Abstract
Recent studies in cancer patients and animal models demonstrate that intestinal microbiota influence the therapeutic efficacy of cancer treatments, including immune checkpoint inhibition. However, no studies to-date have investigated relationships between gastrointestinal microbiota composition and response to checkpoint inhibition in advanced metastatic castrate resistant prostate cancer (mCRPC). We performed 16S rRNA gene sequencing of fecal DNA from 23 individuals with mCRPC progressing on enzalutamide and just prior to treatment with anti-PD-1 (pembrolizumab) to determine whether certain features of the microbiome are associated with treatment response (defined as serum PSA decrease >50% at any time on treatment or radiographic response per RECIST V.1.1). Global bacterial composition was similar between responders and non-responders, as assessed by multiple alpha and beta diversity metrics. However, certain bacterial taxa identified by sequencing across multiple 16S rRNA hypervariable regions were consistently associated with response, including the archetypal oral bacterium Streptococcus salivarius. Quantitative PCR (qPCR) of DNA extracts from fecal samples confirmed increased Streptococcus salivarius fecal levels in responders, whereas qPCR of oral swish DNA extracts showed no relationship between oral Streptococcus salivarius levels and response status. Contrary to previous reports in other cancer types, Akkermansia muciniphila levels were reduced in responder samples as assessed by both 16S rRNA sequencing and qPCR. We further analyzed our data in the context of a previously published “integrated index” describing bacteria associated with response and non-response to checkpoint inhibition. We found that the index was not reflective of response status in our cohort. Lastly, we demonstrate little change in the microbiome over time, and with pembrolizumab treatment. Our results suggest that the association between fecal microbiota and treatment response to immunotherapy may be unique to cancer type and/or previous treatment history.
Keywords: 16S rRNA, Microbiome, Prostate cancer, Immune checkpoint inhibition, Enzalutamide
Introduction
Treatment options for individuals with advanced metastatic castrate resistant prostate cancer (mCRPC) are limited. While second line androgen receptor axis-targeted therapies can be initially effective, resistance often develops, leading to disease progression. Immune checkpoint inhibition (ICI) has been investigated as an alternative treatment for advanced prostate cancer, however clinical trials show limited efficacy for the majority of patients [1], [2], [3]. Much attention has been devoted to identifying biomarkers predictive of response to ICI and recent studies have shown that cancers that respond best to checkpoint inhibition are those that have high tumor mutational burdens (TMB), often resulting from defects in DNA damage response pathways [4]. While metastatic prostate cancers have been shown to have higher mutational loads than primary prostate cancers [5], the TMB is still much lower than that of melanoma and non-small cell lung cancer which typically respond favorably to checkpoint inhibition [6]. However, some cancers with high TMB do not respond to checkpoint inhibition and vice versa, suggesting that genomic alterations alone are inadequate in prediction of cancer immunotherapy response [7].
In addition, the expression of immune checkpoint receptors and ligands in prostate cancer does not always correlate with response [8]. However, recent studies show that some individuals with mCRPC who are progressing on the androgen receptor inhibitor enzalutamide have durable responses to subsequent use of the anti-PD-1 agent pembrolizumab [9], [10], [11], [12]. A prior study demonstrated that progression on enzalutamide is associated with increased circulating PD-L1/2+ dendritic cells compared to those naïve or responding to treatment, and a high frequency of PD-1+T cells [13]. Likewise, a subset of individuals with mCRPC progressing on enzalutamide respond to anti-PD-1, sometimes irrespective of TMB and MMR deficiency status [9,14].
A growing body of evidence indicates that gastrointestinal (GI) microbiota influence the efficacy of cancer treatments, including immunotherapy. For example, in animal models, intestinal microbiota are essential for the anti-tumor effect of agents such as cyclophosphamide [15], platinum chemotherapy (oxaliplatin) [16], CTLA-4 inhibition [17] and anti-PD-L1 immunotherapy [18]. Efficacy of these agents in multiple tumor models is decreased in germ free animals and conventionally housed mice with eradicated commensal intestinal flora compared to conventionally housed mice with normal flora. More recently, studies in human cancer patients have confirmed that abnormal gut microbiome composition contributes to checkpoint inhibition resistance in epithelial tumors [19] and that melanoma patients who respond to anti-PD-1 checkpoint inhibition have distinct fecal microbiome and metagenome makeups compared to non-responders [20]. Based on these findings, we hypothesized that commensal GI microbiota composition may be an influential component of tumor response to checkpoint inhibition and should be investigated in the context of prostate cancer.
In the present study, we performed 16S rRNA gene sequencing of longitudinally collected fecal samples from a cohort of individuals with mCRPC progressing on enzalutamide and initiating treatment with pembrolizumab to determine whether certain features of the microbiome are associated with treatment response. Our findings, such as increased Bacteroides thetaiotaomicron [21] Enterococcaceae [22], and Enterococcus [22] in checkpoint inhibition responders corroborate previous findings in other cancer types. However, unique to previous reports we find that increased Collinsella aerofaciens [18] and Akkermansia muciniphila [19] are associated with non-responders, suggesting that the influence of the GI microbiota on immunotherapy response may be unique to cancer type or previous medical treatment.
Materials and methods
Study cohort
All protocols were approved by Oregon Health Sciences University (OHSU) and Johns Hopkins University (JHU) institutional review boards and patient data were anonymized (trial registration number, clinicaltrials.gov, NCT02312557). Study design, inclusion criteria, and relevant clinical data are previously described [1,12]. Briefly, all participants had mCRPC, had progressed on enzalutamide by prostate specific antigen (PSA) testing and/or imaging, and continued receiving enzalutamide during the study. Patients received 200 mg of pembrolizumab intravenously every three weeks for four cycles (Supplementary Fig. 1A). Body mass index (BMI), age, and baseline PSA levels were statistically similar between responders and non-responders (Supplementary Fig. 1B-D). Treatment response was defined as serum PSA decrease >50% at any time on treatment or radiographic response per RECIST V.1.1. Patient 12 was classified as a responder per these criteria, however it should be noted that response in this individual could have been due to targeted radiation as opposed to study treatment.
Sample collection
All patient-related study procedures were done at OHSU, the site of the interventional clinical trial. Rectal swabs were collected at each treatment visit with a sterile swab (Cat. No. 552C, Copan Diagnostics, Murrieta, CA). The swab was placed in a 3 ml sterile external thread cryovial (Cat. No. T310-3A, Simport Scientific, Beloeil QC Canada) and stored at -80°C. An oral swish was collected at each treatment visit as follows: 10 mL of sterile 0.9% NaCl (Cat. No. 200-59, Teleflex Medical, Morrisville, NC) was swished in the patient's mouth for 30 seconds, the solution was collected, aliquoted into 3 mL sterile external thread cryovials and stored immediately at -80°C. Frozen samples were shipped to JHU on dry ice, and stored at -80°C until DNA was isolated. Supplementary Table 1A-D summarizes the samples collected at each visit for each patient.
Cohort assignment
To account for variable numbers of samples and sample time points per person (Supplementary Table 1), we constructed three data analysis cohorts (Pre-Treatment, First Timepoint, and All Samples) which are outlined in Supplementary Table 1A-D. The “Pre-Treatment” cohort includes only samples collected prior to pembrolizumab treatment at Cycle 1 for a total of 16 individuals (4 responder and 12 non-responder, Supplementary Table 1B). The “First Timepoint” cohort includes the first fecal sample collected from every person, regardless of treatment time point, for a total of 23 individuals (9 responder, 14 non-responder, Supplementary Table 1C). The “All Samples” cohort includes all samples (including longitudinal samples) collected from all 23 individuals, for a total of 136 samples (57 responder, 79 non-responder, Supplementary Table 1D). The results in the main manuscript are presented for the First Timepoint cohort, with results for the Pre-Treatment and All Samples cohorts given in the supplementary material. We highlight the data from the First Timepoint cohort because this sample set includes data from all participants and is therefore the most representative data, yet normalizes the cohort to one sample per person.
DNA isolation
All procedures were performed using sterile technique in a tissue culture hood. Fecal material from the rectal swabs was resuspended in a 1.5 mL microcentrifuge tube containing 500 µl sterile PBS (Cat. No. 21-031-CV, Corning, Manassas, VA) by vigorously swishing the swab in the microcentrifuge tube for 60 seconds. For oral swishes, one 3 mL vial was thawed on ice and contents were divided into two, 2 mL microcentrifuge tubes, and spun at 5000 rpm for 5 minutes. Supernatant was removed and the pellets were combined and resuspended in 500 µl TE50 Buffer (Cat. Nos. 15575-038 and 15568025, Invitrogen, Grand Island, NY).
An enzyme cocktail was made by combining 50 µl of lysozyme (10 mg/ml, Cat. No. L7773, Sigma-Aldrich, St. Louis, MO), 6 µl of mutanolysin (25 KU/ml, Cat. No. M4782, Sigma-Aldrich, St. Louis, MO), and 44 µl of TE50 Buffer per sample. 100 µl of enzyme cocktail was added to each sample. Samples were incubated on a heat block for 1 hr at 37°C. Samples were removed from the heat block and transferred from a microcentrifuge tube into a FastPrep™ Lysing Matrix B tube (Cat. No. 6911050, MP Biomedicals, Santa Ana, CA). 210 µl of 20% SDS (Cat. No. 05030, Sigma-Aldrich, St. Louis, MO) and 810ul of phenol:chloroform:isoamyl alcohol (25:24:1, Cat. No. 108-95-2, ThermoFisher Scientific, Waltham, MA) were added to each sample. Samples were homogenized for 30 seconds at 6.0m/sec twice in an MP FastPrep-24 and then allowed to sit at room temperature for 5 minutes. Samples were centrifuged at 14000 rpm for 5 min at room temperature. The aqueous layer was transferred to a sterile 1.5 ml LoBind microcentrifuge tube (Cat. No. 022431021, Eppendorf, Hamburg, Germany) and volume was estimated. One sample volume of isopropanol (Cat. No. 19516-25ML, Sigma-Aldrich, St. Louis, MO) and 0.1x sample volume of sodium acetate (Cat. No. R1181, ThermoFisher Scientific, Waltham, MA) was added to each sample and gently mixed. Samples were incubated overnight at -20°C, centrifuged at 14000 rpm for 15 minutes at 4°C, and the supernatant was removed. The pellet was rinsed with 1 ml ice cold 70% ethanol (Cat. No. BP2818-500, Fisher Scientific, Fair Lawn, NJ), centrifuged at 14000 rpm for 5 minutes at 4°C, and the supernatant was removed. The pellet was dried in the hood for 5-10 minutes and resuspended in 50-100 µl of DNA-free PCR grade water (Cat. No. P-020-0003, Molzym, Bremen, Germany). Samples were quantitated with the QuBit dsDNA HS (high sensitivity) kit (Cat. No. Q32851, Life Technologies, Carlsbad, CA).
Real-time PCR
All procedures were performed in a sterile hood using sterile reagents and materials. Samples were diluted to 10 ng/µl in DNA-free water (Cat. No. P-020-0003, Molzym, Bremen, Germany). For each 20 µl reaction, the following reagents were combined: 10 µl of iQ SYBR Green Mix (Cat. No. 1708882, Bio Rad Laboratories, United States) 2 µl of 10 mM Forward/Reverse Primer set, 6 µl DNA-free water, 1 µl of 2 µg/µl BSA (Cat. No. B9000S, New England Biolabs Inc, Ontario, Canada), 1 µl of 10 ng/µl DNA. Real-time PCR (qPCR) conditions and primers [23], [24], [25] are outlined in Supplementary Table 2. Total copies of Akkermansia muciniphila and Streptococcus salivarius were estimated using standard curves with A. muciniphila and S. salivarius genomic DNA (Cat. No. BAA-835D-5 and BAA-1024D-5, respectively, ATCC, Manassas, VA). The qPCR efficiency of all qPCR assays was determined to be between 90 - 110%.
Library preparation
Targeted amplification of seven 16S rRNA hypervariable regions (V2, V3, V4, V6-7, V8, V9) was achieved using the Ion Torrent 16S Metagenomics Kit (Cat. No. A26216, ThermoFisher, Waltham, MA) following the manufacturer's instructions. Briefly, 10 ng of each sample was aliquoted in duplicate into PCR strip tubes. Environmental master mix and 16S primer set (10x, Pool 1 or Pool 2) were added to each tube, which was mixed and underwent PCR. Samples were cleaned with AMPure XP beads (Cat. No. A63881, Beckman Coulter, Pasadena, CA) and run on a Bioanalyzer to determine amplicon sizes and concentration. 50 ng of DNA from each pool were combined with water, 5x End Repair Buffer, and End Repair Enzyme, and incubated at room temperature for 20 min. Samples were immediately cleaned with AMPure XP beads. Ligation and nick repair was performed by adding 10x Ligase Buffer, Ion P1 Adaptor, Ion Xpress Barcode, dNTP mix, nuclease free water, DNA ligase, and nick repair polymerase to the sample, incubating per manufacturer's instructions. Samples were cleaned with AMPure XP beads, and a final amplification of the sample was performed by adding Platinum PCR Supermix High Fidelity and Library Amplification Primer Mix to the sample and undergoing PCR according to manufacturer's instructions. Samples were cleaned with AMPure XP beads prior to resuspension in 20 µl of low TE and the final sample was quantitated using the Bioanalyzer High Sensitivity DNA Kit (Cat. No. 5067-4626, Agilent Technologies, Santa Clara, CA). Samples were diluted to 26 pM, pooled to a final volume greater than 25 µl, stored at 4°C and sequenced within 48 hours.
Sequencing
Sequencing was performed by the Sidney Kimmel Comprehensive Cancer Center (SKCCC) Experimental and Computational Genomics Core (ECGC). Pooled libraries and reagents were loaded onto chips using the Ion Chef Instrument and Ion 520TM and Ion 530TM Kit-Chef (Cat. No. A34461, ThermoFisher Scientific, Waltham, MA), which in an automated manner, loads libraries onto beads, performs emulsion PCR, purifies DNA post-PCR, and loads purified templated beads onto the Ion S5 chip. Loaded chips were transferred to the Ion GeneStudio S5 System and were sequenced across three separate sequencing runs using 400 bp sequencing kits and Ion S5 Sequencing Kit reagents (Cat. No. A3580, ThermoFisher Scientific, Waltham, MA). Sequences were demultiplexed by sample using the S5 device software, and then separated per hypervariable region by ThermoFisher prior to downstream analysis.
Data and code availability
All sequence files are available in the NCBI Sequence Read Archive (SRA) under Bioproject ID PRJNA793907. All codes used can be found on the public GitHub repository mCRPC-pembro (http://github.com/Sfanos-Lab-Microbiome-Projects/mCRPC-pembro/).
Bioinformatics
Sequences were analyzed as previously described [26]. Briefly, sequences were sent to ThermoFisher for separation by primer using cutprimer. Sequences less than 100 bp were filtered, and data was imported into QIIME2 v 2020.6 [27]. Sequences were separated by hypervariable region and sequencing run for denoising (DADA2 denoise-pyro, reads neither trimmed nor truncated, max-ee of 5) [28], at which point results from each sequencing run were pooled, keeping variable regions separate. Open reference operational taxonomic unit (OTU) clustering of the amplicon sequence variants (ASVs) generated with DADA2 (vsearch cluster-features-open-reference; 97% identity, 4 threads), mafft tree generation (align-to-tree-masfft-fasttree; 4 threads) [29,30], and taxonomic classification (classify-consensus-vsearch; default settings) were performed against our lab-curated SILVA database [26]. Sequences were rarefied to the lowest number of reads (1,000 reads) to include all samples in downstream analysis. Good's coverage analysis [31] demonstrated average retention of ≥98% observed species for all regions in samples rarefied to 1,000 reads (Supplementary File 1).
The following alpha and beta diversity metrics were performed using qiime diversity core-metrics-phylogenetic: Faith's phylogenetic diversity [32], Shannon's, evenness, observed features, Bray Curtis [33], Jaccard [34], Unweighted Unifrac [35], and Weighted Unifrac [36]. Results were imported into Rstudio; the generalized linear model function was used to incorporate results from multiple hypervariable regions (V2, V3, V4, V6-7) in the statistical analysis of alpha diversity metrics and taxonomic abundance. Sequencing results from the V8 and V9 hypervariable regions were excluded per our previous findings of low taxonomic sensitivity of amplicons targeting these regions [26]. Results from beta diversity metrics were used to perform PERMANOVA individually for results from each hypervariable region.
Taxonomic classification results were mined for sequences from the 17 bacteria included in integrated index from Shaikh et al. [37]. Due to differences in classification, not all species were represented in our data. Therefore, some species were not included in the current analysis and others were substituted with a proxy (summarized in Supplementary File 2). The index was then calculated according to Shaikh et al. [37].
Results
Responder status in relation to alpha diversity
We first investigated whether alpha diversity of responder samples was different from non-responders by performing an analysis of four alpha diversity metrics (Shannon diversity, evenness, observed OTUs, and Faith's phylogenetic diversity) on sequencing results obtained from each 16S rRNA hypervariable region. We then integrated alpha diversity results from hypervariable regions using a generalized linear mixed effects model (GLM), which produces a coefficient associated with response status and a corresponding p-value. A positive GLM coefficient indicates increased diversity in responders and a negative GLM coefficient indicates decreased diversity. Overall, there were no significant differences in alpha diversity in the First Timepoint cohort, and results from individual hypervariable regions correlated with the overall GLM results (Fig. 1). In the All Samples cohort, Faith's phylogenetic diversity and observed features (both measures of richness) were significantly decreased in responders across multiple hypervariable regions, whereas evenness was increased in responders (Supplementary Fig. 2, Supplementary File 3). Faith's phylogenetic diversity of the Pre-Treatment cohort was also significantly decreased in responders (Supplementary Fig. 2, Supplementary File 3).
Fig. 1.
Alpha Diversity of fecal DNA in responders verses non-responders by hypervariable region (First Timepoint cohort). When statistical results from four hypervariable regions are incorporated using a generalized linear mixed effects model (GLM), there are no significant differences between responders and non-responders. (A) Shannon diversity (B) Evenness (C) Observed features (D) Faith's phylogenetic diversity.
Responder status in relation to beta diversity
We next investigated whether beta diversity of responder samples was different from non-responders by performing four beta diversity metrics (Bray Curtis, Jaccard, Unweighted Unifrac, and Weighted Unifrac) on results from each hypervariable region. PERMANOVA was used to determine how much variability between samples could be attributed to response status. In the First Timepoint and Pre-Treatment cohorts, we found no measurable differences in microbiome composition as assessed by beta diversity metrics between responders and non-responders after adjusting for individual person and sample time point. In the All Samples cohort, the majority of differences in microbiome composition were attributable to individual people (∼50%), approximately 3% was attributed to response status, and 3% to time point status. Results with PERMANOVA statistics for all three cohorts are summarized in Supplementary File 4.
Different bacterial taxa are enriched in responders versus non-responders
We performed taxonomic classification and calculated the percent abundance for all bacterial sequences obtained from each individual hypervariable region before integrating the results in a GLM. As with alpha diversity, a positive GLM coefficient corresponds to increased abundance in responders and a negative GLM coefficient corresponds to decreased abundance in responders. The results for the top thirty most differentially abundant bacterial genera and species between responders and non-responders in the First Timepoint cohort are summarized in Fig. 2 and the results for the Pre-Treatment and All Timepoints cohorts are summarized in Supplementary Fig. 3. All taxonomic results are listed in Supplementary File 5. We compared our taxonomic abundance results to seven prior studies examining gastrointestinal microbiomes in association with response to checkpoint inhibition (Table 1). Of interest, our study in prostate cancer demonstrates the opposite association between several previously reported taxa and response to checkpoint inhibition in other types of cancer.
Fig. 2.
Taxonomic differential abundance for the First Timepoint cohort. Volcano plot (A) and histogram (B) of the 30 most differentially abundant bacterial species between responders and non-responders. Taxonomic percent abundance was calculated for each hypervariable region and results were combined using a GLM to produce a single GLM coefficient. A positive GLM coefficient indicates enrichment in responders, whereas a negative GLM coefficient indicates depletion in responders and enrichment in non-responders. The magnitude of the GLM coefficient corresponds with the magnitude of enrichment. p values are log transformed and multiplied by negative one so that more significant p-values are higher in value. In B, species are listed in order of magnitude of GLM coefficient from top to bottom.
Table 1.
Summary of previously reported associations between taxonomic abundance and response to checkpoint blockade, and how they compare to results from this data set. np = not present in our dataset, ns = not significant, x = present and significant.
| Author | Bacteria | Published correlation | Our correlation | First Timepoint | All Samples | Pre treatment |
|---|---|---|---|---|---|---|
| Frankel, et al | Bacteroides caccae | Increased in R | np | np | np | |
| Streptococcus parasanguinis | Increased in R | Increased in R | ns | x | ns | |
| Faecalibacterium prausnitzii | Increased in R | np | np | np | ||
| Holdemania filiformis | Increased in R | np | np | np | ||
| Bacteroides thetaiotaomicron | Increased in R | Increased in R | x | ns | x | |
| Routy, et al | Akkermansia muciniphila | Increased in R | Increased in NR | ns | x | ns |
| Enterococcus hirae | Increased in R | np | np | np | ||
| Chaput, et al | Faecalibacterium spp. | Increased in R | Increased in NR | ns | x | ns |
| Gemmiger spp. | Increased in R | np | np | np | ||
| Matson, et al | Bifidobacterium longum | Increased in R | Increased in NR | ns | x | ns |
| Collinsella aerofaciens | Increased in R | Increased in NR | x | x | x | |
| Enterococcus faecium | Increased in R | ns | ns | np | ||
| Gopalakrishnan, et al | Faecalibacterium spp. | Increased in R | Increased in NR | ns | x | ns |
| Faecalibacterium prausnitzii | Increased in R | np | np | np | ||
| Ruminococcaceae | Increased in R | Increased in NR | ns | x | ns | |
| Baruch, et al | Enterococcaceae | Increased in R | Increased in R | x | x | x |
| Enterococcus spp. | Increased in R | Increased in R | x | x | x | |
| Streptococcus australis | Increased in R | ns | ns | ns | ||
| Veillonella atypica | Increased in NR | Increased in R | ns | x | np | |
| Wind, et al | Streptococcus parasanguinis | Increased in R | Increased in R | ns | x | ns |
| Bacteroides massiliensis | Increased in R | np | np | np | ||
| Peptostreptococcaceae | Increased in NR | Increased in NR | ns | ns | x |
The oral bacterium Streptococcus salivarius is elevated in responders to pembrolizumab
Streptococcus salivarius (S. salivarius) was the most differentially abundant species between responders and non-responders in the First Timepoint cohort (Fig. 2). Furthermore, this species was consistently elevated in responders across the sequencing results from multiple hypervariable regions (Fig. 3A) in all three cohorts (Supplementary Fig. 4). Therefore, we aimed to verify these results using qPCR. qPCR of fecal DNA from the First Timepoint cohort corroborated the sequencing results and showed elevated S. salivarius in responders compared to non-responders (Fig. 3B, 3C). Since S. salivarius is a commensal of the oral cavity, we also investigated the levels of S. salivarius in DNA from oral swish samples by qPCR in the First Timepoint cohort. Interestingly, qPCR of oral DNA samples showed no difference in S. salivarius levels between responders and non-responders (Fig. 3D). Likewise, S. salivarius levels in oral DNA did not correlate with levels in fecal DNA measured by qPCR (Fig. 3E).
Fig. 3.
Fecal Streptococcus salivarius levels are increased in responders. (A) Percent abundance of fecal Streptococcus salivarius by Ion Torrent 16S rRNA gene sequencing (First Timepoint cohort). GLM coefficient = 0.967, p value = 0.00001 (FDR adjusted p value = 0.004). (B) Percent abundance of fecal Streptococcus salivarius relative to total 16S rRNA gene copies by qPCR (First Timepoint cohort, fecal DNA). P value = 0.055, Mann Whitney two tailed t test. (C) Correlation between fecal DNA percent abundance of Streptococcus salivarius by qPCR (x axis) and 16S rRNA gene sequencing (V3 region, y axis), R2 = 0.9337. (D) Percent abundance of oral Streptococcus salivarius relative to total 16S rRNA gene copies by qPCR (First Timepoint cohort, oral swish). No significant difference, Mann Whitney two tailed t test. (E) Correlation between qPCR percent abundance of Streptococcus salivarius in rectal swab (x axis) versus oral swish (y axis), R2 = 0.087.
Akkermansia muciniphila is depleted in responders to pembrolizumab
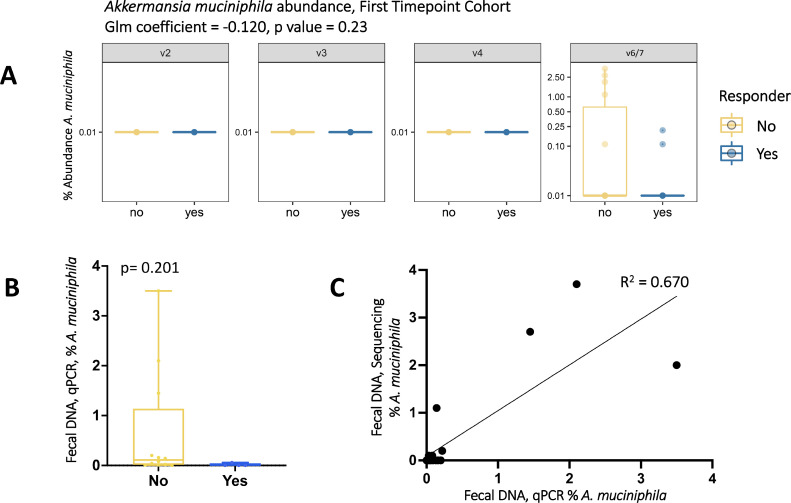
Elevated Akkermansia muciniphila (A. muciniphila) levels have been associated with response to anti-PD-1 in melanoma patients [19] and consumption of oral androgen receptor axis-targeted therapies in prostate cancer patients [38,39]. We therefore investigated whether A. muciniphila levels correlated with response to pembrolizumab in our cohorts. A. muciniphila was not initially detected in our sequencing data, but the OTU classified as Family Verrucomicrobiaceae had 100% identity with A. muciniphila through the Basic Local Alignment Search Tool (BLAST) [40] and is therefore used as a proxy for A. muciniphila throughout this study. A. muciniphila was only detected using primers targeting the V6-7 hypervariable region, which showed decreased levels in responders compared to non-responders (Fig. 4A First Timepoint cohort, Supplementary Fig. 5A,C Pre-Treatment and All Sample cohorts). This trend was corroborated by A. muciniphila-specific qPCR of fecal DNA (Fig. 4B,C, Supplementary Fig. 5B,D).
Fig. 4.
Percent abundance of Akkermansia muciniphila is decreased in responders compared to non-responders in fecal samples from the First Timepoint cohort. (A) Percent abundance of Akkermansia muciniphila copies by Ion Torrent 16S rRNA gene sequencing. Glm coefficient = -0.120, p value = 0.227 (FDR adjusted p value = 0.425). (B) Percent abundance of Akkermansia muciniphila copies relative to total 16S rRNA gene copies by qPCR. P value = 0.201, Mann Whitney two tailed t test. (C) Correlation between rectal swab percent abundance of Akkermansia muciniphila copies by qPCR (x axis) and 16S rRNA gene sequencing (V6-7 region, y axis), R2 = 0.670.
Integrated index analysis applied to prostate cancer
Recently, Shaikh et al. combined sequencing data from multiple checkpoint inhibition microbiome studies to develop an “integrated index” describing bacteria associated with response and non-response to checkpoint inhibition [37]. In order to further investigate how our results compare to previous published reports, we ran our data through a modified integrated index, optimizing for differences in bacterial classification between the two data sets (Supplementary File 2). Due to differences in taxonomic identification between studies, we did not include six of the seventeen bacteria in our analyses, substituted four species with a representative genus or species, and substituted Family Verrucomicrobiaceae for A. muciniphila as previously discussed. Positive index values correlate with bacteria associated with response, whereas negative index values correlate with bacteria associated with non-response. In all, we saw no significant differences for integrated results between responders and non-responders (Fig. 5A, Supplementary File 2), but using the index highlighted interesting signals at the individual taxon levels. Bacteroides thetaiotaomicron, Bacteroides uniformis, Bacteroides fragilis, Faecalibacterium unassigned, and Roseburia unassigned were the five taxa contributing the most signal to this analysis. Of these five taxa, three were significantly different between responders and non-responders in our data, all of which were increased in responders: B. thetaiotaomicron, B. fragilis, and Roseburia unassigned (Fig. 5B-D).
Fig. 5.
Evaluation of the integrated microbiome prediction index adapted from Shaikh et al. (A) Neither bacteria associated with non-response (top) or response (bottom) according to the index are differentially abundant between responders and non-responders in our data set. (B-D) Individual plots of three of the bacteria contributing the most signal to the integrated index.
Pembrolizumab treatment does not elicit consistent changes to fecal bacterial composition
Gopalakrishnan et al. reported that the GI microbiome is stable over time with checkpoint inhibition treatment [20] however overall, the data on this topic is limited. To further investigate the stability of the GI microbiome over the course of checkpoint inhibition treatment, we performed longitudinal analysis on data from nine individuals (Patient ID 11, 13, 14, 16, 18, 20, 21, 22, 23) for which longitudinal samples were collected across the entirety of the pembrolizumab treatment cycles (Cycle 1, Cycle 2, Cycle 3, Cycle 4, and Monitoring Phase). For each of these individuals, we plotted alpha diversity and taxonomic abundance values over time, in addition to performing PERMANOVA analysis. There were no statistically significant differences in community-wide alpha and beta diversity analyses over time. Specifically, there was no difference between alpha diversity of Cycle 1 (pre-treatment) and Cycle 2 (on-treatment) by any of the four alpha diversity metrics (statistics available in Supplementary File 6). Additionally, beta diversity analysis revealed that different time point samples from the same patient cluster together (Fig. 6A), and that time point does not significantly contribute to variability by PERMANOVA in these nine individuals with full treatment cycle samples available (statistics available in Supplementary File 6). There was a subset of seven taxa present in samples n ≥ 6 that showed statistically significant differences between Cycle 1 and Cycle 2 (Supplementary Fig. 6, Supplementary File 6), however the direction of abundance (e.g. increase or decrease) was not consistent for all individuals (Fig. 6B) or consistent over time (Fig. 6C). None of these bacteria were identified as being members of the integrated index, and none of them were differentially abundant between responders and non-responders aside from Corynebacterium tuberculostearicum.
Fig. 6.
Longitudinal analysis of bacterial composition during treatment. (A) Beta diversity of 5 longitudinal samples from 9 individuals demonstrates that different samples from the same person cluster together, indicating little variation in global microbiome composition over time. (B) Percent abundance of Escherichia coli (E. coli) is significantly higher in Cycle 1 compared to Cycle 2 (Mann Whitney two-tailed t test, GLM coefficient -4.295, p value 0.005), demonstrating that certain taxa decrease after starting pembrolizumab treatment. (C) Percent abundance of E. coli varies over time in individual patients.
Discussion
Gopalakrishnana et al. [20] and Jin et al. [41] previously reported increased alpha diversity in responders to checkpoint inhibition therapy. In the present study, we observed no difference in alpha diversity between responders and non-responders in the First Timepoint cohort (Fig. 1). However, both metrics of richness were decreased in responders in the All Samples cohort, and one was decreased in the Pre-Treatment cohort (Supplementary Fig. 2). Alternatively, evenness was increased in responders in the All Samples cohort (Supplementary Fig. 2). While some studies have reported relationships between alpha diversity and response to checkpoint inhibition, other results are conflicting. In a retrospective meta-analysis, Shaikh et al. [37] reported that alpha diversity is not consistently predictive of response to checkpoint inhibition. However, it should be noted that this analysis was performed using data from different types of cancer patients, and it could be that disease type has an important influence on GI microbiome characteristics.
Across all cohorts, the majority of compositional changes (∼50%) were attributed to the donor (between-person variation). After adjusting for donor and treatment time point, there were minimal differences in global compositional variability (up to 3%) attributable to response status (Supplementary File 4). This indicates that response is not associated with global alteration to an individual's GI microbial composition, but likely associated with more granular differences such as individual bacteria abundance or metabolite production. As such, we next investigated whether certain bacterial species were differentially abundant in responders verses non-responders to pembrolizumab. Not only did we find several differentially abundant taxa, but our study also demonstrates the opposite association between several previously reported taxa and response to checkpoint inhibition (Table 1). These conflicting results could be for many reasons, including differences in study design (type of sequencing and taxonomic databases used), variability in clinical metadata (ethnicity, age, sex), exposure of each cohort to different medications (androgen deprivation therapy), or physiologic differences in cancer type (e.g., melanoma versus prostate cancer). In addition, it should be noted that diet and other nutrient-specific factors play a significant role in the composition and metabolic output of the GI microbiome. For example, high dietary fiber diet was associated with significantly improved progression-free survival in melanoma patients treated with ICI [42]. As previously reported studies varied geographically, it is plausible that differences in diet and/or other non- clinicodemographic factors influenced differing results. Due to differences between our results and previously published reports, we further investigated two bacteria of interest in our cohort: S. salivarius and A. muciniphila.
In the First Timepoint cohort, S. salivarius was increased in fecal DNA of responders by both 16S rRNA sequencing and qPCR. S. salivarius is a commensal of the human oral cavity that is also reported at other sites along the GI tract, namely the stomach [43], jejunum [44], and ileum [45]. S. salivarius possesses antimicrobial activities due to the production of lantibiotic bacteriocins, and thus has been used as an oral probiotic [46]. Its close relative, Streptococcus thermophilus (S. thermophilus), is a common probiotic and has several perceived health benefits including stimulation of the gut immune system [47]. Recent evidence indicates that studies investigating the effects of S. thermophilus as a probiotic may have misidentified S. salivarius for S. thermophilus, further suggesting that S. salivarius may be play a role in intestinal health [48].
Accordingly, we next investigated whether S. salivarius levels in oral swish samples correlate with levels in rectal swab samples. Interestingly, S. salivarius levels in oral swish samples did not correlate with levels in rectal swabs (Fig. 3), suggesting that association between S. salivarius levels and pembrolizumab response in our cohorts was site-specific. S. salivarius has been found to modulate PPAR gamma expression of intestinal epithelial cells [49], indicating that it may have immunomodulatory functions within the GI tract relevant to treatment with checkpoint inhibition. However, a recent report found a correlation between increased levels of fecal S. salivarius and non-response to anti-PD-1 in melanoma patients [50], suggesting that this correlation may not transcend cancer types.
We were specifically interested in examining levels of A. muciniphila because we and others previously reported increased A. muciniphila in individuals receiving oral androgen receptor axis-targeted therapies such as enzalutamide and abiraterone acetate [38,39], and because this species has been reported to be increased in responders to PD-1 inhibition [19]. Therefore, we expected increased A. muciniphila levels in responders. However, both our qPCR data and 16S rRNA sequencing data show decreased A. muciniphila in responders. A proposed mechanism of A. muciniphila improving ICI response is through production of inosine. Mager et al. propose that disruption of the gut barrier by ICI allows translocation of inosine produced by gut bacteria into systemic circulation, which thereby activates T cells via the adenosine A2A receptor [51]. However, preliminary studies suggest that inosine may facilitate metastasis via decreased cAMP and thereby cell-extracellular matrix adhesion in prostate cancer [52]. Interestingly, another major inosine producer, Bifidobacterium pseudolongum, was significantly increased in responders in both the First Timepoint Cohort (Fig. 2B) and the All Samples Cohort (Supplementary File 5). Inosine production by B. pseudolongum has been previously shown to enhance the efficacy of ICI in mouse models of cancer [53].
Recently, Shaikh et al. reported on the lack of consensus regarding the microbial signals associated with clinical response to checkpoint inhibition. By re-analyzing combined data from multiple studies, they developed an “integrated index” describing bacteria associated with response and non-response to checkpoint inhibition [37]. We therefore aimed to determine whether our data would be predictive of response and non-response using this index, so we ran our data through a modified integrated index. Firstly, we found discrepancies between taxonomic classification of our dataset and the Shaikh et al. dataset, necessitating us to amend the index by removing some unidentified taxa and substituting others. This is a common problem in the microbiome field, and argues the need for a better annotated, more complete, uniform taxonomic classification database and uniform pipelines for microbial analysis.
When we integrated the signals across all bacteria in the index, we saw no significant differences between responders and non-responders (Fig. 5A, Supplementary File 2), however using the index highlighted interesting signals for three taxa: Bacteroides thetaiotaomicron, Bacteroides fragilis, and Roseburia unassigned (Fig. 5B-D). Interpretation of our results compared to previous reports is complex. For example, B. thetaiotaomicron has been reported as both increased in responders [21] and increased in non-responders [20,37]. As another example, Roseburia intestinalis was previously documented as increased in non-responders [18], whereas Shaikh et al. found Roseburia hominis enriched in responders [37]. Of note, sequences assigned to Roseburia in our data set were only able to be classified to the genus level. Upon BLAST analysis of a subset of the sequences, they were not definitively traceable to a particular Roseburia species, likely due to sequence homology between species and because the database contains sequences of uncultured species.
A complicating factor in GI microbiome research is that bacteria live in multispecies communities within the gut; therefore, results cannot always be interpreted on the single species level [54]. Notably, A. muciniphila and B. thetaiotaomicron inhabit a similar niche of the GI microbiome in that they are both part of a small group of bacteria with the ability to degrade mucin [55]. If mucin degradation is in a state of equilibrium and these two bacteria both play the role of mucin degradation, this may at least partially explain why we see an inverse relationship compared to expected (A. muciniphila enriched in non-responders and B. thetaiotaomicron enriched in responders).
Finally, it has been demonstrated that the abundance of certain bacterial taxa is influenced by use of medications [56], therefore we investigated whether pembrolizumab caused a shift in bacterial makeup over time. We performed alpha diversity analysis and taxonomic abundance longitudinally over time for nine individuals with samples at identical time points (Cycle 1 (pre-treatment), Cycle 2, 3, 4 (on treatment), and Monitoring Phase (post treatment)) (Supplementary File 6). There were seven taxa present in n ≥ 6 samples that varied significantly between Cycle 1 and Cycle 2 (Supplementary Fig. 6). Of those seven taxa, Blautia coccoides and Clostridium algidixylanolyticum were both consistently decreased in Cycle 2, however directionality of variation compared to Cycle 1 for the other taxa was inconsistent between patients (Fig. 6B,C, Supplementary Fig. 6), suggesting that pembrolizumab did not have a consistent effect on bacterial composition for the majority of taxa. Importantly, none of these seven taxa were differentially abundant between responders and non-responders aside from C. tuberculostearicum (Fig. 2B, Supplementary Fig. 3C). Additionally, beta diversity analysis comparing Cycle 1 samples to Cycle 2 samples showed no significant variation in composition due to treatment time point (Supplementary File 6). Fig. 6A illustrates how longitudinal samples from the same patient cluster together, further demonstrating minimal changes to an individual's microbiome on and off pembrolizumab treatment.
Conclusions
The interplay between a person's gastrointestinal microbiome and their response to immune checkpoint inhibition is a recognized but poorly characterized phenomenon. Herein, we provide the first report of microbiome profiling of individuals with advanced prostate cancer in the context of immune checkpoint inhibition. We identified a number of differentially abundant taxa between responders and non-responders, however we find the opposite association between several previously reported taxa and response to checkpoint inhibition. These findings highlight that the effect of the microbiome on response to ICI may potentially be cancer-type specific, population-specific, or influenced by other clinical and/or non- clinicodemographic factors. We likewise highlight the importance of considering microbial community interactions when interpreting results.
Funding
This work was supported by Prostate Cancer Foundation Challenge Awards 16CHAL13 (to K.S.S.) and 18CHAL01 (to L.B.P, A.E.M., J.N.G., and K.S.S.), Department of Defense Prostate Cancer Research Award W81XWH-18-1-0545 (to L.B.P. and K.S.S.), and NCI SPORE award P50CA058236 (to K.S.S). Merck Sharpe & Dohme provided funding for the clinical trial.
CRediT authorship contribution statement
Lauren B. Peiffer: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft. James R. White: Formal analysis, Writing – review & editing. Carli B. Jones: Data curation, Formal analysis. Rachel E. Slottke: Data curation, Formal analysis, Project administration. Sarah E. Ernst: Investigation, Methodology. Amy E. Moran: Formal analysis, Methodology, Writing – review & editing. Julie N. Graff: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – review & editing. Karen S. Sfanos: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Supervision, Writing – original draft.
Acknowledgements
We would like to thank Jennifer Meyers, Hai Xu, and Kornel Schuebel from the JHU SKCCC Experimental and Computational Genomics Core for their help in generating the sequencing data. We would also like to thank Dr. Cynthia Sears and Dr. Drew Pardoll for helpful advice and discussion. Thank you to Bradley Toms and Leonardo Varuzza from ThermoFisher for their assistance with library preparation and data analysis.
Footnotes
J.N.G. is an employee of the US Government. The contents do not represent the views of the US Department of Veterans Affairs or the United States Government. J.R.W. has financial and/or other relationship with Resphera Biosciences. A.E.M. received research funding from AstraZeneca.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100822.
Contributor Information
Julie N. Graff, Email: graffj@ohsu.edu.
Karen S. Sfanos, Email: ksfanos@jhmi.edu.
Appendix. Supplementary materials
References
- 1.Beer TM, Kwon ED, ke CG, Fizazi K, Logothetis C, Gravis G, Ganju V, Polikoff J, Saad F, Humanski P, et al. Randomized, double-blind, Phase III trial of Ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J Clin Oncol. 2017;35:40–47. doi: 10.1200/JCO.2016.69.1584. [DOI] [PubMed] [Google Scholar]
- 2.Kwon EDP, Drake CGMD, Scher HIP, Fizazi KP, Bossi AMD, van den Eertwegh AJMMD, Krainer MMD, Houede NMD, Santos RMD, Mahammedi HMD, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15:700–712. doi: 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powles T, Yuen KC, Gillessen S, Kadel EE, Rathkopf D, Matsubara N, Drake CG, Fizazi K, Piulats JM, Wysocki PJ, et al. Atezolizumab with enzalutamide versus enzalutamide alone in metastatic castration-resistant prostate cancer: a randomized phase 3 trial. Nat Med. 2022;28:144–153. doi: 10.1038/s41591-021-01600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abeshouse A, Akbani R, Ally A, Aprikian A, Auman JT, Beroukhim R, Bradley Robert K, Bryce Alan H, Button M, Canes D, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, Rathkopf D, Morris MJ, Danila DC, Slovin SF, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 2019;5:471–478. doi: 10.1001/jamaoncol.2018.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haffner MC, Guner G, Taheri D, Netto GJ, Palsgrove DN, Zheng Q, Guedes LB, Kim K, Tsai H, Esopi DM, et al. Comprehensive evaluation of programmed death-ligand 1 expression in primary and metastatic prostate cancer. Am J Pathol. 2018;188:1478–1485. doi: 10.1016/j.ajpath.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graff JN, Alumkal JJ, Drake CG, Thomas GV, Redmond WL, Farhad M, Cetnar JP, Ey FS, Bergan RC, Slottke R, et al. (2016). Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancerOncotarget7. [DOI] [PMC free article] [PubMed]
- 10.Lin H, Liu Q, Zeng X, Yu W, Xu G. Pembrolizumab with or without enzalutamide in selected populations of men with previously untreated metastatic castration-resistant prostate cancer harbouring programmed cell death ligand-1 staining: a retrospective study. BMC Cancer. 2021;21:399. doi: 10.1186/s12885-021-08156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tucker MD, Zhu J, Marin D, Gupta RT, Gupta S, Berry WR, Ramalingam S, Zhang T, Harrison M, Wu Y, et al. Pembrolizumab in men with heavily treated metastatic castrate-resistant prostate cancer. Cancer Med. 2019;8:4644–4655. doi: 10.1002/cam4.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graff JN, Beer TM, Alumkal JJ, Slottke RE, Redmond WL, Thomas GV, Thompson RF, Wood MA, Koguchi Y, Chen Y, et al. A phase II single-arm study of pembrolizumab with enzalutamide in men with metastatic castration-resistant prostate cancer progressing on enzalutamide alone. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop JL, Sio A, Angeles A, Roberts ME, Azad AA, Chi KN, Zoubeidi A. PD-L1 is highly expressed in enzalutamide resistant prostate cancer. Oncotarget. 2015;6:234–242. doi: 10.18632/oncotarget.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan X, Polesso F, Wang C, Sehrawat A, Hawkins RM, Murray SE, Thomas GV, Caruso B, Thompson RF, Wood MA, et al. Androgen receptor activity in T cells limits checkpoint blockade efficacy. Nature. 2022 doi: 10.1038/s41586-022-04522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 20.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19:848–855. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 23.Kim Seow W, Lam JHC, Tsang AKL, Holocombe T, Bird PS. Oral Streptococcus species in pre-term and full-term children – a longitudinal study. Int J Paediatr Dent. 2009;19:406–411. doi: 10.1111/j.1365-263X.2009.01003.x. [DOI] [PubMed] [Google Scholar]
- 24.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shrestha E, White JR, Yu SH, Kulac I, Ertunc O, De Marzo AM, Yegnasubramanian S, Mangold LA, Partin AW, Sfanos KS. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J Urol. 2018;199:161–171. doi: 10.1016/j.juro.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones CB, White JR, Ernst SE, Sfanos KS, Peiffer LB. Incorporation of data from multiple hypervariable regions when analyzing bacterial 16S rRNA gene sequencing data. Front Genet. 2022;13 doi: 10.3389/fgene.2022.799615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith G, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price MN, Dehal PS, Arkin AP (2010). FastTree 2–approximately maximum-likelihood trees for large alignmentsPLoS One5, e9490. [DOI] [PMC free article] [PubMed]
- 30.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 32.Faith DP, Minchin PR, Belbin L. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio. 1987;69:57–68. [Google Scholar]
- 33.Sorensen TA (1948). A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commonsBiolSkar5, 1-34.
- 34.Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;44:223–270. [Google Scholar]
- 35.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaikh FY, White JR, Gills JJ, Hakozaki T, Richard C, Routy B, Okuma Y, Usyk M, Pandey A, Weber JS, et al. A uniform computational approach improved on existing pipelines to reveal microbiome biomarkers of nonresponse to immune checkpoint inhibitors. Clin Cancer Res. 2021;27:2571–2583. doi: 10.1158/1078-0432.CCR-20-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daisley BA, Chanyi RM, Abdur-Rashid K, Al KF, Gibbons S, Chmiel JA, Wilcox H, Reid G, Anderson A, Dewar M, et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat Commun. 2020;11:4822. doi: 10.1038/s41467-020-18649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sfanos KS, Markowski MC, Peiffer LB, Ernst SE, White JR, Pienta KJ, Antonarakis ES, Ross AE. Compositional differences in gastrointestinal microbiota in prostate cancer patients treated with androgen axis-targeted therapies. Prostate Cancer Prostatic Dis. 2018;21:539–548. doi: 10.1038/s41391-018-0061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S (2019). The diversity of gut microbiome is associated with favorable responses to anti–programmed death 1 immunotherapy in Chinese patients with NSCLCJ Thorac Oncol14, 1378-1389. [DOI] [PubMed]
- 42.Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, Khan MAW, Zhang X, White MG, Peterson CB, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. 2021;374:1632–1640. doi: 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hakalehto E, Vilpponen-Salmela T, Kinnunen K, von Wright A. Lactic acid bacteria enriched from human gastric biopsies. ISRN Gastroenterol. 2011;2011 doi: 10.5402/2011/109183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 45.den Bogert Bv, Erkus O, Boekhorst J, Md Goffau, Smid EJ, Zoetendal EG, Kleerebezem M. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol. 2013;85:376–388. doi: 10.1111/1574-6941.12127. [DOI] [PubMed] [Google Scholar]
- 46.Burton JP, Wescombe PA, Moore CJ, Chilcott CN, Tagg JR. Safety assessment of the oral cavity probiotic Streptococcus salivarius K12. Appl Environ Microbiol. 2006;72:3050–3053. doi: 10.1128/AEM.72.4.3050-3053.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perdigon G, Nader de Macias ME, Alvarez S, Oliver G, Pesce de Ruiz Holgado AA. Enhancement of immune response in mice fed with Streptococcus thermophilus and Lactobacillus acidophilus. J Dairy Sci. 1987;70:919–926. doi: 10.3168/jds.S0022-0302(87)80095-4. [DOI] [PubMed] [Google Scholar]
- 48.Martinović A, Cocuzzi R, Arioli S, Mora D. Streptococcus thermophilus: to survive, or not to survive the gastrointestinal tract, that is the question. Nutrients. 2020;12:2175. doi: 10.3390/nu12082175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couvigny B, de Wouters T, Kaci G, Jacouton E, Delorme C, Doré J, Renault P, Blottière HM, Guédon E, Lapaque N. Commensal Streptococcus salivarius modulates PPARγ transcriptional activity in human intestinal epithelial cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wind TT, Gacesa R, Vich Vila A, de Haan JJ, Jalving M, Weersma RK, Hospers GAP. Gut microbial species and metabolic pathways associated with response to treatment with immune checkpoint inhibitors in metastatic melanoma. Melanoma Res. 2020;30:235–246. doi: 10.1097/CMR.0000000000000656. [DOI] [PubMed] [Google Scholar]
- 51.Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 52.Charles C, Gohlke J, Lloyd S, Rasaily U, Henderson J, Karanam B, Simons B, Navone N, Kittles R, Ambs S, et al. Abstract PR17: metabolic re-wiring in African-American prostate cancer: a role of adenosine-inosine axis. Cancer Epidemiol Biomarkers Prev. 2020;29:PR17. [Google Scholar]
- 53.Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369:1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 54.Stubbendieck RM, Vargas-Bautista C, Straight PD. Bacterial communities: interactions to scale. Front Microbiol. 2016;7:1234. doi: 10.3389/fmicb.2016.01234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Herreweghen F, De Paepe K, Roume H, Kerckhof F-M, Van de Wiele T. Mucin degradation niche as a driver of microbiome composition and Akkermansia muciniphila abundance in a dynamic gut model is donor independent. FEMS Microbiol Ecol. 2018;94:1–13. doi: 10.1093/femsec/fiy186. [DOI] [PubMed] [Google Scholar]
- 56.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, Mujagic Z, Jonkers DMAE, Masclee AAM, Fu J, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11:362. doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence files are available in the NCBI Sequence Read Archive (SRA) under Bioproject ID PRJNA793907. All codes used can be found on the public GitHub repository mCRPC-pembro (http://github.com/Sfanos-Lab-Microbiome-Projects/mCRPC-pembro/).