Abstract
Cellular carbohydrates or glycans are critical mediators of biological function. Their remarkably diverse structures and varied activities present exciting opportunities for understanding many areas of biology. In this Primer, we discuss key methods and recent breakthrough technologies for identifying, monitoring, and manipulating glycans in mammalian systems.
Introduction
Glycans are ubiquitous and play important roles in fundamental processes ranging from neural development and cell signaling to immune regulation and host-pathogen interactions (Varki, 2017). All living cells are coated with a complex array of glycans that adorn the proteins, lipids, and even RNAs in cell membranes and cell walls. Glycosylation is one of the most common post-translational modifications. The majority of all human proteins are glycosylated, and many glycans become altered and contribute to the pathophysiology of major diseases, including cancer (Pinho and Reis, 2015), diabetes (Reily et al., 2019), Alzheimer’s disease (Haukedal and Freude, 2020), and infectious diseases such as COVID-19 (Clausen et al., 2020; Zhou and Cobb, 2021). With the advent of new technologies for studying glycans, there is a growing ability to understand glycosylation and its myriad of functions.
Glycans have historically been challenging to study compared to other major classes of biomolecules. Unlike DNA, RNA, and proteins, glycans are not produced using a specific template. Rather, their biosynthesis relies on the intrinsic specificities and spatiotemporal expression of a series of glycosyltransferases (GTs), which ultimately leads to families of related but non-equivalent glycan structures. Glycans can also be assembled into both linear and branched structures, wherein the monosaccharide building blocks are joined at one of several positions around the sugar ring with α- or β- stereochemistry at the anomeric carbon (Figure 1A). Thus, the potential structural complexity of glycans vastly exceeds that of DNA, RNA, and proteins. Due to the decentralized biosynthetic machinery that may have purposefully evolved to generate microheterogeneity and the resulting structural diversity of glycans, standardized methods used for other biopolymers like polymerase chain reaction (PCR) amplification and sequencing are not directly applicable to glycans. Moreover, the linear, stepwise nature of glycan biosynthesis limits the power of genetic methods, which disrupt not only the target glycan on multiple glycoconjugates but also other glycan structures produced in subsequent biosynthetic steps.
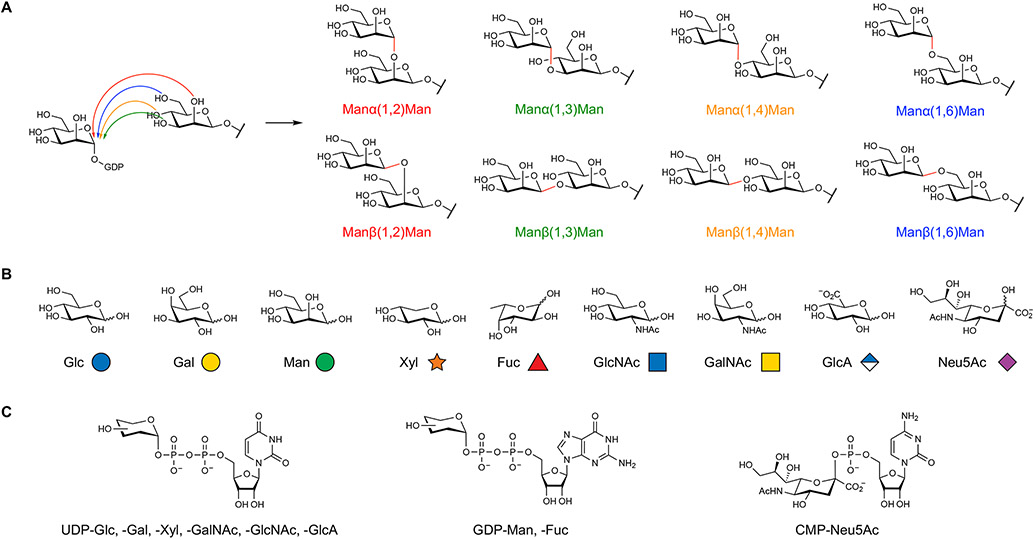
Figure 1. Chemical diversity of mammalian glycans.
(A) The native assembly of two monosaccharides leads to eight potential disaccharides, depending on the regioselectivity (e.g., 1,2 versus 1,3) and stereoselectivity (α versus β) of the newly formed glycosidic bond. In contrast, only a single structure is produced from the native assembly of dinucleotides or dipeptides.
(B) Mammalian glycans are composed of nine monosaccharides, which are pictorially represented by specific symbols from the Symbol Nomenclature for Glycans.
(C) Glycosyltransferases utilize activated nucleotide sugar donors to transfer monosaccharide units onto growing glycan chains.
Glycoscientists have addressed many of these challenges through the development of new technologies for understanding glycan function that often embrace and even exploit the structure-specific differences across glycan classes. This approach has yielded a wide-ranging set of customized tools, which have been accompanied by an enthusiasm from glycoscientists to collaborate with the broader scientific community. In this Primer, we aim to provide a critical overview and interpretation of current methods in mammalian glycobiology. Because of the sheer breadth of glycans and approaches, this Primer is by no means comprehensive. Instead, we have selected important techniques that can be used to address major questions in glycoscience research. Throughout the Primer, we will draw examples from four major classes of mammalian glycans: O-linked/N-acetylglucosamine (O-GlcNAc), O- and N-glycans, and glycosaminoglycans (GAGs). These classes exemplify both the structural diversity of carbohydrates as well as the range of approaches that can be used to study them. When possible, we also highlight other notable reviews for further reading. Glycoscience is a central field with links to all areas of biology. We hope that this Primer on the current glycoscience toolkit will inspire the broader scientific community to take on the exciting challenge of understanding glycan function and help define new roles for these exquisitely varied and complex molecules.
A Brief Guide to Glycans and Glycosylation
Glycans are a wide-ranging group of biomolecules that vary significantly in size, composition, localization, and attachment. Their structures span from single monosaccharides to elaborately branched oligosaccharides as well as long polysaccharide polymers with molecular masses >1000 kDa. In vertebrates, glycans are composed of nine main monosaccharide building blocks: glucose (Glc), galactose (Gal), xylose (Xyl), mannose (Man), fucose (Fuc), N-acetylglucosamine (GlcNAc), N-acetylgalactosamine (GalNAc), glucuronic acid (GlcA), and N-acetylneuraminic acid (Neu5Ac) (Figure 1B) and are synthesized by GT enzymes using activated nucleotide sugar donors (Figure 1C). Unlike nucleic acids and proteins, glycans are not linear polymers with a conserved backbone and functional side groups. Rather, monosaccharides form the backbone of glycan structures and are joined by various regioisomeric and stereoisomeric linkages. Further diversification of glycan structure arises through the post-glycosylational modification of certain glycans by sulfation, acetylation, methylation, phosphorylation, and epimerization. To streamline and standardize the depiction of glycans, the glycoscience community has adopted the modern Symbol Nomenclature for Glycans (SNFG) (Neelamegham et al., 2019), which represents monosaccharides as color-coded shapes with text abbreviations to indicate glycosidic linkages and post-glycosylational modifications. Glycan diagrams are conventionally drawn from their non-reducing end on the left or top to their reducing end on the right or bottom to rapidly convey and compare complex structural information across related families. We will use the SNFG standard throughout the Primer and strongly encourage its general use by all scientists to facilitate accurate communication regarding glycans.
Many mammalian glycans can be categorized into one of four structurally diverse classes: O-GlcNAc, O-and N-glycans, and GAGs. O-linked N-acetylglucosamine or O-GlcNAc, is a single GlcNAc monosaccharide that is attached to serine or threonine residues of proteins (Figure 2A). Unlike nearly all other forms of glycosylation, the O-GlcNAc sugar is not further elaborated, and O-GlcNAc glycosylation (also known as O-GlcNAcylation) is a dynamic, inducible modification that occurs primarily on intracellular proteins. O-GlcNAcylation has been identified on thousands of proteins yet is mediated by only a single pair of enzymes: O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA). OGT, like other GTs, uses a nucleotide sugar donor, uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), to modify substrate proteins. As UDP-GlcNAc biosynthesis incorporates metabolites from central carbon, lipid, and nucleotide production, O-GlcNAcylation often serves as a nutrient and stress sensor that links the global metabolic status of the cell to the regulation of fundamental processes such as transcription, translation, and signal transduction (Yang and Qian, 2017), with aberrant O-GlcNAcylation events associated with metabolic and aging disorders (Bond and Hanover, 2013).
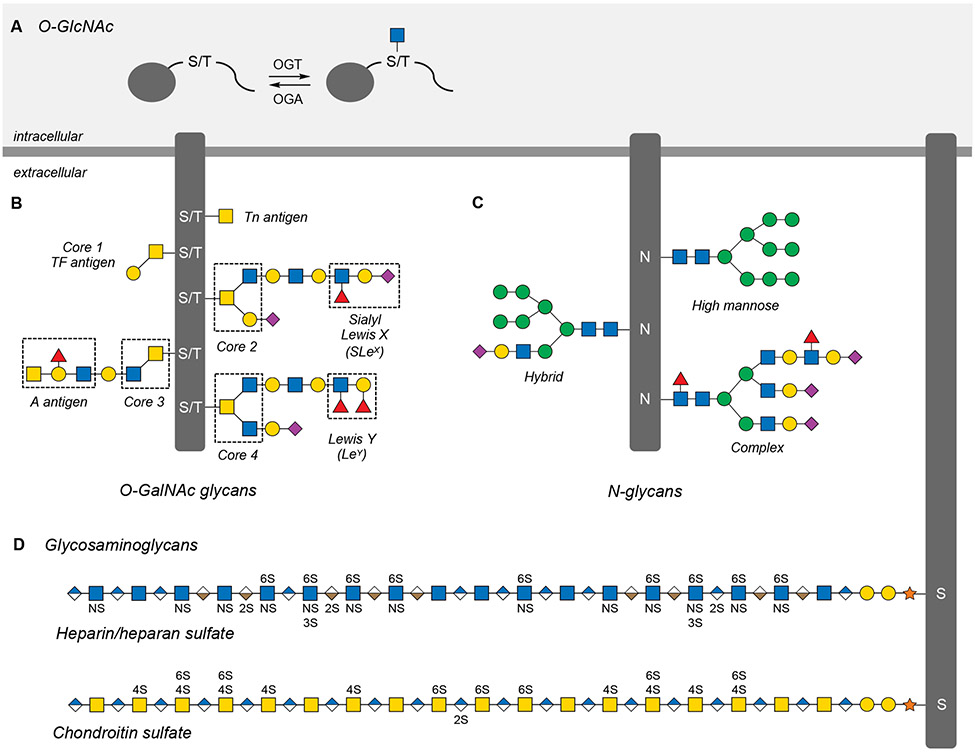
Figure 2. Major glycan classes in mammals.
(A) O-GlcNAcylation is the dynamic and reversible addition of N-acetylglucosamine to Ser and Thr residues on thousands of intracellular proteins.
(B) O-GalNAc or mucin-like glycans are a broad class of O-linked extracellular glycans categorized by one of eight core structures that can be elaborated with a number of glycan antigens.
(C) N-glycans are branched glycans attached to Asn residues of extracellular proteins and are categorized by the number and composition of their antennae branching from a conserved core structure.
(D) Glycosaminoglycans (GAGs) are linear extracellular polysaccharides that can be sulfated at different hydroxyl and amine positions along the length of the glycan chain.
Glycans are also often attached to cell-surface or secreted extracellular proteins. These O- and N-glycans are named based on their site of attachment: serine or threonine for O-glycans and asparagine for N-glycans. Although similar to O-GlcNAc in terms of attachment, O-glycans contain a much broader range of sugars, encompassing O-linked GalNAc, GlcNAc, Fuc, Glc, Xyl, and Man mono- and oligosaccharides, and typically range in size from one to more than twelve sugar residues. Mucin-type O-glycans are the most abundant class of these structures and are characterized by an O-linked GalNAc residue, also known as the Tn antigen (Figure 2B) (Rangel-Angarita and Malaker, 2021). The first GalNAc residue can be extended with multiple monosaccharides to produce eight core structures, which in turn are further elaborated through the activities of other GTs to contain other motifs such as fucosylation and sialylation. Hundreds of these modifications are found on the mucin proteins that line mucosal surfaces and form a physical barrier between host and the environment. Moreover, mucin and its glycans are highly dysregulated across many cancers and have been explored extensively for the development of cancer vaccines (Pinzon Martin et al., 2019).
N-glycans are also built on a conserved core (the branched pentasaccharide Man3GlcNAc2), which is modified with glycan “antennae,” generating structures that are classified into three broad groups: high mannose, complex, and hybrid (Figure 2C). During N-glycan synthesis, a large N-glycan precursor is attached to a dolichol phosphate lipid anchor and then transferred by oligosaccharyltransferase (OST) onto targeted Asn residues. The glycan undergoes trimming by glycosidases and further elaboration by GTs. Under normal conditions, N-glycans are first produced in the endoplasmic reticulum and O-glycans in the Golgi apparatus. As protein substrates transit the Golgi, nascent O- and N-glycans encounter a network of GTs that extend and cap the growing oligosaccharide in an incompletely understood process dictated by the expression levels, localization, and activation of each enzyme, as well as nucleotide sugar donor levels. Unlike other biosynthetic processes like transcription and translation where low fidelity can be detrimental, the imprecise process of O- and N-glycan biosynthesis is likely an evolutionary benefit. The resulting divergent glycans may allow cells to finely tune protein structure, folding, and function by sampling multiple combinations of glycan structures or “glycoforms” on individual proteins.
Mammals can also produce polysaccharides, which are represented by the ubiquitous, abundant linear polymers known collectively as glycosaminoglycans (GAGs) (Figure 2D). GAGs such as heparin/heparan sulfate (HS), chondroitin sulfate (CS), and dermatan sulfate (DS) are assembled from disaccharide units consisting of a glucuronic acid (GlcA) and a hexosamine sugar (GlcNAc or GalNAc). The iduronic acid (IdoA) residues found at irregular intervals in both HS and DS are generated through modification of GlcA by epimerase enzymes. Each disaccharide is also differentially sulfated along the polysaccharide chain by sulfotransferases, resulting in diverse patterns of sulfation. Consequently, GAGs have remarkable structural complexity in the form of many sulfation sequences, as well as domains of high and low sulfation density, which serve as docking sites for more than 800 proteins (Vallet et al., 2021; Xu and Esko, 2014). Accordingly, GAGs regulate a wide array of biological processes, ranging from animal development and immune regulation to infection and neuroregeneration (Mikami and Kitagawa, 2013; Xu and Esko, 2014). In the case of HS, CS, and DS, the polysaccharide chains are attached to serine residues of proteins through a xylose-containing tetrasaccharide linker. Defects in the enzymes that generate this linker region are associated with severe congenital disorders of glycosylation (CDGs) (Ng and Freeze, 2018). Proteoglycans, the cell-surface or secreted proteins to which GAGs are attached, generally possess between one and five polysaccharide chains from one or more GAG classes. Thus, the multiple levels of structural diversity found in GAGs, from sulfation motif to charge density to protein anchor, provide ample means to engage and direct proteins and cellular activity.
Beyond the glycans described here, many other carbohydrate structures exist in mammalian systems. These include unique modifications to other protein residues including C-mannosylation of tryptophans within thrombospondin type I repeats and galactosylation of hydroxyproline and hydroxylysine residues within collagen. In addition to protein anchors, glycans are attached to lipids on the cell surface such as sphingo- or glycerolipids. Specialized glycosylphosphatidylinositol (GPI) lipids are used to anchor over 100 different proteins to the cell surface. Very recently, small RNAs on the cell surface were also found to be decorated with N-glycans (Flynn et al., 2021), extending the scope of glycan modifications to nucleic acids. More detailed information about glycan structures and their biosynthesis can be found in Essentials of Glycobiology (https://www.ncbi.nlm.nih.gov/books/NBK579918/), an authoritative and freely available textbook written by leading experts in the field.
The functions and mechanisms of mammalian glycans mirror their wide range of structures. Small modifications such as O-GlcNAcylation can dynamically modulate proteins similar to phosphorylation and other post-translational modifications, altering tertiary structure, blocking ligand interactions, competing with other post-translational modifications, and/or controlling enzyme activity (Yang and Qian, 2017). As the glycan size increases, modifications like O- and N-glycans can alter the physicochemical properties of proteins such as solubility and folding (Xu and Ng, 2015), while also directly serving as binding partners for cell-surface receptors on nearby cells or for microorganisms such as viruses or bacteria (Raman et al., 2016). For larger polymeric glycans like GAGs, the polysaccharides can act independently or in concert with their protein anchors, recruiting soluble ligands to the cell surface to control protein diffusion and establish protein gradients or engaging cell-surface receptors directly to initiate signaling independently of canonical protein ligands (Kjellen and Lindahl, 2018).
Considering the many diverse roles of glycans, key questions arise when aiming to connect glycan structure or “glycotype” to phenotype. For example, which glycan structures are present on which cell types, and how do these glycan populations and their interactions change during development, normal physiology, and disease? With the advent of single-cell technologies, the ability to identify and quantify levels of glycans and glycoconjugates with increasing cellular resolution will be critical for advancing an understanding of glycan function. While some glycans correlate well with a given cell type, state, or disease and may serve as good biomarkers, others may directly influence function. How can one establish causation for specific glycan-associated phenomena? Questions such as these require robust methods to detect, quantify, and manipulate individual glycans or glycosylation events both in vitro and in vivo. Here, we will provide a broad overview of state-of-the-art methods in glycoscience, as well as a guide to interpreting results and the potential limitations of each approach. Our goal is to inspire new testable hypotheses for glycan function, provide practical guidance, and connect the broader scientific community with glycoscientists to address these central questions across various biological fields.
Tools of the Trade
Identifying relevant glycan structures or “glycotypes”
The diversity of glycan structures may initially seem daunting when investigating their biological functions. A crucial first step is to identify the relevant glycan class, the specific glycan structure if possible, and its mode of attachment to proteins or lipids. Bioinformatic resources are often helpful for determining glycotypes and guiding experimental designs. Three large-scale web-based glycoinformatics resources include: (1) GlyGen (https://glygen.org/) (York et al., 2020), (2) Glycomics@ExPASy (https://glycoproteome.expasy.org/) (Mariethoz et al., 2018), and (3) GlyCosmos (https://glycosmos.org) (Yamada et al., 2020). Coordinated by the GlySpace Alliance and supported by national scientific funding agencies, these three bioinformatic organizations integrate data regarding glycan structure, biosynthesis, gene, organism, and disease. Although the databases are relatively new and still undergoing expansion, these resources can help clarify glycan-related hits from genetic screens or proteomics experiments, and they provide key infrastructure for the collection and dissemination of data in the field.
A variety of experimental reagents and approaches can be used to identify the relevance of individual glycotypes in specific biological contexts. Pharmacological inhibition can be a good starting point to link glycans to a particular function in systems with well-defined phenotypes. Commonly used inhibitors of glycosylation target broad classes of glycans by impeding their biosynthesis or preventing glycan attachment to proteins. For example, OSMI-1 acts directly on OGT to prevent transfer of the GlcNAc sugar to O-GlcNAcylated proteins (Figure 3A) (Ortiz-Meoz et al., 2015). Modified sugar donors have also been employed as mechanism-based GT inhibitors. In the case of O-GlcNAc, the sugar analog 5SGlcNAc, in which the endocyclic ring oxygen of GlcNAc is replaced with a sulfur atom, is converted by biosynthetic pathways to the corresponding nucleotide 5-thiosugar donor and transferred by OGT less efficiently than the natural UDP-GlcNAc donor, resulting in OGT inhibition and decreasing cellular O-GlcNAc levels (Gloster et al., 2011). Conversely, global O-GlcNAc levels can be increased by OGA inhibitors such as the widely used Thiamet-G (Yuzwa et al., 2008).
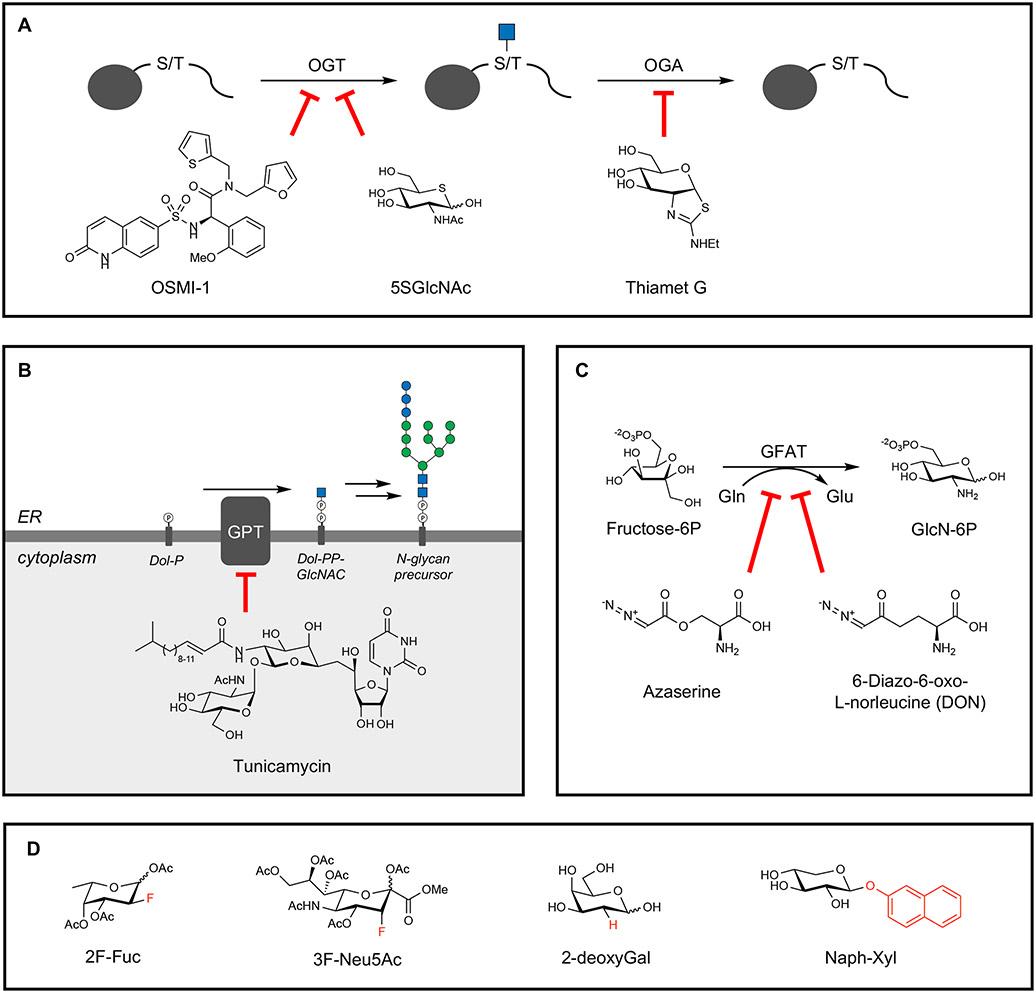
Figure 3. Pharmacological inhibitors of glycosylation.
(A) Protein O-GlcNAcylation can be blocked through the OGT inhibitors OSMI-1 and 5SGlcNAc, and the removal of O-GlcNAc can be targeted through the OGA inhibitor Thiamet G.
(B) N-glycosylation can be broadly inhibited using tunicamycin, which inhibits the attachment of GlcNAc-1-phosphate to dolichol phosphate (Dol-P) by GlcNAc-1-phosphate transferase (GPT).
(C) Glycans containing GlcNAc and GalNAc can be targeted using the glutamine mimics azaserine and DON, which target GFAT activity in hexosamine biosynthesis.
(D) “Look-alike” mimics of monosaccharides can inhibit specific modifications such as fucosylation (2F-Fuc and 2-deoxyGal) and sialylation (3F-Neu5Ac) or act as decoys for GTs in GAG biosynthesis (Naph-Xyl).
Despite these successes, cell-permeable inhibitors selective for specific GTs or glycosyl hydrolases have been generally difficult to obtain, due partly to similar substrate binding sites across the protein families. Thus, natural toxins are commonly used to alter glycosylation levels more globally. Originally isolated as a class of antibiotics from Streptomyces species, the natural product tunicamycin blocks N-glycosylation by inhibiting transfer of the initial GlcNAc residue onto dolichol phosphate (Figure 3B) (Duksin and Mahoney, 1982). Azaserine and DON (6-diazo-5-oxo-L-norleucine), other Streptomyces products, inhibit hexosamine biosynthesis by blocking glutamine fructose-6-phosphate amidotransferase (GFAT), thereby reducing O-GlcNAcylation and other forms of glycosylation (Figure 3C) (Brimble et al., 2010). However, these molecules act as glutamine mimics that broadly inhibit amidotransferases, causing pleiotropic effects. Synthetic “look-alike” monosaccharides such as fluorinated and deoxy sugar analogues antagonize glycosylation through various mechanisms (Figure 3D). For example, fluorinated Fuc and Neu5Ac analogues have been shown to prevent fucosylation and sialylation of glycans, respectively, through competitive inhibition of the respective GTs (Rillahan et al., 2012). Although the mechanisms underlying their inhibitory activity remain relatively unclear and likely involve multiple pathways, deoxysugars like 2-deoxyglucose have been used to prevent N-glycosylation (Kurtoglu et al., 2007). For Fucα(1,2)Galcontaining glycans, 2-deoxygalactose has been employed to prevent fucosylation due to lack of the 2-hydroxyl group on galactose required for Fuc attachment (Bullock et al., 1990). Hydrophobic xylosides can partially prevent GAG assembly by acting as competitive “decoy” substrates for GTs and diverting enzymatic activity away from natural proteoglycan substrates (Chua and Kuberan, 2017). GAG sulfation can also be inhibited by using sodium chlorate, a sulfate analogue that reduces global production of the sulfate donor phosphoadenosine 5’-phosphosulfate (PAPS) and affects all forms of sulfation including protein sulfation (Greve et al., 1988), or by using specific inhibitors of GAG sulfotransferases (Cheung et al., 2017). Nevertheless, results obtained using pharmacological inhibitors should be interpreted with caution and supported by other independent methods as they typically modulate large classes of glycans across the entire proteome and can result in pleiotropic effects such as ER stress or cytotoxicity that may complicate observable phenotypes.
A complementary approach to chemical inhibitors is the use of recombinant enzymes to modify or cleave glycans of interest. Removal of N-glycans can be accomplished with PNGase F or Endo F1/2/3 (Figure 4A) (Tarentino and Plummer, 1994). PNGase F cleaves at the Asn-GlcNAc bond that attaches N-glycans to the protein, whereas Endo F enzymes hydrolyze N-glycans between the first and second GlcNAc residues. Each enzyme shows a distinct specificity toward N-glycan structures (e.g., number and composition of antennae or core fucosylation). Therefore, the selective use of endoglycosidases (ENGases) individually or in combination can also help to determine the relevant N-glycan structures. In addition to ENGases, exoglycosidases (EXGases) can be employed to cleave terminal glycans. For example, recombinant sialidases and fucosidases were used to study the roles of Neu5Ac and Fuc in cancer cell progression and clearance (Hudak et al., 2014; Yuan et al., 2008). Although deglycosylating enzymes are commonly applied to purified proteins, many of these enzymes have been successfully employed with live cells. For GAGs, chondroitinase ABC and heparinase I/II/III can be used to digest CS and HS polysaccharides, respectively, and have been applied to purified proteins, cultured cells, and organisms in vivo (Bradbury et al., 2002; Brown et al., 2012; Griffin et al., 2021).
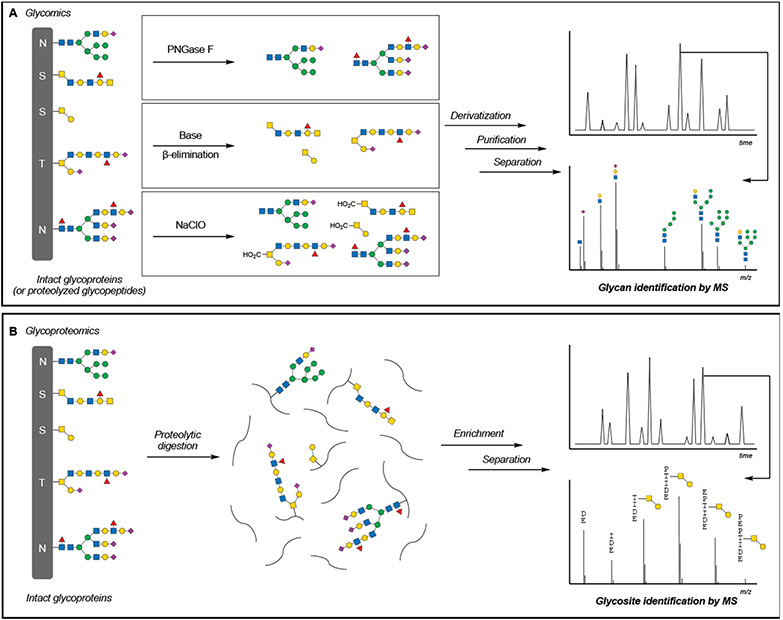
Figure 4. Glycomics and glycoproteomics workflows.
(A) Glycomics, or the analysis of glycan composition, is accomplished through the enzymatic or chemical release of glycans, followed by chemical derivatization, purification, separation, and mass spectrometric characterization of glycan structures.
(B) Glycoproteomics, or the analysis of protein glycosylation, is accomplished through the digestion of glycoproteins and enrichment of glycopeptides, followed by separation and mass spectrometric identification of glycosylated peptides.
The presence or absence of specific glycan structures can often be confirmed using protein-based probes such as lectins and antibodies. Lectins are naturally occurring glycan-binding proteins (GBPs) that are often isolated from plants. Many lectins are commercially available and have moderate affinities for defined glycan motifs. Commonly used lectins include wheat germ agglutinin (WGA) for terminal GlcNAc residues, concanavalin A (ConA) for branching oligomannose residues of N-glycans, Sambucus nigra agglutinin (SNA) for α(2,6)-sialic acids, and Ulex europaeus agglutinin-I (UEA-I)-for α(1,2)-linked Fuc. Potential cross-reactive binding of lectins towards multiple glycan structures must be considered. The Consortium for Functional Glycomics has made microarray data on the binding of lectins to hundreds of defined glycan structures publicly available (https://ncfg.hms.harvard.edu/ncfg-data/microarray-data/lectin-quality-assurancequality-control). Moreover, machine-learning methods were recently applied to these data to annotate the complex binding specificities of 57 commercially available lectins, providing a critical guide to these important reagents (Bojar et al., 2022). As a complement to lectins, antibodies have been generated against all major classes of mammalian glycans, with over 1,000 monoclonal antibodies previously described (Sterner et al., 2016). The Database for Glycan Reagents (DAGR) was established through the Center for Cancer Research at the National Cancer Institute (https://dagr.ccr.cancer.gov) to facilitate the use of anti-carbohydrate antibodies and lectins.
Glycan-binding lectins and antibodies have been leveraged for glycan detection using standard methods, including histology, protein or western blotting, and enzyme-linked lectin/immunosorbent assays. Microarrays of lectins and carbohydrate antibodies have also been constructed for glycomics applications (Dang et al., 2020) and to identify glycans involved in processes such as melanoma metastasis (Agrawal et al., 2017) and viral host response (Heindel et al., 2020). As mentioned above, care must be taken when interpreting data using such reagents as they often bind multiple glycans containing related (and sometimes unrelated) structural motifs. Glycan structure cannot be definitively determined based solely on lectin or antibody binding. The comparative use of multiple glycan-binding reagents should be considered to provide further evidence of glycan identity, along with mass spectrometry (MS) and other methods such as metabolic or chemoenzymatic labeling (discussed in “Detecting and monitoring glycans”).
By far, MS analysis is the most definitive and comprehensive method to identify glycan structures (Ruhaak et al., 2018). Unlike other methods described above, liquid chromatography in conjunction with MS (LC-MS) provides a direct readout of the glycan structure through intact molecular mass determination and sequencing by MS/MS fragmentation (Figure 4A). Although MS cannot easily discriminate between isobaric diastereomers (e.g., GlcNAc vs. GalNAc) without further fragmentation of the monosaccharide, glycomic approaches have now become more standardized, with commercialized kits for sample preparation and internal standards for spectral matching. Kits frequently employ chemical or enzymatic methods to release the targeted glycans, which are then derivatized with UV-active compounds like 2-aminobenzamide (2-AB) to facilitate detection during LC separation. Alternatively, benzyl GalNAc glycosides have also been employed as decoys to produce secreted O-glycans for cell state-dependent O-glycome profiling (Kudelka et al., 2016). For larger polysaccharides like GAGs, the full determination of individual linear sequences remains challenging due to their overall size and multiple levels of structural heterogeneity. While a few examples of GAG sequencing have been reported (Ly et al., 2011; van Kuppevelt et al., 2017), GAGs are typically enzymatically digested and subjected to disaccharide compositional analysis by LC-MS, providing a relative quantification of individual disaccharide motifs at the expense of linear sequence data.
Although glycomics analyses can often determine the glycan structure, critical information regarding their attachment sites to proteins is often lost. O-glycans, including O-GlcNAc, generally lack consensus motifs, and knowledge of the glycosylation sites can be critical for generating and testing hypotheses regarding glycan function. Therefore, the goal of glycoproteomics approaches is not only to identify the glycan structure but also to sequence the underlying peptide (Figure 4B). The mass spectra of complex O- and N-glycans can be severely complicated by partial fragmentation of the glycan. Next-generation bioinformatic approaches and various MS fragmentation methods have been developed to address this complexity and increase the number of identified glycosylated proteins (reviewed in (Chernykh et al., 2021; Oliveira et al., 2021)).
Glycoproteomics has also greatly benefited from new methods to enrich for glycosylated proteins and peptides (Riley et al., 2021). The presence of abundant, nonglycosylated peptides often obscures the detection of rarer, glycosylated peptides. Glycopeptide enrichment can be achieved by lectin affinity chromatography, hydrophilic interaction liquid chromatography, and metabolic or chemoenzymatic labeling of glycans with affinity tags (described in ‘Detecting and monitoring glycosylation’). Moreover, specialized methods for enriching and mapping O-GalNAc sites have also been developed using bacterial proteases that cleave specific peptide sequences proximal to mucin-type O-glycans (Rangel-Angarita and Malaker, 2021). When used successfully, these methods have enabled large-scale, proteome-wide profiling of glycoproteins in multiple contexts, including the identification of over 1,750 O-GlcNAcylation sites in neuronal synaptosomes (Trinidad et al., 2012), 2,200+ O-glycopeptides in activated T cells (Woo et al., 2018), 600+ N-glycopeptides from human serum (Li et al., 2019a), and full profiling of GAG composition in 20 human cell lines (Li et al., 2015).
Generating chemically defined glycans and glycoconjugates
Just as the automated synthesis of oligonucleotides and peptides revolutionized our understanding of these biomolecules and ushered in a new era of modern molecular biology, access to a broad range of glycan structures will be critical for advancing glycoscience and constitutes an essential part of the glycoscientist’s toolkit. Glycans and glycoconjugates of unique, defined structure serve as invaluable tools for many applications, acting as standards for structure identification, as ligands for studying GBP interactions, and as simplified glycan or glycoprotein mimetics for probing function. Significant advances in carbohydrate chemistry have recently been made toward producing complex oligosaccharides and are reviewed elsewhere (Boltje et al., 2009; Mende et al., 2016). Here, we will highlight the emergence of innovative technologies that facilitate the production of a wide range of glycans and increase availability of these chemical probes for broad distribution.
A major focus of modern carbohydrate chemistry has been the development of methodologies for the automated assembly of oligo- and polysaccharides (Figure 5A) (Panza et al., 2018; Wen et al., 2018). Glycan synthesis is challenging due to the need to control the regioselectivity (which hydroxyl position around the sugar ring) and the stereoselectivity (α or β anomer) of each newly formed glycosidic bond. To achieve this, monosaccharide building blocks with chemical protecting groups at various hydroxyl positions must first be prepared. These protecting groups must be removable to unmask a desired position for coupling with another monosaccharide yet remain inert under other reaction conditions. As the optimal set of protecting groups depends on the glycan structure, there are no universal, “one-size-fits-all” building blocks for glycan synthesis. Thus, the synthesis of a glycan target can take several months and in some cases years to complete.
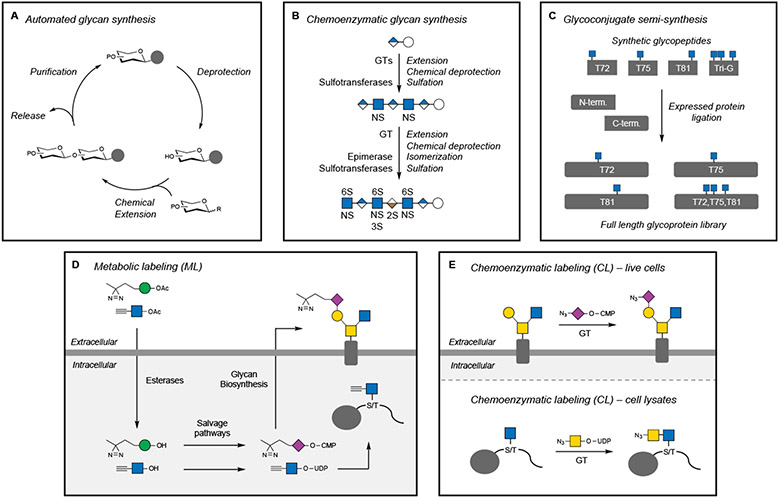
Figure 5. Methods to generate and modify glycans and glycoconjugates.
(A) Automated methods for the chemical synthesis of oligosaccharides use a similar approach as solid-phase peptide or oligonucleotide synthesis. The glycan is elongated through a series of deprotection and coupling steps, followed by release from the resin.
(B) The chemoenzymatic synthesis of an Arixtra biosimilar oligosaccharide employs multiple enzymatic and chemical steps to assemble and functionalize an HS heptasaccharide.
(C) Semi-synthesis of O-GlcNAcylated α-synuclein utilizes synthetic glycopeptide fragments for expressed protein ligation to generate a library of specific protein glycoforms.
(D) Metabolic labeling (ML) utilizes peracetylated (indicated by OAc) nonnatural monosaccharides that cross cell membranes, are deprotected (indicated by OH) by endogenous esterases, converted into nucleotide sugar donors, and then incorporated by GTs into glyconjugates. Nonnatural glycans with diazirine or alkyne functionalities are shown as representative examples.
(E) Chemoenzymatic labeling (CL) employs exogenous GTs and nonnatural nucleotide sugar donors to modify specific glycan structures recognized by the GT. Nonnatural glycans with azide functionalities are shown.
Nonetheless, multiple methods have been developed to expedite glycan assembly. For example, elongation of the growing oligosaccharide chain while attached to Merrifield and other resins used for solid-phase peptide synthesis has enabled the production of various complex glycans, including α-glucan polysaccharides, mycobacterial oligoarabinofuranosides, blood group antigens, and GAGs (Guberman and Seeberger, 2019). Alternatively, installation of a fluorinated tag onto the reducing end of the growing oligosaccharide chain can facilitate automated solution-phase synthesis and purification of reaction intermediates by fluorous solid-phase extraction (Tang and Pohl, 2016). These methods have been incorporated into automation platforms such as the commercially available synthesizer Glyconeer 2.1 (Hahm et al., 2017), HPLC-based systems (Panza et al., 2020), and microwave-assisted peptide synthesizers (Danglad-Flores et al., 2021).
Recent technologies have also advanced the large-scale purification of complex glycans from natural sources (Zhang et al., 2020). A key step in the purification is the release of natural glycans from their pendant protein or lipid conjugates. A mild method employing dilute bleach (NaClO) was developed to oxidatively release O- and N-glycans from glycoproteins and glycan nitriles from glycosphingolipids (Song et al., 2016; Zhu et al., 2018) (Figure 4A). Notably, these chemical methods could be scaled to kilogram quantities of protein or tissue and eliminated the need for expensive enzymes like PNGase F. Although chemical synthesis is still the better choice for lower abundance glycans, the harvesting of oligosaccharides from plant and animal tissue now provides a practical alternative for certain N- and O-glycans.
Enzymes have also been exploited to facilitate glycan production (Figure 5B). These methods often employ purified natural or engineered bacterial GTs along with their nucleotide sugar donor substrates. Powerful multienzyme systems have been developed that combine GTs with upstream biosynthetic enzymes to produce the required nucleotide sugars, providing one-pot syntheses of oligosaccharides (Yu and Chen, 2016). The scope of enzymatic glycan synthesis can be expanded by combining enzymes with chemically modified substrates. Such chemoenzymatic approaches have enabled the production of many bioactive carbohydrates and continue to transform the field. For example, the anticoagulant drug Arixtra is a synthetic heparin-based pentasaccharide used for the treatment of deep vein thrombosis. While Arixtra requires 50 chemical steps to synthesize (~0.1% yield), a biosimilar heptasaccharide containing the Arixtra pentasaccharide motif was obtained chemoenzymatically using GT and sulfotransferase enzymes in 12 steps and 37% overall yield (Xu et al., 2011). In another example, the challenge of producing asymmetrically branched N-glycans was overcome using a chemoenzymatic approach (Wang et al., 2013b). Automation platforms for enzymatic and chemoenzymatic glycan assembly have also been developed (Li et al., 2019c; Wen et al., 2018), laying the foundation for the production and broad distribution of large collections of complex oligosaccharides.
Glycans and GBPs are often presented in multivalent forms in vivo, which enhances the affinity of glycan-protein interactions through avidity. Glycopolymers have been synthesized to mimic these high avidity interactions by presenting multiple copies of individual glycan motifs (Kiessling and Grim, 2013) and are useful tools for modulating biological function. For example, linear polymers containing pendant sulfated Lewis X epitopes or Neu5Ac glycans have been exploited to characterize leukocyte rolling (Sanders et al., 1999) and suppress B cell activation (Courtney et al., 2009), respectively. As GAGs are naturally multivalent, polymers with pendant HS or CS disaccharides are excellent simplified mimetics that can elicit diverse phenotypes, including neurite outgrowth (Rawat et al., 2008), chemokine signaling activation (Sheng et al., 2013), and blood anticoagulation (Oh et al., 2013) in a sulfation motif-dependent manner. Multivalent glycopolymers have also been functionalized with lipid tails to remodel cell surfaces with glycans (described in ‘Modulating glycans to probe function: connecting ‘glycotype’ to phenotype’) or with photocrosslinking functionalities to identify GBPs (described in ‘Discovering and characterizing glycan-protein interactions’).
Glycans often modulate the proteins to which they are attached, affecting physical properties such as folding, stability, and solubility, as well as their biological activities. Thus, the production of homogeneous glycoproteins has been crucial for defining the functions of specific glycans and advancing the development of biologic drugs. Semi-synthesis methods such as native chemical ligation and expressed protein ligation, in which synthetic glycopeptides are ligated to peptide or protein fragments, are highly effective at producing homogeneously glycosylated proteins (Figure 5C). These approaches have been applied to glycoproteins such as the drug erythropoietin (Wang et al., 2013a), a 166-amino acid protein with four glycosylation sites that stimulates erythrocyte production, as well as α-synuclein (Marotta et al., 2015), a 140-amino acid O-GlcNAcylated protein whose aggregation is associated with the pathology of Parkinson’s disease. Alternatively, the glycan profiles of proteins can be tailored by engineering glycosylation pathways in cells (similar to Figure 6A). Genetic engineering to manipulate the glycosylation patterns on purified proteins is commonly performed, and although the resulting proteins are still glycoform mixtures, these facile and scalable production systems reduce structural complexity and remain an industry standard.
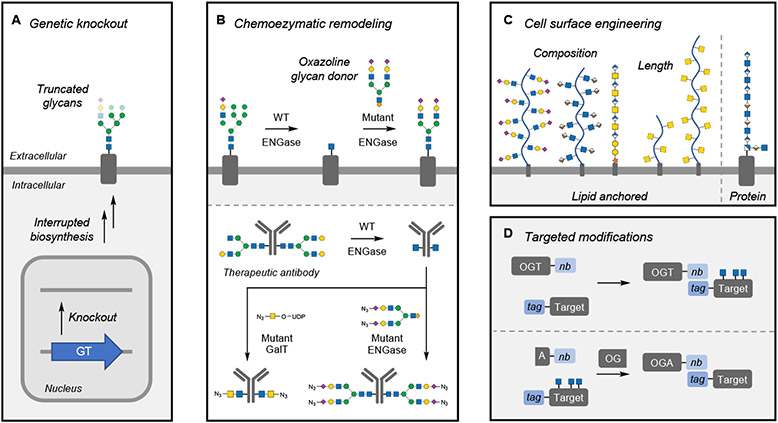
Figure 6. Methods to modulate glycans.
(A) Genetic knockout of individual glycosyltransferases (GTs) and other glycan biosynthetic enzymes globally affects cellular glycan populations, leading to truncated glycan structures.
(B) Chemoenzymatic remodeling using endoglycosidases (ENGase) simplifies N-glycan heterogeneity on the cell surface or on proteins. Installation of specific structures is accomplished using mutant ENGases or GTs that can install specific structures functionalized for further biorthogonal reactions.
(C) Cell-surface engineering has been accomplished with both synthetic and naturally occurring glycopolymers anchored to the cell surface by lipids or proteins. Techniques can modulate the glycocalyx by changing its composition or thickness.
(D) Targeted O-GlcNAc modification utilizes nanobody (nb)-fused OGT or OGA, which directs the enzymes to tagged target proteins. In the case of OGA, this approach is accomplished through a split OGA construct.
Chemical biology approaches provide more precise control over glycoform production in vitro. For example, the N-glycans on therapeutic antibodies can be modified by ENGases, which first trim the conserved N-glycans to a single GlcNAc residue (similar to Figure 6B) (Giddens et al., 2018; Huang et al., 2012). Mutant ENGases then transfer a chemically modified intact N-glycan oxazoline donor onto the glycan stub, producing a homogeneously N-glycosylated antibody. Additional modifications using other enzymes like fucosidases can be added to the process, further defining the specific glycoform of the antibody. Another complementary approach uses genetic code expansion to make defined glycosylated proteins by installing chemically reactive, nonnatural amino acids at specific sites for click chemistry-based functionalization (Machida et al., 2015). A nonnatural dehydroalanine amino acid was employed to produce O-GlcNAc mimics via radical-mediated ligation of alkyl halides, and N-glycan mimics were installed through ENGase-mediated extension of the GlcNAc residue, providing a general method to produce glycoprotein mimetics (Wright et al., 2016).
Many of these defined synthetic glycans and new technologies are currently available through individual laboratories or companies. Through the support of the NIH Common Fund Glycoscience Program, great progress has been made in creating new resources, tools, and methods to render the study of glycans more accessible to the larger research community. A partial listing of these resources can be found at https://commonfund.nih.gov/Glycoscience/programresources.
Discovering and characterizing glycan-protein interactions
Glycans often exert their activities through direct interactions with proteins. Glycoproteins, glycolipids, and polysaccharides form an extramembrane compartment, termed the glycocalyx, which is found on nearly all eukaryotic cells. The glycocalyx is the first site of cellular contact with the environment, and thus glycans play key roles in cell-cell and cell-matrix interactions critical for processes such as immune cell trafficking, embryonic development, and cancer metastasis. GBPs include lectins but also extend to many other proteins not classically defined as lectins. For example, a wide variety of proteins bind to GAGs, including soluble ligands like growth factors and cytokines, transmembrane proteins such as receptor tyrosine kinases and phosphatases, as well as proteins from microbial pathogens like the SARS-CoV-2 spike glycoprotein (Vallet et al., 2021). Therefore, the study of carbohydrate-protein interactions is critical to understanding glycan function.
Glycan molecules of defined structure have greatly facilitated the discovery of novel protein receptors. For example, affinity purification using immobilized glycans followed by mass spectrometry (AP-MS) is a powerful method to enrich and identify GBPs. Because carbohydrate-protein interactions can be low to moderate affinity, methods that capitalize on multivalent interactions to strengthen the interaction or covalently capture proteins using chemical crosslinking agents are highly effective. For example, GBPs have been identified using gold nanoparticles (Sakurai et al., 2016) and synthetic glycopolymers (Wibowo et al., 2014) functionalized with multiple copies of sugar epitopes, along with photocrosslinkers such as benzophenone or nitrophenylazide. Direct conjugation of a bifunctional probe containing a photocrosslinker and an alkyne group for appending a biotin tag to commercially available, natural GAG polysaccharides enabled sulfation motif-specific CS binding proteins to be enriched and identified from neurons (Joffrin and Hsieh-Wilson, 2020).
In some cases, binding may be mediated by interactions not only with the glycan itself but also with the associated glycoprotein. To enable detection of such glycoprotein-protein interactions, metabolic labeling (ML) can be used to install photoaffinity labels onto cellular glycoproteins. ML, also known as metabolic oligosaccharide engineering, exploits the promiscuity of mammalian salvage pathways to convert nonnatural monosaccharides into nucleotide sugar donors, which are then incorporated into cellular glycans by GTs (Figure 5D). Sugar analogs of GlcNAc, ManNAc, and Neu5Ac have been synthesized containing aryl azide and diazirine groups for photocrosslinking (Han et al., 2005; Tanaka and Kohler, 2008; Yu et al., 2012). Cells are treated with membrane-permeable versions of these analogs, which are deacetylated by intracellular esterases and subsequently incorporated into newly formed glycoproteins. Following light-induced crosslinking, immunoprecipitation of the glycoprotein, coupled with MS analysis, can be used to identify putative glycoprotein interactors.
Specific glycoprotein-protein interactions can also be identified by proximity labeling methods. For example, ML was employed to install a non-natural azide group into Neu5Ac glycans on cell-surface glycoproteins (Li et al., 2019b). This modified glycan was then reacted using bioorthogonal chemistry with a cyclooctyne probe containing a coordinated Fe(III) ion. When treated with hydrogen peroxide, Fe(III) generated hydroxyl radicals, which in turn oxidized nearby amino acid residues that could be detected by MS. In another approach, tyramide radicalization was used to identify glycoprotein ligands for Siglecs, a family of Neu5Ac recognition proteins essential for self-nonself discrimination by the immune system (Chang et al., 2017). Siglec-horseradish peroxidase (HRP) complexes were formed using recombinant FLAG-tagged Siglec proteins and anti-FLAG antibodies conjugated to HRP. Incubation of the Siglec-HRP complexes with cells, followed by addition of biotin tyramide and hydrogen peroxide generated short-lived tyramide radicals that biotinylated nearby proteins, allowing for identification of both known and new Siglec ligands. Similarly, glycoprotein ligands for galectin-1 and galectin-3 were identified by conjugating these GBPs to the ascorbate peroxidase APEX2, which enabled labeling of proximal proteins using biotin tyramide and hydrogen peroxide (Joeh et al., 2020; Vilen et al., 2021). As the HRP and APEX methods use a commercially available biotin probe, application to other GBPs should be readily feasible.
Upon identifying a glycoprotein-protein interaction, establishing the glycan structural motif(s) responsible for mediating the interaction can provide crucial insights into its specificity and function. The selectivity of glycan-protein interactions can range from promiscuous affinity for multiple, related carbohydrate structures to high selectivity for individual structures. Glycan microarrays have emerged as a high-throughput technology to determine the binding specificities of GBPs (Rillahan and Paulson, 2011). Similar to traditional DNA microarrays, glycan microarrays containing a wide range of both synthetic and natural glycans have been constructed using robotic printing technologies. Protein binding to specific glycans spotted on the microarray is typically detected using biotinylated or fluorescently labeled antibodies. This miniaturized format permits rapid interrogation of many glycan-protein interactions in parallel, while requiring minimal glycan and protein material. As mentioned above, glycan microarrays have provided vital information regarding lectin specificity and have been applied to a wide range of GBPs (Gao et al., 2019), serum antibodies (Xia and Gildersleeve, 2015), and even intact viruses (Smith and Cummings, 2014).
Despite their ease, glycan microarrays are inherently limited by the diversity of glycans on the array. Current microarrays contain only a fraction of the mammalian glycome and an even smaller proportion of the microbial glycome, highlighting a critical need to expand access to pure, well-defined glycan molecules. As discussed above, the synthesis and biochemical isolation of large panels of glycans is technically challenging. To combat these issues and facilitate the use of glycan microarrays in the broader community, the NIH has supported large-scale efforts to produce glycans and glycan microarrays. Numerous glycan microarrays are currently available upon request from the Consortium for Functional Glycomics (CFG) and National Center for Functional Glycomics (https://ncfg.hms.harvard.edu/microarrays). The widely used CFG array (version 5.2) has approximately 600 mammalian glycans, while the microbial glycan array has over 300 glycans. In addition, the Glycosciences Laboratory at Imperial College London has a large collection of glycans and offers microarray analyses (http://www.imperial.ac.uk/glycosciences/). Several companies also provide glycan microarrays and analysis services for targeted subsets of glycans, including GAGs and common N- and O-glycans found in serum, plasma, and other tissues. Newer approaches with chemically released natural glycans, multiplexing capabilities, and alternative solution-binding assays will continue to advance this cornerstone technology.
As glycan density, spatial arrangement, and presentation on lipids or proteins, as well as the presence of competing glycan structures, can significantly influence glycan recognition (Kiessling and Grim, 2013; Mende et al., 2019), cell-based platforms have also been explored as a complementary approach to glycan microarrays. Knockout cell lines generated by chemical mutagenesis and selected for altered glycosylation have historically been used to characterize glycosylation pathways, identify relevant genes, and elucidate the functional roles of glycans (Patnaik and Stanley, 2006). Modern genetic engineering techniques like zinc finger nuclease (ZFN) and CRISPR/Cas9-directed editing have recently been employed to generate panels of isogenic HEK293 cells with predictable structural changes to O- and N-glycans, as well as GAGs (Briard et al., 2018; Narimatsu et al., 2019). This “glycotopiary” approach to prune cell-surface glycans provides a valuable cell-based array for investigating binding specificities to glycans. Such cell lines have been exploited in conjunction with flow cytometry to determine the binding selectivity of Neu5Ac-binding Siglec proteins and hemagglutinin (HA) proteins from individual strains of influenza. It is worth noting, however, that the cells generated by genetic and chemoenzymatic approaches simultaneously present multiple different glycoforms, necessitating comparative analyses across various cell lines to determine specificity for individual glycans. Nevertheless, cell-based platforms are a powerful complement to traditional glycan microarrays and are being made broadly available to the scientific community.
Another recently developed approach that expands the capabilities of traditional glycan microarrays is the “liquid” glycan array platform. In this approach, defined glycans are covalently attached to DNA-barcoded M13 bacteriophages using click chemistry (Sojitra et al., 2021). The bacteriophage mixture is applied to GBPs in vitro or even in vivo, and bound bacteriophages are then sequenced to identify the structures and valencies of potential interacting glycans. Notably, this liquid glycan array method enables not only the study of GBP specificities, but also other important facets of glycan recognition, such as multivalency, avidity, as well as the potential for crosstalk and dynamic competition between glycans in vivo. Here again, access to defined, azide-functionalized glycans is required, which may limit the diversity of structures that can be used. Nonetheless, this DNA-encoded approach has unique advantages and may become widely utilized as the library of phage-displayed glycans grows.
Glycan microarrays are valuable as a hypothesis-generating discovery tool to rapidly screen putative GBPs and identify potential glycan structures important for recognition. These initial screens should be followed up with conventional binding assays such as enzyme-linked immunosorbent assays, surface plasmon resonance, biolayer interferometry, isothermal titration calorimetry, fluorescence polarization, and frontal affinity chromatography (Nagae and Yamaguchi, 2018). New technologies such as mass photometry are also likely amenable to measuring protein complexes mediated by glycans (Young et al., 2018). Importantly, these assays provide independent validation of structure-dependent binding and can be used to derive kinetic and thermodynamic parameters such as enthalpy and entropy (ΔH, ΔS), dissociation constants (Kd), and Kon and Koff rate constants.
Structural approaches and site-directed mutagenesis help to define further the molecular basis of glycan-protein recognition, including the binding interface, intermolecular forces, and specificity of the interaction. The most widely used techniques, nuclear magnetic resonance (NMR) and X-ray crystallography, require pure, structurally defined glycans, which limits their use to certain accessible carbohydrate classes. Another major challenge to structural glycobiology and high-resolution structure determination is the intrinsic flexibility of glycans, which leads to heterogeneous ensembles of defined conformational states. NMR methods are particularly well suited to studying glycan conformation and dynamics, and various NMR techniques have been utilized to gain insights into glycan-protein interactions, most notably nuclear Overhauser effect spectroscopy (NOESY), saturation transfer difference NMR (STD-NMR), and WaterLOGSY (reviewed in (Gimeno et al., 2020; Nieto, 2018)). X-ray crystallography studies have provided crucial data on conformational features of carbohydrates and their interactions within protein binding clefts. For example, seminal structures of the ternary HS-fibroblast growth factor (FGF)-fibroblast growth factor receptor (FGFR) complex highlighted how GAGs can engage both ligands and receptors in a single complex, likely aiding in receptor activation (Pellegrini et al., 2000; Schlessinger et al., 2000).
Computational modeling methods are often used in combination with experimental structures or protein homology models when structures are unobtainable. For example, a combined computational-experimental approach was used to investigate the specificity of an antibody against the tumor-associated carbohydrate antigen sialyl-Tn (Amon et al., 2018). Antibody specificity was first investigated by glycan microarray, alanine-scanning mutagenesis, and STD-NMR to define the antibody-glycan contact surface. Computational grafting of various sialyl-Tn-related carbohydrates onto the modeled antibody structure using Gly-Spec (www.glycam.org) led to a 3D model of the antibody-glycan complex that was consistent with the experimental data, revealing features important for the high selectivity of the antibody. For GAGs, various computational programs such as GAG-Dock (Griffith et al., 2017), VinaCarb (Nivedha et al., 2016), and GlycoTorch Vina (Boittier et al., 2020) have been developed for docking of GAG oligosaccharides to their binding proteins, providing structural models that closely mimic known crystal structures and guiding investigations into the functional consequences of GAG binding.
Detecting and monitoring glycosylation in vitro and in vivo
The glycome responds dynamically within minutes in response to cellular stimuli or over long periods of time, such as during development or disease progression. Thus, robust methods to detect and monitor specific glycans are critical for establishing the functions of glycans in physiology and disease.
As mentioned above, lectins or antibodies are often used to detect carbohydrates on glycoconjugates by western blotting or immunocytochemistry, and because many recognize the carbohydrate epitope independent of the pendant protein or lipid, they can be used to compare the relative glycosylation levels of a given glycoconjugate across different conditions. Antibodies that selectively recognize glycans at specific sites in proteins have been difficult to generate, although site-specific O-GlcNAc antibodies have been produced against a small number of O-GlcNAcylated targets (Gorelik and van Aalten, 2020). As lectins and antibodies may recognize multiple related glycans with varying affinities, care should be taken in interpreting results using these reagents, and the glycan structure and glycosylation site(s) on a protein should be confirmed by mass spectrometry and/or site-directed mutagenesis in all cases.
Glycans can also be detected by modification of specific glycans with chemical reporters such as fluorescent dyes or biotin. For example, mild periodate oxidation of Neu5Ac-containing glycans generates a terminal aldehyde that can be functionalized with reporter groups using oxime chemistry (Zeng et al., 2009). Metabolic labeling (ML) and chemoenzymatic labeling (CL) methods allow multiple other classes of carbohydrates to be targeted. As described above (‘Discovering and characterizing glycan-protein interactions’ section), ML exploits the substrate promiscuity of biosynthetic enzymes to introduce nonnatural glycans bearing small chemical functionalities into cellular glycoconjugates (Figure 5D) (Wang and Mooney, 2020). In CL, an exogenous GT is used to tag existing glycans on cellular glycoconjugates with a nonnatural sugar modified with a small chemical functionality (Figure 5E) (Lopez Aguilar et al., 2017). Covalent attachment of chemical reporters to these nonnatural sugars using biorthogonal chemistry enables rapid labeling of glycans in cells, tissues, and even whole organisms. For live-cell imaging, the biorthogonal copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction can be cytotoxic, likely due to reactive oxygen species generated by the catalyst (Kennedy et al., 2011), but cytotoxicity can sometimes be avoided with improved copper ligands (Parker and Pratt, 2020) or catalyst-free reactions such as strain-promoted azide-alkyne cycloaddition (SPAAC) or tetrazine ligation (Nguyen and Prescher, 2020).
CL was originally developed as a sensitive method for detecting O-GlcNAcylated proteins. A mutant β-1,4-galactosyltransferase (Y289L GalT) can be used to append a nonnatural azido- or keto-GalNAc sugar onto O-GlcNAc moieties (Clark et al., 2008; Khidekel et al., 2003). The nonnatural sugar can then be reacted to label the glycoproteins with a biotin tag, which allows for both sensitive detection and affinity enrichment for MS analysis. CL has facilitated the identification of O-GlcNAc-modified proteins, MS determination of their glycosylation sites, and quantitative proteomic profiling of O-GlcNAcylated proteins across cells and tissues. In addition to biotin, other reporters have been employed with CL, including polyethylene glycol polymers of defined mass (Rexach et al., 2010). These polymer mass tags shift the molecular weight of the glycoprotein and are visualized by western blotting to determine the stoichiometry of glycosylation. Using this approach, a significant increase in O-GlcNAcylation on phosphofructokinase-1 (PFK1) was quantified in human breast tumor tissue compared to normal tissue (Yi et al., 2012). CL with polymer mass tags has also been used to study the interplay between post-translational modifications, demonstrating that O-GlcNAcylation was induced specifically on the phosphorylated subpopulation of the transcription factor CREB in response to neuronal depolarization (Rexach et al., 2012). Notably, CL has been expanded to the detection of many other glycan motifs, including the O- and N-glycan disaccharide N-acetyllactosamine (LacNAc) (Zheng et al., 2011), the Tn antigen (O-linked GalNAc) (Wu et al., 2016), the TF antigen (O-linked Gal-GalNAc) (Li et al., 2014), Fucα(1,2)Gal (Chaubard et al., 2012), Neu5Acα(2,3)Gal (Wen et al., 2016), and terminal HS residues (Wu et al., 2018). CL has also been developed as a broad labeling method for O- and N-glycans by exploiting the substrate specificities of different sialyltransferases (Mbua et al., 2013; Yu et al., 2016). Labeling kits for O-GlcNAc, as well as many of the enzymes and nonnatural sugar donors, are commercially available or accessible as shared reagents from researchers.
ML has been used in conjunction with fluorescent reporters to image cellular glycans in a variety of contexts. For example, super-resolution imaging of the glycocalyx was achieved through ML of GalNAc-containing glycans and periodate labeling of Neu5Ac (Möckl et al., 2019). The results showed nanoscale organization of glycans in the glycocalyx, along with changes in glycocalyx thickness upon oncogenic KRAS activation. ML has also been applied to image glycans in mammalian organs, including the heart (Rong et al., 2014) and brain (Xie et al., 2016), and in whole organisms such as zebrafish (Laughlin et al., 2008). To improve the selectivity of ML in vivo, “caged” sugars can be employed, in which chemical groups on the nonnatural sugar are cleaved by enzymes in the target tissue, allowing the nonnatural sugar to be incorporated (Chang et al., 2010). “Decaging” of sugars by histone deacetylase and cathepsin L enabled selective labeling of glycoproteins in ectopic tumors in mice (Wang et al., 2017). To enhance the efficiency of ML in vivo, liposomes have been used to encapsulate and deliver the nonnatural sugar analog (Xie et al., 2016). This approach allowed an azide-functionalized Neu5Ac derivative to cross the blood-brain barrier and become incorporated into sialylated glycans in mouse brains.
Glycan labeling via ML and CL can be combined with other protein-specific labeling procedures to monitor the glycosylation status of specific glycoproteins directly. For example, a protein-specific fluorescent donor can be combined with a glycan-specific acceptor dye, appended via ML, for fluorescence resonance energy transfer (FRET) to measure the glycosylation status, occupancy, and cell-surface localization of glycoproteins. A modified in situ proximity ligation assay (PLA) can also be used to detect glycosylation levels on specific glycoproteins of interest (Robinson et al., 2016). In this assay, CL is used to install a biotin tag on the glycan of interest. An antibody that recognizes the target protein and an anti-biotin antibody are then incubated with the samples. Each antibody contains a complementary single-stranded DNA oligonucleotide, which hybridize with one another when in proximity, such as on the same glycoprotein. Hybridization can be detected after DNA ligation using fluorescently tagged complementary oligonucleotides or by quantitative polymerase chain reaction (qPCR).
The selection of reagents for ML is critical to target specific glycans (Parker and Pratt, 2020). ManNAc-based probes (Ac4ManNAz and Ac4ManNAlk) are the preferred method to label Neu5Ac-containing glycans in mammalian systems, in part due to their cell permeability. Fuc-specific probes have been generated by additions to the C-6 position (Ac46AzFuc and Ac46AlkFuc). As GalNAc-based probes have been shown to undergo metabolic crosstalk through UDP-GlcNAc/GalNAc-4-epimerase (GALE), leading to promiscuous labeling of various glycans (Boyce et al., 2011), significant efforts have been undertaken to develop selective probes for both GlcNAc- and GalNAc-containing glycans. The development of C-6 modified GlcNAc metabolic probes (Ac36AzGlcNAc and Ac36AlkGlcAc) has enabled the specific profiling of O-GlcNAcylation (Chuh et al., 2017; Chuh et al., 2014), whereas probes with acetamide modifications (Ac4GlcNAz and Ac4GlcNAlk) will label both O-GlcNAc and GlcNAc-containing cell-surface glycoproteins. Work exploring sterically bulky acetamide derivatives of GalNAc has led to the generation of a caged derivative of N-(2-azidopropanoyl)-GalNAc-1-phosphate that can be incorporated specifically into O-GalNAc glycans in cells expressing a mutant form of the nucleotide sugar donor biosynthetic enzyme AGX1 (Debets et al., 2020). As some ML probes can nonspecifically label cysteine residues (Qin et al., 2018), newly discovered glycoproteins should be further verified by mass spectrometry or other methods.
Ultimately, the choice of ML or CL depends on the specific glycan of interest, the extent of labeling and specificity required, and the experimental question being addressed. As described above, ML works particularly well for imaging glycans in vivo, whereas CL is well suited for analyzing human tissue. ML is typically substoichiometric due to competition with natural substrates and therefore should be used with caution when quantifying glycosylation levels. As CL can allow for stoichiometric tagging of endogenous glycans, CL may be better suited for quantification or when high detection sensitivity is required. In terms of specificity, ML generally labels multiple glycan classes containing the monosaccharide of interest, whereas the selectivity of CL depends on the substrate specificity of the GT employed. Thus, CL can be used to detect specific di- or trisaccharide glycan motifs unlike ML. Several detailed reviews are available that outline the best practices for these methods (Cheng et al., 2021; Lopez Aguilar et al., 2017; Parker and Pratt, 2020).
In the future, advancing an understanding of the structure and function of glycans will require new tools for monitoring the glycome with increasing cellular resolution across different cell types, organs, physiological stimuli, and disease states. Single-cell techniques such as flow cytometry and single-cell RNA-sequencing (scRNA-seq) have provided key insights into cellular diversity and heterogeneity, as well as molecular and cellular states important for health and disease. Methods that employ oligonucleotide-conjugated antibodies such as CITE-seq have been developed to translate protein detection into “sequenceable” readouts (Stoeckius et al., 2017). Recently, glycan detection using DNA-barcoded lectins has been combined with this technology to quantify N-glycans on individual cells (Kearney et al., 2021; Minoshima et al., 2021). Although limited by the cross-reactive nature of lectin binding, this proof-of-principle method suggests that new technologies can be developed to profile glycans more precisely at the single-cell level in the future.
As a complement to single-cell barcoding technologies, MS methods to monitor glycan structures have greatly improved and are becoming more routine (described in ‘Identifying relevant glycan structures or “glycotypes”‘). These methods have elucidated important differences in O- and N-glycan structures between mouse brain regions and between sexes, suggesting that distinct glycan repertoires are expressed in different tissues and are subject to tight regulation (Williams et al., 2022). The localization of glycans in tissues from cancer biopsies has also been studied using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS imaging (MSI) (Powers et al., 2013). This approach has been most extensively applied to N-glycans, which are first released from the tissue using PNGase F and then identified in specific regions targeted by the ionizing laser. Such methods have also been applied to formalin-fixed paraffin embedded tissue samples, allowing new analyses for archival tissue samples (Powers et al., 2014). The continued development of these and other related technologies, such as single-cell proteomics and mass cytometry imaging, should enable glycans to be added to multi-omics analyses, providing an essential, missing element for understanding cellular complexity and function.
Modulating glycans to probe function: connecting ‘glycotype’ to phenotype
The ability to selectively modulate glycans on glycoconjugates is critical for relating specific glycan structures and glycosylation events to particular cellular phenotypes. Both loss-of-function and gain-of-function approaches are available for manipulating glycans. For example, pharmacological inhibitors, glycosidase treatment, or genetic knockdown/knockout approaches often lead to loss of function by removing specific cellular glycans. As these methods typically deplete entire families of glycan structures, the results can sometimes be difficult to interpret. Comparative studies in which multiple genes are systematically deleted within a single biosynthetic pathway can provide better control of glycan populations and be more informative.
With the advent of facile gene knockout and editing by ZFNs, TALENs, and CRISPR/Cas9, libraries of defined mutants have been developed for O- and N-glycans (Narimatsu et al., 2019), GPI anchors (Liu et al., 2021), and GAGs (Chen et al., 2018; Qiu et al., 2018), significantly expanding the available toolkit of cell lines with simplified glycomes (Figure 6A). In addition to cell lines, this loss-of-function approach has been applied to more complex systems such as human organotypic skin models to understand the role of glycans in tissue formation (Dabelsteen et al., 2020). Genetic disruption approaches have also been used to generate secreted mucins with defined O-glycan composition as protein-based probes (Nason et al., 2021). Beyond targeting glycan biosynthetic pathways, genetic manipulation enables the interrogation of specific glycosylation events in various biological settings. For example, viral-mediated expression of a site-specific O-GlcNAc-deficient mutant of CREB in cultured neurons and murine brains in vivo revealed that activity-induced O-GlcNAcylation at Ser-40 modulates dendritic and axonal growth, as well as long-term memory consolidation (Rexach et al., 2012). In addition to alanine mutagenesis, specific O-GlcNAcylation sites on proteins have been mutated to Cys using CRISPR-Cas9 technologies. OGT-mediated GlcNAcylation still occurred at the mutated site, producing a hydrolytically resistant, structural mimic of O-GlcNAc (Gorelik et al., 2019). For example, S405C mutation of OGA led to hyper-S-GlcNAcylation and substantially reduced its cellular half-life, suggesting a role for this site-specific modification in regulating OGA stability.
Knockout animal models have also been extensively used to study the roles of glycans and are particularly helpful for examining biological phenotypes that cannot be recapitulated in vitro. As reviewed elsewhere (Stanley, 2016), many constitutive GT knockouts in mice exhibit embryonic lethality, underscoring the importance of glycans for development and health and prompting the development of conditional knockout models. However, interpretation of knockout animal phenotypes can be complicated by the pleiotropic functions of glycans in vivo, whereby similar but nonequivalent animal models may affect only some of the pleiotropic roles of glycans (Häcker et al., 2005). In addition to disrupting glycan biosynthesis, more precise targeting of specific glycan-binding events in model organisms is possible using gene editing technologies. For example, HS GAGs were found to engage TIE1, an orphan receptor critical for vascular development and homeostasis (Griffin et al., 2021). However, knockout of either the glycan or receptor leads to embryonic lethality, preventing the study of HS-TIE1 interactions in the maturing vasculature. To address this, Cas9-targeted mutations were generated in the HS binding cleft of TIE1, allowing selective ablation of the interaction without loss of the protein or polysaccharide. These mutant animals showed aberrant vasculature and altered vascular survival signaling, highlighting how the functional roles of individual glycan-protein interactions can be teased apart and linked to complex in vivo processes.
Gain-of-function approaches have been developed to install a variety of O- and N-glycans onto proteins and cell surfaces. For example, N-glycans can be remodeled on cells using a two-step enzymatic approach like the remodeling of N-glycans on therapeutic antibodies (described in ‘Generating chemically defined glycans and glycoconjugates’) (Figure 6B). This approach greatly simplifies the overall structural complexity of N-glycosylation on the cell. N-glycan engineering has also been applied to impart new functions onto purified proteins. For example, N-glycan engineering of antibodies using CL methods has been used to attach cytotoxic molecules for antibody-drug conjugates (Tang et al., 2017; van Geel et al., 2015).
O-glycan engineering approaches have exploited artificial scaffolds with defined O-glycan structures (Figure 6C). For example, synthetic glycopolymers displaying Neu5Ac-containing motifs and a terminal phospholipid anchor were incorporated into cell surfaces and shown to engage immune inhibitory Siglec receptors and inhibit NK cell-based cytotoxicity (Hudak et al., 2014). In another study, glycopolymers of varying lengths were exploited to modulate the thickness of the glycocalyx surrounding cancer cells (Paszek et al., 2014). These studies demonstrated that the physical properties of the glycocalyx can drive integrin clustering in cancer cells.
A variety of cell-surface glycan engineering methods have also been developed for GAGs (Figure 6C). Using a similar lipid-anchored approach, the presentation of glycopolymers decorated with specific sulfated HS disaccharides was found to accelerate the differentiation of embryonic stem cells into neuronal precursors (Huang et al., 2014) and facilitate agrin-induced clustering of neurotransmitter receptors at the neuromuscular junction (Huang et al., 2018). In a complementary approach, commercially available CS polysaccharides were conjugated to a simple lipid anchor and incorporated into liposomes (Pulsipher et al., 2014). Addition of these functionalized GAG liposomes to primary neurons remodeled the cell surface and promoted signaling and outgrowth in a sulfation-dependent manner. Lipid anchoring of GAGs is often short-lived, with membrane-inserted probes showing half-lives on the order of several hours, but anchoring GAGs to a transmembrane HaloTag protein can produce longer-lived GAGs that are stably detected on the cell surface for over one week (Pulsipher et al., 2015). HS GAG engineering was shown to potentiate neural differentiation of embryonic stem cells (Pulsipher et al., 2015) and angiopoietin signaling (Griffin et al., 2021) based on the sulfation pattern of the displayed GAG. New methods enable the creation of semi-synthetic proteoglycans, wherein both the core protein and GAG composition can be systematically engineered and displayed on cell surfaces (O'Leary et al., 2022). Such approaches may shed light on the importance of the proteoglycan core protein and architecture in the biology of GAGs.
Finally, intracellular O-GlcNAcylation can be modulated by targeting nanobody-fused OGT or OGA enzymes to O-GlcNAcylated substrates tagged with a nanobody recognition epitope (Figure 6D) (Ge et al., 2021; Ramirez et al., 2020). This approach allows for the control of O-GlcNAcylation stoichiometry on specific overexpressed proteins in cells and will likely be extendable to endogenous substrates using CRISPR/Cas9-mediated gene editing. Analysis of proteins containing multiple modification sites may be complicated as the exact sites and their relative glycosylation stoichiometries may differ using this strategy compared to native O-GlcNAcylation. Nevertheless, this emerging approach has been successfully used to show that O-GlcNAcylation alters the kinase activity of casein kinase 2α (Schwein et al., 2022), enabling new studies to understand O-GlcNAc function and its crosstalk with other post-translational modifications.
Conclusions and Outlook
Glycans are intricately involved in all facets of biology, and new breakthrough technologies have begun to reveal mechanisms by which these biomolecules regulate critical functions. In this Primer, we have outlined current methods to identify, characterize, monitor, and modulate glycans for a variety of applications. These approaches overcome the difficulties of glycan complexity and heterogeneity that have historically challenged glycoscience research, providing a clearer picture of the central features that underlie glycan activity. The glycoscience field is still in its exponential phase of growth, and future work will continue to expand the modern toolkit and improve our ability to decipher glycan function. A key remaining challenge for the future will be to rapidly produce larger collections of diverse, chemically pure glycan and glycoconjugate structures. As methods toward streamlined and automated syntheses advance, we envision that these molecules will one day be as accessible as tailored oligonucleotides and peptides are today. Progress in other fields such as protein structure prediction and directed evolution may help to develop new probes and enzymatic reagents with greater selectivity for specific glycan structures. New techniques to analyze data such as machine learning approaches combined with glycan array and other screening methods could propel novel discoveries by parsing information-rich experiments into actionable structure-function hypotheses. The application of new imaging and structural techniques such as super-resolution microscopy, micro-crystal electron diffraction, as well as cryo-electron microscopy and tomography should reveal key insights into the relationships between glycans and the structural organization of multiprotein complexes and subcellular compartments. Looking forward, the development of single-cell, spatial, and temporal technologies for quantifying glycans will greatly expand an understanding of their functions across multicellular systems. This information, combined with glycomics and glycoproteomics data, will add another crucial dimension to multi-omics experiments, providing important insights into cellular complexity and disease, as well as novel disease biomarkers. Although this Primer has focused specifically on tools to study mammalian glycans, a great diversity of carbohydrates is found across other kingdoms of life, including bacteria, fungi, and archaea. These microbial glycans can act as crucial regulators of host function and thus are key to understanding the human holobiont.
The broad integration of glycoscience across other fields of biology will require access to readily usable tools and expertise. The focus on methods development and dissemination, epitomized by the NIH Common Fund Glycoscience Program and similar international efforts, will enable the democratization of these powerful technologies. When coupled with the proper selection of methods, controls, and data analysis, these tools can uncover new biological mechanisms and therapeutic strategies. The glycoscience community at large has a rich history of collaboration. With a broad repertoire of tools in hand along with the enthusiasm of the community, glycoscience research will continue to expand our understanding of the intricacies of mammalian biology.
Glycans are complex, ubiquitous, heterogeneous and play important roles in fundamental processes broadly across biology. Griffin and Hsieh-Wilson provide a critical overview and discuss current methods to identify, characterize, monitor and modulate mammalian glycans and how technological advances have overcome the difficulties of glycan complexity and heterogeneity to provide a clearer picture of central features of that underlie glycan activity.
Acknowledgments
This work is supported the Hope Funds for Cancer Research (HCFR-19-03-02, M.E.G.), the Melanoma Research Foundation (career development award, M.E.G.), and the National Institutes of Health (U01 GM116262, RF1-AG060540, and RF1-AG062324, L.C.H.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, Davalos V, McDermott M, Nayak S, Darvishian F, et al. (2017). A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell 31, 804–819.e807. 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon R, Grant OC, Leviatan Ben-Arye S, Makeneni S, Nivedha AK, Marshanski T, Norn C, Yu H, Glushka JN, Fleishman SJ, et al. (2018). A combined computational-experimental approach to define the structural origin of antibody recognition of sialyl-Tn, a tumor-associated carbohydrate antigen. Sci Rep 8,10786. 10.1038/s41598-018-29209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boittier ED, Burns JM, Gandhi NS, and Ferro V (2020). GlycoTorch Vina: docking designed and tested for glycosaminoglycans. J Chem Inf Model 60, 6328–6343. 10.1021/acs.jcim.0c00373. [DOI] [PubMed] [Google Scholar]
- Bojar D, Meche L, Meng G, Eng W, Smith DF, Cummings RD, and Mahal LK (2022). A useful guide to lectin binding: machine-learning directed annotation of 57 unique lectin specificities. ACS Chem Biol. 10.1021/acschembio.1c00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltje TJ, Buskas T, and Boons GJ (2009). Opportunities and challenges in synthetic oligosaccharide and glycoconjugate research. Nat Chem 1, 611–622. 10.1038/nchem.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MR, and Hanover JA (2013). O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr 33, 205–229. 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, and Bertozzi CR (2011). Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci U S A 108, 3141–3146. 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, and McMahon SB (2002). Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640. 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Briard JG, Jiang H, Moremen KW, Macauley MS, and Wu P (2018). Cell-based glycan arrays for probing glycan-glycan binding protein interactions. Nat Commun 9, 880. 10.1038/s41467-018-03245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimble S, Wollaston-Hayden EE, Teo CF, Morris AC, and Wells L (2010). The Role of the O-GlcNAc modification in regulating eukaryotic gene expression. Curr Signal Transduct Ther 5, 12–24. 10.2174/157436210790226465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Xia J, Zhuang B, Cho KS, Rogers CJ, Gama CI, Rawat M, Tully SE, Uetani N, Mason DE, et al. (2012). A sulfated carbohydrate epitope inhibits axon regeneration after injury. Proc Natl Acad Sci U S A 109, 4768–4773. 10.1073/pnas.1121318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S, Potter J, and Rose SP (1990). Effects of the amnesic agent 2-deoxygalactose on incorporation of fucose into chick brain glycoproteins. J Neurochem 54, 135–142. 10.1111/j.1471-4159.1990.tb13293.x. [DOI] [PubMed] [Google Scholar]
- Chang L, Chen YJ, Fan CY, Tang CJ, Chen YH, Low PY, Ventura A, Lin CC, Chen YJ, and Angata T (2017). Identification of Siglec ligands using a proximity labeling method. J Proteome Res 16, 3929–3941. 10.1021/acs.jproteome.7b00625. [DOI] [PubMed] [Google Scholar]
- Chang PV, Dube DH, Sletten EM, and Bertozzi CR (2010). A strategy for the selective imaging of glycans using caged metabolic precursors. J Am Chem Soc 132, 9516–9518. 10.1021/ja101080y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubard JL, Krishnamurthy C, Yi W, Smith DF, and Hsieh-Wilson LC (2012). Chemoenzymatic probes for detecting and imaging fucose-alpha(1-2)-galactose glycan biomarkers. J Am Chem Soc 134, 4489–4492. 10.1021/ja211312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Narimatsu Y, Clausen TM, Gomes C, Karlsson R, Steentoft C, Spliid CB, Gustavsson T, Salanti A, Persson A, et al. (2018). The GAGOme: a cell-based library of displayed glycosaminoglycans. Nat Methods 15, 881–888. 10.1038/s41592-018-0086-z. [DOI] [PubMed] [Google Scholar]
- Cheng B, Tang Q, Zhang C, and Chen X (2021). Glycan labeling and analysis in cells and in vivo. Annu Rev Anal Chem 14, 363–387. 10.1146/annurev-anchem-091620-091314. [DOI] [PubMed] [Google Scholar]
- Chernykh A, Kawahara R, and Thaysen-Andersen M (2021). Towards structure-focused glycoproteomics. Biochem Soc Trans 49, 161–186. 10.1042/BST20200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ST, Miller MS, Pacoma R, Roland J, Liu J, Schumacher AM, and Hsieh-Wilson LC (2017). Discovery of a small-molecule modulator of glycosaminoglycan sulfation. ACS Chem Biol 12, 3126–3133. 10.1021/acschembio.7b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua JS, and Kuberan B (2017). Synthetic xylosides: probing the glycosaminoglycan biosynthetic machinery for biomedical applications. Acc Chem Res 50, 2693–2705. 10.1021/acs.accounts.7b00289. [DOI] [PubMed] [Google Scholar]
- Chuh KN, Batt AR, Zaro BW, Darabedian N, Marotta NP, Brennan CK, Amirhekmat A, and Pratt MR (2017). The new chemical reporter 6-alkynyl-6-deoxy-GlcNAc reveals O-GlcNAc modification of the apoptotic caspases that can block the cleavage/activation of caspase-8. J Am Chem Soc 139, 7872–7885. 10.1021/jacs.7b02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuh KN, Zaro BW, Piller F, Piller V, and Pratt MR (2014). Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J Am Chem Soc 136, 12283–12295. 10.1021/ja504063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew J, and Hsieh-Wilson LC (2008). Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc 130, 11576–11577. 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, et al. (2020). SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, 1043–1057 e1015. 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney AH, Puffer EB, Pontrello JK, Yang ZQ, and Kiessling LL (2009). Sialylated multivalent antigens engage CD22 in trans and inhibit B cell activation. Proc Natl Acad Sci U S A 106, 2500–2505. 10.1073/pnas.0807207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabelsteen S, Pallesen EMH, Marinova IN, Nielsen MI, Adamopoulou M, Romer TB, Levann A, Andersen MM, Ye Z, Thein D, et al. (2020). Essential functions of glycans in human epithelia dissected by a CRISPR-Cas9-engineered human organotypic skin model. Dev Cell 54, 669–684 e667. 10.1016/j.devcel.2020.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang K, Zhang W, Jiang S, Lin X, and Qian A (2020). Application of lectin microarrays for biomarker discovery. ChemistryOpen 9, 285–300. 10.1002/open.201900326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danglad-Flores J, Leichnitz S, Sletten ET, Abragam Joseph A, Bienert K, Le Mai Hoang K, and Seeberger PH (2021). Microwave-assisted automated glycan assembly. J Am Chem Soc 143, 8893–8901. 10.1021/jacs.1c03851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debets MF, Tastan OY, Wisnovsky SP, Malaker SA, Angelis N, Moeckl LKR, Choi J, Flynn H, Wagner LJS, Bineva-Todd G, et al. (2020). Metabolic precision labeling enables selective probing of O-linked N-acetylgalactosamine glycosylation. Proc Natl Acad Sci U S A 117, 25293–25301. 10.1073/pnas.2007297117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duksin D, and Mahoney WC (1982). Relationship of the structure and biological activity of the natural homologues of tunicamycin. J Biol Chem 257, 3105–3109. [PubMed] [Google Scholar]
- Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, George BM, Majzoub K, Villalta PW, Carette JE, and Bertozzi CR (2021).Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124.e3122. 10.1016/j.cell.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wei M, McKitrick TR, McQuillan AM, Heimburg-Molinaro J, and Cummings RD (2019). Glycan microarrays as chemical tools for identifying glycan recognition by immune proteins. Front Chem 7, 833. 10.3389/fchem.2019.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Ramirez DH, Yang B, D'Souza AK, Aonbangkhen C, Wong S, and Woo M (2021). Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nat Chem Biol 17, 593–600. 10.1038/s41589-021-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddens JP, Lomino JV, DiLillo DJ, Ravetch JV, and Wang LX (2018). Site-selective chemoenzymatic glycoengineering of Fab and Fc glycans of a therapeutic antibody. Proc Natl Acad Sci U S A 115, 12023–12027. 10.1073/pnas.1812833115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno A, Valverde P, Arda A, and Jimenez-Barbero J (2020). Glycan structures and their interactions with proteins. A NMR view. Curr Opin Struct Biol 62, 22–30. 10.1016/j.sbi.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, and Vocadlo DJ (2011). Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol 7, 174–181. 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik A, Bartual SG, Borodkin VS, Varghese J, Ferenbach AT, and van Aalten DMF (2019). Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation. Nat Struct Mol Biol 26, 1071–1077. 10.1038/s41594-019-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik A, and van Aalten DMF (2020). Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chem Biol 1, 98–109. 10.1039/d0cb00052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve H, Cully Z, Blumberg P, and Kresse H (1988). Influence of chlorate on proteoglycan biosynthesis by cultured human fibroblasts. J Biol Chem 263, 12886–12892. [PubMed] [Google Scholar]
- Griffin ME, Sorum AW, Miller GM, Goddard WA 3rd, and Hsieh-Wilson LC (2021). Sulfated glycans engage the Ang-Tie pathway to regulate vascular development. Nat Chem Biol 17, 178–186. 10.1038/s41589-020-00657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith AR, Rogers CJ, Miller GM, Abrol R, Hsieh-Wilson LC, and Goddard WA 3rd (2017). Predicting glycosaminoglycan surface protein interactions and implications for studying axonal growth. Proc Natl Acad Sci U S A 114, 13697–13702. 10.1073/pnas.1715093115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guberman M, and Seeberger PH (2019). Automated glycan assembly: a perspective. J Am Chem Soc 141, 5581–5592. 10.1021/jacs.9b00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker U, Nybakken K, and Perrimon N (2005). Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol 6, 530–541. 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Hahm HS, Schlegel MK, Hurevich M, Eller S, Schuhmacher F, Hofmann J, Pagel K, and Seeberger PH (2017). Automated glycan assembly using the Glyconeer 2.1 synthesizer. Proc Natl Acad Sci U S A 114, E3385–E3389. 10.1073/pnas.1700141114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Collins BE, Bengtson P, and Paulson JC (2005). Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat Chem Biol 1, 93–97. 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- Haukedal H, and Freude KK (2020). Implications of glycosylation in Alzheimer's disease. Front Neurosci 14, 625348. 10.3389/fnins.2020.625348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel DW, Koppolu S, Zhang Y, Kasper B, Meche L, Vaiana CA, Bissel SJ, Carter CE, Kelvin AA, Elaish M, et al. (2020). Glycomic analysis of host response reveals high mannose as a key mediator of influenza severity. Proc Natl Acad Sci U S A 117, 26926–26935. 10.1073/pnas.2008203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ML, Smith RA, Trieger GW, and Godula K (2014). Glycocalyx remodeling with proteoglycan mimetics promotes neural specification in embryonic stem cells. J Am Chem Soc 136, 10565–10568. 10.1021/ja505012a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ML, Tota EM, Lucas TM, and Godula K (2018). Influencing early stages of neuromuscular junction formation through glycocalyx engineering. ACS Chem Neurosci 9, 3086–3093. 10.1021/acschemneuro.8b00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Giddens J, Fan SQ, Toonstra C, and Wang LX (2012). Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc 134, 12308–12318. 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak JE, Canham SM, and Bertozzi CR (2014). Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol 10, 69–75. 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeh E, O'Leary T, Li W, Hawkins R, Hung JR, Parker CG, and Huang ML (2020). Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc Natl Acad Sci U S A 117, 27329–27338. 10.1073/pnas.2009206117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffrin AM, and Hsieh-Wilson LC (2020). Photoaffinity probes for the identification of sequence-specific glycosaminoglycan-binding proteins. J Am Chem Soc 142, 13672–13676. 10.1021/jacs.0c06046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney CJ, Vervoort SJ, Ramsbottom KM, Todorovski I, Lelliott EJ, Zethoven M, Pijpers L, Martin BP, Semple T, Martelotto L, et al. (2021). SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Sci Adv 7. 10.1126/sciadv.abe3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DC, McKay CS, Legault MC, Danielson DC, Blake JA, Pegoraro AF, Stolow A, Mester Z, and Pezacki JP (2011). Cellular consequences of copper complexes used to catalyze bioorthogonal click reactions. J Am Chem Soc 133, 17993–18001. 10.1021/ja2083027. [DOI] [PubMed] [Google Scholar]
- Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, and Hsieh-Wilson LC (2003). A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc 125, 16162–16163. 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- Kiessling LL, and Grim JC (2013). Glycopolymer probes of signal transduction. Chem Soc Rev 42, 4476–4491. 10.1039/c3cs60097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellen L, and Lindahl U (2018). Specificity of glycosaminoglycan-protein interactions. Curr Opin Struct Biol 50, 101–108. 10.1016/j.sbi.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Kudelka MR, Antonopoulos A, Wang Y, Duong DM, Song X, Seyfried NT, Dell A, Haslam SM, Cummings RD, and Ju T (2016). Cellular O-glycome reporter/amplification to explore O-glycans of living cells. Nat Methods 13, 81–86. 10.1038/nmeth.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtoglu M, Maher JC, and Lampidis TJ (2007). Differential toxic mechanisms of 2-deoxy-D-glucose versus 2-fluorodeoxy-D-glucose in hypoxic and normoxic tumor cells. Antioxid Redox Signal 9, 1383–1390. 10.1089/ars.2007.1714. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Baskin JM, Amacher SL, and Bertozzi CR (2008). In vivo imaging of membrane-associated glycans in developing zebrafish. Science 320, 664–667. 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Li L, Tian F, Zhang L, Xue C, and Linhardt RJ (2015). Glycosaminoglycanomics of cultured cells using a rapid and sensitive LC-MS/MS approach. ACS Chem Biol 10, 1303–1310. 10.1021/acschembio.5b00011. [DOI] [PubMed] [Google Scholar]
- Li Q, Kailemia MJ, Merleev AA, Xu G, Serie D, Danan LM, Haj FG, Maverakis E, and Lebrilla CB (2019a). Site-specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal Chem 91, 5433–5445. 10.1021/acs.analchem.9b00776. [DOI] [PubMed] [Google Scholar]
- Li Q, Li Z, Duan X, and Yi W (2014). A tandem enzymatic approach for detecting and imaging tumor-associated Thomsen-Friedenreich antigen disaccharide. J Am Chem Soc 136, 12536–12539. 10.1021/ja5054225. [DOI] [PubMed] [Google Scholar]
- Li Q, Xie Y, Xu G, and Lebrilla CB (2019b). Identification of potential sialic acid binding proteins on cell membranes by proximity chemical labeling. Chem Sci 10, 6199–6209. 10.1039/c9sc01360a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu L, Wei N, Yang JY, Chapla DG, Moremen KW, and Boons GJ (2019c). An automated platform for the enzyme-mediated assembly of complex oligosaccharides. Nat Chem 11, 229–236. 10.1038/s41557-019-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SS, Liu YS, Guo XY, Murakami Y, Yang G, Gao XD, Kinoshita T, and Fujita M (2021). A knockout cell library of GPI biosynthetic genes for functional studies of GPI-anchored proteins. Commun Biol 4, 777. 10.1038/s42003-021-02337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Aguilar A, Briard JG, Yang L, Ovryn B, Macauley MS, and Wu P (2017). Tools for studying glycans: recent advances in chemoenzymatic glycan labeling. ACS Chem Biol 12, 611–621. 10.1021/acschembio.6b01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly M, Leach FE 3rd, Laremore TN, Toida T, Amster IJ, and Linhardt RJ (2011). The proteoglycan bikunin has a defined sequence. Nat Chem Biol 7, 827–833. 10.1038/nchembio.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida T, Lang K, Xue L, Chin JW, and Winssinger N (2015). Site-Specific glycoconjugation of protein via bioorthogonal tetrazine cycloaddition with a genetically encoded trans-cyclooctene or bicyclononyne. Bioconjug Chem 26, 802–806. 10.1021/acs.bioconjchem.5b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariethoz J, Alocci D, Gastaldello A, Horlacher O, Gasteiger E, Rojas-Macias M, Karlsson NG, Packer NH, and Lisacek F (2018). Glycomics@ExPASy: bridging the gap. Mol Cell Proteomics 17, 2164–2176. 10.1074/mcp.RA118.000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta NP, Lin YH, Lewis YE, Ambroso MR, Zaro BW, Roth MT, Arnold B, Langen R, and Pratt MR (2015). O-GlcNAc modification blocks the aggregation and toxicity of the protein alpha-synuclein associated with Parkinson's disease. Nat Chem 7, 913–920. 10.1038/nchem.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbua NE, Li X, Flanagan-Steet HR, Meng L, Aoki K, Moremen KW, Wolfert MA, Steet R, and Boons GJ (2013). Selective exo-enzymatic labeling of N-glycans on the surface of living cells by recombinant ST6Gal I. Angew Chem Int Ed Engl 52, 13012–13015. 10.1002/anie.201307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende M, Bednarek C, Wawryszyn M, Sauter P, Biskup MB, Schepers U, and Brase S (2016). Chemical synthesis of glycosaminoglycans. Chem Rev 116, 8193–8255. 10.1021/acs.chemrev.6b00010. [DOI] [PubMed] [Google Scholar]
- Mende M, Bordoni V, Tsouka A, Loeffler FF, Delbianco M, and Seeberger PH (2019). Multivalent glycan arrays. Faraday Discuss 219, 9–32. 10.1039/c9fd00080a. [DOI] [PubMed] [Google Scholar]
- Mikami T, and Kitagawa H (2013). Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta 1830, 4719–4733. 10.1016/j.bbagen.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Minoshima F, Ozaki H, Odaka H, and Tateno H (2021). Integrated analysis of glycan and RNA in single cells. iScience 24, 102882. 10.1016/j.isci.2021.102882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckl L, Pedram K, Roy AR, Krishnan V, Gustavsson AK, Dorigo O, Bertozzi CR, and Moerner WE (2019). Quantitative super-resolution microscopy of the mammalian glycocalyx. Dev Cell 50, 57–72.e56. 10.1016/j.devcel.2019.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, and Yamaguchi Y (2018). Biophysical analyses for probing glycan-protein interactions. Adv Exp Med Biol 1104, 119–147. 10.1007/978-981-13-2158-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen YH, Schjoldager KT, Steentoft C, et al. (2019). An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol Cell 75, 394–407 e395. 10.1016/j.molcel.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nason R, Bull C, Konstantinidi A, Sun L, Ye Z, Halim A, Du W, Sorensen DM, Durbesson F, Furukawa S, et al. (2021). Display of the human mucinome with defined O-glycans by gene engineered cells. Nat Commun 12, 4070. 10.1038/s41467-021-24366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lutteke T, O'Boyle N, Packer NH, Stanley P, Toukach P, et al. (2019). Updates to the Symbol Nomenclature for Glycans guidelines. Glycobiology 29, 620–624. 10.1093/glycob/cwz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BG, and Freeze HH (2018). Perspectives on glycosylation and its congenital disorders. Trends Genet 34, 466–476. 10.1016/j.tig.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen SS, and Prescher JA (2020). Developing bioorthogonal probes to span a spectrum of reactivities. Nat Rev Chem 4, 476–489. 10.1038/s41570-020-0205-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto PM (2018). The Use of NMR to Study Transient Carbohydrate-Protein Interactions. Front Mol Biosci 5, 33. 10.3389/fmolb.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivedha AK, Thieker DF, Makeneni S, Hu H, and Woods RJ (2016). Vina-Carb: improving glycosidic angles during carbohydrate docking. J Chem Theory Comput 12, 892–901. 10.1021/acs.jctc.5b00834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary TR, Critcher M, Stephenson TN, Yang X, Hassan AA, Bartfield NM, Hawkins R, and Huang ML (2022). Chemical editing of proteoglycan architecture. Nat Chem Biol 18, 634–642. 10.1038/s41589-022-01023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YI, Sheng GJ, Chang SK, and Hsieh-Wilson LC (2013). Tailored glycopolymers as anticoagulant heparin mimetics. Angew Chem Int Ed Engl 52, 11796–11799. 10.1002/anie.201306968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira T, Thaysen-Andersen M, Packer NH, and Kolarich D (2021). The hitchhiker's guide to glycoproteomics. Biochem Soc Trans 49, 1643–1662. 10.1042/BST20200879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Meoz RF, Jiang J, Lazarus MB, Orman M, Janetzko J, Fan C, Duveau DY, Tan ZW, Thomas CJ, and Walker S (2015). A small molecule that inhibits OGT activity in cells. ACS Chem Biol 10, 1392–1397. 10.1021/acschembio.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza M, Pistorio SG, Stine KJ, and Demchenko AV (2018). Automated chemical oligosaccharide synthesis: novel approach to traditional challenges. Chem Rev 118, 8105–8150. 10.1021/acs.chemrev.8b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza M, Stine KJ, and Demchenko AV (2020). HPLC-assisted automated oligosaccharide synthesis: the implementation of the two-way split valve as a mode of complete automation. Chem Commun 56, 1333–1336. 10.1039/c9cc08876h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CG, and Pratt MR (2020). Click chemistry in proteomic investigations. Cell 180, 605–632. 10.1016/j.cell.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, et al. (2014). The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511, 319–325. 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik SK, and Stanley P (2006). Lectin-resistant CHO glycosylation mutants. Methods Enzymol 416, 159–182. 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Burke DF, von Delft F, Mulloy B, and Blundell TL (2000). Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034. 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- Pinho SS, and Reis CA (2015). Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 15, 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Pinzon Martin S, Seeberger PH, and Varon Silva D (2019). Mucins and pathogenic mucin-like molecules are immunomodulators during infection and targets for diagnostics and vaccines. Front Chem 7, 710. 10.3389/fchem.2019.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers TW, Jones EE, Betesh LR, Romano PR, Gao P, Copland JA, Mehta AS, and Drake RR (2013). Matrix assisted laser desorption ionization imaging mass spectrometry workflow for spatial profiling analysis of N-linked glycan expression in tissues. Anal Chem 85, 9799–9806. 10.1021/ac402108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, Haab BB, and Drake RR (2014). MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 9, e106255. 10.1371/journal.pone.0106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher A, Griffin ME, Stone SE, Brown JM, and Hsieh-Wilson LC (2014). Directing neuronal signaling through cell-surface glycan engineering. J Am Chem Soc 136, 6794–6797. 10.1021/ja5005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher A, Griffin ME, Stone SE, and Hsieh-Wilson LC (2015). Long-lived engineering of glycans to direct stem cell fate. Angew Chem Int Ed Engl 54, 1466–1470. 10.1002/anie.201409258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Qin K, Fan X, Peng L, Hong W, Zhu Y, Lv P, Du Y, Huang R, Han M, et al. (2018). Artificial cysteine S-glycosylation induced by per-O-acetylated unnatural monosaccharides during metabolic glycan labeling. Angew Chem Int Ed Engl 57, 1817–1820. 10.1002/anie.201711710. [DOI] [PubMed] [Google Scholar]
- Qiu H, Shi S, Yue J, Xin M, Nairn AV, Lin L, Liu X, Li G, Archer-Hartmann SA, Dela Rosa M, et al. (2018). A mutant-cell library for systematic analysis of heparan sulfate structure-function relationships. Nat Methods 15, 889–899. 10.1038/s41592-018-0189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, Tharakaraman K, Sasisekharan V, and Sasisekharan R (2016). Glycan-protein interactions in viral pathogenesis. Curr Opin Struct Biol 40, 153–162. 10.1016/j.sbi.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez DH, Aonbangkhen C, Wu HY, Naftaly JA, Tang S, O'Meara TR, and Woo CM (2020). Engineering a proximity-directed O-GlcNAc transferase for selective protein O-GlcNAcylation in cells. ACS Chem Biol 15, 1059–1066. 10.1021/acschembio.0c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Angarita V, and Malaker SA (2021). Mucinomics as the next frontier of mass spectrometry. ACS Chem Biol 16, 1866–1883. 10.1021/acschembio.1c00384. [DOI] [PubMed] [Google Scholar]
- Rawat M, Gama CI, Matson JB, and Hsieh-Wilson LC (2008). Neuroactive chondroitin sulfate glycomimetics. J Am Chem Soc 130, 2959–2961. 10.1021/ja709993p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily C, Stewart TJ, Renfrow MB, and Novak J (2019). Glycosylation in health and disease. Nat Rev Nephrol 15, 346–366. 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Clark PM, Mason DE, Neve RL, Peters EC, and Hsieh-Wilson LC (2012). Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol 8, 253–261. 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, and Hsieh-Wilson LC (2010). Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol 6, 645–651. 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley NM, Bertozzi CR, and Pitteri SJ (2021). A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Mol Cell Proteomics 20, 100029. 10.1074/mcp.R120.002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, and Paulson JC (2012). Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol 8, 661–668. 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan CD, and Paulson JC (2011). Glycan microarrays for decoding the glycome. Annu Rev Biochem 80, 797–823. 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PV, Tsai CT, de Groot AE, McKechnie JL, and Bertozzi CR (2016). Glyco-seek: ultrasensitive detection of protein-specific glycosylation by proximity ligation polymerase chain reaction. J Am Chem Soc 138, 10722–10725. 10.1021/jacs.6b03861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J, Han J, Dong L, Tan Y, Yang H, Feng L, Wang QW, Meng R, Zhao J, Wang SQ, and Chen X (2014). Glycan imaging in intact rat hearts and glycoproteomic analysis reveal the upregulation of sialylation during cardiac hypertrophy. J Am Chem Soc 136, 17468–17476. 10.1021/ja508484c. [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Xu G, Li Q, Goonatilleke E, and Lebrilla CB (2018). Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem Rev 118, 7886–7930. 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Hatai Y, and Okada A (2016). Gold nanoparticle-based multivalent carbohydrate probes: selective photoaffinity labeling of carbohydrate-binding proteins. Chem Sci 7, 702–706. 10.1039/c5sc03275j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders WJ, Gordon EJ, Dwir O, Beck PJ, Alon R, and Kiessling LL (1999). Inhibition of L-selectin-mediated leukocyte rolling by synthetic glycoprotein mimics. J Biol Chem 274, 5271–5278. 10.1074/jbc.274.9.5271. [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, and Mohammadi M (2000). Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 6, 743–750. 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Schwein PA, Ge Y, Yang B, D'Souza A, Mody A, Shen D, and Woo CM (2022). Writing and erasing O-GlcNAc on casein kinase 2 alpha alters the phosphoproteome. ACS Chem Biol 17, 1111–1121. 10.1021/acschembio.1c00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng GJ, Oh YI, Chang SK, and Hsieh-Wilson LC (2013). Tunable heparan sulfate mimetics for modulating chemokine activity. J Am Chem Soc 135, 10898–10901. 10.1021/ja4027727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, and Cummings RD (2014). Investigating virus-glycan interactions using glycan microarrays. Curr Opin Virol 7, 79–87. 10.1016/j.coviro.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojitra M, Sarkar S, Maghera J, Rodrigues E, Carpenter EJ, Seth S, Ferrer Vinals D, Bennett NJ, Reddy R, Khalil A, et al. (2021). Genetically encoded multivalent liquid glycan array displayed on M13 bacteriophage. Nat Chem Biol 17, 806–816. 10.1038/s41589-021-00788-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, and Cummings RD (2016). Oxidative release of natural glycans for functional glycomics. Nat Methods 13, 528–534. 10.1038/nmeth.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P (2016). What have we learned from glycosyltransferase knockouts in mice? J Mol Biol 428, 3166–3182. 10.1016/j.jmb.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner E, Flanagan N, and Gildersleeve JC (2016). Perspectives on anti-glycan antibodies gleaned from development of a community resource database. ACS Chem Biol 11, 1773–1783. 10.1021/acschembio.6b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, and Smibert P (2017). Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868. 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, and Kohler JJ (2008). Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J Am Chem Soc 130, 3278–3279. 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- Tang F, Wang LX, and Huang W (2017). Chemoenzymatic synthesis of glycoengineered IgG antibodies and glycosite-specific antibody-drug conjugates. Nat Protoc 12, 1702–1721. 10.1038/nprot.2017.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SL, and Pohl NLB (2016). Automated fluorous-assisted solution-phase synthesis of beta-1,2-, 1,3-, and 1,6-mannan oligomers. Carbohydr Res 430, 8–15. 10.1016/j.carres.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino AL, and Plummer TH Jr. (1994). Enzymatic deglycosylation of asparagine-linked glycans: purification, properties, and specificity of oligosaccharidecleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol 230, 44–57. 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, and Burlingame AL (2012). Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics 11, 215–229. 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet SD, Clerc O, and Ricard-Blum S (2021). Glycosaminoglycan-protein interactions: the first draft of the glycosaminoglycan interactome. J Histochem Cytochem 69, 93–104. 10.1369/0022155420946403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geel R, Wijdeven MA, Heesbeen R, Verkade JM, Wasiel AA, van Berkel SS, and van Delft FL (2015). Chemoenzymatic conjugation of toxic payloads to the globally conserved N-glycan of native mAbs provides homogeneous and highly efficacious antibody-drug conjugates. Bioconjug Chem 26, 2233–2242. 10.1021/acs.bioconjchem.5b00224. [DOI] [PubMed] [Google Scholar]
- van Kuppevelt TH, Oosterhof A, Versteeg EMM, Podhumljak E, van de Westerlo EMA, and Daamen WF (2017). Sequencing of glycosaminoglycans with potential to interrogate sequence-specific interactions. Sci Rep 7, 14785. 10.1038/s41598-017-15009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A (2017). Biological roles of glycans. Glycobiology 27, 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilen Z, Joeh E, Critcher M, Parker CG, and Huang ML (2021). Proximity tagging identifies the glycan-mediated glycoprotein interactors of galectin-1 in muscle stem cells. ACS Chem Biol 16, 1994–2003. 10.1021/acschembio.1c00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, and Mooney DJ (2020). Metabolic glycan labelling for cancer-targeted therapy. Nat Chem 12, 1102–1114. 10.1038/s41557-020-00587-w. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang R, Cai K, He H, Liu Y, Yen J, Wang Z, Xu M, Sun Y, Zhou X, et al. (2017). Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat Chem Biol 13, 415–424. 10.1038/nchembio.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Dong S, Shieh JH, Peguero E, Hendrickson R, Moore MAS, and Danishefsky SJ (2013a). Erythropoietin derived by chemical synthesis. Science 342, 1357–1360. 10.1126/science.1245095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chinoy ZS, Ambre SG, Peng W, McBride R, de Vries RP, Glushka J, Paulson JC, and Boons GJ (2013b). A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans. Science 341, 379–383. 10.1126/science.1236231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Edmunds G, Gibbons C, Zhang J, Gadi MR, Zhu H, Fang J, Liu X, Kong Y, and Wang PG (2018). Toward automated enzymatic synthesis of oligosaccharides. Chem Rev 118, 8151–8187. 10.1021/acs.chemrev.8b00066. [DOI] [PubMed] [Google Scholar]
- Wen L, Zheng Y, Jiang K, Zhang M, Kondengaden SM, Li S, Huang K, Li J, Song J, and Wang PG (2016). Two-Step chemoenzymatic detection of N-acetylneuraminic acid-alpha(2-3)-galactose glycans. J Am Chem Soc 138, 11473–11476. 10.1021/jacs.6b07132. [DOI] [PubMed] [Google Scholar]
- Wibowo A, Peters EC, and Hsieh-Wilson LC (2014). Photoactivatable glycopolymers for the proteome-wide identification of fucose-alpha(1-2)-galactose binding proteins. J Am Chem Soc 136, 9528–9531. 10.1021/ja502482a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Noel M, Lehoux S, Cetinbas M, Xavier RJ, Sadreyev RI, Scolnick EM, Smoller JW, Cummings RD, and Mealer RG (2022). Mammalian brain glycoproteins exhibit diminished glycan complexity compared to other tissues. Nat Commun 13, 275. 10.1038/s41467-021-27781-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, and Pitteri SJ(2018). Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag). Mol Cell Proteomics 17, 764–775. 10.1074/mcp.RA117.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TH, Bower BJ, Chalker JM, Bernardes GJ, Wiewiora R, Ng WL, Raj R, Faulkner S, Vallee MR, Phanumartwiwath A, et al. (2016). Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 354. 10.1126/science.aag1465. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Huang X, Burton AJ, and Swift KA (2016). Probing sialoglycans on fetal bovine fetuin with azido-sugars using glycosyltransferases. Glycobiology 26, 329–334. 10.1093/glycob/cwv109. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Person AD, Anderson M, Burroughs B, Tatge T, Khatri K, Zou Y, Wang L, Geders T, Zaia J, and Sackstein R (2018). Imaging specific cellular glycan structures using glycosyltransferases via click chemistry. Glycobiology 28, 69–79. 10.1093/glycob/cwx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, and Gildersleeve JC (2015). The glycan array platform as a tool to identify carbohydrate antigens. Methods Mol Biol 133l, 27–40. 10.1007/978-1-4939-2874-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Dong L, Du Y, Zhu Y, Hua R, Zhang C, and Chen X (2016). In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc Natl Acad Sci U S A 113, 5173–5178. 10.1073/pnas.1516524113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, and Ng DT (2015). Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol 16, 742–752. 10.1038/nrm4073. [DOI] [PubMed] [Google Scholar]
- Xu D, and Esko JD (2014). Demystifying heparan sulfate-protein interactions. Annu Rev Biochem 83, 129–157. 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, and Liu J (2011). Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science 334, 498–501. 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada I, Shiota M, Shinmachi D, Ono T, Tsuchiya S, Hosoda M, Fujita A, Aoki NP, Watanabe Y, Fujita N, et al. (2020). The GlyCosmos Portal: a unified and comprehensive web resource for the glycosciences. Nat Methods 17, 649–650. 10.1038/s41592-020-0879-8. [DOI] [PubMed] [Google Scholar]
- Yang X, and Qian K (2017). Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 18, 452–465. 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd, Peters EC, Driggers EM, and Hsieh-Wilson LC (2012). Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980. 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Mazumder R, Ranzinger R, Edwards N, Kahsay R, Aoki-Kinoshita KF, Campbell MP, Cummings RD, Feizi T, Martin M, et al. (2020). GlyGen: computational and informatics resources for glycoscience. Glycobiology 30, 72–73. 10.1093/glycob/cwz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G, Hundt N, Cole D, Fineberg A, Andrecka J, Tyler A, Olerinyova A, Ansari A, Marklund EG, Collier MP, et al. (2018). Quantitative mass imaging of single biological macromolecules. Science 360, 423–427. 10.1126/science.aar5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, and Chen X (2016). One-pot multienzyme (OPME) systems for chemoenzymatic synthesis of carbohydrates. Org Biomol Chem 14, 2809–2818. 10.1039/c6ob00058d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SH, Boyce M, Wands AM, Bond MR, Bertozzi CR, and Kohler JJ (2012). Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc Natl Acad Sci U S A 109, 4834–4839. 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SH, Zhao P, Sun T, Gao Z, Moremen KW, Boons GJ, Wells L, and Steet R (2016). Selective exo-enzymatic labeling detects increased cell surface sialoglycoprotein expression upon megakaryocytic differentiation. J Biol Chem 291, 3982–3989. 10.1074/jbc.M115.700369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Listinsky CM, Singh RK, Listinsky JJ, and Siegal GP (2008). Cell surface associated alpha-L-fucose moieties modulate human breast cancer neoplastic progression. Pathol Oncol Res 14, 145–156. 10.1007/s12253-008-9036-x. [DOI] [PubMed] [Google Scholar]
- Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, and Vocadlo DJ (2008). A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol 4, 483–490. 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Ramya TN, Dirksen A, Dawson PE, and Paulson JC (2009). High-efficiency labeling of sialylated glycoproteins on living cells. Nat Methods 6, 207–209. 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li Z, and Song X (2020). Preparation of complex glycans from natural sources for functional study. Front Chem 8, 508. 10.3389/fchem.2020.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Jiang H, Gros M, del Amo DS, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, and Wu P (2011). Tracking N-acetyllactosamine on cell-surface glycans in vivo. Angew Chem Int Ed Engl 50, 4113–4118. 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JY, and Cobb BA (2021). Glycans in immunologic health and disease. Annu Rev Immunol 39, 511–536. 10.1146/annurev-immunol-101819-074237. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Yan M, Lasanajak Y, Smith DF, and Song X (2018). Large scale preparation of high mannose and paucimannose N-glycans from soybean proteins by oxidative release of natural glycans (ORNG). Carbohydr Res 464, 19–27. 10.1016/j.carres.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]