Abstract
Rapid advances in automated methods for extracting large numbers of quantitative features from medical images have led to tremendous growth of publications reporting on radiomic analyses. Translation of these research studies into clinical practice can be hindered by biases introduced during the design, analysis, or reporting of the studies. Herein, the authors review biases, sources of variability, and pitfalls that frequently arise in radiomic research, with an emphasis on study design and statistical analysis considerations. Drawing on existing work in the statistical, radiologic, and machine learning literature, approaches for avoiding these pitfalls are described.
© RSNA, 2022
Summary
This review highlights biases and inappropriate methods used in radiomic research that can lead to erroneous conclusions; addressing these issues will accelerate translation of research to clinical practice and have the potential to positively impact patient care.
Essentials
■ Many radiomic research studies are hindered by systematic biases.
■ In addition to ongoing initiatives for standardization, improvements in study design, data collection, rigorous statistical analysis, and thorough reporting are needed in radiomic research.
■ Insight into potential problems and suggestions for how to circumvent common pitfalls in radiomic studies are provided.
Introduction
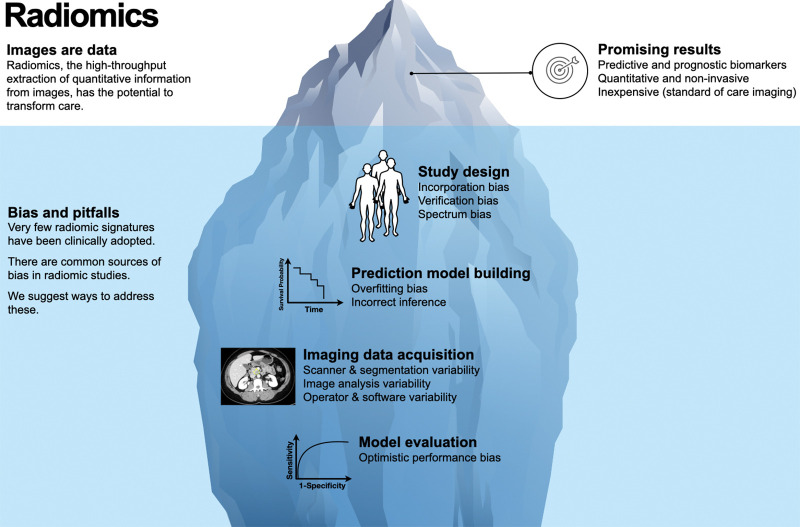
Rapid advances in automated methods for extracting large numbers of quantitative features from medical images have led to an explosion of publications exploring combinations of features as imaging biomarkers for diagnosis, clinical prognosis, treatment selection, or other decision support (1,2). Radiomics is a term used to describe both the automatic conversion of medical imaging data to quantifiable features and the quantified features themselves; these features may include well-known imaging descriptors, such as Hounsfield units, or more exploratory features such as gray-level texture or machine learned features. However, a sobering consideration is that only a small fraction of quantitative imaging biomarkers is clinically adopted. Moreover, to our knowledge, there are no radiomic signatures that are identified by way of a high-throughput pipeline in widespread clinical use (3–5). Despite the great promise it holds, radiomics research is susceptible to hidden obstacles (Fig 1).
Figure 1:
Diagram illustrates how the potential of radiomics can be weighed down by multiple sources of bias and variability that are often overlooked and require careful consideration for the field to be successful. Radiomics analysis has shown promise for generating imaging biomarkers, which is well described in the literature.
While considerable progress has been made in radiomic biomarker taxonomy and standardization (6–8), commensurate attention has not been paid to the design and conduct of radiomic studies for imaging biomarker discovery. As a result, many published studies harbor systematic biases or do not include sufficient information for readers to interpret findings in an appropriate context (9). Herein, we discuss study design and statistical analysis considerations for radiomic studies, drawing from our recent experience as collaborating statisticians and computer scientists and as reviewers for journals such as Radiology. It is not our intention to provide an in-depth review of technical radiomic features (eg, gray scale and bin width); we refer readers elsewhere for that (7,10–14). We do not provide a comprehensive list of sources of bias and variability or a ranking of their impact. Instead, our aim is to highlight common pitfalls that we have observed in the design and statistical analysis of radiomic studies and to suggest ways to potentially circumvent them. Our goal is to facilitate high-quality results with the potential for widespread positive impact on patient care.
Study Design Considerations
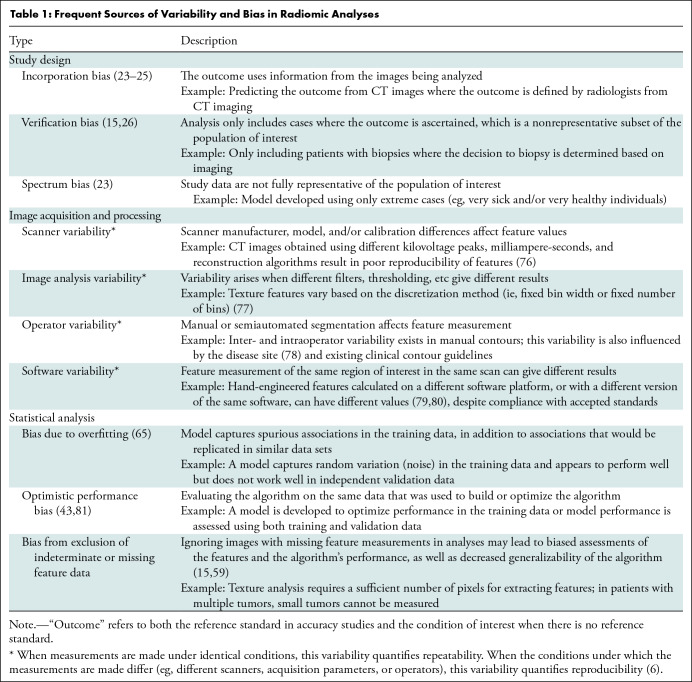
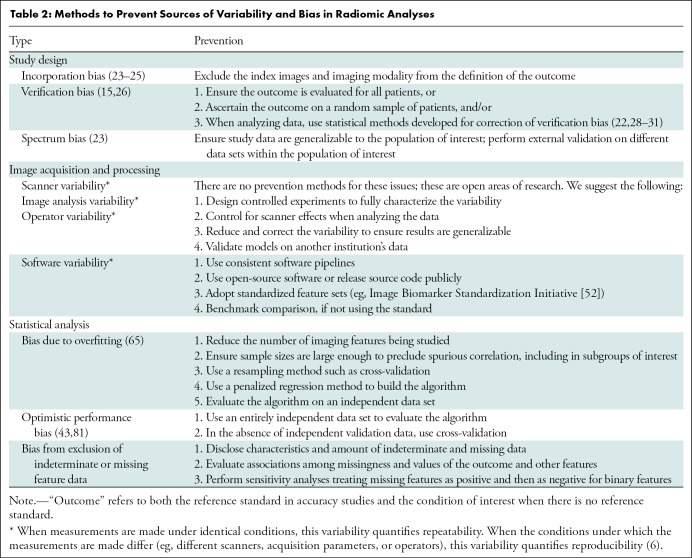
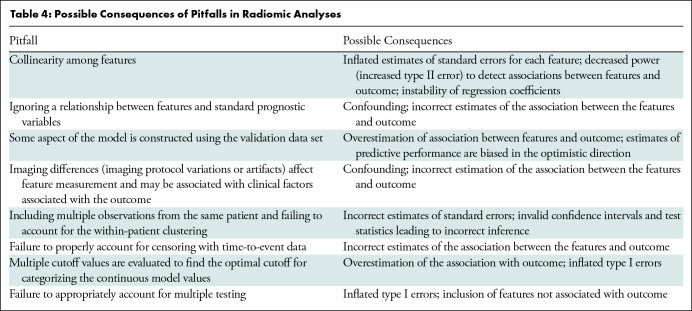
Radiomic analyses require the availability of patient images acquired as part of clinical practice or a clinical trial. These analyses are susceptible to several different biases (Table 1), because systematic error in the research process can lead to erroneous conclusions. Bias may be minimized by careful study design, calculating sample size to yield sufficient power to detect clinically meaningful differences, and prespecification of hypotheses, research objectives, the nature and sources of variables of interest, potential confounders, and appropriate analysis (Tables 2–4). Best practices for design, conduct, and analysis are well developed for diagnostic, prognostic, and predictive models and are applicable to radiomic studies (15–22). In the paragraphs that follow, we alert readers to design choices that occur frequently and lead to bias.
Table 1:
Frequent Sources of Variability and Bias in Radiomic Analyses
Table 2:
Methods to Prevent Sources of Variability and Bias in Radiomic Analyses
Table 4:
Possible Consequences of Pitfalls in Radiomic Analyses
Table 3:
Examples of Potential Pitfalls in Radiomic Analyses
Definition of Outcome
The primary outcome of interest should be defined at study onset. In analyses evaluating accuracy, the outcome is typically an abnormal condition assessed with a reference standard. In radiomic analyses where there may be no reference standard, the outcome may be the presence of an abnormal condition either at the time of imaging or at a future point, such as with overall or progression-free survival. We use the word “outcome” to refer to both the reference standard in accuracy studies and the condition of interest when there is no reference standard.
Outcome assessment should not rely on information from the modality from which the imaging features are extracted to avoid incorporation bias (23–26). This bias is greatest when the outcome is identified solely from the same image from which the features are measured, and one or more of the features are vital for identifying the condition. However, bias may still be present if a subsequent image from the same modality is used or if the features under study play less of a role in identifying the condition. Intuitively, the magnitude of bias will be related to the magnitude of correlation between the features and the condition.
Dercle et al (27) developed a radiomics signature for predicting overall survival using CT images in 667 patients with colorectal cancer who were enrolled in a clinical trial comparing irinotecan, 5-fluorouracil, and leucovorin (FOLFIRI), alone or in combination with cetuximab. Scans from baseline (pretreatment) and 8 weeks after randomization (on-therapy scans) were divided into training and validation data sets. After comparing the signature with known predictors of sensitivity to FOLFIRI plus cetuximab, KRAS mutation status and the 8-week change in tumor burden, the authors concluded that their radiomic signature outperformed the other known predictors. Notably, if they had used progression-free survival as the outcome rather than overall survival, incorporation bias could have been introduced into the analysis. Progression-free survival is determined from CT scans, as are both the radiomic signature and 8-week change in tumor burden, potentially inducing a correlation between the predictors and the outcome, while the KRAS mutation status predictor would not be subject to the same induced correlation.
Selection of Images for Inclusion
The selection of images to be used, both for training and validating a radiomics model, requires careful consideration.
Some outcomes, such as histologic diagnosis, are only assessed for a subset of patients, based in part on clinical interpretation of imaging results. Limiting the study to these patient images results in verification bias (Tables 1, 2), which is a missing data problem that may lead to estimates of sensitivity that are too high and estimates of specificity that are too low, or in extreme cases, the inability to directly estimate sensitivity and specificity (15,26). Different study designs have been suggested to avoid this bias, and there are several proposed bias-correction methods when verification bias is deemed unavoidable (22,28–31).
Kontos et al (32) analyzed mammograms obtained from routine breast cancer screening using unsupervised clustering to identify four radiomic phenotypes of mammographic parenchymal complexity. To evaluate whether these radiomic phenotypes were associated with improved cancer detection, the authors analyzed scans from a separate case-control study that included women at high risk of breast cancer who were diagnosed with breast cancer. If the case-control study included only women for whom a biopsy result was available, their results would be subject to verification bias.
Discovery studies exploring new feature combinations often use a case-control design (33). If the included patients (cases) have severe, overt disease or their health conditions are more obvious, or the healthy patients (controls) are atypically healthy, spectrum bias is likely present. Also referred to as case-mix bias, spectrum bias is not limited to case-control studies and can lead to estimates of accuracy metrics that are too high (23). Because Kontos et al (32) defined their cases to be women at high risk of breast cancer (because of BRCA1 or BRCA2 mutation or a history of chest radiation, for instance), their analysis may be subject to spectrum bias, while the clinical trial data reported by Dercle et al (27) are less likely to overestimate performance of radiomics-derived predictors. (Note, however, that Kontos et al avoid possible spectrum bias in defining the radiomic phenotype by performing unsupervised clustering in the routine screening population without regard for outcome availability.)
We make the distinction here between spectrum bias and spectrum effect, where the latter is defined as the variation in performance in different populations (34). While not necessarily a bias in the statistical sense (where an estimate is biased if its expected value does not match the true value of the corresponding parameter), it is an important aspect of study design to consider, especially in the context of lack of diversity in medical research (35–38). As has been pointed out, analyses of data sets restricted to patients from a single institution may not be generalizable (32,39). Spectrum effects may be related to catchment populations and institutional procedures for treatment and supportive care; radiomics studies may also demonstrate spectrum effects introduced by differences in scanner hardware, scanning protocols, and image analysis protocols (Tables 1, 2).
Finally, when more than one image per patient is available for analysis, the study design should take into account that these images are not completely independent, and, therefore, do not increase the sample size and study power in the same way that adding completely independent images (patients) would. Specialized statistical analysis methods are required to account for within-patient correlation introduced by assessment of patients at multiple time points or with multiple lesions (40–42).
Training and Validation Data Sets
Appropriate division of data for training and validation of models according to radiomic features is needed to avoid optimistic performance bias (Tables 1, 2) (43). There are different ways this division is handled in practice. For instance, Eslami et al (39) divided CT scans from 624 participants in the Framingham Heart Study into training and validation data sets to develop a radiomic-based risk score characterizing coronary artery calcium. Using a composite outcome consisting of all-cause mortality, nonfatal ischemic stroke, or myocardial infarction, the authors found that adding the radiomic-based risk score to a model containing known predictors of cardiovascular events, including a well-established measure of coronary artery calcium, resulted in substantial improvement in identifying high risk individuals.
When designing a radiomics analysis, independent and mutually exclusive data sets should be used as follows:
Training data are used for data exploration, feature selection, hyper-parameter selection, and model development. Training data may be further divided into subsets, such as test data for model discovery and tuning data for model revisions or hyper-parameter selection. Eslami et al (39) used a “discovery” training data set with repeated cross-validation to select parameters in a random forest model.
Validation data are used to assess performance of a model that was “locked down” (no changes in feature selection, data set–specific standardization, or model parameters) using training data. External validation is preferred and uses independent and distinct data (eg, from a different institution). Internal validation uses hold-out data sets from the same source as the training data, although separated from training data by random selection or a different date range (44). When hold-out data sets are impractical due to modest sample size, cross-validation can be used to assess model performance. However, the cross-validation design must be prespecified to avoid selecting a model based on performance of validation with different k-folds. Because feature selection is likely performed within each cross-validation step, another design prespecification should outline how to select final model features when they are not consistently chosen in each cross-validation subset (45).
Image Analysis Considerations
All biomarkers, whether developed from plasma, tissue, imaging, patient-reported data, or in combination, face potential sources of bias due to assay methodology and analysis. Unique to radiomic analyses are the errors and biases that may arise during image acquisition, processing, and imaging feature quantification (Tables 1, 2) (46–51). Standardization of workflow components (7,52) supports the reproducibility needed to move beyond discovery and into clinical practice and was demonstrated by Eslami et al (39) through their use of PyRadiomics, an open-source radiomics platform that follows suggested feature definitions. However, standardization is not always feasible, most notably in clinical image acquisition (7,50,53,54). Within clinical trials, well-defined patient cohorts and a standardized protocol permit precise biomarker development and exploration of causation. Translation to clinical practice will require thorough testing to identify and account for spectrum effects and to ensure that radiomics-derivedmeasures are robust to data acquisition protocols. These efforts will additionally require broad access to large quantities of images available from real-world data, with clinical annotation.
Statistical Analysis Considerations
Statistical analysis of radiomic features often involves multiple steps, including one or more of the following: dimension reduction, feature selection, model building (or classification), selection of a risk-stratification threshold, fine-tuning of model components, internal validation, and external validation (55–58); bias may arise at multiple points in this process (Tables 1–4).
Exclusion of Indeterminate or Missing Data
Exclusion of missing outcome or feature data from analysis can lead to bias, especially when the reason the feature could not be quantified or extracted is directly related to the feature or the outcome (15,59). All three of the aforementioned examples (Dercle et al [27], Kontos et al [32], and Eslami et al [39]) excluded poor-quality images or images with artifacts and are, therefore, susceptible to resultant bias. This type of image exclusion is very common practice across radiomic studies and is an inherent challenge when quantitatively assessing images. Additionally, the impact of image quality on radiomic features is an active and important area of study (60–63).
Overfitting
Overfitting refers to the situation when a model or classifier is highly optimized for a particular data set, consequently captures noise, and then fails to work well in other data sets either by over- or underestimating risk of the patient outcome (64,65). This results in poor model performance metrics, such as low values for the area under the receiver operating characteristic curve. Despite a high level of awareness in the radiomics literature (1), bias due to overfitting is still commonly observed. Overfitting is most likely to occur when the study sample size is small relative to the number of imaging features evaluated and can also occur in cases where only a small number of imaging features are included, particularly if they are only weakly associated with the outcome (64,66). In all three of our stated examples (27,32,39), the authors aimed to minimize overfitting by including a relatively large number of patients and reducing the number of features that were used.
Multiple Testing
In radiomics, multiple testing is widely recognized as problematic and occurs when many radiomic features are examined without prespecified hypotheses or a method for minimizing false discovery (67–69). Hidden multiple testing may arise at any of several analysis steps. For example, testing multiple optimal cutoffs for clinical decision making is also multiple testing and leads to an increased chance of erroneously finding an association (inflated type I error) as well as overestimation of effect sizes (67,70). Additionally, when there are multiple candidate methods for a step (ie, dimension reduction) and none has been shown to be uniformly superior to others (55–57,71), exploration of several methods may lead to selecting a method based on (spurious) fluctuations in performance in the study sample. As for cross-validation, the methodology should be prespecified based on careful consideration of study aims, data characteristics inherent to the study design, and consequences of false-positive and false-negative errors.
Reporting Considerations
It is not always possible to safeguard against all potential sources of study bias in radiomics research. Therefore, it is imperative that researchers thoroughly report on their imaging data (ie, Digital Imaging and Communications in Medicine [DICOM] header information), methodology, limitations, and any other potential sources of variability. Rigorous reporting enables researchers to build on others’ results and protects against failed attempts to replicate spurious and overstated results. For instance, Eslami et al (39) included a detailed description of their methodology in their supplementary material.
Reporting guidelines developed for other research emphases, such as the Standards for Reporting of Diagnostic Accuracy (STARD) initiative for diagnostic accuracy studies (16), the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement for prediction models (72), and the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) for tumor markers (73), are required by some journals for relevant articles and have many elements that are applicable to radiomic research (74). There has been some previous work aiming to establish parallel initiatives in radiomics (46,47,75). It will be key to consider aspects of study design, data collection, and rigorous statistical analysis moving forward.
Discussion
Radiomic analyses are highly susceptible to bias arising from multiple sources. A unifying theme behind the biases and pitfalls we have outlined is that they can all lead to incorrect inference and a model that erroneously includes or excludes imaging features and, ultimately, performs poorly. While not meant to be an all-encompassing list, the issues we have highlighted arise frequently. Some, such as overfitting and lack of adjusting for multiple testing, are particularly relevant in radiomic studies. Others are issues that may arise equally as frequently in other types of studies but have been highlighted here because we have noticed a lack of awareness of these issues among investigators conducting radiomic studies. All are issues that are broadly applicable to many studies, including those where features are derived by the computer using convolution neural network (deep) approaches. In any analysis, the challenge is to identify the most relevant sources of bias and measurement error.
Although software packages to implement analyses are readily available and increasingly user friendly, if they are not implemented with the necessary expertise or correct guidance, there is a high risk that incorrect conclusions will be drawn from the work. The field of radiomics lies at the intersection of medicine, computer science, and statistics (Fig 2). We contend that to produce clinically meaningful results that positively impact patient care and minimize biases and pitfalls, radiomic analysis requires a multidisciplinary approach with a research team that includes individuals with multiple areas of expertise.
Figure 2:
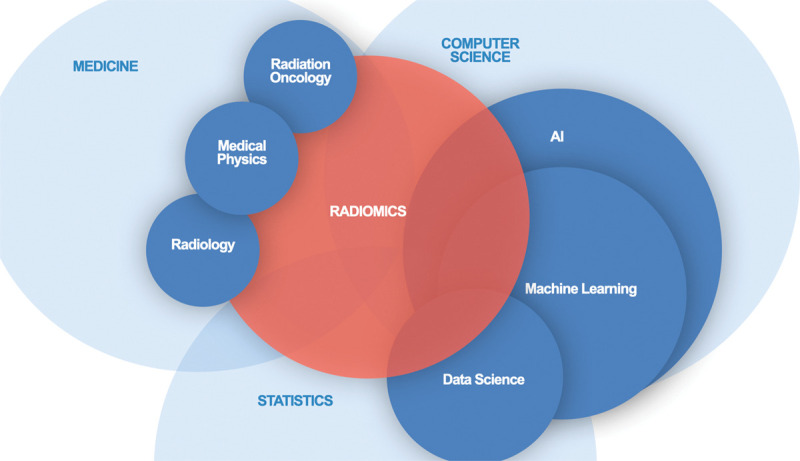
Diagram shows that the expertise required to ensure meaningful results from radiomic analysis spans a variety of disciplines. Collaboration and knowledge sharing is essential.
Footnotes
C.S.M. and M.L.W. contributed equally to this work.
C.S.M. supported by a Cancer Center Support (CORE) grant to Memorial Sloan Kettering (grant P30CA008748). A.L.S. funded in part by the National Institutes of Health (NIH) (R01 CA233888) and the Canada Research Chairs Program. M.A.J. supported in part by NIH Imaging Response Assessment Team grants (5P30CA006973 and U01CA140204-09).
Disclosures of conflicts of interest: C.S.M. Grant from the Foundation for the National Institutes for Health; honoraria from the University of Michigan. M.L.W. No relevant relationships. M.A.J. Grant from the National Institutes for Health; inventor and patent applicant for COVID.AI; Radiology and Medical Physics editorial boards. B.F.K. Consultant, Zionexa and Telix; board member, Randomized Evaluation of Trial Acceptability by Incentive; co-chair, Pharma Imaging Network for Therapeutics and Diagnostics. A.L.S. Charter member of a National Institutes of Health Study Section.
References
- 1. Gillies RJ , Kinahan PE , Hricak H . Radiomics: Images Are More than Pictures, They Are Data . Radiology 2016. ; 278 ( 2 ): 563– 577 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Song J , Yin Y , Wang H , Chang Z , Liu Z , Cui L . A review of original articles published in the emerging field of radiomics . Eur J Radiol 2020. ; 127: 108991. [DOI] [PubMed] [Google Scholar]
- 3. Morin O , Vallières M , Jochems A , et al . A Deep Look Into the Future of Quantitative Imaging in Oncology: A Statement of Working Principles and Proposal for Change . Int J Radiat Oncol Biol Phys 2018. ; 102 ( 4 ): 1074– 1082 . [DOI] [PubMed] [Google Scholar]
- 4. O’Connor JPB , Aboagye EO , Adams JE , et al . Imaging biomarker roadmap for cancer studies . Nat Rev Clin Oncol 2017. ; 14 ( 3 ): 169– 186 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Timmeren JE , Cester D , Tanadini-Lang S , Alkadhi H , Baessler B . Radiomics in medical imaging-“how-to” guide and critical reflection . Insights Imaging 2020. ; 11 ( 1 ): 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sullivan DC , Obuchowski NA , Kessler LG , et al . Metrology Standards for Quantitative Imaging Biomarkers . Radiology 2015. ; 277 ( 3 ): 813– 825 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zwanenburg A , Vallières M , Abdalah MA , et al . The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping . Radiology 2020. ; 295 ( 2 ): 328– 338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kessler LG , Barnhart HX , Buckler AJ , et al . The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions . Stat Methods Med Res 2015. ; 24 ( 1 ): 9– 26 . [DOI] [PubMed] [Google Scholar]
- 9. Park JE , Kim D , Kim HS , et al . Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement . Eur Radiol 2020. ; 30 ( 1 ): 523– 536 . [DOI] [PubMed] [Google Scholar]
- 10. Larue RT , Defraene G , De Ruysscher D , Lambin P , van Elmpt W . Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures . Br J Radiol 2017. ; 90 ( 1070 ): 20160665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuo MD , Gollub J , Sirlin CB , Ooi C , Chen X . Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma . J Vasc Interv Radiol 2007. ; 18 ( 7 ): 821– 831 . [DOI] [PubMed] [Google Scholar]
- 12. Aerts HJWL , Velazquez ER , Leijenaar RTH , et al . Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach . Nat Commun 2014. ; 5 ( 1 ): 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parekh V , Jacobs MA . Radiomics: a new application from established techniques . Expert Rev Precis Med Drug Dev 2016. ; 1 ( 2 ): 207– 226 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Z , Wang S , Dong D , et al . The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges . Theranostics 2019. ; 9 ( 5 ): 1303– 1322 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Begg CB , McNeil BJ . Assessment of radiologic tests: control of bias and other design considerations . Radiology 1988. ; 167 ( 2 ): 565– 569 . [DOI] [PubMed] [Google Scholar]
- 16. Bossuyt PM , Reitsma JB , Bruns DE , et al . STARD 2015: An Updated List of Essential Items for Reporting Diagnostic Accuracy Studies . Radiology 2015. ; 277 ( 3 ): 826– 832 . [DOI] [PubMed] [Google Scholar]
- 17. Pepe MS , Feng Z , Janes H , Bossuyt PM , Potter JD . Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design . J Natl Cancer Inst 2008. ; 100 ( 20 ): 1432– 1438 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sauerbrei W , Taube SE , McShane LM , Cavenagh MM , Altman DG . Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration . J Natl Cancer Inst 2018. ; 110 ( 8 ): 803– 811 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou XH , Obuchowski NA , McClish DK . In: Statistical methods in diagnostic medicine . New York, NY: : Wiley-Interscience; , 2002. . [Google Scholar]
- 20. Schlesselman JJ , Stolley PD . Case-control studies: design, conduct, analysis . New York, NY: : Oxford University Press; , 1982. . [Google Scholar]
- 21. Motulsky HJ . Common misconceptions about data analysis and statistics . Br J Pharmacol 2015. ; 172 ( 8 ): 2126– 2132 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pepe MS . The Statistical Evaluation of Medical Tests for Classification and Prediction . Oxford, England: : Oxford University Press; , 2003. ; 168– 193 . [Google Scholar]
- 23. Ransohoff DF , Feinstein AR . Problems of spectrum and bias in evaluating the efficacy of diagnostic tests . N Engl J Med 1978. ; 299 ( 17 ): 926– 930 . [DOI] [PubMed] [Google Scholar]
- 24. Whiting P , Rutjes AW , Reitsma JB , Glas AS , Bossuyt PM , Kleijnen J . Sources of variation and bias in studies of diagnostic accuracy: a systematic review . Ann Intern Med 2004. ; 140 ( 3 ): 189– 202 . [DOI] [PubMed] [Google Scholar]
- 25. Zhou XH , Obuchowski NA , McClish DK . In: Statistical Methods in Diagnostic Medicine . New York, NY: : Wiley-Interscience; , 2002. ; 73– 74 . [Google Scholar]
- 26. Kohn MA , Carpenter CR , Newman TB . Understanding the direction of bias in studies of diagnostic test accuracy . Acad Emerg Med 2013. ; 20 ( 11 ): 1194– 1206 . [DOI] [PubMed] [Google Scholar]
- 27. Dercle L , Lu L , Schwartz LH , et al . Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway . J Natl Cancer Inst 2020. ; 112 ( 9 ): 902– 912 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Begg CB . Biases in the assessment of diagnostic tests . Stat Med 1987. ; 6 ( 4 ): 411– 423 . [DOI] [PubMed] [Google Scholar]
- 29. de Groot JAH , Bossuyt PMM , Reitsma JB , et al . Verification problems in diagnostic accuracy studies: consequences and solutions . BMJ 2011. ; 343: d4770. [DOI] [PubMed] [Google Scholar]
- 30. Begg CB , Greenes RA . Assessment of diagnostic tests when disease verification is subject to selection bias . Biometrics 1983. ; 39 ( 1 ): 207– 215 . [PubMed] [Google Scholar]
- 31. Zhou XH , Obuchowski NA , McClish DK . In: Statistical Methods in Diagnostic Medicine . New York, NY: : Wiley-Interscience; , 2002. ; 307– 353 . [Google Scholar]
- 32. Kontos D , Winham SJ , Oustimov A , et al . Radiomic Phenotypes of Mammographic Parenchymal Complexity: Toward Augmenting Breast Density in Breast Cancer Risk Assessment . Radiology 2019. ; 290 ( 1 ): 41– 49 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pepe MS , Etzioni R , Feng Z , et al . Phases of biomarker development for early detection of cancer . J Natl Cancer Inst 2001. ; 93 ( 14 ): 1054– 1061 . [DOI] [PubMed] [Google Scholar]
- 34. Usher-Smith JA , Sharp SJ , Griffin SJ . The spectrum effect in tests for risk prediction, screening, and diagnosis . BMJ 2016. ; 353: i3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loree JM , Anand S , Dasari A , et al . Disparity of Race Reporting and Representation in Clinical Trials Leading to Cancer Drug Approvals From 2008 to 2018 . JAMA Oncol 2019. ; 5 ( 10 ): e191870, e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chastain DB , Osae SP , Henao-Martínez AF , Franco-Paredes C , Chastain JS , Young HN . Racial Disproportionality in Covid Clinical Trials . N Engl J Med 2020. ; 383 ( 9 ): e59. [DOI] [PubMed] [Google Scholar]
- 37. Larrazabal AJ , Nieto N , Peterson V , Milone DH , Ferrante E . Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis . Proc Natl Acad Sci U S A 2020. ; 117 ( 23 ): 12592– 12594 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Obermeyer Z , Powers B , Vogeli C , Mullainathan S . Dissecting racial bias in an algorithm used to manage the health of populations . Science 2019. ; 366 ( 6464 ): 447– 453 . [DOI] [PubMed] [Google Scholar]
- 39. Eslami P , Parmar C , Foldyna B , et al . Radiomics of Coronary Artery Calcium in the Framingham Heart Study . Radiol Cardiothorac Imaging 2020. ; 2 ( 1 ): e190119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gönen M , Panageas KS , Larson SM . Statistical issues in analysis of diagnostic imaging experiments with multiple observations per patient . Radiology 2001. ; 221 ( 3 ): 763– 767 . [DOI] [PubMed] [Google Scholar]
- 41. Levine D , Bankier AA , Halpern EF . Submissions to Radiology: Our Top 10 List of Statistical Errors . Radiology 2009. ; 253 ( 2 ): 288– 290 . [Google Scholar]
- 42.Zhou XH, Obuchowski NA, McClish DK. In: Statistical Methods in Diagnostic Medicine. New York, NY:Wiley-Interscience,2002; 104–106, 169–171, 274–303. [Google Scholar]
- 43. Altman DG , Royston P . What do we mean by validating a prognostic model? Stat Med 2000. ; 19 ( 4 ): 453– 473 . [DOI] [PubMed] [Google Scholar]
- 44. Park JE , Park SY , Kim HJ , Kim HS . Reproducibility and generalizability in radiomics modeling: Possible strategies in radiologic and statistical perspectives . Korean J Radiol 2019. ; 20 ( 7 ): 1124– 1137 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krstajic D , Buturovic LJ , Leahy DE , Thomas S . Cross-validation pitfalls when selecting and assessing regression and classification models . J Cheminform 2014. ; 6 ( 1 ): 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lambin P , Leijenaar RTH , Deist TM , et al . Radiomics: the bridge between medical imaging and personalized medicine . Nat Rev Clin Oncol 2017. ; 14 ( 12 ): 749– 762 . [DOI] [PubMed] [Google Scholar]
- 47. Sanduleanu S , Woodruff HC , de Jong EEC , et al . Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score . Radiother Oncol 2018. ; 127 ( 3 ): 349– 360 . [DOI] [PubMed] [Google Scholar]
- 48. Traverso A , Wee L , Dekker A , Gillies R . Repeatability and Reproducibility of Radiomic Features: A Systematic Review . Int J Radiat Oncol Biol Phys 2018. ; 102 ( 4 ): 1143– 1158 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yip SS , Aerts HJ . Applications and limitations of radiomics . Phys Med Biol 2016. ; 61 ( 13 ): R150– R166 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nie K , Al-Hallaq H , Li XA , et al . NCTN Assessment on Current Applications of Radiomics in Oncology . Int J Radiat Oncol Biol Phys 2019. ; 104 ( 2 ): 302– 315 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mackin D , Fave X , Zhang L , et al . Measuring Computed Tomography Scanner Variability of Radiomics Features . Invest Radiol 2015. ; 50 ( 11 ): 757– 765 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. The Image Biomarker Standardisation Initiative Reference Manual . https://ibsi.readthedocs.io/en/latest/#. Accessed March 7, 2021.
- 53. Rosen M , Kinahan PE , Gimpel JF , et al . Performance Observations of Scanner Qualification of NCI-Designated Cancer Centers: Results From the Centers of Quantitative Imaging Excellence (CQIE) Program . Acad Radiol 2017. ; 24 ( 2 ): 232– 245 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buckler AJ , Boellaard R . Standardization of quantitative imaging: the time is right, and 18F-FDG PET/CT is a good place to start . J Nucl Med 2011. ; 52 ( 2 ): 171– 172 . [DOI] [PubMed] [Google Scholar]
- 55. Chandrashekar G , Sahin F . A survey on feature selection methods . Comput Electr Eng 2014. ; 40 ( 1 ): 16– 28 . [Google Scholar]
- 56. Parmar C , Grossmann P , Bussink J , Lambin P , Aerts HJWL . Machine Learning methods for Quantitative Radiomic Biomarkers . Sci Rep 2015. ; 5 ( 1 ): 13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pavlou M , Ambler G , Seaman S , De Iorio M , Omar RZ . Review and evaluation of penalised regression methods for risk prediction in low-dimensional data with few events . Stat Med 2016. ; 35 ( 7 ): 1159– 1177 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Deist TM , Dankers FJWM , Valdes G , et al . Erratum: “Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers” [Med. Phys . 45 (7), 3449–3459 (2018)]. Med Phys 2019;46(2):1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pigott TD . A Review of Methods for Missing Data . Educ Res Eval 2001. ; 7 ( 4 ): 353– 383 . [Google Scholar]
- 60. Park BW , Kim JK , Heo C , Park KJ . Reliability of CT radiomic features reflecting tumour heterogeneity according to image quality and image processing parameters . Sci Rep 2020. ; 10 ( 1 ): 3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Arrowsmith C , Reiazi R , Welch ML , et al . Automated detection of dental artifacts for large-scale radiomic analysis in radiation oncology . Phys Imaging Radiat Oncol 2021. ; 18 : 41– 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wei L , Rosen B , Vallières M , et al . Automatic recognition and analysis of metal streak artifacts in head and neck computed tomography for radiomics modeling . Phys Imaging Radiat Oncol 2019. ; 10 : 49– 54 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao B . Understanding Sources of Variation to Improve the Reproducibility of Radiomics . Front Oncol 2021. ; 11: 633176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harrell FE . Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis . New York, NY: : Springer; , 2001. . [Google Scholar]
- 65. Hawkins DM . The problem of overfitting . J Chem Inf Comput Sci 2004. ; 44 ( 1 ): 1– 12 . [DOI] [PubMed] [Google Scholar]
- 66. Subramanian J , Simon R . Overfitting in prediction models—is it a problem only in high dimensions? Contemp Clin Trials 2013. ; 36 ( 2 ): 636– 641 . [DOI] [PubMed] [Google Scholar]
- 67. Chalkidou A . O’iscovery Rates in PET and CT Studies with Texture Features: A Systematic Review . PLoS One 2015. ; 10 ( 5 ): e0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vallières M , Zwanenburg A , Badic B , Cheze Le Rest C , Visvikis D , Hatt M . Responsible Radiomics Research for Faster Clinical Translation . J Nucl Med 2018. ; 59 ( 2 ): 189– 193 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Krzywinski M , Altman N . Points of significance: Comparing samples—part II . Nat Methods 2014. ; 11 ( 4 ): 355– 356 . [DOI] [PubMed] [Google Scholar]
- 70. Hilsenbeck SG , Clark GM , McGuire WL . Why do so many prognostic factors fail to pan out? Breast Cancer Res Treat 1992. ; 22 ( 3 ): 197– 206 . [DOI] [PubMed] [Google Scholar]
- 71. Deist TM , Dankers FJWM , Valdes G , et al . Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers . Med Phys 2018. ; 45 ( 7 ): 3449– 3459 . [Published correction appears in Med Phys 2019;46(2):1080-1087.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Collins GS , Reitsma JB , Altman DG , Moons KGM . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement . BMJ 2015. ; 350: g7594. [DOI] [PubMed] [Google Scholar]
- 73. McShane LM , Altman DG , Sauerbrei W , et al . REporting recommendations for tumour MARKer prognostic studies (REMARK) . Br J Cancer 2005. ; 93 ( 4 ): 387– 391 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Collins GS , Moons KGM . Reporting of artificial intelligence prediction models . Lancet 2019. ; 393 ( 10181 ): 1577– 1579 . [DOI] [PubMed] [Google Scholar]
- 75. Kocak B , Kus EA , Kilickesmez O . How to read and review papers on machine learning and artificial intelligence in radiology: a survival guide to key methodological concepts . Eur Radiol 2021. ; 31 ( 4 ): 1819– 1830 . [DOI] [PubMed] [Google Scholar]
- 76. Meyer M , Ronald J , Vernuccio F , et al . Reproducibility of CT Radiomic Features within the Same Patient: Influence of Radiation Dose and CT Reconstruction Settings . Radiology 2019. ; 293 ( 3 ): 583– 591 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Duron L , Balvay D , Vande Perre S , et al . Gray-level discretization impacts reproducible MRI radiomics texture features . PLoS One 2019. ; 14 ( 3 ): e0213459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pavic M , Bogowicz M , Würms X , et al . Influence of inter-observer delineation variability on radiomics stability in different tumor sites . Acta Oncol 2018. ; 57 ( 8 ): 1070– 1074 . [DOI] [PubMed] [Google Scholar]
- 79. Fornacon-Wood I , Mistry H , Ackermann CJ , et al . Reliability and prognostic value of radiomic features are highly dependent on choice of feature extraction platform . Eur Radiol 2020. ; 30 ( 11 ): 6241– 6250 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McNitt-Gray M , Napel S , Jaggi A , et al . Standardization in Quantitative Imaging: A Multicenter Comparison of Radiomic Features from Different Software Packages on Digital Reference Objects and Patient Data Sets . Tomography 2020. ; 6 ( 2 ): 118– 128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Harrell FE Jr , Lee KL , Mark DB . Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors . Stat Med 1996. ; 15 ( 4 ): 361– 387 . [DOI] [PubMed] [Google Scholar]