Abstract
Osmoregulation in Saccharomyces cerevisiae involves a multistep phosphorelay system requiring three proteins, SLN1, YPD1, and SSK1, that are related to bacterial two-component signaling proteins, in particular, those involved in regulating sporulation in Bacillus subtilis and anaerobic respiration in Escherichia coli. The SLN1-YPD1-SSK1 phosphorelay regulates a downstream mitogen-activated protein kinase cascade which ultimately controls the concentration of glycerol within the cell under hyperosmotic stress conditions. The C-terminal response regulator domains of SLN1 and SSK1 and full-length YPD1 have been overexpressed and purified from E. coli. A heterologous system consisting of acetyl phosphate, the bacterial chemotaxis response regulator CheY, and YPD1 has been developed as an efficient means of phosphorylating SLN1 and SSK1 in vitro. The homologous regulatory domains of SLN1 and SSK1 exhibit remarkably different phosphorylated half-lives, a finding that provides insight into the distinct roles that these phosphorylation-dependent regulatory domains play in the yeast osmosensory signal transduction pathway.
The osmoregulatory system in Saccharomyces cerevisiae represents a particularly novel signal transduction pathway because it combines widely utilized mechanisms for cellular information processing observed in both prokaryotic and eukaryotic cells. The molecular components of this signal-response pathway have been identified previously by classic yeast genetics (reviewed in references 5, 34, and 54). External changes in osmolarity are detected at the cell surface by a transmembrane autophosphorylating histidine protein kinase, SLN1 (35). Signals are propagated in the cytoplasm by a series of steps involving phosphoryl transfer from SLN1 to YPD1 to SSK1 (Fig. 1), which all share sequence and functional similarity with bacterial “two-component” signal transduction proteins (30, 37). Phosphorylation of SSK1 acts as a molecular switch in controlling downstream effectors, which in the yeast osmosensing pathway consist of a mitogen-activated protein (MAP) kinase cascade. Ultimately, activation of the MAP kinase-dependent pathway leads to an increase in intracellular glycerol concentration (2, 3, 6, 31).
FIG. 1.
His-to-Asp phosphorelay in S. cerevisiae osmoregulation. The four-step phosphorelay system involving the SLN1, YPD1, and SSK1 proteins negatively regulates the downstream MAP kinase cascade. Under hyperosmotic stress conditions, dephosphorylation of SSK1 results in activation of the MAP kinase cascade, which raises the level of glycerol within the cell to restore osmotic balance. Solid bars indicate putative transmembrane regions.
The histidine protein kinase SLN1 is a hybrid protein in which the N-terminal region shares sequence similarity with the bacterial histidine kinase family and the C-terminal region has all the conserved features of the regulatory domain of bacterial response regulator proteins (35). SSK1 is a typical two-domain response regulator protein with the exception that the regulatory domain is located at the C terminus rather than at the N terminus (30). The SLN1 and SSK1 response regulator domains are phosphorylated on an aspartic acid residue. Phosphoryl transfer from SLN1 to SSK1 is dependent upon a histidine-phosphorylated protein intermediate, YPD1 (37). SSK1 is unique among characterized response regulator proteins in that phosphorylation acts as a negative regulator of SSK1. It is the unphosphorylated form of SSK1 that interacts with and activates the downstream MAP kinase cascade (36). Under normal environmental conditions, SSK1 must be maintained in a stably phosphorylated state; however, the mechanism(s) by which this is achieved is not yet thoroughly understood.
In recent years, it has become evident that two-component signaling strategies are utilized across several kingdoms, having recently been discovered in archaebacterial species (26, 38, 44), plants (9, 17, 19, 23, 39, 50, 52), fungal organisms (4, 7, 30, 35, 37, 43), and the slime mold Dictyostelium discoideum (42, 51, 59). Although a number of bacterial two-component signaling proteins have been investigated in kinetic detail, very little biochemical information is available for the growing list of eukaryotic two-component signaling proteins that have been discovered thus far.
Posas and coworkers (37) provided the first biochemical evidence that the eukaryotic two-component signaling proteins SLN1, YPD1, and SSK1 function in a multistep phosphorelay pathway analogous to a minor fraction of known bacterial two-component systems, notably, sporulation in Bacillus subtilis (8), virulence in Bordetella pertussis (49), and anaerobic responses in Escherichia coli (20, 21, 48). It has been postulated that the advantages of incorporating multiple phosphoryl transfer steps along a single signaling pathway are to allow for several regulatory checkpoints and possibly additional input from “cross-talk” between parallel pathways (5, 8, 37, 49).
We have initiated studies aimed at the biochemical characterization of the SLN1-YPD1-SSK1 phosphorelay pathway. In this report, part of the yeast osmoregulation system has been reconstituted in vitro by using purified domains. We have compared and contrasted the roles of the homologous regulatory domains associated with SLN1 and SSK1 by examining phosphoryl transfer from YPD1 and the lifetimes of the phosphorylated regulatory domains. Based on our findings, we propose that under normal osmolarity conditions, SSK1 is maintained in its phosphorylated inactive state, at least in part due to the unusual stability of the phosphoaspartate linkage.
MATERIALS AND METHODS
Materials.
All chemicals and biochemicals used were of ultrapure grade. NdeI and chitin beads were obtained from New England Biolabs. Pfu DNA polymerase was purchased from Stratagene. All other restriction endonucleases, DNA-modifying enzymes, and oligonucleotides were purchased from GIBCO/BRL. H332PO4 (>9,000 Ci/mmol) was purchased from Amersham. Kodak BioMax MS film was used for autoradiography. Chromatography media were purchased from Pharmacia and Sigma. Plasmids pPD2146 and pSSK1220, containing the genes for SLN1 and SSK1, respectively, were a gift from H. Saito (Dana-Farber Cancer Institute). The expression vector encoding Salmonella typhimurium CheY, pME124, was kindly provided by A. M. Stock (University of Medicine and Dentistry of New Jersey).
Construction of plasmid expression vectors.
For expression in bacterial cells, gene fragments corresponding to the regulatory domains of the yeast SLN1 and SSK1 proteins (designated R1 and R2, respectively) were amplified by PCR and subcloned into the pCYB2 vector of the IMPACT system (New England Biolabs) to generate fusion proteins consisting of the target protein located at the N terminus, followed by the yeast VMA1 protein-splicing intein domain and a chitin binding domain (CBD) at the C terminus. The SLN1-R1 and SSK1-R2 gene fragments were obtained by PCR amplification from plasmid DNA templates using Pfu DNA polymerase and synthetic oligonucleotides containing unique restriction enzyme recognition sites at the termini. Specifically, plasmid pFJS17 was constructed by subcloning an NdeI-XhoI PCR fragment containing nucleotides 3250 to 3660 of the coding region of SLN1 (encoding amino acids 1084 to 1220) into pCYB2. Similarly, another plasmid was engineered to contain nucleotides 1483 to 2136 of SSK1 (encoding amino acids 495 to 712) cloned into pCYB2 as an NdeI-SmaI fragment. Since the gene fragments were expressed as N-terminal fusion proteins, an ATG start codon was included as part of the NdeI site at the 5′ end. In order to improve expression of the SSK1-R2 fusion protein, the entire coding region corresponding to the SSK1-R2-intein-CBD fusion was further subcloned and placed under the control of the T7 polymerase promoter in the pET11a expression plasmid (Novagen). This pET derivative was designated pFJS16.
Expression and purification of SLN1-R1.
E. coli DH5α cells transformed with the SLN1-R1 fusion construct (pFJS17) were grown in 1 liter of Luria-Bertani medium in the presence of 100 μg of ampicillin/ml at 37°C. When the optical density (at 600 nm) of the culture reached 0.6, the cells were cooled to room temperature and expression of SLN1-R1 was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM. The cultures were shaken for an additional 8 h at room temperature and then harvested, washed, and resuspended at 5 ml/g (wet weight) of cells in lysis buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1 mM EDTA, 0.1% Triton X-100). Cells were lysed by sonication, and the lysates were clarified by centrifugation at 112,000 × g for 1 h at 4°C. The supernatant was loaded onto a 1.5-ml chitin bead column equilibrated in lysis buffer at 4°C. The column was washed sequentially with 20 ml of lysis buffer, 5 ml of cleavage buffer (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA), 5 ml of cleavage buffer containing 5 mM ATP–10 mM MgCl2, and 5 ml of cleavage buffer. The ATP-containing wash was included to remove any bound heat shock proteins (47). The column was then washed immediately with 5 ml of cleavage buffer containing 30 mM dithiothreitol (DTT), and then buffer flow was stopped. The column was incubated overnight at 4°C to allow thiol-induced cleavage of the fusion protein. The target protein was eluted with cleavage buffer. SLN1-R1 was further purified by separation on a Sephadex G50 gel filtration column (bed volume, 50 ml) equilibrated in 20 mM Tris-HCl (pH 7.5)–50 mM NaCl–1 mM EDTA–1 mM DTT. Fractions containing SLN1-R1 were pooled and concentrated by using a Centricon 10 (Amicon) filter unit. The protein was judged to be ≥99% homogeneous based on analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined by the absorbance at 280 nm by using a calculated extinction coefficient of 4,020 M−1 cm−1. Typical yields were 0.2 mg of pure protein/g of cells. Purified SLN1-R1 protein was stored in gel filtration buffer in the presence of 10% glycerol at −80°C.
Expression and purification of SSK1-R2.
Purification of SSK1-R2 was performed similarly to that of SLN1-R1 with the following modifications. The SSK1-R2 fusion protein construct (pFJS16) was expressed in E. coli BL21(DE3) cells. When the optical density (at 600 nm) of the culture had reached 0.8, expression of the fusion protein was induced by the addition of IPTG to a final concentration of 1 mM. Changes in the buffers were as follows: the cell lysis buffer (pH 8.0) contained 20% glycerol, the cleavage buffer (pH 8.0) contained 10% glycerol, and the gel filtration buffer (pH 8.0) contained 100 mM NaCl and 10% glycerol. Final purification by gel filtration chromatography was performed by using Sephadex G75 resin. The protein was judged to be ≥93% homogenous based on analysis by SDS-PAGE. The protein concentration was determined by the absorbance at 280 nm by using a calculated extinction coefficient of 25,440 M−1 cm−1. Typical yields were 0.18 mg of pure protein/g of cells. Purified SSK1-R2 was stored in gel filtration buffer at −80°C.
Purification of CheY and YPD1.
S. typhimurium CheY was purified from E. coli HB101 cells harboring a CheY overexpression plasmid (pME124) according to the work of Stock et al. (46). The expression and purification of yeast YPD1 have been described elsewhere (56).
Phosphorylation reactions using acetyl phosphate.
[32P]-labeled acetyl phosphate was prepared according to the method of Stadtman (45) and stored in 0.1 M Tris-HCl, pH 7.0, in small aliquots at −20°C. Unless stated otherwise, phosphorylation reaction mixtures (15 μl) contained 2 μM (each) CheY and/or YPD1 and either SLN1-R1 or SSK1-R2 in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–1 mM DTT. Reactions were carried out at room temperature; they were initiated by the addition of acetyl [32P]phosphate to 2 to 5 mM and stopped by the addition of 5 μl of 4× stop buffer (0.25 M Tris-HCl [pH 6.8], 8% SDS, 40 mM EDTA, 40% glycerol, 0.008% bromophenol blue). Samples were analyzed by separation of proteins on an SDS–15% polyacrylamide gel followed by autoradiography.
Chemical stabilities of phosphorylated amino acids.
Protein phosphorylation reactions containing acetyl [32P]phosphate were performed as described above and allowed to proceed for 10 min at room temperature. Reactions were terminated by the addition of stop buffer. To assess the acid and base stability of the phospho-amino acid linkage, HCl or NaOH was added to a final concentration of 0.5 or 1 M, respectively. Reaction mixtures were incubated for 10 min at 37°C, after which the reactions were neutralized. Samples were analyzed by SDS-PAGE as described above.
Preparation of phospho-YPD1.
Phospho-YPD1 was prepared by incubating YPD1 (12 μM) with CheY (3 μM) in the presence of 3 mM acetyl [32P]phosphate in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–1 mM DTT in a total volume of 100 μl for 10 min at room temperature. To separate phospho-YPD1 from CheY, 20 μl of a slurry of Q Sepharose Fast Flow resin (Pharmacia) in resin buffer (50 mM Tris-HCl [pH 7.5]–5 mM MgCl2) was added to the reaction mixture and mixed gently for 5 min. The resin was pelleted by centrifugation at 1,000 × g for 2 min and was then washed four times with 100 μl of resin buffer containing 0.1 M NaCl. Phospho-YPD1 was eluted with 60 μl of resin buffer containing 0.25 M NaCl and was used directly in phosphoryl transfer experiments with SLN1-R1 and SSK1-R2.
Phosphorylation and dephosphorylation of SLN1-R1 and SSK1-R2.
Isolated radiolabeled phospho-YPD1 (0.5 μM) was incubated at room temperature with a 10- to 20-fold molar excess of SLN1-R1 or SSK1-R2 in a total volume of 100 μl. Aliquots were removed at indicated time points, mixed with 4× stop buffer to terminate the reaction, and kept at −20°C until gel analysis. Proteins were separated on an SDS–15% polyacrylamide gel and analyzed by autoradiography. Relative amounts of phosphorylated protein were determined by scanning laser densitometry of the autoradiograph. Rate constants for the dephosphorylation reaction and half-lives of the phosphorylated response regulator domains were determined by least-squares fitting of the data to a single exponential, assuming first-order kinetics.
RESULTS
Expression and purification of the regulatory domains SLN1-R1 and SSK1-R2.
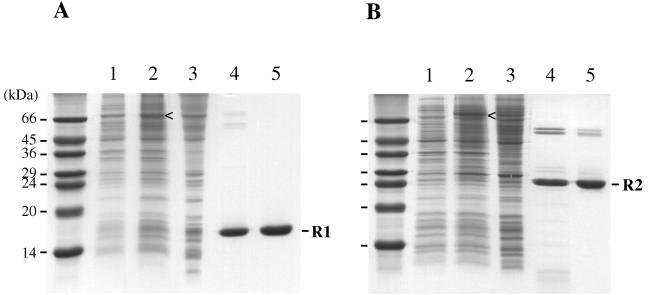
The regulatory domains of the yeast osmoregulation proteins SLN1 and SSK1 were originally identified based on their conserved sequence homology to known bacterial response regulator proteins (30, 35). Each of the two domains, termed SLN1-R1 and SSK1-R2 in this report, is located at the carboxyl terminus of the respective protein. In order to investigate and compare the biochemical properties of these two domains to each other and to other bacterial response regulator proteins, we designed expression plasmids containing only the coding regions for the response regulator domains: amino acids 1084 to 1220 of SLN1 and amino acids 495 to 712 of SSK1. Both domains were overexpressed in E. coli as fusion proteins by using the IMPACT system (Fig. 2). Due to differences in expression levels and solubilities of the resulting fusion proteins, the SLN1-R1 fusion protein was expressed under the control of the Ptac promoter in DH5α cells, whereas the SSK1-R2 fusion protein was expressed under the control of the T7 RNA polymerase promoter in BL21(DE3) cells. Bacterial cultures were grown at room temperature upon induction in order to minimize the formation of inclusion bodies. After overnight incubation with DTT, which induces self-cleavage of the intein fusion protein, the isolated response regulator domains were obtained in relatively pure form (Fig. 2, lanes 4) and were further purified to near-homogeneity by gel filtration chromatography (Fig. 2, lanes 5). As expected, SLN1-R1 and SSK1-R2 migrate on an SDS-polyacrylamide gel with apparent molecular weights of 15,300 and 24,300, respectively (Fig. 2). Both domains were active in terms of phosphoryl transfer activity, as shown below.
FIG. 2.
Purification of SLN1-R1 and SSK1-R2 domains using the IMPACT fusion protein system. Protein expression and purification of SLN1-R1 (A) and SSK1-R2 (B) were analyzed by SDS-PAGE. Lanes 1, whole-cell lysates from uninduced cells; lanes 2, whole-cell lysates of induced cells, grown for 8 h at room temperature; lanes 3, soluble protein fraction after cell lysis; lanes 4, pooled fractions from chitin column following overnight thiol-induced cleavage; lanes 5, pooled fractions after gel filtration. Proteins were denatured in SDS-PAGE sample buffer in the absence of reducing agents, separated on SDS–15% polyacrylamide gels, and stained with Coomassie blue. Sideways carets (<) indicate induced expression of full-length fusion protein. Sizes of molecular markers are given on the left.
Phosphorylation of SLN1-R1 and SSK1-R2 using small-molecule phosphoryl donors.
A number of bacterial response regulator domains can be phosphorylated in vitro by using small-molecule phosphoryl donors (13, 14, 28, 33, 40, 58). For example, the chemotaxis response regulator CheY can be phosphorylated by using acetyl phosphate (Fig. 3, lane 1). In order to assess the activities of the SLN1-R1 and SSK1-R2 response regulator domains, we examined the abilities of the isolated domains to catalyze phosphoryl transfer by using acetyl [32P]phosphate. Both SLN1-R1 and SSK1-R2 were also able to use acetyl phosphate as a phosphoryl donor (Fig. 3, lanes 4 and 7), although with greatly diminished efficiency in comparison to CheY. The weak band representing phosphorylated SLN1-R1 (Fig. 3, lane 4) was observed upon longer exposure of the film.
FIG. 3.
Phosphorylation of YPD1, SLN1-R1, and SSK1-R2 by using a heterologous in vitro system. Phosphorylation reaction mixtures (15 μl) contained 2 μM (each) CheY and/or YPD1, and either SLN1-R1 or SSK1-R2 as indicated in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–1 mM DTT. After the addition of 3 mM acetyl [32P]phosphate, reaction mixtures were incubated for 10 min at room temperature, and reactions were stopped by the addition of 5 μl of 4× stop buffer. Reaction products were separated on an SDS–15% polyacrylamide gel. Immediately following SDS-PAGE, the wet gel was wrapped in plastic film and autoradiography was performed at −80°C. Molecular size markers are indicated on the left.
Phosphorylation of YPD1 using CheY.
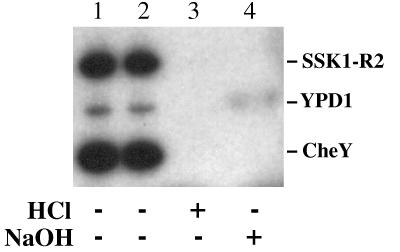
To obtain sufficient quantities of phospho-SLN1-R1 and phospho-SSK1-R2 for biochemical studies, we developed a heterologous system that utilized the bacterial response regulator CheY. In this system, acetyl [32P]phosphate was used to phosphorylate CheY, which in turn served as a phosphoryl donor for the yeast protein YPD1 (Fig. 3, lane 3). Phosphoryl transfer between CheY and YPD1 is reversible (data not shown). To verify proper phosphoryl transfer from the aspartic acid residue of CheY to the histidine residue of YPD1, both phosphorylated proteins were treated with 0.5 M HCl or 1 M NaOH for 10 min at 37°C. Phospho-YPD1 was stable under basic conditions but sensitive to acidic conditions, whereas phospho-CheY was sensitive to both pH extremes (Fig. 4, lanes 3 and 4). This is consistent with an acyl phosphate linkage (aspartyl-phosphate) on CheY and a phosphoramidate linkage (histidinyl-phosphate) on YPD1 (15).
FIG. 4.
Chemical stability of phosphorylated YPD1 and SSK1-R2. Phosphorylation reactions (30 μl) were carried out as described in the legend to Fig. 3 and stopped by the addition of stop buffer. The sample was divided into four aliquots, which were either left untreated (lane 1) or incubated for 10 min at 37°C alone (lane 2) or after the addition of HCl to a final concentration of 0.5 M (lane 3) or NaOH to a final concentration of 1 M (lane 4). Reaction products were separated on an SDS–15% polyacrylamide gel and analyzed by autoradiography.
Phosphoryl transfer from YPD1 to SLN1-R1 and SSK1-R2.
According to the proposed phosphorelay of the yeast osmoregulation pathway, a phosphoryl group is transferred from SLN1 to YPD1, which in turn serves as a phosphoryl donor for SSK1. When either SLN1-R1 or SSK1-R2 was included in a reaction mixture containing acetyl [32P]phosphate, CheY, and YPD1, the phosphoryl group was efficiently transferred to the yeast response regulator domain (Fig. 3, lanes 6 and 9). In the absence of CheY (data not shown) or YPD1 (Fig. 3, lanes 5 and 8), only limited direct phosphorylation of the SLN1-R1 and SSK1-R2 domains with acetyl phosphate was observed. The weak band representing phosphorylated SLN1-R1 (Fig. 3, lane 5) was observed upon longer exposure of the film.
When CheY and YPD1 were preincubated with acetyl [32P]phosphate, phospho-CheY and phospho-YPD1 reached steady-state levels within 10 min (Fig. 5, lane 3). Addition of SSK1-R2 (Fig. 5, lane 5) to this reaction mixture after 30 min resulted in immediate transfer of the phosphoryl group from YPD1 to SSK1-R2 (Fig. 5; compare lanes 4 and 5). Further incubation of the reaction mixture resulted in accumulation of phospho-SSK1-R2 until saturation or steady-state levels were reached, at which time phospho-YPD1 was observed to accumulate (Fig. 5, lanes 8 and 9). The same observation was made when SLN1-R1 was added to the initial reaction mixture containing phospho-CheY and phospho-YPD1 (data not shown). In both phospho-SLN1-R1 (data not shown) and phospho-SSK1-R2 (Fig. 4, lanes 3 and 4), the phospho-amino acid bond was sensitive to acidic and basic pHs, which is consistent with the expected presence of an aspartyl-phosphate on both response regulator domains (15).
FIG. 5.
YPD1-dependent phosphoryl transfer from phospho-CheY to SSK1-R2. CheY (3 μM) and YPD1 (12 μM) were incubated in the presence of 0.7 mM acetyl [32P]phosphate in a total volume of 100 μl in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–1 mM DTT at room temperature. Aliquots (9 μl) were removed at the indicated time points and added to 3 μl of 4× stop buffer (lanes 1 to 4). After 30 min, SSK1-R2 was added to 4 μM, incubation was continued, and aliquots (12 μl) were removed and mixed with 4 μl of 4× stop buffer at indicated time points (lanes 5 to 9). Reaction products were separated on an SDS–15% polyacrylamide gel and analyzed as described in the legend to Fig. 3. Molecular size markers are indicated on the left.
In order to perform biochemical studies on the isolated phosphorylated response regulator domains, we modified the heterologous phosphorylation system by first isolating phospho-YPD1 before the addition of SLN1-R1 or SSK1-R2. Phospho-YPD1 was isolated from the phosphorylation reaction mixture by using anion-exchange chromatography and was subsequently used to phosphorylate SLN1-R1 and SSK1-R2.
Half-lives of the phosphorylated regulatory domains.
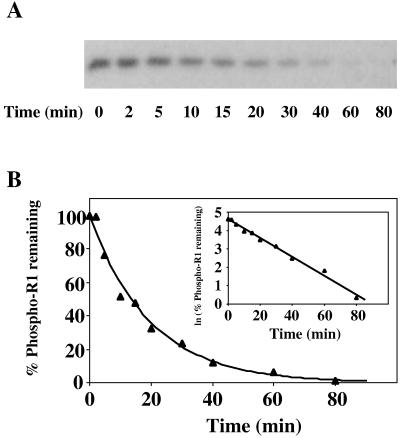
Most phosphorylated bacterial response regulators examined to date have a relatively short half-life in the presence of magnesium ions, ranging from seconds for CheY (16) to about 10 h for the vancomycin resistance protein VanR (53). To address the stability of the phosphorylated form of SSK1-R2, and to compare it to SLN1-R1 and other response regulator proteins, we measured rates of dephosphorylation of phospho-SLN1-R1 and phospho-SSK1-R2. Purified phospho-YPD1 (0.5 μM) was incubated with a molar excess of SLN1-R1 (10 μM) or SSK1-R2 (5 μM) in the presence of MgCl2. Phosphoryl transfer from YPD1 to SLN1-R1 or SSK1-R2 was complete within 1 min. Aliquots were removed at designated time points thereafter, and stop buffer was added. The reaction products were separated on SDS-PAGE gels and analyzed by autoradiography and densitometry (Fig. 6A and 7A). Throughout the time period analyzed, radiolabeled YPD1 was not detectable, indicating that there is no appreciable reverse reaction. The results indicate that the phosphorylated response regulator domains of SLN1 and SSK1 have distinctly different intrinsic stabilities. Phospho-SLN1-R1 exhibited a half-life (t1/2) of 13 ± 1 min (Fig. 6) with a corresponding rate constant (k) of 0.054 min−1, whereas under identical reaction conditions, phospho-SSK1-R2 exhibited a half-life nearly 200-fold greater (t1/2 = 42 ± 10 h; k = 0.017 h−1) (Fig. 7).
FIG. 6.
Dephosphorylation rate of phospho-SLN1-R1. (A) Autoradiograph showing dephosphorylation of phospho-SLN1-R1 as a function of time. Phospho-YPD1 (0.5 μM) was incubated with a 20-fold molar excess of SLN1-R1 in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–1 mM DTT at room temperature for 1 min to allow complete transfer of the phosphoryl group. An aliquot was then removed, mixed with stop buffer, and labeled t0. Subsequently, aliquots were removed at specific time points over the course of 80 min and mixed with stop buffer to terminate the reaction. Proteins were separated on SDS–15% polyacrylamide gels and analyzed as indicated in the legend to Fig. 3. (B) The autoradiographs were further analyzed by scanning laser densitometry to determine the fraction of phospho-SLN1-R1 remaining. Dephosphorylation of phospho-SLN1-R1 followed first-order rate kinetics, and the rate constant and half-life of phospho-SLN1-R1 were determined accordingly. The lines represent computer-generated least-squares fitting to a single exponential or a linear relationship (inset). Data shown are from a representative experiment which was performed multiple times.
FIG. 7.
Dephosphorylation rate of phospho-SSK1-R2. (A) Autoradiograph showing dephosphorylation of phospho-SSK1-R2 as a function of time. Phospho-YPD1 (0.5 μM) was incubated with a 10-fold molar excess of SSK1-R2 as described in the Fig. 6 legend. Subsequent aliquots were removed and mixed with stop buffer over the course of 5 days. Proteins were separated on SDS–15% polyacrylamide gels and analyzed as indicated in the Fig. 3 legend. (B) Autoradiographs were further analyzed by scanning laser densitometry to determine the fraction of phospho-SSK1-R2 remaining. Dephosphorylation of phospho-SSK1-R2 followed first-order rate kinetics, and the rate constant and half-life of phospho-SSK1-R2 were determined as described in the Fig. 6 legend.
DISCUSSION
We expressed and purified the SLN1-R1 and SSK1-R2 domains and YPD1 from E. coli in order to compare and contrast the phosphorylation and dephosphorylation activities of these two response regulator domains that are part of the yeast osmosensing pathway. Although both isolated SLN1-R1 and SSK1-R2 domains could be phosphorylated to a limited extent by using acetyl phosphate, a heterologous system consisting of acetyl phosphate, the bacterial chemotaxis protein CheY, and YPD1 was developed as a more efficient means of phosphorylating SLN1-R1 and SSK1-R2 in vitro.
Histidine-to-aspartate phosphoryl transfer.
Using the heterologous in vitro phosphorylation system, we demonstrated that CheY, phosphorylated by using acetyl phosphate, can serve as a phosphoryl donor to YPD1. This interaction between the bacterial response regulator CheY and the yeast phosphorelay protein YPD1 suggests that molecular recognition elements have been conserved throughout evolution.
YPD1 functions as a phosphoprotein intermediate by receiving a phosphoryl group from SLN1-R1 and transferring it to SSK1-R2. YPD1 is a member of a family of proteins that contain, or consist solely of, a histidine-containing phosphotransfer (HPt) domain; this family includes, among others, ArcB (24), Spo0B (8), and RdeA (10), with which YPD1 shares weak but significant sequence homology. Although there is apparent sequence similarity in the region surrounding the conserved histidine residue that is phosphorylated in YPD1 and two-component sensor kinases (10, 24, 37), YPD1 cannot catalyze autophosphorylation using ATP as a donor and therefore does not itself possess histidine kinase (HK) activity (22). This is consistent with observations made with analogous HPt domains (20, 37, 49).
Posas et al. (37) have demonstrated, using the well-characterized yeast two-hybrid system, that YPD1 physically interacts with the C-terminal response regulator domains of SLN1 and SSK1. Our results show that, at least in vitro, phosphoryl transfer also occurs readily in the “reverse” direction (see Fig. 1), i.e., from YPD1 back to SLN1-R1. Thus, questions regarding molecular recognition and discriminatory interactions of YPD1 with the two response regulator domains SLN1-R1 and SSK1-R2 are raised. We have recently initiated X-ray structural studies of YPD1 (56) in order to examine, in detail, the environment around the conserved histidine and to identify the molecular surface(s) that is available for interaction of YPD1 with the homologous regulatory domains of SLN1 and SSK1.
Interestingly, in addition to activating stress response elements through the HOG1 osmosensing pathway (41), both SLN1 and YPD1 are required for activation of the transcription factors Mcm1 and Skn7 (11), which are involved in growth control and cellular responses to oxidative stress, respectively (7, 12, 25). Thus, YPD1 plays a pivotal role in a branched phosphorelay pathway that is involved in other responses besides osmoregulation. The fact that YPD1 can serve as a phosphoryl acceptor or donor to at least four different response regulator domains indicates that this HPt domain may function in a nondiscriminatory fashion. However, subtle differences in the manner in which YPD1 recognizes and interacts with different response regulator domains may exist, and we are currently investigating this possibility. In light of the participation of YPD1 in other signaling pathways, the mechanism by which SSK1 is phosphorylated via YPD1 and maintained in its inactive phosphorylated state under normal environmental conditions is of fundamental importance, as discussed below.
Stabilities of the phosphorylated response regulator domains.
Studies of several response regulator domains have indicated a much shorter Mg2+-dependent half-life for the phosphorylated state compared to half-lives of small-molecule acyl phosphates (27), ranging from seconds for CheY and CheB (16, 55) to several hours for OmpR and Spo0F (18, 57). These domains thus exhibit an intrinsic phosphatase activity that most likely reflects the duration of the cellular response required for that particular pathway. The response regulator having the longest reported phosphorylated lifetime is VanR (t1/2 = 10 to 12 h) (53). In comparing the in vitro stabilities of the phosphorylated response regulator domains associated with SLN1 and SSK1, we found that while SLN1-R1 exhibited a phosphorylated t1/2 of ∼13 min in the presence of Mg2+, in marked contrast, under identical conditions, the half-life for the phosphorylated SSK1-R2 domain was approximately 200-fold longer (t1/2 = ∼42 h), which to our knowledge is the longest half-life that has been observed for any phosphorylated response regulator domain examined to date. Our preliminary results with full-length SSK1 indicate that the phosphorylated lifetime does not differ significantly from that of the isolated R2 domain, which suggests that the N-terminal effector-like domain does not influence the lifetime of the phosphorylated protein (22).
These results are consistent with the proposed physiological roles for SLN1-R1 and SSK1-R2. The SLN1-R1 domain functions primarily in a phosphorelay capacity, shuttling the phosphoryl group between the sensor kinase SLN1 and YPD1. In contrast, the unusual stability of the phospho-aspartyl bond in SSK1-R2 helps to maintain SSK1 in a phosphorylated inactive state under non-osmotic stress conditions, which is essential to cells because constitutive activation of the HOG1 MAP kinase cascade has lethal consequences (30).
Interestingly, when cells were shifted into high-osmolarity media, activation of the MAP kinase cascade was observed within minutes, as assessed by tyrosine phosphorylation of HOG1 (29). This would necessitate rapid dephosphorylation of SSK1. How is this achieved in light of the inherent stability of the phosphorylated SSK1-R2 domain? One possibility is that the SLN1-HK domain functions as a phosphatase toward SSK1, as has been observed with some prokaryotic sensor kinase-response regulator systems (1, 18, 32, 40). However, studies performed in our laboratory indicate that the presence of the purified SLN1-HK domain does not affect the stability of phospho-SSK1-R2 (22). These observations, therefore, suggest the involvement of an as yet unidentified aspartyl phosphatase that activates SSK1 by catalyzing rapid dephosphorylation of SSK1 when cells are shifted into a high-osmolarity medium. The activation of SSK1 by dephosphorylation is in contrast to the activation of other known characterized bacterial response regulators, which occurs by phosphorylation.
The SLN1-YPD1-SSK1 two-component-based phosphorelay pathway represents a novel form of regulation of a MAP kinase cascade. However, in order to compare features of the two-component signaling strategies that have been retained over evolutionary time and to uncover features that are novel and/or specific to the eukaryotic two-component-regulated pathways, it will be important to more fully investigate the yeast osmoregulatory pathway as a model system.
ACKNOWLEDGMENTS
We are grateful to Ann Stock and Haruo Saito for generously providing plasmid DNAs. We also thank members of the laboratory and Paul F. Cook for critical reading of the manuscript. The helpful comments and suggestions provided by the reviewers are also gratefully acknowledged.
This work was supported by start-up funds provided by the University of Oklahoma to A.H.W.
REFERENCES
- 1.Aiba H, Nakasai F, Mizushima S, Mizuno T. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- 2.Albertyn J, Hohmann S, Prior B A. Characterization of the osmotic-stress response in Saccharomyces cerevisiae: osmotic stress and glucose repression regulate glycerol-3-phosphate dehydrogenase independently. Curr Genet. 1994;25:12–18. doi: 10.1007/BF00712960. [DOI] [PubMed] [Google Scholar]
- 3.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alex L A, Borkovich K A, Simon M I. Hyphal development in Neurospora crassa: involvement of a two-component histidine kinase. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 6.Blomberg A, Adler L. Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J Bacteriol. 1989;171:1087–1092. doi: 10.1128/jb.171.2.1087-1092.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown J L, Bussey H, Stewart R C. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 9.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 10.Chang W-T, Thomason P A, Gross J D, Newell P C. Evidence that the RdeA protein is a component of a multistep phosphorelay modulating rate of development in Dictyostelium. EMBO J. 1998;17:2809–2816. doi: 10.1093/emboj/17.10.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fassler, J., and R. Deschenes. Personal communication.
- 12.Fassler J S, Gray W M, Malone C L, Tao W, Lin H, Deschenes R J. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through the Hog1 osmosensing pathway. J Biol Chem. 1997;272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Atkinson M R, McCleary W, Stock J B, Wanner B L, Ninfa A J. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J Bacteriol. 1992;174:6061–6070. doi: 10.1128/jb.174.19.6061-6070.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher S L, Kim S-K, Wanner B L, Walsh C T. Kinetic comparisons of the specificity of the vancomycin resistance kinase VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- 15.Fujitaki J M, Smith R A. Techniques in the detection and characterization of phosphoramidate-containing proteins. Methods Enzymol. 1984;107:23–36. doi: 10.1016/0076-6879(84)07004-x. [DOI] [PubMed] [Google Scholar]
- 16.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 17.Hua J, Chang C, Sun Q, Meyerowitz E M. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- 18.Igo M M, Ninfa A J, Stock J B, Silhavy T J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- 19.Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishige K, Nagasawa S, Tokishita S, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iuchi S. Phosphorylation/dephosphorylation of the receiver module at the conserved aspartate residue controls transphosphorylation activity of histidine kinase in sensor protein ArcB of Escherichia coli. J Biol Chem. 1993;268:23972–23980. [PubMed] [Google Scholar]
- 22.Janiak-Spens, F., and A. H. West. Unpublished data.
- 23.Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Mizuno T, Shimizu T, Hakoshima T. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell. 1997;88:717–723. doi: 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- 25.Ketela T, Brown J L, Stewart R C, Bussey H. Yeast Skn7p is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol Gen Genet. 1998;259:372–378. doi: 10.1007/s004380050824. [DOI] [PubMed] [Google Scholar]
- 26.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 27.Koshland D E., Jr Effect of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymatic reactions. J Am Chem Soc. 1952;74:2286–2292. [Google Scholar]
- 28.Lukat G S, McCleary W R, Stock A M, Stock J B. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 30.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 31.Mager W H, Varela J C S. Osmostress response of the yeast Saccharomyces. Mol Microbiol. 1993;10:253–258. [PubMed] [Google Scholar]
- 32.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 33.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 34.Morgan B A, Bouquin N, Johnston L H. Two-component signal-transduction systems in budding yeast MAP a different pathway? Trends Cell Biol. 1995;5:453–457. doi: 10.1016/s0962-8924(00)89114-x. [DOI] [PubMed] [Google Scholar]
- 35.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 36.Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SNL1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 38.Rudolf J, Tolliday N, Schmitt C, Schuster S C, Oesterhelt D. Phosphorylation in halobacterial signal transduction. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai H, Hua J, Chen Q G, Chang C, Medrano L J, Bleecker A B, Meyerowitz E M. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schröder I, Wolin C D, Cavicchioli R, Gunsalus R P. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J Bacteriol. 1994;176:4985–4992. doi: 10.1128/jb.176.16.4985-4992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schüller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuster S C, Noegel A A, Oehme F, Gerisch G, Simon M I. The hybrid histidine kinase DokA is part of the osmotic response system of Dictyostelium. EMBO J. 1996;15:3880–3889. [PMC free article] [PubMed] [Google Scholar]
- 43.Shieh J-C, Wilkinson M G, Buck V, Morgan B A, Makino K, Millar J B A. The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-Sty1 MAP kinase pathway and fission yeast cell cycle. Genes Dev. 1997;11:1008–1022. doi: 10.1101/gad.11.8.1008. [DOI] [PubMed] [Google Scholar]
- 44.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stadtman E R. Preparation and assay of acetyl phosphate. Methods Enzymol. 1957;3:228–231. [Google Scholar]
- 46.Stock A M, Mottonen J M, Stock J B, Schutt C E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 47.Thain A, Gaston K, Clarke A R. A method for the separation of GST fusion proteins from co-purifying GroEL. Trends Genet. 1996;12:209–210. doi: 10.1016/s0168-9525(96)90022-0. [DOI] [PubMed] [Google Scholar]
- 48.Tsuzuki M, Ishige K, Mizuno T. Phosphotransfer circuitry of the putative multi-signal transducer, ArcB, of Escherichia coli: in vitro studies with mutants. Mol Microbiol. 1995;18:953–962. doi: 10.1111/j.1365-2958.1995.18050953.x. [DOI] [PubMed] [Google Scholar]
- 49.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 50.Urao T, Yakubov B, Yamaguchi-Shinozaki K, Shinozaki K. Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 1998;427:175–178. doi: 10.1016/s0014-5793(98)00418-9. [DOI] [PubMed] [Google Scholar]
- 51.Wang N, Shaulsky G, Escalante R, Loomis W F. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 1996;15:3890–3898. [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson J Q, Lanahan M B, Yen H-C, Giovannoni J J, Klee H J. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 53.Wright G D, Holman T R, Walsh C T. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993;32:5057–5063. doi: 10.1021/bi00070a013. [DOI] [PubMed] [Google Scholar]
- 54.Wurgler-Murphy S M, Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 55.Wylie D, Stock A, Wong C-Y, Stock J. Sensory transduction in bacterial chemotaxis involves phosphotransfer between Che proteins. Biochem Biophys Res Commun. 1988;151:891–896. doi: 10.1016/s0006-291x(88)80365-6. [DOI] [PubMed] [Google Scholar]
- 56.Xu, Q., V. Nguyen, and A. H. West. Purification, crystallization, and preliminary X-ray diffraction analysis of the yeast phosphorelay protein YPD1. Acta Crystallogr., in press. [DOI] [PubMed]
- 57.Zapf J, Grimshaw C E, Hoch J A, Varughese K I, Whiteley J M. A source of response regulator autophosphatase activity: the critical role of a residue adjacent to the Spo0F autophosphorylation active site. Biochemistry. 1998;37:7725–7732. doi: 10.1021/bi9729615. [DOI] [PubMed] [Google Scholar]
- 58.Zapf J W, Hoch J A, Whiteley J M. A phosphotransferase activity of the Bacillus subtilis sporulation protein Spo0F that employs phosphoramidate substrates. Biochemistry. 1996;35:2926–2933. doi: 10.1021/bi9519361. [DOI] [PubMed] [Google Scholar]
- 59.Zinda M J, Singleton C K. The hybrid histidine kinase dhkB regulates spore germination in Dictyostelium discoideum. Dev Biol. 1998;196:171–183. doi: 10.1006/dbio.1998.8854. [DOI] [PubMed] [Google Scholar]