Abstract
Background
Antimicrobial susceptibility testing (AST) is often needed prior to antimicrobial optimization for patients with gram-negative bloodstream infections (GN-BSIs). Rapid AST (rAST) in combination with antimicrobial stewardship (AS) may decrease time to administration of narrower antibiotics.
Methods
This was a prospective, nonblinded, randomized trial evaluating the impact of a phenotypic rAST method vs conventional AST (cAST) in hospitalized patients with GN-BSI and source control. The primary outcome was time to narrowest effective therapy.
Results
Two hundred seventy-four patients were randomized and 205 underwent analysis (97 cAST, 108 rAST). Median (interquartile range [IQR]) time to susceptibility results was 23 hours shorter in the rAST group (cAST: 62 [59–67] hours vs rAST: 39 [IQR, 35–46] hours; P < .001). Median (IQR) time to narrowest effective therapy was similar between groups (cAST: 73 [44–138] hours vs rAST: 64 [42–92] hours; P = .10). Median (IQR) time to narrowest effective therapy was significantly shorter in a prespecified subgroup of patients not initially on narrowest therapy and during AS working hours (cAST: 93 [56–154] hours vs rAST: 62 [43–164] hours; P = .004). Significant decreases were observed in median (IQR) time to oral therapy (cAST: 126 [76–209] hours vs rAST: 91 [66–154] hours; P = .02) and median (IQR) length of hospital stay (cAST: 7 [4–13] days vs rAST: 5 [4–8] days; P = .04).
Conclusions
In patients with GN-BSI, rAST did not significantly decrease time to narrowest effective therapy but did decrease time to oral antibiotics and length of hospital stay. Rapid AST using existing microbiology platforms has potential to optimize patient outcomes.
Keywords: antimicrobial stewardship, antimicrobial susceptibility testing, bloodstream infection, gram negative, rapid diagnostic testing
This randomized study evaluated rapid antimicrobial susceptibility testing (rAST) in patients with gram-negative bacteremia and source control. Results demonstrated no difference in time to narrowest effective therapy. Time to oral therapy and length of hospital stay were significantly shorter in the rAST group.
Gram-negative (GN) bacilli are responsible for 40% of bloodstream infections (BSIs) and represent a major contributor to healthcare-related morbidity and mortality [1–4]. Increasing incidence of multidrug-resistant GN organisms has made empiric use of broad-spectrum antibiotics standard practice for most institutions [5]. However, judicious use of antibiotics is important to limit the development of resistance [6].
Recent studies have demonstrated that transitioning to oral therapy and shortening durations of therapy are safe strategies for patients with uncomplicated GN-BSI, including the use of oral β-lactams [7–11]. A retrospective study by Tamma and colleagues reported a 2-day decrease in length of hospital stay (LOS) in patients transitioned to oral step-down therapy [9]. However, antimicrobial susceptibility testing (AST) can be a rate-limiting step in de-escalation and transitioning to oral therapy.
Conventional microbiology methods for organism identification and AST require time for organism growth, mass spectrometry (MS) identification of organism, and exposure to antimicrobials to determine phenotypic susceptibility. Shortening time to GN-BSI susceptibility results was associated with decreases in time to antimicrobial changes in multiple retrospective studies and a single randomized controlled trial (RCT) that evaluated a commercially available platform [12–14]. Time to oral therapy and LOS were not affected. A recent Cochrane review [15] also found no evidence that rapid AST (rAST) was associated with decreased mortality or LOS. Previous studies have demonstrated improvements in antibiotic use with faster microbiology results in conjunction with antimicrobial stewardship (AS) programs; however, this is not observed in the absence of AS [9, 16–19].
A phenotypic method for rAST directly from positive blood cultures was used to compare rAST and conventional AST (cAST) in patients with GN-BSI. It was hypothesized that rAST in combination with AS would decrease time to narrowest effective therapy and hospital LOS.
METHODS
Design and Setting
This was a prospective RCT evaluating the impact of rAST on patients with GN-BSI in combination with AS between 1 August 2020 and 5 November 2021. The study was conducted at 2 medical centers in Portland, Oregon (Providence Portland Medical Center and Providence St Vincent Medical Center). The study sites are each approximately 500 beds and part of an 8-hospital health system using a centralized microbiology clinical laboratory and a regional AS program. Resources for the AS program include 2 full-time infectious diseases (ID) pharmacists, a postgraduate year 2 ID pharmacy resident, and 1 full-time ID physician. A rapid, direct-from-blood matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) process was utilized on all study participants and has been part of the institutional standard of care since 2018. This study was approved by the Providence St Joseph Health System Institutional Review Board, and a waiver of informed consent was granted.
Study Participants
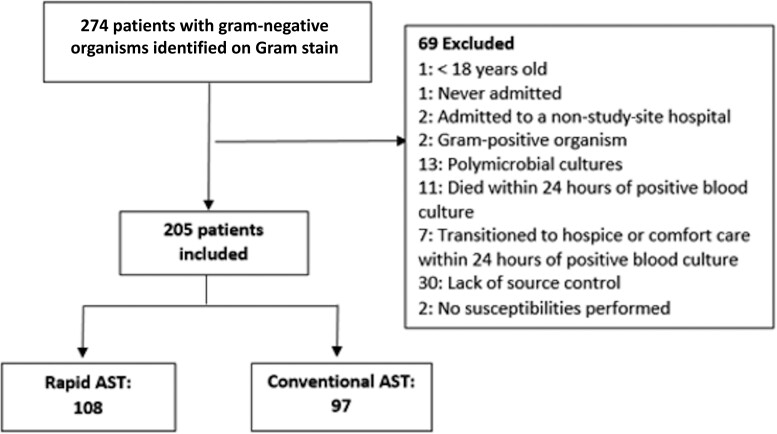
Hospitalized adult (aged ≥18 years) patients were randomized to rAST or cAST when the automated blood culture system (BacT/Alert, bioMérieux, Durham, North Carolina) turned positive and routine Gram stain results demonstrated a GN organism. Only the first positive blood culture per patient encounter was randomized. Patients were excluded after randomization if they had polymicrobial blood cultures, no susceptibility testing performed, organisms eventually identified as gram-positive, died, or transitioned to hospice or comfort care within 24 hours of randomization or lacked source control (Figure 1). Lack of source control was defined as any of the following remaining at discharge: infected prosthetic material, undrained fluid collection, biliary obstruction, urinary obstruction, or deep-seated infection (osteomyelitis or endocarditis).
Figure 1.
Participant screening and randomization. Abbreviation: AST, antimicrobial susceptibility testing.
Procedures
Microbiology
The following laboratory-developed method was successfully validated in the 12 months prior to our study. GN bacilli were rapidly identified to species level by MALDI-TOF MS directly from the positive blood culture bottle [20], similar to methods described previously by Horing and colleagues [21]. In brief, a 5-mL aliquot of blood culture suspension was removed from the blood culture bottle in a biosafety cabinet in a sterile fashion and transferred to a serum separator tube (SST). The SST was spun for 5 minutes at 4000 rpm. Using a sterile bulb transfer pipet, serum supernatant at the top was removed, leaving behind a buffy coat layer, a gel layer, and a red blood cell layer. The SST pellet was washed with 200 µL of nuclease-free water by vigorously mixing the buffy coat layer inside the SST. The mixture was transferred into a 1.5-mL Eppendorf tube. The supernatant was removed, and the pellet used for (1) spotting it on a Vitek MS target slide for identification and (2) inoculating a blood plate with a generous amount of pellet (“smudge plate”), which was then incubated at 5% carbon dioxide for 4 hours. If the patient was randomized to rAST, an aliquot of the pellet was diluted in 0.45% saline to a density of 0.5 McFarland. This suspension was used to inoculate a Vitek 2 susceptibility card (GN72). If the patient was randomized to the cAST, standard procedures for Vitek were followed. If direct identification by MS failed, the smudge plate was used for identification by MALDI-TOF. The above procedures were performed 24 hours per day, 7 days a week as blood culture bottles turned positive.
Antimicrobial Stewardship
Prior to the study initiation, the AS team received real-time pages from the microbiology laboratory when species were identified via rapid-MALDI-TOF procedures. An institutional guidance document existed to guide organism-specific empiric therapy for bacteremic patients. For the duration of the study, ID pharmacists continued to receive real-time pages and no additional notifications were provided when AST resulted. Results and recommendations were reviewed with ID physicians daily Monday through Friday, 7:00 am through 4:00 pm. Pages sent during AS off-hours were reviewed the next working day. Patients were followed until AST returned and recommendations for follow-up blood cultures, antimicrobial changes, transition to oral therapy, and duration of therapy were generated. A note template was created for the purposes of the study and is included in Supplementary Figure 1. For uncomplicated Enterobacterales infections with source control and adequate clinical improvement, AS routinely recommended no follow-up blood cultures, switch to oral therapy, and 7-day duration of therapy. Recommendations were given to the primary team through phone calls and/or communications in the electronic medical record. Changes to antibiotic therapy were made at the discretion of the primary team.
Outcome Measures
The primary outcome was time to narrowest effective therapy, defined as time from blood culture collection to first administration of narrowest-spectrum antimicrobial agent given patient-specific susceptibility results, concomitant infections, antibiotic allergies, and comorbid conditions, plus cessation of unnecessary gram-positive agents. If the narrowest therapy was first administered as a discharge prescription, the presumed intended start of the discharge prescription was used as the time of administration. For patients who did not receive narrowest therapy, the time from blood culture collection to end of antibiotic use was used for the primary outcome. A time of zero was used for patients already on narrowest effective therapy at time of blood culture collection. Narrowest effective therapy was determined retrospectively after blinded review by 2 ID pharmacists (A. B. C. and B. F.) and an ID physician (T. P.) using an agreed-upon definition of narrowest therapy (Supplementary Table 1).
Secondary outcomes included time to susceptibility results, time to oral therapy, infection-related discharge readiness at days 3 and 5, LOS, in-hospital mortality, 30-day mortality, 30-day readmission, and recurrence of bacteremia. Infection-related discharge readiness was defined as meeting all the following: source control achieved, afebrile for 24 hours, Pitt bacteremia score ≤1 [22], improvement in at least 1 local sign or symptom of infection, able to tolerate oral medications, and susceptibility to an oral agent.
Recurrence of bacteremia was defined as identification of the same organism in a blood culture within 30 days of antibiotic completion. Patients were considered immunocompromised if they had any of the following: history of solid organ transplant or stem cell transplant, ≥20 mg/day of prednisone (or equivalent) for ≥14 days in the past 30 days, immunomodulatory medications in the past 90 days, or any daily leukocyte count ≤1000 cells/mL during bacteremia treatment. Additional outcome definitions are described in the Supplementary Methods.
Randomization and Blinding
A randomization key was generated and used to distribute patients between AST groups in equal fashion (with a block randomization scheme and block size randomly picked from 2 and 4) stratified by hospital. Patients were randomized when the Gram stain showed a GN organism.
Patients undergoing rAST had the following comment added to their susceptibility results: “Presumptive susceptibility results. Verification to follow.” AS team members were not blinded to method of AST assigned. Treating clinicians were not informed of the study and were blinded to the randomization group.
Data Collection
Patients were followed for 90 days after the first blood culture result. Age, sex, race, ethnicity, LOS, discharge disposition, and comorbidity data were collected using an internal electronic SAP BI Web Intelligence report. Patient International Classification of Diseases, Tenth Revision diagnoses codes were used to generate a Charlson Comorbidity Index (CCI) score for each patient [23]. All other study data were manually extracted through medical record review and managed using REDCap [24], an electronic data capture tool hosted at Providence St Joseph Health.
Statistical Analysis
Continuous variables were summarized as mean ± standard deviation or median (IQR) as appropriate, whereas categorical variables were summarized as frequency (percentage). Student t test or Wilcoxon rank-sum test was performed to compare continuous variables, and χ2 test or Fisher exact test was performed to compare categorical variables. A prespecified subgroup analysis was planned for the primary outcome, which evaluated the effects of susceptibility methods in patients not initially on narrowest therapy and during AS program hours (Monday–Friday, 7:00 am–4:00 pm) and off-hours, respectively. All analyses were performed using R statistical program (R Foundation for Statistical Computing, Vienna, Austria) [25]. Taking into account a postrandomization exclusion estimate of 30%, a sample size of about 150 per treatment group (yielding 105 per group for the final analysis) was estimated to achieve 80% power in detecting a 10-hour difference in the primary outcome of time to narrowest therapy. A 10-hour difference was chosen based on the smallest expected difference in rAST and cAST results that would be possible. A P value < .05 was considered statistically significant.
RESULTS
A total of 274 GN organisms were identified on Gram stain and corresponding patients were randomized. Medical record review identified 69 patients meeting exclusion criteria as described in Figure 1. The final analysis included 205 patients (108 rAST, 97 cAST). The average age was 69 years, and 53% were female (Table 1). The most common source of infection was urinary (67%). AS progress notes were recorded in the electronic medical record for 77 (37.6%) patients. Pitt bacteremia score and CCI score were similar between groups. Age, source of infection, and immunocompromised status were also similar. ID consultation was more common in the cAST group.
Table 1.
Clinical and Microbiologic Demographics
| Demographic | Overall (N = 205) | cAST (n = 97) | rAST (n = 108) |
|---|---|---|---|
| Age, y, mean ± SD | 68.5 ± 16.4 | 68.3 ± 15.2 | 68.6 ± 17.4 |
| Female sex | 109 (53) | 45 (46) | 64 (59) |
| Race | |||
| Black | 12 (5.9) | 10 (10) | 2 (1.9) |
| White | 48 (24) | 16 (16) | 32 (30) |
| Other/unknown | 144 (71) | 71 (73) | 73 (68) |
| Ethnicity | |||
| Hispanic or Latino | 22 (11) | 6 (6.2) | 16 (15) |
| Not Hispanic or Latino | 175 (86) | 90 (93) | 85 (79) |
| Unknown | 7 (3.4) | 1 (1.0) | 6 (5.6) |
| Missing | 1 | 0 | 1 |
| Hospital | |||
| Hospital 1 | 77 (38) | 35 (36) | 42 (39) |
| Hospital 2 | 128 (62) | 62 (64) | 66 (61) |
| CCI score, mean ± SD | 5.9 ± 3.6 | 6.1 ± 3.9 | 5.7 ± 3.4 |
| Comorbidities | |||
| Diabetes | 112 (54.6) | 55 (56.7) | 57 (52.8) |
| Myocardial infarction | 45 (22.0) | 16 (16.5) | 29 (26.9) |
| Congestive heart failure | 59 (28.8) | 24 (24.7) | 35 (32.4) |
| Peripheral vascular disease | 14 (6.8) | 7 (7.2) | 7 (6.5) |
| CVA or TIA | 18 (8.8) | 10 (10.3) | 8 (7.4) |
| Dementia | 32 (15.6) | 12 (12.4) | 20 (18.5) |
| COPD | 42 (20.5) | 15 (15.5) | 27 (25.0) |
| Connective tissue disorder | 14 (6.8) | 7 (7.2) | 7 (6.5) |
| Peptic ulcer disease | 4 (2.0) | 4 (4.1) | 0 (0) |
| Chronic kidney disease | 62 (30.2) | 34 (35.1) | 28 (25.9) |
| Metastatic solid tumor | 14 (6.8) | 9 (9.3) | 5 (4.6) |
| Leukemia or lymphoma | 15 (7.3) | 2 (2.1) | 13 (12.0) |
| HIV/AIDS | 1 (0.5) | 1 (1.0) | 0 (0) |
| Liver disease | 19 (9.3) | 8 (8.2) | 11 (10.2) |
| Pitt bacteremia score, median (IQR) | 2 (0–3) | 2 (0–3) | 2 (0–3) |
| Temperaturea | |||
| ≤35°C or ≥40°C | 7 (3.4) | 4 (4.1) | 3 (2.8) |
| 35.1°C–36.0°C or 39.0°C–39.9°C | 66 (32) | 26 (27) | 40 (37) |
| 36.1°C–38.9°C | 132 (64) | 67 (69) | 65 (60) |
| Hypotensiona,b | 95 (46) | 43 (44) | 52 (48) |
| Mechanical ventilationa | 8 (3.9) | 4 (4.1) | 4 (3.7) |
| Cardiac arresta | 10 (4.9) | 6 (6.2) | 4 (3.7) |
| Mental statusa | |||
| Alert | 159 (78) | 76 (78) | 83 (77) |
| Comatose | 2 (1.0) | 1 (1.0) | 1 (0.9) |
| Disoriented | 43 (21) | 19 (20) | 24 (22) |
| Stuporous | 1 (0.5) | 1 (1.0) | 0 (0) |
| Immunocompromisedc | 21 (10) | 11 (11) | 10 (9.3) |
| ICU admissiond | 38 (19) | 15 (16) | 23 (22) |
| Organism species | |||
| Acinetobacter | 2 (1.0) | 2 (2.0) | 0 |
| Citrobacter | 3 (1.5) | 0 | 3 (2.8) |
| Escherichia | 139 (67.8) | 61 (62.9) | 78 (74.3) |
| Enterobacter | 6 (2.9) | 2 (2.0) | 4 (3.7) |
| Klebsiella | 29 (14.1) | 14 (14.4) | 15 (13.9) |
| Morganella | 1 (0.5) | 1 (1.0) | 0 |
| Proteus | 13 (6.3) | 10 (10.3) | 3 (2.8) |
| Providencia | 1 (0.5) | 1 (1.0) | 0 |
| Pseudomonas | 10 (4.9) | 6 (6.2) | 4 (3.7) |
| Salmonella | 1 (0.5) | 0 | 1 (0.9) |
| ESBL | 20 (9.8) | 8 (8.2) | 12 (11) |
| Source of infection | |||
| Central line | 2 (1.0) | 1 (1.0) | 1 (0.9) |
| Intra-abdominal | 38 (19) | 17 (18) | 21 (19) |
| Other | 2 (1.0) | 2 (2.1) | 0 (0) |
| Pulmonary | 14 (6.8) | 9 (9.3) | 5 (4.6) |
| Skin | 7 (3.4) | 5 (5.2) | 2 (1.9) |
| Urinary | 137 (67) | 60 (62) | 77 (71) |
| Unknown | 5 (2.4) | 3 (3.1) | 2 (1.9) |
| Follow-up blood culture | 111 (54) | 55 (57) | 56 (52) |
| Follow-up blood culture positive | 10 (9.0) | 5 (9.1) | 5 (8.9) |
| Febrile days, median (IQR) | 1 (0–2) | 1 (0–2) | 1 (0–1) |
| Infectious diseases consult | 28 (14) | 19 (20) | 9 (8.3%) |
| Discharge disposition | |||
| Intermediate care, nonskilled | 3 (1.5) | 2 (2.1) | 1 (0.9) |
| Expired | 3 (1.5) | 2 (2.1) | 1 (0.9) |
| Home or self-care | 110 (54) | 48 (49) | 62 (57) |
| Home with home health | 45 (22) | 25 (26) | 20 (19) |
| Home with home hospice | 7 (3.4) | 2 (2.1) | 5 (4.6) |
| Hospice medical facility | 1 (0.5) | 1 (1.0) | 0 (0) |
| Inpatient rehabilitation | 4 (2.0) | 2 (2.1) | 2 (1.9) |
| Short-term general inpatient | 2 (1.0) | 2 (2.1) | 0 (0) |
| Skilled nursing facility | 30 (15) | 13 (13) | 17 (16) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: cAST, conventional antimicrobial susceptibility testing; CCI, Charlson Comorbidity Index; COPD, chronic obstructive pulmonary disease; CVA, cardiovascular accident; ESBL, extended-spectrum β-lactamase; HIV, human immunodeficiency virus; ICU, intensive care unit; IQR, interquartile range; rAST, rapid antimicrobial susceptibility testing; SD, standard deviation; TIA, transient ischemic attack.
Variables were recorded within 24 hours prior to or 24 hours after blood culture collection individually and as part of the Pitt bacteremia score. The value furthest from normal (either highest or lowest) was used in calculating the Pitt bacteremia score.
Hypotension defined as need for intravenous vasopressor agents, systolic blood pressure (BP) <90 mm Hg, or acute hypotensive event with a drop in systolic BP >30 mm Hg or diastolic BP >20 mm Hg.
Immunocompromised condition defined as any of the following: history of solid organ transplant, hematopoietic stem cell transplant, any daily leukocyte count <1000 cells/mL during treatment, taking prednisone (or equivalent) ≥20 mg per day for ≥14 days, or immunomodulatory medications in past 30 days.
Admitted to an intensive care unit at the time of blood culture organism identification.
Primary Outcome
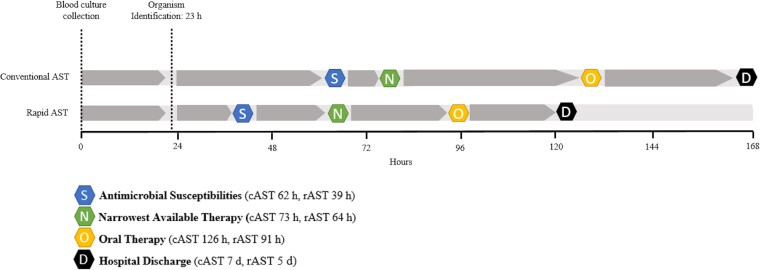
The median (IQR) time to narrowest effective therapy was shorter in the rAST group by 9 hours but was not statistically significant (73 [44–138] vs 64 [42–92] hours; P = .10) (Table 2). When patients already on narrowest therapy at time of organism identification were excluded from the analysis, there was a significant reduction of 21 hours in median (IQR) time to narrowest therapy (89 [58–148] vs 68 [45–95] hours; P = .008) (Table 3). A preplanned subpopulation analysis indicated a 31-hour reduction in time to narrowest therapy within rAST group during AS working hours (median [IQR], 93 [56–154] hours vs 62 [43–91] hours; P = .004), but not during off-hours (median [IQR], 73 [60–138] hours vs 76 [52–115] hours; P = .56).
Table 2.
Outcomes
| Outcome | Overall (N = 205) | cAST (n = 97) | rAST (n = 108) | P Valuea |
|---|---|---|---|---|
| Time to narrowest effective therapy, h | 67 (43–122) | 73 (44–138) | 64 (42–92) | .10 |
| Time to Gram stain, h | 15 (13–18) | 14 (13–19) | 15 (13–18) | .91 |
| Time to species identification, h | 23 (20–31) | 24 (20–33) | 23 (19–27) | .12 |
| Time to susceptibilities, h | 52 (38–63) | 62 (59–67) | 39 (35–46) | <.001 |
| Received oral therapy | 150 (73) | 64 (66) | 86 (80) | .040 |
| Time to oral therapy, h | 97 (68–186) | 126 (76–209) | 91 (66–154) | .022 |
| Length of hospital stay, d | 6 (4–10) | 7 (4–13) | 5 (4–8) | .035 |
| Discharge readiness by day 3 | 98 (48) | 37 (38) | 61 (56) | .012 |
| Discharge readiness by day 5 | 150 (73) | 65 (67) | 85 (79) | .082 |
| Recurrence of bacteremia | 0 | 0 | 0 | |
| 30-d readmission | 22 (11) | 12 (13) | 10 (9.3) | .50 |
| ED visit within 30 d | 13 (6.4) | 7 (7.4) | 6 (5.6) | .78 |
| Mortality | ||||
| In-hospital | 3 (1.5) | 2 (2.1) | 1 (0.9) | .60 |
| 30-d | 6 (2.9) | 5 (5.2) | 1 (0.9) | .10 |
| 90-d | 11 (5.4) | 8 (8.2) | 3 (2.8) | .12 |
| Length of therapyb, d | 10 (8–12) | 10 (8–11) | 10 (7–12) | .49 |
| Days of therapyc | 11 (9–15) | 11 (9–14) | 11 (9–15) | .99 |
Data are presented as No. (%) or median (interquartile range) unless otherwise indicated.
Abbreviations: cAST, conventional antimicrobial susceptibility testing; ED, emergency department; rAST, rapid antimicrobial susceptibility testing.
P values were generated from Wilcoxon rank-sum test for continuous variables and Fisher exact test or χ2 test for categorical variables, between rAST and cAST.
Length of therapy was defined as the number of consecutive days the patient received any antibiotic therapy. Any day in which the patient received a dose of an antibiotic was counted as 1 day.
Days of therapy was defined as the number of days a patient is on any particular antibiotic and is cumulative for all antibiotics the patient received.
Table 3.
Subgroup Analysis of Patients With Time to Narrowest Therapy Exceeding Time to Organism Identification
| Characteristic | Overall (n = 182) | cAST (n = 84) | rAST (n = 98) | P Valuea |
|---|---|---|---|---|
| Time to narrowest therapy, h | 72 (48–126) | 89 (58–148) | 68 (45–95) | .008 |
| Time to oral therapy, h | 95 (68–176) | 121 (76–200) | 90 (66–143) | .017 |
| Length of therapyb, d | 10 (8–11) | 10 (8–11) | 10 (8–12) | .58 |
| Days of therapyc | 11 (9–14) | 11 (9–14) | 11 (9–14) | .82 |
| AS hours (7:00 am–4:00 pm, Mon–Fri) | (n = 101) | (n = 45) | (n = 56) | |
| Time to narrowest therapy, h | 70 (47–126) | 93 (56–154) | 62 (43–91) | .004 |
| Time to oral therapy, h | 94 (66–183) | 121 (77–199) | 86 (63–164) | .035 |
| Length of therapyb, d | 10 (8–11) | 10 (8–11) | 10 (8–11) | .69 |
| Days of therapyc | 12 (9–14) | 12 (10–15) | 11 (9–14) | .28 |
| AS off-hours (4:01 pm–6:59 am, Mon–Fri and all hours Sat–Sun) | (n = 81) | (n = 39) | (n = 42) | |
| Time to narrowest therapy, h | 73 (57–125) | 73 (60–138) | 76 (52–115) | .56 |
| Time to oral therapy, h | 95 (69–174) | 137 (70–207) | 92 (70–135) | .21 |
| Length of therapyb, d | 9 (8–12) | 10 (8–12) | 9 (8–12) | .71 |
| Days of therapyc | 11 (9–14) | 10 (9–13) | 11 (9–15) | .41 |
Data are presented as median (interquartile range) unless otherwise indicated.
Abbreviations: AS, antimicrobial stewardship; cAST, conventional antimicrobial susceptibility testing; rAST, rapid antimicrobial susceptibility testing.
P values were generated from Wilcoxon rank-sum test for continuous variables and Fisher exact test or χ2 test for categorical variables, between rAST and cAST.
Length of therapy was defined as the number of consecutive days the patient received any antibiotic therapy. Any day in which the patient received a dose of an antibiotic was counted as 1 day.
Days of therapy was defined as the number of days a patient is on any particular antibiotic and is cumulative for all antibiotics the patient received.
Secondary Outcomes
Outcomes are summarized in Table 2 and select outcomes are displayed in a timeline in Figure 2. Additional analysis comparing median and mean values of select outcomes are listed in Supplementary Table 2. Time from blood culture collection to susceptibility result was significantly shorter in the rAST group by 23 hours (median [IQR], 62 [59–67] hours vs 39 [35–46] hours; P < .001). Time to Gram stain and identification was similar between groups. Median (IQR) LOS was 2 days shorter in the rAST group (7 [4–13] days vs 5 [4–8] days; P = .04). More patients in the rAST group received oral therapy (66% vs 80%; P = .04) and were discharge ready by day 3 (38% vs 56%; P = .01), with no difference in discharge readiness by day 5. Time to oral therapy was 35 hours shorter in the rAST group (median [IQR], 126 [76–209] hours vs 91 [66–154] hours; P = .02). No differences in mortality, bacteremia recurrence, 30-day emergency department visit without admission, or 30-day hospital readmission were observed. Antibiotic use is described in Supplementary Table 3. The most common oral therapy was cephalexin (45.3%). There was no significant difference in antibiotic consumption or length of therapy.
Figure 2.
Time course of patient outcomes between rapid antimicrobial susceptibility testing (rAST) and conventional antimicrobial susceptibility testing (cAST). Outcomes from the full cohort as reported in Table 2. Time points are reported as median values.
DISCUSSION
In this randomized trial, rAST in the setting of AS resulted in a trend toward shorter time to narrowest effective therapy; however, this result was not statistically significant. When patients who were already on narrowest therapy at the time of organism identification were excluded from the analysis, rAST resulted in a significantly shorter time. A significant reduction in median time to oral therapy was demonstrated in the rAST group.
This is the first study to analyze downstream effects of rAST including time to oral therapy and discharge readiness in a cohort with GN-BSI with source control. The focus of this study was narrowest effective therapy, unlike other studies that have used broader definitions of optimal therapy and generally focused on empiric therapy changes [14, 26–29]. What constitutes narrowest therapy can vary based on local susceptibility patterns. The considerations for narrowest effective therapy (Supplementary Table 1) are similar to agents used in the National Healthcare Safety Network’s Standardized Antimicrobial Administration Ratio narrow category, but made more specific to Gram-negative organisms and our local susceptibility patterns.
Five RCTs have evaluated the impact of rAST [14, 26–30]. Comparison of outcomes between these studies is limited by the variability in rAST method, patient population, and organisms. Nevertheless, these studies demonstrated improvements in antibiotic use with rAST using various definitions of improvement. The RAPIDS-GN study evaluated a rAST platform (Accelerate Pheno) in combination with AS in 448 patients with GN-BSI [14]. The study used a rAST method that reported results in approximately 7 hours, similar to the direct-inoculation method used here. The authors found a significant decrease in time to GN antibiotic change (17.3% vs 42.1%; P < .001). This is the only prospective RCT in addition to the present study to analyze the impact of rAST in a cohort of exclusively GN-BSI.
The subgroup analysis, which excluded patients who were already on narrowest therapy at the time of organism identification and who received AST results during AS off-hours, demonstrated a significant reduction in time to narrowest therapy. This effect was not observed in patients with AST results reported during AS off-hours, suggesting the importance of combining rAST with AS services. This is consistent with previous literature evaluating rapid diagnostic technology (RDT), which demonstrated no impact on outcomes in the absence of AS [16, 17]. AS services were only available Monday through Friday, which may have muted the effect of the intervention during alternative hours. However, a similar study with 24-hour AS coverage reported a low rate of antibiotic changes overnight due to AS preference not to intervene for nonurgent recommendations [26].
In contrast to previous RCTs [15], patients undergoing rAST had a significantly lower LOS. The Rapid Identification and Susceptibility Testing for Gram-negative Bacteremia (RAPIDS-GN) study reported no difference in LOS (8.2 vs 9.8 days). While the authors note the study was not powered to detect decreases in LOS and the sample size was higher compared to the present study (205 vs 448), there are several possible explanations for the decrease observed here. First, patient population differences may have led to differences in severity of illness. Patients in the RAPIDS-GN study had higher rates of mechanical ventilation, intensive care unit admission, and nonurinary sources. However, the Pitt bacteremia score and CCI score (2.4 and 5.9, respectively) were similar to the current study. Second, the present study excluded patients without source control, which may have selected for patients more likely to be impacted by the intervention. A lower proportion of patients in the RAPIDS-GN study had urinary sources of infection (35% vs 67%), highlighting this difference. Last, differences in antibiograms and AS practices can impact antibiotic use. A third-generation cephalosporin resistance rate of only 9.8% among Enterobacterales was observed here, compared to 18.4% in RAPIDS-GN, which can limit the availability of oral therapies often necessary for hospital discharge. High rates of β-lactam–susceptible Escherichia coli (84% susceptible to cefazolin per the 2021 antibiogram, based on a susceptibility breakpoint of ≤4) allows for streamlined transitions to narrow-spectrum oral therapy, with 69% of patients who received oral therapy receiving a β-lactam. A combination of low resistance rates and existing culture of aggressive oral transition practices may have led to a decreased LOS.
All-cause 30-day mortality in our study was 2.9%, which is lower than the reported mortality in the RAPIDS-GN (9.6%); however, that study included patients without source control. Similar to other studies, there was no difference in mortality rates between groups. The only RCT of rAST to have reported a difference in mortality is Doern and colleagues, who reported a 5.7% decrease in attributable mortality [27]. The authors conducted a similar method of rAST; however, comparison of results is limited by the lack of AS intervention and diverse isolate sources and organisms. Retrospective studies of rAST have not demonstrated a difference in mortality but many are not powered to detect differences [31, 32]. A systematic review and meta-analysis of molecular RDT used for BSI found an associated decrease in mortality with RDT when used in combination with AS [17]. While the methods assessed in this study do not include phenotypic rAST, these results are encouraging.
Strengths of this study include the randomized design and a method of rAST, which did not require purchase of a laboratory platform or single-use panels. Many RDTs are supplementary tests that cannot fully replace traditional platforms since they are limited in which bacteria and resistance genes they detect. For example, the Biofire BCID panel used in the study by Banerjee and colleagues detected only 81% of organisms from blood cultures [26]. Similarly, Accelerate Pheno is not currently US Food and Drug Administration approved to report cefazolin, ampicillin, or trimethoprim-sulfamethoxazole susceptibilities, which excludes their use in de-escalation [33]. Second, inclusion of AS intervention was a key component to the study design, which has been shown to enhance outcomes in combination with RDT [17]. Last, patients who never received narrowest therapy were included in the analysis, which reflects intention to treat. In addition, it more closely parallels real-world practice where AS recommendations may not always be accepted. Individual patient factors that would require broader therapy were considered in the determination of the primary outcome, which limits confounding.
There are several limitations to this study. First, this study employed a unique exclusion criteria of infectious source control, which led to postrandomization exclusion of 30 patients. Similar proportions of patients (cAST: 58%, rAST 42%) between groups were excluded for this reason (Supplementary Table 4); however, it is still possible that postrandomization exclusion led to unmeasured confounders between groups. Exclusion of polymicrobial cultures also limits generalizability, though this only accounted for 4.7% of patients. Second, clinicians and AS personnel were not blinded, which could have influenced treatment recommendations. Third, specific types and interventions by AS were not fully captured as many were made via informal channels such as phone calls or internal messaging systems that were not linked to the medical record. Since AS personnel were not blinded and recommendation acceptance rates were not captured, this may have introduced bias that led to observed differences between groups. To mitigate this, AS personnel utilized a standard empiric treatment guideline and intervention (Supplementary Figure 1 and Supplementary Table 1). Although treating clinicians were blinded to the study group and not informed of the study’s existence, they may have observed faster times to AST results based on time stamps in the electronic medical record or the result comment that included the word “presumptive.” Though unlikely, this unblinding could have influenced treatment decisions. Last, this study was performed at a single healthcare system with one AS program, which may limit generalizability. The study institutions also have low resistance rates (E coli extended-spectrum β-lactamase rate: 6%), which limits generalizability.
In this study of patients with GN-BSI, rAST did not significantly decrease time to narrowest effective therapy, but did decrease time to administration of oral antibiotics and decreased LOS. This LOS reduction resulted in an approximate $1.2 million dollar cost-avoidance in the rAST group alone. There was no direct cost of implementing rAST at the study institutions since rapid identification via MALDI-TOF was already in place, although indirect costs such as microbiologist time were not evaluated. The cost-effectiveness of this method likely varies depending on existing laboratory practices, patient populations, and AS resources. In a subgroup analysis excluding patients receiving narrowest therapy prior to organism identification, time to narrowest therapy was significantly shorter in the rAST group, especially when combined with AS. Faster AST reporting in the setting of AS has the potential to facilitate early transitions to oral therapy and hospital discharge. Direct blood culture inoculation of existing microbiology laboratory platforms is an alternative method for optimizing patient outcomes compared to commercial platforms.
Supplementary Material
Contributor Information
Alyssa B Christensen, Department of Pharmacy, Providence St Vincent Medical Center, Portland, Oregon, USA.
Brent Footer, Department of Pharmacy, Providence Portland Medical Center, Portland, Oregon, USA.
Tobias Pusch, Department of Infectious Diseases, Providence St Vincent Medical Center, Portland, Oregon, USA.
Kim Heath, Department of Clinical Microbiology, Providence Oregon Regional Laboratory, Portland, Oregon, USA.
Maha Iqbal, Department of Clinical Microbiology, Providence Oregon Regional Laboratory, Portland, Oregon, USA.
Lian Wang, Center for Cardiovascular Analytics, Research and Data Science, Providence Heart Institute, Providence Research Network, Portland, Oregon, USA.
Gregory Tallman, Department of Pharmacy, Providence St Vincent Medical Center, Portland, Oregon, USA; School of Pharmacy, Pacific University, Hillsboro, Oregon, USA.
Cameron Cover, Department of Infectious Diseases, Providence St Vincent Medical Center, Portland, Oregon, USA.
Jennifer Marfori, Department of Infectious Diseases, Providence St Vincent Medical Center, Portland, Oregon, USA.
Brian Kendall, Department of Infectious Diseases, Providence Portland Medical Center, Portland, Oregon, USA.
Nick Stucky, Department of Infectious Diseases, Providence Portland Medical Center, Portland, Oregon, USA.
Meagan Greckel, Department of Pharmacy, Providence St Vincent Medical Center, Portland, Oregon, USA; Department of Pharmacy, Providence Portland Medical Center, Portland, Oregon, USA.
Ivor L Thomas, Department of Clinical Microbiology, Providence Oregon Regional Laboratory, Portland, Oregon, USA.
Katelynn Tran, Department of Pharmacy, Providence St Vincent Medical Center, Portland, Oregon, USA.
Salena Yip, Department of Pharmacy, Providence St Vincent Medical Center, Portland, Oregon, USA.
Margret Oethinger, Department of Clinical Microbiology, Providence Oregon Regional Laboratory, Portland, Oregon, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Potential conflicts of interest. T. P. reports contracted research support from Gilead Sciences and ViiV Healthcare; reports receiving honoraria as consultant to Gilead Sciences and ViiV Healthcare; and reports being a member of Gilead Sciences’ HIV speaker’s bureau. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 2013; 19:501–9. doi: 10.1111/1469-0691.12195 [DOI] [PubMed] [Google Scholar]
- 2. Kang C-I, Kim S-H, Kim H-B, et al. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis 2003; 37:745–51. doi: 10.1086/377200 [DOI] [PubMed] [Google Scholar]
- 3. Kang C-I, Kim S-H, Park WB, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother 2005; 49:760–6. doi: 10.1128/AAC.49.2.760-766.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diekema DJ, Beekmann SE, Chapin KC, Morel KA, Munson E, Doern GV. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol 2003; 41:3655–60. doi: 10.1128/JCM.41.8.3655-3660.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lodise TP, Kanakamedala H, Hsu WC, Cai B. Impact of incremental delays in appropriate therapy on the outcomes of hospitalized adult patients with gram-negative bloodstream infections: “Every day matters.” Pharmacotherapy 2020; 40:889–901. [DOI] [PubMed] [Google Scholar]
- 6. Bell BG, Schellevis F, Stobberingh E, Goossens H, Pringle M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect Dis 2014; 14:13. doi: 10.1186/1471-2334-14-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yahav D, Franceschini E, Koppel F, et al. Seven versus 14 days of antibiotic therapy for uncomplicated gram-negative bacteremia: a noninferiority randomized controlled trial. Clin Infect Dis 2019; 69:1091–8. doi: 10.1093/cid/ciy1054 [DOI] [PubMed] [Google Scholar]
- 8. Fabre V, Amoah J, Cosgrove SE, Tamma PD. Antibiotic therapy for Pseudomonas aeruginosa bloodstream infections: how long is long enough? Clin Infect Dis 2019; 69:2011–4. doi: 10.1093/cid/ciz223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamma PD, Conley AT, Cosgrove SE, et al. Association of 30-day mortality with oral step-down vs continued intravenous therapy in patients hospitalized with Enterobacteriaceae bacteremia. JAMA Intern Med 2019; 179:316. doi: 10.1001/jamainternmed.2018.6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sutton JD, Stevens VW, Chang N-CN, Khader K, Timbrook TT, Spivak ES. Oral β-lactam antibiotics vs fluoroquinolones or trimethoprim-sulfamethoxazole for definitive treatment of Enterobacterales bacteremia from a urine source. JAMA Netw Open 2020; 3:e2020166-e. doi: 10.1001/jamanetworkopen.2020.20166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heil EL, Bork JT, Abbo LM, et al. Optimizing the management of uncomplicated gram-negative bloodstream infections: consensus guidance using a modified Delphi process. Open Forum Infect Dis 2021; 8:ofab434. doi: 10.1093/ofid/ofab434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robinson ED, Stilwell AM, Attai AE, et al. Implementation of a rapid phenotypic susceptibility platform for gram-negative bloodstream infections with paired antimicrobial stewardship intervention: is the juice worth the squeeze? Clin Infect Dis 2021; 73:783–92. doi: 10.1093/cid/ciab126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walsh TL, Bremmer DN, Moffa MA, et al. Impact of an antimicrobial stewardship program-bundled initiative utilizing Accelerate Phenosystem in the management of patients with aerobic gram-negative bacilli bacteremia. Infection 2021; 49:511–9. doi: 10.1007/s15010-021-01581-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee R, Komarow L, Virk A, et al. Randomized trial evaluating clinical impact of RAPid identification and susceptibility testing for gram-negative bacteremia: RAPIDS-GN. Clin Infect Dis 2021; 73:e39–46. doi: 10.1093/cid/ciaa528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anton-Vazquez V, Hine P, Krishna S, Chaplin M, Planche T. Rapid versus standard antimicrobial susceptibility testing to guide treatment of bloodstream infection. Cochrane Database Syst Rev 2021; 5:CD013235. doi: 10.1002/14651858.CD013235.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pliakos EE, Andreatos N, Shehadeh F, Ziakas PD, Mylonakis E. The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev 2018; 31:e00095-17. doi: 10.1128/CMR.00095-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64:15–23. doi: 10.1093/cid/ciw649 [DOI] [PubMed] [Google Scholar]
- 18. Frye AM, Baker CA, Rustvold DL, et al. Clinical impact of a real-time PCR assay for rapid identification of staphylococcal bacteremia. J Clin Microbiol 2012; 50:127–33. doi: 10.1128/JCM.06169-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beganovic M, Timbrook TT, Wieczorkiewicz SM. Predictors of time to effective and optimal antimicrobial therapy in patients with positive blood cultures identified via molecular rapid diagnostic testing. Open Forum Infect Dis 2019; 6:ofy350. doi: 10.1093/ofid/ofy350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hogan CA, Watz N, Budvytiene I, Banaei N. Rapid antimicrobial susceptibility testing by VITEK®2 directly from blood cultures in patients with gram-negative rod bacteremia. Diagn Microbiol Infect Dis 2019; 94:116–21. doi: 10.1016/j.diagmicrobio.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 21. Horing S, Massarani AS, Loffler B, Rodel J. Rapid antibiotic susceptibility testing in blood culture diagnostics performed by direct inoculation using the VITEK(R)-2 and BD Phoenix platforms. Eur J Clin Microbiol Infect Dis 2019; 38:471–8. doi: 10.1007/s10096-018-03445-3 [DOI] [PubMed] [Google Scholar]
- 22. Al-Hasan MN, Lahr BD, Eckel-Passow JE, Baddour LM. Predictive scoring model of mortality in gram-negative bloodstream infection. Clin Microbiol Infect 2013; 19:948–54. doi: 10.1111/1469-0691.12085 [DOI] [PubMed] [Google Scholar]
- 23. Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 2005; 43:1130–9. doi: 10.1097/01.mlr.0000182534.19832.83 [DOI] [PubMed] [Google Scholar]
- 24. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Core Team R, Core Team R. R: A language and environment for statistical computing. Version 3.6.1. Vienna, Austria: R Foundation for Statistical Computing, 2019. [Google Scholar]
- 26. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction–based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. doi: 10.1093/cid/civ447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doern GV, Vautour R, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol 1994; 32:1757–62. doi: 10.1128/jcm.32.7.1757-1762.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J-H, Kim I, Kang C, et al. Enhanced antimicrobial stewardship based on rapid phenotypic antimicrobial susceptibility testing for bacteraemia in patients with haematological malignancies: a randomized controlled trial. Clin Microbiol Infect 2021; 27:69–75. doi: 10.1016/j.cmi.2020.03.038 [DOI] [PubMed] [Google Scholar]
- 29. Beuving J, Wolffs P, Hansen W, et al. Impact of same-day antibiotic susceptibility testing on time to appropriate antibiotic treatment of patients with bacteraemia: a randomised controlled trial. Eur J Clin Microbiol Infect Dis 2015; 34:831–8. doi: 10.1007/s10096-014-2299-0 [DOI] [PubMed] [Google Scholar]
- 30. Banerjee R, Humphries R. Rapid antimicrobial susceptibility testing methods for blood cultures and their clinical impact. Front Med (Lausanne) 2021; 8:635831. doi: 10.3389/fmed.2021.635831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ehren K, Meißner A, Jazmati N, et al. Clinical impact of rapid species identification from positive blood cultures with same-day phenotypic antimicrobial susceptibility testing on the management and outcome of bloodstream infections. Clin Infect Dis 2020; 70:1285–93. [DOI] [PubMed] [Google Scholar]
- 32. Perez KK, Olsen RJ, Musick WL, et al. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant gram-negative bacteremia. J Infect 2014; 69:216–25. doi: 10.1016/j.jinf.2014.05.005 [DOI] [PubMed] [Google Scholar]
- 33. Accelerate Diagnostics, Inc . Accelerate PhenoTest BC Kit. Available at: https://acceleratediagnostics.com/products/accelerate-phenotest-bc/#panel. Accessed 18 January 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.