Abstract
The International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) COVID-19 dataset is one of the largest international databases of prospectively collected clinical data on people hospitalized with COVID-19. This dataset was compiled during the COVID-19 pandemic by a network of hospitals that collect data using the ISARIC-World Health Organization Clinical Characterization Protocol and data tools. The database includes data from more than 705,000 patients, collected in more than 60 countries and 1,500 centres worldwide. Patient data are available from acute hospital admissions with COVID-19 and outpatient follow-ups. The data include signs and symptoms, pre-existing comorbidities, vital signs, chronic and acute treatments, complications, dates of hospitalization and discharge, mortality, viral strains, vaccination status, and other data. Here, we present the dataset characteristics, explain its architecture and how to gain access, and provide tools to facilitate its use.
Subject terms: Epidemiology, Viral infection
| Measurement(s) | CDISC SDTM Vital Sign Test Name Terminology |
| Technology Type(s) | CDISC Define-XML General Observation Class Terminology |
| Factor Type(s) | Hospital Mortality • COVID-19 |
| Sample Characteristic - Organism | Homo sapiens |
| Sample Characteristic - Environment | hospital |
Background & Summary
The International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC) is a global federation of clinical research networks collaborating to prevent illness and death from infectious disease outbreaks through proficient and agile research response1. In January 2020, ISARIC launched a research response to the emergence of a novel severe acute respiratory syndrome coronavirus (SARS-COV-2), detected weeks earlier in Wuhan, China2,3. The initial focus was on the clinical characterisation of COVID-19, the disease caused by SARS-CoV-2, which mainly affects the respiratory system2. The fatality rate of COVID-19 varies substantially across different locations, which may reflect differences in population age, comorbidities, vaccination status, and other factors4. In June 2022, there were more than 500 million reported cases and more than 6 million deaths. Despite unprecedented success in the rapid generation of vaccines and effective treatments, COVID-19 continues to cause severe and widespread health consequences5,6. Therefore, the continuation of high-quality, globally-representative research is critical – as are the data required to deliver it.
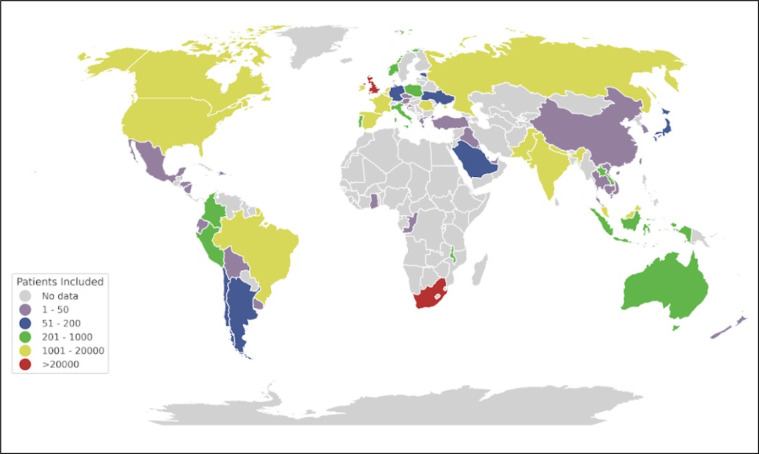
At the beginning of the COVID-19 outbreak, ISARIC adapted the ISARIC-WHO Clinical Characterization Protocol and data tools7 to facilitate global research collaboration and accelerate the understanding of COVID-19 as part of the public health response to the pandemic1,8,9. Between January 2020 and September 2021, information about the clinical presentation, treatment, and outcomes of more than 705,000 patients with COVID-19, hospitalized across 62 countries, was aggregated to form the ISARIC-COVID-19 dataset. Clinical teams in 1,559 participating institutions collected the data. Figure 1 shows the number of patients per country included in the database as of September 20211,4,10. The number of patients included in the dataset continues to grow as data collection continues across the globe.
Fig. 1.
The number of patients per country is included in the ISARIC COVID-19 database.
The objective of the dataset is to accelerate understanding of COVID-19 through access to detailed clinical information on infected patients from a range of settings. Access to data facilitates science, improves scientific transparency and integrity, and has played a substantial role in the generation of knowledge that has led to better patient management and vaccine production for COVID-1911. The diversity of populations, regions, and resource levels from which the data originate increases the generalizability of the evidence generated and supports comparisons across them. By collating, standardizing, and sharing large volumes of disparate data, curation and governance efforts are invested centrally by a specialised team, enabling efficient data access, and analysis by many researchers focused on the questions most relevant to the patients in their settings. This approach accelerates pandemic response by promoting locally-driven, locally-relevant knowledge generation, which is most likely to have an impact on public health policy and drive societal benefits beyond health12,13.
Methods
Data collection
Standardized clinical data of patients with suspected or confirmed COVID-19 are collected on the ISARIC-WHO case report forms (CRFs) (https://isaric.org/research/covid-19-clinical-research-resources/covid-19-crf/) or site-specific iterations of these forms. These forms are available in multiple languages to support accessibility for a global response.
Sites implement data collection contemporaneously to clinical care. Data are collected through direct observation and/or reviewing and extracting electronic health records or patient registries. Data can be submitted to ISARIC by completing the CRF on the Research Electronic Data Capture platform (REDCap version 10.6 Vanderbilt University14) hosted by the University of Oxford. Alternatively, institutions using other data collection forms and/or a different data management system can share patient data in any format to the ISARIC COVID-19 data platform, hosted by the Infectious Diseases Data Observatory (IDDO, www.iddo.org). Data were prospectively collected on patients with clinical suspicion or laboratory confirmation of SARS-CoV-2 infection and admitted to a participating hospital or ward. Recruitment aimed to include all identified patients; however, resource constraints limited enrolment when patient numbers surged and health systems became overwhelmed. In such cases, or in sites where prospective data collection was impossible, data were extracted from electronic health records. Ethics approval and informed consent were obtained according to local regulations, which included a waiver of consent to collect de-identified data at several sites due to the burden on front-line workers and the data protection framework in place. The WHO-ISARIC Clinical Characterization Protocol was approved by the WHO Ethics Committee (RPC571 and RPC572).
Data standardization
The ISARIC COVID-19 dataset is a large, clinically comprehensive, international resource. The diversity of data aggregated to create this resource required a uniform data model to standardize the structures and ontologies to a harmonized format. Thus, all data are standardized to the Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM) to facilitate pooled analyses. While there is no perfect data model, the CDISC SDTM was chosen to allow maximum flexibility to accommodate the diverse data types collected by different groups. This was preferred over other options, such as the Observational Medical Outcomes Partnership (OMOP) model, which was more rigid with a fixed number of possible tables and variables. The use of SDTM also allows for greater interoperability to enable integration with COVID-19 clinical trial data that may be added to the dataset in the future. This data model is designed for data tabulation and storage. Using the dataset requires processing to create an analysis dataset from which results can be derived. Here we present a complete description of the available data, how it is formatted, and describe a generalizable strategy to use and maximize its utility in research.
Data standardization - de-identification
Data entered in the ISARIC REDCap database or uploaded to the IDDO data platform are reviewed to ensure no direct identifiers are included. Direct identifiers, including those listed in the UK General Data Protection Regulation (https://ico.org.uk/for-organisations/guide-to-data-protection/guide-to-the-general-data-protection-regulation-gdpr/) and the US Health Insurance Portability and Accountability Act (https://www.hhs.gov/hipaa/index.html), are permanently deleted before data are curated through various processes.
Data standardisation - pre-mapping
Data and all documentation shared with the data, such as dictionaries, protocols, publications, and data collection forms, are reviewed by the data curator to fully understand the contents of the dataset. Queries are raised with the data contributor when required. Each variable in the dataset is assigned to the appropriate SDTM domain(s), variable(s), and controlled vocabulary (if applicable) according to the rules found within the IDDO SDTM Implementation Manual (https://www.iddo.org/tools-and-resources/data-tools). The implementation manual chronicles each type of data curated to the platform and is consulted and updated with each new dataset to ensure consistency across the repository. An audit trail of the assignments is also recorded in a dataset-specific SDTM mapping guide.
Data standardization - data wrangling
For formatting and coding, the contributed datasets are loaded into Trifacta®, a data wrangling programme. This can include merging files, splitting variables into separate domains, applying controlled terminology to variables, and adding created variables as required. IDDO-defined standardization, conversion, and categorization formulas are also used as described in the IDDO SDTM Implementation Manual. Transformations on the contributed data (in the interests of standardization) are recorded and stored in a form that documents the transformation and enables it to be reproduced.
Data standardization - review and edit checks
Data is run through Pinnacle 21® (community version) software, a CDISC standards compliance-verification tool that checks the standard SDTM implementation guide rules and requirements for regulatory submission. The resulting checks and warnings are assessed for applicability to the individual dataset. The data are also run through standard edit checks to identify possible mapping errors separate from SDTM conformance. The curator adjusts the mapping as needed to make corrections.
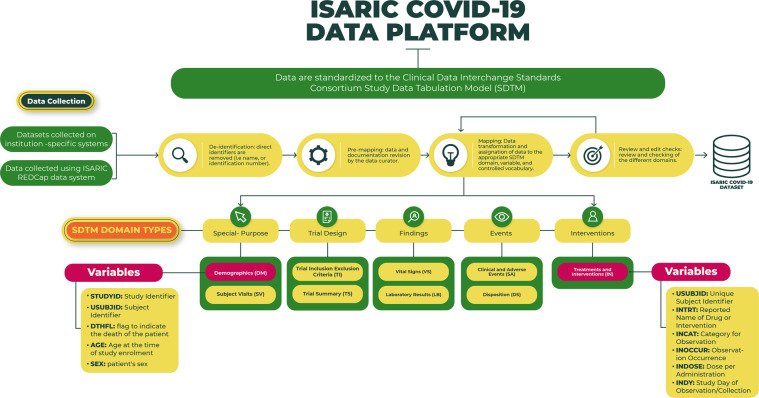
Figure 2 describes the workflow from data acquisition to the final, pooled dataset that researchers can access to conduct their research.
Fig. 2.
Overview of the ISARIC COVID-19 Database.
Data Records
The dataset is available from the Infectious Diseases Data Observatory – IDDO at 10.48688/nx85-bv3015 The ISARIC-COVID-19 dataset is a relational database consisting of 16 tables, each representing a domain of information set out in the CDISC SDTM data model. Unique identifiers link these with the suffix ‘ID.’ For example, USUBJID refers to the subject’s unique identifier, which is the primary key for assessing individual-level data; STUDYID contains the unique identifier for an individual hospital or network of hospitals. Each table defines and tracks different aspects of illness and treatment.
Data tables
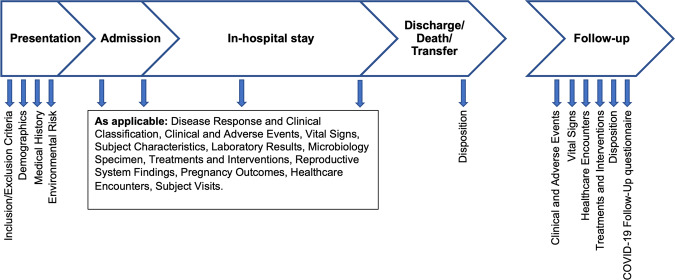
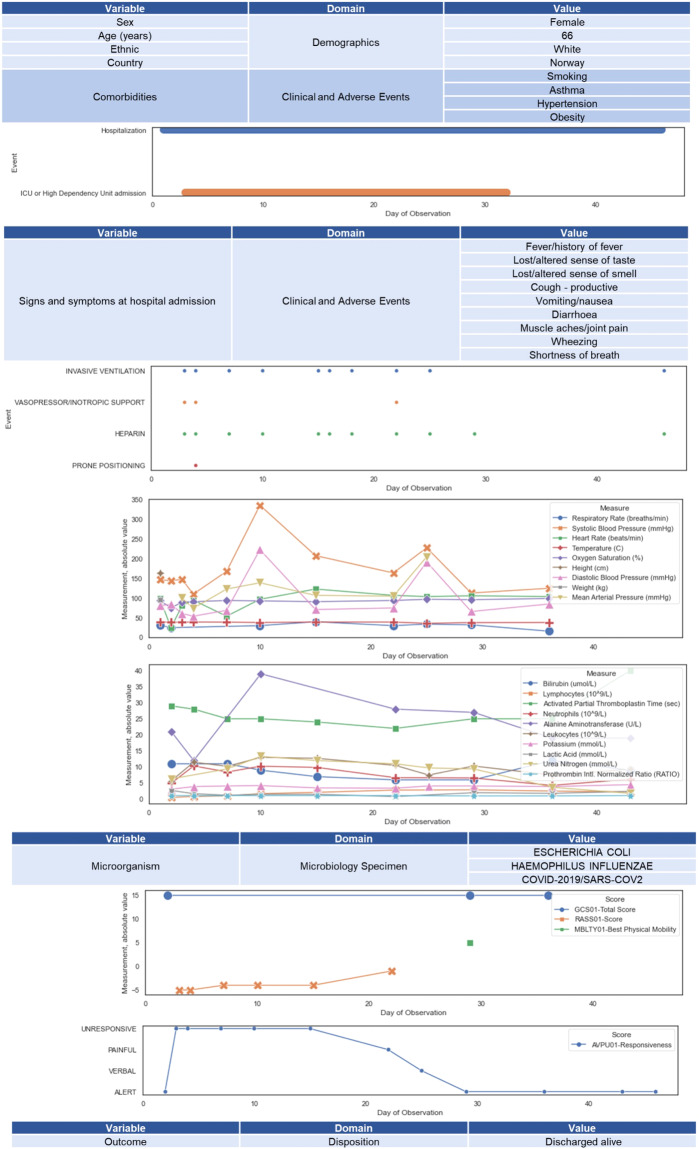
The tables (i.e., domains) currently included in the dataset are Demographics (DM), Disposition (DS), Environmental Risk (ER), Healthcare Encounters (HO), Inclusion/Exclusion Criteria (IE), Treatments and Interventions (IN), Laboratory Results (LB), Microbiology Specimen (MB), Reproductive System Findings (RP), Disease Response and Clinical Classification (RS), Clinical and Adverse Events (SA), Subject Visits (SV), Vital Signs (VS), COVID-19 Follow-Up questionnaire (CQ), Subject Characteristics (SC), and Pregnancy Outcomes (PO) (Supplementary Table 1); The majority of those tables are at a patient level, so it has a subject id (USUBJID) that that relates the information of a single patient distributed in the multiple tables. The Trial Summary (TS), Trial Inclusion Exclusion Criteria (TI), and Device Identifiers (DI) are study-level domains; thus, there is no individual patient-level data in those domains. Instead, there is information about the uniqueness of each institution, for instance, the inclusion/exclusion criteria or the devices used at each hospital. Data collection times for each data type are presented in Fig. 316–18. As an example, we show in Fig. 4 a synthetic, representative subset of the available data for a female patient.
Fig. 3.
Data collection points for each data type.
Fig. 4.
A synthetic, representative subset of the available data for a female patient.
The CDISC SDTM data model has several advantages. For example:
It can adapt to any number of events. Frequently recorded events such as vital signs, laboratory tests, and patient status scores are stored as a series of events. The order is recorded in the variables with the suffix ‘DY,’ which describes the day of the observation relative to the patient’s hospital admission date. For example, the variable ‘VSDY’ indicates the day when a particular vital sign was measured. Events occurring within the same day can be further ordered using the variables with the suffix ‘SEQ’, which captures the sequence of events independently of the day on which they occurred.
It captures whether or not a variable was collected for a given patient (this is critical to count denominators accurately in an aggregated collection of many different datasets). The model enables this by collecting the existence of a variable separately from the occurrence or completion of that variable. E.g., if the CRF for a dataset includes data on fever, the model shows that this question was prespecified as FEVER_PRESP = Yes; if the patient had a fever, it is captured as FEVER_OCCUR = Yes; if the patient was afebrile, it is registered as FEVER_OCCUR = No. Combining these two variables makes it possible to accurately quantify how many patients were evaluated for fever and how many had a fever. This distinction is found in the ER, HE, IN, and SA tables. A full description of how SDTM is implemented for these data, Frequently Asked Questions, and other data tools are available within the IDDO suite of curation and data resources (https://www.iddo.org/tools-and-resources/data-tools) to assist analysts in understanding these nuances. The remaining tables contain study-level data (e.g., Study Inclusion Exclusion Criteria and Device Identifiers); thus, there are no individual-level data in these domains.
The dataset also contains a rich repository of free-text entries that capture more fine-grained information not included in the CRF solicited entries. Such information can be identified by applying simple search functions or Natural Language Processing (NLP) techniques to the **TERM variable. Supplementary Table 1 describes how data is distributed across the domain data tables and how many unique patients are included in each table.
Patient characteristics
Among the 708,158 patients whose data were entered as of September 2021, 552,366 (78%) had laboratory confirmation of SARS-CoV-2 infection, and 50,426 (7,1%) were clinically diagnosed (where testing was not available or results were not reported). Of these patients, the median age (interquartile ranges: first quartile (Q1) and third quartile (Q3)) is 58 (IQR: 44–72) years, 48.9% are male, and 50.9% are female (the sex of 0.1% of the patients is unknown). A total of 126,069 (20.9%) patients were admitted to a critical care unit (ICU or HDU), and in-hospital mortality was 23.5%5. Table 1 provides a breakdown of the population by continent, and Supplementary Table 1 shows the number of unique patients with data reported per each domain.
Table 1.
Details of the ISARIC-COVID-19 patient population by continent.
| Continent | Global | Africa | Europe | Asia | North America | South and Central America | Oceania |
|---|---|---|---|---|---|---|---|
| n = 602792 | n = 369467 | n = 206992 | n = 16019 | n = 6687 | n = 2709 | n = 448 | |
| Critical care admission, counts (%) | 126069 (20.91) | 73095 (19.78) | 35454 (17.13) | 11544 (72.06) | 3619 (54.12) | 1872 (69.10) | 427 (95.31) |
| Age, years, median (Q1-Q3) | 58 (44–72) | 54 (40–66) | 70 (54–82) | 58 (46–68) | 64 (52–76) | 54 (42–66) | 62(51–70) |
| Male, counts (%) | 294928 (48.93) | 165376 (44.76) | 113148 (54.66) | 10366 (64.71) | 3857 (57.68) | 1659 (61.24) | 269 (60.04) |
| In-hospital mortality, counts (%) | 141646 (23.5) | 88737 (24.02) | 46424 (22.43) | 4310 (26.91) | 1672 (25) | 440 (16.24) | 59 (13.17) |
The information presented in the table is based on the raw data, and there is missing data, for instance: 470 patients do not have their country of origin registered; 8143 patients do not have age; 149 do not have their sex registered, and the outcome of 10130 patients is missing.
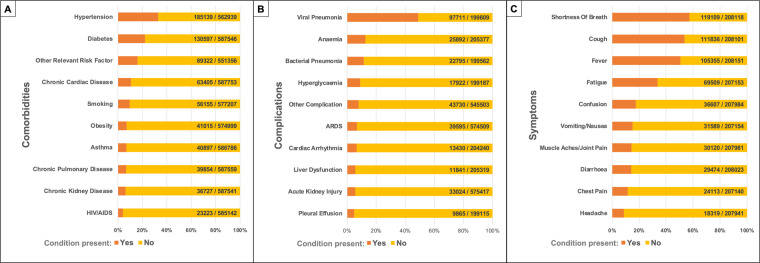
The most frequently reported comorbidities, symptoms at hospital admission, and complications during hospital admission are presented in Fig. 5. Among comorbid conditions, hypertension (30.7%), diabetes mellitus (29.6%), and chronic cardiac disease (10.5%) were the most frequently reported. The top five symptoms at admission were cough (23.7%), shortness of breath (19.8%), fever (17.5%), fatigue (11.5%), and altered consciousness (6.1%). Regarding complications, viral pneumonia (16.2%), acute respiratory distress syndrome (6.6%), acute kidney injury (5.5%), anaemia (4.3%), and bacterial pneumonia (3.8%) were the most frequently identified.
Fig. 5.
Distribution of primary symptoms, comorbidities, and treatments. (A) shows the prevalence of comorbidities; (B) shows the prevalence of symptoms at admission; (C) shows the proportion of patients receiving each treatment.
Technical Validation
Data submitted via the ISARIC REDCap system are subjected to a series of field-specific data quality checks designed by ISARIC. These trigger error alerts inform users of issues based on value limits, validate dates, flag missing variables, and perform logic checks to compare related variables. Data are further reviewed by a data manager who sends data quality reports and queries to sites when critical data are missing or outside expected values. Staff at data collection sites review the alerts and make the necessary corrections to their data in the REDCap system.
Data uploaded to the IDDO platform are verified during the ‘pre-mapping’ and ‘data review and edit checks’ processes described above. Interpretation of the data dictionary (for sites that used a unique data collection tool) and any missing values are queried directly with staff at the data collection sites. Results are charted per variable to identify and query outlier values. Where correction is suggested, the contributing site is contacted and asked to correct the data as needed before re-uploading them to the data platform.
Usage Notes
The utility of the data collected is optimised by issuing regular open-access ISARIC COVID-19 Clinical Data Reports (https://isaric.org/research/covid-19-clinical-research-resources/evidence-reports/) and periodic updates to the ISARIC COVID-19 Dashboard (https://livedataoxford.shinyapps.io/CovidClinicalDataDashboard/). Data are available for analysis through two mechanisms to maximize uptake: a collaborative mechanism for ISARIC partners who contribute data to the dataset and a data-sharing platform for external researchers. The sites that contribute to the data retain ownership and decision-making authority on their data at all times.
It is essential to highlight that more countries are globally transitioning to digital-based healthcare systems. During the transitioning process, quality control measures are necessary to enhance the effectiveness of healthcare-related communication and data quality19. Thus, the ISARIC-COVID-19 dataset can generate insights facilitating quality control measures, especially in developing countries where scarce scientific resources.
Data access
Staff from sites that contribute data to the dataset may access data for collaborative analysis via the ISARIC Partner Analysis scheme (https://isaric.org/research/isaric-partner-analysis-frequently-asked-questions/). Proposals for these analyses are governed and supported by ISARIC and executed with all data contributors’ contributions, oversite, and accreditation4,10,20. ISARIC provides statistical, clinical, and administrative support to promote analyses by partners who contribute the data, especially those based in low-resource settings.
External researchers who have not contributed to the dataset are also welcome to submit a data access and analysis proposal via the IDDO platform (https://www.iddo.org/covid19). An independent Data Access Committee reviews these requests according to the Data Access Guidelines of the platform. (https://www.iddo.org/covid19/data-sharing/accessing-data). Statistical analysis plans and outputs from both types of access can be viewed at: https://www.iddo.org/covid19/research/approved-uses-platform-data.
Data management, curation, governance, and the data-sharing platform are free to use and supported by the ISARIC and IDDO data management teams. When shared through the governed data access mechanisms, the ISARIC COVID-19 database is provided as a collection of comma-separated value (CSV) files (i.e., tables), along with scripts to help import the data into PostgreSQL and codes that enable the reuse of the data. Notably, where data transformations are made during the database construction process, care is taken not to modify raw study data. The teams performing analyses can develop analytic codes based on assumptions they deem appropriate.
Data use
The breadth of analyses published to date demonstrates the diversity of science that can be generated from these data. Examples include identification of unique COVID-19 symptomology at the extremities of age21; to develop the ISARIC 4 C mortality score that outperformed existing scores and showed utility to directly inform clinical decision making22; to identify temporal trends in inpatient journeys and inform resource needs in an evolving pandemic10, and to improve the diagnosis of acute kidney injury23. Further analyses to develop natural language processing, understand neurological outcomes in COVID-19 and develop models that predict a range of outcomes.
The use of such a large and diverse dataset is not without challenges. Robust interpretation of analytic outputs requires an understanding of the variation in recruitment practices between sites and during the course of the outbreak and the availability of treatments and facilities (e.g., ICUs and ventilators) across the range of resource settings. ISARIC’s collaborative approach to research outputs addresses these challenges by involving all staff who contributed to the collection of data in the review of the analysis plans and manuscripts. When designing an analysis plan, researchers must also consider which data are and are not available from each site and account for high levels of missingness, particularly during regional peaks in COVID-19 transmission. The CDISC SDTM data model was selected for harmonisation of these data, specifically because it captures these aspects of data providence. Those using the dataset benefit from the richness of the model; however, they will need to master the challenges of its complexity. Tools to support understanding of the data model can be found at https://www.iddo.org/tools-and-resources/data-tools.
Collaborative research
The ISARIC WHO characterization protocol has proven to be a successful strategy for generating standardized data from multiple sites that international researchers can access for analysis18,21,22,24–27. Having a pre-prepared protocol for clinical investigation of an emerging infectious disease established before the beginning of the COVID-19 pandemic allowed us to gather patient data very early in the pandemic. As a result, contributors benefited from clinical data captured in other regions before they experienced cases and improved confidence in a larger dataset. By implementing systems to harmonize global data, ISARIC and IDDO have made international collaboration more efficient1. The evolution of these systems, including integrating epidemiological and genomic data to address new types of research questions, is in progress. Finally, ISARIC’s data governance model allows members and non-members to propose research questions that could be answered using this dataset, which has helped advance science and empowers scientists worldwide4,10,20. This open and collaborative approach maximizes the scientific utility and public health impact of global data. With a focus on ensuring the representation of patient data and researchers from lower-resourced settings, the ISARIC network has accelerated understanding of COVID-19, advanced preparedness for future pandemics, and raised the bar on global collaboration for health.
Supplementary information
Acknowledgements
The investigators acknowledge: This work is part of the Grand Challenges ICODA pilot initiative, delivered by Health Data Research UK and funded by the Bill & Melinda Gates Foundation and the Minderoo Foundation. The philanthropic support of the donors to the University of Oxford’s COVID-19 Research Response Fund; UK Foreign, Commonwealth and Development Office and Wellcome [215091/Z/18/Z and 220757/Z/20/Z]; the Bill & Melinda Gates Foundation [OPP1209135]; the National Institute for Health Research (NIHR; award CO-CIN-01); the Medical Research Council (MRC; grant MC_PC_19059); the NIHR Health Protection Research Unit (HPRU) in Emerging and Zoonotic Infections at University of Liverpool in partnership with Public Health England (PHE)(award 200907); NIHR HPRU in Respiratory Infections at Imperial College London with PHE (award 200927); Liverpool Experimental Cancer Medicine Centre (grant C18616/A25153); NIHR Biomedical Research Centre at Imperial College London (award IS-BRC-1215-20013); NIHR Clinical Research Network (infrastructure support); CIHR Coronavirus Rapid Research Funding Opportunity OV2170359 and was coordinated out of Sunnybrook Research Institute; the endorsement of the Irish Critical Care- Clinical Trials Group, co-ordinated in Ireland by the Irish Critical Care- Clinical Trials Network at University College Dublin and funded by the Health Research Board of Ireland [CTN-2014-12]; Rapid European COVID-19 Emergency Response research (RECOVER) [H2020 project 101003589]; European Clinical Research Alliance on Infectious Diseases (ECRAID) [965313]; COVID clinical management team, AIIMS, Rishikesh, India; Cambridge NIHR Biomedical Research Centre; the dedication and hard work of the Groote Schuur Hospital Covid ICU Team; the Groote Schuur nursing and University of Cape Town registrar bodies coordinated by the Division of Critical Care at the University of Cape Town; Wellcome Trust fellowship [205228/Z/16/Z]; the Liverpool School of Tropical Medicine; the University of Oxford; the dedication and hard work of the Norwegian SARS-CoV-2 study team; the Research Council of Norway grant no 312780; a philanthropic donation from Vivaldi Invest A/S owned by Jon Stephenson von Tetzchner; Innovative Medicines Initiative Joint Undertaking under Grant Agreement No. 115523 COMBACTE, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies, in-kind contribution; preparedness work conducted by the Short Period Incidence Study of Severe Acute Respiratory Infection; Stiftungsfonds zur Förderung der Bekämpfung der Tuberkulose und anderer Lungenkrankheiten of the City of Vienna, Project Number: APCOV22BGM; Italian Ministry of Health “Fondi Ricerca corrente–L1P6” to IRCCS Ospedale Sacro Cuore–Don Calabria; Australian Department of Health grant (3273191); Gender Equity Strategic Fund at University of Queensland; Artificial Intelligence for Pandemics (A14PAN) at University of Queensland; the Australian Research Council Centre of Excellence for Engineered Quantum Systems (EQUS, CE170100009); the Prince Charles Hospital Foundation, Australia; UK Medical Research Council Clinical Research Training Fellowship MR/V001671/1; Instituto de Salud Carlos III, Ministerio de Ciencia, Spain; Brazil, National Council for Scientific and Technological Development Scholarship number 303953/2018-7; Firland Foundation, Shoreline, Washington, USA; the French COVID cohort (NCT04262921) is sponsored by INSERM and is funding by the REACTing (REsearch & ACtion emergING infectious diseases) consortium and by a grant of the French Ministry of Health (PHRC n°20-0424). This work uses Data/Material provided by patients and collected by the NHS as part of their care and support #DataSavesLives. The Data/materials used for this research were obtained from ISARIC4C. ISARIC4C Investigators collated the COVID-19 Clinical Information Network (CO-CIN) data.
Code availability
Processing codes for the ISARIC COVID-19 database are openly available online, and contributions from the research community to share these codes are encouraged. For this reason, a public code repository has been created along with this manuscript to develop and share code collectively: https://github.com/ISARICDataPlatform/ISARICBasics.git. The content of this repository is under continuous development. Still, it has been seeded with code to generate patient-level datasets suitable for statistics and machine learning research, such as patient demographic, comorbid conditions at the time of admission, application of treatments, and severity scores, among others. It is possible for the research community to directly submit updates, improvements, and additions to the repository via GitHub. Moreover, a Jupyter Notebook containing the code used to generate the tables and descriptive statistics included in this paper is openly available on GitHub.
Competing interests
Allavena, C. declares personal fees from ViiVHealthcare, MSD, Janssen, and Gilead, all outside the submitted work. Andréjak, C. declares personal fees for lectures from Astra Zeneca, outside the submitted work. Antonelli, M. declares unrestricted research grants from GE and Estor/Toray, Board participation from Pfizer and Shionogi. All unrelated to the present work. Beltrame, A. has nothing to declare concerning the current work. A Borie, R. declares personal fees for Roche, Sanofi, and Boehringer Ingelheim’s lectures outside the submitted work. Bosse, Hans Martin is co-investigator for placebo studies in infants and children in clinical trials by Actelion/Janssen (Johnson&Johnson), outside the submitted work. Cheng, M. declares grants from McGill Interdisciplinary Initiative in Infection and Immunity, grants from Canadian Institutes of Health Research, during the conduct of the study; personal fees from GEn1E Lifesciences (as a member of the scientific advisory board), personal fees from nplex biosciences (as a member of the scientific advisory board), outside the submitted work. He is the co-founder of Kanvas Biosciences and owns equity in the company. In addition, M. Cheng reports a patent Methods for detecting tissue damage, graft versus host disease, and infections using cell-free DNA profiling pending, and a patent Methods for assessing the severity and progression of SARS-CoV-2 infections using cell-free DNA pending. Cholley, B. declares personal fees (for lectures and participation to advisory boards) from Edwards, Amomed, Nordic Pharma, and Orion Pharma. Claure-Del Granado, R. declares individual fees (for lectures and participation to advisory boards) from Nova Biomedical, Medtronic, and Baxter all outside the submitted work. Cruz-Bermúdez J.L. declares personal fees from Elsevier for advice outside the submitted work. Cummings, M. and O’Donnell, M. participated as investigators for clinical trials evaluating the efficacy and safety of remdesivir (sponsored by Gilead Sciences) and convalescent plasma (sponsored by Amazon) in hospitalized patients with COVID-19. Support for this work is paid to Columbia University. Dalton, H. declares personal fees for medical director of Innovative ECMO Concepts and honorarium from Abiomed/BREETHE Oxi-1 and Instrumentation Labs. Consultant fee, Entegrion Inc. Dyrhol-Riise, AM, declares grants from Gilead outside this work. Deplanque, D. declares personal fees from Biocodex, Bristol-Myers Squibb, and Pfizer (advisory boards). Donnelly, C.A. declares research funding from the UK Medical Research Council and the UK National Institute for Health Research. Douglas, J.J. declares personal fees from lectures from Sunovion and Merck, consulting fees from Pfizer. Durante-Mangoni, E. declares funding via his Institution from MSD, Pfizer, and personal fees or participation in advisory boards or participation to the speaker’s bureau of Roche, Pfizer, MSD, Angelini, Correvio, Nordic Pharma, Bio-Merieux, Abbvie, Sanofi-Aventis, Medtronic, Tyrx and DiaSorin. Gordon, AC. reports a Research Professorship from NIHR (RP-2015-06-18), consulting fees from GlaxoSmithKline, Bristol Myers Squibb, and 30 Respiratory paid to his institution outside of the submitted work. Grasselli, G. declares personal fees from Getinge, Biotest, Draeger Medical, Fisher & Paykel, MSD, and unrestricted research grants from MSD and Fisher & Paykel, all outside the submitted work. Guerguerian AM. Participated as a site investigator for the Hospital for Sick Children, Toronto, Canada, through the SPRINT-SARI Study via the Canadian Critical Care Trials Group sponsored in part by the Canadian Institutes of Health Research. Hammond, TC declares consulting fees from Regeneron, Pfizer, and Agenus. Ho, A. declares grant funding from Medical Research Council UK, Scottish Funding Council - Grand Challenges Research Fund, and the Wellcome Trust, outside this submitted work. Holter, J. C. reports grants from Research Council of Norway grant no 312780, and Vivaldi Invest A/S owned by Jon Stephenson von Tetzchner, during the conduct of the study. Hulot, J.S. reports grants from Bioserenity, Sanofi, Servier, and Novo Nordisk.; speaker, advisory board or consultancy fees from Amgen, Astra Zeneca, Bayer, Bioserenity, Boehringer Ingelheim, Bristol-Myers Squibb, MSD, Novartis, Novo Nordisk, Vifor (all unrelated to the present work). Kalleberg, K.T. Is a founder and shareholder of the company Age Labs, which develops epigenetic tests, including one for COVID-19 severity. Kimmoun, A. declares personal fees (payment for lectures) from Baxter, Aguettant, Aspen. Kumar, D. declares grants and personal fees from Roche, GSK, and Merck, and personal fees from Pfizer and Sanofi. Kutsogiannis, D.J. declares personal fees for a lecture from Tabuk Pharmaceuticals and the Saudi Critical Care Society. Kutsyna, G. declares the study consulting fee for clinical trial ClinicalTrials.gov Identifier: NCT04762628. Laffey, J. reports that he has received fees for consultancy from GlaxoSmithKline and from Baxter Therapeutics for work outside the scope of this work. Lairez, O. declares grant funding from Pfizer; conference fees from Amicus, GE Healthcare, Novartis, Sanofi-Genzyme, and Takeda-Shire; and consultancy fees from Alnylam, Amicus, Pfizer, Takeda-Shire. Lee, J. reports grants from European Commission PREPARE grant agreement No 602525, European Commission RECOVER Grant Agreement No 101003589 and European Commission ECRAID-Plan Grant Agreement 825715 supporting the conduct, coordination, and management of the work. Lee, T.C. declares research salary support from les Fonds de recherche du Québec – Santé. Lefèvre, B. declares travel/accommodation/meeting expenses from Mylan and Gilead, all outside the submitted work. Lellouche, F. declares grants from CIHR for COVID-19 studies is co-founder and administrator of Oxynov. inc, fees from Fisher&Paykel, Vygon, and Novus. Lemaignen, A. declares personal fees (payment for lectures) from MSD and Gilead; and travel/accommodation/meeting expenses from Pfizer. Leone, M declares personal fees from Gilead, MSD, Aspen, Ambu, and Amomed Lescure, F.X. declares personal fees (payment for lectures) from Gilead, MSD; and travel/accommodation/meeting expenses from Astellas, Eumedica, MSD. Lim, W.S. declares his institution has received unrestricted investigator-initiated research funding from Pfizer for an unrelated multicentre cohort study in which he is the Chief Investigator, and research funding from the National Institute for Health Research, the UK, for various clinical trials outside the submitted work. Liu, K. reports personal fees from MERA and receives a salary from TXP Medical completely outside the submitted work. Maier, Lars S. has nothing to declare with respect to the present work. Mark, P.B. declares grant support from Boehringer Ingelheim, lecture fees and/or travel support from AstraZeneca, Astellas, Vifor, Pharmacosmos, and Napp, and grant funding from British Heart Foundation, Medical Research Council, National Institute for Health Research and Kidney Research UK all outside the submitted work. Martin-Blondel G declares support for attending meetings and personal fees from BMS, MSD, Janssen, Sanofi, Pfizer, and Gilead for lectures outside the submitted work. Martin-Loeches I. declared lectures for Gilead, Thermofisher, Pfizer, MSD; advisory board participation for Fresenius Kabi, Advanz Pharma, Gilead, Accelerate, Merck; and consulting fees for Gilead outside of the submitted work. Martín-Quiros, A. declares consulting fees for Gilead. Mentré F declares consulting fees from IPSEN, Servier, and Da Volterra and reports research grants to her group from Sanofi, Roche, Servier, and Da Voleterra, all outside the submitted work. Montrucchio, G declares personal fees for a lecture from Pfizer Gilead outside the submitted work. Murthy, S declares receiving salary support from the Health Research Foundation and Innovative Medicines Canada Chair in Pandemic Preparedness Research. Nichol, A. declares a grant from the Health Research Board of Ireland to support data collection in Ireland (CTN-2014-012), an unrestricted grant from BAXTER for the TAME trial kidney substudy, and consultancy fees paid to his institution from AM-PHARMA. Nseir S. declares lectures for Gilead, Pfizer, MSD, Biomérieux, Fischer and Paykel, and Bio-Rad, outside the submitted work. Openshaw, P. has served on scientific advisory boards for Janssen/J&J, Oxford Immunotech Ltd, GSK, Nestle, and Pfizer (fees to Imperial College). He is Imperial College lead investigator on EMINENT, a consortium funded by the MRC and GSK. He is a member of the RSV Consortium in Europe (RESCEU) and Inno4Vac, Innovative Medicines Initiatives (IMI) from the European Union. Peltan, I.D. declares grant support from the National Institutes of Health and, outside the submitted work, grant support from Centers for Disease Control and Prevention, National Institutes of Health, and Jannsen and payments to his institution from Regeneron and Asahi Kasei Pharma. Pesenti, A. declares personal fees from Maquet, Novalung/Xenios, Baxter, and Boehringer Ingelheim. Peytavin G. declares consulting fees (for lectures and/or participation in advisory boards) and travel grants from Gilead Sciences, Janssen, Merck, Takeda, Theratechnologies, and ViiV Healthcare. Poissy, J. declares personal fees from Gilead for lectures outside the submitting work. Povoa, P. declares personal fees (for lectures and advisory boards) from MSD, Technophage, Sanofi, and Gilead. Póvoas, D. declares consulting fees (for lectures and/or participation in advisory boards) from Roche and Viiv Healthcare; and travel/accommodation/meeting expenses from Abbvie, Gilead Sciences, Janssen Cilag, Merck Sharp & Dohme, and ViiV Healthcare. Rewa, O. declares honoraria from Baxter Healthcare Inc and Leading Biosciences Inc. Rössler, B. declares grants from CytoSorbent Inc. Rossanese, A. declares consulting fees (for lectures and/or participation to advisory boards) from Emergent BioSolutions and Sanofi Pasteur, but all outside of the frame of the submitted work. Săndulescu, O. has been an investigator in COVID-19 clinical trials by Algernon Pharmaceuticals, Atea Pharmaceuticals, Regeneron Pharmaceuticals, Diffusion Pharmaceuticals, and Celltrion, Inc. and Atriva Therapeutics, outside the scope of the submitted work. Semple, M.G. reports grants from DHSC National Institute of Health Research UK, from the Medical Research Council UK, and from the Health Protection Research Unit in Emerging & Zoonotic Infections, University of Liverpool, supporting the conduct of the study; other interest in Integrum Scientific LLC, Greensboro, NC, USA, outside the submitted work. Serpa Neto, A. declares personal lecture fees from Drager outside the submitted work. Serrano-Balazote, P. declares funding via his Institution from Novartis and Janssen, and personal fees or participation in advisory boards or participation to the speaker’s bureau of Roche, all outside of the submitted work. Shrapnel, S. participated as an investigator for an observational study analysing ICU patients with COVID-19 (for the Critical Care Consortium including ECMOCARD) funded by The Prince Charles Hospital Foundation during the conduct of this study. S. Shrapnel reports in-kind support from the Australian Research Council Centre of Excellence for Engineered Quantum Systems (CE170100009). Streinu-Cercel, Adrian has been an investigator in COVID-19 clinical trials by Algernon Pharmaceuticals, Atea Pharmaceuticals, Regeneron Pharmaceuticals, Diffusion Pharmaceuticals, and Celltrion, Inc., outside the scope of the submitted work. Streinu-Cercel, Anca has been an investigator in COVID-19 clinical trials by Algernon Pharmaceuticals, Atea Pharmaceuticals, Regeneron Pharmaceuticals, Diffusion Pharmaceuticals, and Celltrion Inc. and Atriva Therapeutics, outside the scope of the submitted work. Summers, C. reports that she has received fees for consultancy for Abbvie and Roche relating to COVID-19 therapeutics. She was also the UK Chief Investigator of a GlaxoSmithKline plc sponsored study of therapy for COVID and is a member of the UK COVID Therapeutic Advisory Panel (UK-CTAP). Outside the scope of this work, Dr. Summers’ institution receives research grants from the Wellcome Trust, UKRI/MRC, National Institute for Health Research (NIHR), GlaxoSmithKline, and AstraZeneca to support research in her laboratory. Susanne Dudman reports grants from Research Council of Norway grant no 312780. Tedder, R. reports grants from MRC/UKRI during the conduct of the study. In addition, R. Tedder has a patent United Kingdom Patent Application No. 2014047.1 “SARS-CoV-2 antibody detection assay” issued. Terzi, N. reports personal fees from Pfizer outside the submitted work. Timsit, J.F. participated in an advisory board for MSD, Pfizer, nabriva, Gilead, Shionoghi, Medimune outside the submitted work. JF Timsit declared lecture fees from MSD, Biomerieux, Pfizer, Shionoghi. Turtle, L. reports grants from MRC/UKRI during the conduct of the study and fees from Eisai for delivering a lecture related to COVID-19 and cancer, paid to the University of Liverpool. Ullrich, R. reports grant funding to his institution from Apeptico, APEIRON, Biotest, Bayer, CCORE, and Philips and personal fees from Biotest. He holds European patent EP15189777.4 “Blood purification device” and equity in CCORE Technology GesmbH, medical device research, and development company. Visseaux B. declares personal fees from BioMérieux, Qiagen, and Gilead and research grants from Qiagen, all outside the submitted work. West, E. reports grant funding from the Firland Foundation, the US CDC, and the Bill and Melinda Gates Foundation for studies of COVID-19, and grant funding from the US NIH for studies of other respiratory infections. Couffin-Cadiergues S. has nothing to declare relating to the present work. Søraas, A. Is a founder of the company Age Labs, which develops epigenetic tests, including COVID-19 severity.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Laura Merson, Email: laura.merson@ndm.ox.ac.uk.
Luis Felipe Reyes, Email: luis.reyes5@unisabana.edu.co.
ISARIC Clinical Characterization Group:
Ali Abbas, Sheryl Ann Abdukahil, Nurul Najmee Abdulkadir, Ryuzo Abe, Laurent Abel, Lara Absil, Kamal Abu Jabal, Hiba Abu Zayyad, Subhash Acharya, Andrew Acker, Shingo Adachi, Elisabeth Adam, Enrico Adriano, Diana Adrião, Saleh Al Ageel, Shakeel Ahmed, Marina Aiello, Kate Ainscough, Eka Airlangga, Tharwat Aisa, Ali Ait Hssain, Younes Ait Tamlihat, Takako Akimoto, Ernita Akmal, Eman Al Qasim, Tala Al-dabbous, Abdulrahman Al-Fares, Razi Alalqam, Angela Alberti, Senthilkumar Alegesan, Cynthia Alegre, Marta Alessi, Beatrice Alex, Kévin Alexandre, Huda Alfoudri, Adam Ali, Imran Ali, Naseem Ali Shah, Naseem Ali Sheikh, Kazali Enagnon Alidjnou, Jeffrey Aliudin, Qabas Alkhafajee, Clotilde Allavena, Nathalie Allou, Aneela Altaf, João Alves, João Melo Alves, Rita Alves, Maria Amaral, Nur Amira, Heidi Ammerlaan, Phoebe Ampaw, Roberto Andini, Claire Andrejak, Andrea Angheben, François Angoulvant, Séverine Ansart, Sivanesen Anthonidass, Massimo Antonelli, Carlos Alexandre Antunes de Brito, Kazi Rubayet Anwar, Ardiyan Apriyana, Yaseen Arabi, Irene Aragao, Francisco Arancibia, Carolline Araujo, Antonio Arcadipane, Patrick Archambault, Lukas Arenz, Jean-Benoît Arlet, Christel Arnold-Day, Ana Aroca, Lovkesh Arora, Rakesh Arora, Elise Artaud-Macari, Diptesh Aryal, Motohiro Asaki, Angel Asensio, Elizabeth Ashley, Muhammad Ashraf, Namra Asif, Mohammad Asim, Jean Baptiste Assie, Amirul Asyraf, Anika Atique, A. M. Udara Lakshan Attanyake, Johann Auchabie, Hugues Aumaitre, Adrien Auvet, Eyvind W. Axelsen, Laurène Azemar, Cecile Azoulay, Benjamin Bach, Delphine Bachelet, Claudine Badr, Roar Bævre-Jensen, Nadia Baig, J. Kenneth Baillie, J Kevin Baird, Erica Bak, Agamemnon Bakakos, Nazreen Abu Bakar, Andriy Bal, Mohanaprasanth Balakrishnan, Valeria Balan, Firouzé Bani-Sadr, Renata Barbalho, Nicholas Yuri Barbosa, Wendy S. Barclay, Saef Umar Barnett, Michaela Barnikel, Helena Barrasa, Audrey Barrelet, Cleide Barrigoto, Marie Bartoli, Cheryl Bartone, Joaquín Baruch, Mustehan Bashir, Romain Basmaci, Muhammad Fadhli Hassin Basri, Denise Battaglini, Jules Bauer, Diego Fernando Bautista Rincon, Denisse Bazan Dow, Abigail Beane, Alexandra Bedossa, Ker Hong Bee, Husna Begum, Sylvie Behilill, Karine Beiruti, Albertus Beishuizen, Aleksandr Beljantsev, David Bellemare, Anna Beltrame, Beatriz Amorim Beltrão, Marine Beluze, Nicolas Benech, Lionel Eric Benjiman, Dehbia Benkerrou, Suzanne Bennett, Luís Bento, Jan-Erik Berdal, Delphine Bergeaud, Hazel Bergin, José Luis Bernal Sobrino, Giulia Bertoli, Lorenzo Bertolino, Simon Bessis, Adam Betz, Sybille Bevilcaqua, Karine Bezulier, Amar Bhatt, Krishna Bhavsar, Isabella Bianchi, Claudia Bianco, Farah Nadiah Bidin, Moirangthem Bikram Singh, Felwa Bin Humaid, Mohd Nazlin Bin Kamarudin, François Bissuel, Patrick Biston, Laurent Bitker, Jonathan Bitton, Pablo Blanco-Schweizer, Catherine Blier, Frank Bloos, Mathieu Blot, Lucille Blumberg, Filomena Boccia, Laetitia Bodenes, Alice Bogaarts, Debby Bogaert, Anne-Hélène Boivin, Pierre-Adrien Bolze, François Bompart, Patrizia Bonelli, Aurelius Bonfasius, Diogo Borges, Raphaël Borie, Hans Martin Bosse, Elisabeth Botelho-Nevers, Lila Bouadma, Olivier Bouchaud, Sabelline Bouchez, Dounia Bouhmani, Damien Bouhour, Kévin Bouiller, Laurence Bouillet, Camile Bouisse, Thipsavanh Bounphiengsy, Latsaniphone Bountthasavong, Anne-Sophie Boureau, John Bourke, Maude Bouscambert, Aurore Bousquet, Jason Bouziotis, Bianca Boxma, Marielle Boyer-Besseyre, Maria Boylan, Fernando Augusto Bozza, Axelle Braconnier, Cynthia Braga, Timo Brandenburger, Filipa Brás Monteiro, Luca Brazzi, Dorothy Breen, Patrick Breen, Kathy Brickell, Alex Browne, Shaunagh Browne, Nicolas Brozzi, Sonja Hjellegjerde Brunvoll, Marjolein Brusse-Keizer, Nina Buchtele, Christian Buesaquillo, Polina Bugaeva, Marielle Buisson, Danilo Buonsenso, Erlina Burhan, Aidan Burrell, Ingrid G. Bustos, Denis Butnaru, André Cabie, Susana Cabral, Eder Caceres, Cyril Cadoz, Mia Callahan, Kate Calligy, Jose Andres Calvache, Caterina Caminiti, João Camões, Valentine Campana, Paul Campbell, Josie Campisi, Cecilia Canepa, Mireia Cantero, Pauline Caraux-Paz, Sheila Cárcel, Chiara Simona Cardellino, Filipa Cardoso, Filipe Cardoso, Nelson Cardoso, Sofia Cardoso, Simone Carelli, Francesca Carlacci, Nicolas Carlier, Thierry Carmoi, Gayle Carney, Inês Carqueja, Marie-Christine Carret, François Martin Carrier, Ida Carroll, Gail Carson, Leonor Carvalho, Maire-Laure Casanova, Mariana Cascão, Siobhan Casey, José Casimiro, Bailey Cassandra, Silvia Castañeda, Nidyanara Castanheira, Guylaine Castor-Alexandre, Henry Castrillón, Ivo Castro, Ana Catarino, François-Xavier Catherine, Paolo Cattaneo, Roberta Cavalin, Giulio Giovanni Cavalli, Alexandros Cavayas, Adrian Ceccato, Minerva Cervantes-Gonzalez, Anissa Chair, Catherine Chakveatze, Adrienne Chan, Meera Chand, Christelle Chantalat Auger, Jean-Marc Chapplain, Julie Chas, Allegra Chatterjee, Mobin Chaudry, Jonathan Samuel Chávez Iñiguez, Anjellica Chen, Yih-Sharng Chen, Matthew Pellan Cheng, Antoine Cheret, Alfredo Antonio Chetta, Thibault Chiarabini, Julian Chica, Suresh Kumar Chidambaram, Leong Chin Tho, Catherine Chirouze, Davide Chiumello, Hwa Jin Cho, Sung-Min Cho, Bernard Cholley, Danoy Chommanam, Marie-Charlotte Chopin, Ting Soo Chow, Yock Ping Chow, Nathaniel Christy, Hiu Jian Chua, Jonathan Chua, Jose Pedro Cidade, José Miguel Cisneros Herreros, Barbara Wanjiru Citarella, Anna Ciullo, Emma Clarke, Jennifer Clarke, Rolando Claure-Del Granado, Sara Clohisey, Perren J. Cobb, Cassidy Codan, Caitriona Cody, Alexandra Coelho, Megan Coles, Gwenhaël Colin, Michael Collins, Sebastiano Maria Colombo, Pamela Combs, Marie Connor, Anne Conrad, Sofía Contreras, Elaine Conway, Graham S. Cooke, Mary Copland, Hugues Cordel, Amanda Corley, Sabine Cornelis, Alexander Daniel Cornet, Arianne Joy Corpuz, Andrea Cortegiani, Grégory Corvaisier, Emma Costigan, Camille Couffignal, Sandrine Couffin-Cadiergues, Roxane Courtois, Stéphanie Cousse, Rachel Cregan, Charles Crepy D’Orleans, Sabine Croonen, Gloria Crowl, Jonathan Crump, Claudina Cruz, Juan Luis Cruz Berm, Jaime Cruz Rojo, Marc Csete, Alberto Cucino, Ailbhe Cullen, Matthew Cummings, Ger Curley, Gerard Curley, Elodie Curlier, Colleen Curran, Paula Custodio, Federico D’Amico, Frédérick D’Aragon, Eric D’Ortenzio, Ana da Silva Filipe, Charlene Da Silveira, Al-Awwab Dabaliz, Andrew Dagens, John Arne Dahl, Darren Dahly, Heidi Dalton, Jo Dalton, Seamus Daly, Nick Daneman, Corinne Daniel, Emmanuelle A Dankwa, Jorge Dantas, Mark de Boer, Gillian de Loughry, Diego de Mendoza, Etienne De Montmollin, Rafael Freitas de Oliveira França, Ana Isabel de Pinho Oliveira, Rosanna De Rosa, Cristina De Rose, Thushan de Silva, Peter de Vries, Jillian Deacon, David Dean, Alexa Debard, Bianca DeBenedictis, Marie-Pierre Debray, Nathalie DeCastro, William Dechert, Lauren Deconninck, Romain Decours, Eve Defous, Isabelle Delacroix, Eric Delaveuve, Karen Delavigne, Nathalie M. Delfos, Ionna Deligiannis, Andrea Dell’Amore, Christelle Delmas, Pierre Delobel, Corine Delsing, Elisa Demonchy, Emmanuelle Denis, Dominique Deplanque, Pieter Depuydt, Mehul Desai, Diane Descamps, Mathilde Desvallées, Santi Dewayanti, Pathik Dhanger, Alpha Diallo, Sylvain Diamantis, André Dias, Andrea Dias, Juan Jose Diaz, Priscila Diaz, Rodrigo Diaz, Kévin Didier, Jean-Luc Diehl, Wim Dieperink, Jérôme Dimet, Vincent Dinot, Fara Diop, Alphonsine Diouf, Yael Dishon, Félix Djossou, Annemarie B. Docherty, Helen Doherty, Arjen M Dondorp, Andy Dong, Christl A. Donnelly, Maria Donnelly, Chloe Donohue, Sean Donohue, Yoann Donohue, Peter Doran, Céline Dorival, Phouvieng Douangdala, James Joshua Douglas, Renee Douma, Nathalie Dournon, Triona Downer, Joanne Downey, Mark Downing, Tom Drake, Aoife Driscoll, Amiel A. Dror, Murray Dryden, Claudio Duarte Fonseca, Vincent Dubee, François Dubos, Audrey Dubot-Pérès, Alexandre Ducancelle, Toni Duculan, Susanne Dudman, Abhijit Duggal, Paul Dunand, Jake Dunning, Mathilde Duplaix, Emanuele Durante-Mangoni, Lucian Durham, III, Bertrand Dussol, Juliette Duthoit, Xavier Duval, Anne Margarita Dyrhol-Riise, Sim Choon Ean, Marco Echeverria-Villalobos, Giorgio Economopoulos, Michael Edelstein, Siobhan Egan, Linn Margrete Eggesbø, Carla Eira, Mohammed El Sanharawi, Subbarao Elapavaluru, Brigitte Elharrar, Jacobien Ellerbroek, Merete Ellingjord-Dale, Philippine Eloy, Tarek Elshazly, Iqbal Elyazar, Isabelle Enderle, Tomoyuki Endo, Chan Chee Eng, Ilka Engelmann, Vincent Enouf, Olivier Epaulard, Martina Escher, Mariano Esperatti, Hélène Esperou, Marina Esposito-Farese, João Estevão, Manuel Etienne, Nadia Ettalhaoui, Anna Greti Everding, Mirjam Evers, Isabelle Fabre, Marc Fabre, Amna Faheem, Arabella Fahy, Cameron J. Fairfield, Zul Fakar, Komal Fareed, Pedro Faria, Ahmed Farooq, Hanan Fateena, Arie Zainul Fatoni, Karine Faure, Raphaël Favory, Mohamed Fayed, Niamh Feely, Laura Feeney, Jorge Fernandes, Marília Andreia Fernandes, Susana Fernandes, François-Xavier Ferrand, Eglantine Ferrand Devouge, Joana Ferrão, Carlo Ferrari, Mário Ferraz, Benigno Ferreira, Isabel Ferreira, Sílvia Ferreira, Ricard Ferrer-Roca, Nicolas Ferriere, Céline Ficko, Claudia Figueiredo-Mello, William Finlayson, Juan Fiorda, Thomas Flament, Clara Flateau, Tom Fletcher, Letizia Lucia Florio, Brigid Flynn, Deirdre Flynn, Federica Fogliazza, Claire Foley, Jean Foley, Victor Fomin, Tatiana Fonseca, Patricia Fontela, Simon Forsyth, Denise Foster, Giuseppe Foti, Erwan Fourn, Robert A. Fowler, Marianne Fraher, Diego Franch-Llasat, Christophe Fraser, John F Fraser, Marcela Vieira Freire, Ana Freitas Ribeiro, Craig French, Caren Friedrich, Ricardo Fritz, Stéphanie Fry, Nora Fuentes, Masahiro Fukuda, G Argin, Valérie Gaborieau, Rostane Gaci, Massimo Gagliardi, Jean-Charles Gagnard, Nathalie Gagné, Amandine Gagneux-Brunon, Sérgio Gaião, Linda Gail Skeie, Phil Gallagher, Elena Gallego Curto, Carrol Gamble, Yasmin Gani, Arthur Garan, Rebekha Garcia, Noelia García Barrio, Julia Garcia-Diaz, Esteban Garcia-Gallo, Navya Garimella, Federica Garofalo, Denis Garot, Valérie Garrait, Basanta Gauli, Nathalie Gault, Aisling Gavin, Anatoliy Gavrylov, Alexandre Gaymard, Johannes Gebauer, Eva Geraud, Louis Gerbaud Morlaes, Nuno Germano, Praveen Kumar Ghisulal, Jade Ghosn, Marco Giani, Carlo Giaquinto, Jess Gibson, Tristan Gigante, Morgane Gilg, Elaine Gilroy, Guillermo Giordano, Michelle Girvan, Valérie Gissot, Jesse Gitaka, Gezy Giwangkancana, Daniel Glikman, Petr Glybochko, Eric Gnall, Geraldine Goco, François Goehringer, Siri Goepel, Jean-Christophe Goffard, Jin Yi Goh, Jonathan Golob, Rui Gomes, Kyle Gomez, Joan Gómez-Junyent, Marie Gominet, Bronner P. Gonçalves, Alicia Gonzalez, Patricia Gordon, Yanay Gorelik, Isabelle Gorenne, Laure Goubert, Cécile Goujard, Tiphaine Goulenok, Margarite Grable, Jeronimo Graf, Edward Wilson Grandin, Pascal Granier, Giacomo Grasselli, Lorenzo Grazioli, Christopher A. Green, Courtney Greene, William Greenhalf, Segolène Greffe, Domenico Luca Grieco, Matthew Griffee, Fiona Griffiths, Ioana Grigoras, Albert Groenendijk, Anja Grosse Lordemann, Heidi Gruner, Yusing Gu, Fabio Guarracino, Jérémie Guedj, Martin Guego, Dewi Guellec, Anne-Marie Guerguerian, Daniela Guerreiro, Romain Guery, Anne Guillaumot, Laurent Guilleminault, Maisa Guimarães de Castro, Thomas Guimard, Marieke Haalboom, Daniel Haber, Hannah Habraken, Ali Hachemi, Amy Hackmann, Nadir Hadri, Fakhir Haidri, Sheeba Hakak, Adam Hall, Matthew Hall, Sophie Halpin, Jawad Hameed, Ansley Hamer, Raph L. Hamers, Rebecca Hamidfar, Bato Hammarström, Terese Hammond, Lim Yuen Han, Rashan Haniffa, Kok Wei Hao, Hayley Hardwick, Ewen M. Harrison, Janet Harrison, Samuel Bernard Ekow Harrison, Alan Hartman, Mohd Shahnaz Hasan, Junaid Hashmi, Madiha Hashmi, Ailbhe Hayes, Leanne Hays, Jan Heerman, Lars Heggelund, Ross Hendry, Martina Hennessy, Aquiles Henriquez-Trujillo, Maxime Hentzien, Jaime Hernandez-Montfort, Daniel Herr, Andrew Hershey, Liv Hesstvedt, Astarini Hidayah, Dawn Higgins, Eibhilin Higgins, Rita Hinchion, Samuel Hinton, Hiroaki Hiraiwa, Haider Hirkani, Hikombo Hitoto, Antonia Ho, Yi Bin Ho, Alexandre Hoctin, Isabelle Hoffmann, Wei Han Hoh, Oscar Hoiting, Rebecca Holt, Jan Cato Holter, Peter Horby, Juan Pablo Horcajada, Koji Hoshino, Kota Hoshino, Ikram Houas, Catherine L. Hough, Stuart Houltham, Jimmy Ming-Yang Hsu, Jean-Sébastien Hulot, Stella Huo, Iqbal Hussain, Samreen Ijaz, Hajnal-Gabriela Illes, Patrick Imbert, Mohammad Imran, Rana Imran Sikander, Aftab Imtiaz, Hugo Inácio, Carmen Infante Dominguez, Yun Sii Ing, Elias Iosifidis, Mariachiara Ippolito, Sarah Isgett, Tiago Isidoro, Nadiah Ismail, Margaux Isnard, Mette Stausland Istre, Junji Itai, Asami Ito, Daniel Ivulich, Danielle Jaafar, Salma Jaafoura, Julien Jabot, Clare Jackson, Nina Jamieson, Pierre Jaquet, Waasila Jassat, Coline Jaud-Fischer, Stéphane Jaureguiberry, Jeffrey Javidfar, Denise Jaworsky, Florence Jego, Anilawati Mat Jelani, Synne Jenum, Ruth Jimbo-Sotomayor, Ong Yiaw Joe, Ruth N. Jorge García, Silje Bakken Jørgensen, Cédric Joseph, Mark Joseph, Swosti Joshi, Mercé Jourdain, Philippe Jouvet, Jennifer June, Anna Jung, Hanna Jung, Dafsah Juzar, Ouifiya Kafif, Florentia Kaguelidou, Neerusha Kaisbain, Thavamany Kaleesvran, Sabina Kali, Alina Kalicinska, Karl Trygve Kalleberg, Smaragdi Kalomoiri, Muhammad Aisar Ayadi Kamaluddin, Zul Amali Che Kamaruddin, Nadiah Kamarudin, Kavita Kamineni, Darshana Hewa Kandamby, Chris Kandel, Kong Yeow Kang, Darakhshan Kanwal, Dyah Kanyawati, Pratap Karpayah, Todd Karsies, Christiana Kartsonaki, Daisuke Kasugai, Anant Kataria, Kevin Katz, Aasmine Kaur, Tatsuya Kawasaki, Christy Kay, Hannah Keane, Seán Keating, Pulak Kedia, Andrea Kelly, Aoife Kelly, Claire Kelly, Niamh Kelly, Sadie Kelly, Yvelynne Kelly, Maeve Kelsey, Ryan Kennedy, Kalynn Kennon, Sommay Keomany, Maeve Kernan, Younes Kerroumi, Sharma Keshav, Evelyne Kestelyn, Imrana Khalid, Osama Khalid, Antoine Khalil, Coralie Khan, Irfan Khan, Quratul Ain Khan, Sushil Khanal, Abid Khatak, Amin Khawaja, Krish Kherajani, Michelle E. Kho, Denisa Khoo, Ryan Khoo, Saye Khoo, Nasir Khoso, Khor How Kiat, Yuri Kida, Harrison Kihuga, Peter Kiiza, Beathe Kiland Granerud, Anders Benjamin Kildal, Jae Burm Kim, Antoine Kimmoun, Detlef Kindgen-Milles, Alexander King, Nobuya Kitamura, Eyrun Floerecke Kjetland Kjetland, Paul Klenerman, Rob Klont, Gry Kloumann Bekken, Stephen R Knight, Robin Kobbe, Chamira Kodippily, Malte Kohns Vasconcelos, Sabin Koirala, Mamoru Komatsu, Volkan Korten, Caroline Kosgei, Arsène Kpangon, Karolina Krawczyk, Sudhir Krishnan, Vinothini Krishnan, Oksana Kruglova, Deepali Kumar, Ganesh Kumar, Mukesh Kumar, Pavan Kumar Vecham, Dinesh Kuriakose, Ethan Kurtzman, Neurinda Permata Kusumastuti, Demetrios Kutsogiannis, Galyna Kutsyna, Konstantinos Kyriakoulis, Erwan L’Her, Marie Lachatre, Marie Lacoste, John G. Laffey, Marie Lagrange, Fabrice Laine, Olivier Lairez, Sanjay Lakhey, Antonio Lalueza, Marc Lambert, François Lamontagne, Marie Langelot-Richard, Vincent Langlois, Eka Yudha Lantang, Marina Lanza, Cédric Laouénan, Samira Laribi, Delphine Lariviere, Stéphane Lasry, Sakshi Lath, Naveed Latif, Odile Launay, Didier Laureillard, Yoan Lavie-Badie, Andrew Law, Andy Law, Cassie Lawrence, Teresa Lawrence, Minh Le, Clément Le Bihan, Cyril Le Bris, Georges Le Falher, Lucie Le Fevre, Quentin Le Hingrat, Marion Le Maréchal, Soizic Le Mestre, Gwenaël Le Moal, Vincent Le Moing, Hervé Le Nagard, Paul Le Turnier, Ema Leal, Marta Leal Santos, Biing Horng Lee, Heng Gee Lee, James Lee, Su Hwan Lee, Todd C. Lee, Yi Lin Lee, Gary Leeming, Bénédicte Lefebvre, Laurent Lefebvre, Benjamin Lefevre, Sylvie LeGac, Jean-Daniel Lelievre, François Lellouche, Adrien Lemaignen, Véronique Lemee, Anthony Lemeur, Gretchen Lemmink, Ha Sha Lene, Jenny Lennon, Rafael León, Marc Leone, Michela Leone, Quentin Lepiller, François-Xavier Lescure, Olivier Lesens, Mathieu Lesouhaitier, Amy Lester-Grant, Andrew Letizia, Bruno Levy, Yves Levy, Claire Levy-Marchal, Katarzyna Lewandowska, Gianluigi Li Bassi, Janet Liang, Ali Liaquat, Geoffrey Liegeon, Kah Chuan Lim, Wei Shen Lim, Chantre Lima, Bruno Lina, Lim Lina, Andreas Lind, Maja Katherine Lingad, Guillaume Lingas, Sylvie Lion-Daolio, Samantha Lissauer, Keibun Liu, Marine Livrozet, Patricia Lizotte, Antonio Loforte, Navy Lolong, Leong Chee Loon, Diogo Lopes, Dalia Lopez-Colon, Anthony L. Loschner, Paul Loubet, Bouchra Loufti, Guillame Louis, Silvia Lourenco, Lara Lovelace-Macon, Lee Lee Low, Marije Lowik, Jia Shyi Loy, Jean Christophe Lucet, Carlos Lumbreras Bermejo, Carlos M. Luna, Olguta Lungu, Liem Luong, Nestor Luque, Dominique Luton, Nilar Lwin, Ruth Lyons, Olavi Maasikas, Oryane Mabiala, Sarah MacDonald, Moïse Machado, Gabriel Macheda, Juan Macias Sanchez, Jai Madhok, Hashmi Madiha, Guillermo Maestro de la Calle, Jacob Magara, Giuseppe Maglietta, Rafael Mahieu, Sophie Mahy, Ana Raquel Maia, Lars S. Maier, Mylène Maillet, Thomas Maitre, Maria Majori, Maximilian Malfertheiner, Nadia Malik, Paddy Mallon, Fernando Maltez, Denis Malvy, Patrizia Mammi, Victoria Manda, Jose M. Mandei, Laurent Mandelbrot, Frank Manetta, Julie Mankikian, Edmund Manning, Aldric Manuel, Ceila Maria Sant’Ana Malaque, Daniel Marino, Flávio Marino, Samuel Markowicz, Charbel Maroun Eid, Ana Marques, Catherine Marquis, Brian Marsh, Laura Marsh, Megan Marshal, John Marshall, Celina Turchi Martelli, Dori-Ann Martin, Emily Martin, Guillaume Martin-Blondel, Ignacio Martin-Loeches, Alejandro Martin-Quiros, Alessandra Martinelli, Martin Martinot, Ana Martins, João Martins, Nuno Martins, Caroline Martins Rego, Gennaro Martucci, Olga Martynenko, Eva Miranda Marwali, Marsilla Marzukie, Juan Fernado Masa Jimenez, David Maslove, Phillip Mason, Sabina Mason, Sobia Masood, Basri Mat Nor, Moshe Matan, Henrique Mateus Fernandes, Meghena Mathew, Daniel Mathieu, Mathieu Mattei, Romans Matulevics, Laurence Maulin, Michael Maxwell, Javier Maynar, Mayfong Mayxay, Thierry Mazzoni, Natalie Mc Evoy, Lisa Mc Sweeney, Colin McArthur, Aine McCarthy, Anne McCarthy, Colin McCloskey, Rachael McConnochie, Sherry McDermott, Sarah E. McDonald, Aine McElroy, Samuel McElwee, Victoria McEneany, Allison McGeer, Chris McKay, Johnny McKeown, Kenneth A. McLean, Paul McNally, Bairbre McNicholas, Elaine McPartlan, Edel Meaney, Cécile Mear-Passard, Maggie Mechlin, Maqsood Meher, Omar Mehkri, Ferruccio Mele, Luis Melo, Kashif Memon, Joao Mendes, Ogechukwu Menkiti, Kusum Menon, France Mentré, Alexander J. Mentzer, Emmanuelle Mercier, Noémie Mercier, Antoine Merckx, Mayka Mergeay-Fabre, Blake Mergler, Laura Merson, Tiziana Meschi, António Mesquita, Roberta Meta, Osama Metwally, Agnès Meybeck, Dan Meyer, Alison M. Meynert, Vanina Meysonnier, Amina Meziane, Mehdi Mezidi, Giuliano Michelagnoli, Céline Michelanglei, Isabelle Michelet, Efstathia Mihelis, Vladislav Mihnovit, Hugo Miranda-Maldonado, Nor Arisah Misnan, Nik Nur Eliza Mohamed, Tahira Jamal Mohamed, Asma Moin, David Molina, Elena Molinos, Brenda Molloy, Mary Mone, Agostinho Monteiro, Claudia Montes, Giorgia Montrucchio, Sarah Moore, Shona C. Moore, Lina Morales Cely, Lucia Moro, Diego Rolando Morocho Tutillo, Ben Morton, Catherine Motherway, Ana Motos, Hugo Mouquet, Clara Mouton Perrot, Julien Moyet, Caroline Mudara, Aisha Kalsoom Mufti, Ng Yong Muh, Dzawani Muhamad, Jimmy Mullaert, Fredrik Müller, Karl Erik Müller, Daniel Munblit, Syed Muneeb, Nadeem Munir, Laveena Munshi, Aisling Murphy, Lorna Murphy, Marlène Murris, Srinivas Murthy, Himed Musaab, Carlotta Mutti, Himasha Muvindi, Gugapriyaa Muyandy, Dimitra Melia Myrodia, Farah Nadia Mohd-Hanafiah, Dave Nagpal, Mangala Narasimhan, Nageswaran Narayanan, Rashid Nasim Khan, Alasdair Nazerali-Maitland, Nadège Neant, Holger Neb, Coca Necsoi, Nikita A. Nekliudov, Erni Nelwan, Raul Neto, Emily Neumann, Bernardo Neves, Pauline Yeung Ng, Wing Yiu Ng, Anthony Nghi, Jane Ngure, Duc Nguyen, Orna Ni Choileain, Niamh Ni Leathlobhair, Alistair Nichol, Prompak Nitayavardhana, Stephanie Nonas, Nurul Amani Mohd Noordin, Marion Noret, Nurul Faten Izzati Norharizam, Lisa Norman, Alessandra Notari, Mahdad Noursadeghi, Karolina Nowicka, Adam Nowinski, Saad Nseir, Jose I. Nunez, Nurnaningsih Nurnaningsih, Dwi Utomo Nusantara, Elsa Nyamankolly, Anders Benteson Nygaard, Fionnuala O. Brien, Annmarie O. Callaghan, Annmarie O’Callaghan, Max O’Donnell, Sophie O’Halloran, Katie O’Hearn, Conar O’Neil, Linda O’Shea, Miriam O’Sullivan, Giovanna Occhipinti, Derbrenn OConnor, Tawnya Ogston, Takayuki Ogura, Tak-Hyuk Oh, Shinichiro Ohshimo, Agnieszka Oldakowska, João Oliveira, Larissa Oliveira, Piero L. Olliaro, David S. Y. Ong, Jee Yan Ong, Wilna Oosthuyzen, Anne Opavsky, Peter Openshaw, Saijad Orakzai, Claudia Milena Orozco-Chamorro, Andrés Orquera, Jamel Ortoleva, Javier Osatnik, Siti Zubaidah Othman, Paul Otiku, Nadia Ouamara, Rachida Ouissa, Clark Owyang, Eric Oziol, Maïder Pagadoy, Justine Pages, Amanda Palacios, Mario Palacios, Massimo Palmarini, Giovanna Panarello, Prasan Kumar Panda, Hem Paneru, Lai Hui Pang, Mauro Panigada, Nathalie Pansu, Aurélie Papadopoulos, Paolo Parducci, Edwin Fernando Paredes Oña, Rachael Parke, Melissa Parker, Briseida Parra, Vieri Parrini, Taha Pasha, Jérémie Pasquier, Bruno Pastene, Fabian Patauner, Drashti Patel, Mohan Dass Pathmanathan, Luís Patrão, Patricia Patricio, Juliette Patrier, Laura Patrizi, Lisa Patterson, Rajyabardhan Pattnaik, Christelle Paul, Mical Paul, Jorge Paulos, William A. Paxton, Jean-François Payen, Kalaiarasu Peariasamy, Miguel Pedrera Jiménez, Giles J. Peek, Florent Peelman, Nathan Peiffer-Smadja, Vincent Peigne, Mare Pejkovska, Paolo Pelosi, Ithan D. Peltan, Rui Pereira, Daniel Perez, Luis Periel, Thomas Perpoint, Antonio Pesenti, Vincent Pestre, Lenka Petrou, Ventzislava Petrov-Sanchez, Michele Petrovic, Frank Olav Pettersen, Gilles Peytavin, Scott Pharand, Ooyanong Phonemixay, Michael Piagnerelli, Walter Picard, Olivier Picone, Maria de Piero, Carola Pierobon, Djura Piersma, Carlos Pimentel, Raquel Pinto, Catarina Pires, Isabelle Pironneau, Lionel Piroth, Roberta Pisi, Ayodhia Pitaloka, Riinu Pius, Simone Piva, Laurent Plantier, Hon Shen Png, Julien Poissy, Ryadh Pokeerbux, Maria Pokorska-Spiewak, Sergio Poli, Georgios Pollakis, Diane Ponscarme, Jolanta Popielska, Diego Bastos Porto, Andra-Maris Post, Douwe F. Postma, Pedro Povoa, Diana Póvoas, Jeff Powis, Sofia Prapa, Viladeth Praphasiri, Sébastien Preau, Christian Prebensen, Jean-Charles Preiser, Anton Prinssen, Mark G. Pritchard, Gamage Dona Dilanthi Priyadarshani, Lucia Proença, Sravya Pudota, Oriane Puéchal, Bambang Pujo Semedi, Mathew Pulicken, Matteo Puntoni, Gregory Purcell, Luisa Quesada, Vilmaris Quinones-Cardona, Víctor Quirós González, Else Quist-Paulsen, Mohammed Quraishi, Maia Rabaa, Christian Rabaud, Ebenezer Rabindrarajan, Aldo Rafael, Marie Rafiq, Gabrielle Ragazzo, Mutia Rahardjani, Ahmad Kashfi Haji Ab Rahman, Rozanah Abd Rahman, Arsalan Rahutullah, Fernando Rainieri, Giri Shan Rajahram, Pratheema Ramachandran, Nagarajan Ramakrishnan, Kollengode Ramanathan, Ahmad Afiq Ramli, Blandine Rammaert, Grazielle Viana Ramos, Anais Rampello, Asim Rana, Rajavardhan Rangappa, Ritika Ranjan, Elena Ranza, Christophe Rapp, Aasiyah Rashan, Thalha Rashan, Ghulam Rasheed, Menaldi Rasmin, Indrek Rätsep, Cornelius Rau, Francesco Rausa, Tharmini Ravi, Ali Raza, Andre Real, Stanislas Rebaudet, Sarah Redl, Brenda Reeve, Attaur Rehman, Liadain Reid, Dag Henrik Reikvam, Renato Reis, Jordi Rello, Jonathan Remppis, Martine Remy, Hongru Ren, Hanna Renk, Anne-Sophie Resseguier, Matthieu Revest, Oleksa Rewa, Luis Felipe Reyes, Tiago Reyes, Maria Ines Ribeiro, Antonia Ricchiuto, David Richardson, Denise Richardson, Laurent Richier, Siti Nurul Atikah Ahmad Ridzuan, Jordi Riera, Ana L. Rios, Asgar Rishu, Patrick Rispal, Karine Risso, Maria Angelica Rivera Nuñez, Nicholas Rizer, Chiara Robba, André Roberto, Stephanie Roberts, David L. Robertson, Olivier Robineau, Ferran Roche-Campo, Paola Rodari, Simão Rodeia, Julia Rodriguez Abreu, Bernhard Roessler, Claire Roger, Pierre-Marie Roger, Emmanuel Roilides, Amanda Rojek, Juliette Romaru, Roberto Roncon-Albuquerque, Jr, Mélanie Roriz, Manuel Rosa-Calatrava, Michael Rose, Dorothea Rosenberger, Nurul Hidayah Mohammad Roslan, Andrea Rossanese, Matteo Rossetti, Sandra Rossi, Bénédicte Rossignol, Patrick Rossignol, Stella Rousset, Carine Roy, Benoît Roze, Desy Rusmawatiningtyas, Clark D. Russell, Maeve Ryan, Maria Ryan, Mazankowski Heart Institute Ryckaert, Steffi Ryckaert, Aleksander Rygh Holten, Isabela Saba, Luca Sacchelli, Sairah Sadaf, Musharaf Sadat, Valla Sahraei, Nadia Saidani, Maximilien Saint-Gilles, Pranya Sakiyalak, Nawal Salahuddin, Leonardo Salazar, Jodat Saleem, Nazal Saleh, Gabriele Sales, Stéphane Sallaberry, Charlotte Salmon Gandonniere, Hélène Salvator, Olivier Sanchez, Xavier Sánchez Choez, Kizy Sanchez de Oliveira, Angel Sanchez-Miralles, Vanessa Sancho-Shimizu, Gyan Sandhu, Zulfiqar Sandhu, Pierre-François Sandrine, Oana Sandulescu, Marlene Santos, Shirley Sarfo-Mensah, Bruno Sarmento Banheiro, Iam Claire E. Sarmiento, Benjamine Sarton, Ankana Satya, Sree Satyapriya, Rumaisah Satyawati, Egle Saviciute, Parthena Savvidou, Yen Tsen Saw, Justin Schaffer, Tjard Schermer, Arnaud Scherpereel, Marion Schneider, Stephan Schroll, Michael Schwameis, Gary Schwartz, Janet T. Scott, James Scott-Brown, Nicholas Sedillot, Tamara Seitz, Jaganathan Selvanayagam, Mageswari Selvarajoo, Caroline Semaille, Malcolm G. Semple, Rasidah Bt Senian, Eric Senneville, Claudia Sepulveda, Filipa Sequeira, Tânia Sequeira, Ary Serpa Neto, Pablo Serrano Balazote, Ellen Shadowitz, Syamin Asyraf Shahidan, Mohammad Shamsah, Anuraj Shankar, Shaikh Sharjeel, Pratima Sharma, Catherine A. Shaw, Victoria Shaw, Ashraf Sheharyar, Dr. Rajesh Mohan Shetty, Rohan Shetty, Haixia Shi, Nisreen Shiban, Mohiuddin Shiekh, Takuya Shiga, Nobuaki Shime, Hiroaki Shimizu, Keiki Shimizu, Naoki Shimizu, Sally Shrapnel, Pramesh Sundar Shrestha, Shubha Kalyan Shrestha, Hoi Ping Shum, Nassima Si Mohammed, Ng Yong Siang, Jeanne Sibiude, Bountoy Sibounheuang, Atif Siddiqui, Louise Sigfrid, Piret Sillaots, Catarina Silva, Maria Joao Silva, Rogério Silva, Benedict Sim Lim Heng, Wai Ching Sin, Dario Sinatti, Budha Charan Singh, Punam Singh, Pompini Agustina Sitompul, Karisha Sivam, Vegard Skogen, Sue Smith, Benjamin Smood, Coilin Smyth, Michelle Smyth, Morgane Snacken, Dominic So, Tze Vee Soh, Lene Bergendal Solberg, Joshua Solomon, Tom Solomon, Emily Somers, Agnès Sommet, Myung Jin Song, Rima Song, Tae Song, Jack Song Chia, Michael Sonntagbauer, Azlan Mat Soom, Arne Søraas, Camilla Lund Søraas, Alberto Sotto, Edouard Soum, Ana Chora Sousa, Marta Sousa, Maria Sousa Uva, Vicente Souza-Dantas, Alexandra Sperry, Elisabetta Spinuzza, B. P. Sanka Ruwan Sri Darshana, Shiranee Sriskandan, Sarah Stabler, Thomas Staudinger, Stephanie-Susanne Stecher, Trude Steinsvik, Ymkje Stienstra, Birgitte Stiksrud, Eva Stolz, Amy Stone, Adrian Streinu-Cercel, Anca Streinu-Cercel, Samantha Strudwick, Ami Stuart, David Stuart, Decy Subekti, Gabriel Suen, Jacky Y. Suen, Asfia Sultana, Charlotte Summers, Dubravka Supic, Deepashankari Suppiah, Magdalena Surovcová, Suwarti Suwarti, Andrey A. Svistunov, Sarah Syahrin, Konstantinos Syrigos, Jaques Sztajnbok, Konstanty Szuldrzynski, Shirin Tabrizi, Fabio S. Taccone, Lysa Tagherset, Shahdattul Mawarni Taib, Ewa Talarek, Sara Taleb, Jelmer Talsma, Renaud Tamisier, Maria Lawrensia Tampubolon, Kim Keat Tan, Le Van Tan, Yan Chyi Tan, Clarice Tanaka, Hiroyuki Tanaka, Taku Tanaka, Hayato Taniguchi, Huda Taqdees, Arshad Taqi, Coralie Tardivon, Pierre Tattevin, M Azhari Taufik, Hassan Tawfik, Richard S. Tedder, Tze Yuan Tee, João Teixeira, Sofia Tejada, Marie-Capucine Tellier, Sze Kye Teoh, Vanessa Teotonio, François Téoulé, Pleun Terpstra, Olivier Terrier, Nicolas Terzi, Hubert Tessier-Grenier, Adrian Tey, Alif Adlan Mohd Thabit, Anand Thakur, Zhang Duan Tham, Suvintheran Thangavelu, Vincent Thibault, Simon-Djamel Thiberville, Benoît Thill, Jananee Thirumanickam, Shaun Thompson, David Thomson, Emma C. Thomson, Surain Raaj Thanga Thurai, Duong Bich Thuy, Ryan S. Thwaites, Andrea Ticinesi, Paul Tierney, Vadim Tieroshyn, Peter S. Timashev, Jean-François Timsit, Bharath Kumar Tirupakuzhi Vijayaraghavan, Noémie Tissot, Jordan Zhien Yang Toh, Maria Toki, Kristian Tonby, Sia Loong Tonnii, Antoni Torres, Margarida Torres, Rosario Maria Torres Santos-Olmo, Hernando Torres-Zevallos, Michael Towers, Tony Trapani, Théo Treoux, Huynh Trung Trieu, Cécile Tromeur, Ioannis Trontzas, Tiffany Trouillon, Jeanne Truong, Christelle Tual, Sarah Tubiana, Helen Tuite, Jean-Marie Turmel, Lance C. W. Turtle, Anders Tveita, Pawel Twardowski, Makoto Uchiyama, P G Ishara Udayanga, Andrew Udy, Roman Ullrich, Alberto Uribe, Asad Usman, Timothy M. Uyeki, Cristinava Vajdovics, Luís Val-Flores, Piero Valentini, Ana Luiza Valle, Amélie Valran, Ilaria Valzano, Stijn Van de Velde, Marcel van den Berge, Machteld Van der Feltz, Job van der Palen, Paul van der Valk, Nicky Van Der Vekens, Peter Van der Voort, Sylvie Van Der Werf, Marlice van Dyk, Laura van Gulik, Jarne Van Hattem, Carolien van Netten, Gitte Van Twillert, Ilonka van Veen, Noémie Vanel, Henk Vanoverschelde, Pooja Varghese, Michael Varrone, Shoban Raj Vasudayan, Charline Vauchy, Shaminee Veeran, Aurélie Veislinger, Sebastian Vencken, Sara Ventura, Annelies Verbon, James Vickers, José Ernesto Vidal, César Vieira, Deepak Vijayan, Joy Ann Villanueva, Judit Villar, Pierre-Marc Villeneuve, Andrea Villoldo, Nguyen Van Vinh Chau, Gayatri Vishwanathan, Benoit Visseaux, Hannah Visser, Chiara Vitiello, Manivanh Vongsouvath, Harald Vonkeman, Fanny Vuotto, Noor Hidayu Wahab, Suhaila Abdul Wahab, Nadirah Abdul Wahid, Marina Wainstein, Wan Fadzlina Wan Muhd Shukeri, Chih-Hsien Wang, Steve Webb, Jia Wei, Katharina Weil, Tan Pei Wen, Sanne Wesselius, T. Eoin West, Murray Wham, Bryan Whelan, Nicole White, Paul Henri Wicky, Aurélie Wiedemann, Surya Otto Wijaya, Keith Wille, Suzette Willems, Virginie Williams, Evert-Jan Wils, Calvin Wong, Teck Fung Wong, Xin Ci Wong, Yew Sing Wong, Natalie Wright, Gan Ee Xian, Lim Saio Xian, Kuan Pei Xuan, Ioannis Xynogalas, Sophie Yacoub, Siti Rohani Binti Mohd Yakop, Masaki Yamazaki, Yazdan Yazdanpanah, Nicholas Yee Liang Hing, Cécile Yelnik, Chian Hui Yeoh, Stephanie Yerkovich, Touxiong Yiaye, Toshiki Yokoyama, Hodane Yonis, Obada Yousif, Saptadi Yuliarto, Akram Zaaqoq, Marion Zabbe, Kai Zacharowski, Masliza Zahid, Maram Zahran, Nor Zaila Binti Zaidan, Maria Zambon, Miguel Zambrano, Alberto Zanella, Konrad Zawadka, Nurul Zaynah, Hiba Zayyad, Alexander Zoufaly, and David Zucman
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-022-01534-9.
References
- 1.ISARIC Clinical Characterization Group The value of open-source clinical science in pandemic response: lessons from ISARIC. The Lancet. Infectious diseases. 2021;21:1623–1624. doi: 10.1016/S1473-3099(21)00565-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet. Respiratory medicine. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G, et al. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. Jama. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ISARIC Clinical Characterization Group COVID-19 symptoms at hospital admission vary with age and sex: results from the ISARIC prospective multinational observational study. Infection. 2021;49:889–905. doi: 10.1007/s15010-021-01599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes, L. F. et al. Clinical characteristics, risk factors and outcomes in patients with severe COVID-19 registered in the International Severe Acute Respiratory and Emerging Infection Consortium WHO clinical characterisation protocol: a prospective, multinational, multicentre, observational study. ERJ Open Res8, 10.1183/23120541.00552-2021 (2022). [DOI] [PMC free article] [PubMed]
- 6.Reyes LF, et al. Clinical characteristics, systemic complications, and in-hospital outcomes for patients with COVID-19 in Latin America. LIVEN-Covid-19 study: A prospective, multicenter, multinational, cohort study. PloS one. 2022;17:e0265529. doi: 10.1371/journal.pone.0265529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunning JW, et al. Open source clinical science for emerging infections. The Lancet. Infectious diseases. 2014;14:8–9. doi: 10.1016/S1473-3099(13)70327-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom CI, et al. Risk of adverse outcomes in patients with underlying respiratory conditions admitted to hospital with COVID-19: a national, multicentre prospective cohort study using the ISARIC WHO Clinical Characterisation Protocol UK. The Lancet. Respiratory medicine. 2021;9:699–711. doi: 10.1016/S2213-2600(21)00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Docherty AB, et al. Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ. 2020;369:m1985. doi: 10.1136/bmj.m1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ISARIC Clinical Characterization Group. et al. Ten months of temporal variation in the clinical journey of hospitalised patients with COVID-19: An observational cohort. eLife10, 10.7554/eLife.70970 (2021). [DOI] [PMC free article] [PubMed]
- 11.Vuong Q-H, et al. Covid-19 vaccines production and societal immunization under the serendipity-mindsponge-3D knowledge management theory and conceptual framework. Humanities and Social Sciences Communications. 2022;9:22. doi: 10.1057/s41599-022-01034-6. [DOI] [Google Scholar]
- 12.Vuong QH. The (ir)rational consideration of the cost of science in transition economies. Nat Hum Behav. 2018;2:5. doi: 10.1038/s41562-017-0281-4. [DOI] [PubMed] [Google Scholar]
- 13.Jakab, Z., Selbie, D., Squires, N., Mustafa, S. & Saikat, S. Building the evidence base for global health policy: the need to strengthen institutional networks, geographical representation and global collaboration. BMJ Glob Health6, 10.1136/bmjgh-2021-006852 (2021). [DOI] [PMC free article] [PubMed]
- 14.Harris PA, et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.2022. ISARIC (International Severe Acute Respiratory and emerging Infections Consortium. ISARIC COVID-19 Dataset. Exaptive. [DOI]
- 16.Sigfrid L, et al. What is the recovery rate and risk of long-term consequences following a diagnosis of COVID-19? A harmonised, global longitudinal observational study protocol. BMJ Open. 2021;11:e043887. doi: 10.1136/bmjopen-2020-043887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sigfrid L, et al. Long Covid in adults discharged from UK hospitals after Covid-19: A prospective, multicentre cohort study using the ISARIC WHO Clinical Characterisation Protocol. Lancet Reg Health Eur. 2021;8:100186. doi: 10.1016/j.lanepe.2021.100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloom, C. I. et al. Risk of adverse outcomes in patients with underlying respiratory conditions admitted to hospital with COVID-19: a national, multicentre prospective cohort study using the ISARIC WHO Clinical Characterisation Protocol UK. Lancet Respir Med, 10.1016/S2213-2600(21)00013-8 (2021). [DOI] [PMC free article] [PubMed]
- 19.Vuong QH, Le TT, La VP, Nguyen MH. The psychological mechanism of internet information processing for post-treatment evaluation. Heliyon. 2022;8:e09351. doi: 10.1016/j.heliyon.2022.e09351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouziotis, J., Arvanitakis, M., Preiser, J. C. & Group, I. C. C. Association of body mass index with COVID-19 related in-hospital death. Clin Nutr, 10.1016/j.clnu.2022.01.017 (2022). [DOI] [PMC free article] [PubMed]
- 21.ISARIC Clinical Characterization Group. COVID-19 symptoms at hospital admission vary with age and sex: results from the ISARIC prospective multinational observational study. Infection, 10.1007/s15010-021-01599-5 (2021). [DOI] [PMC free article] [PubMed]
- 22.Knight SR, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wainstein M, et al. Use of an extended KDIGO definition to diagnose acute kidney injury in patients with COVID-19: A multinational study using the ISARIC-WHO clinical characterisation protocol. PLoS Med. 2022;19:e1003969. doi: 10.1371/journal.pmed.1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali R, et al. Isaric 4c Mortality Score As A Predictor Of In-Hospital Mortality In Covid-19 Patients Admitted In Ayub Teaching Hospital During First Wave Of The Pandemic. J Ayub Med Coll Abbottabad. 2021;33:20–25. [PubMed] [Google Scholar]
- 25.Gupta RK, et al. Development and validation of the ISARIC 4C Deterioration model for adults hospitalised with COVID-19: a prospective cohort study. The Lancet. Respiratory medicine. 2021;9:349–359. doi: 10.1016/S2213-2600(20)30559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight SR, et al. Prospective validation of the 4C prognostic models for adults hospitalised with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol. Thorax. 2021 doi: 10.1136/thoraxjnl-2021-217629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell CD, et al. Co-infections, secondary infections, and antimicrobial use in patients hospitalised with COVID-19 during the first pandemic wave from the ISARIC WHO CCP-UK study: a multicentre, prospective cohort study. Lancet Microbe. 2021;2:e354–e365. doi: 10.1016/S2666-5247(21)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2022. ISARIC (International Severe Acute Respiratory and emerging Infections Consortium. ISARIC COVID-19 Dataset. Exaptive. [DOI]
Supplementary Materials
Data Availability Statement
Processing codes for the ISARIC COVID-19 database are openly available online, and contributions from the research community to share these codes are encouraged. For this reason, a public code repository has been created along with this manuscript to develop and share code collectively: https://github.com/ISARICDataPlatform/ISARICBasics.git. The content of this repository is under continuous development. Still, it has been seeded with code to generate patient-level datasets suitable for statistics and machine learning research, such as patient demographic, comorbid conditions at the time of admission, application of treatments, and severity scores, among others. It is possible for the research community to directly submit updates, improvements, and additions to the repository via GitHub. Moreover, a Jupyter Notebook containing the code used to generate the tables and descriptive statistics included in this paper is openly available on GitHub.