Abstract
The heat shock response in Escherichia coli depends primarily on the increased synthesis and stabilization of otherwise scarce and unstable ς32 (rpoH gene product), which is required for the transcription of heat shock genes. The heat-induced synthesis of ς32 occurs at the level of translation, and genetic evidence has suggested the involvement of a secondary structure at the 5′ portion (nucleotides −19 to +247) of rpoH mRNA in regulation. We now present evidence for the mRNA secondary structure model by means of structure probing of RNA with chemical and enzymatic probes. A similar analysis of several mutant RNAs with a mutation predicted to alter a base pairing or with two compensatory mutations revealed altered secondary structures consistent with the expression and heat inducibility of the corresponding fusion constructs observed in vivo. These findings led us to assess the possible roles of each of the stem-loop structures by analyzing an additional set of deletions and base substitutions. The results indicated not only the primary importance of base pairings between the translation initiation region of ca. 20 nucleotides (the AUG initiation codon plus the “downstream box”) and the internal region of rpoH mRNA but also the requirement of appropriate stability of mRNA secondary structures for characteristic thermoregulation, i.e., repression at a low temperature and induction upon a temperature upshift.
The heat shock response is a universal, adaptive, and homeostatic cellular response against damage to protein folding under heat and other stresses. In Escherichia coli, the response results primarily from a transient increase in the level of ς32, which is encoded by rpoH and which is specifically required for the transcription of the set of well-conserved heat shock genes (11, 35). The increase in the ς32 level results from both the enhanced synthesis and the stabilization of normally unstable ς32 (12, 30). Whereas the stabilization of ς32 is thought to be triggered by the titration of free DnaK/DnaJ chaperones by stress-induced misfolded proteins (1, 4, 9, 16, 32), the increased synthesis occurs at the level of translation (15, 21, 30) and presumably is regulated via a separate pathway (22, 29, 35). The production of abnormal proteins under various conditions also induces the heat shock response through an increase in the ς32 level (10), but such induction appears to involve only the stabilization and not the increased synthesis of ς32 (16). Furthermore, exposure to extremely high temperatures (e.g., 50°C) can induce ς32 synthesis by enhancing rpoH transcription by activating the second heat shock ς factor, ςE (7, 33), in response to misfolded proteins accumulated in the periplasm (11, 19). Thus, E. coli cells strictly regulate ς32 at various levels to cope with increasing demands for chaperones, ATP-dependent proteases, and other heat shock proteins under a variety of stress conditions (1, 9, 11, 25, 35).
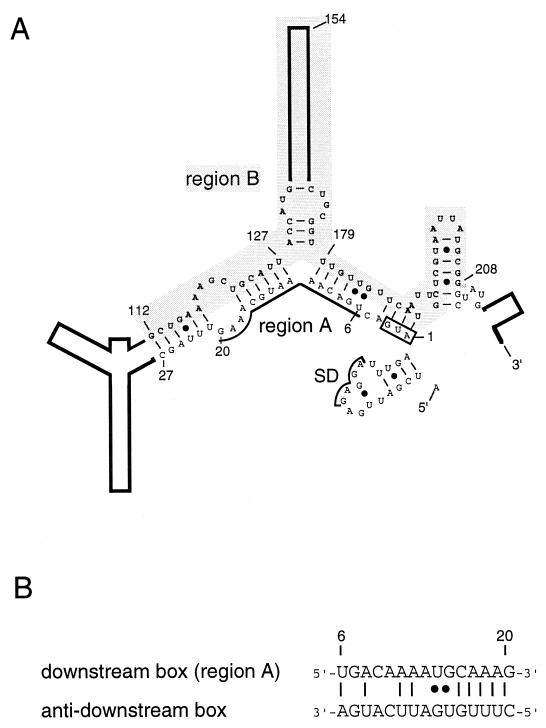
As to the mechanism of translational induction of ς32, extensive deletion analyses of an rpoH-lacZ gene fusion revealed the involvement of positive and negative regulatory regions (regions A and B, respectively) on the 5′ portion of rpoH mRNA (15, 21). Region A (15 nucleotides [nt]), located close to the initiation codon, represents the “downstream box,” (27) which is complementary to part of the 16S rRNA and which potentially enhances translation. Region B, an internal coding segment of ca. 100 nt, is a negative element involved in repressing translation under nonstress conditions. A computer prediction revealed a secondary structure for the 5′ segment (nt −19 to +247) of rpoH mRNA which is fully consistent with the above findings; base pairings between region A and part of region B appeared to negatively modulate rpoH translation (21) (Fig. 1).
FIG. 1.
Schematic representation of the 5′ portion (nt −19 to +247) of E. coli rpoH mRNA as predicted by use of Mulfold (14). (A) Secondary structure thought to be involved in modulating heat-induced synthesis of ς32 (21). Region A (nt +6 to 20), the initiation codon, and the Shine-Dalgarno (SD) sequence are indicated. Region B (nt +112 to 208) is shaded. Numbers refer to the nucleotides of the coding sequence. (B) Putative base pairing between the downstream box (region A) of rpoH and the “anti-downstream box” of 16S rRNA (spanning nt 1469 to 1483). •, G-U pairs.
Mutational analyses of rpoH mRNA deficient in the expression or regulation of a GF364 fusion carrying the initial 364 nt of the rpoH coding region (Fig. 2A) not only substantiated the importance of some of the critical base pairings but also suggested the possible involvement of specific nucleotide sequences in heat induction (36). It was surmised that the translation of rpoH mRNA is restricted by the formation of secondary structure(s) that would limit ribosome entry under nonstress conditions. Upon mild heat shock (e.g., a shift from 30 to 42°C), such mRNA structures were thought to be disrupted to enhance translation, although the mechanism remained unknown (21, 35, 36). In addition, the isolation and characterization of rpoH homologs from a number of gram-negative bacteria revealed evolutionary conservation of both region A and the mRNA secondary structure among the gamma proteobacteria (23). All members of the latter group of bacteria examined seemed to exhibit heat-induced synthesis of ς32 homologs at the translational level, as in E. coli (24).
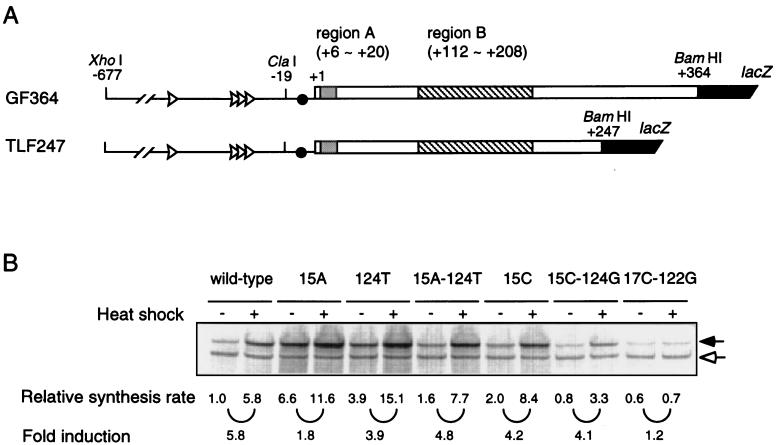
FIG. 2.
Structure and expression of an rpoH-lacZ gene fusion (TLF247). (A) Schematic diagrams of the TLF247 fusion construct and GF364, studied previously (21). The locations of regions A and B are indicated. (B) SDS-PAGE patterns of fusion proteins expressed from the wild-type and mutant forms of TLF247. Cells were grown at 30°C and shifted to 42°C. Pulse-labeling with [35S]methionine was done for 2 min before or 3 min after the temperature shift. The labeled cells were disrupted, and immunoprecipitates obtained with anti–β-galactosidase serum were analyzed by SDS-PAGE as described in Materials and Methods. Closed and open arrows indicate fusion proteins and β-galactosidase ω protein (internal reference), respectively.
The translational induction of ς32 is transient and is followed by a shutoff phase mediated by the DnaK-DnaJ-GrpE chaperones (9, 12, 29). The translational repression and destabilization of ς32 during adaptation periods are part of the feedback regulatory mechanisms (29, 31, 32) mediated by a segment of ς32 protein (18, 22, 35) which contains a highly and uniquely conserved sequence among the rpoH homologs (23, 34). Several lines of evidence suggest that this region plays important roles in the chaperone-mediated negative control of the synthesis and/or stability of ς32 (8, 17, 18). However, the exact regulatory mechanisms of the transient heat induction of ς32 synthesis, including the nature of the sensor(s) and signaling pathway(s), remain largely unresolved.
We now report a structural and functional analysis of the 5′ segment of rpoH mRNA responsible for thermoregulation. We first probed the structures of rpoH RNAs from the wild type and several mutants in vitro and then analyzed their expression in vivo after transcription from a single-copy rpoH-lacZ gene fusion. The data supported some salient features of the predicted mRNA secondary structure and provided the basis for further analysis of each of the component stem-loop structures. The results led us to propose that an mRNA secondary structure with appropriate stability and formed between the translation initiation region (the AUG initiation codon and region A) and the internal coding region is a prerequisite for the thermoregulation of ς32 synthesis.
MATERIALS AND METHODS
Strains, phages, and media.
E. coli K-12 strain MC4100 [araD Δ(argF-lac)U169 rpsL relA flbB deoC ptsF rbsR] (2) was used for all experiments in vivo. The λTLF97-3 vector (28) was used to construct rpoH-lacZ gene fusions. Minimal medium M9 (20) with 0.2% glucose, thiamine (2 μg/ml), and all amino acids except for methionine (20 μg/ml each) was used for pulse-labeling experiments. MacConkey lactose agar (Difco) and L agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (30 μg/ml) were used for isolating λ lysogens containing rpoH-lacZ gene fusions. Recombinant DNA and other general techniques were as described by Sambrook et al. (26) and by Miller (20).
Chemicals, enzymes, and buffers.
1-Cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulfonate (CMCT) and diethyl pyrocarbonate (DEP) were purchased from Sigma. RNase V1 was obtained from Pharmacia, and avian myeloblastosis virus reverse transcriptase was obtained from Life Science. Buffer H was 70 mM HEPES-KOH (pH 7.8) containing 10 mM MgCl2, 270 mM KCl, and 1 mM dithiothreitol, and buffer V1 was 30 mM Tris-HCl (pH 7.8) containing 20 mM MgCl2, 300 mM KCl, and 1 mM dithiothreitol.
Construction of rpoH-lacZ gene fusions.
The gene fusion (translational fusion) designated TLF247 was constructed by in-frame fusion between the XhoI-BamHI fragment of pGF247 (21) containing the rpoH promoters and the 5′ portion of the coding region (nt −677 to +247) and codon 9 of lacZ on the λTLF97-3 vector. The same fragment of pGF247 was also inserted into pBluescript SK(+), yielding pBSK247. Derivatives of TLF247 carrying base substitutions were constructed by PCR with plasmid pFRP103 containing each of the mutations (36) as a template and synthetic oligonucleotide primers that corresponded to the 5′ (nt −46 to −27) and 3′ (nt +227 to 247) portions of the coding region. Seven extra bases containing the BamHI site were added to the latter primer to make in-frame fusions to lacZ (21). A set of 3′ deletions of GFR153 was constructed by PCR with the same 5′ primer as that used above and 3′ primers that corresponded to the end of each deletion (with the same seven extra bases) and with λGFR153 (21) as a template. DNA fragments with the desired sequences containing PCR-amplified products were inserted into pBSK247, and nucleotide sequences were confirmed by dideoxy sequencing. The XhoI-BamHI fragments of the resulting plasmids were then transferred to the λTLF97-3 vector by in vitro packaging. TLF229Δ(stemIII) was constructed from TLF247 by deleting the apical portion of stem II (nt +30 to 110) and all of stem III (nt +128 to 178). Four synthetic oligonucleotides (ca. 60 nt long) were annealed and ligated to create a DNA fragment with 5′ protruding ends for joining with the ClaI or BamHI site at the 5′ or 3′ end, respectively. The resulting fragment was cloned into pBSK247 by replacing the ClaI-BamHI fragment to obtain pBSK229Δ(stemIII).
Determination of rates of synthesis of fusion proteins.
The procedure used for the determination of fusion protein synthesis rates was essentially that described previously (21). Portions (0.1 ml) of log-phase cultures were pulse-labeled with l-[35S]methionine (1,200 Ci/mmol). Extracts were prepared, and portions with equal radioactivity were mixed with a fixed amount of JM103 cell extract (labeled with [35S]methionine) containing β-galactosidase ω protein and treated with antibody against β-galactosidase (Organon Teknika Cappel). The immunoprecipitates were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (7.5% gel), and the intensities of radioactive bands were quantified with a Fujix BAS2000 imaging analyzer to determine the rates of synthesis of fusion proteins after correction for recovery with ω protein as a reference.
RNA preparation.
RNA containing the upstream region and part of the rpoH coding region (nt −60 to +247) was prepared in vitro with T7 RNA polymerase by use of an RNA transcription kit (Stratagene). The AflII-BamHI fragments of pBSK247 were placed under the control of the T7 promoter of vector pSP72, and the resulting plasmids were digested with BamHI and used as templates for RNA synthesis. The RNA obtained was treated at 65°C for 3 min, followed by slow cooling to room temperature prior to use.
Structure probing of RNA.
The procedures used for RNA structure probing were essentially those described by Christiansen et al. (3). Prior to treatment with CMCT, DEP, or RNase V1, RNA (4 μg) was renatured (heating and slow cooling) in 20 μl of buffer H, 200 μl of buffer H, or 20 μl of buffer V1, respectively. RNA was treated with CMCT (50 mM) or DEP (96 μM), and the reaction was terminated by the addition of ethanol on dry ice. For RNase V1 treatment, 6 μl of RNA was mixed with an equal volume of buffer V1 containing enzyme on ice for 30 min, treated with phenol, and precipitated with ethanol. RNA incubated without probes served as a control in all experiments. The identification of modified bases was carried out by primer extension analysis: 0.3 pmol of modified RNA and 3 pmol of 5′-fluorescence-labeled primer complementary to the 5′ (nt +79 to 101) or 3′ (nt +227 to 247) region were incubated with avian myeloblastosis virus reverse transcriptase. Portions of primer extention products were loaded on a 5% polyacrylamide–8 M urea sequencing gel and electrophoresed at 1,400 V for 2.5 h. The bands were detected with Fluorescence BioImage Analyzer FMBIO II Multi-View (Hitachi), and the modified bases were identified by comparison with sequence ladders simultaneously run with the same end-labeled primers.
RESULTS
Thermoregulation of a TLF247 gene fusion mediated by an mRNA secondary structure.
To further understand the rpoH translational control mechanisms, it was important to analyze structural features of the “minimal” mRNA segment(s) essential for thermoregulation. Based on our previous work (21), we constructed a new rpoH-lacZ gene fusion carrying only the first 247 nt of rpoH (TLF247) (Fig. 2A) and reexamined the effects of mutations previously characterized with GF364 carrying the first 364 nt (36). Cells of MC4100 carrying TLF247 or its mutant derivatives were grown at 30°C, shifted to 42°C, and pulse-labeled with [35S]methionine to determine fusion protein synthesis rates (Fig. 2B). Wild-type TLF247 exhibited five- to sixfold induction upon the temperature upshift. In contrast, the mutant fusions (15A, 15C, or 124T) carrying a base substitution predicted to disrupt the 15G-124C base pairing showed enhanced expression at 30°C and reduced induction upon the shift to 42°C (Fig. 3B), consistent with our previous results obtained with GF364 (we discuss below the relatively small effect observed with 15C).
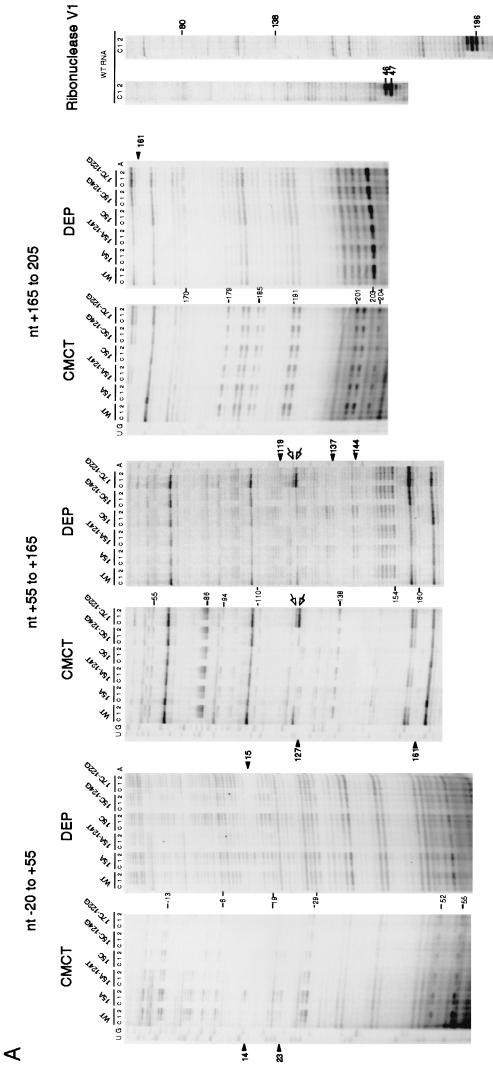
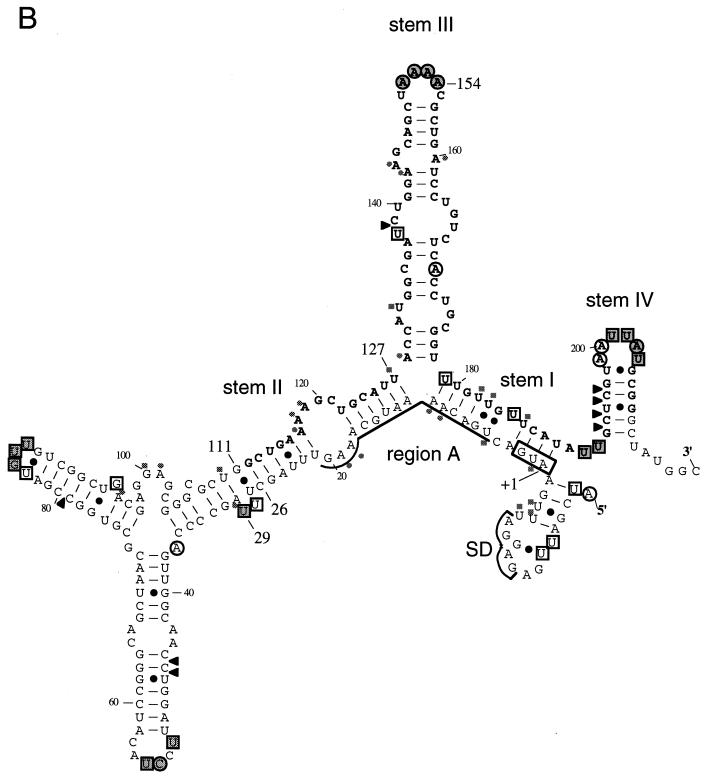
FIG. 3.
Structure probing of rpoH mRNA. (A) Urea-PAGE patterns of primer extension products. A 5′ segment (nt −60 to +247) of wild-type (WT) or mutant RNAs prepared in vitro was treated with CMCT, DEP, or RNase V1, and modified bases were identified by reverse transcription analysis as described in Materials and Methods. Treatment with CMCT was carried out at 30°C for 0 min (lanes c), 10 min (lanes 1), or 30 min (lanes 2), whereas treatment with DEP was done at 30°C for 0 min (lanes c), 5 min (lanes 1), or 10 min (lanes 2). Digestion with RNase V1 was done at 0°C for 30 min with 0 U (lanes c), 0.0005 U (lanes 1), or 0.001 U (lanes 2) of enzyme. Only some of the bases that were clearly modified by each treatment are indicated by nucleotide numbers. Relevant sequence ladders (wild type) are shown to the side as a reference. When comparing the observed bands with the sequence ladders, one should note that cDNA synthesis stops one residue before the modified base. Arrowheads indicate some of the modifications uniquely found in mutant RNA(s), whereas arrows indicate positions at which reverse transcriptase was arrested. Only the data for wild-type RNA are shown for experiments with RNase V1. (B) Modified bases identified with wild-type RNA in panel A are indicated on the mRNA secondary structure. The bases modified by CMCT or DEP are represented by squares or circles, respectively. The relative extents of modification are indicated by outlined stippled, outlined open, and nonoutlined shaded symbols for strong, modest, and weak reactivities, respectively. Arrowheads indicate sites cleaved by RNase V1. C-53 was fortuitously modified by DEP. Stems I, II, III, and IV are indicated, as are the initiation codon, region A, and the Shine-Dalgarno (SD) sequence. Region B (nt +112 to 208) is shown in boldface letters.
As predicted if base pairing were important, the 15A-124T double mutant carrying a compensatory mutation to restore the base pairing exhibited almost normal heat induction. The generally increased expression at both temperatures was presumably due to the relative instability of A-U base pairing compared to G-C pairing. Similarly, the 15C-124G double mutant showed slightly reduced but nearly normal expression and heat induction, contrary to the previous results obtained with the same mutant in the GF364 fusion, which showed no detectable induction at 42°C (36). Reexamination of the latter mutant revealed almost normal induction (data not shown); the 15C-124G mutants in the GF364 and TLF247 constructs thus gave identical results. On the other hand, the 17C-122G double mutant, predicted to form a more stable C-G pairing (than the parental U-A pairing), showed reduced expression at 30°C and no significant induction at 42°C, confirming the previous results (36).
It should be noted that a mutation within region A can affect expression by altering complementarity to 16S rRNA (Fig. 1B) (21). The marked or slight increase found in the expression of 15A or 15C, respectively, relative to that of the wild type (Fig. 2B) was well correlated with the increased or decreased complementarity to 16S rRNA, respectively. Moreover, the higher expression of the 15A-124T double mutant than of the 15C-124G double mutant as well as the lower expression of the 17C-122G double mutant may be partially explained on the same basis. Taken together, these results confirmed the validity of the regulatory model based on the rpoH mRNA secondary structure. However, to support this model, it was necessary to demonstrate the existence of the proposed structure.
Structure probing of the rpoH mRNA secondary structure.
The structure of the 5′ segment of rpoH mRNA (nt −60 to +247) used for construction of the TLF247 fusion was examined with two chemical probes (CMCT and DEP) and an enzymatic probe (RNase V1) (Fig. 3A). RNA prepared by in vitro transcription with T7 RNA polymerase was heated at 65°C for 3 min, slowly cooled to room temperature, and treated with CMCT or DEP, which specifically modifies single-stranded U/G or A, respectively, or with RNase V1, which cleaves double-stranded RNA with no apparent sequence specificity. The modified bases were identified by reverse transcription analysis, and the relative reactivities of individual bases to each of the probes used are illustrated semiquantitatively on the predicted RNA secondary structure (Fig. 3B). The region upstream of nt −19 is not shown here, since it is known not to be involved in thermoregulation (21).
In general, bases strongly modified by chemical probes were found in terminal loops, whereas bases in internal loops, bulges, and branching points were modified less markedly. Some of the A’s and U’s in stem I predicted to form pairings were modified, albeit very weakly, suggesting that the secondary structure in this region might be relatively unstable. With respect to stems II, III, and IV, locations of modified bases were in good agreement with the predicted RNA structure, with a few exceptions. These results thus provided strong evidence for an rpoH mRNA secondary structure with several major stem-loops (21), which has so far been supported by mutational analyses of the expression of rpoH-lacZ gene fusions (36) and by evolutionary conservation of the predicted RNA secondary structure among the rpoH homologs (23). It should be noted, however, that the structure shown in Fig. 3B probably represents a major but not the only structure found under the set of conditions used.
Altered secondary structure of some mutant rpoH RNAs.
To determine the effects of base changes on the mRNA secondary structure, RNAs prepared from five mutants examined above (Fig. 2B) were subjected to similar structural analyses (Fig. 3A). The results obtained, combined with those for the wild-type RNA, revealed certain interesting differences as well as similarities (Table 1). Evidently, stems II and III in the 15A and 15C mutant RNAs were modified to greater extents than those in the wild-type RNA. A-15 was clearly modified by DEP in 15A RNA; in contrast, G-15 in wild-type RNA was not modified by CMCT as expected. More importantly, U-14 was also modified strongly in 15A RNA but not in wild-type RNA, indicating that the neighboring structure was affected by the G-to-A mutation at +15. Since C should not be modified by either probe, the change at +15 could not be seen in 15C RNA. However, U-14 was not modified in 15C RNA, indicating that the change in the neighboring structure was more pronounced in 15A RNA than in 15C RNA. Such differential base modifications, which presumably reflected differences in local secondary structures between the two RNAs, were well correlated with differential fusion protein expression levels at 30°C, namely, higher expression of the 15A mutant than of the 15C mutant (Fig. 2B).
TABLE 1.
Effects of mutations on RNA base modifications by CMCT or DEPa
| Stem | Position(s)b | Effect of mutation detected with RNA fromc:
|
||||
|---|---|---|---|---|---|---|
| 15A | 15A-124T | 15C | 15C-124G | 17C-122G | ||
| II | 14 (U) | ++ | 0 | 0 | 0 | 0 |
| 15 (G) | ++ (A) | 0 | 0 (C)e | 0 | 0 | |
| 17 (A) | + | 0 | + | 0 | 0 (C)e | |
| 18 (A) | + | 0 | + | 0 | 0 | |
| 19 (A) | + | 0 | + | 0 | 0 | |
| 22 (U) | + | 0 | + | 0 | 0 | |
| 23 (U) | + | 0 | + | 0 | 0 | |
| 119 (A) | − | 0 | − | 0 | 0 | |
| 126 (U) | + | 0 | + | 0 | 0 | |
| 127 (U) | + | 0 | + | 0 | − | |
| III | 135 (C) | +d | 0 | +d | 0 | 0 |
| 136 (G) | + | 0 | + | 0 | 0 | |
| 137 (A) | + | 0 | + | 0 | 0 | |
| 144 (A) | + | 0 | + | 0 | 0 | |
| 150 (U) | − | 0 | − | 0 | 0 | |
| 151–154 (A) | − | 0 | − | 0 | 0 | |
| 161 (U) | + | 0 | + | 0 | 0 | |
A 5′ segment (nt −60 to +247) of rpoH RNA was treated with CMCT or DEP, which specifically modifies single-stranded U/G or A, respectively, and the extents of modification relative to that of the wild-type RNA were determined (++, marked increase; +, increase; 0, no change; −, decrease).
The wild-type base at each position is shown in parentheses.
The mutated base is shown in parentheses.
C-135 was fortuitously modified by DEP under these conditions.
The C in these mutant RNAs was refractory to modification by either reagent.
In sharp contrast, RNAs containing the compensatory 15A-124T or 15C-124G mutations exhibited modifications very similar to those of wild-type RNA, consistent with their observed capacity for almost normal regulation of fusion protein synthesis (Fig. 2B). Also, there were few or no changes in modifications specific to the 17C-122G mutant RNA; however, reverse transcriptase tended to stall at the branch point of stems II and III (nt +126 and +127), even without treatment with chemical probes (Fig. 3A). Such an arrest of reverse transcriptase is consistent with the predicted increased stability of stem II in the 17C-122G mutant compared to the wild type (17A-122U) and with the observed inability to be heat induced in vivo (Fig. 2B). These results are therefore in line with the expectations of the mRNA secondary structure model and lend strong support to the notion that the structure of the 5′ segment of rpoH mRNA (nt −19 to +247) plays a major role in modulating translation initiation.
Further deletion analysis of critical regulatory regions.
To further define the rpoH regions critical for thermoregulation, we constructed and examined a set of 5′ and 3′ deletions of the rpoH-lacZ fusion on λTLF247. It had been shown that the internal segment of 127 nt that contains the 5′ half of region B (nt +27 to 153) (Fig. 2) could be deleted from GF364 without affecting regulation, despite the drastic alteration of the predicted mRNA secondary structure; part of stem III formed base pairings with region A (nt +12 to 17) and with seven artificial nucleotides inserted during construction (21) (see Fig. 5). Similarly, the newly constructed fusion TLF247Δ(27–153), which lacked the same segment, exhibited essentially normal expression at 30°C and induction at 42°C, indicating that the altered secondary structure due to the Δ(27–153) deletion fortuitously gained the capacity for thermoregulation (Fig. 4A, line 2). In spite of this anomaly, however, when some of the above mutations (15A, 15A-124T, and 17C-122G) were introduced into the latter construct, they had similar effects on thermoregulation, although less striking than those obtained with the TLF247 derivatives (data not shown). Thus, the thermoregulation observed with TLF247Δ(27–153) appeared to involve mechanisms similar to that found with the parental TLF247 fusion. These results also suggested that the apical portion of stem II and the intact form of stem III were not essential for regulation.
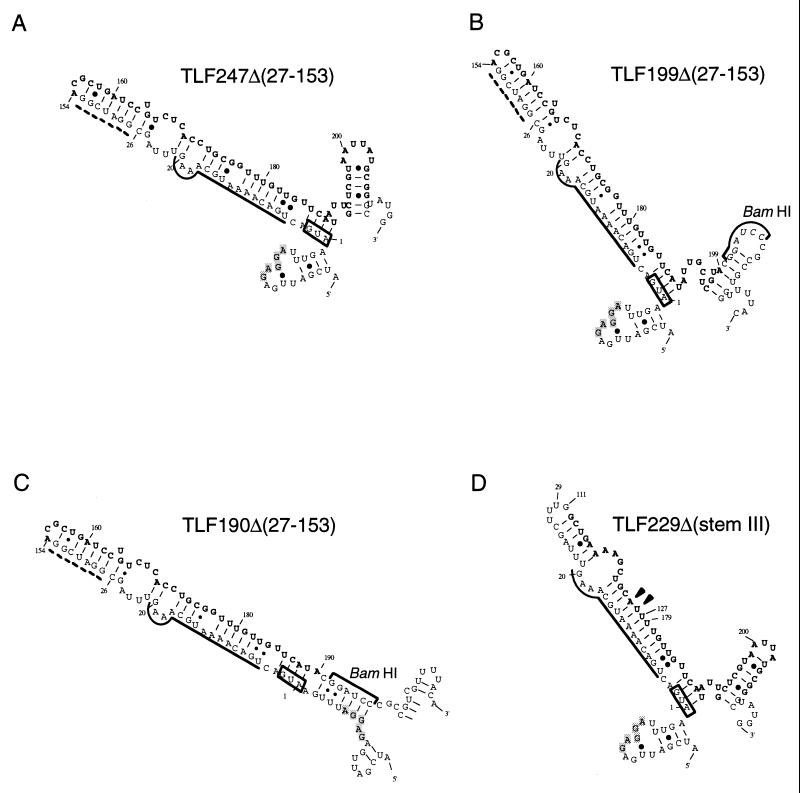
FIG. 5.
Predicted mRNA secondary structures for some of the deletion derivatives used. Structures predicted for RNA of 150 nt (starting from nt −19 for each; the sequences may include a BamHI junction and part of lacZ) and that have minimum free energy are shown for some representative constructs examined in Fig. 4. Only relevant portions are presented. The initiation codon, region A, and region B are indicated as described in the legend to Fig. 3B. The Shine-Dalgarno sequence is shown by shaded letters. Arrowheads indicate the positions where two G’s were replaced in constructing TLF229Δ(stemIII)GG (Fig. 4B). The broken line indicates extra bases inserted during construction (21).
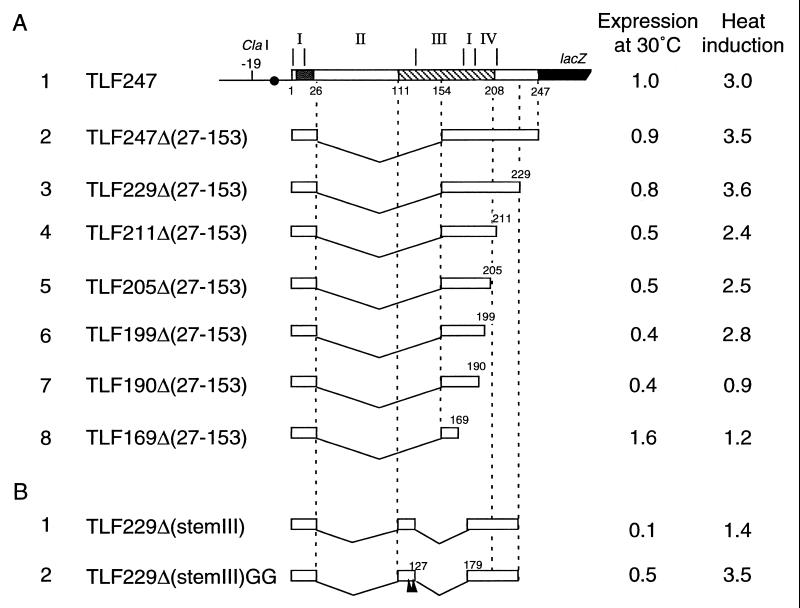
FIG. 4.
Structure and expression of fusion proteins from deletion derivatives of TLF247. (A) A series of 3′ deletions derived from the internal deletion TLF247Δ(27–153). Segments of mRNA that correspond to each of the stem structures (I to IV) are shown above the diagram, and nucleotide numbers are shown below. Regions A and B are indicated by stippled and hatched boxes, respectively. (B) A pair of deletions lacking stem III. Arrowheads indicate the positions of two G’s derived from the Δ(27–153) deletion. Cells were grown at 30°C and shifted to 42°C. Samples taken at time 0 (30°C) and 3 min after the shift were pulse-labeled with [35S]methionine for 1 min, followed by a 3-min chase. The labeled proteins were analyzed by immunoprecipitation, followed by SDS-PAGE as described in the legend to Fig. 2. Synthesis rates were normalized to that for the wild type (TLF247) labeled at 30°C. The apparently lower extents of induction observed here compared to those shown in Fig. 2B were due to slightly different procedures and not to instability of the fusion proteins examined (data not shown).
We next examined a set of 3′ deletions derived from TLF247Δ(27–153). Deletion to nt +229 had little effect on heat induction (Fig. 4A, line 3), but deletion to near the 3′ end of stem IV (and region B) (nt +211) reduced basal expression appreciably (line 4) (see Discussion). Deletions extending into stem IV (nt +205 or +199) affected heat induction only slightly (Fig. 4A, lines 5 and 6), whereas further deletion to nt +190 or +169 abolished induction completely, with a concomitant increase in the expression at 30°C of the latter construct (lines 7 and 8). These results appeared to indicate the importance of stem I but not stem IV for thermoregulation within the limitations of these experiments.
All of the above deletions except for TLF169Δ(27–153) are predicted to form a structure with a single major stem-loop structure that primarily consists of base pairings between the initiation codon plus region A (nt +1 to 21) and part of region B (nt +170 to 189) (Fig. 5A to C). However, the TLF190Δ(27–153) deletion lacking stem IV is predicted to form fortuitous base pairings between part of the Shine-Dalgarno sequence (nt −11 to −6) and the deletion junction with the BamHI site that would hyperstabilize the local secondary structure (Fig. 5C); this prediction probably explains the observed failure to respond to heat shock despite the identity of the major stem-loop structure with those of some of the other constructs. The basal portion of the major stem-loop structure of (27–153) constructs consists of base pairings between the initiation codon plus most of region A (nt +1 to 17) and part of region B (nt +173 to 189), as in authentic rpoH except for the A-13/G-177 mismatch and the U-14·G-176 pairing. In contrast, the apical portion contains A-18 to C-26, A-154 to C-172, and seven extra bases inserted during construction (Fig. 5B, broken line) and differs drastically from that of the parental TLF247 fusion (Fig. 3B). Thus, although the basal portion around the translation initiation region appeared to be most important, it was not sufficient for effective thermoregulation.
A minimal gene fusion that can respond to heat shock.
To construct a minimal fusion with maximum structural similarity to TLF247, the apical portion of the major stem, including seven extra bases of TLF229Δ(27–153), was replaced with part of region B (nt +111 to 127). The resulting fusion, called TLF229Δ(stemIII) (Fig. 5D), lacked the apical half of stem II and all of stem III, in comparison with the parental TLF247 fusion (Fig. 3B). Despite the structural similarity, this construct exhibited extremely low expression at 30°C and was induced little at 42°C (Fig. 4B, line 1). These results were not unexpected, because GF364 lacking all of stem III was unable to be induced upon heat shock, presumably due to the hyperstabilization of stems I and II (36). Thus, to keep the appropriate instability of the RNA secondary structure, the A-13/G-126 mismatch and the U-14·G-125 pairing derived from A-13/G-177 and U-14·G-176, respectively, of TLF247Δ(27–153) (Fig. 5A) were introduced into TLF229Δ(stemIII). The resulting construct, TLF229Δ(stemIII)GG, showed essentially normal thermoregulation, although expression at 30°C was significantly reduced (Fig. 4B, line 2).
It thus seemed evident that a much shorter version (116 bases) of rpoH mRNA present in TLF229Δ(stemIII)GG was sufficient for exhibiting the characteristic thermoregulation of the rpoH-lacZ fusion. These results, combined with the RNA secondary structure prediction for various constructs, suggested that the mRNA secondary structure involving the translation initiation region (the initiation codon plus region A) with appropriate stability or instability may be a primary requirement for the thermoregulation of rpoH translation.
DISCUSSION
The efficiency of translation in E. coli is determined primarily at the stage of initiation, which includes binding of the 30S ribosome to 5′ segments (from approximately nt −20 to +15) of mRNA spanning the Shine-Dalgarno sequence and the initiation codon (6). Thus, the secondary structure of such mRNA segments can play an important role in modulating translation efficiency (5). In the case of rpoH, part of the ribosome binding site (nt +1 to 20) including the AUG codon and region A (downstream box) was thought to be masked through the formation of base pairs with the internal region (region B). This idea was initially suggested by computer prediction (21) and subsequently supported by mutational analyses (21, 36) and structural conservation among the rpoH homologs (23). Such a structure seemed most likely to restrict translation by preventing ribosome entry under nonstress conditions. The present results of structure probing of rpoH mRNA directly supported this model (Fig. 3). Based on the structural information, possible roles of each of the major stems that constitute the whole structure were assessed by further deletion analyses.
The structure probing analyses revealed that mutations within stem II affecting translational repression (15A and 15C) affect not only the neighboring structures of stem II but also the structures of stem III (Table 1). The simultaneous recovery of both of these effects of compensatory mutations (15A-124T and 15C-124G) was well correlated with the expression and regulation of fusion proteins in vivo (Fig. 2B). This finding was not unexpected, because some of the partially constitutive mutations previously isolated from GF346 (133A, 136A, and 142A) (36) were actually localized within stem III. All of these results indicated that the stabilities of stems II and III are interdependent and that changes in stem II stability, at least those involving the mutations analyzed in this study, have particularly striking effects on thermoregulation.
The results of deletion analyses indicated that most of stem II (nt +27 to 111) and stem III were not indispensable for thermoregulation (36) (Fig. 4). However, as discussed below, appropriate stability or instability of the mRNA secondary structure was an essential requirement for normal regulation. In the fusion construct TLF247Δ(27–153), which lacked an appreciable portion of the internal segment, part of stem III (nt +165 to 178) was predicted to form several fortuitous base pairings (as well as some mismatches) with part of stem II (nt +12 to 26) (Fig. 5A). Remarkably, the minimal fusion construct TLF229Δ(stemIII)GG, with much greater similarity to the parental TLF247 fusion, had to retain two mismatches (two G’s) derived from TLF247Δ(27–153) to be heat inducible (Fig. 4B); a similar construct with the parental sequence but lacking the mismatches [TLF229Δ(stemIII)] failed to show heat induction. In this connection, the inability to respond to heat shock was previously observed when stem III was totally deleted from the GF364 fusion (36). Although stem III was not essential, when it was absent, certain mismatches had to be introduced to the remaining segment of RNA to substitute for its function. In other words, stem III appeared to serve as a “wedge” between stems I and II, conferring appropriate instability to the mRNA secondary structure.
3′ Deletions extending into stem IV reduced basal expression significantly and slightly affected heat induction (Fig. 4A, lines 5 and 6). TLF211Δ(27–153) retained intact stem IV, but the sequence immediately downstream at the lacZ junction (BamHI site) was predicted to form base pairings with the Shine-Dalgarno sequence and reduce basal-level expression, as was actually observed (Fig. 4A, line 4), although not as strikingly as in TLF190Δ(27–153) (line 7). These combined results suggested that stem IV was not essential for thermoregulation but would serve to keep the upstream Shine-Dalgarno and adjacent regions “open” for ribosome entry.
The internal deletion TLF247Δ(27–153), like GFR153 studied previously (21), exhibited essentially normal heat induction despite the drastic alteration from the parental TLF247 fusion in the apical (but not the basal) portion of the predicted RNA secondary structure (Fig. 5A). This result indicated that the basal portion containing the translation initiation region (stem I) was most critical for thermoregulation. However, stem I by itself was not sufficient, since TLF190Δ(27–153) containing stem I failed to be heat induced (Fig. 4A, line 7). Also, TLF229Δ(stemIII), which retained intact stem I and part of stem II, was not heat induced (Fig. 4B, line 1). The facts that a drastic alteration in stems II and III did not affect thermoregulation and that the two-G substitution could restore the regulation of TLF229Δ(stemIII) (Fig. 4B, line 2) strongly suggested that the stability of the translation initiation region of rpoH mRNA rather than other structural features was primarily important for thermoregulation. We conclude that there are two major requirements for normal rpoH thermoregulation. First, the translation initiation region (initiation codon plus region A) must be masked through formation of base pairs with part of the internal coding sequence (region B); this factor is crucial for translational repression at a low temperature (30°C). Second, the secondary structure involving the initiation region that includes the Shine-Dalgarno sequence must retain appropriate instability; this factor is essential for heat induction to be observed upon the temperature upshift (42°C).
Besides the mRNA secondary structure, previous results suggested the possible involvement of a specific sequence which may provide a site for protein binding in modulating the heat induction of rpoH translation. This suggestion was based mainly on the noninducible and barely inducible phenotypes of the 15C-124G and 16G-123C mutants, respectively, each containing two compensatory mutations (36). Although we confirmed the results for the latter mutant (16G-123C; data not shown), the 15C-124G mutant actually exhibited slightly reduced but appreciable heat induction (Fig. 2B), eliminating the major basis for suggesting the above possibility. It seemed possible that the subnormal heat induction observed with both of these mutants carrying an alteration in region A (G to C at +15 or C to G at +16) came from the differential effects of decreased complementarity for the anti-downstream box of 16S rRNA (G-15·U to C-15/U or C-16–G to G-16/G) on the translational efficiency at the two temperatures used (30 and 42°C). At present, the involvement of a trans-acting factor(s) in thermoregulation appears unlikely, although it cannot be excluded.
The fact that some of the nucleotides expected to form a stem I structure were modified by chemical probes, albeit weakly (Fig. 3B), suggested that this region was relatively unstable, presumably permitting the limited entry of ribosomes at a low temperature. Moreover, transcription-translation coupling may facilitate a productive interaction between rpoH mRNA and ribosomes because of a delay in forming the stem I structure due to the distance (ca. 180 nt) between the AUG codon and the internal region presumably required for base pairings. In any event, such a dynamic mRNA secondary structure should ensure the production of low but essential basal levels of ς32 at physiological temperatures under nonstress conditions. Mutations such as 15A or 15C may decelerate the formation of an inhibitory RNA structure, thereby permitting ribosome entry and constitutively high expression even at low temperatures.
Finally, ςS encoded by the rpoS gene is another global regulator for a set of genes induced at the stationary phase or upon hyperosmotic stress. Interestingly, ςS itself is regulated primarily at the posttranscriptional level, and recent work indicated the involvement of some specific gene products in the translational control of ςS synthesis (13). In addition, the rpoS mRNA secondary structure was suggested to play a regulatory role, although the mechanism remains unknown. Thus, translational control of global transcription factors such as ς32 and ςS appears to be mediated by an mRNA secondary structure and confers an efficient means for a rapid response to heat or other stress. The results reported here also raise the intriguing possibility that a 5′ portion of the rpoH mRNA secondary structure is involved in direct sensing and responding to high temperatures by enhancing ribosome entry and translation initiation, leading to a rapid increase in the ς32 level and the induction of heat shock proteins. Further work is in progress to examine such possibilities.
ACKNOWLEDGMENTS
We are grateful to T. Linn for the kind gift of the λTLF97-3 vector and to M. Nakayama, H. Kanazawa, and M. Ueda for technical assistance.
This work was supported in part by grants from the Japan Health Sciences Foundation, Tokyo.
REFERENCES
- 1.Bukau B. Regulation of the Escherichia coli heat shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 2.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 3.Christiansen J, Egebjerg J, Larsen N, Garrett R. Analysis of rRNA structure: experimental and theoretical considerations. In: Spedding G, editor. Ribosome and protein synthesis: a practical approach. Oxford, England: IRL Press; 1991. pp. 229–252. [Google Scholar]
- 4.Craig E A, Gross C A. Is hsp70 the cellular thermometer? Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 5.de Smit M H, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draper D E. Translational initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 902–908. [Google Scholar]
- 7.Erickson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternate ς factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 8.Gamer J, Bujard H, Bukau B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor ς32. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 9.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 10.Goff S A, Goldberg A L. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 11.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 12.Grossman A D, Straus D B, Walter W A, Gross C A. ς32 synthesis can regulate the synthesis of heat shock proeins in Escherichia coli. Genes Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 14.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 15.Kamath-Loeb A S, Gross C A. Translational regulation of ς32 synthesis: requirement for an internal control element. J Bacteriol. 1991;173:3904–3906. doi: 10.1128/jb.173.12.3904-3906.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanemori M, Mori H, Yura T. Induction of heat shock proteins by abnormal proteins results from stabilization and not increased synthesis of ς32 in Escherichia coli. J Bacteriol. 1994;176:5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberek K, Galitski T P, Zylicz M, Georgopoulos C. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the ς32 transcription factor. Proc Natl Acad Sci USA. 1992;89:3516–3520. doi: 10.1073/pnas.89.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarty J S, Rudiger S, Schonfeld H-J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. Regulatory region C of the E. coli heat shock transcription factor, ς32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 19.Mecsas J, Rouviere P E, Erickson J W, Donohue T, Gross C A. The activity of ςE, an Escherichia coli heat-inducible ς-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2619–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 20.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 21.Nagai H, Yuzawa H, Yura T. Interplay of two cis-acting mRNA regions in translational control of ς32 synthesis during the heat shock response of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai H, Yuzawa H, Kanemori M, Yura T. A distinct segment of the ς32 polypeptide is involved in DnaK-mediated negative control of the heat shock response in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding ς32 homologs from gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23:4383–4390. [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahigashi K, Yanagi H, Yura T. Regulatory conservation and divergence of ς32 homologs from gram-negative bacteria: Serratia marcescens, Proteus mirabilis, Pseudomonas aeruginosa, and Agrobacterium tumefaciens. J Bacteriol. 1998;180:2402–2408. doi: 10.1128/jb.180.9.2402-2408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neidhardt F C, VanBogelen R A. Heat shock response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1334–1345. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Sprengart M L, Fatscher H P, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16S rRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.St. Pierre R, Linn T. A refined vector system for the in vitro construction of single-copy transcriptional or translational fusions to lacZ. Gene. 1996;169:65–68. doi: 10.1016/0378-1119(95)00787-3. [DOI] [PubMed] [Google Scholar]
- 29.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 30.Straus D B, Walter W A, Gross C A. The heat shock response of E. coli is regulated by changes in the concentration of ς32. Nature. 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 31.Tilly K, McKittrick N, Zylicz M, Georgopoulos C. The dnaK protein modulates the heat-shock response of Escherichia coli. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- 32.Tilly K, Spence J, Georgopoulos C. Modulation of stability of the Escherichia coli heat shock regulatory factor ς32. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Kaguni J M. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J Bacteriol. 1989;171:4248–4253. doi: 10.1128/jb.171.8.4248-4253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yura T. Regulation and conservation of the heat-shock transcription factor ς32. Genes Cells. 1996;1:277–284. doi: 10.1046/j.1365-2443.1996.28028.x. [DOI] [PubMed] [Google Scholar]
- 35.Yura T, Nagai H, Mori H. Regulation of heat shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 36.Yuzawa H, Nagai H, Mori H, Yura T. Heat induction of ς32 synthesis mediated by mRNA secondary structure: a primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 1993;21:5449–5455. doi: 10.1093/nar/21.23.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]