Abstract
Background
After severe traumatic brain injury (sTBI), physicians use long-term prognostication to guide acute clinical care yet struggle to predict outcomes in comatose patients.
Purpose
To develop and evaluate a prognostic model combining deep learning of head CT scans and clinical information to predict long-term outcomes after sTBI.
Materials and Methods
This was a retrospective analysis of two prospectively collected databases. The model-building set included 537 patients (mean age, 40 years ± 17 [SD]; 422 men) from one institution from November 2002 to December 2018. Transfer learning and curriculum learning were applied to a convolutional neural network using admission head CT to predict mortality and unfavorable outcomes (Glasgow Outcomes Scale scores 1–3) at 6 months. This was combined with clinical input for a holistic fusion model. The models were evaluated using an independent internal test set and an external cohort of 220 patients with sTBI (mean age, 39 years ± 17; 166 men) from 18 institutions in the Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) study from February 2014 to April 2018. The models were compared with the International Mission on Prognosis and Analysis of Clinical Trials in TBI (IMPACT) model and the predictions of three neurosurgeons. Area under the receiver operating characteristic curve (AUC) was used as the main model performance metric.
Results
The fusion model had higher AUCs than did the IMPACT model in the prediction of mortality (AUC, 0.92 [95% CI: 0.86, 0.97] vs 0.80 [95% CI: 0.71, 0.88]; P < .001) and unfavorable outcomes (AUC, 0.88 [95% CI: 0.82, 0.94] vs 0.82 [95% CI: 0.75, 0.90]; P = .04) on the internal data set. For external TRACK-TBI testing, there was no evidence of a significant difference in the performance of any models compared with the IMPACT model (AUC, 0.83; 95% CI: 0.77, 0.90) in the prediction of mortality. The Imaging model (AUC, 0.73; 95% CI: 0.66–0.81; P = .02) and the fusion model (AUC, 0.68; 95% CI: 0.60, 0.76; P = .02) underperformed as compared with the IMPACT model (AUC, 0.83; 95% CI: 0.77, 0.89) in the prediction of unfavorable outcomes. The fusion model outperformed the predictions of the neurosurgeons.
Conclusion
A deep learning model of head CT and clinical information can be used to predict 6-month outcomes after severe traumatic brain injury.
© RSNA, 2022
Online supplemental material is available for this article.
See also the editorial by Haller in this issue.
Summary
Deep learning prognostic models using both admission CT scans and clinical information can predict 6-month mortality and unfavorable outcomes after severe traumatic brain injury and outperformed the predictions of neurosurgeons.
Key Results
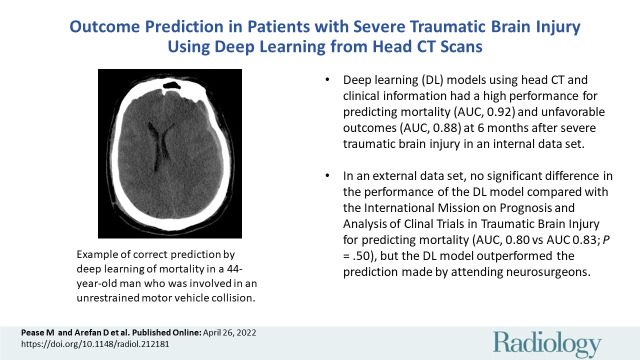
■ The deep learning models using head CT and clinical information had good performance for predicting mortality (area under the receiver operating characteristic [AUC] curve, 0.92) and unfavorable outcomes (AUC, 0.88) at 6 months after severe traumatic brain injury (sTBI) in an internal data set.
■ In an external data set, there was no significant difference in the performance of the deep learning model compared with the International Mission on Prognosis and Analysis of Clinical Trials in TBI (IMPACT) for predicting mortality (AUC, 0.80 vs 0.83; P = .50), but the deep learning model outperformed the predictions made by attending neurosurgeons.
Introduction
Traumatic brain injury (TBI) is a condition that disrupts normal brain function and can lead to permanent neurologic, emotional, and occupational disability. Disability from TBI is estimated to affect nearly 55 million people worldwide and 5 million people in the United States alone (1,2). Patients with severe TBI (sTBI), defined as a postresuscitation Glasgow Coma Scale score of 8 or less, have mortality rates approaching 40% (3,4). Prediction of long-term clinical outcomes is challenging in these patients because of their comatose status and concerning imaging features, including cerebral edema and intracranial hemorrhage (3,5,6). However, despite their early moribund status many patients have the potential to make a favorable recovery (7,8).
Clinicians may not accurately assign a prognosis when assessing sTBI in the acute postinjury phase, and neurosurgeons are frequently pessimistic (9–11). Despite this, neurotrauma practitioners often use their subjective versions of prognostication to guide life or death decisions, including whether to provide life-saving surgical procedures, such as decompressive craniectomy. These decisions must often be made rapidly and early during care, as delays in treatment are associated with worse outcomes (12). Multivariate models, such as the International Mission on Prognosis and Analysis of Clinical Trials in TBI (IMPACT), attempted to assign outcomes to patients using information available in the emergency department. IMPACT, however, was designed to guide clinical trials, rather than assign outcomes to individual patients, and it is not used widely in clinical practice (13,14). In fact, no current national guidelines recommend prognostic models in the care decision for patients with a TBI (15).
Recently, deep learning has transformed medical imaging diagnosis and prognostication (16–18). Deep convolutional neural network (CNN) models can identify abnormalities in radiologic images to assist computer-aided diagnosis of various diseases (19), but these techniques have not been widely adapted for prognostication of neurosurgical conditions. We hypothesized that imaging-based deep learning models could be adapted to predict long-term outcomes after sTBI. The purpose of this study was to develop and evaluate a prognostic model combining deep learning of head CT and clinical information to predict long-term outcomes after sTBI.
Materials and Methods
Study Cohorts and Data Collection
This study received the approval of the institutional review board at our institution, as well as at each institution participating in the Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) study. All institutions complied with Health Insurance Portability and Accountability Act requirements. Written consent was obtained from legally appointed representatives. We adhered to the Standards for Reporting Diagnostic Accuracy Studies (20) and Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (21). We built and tested our prediction model on an internal cohort of patients from the University of Pittsburgh Medical Center (UPMC) and externally tested our model with patients from the TRACK-TBI consortium.
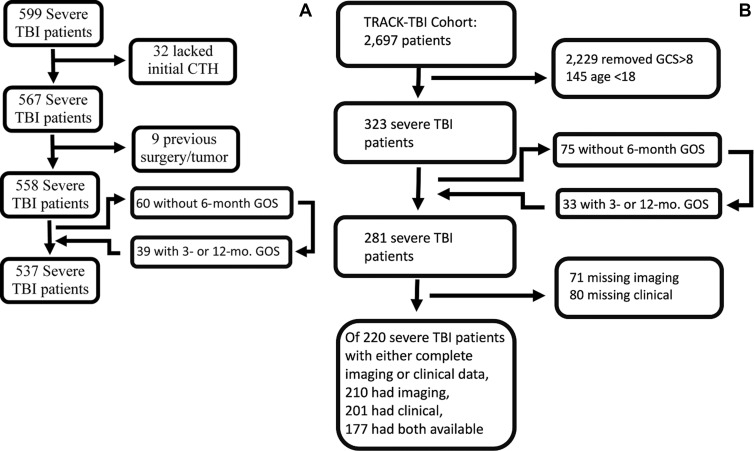
UPMC has a prospectively collected database of patients with sTBI admitted to a level 1 trauma center from November 2002 to December 2018 (Fig 1). This database has previously been described (22,23) and includes patients aged 16–80 years with sTBI. TRACK-TBI is a prospective multicenter study recruiting participants from 18 sites across the United States, enrolling nearly 3000 patients from February 2014 through April 2018 (NCT02119182) (24). We selected consecutive patients with sTBI and excluded patients coenrolled at UPMC. For both cohorts, we excluded patients with pre-existing neurosurgical disease, those without an admission CT head scan prior to neurosurgical intervention, and those who had substantial motion artifacts. Neurologic outcomes were assessed at 3, 6, and 12 months through a structured interview by trained neuropsychologists using the Glasgow Outcomes Scale (1 = death, 2 = persistent vegetative state, 3 = severe disability, 4 = moderate disability, 5 = low disability).
Figure 1:
Consolidated Standards of Reporting Trials diagram for (A) University of Pittsburgh Medical Center and (B) Transforming Research and Clinical Knowledge in Traumatic Brain Injury cohorts. For patients who were missing 6-month outcomes, the 3- or 12-month outcome was substituted in place of the 6-month outcome, if available for model prediction. CTH = CT of the head, GCS = Glasgow Coma Scale, GOS = Glasgow Outcome Scale, TBI = traumatic brain injury.
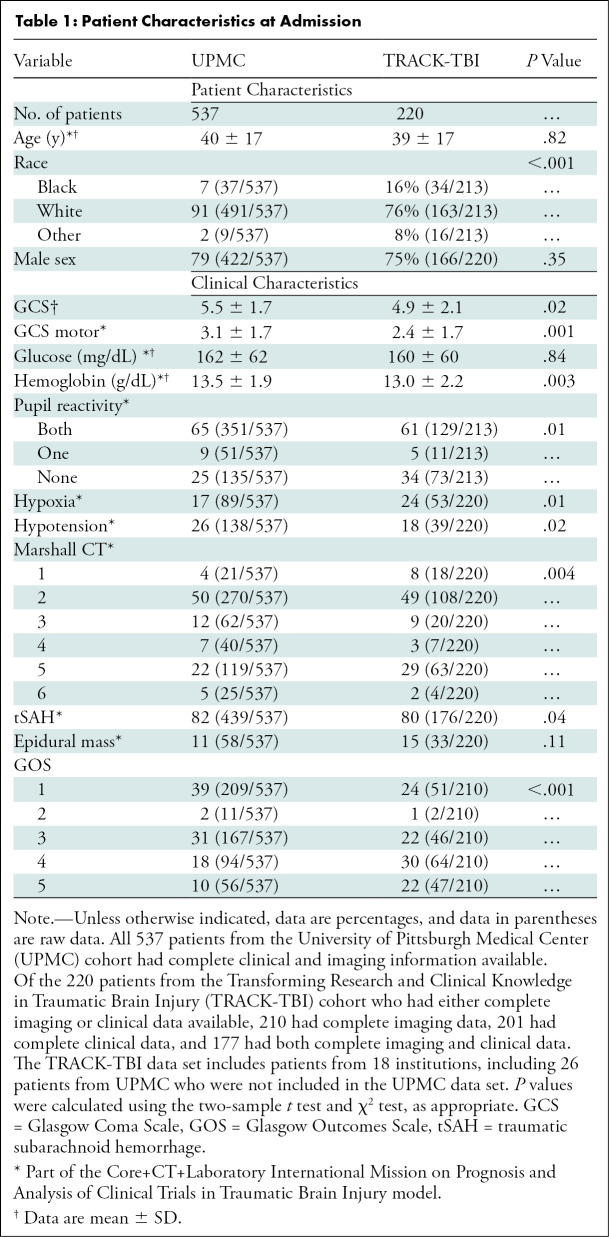
Demographic, clinical, and qualitative imaging variables in IMPACT were collected from the prospective databases (Table 1). We also collected sex, race, and mechanism of injury, which were not included in IMPACT. Data missing from the prospective collection of the UPMC database, including head CT data, were retrospectively collected without blinding to outcomes. For both cohorts, patients with missing 6-month outcomes had 3- or 12-month outcomes substituted, if available (Appendix E1 [online]). All remaining patients with incomplete data or who lacked sufficient follow-up data were removed from the study. Similar to IMPACT, we predicted mortality and unfavorable (Glasgow Outcomes Scale 1–3) or favorable (Glasgow Outcomes Scale 4–5) outcomes at 6 months.
Table 1:
Patient Characteristics at Admission
CT Imaging
Appendix E2 (online) describes details of image acquisition. For the UPMC database, all CT images were obtained with a GE Lightspeed scanner (GE Healthcare) with 5-mm section thickness. For the TRACK-TBI database, CT images were obtained with various scanners and with a section thickness of 2–6 mm. A representative subvolume of the head CT, spanning the midpoint of the body of the lateral ventricles to the midbrain and selected by an expert physician (M.P.), was used for modeling.
Machine Learning and Model Development
We built four machine learning models using various inputs to make 6-month predictions for mortality and unfavorable outcomes (Appendix E3 [online]).
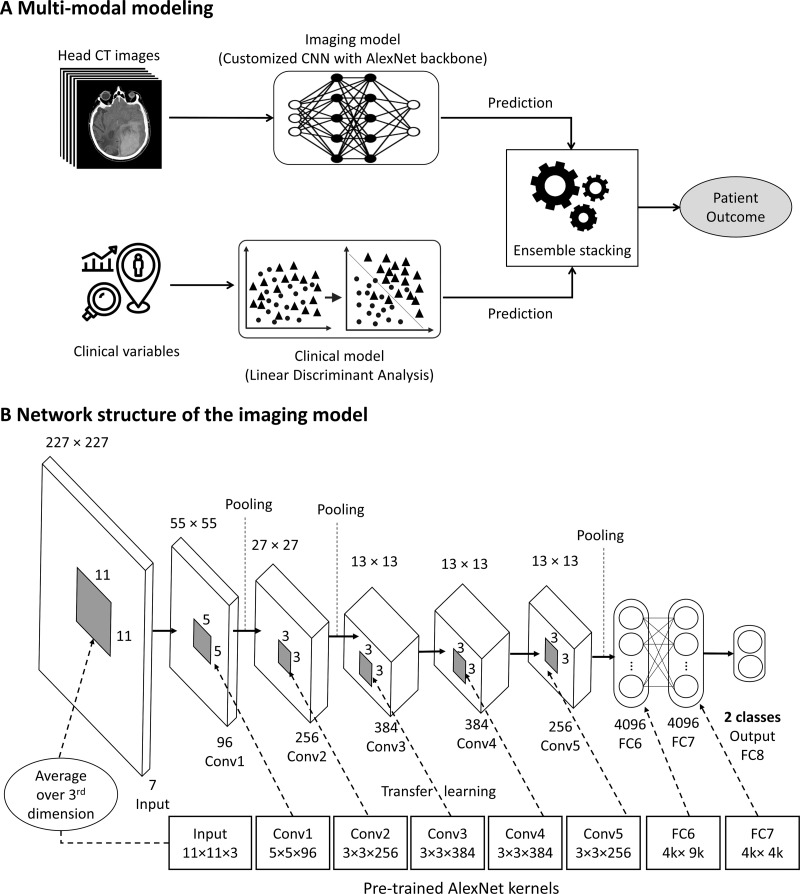
Imaging model.—We used head CT scans as inputs and built a customized CNN model with an AlexNet backbone pretrained with ImageNet data set (http://www.image-net.org/) (Fig 2). The CNN model was designed to analyze the subvolume of each CT scan, spanning from the midbrain to the lateral ventricle. To enhance the training of the CNN Imaging model on heterogenous CT scans from different vendor platforms and imaging reconstruction kernels, we developed a tailored curriculum learning technique. We first started to train the CNN model using a selected subset of homogeneous data (ie, images with the same reconstruction kernel, the so-called easy task) and then gradually increased the capacity of learning by involving a subset of heterogeneous images with a different reconstruction kernel (the so-called difficult task). This novel approach improved model performance through accounting for different image acquisition techniques.
Figure 2:
Outline of deep learning modeling to predict long-term outcomes in patients with severe traumatic brain injury based on radiographic and clinical information available in the emergency department. (A) An analysis of multimodal data, including a customized convolutional neural network (CNN) structure for modeling CT imaging data (imaging model) and a clinical model, was performed to generate a holistic prediction of the long-term outcomes (fusion model). (B) The customized CNN imaging model was structured using AlexNet backbone. The size of kernels in the input layer was changed from 11 × 11 × 3 in AlexNet to 11 × 11 × 7 in the customized model. Transfer learning was applied for all learnable layers, except for the last fully connected layer (FC8). For the input layer with seven channels, each of the available 96 kernels in the input layer of a pretrained AlexNet were averaged over the third dimension (three red-green-blue channels), and then the weights to each of the seven channels of the kernels in the input layer of the CNN model were transferred. Conv = convolutional layer, FC = fully connected layer..
Clinical model.—We built a linear discriminant analysis model using the same inputs as IMPACT with race, sex, and mechanism of injury added. The categorical variables (eg, race and mechanism of injury) were converted to dummy variables before feeding to the linear discriminant analysis model.
Fusion model.—We combined the imaging model with the clinical model using the ensemble stacking technique.
IMPACT-fusion model.—Similar to the fusion model, we fused our CNN imaging model with IMPACT using the ensemble stacking technique. This allowed us to evaluate if the CNN Imaging model provides additional prognostic information to IMPACT.
Attending Neurosurgeon Predictions
We developed a shadow clinical environment to assess how attending neurosurgeons at UPMC predicted outcomes in patients with sTBI. We selected three attending neurosurgeons, all of whom take neurotrauma calls, with 1, 5, and 25 years of experience. The neurosurgeons with 5 and 25 years of experience had subspecialty training in neurovascular surgery, and the other neurosurgeon had subspecialty training in neurotrauma. We used neurosurgeons for the reader study for two reasons. First, neurosurgeons are often the gatekeepers in sTBI care through decisions to offer life-saving surgical procedures. Second, many neurosurgeons care for patients with a TBI for many months after their initial injury, positioning them to better observe long-term outcomes. In our study, 50 patients were randomly selected from the UPMC test cohort of 107 patients. The neurosurgeons were given access to the same clinical information used in our fusion model and had access to the CT scans. For each patient, the neurosurgeon made binary predictions for mortality (alive or dead) and unfavorable (favorable or unfavorable) outcomes at 6 months.
Statistical Analyses
The prediction model was evaluated using both an internal test cohort (UPMC) and an external test cohort using patients from TRACK-TBI. For the UPMC evaluation, we used a stratified random sampling procedure on Glasgow Outcomes Scale and different reconstruction kernels to split the internal cohort into 70%, 10%, and 20% for training, validation, and testing, respectively. For the TRACK-TBI evaluation, we used the entire UPMC cohort to train the prediction model, using the parameters learned in the internal evaluation, and tested this model using the TRACK-TBI cohort. TRACK-TBI provided the CT imaging and clinical inputs to the model development team, who were blinded to the long-term outcomes of patients in the TRACK-TBI study. The TRACK-TBI consortium independently analyzed the model’s prediction results.
Model performance was evaluated using area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, and specificity. Two-sided DeLong test was used to assess the significance of differences between two AUCs (25). For multiple testing correction, we used the Benjamini-Hochberg procedure to control the false detection rate at 0.05 (26). We estimated the sensitivity at fixed specificity using the Pepe method (27). The 95% CI for sensitivity was estimated using naive exact binomial and Linnet adjusted techniques (27). Neurosurgeon predictions were reported as accuracy and their overall sensitivity and specificity for mortality and unfavorable outcome predictions. A positive prediction for mortality or unfavorable outcomes was a prediction probability of the event greater than 50%. All the statistical analyses were performed using R software (version 0.0.456; R Foundation for Statistical Computing) and SAS software (version 0.04 for Linux (SAS Institute). The code for building the models and data analysis can be found at https://github.com/Pitt-ICCI/TBI-study.
Results
Patient Overview for UPMC Cohort
For the UPMC cohort, 599 patients with sTBI were initially enrolled, and 537 remained after exclusion (Fig 1). Admission data are found in Table 1 (mean age, 40 years ± 17 [SD]; 422 men).
Model Testing on the Internal Test Cohort
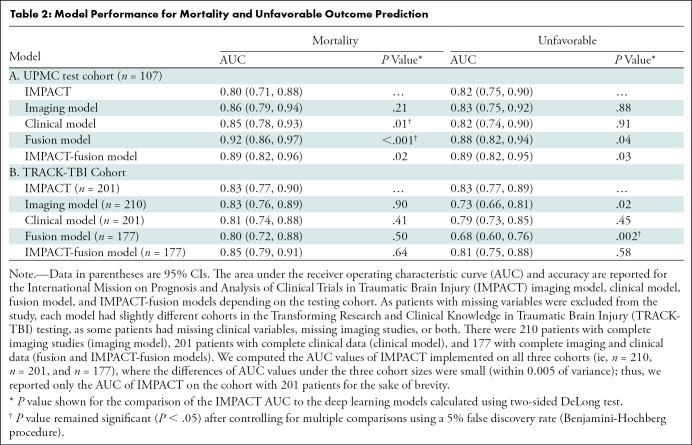
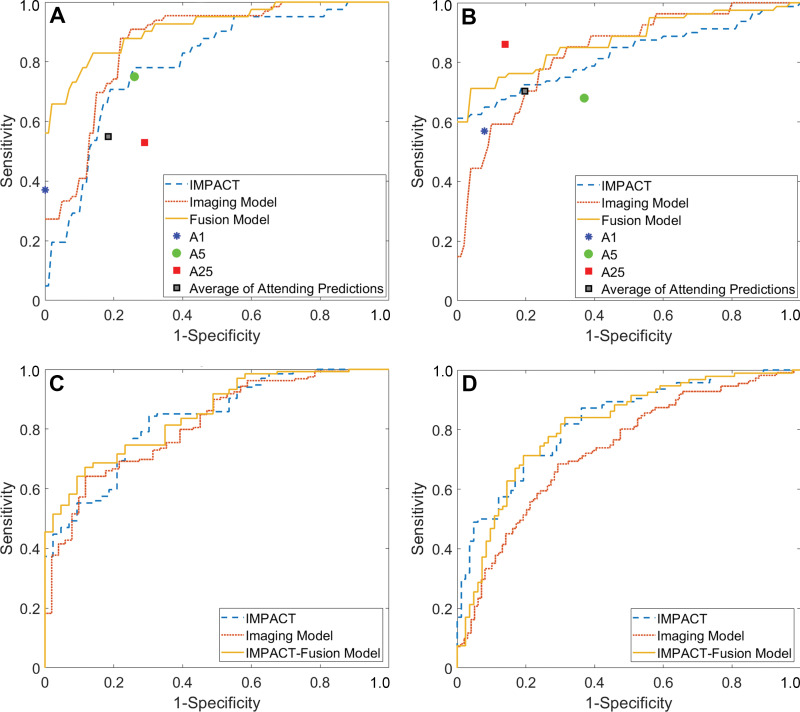
In the UPMC test cohort, IMPACT had an AUC of 0.80 (95% CI: 0.71, 0.88) for mortality and 0.82 (95% CI: 0.75, 0.90) for unfavorable outcomes (Table 2; Figs 3, 4). The imaging model using only CT scans showed no evidence of a significant difference from IMPACT for predicting mortality (AUC, 0.86; 95% CI: 0.79, 0.94; P = .21) or unfavorable outcomes (AUC, 0.83; 95% CI: 0.75, 0.92; P = .88). The clinical model had a better performance for predicting mortality, with an AUC of 0.85 (95% CI: 0.78, 0.93; P = .01), whereas no evidence of a significant difference was found for predicting unfavorable outcomes, with an AUC of 0.82 (95% CI: 0.74, 0.90; P = .91) as compared with IMPACT. The best-performing model was the fusion model, which combined head CT scans and clinical information. It had a better performance than IMPACT for predicting both mortality (AUC, 0.92; 95% CI: 0.86, 0.97; P < .001) and unfavorable outcomes (AUC, 0.88; 95% CI: 0.82, 0.95; P = .04).
Table 2:
Model Performance for Mortality and Unfavorable Outcome Prediction
Figure 3:
Comparison of performance of imaging, fusion, and International Mission on Prognosis and Analysis of Clinal Trials in Traumatic Brain Injury (IMPACT)-fusion models with IMPACT for survival and unfavorable outcomes. Receiver operating characteristic curves compare (A) mortality and (B) unfavorable outcomes for the University of Pittsburgh Medical Center data set and (C) mortality and (D) unfavorable outcomes for the Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) validation. Sensitivity and specificity for attending neurosurgeon predictions are reported in A and B, both as an average and for the individual attending neurosurgeon with 1 (A1), 5 (A5), and 25 (A25) years of experience.
Figure 4:
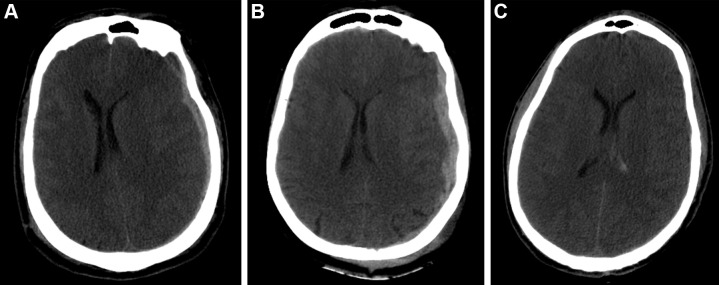
Example predictions by fusion model on University of Pittsburgh Medical Center patients. (A) Correct prediction in a 44-year-old man who was involved in an unrestrained motor vehicle collision. He underwent emergent decompressive hemicraniectomy (DHC), had bilateral lung injuries, and ultimately developed a pulmonary embolism with difficulty oxygenating on posttrauma day 6. His care was withdrawn, and he died. The model correctly predicted mortality. (B) Incorrect prediction in a 57-year-old woman who was in a motor vehicle collision and underwent DHC. The model predicted she would die, but she had a Glasgow Outcomes Scale of 3 at 2 years after trauma. She lived in a nursing home and was dependent on others for most daily living activities. (C) Incorrect prediction in a 28-year-old man who was in a motorcycle collision and had a minor head injury with intraventricular hemorrhage. Several weeks after trauma, he developed Klebsiella ventriculitis and pneumonia that led to an episode of severe hypotension. He subsequently developed malignant cerebral edema and died by brain death criteria. While the model predicted this patient would survive, this scenario highlights the difficulty of predicting outcomes based on information available in the emergency department, as events later in the patient’s course affect outcomes.
Patient Overview for TRACK-TBI
In the TRACK-TBI cohort, 323 patients with sTBI were identified. Of the 281 patients remaining after exclusion, 71 were missing CT head scans, and 80 had missing clinical information. In total, out of 220 patients who had either complete imaging data or complete clinical information, 210 had complete imaging data, 201 had complete clinical information, and 177 had both. When compared with the UPMC cohort, patients in the TRACK-TBI cohort had several markers of more severe injury, including higher rates of nonreactive pupils (P = .01), lower Glasgow Coma Scale scores (P = .02), lower levels of hemoglobin (P = .003), and lower rates of hypoxia P = .01). Despite this, patients in the TRACK-TBI cohort had improved 6-month outcomes, with lower mortality and higher rates of favorable outcomes (P < .001), as compared with the UPMC cohort.
Model Testing on the TRACK-TBI Cohort
In the TRACK-TBI testing cohort, IMPACT had an AUC of 0.83 (95% CI: 0.77, 0.90) for predicting mortality and an AUC of 0.83 (95% CI: 0.77, 0.89) for unfavorable outcomes. There was no evidence of a significant difference in the performance of any models compared with IMPACT for predicting mortality. The imaging model had an AUC of 0.83 (95% CI: 0.76, 0.89; P = .90) and the IMPACT-fusion model had an AUC of 0.85 (95% CI: 0.79, 0.91; P = .64) for predicting mortality. Both the imaging model (AUC, 0.73; 95% CI: 0.66, 0.81; P = .02) and the fusion model (AUC, 0.68; 95% CI: 0.60, 0.76; P = .002) had a lower performance than IMPACT for predicting unfavorable outcomes.
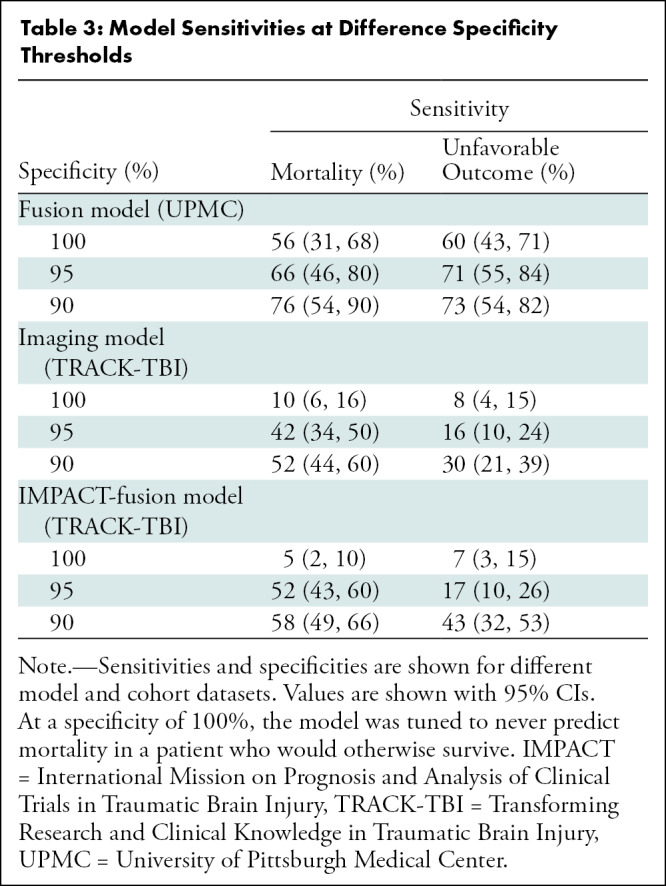
Table 3 reports the sensitivity for mortality and unfavorable outcomes for various specificity levels. For the UPMC test set, the sensitivity for mortality was 56% (95% CI: 31, 68) when the specificity was set to 100% (ie, never recommending withdrawal of care in a patient who would otherwise survive). Lowering the specificity to 90% resulted in a sensitivity of 76% (95% CI: 54, 90). For predicting mortality, when the CNN Imaging model was set to 100%, the sensitivity was 10% (95% CI: 6, 16), and when it was set to 90%, the sensitivity was 52% (95% CI: 44, 60).
Table 3:
Model Sensitivities at Difference Specificity Thresholds
Attending Neurosurgeon Predictions
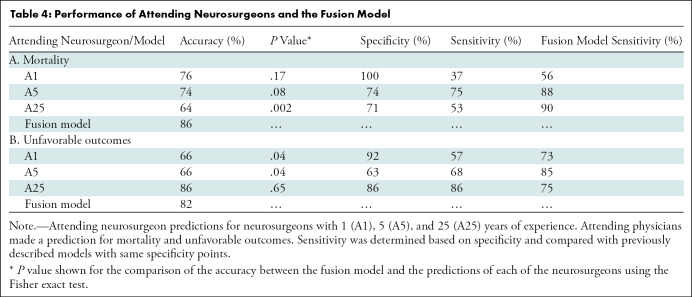
As shown in Table 4, neurosurgeons with 1, 5, and 25 years of experience had varying performance for mortality (accuracies of 76%, 74%, and 64%, respectively) and unfavorable outcomes (accuracies of 66%, 66%, and 86%, respectively). The most experienced neurosurgeon performed worse (64%) than the others for mortality prediction. The neurosurgeons with 5 and 25 years of experience incorrectly predicted that nine and 10 patients, respectively, would have died, even though these patients ultimately survived. The neurosurgeon with 1 year of experience made no mortality predictions in patients who ultimately survived. For comparison, the machine learning model (fusion model) had an accuracy of 86% for mortality and 82% for unfavorable outcome, which is comparable or significantly higher than the accuracy of the predictions made by the three neurosurgeons (Table 4).
Table 4:
Performance of Attending Neurosurgeons and the Fusion Model
Discussion
Neurotrauma practitioners frequently make time-sensitive decisions to provide life-saving surgery upon admission of patients with a severe traumatic brain injury (sTBI). To assist with these decisions, we built a deep learning model integrating head CT images with clinical data to predict long-term outcomes for patients with sTBI. Internal testing of our model outperformed the International Mission on Prognosis and Analysis of Clinical Trials in TBI (IMPACT) for both mortality (area under the receiver operating characteristic curve [AUC], 0.92 vs 0.80; P = <.001) and unfavorable outcome (AUC, 0.88 vs 0.82; P = .04) predictions at 6 months. In the Transforming Research and Clinical Knowledge in Traumatic Brain Injury test cohort, our models maintained discriminatory ability for mortality and unfavorable long-term outcomes. Our imaging model, built using only head CT, had noninferior performance to IMPACT for mortality (AUC, 0.83 vs 0.83; P = .90). This model could potentially function in a near-automated and time-efficient manner, obviating burdensome data collection required by IMPACT. This study shows that deep learning analysis of head CT can yield prognostic information to guide the care of patients with sTBI.
Although the trajectory of recovery from coma after TBI is unknown, recent work suggests great potential for recovery among TBI survivors (7). Despite this, most deaths (range, 55%–72%) after TBI are from the withdrawal of life-sustaining treatment, usually within 72 hours of injury, and are based on physician perception of a poor prognosis (28,29). Withdrawal of life-sustaining treatment is associated more strongly with the facility providing care than with underlying patient characteristics, suggesting that local practice patterns for withdrawing care may matter more than the injury itself (28). Recognizing the high cost of errors for models designed to guide life-or-death decisions, we assessed the performance when tuned for a zero false-positive rate (ie, never inappropriately withdrawing life-sustaining therapies in a patient who would survive). Under these stringent conditions, our sensitivity is 56% for mortality in the internal cohort and 10% in the TRACK-TBI cohort. This increased to 42% in the TRACK-TBI cohort when we reduced the parameters to a 5% false-positive rate. These numbers show that quantitative analysis of head CT imaging data early in the course of TBI may allow for more effective care of patients with sTBI by avoiding inappropriate withdrawal of life-sustaining treatment.
Previous modeling efforts predominantly used standard statistical techniques with moderate effect sizes (30). IMPACT is a multivariate model that has been externally validated on a large data set, but it has not gained widespread adoption (14), partly because clinicians mistrusted models designed to guide clinical trials rather than prognose individual patient outcomes. In clinical practice, physicians relied on their own prognostication, which historically lacked sufficient accuracy to guide withdrawal of life-sustaining therapy decisions (10,31). Our model is positioned to potentially serve as a rapid point-of-care test in the emergency room to guide personalized care decisions early, before surgical intervention. Our study did not include other diagnostic tests, such as electroencephalography or MRI, as performing these tests is not feasible before emergent neurosurgery, where treatment decisions must be made rapidly (12).
Overall, our results support the belief that deep CNN image analysis is able to identify abnormalities in brain imaging to predict long-term outcomes. In both the UPMC and TRACK-TBI testing cohorts, our imaging model performed well for predicting long-term outcomes compared with the predictions of attending neurosurgeons and IMPACT. The performance of the fusion model decreased when our models were tested on the independent TRACK-TBI cohort. A potential reason could be due to the characteristic differences of the TRACK-TBI cohort compared with the UPMC cohort (Table 1), where the TRACK-TBI cohort had markers of more severe injuries, such as higher rates of nonreactive pupils, lower Glasgow Coma Scale scores, lower levels of hemoglobin, and lower rates of hypoxia, yet superior outcomes compared with those of UPMC. If we combine the UPMC data and the TRACK-TBI data to train an updated prediction model, it is expected that the updated model would exhibit more robust performance than is currently seen, and we plan to explore this in the next steps of our work.
The comparisons of our models to the predictions of the attending neurosurgeons reveal important insights. The most experienced neurosurgeon had the lowest accuracy for mortality prediction but the highest accuracy for unfavorable outcome prediction, which reflects the difficulty and qualitative nature of the predictions made by human experts. To guide decisions about life-saving surgery, neurosurgeons must make critical clinical decisions as to whether a patient can survive an injury. Our models showed improved accuracy compared with the predictions of the neurosurgeons, suggesting that our model may improve TBI prognostication over the qualitative assessments made by neurosurgeons. After thorough validation, our model could provide quantitative prognostic information to better enable neurosurgeons to make rapid, reproducible, and more accurate decisions to guide the care of patients with sTBI.
Our study had several limitations. First, our model used a subvolume of CT head scans from the midbrain to the body of the lateral ventricles (Appendix E2 [online]). Without using the entire CT volume, rare findings, such as a cerebellar or pontine contusion, may be missed. Further comparisons with models using the entire CT volume may be conducted in future work. Second, our retrospective attending neurosurgeon prediction study may not capture the full ability of human experts to predict long-term outcomes. Although neurosurgeons had access to imaging and clinical patient descriptors, evaluating the patient and performing a physical examination is a key portion of a physician’s assessment.
In conclusion, we developed and evaluated a deep learning model combining head CT and clinical information for prognosing 6-month outcomes early after severe traumatic brain injury (sTBI). We demonstrated that quantitative analysis of head CT images improves the prediction of outcomes. Because of its ease of implementation, our model could be deployed as a fast and automated point-of-care tool to help physicians prognose long-term outcomes in patients with sTBI.
Acknowledgments
Acknowledgments
We thank Amy Klym for assistance with data collection and maintenance at the University of Pittsburgh. We thank Amy Markowitz for editorial support and Brian Fabian for administrative support with TRACK-TBI.
M.P. and D.A. contributed equally to this work.
D.O.O. and S.W. are co-senior authors.
Supported in part by the National Institutes of Health and National Cancer Institute (R01CA218405), National Institutes of Health and National Institute of Neurologic Disorders and Stroke (U01 NS1365885, U01 NS086090), and U.S. Department of Defense (W911QY-14-C-0070, W81XWH-18–2-0042, W81XWH-15–9-0001, W81XWH-14–2-0176). Supported by a Radiological Society of North America Research Scholar grant (RSCH1530), an Amazon Machine Learning Research Award, and the Congress of Neurologic Surgeons Data Science Fellowship grant. This work used the Extreme Science and Engineering Discovery Environment, which is supported by the National Science Foundation (ACI-1548562). Specifically, it used the Bridges system, which is supported by the National Science Foundation (ACI-1445606), at the Pittsburgh Supercomputing Center. TRACK-TBI received support from the National Football League Scientific Advisory Board, the U.S. Department of Energy for Precision Medicine, One Mind for TRACK-TBI patients’ stipends and support to clinical sites, NeuroTrauma Sciences to support data curation efforts, and Abbott Laboratories for add-in clinical studies.
TRACK-TBI Investigators: The group authorship from TRACK-TBI includes the following investigators who agreed to the group authorship: Neeraj Badjatia, MD, University of Maryland; Yelena Bodien, PhD, Massachusetts General Hospital; Ann-Christine Duhaime, MD, Massachusetts General Hospital for Children; V. Ramana Feeser, MD, Virginia Commonwealth University; Adam R. Ferguson, PhD, University of California, San Francisco; Brandon Foreman, MD, University of Cincinnati; Raquel Gardner, MD, University of California, San Francisco; Shankar Gopinath, MD, Baylor College of Medicine; C. Dirk Keene, MD, PhD, University of Washington; Christopher Madden, MD, UT Southwestern; Michael McCrea, PhD, Medical College of Wisconsin; Pratik Mukherjee, MD, PhD, University of California, San Francisco; Laura B. Ngwenya, MD, PhD, University of Cincinnati; David Schnyer, PhD, UT Austin; Sabrina Taylor, PhD, University of California, San Francisco; and John K. Yue, MD, University of California, San Francisco.
Disclosures of Conflicts of Interest: M.P. Fellowship grant from the Congress of Neurological Surgeons Data Science, Member of the Congress of Neurological Surgeons Data Science Committee and Quality Committee. D.A. No relevant relationships. J.B. Licensing payments for U.S. patent 11,200,664. E.Y. No relevant relationships. A.P. No relevant relationships. K.H. No relevant relationships. E.N. No relevant relationships. S.R. No relevant relationships. S.C. No relevant relationships. N.T. Consulting fees from the U.S. Department of Energy. D.O.O. Chair of the AANS/CNS Section on Neurotrauma and Critical Care. S.W. Grants from Stanly Marks Research Foundation, UPMC Hillman Cancer Center, University of Pittsburgh, NSF, and NSF/NIH joint program; consulting fees from COGNISTX; patent for a real-time artificial intelligence–enabled analysis device and method for use in nuclear medicine imaging is under professional review; past member of the Breast Cancer Early Detection, Prevention, and Risk Assessment Scientific Committee of the Wu Jie-Ping Medical Foundation; member of the RSNA Scientific Program Committee for Breast Imaging; member of the ECOG-ACRIN Cancer Research Group Radiomics Imaging Committee; member of the SPIE Medical Imaging Conference Technical Committee; member of the Computer-aided Diagnosis Conference SPIE Medical Imaging Conference Technical Committee; member of the Imaging Informatics for Healthcare, Research, and Applications; member of the Council of Early Career Investigators in Imaging of the Academy of Radiology Research and Coalition for Imaging and Bioengineering Research; member of the RSNA R&E Foundation Fund Development Committee–Corporate Giving Subcommittee; stock in Cognistx; computational devices from Nvidia; Journal of Digital Imaging editorial board.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- CNN
- convolutional neural network
- IMPACT
- International Mission on Prognosis and Analysis of Clinical Trials in TBI
- sTBI
- severe TBI
- TBI
- traumatic brain injury
- TRACK-TBI
- Transforming Research and Clinical Knowledge in Traumatic Brain Injury
- UPMC
- University of Pittsburgh Medical Center
Contributor Information
Shandong Wu, Email: wus3@upmc.edu.
Neeraj Badjatia, University of Maryland.
Yelena Bodien, Massachusetts General Hospital.
Ann-Christine Duhaime, Massachusetts General Hospital for Children.
V. Ramana Feeser, Virginia Commonwealth University.
Adam R. Ferguson, University of California, San Francisco.
Brandon Foreman, University of Cincinnati.
Raquel Gardner, University of California, San Francisco.
Shankar Gopinath, Baylor College of Medicine.
C. Dirk Keene, University of Washington.
Christopher Madden, UT Southwestern.
Michael McCrea, Medical College of Wisconsin.
Pratik Mukherjee, University of California, San Francisco.
Laura B. Ngwenya, University of Cincinnati.
David Schnyer, UT Austin.
Sabrina Taylor, University of California, San Francisco.
John K. Yue, University of California, San Francisco.
Collaborators: Neeraj Badjatia, Yelena Bodien, Ann-Christine Duhaime, V. Ramana Feeser, Adam R. Ferguson, Brandon Foreman, Raquel Gardner, Shankar Gopinath, C. Dirk Keene, Christopher Madden, Michael McCrea, Pratik Mukherjee, Laura B. Ngwenya, David Schnyer, Sabrina Taylor, and John K. Yue
References
- 1. Centers for Disease Control and Prevention . Report to Congress on Traumatic Brain Injury in the United States: Epidemiology and Rehabilitation . National Center for Injury Prevention and Control; Division of Unintentional Injury Prevention. Atlanta, Ga: : Centers for Disease Control and Prevention, 2015. . [DOI] [PubMed] [Google Scholar]
- 2. GBD . 2016 Traumatic Brain Injury and Spinal Cord Injury Collaborators. Global, regional, and national burden of traumatic brain injury and spinal cord injury, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 2019;18(1):56–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Myburgh JA, Cooper DJ, Finfer SR, et al. Epidemiology and 12-month outcomes from traumatic brain injury in Australia and New Zealand. J Trauma 2008;64(4):854–862. [DOI] [PubMed] [Google Scholar]
- 4. Jha RM, Elmer J, Zusman BE, et al. Intracranial pressure trajectories: A novel approach to informing severe traumatic brain injury phenotypes. Crit Care Med 2018;46(11):1792–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maeda Y , Ichikawa R , Misawa J , et al . External validation of the TRISS, CRASH, and IMPACT prognostic models in severe traumatic brain injury in Japan . PLoS One 2019. ; 14 ( 8 ): e0221791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Panczykowski DM, Puccio AM, Scruggs BJ, et al. Prospective independent validation of IMPACT modeling as a prognostic tool in severe traumatic brain injury. J Neurotrauma 2012;29(1):47–52. [DOI] [PubMed] [Google Scholar]
- 7. Kowalski RG, Hammond FM, Weintraub AH, et al. Recovery of Consciousness and Functional Outcome in Moderate and Severe Traumatic Brain Injury. JAMA Neurol 2021;78(5):548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puffer RC, Yue JK, Mesley M, et al. Long-term outcome in traumatic brain injury patients with midline shift: a secondary analysis of the Phase 3 COBRIT clinical trial. J Neurosurg 2018;131(2):596–603. [DOI] [PubMed] [Google Scholar]
- 9. Edlow BL , Chatelle C , Spencer CA , et al . Early detection of consciousness in patients with acute severe traumatic brain injury . Brain 2017. ; 140 ( 9 ): 2399– 2414 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaufmann MA , Buchmann B , Scheidegger D , Gratzl O , Radü EW . Severe head injury: should expected outcome influence resuscitation and first-day. decisions? Resuscitation 1992. ; 23 ( 3 ): 199– 206 . [DOI] [PubMed] [Google Scholar]
- 11. Bonds B , Dhanda A , Wade C , Diaz C , Massetti J , Stein DM . Prognostication of Mortality and Long-Term Functional Outcomes Following Traumatic Brain Injury: Can We Do Better? J Neurotrauma 2021. ; 38 ( 8 ): 1168– 1176 . [DOI] [PubMed] [Google Scholar]
- 12. Seelig JM , Becker DP , Miller JD , Greenberg RP , Ward JD , Choi SC . Traumatic acute subdural hematoma: major mortality reduction in comatose patients treated within four hours . N Engl J Med 1981. ; 304 ( 25 ): 1511– 1518 . [DOI] [PubMed] [Google Scholar]
- 13. Steyerberg EW , Mushkudiani N , Perel P , et al . Predicting outcome after traumatic brain injury: development and international validation of prognostic scores based on admission characteristics . PLoS Med 2008. ; 5 ( 8 ): e165; discussion e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letsinger J , Rommel C , Hirschi R , Nirula R , Hawryluk GWJ . The aggressiveness of neurotrauma practitioners and the influence of the IMPACT prognostic calculator . PLoS One 2017. ; 12 ( 8 ): e0183552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carney N , Totten AM , O’Reilly C , et al . Guidelines for the Management of Severe Traumatic Brain Injury, Fourth Edition . Neurosurgery 2017. ; 80 ( 1 ): 6– 15 . [DOI] [PubMed] [Google Scholar]
- 16. Mei X, Lee HC, Diao KY, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med 2020;26(8):1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Courtiol P, Maussion C, Moarii M, et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat Med 2019;25(10):1519–1525. [DOI] [PubMed] [Google Scholar]
- 18. Ardila D , Kiraly AP , Bharadwaj S , et al . Author Correction: End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography . Nat Med 2019. ; 25 ( 8 ): 1319. [DOI] [PubMed] [Google Scholar]
- 19. Titano JJ, Badgeley M, Schefflein J, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med 2018;24(9):1337–1341. [DOI] [PubMed] [Google Scholar]
- 20. Bossuyt PM , Reitsma JB , Bruns DE , et al . STARD 2015: An Updated List of Essential Items for Reporting Diagnostic Accuracy Studies . Radiology 2015. ; 277 ( 3 ): 826– 832 . [DOI] [PubMed] [Google Scholar]
- 21. Moons KGM , Altman DG , Reitsma JB , et al . Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration . Ann Intern Med 2015. ; 162 ( 1 ): W1– W73 . [DOI] [PubMed] [Google Scholar]
- 22. Goldschmidt E , Deng H , Puccio AM , Okonkwo DO . Post-traumatic hydrocephalus following decompressive hemicraniectomy: Incidence and risk factors in a prospective cohort of severe TBI patients . J Clin Neurosci 2020. ; 73 : 85– 88 . [DOI] [PubMed] [Google Scholar]
- 23. Mellett K , Ren D , Alexander S , et al . Genetic Variation in the TP53 Gene and Patient Outcomes Following Severe Traumatic Brain Injury . Biol Res Nurs 2020. ; 22 ( 3 ): 334– 340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Manley GT , Robertson CS , Okonkwo DO , et al . (March 2014 - June 2019) Transforming Research and Clinical Knowledge in Traumatic Brain Injury. Identifier NCT02119182. https://clinicaltrials.gov/ct2/show/NCT02119182 Accessed June 6, 2020 . [Google Scholar]
- 25. DeLong ER , DeLong DM , Clarke-Pearson DL . Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach . Biometrics 1988. ; 44 ( 3 ): 837– 845 . [PubMed] [Google Scholar]
- 26. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc B 1995;57(1):289–300. [Google Scholar]
- 27. Pepe MS . The Statistical Evaluation of Medical Tests for Classification and Prediction . New York, NY: : Oxford University Press, 2003. . [Google Scholar]
- 28. Turgeon AF , Lauzier F , Simard JF , et al . Mortality associated with withdrawal of life-sustaining therapy for patients with severe traumatic brain injury: a Canadian multicentre cohort study . CMAJ 2011. ; 183 ( 14 ): 1581– 1588 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tisherman SA, Schmicker RH, Brasel KJ, et al. Detailed description of all deaths in both the shock and traumatic brain injury hypertonic saline trials of the Resuscitation Outcomes Consortium. Ann Surg 2015;261(3):586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dijkland SA, Foks KA, Polinder S, et al. Prognosis in moderate and severe traumatic brain injury: A systematic review of contemporary models and validation studies. J Neurotrauma 2020;37(1):1–13. [DOI] [PubMed] [Google Scholar]
- 31. Perel P, Wasserberg J, Ravi RR, Shakur H, Edwards P, Roberts I. Prognosis following head injury: a survey of doctors from developing and developed countries. J Eval Clin Pract 2007;13(3):464–465. [DOI] [PubMed] [Google Scholar]