Abstract
Patients with high-risk diffuse large B-cell lymphoma have poor outcomes following first-line cyclophosphamide, doxorubicin, vincristine, prednisone, and rituximab (R-CHOP); thus, treatment of this fatal disease remains an area of unmet medical need and requires identification of novel therapeutic approaches. Dysregulation of protein translation initiation has emerged as a common downstream node in several malignancies, including lymphoma. Ubiquitination, a prominent posttranslational modification associated with substrate degradation, has recently been shown to be a key modulator of nascent peptide synthesis by limiting several translational initiation factors. While a few deubiquitinases have been identified, the E3 ligase responsible for the critical ubiquitination of these translational initiation factors is still unknown. In this study, using complementary cellular models along with clinical readouts, we establish that PARK2 ubiquitinates eIF4B and consequently regulates overall protein translational activity. The formation of this interaction depends on upstream signaling, which is negatively regulated at the protein level of PARK2. Through biochemical, mutational, and genetic studies, we identified PARK2 as a mTORC1 substrate. mTORC1 phosphorylates PARK2 at Ser127, which blocks its cellular ubiquitination activity, thereby hindering its tumor suppressor effect on eIF4B's stability. This resultant increase of eIF4B protein level helps drive enhanced overall protein translation. These data support a novel paradigm in which PARK2-generated eIF4B ubiquitination serves as an anti-oncogenic intracellular inhibitor of protein translation, attenuated by mTORC1 signaling.
Implications:
Our data implicate the FASN/mTOR-PARK2-eIF4B axis as a critical driver of enhanced oncogene expression contributing to lymphomagenesis.
Introduction
Rapid protein degradation by the proteolytic system comprising ubiquitin ligases can impact various pathologic events, including tumor development and progression (1). Aberrant protein stability secondary to defects in the ubiquitin-proteasome system is an emerging key driver for both tumor induction and chemoresistance in several cancers (2). Owing to their pivotal role, ubiquitin ligases are often deregulated in several cancers altering their substrate availability or activity in a manner that can promote cellular transformation (3). This deregulation hampers the homeostasis of essential cellular regulatory proteins resulting in several pathologic conditions, including cancer.
The contribution of protein ubiquitination underlying the development of human lymphoid malignancies is established. Over the past two decades, several E3 ligases and E2 ligases were identified that have modulated cellular events promoting lymphoid malignancies. Enzymes such as TRAF6 (4), FBXO10 (5), and c-IAP1 (6) are reported to be inactivated either by mutations or deletion/suppression by epigenetic modifications in some hematologic malignancies. Conversely, enzymes like RNF31 having a rare germline mutation are known to have enhanced function in activated B-cell-like diffuse large B-cell lymphoma (ABC-DLBCL; ref. 7). Collectively, these enzymes, either through monoubiquitination or polyubiquitination, regulate key signaling events, including B-cell receptor signaling, NFκB pathway, and transcriptional regulation. Surprisingly, few published reports evaluated the direct impact of E3 ligase on translational apparatus proteins in lymphoid malignancies or other tumors.
There is ample literature indicating a direct link between deregulation of the ribosome recruitment phase of translation initiation with cellular transformation (8). Consequently, therapeutic targeting of translation initiation factors has emerged as attractive candidates for cancer therapy. Indeed, preclinical studies using mouse models revealed that specific suppression of translational initiation factors restores chemosensitivity in lymphoma (9). Emerging data from mechanistic studies defining the regulation of translation initiation underscore protein stability as a critical event in augmenting the translational output. Recently, Li and colleagues reported that USP9X is a novel binding partner of eIF4A1, which modulates its ubiquitination attenuating the cap-dependent translational initiation (10). Similarly, the Gartenhaus lab using both pharmacologic and genetic approaches, reported that FASN induced S6Kinase signaling to phosphorylate USP11 enhancing its interaction and stability of eIF4B in DLBCL consequently promoting oncogenic translation (11). As we gain deeper insights into the regulation of translation initiation, it is becoming clear that E3 ligases are major effectors of these critical cellular processes. In this study, we identified eIF4B as a PARK2 E3 ligase substrate. Furthermore, PARK2 expression is sensitive to metabolic and proliferative signaling. An unexpected finding was that mTOR phosphorylates PARK2. This posttranslational modification of PARK2 driven by mTOR signaling hinders its E3 ligase activity, which in part, contributes to the oncogenic transformation of DLBCL.
Materials and Methods
Cell lines, culture, and generation of stable cell lines
All cells were obtained from ATCC (DS, RC, Toledo, Farage, SUDHL-2, SUDHL-4) and maintained at 37 °C with 5% CO2, except HLY-1 cells (gracious gift from Dr. Ari Melnick, Weill Cornell Medicine, New York, NY) and OCI-Ly3 (purchased from DSMZ). HEK293T/17 were cultured in DMEM supplemented with 10% FBS (Atlanta Biologicals). GM B cells were procured from the National Institutes of General Medical Sciences Human Genetic Mutant Cell Repository (Coriell Institute for Medical Research, Camden, NJ). All other cells were grown in RPMI1640, 10% FBS (GM B cells were maintained on 15% FBS). OCI-Ly3 were grown in IMDM with 20% FBS. Platinum A cells were procured from Cell Biolabs. The quality and authenticity of cell lines were performed on a regular basis using regular Mycoplasma testing and short tandem repeat (STR) profiling through the Nucleic Acid Research Facilities (NARF) at VCU (Richmond, VA), compared against known STR profiles. Cells were regularly passaged according to prescribed guidelines. Exponentially growing cells were treated with selected inhibitors and maintained at 37°C, harvested at indicated timepoints for further analysis. Stable cells were generated by lentivirus and retroviral infection, selected, and maintained with puromycin (1 mg/mL). After selection, cells were cultured in their respective complete media in the absence of puromycin. All the chemical inhibitors were purchased from Selleckchem.
Plasmids, lentiviral production, and infection
PARK2-HA [wild type (WT) and catalytically inactive (CS)] and PARK2-GFP constructs were a generous gift from Drs. Toshio Kitamura, Ted Dawson, and Edward Fon (Addgene plasmids #38248, 17613, 45875, 45877). pMX-IP-PARK2-HA (S127A, C431S) mutants were generated by using a site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's recommendations. Retroviral constructs for overexpression of encoding plasmids were transfected in Platinum-A cells, and the particles were concentrated as reported earlier. Lentiviral constructs for knockdown of PARK2 and FASN were purchased from Sigma. For lentiviral packaging, HEK293T/17 cells were seeded at 50% confluence and transfected using packing plasmids psPAX2 and pMD2.G (a generous gift from Dr. Didier Trono; Addgene # 12260 and #12259). Lentiviral particles were concentrated as defined earlier. DLBCLs were treated with polybrene (4 mg/mL, American Bioanalytical) and centrifuged (35 minutes, 300 × g, 25°C). Cell pellets were resuspended in the virus-containing medium for 48 hours, then selected and maintained with puromycin (1 mg/mL).
B-cell sorting
Human B cells from otherwise healthy motor vehicle collision patients were provided by the R. Adams Cowley Shock Trauma Center and UMGCC Pathology Biorepository and Research Core following the University of Maryland Medical School Institutional Review Board's guidelines conform to the Declaration of Helsinki. Primary human tissue was minced on ice, and mononuclear cells were isolated using density centrifugation (lymphocyte separation medium). B cells were isolated by magnetic bead separation (B-CLL cell isolation kit, human, Miltenyi Biotec, per manufacturer protocol) before further use (11).
Puromycin incorporation assay
Puromycin incorporation assay (SUnSET) assay was performed as per the manufacturer's recommendations (Kerafast) as reported previously (11). In brief, cells were pulse labeled with puromycin [normal cells (noninfected): 1 μg/mL, infected cells: 3 μg/mL] for 30 minutes. Posttreatment, cells were washed with ice-cold PBS, lysed, and probed (10 μg of protein) with an anti-puromycin antibody. Signals were normalized with investigating GAPDH (loading control).
Immunoblotting and immunoprecipitation
Cells were lysed in RIPA buffer [50 mmol/L TRIS pH 7.5, 150 mmol/L NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% triton X-100, 1 mmol/L EDTA, and 1 mmol/L EGTA, 1 mmol/L sodium orthovanadate, 1 mmol/L sodium fluoride, 1× protease inhibitor (Sigma-Aldrich), phosphatase inhibitor cocktails #2 and #3 (Sigma-Aldrich), and 1 mmol/L phenylmethylsulfonylfluoride]. Cells lysate were quantified using Bradford reagents and equal amount of protein was separated on a BOLT 4%–12% Bis-Tris gradient gel (Life Technologies) and probed with the following antibodies: eIF4B (SCBT, sc-376062, 1:1,000), PARK2 (Cell Signaling Technology, 4211, 1:1,000 or SCBT, sc-32282, 1: 1,000), eIF4A (sc-377315 or sc-14211, 1:1,000), eIF4E (SCBT, sc-9976, 1:1,000), cMYC (SCBT, sc-373712 or SC-764, 1:1,000), Bcl2 (Cell Signaling Technology #2870 or SCBT, sc-509, 1:1,000), Bcl6 (Cell Signaling Technology, #5650, 1:1,000), PARP-1 (SCBT, sc-7150, 1:1,000), XIAP (Cell Signaling Technology #2042 or #2045, 1:1,000), p-70S6kinase (T421, S424; Cell Signaling Technology, #9204, 1:1,000), Total 70S6kinase (Cell Signaling Technology, #9202, 1:1,000), SMURF1 (SCBT, sc-100616, 1:1,000) Actin (SCBT, sc-1615, 1:1,000), GAPDH (Abcam, ab8245, 1:10,000), Flag (Sigma, F3165, 1:10,000), HA (BioLegend, MMS-101P, 1:10,000), Ubiquitin (SCBT, sc-8017, 1:2,000), His (SCBT, sc-803, 1:5,000), SOCS1 (Cell Signaling Technology #3950, 1:1,000), A20 (Cell Signaling Technology #4625, 1:1,000), BLIMP1 (SCBT, sc-130917, 1:1,000), DUSP4 (SCBT, sc-135991, 1:1,000), Vinculin (Sigma, V9131, 1:10,000), mTOR (Cell Signaling Technology, #2983, 1:1,000), Akt (Cell Signaling Technology, #4691, 1:1,000), pAkt-Substrate (Cell Signaling Technology, #9614, 1:1,000). Densitometry analyses were performed using Image Studio (LI-COR Biosciences) and presented as ratio of target band signal intensity to GAPDH/Actin/Vinculin band signal intensity. For immunoprecipitation, 500 μg of protein lysates were precleared with protein A Sepharose beads (50 μL/500 μL of sample) for 1 hour and processed as described earlier. Input (50 μg) was 10% of amount used for immunoprecipitation (IP).
In vitro mTOR kinase assay and cellular phosphorylation assay
mTORC1 kinase assay was performed as described previously (12). Active mTOR protein was procured from Sigma. Immunoprecipitated PARK2-HA (wt and alanine mutant) was washed three times and incubated with kinase assay buffer (20 mmol/L HEPES, 50 mmol/L KCl, 10 mmol/L MgCl2), and set on ice. The reaction was primed with the addition of 100 μmol/L ATP along with incubation at 30°C for 10 minutes. The reaction was terminated by adding 2× Laemmli buffer and boiled at >90 °C for 5 minutes. Samples were separated on a 4%–12% SDS page and probed with a pAKT substrate antibody. The blots were stripped and examined for mTOR and HA, respectively. Cellular phosphorylation assay was performed as described earlier. Here, 293T cells were transfected with PARK2 (wt) or with mutant and cultured in the presence of 20% FBS to increase basal signaling. Cells transfected with PARK2 alone (Fig. 5D) were treated for 2 hours with Rapamycin (50 nmol/L). Cells were subsequently washed, lysed, and PARK2 was precipitated using HA antibody or beads. Phospho-signal of PARK2 was quantified by pAKT substrate antibody.
Figure 5.
mTOR-modified PARK2 fails to ubiquitinate eIF4B. A, Indicated cells were stably infected with the retroviral particles expressing GFP or PARK2 mutants and probed for the defined antibodies. B, Densitometric quantification of the immunoblots in A. Values were normalized with their corresponding loading controls and neutralized with GFP-expressing cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Bonferroni post hoc analysis. b, P < 0.01; d, P < 0.001 versus GFP-infected cells. A, P < 0.05; C, P < 0.005 versus PARK2-W–infected cells. C, Transiently infected HLY1 cells were lysed, and eIF4B was immunoprecipitated, followed by the probing with ubiquitin. D, Indicated cells stably expressing the genes were challenged with puromycin. Posttreatment, cells were lysed, and the puromycin incorporation was studied using an anti-puromycin antibody. E, Densitometric quantification of the immunoblots in D. Values were normalized with their corresponding loading controls and neutralized with GFP expressing cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Bonferroni post hoc analysis. b, P < 0.01; d, P< 0.001 versus GFP-infected cells. D, P < 0.001 versus PARK2-W–infected cells. F and H, Indicated cells were stably infected with the retroviral particles expressing GFP or PARK2 mutants and probed for the defined antibodies. G and I, Densitometric quantification of the immunoblots in F and H, respectively. Values were normalized with their corresponding loading controls and neutralized with GFP-expressing cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Bonferronis post hoc analysis. a, P < 0.05; b, P < 0.01; c, P < 0.005; d, P < 0.001 versus GFP-infected cells. B, P < 0.01; C, P < 0.005; D, P < 0.001 versus PARK2-W–infected cells. J, Indicated PARK2 (wild type and mutants) stable cells were seeded 1 million per well in 6-well plates. GFP-infected were used as the internal control. After 12 hours, the cells were collected and counted using trypan blue. Values are expressed as mean ± SD (n = 3); *, P < 0.05 versus GFP-infected corresponding cells; a, P < 0.05 versus PARK2-W–infected corresponding cells. K, The total number of colonies grown in PARK2-expressing (wild type and mutants) cells in methylcellulose culture. Colony counts were performed on the 15th day of methylcellulose culture. ***, P < 0.005; ****, P < 0.001 versus corresponding GFP-infected control cells; c, P < 0.005; d, P < 0.001 versus corresponding PARK2-W–infected control cells.
In vitro PARK2 ubiquitination assay and cellular ubiquitinylation assay
In vitro ubiquitination assay was performed using commercially procured reagents: E1 ligase, E2 ligase (Ubc7), PARK2, E3 ligase, Ubiquitin, HA-Ubiquitina, and pS65-Ubiquitin, all procured from the R&D system. His-tagged eIF4B was purified from the bacterial source (BL21*DE3). PARK2 (100 nmol/L) was incubated with 1 μmol/L E1 ligase, 2.5 μmol/L E2 ligase along with Ubiquitin (1 μmol/L) and pS65 Ubiquitin (100 nmol/L) at 30°C for 10 minutes for priming in ubiquitinylation assay buffer (10× buffer, 5 mmol/L ATP, 10 mmol/L MgCl2, 500 mmol/L Tris pH7.5, 1 mmol/L DTT). After priming, His-tagged eIF4B was added along with 2.5 μmol/L of HA-ubiquitin and was incubated at 30°C for 20 minutes. The reaction was terminated by the addition of 2× Laemmli sample buffer and analyzed by Western blotting. In a separate reaction, to evaluate the in vitro ubiquitination activity of PARK2 (wt and mutants), PARK2 was enriched by immunoprecipitation from 293T lysates (10-cm dish, 8 μg DNA), washed stringently, and resuspended in ubiquitination assay buffer. After priming, as discussed above, the complex was mixed with His-tagged eIF4B for in vitro eIF4B ubiquitination reaction. Ubiquitination reactions were resolved by SDS-PAGE and subjected to Western blot analysis with defined antibodies.
For analysis of endogenous eIF4B ubiquitination under cellular conditions, lysates were immunoprecipitated with eIF4B. A total of 500 μg of protein lysates were precleared by the addition of 50 μL of washed Protein A agarose bead slurry. The agarose beads were collected by pulsing 5 seconds in the microcentrifuge at 5,000 g and discarded. Sample supernatants were incubated with 10 μL of anti-eIF4B at 4°C for 2 hours. A total of 50 μL of washed Protein A agarose bead slurry were added to the reaction mixture and gently rocked at 4°C for an additional 2 hours. The agarose beads were then collected by pulsing 5 seconds in the microcentrifuge at 5,000 g and washed three times with ice-cold washing buffer [20 mmol/L Tris, pH 7.5, 500 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 0.1% Triton X-100, protease inhibitors (Roche), 50 mmol/L MG132). The beads were resuspended in 40 μL of 2× Laemmli sample buffer, boiled, analyzed by SDS-PAGE, and developed with the indicated antibodies.
IHC
IHC was performed as discussed earlier (11). Briefly, tissue microarray (TMA) slides (US Biomax, # LY800a, b) were dewaxed, washed once in absolute ethanol, and incubated in 3.3% hydrogen peroxide in methyl alcohol for 40 minutes in the dark at room temperature to inactivate endogenous peroxidase. The sections were hydrated in graded alcohol followed by three washes in PBS for 5 minutes. Sections were incubated in blocking solution containing 5% BSA and 2% horse serum for 2 hours, followed by overnight incubation at 4°C with eIF4B, FASN, and PARK2 antibody. Sections were then washed three times with PBS, incubated in a biotinylated goat anti-rabbit IgG (diluted 1:200) for 1 hour, washed again in PBS, and incubated in Vectastain ABC kit (Vector Labs Inc.) for 1 hour. After three washes in PBS, immunolabeling was revealed with 0.05% diaminobenzidine. Sections were then dehydrated in ethanol, cleared in xylene, and cover slipped with a mounting medium. On the basis of the staining intensity, samples were scaled between 0 and 4, as discussed earlier (0 means low staining intensity while 4 represents the highest staining intensity).
Extraction and analysis of The Cancer Genome Atlas datasets
UACLAN (http://ualcan.path.uab.edu/) is a comprehensive resource for investigating cancer data, primarily The Cancer Genome Atlas (TCGA). The expression of PARK2 and other transcripts in TCGA DLBCL samples (n = 41) was extracted using the UACLAN database. The database computes the overall survival (OS) analysis (Kaplan–Meier survival plot) of PARK2 using survival data of patients with high gene expression [transcript per million (TPM) values above upper quartile] and low/medium gene expression (TPM values below upper quartile). We obtained PARK2 and other transcript expressions in naïve B cells from healthy individuals (n = 91) from the DICE [Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs), and Epigenomics] database (https://dice-database.org/), a critical interactive resource of the transcriptional and epigenomic background of several types of human immune cells.
Assessment of viable cells
Exponentially growing stable cells were seeded in equal density (1 million). After 12 hours of seeding, cells were collected till the indicated timepoints and stained with trypan blue. The number of nonviable cells and viable cells were counted depending upon the intake of trypan blue in the hemocytometer.
Clonogenic methylcellulose assays
Genetically modified GMO and DLBCL cells (25–50 × 103) were plated in methylcellulose (RnD). Colonies were visualized and counted after 15 days.
Statistical analysis
Values were expressed as mean ± SD. For comparison between two groups, the unpaired Student t test was used. One-way ANOVA followed by either Dunnet or Bonferroni post hoc analysis was used to compare more than two groups as mentioned in figure legends. Wilcoxon signed-rank test was used to compare the datasets between normal and DLBCL samples, P < 0.05 was considered as significant. Results represent means ± SD of a minimum of three independent experiments.
Data availability
All the supporting data for this study are available from the corresponding author on reasonable request.
Results
PARK2 interacts with eIF4B
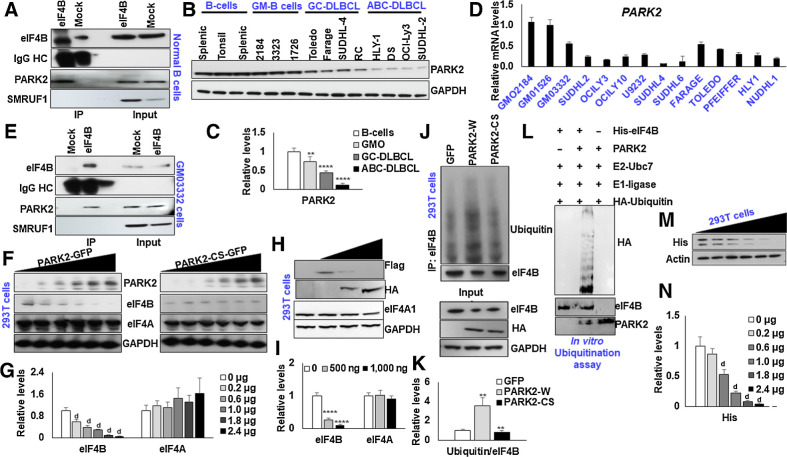
In an attempt to identify the E3 ligase interacting protein partners of eIF4B, we interrogated the publicly available databases, BioGRID (13). Among these, we found PARK2 and SMURF1 as the potential interacting partners with eIF4B (Supplementary Fig. S1). To experimentally validate these interactions, we performed co-immunoprecipitation with eIF4B in normal B cells. PARK2, but not SMURF1, was readily detected in the immunoprecipitants of eIF4B (Fig. 1A). PARK2, initially identified to be associated with Parkinson disease, is emerging as a potential tumor suppressor in several cancers, including DLBCL (14, 15). PARK2 belongs to the RING between RING family of the E3 ligase, which is part of the multiprotein ubiquitin-proteasome system that mediates the targeting protein for degradation (16). Next, we analyzed the expression of PARK2 and eIF4B in a panel of normal B cells, lymphoblastoid cells (GM cells), and DLBCL [ABC- and Germinal Center (GC)-DLBCL]. Immunoblot analysis revealed a significant decrease in the PARK2 protein in both ABC and GC-DLBCLs compared with normal B cells, while the expression of eIF4B, consistent with the literature, was noted to be enhanced (Supplementary Fig. S2A and S2B), suggesting a negative correlation between levels of the two proteins. Notably, the expression of PARK2 in GM-B cells (lymphoblastoid cells) was almost comparable with that of normal B cells (Fig. 1B–D). It should be pointed out that Epstein-Barr virus (EBV) infected and subsequently immortalized normal B cells (GM-B cells) harbor minor genetic and phenotypic alterations (17). Next, we checked the interaction between eIF4B and PARK2 in GM-B cells. Consistent with our previous observation, enrichment of PARK2, but not SMURF1, was readily detected in eIF4B precipitated complex (Fig. 1E).
Figure 1.
PARK2 ubiquitinates eIF4B. A, Lysates from naïve B cells were immunoprecipitated with eIF4B antibody and probed for PARK2 and SMURF1. B, The protein levels PARK2 were shown in naïve B cells (three cases of different donors), GMO B cells (2184, 3323, 1726), and DLBCL (ABC-HLY-1, DS, OCI-Ly3, SUDHL-2 and GC-Toledo, Farage, SUDHL-4 and RC) cells. C, Densitometric quantification of the immunoblots in B. Values were first normalized with their respective loading controls and neutralized with the average of naïve B cells and expressed as mean ± SD, which was set at 1. Statistical analysis was performed using two-way ANOVA followed by Dunnett post hoc analysis. **,P < 0.01; ***, P < 0.005 versus naïve B cells. D, qRT-PCR analysis for checking mRNA expression of PARK2 in indicated cells. Results were normalized with GMO2184 cells, which were set at 1 and expressed as mean ± SD (n = 3). E, Lysates from GMO3323 were immunoprecipitated with eIF4B antibody and probed for PARK2 and SMURF1. F, 293T cells were transfected with increasing concentration of PARK2-GFP or PARK2-CS-GFP, and levels of endogenous eIF4B was confirmed by immunoblotting. G, Densitometric quantification of the immunoblots in F. Values were normalized with corresponding loading control and neutralized with GFP (0 μg) transfected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. d, P < 0.001 versus GFP transfected cells. H, 293T cells were transfected with SFB-eIF4B along with the increasing concentration of PARK2-HA, and levels of ectopic levels of eIF4B was confirmed by immunoblotting. I, Densitometric quantification of the immunoblots in H. Values were normalized with their respective loading control and neutralized with SFB-eIF4B (2 μg) transfected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. ****,P < 0.001 versus SFB-eIF4B transfected cells. J, 293T cells were transfected with indicated constructs. After 36 hours of transfection, endogenous eIF4B was enriched from the lysates and ubiquitin levels were detected by immunoblotting. Lysates were probed for the indicated antibodies. K, Densitometric quantification of the immunoblots in J. Values were normalized with their corresponding eIF4B levels and neutralized with GFP transfected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Bonferroni post hoc analysis. **, P < 0.01 versus GFP transfected cells, b, P < 0.01 versus PARK2-HA transfected cells. L,In vitro ubiquitination assay of His-eIF4B using recombinant E1 (human ubiquitin-activating enzyme E1), recombinant E2 (UbcH7), HA-ubiquitin, and recombinant PARK2. His-eIF4B was ubiquitinated in the presence recombinant PARK2. M, 293T cells were transfected with pYIC and indicated the amount of PARK2-HA and/or GFP. Posttransfection, cell lysates were resolved and probed with His antibody. Actin was used as the loading control. N, Densitometric quantification of the immunoblots in M. Values were normalized with their corresponding actin levels and neutralized with GFP (0 μg) transfected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. d, P < 0.001 versus GFP transfected cells.
Given the observation that PARK2 is a potential ligase interacting with eIF4B, we performed a classical protein stability assay to determine the functional impact of PARK2 on eIF4B stability. First, we transfected 293T cells with increasing concentrations of PARK2-GFP or catalytically inactive PARK2-GFP (PARK2-GFP) and determined the protein levels of eIF4B by immunoblots. As shown in Fig. 1F, a dose-dependent increase in the concentration of PARK2 decreased the protein levels of eIF4B (Fig. 1G); however, transient transfection of PARK2-CS had minimal impact on eIF4B protein content. Next, we cotransfected 293T cells with SFB-eIF4B with an increasing concentration of PARK2-HA. Increasing the concentration of PARK2-HA expression significantly reduced the flag, further supporting our hypothesis that PARK2-HA destabilizes the eIF4B protein (Fig. 1H and I). Examining the protein turnover using the cycloheximide assay, we observed that ectopic expression of PARK2-HA (wild type) but not PARK2-CS (catalytically inactive) or GFP transfected cells destabilizes the protein (Supplementary Fig. S2C–S2F). Next, we investigated whether PARK2 induces eIF4B ubiquitination in destabilizing the protein levels. Here 293T cells were transfected with PARK2-HA or PARK2-CS, GFP transfected cells were used as an internal control. After 36 hours of transfection, endogenous eIF4B was enriched, and immunoblots detected ubiquitination levels. Forced expression of PARK2 significantly enhanced the ubiquitination of eIF4B compared with GFP transfected cells; however, the ubiquitin signals of PARK2-CS were almost comparable with that of GFP (Fig. 1J and K). To further bolster over observation, we transfected 293T cells with increasing concentrations of PARK2-HA. After 48 hours of transfection, cells were treated with MG132, a potent, reversible, and cell-permeable proteasome inhibitor, for 4 hours. Posttreatment cells were lysed, and protein levels of eIF4B were detected by Western blotting. As anticipated, treatment with MG132 partially restored the eIF4B protein level indicating that the protein is channeled to ubiquitin-mediated degradation upon enhanced PARK2 activity (Supplementary Fig. S2G and S2H). To determine whether PARK2 directly ubiquitinates eIF4B, we incubated purified His-tagged eIF4B with commercially procured recombinant PARK2 in ubiquitination assay buffer. Our results indicate that PARK2 directly ubiquitinates eIF4B in the presence of ubiquitin-activating enzyme E1 and the ubiquitin-conjugating enzyme E2 along with ATP (Fig. 1L). To further expand our observation, we next asked whether PARK2 plays any role in regulating protein translation. Given the central role of eIF4B in regulating both cap-dependent and IRES-dependent translation, we challenged 293T cells with pYIC and the increasing concentration of PARK2. pYIC is a bicistronic fluorescent reporter gene with cap-dependent eYFP and IRES-dependent eCFP translation under the control of the cytomegalovirus promoter (18). Consistent with our dose-dependent study of eIF4B (11), PARK2 overexpression displayed a dose-dependent decrease in both cap-dependent as well as IRES-dependent translation (Fig. 1M and N). Thus, we demonstrated that PARK2 is a bona fide ubiquitin E3 ligase of eIF4B.
Downregulation of PARK2 activates eIF4B-sensitive gene expression.
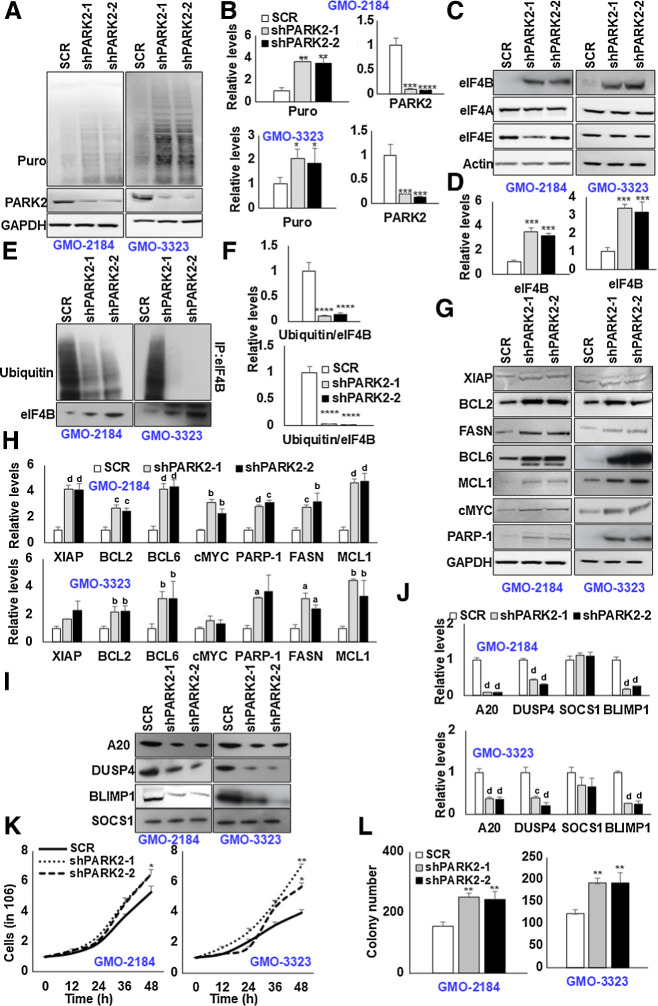
To explore the functional significance of PARK2 expression in B cells, we examined the downregulation of PARK2 in four different GM-B cells. GM-B cells (lymphoblastoid cell lines) are commonly used as a surrogate for resting B cells from peripheral blood with EBV providing a continuous replicating source, bearing minor genetic and phenotypic alterations. To address the potential confounding issue of off-target effects, we selected two independent short hairpin RNAs (shRNA). Non-specific target (SCR)-infected cells were used as an internal control. Having observed that PARK2-HA decreases both cap-dependent as well as IRES-dependent translation, we first challenged PARK2-depleted GM-B cells with SUnSET assay (19). As shown in Fig. 2A and B, depletion of PARK2 significantly enhanced the overall translational capacity of the cells (Supplementary Fig. S3A and S3B). Depletion of PARK2 was confirmed by Western blot and qPCR analysis (Fig. 2A and B; Supplementary Fig. S3A, S3B, and S4A). Next, we assessed the expression of translation initiation factors. In coherence with our previous observation, depletion of PARK2 significantly enhanced the eIF4B protein levels. However, the protein content of eIF4A and eIF4E were minimally modified (Fig. 2C and D; Supplementary Fig. S3C and S3D). More importantly, PARK2 depletion had a nonsignificant impact on the eIF4B transcript, revealing the enhanced protein levels of eIF4B are independent of its transcriptional regulation (Supplementary Fig. S4A). To assess whether the protein stability of eIF4B is altered because of posttranslational ubiquitinylation, we challenged genetically modified GM-B cells with C75, an established FASN inhibitor, for 6 hours. Posttreatment, cells were lysed, and eIF4B protein content was enriched by immunoprecipitation. Consistent with our previous observation, treatment with C75 significantly induced eIF4B ubiquitination. Notably, the ubiquitin signals were shunted upon PARK2 depletion, further supporting our hypothesis that PARK2 ubiquitinates eIF4B upon FASN inhibition (Fig. 2E and F).
Figure 2.
Depletion of PARK2 augments eIF4B signaling. A, PARK2-depleted stable GMO cells (2184, 3323) were cultured in the presence of puromycin (3 μg/mL) for 30 minutes, and lysates were probed for defined antibodies. GAPDH was used as the loading control. B, Densitometric quantification of the immunoblots in A. Values were normalized with their corresponding loading controls and neutralized with corresponding SCR-infected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.005 versus SCR-infected cells. C, G, and I, PARK2-depleted stable GMO cells (2184, 3323) were probed for defined antibodies. Actin was used as the loading control. D, H, and J, Densitometric quantification of the immunoblots in C, G, and I, respectively. Values were normalized with their corresponding loading controls and neutralized with corresponding SCR-infected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. a, P < 0.05; b, P < 0.01; c, P < 0.005; d, P < 0.001 versus SCR-infected cells. E, PARK2-depleted stable GM-B cells (2184, 3323) were treated with C75 (10 μmol/L) for 8 hours. Posttreatment, eIF4B was enriched from the cellular lysates, and ubiquitin levels were determined by immunoblotting. F, Densitometric quantification of the immunoblots in E. Values were normalized with corresponding eIF4B levels and neutralized with their corresponding SCR-infected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. ****, P < 0.001 versus SCR-infected cells. K, Indicated PARK2-depleted stable cells were seeded 1 million per well in 6-well plates. After 12 hours, the cells were collected and counted using trypan blue. Values are expressed as mean ± SD (n = 3), *, P < 0.05; **, P < 0.01 versus SCR-infected corresponding cells. L, The total number of colonies grown in PARK2-depleted cells in methylcellulose culture. Colony counts were performed on the 15th day of methylcellulose culture. **, P < 0.01 versus corresponding SCR-infected control cells.
To further expand our observation of PARK2 regulation on eIF4B activity, we next investigated the effects of PARK2 depletion on eIF4B-sensitive oncogene levels. In agreement with our previous findings (11), we noted that depletion of PARK2 significantly enhanced the protein levels of XIAP, BCL2, BCL6, cMYC, PARP-1, MCL1, and FASN (Fig. 2G and H; Supplementary Fig. S3E and S3F). In addition, the transcripts of cMYC, PARP-1, and FASN were noted to be upregulated, minimal changes were observed in other oncogenes (Supplementary Fig. S4B). More importantly, the expression of tumor suppressor genes: A20, BLIMP1, and DUSP4 were depleted at protein and mRNA levels (Fig. 2I and J; Supplementary Fig. S3G, S3H, and S4C). However, the expression of SOCS1 was noted to be modestly altered. Taken together, depletion of PARK2 not only enhances eIF4B protein levels but also enhances the downstream eIF4B-sensitive target expression. Given the positive impact on oncogene expression upon PARK2 depletion, we next aimed to evaluate the proliferative and transformative effect of PARK2-depleted B cells. First, we checked the cellular proliferative capacity upon PARK2 depletion. Consistent with the literature (20, 21), the depletion of the PARK2 resulted in enhanced cellular proliferation (Fig. 2K; Supplementary Fig. S5A). Next, we studied the transformative impact of PARK2 using the clonogenic assay (colony formation assay). It is a widely used experimental approach to assess the transformed characteristics of cells in vitro (11, 22–24). In agreement with our proliferative and oncogenic studies, decreased PARK2 expression enhanced colony formation in GM-B cells (Fig. 2L; Supplementary Fig. S5B). These observations support our hypothesis that a decrease in PARK2 promotes lymphomagenesis.
Proliferative signals modulate PARK2 expression
We previously reported that FASN regulates S6Kinase-mediated USP11-dependent eIF4B stability (11). We noted that PARK2-mediated ubiquitination of eIF4B induces proteasomal degradation (Fig. 1J and K). Moreover, PARK2-depleted cells failed to ubiquitinate eIF4B upon C75 (FASN inhibitor) treatment (Fig. 2E and F). Given the intriguing cross-talk between FASN, eIF4B, and PARK2, we were tempted to address whether FASN activity regulates PARK2. Here, DLBCL cells (4 of ABC origin, 3 of GC origin) were exposed to C75, and protein levels of PARK2 were determined by immunoblot analysis. As shown in Fig. 3B, increasing the concentration of C75 significantly induced the protein content of PARK2 in ABC-DLBCL (Supplementary Fig. S6A). However, consistent with our previous findings (11), treatment of GC-DLBCL with C75 failed to elicit any changes in PARK2 expression (Supplementary Fig. S7A). To ascertain whether the observed effect is due to FASN activity and not off-target effects, FASN-depleted DLBCL cell lysates (4 of ABC origin, 3 of GC origin, one double-hit lymphoma) were probed for PARK2 protein levels. In agreement with C75 treatment, PARK2 expression was noted to be upregulated in FASN depletion in ABC-DLBCL, but not in GC-DLBCL as well as double-hit lymphoma cells (Fig. 3C; Supplementary Fig. S6B and S7B). Our previous report (11) noted that FASN activated PI3K signaling tethered USP11-eIF4B interactions. Thus, to assess whether PI3K signaling plays a role in PARK2 expression, we treated DLBCLs with PI3K signaling inhibitor (Ly294002: PI3K inhibitor, Rapamycin: mTOR inhibitor, Torin 1: mTORC1 inhibitor MK2206 or AZD5363: Akt inhibitor, Pf-4708671: p70S6Kinase inhibitor). Consistent with the FASN inhibition, treatment with PI3K (Fig. 3D; Supplementary Fig. S6C), Akt (Fig. 3E; Supplementary Fig. S6D), and mTOR inhibitors (Fig. 3F; Supplementary Fig. S6E) in ABC-DLBCL induces PARK2 protein levels. Furthermore, the compound treatment failed to induce PARK2 expression in non–ABC-DLBCLs, an observation in agreement with our previous findings (Supplementary Fig. S7C–S7F). However, to our surprise, treatment with p70-S6Kinase inhibitor failed to provoke any modification in PARK2 expression either in ABC-origin, GC-origin, or double-hit DLBCL lymphoma cells (Fig. 3G; Supplementary Fig. S6F and S7F). Having observed that inhibition of oncogenic signals in ABC-DLBCL, but not in GC-DLBCL, enhances PARK2 protein content, we next assessed the impact of PARK2 protein levels upon cotreatment in GC-DLBCL. Taking a cue from our previous report (11), we exposed GC-DLBCL cells with C75 in combination with PI3K signaling inhibitors. As shown in Figs. 3H–K, inhibition of PI3K signaling in combination with FASN significantly enhanced the protein levels of PARK2 in Toledo and Farage GC-DLBCL cells. Collectively, the data establish that PARK2 protein levels are inversely correlated with proliferative/oncogenic stimuli, which entirely aligns with our other observations and the literature (25).
Figure 3.
Inhibition of proliferative signaling increases PARK2 protein levels. A, Schematic presentation showing proliferative signaling pathway and inhibitors. Indicated cells were cultured with either C75 (B), Ly284002 (D), Rapamycin (E), MK2206 (F), or PF470851 (G) for 16 hours or infected with shRNA against FASN (C) and lysed. Posttreatment/infection, cell lysates were probed for PARK2 expression. GAPDH was used as the loading control. The bar diagram beneath each figure represents densitometric quantification of the immunoblots. Values were normalized with corresponding loading control and neutralized with corresponding DMSO (0.1%) or SCR-infected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 versus corresponding cells. H and J, Toledo and Farage were treated with indicated PI3K signaling inhibitors [LY294002: PI3K inhibitor (1 μmol/L), AZD5363: Akt inhibitor (500 nmol/L), Rapamycin: mTOR inhibitor (50 nmol/L), Torin 1: mTOR inhibitor (250 nmol/L), PF-4708671: S6Kinase inhibitor (1 μmol/L)] in presence or absence of C75. Posttreatment, cell lysates were probed for PARK2. GAPDH was used as the loading control. I and K, Densitometric quantification of the immunoblots in H and J. Values were normalized with corresponding GAPDH and neutralized with DMSO-treated corresponding cells, which was set to 1. Values were represented as mean ± SD for n = 3. Statistical analysis was performed using Student t test (unpaired two-tailed) *, P < 0.05; **, P < 0.01, versus corresponding treated cells.
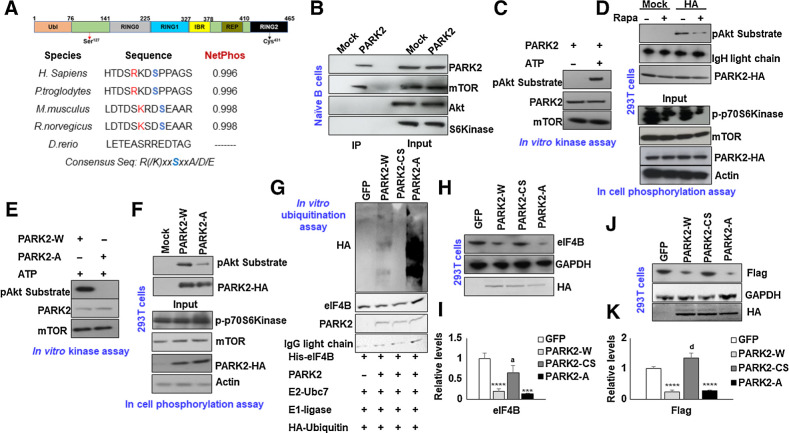
mTOR phosphorylates PARK2
To further verify the association between eIF4B, PARK2, FASN, and the oncogenic stimuli, we next assessed the possibility of PARK2 as a PI3K signaling substrate. We previously reported that USP11, a bona fide eIF4B deubiquitinase, is phospho-modified by p70-S6Kinase and this posttranslational event enhances USP11–eIF4B interactions (11). Furthermore, the protein levels of PARK2 were noted to be increased upon pharmacologic inhibition of the PI3K signaling cascade. Surprisingly, the chemical inhibition of p70-S6kinase displayed minimal impact on PARK2 protein levels. To interrogate the mechanism(s) controlling PARK2 activity, we performed in silico analysis to address whether oncogenic PI3K signaling phosphorylates PARK2 protein. This analysis revealed the presence of one consensus site of phosphorylation at Ser127 (R/KxxS). Interestingly, the site was observed to be evolutionary conserved across different species (Fig. 4A). To identify the potential phospho-modifying kinase, we precipitated PARK2 from normal B cells and probed for the PI3K signaling kinase. As shown in Fig. 4B, mTORC1 was readily detected in enriched PARK2 samples. However, other kinases (Akt and p70S6Kinase) were not detected, suggesting that mTORC1 may modify PARK2. To test our hypothesis, we incubated HA-tagged PARK2 enriched from 293T cells with commercially procured catalytically active mTORC1 complex in the presence of ATP. After in vitro kinase reaction, samples were separated on SDS-PAGE, and the phosphorylation signals were detected using pAKT substrate antibody (26). As shown in Fig. 4C, incubating active mTOR complex in the presence of ATP modified PARK2 under in vitro conditions. To evaluate whether mTORC1 can phosphorylate PARK2 under cellular conditions, we treated 293T cells overexpressing PARK2-HA with Rapamycin for two hours. Posttreatment, cells were lysed, and PARK2 protein levels were enriched using anti-HA agarose beads. Consistent with our in vitro reaction, under basal conditions, the PARK2 phospho-signals were readily detected; however, treatment with Rapamycin significantly reduced the cellular phospho-signals of PARK2, establishing that PARK2 is indeed phosphorylated by mTORC1 kinase (Fig. 4D). To evaluate whether the predicted Ser127 is certainly the phospho-acceptor site on PARK2, we incubated PARK2-HA [wild type (W) and Ser127Ala (A)] with an active mTOR complex for the in vitro kinase reaction. Consistent with our previous data, mTOR was able to modify the wild-type protein. Still, the phospho-signals were lost upon mutation of Ser127 to Alanine, confirming that Ser127 is the mTOR-driven phospho-acceptor site on PARK2 (Fig. 4E). Further to evaluate under cellular conditions, 293T cells were transfected with PARK2-HA (W and A) and cultured with 20% FBS to activate the mTOR signals. After transfection, cells were lysed, and PARK2 was enriched using an anti-HA agarose bead and separated on SDS-PAGE. As shown in Fig. 4F, phospho-signals were detected in wild-type PARK2, but the intensity was significantly diminished upon point mutation to Alanine.
Figure 4.
mTOR phosphorylates PARK2 at Ser127. A, Schematic presentation of protein architecture of PARK2. Sequence alignment of AGC substrate consensus sequences within PARK2 across different species. B, Lysates from naïve B cells were immunoprecipitated with PARK2 antibody and probed for mTOR, AKT, and p70S6Kinase. C and E, HA-enriched PARK2 (wt and mutants) were incubated with active mTOR protein in the presence and absence of ATP as indicated, and phospho-signals of modified PARK2 were captured using pAkt substrate antibody. 293T cell transfected with PARK2-HA (D) and/or its mutant (F) and cultured in 20% FBS. Cells were treated with Rapamycin for 2 hours. D, Posttransfection/posttreatment, PARK2 was enriched with HA beads, and phospho-signals were captured using pAkt substrate antibody. Lysates were probed for the indicated antibodies. G,In vitro ubiquitination assay of His-eIF4B using recombinant E1 (human ubiquitin-activating enzyme E1), recombinant E2 (UbcH7), HA-ubiquitin and PARK2 (wt and mutants). His-eIF4B was ubiquitinated in the presence PARK2-W, PARK2-A significantly increased eIF4B ubiquitination. PARK2-CS was used as internal control. H and J, 293T cells were transfected with PARK2-HA (wt and mutants) or GFP either in the presence (J) or absence (H) of SFB-eIF4B, and levels of eIF4B (endogenous or ectopic) were determined by immunoblotting. GAPDH was used as the loading control. I and K, Densitometric quantification of the immunoblots in H and J, respectively. Values were normalized with GFP transfected cells and were represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Bonferroni post hoc analysis. ***, P < 0.005; ****, P < 0.001 versus GFP transfected cells, a, P < 0.05; d, P <0.001 versus PARK2-W transfected cells.
We next inquired what is the impact of mTOR-mediated PARK2 phosphorylation on its ubiquitinylation activity. To address this question, we enriched PARK2 (wt and mutants) and subjected them to in vitro ubiquitination assay using His-tagged eIF4B as a substrate. Consistent with our previous observation (Fig. 1L), PARK2-W was able to ubiquitinate eIF4B directly. Notably, the ubiquitination of eIF4B with PARK2-A was enhanced, indicating that the mTORC1 mediated phosphorylation may hamper the PARK2 enzymatic activity (Fig. 4G). PARK2-CS was used as an internal control. Having observed that mTORC1 phosphorylates PARK2 and may hinder its activity, we next inquired about the physiologic relevance of this phospho-event. Here, 293T cells were transfected with either GFP or PARK2-HA (wild type and mutants) and measured the protein levels of endogenous eIF4B levels. As anticipated, forced expression of PARK2, but not catalytically inactive PARK2 (PARK2-CS), robustly reduced the endogenous eIF4B protein levels compared with GFP transfected cells. Significantly, ectopic expression of PARK2 Alanine mutant (PARK2-A) also decreased the eIF4B protein levels, almost comparable with WT PARK2 (Fig. 4H and I). To further support our observations, we transfected 293T cells with SFB-eIF4B along with either GFP or PARK2 constructs. After 48 hours of transfection, the ectopically expressed eIF4B levels were determined by flag antibody. Concurrence with our endogenous data, the flag signals was also found to decline with coexpression of PARK2 (W and A); however, the overexpression of PARK2-CS failed to reduce the flag levels (Fig. 4J and K). Collectively, the data indicate the PARK2 is mTORC1 substrate, and mTORC1 mediated PARK2 phosphorylation at Ser127 may modulate its ubiquitin activity.
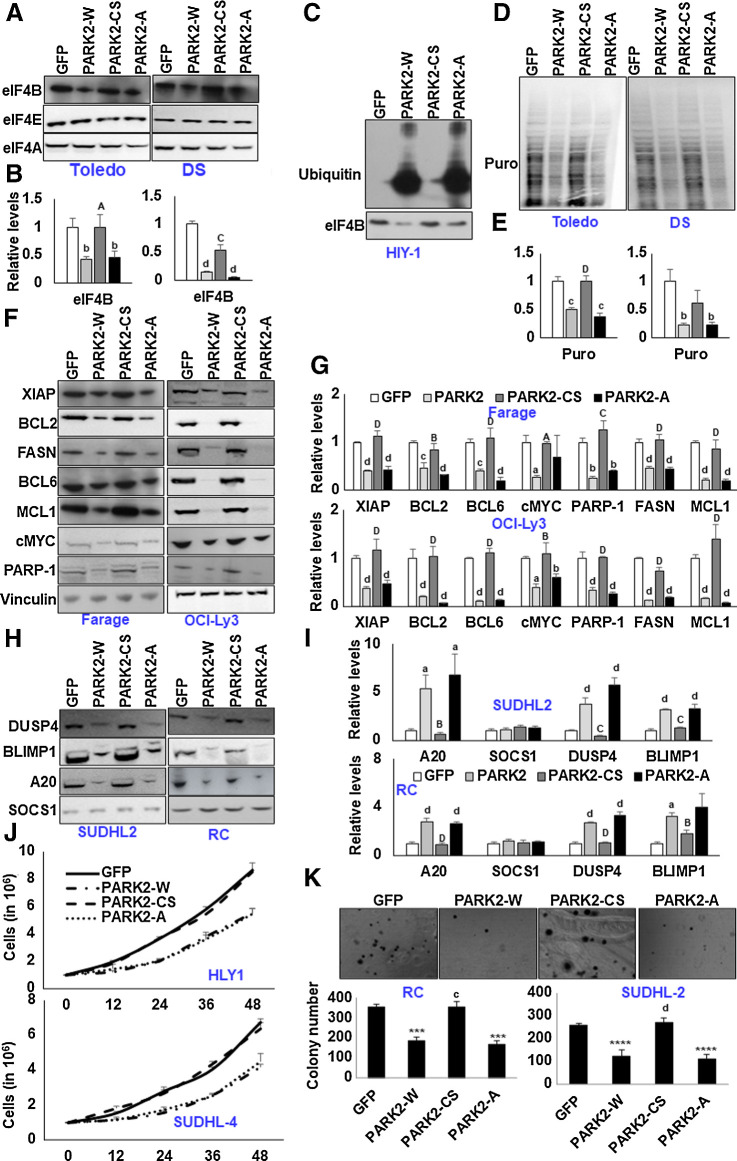
mTORC1-mediated PARK2 phosphorylation enhances eIF4B oncogenic activity
To study the effect of mTORC1-mediated PARK2 phosphorylation on eIF4B activity, we infected DLBCLs (4 ABC origin, 3 GC, and one double-hit lymphoma) with PARK2 (wild type and mutants). In our assay, GFP-infected cells were used as internal control, while PARK2-CS–overexpressing cells served as a negative control. Ectopic expression of PARK2-W, but not PARK2-CS, significantly depleted eIF4B protein levels in DLBCLs. Significantly overexpressing PARK2-A also hampered the protein levels of eIF4B in DLBCLs. eIF4B transcripts in PARK2-modulated DLBCL cells were minimally modified. Significantly, the protein levels of eIF4A and eIF4E were minimally altered (Fig. 5A and B; Supplementary Fig. S8A and S8B). Furthermore, eIF4B transcripts were noted to be modestly changed upon genetic manipulation of PARK2 in DLBCL cells (Supplementary Fig. S9). To assess whether PARK2 depletes eIF4B in a ubiquitin-dependent manner in DLBCLs, we transiently infected HLY1 cells with PARK2 constructs. After 48 hours of infection, cells were lysed, and eIF4B was enriched using an eIF4B-specific antibody. As shown in Fig. 5C, forced expression of PARK2 (W or A) robustly induced the ubiquitin signals with a substantial decrease in eIF4B protein levels. Because eIF4B is critical for augmenting overall translation, we examined the PARK2-modified DLBCLs with the SUnSET assay. Consistent with our other observations, puromycin pulse labeling of the nascent peptide was significantly reduced in PARK2-W–infected DLBCLs, but not in PARK2-CS–infected cells, revealing catalytically active PARK2 as a potent regulator of the transitional apparatus. Importantly, overexpression of PARK2-A, mTORC1 phospho-deficient PARK2 point mutant, displayed a significant decrease in anti-puromycin signals, almost comparable with the PARK2-W, revealing that mTORC1-mediated PARK2 phosphorylation hampers its activity in translational regulation (Fig. 5D and E, Supplementary Fig. S8C and S8D).
Having noticed a significant reduction in overall translation linked to eIF4B protein upon PARK2 expression, we next evaluated the expression of eIF4B-sensitive genes in these cells. Consistently, overexpression of PARK2 (W and A), but not PARK2-CS, significantly reduced the protein levels of oncogenes, namely cMYC, MCL1, XIAP, BCL2, BCL6, PARP-1, and FASN compared with GFP-infected cells (Fig. 5F and G; Supplementary Fig. S10A and S10B). Furthermore, in concert with our previous report, the transcripts of cMYC, PARP-1, and FASN were noted to be depleted upon PARK2 (W and A), but not PARK2-CS, compared GFP-infected cells (Supplementary Fig. S11). Likewise, the protein and the transcript of tumor suppressor genes like A20, DUSP4, and BLIMP1 were reduced upon infection with constructs expressing PARK2 (W and A), but not PARK2-CS in comparison with GFP-expressing cells. Significantly, expression of SOCS1, a Wnt signaling sensitive tumor suppressor gene (27), was minimally modified upon PARK2 modulation (Fig. 5H and I; Supplementary Fig. S12A, S12B, and S13). Taken together, the current data strongly endorse the potential role of PARK2 in translational regulation. Next, we aimed to evaluate the oncogenic properties of PARK2 and its mutant in DLBCL. First, we checked the cellular proliferative capacity. Consistent with the extant literature, ectopic expression of the PARK2, but not PARK2-CS, resulted in a significant reduction of cellular proliferation. Moreover, forced expression of PARK2-A in DLBCL displayed a substantial reduction in cellular proliferation, almost comparable with PARK2-W (Fig. 5K; Supplementary Fig. S14). Next, we studied the impact of PARK2 on the clonogenic assay. In agreement with our proliferative and oncogenic studies, PARK2 (W and A) overexpression, but not PARK2-CS, decreased the colony-forming capacity of DLCBLs (Fig. 7K). Taken together, PARK2 acts as a potent tumor suppressor, and mTOR-mediated PARK2 phosphorylation depletes its ligase activity and thus its tumor-suppressive capacity.
Figure 7.
PARK2 in eIF4B regulation and lymphomagenesis. Functional PARK2 targets eIF4B for ubiquitination- and proteasome-mediated degradation in maintaining B-cell homeostasis. Loss of expression or activity of PARK2 enhances eIF4B protein levels resulting in aberrant oncogene translation which contributes to lymphomagenesis.
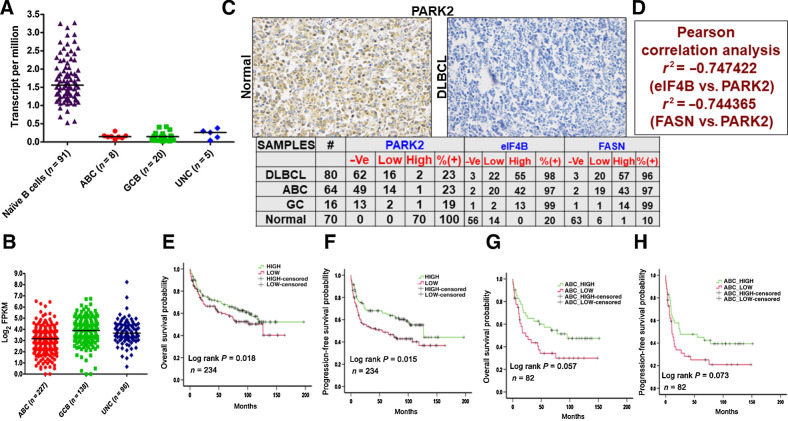
PARK2 expression is depleted in DLBCL
To elucidate the clinical relevance of PARK2 in DLBCL, we first examined the expression of PARK2 using the DICE and TCGA datasets (28, 29). The expression of PARK2 was significantly reduced in DLBCL samples compared with those of naïve B cells, suggesting that the protein may have a tumor suppressor role in lymphomagenesis. Furthermore, the transcripts of PARK2 in ABC-DLBCL was significantly lower compared with GC-DLBCL (Fig. 6A). To further corroborate our findings, gene expression of PARK2 was analyzed from the publicly available transcriptomic dataset (30). Evaluating more than 450 DLBCL samples, the expression of PARK2 was noted to be significantly reduced in ABC compared with GC and unclassified DLBCL samples (Fig. 6B). To corroborate this observation, we stained primary DLBCL specimens from commercially obtained TMA slides (US Biomax). IHC analysis of PARK2 expression was performed in 80 DLBCL as well as 70 reactive B-cell samples. Staining naïve B cells displayed 100% expression of PARK2, while that of the B-cell tumor was around 23% (Fig. 6C). Collectively, PARK2 appears to be depleted in the vast majority of DLBCL, independent of its COO, a similar expression profile was also observed from DLBCL cultured cells (Fig. 1B)
Figure 6.
Clinicopathologic evaluation of PARK2. A, Representative plots show expression profiles of PARK2 in naïve B cells (obtained from DICE database https://dice-database.org/) compared with molecular subgroups of patients with DLBCL in TCGA dataset. PARK2 showed significantly lower expression in tumor samples compared with control. B, Comparison of PARK2 in molecular subgroups using a publicly available large dataset of patients with DLBCL (https://gdc.cancer.gov/about-data/publications/DLBCL-2018). PARK2 showed significantly lowest expression in ABC-DLBCL subgroups compared with GCB-DLBCL and UNC-DLBCL. C, Representative IHC image of commercially procured TMA slides stained with PARK2 antibody. Summary of the PARK2, eIF4B, and FASN stained slides for DLBCL and normal reactive lymph node samples. -Ve: no staining detected, low: 1–2 staining density, high: 3–4 staining density (D) Pearson correlation evaluation of the stained slides. E, PARK2 expression was found to be significantly (P 0.05) associated with OS of patients with DLBCL in the publicly available dataset. Patients with a higher median expression of PARK2 showed better prognosis than patients having lower than median expression. F, PARK2 expression was also found to be significantly (P 0.05) associated with the PFS in the same cohort of patients with DLBCL having a similar outcome. G, The same cohort was segregated into molecular subgroups, and higher median PARK2 expression in patients with ABC-DLBCL showed significantly (P 0.05) better prognosis than patients having lower than median expression. H, PFS analysis in patients with ABC-DLBCL showed a similar trend, albeit not statistically significant (P 0.05).
Next, replicate IHC slides used for PARK2 staining were also examined for eIF4B and FASN expression. In parallel with our previous report (11), expression of eIF4B and FASN were noted to be upregulated in more than 95% of all the samples assayed (Fig. 6C). Performing Pearson correlation analysis, we found that there is a strong negative correlation in the expression pattern of all three proteins (Fig. 6D). Subsequently, the survival of the patients was evaluated using Kaplan–Meier analysis (31). PARK2 expression was significantly (P = 0.018) associated with the OS and progression-free survival (PFS; P = 0.015) of patients with DLBCL in the publicly available dataset (Fig. 6E and F; ref. 28). Patients with a higher median expression of PARK2 showed a better prognosis than patients with lower median expression. Similar analysis upon molecular subgroup classifications, we found that patients with ABC-DLBCL with lower PARK2 expression (less than the median of PARK2 level) showed shortened survival periods than the high expression of PARK2 (P = 0.057; Fig. 6G). Likewise, the expression profile of PARK2 in GC and unclassified DLBCL tended toward a poorer prognosis, but the difference was not statistically significant in our analysis (Supplementary Fig. S15C and S15D). Significantly, PARK2 expression displayed a positive correlation with PFS in ABC-DLBCL (P = 0.072; Fig. 6F), along with a similar trend in GC and unclassified (Supplementary Fig. S15E and S15F). While a more detailed analysis including additional clinicopathologic parameters will shed more light on our findings, our results do suggest a putative correlation between PARK2 levels and clinical outcomes. Together, the above clinical data reveals lower expression of PARK2 being associated with a decrease in both OS and PFS, which supports our contention that PARK2 protein acts as a tumor suppressor in DLBCL.
Discussion
Translational control of cancer is a multifaceted process involving alterations in translation factor levels culminating in a sustained expression of oncogenes supporting the survival and proliferation of cancer cells under challenging conditions (8, 32). There is greatly expanding interest in exploring the regulatory cascades of translational control in cancer. Ubiquitination, an essential type of covalent ligation, plays a crucial role in controlling substrate degradation and subsequently mediates the “quantity” and “quality” of various proteins, serving to ensure robust regulation of cellular balance and activity (33–36). While much progress has been made in characterizing the quality of newly synthesized peptides as well as quality control of stalled ribosomes via ubiquitin-mediated pathways, elucidating the fine details of this critical regulatory process in controlling the vital translational initiation proteins is still an area ripe for discovery (37–39). Herein, we identify PARK2 as a novel E3 ligase factor critical to regulating eIF4B stability. Our data suggest that PARK2 is positioned as a tumor suppressor downstream of the mTORC1-mediated pathway that regulates eIF4B stability in conjunction with external (PI3K) and internal metabolic signaling (FASN). They converge downstream in determining the eIF4B-dependent translational output, both in normal and disease states.
Despite the importance of eIF4B's regulatory capacity on the expression of crucial oncogenes associated with the progression and survival of cancer, a detailed molecular understanding of what drives the enhancement of the protein's stability is still unclear (11, 40, 41). We previously reported that metabolic signaling induced by enhanced expression of FASN increases the stability of the protein in DLBCL. Furthermore, while embarking on the molecular characterization, we noted that USP11 in a FASN-S6kinase-dependent manner deubiquitinates eIF4B to enhance its protein level (11). In this study, we demonstrate that PARK2, an established tumor suppressor, is a bona fide E3 ligase of eIF4B in a FASN-dependent manner lends a mechanistic basis to our understanding of eIF4B stability and activity of DLBCL.
PARK2 was initially identified as a spontaneous element in Parkinson disease (14). Subsequent studies reported that the gene is located on the common fragile site region in FRA6E, which is reported to be deleted/suppressed in several cancers (42, 43). Furthermore, PARK2 knockout mice are highly susceptible to γ-irradiation–induced lymphoma (14, 44). Previous studies reported that loss/mutation of PARK2 predisposes the cells toward a transformed state by augmenting cellular proliferation, increased cell cycle, and mitochondrial biosynthesis (14). Furthermore, the genetic loci of PARK2 are within the region of 6q26q27, which was noted to be lost/silenced in more than 50% of patients with DLBCL (15). This mechanistic study establishes that PARK2 directly impacts the overall translational output regulating both cap-dependent and IRES-dependent translational apparatus, an essential cellular activity for sustained cell growth and viability. Significantly, the clinical evaluation of PARK2 protein levels in DLBCL samples evaluated by Carreras and colleagues demonstrated the absence of PARK2 expression in lymphoid tissues (15). In accord with their observation, our expression analysis of PARK2 studied by IHC staining in commercially procured TMA slides displayed a robust reduction in the protein expression under various lymphoma types. These data are further supported by the depletion of PARK2 transcripts in DLBCL evaluated from TCGA, GDC, and DICE datasets.
Owing to its critical role in translational regulation, eIF4B has been proposed as the convergent target of the PI3K/mTOR pathway (40, 41, 45). PARK2 is a well-established intracellular PI3K/Akt signaling inhibitor (25). Gupta and colleagues using the genomic profile from publicly available databases, reported that PARK2 enhances PI3K/Akt activation (25). Park and colleagues noted that under mitochondrial stress, PARK2 ubiquitinates mTORC1 on Lys2066 and Lys2306 for maintaining the kinase activity for cellular survival (46). Furthermore, Lin and colleagues demonstrated that PARK2 hampers the Akt-driven tumorigenic events in glioma cells (47). Precisely how PI3K/Akt signaling regulates PARK2 activity is unclear, although its expression is known to be suppressed by epigenetic regulation (48). To this end, we observed that PARK2 is a mTORC1 substrate, and mTORC1 mediated PARK2 phosphorylation at Ser127 suppressing its E3 ligase activity. It is well established that PINK1-mediated PARK2 phosphorylation at Ser65 exponentially enhances its E3 ligase activity (49). Depletion of PTEN (a potent regulator or PI3K signaling) is known to downregulate PINK1 expression (25). Conversely, PINK1 can directly activate Akt via activation of mTORC2 to enhance cancer cell invasiveness (50). It will be intriguing to dissect the molecular events regulating PARK2 by PINK1 and mTORC1 in lymphoid malignancies. In the same vein, the impact of fatty acid synthase on PARK2 is likely intertwined with the PI3K/mTOR pathway. In addition, Kim and colleagues noted that PARK2 expression is lipid-dependent, and the protein may be involved in fatty acid metabolism (51). Interestingly, by blocking FASN using chemical modulators, Verstreken and colleagues overcame the PINK1 deficiency in flies (52).
Overall, the PARK2-eIF4B axis affects B-cell lymphoma proliferation through regulating translational output and cellular signaling (Fig. 7). This novel molecular link between the ubiquitin-proteasome system and the mRNA translational apparatus reinforces the convergence of these two critical pathways in the oncogenic events driving lymphoid malignancies. Further studies will be required to address the cross-talk between PINK1- and mTOR-mediated phosphorylation of PARK2 to elucidate the signaling that regulates the translational output and cellular proliferation. In the aggregate, our findings reveal a complex network driven by FASN involving p70S6Kinase/USP11 (11) and PARK2/mTORC1 signaling in the regulation of eIF4B, providing additional candidate therapeutic targets in DLBCL and related malignancies.
Supplementary Material
Acknowledgments
The authors would like to thank Didier Trono, Toshio Kitamura, Ted Dawson, and Edward Fon for generously providing reagents. This work was supported in part by a Merit Review Award from the Department of Veterans Affairs (R.B. Gartenhaus), R01CA164311 (R.B. Gartenhaus) from the NIH and shared resource and funding Massey NIH-NCI Cancer Center Support Grant P30 CA016059.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
B.B. Kapadia: Conceptualization, validation, methodology, writing–original draft, writing–review and editing. A. Roychowdhury: Data curation, formal analysis. F. Kayastha: Formal analysis, writing–review and editing. N. Nanaji: Resources, formal analysis. R.B. Gartenhaus: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 2006;17:1807–19. [DOI] [PubMed] [Google Scholar]
- 2. Di Costanzo A, Del Gaudio N, Conte L, Altucci L. The Ubiquitin proteasome system in hematological malignancies: new insight into its functional role and therapeutic options. Cancers 2020;12:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qi J, Ronai ZA. Dysregulation of ubiquitin ligases in cancer. Drug Resist Updat 2015;23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qu C, Kunkalla K, Vaghefi A, Frederiksen JK, Liu Y, Chapman JR, et al. Smoothened stabilizes and protects TRAF6 from degradation: a novel non-canonical role of smoothened with implications in lymphoma biology. Cancer Lett 2018;436:149–58. [DOI] [PubMed] [Google Scholar]
- 5. Masle-Farquhar E, Russell A, Li Y, Zhu F, Rui L, Brink R, et al. Loss-of-function of Fbxo10, encoding a post-translational regulator of BCL2 in lymphomas, has no discernible effect on BCL2 or B lymphocyte accumulation in mice. PLoS One 2021;16:e0237830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akyurek N, Ren Y, Rassidakis GZ, Schlette EJ, Medeiros LJ. Expression of inhibitor of apoptosis proteins in B-cell non-Hodgkin and Hodgkin lymphomas. Cancer 2006;107:1844–51. [DOI] [PubMed] [Google Scholar]
- 7. Yang Y, Schmitz R, Mitala J, Whiting A, Xiao W, Ceribelli M, et al. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov 2014;4:480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 2013;5:a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor J, Yeomans AM, Packham G. Targeted inhibition of mRNA translation initiation factors as a novel therapeutic strategy for mature B-cell neoplasms. Explor Target Antitumor Ther 2020;1:3–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Z, Cheng Z, Raghothama C, Cui Z, Liu K, Li X, et al. USP9X controls translation efficiency via deubiquitination of eukaryotic translation initiation factor 4A1. Nucleic Acids Res 2018;46:823–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kapadia B, Nanaji NM, Bhalla K, Bhandary B, Lapidus R, Beheshti A, et al. Fatty Acid Synthase induced S6Kinase facilitates USP11-eIF4B complex formation for sustained oncogenic translation in DLBCL. Nat Commun 2018;9:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dodd KM, Tee AR. In vitro mTORC1 kinase assay for mammalian cells protocol. Bio Protoc 2016;6:e1827. [Google Scholar]
- 13. Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34:D535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu J, Zhang C, Hu W, Feng Z. Parkinson's disease-associated protein Parkin: an unusual player in cancer. Cancer Commun 2018;38:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carreras J, Kikuti YY, Bea S, Miyaoka M, Hiraiwa S, Ikoma H, et al. Clinicopathological characteristics and genomic profile of primary sinonasal tract diffuse large B cell lymphoma (DLBCL) reveals gain at 1q31 and RGS1 encoding protein; high RGS1 immunohistochemical expression associates with poor overall survival in DLBCL not otherwise specified (NOS). Histopathology 2017;70:595–621. [DOI] [PubMed] [Google Scholar]
- 16. Chaugule VK, Walden H. Specificity and disease in the ubiquitin system. Biochem Soc Trans 2016;44:212–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hussain T, Mulherkar R. Lymphoblastoid cell lines: a continuous in vitro source of cells to study carcinogen sensitivity and DNA repair. Int J Mol Cell Med 2012;1:75–87. [PMC free article] [PubMed] [Google Scholar]
- 18. Nie M, Htun H. Different modes and potencies of translational repression by sequence-specific RNA-protein interaction at the 5'-UTR. Nucleic Acids Res 2006;34:5528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009;6:275–7. [DOI] [PubMed] [Google Scholar]
- 20. Duan H, Lei Z, Xu F, Pan T, Lu D, Ding P, et al. PARK2 suppresses proliferation and tumorigenicity in non-small cell lung cancer. Front Oncol 2019;9:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lei Z, Duan H, Zhao T, Zhang Y, Li G, Meng J, et al. PARK2 inhibits osteosarcoma cell growth through the JAK2/STAT3/VEGF signaling pathway. Cell Death Dis 2018;9:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang KC, Chen RY, Wang YC, Hung LY, Medeiros LJ, Chen YP, et al. Stem cell characteristics promote aggressiveness of diffuse large B-cell lymphoma. Sci Rep 2020;10:21342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pichard A, Marcatili S, Karam J, Constanzo J, Ladjohounlou R, Courteau A, et al. The therapeutic effectiveness of (177)Lu-lilotomab in B-cell non-Hodgkin lymphoma involves modulation of G2-M cell cycle arrest. Leukemia 2020;34:1315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A 2012;109:20047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gupta A, Anjomani-Virmouni S, Koundouros N, Dimitriadi M, Choo-Wing R, Valle A, et al. PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-nitrosylation. Mol Cell 2017;65:999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walia V, Cuenca A, Vetter M, Insinna C, Perera S, Lu Q, et al. Akt regulates a Rab11-effector switch required for ciliogenesis. Dev Cell 2019;50:229–46 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trinath J, Holla S, Mahadik K, Prakhar P, Singh V, Balaji KN. The WNT signaling pathway contributes to dectin-1-dependent inhibition of Toll-like receptor-induced inflammatory signature. Mol Cell Biol 2014;34:4301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell 2018;175:1701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 2018;378:1396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer 2010;10:254–66. [DOI] [PubMed] [Google Scholar]
- 33. Kapadia BB, Gartenhaus RB. DUBbing down translation: the functional interaction of deubiquitinases with the translational machinery. Mol Cancer Ther 2019;18:1475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther 2020;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002;82:373–428. [DOI] [PubMed] [Google Scholar]
- 36. Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem 2017;86:193–224. [DOI] [PubMed] [Google Scholar]
- 37. Wang F, Canadeo LA, Huibregtse JM. Ubiquitination of newly synthesized proteins at the ribosome. Biochimie 2015;114:127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brandman O, Hegde RS. Ribosome-associated protein quality control. Nat Struct Mol Biol 2016;23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karamyshev AL, Karamysheva ZN. Lost in translation: ribosome-associated mRNA and protein quality controls. Front Genet 2018;9:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shahbazian D, Parsyan A, Petroulakis E, Hershey J. Sonenberg N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 2010;9:4106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen K, Yang J, Li J, Wang X, Chen Y, Huang S, et al. eIF4B is a convergent target and critical effector of oncogenic Pim and PI3K/Akt/mTOR signaling pathways in Abl transformants. Oncotarget 2016;7:10073–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Glover TW, Wilson TE, Arlt MF. Fragile sites in cancer: more than meets the eye. Nat Rev Cancer 2017;17:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Denison SR, Callahan G, Becker NA, Phillips LA, Smith DI. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer 2003;38:40–52. [DOI] [PubMed] [Google Scholar]
- 44. Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A 2011;108:16259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horvilleur E, Sbarrato T, Hill K, Spriggs RV, Screen M, Goodrem PJ, et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia 2014;28:1092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park D, Lee MN, Jeong H, Koh A, Yang YR, Suh PG, et al. Parkin ubiquitinates mTOR to regulate mTORC1 activity under mitochondrial stress. Cell Signal 2014;26:2122–30. [DOI] [PubMed] [Google Scholar]
- 47. Lin DC, Xu L, Chen Y, Yan H, Hazawa M, Doan N, et al. Genomic and functional analysis of the E3 ligase PARK2 in glioma. Cancer Res 2015;75:1815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Agirre X, Roman-Gomez J, Vazquez I, Jimenez-Velasco A, Garate L, Montiel-Duarte C, et al. Abnormal methylation of the common PARK2 and PACRG promoter is associated with downregulation of gene expression in acute lymphoblastic leukemia and chronic myeloid leukemia. Int J Cancer 2006;118:1945–53. [DOI] [PubMed] [Google Scholar]
- 49. Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J 2014;460:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Flanagan CH, Morais VA, Wurst W, De Strooper B, O'Neill C. The Parkinson's gene PINK1 regulates cell cycle progression and promotes cancer-associated phenotypes. Oncogene 2015;34:1363–74. [DOI] [PubMed] [Google Scholar]
- 51. Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest 2011;121:3701–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vos M, Geens A, Bohm C, Deaulmerie L, Swerts J, Rossi M, et al. Cardiolipin promotes electron transport between ubiquinone and complex I to rescue PINK1 deficiency. J Cell Biol 2017;216:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the supporting data for this study are available from the corresponding author on reasonable request.



![Figure 3. Inhibition of proliferative signaling increases PARK2 protein levels. A, Schematic presentation showing proliferative signaling pathway and inhibitors. Indicated cells were cultured with either C75 (B), Ly284002 (D), Rapamycin (E), MK2206 (F), or PF470851 (G) for 16 hours or infected with shRNA against FASN (C) and lysed. Posttreatment/infection, cell lysates were probed for PARK2 expression. GAPDH was used as the loading control. The bar diagram beneath each figure represents densitometric quantification of the immunoblots. Values were normalized with corresponding loading control and neutralized with corresponding DMSO (0.1%) or SCR-infected cells, which was set as 1; represented as mean ± SD for n = 3. Statistical analysis was performed using one-way ANOVA followed by Dunnett post hoc analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001 versus corresponding cells. H and J, Toledo and Farage were treated with indicated PI3K signaling inhibitors [LY294002: PI3K inhibitor (1 μmol/L), AZD5363: Akt inhibitor (500 nmol/L), Rapamycin: mTOR inhibitor (50 nmol/L), Torin 1: mTOR inhibitor (250 nmol/L), PF-4708671: S6Kinase inhibitor (1 μmol/L)] in presence or absence of C75. Posttreatment, cell lysates were probed for PARK2. GAPDH was used as the loading control. I and K, Densitometric quantification of the immunoblots in H and J. Values were normalized with corresponding GAPDH and neutralized with DMSO-treated corresponding cells, which was set to 1. Values were represented as mean ± SD for n = 3. Statistical analysis was performed using Student t test (unpaired two-tailed) *, P < 0.05; **, P < 0.01, versus corresponding treated cells.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/2e28/9381106/078c0aafbb81/735fig3.jpg)