Abstract
The pandemic caused by severe acute respiratory Coronavirus-2 (SARS-CoV-2) has been ongoing for over two years, and treatment for COVID-19, other than monoclonal antibodies, is urgently required. Accordingly, we have investigated the inhibitors of SARS-CoV-2 protein targets by high-throughput virtual screening using a marine natural products database. Considering the calculated molecular properties and availability of the compounds, (+)-usnic acid was selected as a suitable hit. In the in vitro antiviral assay of (+)-usnic acid by the immunofluorescence method, IC50 was 7.99 μM, which is similar to that of remdesivir used as a positive control. The generalized Born and surface area continuum solvation (MM/GBSA) method was performed to find the potent target of (+)-usnic acid, and the Mpro protein showed the most prominent value, −52.05 kcal/mol, among other SARS-CoV-2 protein targets. Thereafter, RMSD and protein–ligand interactions were profiled using molecular dynamics (MD) simulations. Sodium usnate (NaU) improved in vitro assay results with an IC50 of 5.33 μM and a selectivity index (SI) of 9.38. Additionally, when (+)-usnic acid was assayed against SARS-CoV-2 variants, it showed enhanced efficacy toward beta variants with an IC50 of 2.92 μM and SI of 11.1. We report the in vitro anti-SARS-CoV-2 efficacy of (+)-usnic acid in this study and propose that it has the potential to be developed as a COVID-19 treatment if its in vivo efficacy has been confirmed.
Subject terms: Infectious diseases, Computational chemistry, Drug discovery, Virtual screening
Introduction
The outbreak of the pandemic coronavirus, SARS-CoV-2, in 2019 has enormously changed our way of living. SARS-CoV-2 causes COVID-19, which is a respiratory disease1. Because of the rapid spread of the virus and the lack of effective medication, SARS-CoV-2 has infected 500 million people worldwide with a mortality of approximately 6 million2. It is the third most pathogenic human coronavirus, which causes severe infection3,4.
SARS-CoV-2 is a single-stranded RNA virus with a genome comprising 14 open reading frames (ORFs) that encode four structural proteins, 16 nonstructural proteins (NSPs), and nine accessory proteins5. The structural proteins include spike (S), membrane (M), envelope (E) and nucleocapsid (N). The S glycoprotein is the main factor for SARS-CoV-2 entry into host cells. The binding of the S protein to the human ACE2 receptor cleaves the S into S1 and S2 subunits using proteases, and afterward, they are internalized through either clathrin-mediated endocytosis or plasma membrane fusion6–8. As the virus enters the host, the viral genome ORF1a/b is translated to pp1a and pp1ab polyproteins9. 16 NSPs are released by the cleavage of the proteases using papain-like protease (PLpro) and 3C-like protease (3CLpro), eventually forming the replication transcription complex (RTC)10,11. RTC with NSPs undergoes viral genomic RNA replication, and the transcription of subgenomic RNA produces structural proteins10,12. Finally, the virion is assembled through the endoplasmic reticulum (ER), ER-Golgi intermediate compartment (ERGIC), and Golgi apparatus, and thereafter, the virion is released by exocytosis10,13,14. Meanwhile, the point mutations in the receptor binding domain (RBD) of the S in SARS-CoV-2 yielded several deadly variants with increased susceptibility and transmissibility15–17.
From the perspective of the mechanism of the SARS-CoV-2 infection, the target proteins for drug discovery can be mainly categorized into two classes. The first class is the viral protein NSPs, including NSP3, NSP5, NSP12 and NSP13. The second class of target proteins is involved in the influx of viruses into the cells, originally presents in human body18.
NSPs play an essential role in viral replication and transcription. Of the 16 NSPs, the above-mentioned four NSPs are known as major target proteins. NSP3, also called papain-like protease (PLpro), cleaves NSP1 and NSP2 from the polyproteins pp1a and pp1ab. GRL0617, a SARS-CoV inhibitor, has been reported to exhibit in vitro antiviral efficacy against SARS-CoV-2 with IC50 of 2.1 μM by binding to PLpro19,20. NSP5, also called 3CLpro or Mpro, is the main protease involved in the maturation of other NSPs and the cleavage of polyproteins pp1a and pp1ab. MI-09 and MI-30, synthetic molecules with a five-membered bicycloproline moiety and γ-lactam ring, have shown potent inhibitory activities against SARS-CoV-2 Mpro with an IC50 of 15.2 and 17.2 nM respectively. Further, these compounds have shown in vivo antiviral efficacy with good PK properties and no significant toxicity21. NSP12, also called RNA-dependent RNA polymerase (RdRP), catalyzes the replication and transcription of the SARS-CoV-2 RNA genome. Remdesivir is an FDA-approved drug only for emergency use in COVID-19 treatment. The incorporation of remdesivir into the growing RNA product induces RdRp stalling and creates a barrier to RNA translocation22. NSP13 is an RNA helicase that interacts with NSP12 to enhance viral replication efficiency23. Posaconazole and grazoprevir have recently been identified as potential repurposing drugs that target helicase by in silico studies24.
The influx of the SARS-CoV-2 can be prevented by inhibiting the host proteins involved in the viral infection. Furin is the protease responsible for the proteolytic activation of the spike at the S1/S2 site25. Although many studies point out that furin is not the only protease involved in the cleavage of the S1/S2 site, treatment with a furin inhibitor, decanoyl-RVKR-chloromethylketone (CMK), blocks the entry of the virus26. TMPRSS2, a crucial protease in SARS-CoV-2 entry into the host cell, is inhibited by camostat mesilate and nafamostat, both of which are approved treatments for other medical uses. The in vitro antiviral IC50 of camostat mesilate is measured as 4.2 nM27. Further, camostat mesilate has shown in vivo antiviral activity against SARS-CoV with a survival level of above 60% when dosed in mice at 30 mg/kg28. Furthermore, camostat mesilate has undergone clinical trials for the treatment of COVID-19; however, it did not show any particular efficacy29. Cathepsin L, a lysosomal protease and a marker for various cancers, plays a crucial role in SARS-CoV-2 infection by cleaving the S protein at a position different from the furin cleavage site30,31. Aloxistatin (E-64d), a broad cysteine protease inhibitor, reduces the cell entry of SARS-CoV-2 pseudovirions by 92.5% at 30 μM32. AAK1, the numb-associated kinase (NAK) family, is responsible for clathrin-mediated endocytosis. Barcitinib is a selective Janus kinase 1/2 inhibitor that shows a high affinity for AAK1. The phase 3 clinical trial for the treatment of COVID-19 has been conducted using barcitinib and the 28-day mortality rate has been reduced by 38.2% compared to that of the placebo33.
Natural products have long been used to treat various diseases including cancer, metabolic diseases and infectious diseases. Natural products that hang down through folk remedies are profiled along with advances in chemistry and are regarded as useful resources for drug development34,35. Over 300,000 natural products obtained from various resources including plants, animals, microorganisms and marine organisms have been reported and new compounds with distinct chemical scaffolds are continually being reported34,36. For marine natural products that have not been thoroughly studied compared to terrestrial compounds, more than 1,000 compounds with novel skeletons have been reported every year37. Natural products are structurally diverse and many of them exhibit characteristic bioactivities. Among the natural products, there are many substances with antiviral effects35. Spirooliganone, uncinoside, baicalein and curcumin are natural products with antiviral effects38. Specifically, Tamiflu, a treatment for influenza A and B, is based on a natural product derived from the Chinese spice, star anise (Illicium verum)39. Thus, the use of natural products as a starting point for new drug discovery will be a viable strategy for the successful development of therapeutics that can effectively control COVID-19.
In this study, the hit compound, (+)-usnic acid, was identified by the in silico molecular docking of more than 20,000 marine natural products against eight SARS-CoV-2 targets. The antiviral efficacy of the hit compound was evaluated using immunofluorescence. To rationalize the antiviral efficacy of the hit compound, MM-GBSA and molecular dynamics (MD) approach were employed. Finally, the potency of (+)-usnic acid was examined under various conditions including the salt formation of (+)-usnic acid (sodium and potassium salts) and in vitro assays against some SARS-CoV-2 variants. Through this study, we have demonstrated that (+)-usnic acid could be a good drug lead for the treatment of the deadly COVID-19.
Methods
Hit discovery through high throughput virtual screening (HTVS)
An in-house database containing natural products derived from marine sources (animals, plants, bacteria, and fungi) was selected as the HTVS ligand for the SARS-CoV-2 protein targets. The in-house database contained 24,669 marine natural products. Ligands were prepared using the LigPrep software offered by Schrödinger40. The OPLS3e force field was applied to the ligands generating tautomers and possible states at a target pH of 7.0 ± 2.0.
The grids were generated based on the eight SARS-CoV-2 targets listed in Table 1. For the protein grid generation, the grid box was located within the centroid of the selected binding site residues. The size of the grid box was set to dock the ligands within 20 Å. The ligands were docked using the Glide module from Schrödinger with HTVS precision41. The molecular properties such as absorption, distribution, metabolism and excretion (ADME) of the ligands were calculated using QikProp software42.
Table 1.
The SARS-CoV-2 target protein PDBs and the binding site amino acid used in grid generation.
| No | Target protein | PDB ID | Binding site amino acids | Ref |
|---|---|---|---|---|
| 1 | PLpro | 6WX4 | Met208, Pro247, Tyr264, Tyr268 | 43 |
| 2 | Mpro | 7AKU | His41, Gly143, Gln189 | 44,45 |
| 3 | RdRp | 7BV2 | Asp618, Asp760, Asp761 | 22,46 |
| 4 | Helicase | 7NNG | Gln404, Arg443, Arg567 | 23 |
| 5 | Furin | 6EQW | Leu227, Glu257 | 47 |
| 6 | TMPRSS2 | 7MEQ | His296, Ser441, Ser460 | 48 |
| 7 | CathepinL | 1MHW | Gln21, Gly23, Asp162 | 49 |
| 8 | AAK1 | 4WSQ | Asp127, Glu180, Asn181 | 50 |
Compound isolation and structure confirmation
The (+)-usnic acid was isolated from the extracts of marine fungus, Mycosphaerella sp., which was collected from a marine sediment at Donghae-si, Korea. The fungus was cultivated in 6 L of potato dextrose broth (PDB) dissolved in seawater in 27 °C, at 140 rpm shaking incubator for 7 days. The broth was extracted with ethyl acetate and yielded 4.01 g extract. The crude extract was fractionated into 3 fractions with a silica gel column chromatography using CH2Cl2 and MeOH as solvent. Fractions were further purified by C18 HPLC (Phenomenex luna C18 column, 250 mm × 10 mm, 5 μm) using 65% CH3CN in H2O to yield 6.8 mg (+)-usnic acid. NMR spectra were measured using a Bruker Ascend 700 MHz spectrometer. Electrospray ionization source (ESI) mass data was obtained using Agilent Technologies 6120 quadrupole mass spectrometer coupled with Agilent Technologies 1260 series HPLC. The optical rotation of (+)-usnic acid was measured in chloroform using a 10 mm path length cell on a Digital Polarimeter P-2000, Jasco Inc.
In vitro SARS-CoV-2 inhibition evaluation by immunofluorescence method
The efficacy of antiviral activity against SARS-CoV-2 was verified by an immunofluorescence method. The SARS-CoV-2 virus was obtained from the Korea Centers for Disease Control and Prevention (KCDC, βCoV/Korea/KCDC/2020) and vero cells were obtained from the ATCC (ATCC-CCL81). The specific primary antibody for anti-SARS-CoV-2 nucleocapsid (N) protein was purchased from Sino Biological and the secondary antibodies, Alexa Fluor 488 goat anti-rabbit IgG and Hoechst 3342, were purchased from Molecular Probes. All the experiments using SARS-CoV-2 were carried out in biosafety level 3 (BSL-3) containment at Institut Pasteur Korea with strict accordance of the standard protocol for Korea National Institute of Health (KNIH) and Seoul National University (SNU).
For the cell preparation, the 384-tissue culture plates were seeded with 1.2 × 104 vero cells per well. After 24 h, compounds were prepared at 10 points by twofold serial dilutions in dimethyl sulfoxide (DMSO) to 50 μM and treated to the cells. One hour after the compound treatment, cells were transferred to BSL-3 facility and the viruses were infected at MOI of 0.008. The plates were incubated at 37 ℃ for 24 h. After fixing the cells with 4% paraformaldehyde (PFA), permeabilization was performed. Anti-SARS-CoV-2 N primary antibody was treated and afterward, cells were stained with Alexa Fluor 488-conjugated goat anti-rabbit IgG secondary antibody and Hoechst 33342. The fluorescent images of the infected cells were obtained using a large capacity image analysis instrument, Operetta (Perkin Elmer).
Computational calculation of MM-GBSA and MD simulations
The extra precision (XP) docking of (+)-usnic acid was performed against eight selected SARS-CoV-2 receptors. Based on the docking studies, the Prime MM-GBSA offered by Schrödinger was used to calculate the binding-free energy of the docked ligand for each protein target51. The binding free energy was calculated using the following formula:
∆G(Solv) is the difference in the GBSA solvation energy of the (+)-usnic acid-protein complex compared to the sum of the energies of the uncomplexed protein and ligand. ∆E(MM) is the difference in the minimized energy of the (+)-usnic acid-protein complex compared to the sum of the energies of the uncomplexed protein and ligand. ∆G(SA) is the difference in the surface area energy compared to the sum of the energies of the uncomplexed protein and ligand. Prime MM-GBSA was operated in the OPLS3e force field using the variable dielectric surface generalized Born (VSGB) solvation model.
The thermodynamic behavior, stability and interaction of (+)-usnic acid with Mpro were assessed by MD simulations. The MD calculations were performed using Desmond software52,53. The MD system was built using the TIP3P 3-site water model. The system was neutralized by adding 3 sodium ions and the salt concentration was adjusted to 0.15 M using the sodium and chloride ions. For the MD simulations, an isothermal-isobaric (NPT) ensemble class was applied with the temperature and pressure set to 300 K and 1.01325 bar respectively. The Nose–Hoover chain method was employed for the thermostat method and the Martyna–Tobias–Klein method was applied to the barostat method. The total simulation time was 100 ns, at the recording interval of 100 ps. The analysis of the MD simulation was executed using the simulation interaction diagram enclosed in the Desmond module.
Anti-SARS-CoV-2 assay of the (+)-usnic acid salts
The anti-SARS-CoV-2 assay of the salt form of (+)-usnic acid with the counter-ions, Na+ or K+, was evaluated. The salt form of (+)-usnic acid was synthesized by titration with sodium hydroxide and potassium hydroxide and, thereafter, it was lyophilized. The structure was confirmed by NMR. The efficacy of the (+)-usnic acid salts was evaluated using the immunofluorescence as described above.
In vitro SARS-CoV-2 variants inhibition evaluation
For the in-vitro evaluation of the (+)-usnic acid against the SARS-CoV-2 variants, three major variants of concern were selected for the assessment. The three variants included alpha (UK, B.1.1.7) variant, beta (South Africa, B.1.351) and delta (India, B.1.617.2) variants. All three strains were obtained from the Korea Disease Control and Prevention Agency (KDCA). The serial codes of each variant were hCoV-19/Korea/KDCA51463/2021, hCoV-19/Korea/KDCA55905/2021 and hCoV19/Korea/KDCA119861/2021 respectively. The protocol for in vitro evaluation was identical to that of the immunofluorescence method described above.
Results
Analysis of the HTVS of marine natural products against SARS-CoV-2 proteins
The HTVS for marine natural products was performed using the Glide software. An in-house database containing 24,669 marine natural product structures was analyzed. The docking scores of eight SARS-CoV-2 target proteins (PLpro, Mpro, RdRp, helicase, furin, TMPRSS2, cathepin L, and AAK1) were profiled. The number of compounds that were identified as possible hits was 2,251, with a docking score below –7. Further, the compounds were filtered with ADME properties calculated by Qikprop, following the criteria suggested by Schrödinger. Twenty-seven compounds were passed as potential hits. Considering the availability and ease of supply, four natural products were analyzed at a single dose (10 μM), and (+)-usnic acid showed efficacy against SARS-CoV-2 in an in vitro antiviral assay (Fig. S3). (+)-Usnic acid was selected as the hit compound for the further analysis. The calculated properties of the (+)-usnic acid are shown in Table 2.
Table 2.
The calculated molecular property of the selected compound, (+)-usnic acid. The molecular property includes docking scores of protein targets and predicted ADME properties (dipole, PSA, volume, SASA, QplogS, %HumanOralAbsorption).
| Compound | Docking score (Glide) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (+)-Usnic acid | PLpro | Mpro | RdRp | Helicase | Furin | TMPRSS2 | CathepsinL | AAK1 | |||||||
| −6.00 | −7.70 | −6.24 | −4.72 | −4.77 | −5.53 | −5.40 | −9.34 | ||||||||
| Predicted ADME properties (QikProp) | |||||||||||||||
| MW | Dipole | PSA | volume | SASA | QPlogS | %Human OralAbsorption | Rule of Five | ||||||||
| 344.3 | 4.9 | 144.6 | 1000.3 | 553.1 | −2.5 | 66.1 | 0 | ||||||||
The (+)-usnic acid used in the experiment was structurally confirmed by NMR (Fig. S7). The purity was monitored by LC–MS and confirmed that it was over 99% (Fig. S9). The optical rotation was 500.4 ([α]25D) and it was considered optically pure by comparing with standard compound.
Evaluation of the in vitro antiviral activities of (+)-usnic acid against SARS-CoV-2
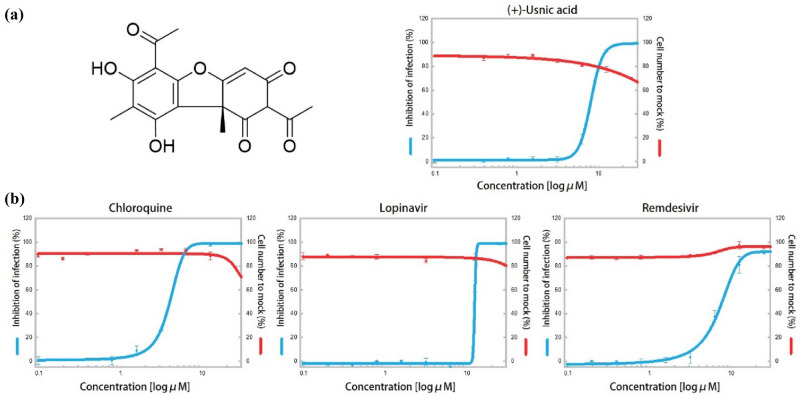
The SARS-CoV-2 antiviral efficacy of the selected compound, (+)-usnic acid, was evaluated by immunofluorescence. The dose response curves of (+)-usnic acid with three control compounds (chloroquine, lopinavir, and remdesivir) are shown in Fig. 1. The IC50 value of (+)-usnic acid was 7.99 μM and CC50 was over 50 μM. The selectivity index (SI) is calculated to be 6.26. The IC50 values of the controls, chloroquine, lopinavir and remdesivir, were 11.48, 11.80, and 7.42 μM, respectively. The CC50 values of the controls were all above 50 μM; the CC50 of lopinavir was higher than 150 μM. The SI of chloroquine, lopinavir and remdesivir was 13.07, 4.24, and 6.74 respectively. The efficacy of (+)-usnic acid against SARS-CoV-2 is similar to that of remdesivir, which is approved by the FDA for the treatment of emergency patients with COVID-19. The (+)-usnic acid showed a lower IC50 value compared to those of the two other controls, chloroquine and lopinavir.
Figure 1.
The anti-SARS-CoV-2 efficacy of (+)-usnic acid and controls; (a) The structure of (+)-usnic acid on the left and dose response curve of (+)-usnic acid against SARS-CoV-2 on the right; (b) The dose response curve of controls (chloroquine, lopinavir and remdesivir); the inhibition of infection and cell number to mock is shown using blue and red lines, respectively.
MM-GBSA and MD analysis of (+)-usnic acid against Mpro
The MM-GBSA method was used to identify the most preferable SARS-CoV-2 target proteins for (+)-usnic acid. The binding free energies are shown in Fig. 2. The target protein that showed the highest difference in the binding-free energy is Mpro, –52.05 kcal/mol. The binding free energies of remaining proteins were in the range of –39 and –29 kcal/mol, which had at least 10 kcal/mol difference with that of Mpro.
Figure 2.
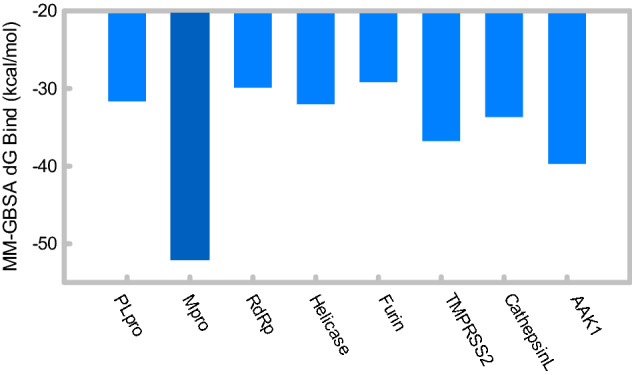
Predicted binding-free energies (kcal/mol) of 8 SARS-CoV-2 target proteins bound with (+)-usnic acid, calculated by Prime MM-GBSA.
The detailed contributions of the binding-free energies are shown in Table 3. The binding free energy (dGbind) was calculated from the sum of dGcoloumb, dGcovalent, dGHbond, dGLipo, dGPacking, dGSolvGB and dGvdW. The main contributor to Mpro, the protein with the lowest binding-free energy when complexed with (+)-usnic acid, was dGvdW with a value of –45.30 kcal/mol. Further, Mpro had relatively low dGLipo compared to those of other proteins; –7.81 kcal/mol.
Table 3.
Detailed values of the binding-free energies (kcal/mol) and binding energy states of the 8 SARS-CoV-2 target proteins that bound with (+)-usnic acid.
| Protein | dGBind | dGCoulomb | dGCovalent | dGHbond | dGLipo | dGPacking | dGSolvGB | dGvdW |
|---|---|---|---|---|---|---|---|---|
| PLpro | −31.66 | −15.02 | 4.41 | −1.05 | −3.57 | 0.00 | 12.68 | −29.11 |
| Mpro | −52.05 | −19.16 | 1.43 | −1.93 | −7.81 | −2.35 | 23.07 | −45.30 |
| RdRp | −29.88 | −11.91 | 2.38 | −0.69 | −3.80 | −3.52 | 24.06 | −36.41 |
| Helicase | −32.00 | −26.05 | 6.96 | −2.66 | −3.23 | −3.18 | 27.27 | −31.10 |
| Furin | −29.16 | −12.82 | 1.18 | −1.27 | −5.57 | −1.08 | 18.86 | −28.46 |
| TMPRSS2 | −36.75 | −15.91 | −0.34 | −1.13 | −5.46 | 0.00 | 18.08 | −31.99 |
| CathepsinL | −33.66 | −12.27 | 0.34 | −0.94 | −6.36 | −2.44 | 20.62 | −32.62 |
| AAK1 | −39.70 | −15.43 | 2.05 | −1.31 | −8.17 | 0.00 | 21.70 | −38.54 |
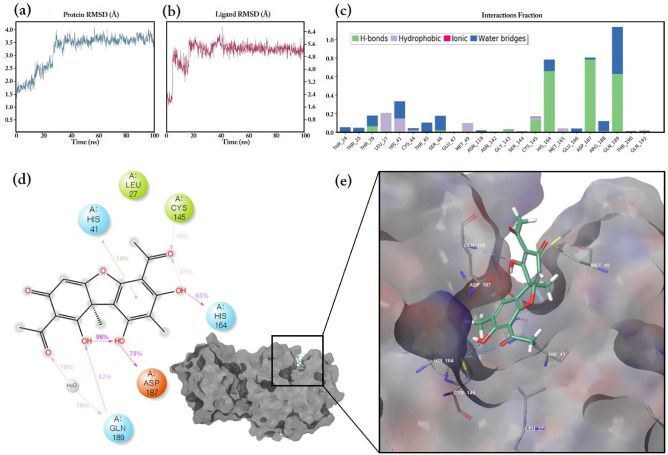
The MD simulation of Mpro complexed with (+)-usnic acid was monitored and is shown in Fig. 3. Protein and ligand RMSD are shown in Fig. 3a,b. The RMSD of the alpha-carbon (Cα) of the Mpro protein reached up to 3.5 Å in 30 ns and then remained equilibrated for the rest of measurement times. The RMSD fluctuation of Cα remained within 1–3 Å, which is acceptable for small and globular proteins. The ligand RMSD rapidly rose to 5 Å in 20 ns. Then, the fluctuation of ligand RMSD remained in a steady-state between 4.8 and 5.6 Å for the other monitoring period.
Figure 3.
The MD simulation of (+)-usnic acid bound to the SARS-CoV-2 Mpro protein; (a) RMSD of the alpha-carbon of Mpro; (b) The ligand RMSD of (+)-usnic acid bound to Mpro, simulated for 100 ns; (c) The Mpro amino acid interaction with (+)-usnic acid showing H-bonds, hydrophobic interactions and water bridges; (d) The 2D % interaction illustration of (+)-usnic acid in tautomeric form; (e) The 3D illustration of (+)-usnic acid bound to Mpro showing interacting amino acids and hydrogen bonds.
Through the MD simulation, the amino acid of Mpro protein, that (+)-usnic acid mainly interacts with, was profiled and shown in Fig. 3c-d. The interaction through the H-bond and water bridges was predominant and there were slight hydrophobic interactions as well; however, ionic interaction was not detected. His164, Asp187 and Gln189 were the amino acids with the interaction fractions over 0.8, in which most of them were accounted by H-bonds and water bridges. According to the percentage interaction illustration, (+)-usnic acid interacted with Asp187 at most with 78%, followed by 65% of His164. At the 3D model in Fig. 3e, (+)-usnic acid bound to the Mpro binding pocket and interacted with His41, Cys145, Asp187 and Gln189 through hydrogen bonds.
Anti-SARS-CoV-2 efficacy of (+)-usnic acid and its salts
The anti-SARS-CoV-2 efficacy of (+)-usnic acid salts, sodium usnate (NaU) and potassium usnate (KU), was evaluated. NaU showed a slightly improved efficacy compared to those of the (+)-usnic acid and KU. The IC50 of NaU and KU was 5.33 and 7.57 μM respectively, which were lower than that of (+)-usnic acid. The CC50 of all compounds remained over 50 μM. The SI of (+)-usnic acid salts was 9.38 for NaU and 6.60 for KU, both showing slightly improved values than (+)-usnic acid (Table 4).
Table 4.
Anti-SARS-CoV-2 efficacy of (+)-usnic acid and its salts (NaU and KU).
| Compound | IC50 (μM) | CC50 (μM) | SI |
|---|---|---|---|
| (+)-Usnic acid | 7.99 (SE ± 0.23) | > 50 | 6.26 |
| NaU | 5.33 (SE ± 0.01) | > 50 | 9.38 |
| KU | 7.57 (SE ± 1.27) | > 50 | 6.60 |
Inhibition of the SARS-CoV-2 variants by (+)-usnic acid
The antiviral efficacy of (+)-usnic acid against SARS-CoV-2 differs depending on the variants. The IC50 of (+)-usnic acid against alpha and delta variants were 6.05 μM and 7.17 μM respectively, which were similar to the efficacy against the original strain. However, usnic acid showed enhanced efficacy against the beta variant where the IC50 was 2.92 μM. The SI for the beta variant was 11.1, indicating more than twice the SI of the other two variants assayed (Table 5).
Table 5.
The antiviral efficacy of (+)-usnic acid and controls against the SARS-CoV-2 variants [Alpha (B.1.1.7), Beta (B.1.351), and Delta (B.1.617.2)].
| Compounds | (+)-Usnic Acid | Chloroquine | Remdesivir | Lopinavir | |||||
|---|---|---|---|---|---|---|---|---|---|
| Variants | IC50 (μM) |
CC50 (μM) |
SI | IC50 (μM) |
CC50 (μM) |
IC50 (μM) |
CC50 (μM) |
IC50 (μM) |
CC50 (μM) |
|
SARS-CoV-2 Alpha var (B.1.1.7) |
6.05 (SE ± 0.33) |
34.8 (SE ± 1.9) |
5.8 |
6.16 (SE ± 0.30) |
> 150 |
2.25 (SE ± 0.16) |
> 50 |
10.8 (SE ± 0.9) |
> 50 |
|
SARS-CoV-2 Beta var (B.1.351) |
2.92 (SE ± 0.06) |
32.4 (SE ± 1.0) |
11.1 |
2.64 (SE ± 0.49) |
> 150 |
1.47 (SE ± 0.16) |
> 50 |
11.8 (SE ± 0.5) |
> 50 |
|
SARS-CoV-2 Delta var (B.1.617.2) |
7.17 (SE ± 0.59) |
> 50 | 6.97 |
6.22 (SE ± 0.34) |
> 150 |
6.48 (SE ± 0.40) |
> 50 |
15.3 (SE ± 5.6) |
> 50 |
Discussion
From a marine natural products in-house database containing 24,669 natural products, (+)-usnic acid, a substance effective against SARS-CoV-2, was discovered as a hit compound through virtual screening and in vitro assays. The thermodynamic properties and amino acid interactions of (+)-usnic acid were analyzed using MD simulation. Furthermore, the efficacy of (+)-usnic acid and its salts against the SARS-CoV-2 variants was evaluated in this study.
The HTVS of natural product libraries allowed the profiling of docking energies for eight SARS-CoV-2 target proteins within a few days. The docking energy was the priority in the selection and compounds exceeding the permitted range of calculated PK properties were removed. Twenty-seven natural products were identified as potential hits, and considering the ease of securing the compounds, four natural products were selected for in vitro evaluation at a single dose. The (+)-usnic acid showed potent efficacy against SARS-CoV-2 and we conducted further experiments with this compound.
Usnic acid (UA) was first reported in 1844, since then many bioactivities have been studied for this dibenzofuran compound and its derivatives54. Usnic acids are found mostly in lichens such as Alectoria, Cladonia, Evernia, Lecanora, Ramalina and Usnea55. The usnic acid shows wide-ranging bioactivities including antimicrobial, antifungal, antiprotozoal, insecticidal, anti-inflammatory, phytotoxic, anti-inflammatory, and antimitotic activities56. Moreover, the usnic acid shows weak antiviral activity against Herpes simplex type1, Polio type1, papiloma and influenza virus A (H1N1)56,57. Recently, usnic acid was produced from the marine fungus Mycosphaerella sp., which was isolated from marine sediment in Korea with several compounds in similar scaffolds58.
The in vitro immunofluorescence test against the SARS-CoV-2 showed that (+)-usnic acid showed efficacy with IC50, CC50 and SI corresponding to 7.99 μM, more than 50 μM and 6.26 respectively. Compared to the three compounds (remdesivir, lopinavir and chloroquine) used as positive controls, (+)-usnic acid showed a similar profile to remdesivir, which was approved as a treatment for COVID-19 only for an emergency use. (+)-Usnic acid showed lower IC50 values than those of lopinavir and chloroquine.
Although the antiviral efficacy of (+)-usnic acid was validated, we did not know the exact protein target. Therefore, we attempted to identify it using MM-GBSA. MM-GBSA was applied to the energy-minimized pose of docked (+)-usnic acid with eight SARS-CoV-2 protein targets. The binding free energy of Mpro protein was the lowest at –52.05 kcal/mol and the energy difference among those of other proteins was at least 10 kcal/mol. Thus, we determined that Mpro was the most plausible SARS-CoV-2 protein target for (+)-usnic acid. For further analysis, the MD simulations of Mpro with (+)-usnic acid were performed for 100 ns. The protein RMSD increased up to 3.5 Å for 30 ns and ligand RMSD rose to 5 Å for same period as well. The ligand moved slightly to inner pocket of Mpro compared to the binding pose of docking analysis. Instead of a hydrogen bond with Gly143, (+)-usnic acid became more stabilized by H-Bonds with Asp187 and Gln189. Afterwards, (+)-usnic acid was remained stabilized for other 100 ns monitoring period. As a result, the most likely target protein of SARS-CoV-2 for (+)-usnic acid is Mpro.
Subsequently, the antiviral efficacy of (+)-usnic acid under various conditions was evaluated. First, the sodium and potassium salts of (+)-usnic acid were prepared. The IC50 and SI of NaU were slightly improved, while those of KU remained almost the same as that of (+)-usnic acid. This improvement in the salts could be explained by the enhanced water solubility. The CC50 of both salts was greater than 50 μM. Next, the antiviral efficacy of (+)-usnic acid against the SARS-CoV-2 variants, which are causing serious pandemic worldwide, was evaluated. The (+)-usnic acid against the original SARS-CoV-2 strain showed profiles similar to those of the alpha and delta variants, while it exhibited a superior effect on the beta variant. The reason for this is still unclear and further research is needed to understand why (+)-usnic acid exhibits better efficacy against the beta variant, but it is hypothesized that the K90R mutation on Mpro, 95% prevalent in beta affected the protein stability of the Mpro59,60. As lys90 is located on the opposite side of the binding site, it does not significantly affect the binding of (+)-usnic acid. But as a result, protein folding of Mpro becomes destabilized and the overall efficacy of (+)-usnic acid against beta variant seem enhanced.
Usnic acid has been widely used as an ingredient for daily necessities including toothpastes and mouthwashes55. It has also been used as a dietary supplement for weight loss61. Unfortunately, usnic acid exhibits hepatotoxicity because of the uncoupling of oxidative phosphorylation, resulting in the destruction of mitochondrial respiration at very high concentrations62. Nevertheless, usnic acid is expected to have sufficient efficacy and safety for the treatment of COVID-19. The maximum period required for COVID-19 recovery is usually 7 days and paxlovid and molnupiravir, approved for emergency use, are employed for five days only. The use of usnic acid within five days may not show any significant toxicity since there are records of human consumption for a relatively long period of over two weeks63. Therefore, it is suggested that usnic acid could be used in emergency situations for the treatment of COVID-19, if its in vivo efficacy is proven. Additionally, oral administration is expected to be possible based on a study showing the improved bioavailability of KU in a mouse tumor xenograft model64.
In conclusion, this study identified (+)-usnic acid as a potential drug lead against COVID-19 by the HTVS of a marine natural products database. Further, the in vitro efficacy of (+)-usnic acid has been validated in several SARS-CoV-2 variants. The protein target of (+)-usnic acid was explored using MM-GBSA and MD approaches. SARS-CoV-2 Mpro was identified as a potent protein target of (+)-usnic acid. Although there is a toxicity issue on usnic acid, it is easy to be obtained and has a record of its human use. Therefore, we propose (+)-usnic acid as a potential therapeutic tool for COVID-19 treatment, considering the desperate need to end the deadly pandemic.
Supplementary Information
Acknowledgements
This work was supported by the Basic Science Research Program funded by the Ministry of Science and ICT (2019R1A2C2005492).
Author contributions
E.O., W.W. and H.K. wrote the main manuscript text. E.O., K.P., C.P., Y.C., J.L. and E.K. performed the experiments. All authors reviewed the manuscript.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-17506-3.
References
- 1.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (2021).
- 3.Ruan, L. & Zeng, G. in Emerging Infections in Asia, 75–96 (Springer, 2008).
- 4.Uyeki TM, et al. Development of Medical Countermeasures to Middle East Respiratory Syndrome Coronavirus. Emerg. Infect. Dis. 2016;22:1. doi: 10.3201/eid2207.160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng R, Wu LA, Wang Q, Qi J, Gao GF. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021;46:848–860. doi: 10.1016/j.tibs.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2021 doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, et al. SARS-CoV-2: Mechanism of infection and emerging technologies for future prospects. Rev. Med. Virol. 2021;31:e2168. doi: 10.1002/rmv.2168. [DOI] [PubMed] [Google Scholar]
- 10.V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krichel B, Falke S, Hilgenfeld R, Redecke L, Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr AR, Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses. 2015;1:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein S, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saraste J, Prydz K. Assembly and cellular exit of coronaviruses: Hijacking an unconventional secretory pathway from the pre-golgi intermediate compartment via the golgi ribbon to the extracellular space. Cells. 2021;10:1. doi: 10.3390/cells10030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang, T. J. et al. Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function. Nat Struct Mol Biol28, 731–739, doi:10.1038/s41594-021-00652-z (2021). [DOI] [PubMed]
- 16.La Rosa G, et al. Key SARS-CoV-2 mutations of alpha, gamma, and eta variants detected in urban wastewaters in Italy by long-read amplicon sequencing based on nanopore technology. Water. 2021;13:2503. doi: 10.3390/w13182503. [DOI] [Google Scholar]
- 17.Han P, et al. Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-020-20314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haque, S. M., Ashwaq, O., Sarief, A. & Azad John Mohamed, A. K. A comprehensive review about SARS-CoV-2. Future Virol15, 625–648. 10.2217/fvl-2020-0124 (2020). [DOI] [PMC free article] [PubMed]
- 19.Ratia K, et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Z, et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021;12:488. doi: 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao J, et al. SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokic G, et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman JA, et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12:4848. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abidi SH, et al. Repurposing potential of posaconazole and grazoprevir as inhibitors of SARS-CoV-2 helicase. Sci. Rep. 2021;11:10290. doi: 10.1038/s41598-021-89724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang T, et al. Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: Potential role of proteases beyond Furin. ACS Infect Dis. 2021;7:264–272. doi: 10.1021/acsinfecdis.0c00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng YW, et al. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33:108254. doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann M, et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65:103255. doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, et al. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunst JD, et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. EClinicalMedicine. 2021;35:100849. doi: 10.1016/j.eclinm.2021.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao MM, et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target Ther. 2021;6:134. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes CP, et al. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Front. Cell Infect. Microbiol. 2020;10:589505. doi: 10.3389/fcimb.2020.589505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marconi VC, et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021 doi: 10.1016/S2213-2600(21)00331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 35.Omokhua-Uyi AG, Van Staden J. Natural product remedies for COVID-19: A focus on safety. S. Afr. J. Bot. 2021;139:386–398. doi: 10.1016/j.sajb.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorokina M, Steinbeck C. Review on natural products databases: where to find data in 2020. J. Cheminform. 2020;12:20. doi: 10.1186/s13321-020-00424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat. Prod. Rep. 2021;38:362–413. doi: 10.1039/d0np00089b. [DOI] [PubMed] [Google Scholar]
- 38.Lin LT, Hsu WC, Lin CC. Antiviral natural products and herbal medicines. J. Tradit. Complement Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patra JK, et al. Star anise (Illicium verum): Chemical compounds, antiviral properties, and clinical relevance. Phytother. Res. 2020;34:1248–1267. doi: 10.1002/ptr.6614. [DOI] [PubMed] [Google Scholar]
- 40.Schrödinger Release 2021–3: LigPrep, Schrödinger, LLC, New York, NY, (2021).
- 41.Schrödinger Release 2021–3: Glide, Schrödinger, LLC, New York, NY, (2021).
- 42.Schrödinger Release 2021–3: QikProp, Schrödinger, LLC, New York, NY, (2021).
- 43.Rut W, et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci. Adv. 2020;6:1. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunther S, et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshino R, Yasuo N, Sekijima M. Identification of key interactions between SARS-CoV-2 main protease and inhibitor drug candidates. Sci. Rep. 2020;10:12493. doi: 10.1038/s41598-020-69337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin W, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahms SO, Hardes K, Steinmetzer T, Than ME. X-ray structures of the proprotein convertase furin bound with substrate analogue inhibitors reveal substrate specificity determinants beyond the S4 Pocket. Biochemistry. 2018;57:925–934. doi: 10.1021/acs.biochem.7b01124. [DOI] [PubMed] [Google Scholar]
- 48.Hussain M, et al. Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: Conformation and intermolecular interactions. AIMS Microbiol. 2020;6:350–360. doi: 10.3934/microbiol.2020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chowdhury, S. F. et al. Design of noncovalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides. J Med Chem45, 5321–5329, 10.1021/jm020238t (2002). [DOI] [PubMed]
- 50.Sorrell, F. J., Szklarz, M., Abdul Azeez, K. R., Elkins, J. M. & Knapp, S. Family-wide Structural Analysis of Human Numb-Associated Protein Kinases. Structure24, 401–411, doi:10.1016/j.str.2015.12.015 (2016). [DOI] [PMC free article] [PubMed]
- 51.Schrödinger Release 2021–3: Prime, Schrödinger, LLC, New York, NY, 2021.
- 52.Bowers, K. J. et al. in SC'06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. 43–43 (IEEE).
- 53.Schrödinger Release 2021–3: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2021. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2021.
- 54.Knop W. Chemisch-physiologische Untersuchung über die Flechten. Justus Liebigs Ann. Chem. 1844;49:103–124. doi: 10.1002/jlac.18440490202. [DOI] [Google Scholar]
- 55.Ingolfsdottir K. Usnic acid. Phytochemistry. 2002;61:729–736. doi: 10.1016/S0031-9422(02)00383-7. [DOI] [PubMed] [Google Scholar]
- 56.Galanty A, Paśko P, Podolak I. Enantioselective activity of usnic acid: A comprehensive review and future perspectives. Phytochem. Rev. 2019;18:527–548. doi: 10.1007/s11101-019-09605-3. [DOI] [Google Scholar]
- 57.Sokolov DN, et al. Anti-viral activity of (-)- and (+)-usnic acids and their derivatives against influenza virus A(H1N1)2009. Bioorg Med Chem Lett. 2012;22:7060–7064. doi: 10.1016/j.bmcl.2012.09.084. [DOI] [PubMed] [Google Scholar]
- 58.Lee, J. et al. Mycousfurans A and B, Antibacterial Usnic Acid Congeners from the Fungus Mycosphaerella sp., Isolated from a Marine Sediment. Mar Drugs17. 10.3390/md17070422 (2019). [DOI] [PMC free article] [PubMed]
- 59.Ullrich, S., Ekanayake, K. B., Otting, G. & Nitsche, C. Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg Med Chem Lett62, 128629. 10.1016/j.bmcl.2022.128629 (2022) [DOI] [PMC free article] [PubMed]
- 60.Sokalingam, S., Raghunathan, G., Soundrarajan, N. & Lee, S. G. A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PLoS One7, e40410, doi:10.1371/journal.pone.0040410 (2012) [DOI] [PMC free article] [PubMed]
- 61.Durazo, F. A. et al. Fulminant liver failure due to usnic acid for weight loss. Official journal of the American College of Gastroenterology| ACG99, 950–952 (2004). [DOI] [PubMed]
- 62.Pramyothin P, Janthasoot W, Pongnimitprasert N, Phrukudom S, Ruangrungsi N. Hepatotoxic effect of (+)usnic acid from Usnea siamensis Wainio in rats, isolated rat hepatocytes and isolated rat liver mitochondria. J. Ethnopharmacol. 2004;90:381–387. doi: 10.1016/j.jep.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 63.Hsu LM, Huang YS, Chang FY, Lee SD. 'Fat burner' herb, usnic acid, induced acute hepatitis in a family. J. Gastroenterol. Hepatol. 2005;20:1138–1139. doi: 10.1111/j.1440-1746.2005.03855.x. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, et al. Potassium usnate, a water-soluble usnic acid salt, shows enhanced bioavailability and inhibits invasion and metastasis in colorectal cancer. Sci. Rep. 2018;8:16234. doi: 10.1038/s41598-018-34709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.