Abstract
N6-methyladenosine (m6A) is the most prevalent and internal modification that occurs in the messenger RNAs of eukaryotes. However, knowledge of the impact of these modifications on gene expression regulation remains limited. By using the in vitro MeRIP-seq and RNA-seq assays, we discovered that the mRNA demethylase FTO was significantly up-regulated in esophageal squamous cell carcinoma (ESCC) tissues and cells. Knockdown of FTO drastically suppressed the proliferation, migration, and invasion of ESCC cells. Furthermore, by using transcriptome-wide m6A-seq and RNA-seq assays, we identified ERBB2 is the target of FTO, which acts in concert in ESCC tumorigenesis and metastasis. Moreover, loss and gain functional studies suggested that the m6A reader YTHDF1 stabilizes ERBB2 mRNA via decoding the m6A modification. All these results uncovered a new signaling cascade, including FTO, YTHDF1, and ERBB2, which finely regulates the ESCC progression.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10585-022-10169-4.
Keywords: Esophageal squamous cell carcinoma, N6-methyladenosine (m6A) modification, FTO, ERBB2, YTHDF1
Introduction
Although post-transcriptional modifications are known to occur in RNAs, the biological functions of these modifications has only recently begun to be explored [1]. The mRNA m6A modification was first identified in the 1970s [2–5]. Since then, the accumulating studies have demonstrated this modification is most abundant in eukaryotic mRNAs with unique distribution patterns [6, 7]. In addition, m6A modification also exists in different types of RNAs, including ribosomal RNAs, small nuclear RNAs, and transfer RNAs [8–11]. Despite its widespread distribution in mammalian transcriptome, functional studies remain limited, possibly due to the low abundance of m6A mRNA and technical difficulties in global detection.
In mammalian cells, the m6A modification is catalyzed by a methyltransferase complex (“writers”) consisting of the proteins methyltransferase-like 3 (METTL3) and METTL14, which form a stable heterodimer complex [12, 13]. Then the mammalian splicing factor WTAP (Wilms tumor 1 associated protein) was identified as the third auxiliary factor of the core methyltransferase complex that affects cellular m6A methylation [12, 14]. Notably, the first RNA demethylase, fat mass and obesity-associated protein (FTO), was identified to function as an m6A “eraser” to selectively reverse m6A to adenosine in nuclear RNA, suggesting that RNA m6A modification is reversible [15]. Afterwards, the alkylation repair homolog protein 5 (ALKBH5) was proved to be another “eraser” of m6A modification [16, 17], indicating a dynamic nature of m6A methylation. Since then, the m6A-binding proteins with YTH domain, including cytoplasmic protein YTHDF1, YTHDF2, YTHDF3, and nuclear protein YTHDC1, have been identified to be the “readers” of m6A, which preferentially recognizes m6A-containing mRNA to modulate its stability and translation [7, 18–20].
Numerous studies have demonstrated that mRNA m6A modifications play important roles in various biological processes, such as heat-shock response [19], DNA damage response [21, 22], mRNA clearance [23], neuronal functions [24], cortical neurogenesis [25], progenitor cell specification [26], and T-cell homeostasis [27]. Moreover, the m6A modifications are commonly occurred in eukaryotes, which involved in cancer progression under multiple regulatory mechanisms [28]. As the first identified demethylase of mRNA m6A modification, FTO with single nucleotide polymorphism (SNP) is associated with cancer susceptibility and tumorigenesis [29–31]. In addition, the increasing evidences showed that overexpression of FTO in a variety of tumor tissues was highly correlated with the prognosis of tumors [32]. In vivo experiments showed that FTO promoted the invasion capability of AML cells and enhanced the cell transformation mediated by leukemia oncogenes [33]. Owing to the complicated regulatory mechanisms, the FTO targets and their definitive roles in cancer remained limited.
In this study, we investigated the role of m6A modification in the tumorigenesis of ESCC, and found a significant increased level of FTO in ESCC. In vitro assays showed that the epidermal growth factor receptor ERBB2 (Erb-B2 Receptor Tyrosine Kinase 2) [34] is a downstream target of FTO. Further studies demonstrated that FTO and ERBB2 act in concert to regulate the tumorigenesis and metastasis of ESCC. Our findings suggested that FTO mediated ERBB2 m6A modification might be applied to offer a new therapeutic strategy to treat ESCC.
Materials and methods
The ESCC specimens and cell lines
A total of 28 cases of ESCC and its paracancerous tissues with histologically confirmed ESCC were obtained by tumor surgical resection at Anhui Provincial Cancer Hospital, West Branch of the First Affiliated Hospital of University of Science and Technology of China (USTC). All experimental procedures were conducted by Biomedical Ethics Committee of USTC. All participants have signed informed consent, and their clinicopathological information is shown in Table S1 of the SI Appendix.
Human normal esophageal epithelial cell lines HEEC and esophageal squamous cell carcinoma (ESCC) cell lines KYSE140, KYSE180, KYSE450, KYSE30 and KYSE150 were acquired from China Cell Resource Center (Shanghai, China) and cultured in RPMI 1640 (BI) medium supplemented with 10% fetal bovine serum (PAN) at 37 °C and 5% CO2. All cell lines had been tested negative for mycoplasma.
Plasmids and transfection
Stable knockdown and overexpression of FTO and ERBB2 were achieved by lentivirus which were purchased from Hanbio (Shanghai, china) and carried short-hairpin RNA (shRNA) and open reading frames of the genes (ORF). The flag labeled YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2 plasmids were constructed by pcDNA3.1-3XFlag vector. TheYTHDF1 ORF was inserted into PCDH vector to construct YTHDF1 overexpression plasmid. The PLKO.1 plasmid carrying YTHDF1 short hairpin RNA (shRNA) was constructed to knockout YTHDF1expression. Small interfering RNA (siRNA) targeting ERBB2-specific regions were synthesized by Ruibo (Guangzhou, China). Transfections were carried out using the Lipofectamine2000 (Invitrogen, USA) following the manufacturer’s instructions. The siRNAs and shRNAs sequences were listed in Table S2 of the SI Appendix.
RNA extraction and real-time PCR
Total RNA was isolated from cell lines by TRIzol reagent (Vazyme). cDNA was synthesized using HiScript®II 1st Strand cDNA Synthesis Kit (Vazyme). The relative target gene mRNA expression was analyzed using the comparative CT method, with GAPDH as the endogenous control. All the qPCR primer sequences were shown in SI Appendix, Table S2.
Western blot assays
ESCC tissues, paracancerous tissues, cultured cells were lysed by lysis buffer (60 mM Tris–HCl, pH 6.8, 2% SDS, 20% Glycerol, 0.25% bromophenol blue and 1.25% 2-mercaptoethanol). The proteins were separated by SDS-PAGE and transferred to PVDF membrane (Millipore). The membrane was blocked in 5% skimmed milk for 1 h and then incubated overnight with specific antibodies at 4 °C, followed by treatment of horseradish peroxidase-conjugated secondary antibody (Proteintech). The signal bands were detected with ECL kit (Thermo). All antibodies used in this paper are from Proteintech. Detailed information regarding the full-length gels is depicted in SI Appendix, Figs. S2–S11.
Cell proliferation assays
A total of 3000 transfected cells were cultured into 96-well plates, the cells were added with CCK-8 solution (Bimake) at 0, 24, 48, 72, and 96 h after cell adherent according to the manufacturer’s instructions and incubated for additional 2 h. The absorbance values were measured at 450 nm by Universal Microplate Spectrophotometer.
Colony formation assays
Different treated cells were resuspended and seed into each well of the six-well plate with a density of 500 cells. After 2 weeks, the clones were fixed with alcohol and then stained with crystal violet, and the number of cells was counted.
In vitro migration and invasion assays
The treated cells in 200 μl serum-free medium (Cell density is 2.5 × 105 for migration assays, and 8 × 104 for invasion assays) were loaded in a 24-well transwell inserts upper compartment with an 8 μm diameter pores (Corning). Transwell chamber was covered with matrigel (BD) for invasion assay, but no matrix glue for migration assays. The medium of 600 μl containing 20% serum was added to the lower compartment. After 40 h, the non-migrating and non-invasive cells were scraped off, the migrating and invasive cells were fixed on the lower surface, stained with crystal violet solution, counted and photographed.
Methylated RNA immunoprecipitation sequencing (MeRIP-seq) and RNA-seq
Total RNA was first lyextracted from stably knockdown FTO KYSE150 cells and their corresponding controls. Then the mRNA was isolated from the total RNA by PolyATtract® mRNA Isolation Systems (Promega). M6A methylated mRNA fragments were enriched using immunomagnetic beads with m6A antibody. The enriched mRNA fragments were purified, constructed a high-throughput sequencing library using Illumina’s Nebnext Ultra RNA library preparation kit and sequenced by Illumina HiSeq 2000. At the same time, a common transcriptome library should be constructed separately as a control. Library preparation and high-throughput sequencing were performed by Novo gene. Reads Mapping, m6A Peak Calling, Motif Search, and Following analysis were performed as described previously [35].
Quantification of RNA m6A modification
RNA m6A methylation status in total RNAs was detected using the m6A RNA Methylation Quantitative Kit (Epigentek) according to the manufacturer’s instructions. In short, 200 ng RNA was bound to the strip wells. Then, m6A RNA capture antibody and detection antibody were added respectively, and the detection signal was enhanced with a signal enhancer. Finally, the absorbance value at 450 nm was recorded by the microplate reader, and the percentage of m6A in the total RNA was calculated.
Dot blot assays
The mRNA was isolated from the total RNA using PolyATtract®mRNA Isolation Systems (Promega) Kit and denaturated, fixed on a positively charged nylon membrane (Beyotime), and then sealed with 5% skimmed milk. The m6A antibody (Active motif) was incubated overnight at 4 °C, and then the membrane was cleaned and incubated at room temperature for 2 h with rabbit secondary antibody (Proteintech). The signal on the nylon membrane is captured by an Image Reader. Finally, the nylon membrane was stained with 0.2% methylene blue to determine whether the sample loading quantity was the same. Detailed information regarding the full-length gels is depicted in SI Appendix, Fig. S12.
RNA-binding protein immunoprecipitation (RIP) assays
RIP detection was performed using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Briefly, the cell lysates prepared were incubated with magnetic beads-complex containing positive control antibody (Millipore), negative control antibody IgG (Millipore), and Flag antibody at 4 °C for 3 h or overnight. Then the RNA–protein complex was incubated at 55 °C for 30 min in a buffer solution with protease K. Finally, the RNA was extracted with phenol/chloroform/isoamyl alcohol. The interaction between ERBB2 and m6A reader proteins was identified by PCR and normalized to the input. The ERBB2 primers for PCR are detailed in the Appendix SI, Table S2.
M6A-RNA immunoprecipitation(MeRIP) qPCR assay
The Magna MeRIP™ m6A Kit (Millipore) is used for quantitative analysis of RNA methylation levels of specific genes. Briefly, the mRNA was isolated and acquired from the total RNA using PolyA Ttract® mRNA Isolation Systems (Promega) Kit. Then, the mRNA was chemically segmented to a nucleotide fragment of about 100 bp and the mRNA containing m6A was enriched by immunoprecipitation reaction with m6A antibody. Finally, the enrichment of mRNA was detected by qPCR with m6A primers for ERBB2. The specific primers sequences for ERBB2 were shown in Table S2 of the SI Appendix [36, 55].
RNA pull-down assays
Lysates from 3 × 107 KYSE150 cells were collected and incubated with 4ug biotin-labeled sense or antisense ERBB2 probes. Followed by, after overnight at 4 °C of incubation, streptavidin magnetic beads were added to the reaction mix and incubated at 4 °C for 4 h. The magnetic beads were washed by pull-down buffer, added to loading buffer and boiled for western blot analysis.
RNA stability
In order to detect the mRNA stability of knock-down FTO cells, sh-FTO and control cells were implanted into 12-well plates overnight, and then treated with Actinomycetes D (ApexBio, 6 μg/ml) for corresponding time (0, 2, 4, 6, and 8 h) to block RNA transcription. Finally, RNA was extracted with Trizol and analyzed by qPCR. The mRNA expression level of each group at the specified time was calculated and normalized with β-actin.
Luciferase reporter assays
The wild-type or mutant PGL plasmid containing the 3′UTR fragment (in which adenosine at the m6A site was replaced by cytosine) of ERBB2 was transfected into the stable FTO knockdown KYSE150 with Lipofectamine 2000, respectively. Luciferase activity was detected using the dual luciferase reporting system kit (Promega) by Promega Glomax 20/20 Luminomete at 24 h after transfection, and was normalized with Renilla fluorescence. Detailed information regarding the primers used for plasmid construction is depicted in SI Appendix, Table S2.
In vivo xenograft and metastasis model
Female BALB/c nude mice (4-week-old) was purchased from Shanghai SLAC Experimental Animal Co., Ltd. For the subcutaneous transplantation model, sh-control, sh-FTO, NC-OE and ERBB2-OE KYSE150 stable cells (6 × 106 per mouse, n = 3 for each group) were diluted to 100 μl PBS + 100 μl Matrigel (BD) and subcutaneously injected to two points in the middle and upper groin of immunodeficient mice to study tumor growth. When the tumor volume reached ~ 1000 mm3 in each group, the nude mice were killed, and the tumors were excised and weighed for histology and further study. The tumor volume was calculated by the formula V = 1/2 × large diameter × (small diameter)2. For a model of lung metastasis in vivo, nude mice were injected with 100 μl WT (wide type), sh-FTO, ERBB2-OE and sh-FTO+ERBB2-OE KYSE150 stable cells (1 × 106 cells per mouse, n = 3 for each group) through tail vein, respectively. Six weeks after injection, the mice were killed and analyzed for metastatic lung tumors. All animal research procedures are carried out under a program approved by the Animal Laboratory Center of University of Science and Technology of China.
Immunohistochemistry assays (IHC)
Tissue arrays were constructed using 28 pairs of human ESCC and corresponding paracancerous tissues, as well as nude mouse specimens. Paraffin-embedded sections and immunohistochemical staining were performed on all specimens to detect the expression of FTO for human ESCC and paracancerous tissues, and the expression of Vimentin and E-cadherin, for nude mouse specimens. In short, the slides were incubated overnight with the appropriate antibody at 4 °C and the secondary antibody at room temperature for 2 h. Then, Subsequent steps were performed using the immunohistochemical kit (Beyotime, FD008) and peroxidase (Beyotime, P0202). The intensity of staining was the staining intensity was detected using the IMAGE-PRO PLUS 6.0 software (Media Cybernetics, Silver Spring, MD, USA). Specific steps: (1) finding and measuring the section of interest; (2) adjusting the optical density; (3) acquiring, converting and saving the image; (4) correcting the background and background staining; (5) configuring the section of interest to determine the light density; (6) Checking the optical density. The positive staining in images was quantified as integral optical density (IOD)/area, that is, mean density = density sum/area sum [36].The relevant clinical information was provided in Table S1.
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis (GEPIA) database was used to analyze the total expression level of FTO ESCC and normal esophageal epithelial samples. In addition, the correlation between YTHDF1 and ERBB2 expression was calculated. In addition, the protein–protein interaction among FTO and m6A writers was predicted using the STRING database.
Vector and m6A mutation assays
Based on MeRIP-seq and mRNA-seq data, the 3′-terminal untranslated region (3′UTR) of potential m6A site ERBB2 containing wild and mutated m6A motif was cloned into vector pGL3 for luciferase reporter gene analysis. The sequences was provided in SI Appendix, Table S2.
Statistical analysis
All values are expressed as mean ± SD from at least three independent trials. Statistical analysis was performed using GraphPad Prism 7 software. Two-tailed Student’s t-test, One-way ANOVA, and Two-way ANOVA were used to determine the statistical significance between the groups. Spearman analysis was used for the correlation between the two independent groups. The coefficient R > 0.3 indicates a moderate positive correlation. Survival curves were plotted using the Kaplan–Meier method with the log-rank test. p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001) was considered statistically significant.
Results
FTO was significantly up-regulated in ESCC and esophageal carcinoma cells
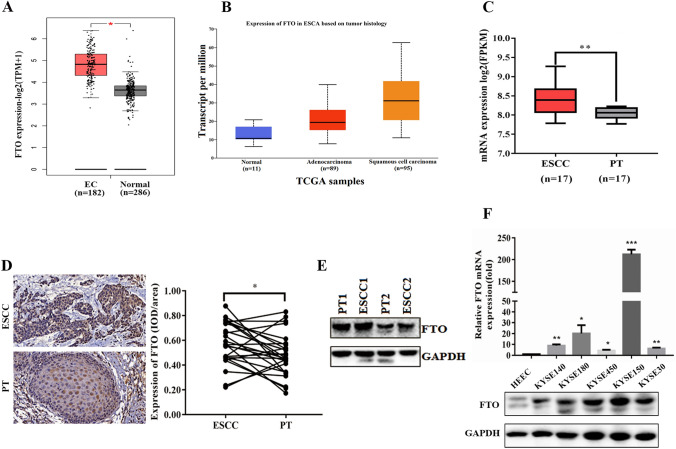
To explore the potential role of FTO in ESCC, we analyzed the expression level of FTO in ESCC patient tissues. Data from the GEPIA database showed that FTO was significantly upregulated in 182 esophageal cancer tissue samples compared to 286 normal esophageal epithelial tissue samples (Fig. 1A). To further identify whether FTO expression is associated with esophageal cancer histology, we analyzed The Cancer Genome Atlas (TCGA) data through the UALCAN database and found that FTO expression was significantly upregulated in 95 ESCC specimens compared to 11 normal esophageal epithelial cells. In addition, FTO expression was also up-regulated in 89 adenocarcinoma specimens, but without significant difference (Fig. 1B). In addition, ESCC has been reported to account for approximately 90% of esophageal cancers [37]. In the next step, FTO was also significantly upregulated in ESCC tissues compared to normal esophageal epithelium tissue based on GSE20347 dataset that microarray analysis of the gene expression profiles of 17 paired ESCC specimens and normal esophageal epithelium tissues (Fig. 1C).Consistently, the immunohistochemical (IHC) staining of the tissue arrays of 28 pairs of ESCC specimens and adjacent normal esophageal epithelial tissues also showed that the expression level of FTO was significantly higher than that of normal tissues (Fig. 1D). Furthermore, western blotting of FTO in two ESCC samples showed that FTO expression in primary tumor samples was significantly higher than that in the corresponding paracancerous tissues (Fig. 1E). Next, we examined FTO mRNA and protein levels in five ESCC cell lines, with the normal esophageal epithelial cell line (HEEC) as a control. The results showed that mRNA levels were significantly up-regulated in all five ESCC cells, with KYSE150 of the highest level (Fig. 1F). In addition, the protein levels were also greatly increased in each of the five ESCC cell compared to normal cells (HEECs) (Fig. 1F). All these results clearly indicated that FTO is highly expressed in ESCC patients, which may be a potential therapeutic biomarker for ESCC.
Fig. 1.
FTO was significantly up-regulated in ESCC and esophageal carcinoma cells. A FTO was significantly up-regulated in Esophageal carcinoma (EC) compared with normal Esophageal epithelial tissues from the GEPIA database data. B The expression levels (transcript per million) of FTO were analyzed in Esophageal adenocarcinoma (n = 89), Esophageal squamous cell carcinoma tissues (n = 95) and normal Esophageal epithelial tissues (n = 11) in the TCGA cohort. C The relative mRNA expression levels of FTO in 17 ESCC tissues and their adjacentnormal epithelial tissues (from the GSE20347 dataset) were analyzed. D Representative image of immunohistochemical staining by FTO in 400-fold magnified ESCC tissues and paired normal tissues in human samples (above).Immunohistochemical expression of FTO in ESCC tumor tissue and paired paracancerous tissues (n = 28) was quantitatively analyzed using IMAGE-PRO PLUS 6.0 software (below). E Western blotting assay of FTO expression in three paired in 2 pairs of ESCC primary tumor samples and their matched paracancerous tissues. F Real-time PCR (n = 3) and western blotting analysis of FTO expression in five ESCC cell lines and one normal esophageal cell line. Two-tailed Student’s t-test and one-way ANOVA were used to perform comparison between two groups and more groups, respectively. *p < 0.05,**p < 0.01; ***p < 0.001.
FTO involves in the cell proliferation, migration and invasion of ESCC
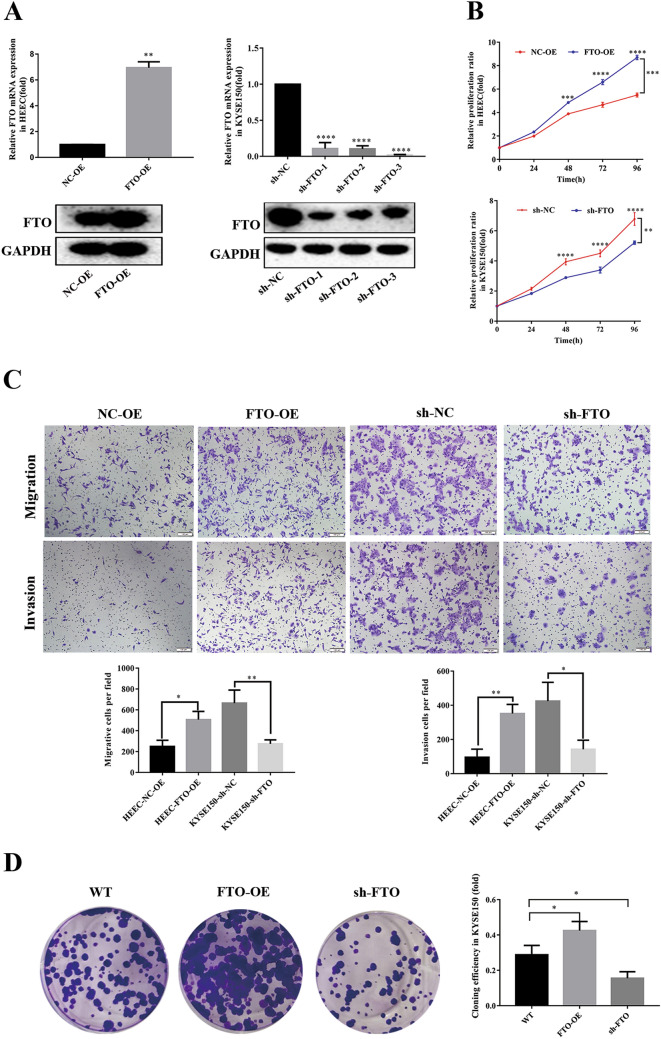
To investigate the potential roles of FTO in ESCC, we performed a series of in vitro functional assays. First, we attempted to up-regulate or down-regulate FTO expression in KYSE150 cells. The qRT-PCR and western blotting analysis showed a significantly higher expression of FTO in both mRNA and protein levels by transfecting FTO-overexpressing lentivirus in HEEC cells (Fig. 2A). Moreover, transfection of each of the three different shRNAs of FTO in KYSE150 dramatically decreased the FTO expression in both mRNA and protein levels (Fig. 2A). The following CCK-8 assays in 96 h showed that HEEC cells with FTO overexpression possess a higher cell proliferation capability compared to the control cells, whereas silence of FTO expression significantly decreased the cell proliferation ratio in KYSE150 cells (Fig. 2B).
Fig. 2.
FTO regulates ESCC cell proliferation and migration, invasion and colony formation. A The over-expression efficiency of FTO was identified at RNA and protein levels using PCR (top) and western blotting (bottom). The interference efficiency of FTO was identified at RNA and protein levels by PCR (top) and western blotting (bottom). B CCK-8 assay was used to analyze the proliferation efficiency of cells after interfering with the expression efficiency of FTO. Overexpression of FTO (FTO-OE) significantly increased the proliferation efficiency of HEEC cells, while inhibition of FTO (sh-FTO) significantly decreased the proliferation efficiency of KYSE150 cells. C Transwell assay was used to assay the invasion and migration ability of HEEC cells. Overexpression of FTO (FTO-OE) significantly increased the invasion and migration ability of HEEC cells, while inhibition of FTO (sh-FTO) significantly decreased the invasion and migration ability of KYSE150 cells. D Colony formation assay was used to analyze the clone formation ability of KYSE150 cells after inhibition or overexpression of FTO. Results were presented as means ± SD (n = 3 per group). The two-tailed Student’s t-test, one-way ANOVA and two-way ANOVA were used to perform comparison between two groups and more groups, respectively. *p < 0.05, ***p < 0.001
Afterwards, we performed the transwell assays to detect the invasion and migration capabilities upon up-regulating FTO in the normal HEEC cell or down-regulating FTO in the ESCC cell line KYSE150. The results showed that FTO overexpression in HEEC cells increased the cell migration and invasion capability compared to that of the control cells (Fig. 2C). In contrast, FTO down-regulation in KYSE150 cells inhibited the cell migration and invasion to a great extent (Fig. 2C).
Then, the colony formation assays were performed in KYSE150 cells to further compare the cell proliferation capability. As shown in Fig. 2D, more colonies were formed upon overexpression of FTO in KYSE150 cells, compared to the much less colonies formed in FTO-silenced KYSE150 cells (Fig. 2D). The results also confirmed that FTO overexpression promoted the ESCC cell proliferation. All these results indicated an oncogenic role of FTO, which involved in the cell proliferation, migration and invasion in ESCC.
FTO regulates the level of m6A modification in ESCC cells
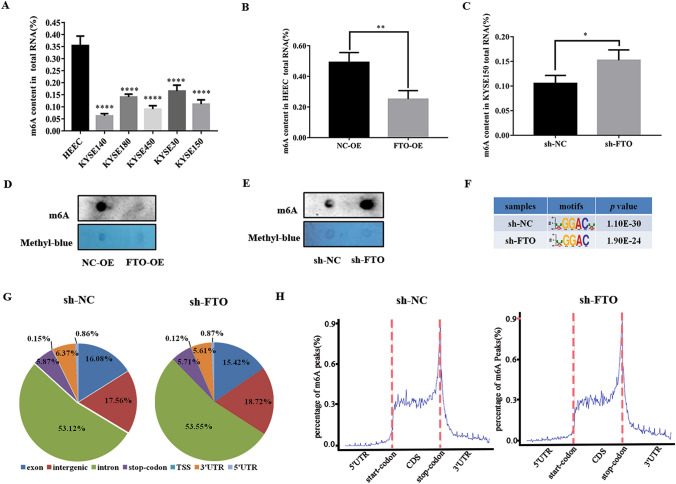
As FTO is the well-investigated mRNA m6A demethylase, we therefore further analyze the m6A modification in five ESCC cell lines and one normal esophageal epithelial cell. The m6A RNA methylation kit was used to quantitatively measure the relative content of m6A modification. As expected, these cell lines with a higher FTO expression have a much lower content of RNA m6A modification, ranging from 5 to 20%, compared to the control cells (Fig. 3A). To further test the role of FTO on RNA m6A modification, we compared the m6A content with the control cells upon either FTO overexpression in HEEC cells or silencing in KYSE150 cells. As expected, upon the up-regulation of FTO in HEEC cells, the content of m6A modification among the total RNA greatly decreased from ~ 50 to ~ 30% (Fig. 3B). In contrast, down-regulation of FTO in KYSE150 cells led to an elevated level of m6A modification from ~ 10 to ~ 15% (Fig. 3C). In addition, the dot blot assays also showed a similar effect on the m6A content upon FTO overexpression in HEEC cells or FTO knockdown in KYSE150 cells (Fig. 3D, E). Sequence analysis of the m6A-seq data revealed the predominant consensus motif GGAC in KYSE150 cells (Fig. 3F). These results clearly showed that FTO indeed functions as a demethylase to remove mRNA m6A modification in ESCC cells.
Fig. 3.
FTO regulates the level of m6A modification in ESCC cells. A The content of m6A in total mRNA in five ESCC cell lines and normal esophageal epithelial cell (HEEC) was determined by the m6A RNA methylation quantitative kit. B, C The RNA m6A content was determined with the m6A methylation kit after overexpression FTO (FTO-OE) in HEEC (B) and FTO-knockdown (sh-FTO) in KYSE150 cells (C). D, E Dot blot assay was used to analyze the methylation of m6A RNA after overexpression FTO (FTO-OE) in HEEC (D) and FTO-knockdown (sh-FTO) in KYSE150 cells (E). F Predominant consensus motif GGAC was detected in both the control and sh-FTO KYSE150 cells in m6A-seq. G Proportion of m6A peak distribution in the exon, intergenic, intron, stop-codon, transcription start site (TSS), 3′-untranslated region (3′UTR), 5′-untranslated region (5′UTR) across the entire set of mRNA transcripts. H Density distribution of m6A peaks across mRNA transcripts. Regions of the 5′UTR, coding region (CDS), and 3′UTR were split into 100 segments, the percentage of m6A peaks in each segment was determined. The data was presented as the mean ± SDs (n = 3). The two-tailed Student’s t-test and one-way ANOVA were used to perform comparison between two groups and more groups, respectively. *p < 0.05,**p < 0.01; ****p < 0.0001
Next, we analyzed the m6A peak distribution in the different regions of the mRNA transcripts in the KYSE150 cells. The results showed that the m6A peaks are mostly enriched in the intron (> 50%), followed with a large proportion present in the intergenic (~ 18%) and exon (~ 16%) regions (Fig. 3G). In contrast, there are only minor proportions of m6A peaks in the stop-codon, transcription start site (TSS), 3′-untranslated region (3′UTR), or 5′-untranslated region (5′UTR) (Fig. 3G). Notably, no significant difference was found in the KYSE150 cells with or without FTO knockdown, thus FTO did not alter the m6A distribution in the mRNA transcripts. We then analyzed the density distribution of m6A peaks across mRNA transcripts by splitting the corresponding regions into 100 segments. The data showed that the m6A modification mainly distributed in the coding region of mRNA transcripts, with the highest peak near by the stop-codon region (Fig. 3H).
ERBB2 is the target of FTO in ESCC cells
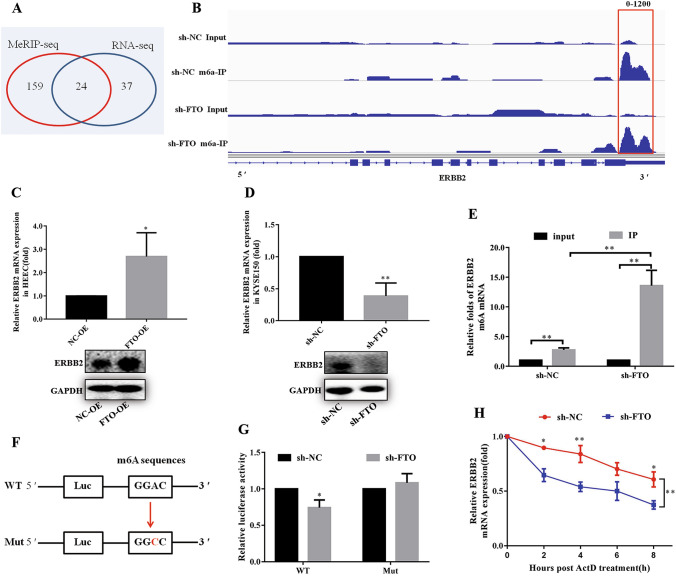
To identify the molecular mechanism of FTO-regulated ESCC metastasis, we attempted to identify the potential targets of FTO in ESCC by analyzing the m6A contents in total mRNA using MeRIP-seq and RNA-seq in FTO-silenced KYSE150 cells and the control cells (Fig. 4A). The MeRIP-seq identified 183 genes (fold change > 2, p < 0.05) with m6A peak enriched in the stop codon and 3′UTR regions after FTO knockdown (Fig. 4A; Table S1). In addition, the RNA-seq data showed that totally 61 transcripts were markedly changed upon FTO knockdown (Fig. 4A; Table S2). Notably, 24 transcripts were shared in RNA-seq and MeRIP-seq data, in which 12 transcripts were up-regulated, whereas the other 12 transcripts were down-regulated (Fig. 4A).
Fig. 4.
ERBB2 was identified as a downstream target of FTO. A Venn diagram showing identified differentially expressed genes using MeRIP-seq and RNA-seq in FTO stable knockdown KYSE150 cell compared with corresponding control. B m6A peaks were enriched in stop-codon and 3′UTRs of ERBB2 genes from m6A RIP-seq data. Squares marked increases of m6A peaks in ESCC cells with stable FTO down regulation compared with control cells. C, D The mRNA and protein levels of ERBB2 in FTO up-regulated cell (C) and down-regulated (D) cell were detected by q-PCR and western blot. D The mRNA and protein levels of ERBB2 in FTO down-regulated cells were detected by q-PCR and western blot. E m6A RIP-qPCR analysis of ERBB2 mRNA in the control and knockdown FTO cells. F Schematic representation that ERBB2 3′UTR wild (GGAC) and mutated (GGCC) were inserted into PGL vector. G Luciferase assay showed that knockdown of FTO repressed the expression of wide-type ERBB2 reporter. H The mRNA level of ERBB2 in KYSE150 cells with FTO knockdown and then treated with Actinomycete D (6 μg/ml) in 0, 2, 4, 6, and 8 h measured by real-time PCR. The data was presented as the mean ± SD (n = 3). The two-tailed Student’s t-test, one-way ANOVA and two-way ANOVA were used to perform comparison between two groups and more groups, respectively. *p < 0.05, **p < 0.01
Among the 12 shared down-regulated transcripts, we selected the top seven most differentially expressed members and detected their expression in KYSE150 cells by real-time PCR. The results showed that METTL1, CAMKK1, PKN2, and TGFBR1 were upregulated in the FTO-silenced KYSE150 cells, whereas ERBB2, AKT3 and PAK1P1 were downregulated. We selected the ERBB2 gene for further studies, which encodes the epidermal growth factor receptor that was previously found to be involved in the tumorigenesis of many different types of cancers [34, 38, 39]. The m6A RIP-seq data showed that the m6A peaks were enriched in the stop-codon and 3′UTRs of ERBB2 transcript in KYSE150 cells with or without FTO knockdown. Compared to the control, m6A methylation was up-regulated in the vicinity of the stop codon of ERBB2 after FTO knockdown (Fig. 4B).
To further investigate the correlation between ERBB2 and FTO, we detected the expression level of ERBB2 in FTO-overexpressed HEEC cells or FTO-silenced KYSE150 cells. The q-PCR and western blot analysis showed that accompanied with the up-regulation of FTO in HEEC cells, expression of ERBB2 is also significantly increased in both mRNA and protein levels (Fig. 4C). In contrast, silence of FTO in KYSE150 cells led to a dramatic decrease of ERBB2 in both mRNA and protein levels (Fig. 4D). The following m6A RIP-qPCR analysis showed that FTO down-regulation results in a higher content of m6A modification of ERBB2 transcript compared to the control cells. These results indicated that the demethylation of ERBB2 m6A might be catalyzed by FTO.
Therefore, we generated luciferase reporters containing a firefly luciferase and 3′UTR regions of wild-typeERBB2 or mutant (Fig. 4F). The results showed that knockdown of FTO in KYSE150 cells repressed the expression of wild-type ERBB2 reporter, whereas the mutant has a minor effect on ERBB2 expression (Fig. 4G). The results indicated that FTO demethylates the m6A modification of ERBB2 mRNA, which might affect its stability. To further prove this, we tested the ERBB2 mRNA levels in KYSE150 cells with FTO knockdown after the treatment with Actinomycete D, which is metabolic inhibitor [40, 41]. The results showed that the mRNA level is dramatically decreased along the time with FTO knockdown (Fig. 4H), indicating that FTO might increase the stability of ERBB2 mRNA, probably via regulating the m6A modification of ERBB2 mRNA.
The inhibition effects of loss FTO are reversed by overexpression of ERBB2
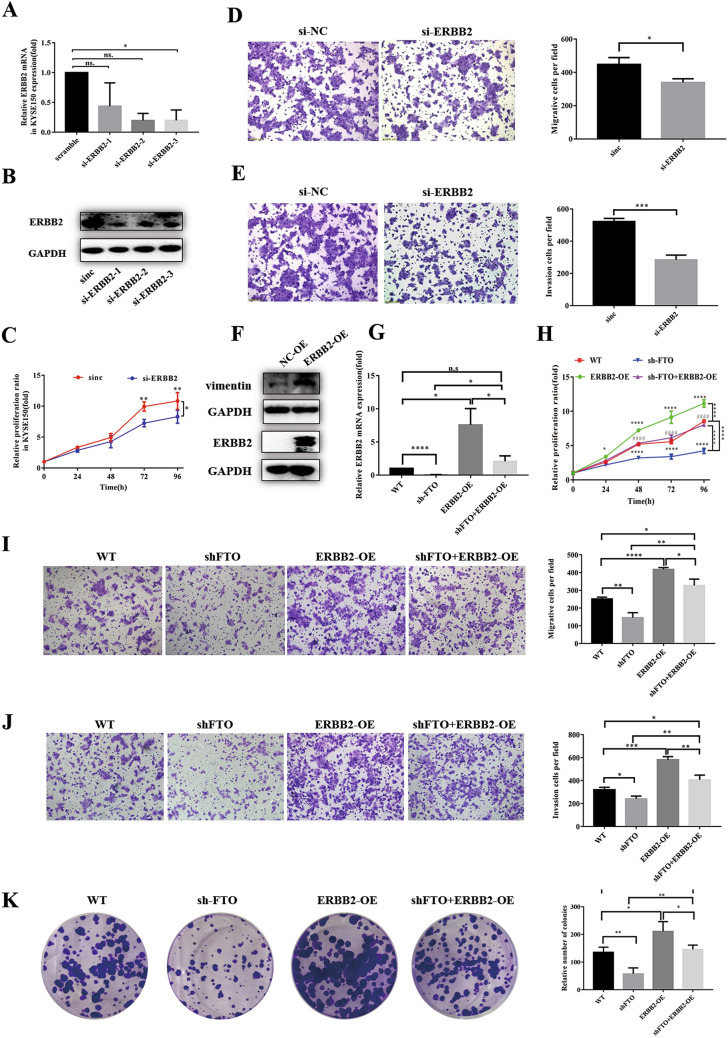
We then characterized the roles of ERBB2 on ESCC cellular functions by several in vitro experiments. We first down-regulated the ERBB2 expression in KYSE150 cells via individually transfecting three ERBB2 siRNAs. The following qRT-PCR and western blot analysis showed that either of the three siRNAs could significantly inhibit the ERBB2 expression in both mRNA and protein levels (Fig. 5A, B). Accompanied with the decrease of ERBB2 level by si-ERBB2 transfection in KYSE150 cells, the cell proliferation ratio is significantly lower than that of the control cells (Fig. 5C). Moreover, the invasion and migration capabilities of ERBB2-slienced KYSE150 cells were largely decreased compared to the control cells (Fig. 5D, E). Notably, overexpression of ERBB2 significantly increased the expression of Vimentin (Fig. 5F), which is a major constituent of the intermediate filament family of proteins, and is known to maintain cellular integrity and provide resistance against stress. In addition, Vimentin is overexpressed in various epithelial cancers, including prostate cancer, gastrointestinal tumors, tumors of the central nervous system, breast cancer, malignant melanoma, and lung cancer [42–44], indicating that ERBB2 promotes ESCC cell migration. All these results indicated that ERBB2 involves in the tumorigenesis of ESCC progression.
Fig. 5.
ERBB2 regulates the invasion and metastasis of ESCC cells and reverses biological effects of FTO. A, B RT-PCR and western blot was used to analyze the mRNA and protein level of ERBB2 in KYSE150 cells. C The CCK-8 assays showed that down-regulation of ERBB2 could inhibit the proliferation of KYSE150 cells. D, E Transwell assay was used to analyze the ability of down-regulation of ERBB2 to inhibit KYSE150 cell invasion and migration. The bar chart shows the corresponding quantitative analysis (right). F Western blot experiment verified the efficiency of the up-regulation ERBB2 expression and the promotion of EMT by the up-regulation ERBB2 expression. G The up-regulation of ERBB2 expression restored the down-regulation of FTO to the inhibition of ERBB2 expression using RT-PCR. H CCK-8 assay confirmed that up-regulation ERBB2 expression restored down-regulated FTO to KYSE150 cell proliferation inhibition. I, K and H Migration (I) invasion (K) and colony formation (H) assays showed that up-regulation of ERBB2 expression restored down-regulated FTO to KYSE150 cell migration, invasion and proliferation inhibition. The bar chart shows the corresponding quantitative analysis (right). The data was presented as the mean ± SD (n = 3). The two-tailed Student’s t-test, one-way ANOVA and two-way ANOVA were used to perform comparison between two groups and more groups, respectively. n.s no statistical significance. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
To further investigate the correlation between FTO and ERBB2 on ESCC tumorigenesis, we overexpressed ERBB2 in KYSE150 cells accompanied with the slience of FTO. As shown in Fig. 5G, compared to the transfection of ERBB2-overexpressed vector (ERBB2-OE) that increased the ERBB2 mRNA expression to ~ tenfold, the ERBB2 expression in FTO-silenced KYSE150 cells with transfection of ERBB2-overexpressed vector (sh-FTO+ERBB2-OE) increased to ~ 2.5-fold, which largely compromises the effect of ERBB2 up-regulation. It also coincides with the notion that FTO-regulated m6A demethylation promotes the ERBB2 mRNA stability (Fig. 4H). As a result, the ERBB2 protein levels are up-regulated in the ERBB2-OE and sh-FTO+ERBB2-OE KYSE150 cells (Fig. 5G), but is much lower in the sh-FTO KYSE150 cells. To test whether ERBB2 could restore the effect of FTO knockdown, we tested the proliferation ratio of KYSE150 cells using CCK-8 assays. The results showed that FTO knockdown decreased the proliferation ratio, which could be restored by ERBB2 overexpression in the sh-FTO+ERBB2-OE cells (Fig. 5H). Similarly, the migration, invasion and colony formation assays showed that the effects of FTO knockdown in KYSE150 cells could also be restored by ERBB2 overexpression (Fig. 5I–K). These results suggested that FTO-regulated m6A demethylation on ERBB2 is associated with the tumorigenesis and metastasis in ESCC cells.
YTHDF1 stabilizes ERBB2 mRNA via recognizing the m6A modification
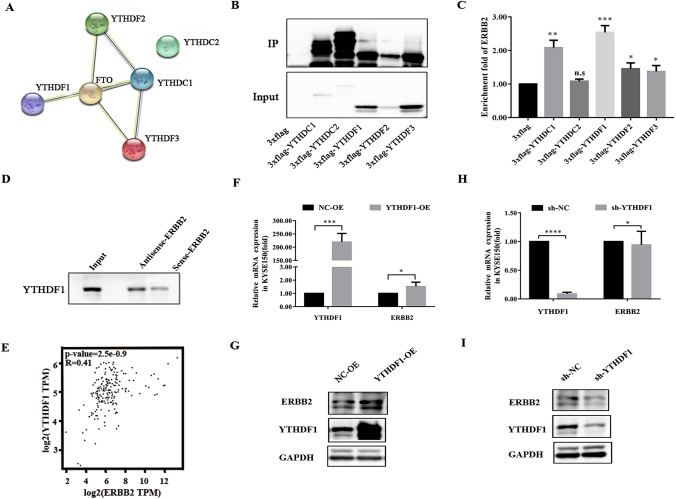
Previous studies had identified two major families of m6A “readers”, including the YTH family and the IGF2BP family, which might play roles in regulating the fate of methylated mRNA [45–47]. The STRING database indicated that FTO has potential interactions with the m6A readers of YTHDC1, YTHDF1, YTHDF2 and YTHDF3, but not YTHDC2 (Fig. 6A). To further identify the specific m6A readers of FTO, we performed the FLAG RNA pull-down assays in KYSE150 cells to screen the ERBB2-related m6A readers. Notably, ERBB2 was enriched by all members in the YTH family, with YTHDF1 of the greatest extent (Fig. 6B, C). Indeed, the biotin-based pull-down assays confirmed the direct interactions of ERBB2 mRNA with YTHDF1 (Fig. 6D). Furthermore, the bioinformatics analysis from the GEPIA database also indicated YTHDF1 negatively regulate the expression of ERBB2 (Fig. 6E).
Fig. 6.
YTHDF1 preferentially decoded the m6A residue of ERBB2. A Protein–protein interaction network between FTO and m6A readers was analyzed using the STRING database. B, C Western blot (the left) and RIP-PCR (the right) were used to analyze the enrichment of 5 m6A recognition proteins in KYSE150 cells for ERBB2. The results showed that all proteins enriched the expression of ERBB2, however, ERBB2 was enriched to the greatest extent by YTHDF1. D The biotin-based pull down assays were used to further analyze the enrichment of YTHDF1 by ERBB2. The results showed a direct interaction between ERBB2 mRNA and YTHDF1. E The correlation between the expression of YTHDF1 and ERBB2 in ESCC was analyzed using GEPIA database and the results showed positive correlation. F, G The real-time PCR and the western blot analyzed showed that the expression of YTHDF1 and ERBB2 were detected in overexpression YTHDF1 (YTHDF1-OE) KYSE150 cells. H, I The real-time PCR and the western blot analyzed showed that the expression of YTHDF1 and ERBB2 were detected in knock-down YTHDF1 (sh-YTHDF1) KYSE150 cells. The result that The ERBB2 expression is related to YTHDF1 positively. The data in C, F and H was presented as the mean ± SD (n = 3). The two-tailed Student’s t-test and one-way ANOVA were used to perform comparison between two groups and more groups, respectively. n.s no statistical significance. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
To further test the role of YTHDF1 on ERBB2 expression and stability, we inhibited or increased the expression of YTHDF1 in KYSE150 cells, and detected the ERBB2 mRNA and protein levels. Accompanied with the up-regulated levels of YTHDF1 in KYSE150 cells, the ERBB2 mRNA and protein levels were significantly up-regulated (Fig. 6F, G). In contrast, ERBB2 mRNA and protein levels were somewhat down-regulated when YTHDF1 was silenced in KYSE150 cells (Fig. 6H, I). Taken together, our results suggested that the methylated ERBB2 transcripts might directly recognized by YTHDF1, which maintained the stability of the ERBB2 transcripts.
FTO and ERBB2 promote in vivo ESCC progression
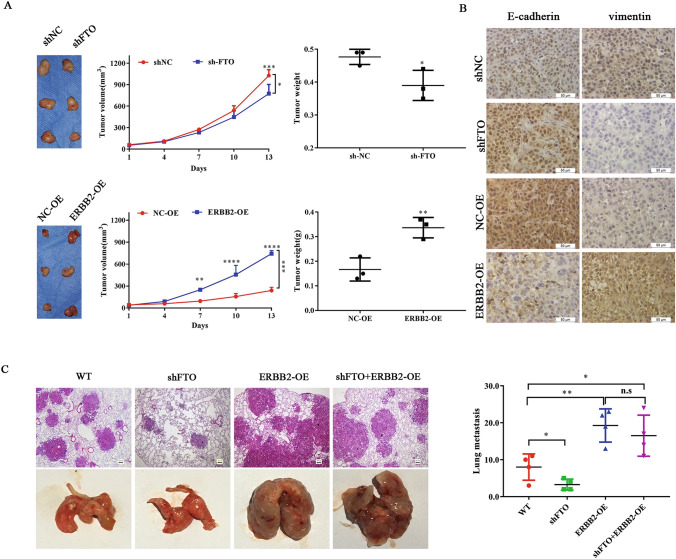
To test the potential role of FTO and ERBB2 on ESCC biogenesis in vivo, we injected the sh-FTO or ERBB2-OE KYSE150 cells subcutaneously into the nude mice. Compared to the control groups, FTO knockdown in KYSE150 cells effectively inhibited the tumor formation and growth (Fig. 7A). In contrast, overexpression of ERBB2 in KYSE150 cells enhanced the tumor growth (Fig. 7A). Further immunohistochemistry assays showed that FTO knockdown inhibited the expression of Vimentin and E-cadherin, indicating an inhibiting effect of tumor metastasis, whereas ERBB2 has a reverse effect (Fig. 7B). To further determine the impacts of m6A methylation on in vivo metastasis, sh-FTO, ERBB2-OE or sh-FTO&ERBB2-OE KYSE150 cells were injected into the nude mice, respectively. As shown in Fig. 7C, the lung metastasis was significantly inhibited upon injection of FTO silenced KYSE150 cells, accompanied with a decreased number of lung tumors. However, ERBB2-OE or sh-FTO & ERBB2-OE significantly promoted the lung metastasis and increased number of lung tumors compared with control cells (Fig. 7C). These results suggested that ERBB2 overexpression promoted tumor metastasis in vivo, which also confirmed the roles of FTO and ERBB2 involved in ESCC progression.
Fig. 7.
FTO and ERBB2 regulates in vivo ESCC progression. A The effects of FTO and ERBB2 on tumor formation, tumor weight and tumor volume change in nude mouse KYSE150-derived xenograft model. Representative images of tumors from FTO knockdown (sh-FTO) and up regulation expression ERBB2 (ERBB2-OE) versus the negative control sh-NC and NC-OE, respectively (n = 3 for each group).FTO knockdown effectively decreased ESCC subcutaneous tumor formation and growth. Overexpression of ERBB2 effectively increased ESCC subcutaneous tumor formation and growth. B The expression of Vimentin and E-cadherin, were analyzed from sh-NC, sh-FTO, NC-OE and ERBB2-OE KYSE150-derived xenograft by IHC. C Wide type (WT), FTO knockdown (sh-FTO), stable up regulation expression ERBB2 (ERBB2-OE) and FTO knockdown simultaneously stable up regulation expression ERBB2 (sh-FTO+ERBB2-OE) from KYSE150 cells were injected into the nude mice by tail vein injection. Representative images of metastatic lung tumors and the H&E staining results were shown (left), and the number of lung tumors was quantitatively analyzed (right). Results were presented as means ± SD (n = 4 per group). The two-tailed Student’s t-test and one-way ANOVA were used to compare the difference between two groups and more groups, respectively. n.s no statistical significance. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Discussion
In this study, we uncovered a significant oncogenic role form6A modification in the tumorigenesis of ESCC. In brief, the decreased levels of mRNA m6A modification in ESCC is correlated with a higher level of FTO. A reversal change of FTO levels largely affects the in vitro proliferation, migration, and invasion of ESCC cells. Further investigations identified that ERBB2, an epidermal growth factor (EGF) receptor which belongs to subclass 1 of the receptor tyrosine kinase (RTK) superfamily [34, 48], is one of the targets of FTO. As found previously, ERBB2 was implicated in the development of human cancers [49]. For example, the ERBB2 alterations have been identified as oncogenic drivers and potential therapeutic targets in lung cancers [50, 51]. Here we showed that ERBB2 is also involved in the progression of ESCC, which provide the basis for further targeting ERBB2 pathway for clinical therapy of ESCC.
To date, the knowledge on the roles of mRNA modification in regulating cancer progression remains limited. As the first characterized m6A demethylase, FTO has been reported to regulate the tumorigenesis in different types of cancers. FTO was found to enhance leukemic oncogene-mediated cell transformation and leukemogenesis via reducing the m6A levels of its targets [33]. In addition, pharmaceutical inhibition of FTO by a chemical inhibitor suppresses tumor progression and substantially prolongs the lifespan of glioblastoma stem cell-grafted mice [52]. In this study, we provided the direct evidence of FTO in regulating the m6A methylation of ERBB2 mRNA. We found that FTO regulates multiple aspects of ESCC progression, including migration, invasion, proliferation, and tumorigenesis. Our results emphasized the roles of FTO-regulated m6A modification in cancer progression, and also provided hints to develop therapeutic strategies against ESCC metastasis by targeting m6A modification and its related targets.
The m6A modification modulates almost all stages in the life cycle of RNA, such as RNA processing, nuclear export, and translation modulation [20, 53]. Previous studies showed that the reader protein YTHDF1 associates with the progression of various cancers, including non-small cell lung cancer [54, 55], and ovarian cancer [56]. In our study, we found that knockdown of YTHDF1 decreased the ERBB2 expression in both mRNA and protein levels (Fig. 6), whereas YTHDF1 overexpression significantly enhanced the ERBB2 mRNA and protein levels. These data support the notion that YTHDF1 directly targets ERBB2, which enhanced the stability of ERBB2 mRNA transcripts. The results also indicated that YTHDF1 might regulate the transcription and translation of ERBB2. However, more investigations are needed to elucidate the fine mechanism of YTHDF1-regulated ERBB2 functions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Prof. YongLiang Jiang on proofreading the manuscript and the staff who participated in this study.
Abbreviations
- ESCC
Esophageal squamous cell carcinoma
- m6A
N6-methyladenosine
- MeRIP-seq
Methylated RNA immunoprecipitation sequencing
- RNA-seq
Transcriptomic RNA sequencing
- FTO
Fat-mass and obesity-associated protein
- ERBB2
Erb-B2 receptor tyrosine kinase 2
- YTHDF1
YTH N6-methyladenosine RNA binding protein 1
- YTHDF2
YTH N6-methyladenosine RNA binding protein 2
- IGF2BP1
Insulin like growth factor 2 mRNA binding protein 1
- IGF2BP2
Insulin like growth factor 2 mRNA binding protein 2
- ActD
Actinomycetes D
- E-cad
E-cadherin
- VIM
Vimentin
- WT
Wide-type
- IHC
Immunohistochemical
- RT-qPCR
Quantitative real-time PCR
- RIP
RNA Immunoprecipitation
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- 3′UTR
Three prime untranslated region
- CDS
Coding sequence
- TCGA
The Cancer Genome Atlas
Author contributions
YAT and XCZ carried out the structure of experiments and revised the manuscript; FFZ, FFG and MHX drafted initial manuscript; MHX, YAT, ZYL, CBZ, LSK, YGP conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities granted to FFZ (WK9110000008), XCZ (WK9110000136), LSK (WK9110000090) and CBZ (WK9110000132), respectively. The Youth Fund of Anhui Cancer Hospital granted to FFZ and LSK, respectively. The Youth Technical Backbone Fund of West Branch of the First Affiliated Hospital of USTC granted to CBZ and LSK, respectively. And the Bethune Medical Science Research Foundation (B19393ET) granted to XCZ.
Data availability
Not applicable.
Material availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethical approval
The animal study proposal was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Science and Technology of China (2021-N(A)-139). All of the mouse experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fangfang Zhao, Fangfang Ge, and Minghua Xie have contributed equally to this work.
Contributor Information
Xucai Zheng, Email: zhengxucai@ustc.edu.cn.
Yiao Tan, Email: tanyiaoa@ustc.edu.cn.
References
- 1.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6(12):863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 2.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavi S, Shatkin AJ. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc Natl Acad Sci USA. 1975;72(6):2012–2016. doi: 10.1073/pnas.72.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977;115(4):695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 5.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 6.Holzer H. Chemistry and biology of macromolecular inhibitors from yeast acting on proteinases A and B, and carboxypeptidase Y. Adv Enzyme Regul. 1975;13:125–134. doi: 10.1016/0065-2571(75)90011-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-Methyladenosine modulates Messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77(1–2):22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 9.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/S0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 10.Shimba S, Bokar JA, Rottman F, Reddy R. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 1995;23(13):2421–2426. doi: 10.1093/nar/23.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366(1):1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG, et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15(1):88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bland RD, Clarke TL, Harden LB. Rapid infusion of sodium bicarbonate and albumin into high-risk premature infants soon after birth: a controlled, prospective trial. Am J Obstet Gynecol. 1976;124(3):263–267. doi: 10.1016/0002-9378(76)90154-X. [DOI] [PubMed] [Google Scholar]
- 17.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. Corrigendum: RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;552(7685):430. doi: 10.1038/nature24007. [DOI] [PubMed] [Google Scholar]
- 22.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543(7646):573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature. 2016;540(7632):242–247. doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 25.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 2017;171(4):877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 27.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, Bailis W, Cao G, Kroehling L, Chen Y, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng X, Su R, Feng X, Wei M, Chen J. Role of N(6)-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soderberg KC, Kaprio J, Verkasalo PK, Pukkala E, Koskenvuo M, Lundqvist E, Feychting M. Overweight, obesity and risk of haematological malignancies: a cohort study of Swedish and Finnish twins. Eur J Cancer. 2009;45(7):1232–1238. doi: 10.1016/j.ejca.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Chen Q, Wang L, Ke D, Yuan Z. Association between FTO gene polymorphism and cancer risk: evidence from 16,277 cases and 31,153 controls. Tumour Biol J Int Soc Oncodev Biol Med. 2012;33(4):1237–1243. doi: 10.1007/s13277-012-0372-9. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez-Caballero ME, Sierra-Ramirez JA. Single nucleotide polymorphisms of the FTO gene and cancer risk: an overview. Mol Biol Rep. 2015;42(3):699–704. doi: 10.1007/s11033-014-3817-y. [DOI] [PubMed] [Google Scholar]
- 32.Zheng QK, Ma C, Ullah I, Hu K, Ma RJ, Zhang N, Sun ZG. Roles of N6-methyladenosine demethylase FTO in malignant tumors progression. Onco Targets Ther. 2021;14:4837–4846. doi: 10.2147/OTT.S329232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19(13):3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng J, Lu Z, Liu H, Zhang L, Zhang S, Chen Y, Rao MK, Huang Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69(3):274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang CJ, Zhou ZG, Holmqvist A, Zhang H, Li Y, Adell G, Sun XF. Survivin expression quantified by Image Pro-Plus compared with visual assessment. Appl Immunohistochem Mol Morphol AIMM. 2009;17(6):530–535. doi: 10.1097/PAI.0b013e3181a13bf2. [DOI] [PubMed] [Google Scholar]
- 37.Zeng H, Zheng R, Zhang S, Zuo T, Xia C, Zou X, Chen W. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thoracic Cancer. 2016;7(2):232–237. doi: 10.1111/1759-7714.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ursini-Siegel J, Schade B, Cardiff RD, Muller WJ. Insights from transgenic mouse models of ERBB2-induced breast cancer. Nat Rev Cancer. 2007;7(5):389–397. doi: 10.1038/nrc2127. [DOI] [PubMed] [Google Scholar]
- 39.Lassus H, Leminen A, Vayrynen A, Cheng G, Gustafsson JA, Isola J, Butzow R. ERBB2 amplification is superior to protein expression status in predicting patient outcome in serous ovarian carcinoma. Gynecol Oncol. 2004;92(1):31–39. doi: 10.1016/j.ygyno.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, Luo G, Tauler J, Du J, Lin S, et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Dashti Y, Grkovic T, Abdelmohsen UR, Hentschel U, Quinn RJ. Actinomycete metabolome induction/suppression with N-acetylglucosamine. J Nat Prod. 2017;80(4):828–836. doi: 10.1021/acs.jnatprod.6b00673. [DOI] [PubMed] [Google Scholar]
- 42.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci CMLS. 2011;68(18):3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strouhalova K, Prechova M, Gandalovicova A, Brabek J, Gregor M, Rosel D. Vimentin intermediate filaments as potential target for cancer treatment. Cancers. 2020;12(1):184. doi: 10.3390/cancers12010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson AM, Havel LS, Koyen AE, Konen JM, Shupe J, Wiles WGT, Martin WD, Grossniklaus HE, Sica G, Gilbert-Ross M, et al. Vimentin is required for lung adenocarcinoma metastasis via heterotypic tumor cell-cancer-associated fibroblast interactions during collective invasion. Clin Cancer Res Off J Am Assoc Cancer Res. 2018;24(2):420–432. doi: 10.1158/1078-0432.CCR-17-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silen W, Machen TE, Forte JG. Acid-base balance in amphibian gastric mucosa. Am J Physiol. 1975;229(3):721–730. doi: 10.1152/ajplegacy.1975.229.3.721. [DOI] [PubMed] [Google Scholar]
- 46.Kidder GW, Montgomery CW. Oxygenation of frog gastric mucosa in vitro. Am J Physiol. 1975;229(6):1510–1513. doi: 10.1152/ajplegacy.1975.229.6.1510. [DOI] [PubMed] [Google Scholar]
- 47.Poole-Wilson PA, Langer GA. Effect of pH on ionic exchange and function in rat and rabbit myocardium. Am J Physiol. 1975;229(3):570–581. doi: 10.1152/ajplegacy.1975.229.3.570. [DOI] [PubMed] [Google Scholar]
- 48.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 49.Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2005;23(11):2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 50.Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, Paik PK, Zakowski MF, Kris MG, Ladanyi M. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res Off J Am Assoc Cancer Res. 2012;18(18):4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Correction A. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2018;559(7715):E12. doi: 10.1038/s41586-018-0228-6. [DOI] [PubMed] [Google Scholar]
- 52.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, Shen Q, Xu P, Zeng L, Zhou Y, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10(1):4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.
Not applicable.