Abstract
We have identified a novel gene, gtcA, involved in the decoration of cell wall teichoic acid of Listeria monocytogenes serotype 4b with galactose and glucose. Insertional inactivation of gtcA brought about loss of reactivity with the serotype 4b-specific monoclonal antibody c74.22 and was accompanied by a complete lack of galactose and a marked reduction in the amounts of glucose on teichoic acid. Interestingly, the composition of membrane-associated lipoteichoic acid was not affected. Complementation of the mutants with the cloned gtcA in trans restored galactose and glucose on teichoic acid to wild-type levels. The complemented strains also recovered reactivity with c74.22. Within L. monocytogenes, sequences homologous to gtcA were found in all serogroup 4 isolates but not in strains of any other serotypes. In serotype 4b, gtcA appears to be the first member of a bicistronic operon which includes a gene with homology to Bacillus subtilis rpmE, encoding ribosomal protein L31. In contrast to gtcA, the latter gene appears conserved among all screened serotypes of L. monocytogenes.
Listeria monocytogenes is a gram-positive, facultative intracellular pathogen that is widespread in the environment and capable of causing severe infections in humans and animals. Populations at risk include pregnant women, newborns, and immunocompromised individuals, and infections are commonly food-borne. Although several serotypes of L. monocytogenes are present in the environment and in foods, the majority of infections are caused by strains of serogroup 1/2 (mostly serotypes 1/2a and 1/2b) and 4 (almost always serotype 4b) (6, 35). Both flagellar and somatic (mostly carbohydrate) surface antigens contribute to the serotypic designation of the organism (36).
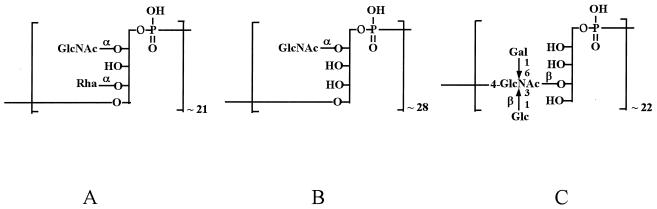
As for many other gram-positive bacteria, the cell wall of L. monocytogenes contains large amounts of the anionic polymer teichoic acid (TA), covalently linked to peptidoglycan. In Bacillus subtilis, TAs have been shown to be essential for viability (22, 28). The TA of B. subtilis 168 consists of polyglycerol phosphate, in contrast to polyribitol phosphate in B. subtilis W23 (1, 5, 28). In L. monocytogenes, a polyribitol phosphate type of TA appears to be the prevalent accessory cell wall polymer (7, 14, 40). Pronounced diversity in TA structure and antigenicity is conferred by glycosidic substitution(s) of the ribitol phosphate units (7, 10, 40). TA of strains of serogroup 1/2 and 3 consists of polyribitol phosphate, with N-acetylglucosamine (Glc-NAc) and rhamnose (in the case of serogroup 1/2) substituents on ribitol (Fig. 1). In contrast, in serogroup 4 strains, Glc-NAc is incorporated in the TA chains, and, depending on the serotype (4a, 4b, etc.), bears galactose and/or glucose substituents. Serotype 4b strains are unique in bearing both galactose and glucose substituents on the Glc-NAc of TA (Fig. 1).
FIG. 1.
Diagrammatic representation of TA composition in different L. monocytogenes serotypes. (A) Type 1/2 (serotypes 1/2a, 1/2b, and 1/2c); (B) type 3 (serotypes 3a, 3b, and 3c); (C) serotype 4b. Modified from the work of Uchikawa et al. (40).
Genetic studies of TA biosynthesis in gram-positive bacteria have mostly involved B. subtilis (19, 28). Although several structural and immunochemical studies of TA in cell walls of different L. monocytogenes serotypes have been performed (7, 10, 14, 39), the molecular genetic mechanisms underlying TA biosynthesis and diversity in Listeria remain to be elucidated. In this communication, we describe gtcA, a novel, serogroup 4-specific gene, which is essential for the presence of normal amounts of galactose and glucose on the TA of L. monocytogenes serotype 4b.
MATERIALS AND METHODS
Bacterial strains and growth media.
Bacteria were grown in brain heart infusion broth (BHI; Difco) at the indicated temperature as previously described (16). Growth on agar was on tryptic soy agar (Difco) supplemented with yeast extract (0.7%) (TSA-YE). All Listeria strains were preserved in BHI at −70°C. When appropriate, antibiotics used for Listeria were streptomycin (1,200 μg/ml), erythromycin (10 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (7 μg/ml). Antibiotics used for Escherichia coli were ampicillin (50 μg/ml) and kanamycin (50 μg/ml). Antibiotics were purchased from Sigma.
Generation and screening of mutants.
Transposon mutants of the serotype 4b strains 4b1 and 2381L were produced as described previously (15) by using filter-mating conjugations with Enterococcus faecalis CG110 or E. faecalis RH110, donors of Tn916 and Tn916ΔE, respectively. Strains 4b1 and 2381L were streptomycin-resistant derivatives of NCTC 10527 (sporadic case isolate) and of strain F2381 (implicated in the Jalisco cheese outbreak), respectively (44). The mutants were grown in 96-well microtiter plates with the appropriate antibiotics (streptomycin-tetracycline and streptomycin-erythromycin for Tn916 and Tn916ΔE mutants, respectively). The mutant banks were kept frozen at −70°C.
Monoclonal antibodies (MAbs) c74.22, c74.33, and c74.180, which react with serotypes 4b, 4d, and 4e but not with other L. monocytogenes serotypes, have been described before (16). For colony immunoblots with these MAbs, the bacteria were grown at 22°C, transferred to nitrocellulose, and processed as described previously (20). An enzyme-linked immunosorbent assay (ELISA) using these MAbs was done as described before (16). Mutant M44 was derived from an earlier Tn916ΔE mutant bank of strain 4b1. Its lack of reactivity with c74.22 but reactivity with c74.33 and c74.180 have been described (16).
Biochemical analysis of cell wall composition.
Cell wall composition was determined as described by Fiedler et al. (7). Preparation and analysis of TA and lipoteichoic acid (LTA) from Listeria were done as previously described (7, 8, 14).
Isolation of transposon-flanking sequences in the mutants.
Procedures for extraction of plasmid DNA from E. coli and genomic DNA from listeriae were previously described (20). Unless otherwise indicated, restriction enzymes and other molecular biologic reagents were purchased from Promega (Madison, Wis.).
The thermostable polymerase used was Tfl (Epicenter, Madison, Wis.). The transposon copy number in the mutants was determined by using Southern blots as previously described (15, 20) with the Genius digoxigenin labeling and detection system (Boehringer-Mannheim). The single-copy transposon mutant M44 was used to amplify a transposon-flanking fragment by means of the single specific primer PCR (SSP-PCR) (37) using EcoRI-digested M44 DNA and pUC18, which was digested by EcoRI and dephosphorylated (Pharmacia) as described previously (43). A 500-bp product was amplified, cloned in pCR1000 (Invitrogen), and sequenced. Primers internal to this fragment (PALN, 5′ TGG GTT ACT ACA AGA AGA G 3′, and PALT, 5′ AGT ACT GAT GCG ATA AAA GCA 3′) were used to amplify a 300-bp fragment which was cloned in pCR1000, resulting in plasmid pNP1. Transposon-flanking fragments from the other mutants were obtained by PCR using primer OTL (5′-CGG AAT TCC GTG AAG TAT CTT CCT ACA G-3′) derived from one of the Tn916 terminal sequences and primer PLNEC (5′ CGG AAT TCC TGA GTT ACT ACA AGA AGA G 3′). Both primers contained EcoRI sites (underlined) at their 5′ end.
Plasmid constructions (pNP500, pNP21, and pNP95).
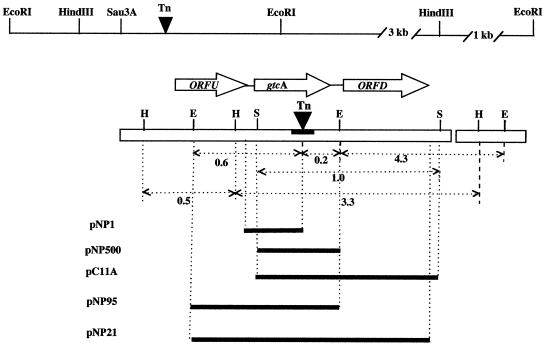
A 4b1 genomic library (5,000 clones; average insert size, 4 to 10 kb) was constructed as described previously (43). This library was screened with pNP1 as a probe, but no positive clones were identified. Another 4b1 library (2,000 clones) was constructed similarly, except that inserts were in the 2- to 4-kb range. One positive clone, pC11A, was identified in this library following screening with pNP1. Plasmid pNP500 was constructed by digesting pC11A with EcoRI (one EcoRI site was in the multiple cloning site of the vector, and the other was in the cloned insert) and subcloning the EcoRI fragment into EcoRI-digested and dephosphorylated pUC18. The locations of the fragments cloned in pNP1, pNP500, and pC11A are indicated in Fig. 3A (see below).
FIG. 3.
Organization of the gtcA genomic region. Arrows indicate the direction of transcription. The solid bar in gtcA indicates the location of the transposon insertion sites (Tn) in M44, B6S, and 27E8. The thick lines indicate the DNA fragments cloned in the indicated plasmids. Numbers indicate sizes of fragments in kilobases. Recognition sites for HindIII (H), Sau3A (S), and EcoRI (E) are indicated.
To construct plasmid pNP21, we used primers RhoBam (5′ GCG CGG ATC CAA AGG GAC AGG CAA CA 3′) and SmeBam (5′ GCG CGG ATC CAC TTG AAA ACC TCC T 3′) corresponding to nucleotides (nt) 6 to 22 and 1302 to 1319, respectively, and containing BamHI recognition sites (underlined) at their 5′ ends. The PCR fragment was digested with BamHI and ligated to the L. monocytogenes-E. coli shuttle vector pKSV7 (38), which had been digested with BamHI and dephosphorylated. To construct pNP95, pNP21 was digested with EcoRI, and the 860-bp EcoRI fragment internal to the cloned insert was cloned into EcoRI-digested and dephosphorylated pKSV7. The fragments cloned in pNP21 and pNP95 are shown in Fig. 3A.
To amplify the gtcA gene from strain 2381L, we used primers RhoBam and SmeBam (see above) and genomic DNA from 2381L as the template. The PCR fragment was purified and sequenced directly.
Preparation of L. monocytogenes electrocompetent cells and electroporation.
To prepare electrocompetent cells of L. monocytogenes, we used the penicillin treatment protocol of Park and Stewart (24). Electrocompetent cells were used immediately or frozen at −70°C. For electroporation, 100 μl of cells (thawed on ice if kept frozen) was mixed gently with 1 μg of plasmid DNA (Wizard minipreps; Promega). The cells were then transferred into 1-mm prechilled electroporation cuvettes (Eppendorf, Madison, Wis.). The electroporator (Eppendorf) was set to 1.5 kV to obtain the field strength of 12.5 kV/cm, as suggested by the vendor. After application of the electrical pulse, 1 ml of BHI with 0.5 M sucrose was added, and the cuvette was incubated at 30°C for 1 h. The cells (100 μl) were then plated on TSA-YE plates containing chloramphenicol (7 μg/ml) and incubated at 30°C for 2 to 3 days.
Northern blotting and reverse transcriptase PCR (RT-PCR).
RNA for Northern blots was prepared from mid-logarithmic-phase cultures. Following treatment with lysozyme and pronase as for DNA extraction, a 10× volume of Tri Reagent (Molecular Research Center, Inc.) was added and the mixture was incubated at room temperature for 5 min to allow complete dissociation of nucleoprotein complexes. Extraction and precipitation of RNA were done in accordance with the suggestions of the vendor, and Northern transfer was done as described before (2) by using positively charged membranes (Magnacharge Micron Separations Inc.). Radioactive probe labeling and signal detection were done as described previously (2).
4b1 RNA for RT-PCR was prepared by the procedures of Sato (34) and Roels and Losick (32). RNA was resuspended in 100 μl of diethylpyrocarbonate-treated water and stored at −70°C. Genomic DNA that might be contaminating the RNA preparation was destroyed by DNase I (Promega) (1 U/μl, 37°C, for 2 to 3 h), and the DNase was then inactivated by boiling for 10 min. The absence of contaminating DNA was confirmed by PCR using the DNase-treated RNA as the template.
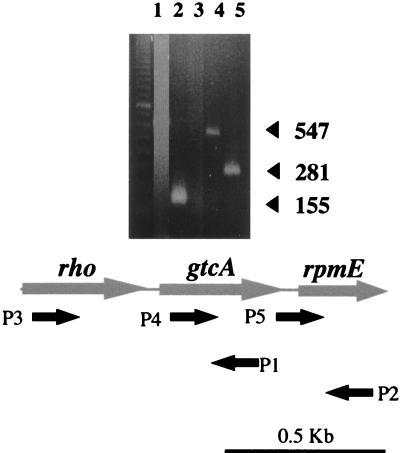
For RT-PCR, 15 μl of the RNA was used to synthesize cDNA in a 33-μl reaction mixture, including 11 μl of first-strand reaction mix (Pharmacia Biotech), 1 mM dithiothreitol, 1 μl of antisense primer, and 5 μl of RNase-free water at 37°C for 1 h. The samples were then heated (95°C, 5 min) to destroy the RT, and 2 to 3 μl of the cDNA was used as the template in a subsequent 25-μl PCR (94°C, 1 min; 50°C, 1 min; 70°C, 2 min; for a total of 30 cycles in a Perkin-Elmer thermocycler). Antisense primers used for cDNA synthesis were P1 (5′ CTG ATT AAC AGA ATC ATG AC 3′, nt 621 to 640) and P2 (5′ TGT TTA CCA GTA TAG AAC GG 3′, nt 1013 to 1032). The subsequent PCRs included one of these primers as well as one of the following primers, as indicated in Fig. 7: P3 (5′ AAA GGG ACA GGC AAC A 3′, nt 6 to 21), P4 (3′ CGC ATC AGT ACT TTT TGC C 3′, nt 484 to 503), and P5 (5′ GCT TGT TGC CTT CAA CAT TC 3′, nt 751 to 770).
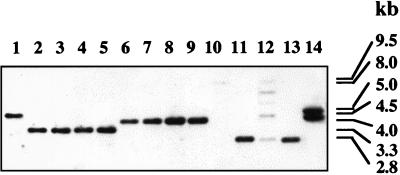
FIG. 7.
Southern blot of HindIII-digested genomic DNA from different listerial strains with pC11A as the probe. Lanes 1 to 11, DNA from L. monocytogenes serotype 1/2b (strain F4233 [lane 1]), serotype 4b (strains F2381, 33, 15U, and 18 [lanes 2 to 5, respectively]), and serotype 1/2a (strains Mack, G2228, G2295, and G3412 [lanes 6 to 9, respectively]). Lanes 10 to 14 contain DNA from L. grayi, Listeria welshimeri, L. seeligeri, L. innocua, and Listeria ivanovii, respectively.
DNA sequencing and sequence analysis.
DNA was sequenced on both strands either manually (Silver Sequence; Promega) or by automated sequencing at the Biotechnology Core Facility (University of Hawaii).
For DNA and protein database searches and analyses, we used FASTA (University of Wisconsin GCG package; Genetics Computer Group, Madison, Wis.) and the BLAST protein database (http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession number.
The nucleotide sequence data determined in this study have been deposited in the GenBank database under accession no. AFO 72894.
RESULTS
Mutants negative for serotype-specific MAb c74.22 have impaired TA glycosylation.
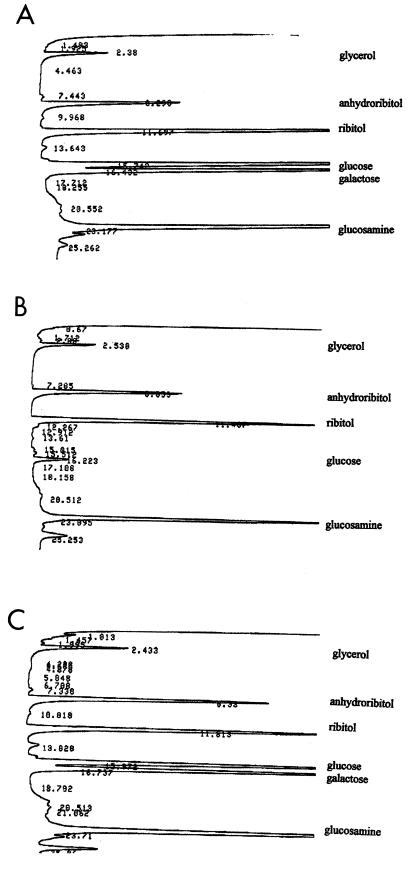
Within L. monocytogenes, MAbs c74.22, c74.33, and c74.180 have been shown to react exclusively with strains of serotypes 4b, 4d, and 4e (16). We have identified three serotype 4b transposon mutants, M44, B6S (also termed XL1), and 27E8 (also termed XL3), which were completely negative for c74.22 but retained reactivity with the other two MAbs (c74.33 and c74.180) on the basis of ELISA and colony immunoblots (16, 20). Mutant M44 has been shown to harbor a single copy of Tn916ΔE (20). Western blots of total cellular and extracellular proteins of M44 and its parental strain 4b1 with MAb c74.22 did not identify any specific protein band(s) reactive with this MAb (data not shown). Biochemical analysis of cell wall composition was then undertaken to determine whether the mutant was altered in components of TA, which, on the basis of biochemical and immunological studies, have been shown to vary among different serotypes of L. monocytogenes (7, 10, 40, 41). It was found that cell wall TA from M44 lacked galactose and had only trace amounts of glucose (Fig. 2B). In contrast, cell wall TA from a wild-type isogenic strain (4WT) contained both galactose and glucose (Fig. 2A), as is typical of serotype 4b strains. Other cell wall TA components (phosphate, ribitol, Glc-Nac) remained normal (Fig. 2A and B), as did the amount and composition of peptidoglycan (data not shown). Interestingly, LTA composition was also completely normal, with both galactose and glucose being present at wild-type levels (data not shown). Results identical to those obtained with M44 were also obtained with the independently derived c74.22-negative mutants B6S and 27E8 (data not shown).
FIG. 2.
TA composition in wild-type strain 4WT (A), gtcA mutant M44 (B), and M44 complemented with wild-type gtcA in trans (pNP95). TA preparation and analysis were done as described previously (7, 14).
Cloning and molecular characterization of gtcA.
SSP-PCR was used as described in Materials and Methods to amplify a transposon-adjacent DNA fragment from mutant M44. The amplified flanking fragment (500 bp) was cloned in pCR1000, and the resulting plasmid was used as a probe in Southern blots to determine the sizes of homologous fragments in the wild type (strain 4b1) and in mutant M44. Such blots confirmed that the amplified fragment was indeed from the transposon-flanking region of M44 and localized the transposon insertion in a ca. 0.85-kb EcoRI and a 3.5-kb HindIII genomic DNA fragment. Identical Southern blot results were obtained with mutants B6S and 27E8 (data not shown), suggesting that these independently derived mutants harbored transposon insertions in the same EcoRI and HindIII fragments.
The cloned transposon-flanking fragment was used to screen a genomic library of strain 4b1 (4- to 10-kb fragments) with no success. A single positive clone was identified from the screening of another 4b1 library that contained smaller (2- to 4-kb) inserts. Southern blot analysis suggested that this clone (pC11A) contained 1.2 kb of DNA within the 3.5-kb HindIII fragment which harbored the transposon in M44 (Fig. 3). DNA sequencing of the original SSP-PCR fragment and of the fragment cloned in pC11A revealed that in M44 the transposon was inserted within an open reading frame (ORF), termed gtcA (for galactose-glucose in TA). Nucleotide sequences of transposon-flanking fragments of mutants B6S and 27E8 (amplified as described in Materials and Methods) showed that the transposon had inserted in a common target site within gtcA. In M44, Tn916ΔE was inserted between nt 512 and 513, whereas Tn916ΔE in B6S and Tn916 in 27E8 were both inserted between nt 514 and 515. The transposon target site in these mutants, TTTTCGAATAAAAA, was in close agreement with the Tn916 preferred target sites reported for other gram-positive bacteria (21).
ORF analysis.
Sequence analysis revealed two complete ORFs (gtcA and ORFD) and one incomplete ORF (ORFU) in this region, all transcribed in the same direction (Fig. 3). An inverted repeat followed by a series of T’s after the end of ORFD may represent a rho-independent transcription terminator (estimated free energy of formation, −25 kcal/mol). The transposon-harboring ORF (gtcA) was preceded by an incomplete ORF (ORFU) with 60 to 74% identity to genes encoding the transcription termination factor rho in B. subtilis, E. coli, Salmonella typhimurium, Haemophilus influenzae, Neisseria gonorrhoeae, and other bacteria. The available sequence of this ORF would be expected to correspond to the 275 bp in the 3′ portion of the putative rho gene in L. monocytogenes, a portion of the coding sequence known to be especially conserved among rho sequences (23).
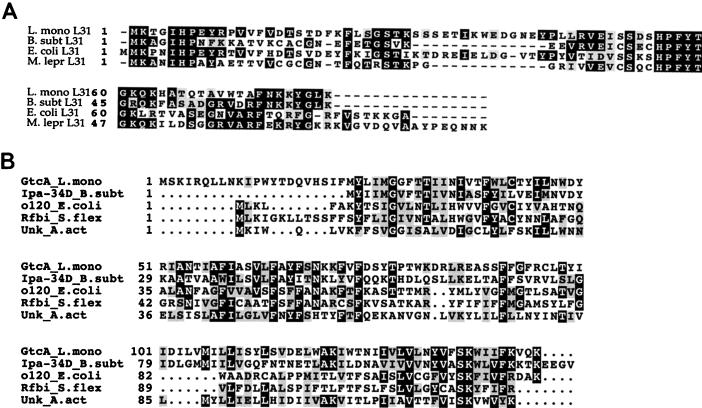
The ORF immediately downstream of gtcA (ORFD) showed 55% homology to the rpmE gene of B. subtilis (31) encoding ribosomal protein L31. Homology was also detected with the rpmE gene sequences of Mycobacterium leprae (41%), E. coli (32%), and other bacteria. BLAST analysis suggested significant similarity between the deduced ORFD product and the L31 proteins from B. subtilis and other bacteria (Fig. 4A shows a multiple sequence alignment). It appears therefore likely that ORFD represents the coding sequence of the L. monocytogenes rpmE gene. The hydrophobicity plot of the putative RpmE suggested that the protein was hydrophilic (data not shown). A putative ribosomal binding site was identified 5 nt upstream of the start codon of this gene.
FIG. 4.
Multiple sequence alignments (CLUSTAL) of the deduced ORFD (putative L31) sequence (A) and the deduced GtcA sequence (B). Abbreviations: B. subt L31 (accession no. M97678), E. coli L31 (accession no. U00096), and M. lepr L31 (accession no. Z73419), L31 sequences from B. subtilis, E. coli, and M. leprae, respectively; Ipa-34D B. subt (accession no. X73124), o120 E. coli (accession no. U000323), Rfbi S. flex (accession no. X71970), and Unk A. act (accession no. AB002668), sequences with similarity to the deduced gtcA product and derived from B. subtilis, E. coli, S. flexneri, and A. actinomycetemcomitans, respectively, as described in the text.
The coding sequence of gtcA (434 bp) was separated from ORFU and ORFD by intergenic regions of 32 and 101 bp, respectively. A putative ribosomal binding site was identified 5 nt upstream of the gtcA start codon, although its similarity with the consensus was noticeably lower than that of the putative ribosomal binding site preceding ORFD. The deduced gene product (17.4 kDa) was calculated to have a pI of 10.17. The gtcA coding sequence was similar (55% identity) to a gene of B. subtilis (ipa34-d) which was identified during the B. subtilis genome sequencing project and which appears to be a member of an operon involved in galactose metabolism (11). The deduced product of gtcA was found to be similar to the deduced product of ipa34-d (46% identity over 123 amino acids). The deduced gtcA product had lower similarity with several other proteins in the database, i.e., a hypothetical protein of Actinobacillus actinomycetemcomitans (33% identity over 140 amino acids) and the deduced products of the E. coli gene yeij (30% identity over 125 amino acids) and of the Shigella flexneri gene rfbI (27% identity over 122 amino acids). The latter two genes are in genomic regions dedicated to surface antigen biosynthesis in E. coli and S. flexneri, respectively. Figure 4B shows the alignment between these deduced sequences. It is noticeable that the 22-amino-acid N-terminal region of the deduced gtcA product does not have a counterpart in the deduced ipa34-d sequence of B. subtilis. In fact, an alternative start codon (base position 375, residue 23) could be identified 12 nt downstream of a putative ribosomal binding site (AGATCAGGTA). The 23-amino-acid N-terminal region of the deduced gtcA product also lacked significant similarity with the hypothetical proteins of A. actinomycetemcomitans, E. coli, and S. flexneri. N-terminal sequencing of the protein will be required for determination of the actual start codon. Hydropathy analysis of the deduced gtcA gene product indicated four putative membrane-spanning domains, suggesting that the protein may be membrane associated. The G+C content of gtcA was found to be ca. 30%, noticeably lower than the average G+C content of L. monocytogenes (38%). In contrast, the G+C contents of the putative rpmE and rho (partial) sequences were 41 and 39%, respectively.
Interestingly, in B. subtilis, rho and rpmE were located adjacent to each other on the chromosome (31), whereas ipa34-d was located in a different genomic region (11). In L. monocytogenes serotype 4b, gtcA was positioned exactly between the putative rho and rpmE genes of this organism (ORFU and ORFD, respectively) (Fig. 3).
Determination of gtcA sequence in epidemic-associated strain F2381.
Strain F2381 is a representative of a clonal lineage implicated in numerous outbreaks of listeriosis and known to be genetically distinct from other serotype 4b strains (3, 25, 44). Sequencing of the gtcA region (nt 289 to 1045) showed that the coding sequence of gtcA was identical for strains 4b1 and F2381. The two sequence differences identified in this region were both in the gtcA-rpmE intergenic region (at nt 756, strain F2381 had an insertion of a T; at nt 819, strain 4b1 had a G that was absent in F2381) (Fig. 3B). The sequenced portion of the coding sequence of rpmE (up to and including nt 1045) was identical for strains 4b1 and F2381.
Transcriptional analysis of gtcA.
Northern blots using the gtcA-internal fragment cloned in pNP1 as probe suggested the presence of a ca. 700-nt transcript (data not shown). Signal intensity was quite low in Northern blots, suggesting a low abundance of the message. RT-PCR was therefore used to determine whether gtcA was cotranscribed with its flanking ORFs. The results of a typical RT-PCR experiment are shown in Fig. 5. cDNA synthesis was done by using primers P1 (nt 621 to 640, complementary to the expected gtcA message) and P2 (nt 1013 to 1032, complementary to the expected rpmE message). The RT-PCR results suggest that gtcA was cotranscribed with ORFD (Fig. 5, lane 4) but not with ORFU (lanes 1 and 3). Since only 32 bp separated the gtcA start codon from the ochre codon of ORFU, the gtcA promoter may be located in the 3′-end portion of ORFU. RT-PCR data suggest that the transcript initiation site was within the last 30 to 40 nt of ORFU (16).
FIG. 5.
RT-PCR to determine transcriptional status of ORFU, gtcA, and ORFD in L. monocytogenes serotype 4b. Primers P1 and P2 were used for cDNA synthesis. Subsequent PCR was done with primers P3 and P1 (lane 1), P4 and P1 (lane 2), P3 and P2 (lane 3), P2 and P4 (lane 4), and P2 and P5 (lane 5). No PCR product was observed in lanes 1 and 3. The leftmost lane contains molecular size markers. The location and orientation of the primers are shown at the bottom of the figure. Precise location and sequence of the primers as well as procedures for RT-PCR were as described in Materials and Methods. Templates for negative control were RNA with the same primer sets. None of the negative control reactions produced detectable PCR products (data not shown).
TA defects of gtcA mutants are complemented by wild-type gtcA in trans.
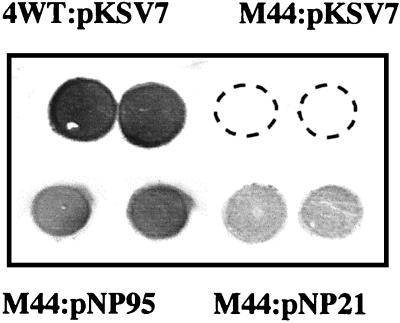
Plasmids pNP21 and pNP95, harboring gtcA with or without ORFD (putative rpmE), respectively (Fig. 3), were electroporated into mutant M44. Both plasmids include the ORFU-gtcA intergenic region and the 3′-end region of ORFU, since the gtcA promoter may be contained within this region. The resulting strains were grown in the presence of chloramphenicol at 30°C, a temperature which permits both replication of the temperature-sensitive plasmid (38) and optimal expression of the serotype 4b-specific surface antigens (16). Reactivity of the mutant with c74.22 was restored by both plasmids. Reactivity was relatively low when pNP21 was used but stronger (albeit still somewhat lower than that of the wild type) with pN95 (Fig. 6). Identical results were obtained with mutant 27E8 (data not shown). Interestingly, introduction of either plasmid completely restored TA composition. The TA of M44 complemented with pNP95 contained both galactose and glucose, at levels indistinguishable from those of the wild type (Fig. 2C); identical results were obtained with M44 complemented with pNP21 (data not shown).
FIG. 6.
Complementation of surface antigen expression of M44 with wild-type gtcA in trans. Colony immunoblotting with MAb c74.22 was done as described in Materials and Methods. Top left (duplicate spots), positive control (wild-type strain 4WT harboring the cloning vector pKSV7); top right (duplicate spots), mutant M44 harboring the cloning vector pKSV7; bottom left and bottom right (duplicate spots in each), M44 harboring pNP95 and pNP21, respectively. No reaction was seen in the top right spots (duplicate spots of M44 harboring pKSV7). Fragments contained in pNP95 and pNP21 were as shown in Fig. 3A.
Within L. monocytogenes, only serogroup 4 strains harbor gtcA sequences whereas ORFD appears to be conserved among Listeria spp.
Recently, we showed that the region adjacent to the transposon insertion in mutant M44 was present in serogroup 4 (serotypes 4a, 4b, 4c, 4d, and 4e) and in certain strains of Listeria innocua (which, in contrast to all other screened L. innocua strains, were also positive for MAbs c74.22, c74.33, and c74.180 [16]) but not in other serotypes of L. monocytogenes or other Listeria species (20). Southern blots using pNP1 as the probe confirmed these findings (data not shown). In addition, Southern blots with pC11A, which includes gtcA as well as ORFD, suggested that the latter gene (putative rpmE) was present in other serotypes of L. monocytogenes as well as in all other Listeria species (hybridizing signals were weak in Listeria grayi and Listeria seeligeri) (Fig. 7). Such Southern blots also revealed HindIII restriction fragment length polymorphisms (RFLPs) which could differentiate L. monocytogenes strains of serotype 4b (Fig. 7, lanes 2 to 5) from those of serotypes 1/2a and 1/2b (lanes 1 and 6 to 9). Similar differentiation could be obtained with EcoRI RFLPs (data not shown). Southern blots of EcoRI-cut DNA with probe pNP500, which includes gtcA and only the gtcA-ORFD intergenic region, furthermore indicated that the intergenic region was present in strains of other serotypes. In serogroup 4 (serotypes 4a, 4b, 4c, 4d, and 4e), a hybridizing band of 0.8 kb was observed, whereas a 2-kb hybridizing band was observed with serogroup 1/2 (serotypes 1/2a, 1/2b, and 1/2c), serogroup 3 (serotypes 3a, 3b, and 3c), and serotype 7 (data not shown).
DISCUSSION
The gene described in this report, gtcA, appears to be essential for presentation of galactose and glucose substituents on serotype 4b TA. Support for this was provided by the findings that (i) insertional inactivation of gtcA in several independently obtained mutants abolished galactose and markedly reduced glucose levels in TA and (ii) the amounts of both sugars were restored to wild-type levels by the wild-type gtcA gene in trans. These results suggest that the phenotype of gtcA insertion mutants was due to the transposon insertion and not to an unrelated (e.g., spontaneous) mutation. In addition, complementation by the wild-type gtcA in trans strongly suggested that the impact of the gtcA mutations on TA composition was indeed due to inactivation of gtcA itself and not to a possible polar effect of the transposon insertion on a downstream gene(s).
Cell wall compositional analysis of the gtcA mutants led to two unexpected observations: in contrast to its impact on TA, the mutation was found to have no effect on LTA composition, suggesting genetic and enzymatic independence of glycosylation between TA and LTA. Furthermore, TA of gtcA mutants was found not only to lack galactose but also to have markedly reduced glucose. A possible explanation for this may be that the enzymatic step(s) mediating incorporation of glucose onto TA may require the prior presence of galactose. Although the precise function of the gtcA gene product remains to be determined, our genetic, immunological, and biochemical data suggest that it is involved in glycosylation of TA domains and that such domains serve as serotype-specific surface antigens on bacteria of serotype 4b. Key supporting data include the impaired TA glycosylation of the gtcA mutants and their lack of reactivity with MAb c74.22, as well as the finding that normal TA glycosylation and reactivity were restored when gtcA was provided in trans. The presence of hydrophobic, putative membrane-spanning domains in the deduced gtcA product would be in agreement with its involvement in TA biosynthesis. In B. subtilis, several TA biosynthesis gene products have been shown to have membrane-spanning and/or membrane-anchoring domains (18, 19), and enzymes involved in TA biosynthesis have been shown to be membrane associated in Staphylococcus aureus and Staphylococcus xylosus (8, 9). Additional insight into the function of the gtcA product will be provided by biochemical characterization of the protein and, perhaps, by characterization of homologous genes or gene products in other bacteria. In this respect, it is of interest that the gtcA homologue of B. subtilis (ipa34-d) appears to be in an operon dedicated to galactose metabolism (11). However, the precise function of ipa34-d has not yet been determined, and B. subtilis is not known to have galactose in its cell wall TA. In B. subtilis, two glucosyl transferases involved in the incorporation of glucose onto TA have been identified, the products of the gtaB and tagE (rodD), respectively (4, 12, 26, 27, 39). B. subtilis, however, uses glycerol as the glucose acceptor in its TA, whereas in L. monocytogenes serotype 4b, N-acetylglucosamine serves as the galactose and glucose acceptor. It is possible that the gtcA product may have a glycosyl transferase activity, although no homologies with similar transferases were detected by the protein database searches. Another possibility that cannot be excluded at this point is that gtcA may encode a regulatory protein, essential for expression of a gene(s) involved in glycosylation of TA in L. monocytogenes serotype 4b. The potential of gtcA to confer c74.22 reactivity to heterologous serotypes (e.g., serogroup 1/2 or 3) remains to be determined. It must be kept in mind, however, that these strains (e.g., serogroup 1/2 or 3) differ in their TA backbone structure and in TA glycosylation sites from serotype 4b strains (Fig. 1). A more likely candidate for heterologous expression would be c74.22-negative strains of L. innocua, which lack galactose and glucose but have a TA backbone structure identical to that of serotype 4b strains. We were, however, unable to introduce gtcA (cloned in pKSV7) into such an L. innocua strain (serotype 6a), perhaps because of a restriction barrier (data not shown).
It is of interest that in serogroup 4 strains of L. monocytogenes, the gtcA gene was positioned between ORFs with homology to rho and rpmE genes, whereas rho and rpmE were directly adjacent to each other in B. subtilis. Our data suggested that gtcA was the first gene of a bicistronic operon which included the putative rpmE. This was surprising since no apparent commonality in the function of these two genes was readily evident. Our data also indicated that of the two genes, only gtcA was unique to serogroup 4, whereas the putative rpmE (as well as the gtcA-rpmE intergenic region) was conserved among listeriae. It is tempting to speculate that the rho-rpmE genomic arrangement has been preserved in L. monocytogenes strains other than those of serogroup 4 (e.g., 1/2 and 3, etc.) which lack gtcA and that, in the course of evolution of serogroup 4 strains, gtcA was introduced in its present location, conferring serogroup-specific expression of a TA-associated, serotype-specific surface antigen(s). The gene may have been transferred between L. monocytogenes serogroup 4 and a lineage of L. innocua, a species which is nonpathogenic but otherwise closely related genetically and antigenically to L. monocytogenes. The gtcA gene may have originated in an organism other than Listeria, and its unusually low G+C content (30%, in contrast to 38% for L. monocytogenes and other Listeria spp.) may be indicative of this. Introduction of gtcA could have been mediated by a phage or transposon, although the currently available sequence data do not reveal any discernible traces or evidence of such elements (inverted repeats, tRNA sequences, or sequences with possible integrase or transposase functions). An alternative possibility is that the gtcA sequences were present in an ancestral L. monocytogenes-L. innocua lineage but were preserved only in serogroup 4 strains and in certain L. innocua strains. We are currently in the process of further characterizing the genomic region that harbors the putative rho and rpmE genes in strains of L. monocytogenes other than those of serogroup 4, in order to gain further insight on this issue.
Extensive molecular studies of virulence determinants have focused on a select group of extracellular and surface-associated proteins of L. monocytogenes (reviewed in references 13 and 29). In contrast, the possible involvement of carbohydrate-based surface antigens in pathogenesis remains poorly understood. Earlier studies suggested that glycosylated TA components were important antigenic determinants in L. monocytogenes (14, 41). We have obtained data, to be described in a separate presentation, which strongly suggest that galactose on TA of L. monocytogenes serotype 4b was essential as a receptor for serotype-specific phages and, in addition, for invasion of several mammalian cell lines by the bacteria (30). Such data are in agreement with findings from other investigations which showed that in serotype 1/2, rhamnose and Glc-NAc on TA served as receptors for serotype-specific phages (42). However, genes involved in the decoration of serotype 1/2 TA with rhamnose and Glc-NAc have not yet been identified, and information regarding their possible serotype-specific distribution in L. monocytogenes is lacking. Further investigations are needed to enhance our understanding of the evolution of genetic systems dedicated to TA glycosylation and of the possible function(s) of the different types of TA glycosylation in the adaptive physiology and pathogenesis of the microorganism.
ACKNOWLEDGMENTS
This research was partially supported by U.S. Department of Agriculture Competitive Research Initiative AAFS grant 92-37 201-8095, by ILSI—North America, and by a seed grant from the University of Hawaii.
We thank Vladimir Lazarevic for helpful discussions and exchange of information related to TA biosynthesis. We also thank Nikhat Parveen for assistance with Northern blots, and we are grateful for the valuable participation of Timothy Rodwell, Thong Truong, Xiang-He Lei, and Wei Zheng in certain aspects of this work. We thank the members of our laboratories for valuable feedback and support throughout the course of this work.
REFERENCES
- 1.Armstrong J J, Baddiley J, Buchanan J G. Structure of the ribitol teichoic acid from walls of Bacillus subtilis. Biochem J. 1960;76:610–621. doi: 10.1042/bj0760610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bibb W F, Gellin B G, Weaver R, Schwartz B, Plikaytis B D, Reeves M W, Pinner R W, Broome C V. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiological investigations. Appl Environ Microbiol. 1990;56:2133–2141. doi: 10.1128/aem.56.7.2133-2141.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks D, Mays L L, Hatefi Y, Young F E. Glucosylation of teichoic acid: solubilization and partial characterization of the uridine diphosphoglucose:polyglycerolteichoic acid glucosyl transferase from membranes of Bacillus subtilis. J Bacteriol. 1971;107:223–229. doi: 10.1128/jb.107.1.223-229.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin T, Burger M M, Glaser L. Synthesis of teichoic acids. VI. The formation of multiple wall polymers in Bacillus subtilis W23. Arch Biochem Biophys. 1966;116:358–367. doi: 10.1016/0003-9861(66)90042-7. [DOI] [PubMed] [Google Scholar]
- 6.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiedler F, Seger J, Schrettenbrunner A, Seeliger H P R. The biochemistry of murein and cell wall teichoic acids in the genus of Listeria. Syst Appl Microbiol. 1984;5:360–376. [Google Scholar]
- 8.Fiedler F, Glaser L. The synthesis of polyribitol phosphate. I. Purification of polyribitol phosphate polymerase and lipoteichoic acid carrier. J Biol Chem. 1974;245:2684–2689. [PubMed] [Google Scholar]
- 9.Fiedler F, Steber J. Structure and biosynthesis of teichoic acids in the walls of Staphylococcus xylosus DSM 20266. Arch Microbiol. 1984;138:321–328. doi: 10.1007/BF00410898. [DOI] [PubMed] [Google Scholar]
- 10.Fujii H, Kamisango K, Nagaoka M, Uchikawa K, Sekikawa I, Yamamoto K, Azuma I. Structural study of teichoic acids of Listeria monocytogenes types 4a and 4d. J Biochem. 1985;97:883–891. doi: 10.1093/oxfordjournals.jbchem.a135130. [DOI] [PubMed] [Google Scholar]
- 11.Glaser P, Kunst F, Arnaud M, Goudart M P, Gonzales W, Hullo M F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweizer J, Vertes A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97kb region from 325 degrees to 333 degrees. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 12.Honeyman A L, Stewart G C. The nucleotide sequence of the rodC operon of Bacillus subtilis. Mol Microbiol. 1989;3:1257–1268. doi: 10.1111/j.1365-2958.1989.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 13.Ireton K, Cossart P. Host-pathogen interactions during entry and actin-based movement of Listeria monocytogenes. Annu Rev Genet. 1997;31:113–138. doi: 10.1146/annurev.genet.31.1.113. [DOI] [PubMed] [Google Scholar]
- 14.Kamisango K, Fujii H, Okumura H, Saiki I, Araki Y, Yamamura Y, Azuma I. Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J Biochem. 1983;93:1401–1409. doi: 10.1093/oxfordjournals.jbchem.a134275. [DOI] [PubMed] [Google Scholar]
- 15.Kathariou S, Metz P, Hof H, Goebel W. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J Bacteriol. 1987;169:1291–1297. doi: 10.1128/jb.169.3.1291-1297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathariou S, Mizumoto C, Allen R D, Fok A K, Benedict A A. Monoclonal antibodies with a high degree of specificity for Listeria monocytogenes serotype 4b. Appl Environ Microbiol. 1994;60:3548–3552. doi: 10.1128/aem.60.10.3548-3552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan, Z., and S. Kathariou. Unpublished data.
- 18.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 19.Lazarevic C, Mauel C, Soldo B, Freymond P-P, Argot P, Karamata D. Sequence analysis of the 308° to 311° segment of the Bacillus subtilis 168 chromosome, a region devoted to cell wall metabolism containing noncoding gray holes which reveal chromosomal rearrangements. Microbiology. 1995;141:329–335. doi: 10.1099/13500872-141-2-329. [DOI] [PubMed] [Google Scholar]
- 20.Lei X-H, Promadej N, Kathariou S. DNA fragments from regions involved in surface antigen expression specifically identify Listeria monocytogenes serovar 4 and a subset thereof: cluster IIB (serotypes 4b, 4d, and 4e) Appl Environ Microbiol. 1997;63:1077–1082. doi: 10.1128/aem.63.3.1077-1082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu F, Churchward G. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J Bacteriol. 1995;177:1938–1946. doi: 10.1128/jb.177.8.1938-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauel C, Young M, Margot P, Karamata D. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol Gen Genet. 1989;215:388–394. doi: 10.1007/BF00427034. [DOI] [PubMed] [Google Scholar]
- 23.Opperman T, Richardson J P. Phylogenetic analysis of sequences from diverse bacteria with homology to the Escherichia coli rho gene. J Bacteriol. 1994;176:5033–5043. doi: 10.1128/jb.176.16.5033-5043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park S F, Stewart G S A B. High efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene. 1990;94:129–134. doi: 10.1016/0378-1119(90)90479-b. [DOI] [PubMed] [Google Scholar]
- 25.Piffaretti J C, Kressebuch H, Aeschbacher M, Bille J, Bannerman E, Musser J M, Selander R K, Rocourt J. Genetic characterization of clones of the bacterium L. monocytogenes causing epidemic disease. Proc Natl Acad Sci USA. 1989;86:3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pooley H M, Paschoud D, Karamata D. The gtaB marker in Bacillus subtilis 168 is associated with a deficiency in UDP-glucose pyrophosphorylase. J Gen Microbiol. 1987;133:3481–3493. doi: 10.1099/00221287-133-12-3481. [DOI] [PubMed] [Google Scholar]
- 27.Pooley H M, Abellan F-X, Karamata D. CDP-glycerol:poly(glycerolphosphate) glycerophosphotransferase, which is involved in the synthesis of the major wall teichoic acid in Bacillus subtilis 168, is encoded by tagF (rodC) J Bacteriol. 1992;174:646–649. doi: 10.1128/jb.174.2.646-649.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pooley H M, Karamata D. Teichoic acid synthesis in Bacillus subtilis: genetic organization and biological roles. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 187–198. [Google Scholar]
- 29.Portnoy D A, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. J Bacteriol. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Promadej, N., F. Fiedler, and S. Kathariou. Unpublished data.
- 31.Quirk P G, Dunkley E A, Jr, Lee P, Krulwich T A. Identification of a putative Bacillus subtilis rho gene. J Bacteriol. 1993;175:647–654. doi: 10.1128/jb.175.3.647-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roels S, Losick R. Adjacent and divergently oriented operons under the control of the sporulation regulatory protein GerE in Bacillus subtilis. J Bacteriol. 1995;177:6263–6275. doi: 10.1128/jb.177.21.6263-6275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruhland G J, Fiedler F. Occurrence and biochemistry of lipoteichoic acids in the genus Listeria. Syst Appl Microbiol. 1987;9:40–46. [Google Scholar]
- 34.Sato N. A family of cold-regulated RNA-binding protein genes in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 1995;23:2161–2167. doi: 10.1093/nar/23.12.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seeliger H P R, Hoehne K. Serotypes of Listeria monocytogenes and related species. Methods Microbiol. 1979;13:31–49. [Google Scholar]
- 37.Shyamala V, Ferro-Luzzi Ames G. Genome walking by single-specific primer polymerase chain reaction: SSP-PCR. Gene. 1989;84:1–8. doi: 10.1016/0378-1119(89)90132-7. [DOI] [PubMed] [Google Scholar]
- 38.Smith K, Youngman P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 39.Soldo B, Lazarevic V, Margot P, Karamata D. Sequencing and analysis of the divergon comprising gtaB, the structural gene of UDP-glucose pyrophosphorylase of Bacillus subtilis 168. Gen Microbiol. 1993;139:3185–3195. doi: 10.1099/00221287-139-12-3185. [DOI] [PubMed] [Google Scholar]
- 40.Uchikawa K, Sekikawa I, Azuma I. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J Biochem. 1986;99:315–327. doi: 10.1093/oxfordjournals.jbchem.a135486. [DOI] [PubMed] [Google Scholar]
- 41.Ullman W W, Cameron J A. Immunochemistry of the cell walls of Listeria monocytogenes. J Bacteriol. 1969;98:486–493. doi: 10.1128/jb.98.2.486-493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wendlinger G, Loessner M J, Scherer S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology. 1996;142:985–992. doi: 10.1099/00221287-142-4-985. [DOI] [PubMed] [Google Scholar]
- 43.Zheng W, Kathariou S. Transposon-induced mutants of Listeria monocytogenes incapable of growth at low temperature (4°C) FEMS Microbiol Lett. 1994;121:287–292. doi: 10.1111/j.1574-6968.1994.tb07114.x. [DOI] [PubMed] [Google Scholar]
- 44.Zheng W, Kathariou S. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl Environ Microbiol. 1997;63:3085–3089. doi: 10.1128/aem.63.8.3085-3089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]