Abstract
Misalignment of repeated sequences during DNA replication can lead to deletions or duplications in genomic DNA. In Escherichia coli, such genetic rearrangements can occur at high frequencies, independent of the RecA-homologous recombination protein, and are sometimes associated with sister chromosome exchange (SCE). Two mechanisms for RecA-independent genetic rearrangements have been proposed: simple replication misalignment of the nascent strand and its template and SCE-associated misalignment involving both nascent strands. We examined the influence of the 3′ exonuclease of DNA polymerase III and exonuclease I on deletion via these mechanisms in vivo. Because mutations in these exonucleases stimulate tandem repeat deletion, we conclude that displaced 3′ ends are a common intermediate in both mechanisms of slipped misalignments. Our results also confirm the notion that two distinct mechanisms contribute to slipped misalignments: simple replication misalignment events are sensitive to DNA polymerase III exonuclease, whereas SCE-associated events are sensitive to exonuclease I. If heterologies are present between repeated sequences, the mismatch repair system dependent on MutS and MutH aborts potential deletion events via both mechanisms. Our results suggest that simple slipped misalignment and SCE-associated misalignment intermediates are similarly susceptible to destruction by the mismatch repair system.
Repeated DNA sequences are prone to rearrangements at relatively high frequencies. Both deletion and duplication of genomic DNA at repeated sequences are responsible for several human diseases (12, 13). Expansion of trinucleotide repeat arrays forms the basis of a growing number of diseases in humans (30). To understand the factors that govern genomic stability, it is therefore important to define the molecular mechanisms of repeated sequence rearrangements.
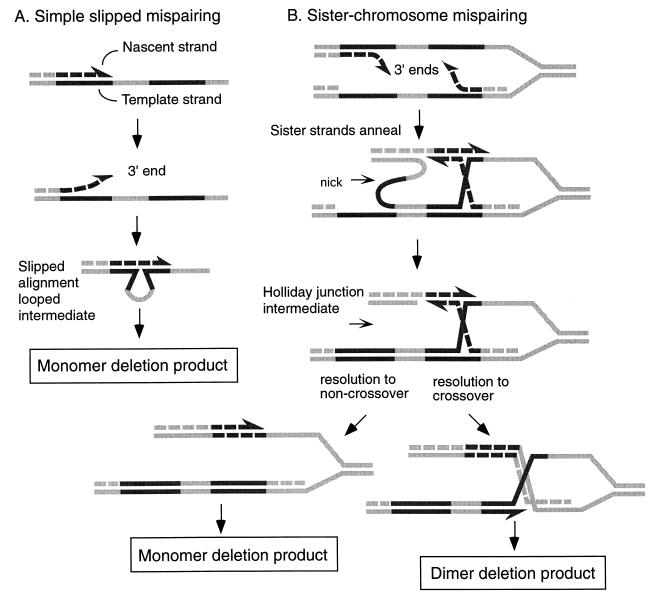
Deletion between repeated sequences in the bacterium Escherichia coli has been studied systematically and has provided evidence for the mechanisms underlying rearrangements of repeated sequences. Sufficiently large homologies (>200 bp) rearrange, in part, via homologous recombination, dependent on the RecA strand transfer protein of E. coli (3). However, rearrangements can also occur efficiently by a RecA-independent “nonrecombinational” mechanism. These rearrangements are dependent on the close proximity of the repeated sequences (3, 5, 19) but can occur between repeats ranging from several to thousands of nucleotides in length (3, 7, 20). It has been proposed that these nonrecombinational rearrangements may occur by slipped misalignment of the repeated sequences during DNA replication (1, 9, 28). The replication slipped misalignment model (Fig. 1A) nicely accounts for the proximity dependence and RecA independence of these events. A replicational mechanism for RecA-independent rearrangements is supported by experimental evidence (18, 29). Furthermore, mutations in many replication components of E. coli stimulate such rearrangements (4, 27).
FIG. 1.
Replication misalignment models for deletion formation. Newly replicated DNA is denoted with dashed lines. (A) Simple slipped mispairing involves the dislocation of a nascent strand to mispair with a second copy of a repeated sequence on its template, forming a looped misaligned intermediate. A 3′ end may be transiently unpaired and susceptible to 3′ exonucleases during this process. If replication is completed, a monomeric deletion product will result. (B) Sister chromosome mispairing involves the displacement and mispairing of both nascent strands in a stalled replication fork. This mispairing produces a Holliday junction-like intermediate which may resolve as a crossover between sister chromosomes, producing a dimeric replicon. Alternative resolution may produce monomeric deletion chromosomes.
However, there is evidence for a second mechanism of RecA-independent rearrangements, associated with replication but distinguished by its molecular products. Analysis of repeat rearrangements occurring on circular molecules suggests that a crossover event involving sister chromosomes is sometimes associated with repeat rearrangements (17, 21). Such sister-chromosome exchange (SCE) events on circular plasmids are detected as dimeric plasmid molecules. These dimeric molecules are not easily explained by a simple replication slippage model, and we have proposed a slipped misalignment mechanism involving sister strands across the replication fork (10, 17). This mechanism (Fig. 1B) resembles recombination in certain respects but is independent of recombination strand transfer protein RecA. This contrasts with rearrangements on plasmids which are recovered as monomeric products, which are consistent with the simple slipped mispairing model where a nascent DNA strand is dislocated relative to its template.
Both the simple replication slippage model and the SCE-associated slippage model (Fig. 1) have several common features. Both types of rearrangements may be promoted by stalled replication. Both models invoke rearrangements promoted by a displaced 3′ strand which subsequently mispairs with a second copy of the sequence. However, SCE-associated slippage involves displacement of both leading and lagging nascent strands; simple slippage can occur by displacement of one or the other nascent strand. In addition, the simple slippage model invokes a looped intermediate that may not exist in SCE-associated slippage.
It is therefore of interest to determine which genetic factors may differentially influence simple slippage or SCE-associated slippage events. In this work, we examined the sensitivity of these events to DNA nucleases. Mutations abolishing the 3′ exonuclease associated with DNA polymerase III (dnaQ) and the 3′ single-stranded DNA exonuclease, exonuclease I (sbcB), were examined for effects on the rate of deletion between 101- and 787-bp tandem repeats. Plasmid deletion products were examined to determine whether simple slippage (producing monomer products) or SCE-associated slippage (producing dimeric products) was affected. Our experiments confirm that displaced 3′ ends are a common intermediate in slipped misalignments. In addition, we show that SCE-associated deletion events are differentially susceptible to exonuclease I whereas the simple slippage events are sensitive to DnaQ. We also compared the susceptibility of simple slippage and SCE-associated deletions to the mismatch repair system. We have previously shown that heterologies between repeated sequences elicit mismatch repair, thereby resulting in a reduced deletion rate between heterologous repeats (18). We show here that both simple slippage and SCE-associated tandem repeat deletions are aborted by the mismatch repair system. This exclusion of deletion via simple slippage and SCE-associated slippage requires MutS and partially requires the MutH endonuclease. Therefore, simple slippage and SCE-associated slippage intermediates are similarly accessible to mismatch repair.
MATERIALS AND METHODS
Bacterial strains and growth.
All strains used are derived from the E. coli K-12 strain AB1157 [F− thi-1 hisG4 Δ(gpt-proA)62 argE3 thr-1 leuB6 kdgK51 rfbD1 ara-14 lacY1 galK2 xyl-5 mtl-1 tsx-33 supE44 rpsL31 Rac− λ− (2)]. Strain JC10287 [AB1157 Δ(srlR-recA)304] was obtained from R. Kolodner. Strain STL1671 [AB1157 sbcB15 Δ(srlR-recA)304] was constructed by P1 virA transduction using donor strain JC10287 and recipient strain STL4477 (AB1157 sbcB15 cysC95::Tn10) and selecting for Cys+ and UV. Strains STL2314 [AB1157 dnaQ49 Δ(srlR-recA)304] and STL2172 [AB1157 mutS201::Tn5 Δ(srlR-recA)304] have been described previously (18, 27). Strain STL3926 (mutH34 recA::cam) was constructed by P1 virA transduction using donor strain JJC432 (leuB6 hisG4 argE3 lacY1 galK2 ara-14 xyl-5 mtl-1 tsx-33 rpsL31 supE44 hsdR recD1901::Tn10 recA::cam) obtained from B. Michel and recipient strain ES1582 (AB1157 mutH34) obtained from M. Marinus and selecting for chloramphenicol resistance.
Strains were grown on Luria-Bertani (LB) or 56/2 minimal medium (31). Strain STL2314 was grown at 30°C and assayed at 37°C. All other strains were grown and assayed at 37°C. Transductions were performed on LCG medium, which consisted of LB medium supplemented with 1% glucose and 2 mM calcium chloride. Antibiotics used were ampicillin at 100 μg/ml, tetracycline at 15 μg/ml, and chloramphenicol at 15 μg/ml.
Deletion assays and analysis of plasmid deletion products.
Deletion was assayed using previously described plasmids pSTL55 (17), pSTL57 (19), and pSTL113 (18). All plasmids used are pBR322 derived and contain a functional copy of bla, which confers ampicillin resistance, and various repeated sequences in the tetA gene. Plasmid pSTL55 contains a 787-bp exact sequence duplication in tetA, and plasmid pSTL57 contains a 101-bp exact sequence duplication in tetA. Plasmid pSTL113 contains an imperfect 101-bp repeat sequence disrupting tetA, the two repeats differing by four bases positioned at intervals of 21 bp (18). These plasmids were introduced into respective strains by electroporation (8) or TSS transformation (6).
Deletion was assayed as described previously (17) for a total of 31 to 64 independent isolates. Briefly, independent cultures were prepared in liquid media, diluted, and plated. The number of Tcr colonies was compared to the total number of Apr colonies, and deletion rates were calculated by the method of the median (15), using the following formula: deletion rate = M/N, where M is the calculated number of deletion events and N is the final average number of Apr cells in the 1-ml cultures. M is determined by interpolation from experimental determination of r0, the median number of Tcr cells, determined by using the formula r0 = M(1.24 + ln M). A 95% confidence interval was determined as described previously (27).
Dimer and monomer deletion products were examined as described previously (27). Briefly, plasmids were purified by phenol-chloroform extraction and subjected to electrophoresis on a 0.8 to 1% agarose gel for size determination. Independently isolated deletion products were analyzed.
RESULTS
Mutations in 3′ exonucleases stimulate deletion formation via different mechanisms.
A proposed intermediate of either a simple template slippage or an SCE-associated deletion event involves a stalled replication fork (Fig. 1). Since stalled replication liberates 3′ DNA ends whose realignment may initiate deletion formation, we wanted to examine the status of 3′ DNA ends during the deletion event. We tested the effects of two genes encoding 3′-specific exonucleases, sbcB encoding exonuclease I and dnaQ encoding the DNA polymerase III proofreading exonuclease, on tandem repeat deletion. Strains were constructed to combine mutations in these exonuclease genes with a deletion of the recA gene, so that RecA-dependent homologous recombination events would be prohibited and only RecA-independent slipped misalignment events could be detected using our deletion assays.
Two plasmids were used to detect deletions of homologous repeated sequences: pSTL55 and pSTL57. Plasmid pSTL55 (17) carries a 787-bp duplication in tetA which disrupts the gene. Deletion of one copy of the repeat restores the integrity of the tetA gene, and can be selected for by tetracycline resistance. Plasmid pSTL57 (19) carries an analogous but shorter 101-bp duplication in tetA. Deletion rates were calculated by fluctuation analysis of independent cultures. In addition, the products of independent deletion events were examined by plasmid purification and agarose gel electrophoresis to determine whether replicon dimerization had accompanied the selected deletion. This allowed us to calculate the rates of monomer-producing and dimer-producing deletion events. Dimer-producing deletion events are presumed to be the result of SCE-associated slipped misalignments. Monomer-producing deletion events may be simple slippage events or SCE-associated events that are resolved as monomer products. Holliday junction intermediates, proposed for the SCE-associated deletion pathway, may be theoretically resolved in one of two ways (11), producing, in the case of circular molecules, either monomeric or dimeric products (Fig. 1B).
In recA strains, the deletion rates determined using the two assay plasmids were similar, although deletion of the larger duplication in pSTL55 yielded a higher proportion of dimeric products (Table 1). A mutation in the exonuclease I gene, sbcB15, caused a dramatic increase, six- to eightfold, in the rate of dimeric (SCE-associated) deletion products formed from pSTL55 or pSTL57. Because pSTL55 yielded proportionately more dimeric products, sbcB15 also caused a concomitant increase in its overall deletion rate; it merely shifted the dimer distribution for pSTL57. The rate of the monomeric deletion products was also increased sixfold for pSTL55, but only a slight increase was seen for pSTL57. This suggests that many SCE-associated slippage events are aborted by the 3′ single-stranded DNA exonuclease, exonuclease I. Simple slippage intermediates may be similarly susceptible to exonuclease I; however, the stimulation of monomer deletion products could be due to SCE events resolved as monomer products (Fig. 1B). The assumption that 40 to 60% of SCE events are resolved to monomers explains the increase in monomer rate for both deletion assay plasmids.
TABLE 1.
Deletion rates and distribution of products in 3′ exonuclease-deficient backgrounds
| Strain | Plasmida | Deletion rate, 105 (CI)b | % Dimer products (nc) | Rate (monomer), 105 | Rate (dimer), 105 |
|---|---|---|---|---|---|
| recA | pSTL57 | 4.9 (3.6–6.7) | 4.2 (118) | 4.7 | 0.21 |
| sbcB recA | pSTL57 | 7.6 (5.9–11) | 21 (168) | 6.0 | 1.6 |
| dnaQ recA | pSTL57 | 6.9 (3.3–9.4) | 5.2 (115) | 6.5 | 0.36 |
| recA | pSTL55 | 5.5 (4.0–6.1) | 39 (82) | 3.3 | 2.1 |
| sbcB recA | pSTL55 | 36 (26–42) | 33 (177) | 24 | 12 |
| dnaQ recA | pSTL55 | 26 (16–30) | 16 (93) | 22 | 4.1 |
pSTL57 carries a 101-bp duplication in tetA; pSTL55 carries a 787-bp duplication.
Deletion rates were calculated by the method of the median, and the 95% confidence intervals (CI) for the individual rate calculations are shown.
n, number of independent deletion isolates analyzed.
In contrast, a mutation in the gene for the 3′ exonuclease associated with DNA polymerase III, dnaQ, caused an increase only of the monomeric products. Again, as above, a significant sixfold increase was seen for deletion of the larger 787-bp duplication in pSTL55, but only minor effects were detected on deletion of the 101-bp repeat in pSTL57. This suggests that DNA polymerase III 3′ exonuclease aborts simple slippage events (as previously observed by us [27]), although its effect is more pronounced for the 787-bp repeat deletion.
Mismatch repair genes abort deletion between homeologous repeats via both misalignment mechanisms.
To examine the accessibility of deletion intermediates to mismatch repair, we used a deletion assay plasmid, pSTL113, containing an imperfect 101-bp repeat with four silent heterologies. This “homeologous” deletion can be compared to the deletion assayed by plasmid pSTL57, containing perfect 101-bp repeats. We examined recA mutant strains with an intact mismatch repair system and those carrying mutations in MutS, the mismatch repair recognition protein, or MutH, the endonuclease which cleaves at hemimethylated GATC sites to initiate mismatch correction. Previously, we have shown that mismatch repair dependent on MutHLS aborts deletion of homeologous repeats but has no effect on perfectly homologous repeats (18). The presence of mismatches in the heteroduplex intermediate of slipped misalignment presumably elicits excision and hence leads to the destruction of deletion intermediates. However, in this previous study, we did not examine the products of the deletion events and therefore could not ascertain differential effects of mismatch repair on simple slipped misalignment versus SCE-associated misalignment events. SCE-associated deletion events involve a recombination-like intermediate (Fig. 1B) and may be more accessible to mismatch repair than replicational simple slippage events. Specifically, we wondered whether the SCE-associated events might be excised without the need for the MutH endonuclease because of the presence of DNA ends in its intermediate. Indeed, homeologous RecA-dependent recombination is aborted by mismatch repair without the need for MutH endonuclease (23, 26).
The rate of deletion between the perfect repeats (without heterology) was unchanged in mismatch repair-proficient or -deficient strains (Table 2). Approximately 4 to 7% of the recovered deletion events were accompanied by SCE, producing dimeric products. The rate of deletion between the two heterologous repeats in a recA mutant strain was decreased by 400-fold as compared to the rate of deletion of fully homologous repeats. Examination of products showed that the SCE-associated homeologous deletion events were also susceptible to exclusion by mismatch repair, showing a 100-fold decrease relative to that seen for homologous repeats. (Dimeric products were more abundant among deletion events for the homeologous repeats [16%] than for the homologous repeats [4%], but this is not a consequence of mismatch repair as MutS− and MutH− strains show similar dimer frequencies.) A mutation in MutS, the recognition component of mismatch repair, almost fully restored deletion between heterologous repeats, by both monomer- and dimer-producing deletion pathways. A mutation in the MutH endonuclease partially restored deletion rates between homeologous repeats to about 25% of the level seen in the MutS− strain. Contrary to our expectation, exclusion of SCE-associated homeologous deletion events, producing dimeric products, was not more independent of MutH. This indicates that MutH cleavage is important in allowing access to the mismatch sites in intermediates of both simple slippage and SCE-associated slippage intermediates. Presumably, some fraction of the intermediates of both types can initiate mismatch repair without MutH incision, causing deletion rates to be restored only partially by a mutation in MutH as compared to full restoration by a mutation in MutS. Previously, we had observed full rather than partial restoration of homeologous deletion by mutations in MutH (18); this difference may be due to the presence of RecA+ in strains used in the former experiments, which may inhibit MutH-independent mismatch excision.
TABLE 2.
Deletion rates and distribution of products in mismatch repair-deficient backgrounds
| Strain | Heterol-ogy | Deletion rate, 106 (CI)a | % Dimer products (nb) | Rate (monomer), 106 | Rate (dimer), 106 |
|---|---|---|---|---|---|
| recA | − | 49 (36–67) | 4.2 (118) | 47 | 2.1 |
| mutS recA | − | 52 (20–134) | 7.3 (96) | 48 | 3.8 |
| mutH recA | − | 30 (16–64) | 4.2 (112) | 28 | 1.3 |
| recA | + | 0.12 (0.08–1.8) | 16 (103) | 0.11 | 0.019 |
| mutS recA | + | 26 (17–43) | 16 (175) | 22 | 4.2 |
| mutH recA | + | 6.8 (2.8–14) | 10 (102) | 6.1 | 0.7 |
Deletion rates were calculated by the method of the median, and the 95% confidence intervals (CI) for the individual rate calculations are shown. Deletion assay plasmids pSTL57 (carrying 101-bp perfect repeats) and pSTL113 (carrying 101-bp repeats with four silent heterologies) were used in these experiments.
n, number of independent deletion isolates analyzed.
DISCUSSION
Susceptibility of slipped misalignment intermediates to 3′ exonucleases.
A deletion between repeated sequences may proceed through replication misalignment, during which a newly replicated single strand of DNA is initially displaced from the template and subsequently reannealed to it at another homologous site. During or after the displacement of the nascent strand, the replication misalignment models (Fig. 1) predict that the 3′ end of the single-stranded DNA should be vulnerable to degradation by 3′ exonucleases. Our finding that deletion events are stimulated by mutations in 3′ exonucleases supports these models in which deletion formation is initiated via displaced 3′ ends. Moreover, the differential effects of exonuclease I and polymerase III exonuclease on monomeric and dimeric deletion products support the idea that two distinct misalignment mechanisms contribute to deletion formation.
Our results show that a mutation in exonuclease I, the major 3′ single-stranded DNA exonuclease of E. coli (14, 16), increases the rate of SCE-associated deletion of tandem repeated sequences. Therefore, SCE-associated deletion intermediates are substrates for exonuclease I-mediated degradation, or, more precisely, a 3′ single-stranded DNA end is present and accessible to degradation by that enzyme during the deletion event. Because exonuclease I is highly specific for single-stranded DNA (16, 25), this argues that the 3′ end is substantially unpaired at some time during the deletion process. SCE-associated deletion may occur when DNA polymerase III has dissociated from its template, thereby freeing both 3′ nascent strands and rendering them vulnerable to exonuclease I if displaced. A mutation in the gene for the polymerase subunit of DNA polymerase III, dnaE486, also differentially increases SCE-associated deletion (27), perhaps because it encourages polymerase dissociation. Mutations in exonuclease I may also increase the recovery of simple replicational misalignments, which produces monomeric products in our assays. However, it is possible that some SCE-associated deletion events are resolved as monomeric products. A random resolution of the proposed Holliday junction in SCE-associated misalignment to monomer or dimer products accounts for the observed increase brought about by sbcB in our assays.
A mutation in the gene for the DNA polymerase III 3′ proofreading exonuclease, dnaQ, also stimulated deletion but specifically affected the monomeric products. Our results (previously reported for the 787-bp repeats [27]) may imply that simple slippage deletion intermediates are destroyed by the polymerase III exonuclease. However, because a dnaQ mutation did not significantly stimulate the dimeric products of either construct, SCE-associated deletion intermediates may be resistant to the polymerase III exonuclease. In addition, we observed that deletion of the larger 787-bp repeated sequence was more sensitive to polymerase III exonuclease than the smaller 101-bp repeat. This may be a property of the repeat length or may be a sequence-specific effect.
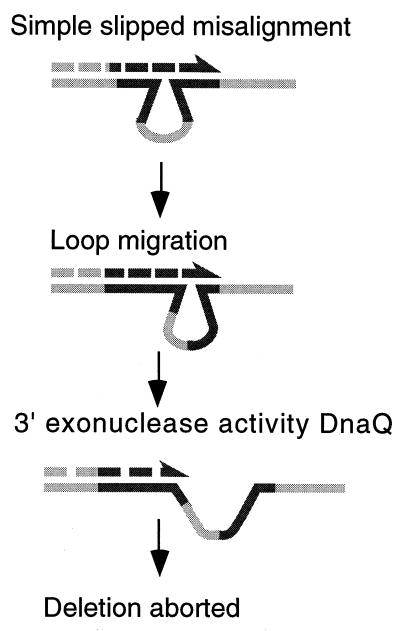
The sensitivity of simple slippage to DnaQ may result from the fact that polymerase III remains associated with the fork during these events and would be in place to attack any displaced 3′ strand. This effect may be more pronounced for misalignment of the larger repeats because more extensive displacement is necessary to effect realignment. (For SCE-associated events, the polymerase may have dissociated and so is not on hand to degrade the displaced 3′ intermediates.) An alternative explanation for our results is that the looped intermediate formed during simple slippage, by realignment of the nascent strand with its template, may be especially vulnerable to polymerase exonucleolytic degradation. The template loop may be free to migrate, and its approach to the 3′ nascent end will destabilize the heteroduplex intermediate (Fig. 2). The rationale for the lesser effect of dnaQ on the smaller repeat is unclear, although it may be that sequence-specific effects stabilize the looped intermediate in the 101-bp region. The intermediate for SCE-associated deletion does not contain a loop and may therefore be inert to destabilization by polymerase III exonuclease.
FIG. 2.
Instability of the looped intermediate of simple slipped misalignment. During deletion formation, the mispairing of nascent and template strands produces a loop on the template strand. Migration of the loop within the repeat can destabilize the intermediate by shortening the heteroduplex pairing region adjacent to the nascent 3′ end. Degradation of the 3′ end by DNA polymerase III (DnaQ) can also destabilize the intermediate and abort potential deletion events.
Alternatively, dnaQ49 mutations may more actively promote simple slippage. As the DnaQ subunit, ɛ, is a component of the core polymerase, it may result in destabilization of the entire polymerase complex. If this is the case, it is difficult to reconcile dnaQ’s effect strictly on simple slippage with the observation that mutations in the gene for the α polymerase subunit itself, dnaE486, preferentially stimulate the SCE-associated deletion pathway (27). A more attractive possibility is that misincorporations by the polymerase in the absence of the proofreading subunit block polymerase extension, providing increased opportunity for simple slipped misalignments. Such a “misincorporation plus slippage” model has been proposed for −1 frameshift mutations produced by polymerase III in vitro (24). The polymerase misincorporation rates in vitro and in vivo, in the absence of proofreading, are sufficiently high (approximately 10−6/base [24]) to account for our observed rates of deletion in dnaQ49 mutants. After the looped slippage intermediate has been formed (as in Fig. 1A), the terminal mismatched base may be removed by the proofreading activities of polymerase I or polymerase II or by other 3′ single-stranded DNA exonucleases, followed by polymerization to form the simple deletion product. (In sequence analysis of deletion products, we have not observed base substitution mutations associated with deletion in dnaQ49 mutS recA strains.) The larger repeat presents a larger target for misincorporation and, for this reason, may be affected by dnaQ49 more severely than the small repeat. If this model is correct, then SCE-associated deletion events must not be stimulated by such misincorporations, perhaps because they are too transient or do not cause polymerase dissociation necessary to elicit strand exchange.
Mismatch repair access to slipped misalignment intermediates.
Mismatched bases, such as may result from polymerase errors, are excised by the MutHLS mismatch repair system in E. coli. A MutSL complex recognizes the mismatch in duplex DNA. MutH provides a nick to the unmethylated strand of a mismatched duplex; this nick serves as an entry point allowing degradation (and subsequent correction) of the newly synthesized strand (22). The mismatch repair system also aborts genetic rearrangements when heterologies are present between otherwise homologous sequences. Mismatch repair that aborts homeologous RecA-dependent recombination does not require MutH (23, 26). This may be because the mismatch excision proteins are able to utilize an already present DNA end of the recombinational intermediate as an entry site to initiate degradation. In contrast, RecA-independent deletion of homeologous tandem repeats is sensitive to MutH and Dam methylation presumably because the heteroduplex intermediate forms during the context of normal DNA replication and is not accessible without incision (18).
To see if the recombination-like RecA-independent SCE intermediate allows for a bypass of MutH function, we analyzed the efficiency of homeologous repeat deletion in mismatch repair-deficient strains and analyzed the products of such deletion events to determine whether deletion had occurred by simple slippage or by SCE-associated misalignment. The SCE-associated misalignment intermediate in our model (Fig. 1B) predicts a mismatched heteroduplex formed by newly replicated strands, which because they are unmethylated should be susceptible to incision by MutH. Subsequent excision of either strand would lead to destabilization and loss of the critical intermediate. Our data indicated that the rate of SCE-associated deletion events was depressed in strains with an intact mismatch repair system when heterologies existed between repeats. SCE-associated homeologous deletion was restored in both mutS and mutH backgrounds, which suggests that MutH plays a significant role in allowing the mismatch excision proteins to access and degrade the mismatched heteroduplex intermediate. Therefore, our results indicate that the intermediates of the SCE-associated deletion pathway are similar to those of the simple slippage pathway in their accessibility to mismatch repair.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants T32 GM07122 (to M.B. and C.J.S.) and RO1 GM43889 and RO1 GM51753.
REFERENCES
- 1.Albertini A M, Hofer M, Calos M P, Miller J H. On the formation of spontaneous deletion: the importance of short sequence homologies in the generation of large deletions. Cell. 1982;29:319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 3.Bi X, Liu L F. recA-independent and recA-dependent intramolecular plasmid recombination: differential homology requirement and distance effect. J Mol Biol. 1994;235:414–423. doi: 10.1006/jmbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 4.Bierne H, Vilette D, Ehrlich S D, Michel B. Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol Microbiol. 1997;24:1225–1234. doi: 10.1046/j.1365-2958.1997.4381795.x. [DOI] [PubMed] [Google Scholar]
- 5.Chedin F, Dervyn E, Dervyn R, Ehrlich S D, Noirot P. Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol Microbiol. 1994;12:561–570. doi: 10.1111/j.1365-2958.1994.tb01042.x. [DOI] [PubMed] [Google Scholar]
- 6.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dianov G L, Kuzminov A V, Mazin A V, Salganik R I. Molecular mechanisms of deletion formation in Escherichia coli plasmids I. Deletion formation mediated by long direct repeats. Mol Gen Genet. 1991;228:153–159. doi: 10.1007/BF00282460. [DOI] [PubMed] [Google Scholar]
- 8.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efstratiadis A, Psalony J W, Maniatis T, Lawn R M, O’Connell C, Spritz R A, DeRiel J K, Forget B G, Weissman S M, Slightom J L, Blechl A E, Smithies O, Baralle F E, Shoulders C C, Proudfoot N J. The structure and evolution of the human β-globin gene family. Cell. 1980;21:653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- 10.Feschenko V V, Lovett S T. Slipped misalignment mechanisms of deletion formation: analysis of deletion endpoints. J Mol Biol. 1998;276:559–569. doi: 10.1006/jmbi.1997.1566. [DOI] [PubMed] [Google Scholar]
- 11.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 12.Hu X, Worton R G. Partial gene duplication as a cause of human disease. Hum Mutat. 1992;1:3–12. doi: 10.1002/humu.1380010103. [DOI] [PubMed] [Google Scholar]
- 13.Krawczak M, Cooper D N. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environments. Hum Genet. 1991;86:425–441. doi: 10.1007/BF00194629. [DOI] [PubMed] [Google Scholar]
- 14.Kushner S R, Nagaishi H, Templin A, Clark A J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 16.Lehman I R, Nussbaum A L. The deoxyribonucleases of Escherichia coli: I. Purification and properties of a phosphodiesterase. J Biol Chem. 1964;235:1479–1487. [PubMed] [Google Scholar]
- 17.Lovett S T, Drapkin P T, Sutera V A, Jr, Gluckman-Peskind T J. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics. 1993;135:631–642. doi: 10.1093/genetics/135.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovett S T, Feschenko V V. Stabilization of diverged tandem repeats by mismatch repair: evidence for deletion formation via a misaligned replication intermediate. Proc Natl Acad Sci USA. 1996;93:7120–7124. doi: 10.1073/pnas.93.14.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lovett S T, Gluckman T J, Simon P J, Sutera V A, Jr, Drapkin P T. Recombination between repeats in Escherichia coli by a recA-independent, proximity-sensitive mechanism. Mol Gen Genet. 1994;245:294–300. doi: 10.1007/BF00290109. [DOI] [PubMed] [Google Scholar]
- 20.Mazin A V, Kuzminov A V, Dianov G L, Salganik R I. Mechanisms of deletion formation in Escherichia coli plasmids. Mol Gen Genet. 1991;228:209–214. doi: 10.1007/BF00282467. [DOI] [PubMed] [Google Scholar]
- 21.Mazin A V, Timchenko T V, Saparbaev M K, Mazina O M. Dimerization of plasmid DNA accelerates selection for antibiotic resistance. Mol Microbiol. 1996;20:101–108. doi: 10.1111/j.1365-2958.1996.tb02492.x. [DOI] [PubMed] [Google Scholar]
- 22.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 23.Petit M-A, Dimpfl J, Radman M, Echols H. Control of large chromosomal duplications in Escherichia coli by the mismatch repair system. Genetics. 1991;129:327–332. doi: 10.1093/genetics/129.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pham P T, Olson M W, McHenry C S, Schaaper R M. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J Biol Chem. 1998;273:23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- 25.Prasher D C, Conarro L, Kushner S R. Amplification and purification of Exonuclease I from Escherichia coli K12. J Biol Chem. 1983;258:6340–6343. [PubMed] [Google Scholar]
- 26.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 27.Saveson C J, Lovett S T. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics. 1997;146:457–470. doi: 10.1093/genetics/146.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye I. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:77–86. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Trinh T Q, Sinden R R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in Escherichia coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 30.Warren S T. The expanding world of trinucleotide repeats. Science. 1996;271:1374–1375. doi: 10.1126/science.271.5254.1374. [DOI] [PubMed] [Google Scholar]
- 31.Willetts N S, Clark A J, Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]