Abstract
The most efficient way to treat tumors is through surgery. However, many cancer patients have a poor prognosis even when they undergo radical excision at an early stage. Micrometastasis is one of the most critical factors that induced this situation. Undetected micrometastasis can lead to the failure of initial treatment. Therefore, preoperative and intraoperative detection of micrometastasis could have a significant clinical influence on the prognosis and optimal therapy for cancer patients. Additionally, to achieve this goal, researchers have aimed to create more effective detection technologies. Herein, we classify the currently reported micrometastasis detection technologies, introduce some representative samples for each technology, including the limitations, and provide future directions to overcome the limitations.
Keywords: tumor, micrometastasis, surface-enhanced Raman scattering, artificial intelligence, liquid biopsy
Introduction
Global Cancer Statistics 2020 estimates that 19.3 million new cancer cases will be diagnosed in 2020, while almost 10.0 million people will die of the disease. 1 With recent advances in the diagnosis and treatment of cancer, the main cause of death among cancer patients is linked increasingly to early metastasis. 2 This early metastasis is often a small collection of tumor cells spread from the original tumor to another through the lymph node or blood. The spread of tumor cells gets together to form a micrometastatic site. This site is too small to detect by conventional medical imaging machines, and only the microscope can identify them. However, we cannot remove all of the lymph nodes or remove the other organ from the body for testing under a microscope. Here, we require some new tools to assist us in detecting micrometastases before, during, or after operations in a noninvasive manner. If micrometastasis can be detected in an early stage cancer patient, it may influence the tumor's staging, thus affecting the treatment plan. Therefore, the detection of micrometastasis in cancer patients has a substantial clinical impact on prognosis and optimal therapy. Given the importance and urgency of detecting micrometastasis, this review introduced emerging detection technologies and their benefits and limitations (Table 1).
Table 1.
Current technologies for the detection of cancer micrometastasis with their benefits and limitation.
| Detection technology | Benefits | Limitation | Reference | |
|---|---|---|---|---|
| Radiology | Noninvasive; rapid; assist staging | Unable to detect micro tumor sites; radiation exposure | 3,4 | |
| Tissue biopsy | Gold standard | Invasive; unable to reflect the entire tumor situation; not easy to obtain the tissue samples | 5–8 | |
| SERS | Rapid; wide detection range; high specificity and sensitivity | Background signal interference; immature clinical application | 9–11 | |
| AI | Diagnosis of complicated metastasis; noninvasive | Lack of multicenter clinical validation | 12–14 | |
| ICG | Intraoperative real-time navigation | Shallow detection depth | 15–17 | |
| Liquid biopsy | Tumor marker (CEA, AFP, CA125, etc.) | Noninvasive; easy to obtain | Low specificity | 18,19 |
| CTCs | Noninvasive; reflect tumor heterogeneity | Hard to isolation; enrichment and identified | 20,21 | |
| ctDNA | Noninvasive; high specificity and sensitivity | Lack of large-scale studies; instability of indicators | 22,23 | |
| Exosomes | Noninvasive; easy to isolation and enrichment | Functional studies are still superficial; no uniform standard | 24–26 | |
Abbreviations: AI, artificial intelligence; CTC, circulating tumor cell; ctDNA, circulating tumor DNA; ICG; indocyanine green; SERS, surface-enhanced Raman scattering.
Technology for Detecting Micrometastasis
Detection by SERS
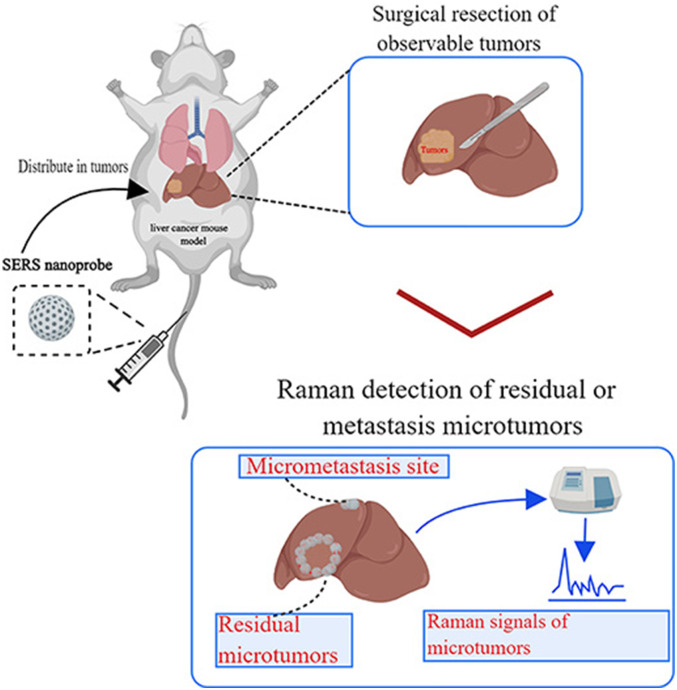
In 1982, an Indian physicist named C.V. Raman first experimentally discovered the Raman effect phenomenon. 27 When light passes through a medium, most photons are scattered by the Rayleigh effect, which means that the reflected photons retain the same energy as the incident photons. 28 However, a small No. of photons are inelastically scattered by the Raman effect. As incident photons hit the medium, they exchange energy, causing the reflected and incident photons to have different wavelengths and frequencies. Therefore, various media scatter light differently, resulting in a Raman spectrum that is unique to the primary substance. This is known as the “Raman fingerprint".10,29 However, traditional Raman scattering has the following disadvantages: weak signals, inadequate anti-interference ability, and inability to quickly identify the composition of substances, thus limiting its application in clinical practice. As research progressed, some researchers discovered that Raman signals from pyridine were significantly enhanced when adsorbed onto a roughened Ag electrode. They called this phenomenon surface-enhanced Raman scattering (SERS).29,30 The emergence of SERS overcomes the low sensitivity of conventional Raman scattering while having all the advantages of Raman signals, such as high specificity and no destruction of specimens. SERS also has the features of a narrow light spectrum, low fluorescence background interference, and near-infrared light excitation for tumor detection. Therefore, researchers have developed a variety of nanoprobes based on SERS to detect tumor lesions. Figure 1 demonstrates the function of SERS in tumor diagnosis.
Figure 1.
Schematic illustration of surface-enhanced Raman scattering (SERS) nanoprobe for intraoperative detection of residual microtumors or micrometastasis sites. Nanoprobe is injected vein before the surgery and waiting their accumulation in tumor tissues. After resection, the tumors detected by conventional methods use the Raman spectrometer to scan the organs again. Finally, the organ additionally detects the residual microtumors and micrometastasis sites. This figure is created with MedPeer (http://image.medpeer.cn/).
Nayak et al 31 developed a novel nanoparticle that combines ALT-863 (an anti-tissue factor monoclonal antibody) with surface-enhanced resonance Raman scattering nanoparticles based on SERS. The team injected this nanoparticle into breast cancer with a lung metastasis mouse model. Then, lung metastasis sites as small as 200 μm were detected by a Raman detector, and all were confirmed by pathology, while no abnormal signals were detected in normal mice. The experiment shows the superior performance of SERS. In addition, Qiu et al 32 developed gap-enhanced Raman tags that the probe has the following advantages: (1) ultrasensitivity to reveal tiny tumor lesions of minimal diameter; (2) superb photostability to maintain an active Raman signal, allowing accurate detection of scattered tumor micrometastasis; and (3) combination with photothermal therapy, allowing intraoperative detection and removal of the remaining microtumor site. Additionally, this team designed cisplatin-loaded gap-enhanced Raman tags. It is specifically used for the intraoperative detection and elimination of unresectable disseminated advanced ovarian tumors. This probe could detect micrometastasis sites as small as 1 mm in the mouse model. 33 Additionally, the probe used thermal chemotherapeutic properties to synergistically eliminate micrometastasis sites and prolong the survival of mice. 33 Another research team constructed a new Raman probe by optimizing gold nanoparticles and accessing Raman reporter molecules, generating high sensitivity and strong signals. Moreover, using this probe can accurately depict tumor boundaries to guide surgical resection and detect tiny micrometastasis sites less than 1 mm in diameter. 34 Harvey et al 35 constructed a Raman spectrometer system for skin measurements and have evaluated more than 1000 cases of skin cancers and other skin diseases. The diagnostic accuracy of the system was comparable to dermatologists. On this basis, they developed a skin cancer diagnostic system (Verisante Aura™) in collaboration with a company. And the diagnostic system has been approved for clinical use in Canada, Australia, and Europe. Using this system may help us detect lesions early that are not visible to the naked eye while reducing the need for unnecessary biopsies. In lung cancer, McGregor et al 36 combined Raman spectroscopy probes with autofluorescence bronchoscopy and white light bronchoscopy to detect cancer in the clinic. The system for localizing high-grade dysplasia and carcinoma in situ shows high sensitivity (90%) and pretty specificity (65%) that have superior capabilities compared to conventional bronchoscopy. That means this machine may improve the rate of early cancer detection and can detect micrometastasis.
According to the above article, this technology demonstrates excellent results in detecting micrometastasis sites. However, it still has several limitations, including interference from background signals and inadequate penetration depth, and most research has focused on animal models. Meanwhile, we need to design multicenter clinical trials to validate its ability to detect micrometastasis. If we can combine its advantages with improvement measures, this technology still has considerable potential in micrometastasis detection.
Application of AI in the Field of Micrometastasis Detection
Recently, with the development of combined modality machines and improved medical devices, image data volumes have increased dramatically. Because of this, the past methods of image data processing and usage for small samples cannot wholly extract the extensive data information contained in images. In 2012, Lambin first introduced the concept of radiomics. Radiomics is a method for extracting a large amount of feature data from medical images using data characteristic algorithms.37–39 These feature data can uncover disease characteristics that the naked eye fails. 40 The different disease forms present distinct imaging features for accurate diagnosis, efficacy assessment, and prognosis prediction.37,38 The radiomics process consists of four main steps. The first step is to acquire high-quality and standardized images. Based on the image, radiologists or automated tools can identify macroscopic tumors. Then, quantitative imaging features are extracted from the tumor region. Finally, the extracted features are analyzed. 37 Nowadays, computed tomograpgy (CT) scan, magnetic resonance imaging (MRI), and positron emission tomogrphy (PET) are commonly to detect metastasis, generating a huge amounts of data. Using radiomics, massive data is collected and analyzed to extract some point features that can be used for classification. And the whole process is time-consuming, laborious, and may generate error. With the advancement of artificial intelligence (AI), especially deep learning technology, in recent years, we have developed a new model that uses end-to-end networks directly from the training data output to attain the final result. The model replaces the traditional method of manually extracting data features and reduces the requirement for manual preprocessing to allow machine learning algorithms to learn from the data and extract features autonomously. 41
At present, several research teams are using AI technology in various areas, including cancer diagnosis, tumor detection, tumor classification, survivability prediction, malignancy prediction, recurrence prediction, and cancer staging. 42 In this field, we focus on applying AI in micrometastasis detection. A team has developed AI-based radiomics to achieve a microperitoneal metastasis diagnosis breakthrough. The team collected clinical and imaging data from 554 gastric cancer patients at four hospitals. These patients' peritoneal metastasis (PM) status was initially negative on CT. However, 122 were later diagnosed as PM-positive by laparoscopy. The research-based venous phase CT images with the largest tumor area and the peritoneal region nearest to the center of the primary tumor extracted 266 features. By analysis, they found two key features from the primary tumor, two key features from the peritoneum, and one clinical factor of the Lauren type, which significantly correlated with peritoneal metastases. Based on these features, researchers constructed a radiomic nomogram and achieved accurate prediction results in the four-hospital validation dataset (all area under curve (AUC)>0.92), especially in 122 patients with positive microperitoneal metastasis, for which the average detection rate of peritoneal metastasis was 85%. 13 Recently, a team obtained a new advance in predicting peritoneal micrometastasis. Using dual-energy computed tomography, they developed a new radiomics model. This model can be used to predict peritoneal micrometastasis of gastric cancer, demonstrating a significantly higher predictive value in terms of peritoneal status than the clinical model and human experts (AUC = 0.7855, p < 0.005; AUC = 0.7322, p <0.001, respectively). 43
As medical machines and computer science progress, researchers focus more on AI. AI technology has an excellent capability of detecting micrometastases. However, there are still some limitations that need to be overcome. Currently, most research involves peritoneal micrometastases. This means that clinical implications are limited and requires us to broaden research directions and cover more cancer types. Second, most research teams included a small No. of patients, which was insufficient. A large sample size, multicenter study, and different ethnicities can improve AI training. Last, the process of defining the entire part will consume considerable time. We may choose a region to detect, omitting potential metastasis in other areas. In summary, AI models have the following advantages: they are noninvasive and easy to use. According to these advantages, it is an effective way of detecting micrometastases.
The Application of ICG Dyes in the Diagnosis and Treatment of Tumor Diseases
Indocyanine green (ICG) is a water-soluble cyanine dye that can bind tightly to plasma proteins but is not readily absorbed by extrahepatic tissues. 44 When illuminated with near-infrared light, it can emit light with a peak wavelength of approximately 840 nm. Based on these features, ICG was initially used for determining hepatic function and for ophthalmic angiography.45,46 Benefits from extensive development and application of fluorescence imaging devices and fluorescence navigation techniques. ICG is no longer limited to evaluating liver function and blood flow. It has also shown significant promise for detecting micro liver tumor lesions and defining tumor boundaries during surgery.16,45
In 2009, Ishizawa et al first reported that ICG fluorescence imaging successfully detected microscopic tumor lesions in the liver during surgery. The principle is that ICG is absorbed by the liver and excreted through the bile duct. Tumors destroy the hepatic normal tissue structure and function, causing the uptake and excretion of ICG to be impaired, resulting in the accumulation of ICG and revealing the lesion in real-time during surgery. 47 Furthermore, we observed 3 patterns of fluorescence intraoperatively based on the degree of differentiation of the tumor tissue: cancerous-type fluorescence, partial fluorescence, and rim-type fluorescence.45,48 On this basis, the team for 276 postoperative patients conducted a retrospective study. In this study, ICG fluorescence imaging revealed 273 of 276 hepatocellular carcinoma cells (HCC) (sensitivity, 99%). Among the 273 HCC patients, 14 lesions were not detected by preoperative imaging, and 3 preoperative imaging confirmed lesions but could not be detected by visual inspection, manual palpation, and intraoperative ultrasound during surgery. Moreover, fluorescence imaging revealed residual fluorescing lesions on the liver's raw surface in 6 patients after liver resection, which were all pathologically proven to be HCCs. 45 Handgraaf et al performed a retrospective multicenter analysis of long-term follow-up after ICG fluorescence imaging-assisted resection of colorectal liver metastases. According to the study, 21 metastases could only be detected by ICG fluorescence imaging. In addition, the tumors detected by ICG fluorescence imaging were significantly smaller (3.2 ± 1.8 mm vs 7.4 ± 2.6 mm, P < .001) than those detected by visual inspection, palpation, and intraoperative ultrasound. 49 Katada et al evaluated 133 pancreatic cancer patients without preoperative evidence of liver metastasis using ICG imaging. They detected 32 abnormal fluorescences among 133 patients, 20 of which were hepatic micrometastases, the minimum metastasis diameter being 2.86 mm. 50 In another study on hepatic micrometastases from pancreatic cancer, 13 of 49 patients detected an abnormal signal with ICG fluorescence imaging. Eight cases were pathologically confirmed to have liver micrometastases. 51
In conclusion, ICG has excellent potential in detecting hepatic microlesions and occult micrometastasis. Nevertheless, this technology still has some limitations. Due to the biological properties of ICG, benign lesions such as cirrhotic nodules can easily be classified as tumor lesions, causing a high false-positive rate. 47 Additionally, the physical characteristics of ICG indicate that the maximum penetration depth is only 5 to 10 mm, which restricts the detection of deep lesions.52,53 ICG fluorescence imaging technology for tumor micrometastasis detection is mainly limited to gastrointestinal tumors, especially liver tumors. We need to do more research to use this technology on other types of tumors.
Liquid Biopsy Using Tumor Diagnosis
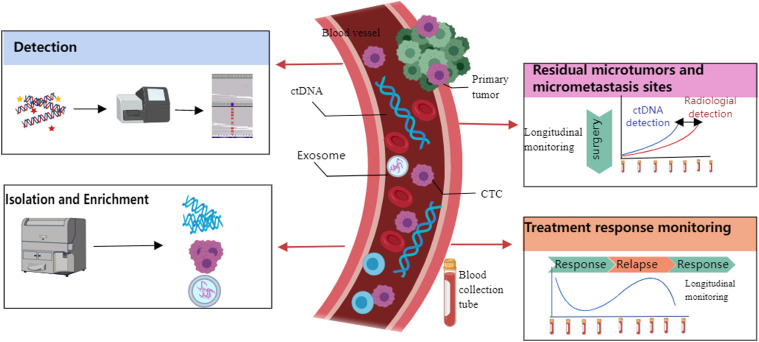
Tumor tissue biopsy is still the gold standard for malignant tumor diagnosis. However, tissue biopsy is invasive and contains a tiny sample that cannot monitor dynamic tumor progression. 54 We need a new technology that can accurately analyze the biological characteristics and heterogeneity of the tumor and be accessible to college samples. 55 In this situation, liquid biopsy might be a valuable addition to current assessment methods. Liquid biopsy is based on detecting tumor-related biomarkers from body fluids (such as peripheral blood). 55 These biomarkers include circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosomes, which have all been proven to be related to tumor metastasis. 56 Figure 2 demonstrates the workflow and functionality of liquid biopsy.
Figure 2.
Schematic illustration of liquid biopsy workflow and functional. (a) Isolation and enrichment circulating tumor cell (CTC)/circulating tumor DNA (ctDNA) from blood according to their biochemical or physical characters. (b) Detecting and longitudinal monitoring of their amount or DNA/RNA/protein changes. Longitudinal monitoring can detect residual microtumors or micrometastasis sites, and monitor treatment response. This figure is created with MedPeer (http://image.medpeer.cn/).
Circulating Tumor Cells
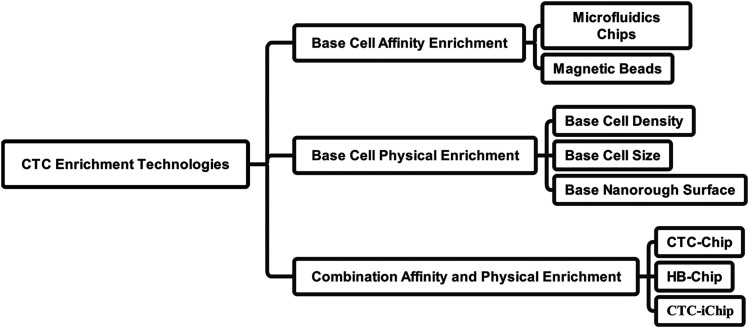
In 1869, Ashworth compared blood cells with tumor cells and first proposed CTCs. 54 CTCs are defined as circulating in peripheral blood tumor cells that shed from primary or metastatic lesions. 57 With the improvement of tumor research in recent years, it has been found that although many patients have clinically undetectable residual tumor lesions after treatment, they ultimately die due to tumor metastasis. The reason may be related to the inability to detect CTCs with conventional imaging technology. In addition, CTCs already exist in the peripheral blood when the cancer is at an early stage. 58 We may use this feature for early diagnosis of metastases. There are still some problems since not all CTCs carry the potential to develop metastasis, and how can we guarantee the CTCs we study are able to form metastasis. Meanwhile, the No. of CTCs is rare in the peripheral blood. 59 Hence, CTC isolation/enrichment/detection is a critical challenge. At present, CTC enrichment technologies are classified as shown in Figure 3. Specifically, they can be divided into affinity-based methods and physical-based methods. Immunomagnetic beads are currently the most commonly used technique for identifying CTCs. The principle is based on the fact that CTCs primarily express epithelial-derived surface markers, and Ag-Ab reactions are used to detect CTCs. This method is divided into positive and negative isolation methods. The former methods isolate CTCs by magnetic beads coupled to epithelial cell adhesion molecules and cytokeratin. The latter method is through magnetic beads coupled to leukocyte markers such as CD45 that indirectly isolate CTCs by removing leukocytes. And the physical-based methods’ principle is CTCs are different from blood cells physical characteristics such as cell size, density, and nanorough surface. 60 Some indicators, such as recovery rate, the limit of detection, and purity, represent the technologies' performance. 57 The following section will describe the application of CTC detection in tumor micrometastasis and early diagnosis.
Figure 3.
A map showing the classification of current circulating tumor cell (CTC) enrichment technologies.
Research proves that most CTCs entering the circulatory system undergo apoptosis under the effect of immune recognition and killing. Only a tiny portion of CTCs survived and developed metastases under certain conditions. Therefore, the detection of CTCs in peripheral blood does not equal metastases. And there is no method that can definitively detect CTCs with the potential to become a metastasis site. At present, the possibility of metastasis is predicted mainly by dynamic monitoring of the No. of CTCs in the peripheral blood. As research progresses, many studies have confirmed the CTC function of tumor micrometastasis and early diagnosis in breast cancer, cervical cancer, and other malignant tumors. Li et al 61 used the CanPatrol system to detect CTCs from 90 patients with early cervical cancer, and the results showed that the positivity rate was 90%. They also found that CTCs with mesenchymal phenotypes are closely associated with lymphatic vascular invasion and pelvic lymph node metastasis in stages I to IIA cervical cancer. Hence, detecting CTC phenotypes may help us diagnose cervical cancer micrometastasis. In breast cancer, many studies are focusing on patients at the early stage without apparent distant metastasis (tumor–node–metastasis [TNM] stage M0). As early as 2010, the American Joint Committee on Cancer (AJCC) included CTCs in the TNM staging system as a new M stage criterion, classified as cM0(i + ). cM0(i + ) refers to no clinical or radiographic evidence of distant metastasis tumor cells in circulating blood, bone marrow, or other nonregional nodal tissue that are no longer than 0.2 mm.20,62 In 2018, the AJCC published the eighth edition cancer staging manual with research improvements. In this manual, they incorporate CTCs, human epidermal growth factor receptor 2, estrogen receptor, and progesterone receptor, grade into the traditional anatomic staging system to evaluate tumor prognosis.63,64 The new manual posits that peripheral blood CTCs ≥5/7.5 mL for advanced breast cancer and ≥1/7.5 mL for early stage suggest poor prognosis, AJCC Level of Evidence II. 65 As the manual gradually improved, CTCs officially entered the tumor staging system, which means that CTC detection technology provides more comprehensive guidance for metastasis and early diagnosis. In clinical practice, Zhang et al 66 examined the No. of CTC in patients with osteosarcoma, and found that patients with ≥2 CTCs compared to <2 CTCs had a shorter median progression-free survival. That means the more CTCs in the peripheral blood, the higher the possibility of metastasis. All this evidence supports the CTC-based detection method, which helped us to diagnose micrometastasis.
Circulating Tumor DNA
Cell-free DNA (cfDNA) exists in the circulatory system and originates from apoptotic cells (mainly lymphocytes).67,68 In 1977, Leon et al first discovered that ctDNA was a part of cfDNA. 69 With the rapid improvement of gene sequencing technology, ctDNA has gained more attention in recent years. ctDNA is fragment DNA released from necrotic and apoptotic tumor cells in the peripheral blood.20,70 Thus, in principle, ctDNA has the same characteristics as their originating tumor cells. However, research shows that the total amount of ctDNA may be less than 0.01% of the total cfDNA concentration.22,71 In addition, the length of ctDNA fragments for many cancers is less than 167bp, and choose between 90bp and 150bp can improve the ability to detect ctDNA. 72 Additionally, many studies found that patients with tumors showed a significant increase in ctDNA levels regardless of the stage and type of tumor. 73 When the tumor load increases, the levels will also be evaluated. 73 Theoretically, ctDNA contains all genetic information about tumor gene mutations, including single-nucleotide variations, copy number variations, and DNA methylation. 25 Thus, ctDNA-based detection may provide more comprehensive genetic variation information than tissue biopsy and present potential value in early diagnosis and individualized treatment. However, the level of ctDNA in the blood is very low, limiting conventional gene sequencing technologies such as the Sanger method. With the rapid development of DNA sequencing technology and molecular diagnostic techniques in recent years, we have more methods to quantify DNA molecules. At present, we often apply polymerase chain reaction, next-generation sequencing, and whole-exome sequencing technologies to detect ctDNA. 74
Tumor diagnosis still relies on tumor markers, tissue biopsy, and radiography in current clinical practice. Although these methods contribute significantly, lower diagnostic efficacy and biopsy damage and radiation remain unavoidable problems. In this situation, ctDNA shows significant advantages of accurate diagnostic capability and minimal invasiveness. Specifically, ctDNA contains all genetic information of tumors. ctDNA can be used as a marker to analyze epigenetic changes in tumors. ctDNA also plays a significant role in early diagnosis, micrometastasis detection or metastasis early diagnosis, and prognosis evaluation. In particular, for metastasis detection and prognostic evaluation, research has proven that early metastasis and micrometastasis may be the main reasons for tumor progression, recurrence, and poor prognosis. Thomas et al found that monitoring ctDNA can detect progression 10 months earlier than the conventional method. 75 This result prompted us to conclude that the tumor may have developed a micrometastasis site a few months ago. Additionally, other scientists have found a similar conclusion. Board et al found that the mutation rate of the PIK3C gene in ctDNA was much higher in grade III breast cancer than in grade I. 76 In a similar study of non-small cell lung and urothelial cancers after using immunotherapy, researchers used targeted sequencing to assess variant allele frequencies of somatic mutations in ctDNA. They found that the changes in VAF occurred earlier than radiographic response. 77 Therefore, we can early diagnose micrometastasis. Additionally, detecting microresidual lesions after surgery is still challenging for doctors. Recent studies show that immediate or serial detection of ctDNA after surgery may solve this problem. 74 Chaudhuri et al used personalized cancer profiling by deep sequencing analysis of ctDNA from patients who received treatment for localized lung cancer. They found that ctDNA was 5.2 months earlier than radiographic progression. In addition, 53% of patients carrying ctDNA mutations have better efficacy with tyrosine kinase inhibitors or immune checkpoint blockade. 78 Ikeda et al detected MET alterations in ctDNA of patients with various tumors and showed that MET ctDNA alterations were highly relevant to bone metastasis. 79 Interestingly, Olsson et al used the approach of combining whole-genome sequencing and droplet digital PCR to identify ctDNA in patients with primary breast cancer. They found that ctDNA-based testing preceded clinical detection of metastasis with an average advance of 11 months through serial monitoring. 80 In addition, Tie et al 81 designed a cohort study that included 230 postoperative stage II colon cancer patients and divided them into two groups. The final results show that the 27-month recurrence rate of the ctDNA-positive group was 79% and that of the ctDNA-negative group was only 9.8%. The above research is also strong evidence that supports ctDNA and can help us diagnose metastasis early.
Exosomes
Exosomes are a subtype of extracellular vesicles with bilayer lipids and are approximately 30 nm to 150 nm in size. 82 In 1986, scientists first discovered exosomes in the medium of sheep reticulocytes. 82 Initial studies considered exosomes as trash bags that help drain nonfunctional cellular components. 83 As research has progressed, exosomes have gradually lifted the veil. Studies show that the formulation of exosomes is a complex and dynamic process. First, the plasma membrane forms intracellular endosomes by budding. 83 Second, intracellular endosomes that continue invagination generate multivesicular bodies (MVBs). 84 MVBs contain DNA fragments, messenger RNAs (mRNAs), microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and proteins.85,86 According to the database ExoCarta (http://www.exocarta.org/) count: 9769 proteins, 3408 mRNAs and 2838 miRNAs were identified in exosomes. Finally, MVBs fuse with the plasma membrane and release their contents into the extracellular space in the form of exosomes or fuse with lysosomes to digest their contents. 87 In addition, exosomes, as storage and transportation of the above biomolecules, can exist in various body fluids, such as blood, urine, cerebrospinal fluid, saliva, and ascites. 88 With increasing research on exosomes, researchers have found that they are involved in targeted messaging and carry much biological information. Research shows that exosomes play an important role in carcinogenesis, metastasis, diagnosis, and treatment. 89 Next, we will describe the relationship between exosomes and tumor metastasis.
Tumor metastasis is a constant topic in treatment because metastasis can occur in any stage of cancer and cannot be avoided. Exosomes promote tumor metastasis mainly through the following mechanisms: enhancing the migration and invasion of tumor cells, establishing a suitable microenvironment for metastasis, and remodeling the extracellular matrix (ECM). 90 Several studies have demonstrated that exosomes are associated with tumor metastasis. Premetastatic niche (PMN) formation joins the regulation of tumor invasion and metastasis. 91 Emmanouilidi et al found that 362 proteins involved in PNM formation are specifically expressed in human pancreatic cancer cells. 92 Likewise, Peinado et al found that exosomes from a metastatic mouse melanoma cell line have higher levels of cMet than exosomes from a less aggressive mouse melanoma cell line. 93 Specifically, exosomes contain Met proteins that affect bone marrow-derived cells (BMDCs). BMDCs can influence the microenvironment, which increases vascular permeability and promotes neovascularization. Then, the change contributes to the metastatic colonization of tumor cells. In the above studies, exosomes from the more highly metastatic cell line included some unique proteins. These proteins can influence the migration and invasion of tumor cells and the microenvironment. In breast cancer, exosomes contain miR-9, which can promote the activation of normal fibroblasts into cancer-associated fibroblasts, thereby remodeling the ECM and affecting metastasis. 94 These studies reflect the close connection between tumor metastasis and exosomes. However, Exosomes still lack solid clinical evidence. The amount of exosomes is huge and isolating tumor-specific exosomes is challenging maybe for a reason.
Others
In addition to the SERS mentioned above and AI technology in microtumor lesion detection, many teams still develop other technologies. Tumor cells have different energy metabolism patterns than normal cells. According to this characteristic, researchers create new fluorescent probes based on nitrogen-doped carbon dots. This probe can identify micron-sized tumor lesions within the choroidal tissue and successfully monitor tumor proliferation and metastasis in an in situ mouse eye tumor model. 95 Zhou et al developed an MRI contrast agent that can bind to fibrin–fibronectin complexes abundant in the tumor microenvironment of fast-growing breast cancer, providing robust contrast enhancement in metastatic tumors enabling the detection of micrometastases 0.5 mm in size. 96 In addition, the researcher was inspired by the mantis shrimp visual system and developed a six-channel color/near-infrared image sensor. This sensor can distinguish diseased from healthy tissue in an estimated 92% of cases. 97 Li et al 98 demonstrated an excellent ability to detect lymph nodes using fluorescent paired-agent imaging. Micrometastases (0.2 mm diameter) can be detected by fluorescent paired-agent imaging with >99% sensitivity and >95% specificity compared to frozen pathology, which has a sensitivity of 20%. In recent years, with the development of computer science, deep learning has become a hot topic of current research. Based on deep learning, Yuan et al 99 built a classifier. This classifier demonstrated an accuracy of 94.11% with an AUC of 0.922 (0.912-0.944), sensitivity of 93.75%, specificity of 94.44%, positive predictive value of 93.75%, and negative predictive value of 94.44%, which shows a great ability to detect peritoneal micometastasis.
Conclusion and Future Directons
The concept of micrometastasis has been proposed for over 60 years. However, research on the clinical significance of micrometastasis has been limited in the past. As research progresses, researchers have found that micrometastasis can affect cancer staging and thus change clinical treatment. However, because microtumor lesions and micrometastasis are too tiny, detecting them is not easy. Therefore, finding microsites has become an urgent problem, and researchers are endeavoring to improve or invent new technologies for detecting micrometastasis.
With the development of medical imaging, the clarity and precision of CT, MR, PET, ultrasound, and other imaging devices have also made significant progress, playing a great role in detecting tumor lesions. However, these devices still cannot detect micrometastasis and thus cannot select better strategies for tumor treatment. The development of SERS and AI technologies has improved the detection accuracy to the millimeter or even micron level, making it possible to detect microtumor lesions while having advantages such as noninvasive and synergistic treatment. However, the above technologies still have some restrictions: background signal interference and insufficient penetration depth. Looking forward to the future, if we can overcome this problem and test it in large-scale clinical experiments, SERS and AI technologies will be a good tool that possesses high resolution and accuracy to help us detect micrometastasis.
Additionally, the emergence of liquid biopsy provides another idea for us to diagnose micrometastasis. However, the clinical application of this technology still needs to solve several limitations. First, the No. of CTCs, ctDNA, and exosomes is small, so the current method cannot solve false positives and false negatives. Second, clinical use cannot solve the issue of standardization. Finally, current detection costs are still high. In the future, we aim to standardize liquid biopsy technologies, automate them, and miniaturize them at low cost.
In summary, we can take advantage of various technologies and apply them in combination. We must make significant progress in the detection of micrometastasis. Moreover, clinical treatment may be based on the detection of micrometastasis in the future.
Abbreviations
- AI
artificial intelligence
- AJCC
American Joint Committee on Cancer
- AUC
Area Under Curve
- BMDCs
bone marrow-derived cells
- CTCs
circulating tumor cells
- CT
Computed Tomography
- ctDNA
circulating tumor DNA
- cfDNA
cell-free DNA
- ECM
extracellular matrix
- HCC
hepatocellular carcinoma cells
- ICG
indocyanine green
- lncRNAs
long noncoding RNAs
- MRI
Magnetic Resonance Imaging
- MVBs
multivesicular bodies
- mRNAs
messenger RNAs
- miRNAs
microRNAs
- PET
Positron Emission Tomograpy
- PM
peritoneal metastasis
- PMN
premetastatic niche
- SERS
surface-enhanced Raman scattering
- TNM
tumor–node–metastasis
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This research was supported by grants from National Natural Science Foundation of China (grant Nos. 81730108, 81973635), Zhejiang Provincial Natural Science Foundation of China for Distinguished Young Scholars (grant No. LR18H160001), and Zhejiang province science and technology project of TCM (grant No. 2019ZZ016).
Ethical Approval: Our study did not require an ethical board approval because it did not contain human or animal trials.
ORCID iD: Xuqing Mao https://orcid.org/0000-0002-7049-5809
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Alix-Panabières C, Riethdorf S. Cancer micrometastases. Nature Reviews Clinical Oncology. 2009;6(6):339‐351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 3.Fass L. Imaging and cancer: a review. Mol Oncol. 2008;2(2):115‐152. doi: 10.1016/j.molonc.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kijima S, Sasaki T, Nagata K, Utano K, Lefor AT, Sugimoto H. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J Gastroenterol. 2014;20(45):16964‐16975. doi: 10.3748/wjg.v20.i45.16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scattoni V, Maccagnano C, Capitanio U, Gallina A, Briganti A, Montorsi F. Random biopsy: when, how many and where to take the cores? World J Urol. 2014;32(4):859‐869. doi: 10.1007/s00345-014-1335-0. [DOI] [PubMed] [Google Scholar]
- 6.VanderLaan PA. Fine-needle aspiration and core needle biopsy: an update on 2 common minimally invasive tissue sampling modalities. Cancer Cytopathol. 2016;124(12):862‐870. doi: 10.1002/cncy.21742. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Li L, Qu C, Liang S, Zeng B, Luo Z. Endoscopic ultrasound-guided fine needle core biopsy for the diagnosis of pancreatic malignant lesions: a systematic review and meta-analysis. Sci Rep. 2016;6:22978. doi: 10.1038/srep22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eskra JN, Rabizadeh D, Pavlovich CP, Catalona WJ, Luo J. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 2019;22(3):362‐381. doi: 10.1038/s41391-019-0127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auner GW, Koya SK, Huang C, et al. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018;37(4):691‐717. doi: 10.1007/s10555-018-9770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher MF. How can we apply the use of surface-enhanced Raman scattering nanoparticles in tumor imaging? Nanomedicine (Lond). 2017;12(3):171‐174. doi: 10.2217/nnm-2016-0385. [DOI] [PubMed] [Google Scholar]
- 11.Vendrell M, Maiti KK, Dhaliwal K, Chang Y-T. Surface-enhanced Raman scattering in cancer detection and imaging. Trends Biotechnol. 2013;31(4):249‐257. doi: 10.1016/j.tibtech.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Lu MY, Chen TY, Williamson DFK, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. 2021;594(7861):106‐110. doi: 10.1038/s41586-021-03512-4. [DOI] [PubMed] [Google Scholar]
- 13.Dong D, Tang L, Li ZY, et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann Oncol. 2019;30(3):431‐438. doi: 10.1093/annonc/mdz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horie Y, Yoshio T, Aoyama K, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89(1):25‐32. doi: 10.1016/j.gie.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 15.Keller DS, Ishizawa T, Cohen R, Chand M. Indocyanine green fluorescence imaging in colorectal surgery: overview, applications, and future directions. Lancet Gastroenterol Hepatol. 2017;2(10):757‐766. doi: 10.1016/S2468-1253(17)30216-9. [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Liu T, Su M, et al. Improving the surgical effect for primary liver cancer with intraoperative fluorescence navigation compared with intraoperative ultrasound. Med Sci Monit. 2019;25:3406‐3416. doi: 10.12659/MSM.916423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim C, Vibert E, Azoulay D, et al. Indocyanine green fluorescence imaging in the surgical management of liver cancers: current facts and future implications. J Visc Surg. 2014;151(2):117‐124. doi: 10.1016/j.jviscsurg.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Michl P, Pauls S, Gress TM. Evidence-based diagnosis and staging of pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20(2):227‐251. [DOI] [PubMed] [Google Scholar]
- 19.Funston G, Hamilton W, Abel G, Crosbie EJ, Rous B, Walter FM. The diagnostic performance of CA125 for the detection of ovarian and non-ovarian cancer in primary care: a population-based cohort study. PLoS Med. 2020;17(10):e1003295. doi: 10.1371/journal.pmed.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479‐491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 21.Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res. 2015;21(21):4786‐4800. doi: 10.1158/1078-0432.CCR-14-1190. [DOI] [PubMed] [Google Scholar]
- 22.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cescon D, Bratman S, Chan S, Siu L. Circulating tumor DNA and liquid biopsy in oncology. Nature Cancer. 2020;1(3):276‐290. doi: 10.1038/s43018-020-0043-5. [DOI] [PubMed] [Google Scholar]
- 24.Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. 2012;44(11):2060‐2064. doi: 10.1016/j.biocel.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics. 2017;15(2):59‐72. doi: 10.1016/j.gpb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766‐769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raman CV, Krishnan KS. A new type of secondary radiation. Nature. 1928;121(3048):501‐502. doi: 10.1038/121501c0. [DOI] [Google Scholar]
- 28.Young AT. Rayleigh Scattering. Appl Opt. 1981;20(4):533‐535. doi: 10.1364/AO.20.000533. [DOI] [PubMed] [Google Scholar]
- 29.Andreou C, Kishore SA, Kircher MF. Surface-Enhanced Raman spectroscopy: a new modality for cancer imaging. J Nucl Med. 2015;56(9):1295‐1299. doi: 10.2967/jnumed.115.158196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombardi JR, Birke RL. The theory of surface-enhanced Raman scattering. J Chem Phys. 2012;136(14):144704. [DOI] [PubMed] [Google Scholar]
- 31.Nayak TR, Andreou C, Oseledchyk A, et al. Tissue factor-specific ultra-bright SERRS nanostars for Raman detection of pulmonary micrometastases. Nanoscale. 2017;9(3):1110‐1119. doi: 10.1039/c6nr08217c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu Y, Zhang Y, Li M, et al. Intraoperative detection and eradication of residual microtumors with gap-enhanced Raman tags. ACS Nano. 2018;12(8):7974‐7985. doi: 10.1021/acsnano.8b02681. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu Z, Thackray BD, et al. Intraoperative Raman-guided chemo-photothermal synergistic therapy of advanced disseminated ovarian cancers. Small. 2018;14(31):e1801022. doi: 10.1002/smll.201801022 [DOI] [PubMed] [Google Scholar]
- 34.Wei Q, Arami H, Santos HA, et al. Intraoperative assessment and photothermal ablation of the tumor margins using gold nanoparticles. Advanced Science. 2021;8(5):2002788. doi: 10.1002/advs.202002788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lui H, Zhao J, McLean D, Zeng H. Real-time Raman spectroscopy for in vivo skin cancer diagnosis. Cancer Res. 2012;72(10):2491‐2500. doi: 10.1158/0008-5472.CAN-11-4061. [DOI] [PubMed] [Google Scholar]
- 36.McGregor HC, Short MA, McWilliams A, et al. Real-time endoscopic Raman spectroscopy for in vivo early lung cancer detection. J Biophotonics. 2017;10(1):98–110. 10.1002/jbio.201500204 [DOI] [PubMed] [Google Scholar]
- 37.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441‐446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nature Reviews Clinical Oncology. 2017;14(12):749‐762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 39.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563‐577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip SSF, Liu Y, Parmar C, et al. Associations between radiologist-defined semantic and automatically computed radiomic features in non-small cell lung cancer. Sci Rep. 2017;7(1):3519. doi: 10.1038/s41598-017-02425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436‐444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 42.Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A, Benali H. From handcrafted to deep-learning-based cancer radiomics: challenges and opportunities. IEEE Signal Process Mag. 2019;36(4):132‐160. doi: 10.1109/MSP.2019.2900993. [DOI] [Google Scholar]
- 43.Chen Y, Xi W, Yao W, et al. Dual-Energy computed tomography-based radiomics to predict peritoneal metastasis in gastric cancer. Front Oncol. 2021;11:659981. doi: 10.3389/fonc.2021.659981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leevy CM, Mendenhall CL, Lesko W, Howard MM. Estimation of hepatic blood flow with indocyanine green. J Clin Invest. 1962;41(5):1169‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishizawa T, Masuda K, Urano Y, et al. Mechanistic background and clinical applications of indocyanine green fluorescence imaging of hepatocellular carcinoma. Ann Surg Oncol. 2014;21(2):440‐448. doi: 10.1245/s10434-013-3360-4. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Kim Y-S, Milenic DE, Baidoo KE, Brechbiel MW. In vitro and in vivo analysis of indocyanine green-labeled panitumumab for optical imaging-a cautionary tale. Bioconjug Chem. 2014;25(10):1801‐1810. doi: 10.1021/bc500312w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishizawa T, Fukushima N, Shibahara J, et al. Real-time identification of liver cancers by using indocyanine green fluorescent imaging. Cancer. 2009;115(11):2491‐2504. doi: 10.1002/cncr.24291. [DOI] [PubMed] [Google Scholar]
- 48.van der Vorst JR, Schaafsma BE, Hutteman M, et al. Near-infrared fluorescence-guided resection of colorectal liver metastases. Cancer. 2013;119(18):3411‐3418. doi: 10.1002/cncr.28203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Handgraaf HJM, Boogerd LSF, Höppener DJ, et al. Long-term follow-up after near-infrared fluorescence-guided resection of colorectal liver metastases: a retrospective multicenter analysis. Eur J Surg Oncol. 2017;43(8):1463‐1471. doi: 10.1016/j.ejso.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katada T, Hashidate H, Yokoyama N, Sudo N, Mitsuma K, Otani T. Initial features of hepatic metastases from pancreatic cancer: histological and radiolographical appraisal of hepatic micrometastases detected by real-time fluorescent imaging. Pancreas. 2017;46(9):1196‐1201. doi: 10.1097/MPA.0000000000000915. [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama N, Otani T, Hashidate H, et al. Real-time detection of hepatic micrometastases from pancreatic cancer by intraoperative fluorescence imaging: preliminary results of a prospective study. Cancer. 2012;118(11):2813‐2819. doi: 10.1002/cncr.26594. [DOI] [PubMed] [Google Scholar]
- 52.Schaafsma BE, Mieog JSD, Hutteman M, et al. The clinical use of indocyanine green as a near-infrared fluorescent contrast agent for image-guided oncologic surgery. J Surg Oncol. 2011;104(3):323‐332. doi: 10.1002/jso.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schols RM, Bouvy ND, van Dam RM, Stassen LPS. Advanced intraoperative imaging methods for laparoscopic anatomy navigation: an overview. Surg Endosc. 2013;27(6):1851‐1859. doi: 10.1007/s00464-012-2701-x. [DOI] [PubMed] [Google Scholar]
- 54.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. doi: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rijavec E, Coco S, Genova C, Rossi G, Longo L, Grossi F. Liquid biopsy in non-small cell lung cancer: highlights and challenges. Cancers (Basel). 2020;12(1):17. 10.3390/cancers12010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2018;19(1):11‐34. doi: 10.1039/c8lc00684a. [DOI] [PubMed] [Google Scholar]
- 57.Shen Z, Wu A, Chen X. Current detection technologies for circulating tumor cells. Chem Soc Rev. 2017;46(8):2038‐2056. doi: 10.1039/c6cs00803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ilie M, Hofman V, Long-Mira E, et al. "Sentinel" circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014;9(10):e111597. doi: 10.1371/journal.pone.0111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235‐1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10(3):374‐394. doi: 10.1016/j.molonc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan L, Yan G, Chen W, Sun L, Wang J, Yang J. Distribution of circulating tumor cell phenotype in early cervical cancer. Cancer Manag Res. 2019;11:5531‐5536. doi: 10.2147/CMAR.S198391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giuliano AE, Connolly JL, Edge SB, et al. Breast cancer-Major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(4):290‐303. doi: 10.3322/caac.21393. [DOI] [PubMed] [Google Scholar]
- 63.Giuliano AE, Edge SB, Hortobagyi GN. Eighth edition of the AJCC cancer staging manual: breast cancer. Ann Surg Oncol. 2018;25(7):1783‐1785. doi: 10.1245/s10434-018-6486-6. [DOI] [PubMed] [Google Scholar]
- 64.Plichta JK, Ren Y, Thomas SM, et al. Implications for breast cancer restaging based on the 8th edition AJCC staging manual. Ann Surg. 2020;271(1):169‐176. doi: 10.1097/SLA.0000000000003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cserni G, Chmielik E, Cserni B, Tot T. The new TNM-based staging of breast cancer. Virchows Arch. 2018;472(5):697‐703. doi: 10.1007/s00428-018-2301-9. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H, Gao P, Xiao X, et al. A liquid biopsy-based method for the detection and quantification of circulating tumor cells in surgical osteosarcoma patients. Int J Oncol. 2017;50(4):1075‐1086. doi: 10.3892/ijo.2017.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abe T, Nakashima C, Sato A, et al. Origin of circulating free DNA in patients with lung cancer. PLoS One. 2020;15(7):e0235611. doi: 10.1371/journal.pone.0235611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem. 2007;53(12):2215. doi: 10.1373/clinchem.2007.092734. [DOI] [PubMed] [Google Scholar]
- 69.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646‐650. [PubMed] [Google Scholar]
- 70.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature Reviews Clinical Oncology. 2013;10(8):472‐484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 71.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548‐554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466):eaat4921. doi: 10.1126/scitranslmed.aat4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madhavan D, Wallwiener M, Bents K, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat. 2014;146(1):163‐174. doi: 10.1007/s10549-014-2946-2. [DOI] [PubMed] [Google Scholar]
- 74.Chae YK, Oh MS. Detection of minimal residual disease using ctDNA in lung cancer: current evidence and future directions. J Thorac Oncol. 2019;14(1):16‐24. doi: 10.1016/j.jtho.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 75.Reinert T, Schøler LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65(4):625‐634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 76.Board RE, Wardley AM, Dixon JM, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120(2):461‐467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 77.Raja R, Kuziora M, Brohawn PZ, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24(24):6212‐6222. doi: 10.1158/1078-0432.CCR-18-0386. [DOI] [PubMed] [Google Scholar]
- 78.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7(12):1394‐1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ikeda S, Schwaederle M, Mohindra M, Fontes Jardim DL, Kurzrock R. MET Alterations detected in blood-derived circulating tumor DNA correlate with bone metastases and poor prognosis. J Hematol Oncol. 2018;11(1):76. doi: 10.1186/s13045-018-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olsson E, Winter C, George A, et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med. 2015;7(8):1034‐1047. doi: 10.15252/emmm.201404913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12(1):133. doi: 10.1186/s13045-019-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208‐1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge Y, Mu W, Ba Q, et al. Hepatocellular carcinoma-derived exosomes in organotropic metastasis, recurrence and early diagnosis application. Cancer Lett. 2020;477:41‐48. doi: 10.1016/j.canlet.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 85.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated metastasis: communication from a distance. Dev Cell. 2019;49(3):347‐360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 86.Zhu L, Sun H-T, Wang S, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13(1):152. doi: 10.1186/s13045-020-00987-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373‐383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255‐289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 89.Kahroba H, Hejazi MS, Samadi N. Exosomes: from carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell Mol Life Sci. 2019;76(9):1747‐1758. doi: 10.1007/s00018-019-03035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8(83):13. 10.1186/s13045-015-0181-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y, Cao X. Characteristics and significance of the Pre-metastatic niche. Cancer Cell. 2016;30(5):668‐681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 92.Emmanouilidi A, Paladin D, Greening DW, Falasca M. Oncogenic and non-malignant pancreatic exosome cargo reveal distinct expression of oncogenic and prognostic factors involved in tumor invasion and metastasis. Proteomics. 2019;19(8):e1800158. doi: 10.1002/pmic.201800158. [DOI] [PubMed] [Google Scholar]
- 93.Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883‐891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baroni S, Romero-Cordoba S, Plantamura I, et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7(7):e2312. doi: 10.1038/cddis.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J, Yang S, Liu Z, et al. Imaging cellular aerobic glycolysis using carbon dots for early warning of tumorigenesis. Adv Mater. 2021;33(1):e2005096. doi: 10.1002/adma.202005096. [DOI] [PubMed] [Google Scholar]
- 96.Zhou Z, Qutaish M, Han Z, et al. MRI Detection of breast cancer micrometastases with a fibronectin-targeting contrast agent. Nat Commun. 2015;6:7984. doi: 10.1038/ncomms8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blair S, Garcia M, Davis T, et al. Hexachromatic bioinspired camera for image-guided cancer surgery. Sci Transl Med. 2021;13(592):eaaw7067. doi: 10.1126/scitranslmed.aaw7067 [DOI] [PubMed] [Google Scholar]
- 98.Li C, Torres VC, He Y, et al. Intraoperative detection of micrometastases in whole excised lymph nodes using fluorescent paired-agent imaging principles: identification of a suitable staining and rinsing protocol. Mol Imaging Biol. 2021;23(4):537‐549. doi: 10.1007/s11307-021-01587-z. [DOI] [PubMed] [Google Scholar]
- 99.Yuan Z, Xu T, Cai J, et al. Development and validation of an image-based deep learning algorithm for detection of synchronous peritoneal carcinomatosis in colorectal cancer. Ann Surg. 2022;275(4):e645–e651. doi: 10.1097/SLA.0000000000004229 [DOI] [PubMed] [Google Scholar]