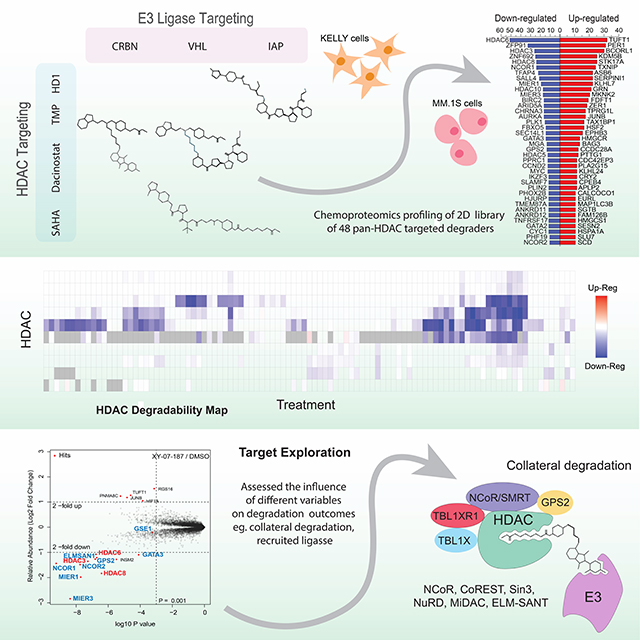
SUMMARY
Targeted protein degradation refers to the use of small molecules that recruit a ubiquitin ligase to a target protein for ubiquitination and subsequent proteasomal degradation. While degraders have been developed for many targets, key questions regarding degrader development and the consequences of acute pharmacological degradation remain, specifically for targets that exist in obligate multi-protein complexes. Here, we synthesize a pan-HDAC degrader library for the chemo-proteomic exploration of acute degradation of a key class of chromatin-modifying enzymes. Using chemo-proteomics, we not only map the degradability of the zinc-dependent HDAC family identifying leads for targeting HDACs 1–8 and 10, but also explore important aspects of degrading epigenetic enzymes. We discover cell line driven target specificity and that HDAC degradation often results in collateral loss of HDAC-containing repressive complexes. These findings potentially offer a new mechanism towards controlling chromatin structure and our open-source resource will facilitate accelerated degrader design and development for HDACs.
Keywords: HDAC, PROTAC, degrader, ubiquitin, E3 ligase, IMiD, targeted degradation, collateral degradation
eTOC Blurb:
Xiong et al. report the design and synthesis of a diverse multi-targeted library of HDAC degraders and assess their activity in cells. The findings provide a roadmap for future design of HDAC targeted degraders and reveal important insights regarding the biological role of this epigenetic family of enzymes.
Graphical Abstract
INTRODUCTION
Targeted protein degradation (TPD) is a therapeutic modality that offers an opportunity to overcome several limitations of traditional inhibitors (Bondeson and Crews, 2017; Burslem and Crews, 2020; Neklesa et al., 2017). TPD utilizes small molecule degraders to hijack the cellular degradation machinery to achieve knockdown of a protein of interest (POI). First demonstrated with a phosphopeptide as the ligase recruiting handle, linked to a small molecule target binder to create a PROteolysis TArgeting Chimera (PROTAC), the resulting heterobifunctional molecule (degrader) was capable of inducing proximity-mediated ubiquitination and subsequent proteasomal degradation in cell lysates (Sakamoto et al., 2001). Over the last two decades, significant improvements have been made to the chemical design of these molecules, starting with the replacement of peptidic recruiters with small molecule recruiters of E3 ubiquitin ligases (Bondeson et al., 2015; Schneekloth et al., 2008; Winter et al., 2015). Degraders now, despite still challenging Lipinski’s rule of 5 (Lipinski et al., 1997), possess more drug-like properties and this has propelled the first degraders into Phase I/II clinical trials for targeting the Androgen Receptor (ARV-110; NCT03888612) and Estrogen Receptor (ARV-471; NCT04072952).
Conceptually, degraders offer advantages such as the potential to target the “undruggable proteome”, which currently makes up about 80% of the human proteome. Due to their catalytic or event-driven mode of action (MoA) enabling sub-stoichiometric degradation, degraders also offer the prospect of sustained efficacy with lower intracellular concentrations of the molecule. Other potential advantages of degraders include: unique pharmacology whereby non-enzymatic functions such as scaffolding functions are targeted (Cromm et al., 2018), high target selectivity through chemical exploitation of the binding pockets and protein-protein interfaces (PPIs) (Farnaby et al., 2019; Gadd et al., 2017; Nowak et al., 2018; Smith et al., 2019), and prolonged efficacy dependent on a target’s turnover rate (Mares et al., 2020; You et al., 2020). Most degraders have been discovered using a bottom-up approach, where potent inhibitors for a POI are selected early in the process and major medicinal chemistry efforts are focused on surveying the linker space between the target inhibitor and different E3 ligase recruiting handles. Optimization is often guided by target-focused assays for measuring target protein levels, and unbiased proteomics analysis is generally used at a later stage to illustrate degradation selectivity of lead molecules. While many degraders have been developed for a variety of targets using this approach (Lai et al., 2016b; Li et al., 2018; Silva et al., 2019; Winter et al., 2015; Wu et al., 2019), they do not sufficiently explore the opportunity of weak target engagement leading to potent degradation, a unique opportunity for degraders (Bondeson et al., 2018; Donovan et al., 2020; Faust et al., 2020; Gadd et al., 2017). The focused bottom-up approach also provides limited data points across a large library of molecules as only final compounds are typically subjected to full characterization. The limited availability of datasets that provide unbiased activity profiling for large libraries of compounds, thereby providing information on off-target degradation and inactive molecules, represents a major limitation for efforts aimed at better understanding the underlying principles of protein degradation. Recently, we pioneered a family-centric top-down approach to explore the degradability of the protein kinase family (Donovan et al., 2020) by combining broad family-wide degrader library generation, with unbiased whole cell degradation profiling using chemo-proteomics. While the commercial success of kinase inhibitors has led to an abundance of selective and pan-kinase inhibitors enabling such efforts, we asked whether a similar approach could be pursued targeting a family with fewer available ligands in which the lack of chemotype diversity would be overcome with more extensive linker exploration. We also wanted to specifically target a family of enzymes, that in contrast to kinases, has strong association with chromatin and whose members typically exist in large multi-protein complexes.
Epigenetic enzymes have a crucial role in regulating gene expression and are frequently dysregulated in human cancers, and this has led to the development of small molecule inhibitors targeting the cancer epigenome (Seto and Yoshida, 2014; Witt et al., 2009b). Histone Deacetylases (HDACs) are one such class of epigenetic enzyme. The abnormal expression of HDACs in diseases including cancers has been found to influence the expression of genes required for promoting cancer cell survival and progression (Chen et al., 2015; Li and Seto, 2016; Seto and Yoshida, 2014). Therefore, therapeutic targeting of HDACs is the focus of numerous drug development efforts and as of 2020, the U.S. Food and Drug Administration (FDA) has approved five HDAC inhibitors for the treatment of T-cell lymphomas and multiple myeloma (Ho et al., 2020). The human HDAC family consists of 18 enzymes, 11 of which contain a divalent zinc cation in the catalytic site and the remaining 7 are Sirtuins with NAD+-dependent activity (Ruijter et al., 2003; Seto and Yoshida, 2014). HDACs can be further classified into four classes: class I (HDAC 1, 2, 3 and 8), class IIa (HDAC 4, 5, 7 and 9), class IIb (HDAC 6 and 10), class IV (HDAC11) and class III (sirtuins) HDACs. HDACs, as chromatin modifying enzymes, are frequently recruited by corepressors as a key component of large histone modifying complexes (Bantscheff et al., 2011; Bradner et al., 2010; Yang and Seto, 2003). In addition to their deacetylase activity, some HDACs are also thought to exert non-enzymatic functions such as a role in scaffolding large complexes (Fischle et al., 2001; Fischle et al., 2002; Magupalli et al., 2020; McKinsey et al., 2000; Nguyen et al., 2020; Verdin et al., 2003). Currently available inhibitors are limited in their isoform selectivity and reports show that achieving complex selectivity is also a major challenge that is thought to be a key source for off-target toxicities and adverse effects associated with HDAC inhibitors (Suraweera et al., 2018).
Targeted protein degradation has proven to be a viable method to overcome limitations of inhibitor isoform selectivity. Moreover, degraders, as tool molecules for chemical knockdown, provide an opportunity to help differentiate the enzymatic and scaffolding functions of HDAC proteins. Although degraders for HDAC6 (An et al., 2019; Dorman et al., 2018; Sinatra et al., 2020; Yang et al., 2020) and class I HDACs (Cao et al., 2020; Smalley et al., 2020; Xiao et al., 2020) have been reported to date, no systematic investigation into the degradability of the HDAC family and the consequences of HDAC degradation on HDAC containing complexes has been pursued. To thoroughly explore the accessibility and degradability of HDACs, we designed and synthesized a comprehensive library of 48 multi-HDAC targeting degrader molecules incorporating four different pan-HDAC binding warheads chemically linked to recruiters of three different ubiquitin E3 ligases – CRBN, VHL and IAP. Using global quantitative chemo-proteomics, we establish an HDAC degradability dataset and demonstrate successful targeting of 9 of the 11 zinc-dependent HDACs. The chemo-proteomics approach not only creates a valuable resource for guiding the development of selective probes, but also sheds light on the consequences of acute HDAC degradation on chromatin-modifying complexes and context dependent activities important to consider for the future development of therapeutics.
RESULTS
HDAC-targeted degrader library design and target identification
We designed and synthesized a library of 48 pan-HDAC heterobifunctional degraders (Figure 1A, Data S1) with emphasis on prioritizing scaffold, linker and ligase diversity together with synthetic tractability to evaluate the degradation of zinc-dependent HDACs. We designed the library to enable broad coverage of the 11 zinc-dependent HDACs using the smallest number of target binding scaffolds by choosing four class-selective, pan-HDAC inhibitors as the HDAC binding ligands: SAHA and dacinostat were selected for targeting class I and IIb HDACs 1, 2, 3, 6, 8, 10 (Bantscheff et al., 2011; Bradner et al., 2010), while TMP269 and NVS-HD1 were selected for targeting class IIa HDACs 4, 5, 7, 9 (Lobera et al., 2013; Luo et al., 2019) (Figure 1A–B, Table S1). To increase the likelihood of HDAC recruitment to a ligase, we sampled the E3 ligase space by selecting four different ligands to recruit the three E3 ligases that are most commonly utilized in degrader development. These include the cereblon (CRL4CRBN) ligand pomalidomide, a von Hippel-Lindau (CRL2VHL) ligand, VH032 analog, with terminal- (VHL1) or internal-positioned (VHL2) linker attachment points, and LCL161 for the recruitment of inhibitor of apoptosis proteins (IAPs). We tethered these target and ligase binding moieties using a variety of linkers including a series of PEG- and hydrocarbon-based linkers with varying lengths and attachment chemistry (Figure 1A–B, Table S1). To enhance the permeability of the molecules contained in our library, we limited the number of hydrogen bond donors and acceptors included in the design by using amine linkers instead of amide linkers. Similarly, we avoided the use of triazole-containing linkers because they often negatively influence cell permeability (Su et al., 2019; Zeng et al., 2019). Individual degrader design elements such as linker attachment point selection with respect to the two protein binders were based on available inhibitor-bound X-ray crystal structures, and for proteins lacking bound crystal structures, structure activity relationships (SAR) for the inhibitor analogs were assessed to establish a linker attachment point that would minimally affect the protein binding activity of the resulting heterobifunctional degraders. Finally, with the aim to build a comprehensive multi-HDAC targeted library, synthetic tractability was a key consideration. For example, in the case of dacinostat, the linker was adapted from its solvent accessible aminoethanol moiety because this synthetic strategy allows for both efficient synthesis of degraders and rapid diversification of linker and ligase at a later stage in the synthetic route.
Figure 1 |. HDAC-targeted degrader library design and target identification.
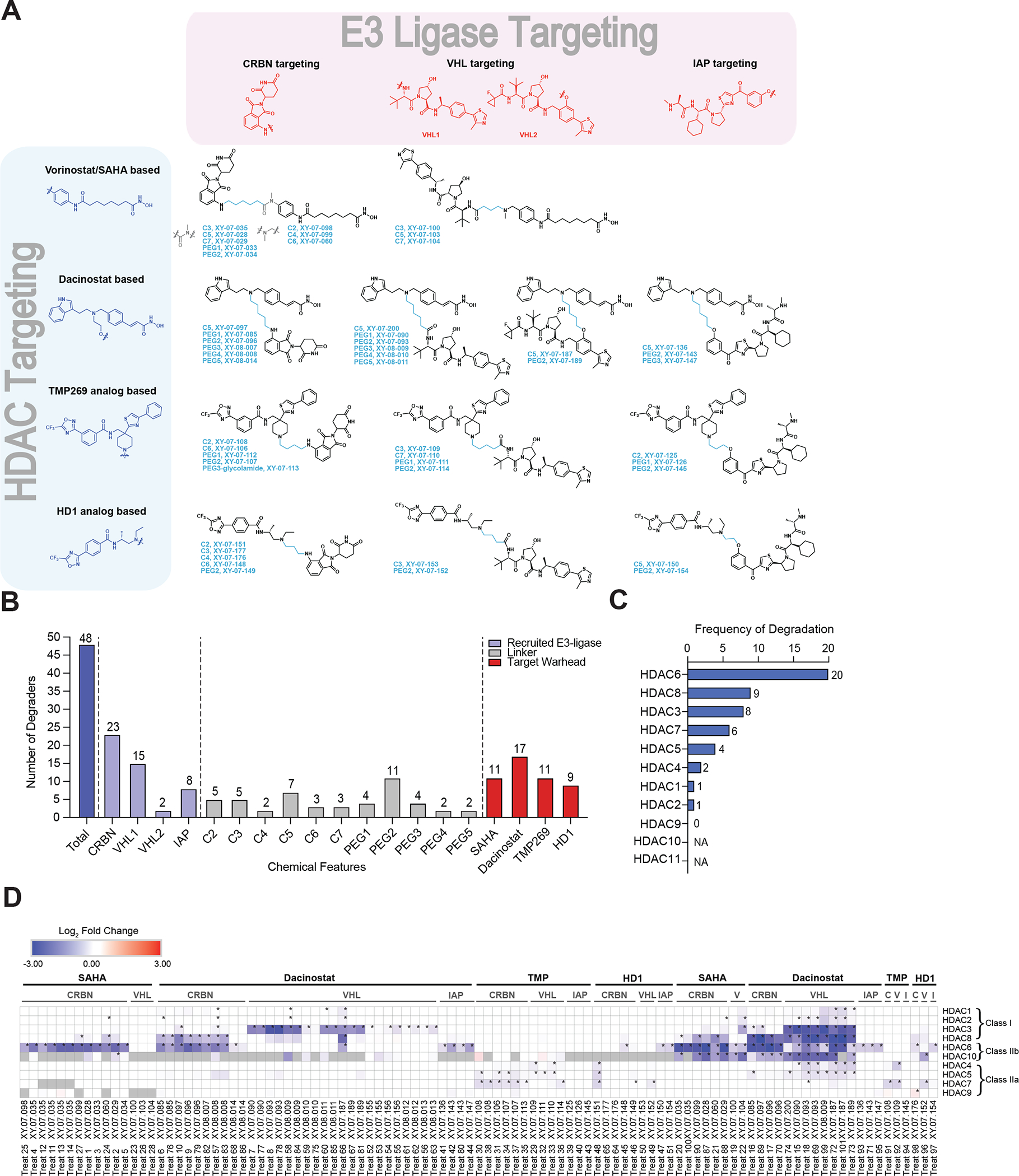
(A) Two-dimensional HDAC-targeting degrader library. (B) Summary of chemical features of the 48 multi-targeted heterobifunctional degraders included in this database. (C) The number of unique degraders that induced degradation of each HDAC in the KELLY-limited dataset. (D) Chemo-proteomic degradation heatmap for each of the treatments included in this study. Proteomics data is from n = 1–2 biological replicates. * on proteomics heatmaps indicates a P-value < 0.001. Corresponding proteomics treatment details can be found in Table S1. See also Table S1–S3, Figure S1–S2.
To assess the degradability of HDACs and effectively map the selectivity profile for each of the 48 pan-HDAC degraders, we employed quantitative proteomics to measure the change in relative abundance of protein expression in response to treatment with each of the HDAC-targeted degraders. Because HDAC inhibitors have demonstrated neuroprotective activity in neurodegeneration models (Chuang et al., 2009; Kim et al., 2007; Lu et al., 2013) and anti-tumoral response in rodent neuroblastoma models (Phimmachanh et al., 2020; Witt et al., 2009a), we decided to explore the effect of HDAC degraders in a neuroblastoma cell line, KELLY cells. We profiled the degrader library in KELLY cells with the default treatment condition as 1 μM for 5 h to assess the degradation profiles of each molecule (Figure 1C–D, Figure S1). The short treatment time was selected to reduce secondary effects (Bushman et al., 2020). Degradation “hits” were called based on a pre-determined threshold of > 1.25 fold change (FC) and P-value < 0.001. Consolidation of the data identified HDAC6 as the most frequently degraded HDAC across our initial profiling database, with 20 independent treatments inducing downregulation of this protein (Figure 1C, Figure S1, Table S2–3). Followed by HDAC8 (9) and HDAC3 (8) as the next most degradable HDACs. These numbers reflect the number of unique degraders that are able to cause degradation of the respective proteins in a limited set of data that includes one treatment per compound in KELLY cells (hereafter referred to as the KELLY-limited dataset). One limitation of this comparison is that not all of the HDACs are reliably quantified in KELLY cells, for example, HDAC10 expression is low in this cell line and this likely limits our ability to detect it in mass spectrometry experiments (Table S1). We validated HDAC degradation by spot checking key degraders for degradation by dose response immunoblot (Figure S2A). To confirm that HDAC degradation was mediated through E3 ubiquitin ligase activity, we performed side-by-side comparisons measuring degradation, degradation rescue by pre-treatment with the neddylation inhibitor MLN4924 and treatment with the respective negative control compound (disrupted ligase engagement) for HDAC6 targeted XY-07-035, and HDAC3, 6 and 8 targeted XY-07-187 (Figure S2B–D).
To expand the dataset, we incorporated additional chemo-proteomics data from an HDAC3-targeted mini-series of degraders and in addition to treatments with variations in treatment concentration and time, we profiled selected degraders in a second cell line of different lineage, multiple myeloma MM.1S cells. Together, these data created an HDAC-targeted chemo-proteomic database incorporating 52 different degraders (Table S1) profiled in over 100 independent treatments (Figure 1D, Figure S1, Table S1–S3) and identifying degraders targeting 9 of the 10 zinc-dependent HDACs quantified across our experiments (Figure 1D, Table S3). Unfortunately, we were unable to quantify HDAC11 in these experiments likely due to low expression levels and important to note is that although HDAC9 was frequently quantified, it was not determined to be a hit under any of the conditions tested in this study (Table S1).
The HDAC family of proteins play critical roles in gene regulation and beyond, but HDAC-substrate relationships are insufficiently understood and many functional roles HDACs play remain unknown. One result of chemo-proteomics that has been underappreciated and underutilized, perhaps because short treatment times drastically reduce secondary effects, are the indirect targets that are up- and down-regulated in response to degrader treatment. To explore the secondary effects of HDAC disruption and assess the combined potential for degraders and chemo-proteomics to expose functions of HDACs, we curated a list of all proteins that were exclusively and consistently up- or down-regulated in response to degrader treatment across the database of >100 treatments (Table S3). We then tabulated this list by calculating the number of degrader treatments that resulted in up- or downregulation of each protein, which allowed us to rank order the proteins to identify those proteins with expression altered most frequently in response to degrader treatment (Figure S2E, Table S3) and found that many of these proteins have reported connections to different HDACs. For example, the most frequently upregulated protein TUFT1 has been reported to be transcriptionally upregulated in response to 6 h treatment with the Class I and IIb HDAC inhibitor SAHA (Nebbioso et al., 2017). Also, the second most frequently upregulated protein PER1, has been linked to HDAC3 and a role in circadian function (Gery et al., 2007), and more specifically in age-related impairment of long-term memory formation (Kwapis et al., 2018). In this study, deletion of HDAC3 was found to rescue age-related impairment of PER1 expression which fits with our data showing increased PER1 expression in response to HDAC disruption.
Collateral degradation of HDAC interacting proteins
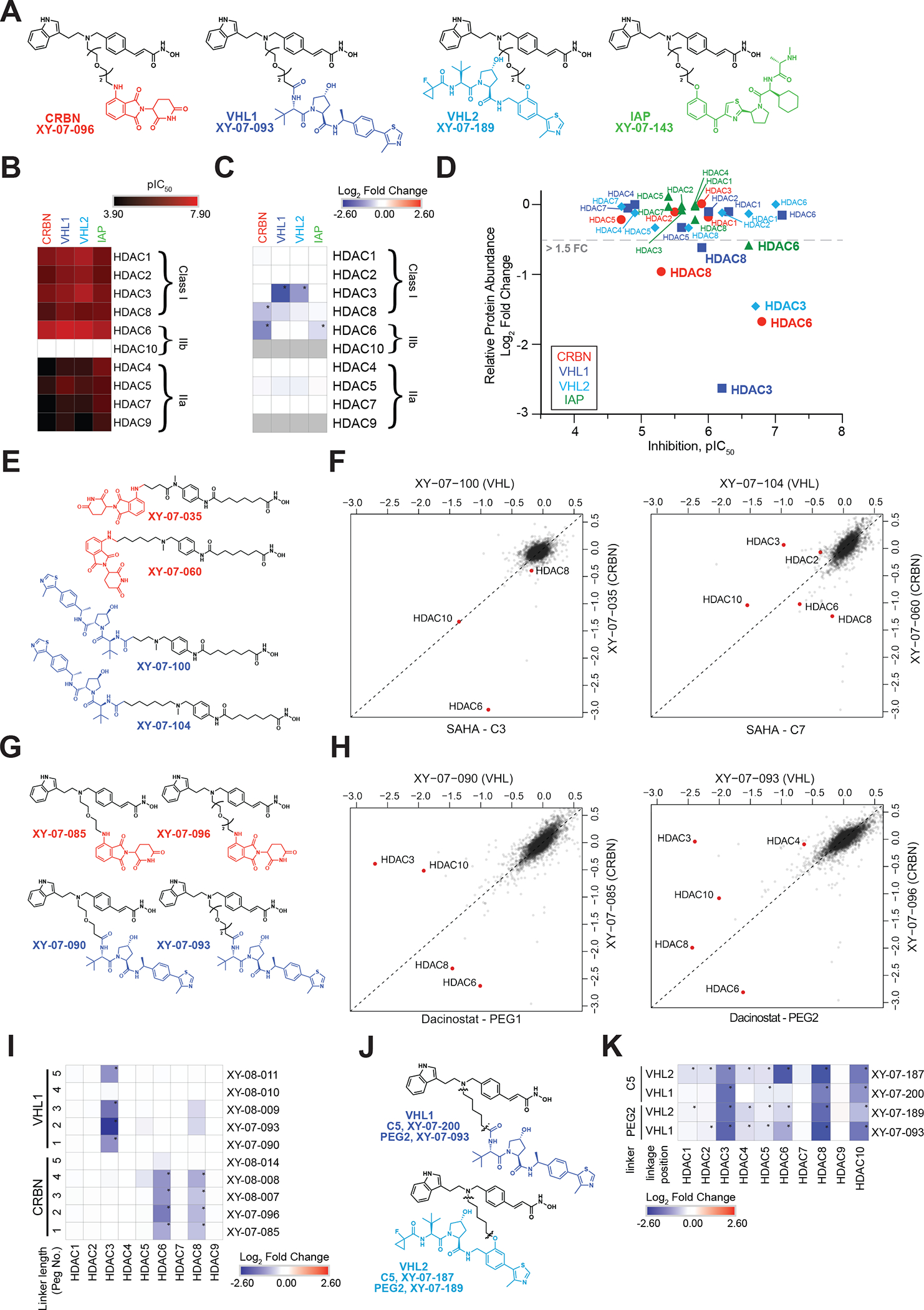
Collateral degradation of protein complex subunits with degraders has been observed for small molecule degraders or covalent fragments targeting the polycomb repressive complex 2 (PRC2) (Hsu et al., 2020), the BAF complex (Farnaby et al., 2019) or the NuRD complex (Vinogradova et al., 2020). But despite the growing numbers of reported degraders, there are fewer examples of collateral degradation described than would be expected by chance. Strikingly, we consistently, across several HDAC-targeted degraders, observe collateral degradation for multiple components of different HDAC-containing protein complexes (Figure 2A, Figure S1, Table S2, S3). We found that the degradation of complex components is influenced by factors such as target binding scaffold, recruited ligase and linker chemistry. Across the dataset we observe that some dacinostat-based degraders show obvious targeting of HDAC3, as well as members of the HDAC3 containing NCoR/SMRT corepressor complex. We also note that degradation of complex components such as GSP2 and NCOR1/2 is more pronounced with dacinostat degraders containing VHL-recruiting moieties (Figure 2A). Other complexes targeted by dacinostat-based degraders include CoREST, MiDAC complexes and ELM/SANT proteins, with different degraders displaying varying abilities to target known binding partners. Studies by Bantscheff and colleagues have shown that HDAC inhibitors not only bind to free HDACs, but also bind to HDACs in the core of multi-subunit complexes which supports the potential for ligase recruitment to complexes (Bantscheff et al., 2011). To investigate whether collateral degradation depends on ligase recruitment, we assessed the global proteome response to a potent multi-targeted degrader, XY-07-187 (Figure S2D), compared to the ligase-inactive control, XY-07-187-NEG. Subtracting the XY-07-187-NEG response from the active degrader treatment eliminates effects that are independent of ligase recruitment. All of the HDAC complex partners remained downregulated indicating that their degradation is strictly dependent on recruitment of the ligase to HDACs. We also confirmed the Cullin RING Ligase (CRL)-dependent mechanism of HDAC and collateral degradation by performing a proteome wide degradation rescue experiment with neddylation inhibitor MLN4924 (Figure S2D). Together these experiments strongly suggest that collateral degradation is a result of HDAC degradation and not inhibition, while additional experiments are needed to elucidate whether complex members get ubiquitinated or HDAC degradation destabilizes binding partners and disintegrates the complex.
Figure 2 |. Collateral degradation and HDAC selectivity with promiscuous binders.

(A) Heatmap displaying the log2FC in relative abundance for HDACs and reported binding partners in response to treatment with indicated degraders. (B) Schematic representation of Reaction Biology’s (www.reactionbiology.com) in vitro HDAC enzymatic inhibition assay. (C) Heatmap displaying in vitro inhibition of HDAC enzymatic activity (pIC50). Inhibition data is from n = 1 ten-point titrations. (D) Scatterplots depicting the log2FC in relative protein abundance of indicated treatments. E) Heatmap displaying the log2FC in relative abundance for quantified HDACs in response to a time course treatment with XY-07-035. (F) As in D, but for XY-07-090 treatment. (G) As in D, but for XY-07-106 and XY-07-151 treatments. HDAC4, 5, 7 and 9 have been colored red to show their location on the scatterplots. Proteomics data is from n = 1–2 biological replicates. * on proteomics heatmaps indicates a P-value < 0.001. See also Table S2–3, Figure S1–S3.
Surprisingly, when switching the recruited ligase of dacinostat-based degraders to IAP, we saw a distinct target profile shift from HDAC3 and NCoR/SMRT complex targeting to HDAC6, CoREST and ELM/SANT targeting with downregulation of GSE1, HMG20B (CoREST) as well as MIER2/3 (ELM/SANT). In some cases, we observe collateral degradation occurring regardless of whether the core HDACs are degraded. For instance, ELM2-SANT domain containing MIER family proteins MIER1/2/3 are often degraded by compounds across our HDAC-focused degrader library. However, while HDAC1/2 are the only HDACs known to be found in complex with MIER family proteins (Derwish et al., 2017), HDAC1 and HDAC2 are not significantly downregulated under the conditions tested. There are a number of possible reasons why MIER1/2/3 are degraded in absence of HDAC1/2 including, for example, HDAC1/2-MIER1/2/3 complex recruitment to the ligase machinery to form a quaternary complex in which only MIER proteins are ubiquitinated and degraded. This could also be true for MIER proteins being recruited through a complex formed with a previously unreported HDAC.
Despite HDACs existing at the core of numerous multiprotein complexes, there are no prior reports of HDAC-targeted degraders inducing collateral degradation of binding partners. There are, however, reports of collateral degradation of BAF (Farnaby et al., 2019), PRC2 (Hsu et al., 2020) and NuRD complexes (Vinogradova et al., 2020). These independent examples of chromatin associated collateral degradation suggest that the collateral degradation phenomenon may be more prevalent for chromatin associated complexes.
HDAC selectivity is attainable with promiscuous binders
Several studies show that cellular degradation has a non-linear dependence on binary binding affinity for kinases and bromodomains (Bondeson et al., 2018; Donovan et al., 2020; Gadd et al., 2017; Huang et al., 2018), which suggests that the target engagement requirement for degradation may be dependent on other factors such as the target, ligase and/or linker chemistry. To investigate whether such non-linear relationships are also present for HDACs, we selected at least one degrader from each of the four target-scaffold series and assessed their ability to engage HDACs in an in vitro fluorescence-based assay (Figure 2B). We found that the SAHA-based degraders retained a similar level of inhibition (pIC50), in addition to similar isoform selectivity as their corresponding parental inhibitor (Figure 2C, S3A, Table S4). When compared to the degradation profile, we found that both XY-07-035 and XY-07-029 showed significant degradation of HDAC6 in line with their strong HDAC6 inhibition profiles (pIC50 ~7.4) (Figure 2C–D). In addition, XY-07-035 also exhibits the ability to bind to and inhibit HDACs 1–3 and 8 in vitro (Figure 2C). But despite multiple chemo-proteomics experiments assessing the targets of this compound, it shows HDAC6 selectivity in KELLY cells. To test the selectivity over time and to generally quantify the temporal cellular response to a representative HDAC degrader, we performed a time course analysis for the HDAC6 selective degrader XY-07-035, treated for 2 to 16 h. The resulting data demonstrate selective and time dependent degradation of HDAC6, while the abundance for other HDACs remains unchanged (Figures 2E). HDAC6 remains the only significantly downregulated target protein up to the 8 h treatment time point with very mild degradation of ZNF692 which is a common off-target of many pomalidomide based degraders (Figure S3B) (Donovan et al., 2018; Nowak et al., 2018; Sievers et al., 2018). At 16 h post treatment, several additional proteins are significantly downregulated, and this is presumably a secondary effect of sustained HDAC6 degradation (Figure S3B–C, Table S2, S3). Similarly, dacinostat-based degraders show stronger inhibition of HDAC6 than other isoforms (Figure 2C, S3A), yet in the KELLY-limited dataset as well as more broadly across the database, HDAC3 is almost always the most prominent degradation target of the VHL-recruiting series of molecules including XY-07-090 (Figure 2F, Table S3). Interesting to note, is that both SAHA- and dacinostat-based degraders retain in vitro inhibition for HDAC1 and HDAC2 (Figure 2C), but it is very rare to see these two isoforms degraded in our library (Table S3). In contrast to SAHA- and dacinostat-based degraders, TMP269-based degrader XY-07-106 showed a significant reduction in inhibition of HDACs 4, 5, 7 and 9, suggesting that the addition of a linker to the piperidine ring may be detrimental to target engagement with this scaffold (Figure 2C). Assessment of the degradation targets of this molecule revealed that although the in vitro selectivity profile clearly favors inhibition of HDAC9, this target was not degraded by the TMP269-based degrader. However, weak engagement of HDAC5 and 7 was sufficient for inducing their degradation (Figure 2G). Interestingly, degrader XY-07-151 which is based on a more potent class IIa inhibitor, NVS-HD1, showed strong inhibition of HDAC4, 5, 7 and 9, but similar to XY-07-106, was only able to initiate significant degradation of HDAC4, 5 and 7 with the treatment conditions tested (Figure 2G). On the other hand, the parental inhibitor NVS-HD1 is reported to have > 200 fold selectivity for class IIa HDACs over other HDACs (Luo et al., 2019), yet when IAP is recruited as the E3 ubiquitin ligase, two different NVS-HD1 based degraders (XY-07-150 and XY-07-154) are able to induce weak and selective degradation of HDAC6 (Figure S1, Figure S3D, Table S3).
Ligase and linker influence degrader selectivity
Targeted protein degradation requires the formation of a productive ternary complex, which includes target protein recruitment to the ligase followed by ubiquitin transfer to an available lysine on the target protein. Many studies have observed that the selectivity profile of degraders towards closely related protein isoforms can be influenced by factors such as the E3 ligase recruited (Lai et al., 2016a; Smith et al., 2019; Zorba et al., 2018), the length of the linker (Cyrus et al., 2011; Nowak et al., 2018; Smith et al., 2019; Zorba et al., 2018), as well as the point of linker attachment on each of the ligands (Smith et al., 2019; Zoppi et al., 2018), and each of these factors is thought to influence the nature of the ternary complex formed. To systematically investigate the impact that the recruited ligase has upon the selectivity of different HDAC isoforms, we evaluated the target scope of a matched series of degraders based on a pan-HDAC inhibitor, dacinostat, which has binding selectivity for HDAC1, 2, 3, 6 and 8 (Bantscheff et al., 2011). We synthesized a set of four degraders containing an identical dacinostat-PEG2 as the target recruiting scaffold-linker, and with each molecule in the series including one of four different ligase recruitment variants designed to recruit three different E3 ligases. The four ligase recruitment variants included CRBN, IAP, and two VHL handles (VHL1 and VHL2) including an exchange of the linker attachment point on the VHL ligand, from the terminal amide group (VHL1) ligand to a phenol linkage on the internal phenyl group (VHL2) ligand (Figure 3A). We first assessed the HDAC inhibition profiles of this tetrad series and confirmed that addition of linker and ligase binder did not interfere with binary target engagement in the in vitro enzymatic binding assay (Figure 3B, Table S3). However, direct comparison of global protein expression levels after degrader treatment revealed that although target engagement was consistent across degraders, recruitment of different ligases does induce distinct preferences in which HDAC isoform is degraded (Figure 3C–D, Table S3). Comparison of the activity of these four molecules in KELLY cells, shows that the CRBN-recruiting compound XY-07-096 preferentially degrades HDAC6 and 8, while IAP-recruiting compound XY-07-143 shows weak but selective degradation of HDAC6. Whereas, both VHL-recruiting molecules show degradation selectivity towards HDAC3, with the VHL1-recruiting compound XY-07-093 being the more potent HDAC3 degrader while also exhibiting weak degradation activity towards HDAC8 (P-value = 0.003), suggesting that altering the VHL ligand linker position could be a useful strategy for tuning HDAC target selectivity (Figure 3C–D). These data highlight the significant impact that recruited ligase can have upon target accessibility.
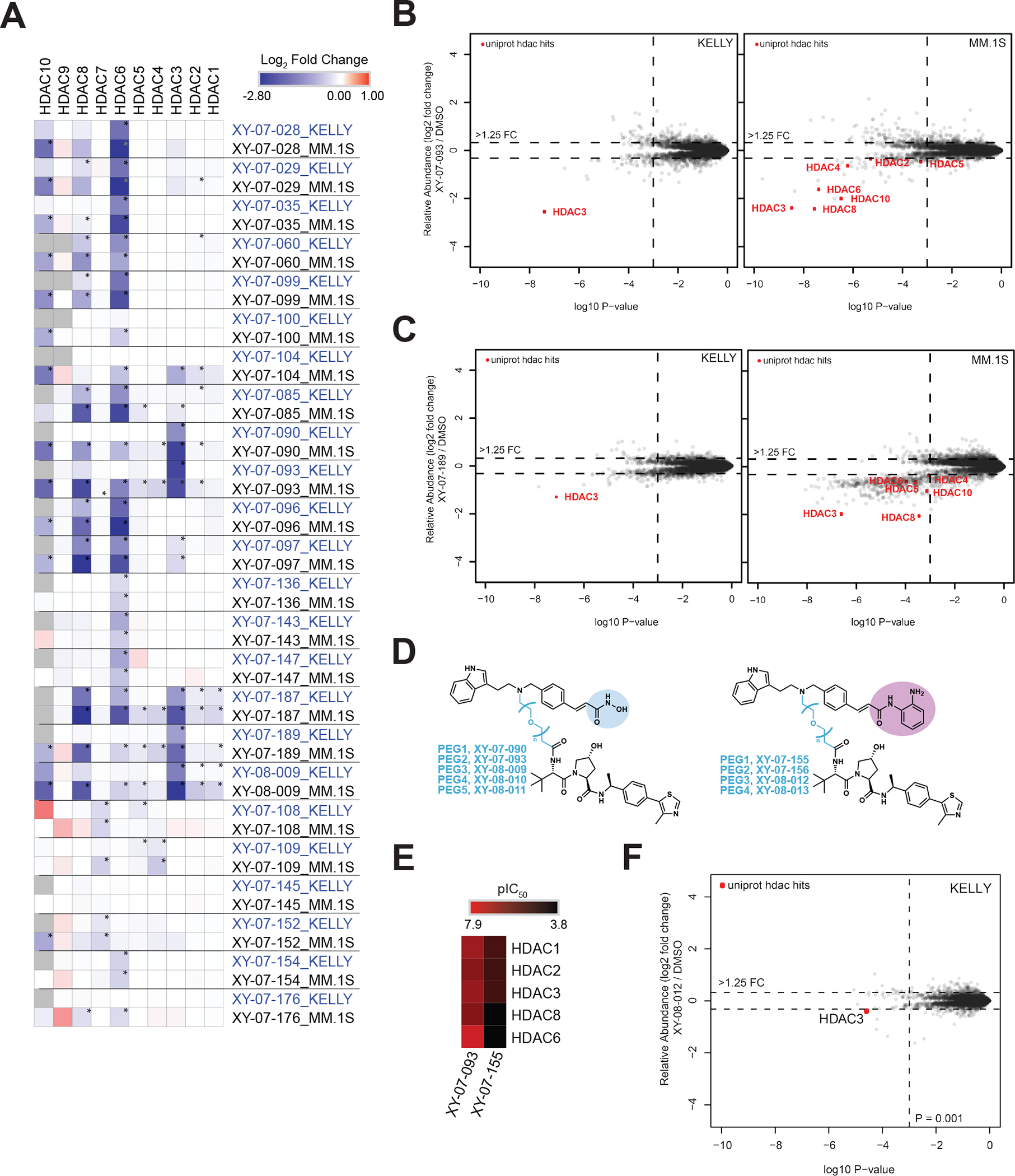
Figure 3 |. Ligase and linker influences degrader selectivity.
(A) Chemical structures for dacinostat-based CRBN/VHL1/VHL2/IAP matched series of degraders (PEG2). (B) In vitro inhibition of HDAC enzymatic activity (pIC50) for the degraders in A. Inhibition data is from n = 1 ten-point titrations. (C) Heatmap displaying the log2FC in relative abundance for quantified HDACs in response to treatment with degraders in A. (D) Scatterplot comparing HDAC engagement B with HDAC degradation C for the matched degraders in A. (E) Chemical structures for two SAHA-based CRBN/VHL1 matched pairs of degraders. (F) Scatterplot comparing HDAC degradation (log2FC) for degraders in E. (G) Chemical structures for two dacinostat-based CRBN/VHL1 matched pairs of degraders. (H) Scatterplot comparing HDAC degradation (log2FC) for degraders in G. (I) Heatmap displaying the log2FC in relative abundance for quantified HDACs in response to indicated degraders assessing linker length. (J) Chemical structures for VHL1 or VHL2-based degraders. (K) Heatmap displaying the log2FC in relative abundance for quantified HDACs in response to indicated degraders. Proteomics data is from n = 1–2 biological replicates. * on proteomics heatmaps indicates a P-value < 0.001. See also Table S2–3, Figure S1, S3, S4.
To further evaluate the influence of the recruited ligase with respect to target space, we decided to focus on CRBN- and VHL1-recruiting degraders because all of the IAP-recruiting degraders in our pilot library showed pronounced cIAP or XIAP self-degradation with limited on-target HDAC degradation (Figure S1, Figure S3D–E) (Sekine et al., 2008). We selected four matched pairs of degraders for direct comparison in MM.1S cells, which have broader HDAC expression: two pairs utilizing SAHA with two different linker lengths (C3 or C7) and two dacinostat-based with two different linker lengths (PEG1 or PEG2) (Figure 3E–H). Direct comparison of the SAHA-C3 pair reveals that HDAC10 has no preference for ligase, whereas HDAC6 shows a strong preference for CRBN as the recruited ligase. Comparison of the short linker data with the longer linker counterpart identifies key differences in target profiles, with the longer linker pair picking up HDAC2 and 3 as weak targets of the VHL-recruiting degrader. In contrast to the strong CRBN preference seen by HDAC6 in the short linker pair, here, HDAC6 looks to be equally targeted by both CRBN and VHL recruiting molecules. Analogous to the SAHA pairs, a longer linker looks to broaden the target profile of the dacinostat-based degraders with the PEG2 pair picking up HDAC4 as a weak hit that was not present in the PEG1 pair. In agreement with the SAHA-C7 data, the two dacinostat pairs both reveal that HDAC3 and 10 have a clear preference for VHL as the recruited ligase and at least for HDAC3, this is in agreement with the database as a whole (Table S3), as not only are more VHL-recruiting degraders able to degrade HDAC3 (VHL = 12/16; CRBN = 2/20), but also, the fold change in degradation is consistently larger with VHL as the recruited ligase. These insights offer valuable information for guiding the prioritization of chemistry efforts for specific targets.
Linker length modifications have proven to be an effective strategy for altering the selectivity profile of degraders (Nowak et al., 2018; Smith et al., 2019; Zorba et al., 2018) and we already see examples of this within our HDAC dataset (Figure 3E–H). To systematically explore the effect of linker length on HDAC degradability, we synthesized two multitargeted sets of degraders based on dacinostat, and capable of recruiting different ligases (CRBN and VHL). Within each of these sets we included molecules spanning PEG1 – 5 linker space and first assessed intracellular ligase engagement to confirm that cell permeability was comparable across linker lengths (Figure S4A–B). Similar to the ligase assessment above, we found that the VHL-recruiting series favored HDAC3 degradation, and the CRBN-recruiting series favored HDAC6 and 8 degradation (Figure 3I, Table S3). Comparison of each series reveals key differences induced by changes in linker length. Remarkably, HDAC degradation activity is completely lost with the PEG4 linker which is surprising because compounds with linker lengths on either side of this (PEG3 and PEG5) are potent and active degraders. Similarly, HDAC degradation activity is lost with the longest linker (PEG5) in the CRBN-set. This data clearly demonstrates how minor changes to the chemical scaffold can have drastic implications to the degradation activity of these molecules (Figure 3I). We noted above that the position of linker attachment on the VHL-recruiting molecule can have some influence upon the degradation profile of the dacinostat series. This initial comparison was performed in KELLY cells and due to low expression levels of a couple of HDACs and the stochastic sampling of global proteomics, we were limited to making comparisons between the 8 HDAC isoforms that were quantified in this experiment. To explore this effect more broadly, we compared two VHL1-linked degraders (C5 and the longer PEG2) with their VHL2-linked matched counterparts (Figure 3J) in MM.1S cells, a cell line that more reliably allows quantification of HDAC isoforms 1 – 10. Direct comparison of the degradation profiles of these molecules showed potent and consistent degradation of HDAC3, 8 and 10 and weak targeting of HDAC5 (0.36 to 0.51 log2FC down) across all four degraders (Figure 3K, Table S3). In addition, all degraders except the VHL1-C5 molecule also show the ability to target HDAC4 and 6 for degradation, but with the C5 degraders showing a stark contrast in their HDAC6 degradation intensity (2.4 log2FC down for VHL2 vs 0.4 log2FC down for VHL1). Overall this data shows some variation in the isoform selectivity profile in response to changes in linker attachment point.
Cell identity influences degrader selectivity profile
Typically, degraders are tested for on-target activity and efficacy in therapeutically relevant cell lines, which can provide a limited view of the full target scope of a discovery-phase molecule because different lineages express different proteins. Studies have also suggested that the local concentration of the two binary binding partners can impact target degradability (Sievers et al., 2018). This is exemplified by the degradation data for the VHL-PEG2 degraders profiled in KELLY cells (Figure 3C) compared to MM.1S cells (Figure 3K), which uncovers a significant difference of the isoform degradation profile across cell lines where these two VHL-PEG2 degraders induce strong and selective HDAC3 degradation in KELLY cells (Figure 3C), but show more broad isoform targeting in MM.1S cells with degradation of HDAC3–6, 8 and 10 (Figure 3K). To our knowledge, there are no prior reports of HDAC degraders having increased multitargeted activity in MM.1S cells, and from a more general sense, there is an absence of data exploring TPD degradation targets that are cell line dependent.
To investigate cell line dependency further, we compared the HDAC degradation profiles of a variety of selected molecules in both KELLY and MM.1S cell lines (Figure 4A) and generally observed more potent HDAC degradation in MM.1S cells. This observation supports published reports that HDAC6 degraders have increased potency in MM.1S cells compared to other tested lineages (An et al., 2019; Xiao and Zhang, 2020). We also noted stark differences in the HDAC target profile between these cell lines, with a significant expansion of the number of HDACs targeted in MM.1S cell experiments for several different molecules. The most striking shift in target profile was observed with compounds XY-07-104 (KELLY-0 targets, MM.1S-4 targets), XY-07-090 (KELLY-1, MM.1S-4), XY-07-093 (KELLY-1, MM.1S-7), XY-07-189 (KELLY-1, MM.1S-6) and XY-08–009 (KELLY-1, MM.1S-7) (Figure 4A, Table S3). A limitation of this comparison is the difference in isoform expression between these two lineages which causes HDAC9 and 10 to be absent from quantification in some KELLY cell experiments (Table S1). However, when these two HDACs are excluded from comparison, the shift in target profile is still apparent. To address whether target expression could be contributing to this difference in isoform degradation, we explored previously published relative protein expression data (Donovan et al., 2020), but found no evidence to suggest that target expression is driving cell line specific degradation. When expanding beyond HDAC degradation to include the global proteome response to these molecules in the two tested cell lines (Figure 4B–C, Figure S1, Table S2, S3), we observed increased proteome disruption in MM.1S cells, especially for those molecules with broad isoform activity, which supports reports of increased potency of these molecules in myeloma (An et al., 2019; Xiao and Zhang, 2020).
Figure 4 |. Cell line dependency and modular design of HDAC3-targeted degraders.
(A) Heatmap displaying the log2FC in relative abundance for HDACs in response to treatment with indicated degraders in KELLY and MM.1S cell lines. (B) Scatterplots depicting the log2FC in relative protein abundance in response to treatment with XY-07-093 in KELLY and MM.1S cells. (C) As in B but treated with XY-07-189. (D) Chemical structures of dacinostat-VHL1 degraders with hydroxamate or benzamide zinc-binding groups. (E) Heatmap displaying HDAC in vitro enzymatic inhibition data (pIC50) comparing HDAC selectivity for hydroxamate- (XY-07-093) and benzamide-based (XY-07-155) molecules. Inhibition data is from n = 1 ten-point titrations. (F) Scatterplot depicting the log2FC in relative protein abundance in response to treatment with XY-08–012. Proteomics data is from n = 1–2 biological replicates. * on proteomics heatmaps indicates a P-value < 0.001. See also Table S2–3, Figure S1, S3, S4.
The increased proteome disturbance in response to pan-HDAC targeting prompted us to perform cell viability assays with selected molecules to investigate whether viability decreases in relation to degradation of specific targets. We found that three of the molecules induced potent cell killing (Figure S4C), but due to the varied target profile including HDAC6 selective (XY-07-143) to multi-targeted (XY-07-187), we were unable to identify a single degraded HDAC that could be responsible for the reduced viability. We do note that the observed cytotoxicity may be due to the HDAC inhibition of essential HDACs due to the pan-HDAC warhead.
Modular design of HDAC3-targeted degraders
Through initial profiling of the pilot HDAC-targeting library, we observed a strong HDAC3 degradation preference for VHL ligase and identified a series of dacinostat-based, VHL-recruiting degraders such as XY-07-093 that were able to induce strong and selective degradation of HDAC3 (Figure 4B). However, this selectivity appeared to be cell line dependent (KELLY cells) as when this degrader was tested in a second cell line, MM.1S cells, the degradation profile revealed additional HDAC targets.
Studies have shown that in addition to the zinc-chelating mechanism of hydroxamate-containing inhibitors, benzamide-based inhibitors also form additional interactions with the pocket adjacent to the zinc binding site (Becher et al., 2014), which is thought to lead to their greater selectivity towards class I HDAC isoforms (Hu et al., 2003; Khan et al., 2008). To improve selectivity, we used the dacinostat-VHL compound series as the basis for design and substituted the hydroxamate group with a benzamide group, and built a miniseries of four degraders with different linker lengths (PEG1 – 4) (Figure 4D). We hypothesized that the degradation preference of VHL (selectivity for HDAC3 over HDAC1 and 2), coupled with a class I selective HDAC binder would result in a selective HDAC3 degrader. Activity profiling of the dacinostat-hydroxamate (XY-07-093) and the dacinostat-benzamide (XY-07-155) degraders revealed a significant difference in the in vitro binding affinity profile, with the benzamide molecule exhibiting about 10-fold higher IC50 for HDAC1, 2 and 3 and no measurable inhibition of HDAC6 and 8, the major degradation off-target for the hydroxamate compound XY-07-093 (Figure 4E, Figure S3A, Table S3). Chemo-proteomic profiling of this series found that although the molecules exhibit HDAC3 isoform selectivity, degradation is weak and only apparent at higher treatment concentrations (5 μM) (Figure 4F, Table S2–3), which could be a consequence of the reduced affinity for HDAC3 compared to the hydroxamates (Figure 4E, Figure S3A, Table S3). Using an HDAC8 reporter assay, we then confirmed that unlike the hydroxamate series, the new benzamide-series of molecules has no degradation activity towards HDAC8 (Figure S4E).
DISCUSSION
Targeted protein degradation is a novel therapeutic modality with rapidly expanding utility in chemical biology and therapeutics development. In this study, we focused on systematically exploring the degradability of an epigenetic family of HDAC proteins.
For decades, the HDAC family has been the focus of many drug discovery campaigns due to their important role in regulating gene expression, but obtaining selective chemical probes and therapeutics has been difficult. Although true selectivity of these degraders includes the selectivity of both inhibition and degradation, degraders offer an opportunity for increased selectivity by taking advantage of PPI for degradation selectivity and sub-stochiometric degradation allowing lower intracellular drug concentrations to reduce inhibition effects of binary target engagement. Degraders also enable drugging of non-enzymatic functions, such as the scaffolding functions that are an important role of some HDACs that act as key components of multiprotein complexes. We thoroughly explored the chemical space surrounding four pan-HDAC inhibitors as the target binding ligands to create a compound matrix capable of mapping the degradability of the HDAC family. Curating the data across the 101 independent proteomics treatments included in this study, we observed degradation of 9 HDACs (HDAC1–8, 10). We identified HDAC6, 3, and 8 as the most frequently degraded HDACs, providing advanced lead molecules for these targets. We identified multiple degraders for novel degradation targets, HDAC5, 7 and 10, confirming isoform degradability while providing valuable initial molecules for the development of more selective probes. We also identified HDAC1, 4 and 2 as infrequently degraded and HDAC9 as not degraded at all, despite the library of degraders having demonstrated broad inhibition profiles covering HDAC1–10. These findings suggests that these isoforms may be more intractable as degradation targets and supports previous studies reporting that binary binding plays an inconsistent role in the efficacy of degraders (Bondeson et al., 2018; Donovan et al., 2020; Gadd et al., 2017; Huang et al., 2018). We assessed the relationship between degradability and reported half-life in cells and we did note that HDAC3, 6 and 8 were consistently amongst the HDACs with the longest half-life and these were also found to be the most degradable HDACs in this study (Savitski et al., 2018; Zecha et al., 2018). However, the lack of clear correlation across all HDACs suggests turnover rate is not responsible for the tractability of targets. Unfortunately, it is difficult to rationalize why specific targets may seem recalcitrant to degradation, and although our chemistry effort encompassed extensive profiling of the accessible degrader space, further experimental investigation into factors such as lysine availability, ternary complex formation, cell line and treatment time, would be required to assess the impact of different factors that could be obstructing degradation of these targets, all of which is beyond the scope of this manuscript.
Since many HDAC isoforms commonly exist within large multi-protein complexes, we explored the use of our HDAC-targeted degraders for targeting the non-HDAC components of these complexes through indirect recruitment to the ligase. We observed a significant number of degraders able to induce downregulation of at least one of the complex components known to bind to the corresponding HDAC, such as the NCoR/SMRT corepressor complex, alongside the degradation of HDAC3. We observed that similar to HDAC degradation, changing the recruited ligase to cIAP, not only changes the HDAC isoform selectivity to HDAC6, but also shifts the complexes degraded to CoREST. We propose that this collateral degradation could be occurring through one of two possible mechanisms. In the first scenario, the ligase is recruited into proximity of the HDAC-containing complex where the binding partners are directly ubiquitinated. Alternatively, the ligase is recruited to the HDAC-containing complex resulting in HDAC degradation which induces destabilization of the complex core and proteasomal clearance. Although multiple components of various different HDAC-containing complexes are degraded across our study, we do note that in some instances, we do not see degradation of the corresponding HDAC isoforms. HDAC1 and 2 are key examples that are known to be major components of multiple different corepressor complexes such as CoREST, MiDAC, NuRD and Sin3. However, we often observe degradation of complex partners in absence of strong HDAC1 or 2 degradation. These data imply that complex degradation may be occurring independent of HDAC degradation, and also offers the possibility that some complex-occupied HDAC isoforms might be less susceptible to methods of TPD due to reduced target binding, inefficient ternary complex formation or even obstruction of available lysines as a result of existing within in a multi-protein complex. While this study provides the first known report of collateral degradation through targeting HDACs, there have been three recent reports confirming bystander degradation of the PRC2 (Hsu et al., 2020), BAF (Farnaby et al., 2019) and NuRD complexes (Vinogradova et al., 2020) complexes which provides four independent examples of collateral degradation that are all linked to chromatin-associated complexes. These examples of bystander targeting shed light on new potential mechanisms of regulating gene expression via differential degradation of corepressor complexes as a mechanism of controlling chromatin structure.
SIGNIFICANCE
HDACs represent a key set of epigenetic enzymes and an important target class for novel therapies. Currently available HDAC inhibitors although effective in approved indications, have strong off-target toxicities and adverse effects which are thought to be a result of the limited selectivity of these molecules. Small molecule mediated degradation can be an effective strategy to overcome the target selectivity limitations of traditional inhibitors. Here, we take a family-centric top-down approach using pan-HDAC inhibitors as the basis of degrader design to explore the degradability of the HDAC family. We collected proteome-wide expression data for 48 pan-HDAC targeting degraders and created a degradability database containing both the positive and negative data required to facilitate predictions and guiding principles for targeting the HDAC family. These findings create a valuable resource for guiding the development of novel degrader-based therapeutics, while also uncovering important information about the biological role of HDACs.
STAR★Methods
RESOURCES AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric S. Fischer (Eric_Fischer@DFCI.HARVARD.EDU).
Materials Availability
Small molecules described in this study will be made available on request, upon completion of a Materials Transfer Agreement.
Data and Code Availability
The raw proteomics datasets generated during this study are available at PRIDE accession: PXD023630; PXD023652; PXD023583; PXD026272; PXD023627; PXD026276; PXD023633; PXD023636; PXD026278; PXD026277; PXD026430; PXD026398; PXD026397 and will be publicly available as of the date of publication.
Proteomics data generated during this study are also available in our custom online database webtool: https://proteomics.fischerlab.org/
The code necessary to reproduce the statistical analysis for quantitative proteomics can be found at DOI: https://doi.org/10.5281/zenodo.5020560
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
All cells were grown in a 37 °C incubator with 5% CO2 and all cell treatments were performed between passage 10 and passage 22. MM.1S cells: Human B lymphoblastic cells from immunoglobulin A lambda myeloma (female, 42 year) were cultured in RPMI-1640 media including 2 mM L-glutamine (Gibco) and supplemented with 10% fetal bovine serum (Gibco). KELLY cells: Human brain cells from neuroblastoma were cultured in RPMI-1640 media including 2 mM L-glutamine (Gibco) and supplemented with 10% fetal bovine serum (Gibco). HEK293T cells: Human embryonic kidney cells (fetus) were cultured in DMEM media including 2 mM L-glutamine (Gibco) and supplemented with 10% fetal bovine serum (Gibco).
METHOD DETAILS
Western blot
KELLY cells were seeded 0.35 × 106 cells/mL in a 12-well cell culture plate and the next day they were treated for 24 hours with DMSO, XY-07-035, XY-07-187, XY-07-097 or XY-07-090 at the indicated concentrations. Cell pellets were lysed with a combination of the addition of 45 μL of M-PER™ Mammalian Protein Extraction Reagent Upon and thermo-mixing before running on a 10% Mini-PROTEAN TGX Precast Protein gels (Bio-rad). Protein was transferred to PVDF membranes using the iBlot 2.0 dry blotting system (Thermo Fisher Scientific). Membranes were blocked with Intercept blocking solution (LiCor) and incubated with primary antibodies overnight at 4 °C, followed by three washes with TBST and incubation with secondary antibodies for 1 hour in the dark. The membrane was washed three final times in TBST and imaged on the Odyssey LCx imaging system (LiCor).
Cell viability assays
Eight thousand KELLY cells were seeded in 50 μL of media per well in a 384-well plate. The next day, cells were treated with DMSO or 10 concentrations of indicated compound. Cells were treated for 24 hours and the cell viability assay was performed according to the manufacturers protocol (CellTiter Glo, Promega). Survival curves were plotted using a nonlinear regression fit in model Prism 8 (GraphPad Software).
Competitive displacement assay for cellular CRBN and VHL engagement
HEK293T cells stably expressing the BRD4BD2-GFP with mCherry reporter were seeded at 30 – 50% confluency in 384-well plates with 50 μL FluoroBrite DMEM media (Thermo Fisher Scientific A18967) containing 10% FBS per well a day before compound treatment. Degrader titrations and 100 nM dBET6 or 250 nM AT1 were dispensed using a D300e Digital Dispenser (HP), normalized to 0.5% DMSO, and incubated with cells for 5 h. Assay plates were imaged using Acumen (TTP Labtech) as described above. Experiments were performed in duplicates and the values for the concentrations that lead to a 50% increase in BRD4BD2-eGFP accumulation (EC50) were calculated using a nonlinear regression fit model in Prism 8 (GraphPad Software).
HDAC8 cellular degradation assays
HEK293T cells stably expressing the full length human HDAC8-EGFP with mCherry reporter in Cilantro2 (Sievers et al., 2018) vector were seeded at 30–50% confluency in 384-well plates with 50 μL FluoroBrite DMEM media (Thermo Fisher Scientific) containing 10% FBS per well a day before compound treatment. Compounds were dispensed using a D300e Digital Dispenser (HP), normalized to 0.5% DMSO, and incubated with cells for 5 h. The assay plate was imaged immediately using an Acumen High Content Imager (TTP Labtech) with 488 nm and 561 nm lasers in 2 μm × 1 μm grid per well format. The resulting images were analyzed using CellProfiler (Carpenter et al., 2006). A series of image analysis steps (‘image analysis pipeline’) was constructed. First, the red and green channels were aligned and cropped to target the middle of each well (to avoid analysis of heavily clumped cells at the edges), and a background illumination function was calculated for both red and green channels of each well individually and subtracted to correct for illumination variations across the 384-well plate from various sources of error. An additional step was then applied to the green channel to suppress the analysis of large auto fluorescent artifacts and enhance the analysis of cell specific fluorescence by way of selecting for objects under a given size, 30 A.U., and with a given shape, speckles. mCherry-positive cells were then identified in the red channel filtering for objects between 8–60 pixels in diameter and using intensity to distinguish between clumped objects. The green channel was then segmented into GFP positive and negative areas and objects were labeled as GFP positive if at least 40% of it overlapped with a GFP positive area. The fraction of GFP-positive cells/mCherry-positive cells in each well was then calculated, and the green and red images were rescaled for visualization. The values for the concentrations that lead to a 50% degradation (DC50) were calculated using the nonlinear fit variable slope model in Prism 8 (GraphPad Software).
HDAC in vitro enzymatic inhibition assay
The in vitro HDAC enzymatic assays were performed by Reaction Biology (Devault, PA) (Howitz, 2015). In summary, HDACs were incubated with indicated HDAC-targeted degraders, HDAC inhibitors or DMSO controls and class-specific acetylated AMC-labeled peptides added to initiate the reaction. The reaction was quenched by addition of a developer containing trypsin to digest deacetylated peptide substrates. As the AMC fluorophore was cleaved, the resulting fluorescence was monitored at 460 nm (λex – 360 nm) (Wang et al., 2017; Wegener et al., 2003). Data was graphed and IC50 calculated using the nonlinear fit variable slope model in Prism 8 (GraphPad Software).
TMT LC-MS3 mass spectrometry
Cells were treated with DMSO (biological triplicate) or degrader at indicated dose and time (Table S1) and cells were harvested by centrifugation. Lysis buffer (8 M Urea, 50 mM NaCl, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (EPPS) pH 8.5, Protease and Phosphatase inhibitors) was added to the cell pellets and homogenized by 20 passes through a 21-gauge (1.25 in. long) needle to achieve a cell lysate with a protein concentration between 1 – 4 mg/mL. A bradford (Bio-Rad) was used to determine the final protein concentration in the cell lysate. 100 – 200 μg of protein for each sample was reduced, alkylated and precipitated using methanol/chloroform as previously described (Donovan et al., 2018) and the resulting washed precipitated protein was allowed to air dry. Precipitated protein was resuspended in 4 M Urea, 50 mM HEPES pH 7.4, followed by dilution to 1 M urea with the addition of 200 mM EPPS, pH 8. Proteins were first digested with LysC (1:50; enzyme:protein) for 12 h at RT. The LysC digestion was diluted to 0.5 M Urea with 200 mM EPPS pH 8 followed by digestion with trypsin (1:50; enzyme:protein) for 6 h at 37 °C. Tandem mass tag (TMT) reagents (Thermo Fisher Scientific) were dissolved in anhydrous acetonitrile (ACN) according to manufacturer’s instructions. Anhydrous ACN was added to each peptide sample to a final concentration of 30% v/v, and labeling was induced with the addition of TMT reagent to each sample at a ratio of 1:4 peptide:TMT label. The 11 or 16-plex labeling reactions were performed for 1.5 h at RT and the reaction quenched by the addition of hydroxylamine to a final concentration of 0.3% for 15 minutes at RT. Each of the sample channels were combined in a 1:1 ratio, desalted using C18 solid phase extraction cartridges (Waters) and analyzed by LC-MS for channel ratio comparison. Samples were then combined using the adjusted volumes determined in the channel ratio analysis and dried down in a speed vacuum. The combined sample was then resuspended in 1% formic acid and acidified (pH 2 – 3) before being subjected to desalting with C18 SPE (Sep-Pak, Waters). Samples were then offline fractionated into 96 fractions by high pH reverse-phase HPLC (Agilent LC1260) through an aeris peptide xb-c18 column (phenomenex) with mobile phase A containing 5% acetonitrile and 10 mM NH4HCO3 in LC-MS grade H2O, and mobile phase B containing 90% acetonitrile and 10 mM NH4HCO3 in LC-MS grade H2O (both pH 8.0). The 96 resulting fractions were then pooled in a non-contiguous manner into 24 fractions and desalted using C18 solid phase extraction plates (SOLA, Thermo Fisher Scientific) followed by subsequent mass spectrometry analysis.
Data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with a Proxeon EASY-nLC 1200 LC pump (Thermo Fisher Scientific) or an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with an UltiMate 3000 RSLCnano System. Peptides were separated on an EasySpray ES803 or ES903 75 μm inner diameter microcapillary column (Thermo Fisher Scientific). Peptides were separated using a 190 min gradient of 6 – 27% acetonitrile in 1.0% formic acid with a flow rate of 300 nL/min.
Each analysis used a MS3-based TMT method as described previously (McAlister et al., 2014). The data were acquired using a mass range of m/z 340 – 1350, resolution 120,000, AGC target 5 × 105, maximum injection time 100 ms, dynamic exclusion of 120 seconds for the peptide measurements in the Orbitrap. Data dependent MS2 spectra were acquired in the ion trap with a normalized collision energy (NCE) set at 35%, AGC target set to 1.8 × 104 and a maximum injection time of 120 ms. MS3 scans were acquired in the Orbitrap with HCD collision energy set to 55%, AGC target set to 2 × 105, maximum injection time of 150 ms, resolution at 50,000 and with a maximum synchronous precursor selection (SPS) precursors set to 10.
LC-MS data analysis
Proteome Discoverer 2.2 or 2.4 (Thermo Fisher Scientific) was used for the processing of .RAW files and controlling peptide and protein false discovery rates (FDR 1%). The MS/MS spectra were searched against a Swissprot human database (September 2017 or December 2019) with both the forward and reverse protein sequences and supplemented with the sequences of common contaminants such as keratin. The database searches were performed with a precursor mass tolerance of 10 ppm and a fragment ion mass tolerance of 0.6 Da. Additional search criteria included: tryptic peptides with two missed cleavages, static alkylation of cysteine (57.02146 Da), static TMT labelling of lysine residues and N-termini of peptides (229.16293 Da for TMT11plex or 304.2071 for TMTpro), and variable oxidation of methionine (15.99491 Da). TMT reporter ion intensities were measured using a 0.003 Da window around the theoretical m/z for each reporter ion in the MS3 scan. Peptide spectral matches with poor quality MS3 spectra were excluded from quantitation (summed signal-to-noise across channels < 100 and precursor isolation specificity < 0.5), and resulting data was filtered to only include proteins that had a minimum of 2 unique peptides quantified. Reporter ion intensities were normalized and scaled using in-house scripts in the R framework (R Development Core Team, 2014), followed by statistical analysis using the limma package within the R framework (Ritchie et al., 2015).
QUANTIFICATION AND STATISTICAL ANALYSIS
All biochemical curves and associated statistical analyses were produced using Prism 8 (GraphPad Software). Details of replicates and data analysis for specific experiments can be found in the figure legends, Table S1 or in the methods section. The reported mean is equivalent to the average of the values determined for each of the replicates and the reported standard deviation is a measure of the variance relative to the determined mean. All proteomics statistical analysis was performed using the limma package within the R framework (Ritchie et al., 2015).
CHEMISTRY GENERAL METHODS
Unless otherwise noted, reagents and solvents were used as received from commercial suppliers. All reactions were monitored using a Waters Acquity UPLC/MS system using an Acquity UPLC® BEH C18 column (2.1 × 50 mm, 1.7 μm particle size), UPLC method A: solvent gradient = 80% A at 0 min, 5% A at 1.8 min; method B: solvent gradient = 100% A at 0 min, 5% A at 1.8 min; solvent A = 0.1% formic acid in H2O; solvent B = 0.1% formic acid in acetonitrile; flow rate: 0.6 mL/min; or an Agilent LC/MS system (Agilent 1200LC/G6130A MS) using SunFire™ C18 column (4.6 × 50 mm, 3.5 μm particle size), LC method: solvent gradient = 95% A to 5% A; solvent A = 0.01% TFA in Water; solvent B = 0.01% TFA in ACN; flow rate: 2.0 mL/min, column temperature 50 °C. Purification of reaction products was carried out by flash chromatography using CombiFlash®Rf with Teledyne Isco RediSep® normal-phase silica flash columns; or Waters HPLC system using SunFireTM C18 column (19 × 100 mm, 5 μm particle size): solvent gradient 0% to 100% acetonitrile or MeOH in H2O (0.035% TFA as additive); flow rate: 20 mL/min, or SunFireTM C18 column (30 × 250 mm, 5 μm particle size): solvent gradient 0% to 100% acetonitrile or MeOH in H2O (0.035% TFA as additive); flow rate: 40 mL/min. All compounds had a resulting purity of over 95% as analyzed by a Waters UPLC system. 1H NMR and 13C NMR spectra were obtained using Bruker Avance III spectrometers (400 MHz or 500 MHz for 1H, and 125 MHz for 13C). Chemical shifts are reported relative to deuterated methanol (δ = 3.31) or dimethyl sulfoxide (δ = 2.50) for 1H NMR. Spectra are given in ppm (δ) and as br = broad, s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet and coupling constants J are reported in Hertz. All compound synthesis and characterization details can be found in Data S1.
Supplementary Material
Table S1 | Chemical library properties and meta-data for chemo-proteomics treatments. Compound names, chemical structures, recruited ligase, linker category, HDAC binder, reported targets of incorporated binder, number and list of degradation targets and SMILES strings provided as well as meta-data for all treatments included in the degradable HDACome study. Related to Figure 1.
Table S2 | Limma tables for 101 chemo-proteomics treatments included in the degradable HDACome dataset. Related to Figure 1.
Table S3 | Degradable HDACome database, degradability data and assessment of cellular and chemical variables on HDAC degradability. Log2 Fold Change and P-value for all proteins in response to each of the 101 treatments included in the degradable HDACome database. In vitro enzymatic inhibition data, ligase-recruiter comparisons, and linker and cell line comparisons included in the degradable HDACome dataset. Related to Figure 1–4.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HDAC3 (1:500) | Abcam | Cat# ab32369 RRID: AB_732780 |
| Anti-HDAC8 (1:1,000) | Abcam | Cat# ab187139 RRID: AB_2715505 |
| Anti-HDAC6 (1:1,000) | Cell Signaling Technology | Cat# 7558 RRID: AB_10891804 |
| Anti-GAPDH (1:10,000) | Millipore-Sigma | Cat# G8795 RRID: AB_1078991 |
| Anti-Vinculin (1:1,000) | Santa Cruz | Cat# sc-73614 RRID: AB_1131294 |
| Anti-Mouse IgG (1:10,000) | LiCor | Cat# 92668072 RRID: AB_10953628 |
| Anti-Rabbit IgG (1:10,000) | LiCor | Cat# 92632211 RRID: AB_621843 |
| Chemicals, peptides, and recombinant proteins | ||
| cOmplete, Mini Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# 11836153001 |
| PhosSTOP Phosphatase Inhibitor Tablets | Sigma-Aldrich | Cat# 04906837001 |
| Halt Protease and Phosphatase Inhibitor Single-Use Cocktail | Thermo Fisher Scientific | Cat# 78442 |
| dBET6 | A gift from Gray Lab, Stanford University | N/A |
| AT1 | Tocris | Cat# 6356 |
| MLN4924 | MedChemExpress | Cat# HY-70062 |
| BRD4BD2-GFP-P2a-mCherry | Fischer Lab, Dana-Farber Cancer Institute (Nowak et al., 2018) | N/A |
| HDAC8-EGFP-IRES-mCherry | Fischer Lab, Dana-Farber Cancer Institute | N/A |
| Critical commercial assays | ||
| Tandem Mass Tag (TMT) Reagents: 11plex | Thermo Fisher Scientific | Cat# A34808 |
| Tandem Mass Tag (TMT) Reagents: 16plex | Thermo Fisher Scientific | Cat# A44520 |
| Bradford Reagent | BioRad | Cat# 500-0205 |
| CellTiter Glo | Promega | Cat# G7570 |
| Histone Deacetylase in vitro inhibition assay | Reaction Biology | www.reactionbiology.com |
| Deposited data | ||
| wp-esf_110 | This paper | PXD023630 |
| wp-esf_120 | This paper | PXD023652 |
| wp-esf_126 | This paper | PXD023583 |
| wp-esf_127 | This paper | PXD026272 |
| wp-esf_130 | This paper | PXD023627 |
| wp-esf_132 | This paper | PXD026276 |
| wp-esf_155 | This paper | PXD023633 |
| wp-esf_162 | This paper | PXD023636 |
| wp-esf_178 | This paper | PXD026278 |
| wp-esf_221 | This paper | PXD026277 |
| wp-esf_229 | This paper | PXD026430 |
| wp-esf_241 | This paper | PXD026398 |
| wp-esf_242 | This paper | PXD026397 |
| Experimental models: Cell lines | ||
| MM.1S (H. sapiens) | ATCC | CRL-2974; RRID:CVCL_8792 |
| KELLY (H. sapiens) | Sigma-Aldrich | 92110411; RRID:CVCL_2092 |
| HEK293T cells (H. sapiens) | ATCC | CRL-3216; RRID:CVCL_0063 |
| Recombinant DNA | ||
| Cilantro2 | (Sievers et al., 2018) | Addgene Cat# 74450 |
| Software and algorithms | ||
| Proteome Discoverer 2.2, 2.4 | Thermo Fisher Scientific | RRID: SCR_014477 https://thermo.flexnetoperations.com/control/thmo/login |
| R Framework | Team RCR: A Language and Environment for Statistical Computing | http://www.R-project.org/ |
| Statistical Analysis Limma Package (R framework) | Ritchie et al. (2015) Nucleic Acids Res. | https://bioconductor.org/packages/release/bioc/html/limma.html |
| Prism 8 | GraphPad | RRID: SCR_002798 http://www.graphpad.com/ |
Highlights:
Pan-HDAC degrader library enabled chemo-proteomics map of HDAC degradability
HDAC degradation results in degradation of HDAC-containing repressive complexes
Cell line dependent HDAC isoform specificity
ACKNOWLEDGEMENTS
Financial support for this work was provided by the Dana-Farber/Novartis Drug Discovery Program (grant to E.S.F).
Abbreviations:
- DIEA
diisopropylethyamine
- DMSO
dimethyl sulfoxide
- HATU
hexafluorophosphate azabenzotriazole tetramethyl uronium
- DCM
dichloromethane
- DMF
dimethylformamide
- EtOAc
ethyl acetate
- THF
tetrahydrofuran
- EDCI
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- HOBt
hydroxybenzotriazole
- DMAP
4-dimethylaminopyridine
- TFA
trifluoroacetic acid
- THP
tetrahydropyranyl ether
Footnotes
DECLARATION OF INTERESTS
K.A.D. is a consultant to Kronos Bio. E.S.F is a founder, science advisory board member and equity holder in Civetta, Jengu (board member), and Neomorph, an equity holder in C4 Therapeutics, and a consultant to Astellas, Novartis, Deerfield, Sanofi and EcoR1 capital. The Fischer lab receives or has received research funding from Novartis, Astellas, Ajax, and Deerfield. K.A.D, Y.X, N.A.E and E.S.F are inventors on 4 patent applications relating to this work owned by DFCI.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An Z, Lv W, Su S, Wu W, and Rao Y (2019). Developing potent PROTACs tools for selective degradation of HDAC6 protein. Protein & cell 10, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, et al. (2011). Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol 29, 255–265. [DOI] [PubMed] [Google Scholar]
- Becher I, Dittmann A, Savitski MM, Hopf C, Drewes G, and Bantscheff M (2014). Chemoproteomics reveals time-dependent binding of histone deacetylase inhibitors to endogenous repressor complexes. ACS Chem Biol 9, 1736–1746. [DOI] [PubMed] [Google Scholar]
- Bondeson DP, and Crews CM (2017). Targeted protein degradation by small molecules. Annual review of pharmacology and toxicology 57, 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson DP, Mares A, Smith IE, Ko E, Campos S, Miah AH, Mulholland KE, Routly N, Buckley DL, and Gustafson JL (2015). Catalytic in vivo protein knockdown by small-molecule PROTACs. Nature chemical biology 11, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson DP, Smith BE, Burslem GM, Buhimschi AD, Hines J, Jaime-Figueroa S, Wang J, Hamman BD, Ishchenko A, and Crews CM (2018). Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell chemical biology 25, 78–87.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, and Mazitschek R (2010). Chemical phylogenetics of histone deacetylases. Nature chemical biology 6, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem GM, and Crews CM (2020). Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell 181, 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman JW, Donovan KA, Schauer NJ, Liu X, Hu W, Varca AC, Buhrlage SJ, and Fischer ES (2020). Proteomics-Based Identification of DUB Substrates Using Selective Inhibitors. Cell Chemical Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, de Weerd S, Chen D, Zwinderman MRH, van der Wouden PE, and Dekker FJ (2020). Induced protein degradation of histone deacetylases 3 (HDAC3) by proteolysis targeting chimera (PROTAC). Eur J Med Chem 208, 112800. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, and Moffat J (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HP, Zhao YT, and Zhao TC (2015). Histone deacetylases and mechanisms of regulation of gene expression. Critical Reviews™ in Oncogenesis 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang D-M, Leng Y, Marinova Z, Kim H-J, and Chiu C-T (2009). Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends in neurosciences 32, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm PM, Samarasinghe KTG, Hines J, and Crews CM (2018). Addressing Kinase-Independent Functions of Fak via PROTAC-Mediated Degradation. Journal of the American Chemical Society. [DOI] [PubMed] [Google Scholar]
- Cyrus K, Wehenkel M, Choi E-Y, Han H-J, Lee H, Swanson H, and Kim K-B (2011). Impact of linker length on the activity of PROTACs. Molecular BioSystems 7, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derwish R, Paterno GD, and Gillespie LL (2017). Differential HDAC1 and 2 Recruitment by Members of the MIER Family. PLoS One 12, e0169338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KA, An J, Nowak RP, Yuan JC, Fink EC, Berry BC, Ebert BL, and Fischer ES (2018). Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KA, Ferguson FM, Bushman JW, Eleuteri NA, Bhunia D, Ryu S, Tan L, Shi K, Yue H, and Liu X (2020). Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman SE, Schumacher SG, Alland D, Nabeta P, Armstrong DT, King B, Hall SL, Chakravorty S, Cirillo DM, Tukvadze N, et al. (2018). Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis 18, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, Zollman D, Steurer S, Karolyi-Oezguer J, and Riedmueller C (2019). BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nature chemical biology 15, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust TB, Yoon H, Nowak RP, Donovan KA, Li Z, Cai Q, Eleuteri NA, Zhang T, Gray NS, and Fischer ES (2020). Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nature chemical biology 16, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, and Verdin E (2001). Human HDAC7 Histone Deacetylase Activity Is Associated with HDAC3in Vivo. Journal of Biological Chemistry 276, 35826–35835. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, and Verdin E (2002). Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Molecular cell 9, 45–57. [DOI] [PubMed] [Google Scholar]
- Gadd MS, Testa A, Lucas X, Chan K-H, Chen W, Lamont DJ, Zengerle M, and Ciulli A (2017). Structural basis of PROTAC cooperative recognition for selective protein degradation. Nature chemical biology 13, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, Marchevsky A, Mckenna R, Taguchi H, and Koeffler HP (2007). Epigenetic silencing of the candidate tumor suppressor gene Per1 in non–small cell lung cancer. Clinical cancer research 13, 1399–1404. [DOI] [PubMed] [Google Scholar]
- Ho TC, Chan AH, and Ganesan A (2020). Thirty years of HDAC inhibitors: 2020 insight and hindsight. Journal of Medicinal Chemistry 63, 12460–12484. [DOI] [PubMed] [Google Scholar]
- Howitz KT (2015). Screening and profiling assays for HDACs and sirtuins. Drug Discovery Today: Technologies 18, 38–48. [DOI] [PubMed] [Google Scholar]
- Hsu JH-R, Rasmusson T, Robinson J, Pachl F, Read J, Kawatkar S, O’Donovan DH, Bagal S, Code E, and Rawlins P (2020). EED-targeted PROTACs degrade EED, EZH2, and SUZ12 in the PRC2 complex. Cell Chemical Biology 27, 41–46. e17. [DOI] [PubMed] [Google Scholar]
- Hu E, Dul E, Sung C-M, Chen Z, Kirkpatrick R, Zhang G-F, Johanson K, Liu R, Lago A, and Hofmann G (2003). Identification of novel isoform-selective inhibitors within class I histone deacetylases. Journal of Pharmacology and Experimental Therapeutics 307, 720–728. [DOI] [PubMed] [Google Scholar]
- Huang HT, Dobrovolsky D, Paulk J, Yang G, Weisberg EL, Doctor ZM, Buckley DL, Cho JH, Ko E, Jang J, et al. (2018). A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell chemical biology 25, 88–99.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, and Carey N (2008). Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochemical Journal 409, 581–589. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Rowe M, Ren M, Hong J-S, Chen P-S, and Chuang D-M (2007). Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: multiple mechanisms of action. Journal of Pharmacology and Experimental Therapeutics 321, 892–901. [DOI] [PubMed] [Google Scholar]
- Kwapis JL, Alaghband Y, Kramár EA, López AJ, Ciernia AV, White AO, Shu G, Rhee D, Michael CM, and Montellier E (2018). Epigenetic regulation of the circadian gene Per1 contributes to age-related changes in hippocampal memory. Nature communications 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, Hines J, and Crews CM (2016a). Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew Chem Int Ed Engl 55, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, Hines J, and Crews CM (2016b). Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew Chem Int Ed Engl 55, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao C, Zhao L, Yuan Z, Chen Y, and Jiang Y (2018). Phthalimide conjugations for the degradation of oncogenic PI3K. European journal of medicinal chemistry 151, 237–247. [DOI] [PubMed] [Google Scholar]
- Li Y, and Seto E (2016). HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harbor perspectives in medicine 6, a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, and Feeney PJ (1997). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23, 3–25. [DOI] [PubMed] [Google Scholar]
- Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, et al. (2013). Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol 9, 319–325. [DOI] [PubMed] [Google Scholar]
- Lu J, Frerich JM, Turtzo LC, Li S, Chiang J, Yang C, Wang X, Zhang C, Wu C, and Sun Z (2013). Histone deacetylase inhibitors are neuroprotective and preserve NGF-mediated cell survival following traumatic brain injury. Proceedings of the National Academy of Sciences 110, 10747–10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Martin SC, Parkington J, Cadena SM, Zhu J, Ibebunjo C, Summermatter S, Londraville N, Patora-Komisarska K, Widler L, et al. (2019). HDAC4 Controls Muscle Homeostasis through Deacetylation of Myosin Heavy Chain, PGC-1alpha, and Hsc70. Cell Rep 29, 749–763 e712. [DOI] [PubMed] [Google Scholar]
- Magupalli VG, Negro R, Tian Y, Hauenstein AV, Di Caprio G, Skillern W, Deng Q, Orning P, Alam HB, and Maliga Z (2020). HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares A, Miah AH, Smith IE, Rackham M, Thawani AR, Cryan J, Haile PA, Votta BJ, Beal AM, and Capriotti C (2020). Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Communications biology 3, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, and Olson EN (2000). Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3-3 to histone deacetylase 5. Proceedings of the National Academy of Sciences 97, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens J, Nees M, Benedetti R, Pallavicini I, and Minucci S (2017). c-Myc modulation and acetylation is a key HDAC inhibitor target in cancer. Clinical Cancer Research 23, 2542–2555. [DOI] [PubMed] [Google Scholar]
- Neklesa TK, Winkler JD, and Crews CM (2017). Targeted protein degradation by PROTACs. Pharmacology & therapeutics 174, 138–144. [DOI] [PubMed] [Google Scholar]
- Nguyen HC, Adlanmerini M, Hauck AK, and Lazar MA (2020). Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature 584, 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, et al. (2018). Plasticity in binding confers selectivity in ligand-induced protein degradation. Nature chemical biology 14, 706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phimmachanh M, Han JZ, O’Donnell YE, Latham SL, and Croucher DR (2020). Histone deacetylases and histone deacetylase inhibitors in neuroblastoma. Frontiers in Cell and Developmental Biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2014). R: A language and environment for statistical computing. (R Foundation for Statistical Computing; ). [Google Scholar]
- Remiszewski SW, Sambucetti LC, Bair KW, Bontempo J, Cesarz D, Chandramouli N, Chen R, Cheung M, Cornell-Kennon S, Dean K, et al. (2003). N-hydroxy-3-phenyl-2-propenamides as novel inhibitors of human histone deacetylase with in vivo antitumor activity: discovery of (2E)-N-hydroxy-3-[4-[[(2-hydroxyethyl)[2-(1H-indol-3-yl)ethyl]amino]methyl] phenyl]-2-propenamide (NVP-LAQ824). Journal of medicinal chemistry 46, 4609–4624. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter A.J.d., GENNIP A.H.v., Caron HN, Kemp S, and KUILENBURG A.B.v. (2003). Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochemical Journal 370, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, and Deshaies RJ (2001). Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences 98, 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Zinn N, Faelth-Savitski M, Poeckel D, Gade S, Becher I, Muelbaier M, Wagner AJ, Strohmer K, Werner T, et al. (2018). Multiplexed Proteome Dynamics Profiling Reveals Mechanisms Controlling Protein Homeostasis. Cell 173, 260–274.e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneekloth AR, Pucheault M, Tae HS, and Crews CM (2008). Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorganic & medicinal chemistry letters 18, 5904–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Takubo K, Kikuchi R, Nishimoto M, Kitagawa M, Abe F, Nishikawa K, Tsuruo T, and Naito M (2008). Small molecules destabilize cIAP1 by activating auto-ubiquitylation. Journal of Biological Chemistry 283, 8961–8968. [DOI] [PubMed] [Google Scholar]
- Seto E, and Yoshida M (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology 6, a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Nagai K, Morita Y, Ujikawa O, Ohoka N, Hattori T, Koyama R, Sano O, Imaeda Y, Nara H, et al. (2018). Development of Protein Degradation Inducers of Androgen Receptor by Conjugation of Androgen Receptor Ligands and Inhibitor of Apoptosis Protein Ligands. Journal of medicinal chemistry 61, 543–575. [DOI] [PubMed] [Google Scholar]
- Sievers QL, Petzold G, Bunker RD, Renneville A, Slabicki M, Liddicoat BJ, Abdulrahman W, Mikkelsen T, Ebert BL, and Thoma NH (2018). Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Ferguson FM, Cai Q, Donovan KA, Nandi G, Patnaik D, Zhang T, Huang H-T, Lucente DE, and Dickerson BC (2019). Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. Elife 8, e45457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinatra L, Bandolik JJ, Roatsch M, Sonnichsen M, Schoeder CT, Hamacher A, Schoeler A, Borkhardt A, Meiler J, Bhatia S, et al. (2020). Hydroxamic Acids Immobilized on Resins (HAIRs): A Toolbox for the Synthesis of Dual-Targeting HDAC Inhibitors and HDAC Degraders (PROTACs). Angew Chem Int Ed Engl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley JP, Adams GE, Millard CJ, Song Y, Norris JKS, Schwabe JWR, Cowley SM, and Hodgkinson JT (2020). PROTAC-mediated degradation of class I histone deacetylase enzymes in corepressor complexes. Chem Commun (Camb) 56, 4476–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, and Crews CM (2019). Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nature communications 10, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Yang Z, Gao H, Yang H, Zhu S, An Z, Wang J, Li Q, Chandarlapaty S, and Deng H (2019). Potent and preferential degradation of CDK6 via proteolysis targeting chimera degraders. Journal of medicinal chemistry 62, 7575–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A, O’Byrne KJ, and Richard DJ (2018). Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Frontiers in oncology 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, and Kasler HG (2003). Class II histone deacetylases: versatile regulators. TRENDS in Genetics 19, 286–293. [DOI] [PubMed] [Google Scholar]
- Vinogradova EV, Zhang X, Remillard D, Lazar DC, Suciu RM, Wang Y, Bianco G, Yamashita Y, Crowley VM, Schafroth MA, et al. (2020). An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell 182, 1009–1026 e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wallach J, Duane S, Wang Y, Wu J, Wang J, Adejare A, and Ma H (2017). Developing selective histone deacetylases (HDACs) inhibitors through ebselen and analogs. Drug design, development and therapy 11, 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener D, Wirsching F, Riester D, and Schwienhorst A (2003). A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chemistry & biology 10, 61–68. [DOI] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, and Bradner JE (2015). Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O, Deubzer H, Lodrini M, Milde T, and Oehme I (2009a). Targeting histone deacetylases in neuroblastoma. Current pharmaceutical design 15, 436–447. [DOI] [PubMed] [Google Scholar]
- Witt O, Deubzer HE, Milde T, and Oehme I (2009b). HDAC family: What are the cancer relevant targets? Cancer letters 277, 8–21. [DOI] [PubMed] [Google Scholar]
- Wu H, Yang K, Zhang Z, Leisten ED, Li Z, Xie H, Liu J, Smith KA, Novakova Z, and Barinka C (2019). Development of multifunctional histone deacetylase 6 degraders with potent antimyeloma activity. Journal of medicinal chemistry 62, 7042–7057. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Wang J, Zhao LY, Chen X, Zheng G, Zhang X, and Liao D (2020). Discovery of histone deacetylase 3 (HDAC3)-specific PROTACs. Chem Commun (Camb) 56, 9866–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, and Zhang X (2020). Recent advances in small molecular modulators targeting histone deacetylase 6. Future Drug Discovery 2, FDD53. [Google Scholar]
- Yang K, Wu H, Zhang Z, Leisten ED, Nie X, Liu B, Wen Z, Zhang J, Cunningham MD, and Tang W (2020). Development of Selective Histone Deacetylase 6 (HDAC6) Degraders Recruiting Von Hippel-Lindau (VHL) E3 Ubiquitin Ligase. ACS Med Chem Lett 11, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, and Seto E (2003). Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev 13, 143–153. [DOI] [PubMed] [Google Scholar]
- You I, Erickson EC, Donovan KA, Eleuteri NA, Fischer ES, Gray NS, and Toker A (2020). Discovery of an AKT Degrader with Prolonged Inhibition of Downstream Signaling. Cell chemical biology 27, 66–73. e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecha J, Meng C, Zolg DP, Samaras P, Wilhelm M, and Kuster B (2018). Peptide level turnover measurements enable the study of proteoform dynamics. Molecular and Cellular Proteomics 17, 974–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Xiong Y, Safaee N, Nowak RP, Donovan KA, Yuan CJ, Nabet B, Gero TW, Feru F, and Li L (2019). Exploring Targeted Degradation Strategy for Oncogenic KRASG12 C. Cell chemical biology. [DOI] [PubMed] [Google Scholar]
- Zoppi V, Hughes SJ, Maniaci C, Testa A, Gmaschitz T, Wieshofer C, Koegl M, Riching KM, Daniels DL, and Spallarossa A (2018). Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von Hippel–Lindau (VHL) based dual degrader probe of BRD9 and BRD7. Journal of medicinal chemistry 62, 699–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorba A, Nguyen C, Xu Y, Starr J, Borzilleri K, Smith J, Zhu H, Farley KA, Ding W, and Schiemer J (2018). Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proceedings of the National Academy of Sciences 115, E7285–E7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 | Chemical library properties and meta-data for chemo-proteomics treatments. Compound names, chemical structures, recruited ligase, linker category, HDAC binder, reported targets of incorporated binder, number and list of degradation targets and SMILES strings provided as well as meta-data for all treatments included in the degradable HDACome study. Related to Figure 1.
Table S2 | Limma tables for 101 chemo-proteomics treatments included in the degradable HDACome dataset. Related to Figure 1.
Table S3 | Degradable HDACome database, degradability data and assessment of cellular and chemical variables on HDAC degradability. Log2 Fold Change and P-value for all proteins in response to each of the 101 treatments included in the degradable HDACome database. In vitro enzymatic inhibition data, ligase-recruiter comparisons, and linker and cell line comparisons included in the degradable HDACome dataset. Related to Figure 1–4.
Data Availability Statement
The raw proteomics datasets generated during this study are available at PRIDE accession: PXD023630; PXD023652; PXD023583; PXD026272; PXD023627; PXD026276; PXD023633; PXD023636; PXD026278; PXD026277; PXD026430; PXD026398; PXD026397 and will be publicly available as of the date of publication.
Proteomics data generated during this study are also available in our custom online database webtool: https://proteomics.fischerlab.org/
The code necessary to reproduce the statistical analysis for quantitative proteomics can be found at DOI: https://doi.org/10.5281/zenodo.5020560
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


