Abstract
The temperate mycobacteriophage L5 integrates site specifically into the genomes of Mycobacterium smegmatis, Mycobacterium tuberculosis, and Mycobacterium bovis bacillus Calmette-Guérin. This integrative recombination event occurs between the phage L5 attP site and the mycobacterial attB site and requires the phage-encoded integrase and mycobacterial-encoded integration host factor mIHF. Here we show that attP, Int-L5, and mIHF assemble into a recombinationally active complex, the intasome, which is capable of attB capture and formation of products. The arm-type integrase binding sites within attP play specialized roles in the formation of specific protein-DNA architectures; the intasome is constructed by the formation of intramolecular integrase bridges between one pair of sites, P4-P5, and the attP core, while an additional pair of sites, P1-P2, is required for interaction with attB.
Establishment of lysogeny by temperate bacteriophages involves site-specific recombination between a phage attachment site (attP) and an attachment site in the bacterial chromosome (attB) (9, 24). Typically, the attP and attB sites have a short segment of DNA sequence identity (the common core) within which strand exchange occurs and which is also part of the attachment junctions, attL and attR, that flank the integrated prophage. Prophage excision involves a second site-specific recombination event between attL and attR to yield attP and attB as products (9, 24). While the mechanism of strand exchange is the same for integrative and excisive recombination, the directionality of these events must be carefully controlled to be in concert with other aspects of the phage life cycle (14).
Site-specific recombinases can be grouped into two main classes on the basis of amino acid similarities, the tyrosine recombinases (including most of the phage integrases) and the serine recombinases (including the resolvases-DNA invertases) (1, 29). Most, but not all, bacteriophages utilize a member of this first group of proteins to catalyze integration and excision (8, 15). While both types of site-specific recombination reactions are utilized across a broad range of biological systems, they must frequently satisfy the demands of directionality, substrate choice, and timing (2, 14). The serine recombinase family generally regulates these events through topological specificity of the DNA substrates (5), while most phage systems use complex DNA sites and additional proteins to control the reactions (9). The complexity of the DNA substrates involved in integrase-mediated recombination reflects the requirement for the assembly of specific protein-DNA architectures within which strand exchange can occur (10, 14).
Mycobacteriophage L5 is a temperate phage that infects Mycobacterium smegmatis, Mycobacterium tuberculosis, and Mycobacterium bovis bacillus Calmette-Guérin (BCG) and forms stable lysogens in which the L5 genome is integrated site specifically into the mycobacterial chromosome (4, 6, 12, 27). Recombination occurs within a 43-bp sequence common to attP and attB and is catalyzed by the phage-encoded integrase Int-L5 (11). The region of L5 phage DNA containing attP includes multiple binding sites for Int-L5 that span a 413-bp segment (19). These integrase binding sites fall into two categories of sequence identity: core-type sites, which overlap the sites of strand exchange within the common core, and arm-type sites (P1-P7), which flank the core (Fig. 1). However, a 246-bp segment is fully active for integrative recombination and the P3 and P6-P7 sites are dispensable (19). While the attP site of the well-studied bacteriophage λ also contains arm- and core-type Int binding sites, the specific locations, arrangements, and orientations of the arm-type sites relative to those of the core are quite different (23).
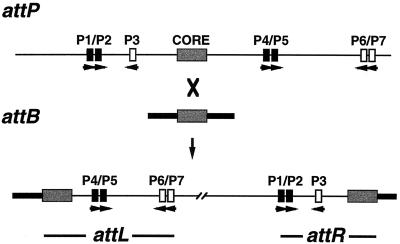
FIG. 1.
Attachment sites in L5 integrative recombination. The arrangements of arm-type (black and white boxes) and core-type (shaded boxes) integrase binding sites in L5 attP and M. smegmatis attB and in the resulting attachment sites attL and attR are shown. The relative orientations of the arm-type sites are indicated by arrowheads. The P1-P2 (spanning coordinates −115 to −135, where the central base pair of the overlap region between the sites of strand exchange is defined as 0) and P4-P5 (coordinates +90 to +110) pairs of sites are required for integrative recombination (black boxes), whereas P3 and the P6-P7 pair are dispensable (white boxes).
The L5 integrase protein is a member of the family of prokaryotic tyrosine recombinases and contains the four catalytic residues conserved throughout this group (11, 15). Although a distant relative of Int-λ, Int-L5 has a similar overall organization, consisting of a small N-terminal domain (residues 1 to 58) which binds sequence specifically to the arm-type sites and a larger C-terminal domain (residues 59 to 371) which binds to the core-type sites and contains the catalytic residues (19, 25). The two domains of Int-L5 can bind to attP DNA simultaneously, producing large complexes that fail to enter a nondenaturing polyacrylamide gel (17). While Int-L5 stimulates recombination in vitro, it requires participation of the mycobacterial integration host factor (mIHF) (17). In contrast to the Escherichia coli integration host factor (IHF) utilized in λ integration (13), the novel mIHF protein does not by itself bind with any preference for attP DNA sequences (17). However, when attP DNA is incubated in the presence of both Int-L5 and mIHF, a tertiary complex (the intasome) is formed which has a well-defined mobility in nondenaturing polyacrylamide gels (17).
It has previously been shown that L5 integrative recombination is strongly stimulated in vitro when the attP substrate is supercoiled (11, 18). Since the intasome complex described above contains linear attP DNA, the question arises as to whether this complex is competent for recombination. Here we demonstrate that the L5 intasome is a noncovalently-associated recombination intermediate and is capable of interaction with attB and strand exchange. We also show that the arm-type sites play highly specialized roles in integration: the P4-P5 pair is required for assembly of the intasome, while the P1-P2 pair is required for association with the recombinational partner, attB.
MATERIALS AND METHODS
DNA fragments.
DNA fragments containing wild-type attP (including sites P1 through P7) were generated by cutting plasmid pMH94 (12) either with BamHI and SalI or with just BamHI to give 612- and 624-bp fragments, respectively, or by cutting plasmid pCP7 (19) with BamHI and EcoRI to give a 634-bp fragment. DNA fragments (all 624 bp) containing substituted arm-type sites were generated by cutting the following plasmids with BamHI: pCP30, pCP31, pCP32, pCP33, pCP34, and pCP35, which have multiple substitutions (at least 8 of 10 bp) in arm-type sites P1, P2, P3, P4, P5, and P4-P5, respectively (19). DNA fragments containing sites P1 to P5 (379 bp) or P3 to P5 (353 bp) were generated by cutting plasmid pCPΔL1 (19) with BamHI and XcmI and plasmid pCPΔL3 (19) with HindIII and XcmI, respectively. attB DNA fragments were generated by annealing pairs of oligonucleotides (to give 29- and 45-bp fragments) as described previously (20) or cut from plasmid pMH12.2 (a pUC119 derivative containing a 1.7-kb SalI attB fragment from M. smegmatis) (12) with AvaII and MseI to give a 126-bp fragment. DNA fragments were radiolabeled on both ends (unless otherwise indicated), as needed, either by phosphorylation or by end-fill with Klenow as described previously (19).
In vitro integrative recombination reactions.
Recombination assays were similar to those described previously (11). Reactions with supercoiled attP substrates were performed in a total volume of 20 μl and contained approximately 0.005 to 0.05 pmol (unless otherwise noted) of supercoiled plasmid containing attP, 0.06 pmol of attB, 0.07 to 0.23 pmol of purified Int-L5, and 3.6 to 12.0 pmol of purified mIHF. Reaction mixtures were incubated for 10 or 30 min (as indicated in Fig. 5C) at 37°C, the reactions were stopped by adding sodium dodecyl sulfate (SDS) to a final concentration of 0.1%, and the mixtures were electrophoresed through a 0.8% agarose gel.
FIG. 5.
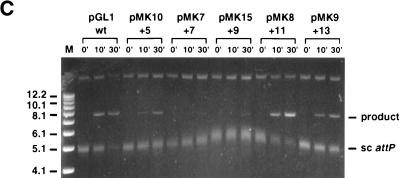
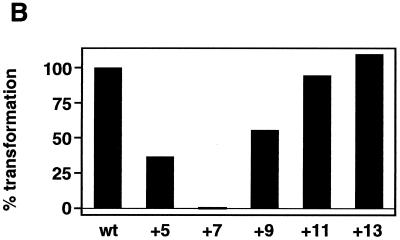
Disruption of recombination by insertion of nonintegral numbers of helical turns into the core–P4-P5 region of attP. (A) Location and sequences of insertion mutants. Insertions were made between bp +50 and bp +51 within attP (the central base pair within the 7-bp strand exchange overlap is designated 0, and the P4-P5 sites span +90 through +110). (B) Integrative transformation assay with attP insertion mutants. Relative frequencies with which the various attP insertion mutant plasmids are able to transform M. smegmatis are shown. The transformation frequency of the wild-type attP plasmid pMH94 is defined as 100%. (C) In vitro recombination assay with attP insertion mutants. Reaction mixtures containing a supercoiled attP plasmid (with insertions as indicated), a short, linear attB DNA, Int, and mIHF were incubated at 37°C for 0, 10, or 30 min as indicated above each lane, stopped by the addition of SDS, and electrophoresed through a 0.8% agarose gel. The positions of the supercoiled attP plasmid (sc attP) and the linear recombination products (sizes ranging from 7,808 to 7,821 bp) are indicated. The slowly migrating band in each lane corresponds to relaxed plasmid DNA. The supercoiled attP band which appears to migrate more slowly in the central lanes is due to a gel artifact (note that Int-L5 does not display topoisomerase activity [18]). Size markers (M) are indicated in kilobases.
Integrative recombination assays with linear attP DNAs contained the same components as described above, with the exceptions that approximately 0.024 pmol of attP was provided as a short, linear DNA fragment, 1 μg of salmon sperm DNA was added to each reaction, and the total reaction volume was 10 μl. The attP DNA was preincubated with Int-L5 and mIHF for 15 to 30 min either at room temperature or on ice, attB was added, and the entire reaction mixture was incubated at room temperature for 1 to 2 h (unless otherwise noted). Reaction mixtures were electrophoresed through a 5% polyacrylamide gel in 1× TBE (100 mM Tris–84 mM borate–1 mM EDTA), and products were visualized by autoradiography. Where indicated, reaction mixtures were treated either by the addition of SDS (final concentration, 0.5%) or proteinase K (final concentration, 1 mg/ml, followed by a 10-min incubation at 55°C) or by heating at 80°C for 10 min.
Two-dimensional gel analysis.
Protein-DNA complexes produced by polyacrylamide gel electrophoresis were denatured by excising the desired lane from a wet gel and soaking it in 0.5% SDS for 10 min. The SDS-treated lane was then laid horizontally across the top of and electrophoresed through a 5% polyacrylamide–0.05% SDS gel in 1× TBE.
In situ DNase I footprinting.
In situ DNase I footprinting was performed as described previously (7). The complexes were formed in reactions identical to those used for linear attP-containing integrative recombination by using a 379-bp P1 to P5 attP DNA 3′ radiolabeled at the P1-proximal end, and mixtures were incubated for 15 min with a 45-bp attB DNA where appropriate. Complexes were separated by electrophoresis through a 1× TBE–5% polyacrylamide gel and visualized by autoradiography; gel slices containing individual complexes were excised and chopped into small pieces. To each footprinting reaction 30 μl of a solution containing 0.5-μg/ml DNase I in 10 mM Tris–0.5-mg/ml bovine serum albumin–2 mM dithiothreitol was added, and the reaction mixtures were incubated for 45 min. The cleavage reaction was started by the addition of 18 μl of 50 mM MgCl2–50 mM CaCl2, and the mixture was incubated for 4 min. The reaction was stopped by adding 30 μl of 0.5 M EDTA, incubating the mixture for 20 min, and then adding 30 μl of 1% SDS. All incubations were at room temperature. The digested DNA was electroeluted from the gel slices, ethanol precipitated, and electrophoresed through a 6 or 10% sequencing gel. Results were visualized by autoradiography and quantified by using NIH Image software (http://rsb.info.nih.gov/nih-image).
Construction of attP insertion mutants.
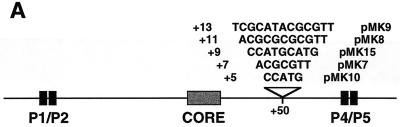
A set of mutants containing insertions in the attP region of the 7,763-bp plasmid pGL1 (19) was constructed as described previously (19) by using the Muta-Gene Phagemid In Vitro Mutagenesis system (Bio-Rad). The mutagenic oligonucleotides were designed to insert 5 bp (to make plasmid pMK10), 7 bp (to make pMK7), or 13 bp (to make pMK9) between attP positions +50 and +51 (between the core and P4), introducing the unique restriction site NcoI (in the +5 and +7 bp insertions) or MluI (in the +13 bp insertion). In order to generate further insertion mutants, plasmids pMK10 and pMK7 were digested with NcoI, 3′ filled to generate blunt ends, and religated, resulting in total insertions of +9 (plasmid pMK15) and +11 bp (pMK8), respectively.
Integrative transformation assays.
In vivo integrative transformation assays were performed as described previously (19). Approximately 0.1 μg (0.025 pmol) of attP-containing plasmid (which also contains L5 int and lacks an origin of replication for mycobacteria) was electroporated into M. smegmatis mc2155 (27, 28) and recovered at 37°C, dilutions were plated on 7H10/ADC plates containing 0.5 μg of tetracycline (attP mutant plasmids pMK7, pMK8, pMK9, pMK10, and pMK15)/ml or 20 μg of kanamycin (wild-type attP plasmid pMH94)/ml, and transformants were scored after a 4- to 5-day incubation at 37°C.
RESULTS
Recombinogenic potential of the L5 intasome.
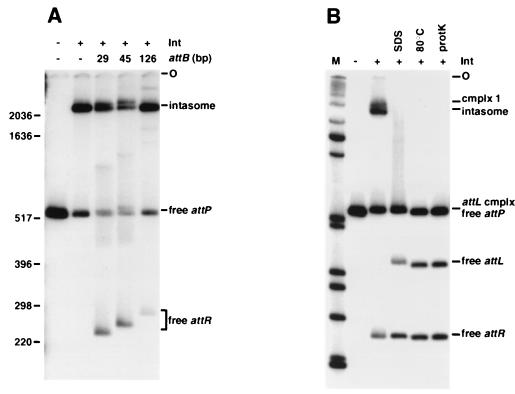
In order to test the recombinogenic potential of the intasome complex, intasomes were formed with radiolabeled attP DNA, Int-L5, and mIHF; attB DNA was added; and the products were analyzed by polyacrylamide gel electrophoresis (Fig. 2A). Use of a 29-bp attB DNA (the minimal functional attB) (20) resulted in the appearance of several new bands, one of the strongest of which had the mobility predicted for a free attR product. The identity of this band as the attR product was confirmed by using attB DNAs of different sizes (45 and 126 bp); as the size of attB increased, the mobility of the putative attR product changed as predicted. This experiment demonstrates that integrative recombination occurs under these conditions.
FIG. 2.
Intasome complexes are active in recombination. (A) Polyacrylamide gel analysis of the products of integrative recombination. Reaction mixtures containing a radiolabeled 612-bp fragment of attP DNA, purified Int-L5 (as indicated), and mIHF were incubated for 30 min, followed by addition of attB DNAs of different sizes (29, 45, or 126 bp, as indicated) and incubation at room temperature for 2 h. The mixtures were then loaded onto a polyacrylamide gel. The predicted sizes for the attR products produced with the 29-, 45-, and 126-bp attB DNAs are 232, 244, and 266 bp, respectively; those for the attL products are 409, 413, and 472 bp, respectively. Size markers are indicated in base pairs. O, origin of electrophoresis. (B) Dissociation of protein-DNA complexes. A radiolabeled 45-bp attB DNA was added to intasome complexes formed with a 624-bp radiolabeled attP DNA, Int-L5 (as indicated), and mIHF as described for panel A. Reactions were either left untreated or were treated with SDS, heat (80°C for 10 min), or proteinase K (protK) as indicated. The predicted sizes for the attR and attL products are 244 and 425 bp, respectively. The positions of free DNAs, the intasome, and complex 1 are shown. M, a radiolabeled 1-kb DNA marker with sizes in bp as follows (from bottom up): 201, 220, 298, 344, 396, 506, 517, 1018, 1636, 2036, and 3054. (C) Two-dimensional gel analysis of recombination products. Reaction products were separated by gel electrophoresis as shown for the untreated sample in panel B. A vertical slice of gel was excised, soaked in SDS, and applied to a polyacrylamide gel containing SDS for the second dimension. The positions of DNAs and complexes in the first dimension are indicated along the top, and the positions of DNAs in the second dimension are indicated on the left. Radiolabeled 1-kb marker fragments are shown at the left. (D) Time course of integrative recombination. Intasomes were formed as described for panel A by using a 634-bp radiolabeled attP DNA. A 126-bp attB DNA was added for different amounts of time (as indicated at top), and products were separated by electrophoresis. Size markers are indicated in base pairs.
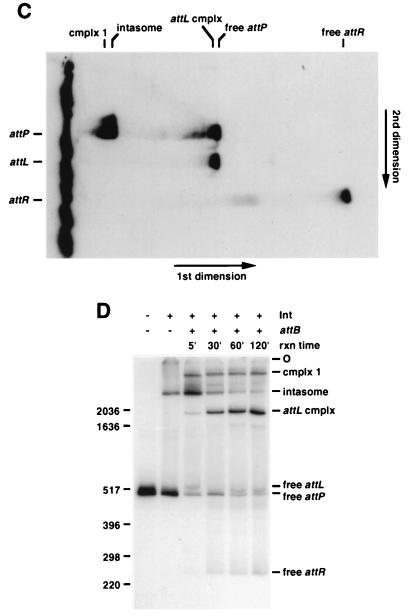
While the predicted attR product DNAs are readily identifiable in this experiment, the corresponding attL DNA fragments are not seen. A simple explanation for this is that the majority of attL DNA remains associated with proteins after strand exchange has occurred. This was confirmed by SDS denaturation of the protein-DNA complexes, which yielded equivalent amounts of attL and attR DNA (Fig. 2B). Since no DNA fragments other than attP, attL, and attR are detected following these treatments, none of the reaction products are likely to be either covalent protein-DNA associations or covalent intermediates in the process of strand exchange (although we cannot rule out the possibility that all treatments could have promoted rapid reversal of any reaction intermediates). By subjecting a sample equivalent to the untreated reaction as shown in Fig. 2B, lane 3, to two-dimensional gel electrophoresis, we determined that the attL protein-DNA complex migrates in a position similar to that of the free attP DNA fragment (Fig. 2C). In addition, small amounts of attR and attP DNAs are seen to dissociate from weak smears and bands elsewhere in the lane (see also Fig. 3A). This experiment also demonstrates that the complex migrating slightly slower than the intasome (complex 1) contains attP DNA but not attL or attR DNA.
FIG. 3.
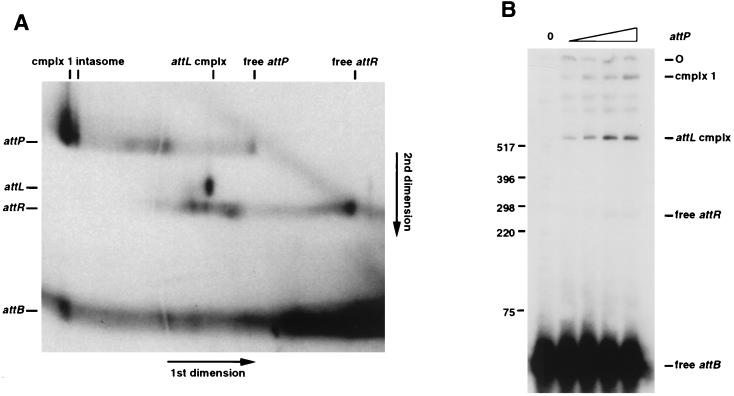
attB is a component of complex 1. (A) Products of recombination were separated by two-dimensional gel electrophoresis as described for Fig. 2C. Both the 624-bp attP and the 45-bp attB DNAs were radiolabeled. The positions of the DNAs and complexes as they ran in the first dimension are indicated along the top. The positions as they ran in the second dimension are indicated on the left. (B) Gel analysis of recombination products formed with a radiolabeled 45-bp attB DNA and increasing amounts (0, 0.012, 0.024, 0.048, and 0.073 pmol) of nonradiolabeled 624-bp attP DNA, Int-L5, and mIHF. At least one of the bands migrating between complex 1 and the attL complex is an attR complex. Size markers are indicated in base pairs.
Incubation of intasomes with attB DNA for various times shows the progression of the integration reaction (Fig. 2D). At early times (5 min after addition of attB DNA) there is an increase in the amount of the intasome band (and the slower-moving complex 1 is clearly visible) even though only small amounts of recombinant products are generated (it is not clear why the increase in the intasome band is seen at early times, although other experiments suggest that this intasome band is not qualitatively different from that seen in the absence of attB; see below). After further incubation, the intasome band diminishes and the amount of product increases. The course of this reaction is consistent with the intasome being a component on the pathway to recombination rather than being an inactive by-product. Quantification of the data shown in Fig. 2D supports this, since the rate of substrate decrease (intasome and free attP DNA) is the same as the rate of product formation (the sum of attL and attR DNAs and their complexes; note that both ends of attP DNA are labeled but that one end, the one that gives rise to attL, has twice as much 32P as the other end). Furthermore, when a gel lane containing products of a reaction with attP, Int, and mIHF (i.e., without attB) was excised, soaked in a solution containing attB DNA and electrophoresed in a second dimension, only the part of the gel containing intasomes gave rise to reaction products, albeit weakly (data not shown). Taken together, these experiments suggest that the intasome complex observed is indeed a noncovalently associated recombinational intermediate, even though the reaction is somewhat slower and less efficient than with supercoiled substrates.
Characterization of an attB-containing complex.
Addition of attB DNA to intasomes promotes the formation of a complex (complex 1) that migrates slightly slower than the intasome (Fig. 2). The mobility of this complex changes with the use of differently sized attB DNAs (Fig. 2A). Since this complex does not contain recombinant products or covalent intermediates (Fig. 2C), complex 1 could represent a quaternary complex containing both attB and attP DNAs (in addition to Int and mIHF). The presence of attB DNA in this complex was confirmed by radiolabeling both DNAs and examining the reaction products by two-dimensional gel electrophoresis (Fig. 3A). Additional support for the presence of attB in this complex and for identification of the recombinant products is provided by analysis of recombination reactions using radiolabeled attB and a nonradiolabeled attP DNA (Fig. 3B). Labeled products migrate in the positions of complex 1, the attL complex, and free attR DNA. These data confirm that complex 1 contains both attP and attB and show that its components are associated through noncovalent interactions. However, this quaternary complex does not behave kinetically as an intermediate in recombination, since it neither significantly accumulates nor decays during the course of the reaction (Fig. 2D).
Arm-type Int binding sites involved in complex formation.
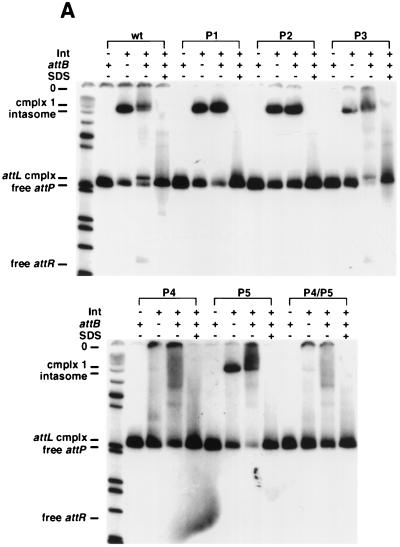
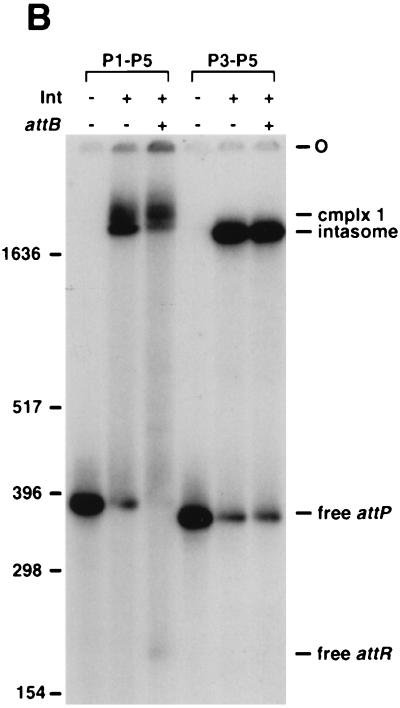
The role of the arm-type Int binding sites in formation of the intasome and complex 1 was investigated by using a series of attP mutants containing multiple substitutions in individual arm-type sites (19). Interestingly, while both the P1 and the P2 mutants fail to undergo recombination, both are fully competent for intasome formation (Fig. 4A); the mobilities of the resulting intasomes are the same as with wild-type attP DNA, suggesting they have the same stoichiometry (Fig. 4A). In fact, both P1 and P2 can be completely removed without affecting intasome assembly (Fig. 4B). Not only are the P1-P2 sites dispensable for intasome formation, but in situ DNase I footprinting of the intasome demonstrates that they are unoccupied in this complex (Fig. 4C). The P4 site clearly is required for assembly of the intasome, and while the P5 mutant forms an intasome (Fig. 4A), it does not appear to be recombinogenic. The three arm-type sites P3, P6, and P7, which were previously shown to be dispensable for integration both in vivo and in vitro (using supercoiled DNA substrates) (19), are not required for the formation of any of the complexes or products shown in Fig. 4A and B.
FIG. 4.
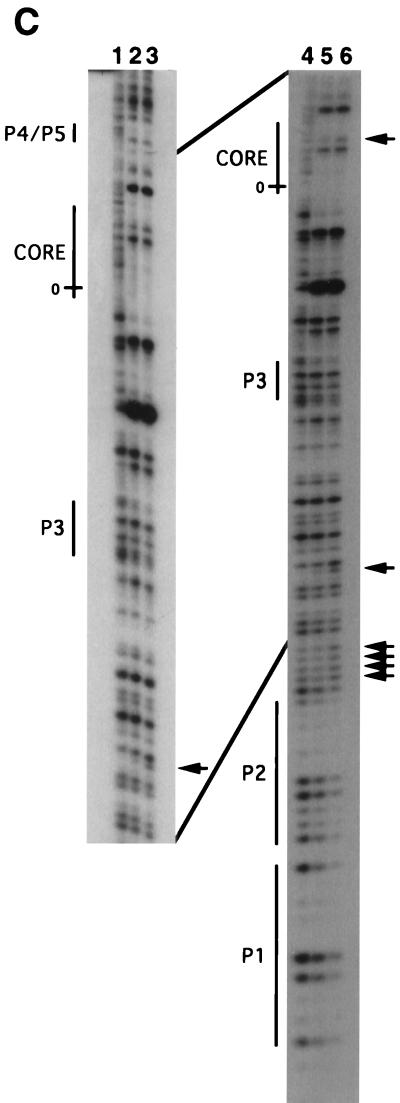
Int binding site requirements for intasome and quaternary complex formation. (A) Formation of protein-DNA complexes with mutant attP DNAs. Reactions were performed as described for Fig. 2A, adding Int-L5 (as indicated), mIHF, a 45-bp attB DNA (as indicated), and attP DNAs (either wild type [wt] or with multiple substitutions in one or more arm-type sites, as indicated), followed by the addition of SDS (as indicated). Since we have shown previously that P3 is dispensable for recombination, it is not surprising that the P3 mutant substrate behaves like wild-type attP in these assays. (B) Formation of the intasome with a P1-P2 deletion in attP. Reactions were performed as described for Fig. 2A with Int-L5 (as indicated), mIHF, an attP DNA (radiolabeled at the left end only) containing either P1 to P5 (a 379-bp fragment) or P3 to P5 (a 353-bp fragment), and a 45-bp attB, as indicated. Size markers are indicated in base pairs. (C) In situ DNase I footprinting assay of attP DNA present in the intasome and complex 1. The intasome and complex 1 were separated by electrophoresis, excised, and treated with DNase I in situ. Use of a relatively small attP fragment provided sufficient separation between the complexes to allow excision with minimal cross-contamination. Lanes 1 and 4, free attP (also excised from the gel); lanes 2 and 5, the intasome; lanes 3 and 6; complex 1. Lanes 1 to 3, electrophoresis through a 10% sequencing gel; lanes 4 to 6, electrophoresis through a 6% sequencing gel. The positions of arm-type and core-type Int binding sites within attP are indicated. 0, the central base pair of the strand exchange overlap region. Lanes were scanned and the intensities of individual bands were measured by using NIH Image software. Enhancements specific to complex 1 are indicated by arrowheads.
While the P1-P2 sites are not involved in intasome formation, they are required for the assembly of complex 1 as shown with radiolabeled attP DNA (Fig. 4A and B) or with radiolabeled attB DNA (data not shown). Moreover, in situ DNase I footprinting of complex 1 shows at least partial protection of P1 and P2 (Fig. 4C), with 50% inhibition of DNase I cutting in this region (as determined by quantification of the lanes in Fig. 4C). It is unclear why less-than-full protection of these sites is seen, since both P1 and P2 are needed for formation of complex 1 (Fig. 4A and B) and the core and P4-P5 sites are fully protected (Fig. 4C). It is possible that this particular interaction is uniquely sensitive to the DNase I cleavage conditions or that simultaneous binding of Int to P1 and P2 is unfavorable in these reactions.
The P4-P5 sites, the core region, and the DNA segment between P4 and the core are occupied in both the intasome (consistent with previous observations [17]) and in complex 1 (Fig. 4C). In addition, both complexes show protection, and an accompanying enhancement of DNase I cleavage (which is the most prominent band in Fig. 4C), of the region immediately to the left of the core (see Fig. 1). This suggests that a protein, presumably mIHF, is bound there. Since mIHF does not bind specifically to attP, and Int is not bound to the left of the core in the intasome (Fig. 4C), mIHF probably binds there through direct interactions with the Int promoters bound at the core. Although the remainder of the region between P2 and the core is not protected in either the intasome or in complex 1, there are a number of DNase I enhancements in this region that are specific to complex 1 (Fig. 4C). These enhancements are spaced approximately 10 bp apart, suggesting that the DNA is bent in this region.
Recombination requires correct phasing of the core and P4-P5 sites.
Formation of the attP intasome requires only the Int binding sites at the core and at the P4-P5 pair of arm-type sites. The sequence of the DNA between the core and the P4 site may not be important for integration but the size and resulting phasing may be critical if the two types of sites participate in Int-mediated protein bridges. This was confirmed by the analysis of a series of attP mutant derivatives in which the spacing was changed between the core and the P4-P5 pair of sites. Insertions were made between positions +50 and +51 in a region that is approximately midway between the P4 site and the crossover point (Fig. 5A) and is poorly protected in DNase I footprinting experiments (17). Five plasmids were generated in which the core-P4 spacing was increased by 5, 7, 9, 11, or 13 bp.
Plasmids containing altered attP DNAs were tested as recombination substrates both in vivo and in vitro (Fig. 5B and C). In both assays, insertion of either 5 or 9 bp reduced recombination significantly but addition of 7 bp eliminated the recombinational potential completely. In contrast, insertion of either 11 or 13 bp had little or no effect on integration. Thus the actual spacing of the core and P4-P5 sites does not appear to be important provided that the appropriate phasing of the sites is maintained. Presumably, the core and P4-P5 sites must be appropriately phased for formation of Int-mediated intramolecular bridges between these sites.
DISCUSSION
Analysis of attP mutants lacking arm-type sites and footprinting analysis of the intasome strongly suggest that the P1-P2 pair of arm-type sites are neither required for nor occupied within the intasome structure. This was somewhat unexpected since the sites required for intasome formation, the core and P4-P5 arm-type sites, are the same as those present in the attachment junction attL. Thus, one of the substrate complexes, the attP intasome, appears to be identical to one of the product complexes, the attL intasome, which is a likely substrate for excision. Moreover, the attP intasome structure appears to be quite stable and remains essentially intact (as an attL complex) following recombination in vitro.
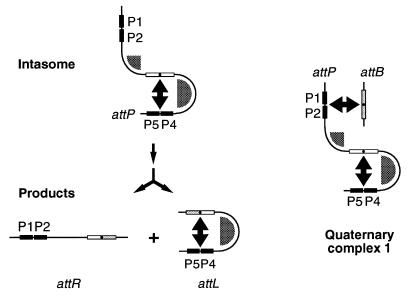
The intasome has two notable aspects to its structure. First, Int-L5 forms intramolecular bridges between the P4-P5 arm-type sites and the core, with mIHF stabilizing a sharp bend in the DNA between these sites (Fig. 6). Support for this structure is provided by the pattern of DNase I protection and the properties of mIHF (17). Further support for the formation of intramolecular protein bridges between the core and P4-P5 during the course of recombination is provided by the observed disruption of recombination upon the insertion of nonintegral numbers of helical turns between the core and P4-P5, while the insertion of approximately one helical turn restores recombination (Fig. 5).
FIG. 6.
Models for the intasome and quaternary complex structures. The figure shows schematic representations of the intasome, recombination products, and quaternary complex 1. Arm-type Int binding sites are represented as black boxes, and core-type sites are shown as white (attP) or shaded (attB) boxes with a black dot indicating bp 0 (central base pair with respect to the sites of strand exchange). Bivalent Int-L5 binding is represented by double-headed arrows, and mIHF binding areas are shaded.
Integrative recombination requires both the P4 and P5 arm-type sites, and DNase I footprinting of the intasome shows that both sites are occupied. However, the P4- and P5-mutant substrates behave differently in intasome formation; the P4 site is required for intasome formation, but the mutant lacking P5 is able to form both the intasome and complex 1 even though no recombination is observed. The difference in the ability to form intasomes could perhaps be accounted for by differences in the affinity of Int for the individual P4 and P5 arm-type sites, or mutant sites, coupled with cooperative interactions between Int subunits. It is possible that the P5-mutant complexes fail to undergo recombination because they lack a critical protein bridge between the P5 site and one-half of the attP core (Fig. 6), although we note that the complexes have mobilities which are identical to those of complexes formed with wild-type attP and there is evidence that Int binds cooperatively to pairs of arm-type sites (19). An alternative explanation is that both P4 and P5 in the P5-mutant are occupied by Int (through cooperative interactions) but that specific interactions with the DNA are necessary, perhaps to ensure proper contacts between Int subunits.
The second feature of the intasome is that the P1-P2 sites are unoccupied. This is rather unusual, since Int-L5 will bind to these sites in the absence of mIHF when used at equivalent concentrations (16, 17). This could be explained by a possible requirement for Int-L5 to bind arm- and core-type sites simultaneously to form stable interactions, perhaps via allosteric communication between the two functional domains (indeed, in mobility shift assays Int-L5 cannot bind either attB DNA or P1-P2 DNA alone but can weakly form bimolecular complexes in an mIHF-independent manner; data not shown). Thus, one consequence of the participation of mIHF in promoting the intramolecular bridge shown in Fig. 5 is the exclusion of core-type sites from bridging with P1 and P2. We also note that the majority of attR, which contains the core and P1-P2 sites, is released as free DNA following recombination in vitro. The inability of attR to form a stable tertiary complex (equivalent to the attL complex shown in Fig. 6) may reflect the phasing of the binding sites, which would position Int protomers bound at the P4-P5 and P1-P2 sites on opposite faces of the DNA helix.
The P1-P2 sites are required for formation of a quaternary attP-attB-containing complex and are at least partially occupied in the structure, indicating that they play a key role in the interaction with attB. One plausible explanation is that an intermolecular Int-L5 bridge is formed between these sites; as with the intasome, we prefer the simple model that both protomers of Int-L5 bound to attB also contact P1 and P2, although other configurations cannot be excluded. The 50% occupancy of P1 and P2 is somewhat puzzling, but raises the intriguing possibility that the lack of coordinate binding of Int to both P1 and P2 could explain the apparent inability of this complex to proceed in the recombination pathway.
Examination of the attP sites of several other phages suggests that the organization of the L5 integration structures which utilize two pairs of directly repeated arm-type sites (one pair on either side of the core region, in the same relative orientation), may be a common motif. The attP sites of phages P2 (30) and φRv2 (21) contain only four arm-type binding sites, placed in this arrangement. The attP site of phage P22 (26) contains five arm-type sites, four arranged in pairs (analogous to the P1-P2 and P4-P5 pairs of sites in L5) and one individual site (with similar placement to the disposable P3 site of L5). This raises the question as to whether all of these phages form similar intasome structures. In contrast, the attP sites of phages λ (9) and HP1 (3) are more complex, containing arm-type sites arranged as both pairs and as individual sites, as direct and indirect repeats, and, in the case of HP1, with multiple core-type binding sites; their corresponding integration complexes may differ from the L5 structures.
Designing a model for L5 intasome function raises further differences between L5 and λ. During λ integration the attB site is not bound by Int-λ independently but instead is captured as naked DNA by integrase molecules which are bound into a preformed intasome via their N-terminal domains and yet contain unsatisfied core-binding domains (22). In contrast, the L5 intasome identified by gel analysis does not appear to contain integrase molecules with unsatisfied binding domains available for the capture of attB (unless they are held in place solely by protein-protein interactions and therefore undetectable by DNase I footprinting assays). Moreover, since Int-L5 does not appear to bind stably to either attB alone or P1-P2 alone, binding of Int-L5 to arm- and core-type sites must be unstable unless both binding domains of Int-L5 are satisfied concurrently, and attB must join the intasome complex by simultaneous binding of Int-L5 to both attB and P1-P2.
Formation of these intasome and quaternary complex structures requires an unusual division of labor among the arm-type integrase binding sites. While little is known about excision of the L5 prophage, these structures allow us to consider possible models for the action of L5 excisionase. For example, excisionase might bind to the region between P2 and the core sites, promoting the formation of an attR tertiary complex which can synapse with an attL intasome (which may be identical to the attL complex identified here as a product of integration and essentially the same as that formed with attP). Alternatively, an excisionase may promote the formation of other protein-DNA complexes utilizing additional Int arm-type binding sites (such as P3 and P6-P7). An excisionase may also function to inhibit integrative recombination by similarly binding to attP DNA, promoting the formation of intramolecular bridges between P1-P2 and core and excluding the necessary P4-P5 interaction.
ACKNOWLEDGMENTS
We thank D. Lever for excellent technical assistance and M. Pedulla, G. Sarkis, and J. Lewis for helpful discussions.
This work was supported by grant GM49647 from the National Institutes of Health.
REFERENCES
- 1.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M J, Kalionis W, Narayana S V L, Pierson L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craig N L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- 3.Hakimi J M, Scocca J J. Binding sites for bacteriophage HP1 integrase on its DNA substrates. J Biol Chem. 1994;269:21340–21345. [PubMed] [Google Scholar]
- 4.Hatfull G F. Mycobacteriophage L5: a toolbox for tuberculosis. ASM News. 1994;60:255–260. [Google Scholar]
- 5.Hatfull G F, Grindley N D F. Resolvases and DNA-invertases: a family of enzymes active in site-specific recombination. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C: American Society for Microbiology; 1988. pp. 357–396. [Google Scholar]
- 6.Hatfull G F, Sarkis G. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 7.Hoeffler W K, Kovelman R, Roeder R G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988;53:907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 8.Kuhstoss S, Rao N J. Analysis of the integration function of the streptomycete bacteriophage phi C31. J Mol Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 9.Landy A. Dynamic, structural and regulatory aspects of λ site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 10.Landy A. Mechanistic and structural complexity in the site-specific recombination pathways of Int and FLP. Curr Opin Genet Dev. 1993;3:699–707. doi: 10.1016/s0959-437x(05)80086-3. [DOI] [PubMed] [Google Scholar]
- 11.Lee M H, Hatfull G F. Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J Bacteriol. 1993;175:6836–6841. doi: 10.1128/jb.175.21.6836-6841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash H A. Bending and supercoiling of DNA at the attachment site of bacteriophage λ. Trends Biochem Sci. 1990;15:222–227. doi: 10.1016/0968-0004(90)90034-9. [DOI] [PubMed] [Google Scholar]
- 14.Nash H A. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments. In: Neid-hardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 2363–2376. [Google Scholar]
- 15.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedulla, M. L., and G. F. Hatfull. Unpublished observations.
- 17.Pedulla M L, Lee M H, Lever D C, Hatfull G F. A novel host factor for integration of mycobacteriophage L5. Proc Natl Acad Sci USA. 1996;93:15411–15416. doi: 10.1073/pnas.93.26.15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peña C E A, Kahlenberg J M, Hatfull G F. The role of supercoiling in mycobacteriophage L5 integrative recombination. Nucleic Acids Res. 1998;26:4012–4018. doi: 10.1093/nar/26.17.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peña C E A, Lee M H, Pedulla M L, Hatfull G F. Characterization of the mycobacteriophage L5 attachment site, attP. J Mol Biol. 1997;266:76–92. doi: 10.1006/jmbi.1996.0774. [DOI] [PubMed] [Google Scholar]
- 20.Peña C E A, Stoner J E, Hatfull G F. Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J Bacteriol. 1996;17:5533–5536. doi: 10.1128/jb.178.18.5533-5536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peña, C. E. A., J. E. Stoner, and G. F. Hatfull. Mycobacteriophage D29 integrase-mediated recombination in vitro: specificity of mycobacteriophage integration. Gene, in press. [DOI] [PubMed]
- 22.Richet E, Abcarian P, Nash H A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988;52:9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 23.Ross W, Landy A. Bacteriophage λ int protein recognizes two classes of sequence in the phage att site: characterization of arm-type sites. Proc Natl Acad Sci USA. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadowski P D. Site-specific genetic recombination: hops, flips and flops. FASEB J. 1993;7:760–767. doi: 10.1096/fasebj.7.9.8392474. [DOI] [PubMed] [Google Scholar]
- 25.Smith, J., and G. F. Hatfull. Unpublished observations.
- 26.Smith-Mungo L, Chan I T, Landy A. Structure of the P22 att site. J Biol Chem. 1994;269:20798–20805. [PubMed] [Google Scholar]
- 27.Snapper S, Lugosi L, Jekkel A, Melton R, Keiser T, Bloom B R, Jacobs W R., Jr Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc Natl Acad Sci USA. 1988;85:6987–6991. doi: 10.1073/pnas.85.18.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snapper S, Melton R, Keiser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 29.Stark W M, Boocock M R, Sherratt D J. Catalysis by site-specific recombinases. Trends Genet. 1992;12:432–439. [PubMed] [Google Scholar]
- 30.Yu A, Haggard-Ljungquist E. Characterization of the binding sites of two proteins involved in the bacteriophage P2 site-specific recombination system. J Bacteriol. 1993;175:1239–1249. doi: 10.1128/jb.175.5.1239-1249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]