Abstract
As the fruit fly, Drosophila melanogaster, progresses from one life stage to the next, many of the enzymes that compose intermediary metabolism undergo substantial changes in both expression and activity. These predictable shifts in metabolic flux allow the fly meet stage-specific requirements for energy production and biosynthesis. In this regard, the enzyme glycerol-3-phosphate dehydrogenase 1 (GPDH1) has been the focus of biochemical genetics studies for several decades and, as a result, is one of the most well-characterized Drosophila enzymes. Among the findings of these earlier studies is that GPDH1 acts throughout the fly lifecycle to promote mitochondrial energy production and triglyceride accumulation while also serving a key role in maintaining redox balance. Here, we expand upon the known roles of GPDH1 during fly development by examining how depletion of both the maternal and zygotic pools of this enzyme influences development, metabolism, and viability. Our findings not only confirm previous observations that Gpdh1 mutants exhibit defects in larval development, lifespan, and fat storage but also reveal that GPDH1 serves essential roles in oogenesis and embryogenesis. Moreover, metabolomics analysis reveals that a Gpdh1 mutant stock maintained in a homozygous state exhibits larval metabolic defects that significantly differ from those observed in the F1 mutant generation. Overall, our findings highlight unappreciated roles for GPDH1 in early development and uncover previously undescribed metabolic adaptations that could allow flies to survive the loss of this key enzyme.
Keywords: Drosophila, metabolomics, glycerol-3-phosphate dehydrogenase, amino acid metabolism
Introduction
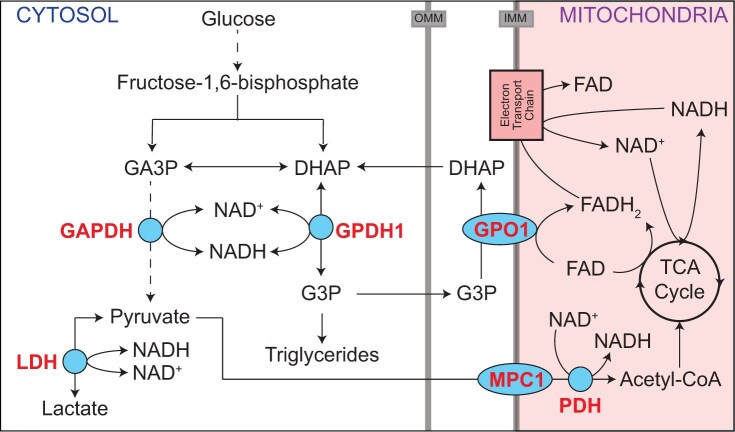
The Drosophila enzyme glycerol-3-phosphate dehydrogenase (GPDH1; encoded by FBgn0001128) is an ideal model for understanding how metabolism adapts to the biosynthetic and energetic requirements of animal development. Although the reaction catalyzed by GPDH1 is relatively simple (Fig. 1), the activity and purpose of the enzyme vary significantly during development. For example, GPDH1 is highly expressed in both the larval fat body and adult flight muscle but serves different purposes within the two tissues. The larval fat body displays high levels of GPDH1 activity and relies on this enzyme to generate glycerol-3-phosphate (G3P; Fig. 1), which is used in triglyceride (TAG) synthesis (Rechsteiner 1970; Sullivan et al. 1983; Merritt et al. 2006; Li et al. 2019). Meanwhile, GPDH1 in adult flight muscle functions in conjunction with the mitochondrial enzyme glycerophosphate oxidase 1 (GPO1) to shuttle electrons into the electron transport chain for ATP synthesis (Sacktor and Dick 1962; O'Brien and MacIntyre 1972a,b; Wojtas et al. 1997; Merritt et al. 2006), thus supporting the intense energy demands of insect flight (Fig. 1).
Fig. 1.
GPDH1 promotes cytosolic redox balance, ATP production, and TAG accumulation. A schematic diagram illustrating the role of GPDH1 in central carbon metabolism. GPDH1 relies on the cofactor NAD+/NADH to interconvert the glycolytic intermediate DHAP to G3P. In Drosophila larvae, the GPDH1-dependent conversion of DHAP to G3P functions in parallel with LDH to maintain redox balance. G3P is used as a precursor for TAG synthesis and functions in the G3P electron shuttle to transfer reducing equivalents to the mitochondria. In adult flight muscle, GPDH1 function in conjunction with GPO1 and the TCA cycle to generate the ATP required for flight. GA3P, glyceraldehyde-3-phosphate; IMM, inner mitochondrial membrane; MPC1, mitochondrial pyruvate carrier 1; OMM, outer mitochondrial membrane; PDH, pyruvate dehydrogenase.
The distinct functions of GPDH1 within the larval fat body and adult flight muscles illustrate why this enzyme serves as a model for understanding how metabolism is coordinately regulated in the context of animal growth, development, and physiology. Nearly 50 years of intensive biochemical and genetic studies revealed that GPDH1 kinetics, stability, and physical characteristics vary dramatically between the larval and adult enzyme pool and that GPDH1 activity fluctuates as a function of developmental time, with activity levels peaking in L3 larvae and in adults (Wright and Shaw 1969; Rechsteiner 1970; O'Brien and MacIntyre 1972a; Sullivan et al. 1983). These biochemical studies, coupled with decades of genetic analysis (O'Brien and MacIntyre 1972b; O'Brien and Shimada 1974; Bewley et al. 1980; Kotarski et al. 1983; Burkhart et al. 1984; Davis and MacIntyre 1988; Gibson et al. 1991; Yamaguchi et al. 1994; Merritt et al. 2006; Carmon et al. 2010; Li et al. 2019), make GPDH1 among the most intensively studied enzymes in developmental biology and provide insight toward how normal animal development relies on dramatic changes in enzyme activity.
Despite Gpdh1 serving as the subject of dozens of biochemical genetic studies, one pool of GPDH1 remains largely overlooked. Drosophila embryos contain a significant amount of maternally loaded GPDH1 that potentially persists into larval development (Wright and Shaw 1969; Wright and Shaw 1970; Casas-Vila et al. 2017). Yet, nearly all Gpdh1 mutant studies only examine zygotic mutants, raising the possibility that novel GPDH1 functions could be discovered by examining mutants lacking the maternal enzyme pool. Here, we address this possibility by examining the metabolic and developmental defects displayed by maternal–zygotic (M/Z) Gpdh1 mutants. Our analysis reveals GPDH1 is not only required for normal oocyte development but also that loss of the maternal GPDH1 pool leads to a significant embryonic lethal phenotype. In contrast, among those Gpdh1 M/Z mutants that survive the embryonic lethal phase, postembryonic development and adult longevity appears similar between zygotic and M/Z Gpdh1 mutants. During our studies, however, we made the unexpected discovery that the homozygous Gpdh1 mutant stock used to study loss of the maternal GPDH1 enzyme pools exhibited striking differences in the steady-state levels of amino acids and tricarboxylic acid (TCA) metabolites when compared with the control strain and F1 mutant generation. These unexpected findings provide insight toward understanding how Drosophila metabolism is rewired to support the complete loss of a major enzyme involved in central carbon metabolism.
Methods
Drosophila melanogaster husbandry and genetics
Fly stocks were maintained on Bloomington Drosophila Stock Center (BDSC) food at 25°C. The Gpdh1A10 mutant allele was described in a previous study from our lab (Li et al. 2019). Briefly, Gpdh1A10 is a CRISPR/Cas9 generated frameshift mutation that deletes 19 bp within exon 3. The mutation is a putative null allele that disrupts both the NAD(P)-binding domain and the C-terminal catalytic domain. For all the experiments, 50 adult virgins were mated with 25 males and the embryos were collected on molasses agar plates with yeast paste for 4 h, as previously described (Li and Tennessen 2017). For larval TAG measurements and GC–MS analysis, animals were allowed to develop on the yeast/molasses agar plate for 60 h at 25°C. For pupariation and eclosion assays, 20 L1 larvae were transferred to individual BDSC food vials and incubated at 25°C. Gpdh1A10 zygotic mutant larvae were collected by crossing Gpdh1A10/CyO, P{w[+mC]=GAL4-twi.G}2.2, P{w[+mC]=UAS-2xEGFP}AH2.2 males and females and selecting for larvae lacking GFP expression. The Gpdh1 maternal–zygotic mutant stock was established by crossing Gdph1A10 zygotic mutant males and females. All experiments described herein were conducted within 1 year of generating the mutant stock.
Ovaries dissection and imaging
Ovaries were dissected from Gpdh1A10/+, Gpdh1A10 (Z), and Gpdh1A10 (M/Z) females that were raised on yeast. Ovaries were fixed with 4% formaldehyde, rinsed twice with phosphate-buffered saline (PBS; pH 7.4), stained with DAPI for 30 min, and mounted on slides using vector shield with DAPI (Vector Laboratories; H-1200-10). Slides were imaged using a Leica SP8 confocal microscope.
Fecundity analysis
Forty virgin females from Gpdh1A10/+ (control), Gpdh1A10 (Z) and Gpdh1A10 (M/Z) were mated with twenty w1118 males in a small mating chamber. Adults were mated for 48 h at 25°C with the egg caps replaced at least once a day. On the morning of day 3, new egg caps were placed in the mating chamber every 2 h. The first and second collections of the day were discarded, and the total number of eggs present during the third 2-h period was quantified for each mating chamber.
Viability assays
Embryonic viability of Gpdh1A10/+, Gpdh1A10 (Z), and Gpdh1A10 (M/Z) genotypes were conducted by crossing 50 female virgins with 25 males in an egg-laying bottle that contained a molasses agar plate partially covered with yeast paste (see Li and Tennessen 2017). The egg-laying plate was replaced every 24 h for 2 days, after which time a fresh egg-laying plate was placed in the bottle and eggs were collected for 2 h. The egg-laying plate was removed, and following an 8h incubation at 25°C to allow for onset of GFP expression from the balancer chromosomes, 30 non-GFP embryos were identified by circling the surrounding agar with a dissecting needle. The number of eggs that hatched to become L1 larvae was recorded 24 h later.
Larval viability was measured by placing 20 synchronized embryos of each genotype on molasses agar plates with yeast paste and measuring time until pupariation. Wandering L3 larvae were subsequently transferred into a glass vial containing BDSC food and monitored until eclosion.
Longevity assay
Virgin female and male adults of the indicated genotypes were separated into glass vials containing BDSC food (n = 10 adults/vial) and maintained at 25°C. Flies were transferred to fresh vials every 3 days without the use of carbon dioxide. The number of dead and alive adults in each vial was recorded daily.
Fat body staining
Mid-L2 larval fat bodies were fixed and stained as previously described (Tennessen et al. 2014a). Briefly, fat bodies were fixed with 4% formaldehyde, rinsed twice with PBS and once with 50% ethanol, and then stained for 2 min at room temperature using filtered 0.5% Solvent Black 3 (CAS Number 4197-25-5; Sigma 199664) dissolved in 75% ethanol. Samples were sequentially rinsed with 50% ethanol, 25% ethanol, and PBS. Stained tissues were mounted on a microscope slide with vector shield with DAPI (Vector Laboratories; H-1200-10).
Gas chromatography–mass spectrometry analysis
Gas chromatography–mass spectrometry (GC–MS) analysis was performed at the Indiana University Mass Spectrometry Facility as previously described (Li and Tennessen 2018). All samples contained 25 mid-L2 larvae and 6 biological replicates were analyzed per genotype. GC–MS data were normalized based on sample mass and an internal succinic-d4 acid standard that was present within the extraction buffer. Data were analyzed using Metaboanalyst version 5.0 following log transformation (base 10) and Pareto Scaling (Pang et al. 2021).
Adult body mass measurements
Ten adult male or female flies were collected 1 day post-eclosion, placed into pretared 1.5-ml microfuge tubes, and the mass was measured using a Mettler XS 105 weighing analytical balance. Six independent samples were measured per genotype.
Statistical analysis
Unless noted, statistical analysis was conducted using GraphPad Prism v9.1. Data are presented as scatter plots, with error bars representing the standard deviation and the line in the middle representing the mean value. Unless noted, data were compared using a Kruskal–Wallis test followed by a Dunn’s multiple comparison test. Longevity data were analyzed using a log-rank (Mantel-Cox) test.
Results
GPDH1 is required for oogenesis and embryonic viability
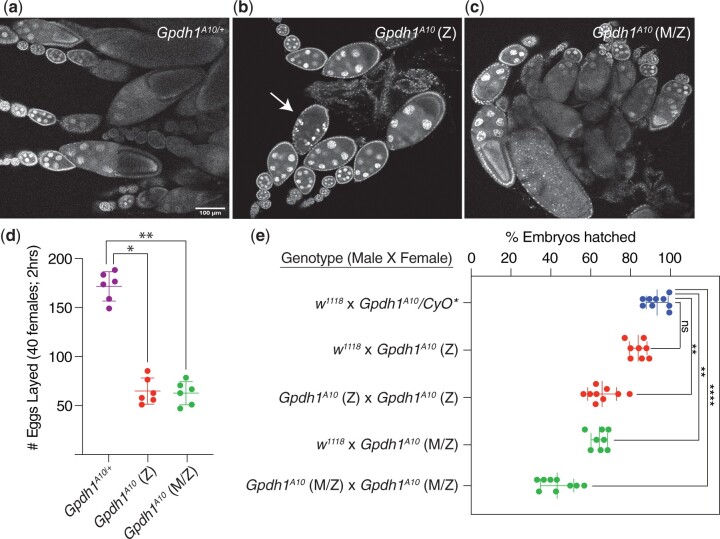
Previous studies indicate that GPDH1 is expressed in the ovary and maternally loaded into the egg (Wright and Shaw 1969, 1970; Casas-Vila et al. 2017). Considering that homozygous Gpdh1 mutant strains are reported to be sub-viable (O'Brien and MacIntyre 1972b; O'Brien and Shimada 1974; Kotarski et al. 1983; Merritt et al. 2006), we examined the possibility that GPDH1 serves essential roles during early development that have been previously overlooked. As a first step toward testing this possibility, we dissected the ovaries from the homozygous Gpdh1A10 mutant flies—both from F1 mutants (generated by crossing Gpdh1A10/CyO, twi-GFP parents and selecting for GFP- larvae), as well as from a homozygous Gpdh1A10 mutant strain (designated M/Z to indicate a complete absence of enzyme during development). We found that the ovaries from both F1 Gpdh1A10 females and Gpdh1A10 M/Z females were smaller than those observed in control females and contained fewer late-stage ovarioles (Fig. 2, a–c). Notably, we regularly observed degenerating egg chambers in mutant females (see Fig. 2b, arrow), indicating that loss of GPDH1 in females limits oocyte production. Consistent with the observed egg chamber defects, both classes of Gpdh1 mutants exhibit significant decreases in egg-laying as compared with controls (Fig. 2d). We would also note the presence of pyknotic nuclei within the mutant follicle cells (see Fig. 2b, arrow, for example), suggesting that future studies should examine the possibility that GPDH1 serves a role in programmed cell death.
Fig. 2.
Gpdh1 is required for oocyte development and embryonic viability. a–c) Ovaries were dissected from 3-day old females and stained with DAPI. When compare to the Gpdh1A10/+ control strain a), the ovaries of b) F1 generation Gpdh1A10 mutants and c) Gpdh1A10 M/Z mutants display b and c) fewer later stage oocytes and b) degenerating egg chambers (see arrow). d) The fecundity of F1 generation Gpdh1A10 mutants and Gpdh1A10 M/Z mutants is significantly decreased when compared with controls. e) Embryos produced by either F1 generation Gpdh1A10 mutants or Gpdh1A10 M/Z mutants exhibit significant mortality when compared with the controls and independent of paternal genotype. *P < 0.05. **P < 0.01. ***P < 0.001. P-values calculated using a Kruskal–Wallis tests followed by a Dunn’s test.
As a complement to our studies of oogenesis, we also examined if the maternal and zygotic GPDH1 enzyme pools are required for embryonic development. Females of a control strain (Gpdh1A10/+), F1 Gpdh1 mutants (Gpdh1A10), and the Gpdh1 mutant strain (Gpdh1A10 M/Z) were crossed with either w1118 controls or Gpdh1A10 mutant males and the resulting offspring were scored for the percentage of embryos that hatched to first instar larvae (L1). We observed that embryos from Gpdh1 mutant mothers of either genetic background died at a significantly higher rate than those produced by heterozygous control mothers, regardless of the parental genotype (Fig. 2e), indicating that both the maternal and zygotic GPDH1 pools are required during embryogenesis. Overall, our findings that Gpdh1 mutants display oogenesis defects and embryonic lethality explains why Gpdh1 mutant females exhibit such low levels of fecundity when compared with control strains.
Zygotic and maternal–zygotic Gpdh1 mutants display similar defects during larval development and adult longevity
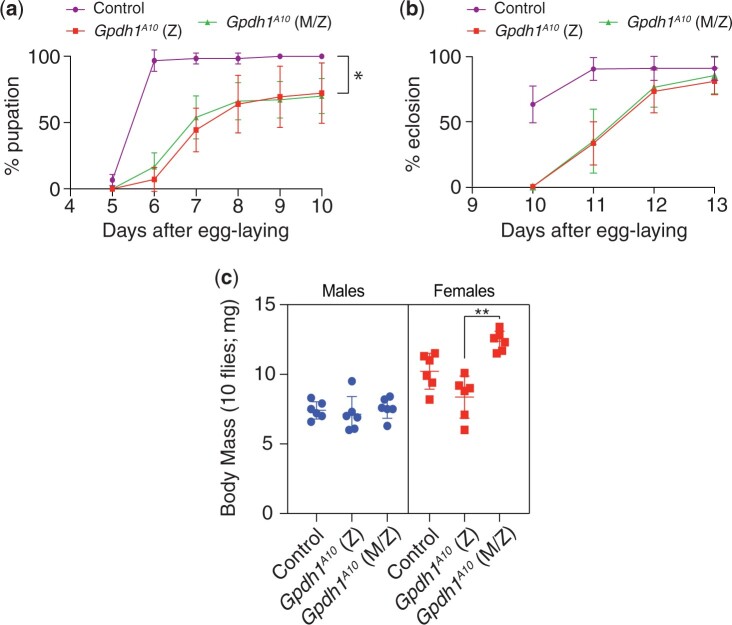
The observed requirement for maternal GPDH1 in embryogenesis led us to reevaluate if loss of this enzyme pool also influences development and lifespan in later life stages. Like previous reports of Gpdh1 zygotic mutants, we found that the Gpdh1 M/Z mutant larvae develop more slowly than heterozygous controls and ∼20% fail to initiate metamorphosis (Fig. 3a). These developmental delay phenotypes, however, are indistinguishable from Gpdh1 zygotic mutants (Fig. 3a). Moreover, Gpdh1 M/Z mutants eclose at the same rate as zygotic mutants (Fig. 3b). The only observable difference between the zygotic and M/Z mutants is that newly eclosed Gpdh1 M/Z mutants females exhibit slightly increased body mass (Fig. 3c); however, we are unable to rule out the possibility that this phenotype is due to differences in genetic background.
Fig. 3.
Gpdh1 zygotic and Gpdh1 maternal–zygotic mutants exhibit similar developmental delays. Gpdh1A10/+ controls, F1 generation Gpdh1A10 mutants, and Gpdh1A10 M/Z mutants were analyzed for a) time from egg-laying to pupariation, b) time from egg-laying to eclosion, and c) body mass 1 day after eclosion. For b), the % eclosion value represents the % of pupae that successfully eclosed and does not include the 20% of larvae that failed to pupariate. **P < 0.01. P-values calculated using a Kruskal–Wallis tests followed by a Dunn’s test.
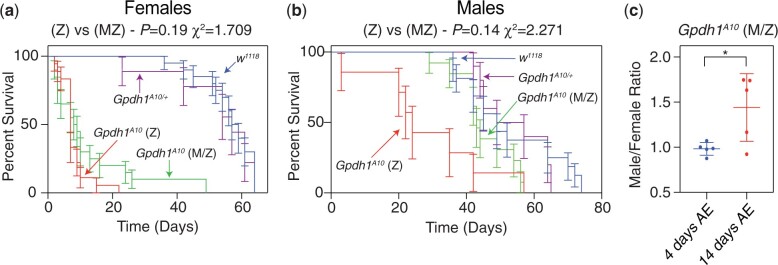
As a complement to our studies of larval development, we also examine the viability of Gpdh1 mutant adults. Here too, we observe little difference between F1 Gpdh1 mutants and M/Z mutants. Consistent with earlier findings (Merritt et al. 2006), we observed that both the Gpdh1A10 zygotic mutant females and M/Z mutant females are short-lived when compared with a heterozygous controls strain, with a majority of mutant animals dying within 2 weeks of eclosion (Fig. 4a). However, we observed that the short lifespan phenotype was milder in F1 Gpdh1A10 mutant males and absent in the Gpdh1A10 M/Z mutant males (Fig. 4b). The observed decrease in female Gpdh1 lifespan is apparent while maintaining the maternal–zygotic stock on Bloomington Drosophila Stock Center food. Although bottles of the Gpdh1A10 M/Z mutant strain contain approximately equal ratios of males: females 2 days after eclosion, this ratio becomes significantly skewed 2 weeks posteclosion due to females dying earlier than male mutants (Fig. 4c). Overall, our findings reveal that nearly all growth, development, and viability phenotypes exhibited by the Gpdh1A10 M/Z mutants are no more severe than those observed in the F1 Gpdh1A10 mutants.
Fig. 4.
Gpdh1 mutant females are short-lived. The lifespan of a) females and b) males from Gpdh1A10/+ heterozygous controls, F1 generation Gpdh1A10 mutants, and a Gpdh1A10 M/Z mutant strain were analyzed for 80 days after eclosion. Longevity data were analyzed using a log-rank (Mantel–Cox) test. c) Ratio of males to females in culture bottles either 2 or 14 days after eclosion. *P < 0.05. P-values calculated using an unpaired t-test.
Gpdh1 zygotic mutants and maternal–zygotic mutants exhibit significant differences in amino acid metabolism
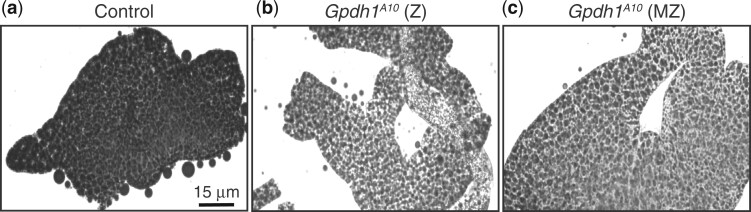
Larval development is able to compensate for loss of zygotic GPDH1 by inducing compensatory changes in central carbon metabolism, rendering Gpdh1 mutants dependent on lactate dehydrogenase (LDH) and GPO1 activity while also inducing significant changes in redox balance and steady-state amino acid levels (Davis and MacIntyre 1988; Li et al. 2019). These earlier metabolic studies were conducted by crossing males and females from a heterozygous Gpdh1-/CyO, twi-GFP stock and selecting for homozygous offspring, thus providing a readout of how loss of zygotic GPDH1 affects larval metabolism. In this regard, our homozygous Gpdh1 M/Z mutant strain provides a unique opportunity to determine if loss of both the maternal and zygotic enzyme pools induce a distinct metabolic profile when compared with zygotic mutants. Toward this goal, we first examined larval TAG levels in the heterozygous control, F1 Gpdh1 mutant, and the homozygous Gpdh1 M/Z strain. Consistent with previous studies (Merritt et al. 2006), we observed similar decreases in fat body TAG levels of the Gpdh1A10 zygotic and Gpdh1A10 M/Z mutant larvae when compared with Gpdh1A10/+ heterozygous controls (Fig. 5, a–c). These results indicate that loss of the maternal GPDH1 pool does not exacerbate the Gpdh1 TAG mutant phenotype.
Fig. 5.
TAG levels are significantly decreased in Gphd1 mutants. Representative images of mid-L2 fat bodies stained with Black Solvent 2 and imaged using bright field microscopy. When compared with a) heterozygous Gpdh1A10/+ control, both b) F1 generation Gpdh1A10 mutants and c) a Gpdh1A10 M/Z mutant strain exhibit a similar decrease in lipid levels. Scale bar in a) applies to b) and c).
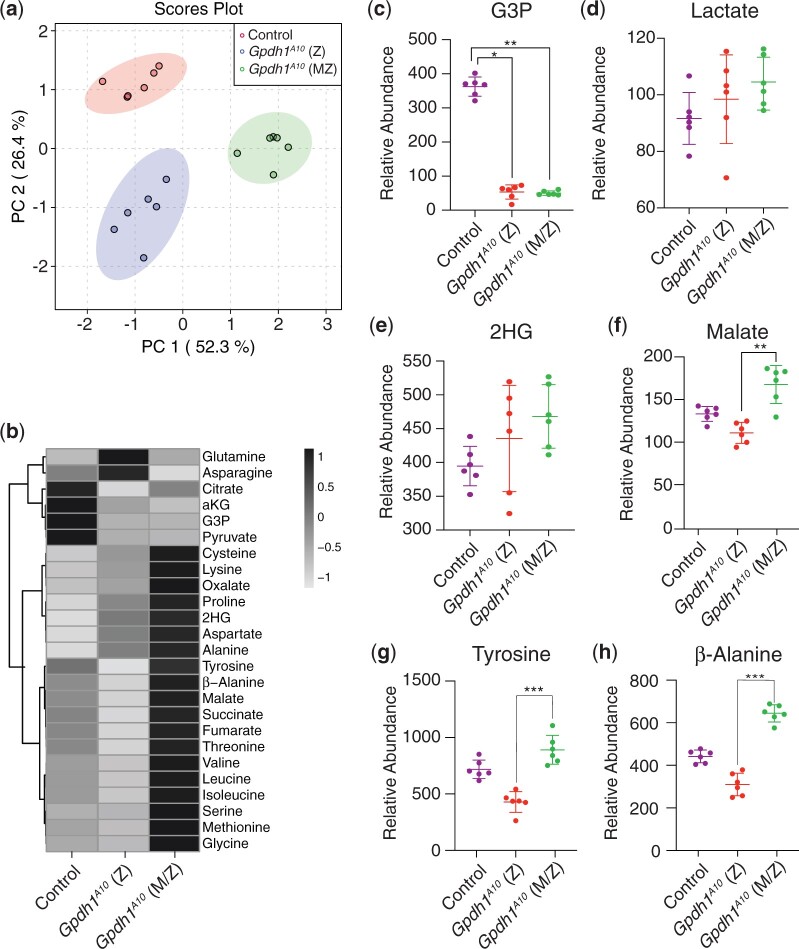
We next used a targeted GC–MS-based approach to compare the levels of amino acids, TCA cycle intermediates, and glycolytic end products in mid-L2 larvae of Gpdh1A10/+ heterozygous controls, Gpdh1A10 zygotic mutants, and Gpdh1A10 M/Z mutants (Supplementary Table 1). When the resulting metabolomics data was analyzed using principal component analysis, the Gpdh1A10 M/Z mutant samples clearly separated from both the Gpdh1A10 zygotic mutants and the Gpdh1A10/+ heterozygous controls (Fig. 6a), suggesting that the 3 genotypes have distinct metabolic profiles. A closer analysis of the datasets revealed that the changes observed in Gpdh1A10 zygotic mutant larvae mimic those observed previously (Li et al. 2019)—G3P levels were significantly decreased in relative to the control strain (Fig. 6, b and c) and both lactate and 2-hydroxyglutarate remained at comparable levels (Fig. 6, b, d, and e). The relative abundance of some amino acids and TCA cycle intermediates were also decreased in the zygotic Gpdh1 mutant larvae compared with Gpdh1A10/+ controls (Fig. 6, b, f, g, and h).
Fig. 6.
Metabolomic analysis of Gpdh1 zygotic and Gpdh1 maternal–zygotic mutants. A targeted GC–MS-based metabolomics method was used to compare the relative abundance of G3P, lactate, 2HG, and amino acids, between heterozygous Gpdh1A10/+ controls, Gpdh1A10 zygotic mutants, and Gpdh1A10 maternal–zygotic mutants. a) PCA plot showing that the control, F1 generation Gpdh1 mutants (Z), and the Gpdh1 M/Z mutant strain separate clearly in their metabolomic profile. b) Heatmap showing the increase in the relative abundance of amino acids in Gpdh1 M/Z mutants when compared to the F1 generation Gpdh1 zygotic mutants and the Gpdh1A10/+ controls. The relative abundance of c) G3P, d) lactate, e) 2HG, f) malate, g) tyrosine, and h) β-alanine are represented as scatter plots with the horizontal lines representing the mean value and standard deviation. *P < 0.05. **P < 0.01. ***P < 0.001. P-values calculated using a Kruskal–Wallis tests followed by a Dunn’s test. Analysis in a) and b) conducted using MetaboAnalyst 5.0 (see Methods).
While the metabolite changes observed in Gpdh1 zygotic mutants largely confirmed previous studies, the metabolic profile of the Gpdh1A10 M/Z mutants exhibited striking differences. Even though G3P, lactate, and 2-hydroxyglutare levels were comparable between the two mutant genotypes (Fig. 6, b–e), many of the same amino acids that were either decreased or unchanged in the zygotic mutants were significantly elevated in the M/Z mutant background (Fig. 6, b and f–h). For example, relative to the control strain, tyrosine and β-alanine were decreased by 40% and 30%, respectively, in zygotic mutants but increased by 40% and 25% in the maternal–zygotic mutant (Fig. 6, g and h). A similar trend was observed with the TCA intermediates succinate, fumarate, and malate (Fig. 6b), with the relative abundance of malate exhibiting a ∼15% decrease in zygotic mutants and a 25% increase in maternal–zygotic mutants when compared to the heterozygous control strain. The results are notable because they support a previously published hypothesis that Gpdh1 mutants experience compensatory changes in malate metabolism (Merritt et al. 2006). Overall, our results reveal that a strain lacking both the maternal and zygotic GPDH1 enzyme pool exhibit changes in the steady-state levels of amino acid and TCA cycle intermediates that are opposite of those observed in mutants lacking only the zygotic enzyme contribution. Future studies should determine if these changes are due to either genetic selection when establishing the homozygous Gpdh1 mutant stock or directly result from loss of the maternal GPDH1 enzyme pool.
Discussion
Here we demonstrate the Drosophila enzyme GPDH1 serves essential roles in both oogenesis and embryogenesis. Our findings expand the known roles for GPDH1 and raise questions as to what function this enzyme serves during early development. Considering that the purpose of GPDH1 activity differs in a context specific manner (e.g. TAG synthesis in fat body, ATP production in flight muscle), our findings raise the question as to the function of GPDH1 in these developmental contexts. Moreover, G3P levels are known to increase over the course of embryogenesis (Tennessen et al. 2014b), suggesting that this compound serves a unique role in the developing embryo.
Our findings also demonstrate that larvae lacking both maternal and zygotic GPDH1 activity exhibit developmental phenotypes that are largely indistinguishable from Gpdh1 zygotic mutants. However, targeted metabolomics analysis indicates that Gpdh1 M/Z mutants exhibit metabolic phenotypes that are more severe than the zygotic mutants, raising the question as to how loss of maternal enzyme pool can influence the larval metabolic program in such a dramatic manner. While we can’t rule out the possibility that maternal GPDH1 activity establishes a metabolic state in the embryo that persists into larval development, a more likely explanation stems from the fact that oogenesis and embryogenesis are energetic processes that impose intense demands on cellular metabolism. As a result, generation of the homozygous Gpdh1 mutant strain would inevitably select for background mutations that compensate for loss of GPDH1 activity in the ovary and embryos—a hypothesis that is supported by previous observations. For example, even though GPDH1 is essential for maintaining ATP production in flight muscle (O'Brien and MacIntyre 1972b; Wojtas et al. 1997; Merritt et al. 2006), Gpdh1 mutants slowly regain the ability to fly when maintained in lab culture (O'Brien and Shimada 1974), indicating that other metabolic processes must be able to compensate for loss of GPDH1 activity. Similarly, Gpdh1 larvae only exhibit slight developmental delays despite displaying a significant disruption in redox balance. This ability of larvae to maintain a somewhat normal growth rate in the absence of GPDH1 depends on the enzymes LDH and GPO1, as loss of either enzyme in a Gpdh1 mutant background enhances the mutant phenotype (Davis and MacIntyre 1988; Li et al. 2019). These earlier studies, combined with our findings and previous observations that Gpdh1 phenotypes are highly dependent on genetic background (Merritt et al. 2006), indicate that GPDH1 functions within a complex and highly adaptable metabolic network that warrants further study.
We would also highlight a few of the limitations imposed by our experimental design. As noted above, the Gpdh1 mutant phenotype is quite sensitive to genetic background effects (Merritt et al. 2006). Since our study relied on a single homozygous mutant line and the control strain consisted of a heterozygous control, there is a distinct possibility that random mutations during establishment of the homozygous strain, as well genetic background differences in the control strain, could have significant effects on the observed phenotypes. In particular, the lifespan phenotypes and metabolomics profiles would likely differ if additional genetic controls would be included. However, we don’t believe that our major findings would change even if additional controls were added to this study. Our major conclusions are that the maternal GPDH1 pool is required for egg production and embryogenesis but has negligible effects on later Gpdh1 mutant phenotypes (apart from the metabolomics profile, which we hypothesize is due to selection for viability; see previous paragraph). We have no doubt that the severity of these phenotypes would differ in a genetic background dependent manner, but our overall findings are consistent with several previous studies. Moreover, the metabolomics profile observed between the zygotic mutant strain and the heterozygous control strain is similar to a previously published study that used a different genetic background (Li et al. 2019).
Regardless of the reason for why Gpdh1 maternal–zygotic mutants exhibit significant metabolic differences when compared with zygotic mutants, our study highlights a poorly understood relationship between G3P and amino acid metabolism. While we are unsure as to the significance of this metabolic relationship in the fly, our findings are consistent with studies of a mouse model of GPD1 deficiency, which induces compensatory amino acid metabolism during fasting in mice (Sato et al. 2016). One intriguing possibility is that the Gpdh1 maternal–zygotic mutants induce changes in one of the central metabolic regulating pathways that control amino acid metabolism. In this regard, the amino acid sensor Tor not only regulates G3P dehydrogenase 1 activity in yeast (Lee et al. 2012), but the GPDH1 substrate, dihydroxyacetone phosphate (DHAP) (see Fig. 1), also activates Tor in mammalian cell culture (Orozco et al. 2020). These correlations between GPDH1 and Tor should be the subject of future investigations.
Our studies also revealed changes in the relative abundance of a subset of TCA acid cycle intermediates, suggesting that mitochondrial metabolism in Gpdh1 maternal–zygotic mutants is fundamentally altered when compared with controls and zygotic mutants. Considering that GPDH1 is an essential enzyme in the G3P electron shuttle (Fig. 1), the observed increases in succinate, fumarate, and malate hint at the possibility that maternal–zygotic mutants exhibit significant changes in mitochondrial metabolism. However, since we observe no changes in the relative abundance of alanine, lactate, and 2-hydroxyglutarate, which are commonly elevated in flies that experience disruption of mitochondrial activity (Feala et al. 2007; Coquin et al. 2008; Campbell et al. 2019; Mahmoudzadeh et al. 2020), the significance of these changes in Gpdh1 maternal–zygotic mutant remain unclear. One explanation is that these metabolomic results hint at a role for maternal GPDH1 that is distinguishable from the zygotic GPDH1 pool. If the primary function of maternal GPDH1 was to drive mitochondrial ATP production during oogenesis and early embryogenesis, loss of GPDH1 would favor selection for genotypes that led to alternative means of mitochondrial energy production. Thus, oocytes and early embryos lacking maternal GPDH1 could compensate by shunting elevated carbon into the TCA cycle, leading to selection for genetic backgrounds with increased TCA flux, and potentially, elevated levels of TCA cycle intermediates. Thus, future studies should examine how loss of maternal GPDH1 interferes with ATP production.
Overall, our results again emphasize that GPDH1 serves a unique role in Drosophila metabolism. The studies presented herein both confirms a large body of literature regarding the role of GPDH1 in physiology, development, and lifespan and reveals new roles for this enzyme in oogenesis, embryogenesis, and amino acid metabolism. Moreover, considering that several studies hint at a key role for human GPD1 in cancer metabolism (Zhou et al. 2017; Rusu et al. 2019; Liu et al. 2021; Xia et al. 2021), our findings highlight the need to better understand how this highly studied enzyme influences gene expression, cell growth and differentiation, and metabolic signaling networks.
Data availability
Gpdh1 mutant strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplemental table.
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
The authors thank the Bloomington Drosophila Stock Center (NIH P40OD018537) for providing fly stocks, the Drosophila Genomics Resource Center (NIH 2P40OD010949) for genomic reagents, and Flybase (NIH 5U41HG000739) and the Indiana University Light Microscopy Imaging Center. Thanks to Lesley N. Weaver for assistance and advice with ovary studies.
Funding
Targeted GC–MS analysis was conducted using instruments housed in the Indiana University Mass Spectrometry Facility, which is supported, in part, by NSF MRI Award 1726633. This publication was made possible with support from the Indiana Clinical and Translational Sciences Institute, which is funded in part by Award Number UL1TR002529 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. JMT is supported by the National Institute of General Medical Sciences of the National Institutes of Health under a R35 Maximizing Investigators’ Research Award (MIRA; 1R35GM119557).
Conflicts of interest
None declared.
Contributor Information
Madhulika Rai, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Sarah M Carter, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Shefali A Shefali, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Nader H Mahmoudzadeh, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Robert Pepin, Department of Chemistry, Indiana University, Bloomington, IN 47405, USA.
Jason M Tennessen, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Literature cited
- Bewley GC, DeZurik JM, Pagelson G.. Analysis of l-glycerol-3-phosphate dehydrogenase mutants in Drosophila melanogaster: complementation for intracellular degradation of the mutant polypeptide. Mol Gen Genet. 1980;178(2):301–308. [DOI] [PubMed] [Google Scholar]
- Burkhart BD, Montgomery E, Langley CH, Voelker RA.. Characterization of allozyme null and low activity alleles from two natural populations of DROSOPHILA MELANOGASTER. Genetics. 1984;107(2):295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JB, Werkhoven S, Harrison JF.. Metabolomics of anoxia tolerance in Drosophila melanogaster: evidence against substrate limitation and for roles of protective metabolites and paralytic hypometabolism. Am J Physiol Regul Integr Comp Physiol. 2019;317(3):R442–R450. [DOI] [PubMed] [Google Scholar]
- Carmon A, Chien J, Sullivan D, MacIntyre R.. The alpha glycerophosphate cycle in Drosophila melanogaster V. molecular analysis of alpha glycerophosphate dehydrogenase and alpha glycerophosphate oxidase mutants. J Hered. 2010;101(2):218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Vila N, Bluhm A, Sayols S, Dinges N, Dejung M, Altenhein T, Kappei D, Altenhein B, Roignant J-Y, Butter F, et al. The developmental proteome of Drosophila melanogaster. Genome Res. 2017;27(7):1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquin L, Feala JD, McCulloch AD, Paternostro G.. Metabolomic and flux-balance analysis of age-related decline of hypoxia tolerance in Drosophila muscle tissue. Mol Syst Biol. 2008;4:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MB, MacIntyre RJ.. A genetic analysis of the alpha-glycerophosphate oxidase locus in Drosophila melanogaster. Genetics. 1988;120(3):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feala JD, Coquin L, McCulloch AD, Paternostro G.. Flexibility in energy metabolism supports hypoxia tolerance in Drosophila flight muscle: metabolomic and computational systems analysis. Mol Syst Biol. 2007;3:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JB, Cao A, Symonds J, Reed D.. Low activity sn-glycerol-3-phosphate dehydrogenase variants in natural populations of Drosophila melanogaster. Heredity. 1991;66(1):75–82. [DOI] [PubMed] [Google Scholar]
- Kotarski MA, Pickert S, Leonard DA, LaRosa GJ, MacIntyre RJ.. The characterization of alpha-glycerophosphate dehydrogenase mutants in Drosophila melanogaster. Genetics. 1983;105(2):387–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jeschke GR, Roelants FM, Thorner J, Turk BE.. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol Cell Biol. 2012;32(22):4705–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Rai M, Buddika K, Sterrett MC, Luhur A, Mahmoudzadeh NH, Julick CR, Pletcher RC, Chawla G, Gosney CJ, et al. Lactate dehydrogenase and glycerol-3-phosphate dehydrogenase cooperatively regulate growth and carbohydrate metabolism during Drosophila melanogaster larval development. Development. 2019;146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tennessen JM.. Methods for studying the metabolic basis of Drosophila development. Wiley Interdiscip Rev Dev Biol. 2017;6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tennessen JM.. Preparation of Drosophila Larval Samples for Gas Chromatography-Mass Spectrometry (GC-MS)-based Metabolomics. J Vis Exp. 2018;136:e57847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Feng Y, Deng Y, Zou Z, Ye J, Cai Z, Zhu X, Liang Y, Lu J, Zhang H, et al. A HIF1alpha-GPD1 feedforward loop inhibits the progression of renal clear cell carcinoma via mitochondrial function and lipid metabolism. J Exp Clin Cancer Res. 2021;40(1):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudzadeh NH, Fitt AJ, Schwab DB, Martenis WE, Nease LM, Owings CG, Brinkley GJ, Li H, Karty JA, Sudarshan S, et al. The oncometabolite L-2-hydroxyglutarate is a common product of dipteran larval development. Insect Biochem Mol Biol. 2020;127:103493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt TJ, Sezgin E, Zhu CT, Eanes WF.. Triglyceride pools, flight and activity variation at the Gpdh locus in Drosophila melanogaster. Genetics. 2006;172(1):293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, MacIntyre RJ.. The -glycerophosphate cycle in Drosophila melanogaster. I. Biochemical and developmental aspects. Biochem Genet. 1972a;7(2):141–161. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, MacIntyre RJ.. The -glycerophosphate in Drosophila melanogaster. II. Genetic aspects. Genetics. 1972b;71(1):127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Shimada Y.. The alpha-glycerophosphate cycle in Drosophila melanogaster. IV. Metabolic, ultrastructural, and adaptive consequences of alphaGpdh-l “null” mutations. J Cell Biol. 1974;63(3):864–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, Lewis CA, Sabatini DM.. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab. 2020;2(9):893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner MC. Drosophila lactate dehydrogenase and alpha-glycerolphosphate dehydrogenase: distribution and change in activity during development. J Insect Physiol. 1970;16(6):1179–1192. [DOI] [PubMed] [Google Scholar]
- Rusu P, Shao C, Neuerburg A, Acikgöz AA, Wu Y, Zou P, Phapale P, Shankar TS, Döring K, Dettling S, et al. GPD1 specifically marks dormant glioma stem cells with a distinct metabolic profile. Cell Stem Cell. 2019;25(2):241–257.e248. [DOI] [PubMed] [Google Scholar]
- Sacktor B, Dick A.. Pathways of hydrogen transport in the oxidation of extramitochondrial reduced diphosphopyridine nucleotide in flight muscle. J Biol Chem. 1962;237:3259–3263. [PubMed] [Google Scholar]
- Sato T, Yoshida Y, Morita A, Mori N, Miura S.. Glycerol-3-phosphate dehydrogenase 1 deficiency induces compensatory amino acid metabolism during fasting in mice. Metabolism. 2016;65(11):1646–1656. [DOI] [PubMed] [Google Scholar]
- Sullivan DT, Donovan FA, Skuse G.. Developmental regulation of glycerol-3-phosphate dehydrogenase synthesis in Drosophila. Biochem Genet. 1983;21(1–2):49–62. [DOI] [PubMed] [Google Scholar]
- Tennessen JM, Barry WE, Cox J, Thummel CS.. Methods for studying metabolism in Drosophila. Methods. 2014a;68(1):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Bertagnolli NM, Evans J, Sieber MH, Cox J, Thummel CS.. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda). 2014b;4(5):839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas K, Slepecky N, von Kalm L, Sullivan D.. Flight muscle function in Drosophila requires colocalization of glycolytic enzymes. Mol Biol Cell. 1997;8(9):1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Shaw CR.. Genetics and ontogeny of alpha-glycerophosphate dehydrogenase isozymes in Drosophila melanogaster. Biochem Genet. 1969;3(4):343–353. [DOI] [PubMed] [Google Scholar]
- Wright DA, Shaw CR.. Time of expression of genes controlling specific enzymes in Drosophila embryos. Biochem Genet. 1970;4(3):385–394. [DOI] [PubMed] [Google Scholar]
- Xia R, Tang H, Shen J, Xu S, Liang Y, Zhang Y, Gong X, Min Y, Zhang D, Tao C, et al. Prognostic value of a novel glycolysis-related gene expression signature for gastrointestinal cancer in the Asian population. Cancer Cell Int. 2021;21(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takano TS, Yamazaki T, Harada K.. Molecular analysis of Gpdh null mutations that arose in mutation accumulation experiments in Drosophila melanogaster. Heredity. 1994;73(4):397–404. [DOI] [PubMed] [Google Scholar]
- Zhou C, Yu J, Wang M, Yang J, Xiong H, Huang H, Wu D, Hu S, Wang Y, Chen X-Z, et al. Identification of glycerol-3-phosphate dehydrogenase 1 as a tumour suppressor in human breast cancer. Oncotarget. 2017;8(60):101309–101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gpdh1 mutant strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplemental table.
Supplemental material is available at G3 online.