Abstract
Occurrence of the hsp70 (dnaK) gene was investigated in various members of the domain Archaea comprising both euryarchaeotes and crenarchaeotes and in the hyperthermophilic bacteria Aquifex pyrophilus and Thermotoga maritima representing the deepest offshoots in phylogenetic trees of bacterial 16S rRNA sequences. The gene was not detected in 8 of 10 archaea examined but was found in A. pyrophilus and T. maritima, from which it was cloned and sequenced. Comparative analyses of the HSP70 amino acid sequences encoded in these genes, and others in the databases, showed that (i) in accordance with the vicinities seen in rRNA-based trees, the proteins from A. pyrophilus and T. maritima form a thermophilic cluster with that from the green nonsulfur bacterium Thermomicrobium roseum and are unrelated to their counterparts from gram-positive bacteria, proteobacteria/mitochondria, chlamydiae/spirochetes, deinococci, and cyanobacteria/chloroplasts; (ii) the T. maritima HSP70 clusters with the homologues from the archaea Methanobacterium thermoautotrophicum and Thermoplasma acidophilum, in contrast to the postulated unique kinship between archaea and gram-positive bacteria; and (iii) there are exceptions to the reported association between an insert in HSP70 and gram negativity, or vice versa, absence of insert and gram positivity. Notably, the HSP70 from T. maritima lacks the insert, although T. maritima is phylogenetically unrelated to the gram-positive bacteria. These results, along with the absence of hsp70 (dnaK) in various archaea and its presence in others, suggest that (i) different taxa retained either one or the other of two hsp70 (dnaK) versions (with or without insert), regardless of phylogenetic position; and (ii) archaea are aboriginally devoid of hsp70 (dnaK), and those that have it must have received it from phylogenetically diverse bacteria via lateral gene transfer events that did not involve replacement of an endogenous hsp70 (dnaK) gene.
The 70-kDa heat-shock protein (HSP70) is a member of a set of proteins (referred to as HSPs) which undergo increased synthesis in response to a variety of physical and chemical stresses (34). Originally identified as inducible proteins, certain HSP70s are constitutively expressed and appear to be essential for physiological cell growth (13, 26). HSP70 has been found in all members of the domains Bacteria and Eucarya investigated until now and in some members of the domain Archaea (20, 22, 37). Certain bacteria (Plantomycetales, Verrucomicrobiales, and Synechococcus spp.; Escherichia coli) contain more than one gene for HSP70 (39, 45, 52), and eucaryotic genomes encode multiple HSP70 versions that are localized to the various cell compartments (cytosol, endoplasmic reticulum, mitochondria, and chloroplasts) (13). In accordance with the proposed bacterial origins of mitochondria and chloroplasts, the (nucleus-encoded) HSP70s of cell organelles are most similar in sequence to homologues from members of the class Proteobacteria and cyanobacteria, respectively (6).
Intriguingly, HSP70-based phylogenies (20–25) contradict both the three-domain division of living organisms inferred from analysis of small-subunit rRNAs (53, 54) and the sisterhood of Archaea and Eucarya evidenced by reciprocally rooted trees of primordially duplicated genes (7, 17, 19, 31, 35). Unlike phylogenies of small-subunit rRNA sequences, the HSP70-based phylogenies predict a close and specific relationship between the archaea and gram-positive bacteria on the one hand and between the eucarya and gram-negative bacteria on the other, rather than between eucarya and archaea (22). These relationships are supported, among other arguments, by the finding of a relatively conserved insert occurring in the same position in all of the HSP70s from gram-negative bacteria and eucarya but absent in all of the homologues from archaea and gram-positive bacteria (21, 22).
The mutual affinities between eucarya, gram-negative bacteria, archaea, and gram-positive bacteria have been taken as evidence that (i) archaea and gram-positive bacteria constitute the two primary (albeit paraphyletic) lines of cellular descent, (ii) gram-negative bacteria are a late offshoot of the primitive gram-positive line, and (iii) the eucaryotic genome arose by chimerism through a unique endosymbiotic event involving the engulfment of an archaeon by a gram-negative bacterium (22).
To study the evolution of the hsp70 (dnaK) gene family with a sample of molecules more representative than that used in earlier works, we examined the sequences currently available in the databases and added two new hyperthermophilic bacteria representing the Aquificales and the Thermotogales. These are considered to be the deepest divergences in the bacterial 16S rRNA phylogenetic tree (3, 10). In addition, we sought the occurrence of the hsp70 (dnaK) gene in various archaea representing the euryarchaeotes and crenarchaeotes.
Here we report (i) the deduced amino acid sequences of the HSP70s from Aquifex pyrophilus and Thermotoga maritima and (ii) results of comparative analyses of these sequences and their homologues in the databases. Based on these results, and on the findings of the distribution of the hsp70 (dnaK) gene among the archaea investigated, we propose an explanation for the anomalous HSP70 phylogenies, which differs from others that are also based on HSP70 sequence comparisons.
MATERIALS AND METHODS
Bacterial and archaeal strains and plasmids.
The bacteria A. pyrophilus (DSM 6858) and T. maritima (DSM 3109) were gifts from K. O. Stetter (Lehrstuhl für Mikrobiologie, Regensburg, Germany). The archaea Desulfurococcus mobilis (DSM 2161), Pyrococcus woesei (DSM 3773), Thermoplasma acidophilum (DSM 1728), and Thermoproteus tenax (DSM 2078) were obtained from W. Zillig (Max Planck Institut für Biochemie, Martinsried, Germany); Methanopyrus kandleri (DSM 6324), Methanothermus fervidus (DSM 2088), and DNA from Archaeoglobus fulgidus (DSM 4139) were gifts from R. Huber (Lehrstuhl für Mikrobiologie, Regensburg, Germany); Methanococcus vannielii was a gift from A. Böck (Lehrstuhl für Mikrobiologie, Munich, Germany); Halobacterium halobium DNA was a gift from F. Pfeiffer (Max Planck Institut für Biochemie, Martinsried, Germany). Sulfolobus solfataricus (formerly Caldariella acidophila MT4) was grown in our laboratories. The Methanosarcina mazei S-6 hsp70 (dnaK) gene (37) was subcloned for this work in a pBluescript-based vector from another construct with a larger insert (2,195 bp) that also contained the other stress genes in the locus (33). PCR products were cloned in pMosBlue vector (Amersham) according to the manufacturer’s instructions. pBluescript SKM13 (Stratagene) was used as the vector, and E. coli TB1 (New England Biolabs) was used as the host. Plasmid-containing strains were grown in LB medium supplemented with ampicillin (75 μg/ml).
DNA preparation, sequencing and digestion.
Genomic DNA was prepared as previously described (4). Isolation and purification of plasmid DNA, recovery of DNA fragments from low-melting-point agarose gels, transformation experiments, and Southern blottings were done according to standard protocols (43). Southern blottings and colony hybridizations using homologous probes were performed at 65°C in the absence of formamide. Southern blottings using heterologous DNA were performed at 37°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% sodium dodecyl sulfate with 25 to 30% formamide, using 7 μg of genomic DNA and 10 to 30 ng of a randomly labeled AccI fragment (1,200 bp) of M. mazei dnaK encompassing codons 169 to 570. All DNA probes were labeled by using [α-32P]dATP (specific activity, 6,000 Ci/mmol) and a random priming labeling kit from Boehringer Mannheim. DNA sequences were determined on both strands by the dideoxynucleotide chain termination method (44) using [35S]dATP (>1,000 Ci/mmol) and both universal and de novo-synthesized primers according to the protocol for the Sequenase sequencing kit (U.S. Biochemical Corp.).
The restriction enzymes used for optimal digestion of genomic DNAs from the species investigated were EcoRI/HindIII (T. maritima), XbaI (S. solfataricus), SacI (H. halobium), EcoRI (M. fervidus), HindIII/XbaI (M. kandleri), and HindIII (D. mobilis, P. woesei, A. pyrophilus, T. tenax, A. fulgidus, and M. vannielii).
PCR-mediated DNA amplification.
dnaK was amplified by PCR in a Perkin-Elmer apparatus according to standard procedures in 3.0 mM MgCl2. Two degenerate primers were used: 5′-CA(AG)GC(ACGT)AC(ACGT)AA(AG)GA(CT)GC(ACGT)GG-3′ (hspI), corresponding to the DNA segment encoding the conserved sequence QATKDAG (E. coli HSP70 residues 152 to 158); and 5′-GC(ACGT)AC(ACGT)GC(CT)TC(AG)TC(ACGT)GG(AG)TT-3′ (hspII), complementary to the DNA segment encoding the conserved sequence NPDEAVA (E. coli HSP70 residues 366 to 372).
Database searches and sequence alignments.
The Genetic Computer Group program suite (15) of the UK MRC Human Genome Mapping Project Resource Centre (Cambridge University, Cambridge, United Kingdom) was used for retrieval of HSP70-related sequences. This was done by probing the DNA and protein databases with the tBLASTn and FASTAp options of the BLAST (1) and FASTA (41) programs. To minimize alignment errors, a preliminary alignment of the full-length HSP70 sequences was generated by CLUSTAL W (49), using default gap penalties. The CLUSTAL W alignment was then locally refined by using the segment-to-segment comparison method implemented in the program DIALIGN (38) with the BLOSUM similarity matrix (27).
Tree-making methods.
Phylogenetic trees were constructed by using maximum-parsimony (MP), evolutionary distance (ED), and maximum-likelihood methods. The MP analyses used the program PROTPARS implemented in PHYLIP version 3.57c (16). The PHYLIP programs SEQBOOT, PROTPARS, and CONSENSE were used sequentially to generate an MP tree which was replicated in 100 bootstraps; on this basis bootstrap confidence levels (BCL) were determined. Evolutionary distances between all pairs of taxa were calculated with the Dayhoff option of the PHYLIP program PROTDIST, which estimates the number of expected amino acid replacements per position, using a substitution model based on the PAM 120 matrix. The resultant pairwise distances were then used to construct a least-squares tree with the program FITCH. The programs SEQBOOT, PROTDIST, FITCH, and CONSENSE were used sequentially to construct a consensus tree based on 100 bootstrap replications of the original alignment. For maximum-likelihood analyses, we used the program PUZZLE version 4.0 (47) with the Jones-Taylor-Thornton (JTT) substitution model and a gamma-distributed model of site-to-site rate variation using eight rate classes to approximate the continuous gamma distribution, as well as a gamma distribution parameter α estimated from the data set.
Nucleotide sequence accession numbers and alignment retrieval.
The A. pyrophilus and T. maritima dnaK sequences are deposited at the EMBL GenBank database with accession no. AJ005800 and AJ005129, respectively. The sequence alignment used in this analysis (file name hsp70.aln) is available at ftp.bce.med.uniroma1.it: dir/cammara.
RESULTS
Cloning of hsp70 (dnaK) homologues.
Fragments of A. pyrophilus and T. maritima dnaK genes were successfully amplified by PCR with two degenerate primers used in the past to clone dnaK genes from a number of bacteria and archaea (20, 52). The amplified DNA fragments, having the expected length of 650 nucleotides, were found to correspond to the sequence encoding residues 152 through 372 of E. coli HSP70 (see Materials and Methods). These fragments were used as probes for isolating genomic clones containing the A. pyrophilus and T. maritima dnaK genes, and dnaK was subcloned and sequenced. In contrast, when PCR was carried out with DNA from a variety of archaea comprising both crenarchaeota (D. mobilis, S. solfataricus, and T. tenax) and euryarchaeota (P. woesei, M. kandleri, M. fervidus, M. vannielii, A. fulgidus, T. acidophilum, and H. halobium), successful amplification of dnaK-related sequences was observed only with T. acidophilum and H. halobium DNA (results not shown). Evidence for dnaK among the archaea listed above was further sought by means of Southern blotting using a 1,200-bp AccI fragment of M. mazei dnaK encompassing codons 169 to 570 (see Materials and Methods). Again, the probe hybridized only with restriction fragments of T. acidophilum and H. halobium; these gave single hybridization bands upon digestion with HindIII (T. acidophilum) and SacI (H. halobium) (results not shown). Although the absence of a gene cannot be asserted with confidence on the basis of negative hybridization results, lack of dnaK in some of the archaea listed above is demonstrated unequivocally by the recent genome sequencing data (see Discussion).
Alignment and sequence comparisons of Aquifex and Thermotoga HSP70s.
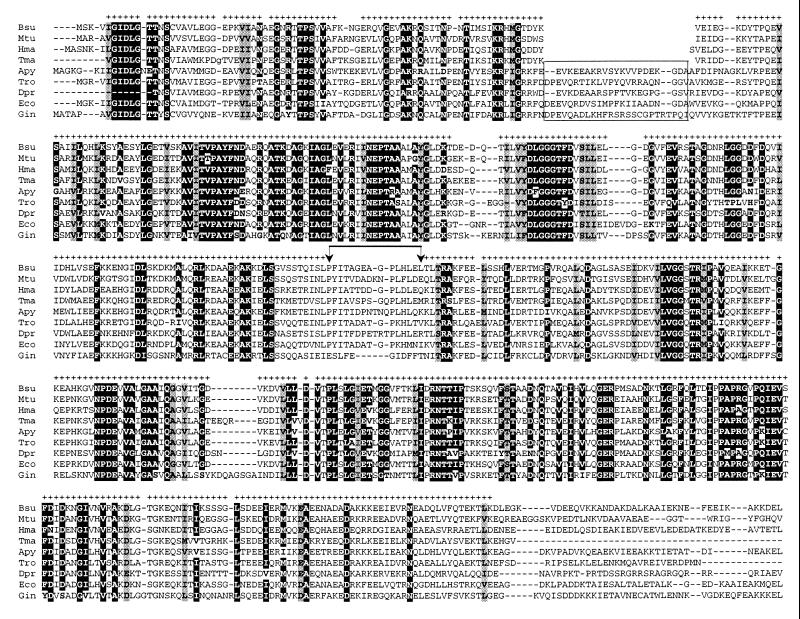
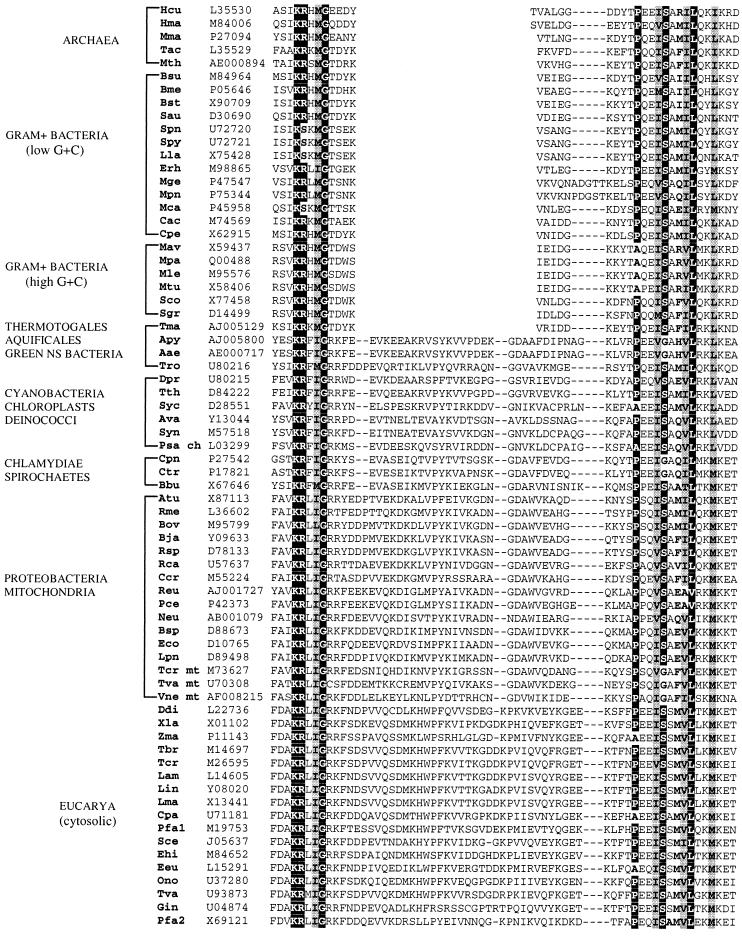
The deduced amino acid sequences of the A. pyrophilus and T. maritima HSP70s were aligned with most of the available homologues by using CLUSTAL W and the DIALIGN segment-to-segment comparison method. In addition to A. pyrophilus and T. maritima, the global alignment included 68 sequences. Of those, 17 were from gram-positive bacteria (of both the low- and high-G+C subdivisions), 5 were from archaea; 29 covered the genera Deinococcus and Thermus, the green nonsulfur bacteria, chlamydiae and spirochetes, α- β-, and γ subdivisions of the class Proteobacteria, mitochondria, cyanobacteria, and chloroplasts; and 17 sequences were eucaryotic cytosolic HSP70s representing a broad sampling of eucaryal diversity. The deduced sequence of the Aquifex aeolicus HSP70 (14) that became available after databank submission of the A. pyrophilus sequence was also used. The complete alignment of 70 HSP70 sequences is retrievable via anonymous ftp as described in Materials and Methods. A subset of the global alignment highlighting strongly conserved sequence motifs constraining the alignment topology is shown in Fig. 1. An excerpt of the alignment focusing on the insertion segment situated in the N-terminal portion of many HSP70 sequences (21, 22) is shown in Fig. 2.
FIG. 1.
Abridged alignment of the A. pyrophilus and T. maritima HSP70 sequences with homologues from the low-G+C gram-positive bacteria (Bsu), the high-G+C gram-positive bacteria (Mtu), the archaea (Hma), the green nonsulfur bacteria (Tro), the deinococci (Dpr), the proteobacteria (Eco), and the eucarya (Gin). Plus signs indicate amino acid positions selected for construction of the phylogenetic trees shown in Fig.s 3 to 5; highlighted blocks indicate positions occupied by an identical amino acid in more than 90% of the 70 aligned sequences; shaded areas indicate positions occupied by similar amino acids (ILVM, DEKRH, ST, GA, FYW, NQ) in 100% of the seventy sequences. For reasons of space, the C-terminal portion of the alignment (comprising residues not selected for tree reconstruction) is not shown. Species abbreviations are listed in the legend to Fig. 2. Vertical arrows delimit a sequence region that could not be confidently aligned between the procaryotic and eucaryotic HSP70s. The boxed region in the top block delimits the insertion segment found in many HSP70s.
FIG. 2.
Excerpt of the HSP70 sequence alignment focusing on the insertion segment which is found in many HSP70 sequences. Highlighting and shading have the same meaning as in Fig. 1. Species abbreviations: Aae (Aquifex aeolicus), Apy (Aquifex pyrophilus), Atu (Agrobacter tumefaciens), Ava (Anabaena variabilis), Bbu (Borrelia burgdorferi), Bja (Bradyrhizobium japonicum), Bme (Bacillus megaterium), Bov (Brucella ovis), Bsp (Buchnera sp.), Bst (Bacillus stearothermophilus), Bsu (Bacillus subtilis), Cac (Clostridium acetobutylicum), Ccr (Caulobacter crescentum), Cpa (Cryptosporidium parvum), Cpe (Clostridium perfringens), Cpn (Chlamydia pneumoniae), Ctr (Chlamydia trachomatis), Ddi (Dictyostelium discoideum), Dpr (Deinococcus proteolyticus), Eco (Escherichia coli), Eeu (Euplotes eurystomus), Ehi (Entamoeba histolytica), Erh (Erysipelothrix rhusiopathiae), Gin (Giardia intestinalis), Hcu (Halobacterium cutirubrum), Hma (Halobacterium marismortui), Lam (Leishmania amazonensis), Lin (Leishmania infantum), Lla (Lactococcus lactis), Lma (Leishmania major), Lpn (Legionella pneumophila), Mav (Mycobacterium avium), Mca (Mycoplasma capricolum), Mge (Mycoplasma genitalium), Mle (Mycobacterium leprae), Mma (Methanosarcina mazei), Mpa (Mycobacterium paratuberculosis), Mpn (Mycoplasma pneumoniae), Mth (Methanobacterium thermoautotrophicum), Mtu (Mycobacterium paratuberculosis), Neu (Nitrosomonas europaea), Ono (Oxytricha nova), Pce (Pseudomonas cepacia), Pfa1 and Pfa2 (Plasmodium falciparum), Psa ch (Pisum sativum, chloroplast), Rca (Rhodobacter capsulatus), Reu (Ralstonia eutropha), Rme (Rhizobium meliloti), Rsp (Rhodopseudomonas sp.), Sau (Staphylococcus aureus), Sce (Saccharomyces cerevisiae), Sco (Streptomyces coelicolor), Sgr (Streptomyces griseus), Spn (Streptococcus pneumoniae), Spy (Streptococcus pyogenes), Syc (Synechococcus sp.), Syn (Synechocystis sp.), Tac (Thermoplasma acidophilum), Tbr (Tripanosoma brucei), Tcr mt (Tripanosoma cruzi, mitochondrion), Tcr (Trypanosoma cruzi), Tma (Thermotoga maritima), Tro (Thermomicrobium roseum), Tth (Thermus thermophilus), Tva mt (Trichomonas vaginalis, mitochondrion), Tva (Trichomonas vaginalis), Vne mt (Vairimorpha necatrix, mitochondrion), Xla (Xenopus laevis), Zma (Zea mays). Databank accession numbers are shown to the right of species designations. NS, nonsulfur.
Consistent with previous reports (22, 37), the HSP70s from the gram-positive bacteria and the five archaea (T. acidophilum, Methanobacterium thermoautotrophicum, M. mazei, and two members of Halobacteriales) lacked the insertion segment seen in the HSP70s from the gram negative organisms. Surprisingly, the T. maritima HSP70, unlike that of the other gram-negative bacteria, including A. pyrophilus, lacked the distinctive insertion.
Phylogenetic analysis of HSP70 sequences.
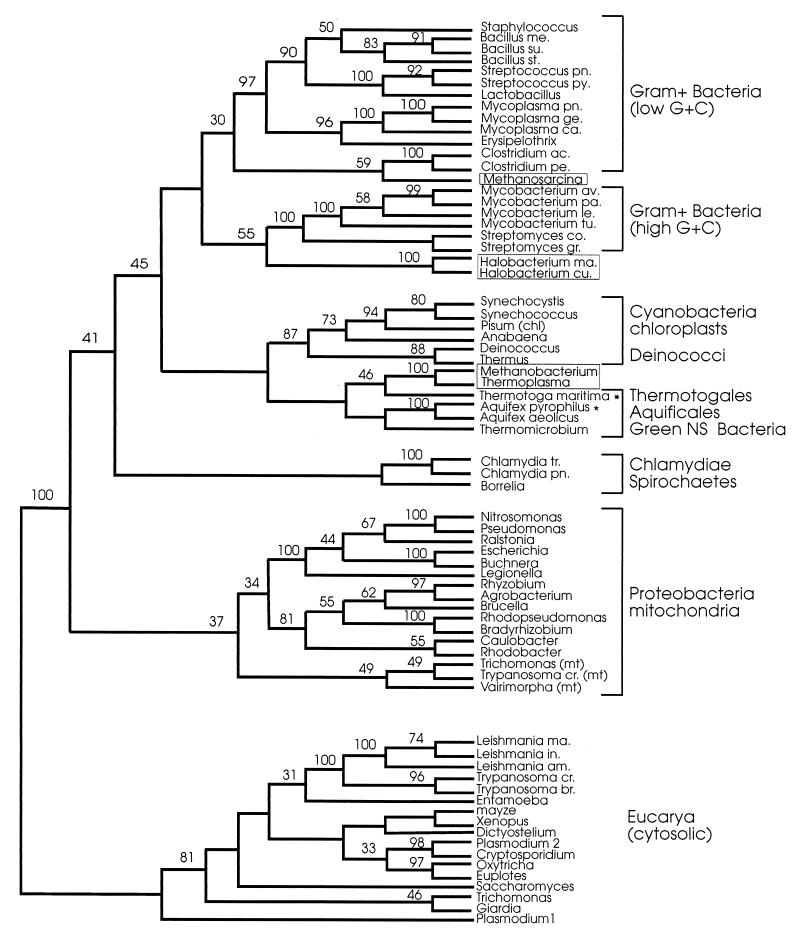
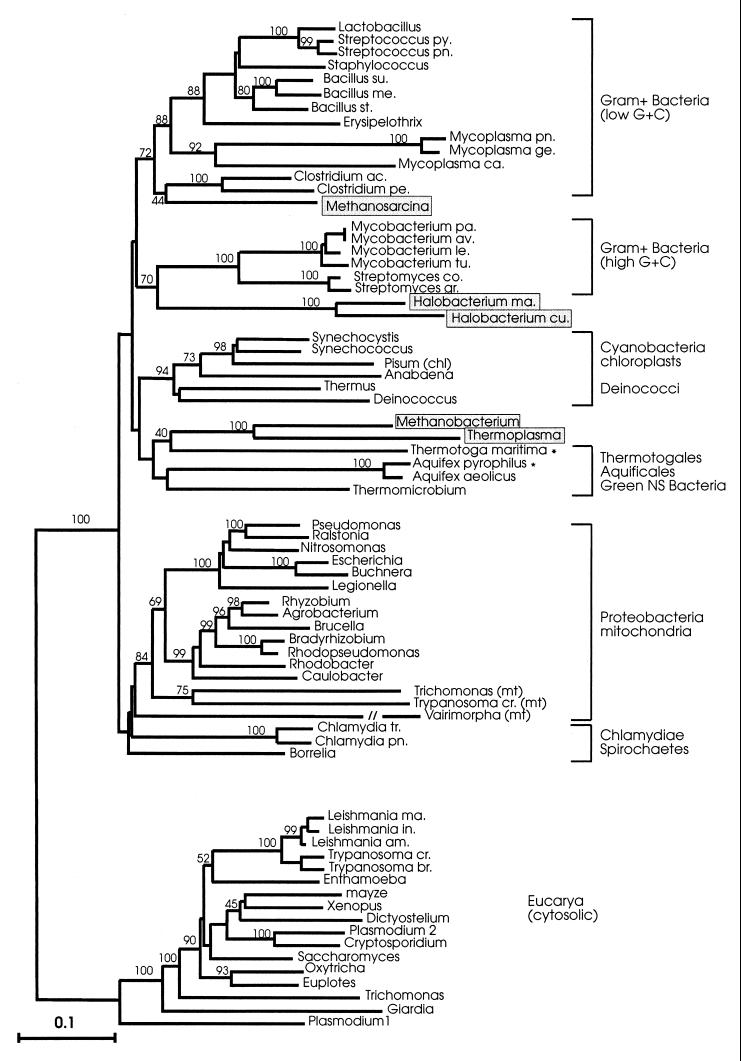
Figures 3 and 4 show MP and ED trees inferred from 492 amino acid positions that could be confidently aligned among all sequences; a short stretch of 14 to 15 residues that was not unambiguously alignable between the procaryotic and eucaryotic sequences was not included in the data set. The ED tree was constructed by the least-squares method using a matrix of evolutionary distances based on the Dayhoff PAM 120 amino acid replacement model. The MP tree was one of two equally parsimonious trees (6,438 steps) that differed in minor details of the branching order.
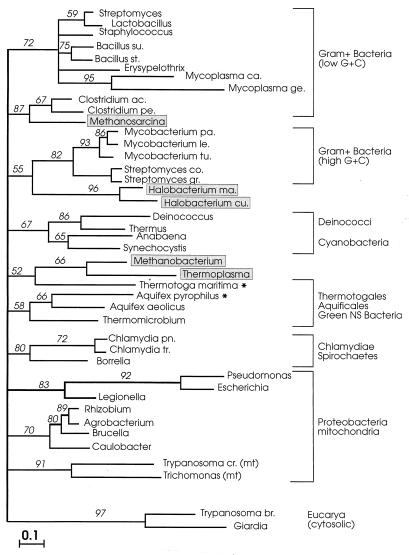
FIG. 3.
MP tree constructed with the program PROTPARS from the 492 amino acid positions marked by + in Fig. 1. Numbers on internal branches are BCL based on 100 bootstrap replications of the original alignment. Only BCL of >30% are shown. Branch lengths do not represent number of steps. Abbreviations: NS, nonsulfur; mt, mitochondria; chl, chloroplasts. Archaeal sequences are boxed; asterisks indicate the two new sequences reported here. For full organism names, see the legend to Fig. 2.
FIG. 4.
ED tree constructed with the program FITCH from the 492 amino acid positions marked by + in Fig. 1. ED matrices were calculated by the PAM 120 amino acid substitution matrix, using the Dayhoff option of the program PROTDIST. Numbers on internal branches are BCL based on 100 bootstrap replications. Only BCL of >50% are shown. The scale bar represents 0.1 amino acid substitution per site. For abbreviations and other details, see the legend to Fig. 3.
In both trees, the deepest divide separated the eucarya from a composite procaryotic cluster showing the archaea interspersed among (and associated with) different bacterial groupings. The topologies of the two trees were basically consistent in reproducing (i) the same clustering of the taxa (a monophyletic gram-positive bacterium clade, the proteobacteria, a chlamydia-spirochete grouping, a strongly supported cyanobacterium-Deinococcus clade); (ii) a similar internal branching order within the clusters with few minor discrepancies; and (iii) the same specific associations of mitochondria with the proteobacteria and of chloroplasts with the cyanobacteria. The specific relationships seen in previous reports (22) between M. mazei and the Clostridium group of low-G+C gram-positive bacteria, and between the Halobacterium cutirubrum/H. marismortui pair and the high G+C gram-positive bacteria, were also confirmed by the analyses. However, due to lack of robustness of the deepest nodes (<50% bootstrap support), the mutual relationships between the principal procaryotic clusters remain statistically indeterminate.
In both trees, the two novel (A. pyrophilus and T. maritima) sequences formed an independent, albeit tenuously supported, grouping of thermophilic organisms with the green nonsulfur bacterium Thermomicrobium roseum and the euryarchaeotes M. thermoautotrophicum and T. acidophilum. The two archaea were strongly related to one another (100% bootstrap support) and appeared to share a last common ancestor with T. maritima. It is noteworthy that the three thermophilic bacterial taxa (Aquifex, Thermotoga, and Thermomicrobium) that clustered together in the HSP70-based trees become resolved into three independent but adjacent lineages in phylogenetic trees of small-subunit rRNA sequences (3, 10). These show Aquificales, Thermotogales, and green nonsulfur bacteria, in that order, as three consecutive offshoots of the bacterial rRNA tree, situated immediately below the Deinococcus-cyanobacterium radiations.
The relationships between the archaeal and bacterial HSP70 sequences seen in Fig. 3 and 4 were further analyzed by quartet puzzling (QP), a maximum-likelihood algorithm that accounts for site-to-site rate variation and gives only fully resolved groupings. As expected from the lack of robustness of the ED and MP trees, the topology of the QP tree (Fig. 5) was largely star-like, indicating that the phylogenetic content of the HSP70 data set does not allow the resolution of the deepest relationships between the largest procaryotic groupings (48). Importantly, however, the tree-like component of the QP phylogeny recovered the association of T. maritima with the Methanobacterium-Thermoplasma pair (52% QP reliability) and confirmed the previously reported relationships between Methanosarcina and the clostridia (87% QP reliability) and between the halobacteria and the high-G+C gram-positive bacteria (55% QP reliability).
FIG. 5.
Maximum-likelihood tree of the HSP70 alignment (the 492 positions marked in Fig. 1). The QP method was used with a gamma-distributed model of site-to-site rate variation using eight rate categories. The gamma distribution parameter α estimated from the data set was 0.71 ± 0.04, and the log-likelihood of the tree was −19,510.8. Numbers above nodes are QP reliability values. The scale bar represents 0.1 amino acid substitution per site. For other details, see the legend to Fig. 3.
Evolutionary distances between HSP70 sequences.
Average maximum-likelihood distances between HSP70s from Archaea, Eucarya, and gram-positive and gram-negative bacterial taxa were calculated by the JTT amino acid substitution model and are shown in matrix form in Table 1. In accordance with results based on the Dayhoff amino acid substitution model (42; see Discussion), the average distances between the eucaryal sequences and the procaryotic homologues are significantly larger than those seen among procaryotes. This finding supports the argument (42) that the most likely rooting of the HSP70 tree lies between the procaryotic cluster and Eucarya, rather than between gram-positive bacteria and Archaea (24), or between a gram-positive/archaeal clade and a gram-negative/eucaryal clade (25).
TABLE 1.
Average evolutionary distances between eucaryotic and procaryotic HSP70 sequencesa
| Group | Pairwise maximum-likelihood distance
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Archaea | G+ | G−
|
Eucarya | ||||||
| P | D/Cy | C/S | Gns | Apy | Tma | ||||
| Archaea | 0.000 | 0.835 | 0.896 | 0.938 | 0.889 | 0.821 | 1.032 | 0.859 | 1.331 |
| G+ | 0.000 | 0.797 | 0.795 | 0.962 | 0.780 | 0.936 | 0.871 | 1.206 | |
| G− | |||||||||
| P | 0.000 | 0.828 | 0.636 | 0.712 | 0.874 | 0.957 | 1.119 | ||
| D/Cy | 0.000 | 0.789 | 0.795 | 0.853 | 0.924 | 1.189 | |||
| C/S | 0.000 | 0.666 | 0.845 | 0.974 | 1.166 | ||||
| Gns | 0.000 | 0.721 | 0.986 | 1.292 | |||||
| Apy | 0.000 | 1.115 | 1.304 | ||||||
| Tma | 0.000 | 1.254 | |||||||
| Eucarya | 0.000 | ||||||||
The pairwise maximum-likelihood distances (JTT amino acid replacement model) used for generating the tree of Fig. 6 were averaged between the taxa comprising the individual groups of organisms. The gram-negative (G−) bacterial group included all of the bacterial taxa shown in Fig. 6 except the low- and high-G+C gram-positive (G+) bacteria. P, proteobacteria; D/Cy, deinococci and cyanobacteria; C/S, chlamydiae and spirochetes; Gns, green nonsulfur bacteria; Apy, A. pyrophilus; Tma, T. maritima.
DISCUSSION
Compared to previous studies of HSP70 sequences, the present data set includes HSP70s from three deep-branching bacteria (A. pyrophilus and T. maritima [this report] and A. aeolicus [14]) and from the archaeon M. thermoautotrophicum (46). The results cast a new light on the evolutionary history of dnaK genes by calling into question such critical issues as (i) the orthology of the dnaK sequences; (ii) the paraphily of Archaea with respect to the gram-positive bacteria; and (iii) the ubiquity of dnaK in the evolutionary spectrum.
Lack of phylogenetic specificity of the HSP70 insertion segment.
According to Gupta and coworkers (18, 20–25), the discrete insertion seen in the N-terminal region of many HSP70 sequences (Fig. 2) is an evolutionary landmark distinguishing Gram-negative bacteria and eucarya (all of which possess the HSP70 insert) from gram-positive bacteria and archaea (which lack the HSP70 insert). Homologues lacking the insert (I−) were assumed to represent the ancestral form of the protein from which all the gram-negative bacterial homologues were derived (21), while eucarya would have obtained the insert-containing form of HSP70 (I+) through the bacterial partner of a postulated endosymbiotic event involving the fusion of a gram-negative bacterium and an archaeon (18, 22–25).
These conclusions are challenged by the evidence in this report that A. pyrophilus and T. maritima, two gram-negative bacteria (29, 30) representing adjacent offshoots in the 16S rRNA tree (3, 10), differ from one another in that A. pyrophilus possesses and T. maritima lacks the insertion segment.
The possibility that the genus Thermotoga is phylogenetically linked to the gram-positive bacteria (8, 12), or that it obtained its dnaK gene from a gram-positive bacterium, is unlikely. First, the HSP70-based phylogenies place Thermotoga outside any of the gram-positive bacterial clusters and close to Aquificales and green nonsulphur bacteria (Thermomicrobium). Second, a similar placement of Thermotoga is predicted by phylogenies of small subunit rRNA (3, 10, 51), elongation factors (EF) (2, 5, 36), and aminoacyl-tRNA synthetases (7). These phylogenies concur in placing the Thermotogales as the deepest or the second deepest grouping in the bacterial tree—somewhat deeper than the green nonsulfur bacteria and the deinococci, and definitely remote from the later-branching gram-positive bacteria and proteobacteria.
Such incongruity as exists between the placement of Thermotoga in trees of molecular sequences, and its linkage to the gram-positives argued from lack of the HSP70 insert, can be rationalized only by positing that (i) different groupings retained either one or the other of two paralogous versions of the dnaK gene or (ii) the insert is an ancestral trait lost more than once in bacterial evolution. Indeed, by taking as a reference the topologies of the small-subunit rRNA and EF trees (2, 3), the distribution of the two HSP70 forms throughout the bacterial phyla would require multiple insertion/deletion events as one moves from Aquificales (I+) to Thermotogales (I−), to green nonsulfur bacteria and deinococci (I+), to cyanobacteria (I+), to gram-positives bacteria (I−), and to proteobacteria (I+).
Clustering of archaea with gram-positive bacteria.
An additional question raised by the phylogenetic placement of the Thermotogales concerns the postulated paraphily of Archaea with respect to gram-positive bacteria (20–25). In contrast to the tenet that archaea are specifically related to (and arose polyphyletically within) the gram-positive bacteria, our results indicate a specific relationship between the archaea T. acidophilum and M. thermoautotrophicum and the bacterium T. maritima. Although the robustness of the ((Thermoplasma, Methanobacterium), Thermotoga) clade is not impressive (52% QP reliability), the association of these three thermophilic taxa is recovered by all the tree-making methods used. Inasmuch as Thermotoga is not affiliated with any of the gram-positive groupings, our observation renders less compelling the general argument of Gupta and coworkers (18, 22–25) that gram-positive bacteria and archaea constitute the two primary (albeit paraphyletic) lines of cellular descent, the gram-negative bacteria being a later offshoot of the primitive gram-positive line.
Absence of dnaK among the archaea.
Several considerations, based on the occurrence of hsp70 (dnaK) among the archaea (Table 2) and the topology of the prokaryotic cluster in the HSP70-based phylogenies (Fig. 3 to 5), support the notion that archaea do not harbor dnaK genes except for taxa that recruited a dnaK sequence from sympatric bacteria.
TABLE 2.
Occurrence of hsp70 (dnaK) among archaeaa
| Organism | PCR amplification,a Southern blottingb (reference)c | Genome sequencing (reference)c |
|---|---|---|
| D. mobilis | − (this report)a,b | ND |
| S. solfataricus | − (this report)a,b | ND |
| Sulfolobus sp. | − (33)b | ND |
| T. tenax | − (this report)a,b | ND |
| M. kandleri | − (this report)a,b | ND |
| P. woesei | − (this report)a,b | ND |
| P. horikoshi | ND | −d |
| M. fervidus | − (33b; this reporta,b) | ND |
| M. thermoautotrophicum ΔH | ND | + (46) |
| M. vannielii | − (this report)a,b | ND |
| M. voltae | − (28)b | ND |
| M. jannaschii | − (33)b | − (9) |
| A. fulgidus | ND | − (32) |
| T. acidophilum | + (22)a,e | ND |
| M. mazei S-6 | + (37)b,e | ND |
| M. thermophila TM-1 | + (33)b,e | ND |
| M. hungatei | − (33)b | ND |
| H. halobium | + (this report)a,b | ND |
| H. cutirubrum | + (22)a,e | ND |
| H. marismortui | + (20)a,e | ND |
PCR amplification using the two degenerate primers for the sequences QATKDAG and NPDEAVA (see Materials and Methods).
Southern blotting with M. mazei dnaK as the probe.
ND, not done; + and −, presence and absence, respectively, of dnaK-related sequences.
Unpublished sequence; retrievable at http://www.bio.nite.go.jp/.
Organism whose dnaK has been cloned and sequenced.
First, dnaK-related sequences are not detectable, by PCR amplification or Southern blotting, in the euryarchaeotes A. fulgidus, M. vannielii, P. woesei, M. kandleri, M. fervidus, Methanococcus jannaschii, Methanococcus voltae, Methanospirillum hungatei, and in the crenarchaeotes T. tenax, D. mobilis, S. solfataricus, and a Sulfolobus species. Secondly, in accordance with the DNA hybridization results, no dnaK homologues have been identified in the completely sequenced genomes of A. fulgidus, M. jannaschii and Pyrococcus horikoshi. Third, the HSP70 sequences from the archaea that possess a dnaK gene (T. acidophilum, M. thermoautotrophicum, M. mazei, H. marismortui, and H. cutirubrum) are distributed among, and clustered with, different bacterial groupings (Fig. 3 to 5) and are not distinguishable from the bacterial homologues by any unique signature. Last, by taking as a reference the 16S rRNA-based phylogeny (40), the dnaK distribution among the archaea is nonsystematic in character: dnaK genes are missing in the crenarcheotes and are haphazardly scattered throughout the major euryarchaeal phyla, regardless of ranking order and phylogenetic kinship. Also, this gene is not shared by members of sister taxa such as M. hungatei (Methanomicrobiales) and M. mazei (Methanosarcinales), or even by members of the same taxon such as M. fervidus and M. thermoautotrophicum (Methanobacteriales).
Taken together, the above observations strongly suggest that archaeal dnaK homologues, whenever they occur, were derived from bacterial donors through lateral gene transfer events. Horizontal transfer of dnaK between the two prokaryotic domains is in fact suggested by the clustering of HSP70 sequences from organisms (Thermotoga, M. thermoautotrophicum, and Thermoplasma) possibly thriving in similar (hot) environments. Also, horizontal transfer of protein-coding genes between the two prokaryotic domains has been reported repeatedly and is evidenced by the clustering of euryarchaeotes and gram-positive bacteria in phylogenetic trees of glutamine synthetase I sequences (8, 50).
A similar interpretation of the anomalous HSP70-based phylogenies had been offered by Roger and Brown (42) before the absence of hsp70 (dnaK) in some archaea became obvious from genome sequencing data. They argued, based on a comparison of evolutionary distances between the HSP70 sequences, that (i) the HSP70 tree should be rooted between eucarya and prokaryotes, rather than between gram-positive bacteria and archaea; and (ii) this rooting is compatible with the canonic Gogarten/Iwabe rooted tree of life (17, 31) if the anomalous placement of archaea is the result of a lateral transfer and replacement of their endogenous hsp70 (dnaK) genes by those from bacteria. The recognition that archaea are aboriginally devoid of dnaK simplifies the argument in that the transfer event does not involve a replacement of the homologous resident gene.
Two explanations can account for the lack of dnaK in archaea; one is based on the Iwabe/Gogarten tree of life, and the other relies on the fusion or chimeric model of cellular evolution advocated by Gupta and coworkers (18, 22–25) and Zillig et al. (55). In the former case, Archaea and Eucarya constitute sister domains sharing a last common ancestor corresponding to the archaeal-eucaryal branch of the universal tree (17, 31, 54). If an ancestral dnaK existed in the last common ancestor of extant organisms, the lack of a dnaK gene in Archaea, and its persistence in Eucarya, could be (most parsimoniously) explained only by positing a unique gene extinction event occurring in the archaeal branch of the tree, provided Archaea represents a monophyletic grouping (see references 2, 11, and 19 for a paraphyletic-Archaea option). The alternative possibilities that dnaK either arose in the bacterial branch, or became extinct in the archaeal-eucaryal branch, can be ruled out, as in both cases the eucaryotic cytosolic HSP70s could have been obtained only through duplication and divergence of the (nucleus-encoded) mitochondrial dnaK, and there is no evidence from the HSP70-based trees (reference 6 and this report) that the cytosolic HSP70s arose from within the proteobacterial/mitochondrial cluster.
The absence of dnaK in various archaea could also be simply explained in the context of the fusion or chimeric model by assuming that one of the two postulated primary lines either contained (Bacteria) or lacked (Archaea) a dnaK sequence. The HSP70 of the hypothetical eukaryotic chimera was thus contributed by the bacterial parent of the fusion, regardless of whether the bacterium (18, 22–25) or the archaeon (55) provided the engulfing partner. However, the fusion model of cellular evolution predicts that reciprocally rooted trees for primordially duplicated genes that are contributed by the bacterial parent of the chimera will show Bacteria and Eucarya, rather than Archaea and Eucarya, as the two sister domains. Given this premise, the fusion model is not a persuasive alternative until such a set of paralogous genes is identified and a sisterhood of Eucarya and Bacteria is convincingly demonstrated.
ACKNOWLEDGMENTS
We thank A. J. L. Macario for his input to this work and for many helpful suggestions during preparation of the manuscript.
This work was supported by grants from Ministero Università e Ricerca Scientifica e Tecnologica, Project Protein-Nucleic Acids Interactions, and from CNR Progetto Finalizzato Biotecnologie.
REFERENCES
- 1.Altschul S F, Gish G, Miller W, Myers E, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf S L, Palmer J D, Doolittle W F. The root of the universal tree and the origin of eucaryotes based on elongation factor phylogeny. Proc Natl Acad Sci USA. 1996;93:7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophily from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9183. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blin N, Stafford D W. A general method for the isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bocchetta M, Ceccarelli E, Creti R, Sanangelantoni A M, Tiboni O, Cammarano P. Arrangement and nucleotide sequence of the gene (fus) encoding elongation factor G (EF-G) from the hyperthermophilic bacterium Aquifex pyrophilus: phylogenetic depth of hyperthermophilic bacteria inferred from analysis of the EF-G/fus sequences. J Mol Evol. 1995;41:803–812. doi: 10.1007/BF00173160. [DOI] [PubMed] [Google Scholar]
- 6.Boorstein W R, Zeigelhoffer T, Craig E A. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 7.Brown J R, Doolittle W F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J R, Masuchi Y, Robb F T, Doolittle W F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38:566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 9.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschi. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 10.Burggraff S, Olsen G J, Stetter K O, Woese C R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- 11.Cammarano, P., R. Creti, A. M. Sanangelantoni, and P. Palm. Splitting the Archaea? A phylogeny of translational elongation factor G(2) sequences inferred from an optimized selection of alignment positions. J. Mol. Evol., in press. [DOI] [PubMed]
- 12.Cavalier-Smith T. Origins of secondary metabolism. Ciba Found Symp. 1992;171:64–87. doi: 10.1002/9780470514344.ch5. [DOI] [PubMed] [Google Scholar]
- 13.Craig E A, Kang P J, Boorstein W. A review of the role of 70k Da heat shock proteins in protein translocation across membranes. Antonie Leeuwenhoek. 1990;58:137–146. doi: 10.1007/BF00548924. [DOI] [PubMed] [Google Scholar]
- 14.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis program for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1993. . (Distributed by the author.) [Google Scholar]
- 17.Gogarten J P, Kibak H, Dittrich P, Taiz L, Bowman E, Bowman M, Manolson M F, Poole R J, Date T, Oshima T, Konishi T, Denda K, Yoshida M. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golding G B, Gupta R S. Protein based phylogenies support a chimeric origin for the eukaryotic genome. Mol Biol Evol. 1995;12:1–6. doi: 10.1093/oxfordjournals.molbev.a040178. [DOI] [PubMed] [Google Scholar]
- 19.Gribaldo S, Cammarano P. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targeting machinery. J Mol Evol. 1998;47:508–516. doi: 10.1007/pl00006407. [DOI] [PubMed] [Google Scholar]
- 20.Gupta R S, Singh B. Cloning of the HSP70 gene from Halobacterium marismortui: relatedness of archaebacterial HSP70 to its eubacterial homologs and a model for the evolution of the HSP70 gene. J Bacteriol. 1992;174:4594–4605. doi: 10.1128/jb.174.14.4594-4605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta R S, Golding G B. Evolution of HSP70 gene and its implications regarding relationships between Archaebacteria, Eubacteria and Eukaryotes. J Mol Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- 22.Gupta R S, Singh B. Phylogenetic analysis of 70 kDa heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol. 1994;4:1104–1114. doi: 10.1016/s0960-9822(00)00249-9. [DOI] [PubMed] [Google Scholar]
- 23.Gupta R S, Aitken K, Falah M, Singh B. Cloning of Giardia lamblia heat shock protein hsp70 homologs: implications regarding origin of eukaryotic cells and of endoplasmic reticulum. Proc Natl Acad Sci USA. 1994;91:2895–2899. doi: 10.1073/pnas.91.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta R S, Golding G B. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- 25.Gupta R S, Bustard K, Falah M, Singh D. Sequencing of heat shock protein 70 (DnaK) homologs from Deinococcus proteolyticus and Thermomicrobium roseum and their integration in a protein-based phylogeny of prokaryotes. J Bacteriol. 1997;179:345–357. doi: 10.1128/jb.179.2.345-357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbert M, Kropinski A M, Jarrel K F. Heat shock response of the archaebacterium Methanococcus voltae. J Bacteriol. 1991;173:3224–3227. doi: 10.1128/jb.173.10.3224-3227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber R, Langworthy T A, Koning H, Thomm M, Woese C R, Sleytr U B, Stetter K O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°C. Arch Microbiol. 1986;144:324–333. [Google Scholar]
- 30.Huber R, Wilharm T, Huber D, Trincone A, Burggraf S, Konig H, Rachel H, Rockinger I, Fricke H, Stetter K O. Aquifex pyrophilus gen. nov. sp. nov. represents a novel group of marine hyperthermophilic hydrogen oxidizing Bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 31.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of Archaea, Bacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klenk H-P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 33.Lange M, Macario A J L, Ahring B, Conway de Macario E. Heat shock response in Methanosarcina mazei S-6. Curr Microbiol. 1997;35:116–121. doi: 10.1007/s002849900222. [DOI] [PubMed] [Google Scholar]
- 34.Lindquist S, Craig E A. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 35.Lawson F S, Charlebois R L, Dillon J A R. Phylogenetic analysis of carbamoylphosphate synthetase genes: evolution involving multiple gene duplications, gene fusions, and insertions and deletions of surrounding sequences. Mol Biol Evol. 1996;13:970–977. doi: 10.1093/oxfordjournals.molbev.a025665. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig W, Weizenegger M, Betzl D, Leidel E, Lenz T, Ludvigsen A, Mollendorf D, Wenzig P, Schleifer K H. Complete nucleotide sequence of seven eubacterial genes coding for the elongation factor Tu: functional, structural and phylogenetic evaluations. Arch Microbiol. 1990;153:241–247. doi: 10.1007/BF00249075. [DOI] [PubMed] [Google Scholar]
- 37.Macario A J L, Dugan C B, Conway de Macario E. A dnaK homolog in the archaebacterium Methanosarcina mazei S-6. Gene. 1991;108:133–137. doi: 10.1016/0378-1119(91)90498-z. [DOI] [PubMed] [Google Scholar]
- 38.Morgenstern B, Dress A, Werner T. Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc Natl Acad Sci USA. 1996;93:12098–12103. doi: 10.1073/pnas.93.22.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nimura K, Yoshikawa H, Takahashi H. Identification of dnaK multigene family in Synechococcus sp. strain PCC 7942. Biochem Biophys Res Commun. 1994;201:466–471. doi: 10.1006/bbrc.1994.1724. [DOI] [PubMed] [Google Scholar]
- 40.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson W R, Lipman D J. Improved tools for biological sequence comparisons. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roger A J, Brown J R. A chimeric origin for eukaryotes re-examined. Trends Biol Sci. 1996;21:370–371. [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seaton B L, Vickery L E. A gene encoding a new Dnak/hsp70 homolog in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:2066–2070. doi: 10.1073/pnas.91.6.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang T Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniles C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strimmer K, Von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 48.Strimmer K, Von Haeseler A. Likelihood mapping: a simple method to visualize the phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiboni O, Cammarano P, Sanangelantoni A M. Cloning and sequencing of the gene encoding glutamine synthetase I from the Archaeum Pyrococcus woesei: anomalous phylogenies inferred from analysis of archaeal and bacterial glutamine synthetase I sequences. J Bacteriol. 1993;175:2961–2969. doi: 10.1128/jb.175.10.2961-2969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van De Peer Y, Neef J M, De Rijk P, De Vos P, De Wachter R. About the order of divergence of the major bacterial taxa during evolution. Syst Appl Microbiol. 1994;17:32–38. [Google Scholar]
- 52.Ward-Rayney N, Rainey F A, Stackebrandt E. The presence of a dnaK (HSP70) multigene family in members of the orders Planctomycetales and Verrucomicrobiales. J Bacteriol. 1997;179:6360–6366. doi: 10.1128/jb.179.20.6360-6366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woese C R, Fox G E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;51:221–271. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zillig W, Palm P, Klenk H-P, Langer D, Hudepohl U, Hain J, Lanzendorfer M, Holz I. Transcription in Archea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of Archaea (Archaebacteria). Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1993. pp. 367–386. [Google Scholar]