Abstract
Although the shapes of organisms are encoded in their genome, the developmental processes that lead to the final form of vertebrates involve a constant feedback between dynamic mechanical forces, and cell growth and motility. Mechanobiology has emerged as a discipline dedicated to the study of the effects of mechanical forces and geometry on cell growth and motility — for example, during cell–matrix adhesion development — through the signalling process of mechanotransduction.
Mechanical forces are controlled by cells and are integrated into tissues to produce the final form of an organism through processes of mechanotransduction that affect cell shape, proliferation, migration and apoptosis (FIG. 1). In the past, “life seemed to have unique properties quite irreducible to the world of physics and chemistry: ‘motion generated from within’, ‘chemistry of a very distinct kind’ ‘replication’ ‘development’, ‘consciousness’ - each of these aspects of life turned into elements that became more and more foreign to the physicist to the extent that many physicists even today look upon biology as something outside their domain” (REF. 1). Many of the early biologists, however, did recognize the importance of physical forces and shape in development and function of organisms, and formed the discipline of ‘physics in biology’ or ‘physiology’ Late in the twentieth century, this discipline fell out of fashion because it was primarily focused on organ-level as opposed to molecular-level phenomena, and because there were few good tools available with which to measure physical parameters of protein function in cells. At the same time, the rapid developments in molecular biology techniques, DNA sequencing and mass spectrometry led to the expansion of our knowledge of the genome and proteome. However, this also led to the realization that the DNA-encoded information was not sufficient to determine the final form of tissues and organs, and that cellular expression profiles could not tell us how complex functions are carried out.
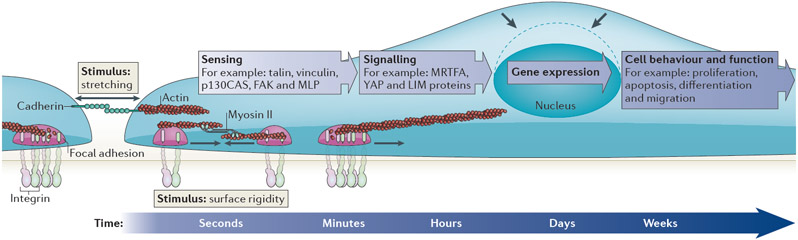
Figure 1 ∣. Mechanotransduction.
Mechanotransduction converts mechanical stimuli — such as substrate rigidity (through contractile units or mature integrin adhesions), stretching (through cell–cell contacts or integrin adhesions) or shear stress (not shown) — into chemical signals to regulate cell behaviour and function. Typically, the pathway involves receptors at focal adhesions or cell–cell contacts (for example, integrins and cadherins), mechanosensors (for example, stretchable proteins such as talin and p130CAS) and nuclear signalling factors to change gene and protein expression profiles. Nuclear deformation (the shape of the nucleus before force is applied is indicated by the dashed line) can also lead to changes in gene expression patterns. The timescale of these events ranges from milliseconds to seconds for the stretching of mechanosensors, hours for altered gene expression, days for changes in cell behaviour and function, and weeks for tissue development. FAK, focal adhesion kinase; MLP, muscle LIM protein; MRTFA, myocardin-related transcription factor A; YAP, Yes-associated protein.
It became increasingly evident that a crucial aspect of organ formation — as well as tissue regeneration, repair and aging — is the dynamic interaction between a cell and its microenvironment, including not only hormones but also neighbouring cells, the extracellular matrix (ECM) and the forces applied to them. The same biochemical components will have different effects on cells when mechanical aspects of their environment are altered. At the heart of these processes are primarily myosin motors that exert forces on actin filaments anchored to cell–cell or cell–matrix adhesions, and mechanosensors that are responsive to counter-forces from matrices, other cells and sometimes membranes. A quasi-steady state in cellular tension is maintained through these actomyosin contractility mechanosensors, which enables cells to define the shape and tension of an organ.
Multidisciplinary approaches are needed to study how organ form and function are affected by actomyosin organization in response to cellular microenvironments. To provide an integrated view of the mechanobiological aspects of cellular function, a new discipline dedicated to the study of subcellular forces, geometries and mechanically responsive complexes is emerging under the rubric of mechanobiology2. Within this discipline, novel nanotechnology techniques and tools provide insights into the functions of various components in sensing and relaying cell–cell and cell–matrix dynamic mechanical forces. Consequently, mechanobiology has become a truly interdisciplinary field of research in which physicists, chemists, engineers and material scientists no longer feel “outside their domain”, and the development of tools, experimental systems and theoretical models is interconnected.
In this Timeline article, we provide a perspective on the history of mechanotransduction research in cell biology and, as a case in point, we present the current working models for matrix adhesion development and mechanosensing.
Milestones in mechanotransduction
The diversity of biological forms that result from mechanotransduction processes (FIG. 1) has interested humans for millennia and early biologists were fascinated by the question of how organisms developed different shapes. In the early twentieth century, D’Arcy Thompson published On Growth and Form (REF. 3) (FIG. 2 (TIMELINE)), which is an archival description of how biological form is shaped by developmental changes. With the advent of cell biology research around the same time, researchers began to explore different ways in which to mechanically manipulate cells (including the use of microneedles or changes in osmotic pressure) and observe their responses. However, the effects of mechanotransduction at the cellular level only drew major attention in the 1950s, when it was first shown that cancer cells can grow in soft agar in an anchorage-independent manner, whereas most non-cancerous cells cannot4,5. This aberrant mechanosensing was described as an in vitro transformation phenotype and has served as an important experimental tool in cancer research. Additional evidence for the existence of cellular mechanosensing came from studies of the coupling between the inner and outer layers of the plasma membrane. The exposure of erythrocytes to anionic and cationic drugs caused differential changes in membrane tension at the intracellular and extracellular surfaces, and consequently led to changes in cell shape6. This indicated the existence of a cellular mechanism for sensing changes in membrane tension, which was later shown to be important for cell spreading and migration7.
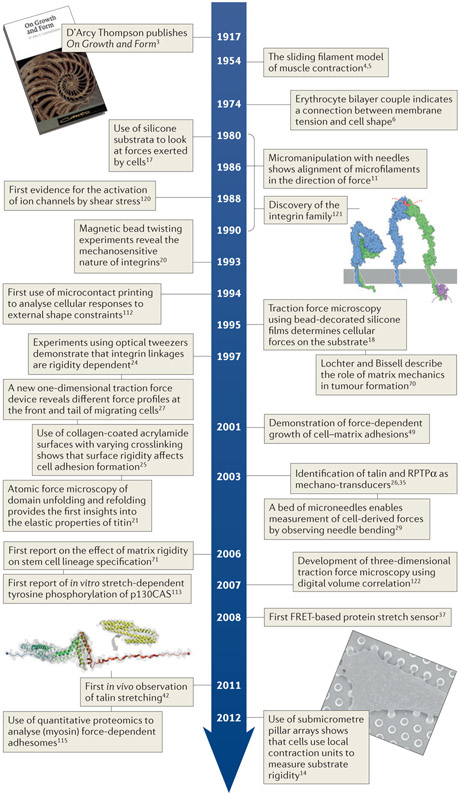
Figure 2 ∣. Timeline of milestones in the history of mechanotransduction research.
Image of the front cover of the 1992 edition of On Growth and Form by D’Arcy Thompson3. Image of integrin protein structure in autoinhibited conformation (left) and active conformation (right) reprinted with permission from David S. Goodsell and the Research Collaboratory for Structural Bioinformatics (RCSB), Protein Data Bank (PDB). Image of vinculin binding to stretched talin from del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009). Reprinted with permission from AAAS. Image of submicrometre pillar array courtesy of Nicolas Biais and Luis Santos, Columbia University, New York, USA.
From that point onwards, a considerable amount of research was dedicated to understanding the mechanisms by which cells communicate with the ECM and their neighbouring cells. This led in the 1970s to the discovery of focal contacts (adhesions), which are multiprotein structures with integrins at their core that provide the mechanical link to the ECM, and that enable inside-out and outside-in mechanosignalling8-10. Soon enough, it was discovered that the actomyosin machinery was attached to cell–matrix adhesions10-12,111 raising the possibility that cellular forces were involved in processes such as cell locomotion. In parallel, the study of muscle biophysics greatly contributed to the rise of the cellular mechanobiology field, starting with the early studies by Hugh Huxley and Andrew Huxley in the 1950s that elucidated the major elements of actomyosin contraction in muscle13,14. These studies led to the groundbreaking sliding filament theory and the swinging crossbridge model of myosin movement on actin filaments15, which have since been confirmed in in vitro studies16.
As many mechanosensory events occur at cell–cell or cell–matrix adhesions, where forces produced by the cytoskeleton are transmitted to the cellular microenvironment8-10, new techniques were needed that were sensitive enough to measure mechanical forces at a subcellular level. The first of such techniques involved the use of elastic silicone surfaces as substrates for cell spreading, and this revealed for the first time that non-muscle cells produced force on their environment17. This early force-sensing tool was the basis for the later development of traction force microscopy, in which the movements of fluorescent beads embedded in elastic gels are used to calculate dynamic cellular forces on continuous surfaces18. Around the same time, ECM patterning at a micrometre scale showed that the form of the matrix contacts was crucial for cell growth and death. In this case, the ability of cells to generate forces over distance seemed to be the key element for downstream signalling19. This was linked to integrin-mediated mechanotransduction (FIG. 1), because a previous study showed that magnetic forces applied to cells through beads coated with integrin ligands led to rapid strengthening of the cytoskeleton20.
Additional tools and methods were introduced in the late 1990s for the application of active and passive forces to cells and molecules (FIG. 3), and these began elucidating the details of the mechanosensing process. In muscles, studies using atomic force microscopy showed that the large muscle protein titin could be mechanically unfolded to unravel its individual immunoglobulin-like domains, which suggested the existence of a mechanism for muscle stabilization against overstretch21. Atomic force microscopy was also used to study the properties of intramolecular bonds between receptors and their ligands — such as biotin and streptavidin — providing important estimates of the energy range of molecular bonds22,23. A study using submicrometre-diameter beads with an optical trap showed that high levels of passive rigidity of matrix molecules were needed to induce strong linkages between the ECM and the cytoskeleton24. In the same year, matrix rigidity was experimentally modified by changing the crosslinking properties of acrylamide gels, and the resulting changes in cell adhesions further reinforced the idea that ECM rigidity affects adhesion size and stability25. Treatment of cells with phosphatase inhibitors later showed that tyrosine phosphorylation and dephosphorylation cycles were involved in sensing rigidity, specifically implicating receptor-type tyrosine-protein phosphatase-α (RPTPα; also known as PTPRA) and the kinase FYN in the case of fibronectin-mediated rigidity sensing26.
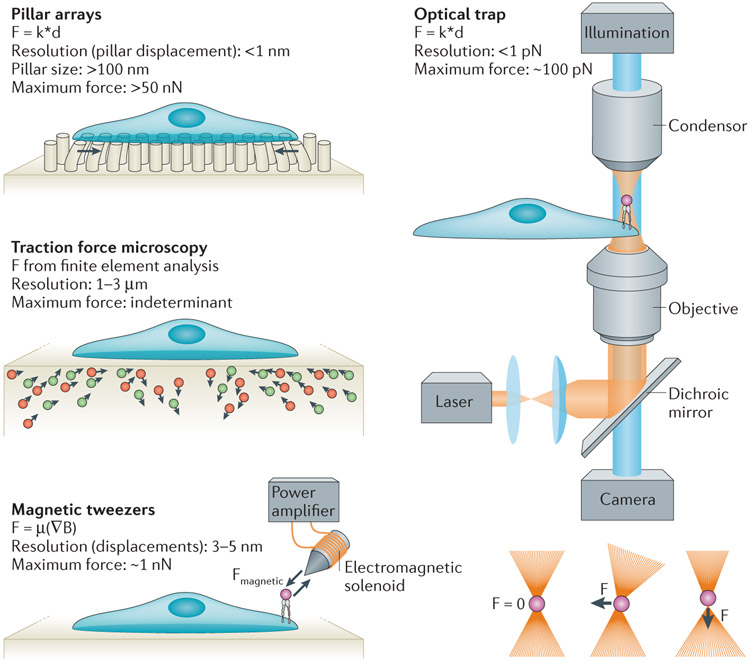
Figure 3 ∣. Experimental tools in mechanobiology.
Pillar arrays can be made to varying dimensions, thereby allowing the determination of substrate rigidity and force resolution. Pillar displacement is measured in live cells and is used to determine cellular forces that are applied to the substrate. Traction force microscopy uses embedded fluorescent beads (often of two colours) and finite element analysis to measure substrate deformations by the cell. Magnetic tweezers create magnetic fields that cause magnetic beads to apply forces to molecules in vitro or in vivo (for example, to integrins with fibronectin-coated beads). Optical traps use a focused laser beam to provide lateral or axial forces onto micrometre-diameter beads and thus apply forces to molecules; dielectric objects such as beads are attracted to the centre of the beam. If the bead is shifted laterally or axially out of the trap centre, the diffraction of the beam results in a restoration force on the bead, pulling it back into the centre. Each tool has advantages in terms of spatial resolution and maximum force that can be applied or experimental data analysis, and thus they are each suitable for specific applications. ∇B, magnetic field gradient; d, distance; F, force; k, spring constant; μ, magnetic moment.
In order to track cellular forces generated by few adhesive contacts in a small subcellular area, a device that used pads suspended on movable cantilevers was developed, which avoided problems in the finite element analysis of continuous surfaces27. Microcontact printing of elastic substrates later provided a simpler solution to finite element analysis and enabled the correlation of cellular traction forces with the localization of fluorescent proteins involved in adhesion assembly and force transmission28,112. Another tool for measuring traction forces was a two-dimensional bed of needles (or pillars)29 (FIG. 3), which raised some concerns that the gaps between pillars would cause altered cell geometry and traction force patterns; however, the pillar arrays generated similar traction force maps to those generated by the previous techniques29. These maps showed that cells were contracting the matrix substrate isometrically; large forces applied from the few focal contacts at the narrow tail of a cell were balanced by many smaller adhesion forces applied across the front of the cell. In the middle of the cell, the densities of adhesions and forces were much lower.
In addition to the study of traction forces, novel tools for measuring the mechanical properties of single cells were developed. A pair of force sensors on micromanipulators enabled the measurement of the elastic and contractile properties of a single cell over time and in response to bioactive substances. For example, sphingosylphosphorylcholine caused an increase in the elasticity of cancer cells, which provided an explanation for how metastatic cells squeeze through membrane pores30-32.
At the molecular level, several different experimental approaches involving ligand valency indicated that crosslinking of three or more integrins was crucial for the sensing and signalling of surface rigidity33. Stretching of exposed cytoskeletons following Triton X-100 treatment revealed force-dependent binding of several adhesion proteins to the cytoskeleton, including paxillin, focal adhesion kinase (FAK; also known as PTK2) and p130CAS (also known as BCAR1)34,113. In related studies of the molecular regulation of force-dependent reinforcement of cell–matrix contacts, several proteins involved in mechanosensing were identified; for example, talin, p130CAS and RPTPα26,35,36,113 (FIG. 1).
Further technical advances improved the temporal, spatial and force-strength resolution of biomechanical cues. Forces that could produce stretching of specific proteins were sensed with fluorescence resonance energy transfer (FRET) at a light microscopy resolution (~250 nm)37,38. To achieve nanometre-level resolution, fluorescence quenching-based tension sensors were developed39-41, and a single-molecule tracking approach revealed dynamic stretch and relaxation cycles of single talin molecules in cells42. In studies of traction forces on pillars, magnetic nickel particles were adhered to the pillars to enable local measurements of the effects of external forces43. The resolution of force measurement was improved by the coating of pillar tips with gold. This enabled covalent labelling of the pillar tips with a wide variety of ligands to specifically measure the contributions of α5β1- and αvβ3-integrins to the cell traction forces44. Arrays of submicrometre-diameter pillars supported more physiological cell spreading than those with larger pillars and provided increased spatial resolution of cellular forces45. Thus, it is now possible to measure cell–matrix forces and their effects from the tissue level to the molecular level, over time and in a physiologically relevant context.
The functions of mechanotransduction
With these tools at hand, the study of integrin-dependent mechanosensing developed rapidly. One particular research catalyst was the realization that cell–ECM contacts are built in a step-wise manner with standard cellular protein assemblies (for example, integrin adhesions or podosomes) and functional modules (defined here as proteins or protein complexes that carry out specific functions, such as those involving motors or kinases). For each well-defined cell state, cell behaviour is very reproducible and thus it is possible to dissect at a nanometre resolution (5–300 nm) the mechanotransduction steps that occur in the sequential transitions from one cell state to the next. Often, dramatic changes in cell state occur abruptly following the inactivation of some functional modules and the activation of others. For example, in the simple case of cell spreading on a surface, a transition from initial rapid isotropic spreading to subsequent contractile spreading involves a momentary increase in membrane tension, which activates periodic contractions to test substrate rigidity through local contraction units (BOX 1). The consequential cellular morphology develops through several steps that may differ slightly in different cell types. However, the steps share many common functional modules that enable the analysis of how the different modules affect cell behaviour. Modern microscopy and nanofabrication techniques now enable us to follow the spatial and temporal distribution of such functional complexes to determine whether the proteins are involved in the initiation, assembly, quasi-steady state behaviour or disassembly of an integrin adhesion or a podosome.
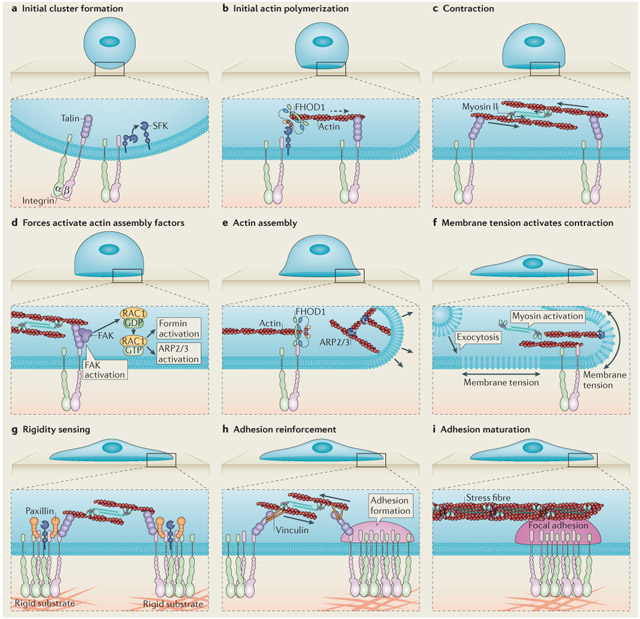
Box 1 ∣. Focal adhesion assembly.
After binding to matrix-coated surfaces, clusters of activated integrins are formed at the cell edge103. Integrin cluster formation leads to the recruitment of talin as well as the activation of SRC family kinases (SFKs)104 (see the figure, part a), which then recruit the formin family protein FH1/FH2 domain-containing protein 1 (FHOD1)to integrin clusters, leading to actin assembly (see the figure, part b; indicated by the dashed arrow)47. The resulting actin polymerization enables clusters to be pulled together by myosin (see the figure, part c)47,48. When nanofabricated barriers (not shown) in the membrane bilayers limit lateral movements, forces are developed on the clusters at the barriers that trigger rapid cell spreading. This presumably involves focal adhesion kinase (FAK), which enhances the activation of the small GTPase RAC1 and targets it to focal adhesions105. RAC1 activates actin assembly through actin-related protein 2/3 (ARP2/3)106, or formins46,105 (see the figure, parts d and e)48,107. Without barriers, cells will round up and often undergo apoptosis. Analysis of membrane dynamics during cell spreading has indicated that rapid isotropic spreading flattens the initially round cell, drawing membrane from the reservoir of folded surface membrane7. Upon the depletion of the folded membrane, tension increases momentarily and signals the activation of exocytosis to increase the membrane surface area by 40% (see the figure, part f) and activate periodic contractions to test substrate rigidity through the local contraction units (see the figure, part g)7,73. Rigidity signalling recruits additional proteins (such as vinculin) and causes adhesion complex reinforcement (see the figure, part h) or disassembly (if the matrix is too soft; not shown), followed by adhesion maturation (see the figure, part i)48,61,65,73,108-110,115. Following adhesion maturation, stress fibres grow from adhesions and will contract to sense matrix rigidity at the whole-cell level76.
The primary functional modules for force generation in animal cells and tissues are myosin II motors that pull on actin filaments, which in turn convey force to actin-anchoring sites (FIG. 1). The major site of actin polymerization was first found at extending cell edges through the incorporation of G-actin into filaments114. However, the rapid turnover of actin filaments — within 1–2 minutes — throughout the cell indicated that there were many different sites of actin polymerization. Actin assembly is a tightly regulated process involving multiple assembly, bundling, crosslinking and severing proteins that are, at least in part, specific for the different types of actin networks46. Recent studies of the early stages of cell adhesion to the ECM showed that the formin protein FH1/FH2 domain-containing protein 1 (FHOD1) was recruited in a force-independent manner to early ligand-bound integrin clusters and stimulated actin assembly from them47,48. Formins such as mammalian diaphanous homologue 1 (mDia1; also known as DIAPH1) and FHOD1 were also found to be important for the maturation of adhesion sites47,49. However, mDia1 and FHOD1 were found to promote the formation of distinct actin structures at different times and thus have distinct roles in this process. Whereas mDia1 was associated with the formation of dorsal stress fibres50, FHOD1 was — together with the actin-related protein 2/3 (ARP2/3) complex — found to be involved in the formation of ventral stress fibres and transverse arcs50,51. Thus, FHOD1 preferentially assembles antiparallel actin structures that probably include contractile units involved in early adhesion formation45,47,51. The alignment of stress fibres in the direction of force during periodic stretching indicated that there is a force-dependent regulation of actin assembly52. A theoretical model suggested that a pulling force in the piconewton range would enhance formin-driven actin assembly from the growing barbed end53, and two recent studies showed that flow forces within a microfluidic device enhanced actin assembly rate from the immobilized formins mDia1 and Bni1p54,55. Thus, adhesion-localized formins, such as FHOD1, could directly couple integrin-sensed forces to alterations in actin assembly47.
The main mechanical link from the ECM to the actin cytoskeleton is, however, provided by proteins such as talin and α-actinin, which can bind to both integrins and to actin filaments56,57. Following the early discovery that talin is involved in force-dependent adhesion reinforcement, it was shown that talin, which links actin to the integrin tail, is periodically stretched in what is best described as a stick-slip mechanism. In this process, the rearward-flowing actin binds to talin (‘stick’) and exerts a force that stretches it until the actin–talin bond breaks (‘slip’) and talin refolds35,42,58-60. The stretching allows binding of vinculin to enhance the coupling to the actin cytoskeleton and reinforcing the adhesion61. Additionally, the actin crosslinkers α-actinin and filamin A compete with talin for integrin binding62-64. Whereas α-actinin recruitment to nascent adhesions enables the transmission of high forces during the maturation of the adhesions65, filamin — like talin — seems to be a genuine mechanosensor, as stretching modulates its affinity for binding partners such as integrins or migfilin (also known as FBLIM1)66-68. Filamin is a dimer, and its dimerization might lead to crosslinking and the stabilization of integrin clusters. The role of filamin in adhesion maturation is also, at least in part, attributed to its binding and ‘buffering’ of the protease calpain, which inhibits the proteolysis of talin66,69.
Sensing substrate rigidity
Of the different forms of mechanosensing (for example, sensing of shear stress or stretching; see FIG. 1), the sensing of matrix rigidity caught recent attention because of its links to cancer and development70,71. The potential to harness the rigidity-sensing pathway — for example, as a therapeutic target or for tissue engineering — led to increasing efforts to characterize its underlying mechanisms. The effects of substrate rigidity on cellular processes were first described in the 1950s4,5, but gaining a better mechanistic understanding relied on the development of experimental tools that allowed the characterization of matrix-rigidity sensing at a nanometre resolution. Matrix-coated beads placed at extending cell leading edges with optical traps (FIG. 3) can develop an increasing force on the cell as the bead moves inwards with the retrograde flowing actin24. With a rigid trap (high beam power), increased resistance of beads to movement by the laser trap indicated a strengthening of the adhesion; with a soft trap (lower beam power), no strengthening was observed and adhesions often broke, causing the beads to jump back to the cell edge. However, if the soft trap was moved rapidly with a piezo to generate high forces on the beads, the adhesions were reinforced72. This indicated that adhesion reinforcement (a rigidity response) was force dependent, and that the adhesion was strengthened when the force exceeded a critical level owing to either cell-driven or piezo-driven displacements in the trap.
At the same time as the above work was carried out, studies of initial cell spreading indicated that cells were sensing matrix rigidity by periodically contracting their leading edge73. Subsequently, cell-spreading assays were developed to enable dissection of the steps that were involved in rigidity-dependent cell spreading and adhesion formation73. On rigid substrates, isotropically spreading cells were found to exhibit a highly reproducible pattern of transitional steps, from initial adhesion formation to the maturation of focal adhesions74. By contrast, spreading assays on lipid bilayers only allowed the analysis of the initial force-independent steps in adhesion formation47,48. Following their spreading on arrays of submicrometre-diameter pillars (which enable traction forces to be measured at a higher resolution), cells produced local contractions with a constant displacement of pillars — which were up to 2 μm apart — independent of substrate rigidity45. The localization of myosin II between the pillars that were displaced by the contractions indicated the formation of bipolar myosin filaments that were possibly organized in contractile units similar to muscle sarcomeres. Furthermore, contraction forces correlated with rigidity sensing; cells pulled the pillars to a constant displacement regardless of pillar stiffness, indicating that the force applied to the pillars was proportional to pillar (matrix) rigidity45. Thus, we suggest that cellular rigidity responses — adhesion formation and further spreading — are triggered by contraction forces when they exceed a certain threshold. How the rigidity sensors are coupled with the tyrosine kinases that mediate mechanosignalling is still a mystery75.
At longer time scales, adhesion formation and disassembly (BOX 1) is a cyclic process, during which the cells actively test the mechanical properties of their microenvironment45,76. The readout of this testing — in the form of protein binding and unbinding, of stretching or of changes in post-translational modifications — is integrated over multiple sites at the cell edge, as well as over multiple cycles, with downstream effects such as changes in cell migration or gene expression (FIG. 1). Notably, the dynamic nature of these processes, as well as the involvement of force in regulating them, has caught the attention of theoretical physicists who began developing models to explain the relationship between force and adhesion dynamics77-81. These models are too complex to be discussed here (for a recent review, see REF. 82), but they can broadly be divided into models involving adhesion-level forces and models involving cellular-level forces. A major challenge is to combine the two levels (both theoretically and experimentally), and also to introduce novel concepts from recent experimental results, which indicate that intra-adhesion forces (that is, forces that are generated within the adhesion, rather than actin flow forces or forces that are generated by actin stress fibres) participate in mechanotransduction as well76.
At a subcellular level, the cell is testing the rigidity of newly bound matrix by contracting it45. There are clear relationships between this subcellular process and rigidity sensing at the tissue level, but the molecular mechanisms connecting them are not understood. Furthermore, recent studies of the relatively simple process of a cell spreading on a matrix-coated surface revealed several sequential steps — that are mechanical in nature — that precede the rigidity-sensing step (see parts a–f of the figure in BOX 1). The sequential nature of these steps indicates that altering any step (for instance, through a different molecular composition of the adhesome in different tissues) will affect subsequent steps and modify cell behaviour, resulting in the use of alternative methods to sense rigidity83,115. Thus, in order to understand the roles that defects in mechanotransduction have in the aetiology of human diseases, it will be important to study the cellular processes that lead to adhesion formation and the alternative processes used by cells that cannot take advantage of a particular pathway to build adhesions.
Other forces and signals
Cells sense other mechanical properties such as forces from shear stress, the contractility of neighbouring cells, or forces generated by different cell and matrix geometries. When considering the sensing of forces by the actomyosin cytoskeleton, the muscle system was historically of primary interest for studying stretch sensing. In the early twentieth century, the Frank–Starling mechanism of the heart described the increase in active force when a muscle is stretched116-119. A.V. Hill84 modelled the muscle as an arrangement containing a contractile element with one elastic element put in series (providing the intrinsic elasticity of the muscle) and another elastic element in parallel (the extracellular matrix). In the 1950s, Huxley and Huxley postulated the sliding filament theory to describe the production of force by the muscle13,14. Later, titin was purified and identified as the elastic component of the muscle, and it is the main contributor to passive tension of the myocardium21,85. Titin also contains an inherent stretch-activated kinase that can regulate muscle gene expression86,87. Other stretch sensors, such as muscle LIM protein (MLP; also known as CSRP3), were discovered in the sarcomeric Z-disc88. Similarly to rigidity sensing, stretch is also sensed by integrins or integrin-linked molecules (for example, the costameres in myofibrils), and the two sensing processes share key elements89.
It is becoming evident that the correlated forces of the cytoskeleton and the matrix contribute substantially to cell growth and tissue morphology and development. Cyclic stretch forces, for example, also cause major changes in the shape and behaviour of non-muscle cells. Initial studies on flexible-bottomed cell culture plates and whole deformable dishes indicated that osteoblasts increase their rate of DNA synthesis and cell division after bi-axial cyclic stretching90,91. By contrast, uniaxial cyclic stretch led to the re-orientation of fibroblasts in the direction of force and provided insights into the molecular basis of this repolarization92-96. Interestingly, a recent study indicates that cyclic stretch forces can stimulate cell spreading and growth on pillar substrates that are otherwise too soft to support spreading and growth (H. Cai and M. P. Sheetz, unpublished observations). In this study, the frequency, magnitude and duration of stretch were all important factors that translated into stress fibre formation and downstream signalling, leading to stretch-dependent growth. Cell growth correlated with the nuclear translocation of myocardin-related transcription factor A (MRTFA; also known as MAL and MKL1), which recently gained attention for its role in epithelial-to-mesenchymal transition, a process that is required for organ development and wound healing, as well as for cancer progression82,97-99. The formation of actin stress fibres is also dependent on cell shape and on ECM rigidity (BOX 1). The formed stress fibres in turn regulate the Hippo signalling pathway to control organ formation100,101. Actin capping and severing leads to the nuclear localization of the Hippo transcription co-activators Yes-associated protein (YAP; also known as YAP1) and TAZ, and subsequently to cell proliferation, differentiation or apoptosis101. Thus, the activity of YAP and TAZ, and of MRTFA, probably contributes to the formation of organ shape by regulating cell proliferation or apoptosis102.
Future perspective and conclusions
It is unlikely that we will be able to fully understand the important processes of cell growth in cancer, cardiovascular diseases and aging without understanding how mechanosensing processes interplay with tyrosine kinases and other signalling pathways. Of particular interest is how cells respond to mechanical forces over longer timescales. It is difficult to study the effect of force signalling over time because cells respond to mechanical and biochemical signals in a timescale of seconds to minutes, whereas tissue formation occurs over weeks to months.
Force is an integral part of the control of cell function by hormones and the ECM; however, we poorly understand how to control or modify its effects. For example, for unknown reasons, wound healing is dramatically increased in the skin following the application of periodic pulses of pressure. It is clear that cells actively test their environment both chemically and mechanically, and adopt specific cell responses depending on matrix stiffness, neighbouring cells, hormone levels, nutrients and other factors. Newly available nanotechnology and advanced light microscopy tools can enable us to map when and how cells make decisions to grow, to undergo apoptosis or to differentiate. When we understand which mechanosensing and signalling pathways are activated, in what sequence and how often, during normal and disease-dependent motile processes, we can hope to mitigate disease-related damage more specifically and to facilitate repair processes.
Acknowledgments
T.I. was supported by a Postdoctoral Fellowship from the American Heart Association. H.W. was supported by a Marie Curie International Outgoing Fellowship within the Seventh European Commission Framework Programme (PIOF-GA-2012-332045). M.P.S. was partially supported by the Mechanobiology Institute, National University of Singapore.
Glossary
- Actomyosin
A basic force-producing or structural unit in cells consisting of myosin motors that bind and pull on actin filaments.
- Adhesome
The combined molecular composition of focal adhesions.
- Dorsal stress fibres
Long parallel actomyosin bundles that are anchored to focal adhesions at one end.
- Finite element analysis
A numerical method of approximation.
- Frank–Starling mechanism
Also known as the Frank–Starling law of the heart; states that there is a direct relationship between the force of cardiac contraction and the volume of blood filling the heart. The stretching of muscle fibres through the increasing blood volume increases calcium sensitivity, thus causing the formation of more actin–myosin crossbridges and hence more force.
- Isotropic spreading
Spreading of cells during which their entire edge (or large parts of it) extends rapidly.
- Ligand valency
The combined effects of the binding of multiple ligands.
- Local contraction units
Multiprotein complexes that are similar to muscle sarcomeres and are used by cells to measure substrate rigidity.
- Microcontact printing
Also known as micropatterning. A form of surface patterning, usually with fluorescent-labelled extracellular matrix proteins.
- Optical trap
Also known as laser tweezers. An appliance that provides force from a highly focused laser beam to hold or move objects such as microspheres.
- Sliding filament theory
A model of muscle contraction postulating that thin filament (mostly actin)-containing I-bands slide past the myosin-containing A-bands to generate force.
- Swinging crossbridge model
The first model of the myosin power stroke, which suggests that ATP-dependent changes in the actin–myosin crossbridge angle would cause the thin filaments to slide past the myosin (see sliding filament theory).
- Transverse arcs
Curved, antiparallel actomyosin bundles that interact with dorsal stress fibres and flow inward towards the cell centre.
- Ventral stress fibres
Antiparallel actomyosin bundles anchored to focal adhesions at both ends.
- Z-disc
A protein complex that defines the boundaries of the muscle sarcomere. It anchors and links actin filaments and titin from adjacent sarcomeres, provides mechanical stability and is a centre of cardiomyocyte signal transduction, including mechanotransduction.
Footnotes
Competing interests statement
The authors declare no competing interests.
Contributor Information
Thomas Iskratsch, Department of Biological Sciences, Columbia University, New York 10027, USA..
Haguy Wolfenson, Department of Biological Sciences, Columbia University, New York 10027, USA..
Michael P. Sheetz, Mechanobiology Institute, National University of Singapore, Singapore 117411; Department of Biological Sciences, Columbia University, New York 10027, USA.
References
- 1.Delbruck M A physicist’s renewed look at biology: twenty years later. Science 168, 1312–1315 (1970). [DOI] [PubMed] [Google Scholar]
- 2.Lim CT, Bershadsky A & Sheetz MP Mechanobiology. J. R. Soc. Interface 7, S291–S293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson DAW On Growth and Form (Cambridge University Press, 1917). [Google Scholar]
- 4.Sanford KK, Likely GD & Earle WR The development of variations in transplantability and morphology within a clone of mouse fibroblasts transformed to sarcoma-producing cells in vitro. J. Natl Cancer Inst 15, 215–237 (1954). [PubMed] [Google Scholar]
- 5.Temin HM & Rubin H Characteristics of an assay for Rous sarcoma virus and Rous sarcoma cells in tissue culture. Virology 6, 669–688 (1958). [DOI] [PubMed] [Google Scholar]
- 6.Sheetz MP & Singer SJ Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc. Natl Acad. Sci. USA 71, 4457–4461 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gauthier NC, Fardin MA, Roca-Cusachs P & Sheetz MP Temporary increase in plasma membrane tension coordinates the activation of exocytosis and contraction during cell spreading. Proc. Natl Acad. Sci. USA 108, 14467–14472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis AS The mechanism of adhesion of cells to glass. A study by interference reflection microscopy. J. Cell Biol 20, 199–215 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izzard CS & Lochner LR Cell-to-substrate contacts in living fibroblasts: an interference reflexion study with an evaluation of the technique. J. Cell Sci 21, 129–159 (1976). [DOI] [PubMed] [Google Scholar]
- 10.Abercrombie M, Heaysman JE & Pegrum SM The locomotion of fibroblasts in culture. IV. Electron microscopy of the leading lamella. Exp. Cell Res 67, 359–367 (1971). [DOI] [PubMed] [Google Scholar]
- 11.Heath JP & Dunn GA Cell to substratum contacts of chick fibroblasts and their relation to the microfilament system. A correlated interference-reflexion and high-voltage electron-microscope study. J. Cell Sci 29, 197–212 (1978). [DOI] [PubMed] [Google Scholar]
- 12.Luduena MA & Wessells NK Cell locomotion, nerve elongation, and microfilaments. Dev. Biol 30, 427–440 (1973). [DOI] [PubMed] [Google Scholar]
- 13.Huxley H & Hanson J Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 173, 973–976 (1954). [DOI] [PubMed] [Google Scholar]
- 14.Huxley AF & Niedergerke R Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature 173, 971–973 (1954). [DOI] [PubMed] [Google Scholar]
- 15.Huxley HE The mechanism of muscular contraction. Science 164, 1356–1365 (1969). [PubMed] [Google Scholar]
- 16.Spudich JA The myosin swinging cross-bridge model. Nature Rev. Mol. Cell Biol 2, 387–392 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Harris AK, Wild P & Stopak D Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208, 177–179 (1980). [DOI] [PubMed] [Google Scholar]
- 18.Oliver T, Dembo M & Jacobson K Traction forces in locomoting cells. Cell. Motil. Cytoskeleton 31, 225–240 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Chen CS, Mrksich M, Huang S, Whitesides GM & Ingber DE Geometric control of cell life and death. Science 276, 1425–1428 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Wang N, Butler JP & Ingber DE Mechanotransduction across the cell surface and through the cytoskeleton. Science 260, 1124–1127 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Rief M, Gautel M, Oesterhelt F, Fernandez JM & Gaub HE Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Moy VT, Florin EL & Gaub HE Intermolecular forces and energies between ligands and receptors. Science 266, 257–259 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Merkel R, Nassoy P, Leung A, Ritchie K & Evans E Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Choquet D, Felsenfeld DP & Sheetz MP Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 88, 39–48 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Pelham RJ Jr & Wang Y Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl Acad. Sci. USA 94, 13661–13665 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Wichert G et al. RPTP-α acts as a transducer of mechanical force on αv/β3-integrin-cytoskeleton linkages. J. Cell Biol 161, 143–153 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galbraith CG & Sheetz MP A micromachined device provides a new bend on fibroblast traction forces. Proc. Natl Acad. Sci. USA 94, 9114–9118 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balaban NQ et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nature Cell Biol. 3, 466–472 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Tan JL et al. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl Acad. Sci. USA 100, 1484–1489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thoumine O & Ott A Time scale dependent viscoelastic and contractile regimes in fibroblasts probed by microplate manipulation. J. Cell Sci 110, 2109–2116 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Micoulet A, Spatz JP & Ott A Mechanical response analysis and power generation by single-cell stretching. Chemphyschem 6, 663–670 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Beil M et al. Sphingosylphosphorylcholine regulates keratin network architecture and visco-elastic properties of human cancer cells. Nature Cell Biol. 5, 803–811 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Coussen F, Choquet D, Sheetz MP & Erickson HP Trimers of the fibronectin cell adhesion domain localize to actin filament bundles and undergo rearward translocation. J. Cell Sci 115, 2581–2590 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Sawada Y & Sheetz MP Force transduction by Triton cytoskeletons. J. Cell Biol. 156, 609–615 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giannone G, Jiang G, Sutton DH, Critchley DR & Sheetz MP Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J. Cell Biol 163, 409–419 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galbraith CG, Yamada KM & Sheetz MP The relationship between force and focal complex development. J. Cell Biol 159, 695–705 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng F, Suchyna TM & Sachs F A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 275, 3072–3087 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grashoff C et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Yehl K, Narui Y & Salaita K Tension sensing nanoparticles for mechano-imaging at the living/nonliving interface. J. Am. Chem. Soc 135, 5320–5323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stabley DR, Jurchenko C, Marshall SS & Salaita KS Visualizing mechanical tension across membrane receptors with a fluorescent sensor. Nature Methods 9, 64–67 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Jurchenko C, Chang Y, Narui Y, Zhang Y & Salaita KS Integrin-generated forces lead to streptavidin-biotin unbinding in cellular adhesions. Biophys. J 106, 1436–1446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margadant F et al. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 9, e1001223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao R, Boudou T, Wang WG, Chen CS & Reich DH Decoupling cell and matrix mechanics in engineered microtissues using magnetically actuated microcantilevers. Adv. Mater 25, 1699–1705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahmouni S et al. Hydrogel micropillars with integrin selective peptidomimetic functionalized nanopatterned tops: a new tool for the measurement of cell traction forces transmitted through αvβ3- or α5β1-integrins. Adv. Mater 25, 5869–5874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghassemi S et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl Acad. Sci. USA 109, 5328–5333 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campellone KG & Welch MD A nucleator arms race: cellular control of actin assembly. Nature Rev. Mol. Cell Biol 11, 237–251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iskratsch T et al. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell 27, 545–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu CH, Law JB, Suryana M, Low HY & Sheetz MP Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc. Natl Acad. Sci. USA 108, 20585–20590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riveline D et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an Mdia1-dependent and ROCK-independent mechanism. J. Cell Biol 153, 1175–1186 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hotulainen P & Lappalainen P Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol 173, 383–394 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulze N et al. FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J. Cell Sci 127, 1379–1393 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Hayakawa K, Sato N & Obinata T Dynamic reorientation of cultured cells and stress fibers under mechanical stress from periodic stretching. Exp. Cell Res 268, 104–114 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Kozlov MM & Bershadsky AD Processive capping by formin suggests a force-driven mechanism of actin polymerization. J. Cell Biol 167, 1011–1017 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Courtemanche N, Lee JY, Pollard TD & Greene EC Tension modulates actin filament polymerization mediated by formin and profilin. Proc. Natl Acad. Sci. USA 110, 9752–9757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jegou A, Carlier MF & Romet-Lemonne G Formin mDia1 senses and generates mechanical forces on actin filaments. Nature Commun. 4, 1883 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Lazarides E & Burridge K α-actinin: immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell 6, 289–298 (1975). [DOI] [PubMed] [Google Scholar]
- 57.Burridge K & Connell L Talin: a cytoskeletal component concentrated in adhesion plaques and other sites of actin-membrane interaction. Cell. Motil 3, 405–417 (1983). [DOI] [PubMed] [Google Scholar]
- 58.Hu K, Ji L, Applegate KT, Danuser G & Waterman-Storer CM Differential transmission of actin motion within focal adhesions. Science 315, 111–115 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Wang YL Flux at focal adhesions: slippage clutch, mechanical gauge, or signal depot. Sci. STKE 2007, e10 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Aratyn-Schaus Y & Gardel ML Transient frictional slip between integrin and the ECM in focal adhesions under myosin II tension. Curr. Biol 20, 1145–1153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.del Rio A et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiema T et al. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell 21, 337–347 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Nieves B et al. The NPIY motif in the integrin β1 tail dictates the requirement for talin-1 in outside-in signaling. J. Cell Sci 123, 1216–1226 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roca-Cusachs P et al. Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc. Natl Acad. Sci. USA 110, E1361–E1370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roca-Cusachs P, Iskratsch T & Sheetz MP Finding the weakest link: exploring integrin-mediated mechanical molecular pathways. J. Cell Sci 125, 3025–3038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lynch CD, Lazar AM, Iskratsch T, Zhang X & Sheetz MP Endoplasmic spreading requires coalescence of vimentin intermediate filaments at force-bearing adhesions. Mol. Biol. Cell 24, 21–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ithychanda SS & Qin J Evidence for multisite ligand binding and stretching of filamin by integrin and migfilin. Biochemistry 50, 4229–4231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rognoni L, Stigler J, Pelz B, Ylanne J & Rief M Dynamic force sensing of filamin revealed in single-molecule experiments. Proc. Natl Acad. Sci. USA 109, 19679–19684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Y et al. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J. Exp. Med 207, 2421–2437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lochter A & Bissell MJ Involvement of extracellular matrix constituents in breast cancer. Semin. Cancer Biol 6, 165–173 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Engler AJ, Sen S, Sweeney HL & Discher DE Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Jiang G, Huang AH, Cai Y, Tanase M & Sheetz MP Rigidity sensing at the leading edge through αvβ3 integrins and RPTPα. Biophys. J 90, 1804–1809 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giannone G et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell 116, 431–443 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Dubin-Thaler BJ et al. Quantification of cell edge velocities and traction forces reveals distinct motility modules during cell spreading. PLoS ONE 3, e3735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prager-Khoutorsky M et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nature Cell Biol. 13, 1457–1465 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Plotnikov SV, Pasapera AM, Sabass B & Waterman CM Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bruinsma R Theory of force regulation by nascent adhesion sites. Biophys. J 89, 87–94 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwarz US et al. Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys. J 83, 1380–1394 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shemesh T, Geiger B, Bershadsky AD & Kozlov MM Focal adhesions as mechanosensors: a physical mechanism. Proc. Natl Acad. Sci. USA 102, 12383–12388 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nicolas A & Safran SA Limitation of cell adhesion by the elasticity of the extracellular matrix. Biophys. J 91, 61–73 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan CE & Odde DJ Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008). [DOI] [PubMed] [Google Scholar]
- 82.De Craene B & Berx G Regulatory networks defining EMT during cancer initiation and progression. Nature Rev. Cancer 13, 97–110 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Zaidel-Bar R & Geiger B The switchable integrin adhesome. J. Cell Sci 123, 1385–1388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hill AV The heat of shortening and the dynamic constants of muscle. Proc. Biol. Sci 126, 136–195 (1938). [Google Scholar]
- 85.Trinick J, Knight P & Whiting A Purification and properties of native titin. J. Mol. Biol 180, 331–356 (1984). [DOI] [PubMed] [Google Scholar]
- 86.Lange S et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308, 1599–1603 (2005). [DOI] [PubMed] [Google Scholar]
- 87.Puchner EM et al. Mechanoenzymatics of titin kinase. Proc. Natl Acad. Sci. USA 105, 13385–13390 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knoll R et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111, 943–955 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Samarel AM Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am. J. Physiol. Heart Circ. Physiol 289, H2291–H2301 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Buckley MJ et al. Osteoblasts increase their rate of division and align in response to cyclic, mechanical tension in vitro. Bone Miner. 4, 225–236 (1988). [PubMed] [Google Scholar]
- 91.Neidlinger-Wilke C, Wilke HJ & Claes L Cyclic stretching of human osteoblasts affects proliferation and metabolism: a new experimental method and its application. J. Orthop. Res 12, 70–78 (1994). [DOI] [PubMed] [Google Scholar]
- 92.Carisey A et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol 23, 271–281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jungbauer S, Gao H, Spatz JP & Kemkemer R Two characteristic regimes in frequency-dependent dynamic reorientation of fibroblasts on cyclically stretched substrates. Biophys. J 95, 3470–3478 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goldyn AM, Rioja BA, Spatz JP, Ballestrem C & Kemkemer R Force-induced cell polarisation is linked to RhoA-driven microtubule-independent focal-adhesion sliding. J. Cell Sci 122, 3644–3651 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu WF, Nelson CM, Tan JL & Chen CS Cadherins, RhoA, and Rac1 are differentially required for stretch-mediated proliferation in endothelial versus smooth muscle cells. Circ. Res 101, e44–e52 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Kaunas R, Nguyen P, Usami S & Chien S Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc. Natl Acad. Sci. USA 102, 15895–15900 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown AC, Fiore VF, Sulchek TA & Barker TH Physical and chemical microenvironmental cues orthogonally control the degree and duration of fibrosis-associated epithelial-to-mesenchymal transitions. J. Pathol 229, 25–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Connor JW & Gomez EW Cell adhesion and shape regulate TGF-β1-induced epithelial-myofibroblast transition via MRTF-A signaling. PLoS ONE 8, e83188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baarlink C, Wang H & Grosse R Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340, 864–867 (2013). [DOI] [PubMed] [Google Scholar]
- 100.Dupont S et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 101.Aragona M et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 102.Nelson CM & Bissell MJ Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol 22, 287–309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Puklin-Faucher E & Sheetz MP The mechanical integrin cycle. J. Cell Sci 122, 179–186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arias-Salgado EG et al. Src kinase activation by direct interaction with the integrin β cytoplasmic domain. Proc. Natl Acad. Sci. USA 100, 13298–13302 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chang F, Lemmon CA, Park D & Romer LH FAK potentiates Rac1 activation and localization to matrix adhesion sites: a role for βPIX. Mol. Biol. Cell 18, 253–264 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miki H, Yamaguchi H, Suetsugu S & Takenawa T IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408, 732–735 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Berrier AL, Martinez R, Bokoch GM & LaFlamme SE The integrin β tail is required and sufficient to regulate adhesion signaling to Rac1. J. Cell Sci 115, 4285–4291 (2002). [DOI] [PubMed] [Google Scholar]
- 108.Kostic A & Sheetz MP Fibronectin rigidity response through Fyn and p130Cas recruitment to the leading edge. Mol. Biol. Cell 17, 2684–2695 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Giannone G & Sheetz MP Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol. 16, 213–223 (2006). [DOI] [PubMed] [Google Scholar]
- 110.Cai Y et al. Cytoskeletal coherence requires myosin-IIA contractility. J. Cell Sci 123, 413–423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kolega J Effects of mechanical tension on protrusive activity and microfilament and intermediate filament organization in an epidermal epithelium moving in culture. J. Cell Biol 102, 1400–1411 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Singhvi R et al. Engineering cell shape and function. Science 264, 696–698 (1994). [DOI] [PubMed] [Google Scholar]
- 113.Sawada Y et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015–1026 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pollard TD & Borisy GG Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 (2003). [DOI] [PubMed] [Google Scholar]
- 115.Schiller HB, Friedel CC, Boulegue C & Fassler R Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12, 259–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Knowlton FP & Starling EH The influence of variations in temperature and blood-pressure on the performance of the isolated mammalian heart. J. Physiol 44, 206–219 (1912). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Markwalder J & Starling EH On the constancy of the systolic output under varying conditions. J. Physiol 48, 348–356 (1914). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Patterson SW & Starling EH On the mechanical factors which determine the output of the ventricles. J. Physiol 48, 357–379 (1914). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Patterson SW, Piper H & Starling EH The regulation of the heart beat. J. Physiol 48, 465–513 (1914). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Olesen SP, Clapham DE & Davies PF Haemodynamic shear stress activates a K+ current in vascular endothelial cells. Nature 331, 168–170 (1988). [DOI] [PubMed] [Google Scholar]
- 121.Hynes RO The emergence of integrins: a personal and historical perspective. Matrix Biol. 23, 333–340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Franck C, Hong S, Maskarinec S, Tirrell D & Ravichandran G Three-dimensional full-field measurements of large deformations in soft materials using confocal microscopy and digital volume correlation. Exp. Mechan 47, 427–438 (2007). [Google Scholar]