Abstract
Septation in Escherichia coli requires several gene products. One of these, FtsQ, is a simple bitopic membrane protein with a short cytoplasmic N terminus, a membrane-spanning segment, and a periplasmic domain. We have constructed a merodiploid strain that expresses both FtsQ and the fusion protein green fluorescent protein (GFP)-FtsQ from single-copy chromosomal genes. The gfp-ftsQ gene complements a null mutation in ftsQ. Fluorescence microscopy revealed that GFP-FtsQ localizes to the division site. Replacing the cytoplasmic and transmembrane domains of FtsQ with alternative membrane anchors did not prevent the localization of the GFP fusion protein, while replacing the periplasmic domain did, suggesting that the periplasmic domain is necessary and sufficient for septal targeting. GFP-FtsQ localization to the septum depended on the cell division proteins FtsZ and FtsA, which are cytoplasmic, but not on FtsL and FtsI, which are bitopic membrane proteins with comparatively large periplasmic domains. In addition, the septal localization of ZipA apparently did not require functional FtsQ. Our results indicate that FtsQ is an intermediate recruit to the division site.
Cell division in the gram-negative bacterium Escherichia coli involves the coordinated invagination of all three layers of the cell envelope—the cytoplasmic membrane, the rigid peptidoglycan layer, and the outer membrane (34). Eventual constriction of the septum severs the cell into two compartments and segregates the replicated genetic material. At least nine essential gene products participate in septation: those of ftsZ, ftsA, ftsQ, ftsL, ftsI, ftsN, ftsK, ftsW, and zipA (18, 23). All of them, except for FtsQ and FtsL, have been localized to the division site by immunolocalization, by tagging with green fluorescent protein (GFP), or by both techniques (1–4, 18, 20, 24, 31, 39, 42, 44). In addition, DivIB, a homolog of FtsQ in Bacillus subtilis, localizes to the septum in that organism (19), implying that FtsQ in E. coli should be no exception. The accumulation of all these essential gene products at the division site suggests that they cooperate intimately in the construction of the septum, possibly as a multiprotein complex. Demonstration of direct physical interactions between FtsZ and FtsA (11, 25, 40) and between FtsZ and ZipA (18) supports the speculation that cell division proteins form such a multimeric complex, the so-called septalsome or divisome (29).
Of the nine known cell division proteins, only FtsZ and FtsI have well-defined enzymatic activities. FtsZ, a homolog of the eukaryotic cytoskeletal protein tubulin (21), has GTPase and polymerization activities (10, 13, 27, 28, 33, 43), and various microscopy studies have indicated that it localizes to the midcell region early in the cell cycle and forms a ring that constricts along with the advancing edge of the invaginating septum (4, 20, 38). These observations suggest that FtsZ contributes to septal constriction by pulling the cell membrane inward (12). FtsI, known also as penicillin-binding protein 3, has a large periplasmic domain with transpeptidase activity specifically required for septal peptidoglycan synthesis (reviewed in references 30 and 36). The precise functions of other cell division proteins, however, remain obscure. All of them are associated with the cell membrane, and some contain large periplasmic domains that suggest that they may have enzymatic functions in septal synthesis.
One such protein is FtsQ, a bitopic membrane protein with a short cytoplasmic amino terminus, a single transmembrane segment, and a relatively large periplasmic domain (6). Results of a genetic analysis in which the cytoplasmic or transmembrane domains of FtsQ were exchanged with analogous domains from other proteins suggested that the cytoplasmic region of FtsQ was necessary for complementation of a null mutation, while the transmembrane segment could be swapped without deleterious effects (but see below) (17). FtsQ lacks extensive sequence homology to other proteins except for FtsQ homologs in other bacterial species. Nevertheless, genetic studies have indicated that FtsQ may functionally interact with other cell division gene products; increases in ftsQ expression are detrimental to strains carrying mutations in ftsA, ftsI, or ftsZ (7), and overexpression of ftsN can partially suppress an ftsQ temperature-sensitive mutation (8).
Recent localization studies have confirmed the functional interaction of Fts proteins originally suggested by genetic analyses. Dependency relationships appear to exist among the cell division proteins for recruitment to the septum. Furthermore, an ordered pathway for recruitment can be formulated from these dependency relationships. FtsZ is the earliest known septal recruit, since it can still form ring structures at potential division sites when other cell division proteins are defective in their function (1, 2, 31, 39, 42, 44). Studies with similar approaches have indicated that FtsA requires only FtsZ for localization (3, 39, 44), while both FtsI and FtsK require FtsZ and FtsA (39, 44) and FtsN requires FtsZ, FtsA, FtsI, and FtsQ (2). Thus, a tentative model is that FtsZ localizes to the septum first, followed immediately by FtsA, then FtsK, FtsQ, and FtsI in an unknown order, and finally FtsN (22, 39). Assuming that the Fts proteins function only at the division site, this pathway may represent the order in which they act on the developing septum. Clarifying the positions of the known cell division proteins in this pathway will facilitate future investigations of their functional significance.
Here we use GFP as a reporter tag to show that FtsQ localizes to the division site. We have exchanged the cytoplasmic or transmembrane domains of FtsQ with analogous domains from MalF, a membrane protein required for maltose transport but not involved in cell division. The results provide evidence that the periplasmic domain is sufficient for septal targeting. In addition, GFP-FtsQ requires FtsZ and FtsA but not FtsL or FtsI for localization. Finally, ZipA fused to GFP can localize to potential division sites in ftsQ filaments.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. NZY medium was used (15); when appropriate, the following antibiotics were added at the indicated concentrations: ampicillin, 200 μg/ml (plasmid) and 25 μg/ml (chromosome); chloramphenicol, 10 μg/ml; kanamycin, 40 μg/ml; and tetracycline, 15 μg/ml. d-Glucose or l-arabinose was used at 0.2% to repress or induce, respectively, the expression of fts alleles cloned under the control of the PBAD promoter (16).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genetic marker(s) or feature(s) | Constructiona | Source or reference |
|---|---|---|---|
| Strains | |||
| Without gfp fusions | |||
| KS272 | F− ΔlacX74 galE galK thi rpsL ΔphoA (PvuII) | 37 | |
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb-5301 deoC7 ptsF25 rbsR | Laboratory collection | |
| LMG194 | KS272 Δara714 leu::Tn10 | 16 | |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen | |
| DRC14 | MC4100 leu::Tn10 ftsZ84(Ts) | D. RayChaudhuri | |
| MM61 | F−araD139 ΔlacU169 Str leu::Tn10 ftsA12(Ts) | Laboratory collection | |
| LMG64 | KS272 recA::cat leu::Tn10 ftsI23(Ts) | 17 | |
| EC433 | MG1655 leu::Tn10 ftsQ1(Ts) | D. Weiss | |
| JOE224 | MC4100 leu::Tn10 ftsQ1(Ts) | P1 (EC433) × MC4100, select Tetr, screen Ts | This study |
| EC548 | KS272 ftsI::TnphoAI173ΔIS50R (Kanr)/pDSW262 | 41 | |
| JM265 | KS272 ftsL::TnphoAL81ΔIS50R (Kanr)/pLD45-33 | J.-M. Ghigo | |
| MJC295 | KS272 ftsQ::TnphoA50 (Kanr)/pLMG161 | M. Carson | |
| JOE170 | KS272 ftsQ::TnphoA50 (Kanr)/pJC10 | From MJC295, replace pLMG161 with pJC10 | This study |
| SM551 | F− Δlac(MS265) λ− λsnalA2 supF58 | S. Michaelis | |
| DHB6521 | SM551 λInCh1 (Kanr) | 5 | |
| With gfp fusions in “wild-type” backgrounds | |||
| EC442 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-ftsQ | From pDSW240 with λInCh | This study |
| EC450 | MC4100 Δ(λattL-lom)::bla lacIq P208-zipA-gfp | 41 | |
| EC452 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp | 41 | |
| EC489 | MC4100 Δ(λattL-lom)::bla lacIq P208-zipA-gfp leu::Tn10 | 41 | |
| JOE99 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-ftsQ leu::Tn10 | P1 (DRC14) × EC442, select Tetr, screen Ti | This study |
| JOE192 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-QQQ | From pJC11 with λInCh | This study |
| JOE193 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-FQQ | From pJC12 with λInCh | This study |
| JOE194 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-QFQ | From pJC14 with λInCh | This study |
| JOE196 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-FFQ | From pJC13 with λInCh | This study |
| JOE257 | MC4100 Δ(λattL-lom)::bla lacIq P207-gfp-QQL | From pJC16 with λInCh | This study |
| With gfp fusions in Ts backgrounds | |||
| JOE95 | P207-gfp-ftsQ leu::Tn10 ftsA12(Ts) | P1 (MM61) × EC442, select Tetr, screen Ts | This study |
| JOE97 | P207-gfp-ftsQ leu::Tn10 ftsZ84(Ts) | P1 (DRC14) × EC442, select Tetr, screen Ts | This study |
| JOE136 | P207-gfp-ftsQ leu::Tn10 ftsI23(Ts) | P1 (LMG64) × EC442, select Tetr, screen Ts | This study |
| JOE165 | P208-zipA-gfp leu::Tn10 ftsQ1(Ts) | P1 (EC433) × EC450, select Tetr, screen Ts | This study |
| JOE225 | P207-gfp leu::Tn10 ftsQ1(Ts) | P1 (EC452) × JOE224, select Ampr | This study |
| JOE226 | P207-gfp-ftsQ leu::Tn10 ftsQ1(Ts) | P1 (EC442) × JOE224, select Ampr | This study |
| JOE228 | P207-gfp-QQQ leu::Tn10 ftsQ1(Ts) | P1 (JOE192) × JOE224, select Ampr | This study |
| JOE229 | P207-gfp-FQQ leu::Tn10 ftsQ1(Ts) | P1 (JOE193) × JOE224, select Ampr | This study |
| JOE230 | P207-gfp-FFQ leu::Tn10 ftsQ1(Ts) | P1 (JOE196) × JOE224, select Ampr | This study |
| JOE231 | P207-gfp-QFQ leu::Tn10 ftsQ1(Ts) | P1 (JOE194) × JOE224, select Ampr | This study |
| JOE259 | P207-gfp-QQL leu::Tn10 ftsQ1(Ts) | P1 (EC433) × JOE257, select Tetr, screen Ts | This study |
| With gfp fusions in depletion backgrounds | |||
| JOE204 | ftsQ::TnphoA50 (Kanr) P207-gfp/pJC10 | P1 (EC452) × JOE170, select Ampr | This study |
| JOE206 | ftsQ::TnphoA50 (Kanr) P207-gfp-ftsQ/pJC10 | P1 (EC442) × JOE170, select Ampr | This study |
| JOE210 | ftsQ::TnphoA50 (Kanr) P207-gfp-QQQ/pJC10 | P1 (JOE192) × JOE170, select Ampr | This study |
| JOE212 | ftsQ::TnphoA50 (Kanr) P207-gfp-FQQ/pJC10 | P1 (JOE193) × JOE170, select Ampr | This study |
| JOE214 | ftsQ::TnphoA50 (Kanr) P207-gfp-FFQ/pJC10 | P1 (JOE196) × JOE170, select Ampr | This study |
| JOE216 | ftsQ::TnphoA50 (Kanr) P207-gfp-QFQ/pJC10 | P1 (JOE194) × JOE170, select Ampr | This study |
| JOE220 | ftsL::TnphoA P207-gfp-ftsQ/pLD45-33 | P1 (EC442) × JM265, select Ampr | This study |
| JOE233 | ftsI::TnphoA P207-gfp-ftsQ/pDSW262 | P1 (EC442) × EC548, select Ampr | This study |
| Plasmids | |||
| pBAD18 | Arabinose regulation; Ampr | 16 | |
| pBAD33 | Arabinose regulation; Cmr | 16 | |
| pLMG161 | ftsQ Ampr | 17 | |
| pJC10 | ftsQ Cmr | See Materials and Methods | This study |
| pET-26b(+) | T7lac Kanr | Novagen | |
| pDSW165 | FtsQ(peri)-His6 | PCR of pLMG161, insert into pET-26b(+) | This study |
| pDSW207 | IPTG regulation, gfp fusion vector; Ampr | 41 | |
| pDSW240 | gfp-ftsQ | PCR of pLMG161, insert into pDSW207 | This study |
| pDSW262 | pBAD33-ftsI | 41 | |
| pLD45-33 | pBAD33-LLL | J.-M. Ghigo | |
| pLD104 | pBAD18-FFQ | 17 | |
| pLD108 | pBAD18-QFQ | 17 | |
| pLD134 | pBAD18-FQQ | 17 | |
| pLD137 | pBAD18-QQQ | 17 | |
| pJC11 | gfp-QQQ | PCR of pLD137, insert into pDSW207 | This study |
| pJC12 | gfp-FQQ | PCR of pLD134, insert into pDSW207 | This study |
| pJC13 | gfp-FFQ | PCR of pLD104, insert into pDSW207 | This study |
| pJC14 | gfp-QFQ | PCR of pLD108, insert into pDSW207 | This study |
| pJC16 | gfp-QQL | See Materials and Methods | This study |
P1 indicates P1 transduction. For example, JOE224 was constructed by infecting MC4100 with a P1 lysate made from EC433. Ts, temperature sensitive; Ti, temperature insensitive (screened for growth at 42°C).
Standard techniques were used for cloning and analysis of DNA, PCR, electroporation, transformation, and P1 transduction (26, 35). Enzymes used to manipulate DNA were from New England BioLabs. DNA sequencing was performed at the Micro Core Facility of the Department of Microbiology and Molecular Genetics, Harvard Medical School.
Plasmid pJC10 was constructed by insertion of the NheI-HindIII fragment containing ftsQ from pLMG161 (17) into pBAD33 (16) digested with XbaI and HindIII.
Construction of strains containing gfp fusions.
Sequence-specific primers were used to amplify ftsQ and the swap constructs from their parent plasmids (Table 1). Oligonucleotide F1 (5′-CGAGAATTCAACAACAACATGGATGTCATTAAAAAGAAACATTGGTGGC-3′) anneals to the 5′ region of malF, Q1 (5′-CGAGAATTCAACAACAACTCGCAGGCTGCTCTGAACACG-3′) anneals to the 5′ region of ftsQ, and Q2 (5′-TGCAAGCTTTCATTGTTGTTCTGCCTGTGC-3′) anneals to the 3′ region of ftsQ; underlining indicates EcoRI or HindIII sites. PCR products were digested with EcoRI and HindIII and ligated into the same sites of the gfp fusion vector pDSW207 (41). Derived from the expression vector pTrc99A (Pharmacia), pDSW207 contains a weakened trc promoter, followed by a Shine-Dalgarno sequence, a bright gfp allele, and a polylinker embedded in an open reading frame (41). The resulting plasmids were confirmed by restriction analysis and DNA sequencing. Fusion proteins containing the cytoplasmic domain of FtsQ have the linker sequence YKEFNNNSQ, where Y and K are the last two residues of GFP and S and Q are the second and third residues of FtsQ. (The initiating methionine of FtsQ is absent in these fusions.) Fusion proteins containing the cytoplasmic domain of MalF have the linker sequence YKEFNNNMD, where Y and K are the last two residues of GFP and M and D are the first two residues of MalF.
A three-letter designation was used to describe the swap constructs: the first letter indicates the cytoplasmic region, the second indicates the transmembrane segment, and the third indicates the periplasmic domain. For example, the construct QFL would contain the cytoplasmic domain of FtsQ, the transmembrane segment of MalF, and the periplasmic domain of FtsL.
Plasmid pJC16, which carries the fusion gfp-QQL, was constructed by replacing the MscI-MscI fragment (containing the 3′ end of gfp and the region encoding the cytoplasmic and transmembrane domains of FtsL) of pDSW207-LLL with the analogous fragment of pJC11 (gfp-QQQ). The gfp-LLL fusion in pDSW207-LLL was constructed by using pLD45 (17) as a template for PCR and inserting the resulting fragment into pDSW207, in the same way that pDSW236 (gfp-ftsL) was constructed (14). Constructs were confirmed by DNA sequencing.
All of the translational fusions to gfp were incorporated into the λInCh phage (5) and integrated into the chromosome in single copies at the λ attachment site (λatt). λInCh picks up plasmid-borne genes by homologous recombination; for this study, the genes included the gfp fusions, bla (conferring ampicillin resistance), and lacIq. Chromosomal insertions made with λInCh were subsequently stabilized by selection for a deletion that removed most of the phage, including its killing functions.
Complementation of ftsQ temperature-sensitive and null mutations.
Complementation of the temperature-sensitive mutation was tested with ftsQ1(Ts) strains carrying gfp fusions in single copies (derivatives of JOE224 or JOE257; Table 1). Cells were streaked onto NZY plates containing tetracycline and ampicillin and incubated at 30 or 42°C. Growth was scored after 16 h. To determine complementation of the null mutation, we used an FtsQ depletion strain (JOE170; Table 1) in which the chromosomal ftsQ allele has been disrupted by a transposon insertion (ftsQ::TnphoA50), leaving the cytoplasmic and transmembrane domains (first 50 amino acids) of FtsQ intact and fused to alkaline phosphatase (6); the only wild-type copy of ftsQ is on a plasmid, under the control of the PBAD promoter (16). This depletion strain prospers when FtsQ expression is induced with arabinose but perishes when expression is repressed with glucose. The gfp fusion alleles were transduced with P1 into this strain by selection for ampicillin resistance (Table 1). Cells were streaked onto NZY plates containing chloramphenicol, kanamycin, ampicillin, and 0.2% arabinose or glucose and incubated at 30, 37, or 42°C. Colony formation was examined after 16 to 24 h.
Growth conditions for fusion protein expression.
For localization of GFP fusions in the wild-type background, cells were grown overnight at 30°C in NZY medium plus ampicillin, subcultured into fresh NZY medium containing several different concentrations (0 to 100 μM) of isopropyl-β-d-thiogalactopyranoside (IPTG), and shaken at 30°C until the optical density at 600 nm (OD600) reached between 0.2 and 0.3. For localization of GFP-FtsQ and ZipA-GFP in temperature-sensitive backgrounds, growth conditions similar to those for the wild-type background were used, except that overnight cultures contained tetracycline as well as ampicillin. After the culture reached the early log phase, 4 ml of cells was transferred to 16 ml of prewarmed NZY medium containing 5 (GFP-FtsQ) or 50 (ZipA-GFP) μM IPTG and shaken vigorously at 42°C for 45 to 60 min; for ftsZ84(Ts) cells, the culture was shifted to NZY medium without NaCl. Cells were harvested immediately before the temperature shift and 45 and 60 min after the shift. For localization of GFP-FtsQ in depletion strains, cells were grown overnight in NZY medium with ampicillin, kanamycin, chloramphenicol, and 0.2% arabinose and subcultured into fresh medium containing chloramphenicol, 0.2% arabinose, and 5 μM IPTG. After the culture reached the desired OD600, aliquots were washed and resuspended in NZY medium plus 5 μM IPTG and 0.2% arabinose or glucose. The resuspended cells were diluted 1:20 to 1:200 in that medium and grown for 3 to 5 h until they became filamentous in the glucose culture. Samples were harvested from both the arabinose and the glucose cultures.
Fluorescence microscopy.
To facilitate observation, we fixed the cell samples by using a procedure similar to that used for immunofluorescence microscopy (31, 32). A 0.5-ml aliquot of the cell culture was added directly to the fixative, which contained 100 μl of 16% paraformaldehyde, 0.4 μl of 25% glutaraldehyde, and 20 μl of 1 M sodium phosphate (pH 7.4). The mixture was incubated for 15 min at room temperature and for 15 min on ice. Cells were then washed three times in PBS (10 mM sodium phosphate [pH 7.4], 150 mM NaCl, 15 mM KCl) and resuspended with 50 or 100 μl of PBS per OD600 unit of 0.1 for the original sample. Fixed cells were allowed to adhere to a 15-well multitest slide (ICN Biomedicals) pretreated with poly-l-lysine (Sigma) by incubation at room temperature for 10 min. We then washed the slide twice by adding PBS and aspirating it to remove free-floating cells. Cells were incubated with 0.2 μg of 4′,6-diamidino-2-phenylindole (DAPI; Sigma) per ml in PBS for 5 min to stain the DNA, and the slide was washed twice again with PBS. Finally, PBS containing 50% glycerol was added prior to sealing with a cover glass. Throughout the microscopy preparation procedure, we minimized the exposure of samples to light to prevent photobleaching.
To examine the cells, we used a Nikon Optiphot 2 microscope equipped with a 60× oil immersion objective (numerical aperture, 1.4), a 100-W mercury lamp, and standard fluorescein isothiocyanate and DAPI filter sets. Images were captured with an Optronics Engineering DEI-750 charge-coupled device video camera system, digitized with a Scion CG7 video card and the public domain NIH Image program (developed at the National Institutes of Health and available on the internet at http://rsb.nih.gov/nih-image/), and processed with Adobe Photoshop version 4.0. Exposures were between 2 and 8 s for GFP images and between 1/60 and 1/15 s for phase-contrast and DAPI images. We measured cell length by using the NIH Image program to compare phase-contrast images of the cells to a calibrated standard. Septal localization was determined from the GFP images. We used the DAPI images to assess whether a cell was healthy prior to fixation, since dead or lysed cells exhibit aberrant nucleoid structures. Only cells with regularly spaced nucleoids were scored.
Antibodies.
Rabbit antibodies against the periplasmic domain of FtsQ were raised at Covance (Denver, Pa.). The protein used as an antigen was obtained as follows. The DNA sequence encoding the periplasmic domain of FtsQ (residues 50 to 276) was amplified by PCR from pLMG161 with the sequences 5′-GAGACCATGGAAGATGCGCAACGCCTG-3′ and 5′-TTCCGCTCGAGTGCTTGTTGTTCTGCCTGTGC-3′ as primers; underlining indicates NcoI or XhoI sites. The PCR product was digested with NcoI and XhoI and ligated into the same sites of pET-26b(+) (Novagen), under the control of a T7lac promoter, to create pDSW165. The insert was fused to two features on plasmid pET-26b(+): a pelB leader sequence, which promotes protein export, and a C-terminal six-histidine (His6) tag, which facilitates protein purification. Plasmid pDSW165 was transformed into strain BL21(DE3). After a 2-h induction with 1 mM IPTG, the transformant overproduced FtsQ(peri)-His6 [where (peri) indicates the periplasmic domain of FtsQ] to approximately 10% of total cell protein, mostly in cytoplasmic inclusion bodies. Insoluble FtsQ(peri)-His6 was purified under denaturing conditions (with 6 M urea) by nickel affinity chromatography (30a). The final preparation was >95% pure, and the yield was about 25 mg per liter of culture.
Polyclonal antibodies against FtsQ were affinity purified against the same protein domain used as an antigen. FtsQ(peri)-His6 was dialyzed into coupling buffer (100 mM sodium phosphate [pH 7.0], 400 mM NaCl, 3 M guanidine), and 2 mg of protein was immobilized on 1 ml of AminoLink resin (Pierce) by reductive amination in accordance with the manufacturer’s instructions. The coupling efficiency was >90%. The column was equilibrated with TBS (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 3 mM KCl). We diluted 5 ml of anti-FtsQ antiserum with 10 ml of TBS, passed the diluted antiserum through the column three times to allow binding of anti-FtsQ antibodies, and washed the column with 15 ml of TBS containing 500 mM NaCl. Bound antibodies were eluted with 0.1 M glycine (pH 2.5), and 1-ml fractions were collected into tubes containing 0.1 ml of 1 M Tris-HCl (pH 8.0). Peak fractions were identified by immunoblotting against purified FtsQ(peri)-His6 that had been spotted onto nitrocellulose membranes. Fraction 2 contained most of the anti-FtsQ activity.
To remove antibodies that recognized soluble E. coli proteins, we passed fraction 2 over a 1-ml column of immobilized E. coli lysate (Pierce) in accordance with the manufacturer’s instructions. Fractions of 0.5 ml were collected and analyzed for protein by the Bradford assay (Pierce). Fractions 2 and 3 contained 75% of the input protein; they were pooled, dialyzed against 10 mM sodium phosphate (pH 7.0)–250 mM NaCl, and concentrated fourfold to a volume of about 250 μl by ultrafiltration in a Centricon 30 apparatus (Amicon). Purified anti-FtsQ antibodies were stored at −20°C at a concentration of 0.2 mg of immunoglobulin G/ml of storage buffer (10 mM sodium phosphate [pH 7.0], 250 mM NaCl, 10 mg of bovine serum albumin per ml, 50% glycerol) and used at a dilution of 1:10,000 for immunoblotting.
Rabbit antibodies against GFP were purchased from Clontech. Nonspecific antibodies were eliminated by incubation with E. coli cell lysate. To prepare the cell lysate, we pelleted 1 liter of an overnight culture of MC4100 by centrifugation and resuspended the cells in 8 ml of PBS; cells were then lysed by repeated sonication and freezing-thawing. We mixed 50 μl of anti-GFP antibody with 500 μl of cell lysate, incubated the mixture on ice for 1 h, and pelleted it by centrifugation at 4°C. The supernatant was mixed with another 500 μl of cell lysate, and the mixture was incubated for 1 h and centrifuged again. The final supernatant (approximately 1 ml) was divided into five aliquots, each corresponding to 10 μl of the original antibody. For Western blotting, an aliquot was diluted into 10 ml of TBS–0.05% Tween 20 so that the final dilution was 1:1,000 of the original antibody.
Rabbit antibodies against FtsL were prepared and purified as described previously (14), similar to the procedures used for anti-FtsQ antibodies. They were used at a dilution of 1:2,500 for immunoblotting.
Western blotting.
Western blotting was done by standard procedures (35). Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (0.2-μm-pore size; Schleicher & Schuell, Inc.). Immunodetection was performed with polyclonal antibody against FtsQ or GFP as the primary antibody, goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Pierce) as the secondary antibody, SuperSignal chemiluminescent substrate (Pierce), and Kodak X-OMAT film. The image on the film was then scanned and processed with Adobe Photoshop version 4.0.
To determine the steady-state levels of the GFP fusion proteins, we harvested cells from the same cultures as those used for fluorescence microscopy. Cells in 2 ml of culture were pelleted by centrifugation and resuspended with 10 or 20 μl of SDS sample buffer (New England BioLabs) per OD600 unit of 0.1. Samples were boiled for 5 min and, when the level of GFP-FtsQ was compared to that of endogenous FtsQ, diluted with SDS sample buffer. Ten to 20 μl of sample was loaded onto a 10 or 12% polyacrylamide-SDS gel.
RESULTS
Construction, characterization, and localization of a GFP-FtsQ fusion.
To study the localization of FtsQ, we constructed a gfp-ftsQ translational fusion such that GFP was attached to the amino terminus (cytoplasmic domain) of FtsQ. The fusion was placed under the control of an IPTG-regulatable promoter and then integrated into the chromosome at the λatt site by use of a newly developed λ phage (5). The resulting strain was a merodiploid containing wild-type ftsQ at the 2-min region and the gfp-ftsQ fusion at the λatt site.
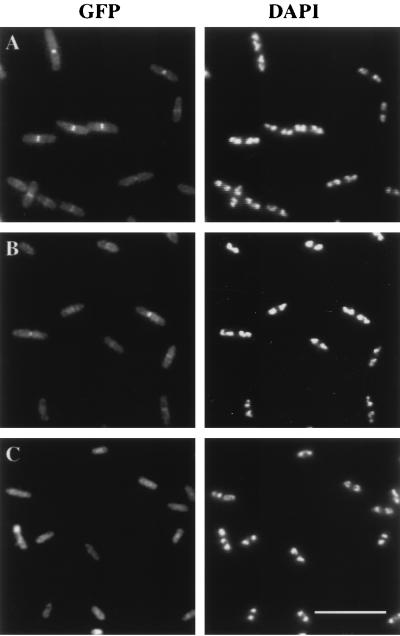
To determine the subcellular location of GFP-FtsQ in this merodiploid strain, we grew cells at different IPTG concentrations to vary the level of expression and then fixed them for examination by fluorescence microscopy. Localization of GFP-FtsQ to the division site was observed over a range of induction levels, from 0 to 50 μM IPTG (Fig. 1A). At higher levels of induction, the background fluorescence throughout the cell became too bright, deterring easy identification of a fluorescent band in the mid-cell region (data not shown). For optimal and consistent observation of GFP-FtsQ localization, we grew cells to the early log phase in the presence of 5 μM IPTG at 30°C. Under these conditions, about 60% of the cells exhibited a band of fluorescence at the division site (Table 2). Bands of fluorescence were not observed at the division sites of control cells expressing GFP alone (Fig. 1C).
FIG. 1.
Subcellular locations of GFP-FtsQ (A), GFP-FFQ (B), and GFP (C). Cells were grown and prepared for microscopy as described in Materials and Methods. Strains used were EC442 (A), JOE196 (B), and EC452 (C). Bar, 10 μm.
TABLE 2.
Localization frequencies of and complementation by GFP-FtsQ and GFP-FtsQ swap protein constructs
| Construct | Structurea | Total no. of cells scored | % Localization frequency (mean ± SD)b | Complementation of the following mutation:
|
|
|---|---|---|---|---|---|
| Tsc | Nulld | ||||
| GFP-FtsQ | 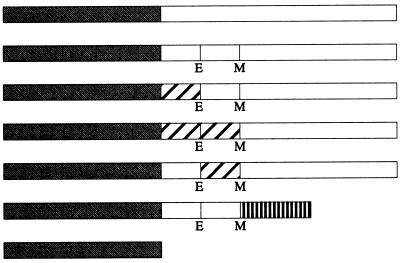 |
715 | 60 ± 3 | + | + |
| GFP-QQQ | 696 | 44 ± 7 | + | + | |
| GFP-FQQ | 703 | 37 ± 8 | + | + | |
| GFP-FFQ | 654 | 35 ± 10 | + | ± | |
| GFP-QFQ | 599 | 30 ± 6 | + | ± | |
| GFP-QQL | 150 | 0 | − | ND | |
| GFP alone | 60 | 0 | − | − | |
Open boxes represent domains of FtsQ. Hatched boxes represent domains derived from MalF. Stippled boxes denote GFP. The box with vertical bars represents the periplasmic domain of FtsL. E and M indicate the borders of the transmembrane domain where the restriction sites for EagI and MscI, respectively, were introduced. These sites caused the following amino acid changes in FtsQ: T23G, W48L, M49G, and E50D. Diagrams of fusion proteins are not to scale. The cytoplasmic and transmembrane domains of FtsQ contained 24 and 25 amino acids, respectively, while those of MalF contained 16 and 25 amino acids. The periplasmic domain of FtsQ contained 227 amino acids, while that of FtsL contained 64 amino acids.
Determined from three independent sets of experiments. Strains EC442, JOE192, JOE193, JOE194, JOE196, and JOE257 were grown in parallel cultures, fixed, and scored for localization as described in Materials and Methods.
Complementation of the ftsQ1(Ts) allele was determined as described in Materials and Methods. The strains used were JOE225, JOE226, JOE228, JOE229, JOE230, JOE231, and JOE259. +, complementation; −, no complementation.
Complementation of the ftsQ::TnphoA50 null allele was determined as described in Materials and Methods. The strains used were JOE204, JOE206, JOE210, JOE212, JOE214, and JOE216. +, complementation; ±, partial complementation; −, no complementation; ND, not determined.
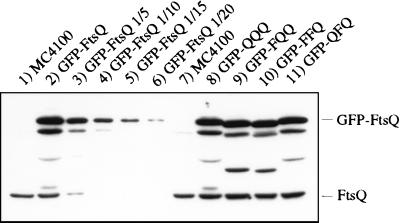
We next estimated the level of expression of GFP-FtsQ by immunoblot analysis with a polyclonal antibody generated against the periplasmic domain of FtsQ. Under the growth conditions used for microscopy, the steady-state level of GFP-FtsQ was about 10 to 15 times that of wild-type FtsQ (Fig. 2). Even in uninduced cells, the level of GFP-FtsQ was about 5 to 10 times that of FtsQ (data not shown). The relatively high level of expression of GFP-FtsQ compared to the expression of endogenous FtsQ was not surprising, because FtsQ is a low-abundance protein (6) and the gfp-ftsQ fusion uses a highly efficient translational initiation sequence from plasmid pDSW207, which was used to construct the fusion (see Materials and Methods). Immunoblotting also revealed three bands between those of FtsQ and GFP-FtsQ that appeared to be degradation products of GFP-FtsQ (Fig. 2). Only a single band, that of full-length GFP-FtsQ, was observed when immunoblotting was performed with a polyclonal anti-GFP antibody (data not shown), suggesting that degradation occurred in the GFP domain of the fusion protein.
FIG. 2.
Immunoblot of FtsQ and GFP-FtsQ fusion proteins. Lanes: 1, MC4100; 2 to 6, serial dilutions of EC442; 7, MC4100; 8, JOE192; 9, JOE193; 10, JOE196; 11, JOE194. Cells were grown at 30°C to the early log phase and boiled in SDS sample buffer as described in Materials and Methods. Equivalent amounts of samples were loaded except that for lanes 3 to 6, the sample in lane 2 was diluted 1/5, 1/10, 1/15, and 1/20, respectively, with SDS sample buffer. The positions of GFP-FtsQ and FtsQ are indicated on the right.
To assess the functionality of the gfp-ftsQ fusion, we tested its ability to complement temperature-sensitive or null mutations in ftsQ. First, gfp alone or gfp-ftsQ was introduced into an ftsQ1(Ts) strain by P1 transduction. Cells expressing gfp-ftsQ formed normal colonies at 42°C, even without IPTG, while those expressing gfp alone failed to grow (Table 2). To test the complementation of a null mutation, we used an FtsQ depletion strain in which the chromosomal ftsQ allele is disrupted by a transposon insertion, leaving intact only the cytoplasmic and transmembrane domains, and a wild-type copy of ftsQ is on a plasmid under the control of an arabinose-dependent promoter (see Materials and Methods). The depletion strain expressing gfp formed colonies on plates containing arabinose but failed to grow on plates containing glucose, while the depletion strain expressing gfp-ftsQ grew well on plates containing either arabinose or glucose (Table 2). Furthermore, GFP-FtsQ continued to localize to the septum in cells depleted of wild-type FtsQ (data not shown). Thus, as far as we can measure, GFP-FtsQ appears to function as well as wild-type FtsQ.
Localization of and complementation by FtsQ swap constructs fused to GFP.
FtsQ is a membrane protein with a simple topology, consisting of a cytoplasmic N-terminal domain, a transmembrane domain, and a periplasmic domain. Previous complementation analysis indicated that the cytoplasmic domain is necessary for function, while the transmembrane domain can be replaced (but see below) (17). To determine whether the cytoplasmic and transmembrane domains of FtsQ are required for localization, we used swap constructs in which these domains were replaced by analogous domains from MalF, a protein not involved in cell division (17). A three-letter designation describes the type of replacement: the first letter indicates the source of the cytoplasmic region, the second indicates that of the transmembrane segment, and the third indicates that of the periplasmic domain. For instance, the swap QFQ contains the cytoplasmic and periplasmic domains of FtsQ and the transmembrane segment of MalF. The QQQ construct differs from wild-type FtsQ in that 4 amino acids at the borders of the membrane-spanning segment have been altered due to the introduction of restriction sites in corresponding regions of the ftsQ gene (Table 2).
GFP was fused to the swap constructs QQQ, FQQ, FFQ, and QFQ, and the fusions were used to generate merodiploid strains similar to those used for the localization of GFP-FtsQ. We found that all of the swap constructs localized to the division site at 30°C, albeit at reduced frequencies compared to that of FtsQ (Fig. 1B and Table 2). Even GFP-QQQ, with only slight amino acid modifications, appeared to localize less well than GFP-FtsQ. The fusion constructs could be ranked, according to their abilities to localize, in the following order, from best to worst: QQQ, FQQ, FFQ, and QFQ. To ensure that differences in localization did not result from differences in expression, we verified by immunoblotting that the steady-state levels of the fusion proteins were equivalent when cells were grown under similar conditions (Fig. 2).
To confirm that the periplasmic domain of FtsQ is required for localization, we fused the swap construct QQL, which contains the cytoplasmic and transmembrane domains of FtsQ and the periplasmic domain of FtsL, to GFP. This fusion protein failed to localize to the septum (Table 2), even though immunoblotting with anti-GFP and anti-FtsL antibodies indicated that the protein was expressed at a level similar to that of GFP-FtsL (data not shown), which did localize to the septum (14).
The localization frequencies of the swap constructs were slightly surprising because they did not show the same pattern as previous complementation results (17). According to the earlier report, FQQ and FFQ failed to complement a null mutation in ftsQ, whereas QFQ did complement such a mutation. We therefore decided to test the complementation of temperature-sensitive and null mutations in ftsQ by using swap constructs fused to GFP. Consistent with previous results, all fusion constructs, except for gfp-QQL, complemented the ftsQ1(Ts) mutation at 42°C (Table 2). However, differences appeared when we tested complementation with the FtsQ depletion strain. All of the swap constructs (except for gfp-QQL, which was not tested) were able to complement the null mutation at 30°C, but cells expressing GFP-FFQ or GFP-QFQ grew more poorly on glucose plates than other cells. At 37°C, the differences in complementation became more pronounced, and at 42°C, GFP-FFQ and GFP-QFQ could not complement the null mutation, while the other fusion constructs could. These complementation results are more consistent with the localization rankings than complementation results from the earlier report: weaker complementation correlates with less frequent localization (Table 2).
The complementation results, in addition, suggested that septal localization of the various GFP-FtsQ swap proteins does not depend on wild-type FtsQ. Indeed, fluorescence microscopy showed that the swap proteins still localized to the division site in cells depleted of wild-type FtsQ at 30°C (data not shown). Thus, the periplasmic domain of FtsQ appears to be sufficient for function.
Dependence of GFP-FtsQ localization on other fts genes.
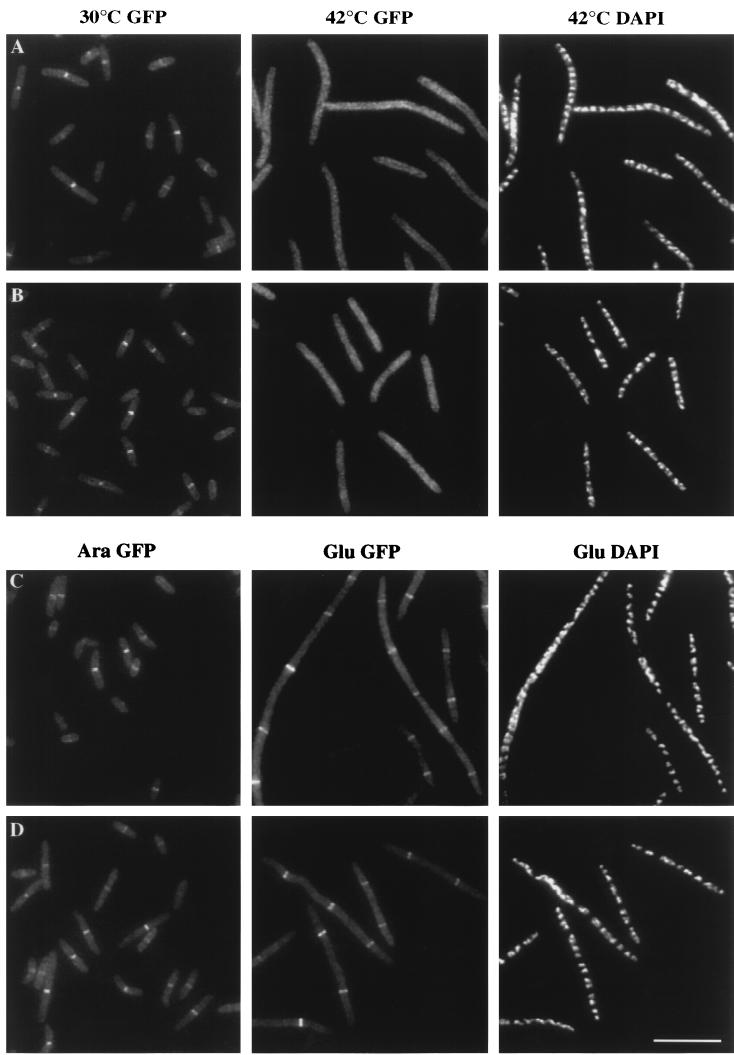
The various cell division proteins appear to be recruited to the developing septum in an ordered pathway. To clarify the order of recruitment, we examined the ability of GFP-FtsQ to localize when other Fts proteins were inactivated. First, we determined whether GFP-FtsQ can localize in ftsZ(Ts) or ftsA(Ts) mutants at the restrictive temperature. Since FtsZ and FtsA, considered early recruits to the division site, can localize in ftsQ1(Ts) filaments (1, 3), they probably function upstream of FtsQ. However, the possibility existed that FtsQ is recruited in parallel and that it can localize without functional FtsZ or FtsA. We transduced an ftsZ(Ts) or ftsA(Ts) allele into the merodiploid strain carrying gfp-ftsQ, grew the cells at 30°C, and then shifted the culture to 42°C to induce filamentation. GFP-FtsQ was recruited to the division site in ftsZ or ftsA mutants at 30°C, but hardly any fluorescent bands were found in ftsZ or ftsA filaments at 42°C (Fig. 3A and B and Table 3). Since, in the control experiment, GFP-FtsQ still localized to the division site when wild-type merodiploid cells were shifted to 42°C (Table 3), the localization ability of GFP-FtsQ was not adversely affected by the temperature shift. Furthermore, the steady-state levels of GFP-FtsQ remained the same before and after the temperature shift, as measured by immunoblot analysis (data not shown). Thus, our results indicate that GFP-FtsQ requires FtsZ and FtsA for localization.
FIG. 3.
Localization of GFP-FtsQ in fts mutants. (A and B) GFP-FtsQ localizes to division sites in ftsZ84(Ts) (A) and ftsA12(Ts) (B) cells at 30°C (left panels) but not at 42°C (middle panels); DAPI images indicate that nucleoid structures are normal in the filaments at 42°C (right panels). (C and D) GFP-FtsQ localizes to potential division sites in FtsI (C) and FtsL (D) depletion strains; the left panels are GFP images of cells growing normally in the presence of arabinose, the middle panels are GFP images of cells grown with glucose and therefore depleted of FtsI or FtsL, and the right panels are corresponding DAPI images of the middle panels. Cells were grown and fixed for fluorescence microscopy as described in Materials and Methods. Strains used were JOE97 (A), JOE95 (B), JOE233 (C), and JOE220 (D). Bar, 10 μm.
TABLE 3.
Localization of GFP-FtsQ in fts mutants
| Expt | Strain background | Strain | Growth condition | No. of cells | Mean ± SD cell length (μm) | Total no. of rings | % of cells with a ring(s) | Avg spacing between rings (μm)a |
|---|---|---|---|---|---|---|---|---|
| Temperature shift | Wild type | JOE99 | 30°C | 124 | 4.0 ± 1.0 | 87 | 70 | 5.7 |
| 42°C | 136 | 3.7 ± 0.9 | 91 | 67 | 5.5 | |||
| ftsZ84(Ts) | JOE97 | 30°C | 97 | 5.0 ± 1.4 | 52 | 54 | 9.3 | |
| 42°C | 51 | 16.1 ± 3.3 | 0 | 0 | >823 | |||
| ftsA12(Ts) | JOE95 | 30°C | 138 | 4.2 ± 1.0 | 97 | 70 | 5.9 | |
| 42°C | 75 | 10.1 ± 2.7 | 3 | 4 | 251 | |||
| ftsI23(Ts) | JOE136 | 30°C | 106 | 9.5 ± 5.4 | 120 | 92 | 8.4 | |
| 42°C | 53 | 16.1 ± 5.8 | 107 | 92 | 8.0 | |||
| Depletion | ftsI::TnphoA | JOE233 | Arabinose | 102 | 4.0 ± 1.1 | 71 | 70 | 5.7 |
| Glucose | 43 | 19.3 ± 9.2 | 98 | 100 | 8.5 | |||
| ftsL::TnphoA | JOE220 | Arabinose | 65 | 6.5 ± 3.0 | 55 | 80 | 7.7 | |
| Glucose | 34 | 17.2 ± 8.8 | 53 | 94 | 11.1 |
Sum of cell lengths divided by total number of rings.
To determine whether FtsQ localization depends on FtsI, we transduced an ftsI(Ts) allele into the gfp-ftsQ merodiploid strain and analyzed the cells before and after the temperature shift. Bands of fluorescence were observed in cells sampled at 30 or 42°C, and the frequencies at which these bands appeared were approximately the same at both temperatures (Table 3). This result suggests that FtsQ does not require functional FtsI for localization. To confirm this result, we introduced gfp-ftsQ into an FtsI depletion strain, which was similar to the FtsQ depletion strain in that it was dependent on arabinose for division; the only intact ftsI gene in this depletion strain was under the control of an arabinose-regulated promoter. Parallel cultures of this strain were grown in rich media containing glucose or arabinose and then fixed for examination. Cell morphology appeared normal in the arabinose culture, and GFP-FtsQ localization was readily observed (Fig. 3C). Discrete bands of fluorescence were also detected in FtsI-depleted filaments in the glucose culture (Fig. 3C). Average spacings between FtsQ rings were about the same in cells grown with arabinose or glucose (Table 3).
Finally, to determine whether FtsQ requires FtsL for septal localization, we transduced gfp-ftsQ into an FtsL depletion strain. The localization results were similar to those obtained with the FtsI depletion strain: bands of fluorescence were readily detected in cells grown with arabinose as well as in filaments from the glucose culture (Fig. 3D), and the average spacing between these bands were similar between the two cultures (Table 3). We concluded that FtsQ does not depend on FtsL for localization to the developing septum.
ZipA-GFP localization in an ftsQ(Ts) mutant.
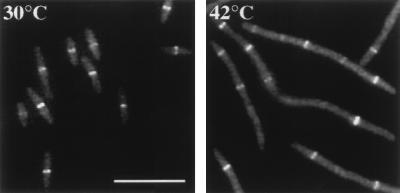
Our localization results suggested that FtsQ is recruited to the division site after FtsZ and FtsA but before FtsL and FtsI. Since ZipA interacts directly with FtsZ and also localizes to the division site (18), we wanted to shed some light on the relative positions of ZipA and FtsQ in this recruitment pathway. To examine the dependence of ZipA localization on FtsQ, we used a merodiploid strain carrying a single copy of zipA-gfp at the att site. We transduced an ftsQ(Ts) allele into the merodiploid strain and conducted temperature shift experiments as before. Fluorescent bands of ZipA-GFP appeared with similar average spacings in cells grown at 30°C and in filaments grown at 42°C (Fig. 4 and Table 4). Thus, ZipA can be recruited to the division site without functional FtsQ.
FIG. 4.
Localization of ZipA-GFP in an ftsQ1(Ts) mutant. (Left) GFP image of JOE165 cells grown at 30°C. (Right) GFP image of JOE165 cells grown at 42°C for 45 min. Cells were fixed for microscopy as described in Materials and Methods. Bar, 10 μm.
TABLE 4.
Localization of ZipA-GFP in wild-type and ftsQ(Ts) backgrounds
| Strain background | Strain | Growth temp (°C) | No. of cells | Mean ± SD cell length (μm) | Total no. of rings | % of cells with a ring(s) | Avg spacing between rings (μm)a |
|---|---|---|---|---|---|---|---|
| Wild type | EC489 | 30 | 102 | 3.2 ± 0.8 | 63 | 62 | 5.3 |
| 42 | 139 | 2.8 ± 0.6 | 99 | 71 | 3.9 | ||
| ftsQ1(Ts) | JOE165 | 30 | 127 | 5.6 ± 1.5 | 104 | 82 | 6.9 |
| 42 | 34 | 25.4 ± 7.7 | 118 | 100 | 7.3 |
Sum of cell lengths divided by total number of rings.
DISCUSSION
In this study, we found that GFP-FtsQ localizes to the division site. Its cytoplasmic and transmembrane domains are not essential for localization, while its periplasmic domain is. In addition, we showed that GFP-FtsQ depends on the cytoplasmic proteins FtsZ and FtsA but not the membrane proteins FtsL and FtsI for septal recruitment. ZipA-GFP does not require FtsQ for localization. Together with results from a parallel study indicating that FtsL also localizes to the division site (14), our findings indicate that all known, essential cell division proteins in E. coli are recruited to the developing septum. Therefore, a criterion for defining a gene product as a direct participant in cytokinesis should be septal localization.
Like other cell division proteins, FtsQ probably localizes as a ring structure. The fluorescent band that we observed is most likely a side view of such a ring. Since FtsQ is a membrane protein, fluorescence should accumulate at the membrane and not in the cytoplasm. Similar to that of other Fts proteins, the septal localization of FtsQ appears to be a timed event. Fluorescent bands tend to be observed in longer, older cells, while shorter, newly born cells generally lack FtsQ at the mid-cell region. Furthermore, once it has been recruited to the division site, FtsQ stays with the invaginating septum until cytokinesis is complete. Thus, FtsQ function in cell division appears to be controlled, at least in part, by timed localization rather than uniform distribution and specific activation at the division site.
Although complementation results indicated that GFP-FtsQ functions as well as wild-type FtsQ, this observation must be treated with caution because Western blotting analysis revealed the presence of GFP-FtsQ breakdown products, one or more of which may be responsible for complementation. This issue, which is often ignored, requires attention whenever complementation by GFP fusions is tested. However, we doubt that the presence of these breakdown products affected our localization conclusions since, when polyclonal anti-GFP antibody was used for immunoblotting, only a single band, that of full-length GFP-FtsQ, appeared, suggesting that GFP-FtsQ produces the fluorescent signal. Even if one of the breakdown products contributes to the localization signal, the domain that allows targeting to the developing septum is still derived from FtsQ.
Studies with other cell division proteins, such as FtsN and FtsK, have indicated that portions of those proteins are sufficient for localization (2, 44). Here, we tested whether the cytoplasmic, transmembrane, or periplasmic domain of FtsQ is required for septal recruitment. We found that the membrane anchor, but not the periplasmic domain, can be swapped without a drastic effect, suggesting that the periplasmic domain is necessary and sufficient for localization. Nevertheless, because these localization experiments were conducted with merodiploid strains, the swap constructs could have interacted with wild-type FtsQ to localize to the division site. To determine whether GFP-FtsQ swap proteins can function by themselves, we assessed their abilities to complement mutations in ftsQ. In agreement with previous results (9, 17), all swap fusions with changes in the membrane anchor complemented the temperature-sensitive mutation. However, our complementation results with the null mutation diverged from those of a previous study (17). All the fusions tested were able to complement the null mutation, but GFP-FFQ and GFP-QFQ complemented less well, suggesting that the transmembrane domain contributes to FtsQ activity. On the other hand, Guzman et al. (17) reported that the cytoplasmic but not the transmembrane domain is required for complementation. The basis of the discrepancy is not known. There are several differences between the complementation tests: distinct techniques were used, expression levels were different, and GFP fusions were analyzed in this study instead of constructs without GFP tags. Moreover, the presence of breakdown products in cells expressing GFP fusions needs to be taken into account when one is considering complementation results. While our results are complex, they indicate that the ftsQ1(Ts) allele is leaky and suggest that only the periplasmic domain of FtsQ is required for function. In support of this conclusion about the periplasmic domain of FtsQ, we found that all of the membrane anchor swap fusions were able to localize to the division site without wild-type FtsQ.
If the periplasmic domain of FtsQ is sufficient for localization, it presumably interacts with factors outside the cytoplasm in order to be recruited to the division site. The most likely candidates for recruiting FtsQ are other cell division proteins. Interestingly, the Fts proteins appear to localize to the developing septum in a linear pathway, and FtsQ seems to be the link between proteins that are predominantly cytoplasmic (FtsZ, FtsA, and ZipA) and proteins that are mainly periplasmic (FtsL, FtsI, and FtsN). Results from this and other studies suggest that FtsZ is the first to arrive at the division site, followed by FtsA, FtsQ, FtsL, FtsI, and FtsN (1–3, 14, 24, 31, 39, 41, 42). So far, there is no evidence for parallel recruitment, in which two proteins can localize without each other, or codependency, in which both proteins fail to localize when one is absent. The position of ZipA in this localization pathway is only partly known, but our results indicate that it acts upstream of FtsQ.
While FtsZ, FtsA, and ZipA all appear to act upstream of FtsQ, none of them is likely to recruit FtsQ by a direct physical interaction. FtsZ and FtsA are both cytoplasmic proteins (23), and ZipA is predicted to be an inner membrane protein without a periplasmic domain (18). FtsQ apparently does not need its membrane anchor for localization. We consider two possible explanations for the mechanism of FtsQ localization: (i) one or more factors act between FtsQ and the other three proteins, or (ii) FtsZ, FtsA, and ZipA alter the inner membrane in a way that allows FtsQ to localize. Whether FtsW, FtsK, or both act upstream of FtsQ and facilitate its localization remains a subject of investigation.
Microscopy studies demonstrating that all known cell division proteins in E. coli localize to the division site lend support to the existence of a protein complex that constructs the developing septum. The recruitment pathway may represent the order in which the cell division proteins assemble into the putative complex. However, caution must be taken in interpreting the localization results. A multimeric complex may not exist at all; instead, the cell division proteins may behave like enzymes in a metabolic pathway, in which an enzyme acts on the product of the previous enzymatic reaction. The cell division proteins may act on distinct septal structures, such as different peptidoglycan and lipid substrates, that form sequentially, and the sequence of formation may be the cause of the linear recruitment pathway. The postulated assembly of a cell division protein complex awaits direct experimental demonstration.
ACKNOWLEDGMENTS
This work was supported by grants from the American Cancer Society and the National Institutes of Health (GM 38922). J.B. is an American Cancer Society Research Professor. J.C.C. was supported by a National Science Foundation predoctoral fellowship. D.S.W. was a DOE Energy Biosciences Fellow of the Life Sciences Research Foundation. J.-M.G. was supported by the Institut Pasteur, Paris, France.
We thank members of the Beckwith laboratory for their general assistance and helpful discussions.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 3.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. Unpublished data.
- 6.Carson M J, Barondess J, Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178:1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer P A J, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 11.Din N, Quardokus E M, Sackett M J, Brun Y V. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol Microbiol. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 12.Erickson H P. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- 13.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghigo, J.-M., D. S. Weiss, J. C. Chen, J. C. Yarrow, and J. Beckwith. Mol. Microbiol., in press. [DOI] [PubMed]
- 15.Guzman L M, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman L M, Weiss D S, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 19.Harry E J, Wake R G. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- 20.Levin P A, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 21.Lowe J, Amos L A. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 22.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 23.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1615–1626. [Google Scholar]
- 24.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Sun Q, Wang R, Singh G, Jonietz E L, Margolin W. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179:6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 27.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen-Disteche M, Fraipont C, Buddelmeijer N, Nanninga N. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol Life Sci. 1998;54:309–316. doi: 10.1007/s000180050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Novagen, Inc. pET system manual. 6th ed. Madison, Wis: Novagen, Inc.; 1995. [Google Scholar]
- 31.Pogliano J, Pogliano K, Weiss D S, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 33.RayChaudhuri D, Park J T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 34.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Spratt B G, Cromie K D. Penicillin-binding proteins of gram-negative bacteria. Rev Infect Dis. 1988;10:699–711. doi: 10.1093/clinids/10.4.699. [DOI] [PubMed] [Google Scholar]
- 37.Strauch K L, Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci USA. 1988;85:1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss, D. S., J. C. Chen, J.-M. Ghigo, D. Boyd, and J. Beckwith. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z-ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508–520. [DOI] [PMC free article] [PubMed]
- 42.Weiss D S, Pogliano K, Carson M, Guzman L M, Fraipont C, Nguyen D M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein Fts1 (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 43.Yu X C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]