Abstract
The frequently emerging SARS-CoV-2 variants have weakened the effectiveness of existing COVID-19 vaccines and neutralizing antibody therapy. Nevertheless, the infections of SARS-CoV-2 variants still depend on angiotensin-converting enzyme 2 (ACE2) receptor-mediated cell entry, and thus the soluble human ACE2 (shACE2) is a potential decoy for broadly blocking SARS-CoV-2 variants. In this study, we firstly generated the recombinant AAVrh10-vectored shACE2 constructs, a kind of adeno-associated virus (AAV) serotype with pulmonary tissue tropism, and then validated its inhibition capacity against SARS-CoV-2 infection. To further optimize the minimized ACE2 functional domain candidates, a comprehensive analysis was performed to clarify the interactions between the ACE2 orthologs from various species and the receptor binding domain (RBD) of SARS-CoV-2 spike (S) protein. Based on the key interface amino acids, we designed a series of truncated ACE2 orthologs, and then assessed their potential affinity to bind to SARS-CoV-2 variants RBD in silico. Of note, we found that the 24-83aa fragment of dog ACE2 (dACE224-83) had a higher affinity to the RBD of SARS-CoV-2 variants than that of human ACE2. Importantly, AAVrh10-vectored shACE2 or dACE224-83 constructs exhibited a broadly blockage breadth against SARS-CoV-2 prototype and variants in vitro and ex vivo. Collectively, these data highlighted a promising therapeutic strategy against SARS-CoV-2 variants.
Keywords: COVID-19, SARS-CoV-2, sACE2, Adeno-associated virus (AAV) vector, Variants
1. Introduction
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, continues to severely challenge the global public health. The frequent outbreaks due to a rapid mutation accumulation of SARS-CoV-2 variants further exacerbated this challenge, including alpha strain (B.1.1.7), beta strain (B.1.351), gamma strain (P.1), kappa (B.1.617.1), delta (B.1.617.2), and mostly recent omicron strain (B.1.1.529) in South Africa etc (Science Brief, 2020). These variants resulted in immune escape to weaken the effectiveness of the existing COVID-19 vaccines and neutralizing antibodies (Planas et al., 2021; Yadav et al., 2022; Wang et al., 2021a). Nevertheless, the cell entry of both SARS-CoV-2 prototype and its variants is still mediated by the angiotensin converting enzyme 2 (ACE2), which can recognize the receptor binding domain (RBD) of the viral spike glycoprotein (S protein) (Scialo et al., 2020). Thus, ACE2 presents a potential therapeutic target for broadly inhibiting the infections of SARS-CoV-2 variants.
Recent studies have demonstrated that human recombinant soluble ACE2 (hrsACE2) effectively blocked the SARS-CoV-2 infections, and its efficacy has been assessed in healthy volunteers and COVID-19 patients in phase I and II trials (Abd El-Aziz et al., 2020; Monteil et al., 2020). Moreover, the optimized forms of sACE2-mediated blockade are extensively exploited, such as random mutagenesis to create the high-affinity sACE2 decoys for binding to the RBD of SARS-CoV-2, fused constructs of the sACE2 with Fc (fragment crystallizable) to prolong its half-life of pharmacokinetics, computational design of the truncated ACE2 functional fragments to decrease its side effects, mRNA-based gene delivery to rapidly generate the circulating sACE2 decay (Feng et al., 2021; Zhang et al., 2021; Chan et al., 2020a). So far, it is of great significance to exploit the sACE2-based therapy as novel agents to control the surge of SARS-CoV-2 infections.
Adeno-associated virus (AAV) has been extensively developed as a gene delivery vector for gene therapy, because of their clinical safety record, broad but tailorable tissue tropism, high-level and long-term gene expression, low immunogenicity, etc (Zheng et al., 2018). Of note, AAVrh10 is a kind of AAV serotype with pulmonary tissue tropism, and thus systemic delivery of AAVrh10 vector was shown to confer an efficient gene expression targeting to the lung system (Watanabe et al., 2010). In the present study, we therefore constructed recombinant AAVrh10-based constructs to express different forms of shACE2 as a novel strategy against SARS-CoV-2 variants infections. We also conducted a comprehensive analysis of the key interface amino acids in the ACE2 of various animal species, and then designed a series of the truncated ACE2 functional domains to increase its affinity to bind the variants of SARS-CoV-2 RBD, and decrease the potential side effects by removing the domain of catalytic activity for angiotensin substrates. Overall, this study highlighted the ACE2-based blockade as a promising therapeutic strategy against SARS-CoV-2 variants infections.
2. Materials and methods
2.1. Cell lines and viruses
2.1.1. Cell lines
Hela cell, 293T cell, BHK-ACE2 cell, CACO-2-N cell, HEK-293T cell, and VeroE6 cell were cultured in complete Dulbecco's modified Eagle's medium (DMEM, Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco), at 37◦C in an incubator containing 5% of CO2.
2.1.2. Viruses
AAVrh10 expressing the different ACE2 fragments, including shACE2, shACE2PD, shACE2(PD + Neck), hACE224-83, dACE224-83, were packaged by PackGene Biotech. The various SARS-CoV-2 pseudovirus of D614G, alpha, beta, gamma, delta, and omicron variants were gifted by VectorBuilder and Dr. Zhongcheng Zhou, and SARS-CoV-2 virus-like-particles (trVLP) was gifted by Dr. Qiang Ding (School of Medicine, Tsinghua University, Beijing, China).
2.2. Western blot analyses
Hela cells were cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin medium, and then transfected with AAV-vector, AAV-shACE2, AAV-shACE2PD and AAV-ACE2(PD + Neck) plasmids according to the manufacturer's manuals using Lipofectamine 2000 CD Transfection Reagent (Cat. No 12566014; Invitrogen, Carlsbad, CA, USA). The medium was replaced with DMEM/High Modified (-Phenol Red) after transfection for 6 h, and the cells were incubated for another 24 h for the expression of the target gene. The Amicon Ultra filtration tube (Cat. UFC801008; Merck millipore, IRL) were used to concentrate the cell supernatant for protein expression level detection. Western blot analysis was performed by standard procedure. DYKDDDDK Tag (D6W5B) Rabbit mAb antibody, GAPDH, Goat Anti-Rabbit IgG H&L and Goat Anti-Mouse IgG H&L were all purchased from Cell Signaling Technology (CST, Danvers, MA, USA).
2.3. Competitive binding experiment
2 × 104 293T cells were transfected with pVAX-codon-optimized-Spike plasmid and cultured in a 96-well plate for 24 h. The supernatant from above cultured Hela cells transfected with different ACE2 fragments was collected and concentrated in an ultrafiltration tube, and mixed with anti-spike-RBD-hIgM antibody (Cat. CM002-0.1A1CoV; CHAMOT) with 1:1 ratio. Then, this mixture was added into above 293T cells expressing spike protein for 1 h. The binding affinity reduction of the anti-spike-RBD-hIgM antibody to the Spike protein was detected by flow cytometry.
2.4. The viral inhibition experiments
2.4.1. Neutralization assay based on SARS-CoV-2 pseudovirus
2 × 104 BHK-ACE2 cells were seeded into 96-well plates per well for 24 h, and these cells were transfected with the AAV-vector, AAV-shACE2, AAV-shACE2PD, and AAV-shACE2(PD + Neck) respectively for 6 h. The medium was replaced with DMEM containing 5% FBS and 1% penicillin/streptomycin, and then 2 × 104 PFU SARS-CoV-2 pseudovirus was added for 24 h. The relative intensity of fluorescence was detected by the Steady-Glo® Renilla Luciferase detection system according to the manufacturer's instructions (Promega, Madison, USA).
2.4.2. Virus inhibition assay based on replication-competent SARS-CoV-2 virus-like-particles (trVLP)
A cell culture system, which consists of the recombinant SARS-CoV-2 virus with a reporter gene (GFP) replacing viral nucleocapsid gene (N) and the CACO-2 cells expressing SARS-CoV-2 N proteins (CACO-2-N), were constructed as previously reported (Ju et al., 2021). CACO-2-N cells were seeded into 24-well plates and transfected with the AAV-vector, AAV-shACE2, AAV-shACE2PD, and AAV-shACE2(PD + Neck) respectively for 6 h. The medium was replaced with DMEM containing 5% FBS and 1% penicillin/streptomycin, and then 2 × 104 PFU SARS-CoV-2 virus-like-particles (trVLP) was added for 24 h. Total RNA of 3 × 105 CACO-2-N cells was extracted with TRIzol reagent (Yeasen, Shanghai, China) and reverse-transcripted to cDNA by HifairTM III 1st Strand cDNA Synthesis SuperMix for qPCR (Yeasen, Shanghai, China). The level of SARS-CoV-2 mRNA was measured using the PerfectStart SYBR Green qPCR supermix (Transgen, Beijing, China) and a QuantStudio 7 Flex quantitative real-time PCR (qPCR) detection system (Applied Biosystems, Foster City, CA, USA). Relative copies of gene expression were calculated by the comparative cycle threshold-value method, and the relative genome copy numbers were calculated based on the normalization with the housekeeping gene β-actin. The results were represented as the fold change in expression level for each transcript. The data were analyzed by the 2− ΔΔCt method.
2.5. Inhibition of SARS-CoV-2 spike-mediated cell–cell fusion
The effector cells expressing S protein were prepared as previously described (Zhang et al., 2021). In brief, 5 × 105 HEK-293T cells were seeded into 6-well plates per well for 24 h, and then transfected with 3 μg pcDNA3.1-SARS-CoV-2 Spike and 2 μg PVAX-GFP simultaneously for 48 h. The effector cells were harvested and incubated with 2 mL supernatant of Hela cells infected by AAVrh10-shACE2, AAVrh10-shACE2(PD + Neck), AAVrh10-hACE224-83, AAVrh10-dACE224-83 respectively at 37 °C for 6 h. In this study, VeroE6 cells were used as target cells. Then, the above-mentioned effector cells and target cells were mixed 1:1 and co-cultured in DMEM for 24 h. The plates were checked for the formation of syncytia under a fluorescence microscope (EVOS digital inverted microscope, Invitrogen). The percent inhibition of cell–cell fusion was calculated using the following formula (number of fused cells/number of the fused and unfused cells) × 100%.
2.6. In silico design of different ACE2 functional domains
2.6.1. Protein sequences analysis of ACE2 derived from multiple species
The ACE2 protein sequences and corresponding accession number of multiple species were obtained from the National Center for Biotechnology Information (NCBI). Multiple sequence-alignment was performed by MAFFT software (version 7.453) with auto strategy. The sequences were analyzed along with the reference strains (Q9BYF1.2) obtained from GenBank using Simplot software (version 3.5.1). The Simplot analysis was performed by setting the window width and the step size to 200 bp and 20 bp based on Kimura 2-parameter model, respectively. The alignment sequences were imported to BioEdit software (version 7.0.9) to calculate the similarity matrix. Heat-map was plotted in R studio (version 4.0.2) using the ggplot2 package.
2.6.2. Analysis of the inter-molecular interactions
The crystal structure of the SARS-CoV-2 spike protein-ACE2 (PDB ID: 6LZG) complex was gained from the Protein Data Bank (https://www.rcsb.org/) for subsequent analysis. The PyMOL software (version 2.3.0) was used to create the figure of crystal structure of 6LZG. The important residues were shown as sticks in the crystal structure whereas other residues were shown as cartoons. The critical inter-molecular interactions, including hydrogen bonds, electrostatic interaction and hydrophobic interactions, were predicted by Protein Interactions Calculator (http://pic.mbu.iisc.ernet.in/job.html) (Tina et al., 2007) and PRODIGY web server (https://bianca.science.uu.nl/prodigy/) (Xue et al., 2016). The measurement tool of PyMOL software was used to determine the distance between the two atoms of inter-molecular interactions, which was shown as the dotted line in the figure. The interactions were identified based on the criteria of the standard hydrogen bonds (Babu, 2003). The interactions between hydrophobic side-chains were identified using a distance cut-off of 5 Å (Tina et al., 2007).
2.6.3. Identification of the hot spot residues through alanine scanning
Based on the PDB file 6LZG, the Computational Interface Alanine Scanning (http://robetta.bakerlab.org/alascansubmit.jsp) was performed to understand the individual roles of the residues and determine which residues contributed mostly to the binding affinity (Kortemme and Baker, 2002; Kortemme et al., 2004). The binding free energy was calculated after the important residues interfacing with SARS-CoV-2 RBD were mutated to alanine. Hot spot residue is defined operationally when the corresponding alanine mutation can have a destabilizing effect on ΔΔG(complex) more than 1 kcal/mol.
2.6.4. Three-dimensional structure prediction and molecular docking
The non-human ACE2 protein sequences were submitted to AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) to predict and build the three-dimensional protein structures (Five structure models have been predicted and we choose the Rank 1 model with the highest pLDDT score for the following study) (Jumper et al., 2021). Molecular docking was performed by HDOCK sever (http://hdock.phys.hust.edu.cn/), which is a hybrid algorithm of template-based modeling and abinitio free docking (Yan et al., 2020a). The predicted PDB file was submitted to HDOCK sever and docked with SARS-CoV-2 RBD (B chain of 6LZG). Then, three-dimensional structure of the truncated ACE2 fragments from different species was aligned with the truncated human ACE2 domain using the align function of PyMol software, and the RMSD value of the protein complexes was calculated. The Protein Interactions Calculator and PRODIGY web server was used to analyze the binding free energy (Gibbs free energy, ΔG), the dissociation constant (Kd) and intermolecular interactions of the docking results. Similarity, the three-dimensional structure model of the RBD region of the SARS-CoV-2 variants based on 6LZG was constructed and docked with the truncated ACE2 fragments from different species.
2.6.5. Assessment of the physicochemical properties, allergenicity and immunogenicity of the designed peptide candidates
The physiochemical properties of the designed peptide candidates were predicted with ProtParam tool (https://web.expasy.org/protparam/). This server helps to compute the various parameters of protein sequence, including theoretical pI, molecular weight, estimated half-life, extension coefficient, instability index, aliphatic and GRAVY index, etc.
To predict the allergenicity of the designed constructs, AllerTOP server (https://www.ddg-pharmfac.net/AllerTOP/) was used. Five machine learning methods were utilized to predict the allergenicity based on the main chemical properties of the peptides (Dimitrov et al., 2013).
To delineate the potential immunogenicity of the designed peptides, these sequences were submitted to the IEDB analysis tool (http://tools.iedb.org/immunogenicity/) with default setting. This tool can score the immunogenicity by determining the position and side chain characteristics of amino acid residues in the peptide-HLA complex that binds to the T cell receptor, and those peptides with low immunogenicity have a lower prediction scores (Calis et al., 2013).
2.7. Ex vivo neutralization assay based on mice experiment
6-8-week-old female BALB/c mice were bred in the Laboratory Animal Center, Sun Yat-sen University. Mice were anesthetized with 30% vol/vol isoflurane diluted in propylene glycol, and then intranasally administered with AAVrh10-GFP, AAVrh10-shACE2, AAVrh10-shACE2(PD + Neck), AAVrh10-hACE224-83, AAVrh10-dACE224-83 with 2 × 1011 genome copies (GC) in 40 μl volume respectively (5 mice per group). After 2 weeks, mouse serum was collected and subjected to SARS-CoV-2 pseudovirus neutralization assay as above mentioned. This study was approved by the Institutional Review Boards and Animal Care and Use Committees of Sun Yat-Sen University (Assurance No: SYSU-IACUC-2022-00081).
2.8. Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software (v.8; GraphPad Software, La Jolla, CA, USA). The data are reported as mean ± SD. The unpaired Student's t-test comparison was performed to assess the significant differences between two groups and One-way ANOVA was used for the comparison among multiple (>2) groups. (*p < 0.05, **p < 0.01, ***p < 0.001).
3. Results
3.1. AAVrh10-vectored shACE2 and shACE2(PD + Neck) to effectively block the SARS-CoV-2 infections
The full-length ACE2 consists of lead peptide (residues 1–18), N-terminal peptidase domain (PD, residues 19–615), neck domain (residues 616–726), transmembrane helix and intracellular segment (residues 727–768). The RBD domain of SARS-CoV-2 S protein directly interacts with the ACE2 PD domain, whereas the neck domain mainly contributes to the dimerization of ACE2 (Yan et al., 2020b). We thus designed a series of truncated ACE2 (Fig. 1 A), including the soluble human ACE2 (shACE2), the shACE2 PD domain (shACE2PD), and the shACE2 PD + neck domain (shACE2(PD + Neck)), to validate its inhibitory effect against SARS-CoV-2. The corresponding genes were cloned into AAVrh10 vector respectively, and these recombinant AAVrh10 viruses could secrete the appropriate proteins in the supernatant (Fig. 1B, and Supplementary Fig. 1A). The competition assay for the S protein-binding antibodies demonstrated that both shACE2 and shACE2(PD + Neck), but not shACE2PD, could effectively bind to S protein of SARS-CoV-2 (Fig. 1C and Supplementary Fig. 1B ). Similarly, neutralizing assay showed that both shACE2 and shACE2(PD + Neck), but not shACE2PD, could effectively inhibit the SARS-CoV-2 pseudovirus infections (Fig. 1D). Importantly, we also assessed that these recombinant AAVrh10 had a significant inhibitory activity against SARS-CoV-2 virus-like-particles (trVLP) replication (Fig. 1E and F), which is a replication-competent SARS-CoV-2 in a complementary CACO-2-N cell line as described in Methods. Thus, these data validated that the recombinant AAVrh10 carrying the truncated shACE2 could effectively block the SARS-CoV-2 infections in vitro.
Fig. 1.
AAVrh10-vectored truncated shACE2 to effectively block the SARS-CoV-2 infections. (A) The sketch map of three truncated ACE2, termed as shACE2, shACE2PD and shACE2 (PD + Neck). (B) After cells were transfected with plasmid-expressing shACE2, shACE2PD, and shACE2(PD + Neck) respectively, the expression of targeted protein in cell lysates (middle) and in the supernatant (lower) was detected by Western blot. (C) The binding ability of shACE2, shACE2PD and shACE2(PD + Neck) with SARS-CoV-2 S protein detected by flow cytometer-based S antibody competition assay. (D and E) The neutralizing activity of plasmid-expressing shACE2, shACE2PD and shACE2(PD + Neck) against SARS-CoV-2 pseudovirus infection (D) and replication-competent SARS-CoV-2 virus-like-particles (trVLP) in CACO-2-N cells(E). (F) The neutralizing activity of AAVrh10-expressing shACE2, shACE2PD and shACE2(PD + Neck) against replication-competent SARS-CoV-2 trVLP in CACO-2-N cells. The data were shown as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001 and ns, no significance.
3.2. In silico design of the functional fragments from different ACE2 orthologs
To elucidate the key interface between ACE2 and SARS-CoV-2 RBD, seventeen ACE2 orthologs were chosen to compare the ACE2 conservation in different species according to the following criteria: (1) species are continually in close contact with human, such as pets and livestock; (2) species are used as animal models in biomedical research such as ferret, rhesus macaque and mouse; (3) species that may serve as potential hosts according to previous studies, such as pangolin, civet, bat. The whole ACE2 gene from other sixteen species maintained over 80% similarity when compared with human ACE2. Of note, the α1 helix, α2 helix and the junction between β3 and β4 hairpin (located at 19–355 aa in human ACE2 protein) in the ACE2 PD domain share over 90% identity with the human ACE2 (Fig. 2 A) (Yan et al., 2020b). In addition, we also performed amino acid sequence multiple alignment between different species, and results showed that rhesus macaque ACE2 had the highest similarity with human ACE2 (95%), while that of chicken and duck were approximately 65% (Fig. 2B). Since the variation in ACE2 orthologs in different species leaded to various susceptibility to SARS-CoV-2 infection (Zhou et al., 2020; Chan et al., 2020b; Chandrashekar et al., 2020; Shi et al., 2020), we further blast the twenty key residues of these ACE2 orthologs that directly interfacing with SARS-CoV-2 RBD, which might be helpful to understand the cross-species transmission of SARS-CoV-2. Except for rhesus macaque's 100% identity, there was some extent difference between these twenty key residues ( Fig. 2C), which may account for the variation of their binding affinity with SARS-CoV-2 RBD.
Fig. 2.
Conservation analysis of ACE2 orthologs and the critical intermolecular interactions between ACE2 and SARS-CoV-2 RBD interfaces. Multiple sequence alignment was performed by MAFFT software (version 7.453) with auto strategy. (A) Simplot analysis of multiple species based on human ACE2 (Q9BYF1.2). The curve with different colors correspond to the amino acid similarity between human ACE2 and different species. The X-axis indicated the amino acid position in the human ACE2 sequence, and the Y-axis indicated amino acid identity between the query sequence and the reference strain. (B) Heat map represents the similarity matrix between these 17 species of ACE2 amino acid sequences. Annotations on right of the heatmap indicated the value of amino acid identity. (C) The key amino acid residues between the interface of ACE2 orthologs and SARS-COV-2 RBD. The mutant residues were highlighted in blue or green. Blue residues represent mutations with similar property (polar to polar or non-polar to non-polar), and green residues represent mutations leading to change in the property of the residue (polar to non-polar or non-polar to polar). (D) Schematic representation of the interactions between SARS-CoV-2 S protein RBD (purple) and human ACE2 binding domain (blue). (E and F) A closeup view of (D), the interface area of SARS-CoV-2 RBD (purple) with human ACE2 binding domain (blue). Key residues at the interface were marked at corresponding position. The intermolecular interactions including hydrogen bonds, electrostatic interaction and hydrophobic interaction are shown in yellow, blue and green, respectively. The panel (D,E,F) was created in PyMOL with PDB file 6LZG.
To further elucidate how these mutations in key residues affect the intermolecular interactions between ACE2 and SARS-CoV-2 RBD, we reconstructed three-dimensional structure of the SARS-CoV-2 spike protein-ACE2 complex (PDB ID: 6LZG) by amino acid substitution experiment ( Fig. 2D–F). In general, the disruption of the critical intermolecular interactions due to mutations in the ACE2 key residues can lead to a decreased binding affinity with SARS-CoV-2 RBD (Table 1 ). For example, hydrogen bonds might be destroyed due to the substitution of Gln 24 by Leu, Glu 35 by Lys or Arg, Tyr 41 and Lys 353 by His; central salt bridge by electrostatic interactions might be affected by the substitution of Asp 30 by Glu, Asn or Ala, Lys31 by Glu. However, mutations in ACE2 residues ASP 30 may increase the binding affinity with SARS-CoV-2 RBD (Table 1), which may account for those susceptible hosts for SARS-CoV-2 infection.
Table 1.
Critical intermolecular interactions between ACE2 and SARS-CoV-2 RBD interface and the potential changes caused by ACE2 mutations.
| Interfacial Contacts | Human ACE2 | SARS-CoV-2 | ACE2 mutation | Change in inter-molecular interactions |
|---|---|---|---|---|
| Hydrogen bonds | SER 19 | ALA 475 | ||
| GLN 24 | ASN 487 | Leu | Destruction | |
| ASP 30 | LYS 417 | |||
| GLU 35 | GLN 493 | Lys/Arg | Destruction | |
| GLU 37 | TYR 505 | |||
| ASP 38 | TYR 449 | |||
| TYR 41 | ASN 501 | His | Destruction | |
| TYR 41 | THR 500 | His | Destruction | |
| GLN 42 | GLN 498 | |||
| GLN 42 | TYR 449 | |||
| TYR 83 | ASN 487 | Phe | Destruction | |
| LYS 353 | GLY 496 | His | Destruction | |
| LYS 353 | ASN 501 | His | Destruction | |
| ASP 355 | THR 500 | |||
| Electrostatic interactions | ASP 30 | LYS 417 | Glu | Enhancement |
| Asn/Ala | Destruction | |||
| LYS 31 | GLU 484 | Glu | Repulsion | |
| Hydrophobic interactions | PHE 28 | TYR 489 | ||
| Leu 79 | PHE 486 | His/Thr/Asn | Destruction | |
| MET 82 | PHE 486 | Asn/Thr/Ser/Arg | Destruction | |
| TYR 83 | PHE 486 |
To rationally design the minimal ACE2 functional fragment, we next performed alanine scanning of the key residues at the ACE2 and RBD binding interface. Results demonstrated that the ΔΔG (complex) value of Gln 24, Tyr 41, Gln 42, Tyr 83 and Asp 355 was greater than 1 kcal/mol and thus considered to be critically important for binding interaction, and Gln 24, Phe 28, Glu 31, Tyr 41, Tyr 83, Asp 355 and Arg 393 also played important role in the stability of the protein complex according to the value of ΔG (partner) (Table 2 ). Given that the α1 and α2 helix from Ser 19 to Tyr 83 contributed approximately 90 percent of the total binding free energy based on the computational analysis by RosettaDock software (Feng et al., 2021), the Gln 24 to Tyr 83 fragments (ACE224-83) in different species was chosen as potential ACE2 fragement to inhibit SARS-CoV-2 infection, and these ACE2 fragements are used in the following function experiments.
Table 2.
Results of computational alanine scanning of ACE2 key residues interfacing with SARS-CoV-2 RBD.a.
| Residue | int_id | Amino acid | ΔΔG (complex) | ΔG (partner) |
|---|---|---|---|---|
| 19 | 1 | S | 0.98 | 0.11 |
| 24 | 1 | Q | 1.94 | 1.12 |
| 27 | 1 | T | 0.67 | −0.05 |
| 28 | 1 | F | 0.24 | 3.22 |
| 30 | 1 | D | 0.36 | −1.28 |
| 31 | 1 | K | 0.18 | −0.45 |
| 34 | 1 | H | 0.67 | −0.71 |
| 35 | 1 | E | 0.34 | −0.30 |
| 37 | 1 | E | 0.13 | 4.79 |
| 38 | 1 | D | 0.82 | −0.99 |
| 41 | 1 | Y | 4.88 | 3.50 |
| 42 | 1 | Q | 1.90 | −0.44 |
| 45 | 1 | L | 0.50 | 0.97 |
| 79 | 0 | L | 0.47 | 0.89 |
| 82 | 1 | M | 0.22 | 0.78 |
| 83 | 1 | Y | 2.91 | 3.61 |
| 330 | 1 | N | 0.18 | 0 |
| 353 | 1 | K | 0.16 | 0.65 |
| 355 | 1 | D | 3.05 | 1.85 |
| 393 | 0 | R | 0 | 7.86 |
a The value of “int_id” is 1 when the distance between one atom of the side chain and one atom of the other partner is within 4 Å, and the value is 0 when the distance is more than 4 Å. ΔΔG (complex): predicted change in binding free energy upon alanine mutation. ΔG (partner): predicted change in protein stability of the mutant complex partner upon alanine mutation.
3.3. Prediction analysis of binding free energy and intermolecular interactions between the designed ACE2 peptides and SARS-CoV-2 variants
To determine whether the designed ACE2 fragments retain the capability to bind to SARS-CoV-2 RBD, three-dimensional structure model was reconstructed using AlphaFold 2 and molecular docking. Results showed that the RMSD values were less than 2 (Fig. 3 A), indicating that there was a similar three-dimensional structure between these different ACE2 ortholog peptides-RBD protein complex and the corresponding human ACE2 peptides-RBD protein complex (Seffernick et al., 2019). Consistent with above data, there was also a similar structure of the protein backbone between these different ACE2 ortholog peptides and the corresponding human ACE2 peptides (Fig. 3B–D). Moreover, we analyzed the critical intermolecular interactions between the designed ACE2 ortholog peptides and SARS-CoV-2 RBD. Interestingly, we found that the RBD binding affinity with ACE2 peptides derived from dog, pig, hamster, ferret, civet, cattle, and sheep and was higher than that of human ACE2 peptides (Table 3 ), implying that these ACE2 ortholog peptides of other species may be a more competitive inhibitor to block SARS-CoV-2 entry than human ACE2.
Fig. 3.
Three-dimension structure reconstruct using AlphaFold2 and molecular docking analysis and the prediction of binding free energy and intermolecular interactions between the designed ACE224-83peptides and SARS-CoV-2 variants. (A) The RMSD value between human ACE2-RBD complex and other species ACE2-RBD complex. Three-dimensional structure alignment between human ACE224-83-RBD complex and cat ACE224-83-RBD complex (B), or bat ACE224-83-RBD complex (C), or chicken ACE224-83-RBD complex (D). The key mutation residues are shown as sticks. (E–I) Prediction of the binding free energy and binding affinity between the designed ACE224-83 of different species and SARS-CoV-2 Wuhan-Hu-1 (E), SARS-CoV-2 variants B.1.351 (K417N, N501Y, E484K) (F), SARS-CoV-2 variants P.1 (K417T, N501Y, E484K) (G), SARS-CoV-2 variants B.1.617 (L452R, E484Q) (H), SARS-CoV-2 variants B.1.1.529 (K417N, S477N, T478K, E484A, Q498R, N501Y) (I). ΔG and Kd value represent the binding free energy and binding affinity, respectively.
Table 3.
Comparison of critical intermolecular interactions and binding affinity between the designed ACE224-83 peptides and SARS‐CoV‐2 RBD.
| Interfacial Contacts | Hydrogen bonds | Electrostatic interactions | Hydrophobic interactions | ΔG (kcal/mol) | Kd (M) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | N 487 | K 417 | Q 493 | Y 505 | Y 449 | N 501 | T 500 | Q 498 | Y 449 | N 487 | K 417 | E 484 | Y 489 | F 486 | F 486 | F 486 | ||
| Human ACE2 | Q 24 | D 30 | E 35 | E 37 | D 38 | Y 41 | Y 41 | Q 42 | Q 42 | Y 83 | D 30 | K 31 | F 28 | L 79 | M 82 | Y 83 | −11.4 | 9.2E-09 |
| rhesus macaque | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | −11.2 | 1.3E-08 |
| hamster | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✕ | ✓ | −11.5 | 8.2E-09 |
| cat | ✕ | E | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✕ | ✓ | −11.3 | 1.1E-08 |
| tiger | ✕ | E | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✕ | ✓ | −11.3 | 1.1E-08 |
| ferret | ✕ | E | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | ✕ | ✕ | ✓ | −11.7 | 6.0E-09 |
| civet | ✕ | E | ✓ | ✕ | E | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✕ | ✓ | ✓ | ✕ | ✓ | −11.9 | 4.4E-09 |
| cattle | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | M | ✕ | ✓ | −11.7 | 6.0E-09 |
| sheep | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | M | ✕ | ✓ | −11.9 | 4.1E-09 |
| rabbit | ✕ | E | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✕ | ✓ | −11.1 | 1.5E-08 |
| dog | ✕ | E | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✕ | ✓ | −12.0 | 3.2E-09 |
| pig | ✕ | E | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | I | ✕ | ✓ | −11.5 | 8.2E-09 |
| pangolin | E | E | ✓ | ✓ | E | ✓ | ✓ | ✓ | ✓ | ✓ | E | ✓ | ✓ | I | ✕ | ✓ | −10.5 | 4.2E-08 |
| bat | E | ✓ | ✕ | ✓ | ✓ | ✕ | ✕ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✕ | ✓ | −10.0 | 8.4E-08 |
| mouse | N | N | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | F | −10.7 | 3.0E-08 |
| chicken | E | ✕ | ✕ | ✓ | ✓ | ✓ | ✓ | E | E | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | F | −10.7 | 2.9E-08 |
| duck | ✓ | ✕ | ✕ | ✓ | ✓ | ✓ | ✓ | E | E | ✕ | ✕ | ✕ | ✓ | ✕ | ✕ | F | −11.0 | 1.6E-08 |
The green ticks and letters represent the presence of interactions, and the red crosses represent the absence of the interactions, respectively. ΔG and Kd value represent the binding free energy and the binding affinity, respectively.
Then, we further predicted the binding free energy and binding affinity between the designed ACE2 peptides and the SARS-CoV-2 variants, including Beta B.1.351 (K417N, N501Y, E484K), Gamma P.1 (K417T, N501Y, E484K), Delta B.1.617 (L452R, E484Q), and Omicron B.1.1.529 (K417N, S477N, T478K, E484A, Q498R, N501Y), by reconstructing the three-dimension structures based on AlphaFold2 algorithm. Results showed that most ACE224-83 peptides retained low binding free energy to different SARS-CoV-2 variants RBD respectively (Fig. 3E–I). Interestingly, compared to that of human ACE224-83, ferret and dog ACE224-83 had an enhanced binding affinity to SARS-CoV-2 variants except Omicron. Of note, all of these ACE224-83 peptides had a reduced binding affinity to the Omicron RBD. Nevertheless, our designed ACE224-83 peptides retained the potential affinity to the SARS-CoV-2 variants.
In addition, we also analyzed the physicochemical properties, allergenicity and immunogenicity of these ACE224-83 peptides as mentioned in Methods. The predicted results showed that ACE224-83 peptides of civet, cattle, chicken, and duck may be allergens; ACE224-83 peptides of ferret, pig, chicken, and duck may be recognized as immunogens; ACE224-83 peptides of rhesus macaque, hamster, cat, tiger, sheep, and dog may have a good safety (Supplementary Table 1).
It is well known that ACE2 is a monocarboxylic peptidase to catalyze the conversion of angiotensin II (Ang II) to angiotensin 1–7 (Ang1–7), and thus plays a key role to regulate the cardiovascular function. Previous studies showed that the amino acid residues Arg169, Trp271, Arg273, His 345, Lys481, and Arg514 were critical to the catalytic activity of ACE2 for angiotensin substrates (Guy et al., 2005; Rushworth et al., 2008). Since all of these residues were not included in our truncated peptides (24-83aa), these truncated peptides could consequently avoid the potential side effects due to the enzymatic activity of the full length ACE2.
After comprehensive consideration of the binding affinity, allergenicity, and immunogenicity, we think that the dog ACE224-83 (dACE224-83) might be a potential therapeutic agent to broadly block SARS-CoV-2 infections.
3.4. AAVrh10-vectored dACE224-83 construct to broadly block the cell entry of SARS-CoV-2 variants in vitro
Next, we generated the recombinant AAVrh10 constructs expressing dog ACE224-83 (dACE224-83), cat ACE224-83 (cACE224-83), mouse ACE224-83 (mACE224-83), human ACE224-83 (hACE224-83), human shACE2, and human shACE2(PD + Neck) respectively for the following functional experiments. Dog and cat are usually in close contact with human daily life, and some studies have showed that they are susceptible to SARS-CoV-2 infections (Calvet et al., 2021; Jairak et al., 2021; Klaus et al., 2021), and therefore we constructed these recombinant AAVrh10 vectors carrying dog ACE2 and cat ACE224-83 for potential therapeutic agents. Mouse is not susceptible to SARS-CoV-2 prototype strain due to the low binding affinity between ACE2 receptor and SARS-CoV-2 RBD, but SARS-CoV-2 variants B.1.351 and P 0.1 had acquired the ability to use mouse ACE2 receptor for cell entry (Wang et al., 2021b). The possible mechanism of this cross-species transmission might be related to RBD N501Y mutation. We therefore constructed the recombinant AAVrh10 vector carrying mouse ACE224-83 to investigate its ability to inhibit SARS-CoV-2 variants. In addition, the recombinant AAVrh10 vector carrying human shACE2, human shACE2(PD + Neck), and human ACE224-83 was constructed as controls in this study.
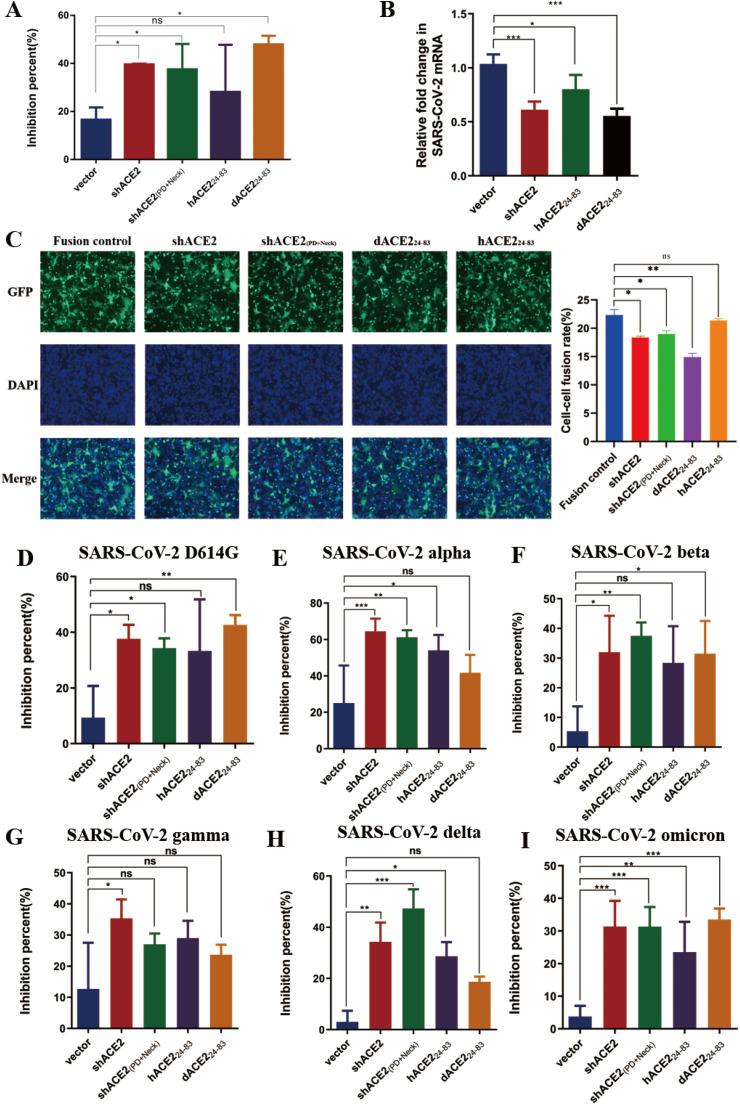
Consistent with above in silico analysis, our in vitro functional experiments demonstrated that human shACE2, human shACE2(PD + Neck), dACE224-83, but not cACE224-83 and mACE224-83, exhibited great potency to block the cell entry of both SARS-CoV-2 pseudovirus (Fig. 4 A) and replication-competent SARS-CoV-2 trVLP (Fig. 4B and Supplementary Fig. 2). We also validated the biological activity of these truncated ACE2 peptides by the inhibition assay of cell-cell fusion, since SARS-CoV-2 showed a potent capacity of plasma membrane fusion by its spike protein-mediated cell-cell fusion. As shown in Fig. 4C, incubation with AAVrh10-vectored dACE224-83 significantly reduced the formation of syncytium, implying that dACE224-83 can effectively block the SARS-CoV-2 spike protein-mediated cell-cell fusion. Moreover, we further detected the blockage breadth of dACE224-83 against emerging SARS-CoV-2 variants of concern, including D614G, Alpha, Beta, Gamma, Delta, and Omicron. Of note, our results showed that AAVrh10-vectored shACE2 or shACE2(PD + Neck) effectively blocked the cell entry of these variants, and AAVrh10-vectored dACE224-83 also potentially blocked the cell entry of D614G, beta, and omicron variants (Fig. 4D–I).
Fig. 4.
AAVrh10-mediated ACE2 functional domain broadly blocked the cell entry of SARS-CoV-2 prototype and variants. (A and B) The neutralizing activity of AAVrh10-shACE2, AAVrh10-shACE2(PD + Neck), AAVrh10-hACE224-83, and AAVrh10-dACE224-83 against SARS-CoV-2 pseudovirus(A) and replication-competent SARS-CoV-2 trVLP(B). The expression level of mRNA was normalized to the expression of β-actin. (C) After treatment with AAVrh10-shACE2, AAVrh10-shACE2(PD + Neck), AAVrh10-hACE224-83 and AAVrh10-dACE224-83 respectively, these AAVrh10-mediated ACE2 functional domain effectively blocked the SARS-CoV-2 spike protein-mediated cell-cell fusion, characteristic with the decreased syncytium formation under a fluorescence microscope. Fusion control means the formation of syncytium by mixing effector cells (pcDNA3.1-SARS-CoV-2 Spike and pVAX-GFP co-transfected HEK293T cells) and target cells (VeroE6 cells) in a 1:1 ratio. Scale bar: 2000 μm. The nuclei were stained with DAPI. The pictures of confocal microscope were analyzed by imageJ software. (D–I) The BHK-ACE2 cells were transfected with the AAV-vector, AAV-shACE2, AAV-shACE2(PD + Neck), AAV-hACE224-83, and AAV-dACE224-83, respectively, and then the inhibitory activity against SARS-CoV-2 variants D614G(D), alpha(E), beta(F), gamma(G), delta(H) and omicron(I) pseudovirus infection was detected. The data from at least triplicates were shown as the mean ± SD.*p < 0.05, **p < 0.01 ***p < 0.001 and ns, no significance.
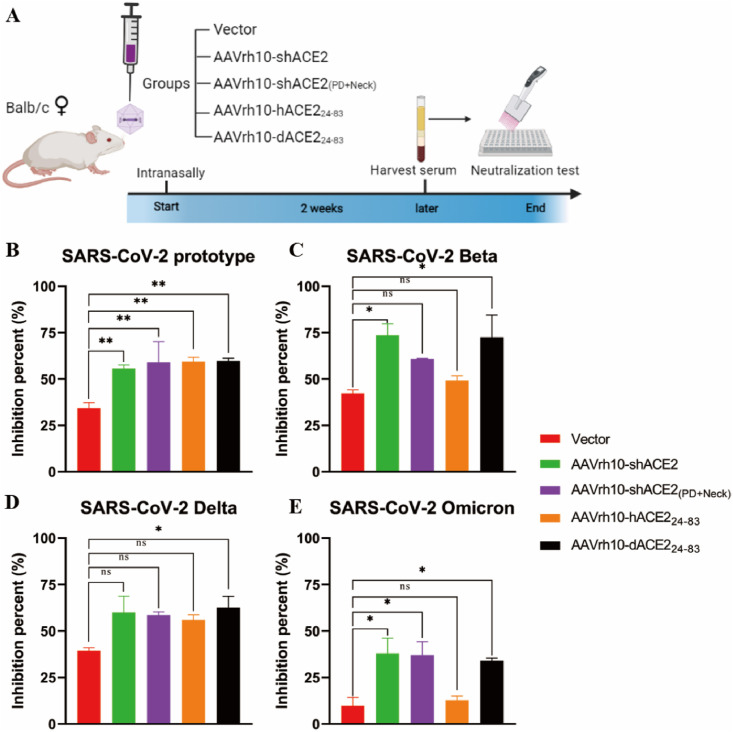
3.5. Intranasal administration of AAVrh10-vectored ACE2 functional domain can broadly block the cell entry of SARS-CoV-2 variants ex vivo
Given that AAVrh10-based vector can infect mice with airway tissue tropism, we then validated the neutralizing ability of these AAVrh10-vectored ACE2 functional domain in mice ex vivo. Mice were intranasally treated with AAVrh10-GFP, AAVrh10-shACE2, AAVrh10-shACE2(PD + Neck), AAVrh10-hACE224-83, and AAVrh10-dACE224-83 respectively, and then the serum was collected to subjected to the neutralizing assays (Fig. 5 A). As shown in Fig. 5B–E, these AAVrh10-vectored truncated ACE2 functional domain had a potent neutralizing ability to inhibit SARS-CoV-2 infections in mice. As expected, the serum from AAVrh10-shACE2-treated mice and AAVrh10-shACE2(PD + Neck)-treated mice had a strong neutralizing activity against SARS-CoV-2 prototype and variants. More importantly, the serum from AAVrh10-dACE224-83-treated mice had effectively blocked the cell entry of SARS-CoV-2 prototype and variants, including Omicron variant (Fig. 5B–E).
Fig. 5.
Intranasal administration for AAVrh10-mediated ACE2 functional domain can broadly block the cell entry of SARS-CoV-2 wild-type and variants through ex vivo serum neutralization assay. (A) Schematic diagram of ex vivo neutralization assay of AAVrh10-vectored truncated ACE2 functional domain in mice serum. 6-8-week-old female BALB/c mice were intranasally administered with 2 × 1011 GC of AAVrh10-GFP, AAVrh10-shACE2, AAVrh10-shACE2(PD + Neck), AAVrh10-hACE224-83, AAVrh10-dACE224-83 respectively (n = 5 mice per gourp). Mice serum was collected after 2 weeks and subjected to the following neutralizing assay. (B–E) The neutralizing activity of above-mentioned mice serum to inhibit cell entry of SARS-CoV-2 prototype (B), Beta (C), Delta (D) and Omicron (E) pseudovirus. Mouse serum samples were tested at a dilution of 1:30. The data from at least triplicates were shown as the mean ± SD.*p < 0.05, **p < 0.01, ***p < 0.001, ns: no significance.
Together, these data demonstrated that AAVrh10-vectored ACE2 functional domain could be a promising therapeutic strategy against SARS-CoV-2 variants infections.
4. Discussion
The emergence of SARS-CoV-2 variants continues to exacerbate the current COVID-19 pandemic and pose a new challenge to current antibody therapies and vaccine inoculation (Sharun et al., 2021). As the key receptor of both SARS-CoV-2 prototype and its variants, ACE2 presents a potential therapeutic target for broadly inhibiting the infections of SARS-CoV-2 variants (Scialo et al., 2020). In physiological conditions, ACE2 contains two forms including full-length membrane-bound ACE2 (mACE2) and soluble ACE2 (sACE2). The mACE2 consists of a transmembrane anchor and an extracellular domain and thus is located on the cell membrane, while the sACE2 is shed into the blood circulation and exerts the function of vasodilation, reduction of vascular permeability, anti-fibrosis, anti-inflammation and other effects (Scialo et al., 2020; Zhong et al., 2011). SARS-CoV-2 enters the cell through membrane fusion, leading to a significant down-regulation of the ACE2 receptor and the loss of the catalytic effect of the ACE2 receptor at the external site of the cell membrane (Zhang et al., 2020; Kuba et al., 2005). Importantly, recent studies demonstrated that sACE2 administration effectively inhibited the cell entry of SARS-CoV-2 through blocking SARS-CoV-2 binding to membrane-anchored ACE2 (Monteil et al., 2020; Feng et al., 2021; Benton et al., 2020). Therefore, increasing the concentration of sACE2 in the blood circulation can serve as a promising therapeutic strategy not only for treatment of cardiovascular diseases, but also for blockade of SARS-CoV-2 infections.
However, the half-life of natural sACE2 in circulation is short (Xie et al., 2021), which raises a question how to increase the level of sACE2 expression in vivo. Compared with other viral vectors, recombinant AAV has been shown as one of the most promising vectors for therapeutic gene delivery, and its efficacy has been demonstrated in extensive clinical trials, ranging from hemophilia B38 to Parkinson's disease (Hu and Lipshutz, 2012; Colella et al., 2018; Ferreira et al., 2014). AAV-based vectors have been reported to possess many advantages, including favorable safety profiles, tailorable tissue tropism, and long-term gene expression (Zheng et al., 2018). Of note, AAVrh10 has been proved as a promising gene delivery vector by increasing pre-clinical and clinical research data (Thwaite et al., 2015; Markmann et al., 2018; Pages et al., 2019). More importantly, AAVrh10 has a great potential for the treatment of various pulmonary diseases because of its pulmonary tissue tropism. For example, intraperitoneal injection of AAVrh10-vectored human α 1-antitrypsin for treatment of α 1-antitrypsin deficiency can reduce progressive destruction of lung parenchyma and early-onset of panacinar emphysema (Chiuchiolo et al., 2013). AAVrh10 is demonstrated as the most effective known AAV vector for intrapleural gene delivery and has the advantage of circumventing human immunity to AAV (De et al., 2006). Intralingual administration of AAVrh10-vectored-miR (superoxide dismutase-1, SOD1) improved respiratory function in a SOD1 mouse model of amyotrophic lateral sclerosis (Lind et al., 2020). In addition, intrapleural administration of an AAVrh10 vector, encoding the bevacizumab monoclonal antibody, effectively suppressed the growth of metastatic lung tumors (Watanabe et al., 2010). In this study, we innovatively used AAVrh10 vector for the first time to deliver different ACE2 domains to target lung tissues, and it is expected to reduce the side effects and to maximize the therapeutic efficacy against respiratory pathogens.
The therapeutic efficacy of full-length rACE2 as a decoy to block the SARS-CoV-2 infection is being assessed in clinical trials (Abd El-Aziz et al., 2020; Zoufaly et al., 2020). However, a recent study demonstrated that sACE2 contributed to SARS-CoV-2 infection and COVID-19 severity, especially in comorbid patients (Rahman et al., 2021). SARS-CoV-2 can exploit the receptor-mediated endocytosis to enhance cell entry through interaction between spike with sACE2-vasopressin via AT1 or AVPR1B (Yeung et al., 2021). Thus, it has raised the potential safety concern regarding to use the full-length rACE2 to treat SARS-CoV-2 infection, which might enhance the SARS-CoV-2 infectivity. As a result, an ideal ACE2 decoy should maintain only the minimum functional domain to interact with SARS-CoV-2 RBD-binding domain, and therefore avoid the redundant domain which can cause the potential side effects.
To address this critical issue, we performed a comprehensive computational analysis to rationally design the minimal functional domain. Given that experimental studies have documented the susceptibility of different animal species to SARS-CoV-2, seventeen of ACE2 orthologs were chosen in this study (Mahdy et al., 2020). Previous study revealed that the emerged SARS-CoV-2 variants, including alpha, beta, gamma and delta, possessed a similar or stronger binding capacity to ACE2 receptor, when compared with that of SARS-CoV-2 prototype (Koehler et al., 2021; Wu et al., 2022), while omicron variant shared a comparable or decreased binding affinity when compared with that of SARS-CoV-2 prototype (Wu et al., 2022; Han et al., 2022). Of note, our results by in silico prediction in binding affinity between ACE2 and SARS-CoV-2 variants were consistent with these findings. In this study, we designed a variety of potential ACE2 functional domain through alanine scanning, structure prediction and molecular docking. Surprisingly, our results suggest that dog ACE224-83 possesses a higher binding affinity with both SARS-CoV-2 prototype and variants than that of human ACE2 and ACE224-83, and the further functional experiments demonstrated that AAVrh10-vectored shACE2 or dACE224-83 construct exhibited a broadly blockage breadth against SARS-CoV-2 wild strain and variants. Due to experimental resources, one limitation for this study is that we did not conduct in vivo experiment in animal infection model to verify the therapeutic efficacy of AAVrh10-dACE224-83 against SARS-CoV-2 infections. In addition, the next study will be more informative if the pharmacokinetics of these truncated ACE2 peptides in serum is assessed and mucosal tissues/samples should be taken to check the local responses. Nevertheless, our findings warrant the further in vivo therapeutic study in the future.
Taken together, this study provide insights to develop the ACE2 functional domain-based blockade as a promising therapeutic strategy against SARS-CoV-2 variants infections.
Author contributions
Project design and supervised by C.S.; experiment performing by M.L., J.C., Y.L., J.Z., Y.H., F.F.; data analysis by M.L., J.C.; picture drawing by M.L., J.C., Y.L.; materials and reagents contributed by C.S., Y.C., L.S., Y.S.; Writing by M.L., J.C., F.F., Y.L.; review and editing by C.S., F.F.,Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2021YFC2300103, 2022YFE0203100), the National Natural Science Foundation of China (82041043), the Natural Science Foundation of Guangdong Province (2019A1515110458), China Postdoctoral Science Foundation (2020T130150ZX), and Shenzhen Science and Technology Program (20190804095916056; JSGG20200225152008136; GXWD20201231165807008; 20200825113322001; RCJC20210706092009004).
Animal and human rights statement
All institutional and national guidelines for the care and use of laboratory animals were followed.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Figure 5A was created with BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105383.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abd El-Aziz T.M., Al-Sabi A., Stockand J.D. Human recombinant soluble ACE2 (hrsACE2) shows promise for treating severe COVID19. Signal Transduct. Targeted Ther. 2020;5(1) doi: 10.1038/s41392-020-00374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M.M. NCI: a server to identify non-canonical interactions in protein structures. Nucleic Acids Res. 2003;31(13):3345–3348. doi: 10.1093/nar/gkg528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588(7837):327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis J.J.A., et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013;9(10) doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet G.A., et al. Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS One. 2021;16(4) doi: 10.1371/journal.pone.0250853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., et al. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuchiolo M.J., et al. Intrapleural administration of an AAVrh.10 vector coding for human alpha1-antitrypsin for the treatment of alpha1-antitrypsin deficiency. Hum. Gene. Ther. Clin. Dev. 2013;24(4):161–173. doi: 10.1089/humc.2013.168. [DOI] [PubMed] [Google Scholar]
- Colella P., Ronzitti G., Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Method. Clin. Dev. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De B.P., et al. High levels of persistent expression of alpha1-antitrypsin mediated by the nonhuman primate serotype rh.10 adeno-associated virus despite preexisting immunity to common human adeno-associated viruses. Mol. Ther. 2006;13(1):67–76. doi: 10.1016/j.ymthe.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Dimitrov I., Flower D.R., Doytchinova I. AllerTOP - a server for in silico prediction of allergens. BMC Bioinf. 2013;14 doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F., et al. Killing two birds with one stone by administration of soluble ACE2: a promising strategy to treat both cardiovascular diseases and SARS-CoV-2 infection. Viruses. 2021;13(11) doi: 10.3390/v13112243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira V., et al. Immune responses to intramuscular administration of alipogene tiparvovec (AAV1-LPL(S447X)) in a phase II clinical trial of lipoprotein lipase deficiency gene therapy. Hum. Gene Ther. 2014;25(3):180–188. doi: 10.1089/hum.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.L., et al. Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis. FEBS J. 2005;272(14):3512–3520. doi: 10.1111/j.1742-4658.2005.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 2022;185(4):630–640 e610. doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Lipshutz G.S. AAV-based neonatal gene therapy for hemophilia A: long-term correction and avoidance of immune responses in mice. Gene Ther. 2012;19(12):1166–1176. doi: 10.1038/gt.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairak W., et al. Transbound Emerg Dis; 2021. First Cases of SARS-CoV-2 Infection in Dogs and Cats in Thailand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X., et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 2021;17(3) doi: 10.1371/journal.ppat.1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus J., et al. SARS-CoV-2 infection in dogs and cats from southern Germany and northern Italy during the first wave of the COVID-19 pandemic. Viruses. 2021;13(8) doi: 10.3390/v13081453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler M., et al. Molecular insights into receptor binding energetics and neutralization of SARS-CoV-2 variants. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-27325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T., Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2002;99(22):14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T., Kim D.E., Baker D. Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004;(219):pl2. doi: 10.1126/stke.2192004pl2. 2004. [DOI] [PubMed] [Google Scholar]
- Kuba K., et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind L.A., et al. Intralingual administration of AAVrh10-miR(SOD1) improves respiratory but not swallowing function in a superoxide dismutase-1 mouse model of amyotrophic lateral sclerosis. Hum. Gene Ther. 2020;31(15–16):828–838. doi: 10.1089/hum.2020.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdy M.A.A., Younis W., Ewaida Z. An overview of SARS-CoV-2 and animal infection. Front. Vet. Sci. 2020;7 doi: 10.3389/fvets.2020.596391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann S., et al. Attenuation of the Niemann-Pick type C2 disease phenotype by intracisternal administration of an AAVrh.10 vector expressing Npc2. Exp. Neurol. 2018;306:22–33. doi: 10.1016/j.expneurol.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Monteil V., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–+. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages G., et al. Intrathecal AAVrh10 corrects biochemical and histological hallmarks of mucopolysaccharidosis VII mice and improves behavior and survival. Hum. Mol. Genet. 2019;28(21):3610–3624. doi: 10.1093/hmg/ddz220. [DOI] [PubMed] [Google Scholar]
- Planas D., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Rahman M.M., Hasan M., Ahmed A. Potential detrimental role of soluble ACE2 in severe COVID-19 comorbid patients. Rev. Med. Virol. 2021;31(5):1–12. doi: 10.1002/rmv.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth C.A., Guy J.L., Turner A.J. Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis. FEBS J. 2008;275(23):6033–6042. doi: 10.1111/j.1742-4658.2008.06733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialo F., et al. ACE2: the major cell entry receptor for SARS-CoV-2. Lung. 2020;198(6):867–877. doi: 10.1007/s00408-020-00408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2020. Science brief: emerging SARS-CoV-2 variants. (CDC COVID-19 Science Briefs). Atlanta (GA) [Google Scholar]
- Seffernick J.T., et al. Predicting protein complex structure from surface-induced dissociation mass spectrometry data. ACS Cent. Sci. 2019;5(8):1330–1341. doi: 10.1021/acscentsci.8b00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharun K., et al. SARS-CoV-2 infection in farmed minks, associated zoonotic concerns, and importance of the One Health approach during the ongoing COVID-19 pandemic. Vet. Q. 2021;41(1):50–60. doi: 10.1080/01652176.2020.1867776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thwaite R., et al. AAVrh.10 immunogenicity in mice and humans. Relevance of antibody cross-reactivity in human gene therapy. Gene Ther. 2015;22(2):196–201. doi: 10.1038/gt.2014.103. [DOI] [PubMed] [Google Scholar]
- Tina K.G., Bhadra R., Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang R., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611–1621. doi: 10.1016/j.immuni.2021.06.003. e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Boyer J.L., Crystal R.G. AAVrh.10-mediated genetic delivery of bevacizumab to the pleura to provide local anti-VEGF to suppress growth of metastatic lung tumors. Gene Ther. 2010;17(8):1042–1051. doi: 10.1038/gt.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.Y., et al. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Targeted Ther. 2022;7(1) doi: 10.1038/s41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., et al. COVID-19 and diabetes: a comprehensive review of angiotensin converting enzyme 2, mutual effects and pharmacotherapy. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.772865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L.C., et al. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32(23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- Yadav P.D., et al. Neutralization of variant under investigation B.1.617.1 with sera of BBV152 vaccinees. Clin. Infect. Dis. 2022;74(2):366–368. doi: 10.1093/cid/ciab411. [DOI] [PubMed] [Google Scholar]
- Yan Y., et al. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- Yan R., et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M.L., et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell. 2021;184(8):2212–2228 e2212. doi: 10.1016/j.cell.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.B., et al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., et al. Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo. Cell Discov. 2021;7(1):65. doi: 10.1038/s41421-021-00302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C.X., et al. Lentiviral vectors and adeno-associated virus vectors: useful tools for gene transfer in pain research. Anat. Rec. 2018;301(5):825–836. doi: 10.1002/ar.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., et al. Prevention of angiotensin II-mediated renal oxidative stress, inflammation, and fibrosis by angiotensin-converting enzyme 2. Hypertension. 2011;57(2):314–322. doi: 10.1161/HYPERTENSIONAHA.110.164244. [DOI] [PubMed] [Google Scholar]
- Zhou P., et al. Addendum: a pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;588(7836):E6. doi: 10.1038/s41586-020-2951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoufaly A., et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8(11):1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.