tillers and low seed-setting rate. Further investigations are needed to decipher the exact underlying mechanisms for these.
The mechanisms underlying CS formation differ between rice and Arabidopsis
In Arabidopsis, CS formation at the endodermis was shown to be coordinately controlled by two distinct signaling pathways: SHR-SCR-AtMYB36 and CIF1/2-SGN3-SGN1 (Drapek et al., 2018; Li et al., 2018; Barbosa et al., 2019). The SHR/SCR pathway activates the transcription of AtMYB36, whose protein product in turn induces the expression of genes involved in CS lignification. Recently, AtMYB36 was also reported to regulate the expression of the suberin biosynthetic genes in roots (Wang et al., 2020a). The SGN3-SGN1 pathway contributes to CS integrity and triggers compensatory lignification and suberization in CS-defective mutants. In Atmyb36, ectopic lignification and an upregulation of suberin are observed, although SGN3 and SGN1 expression was not significantly downregulated, indicating that AtMYB36 is not directly involved in regulating the SGN3-dependent activation of the compensatory mechanism (Kamiya et al., 2015). Homologs for CIF1, CIF2, SGN3, and SGN1 exist in the rice genome (Wang et al., 2019a). Moreover, typical compensatory phenotypes such as ectopic lignification and enhanced suberization at endodermal cells were also observed in rice CS-defective mutants such as Oscasp1, Osmyb36a, and Osmyb36ac, suggesting that the activation of a similar SGN3-dependent pathway might occur in rice upon CS disruption. Yet, different from Atmyb36 and most other CS-defective mutants, the Osmyb36abc triple mutants displayed a delayed and lower accumulation of lignin and suberin at some cortex-facing cell corner/walls between endodermal and cortical cells (Figure�2 and Supplemental Figures S7, S8, S12, and S13). RNA-seq and RT-qPCR analyses showed that the expression levels of some SGN1- and SGN3-like genes are markedly lower in Osmyb36abc mutants, but not in the Osmyb36 single mutants (Figure�7C and Supplemental Data Sets S4 and S5), indicating that all three OsMYB36 transcription factors redundantly regulate the expression of these genes. These data suggest that a strong delay of the compensatory phenotype in Osmyb36abc may be due to a weak activation of the SGN3-dependent compensatory mechanism through downregulating the expression of SGN1- and SGN3-like genes by Osmyb36a/b/c. Therefore, OsMYB36a/b/c may regulate not only CS lignification but also compensatory lignification and suberization at the endodermis in rice, perhaps as regulators of the crosstalk between the CS biosynthetic and CIF1/2-SGN3-SGN1 signaling pathways.
CASPs are key organizers of a membrane scaffold that contributes to CS formation (Roppolo et al., 2011). In Arabidopsis, the expression and localization of CASPs are primarily regulated by AtMYB36. In Atmyb36 plants, AtCASP1-GFP and AtCASP2-GFP did not accumulate at the endodermis (Kamiya et al., 2015; Liberman et al., 2015). When AtCASP1 driven by the SCR promoter was specifically expressed in Atmyb36 endodermal cells, AtCASP1 did not localize to the sites of the CSD between endodermal cells; instead, it uniformly localized to the plasma membrane and cytosol (Kamiya et al., 2015). Compared with AtMYB36, OsMYB36a, OsMYB36b, or OsMYB36c alone has a relatively weak effect on the mRNA accumulation of OsCASP1. The protein abundance of OsCASP1 in endodermal cells was also not obviously altered in the Osmyb36a mutant. However, knockout of OsMYB36a did delay the lateral concentration of OsCASP1 on the anticlinal sides of endodermal cells, whereas overexpressing OsMYB36a but not OsMYB36b promoted the premature localization of OsCASP1 at the endodermis. These results indicate that (1) OsMYB36a regulates the expression of genes involved in the lateral concentration of OsCASP1 into the CSD and (2) OsMYB36a function cannot be replaced by that of OsMYB36b. However, simultaneous knockout of OsMYB36a/b/c resulted in a complete lack of OsCASP1 accumulation at the endodermis, suggesting that all three OsMYB36s regulate the expression and localization of OsCASP1 (Supplemental Figure S25). Like Atmyb36 plants (Kamiya et al., 2015), Osmyb36abc plants showed no CS lignification at the root endodermis. These results indicate that three OsMYB36s and single AtMYB36 play a similar function in the activation of CASP1 expression and CS lignification at the root endodermis. Yet, their expression patterns in roots are quite different. AtMYB36 is specifically expressed in endodermal cells, whereas three OsMYB36s are expressed in both endodermal and nonendodermal cells. In addition, the ectopic expression of AtMYB36 was sufficient to activate the expression of AtCASP1 and cause CS-like structures to form in the epidermis and cortex, whereas we failed to detect either OsCASP1 expression or CS-like structures in OsMYB36a/b/c-expressed nonendodermal cells. Furthermore, overexpressing OsMYB36a or OsMYB36b did not induce the expression of OsCASP1 or the formation of CS-like structures in any nonendodermal root cells. These differences indicate that AtMYB36 is both necessary and sufficient for the activation of CASP1 and CS lignification in Arabidopsis while OsMYB36a/b/c are necessary but not sufficient in rice.
Other unknown factors might also participate in this regulatory cascade in rice. Importantly, the downstream genes regulated by OsMYB36a/b/c versus AtMYB36 are different, with the exceptions of CASP1, CASP2, CASP3, and ESB1 (Supplemental Data Set S6). We showed here that OsMYB36a/b/c regulate many more candidate genes related to CS formation than AtMYB36 does, with some being unique to rice; for instance, Os03g0245500, Os09g0535400, and Os10g0155100 were predicted to encode mannose-specific plant lectins. Os08g0515700 and Os09g0493400 encoded putative proteins containing a domain of unknown function, DUF547, implying that there are additional mechanisms for endodermal CS formation in rice.
Osmyb36s mutations have a distinct effect on the selective uptake of mineral elements from Arabidopsis CS mutants
In Arabidopsis, the compensatory ectopic deposition of lignin and suberin at the endodermis of CS mutants affects both apoplastic and transmembrane ion transport across the endodermis, resulting in complex changes in the shoot ionome (Hosmani et al., 2013; Kamiya et al., 2015; Li et al., 2017). We also observed ectopic deposition of lignin and suberin at the endodermis in the mutants possessing the Osmyb36a mutations (Figure�2, B–B2, C–C2, F–F2, G–G2, H–H2, and I–I2 and Supplemental Figures S12 and S13). When grown in nutrient solution, the Osmyb36a single and Osmyb36ac double mutants exhibited a weak effect on the ionomic changes, while the Osmyb36ab double and Osmyb36abc triple mutants showed a strong effect with higher Ca, Sr, and P contents and lower K, Mn, Fe, Zn, Cu, Ge, and Cd contents in shoots than the WT. However, when grown in soil, Ca was the only element that showed a major increase in concentration in the shoots of the mutants possessing the Osmyb36a mutation versus the WT (Supplemental Figure S17). In contrast, Atmyb36 and other Arabidopsis CS mutants accumulated lower levels of Ca in shoots than the WT.
We previously demonstrated that the CS has distinct effects on controlling mineral element uptake in rice versus Arabidopsis, which was attributed to their different uptake systems and root structures (Wang et al., 2019a). In rice roots, both the exodermis and endodermis contain a CS. Moreover, mature rice roots contain a highly developed aerenchyma between the exodermis and endodermis (Kawai et al., 1998). This root structure underlies the distinct uptake system for mineral uptake in rice; this system is composed of influx and efflux transporters that are polarly localized to the distal and proximal sides of both the exodermis and endodermis (Sasaki et al., 2016; Che et al., 2018). RNA-seq analysis showed that the expression levels of almost all the genes encoding plasma membrane-localized influx/efflux transporters/channels for Ca were not significantly upregulated in Osmyb36s and Oscasp1 roots (Supplemental Data Set S7), indicating that Ca overaccumulation in the shoots of these mutants was not caused by the changes in the membrane transporter/channel-regulated symplastic transport of Ca in roots. In rice, apoplastic Ca uptake mainly occurs in the root tip (less than 10 mm from the root apex), which contains a fully formed CS at the endodermis, but not at the exodermis (Wang et al., 2019a). Similar to Oscasp1, it appears that defects in CS formation at the endodermis of the mutants possessing the Osmyb36a mutation impair the control of the apoplastic flow of Ca to the stele, resulting in the overaccumulation of Ca in shoots (Supplemental Figure S25). This notion is supported by an analysis of the short-term uptake of Sr (a congener of Ca), an apoplastic tracer (Figure�5A). However, Ca levels in the shoots and xylem sap were higher in the Osmyb36ab double and Osmyb36abc triple mutants than in Oscasp1 under the same conditions, likely because knockout of OsMYB36ab or OsMYB36abc has a stronger effect on disrupting CS formation at the endodermis compared with the mutation of OsCASP1. This conclusion is supported by the finding that the absence of CS and compensatory lignification occurred between neighboring endodermal cells in Osmyb36ab and Osmyb36abc, as revealed by lignin staining (Supplemental Figure S7).
Unlike Arabidopsis, rice is a Si- and Mn-accumulating plant. The uptake of Si/Ge and Mn in rice roots is mediated by two pairs of polarly localized influx/efflux transporters: Lsi1 (influx) and Lsi2 (efflux) for Si/Ge (Ma et al., 2006, 2007) and OsNramp5 (influx) and OsMTP9 (efflux) for Mn (Sasaki et al., 2012; Ueno et al., 2015). OsNramp5 also functions as the major pathway for Cd entry to the rice roots (Sasaki et al., 2012). Like Oscasp1, Osmyb36abc contained altered Ge (a chemical analog of Si), Mn, and Cd levels in shoots. The significant reduction in Si and Ge uptake in Oscasp1 mutants is caused by decreased Lsi1 protein abundance, which is likely triggered by the CIF1/2-SGN3-SGN1 signaling pathway (Wang et al., 2019a). Therefore, perhaps the loss of function of OsMYB36abc affected the protein abundance of Lsi1 due to its stronger defects in the CS. Indeed, immunostaining showed no accumulation of Lsi1 at the endodermis in the Osmyb36abc plants (Supplemental Figure S24). Decreased Mn and Cd accumulation in Osmyb36abc might also be due to altered OsNramp5 mRNA or OsNramp5 protein abundance in this mutant. Indeed, the transcript abundance of OsNramp5 was significantly reduced in the Osmyb36abc roots relative to the WT (Supplemental Data Set S7).
In conclusion, three OsMYB36 transcription factors are critical regulators of CS formation at the endodermis of rice roots and redundantly regulate multiple CS-associated genes. Functional analysis of MYB36s revealed that rice controls CS formation via a mechanism at least partly distinct from that of Arabidopsis. In addition, compared with that of Arabidopsis, the rice CS defect has distinct effects on the selective uptake of mineral elements, especially Ca, in roots.
Materials and methods
Plant materials and growth conditions
WT rice (cv. Nipponbare), single- and multiple-knockout lines for three OsMYB36 genes, OsMYB36a- and OsMYB36b-overexpression lines, and the Oscasp1-1 mutant (Wang et�al, 2019a) were used in this study. The WT rice was obtained from rice resources conservation center of Guangxi University. The Oscasp1-1 mutant, OsMYB36-knockout and OsMYB36-overexpression lines were constructed in our laboratory. The seeds were soaked in water for 2 days in the dark at 28�C and transferred to a net floating on 0.5 mM CaCl2 solution. After 7 days, the seedlings were transferred to a 4.0-L plastic pot filled with half-strength Kimura B solution (pH 5.6) as described previously (Yamaji and Ma, 2007) and grown in a greenhouse at 25�C to 30�C. The solution was changed every 2 days.
Phylogenetic analysis
The amino acid sequences of MYB36 proteins in different species were obtained from Genbank (https://www.ncbi.nlm.nih.gov) and aligned using the ClustalW program. The MYB36 sequence alignment is provided as Supplemental Data Set S1. The approximately maximum-likelihood phylogenetic trees were constructed using Jones–Taylor–Thornton model and Gamma distributed with Invariant sites (G + I) on MEGA version 6.
Generation of transgenic rice
Various CRISPR–Cas9 knockout mutants of OsMYB36s were generated as described by Ma et al. (2015). The target sequences of each gene (Supplemental Data Set S8) were introduced into sgRNA expression cassettes via two rounds of PCR with four primers (primer sequences are shown in Supplemental Data Set S8). The amplified fragments were cloned into pYLCRISPR/Cas9Pubi (Ma et al., 2015) in the BsaI restriction enzyme site, producing the pCRISPR-OsMYB36s constructs.
To generate the OsMYB36a or OsMYB36b overexpression lines, the coding region of OsMYB36a or OsMYB36b was amplified by RT-PCR from total RNA from rice using specific primers. The amplified fragment was inserted into the KpnI/HindIII sites of the pYLox vector (Yu et al., 2010) under the control of the maize Ubi promoter, producing the ProUbi-OsMYB36a or ProUbi-OsMYB36b construct. All resulting vectors were introduced into Agrobacterium tumefaciens (strain EHA101), followed by introduction into rice (Nipponbare) by Agrobacterium-mediated transformation.
Phenotypic analysis of the knockout lines
For hydroponic culture, single- and multiple-knockout lines of OsMYB36s, Oscasp1-1, and the WT were grown in half-strength Kimura B solution for 30 days. One day before harvest, the plants were transferred to a solution containing 0.2 �M As and Cd and 1 �M Ge, Rb, and Sr. The roots and shoots were cut from the plants with a razor, separately collected, and used to measure mineral elements as described below.
For soil culture, 2-week-old seedlings of the lines described above were transplanted to a 6-L plastic pot containing paddy soil. The plants were grown in flooded or upland soil supplied with tap water under natural light in a greenhouse. After 14 days, the plants were photographed and the shoots collected for mineral element measurements as described below.
Fourteen-day-old WT and Osmyb36s mutant seedlings were also grown under flooded conditions in a 6.0-L pot containing soil collected from Nanning, Guangxi province, China, in a greenhouse. The plants were photographed at the vegetative and reproductive growth stages. At harvest, the agronomic traits were recorded.
Complementation test
For the complementation test of Osmyb36a-2, we amplified the full-length genomic sequence of OsMYB36a containing the 2.027-kb promoter sequence, 1.25-kb coding sequence, and 0.5-kb downstream sequence (after the translational stop site) from WT Nipponbare genomic DNA using primer sets POsMYB36-F and UOsMYB36-R (primer sequences are listed in Supplemental Data Set S8). The amplified fragment was ligated into binary vector pCAMBIA1300 using a ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) according to the manufacturer’s protocol. This construct was introduced into Agrobacterium strain EHA101 and transformed into rice calli derived from the Osmyb36a-2 mutant lacking the pCRISPR-OsMYB36a construct, which was obtained from the T2 population.
Growth at different Ca concentrations
For phenotypic analysis at different Ca concentrations, the WT, single- and multiple-knockout lines of OsMYB36s, and the Oscasp1-1 mutant were grown in half-strength Kimura B solution under natural light in a greenhouse for 14 days, followed by growth in a solution containing different concentrations of Ca (0.18, 1, and 5 mM) as CaCl2 for 12 days. The plants were then photographed. The root and shoot tissues were separately sampled for Ca measurement as described below.
Measuring Ca content in xylem sap
The WT, single- and multiple-knockout lines of OsMYB36s, and Oscasp1-1 were grown hydroponically for 40 days and exposed to nutrient solution containing 5 mM Ca for 6 h. The plants were cut to a height of 2 cm above the roots, and xylem sap was continuously collected for 1 h with a micropipette. The Ca concentration of the xylem sap was determined as described below.
Elemental analysis
The dried samples were digested with 65% HNO3. The metal concentrations in the digested solution and xylem sap were determined by ICP-MS (Plasma Quant MS; Analytik Jena AG).
Gene expression analysis
To investigate the expression pattern of OsMYB36s, we sampled diverse tissues including root, stem, leaf blade, and leaf sheath tissue. The spatial expression pattern of OsMYB36s in roots was examined using different root segments (0–1, 1–2, 2–3, and 3–4 cm from the root apex) that were separately harvested from 5-day-old seedlings. Total RNA was extracted from the samples using a TRIzol reagent kit (Life Technologies, Carlsbad, CA, USA) and reverse-transcribed using a PrimeScript II 1st Strand cDNA Synthesis kit (Takara, Shiga, Japan) according to the manufacturer’s instructions. The expression of OsMYB36s was determined using ChanQ SYBR Color qPCR Master Mix (Vazyme) in a qTOWER version 2.0 system (Analytik Jena, Jena, Germany). Three biological replicates were analyzed per sample. Histone H3 was used as an internal control. The primers used for RT-qPCR are shown in Supplemental Data Set S8.
Immunostaining with GFP antibody
To examine the cellular localization of OsMYB36s, we generated transgenic plants carrying the ProOsMYB36s:OsMYB36scDNA-GFP or ProOsMYB36a:OsMYB36agenomic-GFP fusion construct. The native promoter (∼2.0-kb region upstream of the start codon) of each OsMYB36 gene was amplified from Nipponbare genomic DNA with the primer set POsMYB36s-GFP-F and POsMYB36s-atg-R, and the OsMYB36s ORF without the stop codon was amplified from cDNA from Nipponbare roots with the primer set POsMYB36s-atg-F and POsMYB36s-GFP-R. We also amplified a 3.274-kb genomic DNA fragment containing the 2.0-kb OsMYB36a promoter sequence and coding sequence without the stop codon from WT genomic DNA by a PCR approach. The amplified fragments were cloned into pCAMBIA1301-GFP using a ClonExpress II One Step Cloning Kit (Vazyme) according to the manufacturer’s protocol, generating the ProOsMYB36s:OsMYB36s-GFP or ProOsMYB36a:OsMYB36agenomic-GFP construct. Using a similar method, we also generated the ProLsi1:Lsi1genomic-GFP construct. To obtain the Osmyb36abc triple mutant carrying the ProOsCASP1:OsCASP1-GFP or ProLsi1: Lsi1genomic-GFP construct, sgRNA expression cassettes for three OsMYB36 genes, together with ProOsCASP1:OsCASP1-GFP or ProLsi1:Lsi1genomic-GFP expression cassettes, were cloned into pYLCRISPR/Cas9Pubi in the BsaI restriction enzyme site, producing the pCRISPR-OsMYB36s-ProOsCASP1:OsCASP1-GFP or pCRISPR-OsMYB36s-ProLsi1:Lsi1genomic-GFP construct. These constructs were transformed into calli derived from Nipponbare or Osmyb36a-2 lacking the pCRISPR-OsMYB36a construct via Agrobacterium (strain EHA101)-mediated transformation. The primer sequences are listed in Supplemental Data Set S8. The Osmyb36a-1, OsMYB36a-, or OsMYB36b-overexpressing line carrying the ProOsCASP1:OsCASP1-GFP construct was obtained from the F2 population of the Osmyb36a-1, OsMYB36a- or OsMYB36b-overexpressing line crossed with the transgenic plant carrying the ProOsCASP1:OsCASP1-GFP construct.
To observe the localization of OsMYB36s-GFP, OsCASP1-GFP, or OsLsi1-GFP in roots, the roots of WT rice, and transgenic plants carrying ProOsMYB36s:OsMYB36s-GFP, ProOsCASP1:OsCASP1-GFP, or ProLsi1:Lsi1genomic-GFP were immunostained using an antibody against GFP (1:1,000 dilution; A11122; Molecular Probes). Following immunostaining as described previously (Wang et al., 2019a), GFP fluorescence was observed under a confocal laser-scanning microscope (TCS SP8; Leica, Wetzlar, Germany).
Subcellular localization of OsMYB36 proteins
To construct the OsMYB36s-GFP fusion protein, each OsMYB36 cDNA without the stop codon was amplified by PCR using the primer set OsMYB36s-GFP-F and OsMYB36s-GFP-R (Supplemental Data Set S8). The restriction sites for subcloning were included in these primer sets. The amplified cDNA fragment for each OsMYB36 gene was inserted into the pYL322-GFP vector upstream of the GFP coding region, producing the OsMYB36s-GFP construct.
The OsMYB36s-GFP construct, together with a nuclear marker (Ghd7-RFP), was introduced into rice leaf protoplasts by PEG-mediated transformation as previously described (Fu et al., 2019). The transformed cells were cultured at room temperature in the dark for 12–16 h. The fluorescence images were captured using a confocal laser-scanning microscope (TCS SP8; Leica).
Histochemical staining of roots
Five-day-old seedlings were used to prepare transverse or longitudinal cross-sections of roots. The roots were embedded in 5% (w/v) agar. Root samples at different positions were sectioned (100-�m thick) with a Microslicer (VT1000 S; Leica). The sections were treated with 0.2% (w/v) Basic Fuchsin for lignin staining or 0.1% (w/v) Calcofluor White for cellulose staining according to Wang et al. (2019a, 2019b). To observe suberin in the samples, Fluorol Yellow 088 fluorescence staining was performed as previously described (Lux et al., 2005). A PI penetration assay was performed as previously described (Wang et al., 2019a, 2019b).
Microscopy observation
Confocal laser-scanning microscopy was performed with a Leica SP8. The following excitation and emission settings were used: GFP, 488 nm/500–550 nm; Basic Fuchsin, 580 nm/600–615 nm; Calcofluor White, 405 nm/415–440 nm; Fluorol Yellow 088, 488 nm/510–525 nm; PI, 488 nm/600–650 nm. The z-stack magnified images of CS and OsCASP1-GFP were taken (∼20 optical sections with 0.3–0.4 �m intervals per sample) and the 3D images were constructed with LAS AF Lite software (Leica Microsystems). Confocal images were processed and analyzed using the Leica Application Suite.
RNA-seq
The roots of 21-day-old WT rice cv. Nipponbare, Osmyb36a-1, Osmyb36ab, Osmyb36abc-1, and oscasp1 seedlings were sampled for RNA-seq analysis. RNA purification and cDNA library preparation were performed as described by Fu et al. (2019). The libraries were sequenced as 150-bp paired-end reads using the Illumina NovaSeq platform (Illumina, Inc., San Diego, CA, USA). Sequence annotation was performed using Hisat2 version 2.0.5 (https://daehwankimlab.github.io/hisat2/) based on the released reference genome downloaded from the Rice Annotation Project Database directly (https://rapdb.dna.affrc.go.jp/download/archive/irgsp1/IRGSP-1.0_genome.fasta.gz). DEGs between the WT and mutants were analyzed by DESeq2 (Love et al., 2014). The standard of log2 fold change ≤−1 and P-value ≤0.05 was used to select downregulated genes between two groups, while the standard of log2 fold change ≥1 and P-value ≤0.05 was used to select upregulated genes. Normalized RPKM (log2 values) of gene expression was used to generate heatmap and hierarchical clustering by R package (https://cran.r-project.org/web/packages/pheatmap/index.html).
To verify the RNA-seq data, RT-qPCR analysis was performed using RNA samples from roots as described above. We examined the expression of six genes with no known homologs in Arabidopsis and 21 genes homologous to known CS-related genes in Arabidopsis. A heatmap of the gene expression levels in various CS defective mutants relative to the WT was generated using MeV software (https://sourceforge.net/projects/mev-tm4/files/mev-tm4/). The primer sequences are listed in Supplemental Data Set S8.
Dual-LUC transient expression assays
Dual-LUC transient expression assays in rice protoplasts were performed according to (Su et al., 2016). The promoter sequences (∼2,100-bp from the start codon) of eight tested genes were amplified from Nipponbare genomic DNA and inserted into pGreen II 0800-LUC upstream of the firefly LUC gene, producing the ProOsCASP1:Luc, ProOsESB1:LUC, ProOs03g0245500:LUC, ProOs08g0515700:LUC, ProOs09g0493400:LUC, ProOs09g0535400:LUC, ProOs10g0155100:LUC, or ProOs11g0499600:LUC vectors. The reporter vector contained the Renilla LUC (REN) gene driven by the CaMV 35S promoter as an internal control. To generate the effector vector, the full-length coding sequence of each OsMYB36 gene was amplified and cloned into the pGreen II 62-SK vector downstream of the CaMV 35S promoter. The resulting vectors were introduced into rice protoplasts as previously described (Fu et al., 2019). LUC and REN LUC activities were detected using a Dual-LUC Reporter Assay System (Promega, Madison, WI, USA). The primer sets used for amplification are listed in Supplemental Data Set S8. All experiments were performed with three biological replicates.
Transactivation activity and Y1H assay
The full-length or truncated coding region of each OsMYB36 gene was amplified and cloned into the NdeI/EcoRI or EcoRI/BamHI sites of the pGBKT7 vector (Clontech, Shiga, Japan) to generate the pGBK-OsMYB36s, pGBK-OsMYB36s-N, and pGBK-OsMYB36s-C constructs. The resulting constructs or a control vector were transformed into yeast strain AH109. The yeast colonies were spotted on SD/Trp− and SD/Trp−/His−/Ade− medium containing 40 mg/L−1 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside, which was used to detect the activity of the MEL1 reporter gene. The plates were incubated at 30�C for 3 days. The primer sets used for amplification are listed in Supplemental Data Set S8.
To investigate the interaction between OsMYB36s and the OsCASP1 and OsESB1 promoters, we amplified the promoter sequences of OsCASP1 (2,082-bp fragment directly upstream of the start codon) and OsESB1 (2,126-bp fragment directly upstream of the start codon) from Nipponbare genomic DNA by PCR. The amplified promoter fragments were cloned into pHIS2.1 upstream of the HIS3 reporter gene in the pHIS2 vector (Clontech), producing the pHIS-ProOsCASP1 and pHIS-ProOsESB1 constructs. The truncated OsMYB36s ORF (1–360 bp) containing the MYB domain was cloned in-frame after the transcriptional activation domain of the yeast GAL4 transcription factor (without the DNA-binding domain) in pGADT7 (Clontech) to generate pGAD-MYB36s-N. A pair of these plasmids (pHIS2-ProOsCASP1 and pGAD-MYB36s-N, pHIS2-ProOsESB1 and pGAD-MYB36s-N, or control pHIS2 and pGADT7) was transformed into yeast strain AH109. The yeast colonies were cultured on SD/-Leu/-Trp and SD/-Leu/-Trp/-His medium containing 0–100 mM 3-amino-1,2,4-triazole at 30�C for 3 days.
EMSA
For the EMSA, the truncated ORF (1–360 bp) of each OsMYB36 gene was cloned in-frame after the His tag in the pRSFDuet-1 vector (Novagen, Beijing, China), generating the pRSFDuetI-His-OsMYB36s-N construct. The resulting construct was transformed into Escherichia coli (BL21). The His-OsMYB36s-N fusion protein was purified using Ni Sepharose 6 Fast Flow columns (GE) according to the manufacturer’s instructions. 5′-carboxyfluorescein (FAM)-labeled oligonucleotide probes containing the MYB-binding motif were synthesized. Probes with the same sequences or without the MYB-binding motif that were not labeled with FAM were used as competitors. EMSAs were performed using a Light Shift Chemiluminescent EMSA Kit following the manufacturer’s protocol. The FAM-labeled probes were detected using the ChemDoc XRS imaging system (Bio-Rad, Hercules, CA, USA). The probe sequences for EMSA are listed in Supplemental Data Set S8.
Statistical analysis
GraphPad Prism version 8 software was used for statistical analysis. Data were analyzed using one-way ANOVA followed by Tukey’s test (Supplemental Data Set S9). Significance of differences at P < 0.05 are indicated by different letters. All experiments were carried out with at least three biological replicates and independently repeated at least twice.
Accession numbers
Sequence data from this article can be found MSU Rice Genome Annotation Project data libraries under accession numbers: OsMYB36a, LOC_Os08g15020; OsMYB36b; LOC_Os02g54520; OsMYB36c, LOC_Os03g56090. The RNA-sequencing raw data can be found in the Sequence Read Archive at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP345199.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis and sequence alignment of OsMYB36 proteins.
Supplemental Figure S2. Subcellular localization of OsMYB36 proteins.
Supplemental Figure S3. Cellular localization of OsMYB36s in rice.
Supplemental Figure S4. The expression pattern of OsMYB36 proteins in roots.
Supplemental Figure S5. Phenotypes of various OsMYB36 CRISPR/Cas9 mutants at the early seedling stage.
Supplemental Figure S6. Observation of CS at the exodermis of rice roots.
Supplemental Figure S7. Transerve observation of CS formation at the endodermis of rice roots.
Supplemental Figure S8. Longitudinal observation of CS formation at the endodermis of rice roots.
Supplemental Figure S9. Effect of overexpressing OsMYB36a or OsMYB36b on root growth at the early seedling stage.
Supplemental Figure S10. Observation of CS formation in WT rice and OsMYB36a-overexpressing line.
Supplemental Figure S11. Observation of CS formation in WT rice and OsMYB36b-overexpressing line.
Supplemental Figure S12. Transerve observation of suberin stain pattern at the roots of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant.
Supplemental Figure S13. Longitudinal observation of suberin stain pattern at the roots of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant.
Supplemental Figure S14. Growth phenotype of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant at the vegetative stage.
Supplemental Figure S15. Complementation of Osmyb36a mutant phenotype.
Supplemental Figure S16. Comparison of the root ionome of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant.
Supplemental Figure S17. Mineral analysis of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant grown under flooded and upland conditions.
Supplemental Figure S18. Root Ca concentration of WT rice, OsMYB36s-knockout lines, and Oscasp1 mutant under different Ca concentrations.
Supplemental Figure S19. Transcriptional activation analysis of OsMYB36 proteins in yeast.
Supplemental Figure S20. Activation of the promoters of six candidate CS-related genes by OsMYB36s in rice protoplast.
Supplemental Figure S21. Mutated sequences in the Osmyb36abc mutants carrying the OsCASP1-GFP or OsLsi1-GFP construct.
Supplemental Figure S22. Observation of OsCASP1 localization in the roots of WT rice and OsMYB36s transgenic lines.
Supplemental Figure S23. Observation of OsCASP1 localization in the roots of WT rice and OsMYB36b overexpressing line.
Supplemental Figure S24. Localization of the Si transporter Lsi1 in WT rice, Osmyb36a, and Osmyb36abc mutant.
Supplemental Figure S25. OsMYB36s-mediated regulation of spatial development of CSs, suberin deposition, and mineral uptake at different root regions.
Supplemental Data Set S1. The sequence alignment and homology analysis of MYB36 in rice and other plants.
Supplemental Data Set S2. DEGs between WT rice and Osmyb36s mutants.
Supplemental Data Set S3. Candidate CS-associated genes regulated by OsMYB36a/b/c.
Supplemental Data Set S4. Homologs of Arabidopsis CS associated genes.
Supplemental Data Set S5. Expression levels of 27 candidate downstream genes in Osmyb36s mutants and Oscasp1 by RT-qPCR.
Supplemental Data Set S6. Comparison of downstream genes regulated by OsMYB36a/b/c and AtMYB36.
Supplemental Data Set S7. Expression levels of OsNramp5 and genes encoding plasma membrane-localized transporters/channels for Ca in Osmyb36s mutants and Oscasp1 in RNA sequencing data.
Supplemental Data Set S8. List of primers used in this study.
Supplemental Data Set S9. Statistical analysis in this study.
Funding
This work was supported by the National Natural Science Foundation of China (32070275), Guangxi Natural Science Foundation (2021GXNSFDA220002), Innovation Project of Guangxi Graduate Education (YCBZ2020022), State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-a202005 and SKLCUSA-a201915), and Hundred-Talent Program of Guangxi (2014).
Conflict of interest statement.�The authors declare no conflict of interest.
J.X. designed the research. Z.W., B.Z., Z.C., M.W., D.C., Q.W., Y.X., L.L., Z.M., and J.X. performed the experiments. Z.W., B.Z., and J.X. analyzed the data. J.X. wrote the article.
The author(s) responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Jixing Xia (xiajx@gxu.edu.cn).
Supplementary Material
Contributor Information
Zhigang Wang, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Baolei Zhang, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Zhiwei Chen, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Mingjuan Wu, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Dong Chao, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Qiuxing Wei, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Yafeng Xin, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Longying Li, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Zhenhua Ming, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
Jixing Xia, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi University, Nanning 530004, China.
References
- Alassimone J, Fujita S, Doblas VG, van Dop M, Barberon M, Kalmbach L, Vermeer JE, Rojas-Murcia N, Santuari L, Hardtke CS, et al. (2016) Polarly localized kinase SGN1 is required for Casparian strip integrity and positioning. Nat Plants 2: 16113. [DOI] [PubMed] [Google Scholar]
- Barbosa I, Rojas-Murcia N, Geldner N (2019) The Casparian strip-one ring to bring cell biology to lignification? Curr Opin Biotechnol 56: 121–129 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Che J, Yamaji N, Ma JF (2018) Efficient and flexible uptake system for mineral elements in plants. New Phytol 219: 513–517 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Drapek C, Sparks EE, Marhavy P, Taylor I, Andersen TG, Hennacy JH, Geldner N, Benfey PN (2018) Minimum requirements for changing and maintaining endodermis cell identity in the Arabidopsis root. Nat Plants 4: 586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enstone DE, Peterson CA, Ma F (2002) Root endodermis and exodermis: structure, function, and responses to the environment. J Plant Growth Regul 21: 335–351 [Google Scholar]
- Fageria NK, Carvalho GD, Santos AB, Ferreira EPB, Knupp AM (2011) Chemistry of lowland rice soils and nutrient availability. Connun Soil Sci Plan 42: 1913–1933 [Google Scholar]
- Fu S, Lu Y, Zhang X, Yang G, Chao D, Wang Z, Shi M, Chen J, Chao DY, Li R, et al. (2019) The ABC transporter ABCG36 is required for cadmium tolerance in rice. J Exp Bot 70: 5909–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N (2013) The endodermis. Annu Rev Plant Biol 64: 531–558 [DOI] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA 110: 14498–14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmbach L, H�maty K, De Bellis D, Barberon M, Fujita S, Ursache R, Daraspe J, Geldner N (2017) Transient cell specific EXO70A1 activity in the CASP domain and Casparian strip localization. Nat Plants 3: 17058. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Borghi M, Wang P, Danku JM, Kalmbach L, Hosmani PS, Naseer S, Fujiwara T, Geldner N, Salt DE (2015) The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci USA 112: 10533–10538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Samarajeewa PK, Barrero RA, Uchimiya MN (1998) Cellular dissection of the degradation pattern of cortical cell death during aerenchyma formation of rice roots. Planta 204: 277–287 [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Li B, Kamiya T, Kalmbach L, Yamagami M, Yamaguchi K, Shigenobu S, Sawa S, Danku JM, Salt DE, Geldner N, et al. (2017) Role of LOTR1 in nutrient transport through organization of spatial distribution of root endodermal barriers. Curr Biol 27: 758–765 [DOI] [PubMed] [Google Scholar]
- Li P, Yu Q, Gu X, Xu C, Qi S, Wang H, Zhong F, Baskin TI, Rahman A, Wu S (2018) Construction of a functional Casparian strip in non-endodermal lineages is orchestrated by two parallel signaling systems in Arabidopsis thaliana. Curr Biol 28: 2777–2786 [DOI] [PubMed] [Google Scholar]
- Liberman LM, Sparks EE, Moreno-Risueno MA, Petricka JJ, Benfey PN (2015) MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc Natl Acad Sci USA 112: 12099–12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux A, Morita S, Abe J, Ito K (2005) An improved method for clearing and staining free-hand sections and whole-mount samples. Ann Bot 96: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Tamai K, Yamaji N, Mitani N, Konishi S, Katsuhara M, Ishiguro M, Murata Y, Yano M (2006) A silicon transporter in rice. Nature 440: 688–691 [DOI] [PubMed] [Google Scholar]
- Ma JF, Yamaji N, Mitani N, Tamai K, Konishi S, Fujiwara T, Katsuhara M, Yano M (2007) An efflux transporter of silicon in rice. Nature 448: 209–212 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y (2017) A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Fujita S, Moretti A, Hohmann U, Doblas VG, Ma Y, Pfister A, Brandt B, Geldner N, Hothorn M (2020) Molecular mechanism for the recognition of sequence-divergent CIF peptides by the plant receptor kinases GSO1/SGN3 and GSO2. Proc Natl Acad Sci USA 117: 2693–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A, Barberon M, Alassimone J, Kalmbach L, Lee Y, Vermeer JE, Yamazaki M, Li G, Maurel C, Takano J, et al. (2014) A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3: e3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnamperuma FN (1972) The chemistry of submerged soils. Adv Agron 24: 29–96 [Google Scholar]
- Reyt G, Chao Z, Flis P, Salas-Gonz�lez I, Castrillo G, Chao DY, Salt DE (2020) Uclacyanin proteins are required for lignified nanodomain formation within Casparian strips. Curr Biol 30: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins NN, Trontin C, Duan L, Dinneny JR (2014) Beyond the barrier: communication in the root through the endodermis. Plant Physiol 166: 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Murcia N, H�maty K, Lee Y, Emonet A, Ursache R, Fujita S, De Bellis D, Geldner N (2020) High-order mutants reveal an essential requirement for peroxidases but not laccases in Casparian strip lignification. Proc Natl Acad Sci USA 117: 29166–29177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Denervaud TV, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Ma JF (2016) Transporters involved in mineral nutrient uptake in rice. J Exp Bot 67: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Shan JX, Gao JP, Lin HX (2016) OsHAL3, a blue light-responsive protein, interacts with the floral regulator Hd1 to activate flowering in rice. Mol Plant 9: 233–244 [DOI] [PubMed] [Google Scholar]
- Ueno D, Sasaki A, Yamaji N, Miyaji T, Fujii Y, Takemoto Y, Moriyama S, Che J, Moriyama Y, Iwasaki K, et al. (2015) A polarly localized transporter for efficient manganese uptake in rice. Nat Plants 1: 15170. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang H, Li P, Li H, Xu C, Cohen H, Aharoni A, Wu S (2020a) Developmental programs interact with abscisic acid to coordinate root suberization in Arabidopsis. Plant J 104: 241–251 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang G, Shi M, Hao D, Wei Q, Wang Z, Fu S, Su Y, Xia J (2019b) Disruption of an amino acid transporter LHT1 leads to growth inhibition and low yields in rice. BMC Plant Biol 19: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Shi M, Wei Q, Chen Z, Huang J, Xia J (2020b) OsCASP1 forms complexes with itself and OsCASP2 in rice. Plant Signal Behav 15: 1706025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yamaji N, Huang S, Zhang X, Shi M, Fu S, Yang G, Ma JF, Xia J (2019a) OsCASP1 is required for Casparian strip formation at endodermal cells of rice roots for selective uptake of mineral elements. Plant Cell 31: 2636–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Jiang J, Zhang C, Jiang L, Ye N, Lu Y, Yang G, Liu E, Peng C, He Z, et al. (2010) Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J Exp Bot 61: 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Zuo D, Tao Y, Cai H, Li L (2020) Laccase3-based extracellular domain provides possible positional information for directing Casparian strip formation in Arabidopsis. Proc Natl Acad Sci USA 117: 15400–15402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.