Significance
Lassa virus (LASV), the causative agent of Lassa fever (LF), represents an important public health problem in Western Africa. There is no Food and Drug Administration (FDA)-approved therapeutic intervention to treat LF. Because of their limited genome coding capacity, LASV proteins are often multifunctional and orchestrate complex interactions with cellular factors to execute steps required to complete the viral life cycle. LASV polymerase is essential for replication and expression of the viral genome and, thus, is an attractive target for antiviral intervention. Here, we present the host interactome of LASV polymerase that can guide identification of novel druggable host cellular targets for the development of cost-effective antiviral therapies for LF.
Keywords: viral replication, proximity proteomics, host-virus interactions, host-directed antivirals, arenaviruses
Abstract
Completion of the Lassa virus (LASV) life cycle critically depends on the activities of the virally encoded, RNA-dependent RNA polymerase in replication and transcription of the viral RNA genome in the cytoplasm of infected cells. The contribution of cellular proteins to these processes remains unclear. Here, we applied proximity proteomics to define the interactome of LASV polymerase in cells under conditions that recreate LASV RNA synthesis. We engineered a LASV polymerase-biotin ligase (TurboID) fusion protein that retained polymerase activity and successfully biotinylated the proximal proteome, which allowed the identification of 42 high-confidence LASV polymerase interactors. We subsequently performed a small interfering RNA (siRNA) screen to identify those interactors that have functional roles in authentic LASV infection. As proof of principle, we characterized eukaryotic peptide chain release factor subunit 3a (eRF3a/GSPT1), which we found to be a proviral factor that physically associates with LASV polymerase. Targeted degradation of GSPT1 by a small-molecule drug candidate, CC-90009, resulted in strong inhibition of LASV infection in cultured cells. Our work demonstrates the feasibility of using proximity proteomics to illuminate and characterize yet-to-be-defined host-pathogen interactome, which can reveal new biology and uncover novel targets for the development of antivirals against highly pathogenic RNA viruses.
Lassa virus (LASV) is a mammarenavirus that is highly prevalent in Western Africa, where it infects several hundred thousand individuals annually; these infections result in a high number of Lassa fever (LF) cases, a hemorrhagic fever disease associated with high morbidity and significant mortality among hospitalized LF patients (1). Although rodent-to-human transmission is the main mode of transmission leading to LASV infections in the human population, a significant number of LF cases can arise from human-to-human transmission (2). Moreover, increased travel has resulted in exported LF cases from endemic Western African countries to nonendemic countries (3). To date, no U.S. Food and Drug Administration (FDA)-licensed countermeasures are available to prevent or treat LASV infections, and current anti-LASV therapy is limited to an off-label use of ribavirin that has limited efficacy and can cause significant side effects. Hence, there is an unmet need for cost-effective therapeutics to combat LASV infections. Development of effective therapeutics would be facilitated by a better understanding of virus-host cell interactions that modulate replication and gene expression of LASV in infected cells.
Like other mammarenaviruses (Bunyavirales: Arenaviridae), LASV is an enveloped virus with a bisegmented, single-stranded RNA genome. Each viral genome segment uses an ambisense coding strategy to direct the synthesis of two viral proteins from open reading frames (ORFs) separated by noncoding intergenic region (IGR). The small (S) segment encodes the nucleoprotein (NP), which is responsible for genome encapsidation and immune evasion, and the glycoprotein precursor (GPC), which is co- and posttranslationally processed to generate the mature GP that mediates virion cell entry via receptor-mediated endocytosis (4, 5). The large (L) segment encodes the large (L) protein that functions as a viral RNA-directed RNA polymerase and the matrix Z protein. NP encapsidates the viral genome and antigenome RNA species to form the viral nucleocapsid to which L associates to form the two viral ribo-NP complexes (vRNPs) (of L and S segments). The resulting vRNPs are responsible for directing replication and transcription of the viral RNA genome (6, 7). LASV polymerase initiates viral transcription from the genome promoter located at the 3′ end of the viral genome, which is primed by a small host-cell-derived, capped RNA fragment via a mechanism called cap-snatching. Primary transcription results in synthesis of NP and L mRNAs from the S and L segments, respectively. The virus polymerase can also adopt a replicase mode and moves across the IGR to generate a copy of the full-length antigenome (cRNA) for each segment. These cRNAs serve as templates for synthesis of the GPC and Z mRNAs from the S and L segments, respectively, as well as templates for amplification of the corresponding viral genome (vRNA) (8–10).
Coordination of multiple domains with distinct enzymatic functions through conformational rearrangement and the engagement of vRNA promoter elements with LASV polymerase have been proposed to mediate the functional transitioning of LASV polymerase between a replicase and transcriptase (11). Here, we investigated whether host cell factors can critically contribute to distinct steps of viral RNA synthesis that are driven by LASV polymerase. We applied the recently developed proximity-labeling technology to characterize the cellular interactome of LASV L polymerase in the context of viral RNA synthesis in living cells. We fused LASV polymerase to the engineered promiscuous biotin ligase TurboID to generate a fusion protein that retained polymerase activity and the ability to biotinylate its cellular interactors in situ. This approach allowed direct affinity capture and identification of biotinylated interactors, leading to the discovery of high-confidence LASV polymerase interactors. To validate the functional role of the identified interactors, we implemented a high-content imaging-based siRNA screen using a human hepatocyte-derived cell line, Huh7 cells, infected with authentic LASV. Among the functional polymerase interactors, we identified GSPT1 as a proviral factor. Pharmacological targeting of GSPT1 with the preclinical drug CC-90009 had a strong inhibitory effect against LASV in cultured cells. Our results have improved our understanding of the cellular proteins and pathways involved in LASV RNA synthesis and uncover potential therapeutic strategies against a lethal human pathogen.
Results
Generating LASV L-HA-TurboID with Polymerase and Proximity-Labeling Activities.
For proximity labeling, we selected TurboID, an engineered biotin ligase that has promiscuous labeling activity and optimized labeling efficiency. TurboID labels exposed lysine residues on neighboring proteins within a ∼10-nm radius and is highly active, requiring <10 min of labeling time for detectable activity (12). We incorporated TurboID with an HA epitope tag into the LASV L protein at a site (after residue 407) previously shown to tolerate incorporation of epitope tags without severely affecting polymerase activity (13) (Fig. 1A). The HA tag facilitated L-HA-TurboID detection and biochemical characterization, as there is no commercially available antibody against LASV L.
Fig. 1.
Generation of a functional LASV polymerase with proximity-labeling activity. (A) Schematic of LASV L-HA-TurboID construct and proximity biotinylation of L-interacting host factors. (B) Polymerase activity and expression of LASV L-TurboID compared to L-WT and L-HA in a cell-based LASV MG assay. MG activity (fluorescent reporter intensity) was quantified in each sample and normalized to the L-WT control. Representative western blots show expression of LASV L-HA-TurboID, L-HA, NP, and cellular β-actin in HEK 293T whole-cell lysates. MG activity was evaluated in three independent biological replicates with triplicated wells for each condition. Bar graphs containing nine data points (white circles) for each condition are displayed, with means plus SEM (error bars). One-way ANOVA with Dunnett’s multiple comparisons test was performed to compare the activity of LASV L-HA-TurboID and L-HA to L-WT (****P < 0.0001). (C) SA blot showing biotinylations mediated by a control HA-TurboID in the absence of LASV proteins (Left) or by LASV L-HA-TurboID in cells coexpressing LASV MG components (Right). (D) Confocal immunofluorescence images of LASV L-HA-TurboID-mediated proximity biotinylation in cells coexpressing LASV MG components. LASV L-HA and L-TurboID were detected by using an anti-HA antibody. Biotinylated proteins were detected by using the SA-AF594. Nuclei were stained with Hoechst and are shown in merged images. Scale bar: 5 μm. Representative images from two independent experiments acquired by Zeiss LSM 880 with Airyscan are shown. (Bottom) Zoomed-in view of the area outlined by (Top) a white dashed line. Each experiment includes at least three different fields of view.
We used a cell-based, LASV minigenome (MG) system to confirm the polymerase activity of the resulting L-HA-TurboID. This MG system recapitulates LASV RNA synthesis using an intracellular reconstituted LASV vRNP expressing a fluorescent reporter, ZsGreen. Reconstitution of LASV vRNP requires coexpression of the LASV L and NP proteins, as well as the LASV MG vRNA. Thus, we performed all experiments requiring reconstituted LASV MG in human embryonic kidney cells (HEK 293T) due to their high transfection efficiency. In these experiments, expression levels of the MG reporter served as a comprehensive measurement of LASV MG replication, transcription, and translation of the MG reporter transcript (14) (SI Appendix, Fig. S1).
We used a wild-type LASV L (L-WT) and an N-terminal HA-tagged L (L-HA) as controls to compare the polymerase activity and protein expression levels with L-HA-TurboID. L-HA-TurboID retained 70% of WT activity, with slightly lower expression levels compared to those of the L-HA control (Fig. 1B). We validated the proximity-labeling activity of L-HA-TurboID by detecting biotinylated proteins in cells reconstituted with LASV MG. In the presence of exogenous biotin, L-HA-TurboID produced a broad range of biotinylated proteins in a time-dependent manner. In comparison, a different group of cellular proteins was biotinylated by an N-terminal HA-tagged TurboID control (Fig. 1C).
We next used confocal fluorescent microscopy to detect the subcellular distribution of L-HA-TurboID-mediated biotinylation in cells. We transfected HEK 293T cells with a LASV MG system containing either L-HA-TurboID or L-HA control, and at 24 h posttransfection, we conducted 1-h biotin labeling. LASV L-HA-TurboID localized to discrete puncta connecting patches in the cytoplasm, similar to the L-HA control. Further, the biotinylation signal overlapped with the location of L-HA-TurboID, supporting efficient and localized biotinylation mediated by the L-HA-TurboID (Fig. 1D).
Defining LASV Polymerase Interactomes by Proximity Proteomics.
We carried out a proximity proteomics experiment using similar conditions to those previously published (12). We transfected HEK 293T cells with the LASV MG system, including either L-WT or L-HA-TurboID followed by biotin labeling. Transfected cells were lysed, and biotinylated proteins were captured with streptavidin (SA) beads. Proteins enriched by SA beads were washed and subjected to on-bead trypsin digestion. The digested peptides in solution were then labeled with unique tandem-mass tags for quantitative proteomic analysis (Fig. 2A).
Fig. 2.
Proximity proteomics of LASV polymerase in living cells. (A) Sample preparation workflow for proximity proteomic assays. (B) Validation of SA enrichment of LASV L-HA-TurboID-mediated biotinylation. Harvested cell lysates are indicated as “Input,” and biotinylated proteins enriched with SA beads are indicated as “SA pulldown.” Representative blots and a silver-stained gel from three biological replicates are presented. (C) Scatter plot showing LASV polymerase-interacting proteins in HEK 293T cells determined by quantitative proteomic analysis. The degree of enrichment for each protein is shown as a fold change, and the statistical confidence of enrichment is shown as adjusted P values that are numerated and log transformed.
To assess the efficiency of the enrichment, biotinylated material bound to SA beads (SA pulldown) from equal amounts of starting material (input) was eluted using sodium dodecyl sulfate (SDS) loading buffer containing dithiothreitol (DTT) and biotin and analyzed by western blot. We observed enrichment of biotinylated proteins specific to samples with L-HA-TurboID expression. Only a small fraction of LASV NP was biotinylated, which may reflect that only a low percentage of the total NP participates in the formation of a functional vRNP or that in the vRNP, the majority of NP was not accessible to biotinylation or remained insoluble under the lysis conditions we used to prepare the samples. We detected L-HA-TurboID in the SA pulldown fraction, suggesting self-biotinylation (Fig. 2B).
To analyze the LASV polymerase interactome, host proteins identified as LASV polymerase interactors were scrutinized by two criteria: 1) degree of enrichment compared to the control proteome and 2) the corresponding statistical confidence for each comparison. We used L-WT to generate the control proteome. For every identified protein, the abundance ratio in the L-HA-TurboID sample was normalized to that in the L-WT sample to obtain a fold-change value. Multiple comparisons were performed across three biological replicates, and adjusted P values (adjusted to false discovery rate of 0.05) were determined to identify proteins that were enriched in the L-HA-TurboID interactome. A threshold of adjusted P < 0.05 and log2(fold change) > 1 was used to identify 42 high-confidence hits corresponding to proteins that interact with LASV polymerase (Fig. 2C).
Effect of siRNA Targeting High-Confidence LASV Polymerase Interactors on Viral Infection.
Next, we assessed the biological importance of the 42 high-confidence hits in the context of infection with live LASV. We used a siRNA-based functional screen based on well-established experimental procedures (15) and performed the screen using human hepatocyte-derived Huh7 cells, as hepatocytes are relevant to LASV-induced pathology (16). Briefly, we transfected Huh7 cells with a library of individual siRNAs targeting each of the 42 high-confidence hits, and at 48 h posttransfection, we infected these cells with the recombinant LASV-eGFP (emerald green fluorescent protein) using a multiplicity of infection (MOI) of 0.5 plaque forming unit per cell (PFU/cell) (17). Using a high-content imaging system, we quantified the percentage of LASV-infected cells with each siRNA knockdown (KD) based on the number of GFP-positive cells and the total cell count (with nuclear stain) (Fig. 3A). The siRNA screen was repeated using a MOI of 1.
Fig. 3.
Identification of functional polymerase interactors by siRNA screen in LASV-infected cells. (A) Workflow of siRNA screen by high-content imaging of Huh7 cells infected with rLASV-eGFP. The raw percentage of infection and total cell count for each siRNA treatment were calculated and normalized to those of NSCs. (B) Heat maps of relative percentage of infection and relative total cell count for LASV. Each value is the mean of technical triplicates. Multiple unpaired t tests were performed to determine the statistical significance of siRNA-mediated changes in the percentage of infection compared to that for NSC. (C) The P values were log-transformed and displayed as the size of each data point on a scatter plot. Data points in the heatmap with P > 0.05 were eliminated from the scatter plot. Names of genes for which multiple siRNAs significantly affected infection, but not cell counts, are highlighted on the corresponding data points. For each gene, only one label is highlighted in red for display purposes. (D) Representative fluorescence images of Huh7 cell monolayers at 48 h.p.i. are shown for NSC and two selected siRNAs. Merged images are composed of green (rLASV-eGFP) and blue (nuclei) channels. Results of the siRNA screen with MOI = 0.5 are shown.
To control potential nonspecific effects of siRNA transfection on LASV infection and total cell count, we normalized these two parameters against values for nonsilencing control (NSC) siRNA. We excluded any hits for which siRNA-KD altered the total cell count by one SD from the average cell count or for which siRNA-KD failed to significantly change the percentage of LASV-infected cells compared to NSC. From the remainder, we considered as true hits those with at least two out of four independent siRNA-KDs resulted in the same infection phenotype.
In total, 19 high-confidence hits exhibited an antiviral role, as depletion of the hit by more than two independent siRNAs increased LASV infection (MOI = 0.5) (Fig. 3B). Seven of these antiviral hits were validated in the repeated siRNA screen (MOI = 1) (SI Appendix, Fig. S2). Limited (10 to 20% of cells) infection of NSC siRNA-transfected cells (SI Appendix, Fig. S3) may have favored the detection of enhancement over reduction in infection. Top hits in the siRNA screen were selected by stringent statistical metrics (two biological replicates, four independent siRNAs per target, technical triplicate per siRNA) and were guided by quantitative parameters of the resulting phenotype (i.e., relative percentages of infection and cell count). These top hits included six antiviral factors (EIF3CL, EIF4G2, TAGLN2, TUBA1B, PSMC5, and upstream frameshift 1 [UPF1]) and one proviral factor (GSPT1).
The Proximity Interactome of LASV L Polymerase.
We summarized the LASV L polymerase interactome in a network representation, in which we highlighted hits that were functionally validated in our siRNA screen. We clustered all interactors based on their STRING (Search Tool for the Retrieval of Interacting Genes/Proteins)-functional classification (18) to identify distinct cellular pathways relevant to LASV infection (Fig. 4A).
Fig. 4.
The proximity interactome of LASV L polymerase. (A) Network of the LASV polymerase interactome identified by proximity proteomics. Each node represents one high-confidence proteomic hit. Each edge represents an identified protein-protein interaction. Nodes that were functionally validated by siRNA screen are highlighted in pink (by two siRNA screens) or in maroon (by one siRNA screen). Nodes shown in heptagons have been previously reported for other virus-host interactomes. Selected nodes are clustered based on enriched biological processes or protein domains by functional enrichment analyses via STRING. (B) Validation of selected proteomic hits by coIP of LASV L-HA transiently expressed in HEK 293T cells. HA-tagged GFP-Halotag fusion protein was used as a control. Representative blots of three experiments are shown. Asterisks indicate band(s) of target protein being detected, including different isoforms. For some blots, a longer-exposure image was used to show visible IP signal, and the same blot with shorter-exposure is shown on the right.
To confirm the LASV L interactome, we selected eight interactors for biochemical validation. We coimmunoprecipitated (coIP) endogenous LASV L interacting proteins in HEK 293T cells using an N-terminal HA-tagged L as the bait (Fig. 4B). Among the eight selected interactors, we validated RARS, AIMP2, RPS3, PSMC5, EIF4G2, UPF1, and GSPT1. AMOT was the only interactor that we could not successfully coIP, likely due to its low expression levels. To control nonspecific binding, we performed a parallel coIP using an HA-tagged GFP-Halotag fusion protein as the bait. These results support the validity of proximity-labeling-based proteomics to identify bona fide LASV L interactors.
We also compared the LASV L interactome described here with previously reported host interactomes of viral proteins derived from the prototypic Old World (OW) arenavirus, lymphocytic choriomeningitis virus (LCMV) (SI Appendix, Fig. S4). We found 11 common host factors shared by multiple interactomes, which likely reflects conserved molecular recognition interfaces between host cells and OW mammarenaviruses.
Role of GSPT1 in LASV Gene Expression.
Here, we focused on the only proviral hit, eukaryotic peptide chain release factor subunit 3a (eRF3a/GSPT1), which has not been reported to play any specific role in viral infection.
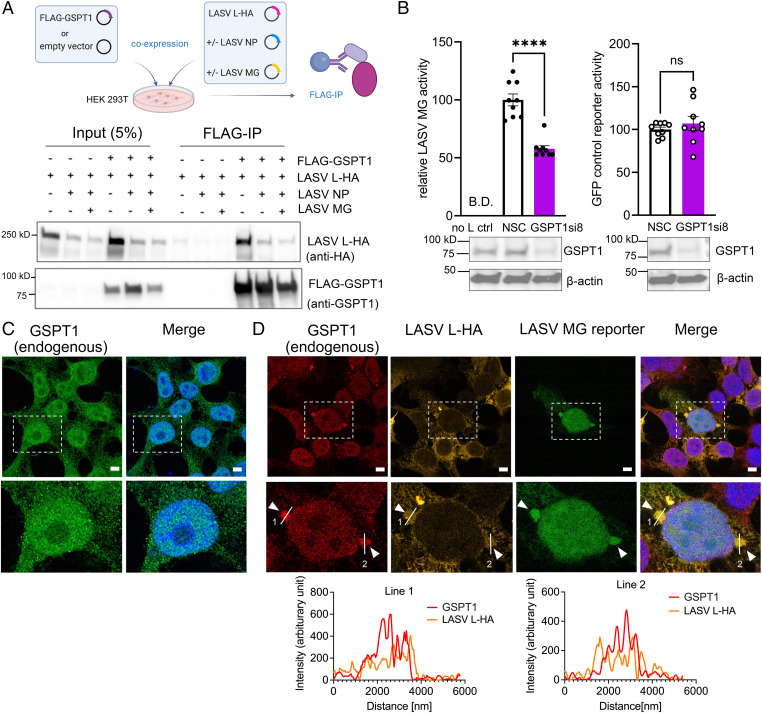
First, we used coIP to verify that GSPT1 physically interacts with LASV L protein. To ensure the level of GSPT1 protein was sufficient for coIP, we cotransfected HEK 293T cells with plasmids expressing an N-terminal FLAG-tagged GSPT1 (long isoform, 68.7 kDa) and LASV L-HA in the presence or absence of other LASV MG components. We found that LASV L-HA consistently IP FLAG-GSPT1 in the presence or absence of LASV NP or LASV NP and the LASV MG (Fig. 5A). We next examined the functional consequence of GSPT1 association with LASV L by the LASV MG assay. We found that depletion of GSPT1 protein by one experimentally validated siRNA, GSPT1si8 (SI Appendix, Fig. S5), significantly suppressed LASV MG activity but did not affect expression of a control eGFP reporter (Fig. 5B).
Fig. 5.
GSPT1 physically and functionally associates with LASV polymerase. (A) Western blot analyses of FLAG-IP from cells coexpressing FLAG-GSPT1 and LASV L-HA with or without LASV MG components. Representative results from three biological replicates are shown. (B) LASV MG activity and expression level of a control GFP reporter in HEK 293T cells upon GSPT1-KD. GSPT1-KD by siRNA was validated by western blot analysis. β-actin served as the loading control. MG activity and expression level of the control GFP reporter in GSPT1si8-transfected cells were normalized to those of NSC transfected cells. The LASV MG experiment was repeated in three independent replicates with triplicate wells for each condition. Nine individual data points for each condition are displayed in the bar graphs, with means ± SEM (error bars) shown. Two-tailed unpaired t tests were performed to determine whether depleting GSPT1 significantly changed activities of LASV MG and the control reporter (ns, not significant; ****P < 0.0001). B.D., below detection limits. Confocal immunofluorescent analysis of the subcellular localization of endogenous GSPT1 alone (C) or with transiently expressed LASV L-HA in LASV MG reconstituted HEK 293T cells (D). Nuclei were stained with Hoechst (blue) and are shown in merged images. Arrowheads show sites where endogenous GSPT1 localize proximal to LASV proteins. Representative images from four independent experiments that were acquired with Zeiss-LSM880 Airyscan are shown, and each experiment includes at least three different fields of view. (Bottom) Zoomed-in view of the area outlined by (Top) a white dashed line. Colocalization analysis of LASV L-HA and endogenous GSPT1 was performed along two white lines for each channel. The line profile plots of the fluorescence intensity of both channels in the same focal plane are shown. Scale bar: 5 μm.
We also examined whether endogenous GSPT1 interacts with LASV polymerase in a cellular context that recapitulates viral RNA synthesis. In HEK 293T control cells, endogenous GSPT1 exhibited a diffuse nucleocytoplasmic distribution (Fig. 5C). However, in HEK 293T cells that successfully reconstituted a functional LASV vRNP, as determined by the ZsGreen reporter signal, we detected a fraction of cytoplasmic GSPT1 that coclustered with LASV L-HA (Fig. 5D), suggesting that GSPT1 interacts with LASV L in cells expressing a functional LASV vRNP.
Role of GSPT1 in LASV Multiplication.
To validate the role of GSPT1 on LASV multiplication, we examined the phenotype of siRNA-mediated GSPT1-KD in multistep LASV growth kinetics in Huh7 cells. Consistent with the siRNA screen results, GSPT1 depletion significantly impaired LASV growth in Huh7 cells starting at 24 h postinfection (h.p.i.) with at least one log reduction in viral titer at 48 h.p.i. (Fig. 6A). The phenotype of impaired LASV growth in GSPT1-KD cells was observed in three biological replicates (Fig. 6A and SI Appendix, Fig. S6A). Upon GSPT1-KD, none of the LASV RNA species were significantly altered (Fig. 6B and SI Appendix, Fig. S6B). Notably, in GPST1-depleted cells at 72 h.p.i., LASV GP2 protein levels were decreased, which might impede production of infectious virions (Fig. 6C and SI Appendix, Fig. S6C). This finding can be related to GSPT1 being a critical component of the cellular translation termination complex (eRF1/eRF3) (19–21). Given the subtle impact on LASV RNA accumulation but decrease in LASV protein levels, we reason that GSPT1 may support LASV infection by assisting viral protein translation.
Fig. 6.
GSPT1 is a druggable host factor required for LASV multiplication. (A) Effect of GSPT1-KD on LASV viral growth kinetics in Huh7 cells. Viral titers from two independent experiments with technical triplicates were log-transformed and plotted as mean ± SD (error bars). Two-way ANOVA analysis on log-transformed titers was performed to determine the statistical significance of the effect of GSPT1-KD on viral growth kinetics (****P < 0.0001). (B) Effect of GSPT1-KD on LASV vRNA and c/mRNA accumulation. Six individual data points from two independent experiments with technical triplicates are displayed, and the mean ± SEM (error bars) is shown. Welch’s t test was performed to determine the statistical significance of the effect of GSPT1-KD on LASV RNA accumulation. (C) Western blot analysis of endogenous GSPT1 protein and LASV GP2 levels in lysates from LASV-infected Huh7 cells. β-actin served as the loading control. Lysates from triplicate wells in the growth kinetic experiments were pooled and analyzed. (D) Representative fluorescence images of Huh7 monolayers at 72 h.p.i. are shown with nuclei (blue) and virus infected cells (green). Representative blots and images from one experiment are shown. (E) Schematic diagram CC-90009-mediated GSPT1 degradation. (F) Effect of CC-90009 on LASV growth kinetics in Huh7 cells. Results of one experiment with technical triplicates were plotted as mean ± SD (error bars). ND, not detectable. Two-way ANOVA analyses on log-transformed viral titers were performed to determine the statistical significance of the effect of CC-90009 treatments compared to DMSO on LASV growth kinetics (***P < 0.001). (G) Effect of CC-90009 treatment on LASV vRNA and c/mRNA accumulation in Huh7 cells. Three individual data points from one experiment with technical triplicates are displayed as mean ± SEM (error bars). Brown-Forsythe and Welch ANOVA with multiple comparisons test were used to determine values that differed significantly from the controls (*P < 0.05; **P < 0.01). (H) Western blot analysis of endogenous GSPT1 protein and LASV GP2 levels in LASV-infected Huh7 lysates. Lysates from triplicate wells in the growth kinetic experiments were pooled and analyzed. (I) Percentage of LASV infection in Huh7 cell (MOI = 0.5) after CC-90009 treatment at 48 h.p.i. Raw values of mean fluorescence intensity of the infected monolayer with drug treatment are normalized to a DMSO control. (J) Percentage of Huh7 cells viability after CC-90009 treatment was determined by Cell titer Glo 2.0. Raw values for cell viability in drug-treated samples are normalized to a DMSO control. For I and J, one experiment with technical triplicates is indicated as mean ± SD (error bars).
Based on the effect of GSPT1 KD on LASV growth kinetics, we predicted that pharmacological inhibition of GSPT1 would suppress LASV multiplication. We therefore tested the antiviral activity of CC-90009, an E3 ubiquitin ligase modulator that selectively tethers GSPT1 to the CRL4CRBN E3 ubiquitin ligase to induce targeted ubiquitination and degradation of GSPT1 (22, 23) (SI Appendix, Fig. S5C). We treated Huh7 cells with CC-90009 immediately following LASV infection and measured the effect of CC-90009 treatment on LASV growth kinetics, viral RNAs, and viral protein accumulation. At 0.1 and 1 μM, CC-90009 effectively inhibited LASV growth (Fig. 6E), which correlated with the dose-dependent reduction in accumulation of both vRNA and c/mRNA accumulation within 6 h of viral infection (Fig. 6F) and diminished viral protein accumulation compared to the dimethyl sulfoxide (DMSO)-treated control (Fig. 6G). CC-90009 had 50% inhibitory concentration (EC50) of 16.86 nM (Fig. 6I) and 50% cytotoxic concentration (CC50) > 10 μM (Fig. 6J), with a selectivity index (CC50/EC50) > 593.
We noticed that unlike GSPT1 depletion mediated by siRNA, CC-90009 treatment equivalently inhibited accumulation of both LASV vRNA and c/mRNA at very early time points after infection (cf. Fig. 6H and 6B), suggesting a fundamental defect in LASV RNA synthesis. This finding, together with the physical association between GSPT1 and LASV L polymerase, suggests that CC-90009 tethering of the E3 ligase to GSPT1 might facilitate ubiquitination and degradation of LASV L. As CC-90009 is able to modulate the spectrum of substrates targeted by the CRL4CRBN E3 ligase (24, 25), LASV L itself could be turned into a neosubstrate and targeted for degradation upon CC-90009 treatment. Thus, a reduced level of LASV L could entail a global decrease in LASV RNA synthesis, although this hypothesis remains to be examined. Together, our results demonstrated that CC-90009 has potent antiviral activity against LASV in human hepatocytes.
Effect of GSPT1 Depletion on Other Mammarenaviruses.
To determine whether the antiviral activity of CC-90009 could be expanded to other OW mammarenaviruses, we examined the effect of CC-90009 on the growth kinetics of LCMV, an OW mammarenavirus related to LASV, in Huh7 cells. Delayed LCMV growth was seen in Huh7 cells infected with LCMV and then treated with CC-90009 (0.1 μM) following infection, and with 1 μM CC-90009, LCMV titers were sustainably reduced by more than one log (Fig. 7A). Similar to the phenotype observed in LASV-infected cells, accumulation of LCMV RNAs (Fig. 7B) and proteins (Fig. 7C) was reduced in CC-90009-treated cells, and fewer cells were infected with LCMV compared to vehicle-treated cells (Fig. 7D). Comparable to the antiviral activity against LASV infection, the EC50 of CC-90009 for LCMV was 14.85 nM (Fig. 7E).
Fig. 7.
Inhibition of LCMV growth by CC-90009-mediated degradation of GSPT1. (A) Effect of CC-90009 on LCMV growth kinetics in Huh7 cells. Results of two independent experiments with technical duplicates are plotted as mean ± SD (error bars). Two-way ANOVA analyses of log-transformed viral titers were performed to determine the statistical significance of the effect of CC-90009 treatments compared to DMSO on LCMV growth kinetics (****P < 0.0001). (B) Effect of CC-90009 treatment on LCMV RNA synthesis in Huh7 cells. Three individual data points from one experiment with technical triplicates are displayed and indicated as mean ± SEM (error bars). Values significantly different from the controls were determined by Brown-Forsythe and Welch ANOVA with multiple comparisons test (*P < 0.05). (C) Western blot analysis of endogenous GSPT1 protein, LCMV GPC, and GP2 levels in LCMV-infected Huh7 lysates. β-actin was used as the loading control. Lysates from duplicated wells in the growth kinetic experiments were pooled and analyzed. (D) Representative fluorescence images of Huh7 monolayers infected with rLCMV-GFP at 72 h.p.i. are shown with nuclei (blue) and virus-infected cells (green). Representative blots and images from one experiment are shown. (E) Percentage of LCMV infection in Huh7 cell (MOI = 0.5) after CC-90009 treatment at 48 h.p.i. Raw values of LCMV infection rate with drug treatment are normalized to a DMSO control.
Discussion
Here, we have presented a host interactome of LASV polymerase in the context of viral RNA synthesis directed by an intracellularly reconstituted functional LASV vRNP. We applied proximity proteomics to identify the LASV polymerase interactors in situ. We then performed a siRNA screen in human hepatocytes infected with live LASV to identify polymerase interactors functionally important during LASV infection. Our finding that most identified polymerase interactors exhibited an antiviral, rather than a proviral, phenotype could reflect that the viral polymerase or viral RNA products, or both, are targets of host innate defense mechanisms. Nevertheless, we validated one proviral factor, GSPT1, and demonstrated that it is a potentially druggable target for the development of therapeutics against LASV.
Previously reported host interactomes on LCMV proteins were based on the affinity purification-mass spectrometry (AP-MS) approach in the context of viral infection (26, 27), which may fail to capture nonstable transient protein-protein interactions. Moreover, the use of live LASV infection requires BSL4 containment, which complicates proteomic experiments using LASV-infected cells. To overcome these obstacles, we used the TurboID-based, proximity-labeling approach to capture both stable and dynamic protein-protein interactions in the context of a cell-based MG system that recapitulates the biological activities of the LASV vRNP. Detection of endogenous host proteins expressed at low levels can be difficult to capture in AP-MS-based interactomes. Nevertheless, in proximity proteomics, direct enrichment of biotinylated interactors to the bait protein offers a higher likelihood of detecting host factors that have low abundance yet important roles such as GSPT1. This technological advantage may have contributed to the identification of 31 cellular proteins not identified by the LCMV L interactome.
Many RNA viruses have evolved strategies to manipulate the host translational machinery to favor translation initiation of viral mRNAs (28–30). We identified two translation initiation factors in our LASV polymerase interactome, EIF4G2 and EIF3CL, as repressors of LASV infection. EIF4G2 is a functional homolog of EIF4G1 (31), which is known to bridge the high-affinity cytoplasmic cap-binding protein EIF4E and the scaffolding protein EIF3, which binds to the small ribosomal subunit. EIF4G2 can also enhance the interaction between EIF4E and cap-containing mRNAs (32). As with other mammarenaviruses, transcription of LASV mRNAs uses a “cap-snatching” mechanism by which viral mRNAs hijack 5′ cap structures from host mRNA species (33), which primes viral transcription and enables cap-dependent translation (34). Although still controversial, one mechanism by which mammarenaviruses could accomplish cap-snatching involves binding of virus NP to m7GpppN cap structures (35, 36), together with the activity of an endonuclease motif (EndoN) present within the N-terminal region of the LASV polymerase (37) that cleaves cellular mRNAs to liberate the 5′ cap structure. Importantly, our LASV L interactome included EIF4G2 and EIF3CL, but not EIF4E, suggesting that LASV polymerase may not hijack the canonical host cap-binding complex. Our results could indicate that L polymerase instead competes for 5′ capped cellular mRNAs with the canonical host cap-binding complex, which contains EIF4E, and L may bind to EIF4G2 as a decoy that interferes with stable complex formation between eIF4E and capped mRNAs. This binding, in turn, would release capped mRNAs that can then serve as substrates for the L EndoN to hijack the 5′ cap structure for LASV transcription initiation. Additional support for this hypothetical model stems from the observation that cap-binding affinity of EIF4E was reduced upon binding of LASV Z protein to EIF4E (38), which further lowers the affinity barrier for a LASV-encoded cap-binding protein, or another proviral factor, to capture fragments of capped cellular mRNAs for priming LASV transcription (SI Appendix, Fig. S7). This model is based on the assumption that EIF4G2 and EIF3CL, but not EIF4E, are bona fide LASV polymerase interactors, which needs to be experimentally validated.
Our siRNA-based functional screen also revealed a LASV polymerase interactor, ATP-dependent RNA helicase UPF1, as an antiviral factor. UPF1 is a key player in multiple cellular RNA decay pathways, typically through binding to the 3′ UTR of a target transcript (39), suggesting that UPF1 might play an antiviral role in LASV infection by mediating LASV mRNA decay. LASV mRNAs could trigger UPF1-dependent mRNA decay through the nonsense-mediated mRNA decay (NMD) pathway. In the NMD pathway, UPF1 degrades aberrant RNA transcripts by joining the translation-termination complex (GSPT1/eRF3-eRF1) upon ribosomal recognition of a premature termination codon (PTC) in the mRNA (40). After recruitment to the termination complex, UPF1 is phosphorylated, thereby triggering a downstream cascade of nucleases and decapping enzymes to dismantle the target transcript (41). LASV mRNAs are likely to incorporate PTCs owing to the presence of an upstream ORF, which can be acquired by viral transcripts as byproducts of the cap-snatching mechanism (42). Future experiments are needed to determine whether the UPF1-LASV L interaction is RNA dependent and whether UPF1 directly targets LASV mRNAs for degradation.
Intriguingly, we identified another host NMD factor, G1-to-S-phase transition 1(GSPT1)/eukaryotic peptide chain release factor GTP-binding subunit A(eRF3a), as a LASV L interactor with proviral activity. GSPT1 is a core component of the cellular translation termination machinery (20, 43) and has been shown to regulate cell-cycle progression (44) and promote apoptosis (45). As GSPT1 participates in multiple cellular pathways, it may regulate viral infection via different mechanisms in a context-dependent manner. We confirmed a physical association between LASV L polymerase and GSPT1. Further, we showed that GSPT1 positively regulates LASV MG activity, arguing against the role of GSPT1 in the NMD being linked to its proviral activity. In the context of a multistep growth kinetics experiment, GSPT1 KD resulted in one-log reduction of the number of infectious LASV progeny, as well as reduced levels of both viral RNA and protein. These results point to a supporting role of GSPT1 in LASV gene expression, which could reflect a critical dependency of LASV on cellular translation termination machinery for its propagation in host cells. Whether the proviral phenotype we observed for GSPT1 is indirectly mediated by its involvement in regulating the cell cycle or apoptosis remains to be determined. Future studies are needed to resolve the functional consequence of the physical association between GSPT1 and LASV polymerase in LASV gene expression and to verify whether GSPT1 contributes to any step of LASV RNA synthesis independent from its potential impact on LASV protein translation.
CC-90009, a CRL4CRBN E3 ligase-modulating drug that specifically targets GSPT1 for proteasomal degradation (22), exhibited antiviral activity against both LASV and LCMV without appreciable cytotoxicity. For both viruses, CC-90009 treatment reduced accumulation of viral RNA and protein in infected cells, suggesting that similar mechanisms drive CC-90009 antiviral activity against LASV and LCMV. Based on the physical interaction between LASV L and GSPT1, it is plausible that LASV L protein might have a higher turnover rate upon CC-90009 treatment. However, due to the lack of access to an antibody to LASV L, we were not able to directly monitor L protein in our viral growth curve experiment. Further validation of this hypothesis can be enabled by monitoring the turnover rate of an epitope or reporter-tagged LASV L in CC-90009-treated cells in the presence or absence of GSPT1.
CC-90009 had EC50 values of 16.86 and 14.85 nM for LASV and LCMV, respectively, in Huh7 cells. We did not observe any significant reduction in cell viability even at the highest (10 µM) concentration of CC-90009 used (CC50 > 10 µM). Importantly, our recent work has shown a potent inhibitory effect of CC-90009 against Zaire Ebola virus growth in cell culture (100-fold reduction in viral titer with 1 μM CC-90009 treatment postinfection) (46). As a comparison, the SARS-CoV-2 polymerase inhibitor molnupiravir, which has been recently issued with an FDA Emergency Use Authorization for COVID-19 treatment, was reported to have an EC50 = 300 nM against SARS-CoV2 in cultured cells (47). CC-90009 is currently in phase 1 clinical development for the treatment of acute myeloid leukemia, raising the possibility of its repurposing as a broad-spectrum antiviral therapy against LASV and Ebola virus infections.
There are several limitations in the current work to be discussed. First, the use of LASV L-WT as the control in our proximity proteomic experiment does not control for nonspecific interactions associated with the TurboID tag in the LASV L-HA-TurboID fusion protein. This might have resulted in some false positives in our list of proteomic hits. However, a control proximity proteome generated by TurboID alone could have resulted in the elimination of cellular proteins that interact with LASV L, because both LASV L and TurboID are cytoplasmic proteins that have no clear physical boundary that would constrain their localizations. To support the specificity of our interactome, we biochemically validated a subset of the identified interactors via coIP using an HA-tagged L that lacks TurboID. We confirmed that 7/8 (87%) tested hits interacted with LASV L. We acknowledge that efficiency of proximity proteomics using TurboID is influenced by the accessibility of lysine residues on the bait-interacting proteins. The list of LASV L-interacting proteins we revealed with proximity proteomics could be complemented by orthogonal proteomic approach to recapitulate the full picture of host-LASV L interactions. Moreover, we utilized the siRNA-based functional screen to support the relevance of LASV L interactome. We confirmed 21 functional hits among 42 interactors (50%) in at least one siRNA screen, including 7 top hits in two independent siRNA screens using different infection conditions. It is possible that some of LASV L interactors we identified in HEK 293T cells are cell-type specific, whereas we performed our siRNA screen in Huh7 cells. We used HEK 293T cells in several experiments because of their high transfection efficiency, and we used Huh7 cells in all viral infection experiments, as liver is a major target organ during LASV infection. Future work should confirm the functional role of LASV L interactors in other primary target cells including macrophage and dendritic cells, as well as in suitable animal models. Taken together, we presented here a cellular interactome of LASV L polymerase, which illuminated mechanisms of host regulation of LASV replication and revealed a landscape of targets for host-directed antiviral drug development.
Materials and Methods
Plasmids, siRNAs, Antibodies, Primers, and Compound.
See SI Appendix, Tables S1–S3 and Dataset S2.
Cell Cultures and Viruses.
See SI Appendix.
LASV MG Assay.
See SI Appendix.
Biotinylation with TurboID Fusion Proteins.
See SI Appendix.
Immunofluorescent Analysis Using Confocal Microscopy.
See SI Appendix.
Proximity-Labeling-Based Proteomics.
See SI Appendix.
siRNA Functional Screening.
Huh7 cells were transfected with individual gene-targeting siRNA and infected with rLASV-eGFP (MOI = 0.5 or 1). Percentage of LASV-infected cells and total cell counts were quantified at 48 h.p.i. See SI Appendix for details.
LASV Growth Kinetics.
Huh7 cells were transfected with siRNA targeting GSPT1 and infected with rLASV-eGFP (MOI = 0.5) 48 h posttransfection or were infected with rLASV-eGFP (MOI = 0.5) and treated with CC-90009 at the specified concentration 1 h.p.i. At indicated time points, LASV vRNAs or c/mRNAs, proteins, and titers were quantified. See SI Appendix for details.
LCMV Growth Kinetics.
rLCMV-GFP-infected Huh7 cells were treated with CC-90009 at the specified concentration 1 h.p.i. At indicated time points, LCMV RNAs, proteins, and titers were quantified. See SI Appendix for details.
Determination of EC50 and CC50 for CC-90009.
See SI Appendix.
CoIP.
See SI Appendix.
RT-qPCR.
See SI Appendix.
Supplementary Material
Acknowledgments
We thank Beatrice Cubitt from the de la Torre Lab (Scripps Research, CA) for helping with the cloning of pCI-FLAG-GSPT1 plasmid and preparation of LASV MG plasmid stocks. We thank Sharon Schendel at La Jolla Institute for Immunology (LJI) for manuscript editing, Zbigniew Mikulski of the Microscopy Core Facility (LJI) for microscopy training, and NIH S10OD021831 for sponsoring the Zeiss laser-scanning microscopy (LSM) 880 microscope. We thank Paul Schimmel (Scripps Research, CA) for providing us with anti-RARS and anti-AIMP2 antibodies and Tianying Zhang (Scripps Research, CA) for the mouse monoclonal antibody to LCMV GP2. This research was supported by institutional funds of LJI (E.O.S.) and NIH/NIAID grants AI125626 and AI128556 (J.C.d.l.T.). J.F. was supported by the Donald E. and Delia B. Baxter Foundation Fellowship. This is publication # 30107 from Scripps Research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.P.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201208119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Proteomic data are shown as Dataset S01 in SI Appendix.
References
- 1.Günther S., Lenz O., Lassa virus. Crit. Rev. Clin. Lab. Sci. 41, 339–390 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Asogun D. A., Günther S., Akpede G. O., Ihekweazu C., Zumla A., Lassa fever: Epidemiology, clinical features, diagnosis, management and prevention. Infect. Dis. Clin. North Am. 33, 933–951 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kofman A., Choi M. J., Rollin P. E., Lassa fever in travelers from West Africa, 1969-2016. Emerg. Infect. Dis. 25, 245–248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rojek J. M., Kunz S., Cell entry by human pathogenic arenaviruses. Cell. Microbiol. 10, 828–835 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Grande-Pérez A., Martin V., Moreno H., de la Torre J. C., Arenavirus quasispecies and their biological implications. Curr. Top. Microbiol. Immunol. 392, 231–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuller-Pace F. V., Southern P. J., Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: In vitro synthesis of full-length viral RNA species. J. Virol. 63, 1938–1944 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyle J. D., Whelan S. P. J., Isolation of reconstructed functional ribonucleoprotein complexes of Machupo virus. J. Virol. 95, e0105421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortín J., Martín-Benito J., The RNA synthesis machinery of negative-stranded RNA viruses. Virology 479-480, 532–544 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Morin B., Kranzusch P. J., Rahmeh A. A., Whelan S. P., The polymerase of negative-stranded RNA viruses. Curr. Opin. Virol. 3, 103–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferron F., Weber F., de la Torre J. C., Reguera J., Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 234, 118–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng R., et al. , Structural insight into arenavirus replication machinery. Nature 579, 615–619 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Branon T. C., et al. , Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel D., Rosenthal M., Gogrefe N., Reindl S., Günther S., Biochemical characterization of the Lassa virus L protein. J. Biol. Chem. 294, 8088–8100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Sobrido L., Paessler S., de la Torre J. C., Lassa virus reverse genetics. Methods Mol. Biol. 1602, 185–204 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Fang J., et al. , Staufen1 interacts with multiple components of the Ebola virus ribonucleoprotein and enhances viral RNA synthesis. MBio 9, e01771-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun N. E., Walker D. H., Pathogenesis of Lassa fever. Viruses 4, 2031–2048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caì Y., et al. , Recombinant Lassa virus expressing green fluorescent protein as a tool for high-throughput drug screens and neutralizing antibody assays. Viruses 10, 655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chauvin C., et al. , Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell. Biol. 25, 5801–5811 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhouravleva G., et al. , Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14, 4065–4072 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauvin C., Jean-Jean O., Proteasomal degradation of human release factor eRF3a regulates translation termination complex formation. RNA 14, 240–245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen J. D., et al. , CC-90009: A cereblon E3 ligase modulating drug that promotes selective degradation of GSPT1 for the treatment of acute myeloid leukemia. J. Med. Chem. 64, 1835–1843 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Baradaran-Heravi A., et al. , Effect of small molecule eRF3 degraders on premature termination codon readthrough. Nucleic Acids Res. 49, 3692–3708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surka C., et al. , CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood 137, 661–677 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krönke J., et al. , Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khamina K., et al. , Characterization of host proteins interacting with the lymphocytic choriomeningitis virus L protein. PLoS Pathog. 13, e1006758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasaki M., et al. , Interactome analysis of the lymphocytic choriomeningitis virus nucleoprotein in infected cells reveals ATPase Na+/K+ transporting subunit Alpha 1 and prohibitin as host-cell factors involved in the life cycle of mammarenaviruses. PLoS Pathog. 14, e1006892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leen E. N., et al. , A conserved interaction between a C-terminal motif in norovirus VPg and the HEAT-1 domain of eIF4G is essential for translation initiation. PLoS Pathog. 12, e1005379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgui I., Yángüez E., Sonenberg N., Nieto A., Influenza virus mRNA translation revisited: Is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 81, 12427–12438 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connor J. H., Lyles D. S., Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76, 10177–10187 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gradi A., et al. , A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18, 334–342 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross J. D., et al. , Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115, 739–750 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Lelke M., Brunotte L., Busch C., Günther S., An N-terminal region of Lassa virus L protein plays a critical role in transcription but not replication of the virus genome. J. Virol. 84, 1934–1944 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olschewski S., Cusack S., Rosenthal M., The cap-snatching mechanism of bunyaviruses. Trends Microbiol. 28, 293–303 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Brunotte L., et al. , Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy. J. Biol. Chem. 286, 38748–38756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi X., et al. , Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468, 779–783 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reguera J., et al. , Comparative structural and functional analysis of bunyavirus and arenavirus cap-snatching endonucleases. PLoS Pathog. 12, e1005636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volpon L., Osborne M. J., Capul A. A., de la Torre J. C., Borden K. L., Structural characterization of the Z RING-eIF4E complex reveals a distinct mode of control for eIF4E. Proc. Natl. Acad. Sci. U.S.A. 107, 5441–5446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y. K., Maquat L. E., UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurosaki T., Maquat L. E., Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 129, 461–467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho J. S. Y., et al. , Hybrid gene origination creates human-virus chimeric proteins during infection. Cell 181, 1502–1517; e1523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frolova L., et al. , Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2, 334–341 (1996). [PMC free article] [PubMed] [Google Scholar]
- 44.Chauvin C., Salhi S., Jean-Jean O., Human eukaryotic release factor 3a depletion causes cell cycle arrest at G1 phase through inhibition of the mTOR pathway. Mol. Cell. Biol. 27, 5619–5629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hegde R., et al. , The polypeptide chain-releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein. J. Biol. Chem. 278, 38699–38706 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Fang J., et al. , Functional interactomes of the Ebola virus polymerase identified by proximity proteomics in the context of viral replication. Cell Rep. 38, 110544 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheahan T. P., et al. , An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12, eabb5883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Proteomic data are shown as Dataset S01 in SI Appendix.