Abstract
The outbreak of novel coronavirus (COVID-19) disease has infected more than 135.6 million people globally. For its early diagnosis, researchers consider chest X-ray examinations as a standard screening technique in addition to RT-PCR test. Majority of research work till date focused only on application of deep learning approaches that is relevant but lacking in better pre-processing of CXR images. Towards this direction, this study aims to explore cumulative effects of image denoising and enhancement approaches on the performance of deep learning approaches. Regarding pre-processing, suitable methods for X-ray images, Histogram equalization, CLAHE and gamma correction have been tested individually and along with adaptive median filter, median filter, total variation filter and gaussian denoising filters. Proposed study compared eleven combinations in exploration of most coherent approach in greedy manner. For more robust analysis, we compared ten CNN architectures for performance evaluation with and without enhancement approaches. These models are InceptionV3, InceptionResNetV2, MobileNet, MobileNetV2, Vgg19, NASNetMobile, ResNet101, DenseNet121, DenseNet169, DenseNet201. These models are trained in 4-way (COVID-19 pneumonia vs Viral vs Bacterial pneumonia vs Normal) and 3-way classification scenario (COVID-19 vs Pneumonia vs Normal) on two benchmark datasets. The proposed methodology determines with TVF + Gamma, models achieve higher classification accuracy and sensitivity. In 4-way classification MobileNet with TVF + Gamma achieves top accuracy of 93.25% with 1.91% improvement in accuracy score, COVID-19 sensitivity of 98.72% and F1-score of 92.14%. In 3-way classification our DenseNet201 with TVF + Gamma gains accuracy of 91.10% with improvement of 1.47%, COVID-19 sensitivity of 100% and F1-score of 91.09%. Proposed study concludes that deep learning modes with gamma correction and TVF + Gamma has superior performance compared to state-of-the-art models. This not only minimizes overlapping between COVID-19 and virus pneumonia but advantageous in time required to converge best possible results.
Keywords: Deep learning, Chest X-ray, COVID-19 analysis, Image enhancement, Image denoising, Pneumonia classification
Introduction
Coronavirus disease 2019 (COVID-19) [49], has been declared a global epidemic by WHO within fewer than four months when 3.3 million confirmed and 238,000 deaths were reported as of 2nd May 2020. COVID-19 disease was instigated by SARS CoV-2 and till 12 April, 2021, WHO reports depict, there have been 135,646,617confirmed cases with 2,930,732 confirmed deaths globally [72]. Due to absence of adequate knowledge related disease and its enormously contagious behaviour, it is extremely important to stop its spreading and to explore diverse methodologies that can help in the early classification of COVID-19. Globally, many researchers of Artificial Intelligence, medicine, clinical study and others have been trying to find out different methods for classification that helps to prevent spreading of COVID-19 virus and any such epidemic in future. Preliminary testing approach RT-PCR (“Reverse-transcriptase polymerase chain reaction”) shows less sensitivity [69] for COVID-19 but on the other hand radiological examinations were found very helpful in diagnosis and assessment of disease progression. Most scientific studies worldwide revealed that symptoms of COVID-19 can be clearly seen in CXR and CT images of lungs [5]. CT (Chest-computed tomography) is most popular and effective technique for lung related infection, but it is expensive. Screening of CT images for COVID-19 diagnosis shows higher sensitivity [34] compared to preliminary testing procedure RT-PCR [29, 69]. But, due to unpredicted increase in COVID-19 prevalence it is tough to make regular use of CT because of its portability and expensiveness. Thus, CXR images are usually preferred to detect infection attacks of COVID-19. Earlier CXR images were commonly used to diagnose pneumonia, abscesses, Tuberculosis [7], lung inammation and enlarged lymph nodes [26]. Radiological studies presented that due to similarity in pneumonia and COVID-19 [5] virus, most patients of COVID-19 were diagnosed with pneumonia. So, it is important to develop techniques that can easily distinguish between viral pneumonia, COVID-19 pneumonia and bacterial pneumonia [1] as crucial for future preventive measure against such deadly epidemic.
To make healthcare systems more proficient to deal with such epidemic situations, Biomedical image analysis is a significant research field. Recent radiological studies depicted certain abnormalities [5] in CXR of patients diagnosed COVID-19 and pneumonia. Even CXR analysis can help in epidemic situation to determine the patients having high risk to give priority treatment. Accordingly, many deep learning models have been used by researchers that help in classification of CXR [3, 6, 9, 15, 58] and CT images [51, 74, 75] better than radiologists. These models have very powerful feature extraction and learning capabilities for numerous image processing tasks. However, analysis of CT images and video endoscopy [75] have been successfully achieved by convolutional neural networks more efficiently. Various comparative studies amongst radiologists and deep learning models have been proposed where DL models performed exceptionally well in image analysis tasks [56, 70]. Numerous convolutional neural network models have been proposed and improved time to time that helps in extraction of more useful features to analyse large [31, 36, 41, 60] volume of images. VGG16 model was proposed by Wang et al. [75] particularly for diagnosis of various types of pneumonia in lung regions. Dense CNN based [56] 121 layers model, proposed by Rajpurkar et al. to differentiate pneumonia amongst other pathology classes using CXR images.
CheXNet is very popular model proposed using 121 layered CNN architecture trained with publicly available ChestX-ray14 dataset [55] containing more than 100,000 X-ray frontal-view images. The performance of the model is compared to radiologists. This study claims that performance of CheXNet model is superior to that of radiologist’s results based on F-measure metric. CheXpert [26] is another common study on chest x-ray images driven by deep learning approaches to gain expert label performance. This study practices on dataset of 224,316 chest x-ray radiographs obtained from 65,240 patients to distinguish the presence of 14 annotations in radiological reports. The results obtained on test set were also evaluated by three radiologists. Deep learning models outperformed the radiologists in diagnosis of several pathologies like edema, cardiomegaly, and pleural effusion etc. in CXR images [26, 29]. Since, deep learning approaches have been used on various labelled datasets to gain expert-level performance on various types of medical images. Initially only COVID-Net was proposed deep learning model by Wong and Wang [71] for diagnosis of COVID-19 cases in CXR with 80% sensitivity. Various deep CNN models including AlexNet, VGG19, Inception, ResNet50, MobileNetV2, InceptionResNetV2, VGG16, GoogleNet, MobileNet, DenseNet121 and many more have been used for drawing conclusions from CXR images.
Although, early achievements of deep learning approaches in finding certain irregularities successfully in CXR images [30] motivated us to examine more about deep CNN architectures for feasibility analysis of COVID-19 [55] among other pathology classes. In such epidemic situations, even a small contribution may assist a lot to handle such data in future. Table 1, represents the abbreviations used in this study. Therefore, primary aim and major contributions of this study is explained as follows:
In the best of author’s knowledge this is first study that cumulatively analysed the effects of image denoising and image enhancement techniques on benchmark datasets used in majority of research studies.
To find best image pre-processing pipeline using image denoising and image enhancement techniques that improve performance and interpretation of the deep learning models.
To find a novel convolutional deep neural network architecture appropriate for finding COVID-19 cases using CXR images with higher COVID-19 sensitivity.
To study the effect of image enhancement techniques on sensitivity of individual classes and confidence score based on different threshold values.
To analyse the effect of image enhancement approaches on interpretation of various models by building visualization attention maps by gradient weighted class activation mapping.
Table 1.
Abbreviations
| Abbreviations | Full form |
|---|---|
| HE | Histogram equalization |
| CLAHE | Contrast limited adaptive histogram equalization |
| BCET | Balance contrast enhancement technique |
| AMF | Adaptive median filter |
| TVF | Total variation filter |
| CAM | Class activation mapping |
| Grad-CAM | Gradient weighted class activation mapping |
The whole structure of this article is defined as; In Section 2, we discuss critical review of existing methodologies for COVID-19 diagnosis. Section 3 discuss about making of dataset by fusion of different data resources. Section 4 describes proposed methodology to achieve desirable task followed by class imbalance, classification process and evaluation metrics. In Section 5 we discuss results obtained by proposed study. Finally, Section 5 ends with a conclusion.
Literature review
With progression in machine intelligence and deep learning approaches in the field of healthcare assist radiologists in more accurate diagnosis of disease in early stage. In last decade, deep learning approaches has been widely applicable in diagnosing respiratory diseases using CXR images. Motivated by early achievements of radiological studies successfully, we explore deep learning approaches for classification of COVID-19 infected via CXR images [9, 51, 55, 58]. For instance, research study proposed in [15], author proposed deep learning approaches with multi objective optimization technique for diagnosis of COVID-19 patients. Decision tree based J48 approach efficiently identify COVID-19 features on CXR images. This study performs comparative analysis of 11 CNN models including DenseNet, ResNet, VGG16, AlexNet and InceptionV3. ResNet101 outperforms others with overall accuracy and sensitivity of 100% with hyena optimizer for parameter optimization. In study [73], author proposed anomaly detection method for finding COVID-19 by 100 COVID samples and 1431 normal samples. This study involves 70 and 1008 COVID-19 and control participants. The obtained results gain sensitivity of 96 and 76% for COVID-19 and control examples with EfficientNet as baseline model. Weak pre-processing and class imbalance are series limitations of study. Another study [12] uses pretrained transfer learning VGG16 model on balanced dataset for COVID-19 classification against normal and pneumonia CXR samples. This Study containing corpus of 132 COVID-19, pneumonia and controls samples each. Obtained results gain 100% sensitivity for diagnosing COVID-19.
In [24], author used AlexNet model to differentiate COVID-19 from control, viral and bacterial pneumonia cases. This Study considers 2 way, 3-way and 4-way classification scenarios. In 2-way classification strategy comparison of normal vs COVID-19, bacterial pneumonia vs COVID-19, normal vs bacterial pneumonia, normal vs viral pneumonia and in 3-way (COVID-19 vs bacterial pneumonia vs normal) are compared whereas in 4-way classification scenario all classes are compared to diagnose COVID-19. Proposed model achieved an accuracy score 91.30% and 89.18% sensitivity in 4-way classification strategy. In another study [14], author pick Xception model as baseline architecture for finding COVID-19. Transfer learning model applied on of 500 pneumonia, 500 normal, 127 COVID-19 samples. Proposed model gains an accuracy score 97% for COVID-19 detection. In [22], author experimented using seven CNN models with limited imbalanced training dataset comprises of 25 COVID-19 and 50 normal samples. Pretrained models VGG19 and DenseNet121 achieved best score with F1-measure 0.91 and 0.89 for covid-19 and normal CXR samples. In Another study [33], Xception architecture is used as baseline for classification by using two multiclass datasets. First task contains 4 classes: i) COVID-19 vs. controls vs. bacterial pneumonia vs viral pneumonia; ii) Second task includes COVID-19 vs normal vs. pneumonia. Also, under-sampling is used to deal with class imbalance. This study contains corpus of 310 Normal, 290 COVID, 330 and 327 bacterial and viral pneumonia CXR samples. Moreover, model gain accuracy of 89% and 94% for 4-class and 3-class respectively.
In article [27], hybrid of CNN and LSTM is applied for COVID-19 detection purpose. CNN is used as feature extractor whereas LSTM plays a vital role for classification purpose. This experimental study contains 4575 CXR images and three classes (COVID-19, normal and Pneumonia). Study uses balanced dataset with each class contributes 1525 samples whereas COVID-19 class contains 912 augmented examples. Hybrid model gain an accuracy score 99% by using 5-fold cross validation. In [40], four CNN models i.e., ResNet50, DenseNet-121, SqueezeNet, and ResNet18 were used to enable transfer learning. Experimental analysis involves 5000 no-finding, pneumonia samples and 184 COVID-19. COVID-19 images are collected using COVID-19 chest-Xray repository. Results indicate 98% sensitivity and 93% specificity. In [52], again five CNN models i.e., ResNet, Inception, NASNetLarge, DenseNet169, ResNet-v2 and InceptionV3 as baseline architectures. Study collects subset of samples from RSNA dataset and compares COVID-19 with normal and pneumonia cases. Class imbalance is resolved using resampling and entropy-based approach. Study depicts NesNetLarge gain an accuracy score of 98% and 96% and sensitivity of 90 and 91% in 2-way and 3-way classification problem. Due to limited availability of COVID-19 examples models may lead to biased results because of class imbalance. So, another approach was used in [68] for generating identical images for better training purposes. With support of GAN (generative adversarial network) artificial examples of CXR images has been generated. This approach resulted in corpus of 1124 control and 403 COVID-19 images. By data augmentation technique, author claimed 10% improvement in accuracy from 85% to 95% via VGG16 as backbone.
Another, comparative study [50] for diagnosis of COVID-19 in 2-class and multiclass problem used similar dataset used in [14] but compares among COVID-19, normal and COVID-19, pneumonia and normal cases. In this study DarkNet model is modified, used as baseline and trained using cross validation technique with K = 5. It gains of 98% and 87% accuracy in 2-way and 3-way classification. In another approach [67], CapsNet (capsule networks) model employed for 3-way classification of COVID-19 vs. normal vs pneumonia and binary classification (normal vs COVID-19). Experimental analysis contains corpus of pneumonia (1050), COVID (231), and normal (1050) CXR images. To reduce class imbalance augmentation operations like rotation and shifting are used to increase COVID images to 1050. Results depict an accuracy score of 97% and 84% for binary and multiclass classification strategy. In [5], similar transfer learning technique applied using five baseline CNN models – MobileNetV2, InceptionResNetV2, InceptionV3, VGG19, Xception to distinguish COVID-19 from pneumonia and control examples. Study was carried out in two stages: one containing COVID (224), control (504) and bacterial pneumonia (700) images and in stage 2 same COVID and normal data used but contained with 714 new viral and bacterial pneumonia samples. MobileNetV2 achieves top result with 94% and 96% accuracy score in 3 and 2-class classification scenario.
Another model is proposed named CovXNet [38] for detection of COVID-19 infection against other classes by using depth wise CNN model. In phase 1 model is trained using bacterial, viral pneumonia and control images. Then, transfer learning is used to obtain new model for COVID-19 training. Study was carried out using two datasets to compare normal vs COVID-19 cases and COVID-19 vs normal vs viral and bacterial images. CovXNet gains 90% and 97% accuracy score in 4 and 2-class classification scenario. In study [37], GAN architecture was implemented to increase the size of dataset using augmentation technique. Dataset collected for study comprises 307 images having four different classes: COVID-19, normal, viral and bacterial pneumonia. AlexNet, Restnet18, GoogleNet were the CNN baseline models used for classification. GoogleNet model gains best accuracy of 99%. In article [49], patch-wise processing strategy was introduced to handle limited training data using. Similar, transfer learning based ResNet-18 was employed for classification network and for segmentation purpose FCDenseNet model is used. The dataset collected under study includes 180 COVID-19, bacterial (54) and viral pneumonia (20), Normal (191) and Tuberculosis (57) cases. This patch-based study gain accuracy of 89%. Especially wong and wang et al. [71] in early studies introduced a deep learning COVID-Net architecture. Experiments performed on benchmark dataset contains corpus of 13,975 CXR images having pneumonia, control and only 266 COVID examples. Proposed COVID-Net architecture gains improved sensitivity and accuracy score of 91% and 93.3% for COVID cases in comparison to ResNet50 and VGG19. Another popular COVID-19 detector COVID-AID [39] based on DenseNet model having 121 layers experimented on covid-chestxray-dataset for [13] COVID cases. It detects COVID infection in CXR images with 100% sensitivity score among viral, bacterial pneumonia and control cases.
In study [6], author performed three individual experiments to differentiate COVID-19 samples from pneumonia and control pathology classes. These experiments were performed on whole slides, cropped images corresponding to lungs and on lung area segmented using U-Net. The study includes more than 8573 images of COVID including augmented samples collected from different repositories. The model achieves top accuracy score of 91.75%. In article [1], DeTrac model (decompose, transfer and compose) grounded on shallow and deep transfer learning i.e., GoogLeNet, SqueezeNet, VGG19, ResNet and AlexNet to differentiate COVID-19 (105) against SARS (11) and normal (80) cases. DeTrac gain an accuracy score of 93.1% for COVID-19 diagnosis. In [53], InceptionResNetV2, Xception, AlexNet and DenseNet-201, were baseline architectures studied to categorise COVID-19 against normal and pneumonia cases. DenseNet-201 claimed to be the best baseline model that classify COVID-19 with accuracy, specificity and sensitivity score as Acc = 98.16%, Sp = 98.77% and Se = 98.93% respectively. In study [21], author aim to classify CXR images of COVID-19, pneumonia and normal patients using Optimized CNN. Optimization of hyperparameters is performed using Greywolf Optimizer algorithm (GWO). The proposed approach contains a total of 2700 samples including 900 COVID -19 generated with augmentation. It achieves best accuracy and sensitivity of 97.78% and 97.75%. In another study [48], author used shallow-CNN for outbreak screening of COVID-19. This model achieved top accuracy score of 99.69%, sensitivity and AUC score of 100% and 99.95% respectively. In another study [16], author used Meta heuristics approach combined with CNN. This study uses ResNet-50 for features extraction and ASSOA (Advanced Squirrel Search Optimization Algorithm) used for selection of better features and finally compared with Genetic Algorithm and GreyWolf Optimizer. Experiments were performed on 5863 samples collected form Kaggle containing pathology classes COVID, control, viral and bacterial pneumonia. It achieves best mean accuracy score of 99.26%.
Specifically, to resolve the issue of COVID-19 data collection, another model developed named MAG-SD [35] for automatic classification of COIVD-19 from pneumonia cases. In this, more relevant features vector is extracted by multiscale attention guided network. It is advantageous because of soft distance regularization, generation of attention guided synthetic data, attention pooling and CLAHE. Proposed model is tested on three dissimilar datasets comprised of 2, 3 and 4 classes. In addition, proposed model is superior to VGG16, ResNet, InceptionV3 and COVID-Net model. Because of data collection from different repositories, there exists certain uncertainty at level of diagnosis. A semi-supervised uncertainty estimation framework [8] was introduced for improving uncertainty of COVID-19 diagnosis using unlabelled data. Various popular approaches like deterministic uncertainty quantification, Monte-Carlo dropout, Softmax scores have been utilized for uncertainty estimation. For reliability comparison of these estimation approaches Jensen-Shannon distance function is used. However, Monte Carlo dropout algorithm gives better results compared to others. Another model named MSRCovXNet [19] was proposed for purpose of efficient feature extraction form limited CXR data. This multi-stage residual network model extracts initial features based on ResNet-18 model and perform feature optimization with two FEM modules i.e., high level and low-level feature maps. Advanced single and multi-stage proposed feature fusion approach enhances the representation of high and low-level features. MSRCovXNet achieves recall and precision score of 94% and 98.9%. This model is compared with COVID-Net and ResNet models. However, it uses only 100 samples from each class for testing. Majority of proposed DL models suffer from generalization errors, high variance and overfitting caused limited corpus of data samples. EDL-COVID, an ensemble model [66] was proposed to combine prediction ability of multiple COVID-Net snapshot models. It claims to improve generalization, interpretation and performance measures. It outperforms accuracy of COVID-Net model i.e., 93.5% on similar COVIDx dataset with improved accuracy of 95% and COVID-19 sensitivity of 94.1%.
In Table 2, we listed the summary of contributions added to the literature with features as reference of article, deep learning models used followed by number of classes, number of samples of each class, performance metrics in terms of overall accuracy (Acc.), COVID-19 sensitivity (Sencov), other metrics including F1-score, precision (PPV), specificity and image enhancement technique applied if any. Interpretation from the related work concludes that transfer learning models found very useful in identification of COVID-19 even with limited availability of training COVID-19 images among other pathology classes.
Table 2.
Proposed contributions of related work
| Ref. | CNN Architecture | # Of Classes | # Of samples | Performance metrics (%) | Image Enh. (IE) | ||||
|---|---|---|---|---|---|---|---|---|---|
| COVID | Normal | Other | Acc. | Sencov | Other | ||||
| [68] | VGG16 | 2 | 403 | 1124 | – | 95 | 90 | PPV - 95 | ☒ |
| [22] | DenseNet121, VGG19, InceptionV3 | 2 | 25 | 25 | – | 90 | – | F1cov - 91 | ☒ |
| [73] | EfficientNet | 2 | 100 | 1431 | – | – | 96 | Sp - 70 | ☒ |
| [21] | CNN + GWO | 3 | 900 | 900 | 900 | 97.78 | 98.5 | PPV - 92.8 | HE |
| [12] | VGG16 | 3 | 132 | 132 | 132 | 85 | 100 | F1–85 | ☒ |
| [27] | CNN, LSTM | 3 | 1525 | 1525 | 1525 | – | 99.3 | F1cov - 98.9 | ☒ |
| [14] | Xception | 3 | 127 | 500 | 500 | – | 97 | F1cov - 96.9 | ☒ |
| [33] | Xception (CoroNet) | 3 | 290 | 310 | 657 | 90.21 | 89 | F1–91 | ☒ |
| [5] | MobileNetV2, Inception InceptionResNetV2, VGG19 | 3 | 224 | 504 | 700 | 92.85 | 98.66 | Spcov − 96.46 | ☒ |
| [50] | DarkNet | 3 | 127 | 500 | 500 | 87.02 | 97.9 | F1–87.37 | ☒ |
| [67] | CapsNet | 3 | 231 | 1050 | 1050 | 84 | 94.57 | F1–84.21 | ☒ |
| [40] | DenseNet-121, ResNet50, ResNet18, SqueezeNet | 3 | 184 | 2400 | 2600 | – | 98 | Spcov - 93 | ☒ |
| [52] | DenseNet169, Inception ResNet, NASNetLarge | 3 | 108 | 533 | 515 | 94 | – |
F1 = 90 Sp - 89 |
TVF |
| [1] | VGG19, ResNet, AlexNet, GoogLeNet and squeezeNet | 3 | 105 | 80 | 11 | 93.1 | 100 | Sp - 85.1 | ☒ |
| [15] | J48 + 11 CNN models | 2 | 50 | 50 | – | 100 | 100 | F1–100 | ☒ |
| [37] | Restnet18, GoogleNet and AlexNet | 4 | 69 | 79 | 158 | 80.6 | 100 | F1–82.32 | ☒ |
| [38] | CovXNet | 4 | 305 | 305 | 610 | 90.2 | 95 | F1–90.4 | ☒ |
| [49] | ResNet18 | 4 | 180 | 191 | 131 | 88.9 | 92.5 | F1–84.4 | HE+G |
| [71] | VGG19, ResNet50, COVID-NET | 3 | 268 | 8066 | 5538 | 93.3 | 91 | PPVcov-98.9 | ☒ |
| [39] | DenseNet121(COVID-AID) | 4 | 155 | 1583 | 4273 | 90.5 | 100 | F1–92.30 | ☒ |
| [53] | AlexNet, DenseNet201, InceptionResNetV2, Xception | 3 | 683 | 2924 | 4272 | – | 98.93 | Sp - 98.77 | ☒ |
| [24] | AlexNet | 4 | 371 | 2017 | – | 93.42 | 89.18 | Sp - 98.9 | ☒ |
| [48] | Shallow-CNN | 3 | 321 | 1583 | 4273 | – | 100 | PPVcov - 99.38 | ☒ |
| [16] | ASSOA | 4 | – | 1227 | 3445 | 99.23 | – | – | ☒ |
| [35] | MAG-SD, InceptionV3, VGG16, ResNet50 | 3 | 462 | 1602 | 1567 | 95.85 | 95 | F1–95.54 | CLAHE |
| 4 | 462 | 1602 | 4265 | 87.12 | 91 | F1–86.98 | |||
| [8] | WideResNet, MinMax UE | 4 | 99 | 1587 | 4273 | – | – | – | ☒ |
| [19] | MSRCovXNet | 3 | 386 | 8066 | 5551 | 95 | 94 | F1–95.33 | ☒ |
| [66] | EDL-COVID | 3 | 573 | 8851 | 6053 | 95 | 94.1 | – | ☒ |
*Bold represents best model; IE – Image enhancement, HE – histogram equalization, TVF - Total variation filter and G – gamma correction, UE- uncertainty estimation
Existing methodologies and results analysis
In this Analysis, we reviewed quality literature from IEEE, Springer and Elsevier databases concerning deep learning for COVID-19 application. Almost thirty papers have been considered in review process regarding diagnosis of COVID-19 from CXR images. Majority of the publications concerning deep learning networks, including VGG [1, 5, 12, 15, 22, 35, 71], NASNetLarge [52], AlexNet [1, 15, 24, 37, 53], DenseNet [15, 16, 22, 39, 40, 53], MobileNet [5, 22], Inception [1, 5, 15, 22, 35, 37, 52, 53], Xception [5, 14, 22, 33, 53], EfficientNet [73], ResNet [1, 15, 16, 22, 35, 37, 40, 49, 52, 71] and with four involving custom CNN [21, 38, 67, 71] architectures. Most of research studies involves two classification scenarios. First scenario, CXR images are classified into one of these, that is COVID-19, normal and pneumonia whereas second involves more specific classification of pneumonia class into viral and bacterial pneumonia in addition to COVID and normal. DenseNet, MobileNet and ResNet models presented better results compared to other DL models with accuracies ranging from 88% to 99%. Similarly, COVID-19 sensitivity is maximum with DenseNet models and ranges from 90% to 100%. However, DenseNet model extract more relevant features and has better interpretation because of more depth and variation in connections as compared to other models. Major drawbacks of majority of studies lies regarding better pre-processing of images, class imbalance, size of dataset, and interpretation of CXR images. Also, some studies balance these classes by generating synthetic data using GAN [37, 68] that further improves performance of models. Usage of advanced meta-heuristic [16] approaches for relevant feature selection also improves performance and confidence score of models. ASSOA optimization approach improves feature selection process compared to grey wolf optimizer (GWO), genetic approach and ant colony algorithm. In addition, hyperparameters selection [21] with GWO improves performance of models. Regarding advanced image enhancement only [12, 35, 49, 52] have randomly selected one of these; Histogram equalization, Total variation filter, CLAHE and gamma correction. Also, traditional machine learning approaches have been applicable on features extracted with VGG, ResNet50 and DenseNet but less effective compared to classification with deep learning approaches.
Two-way classification process
In Fig. 1 we have shown the block diagram showing two ways for analysis of CXR and other sequences of steps required to follow for better classification and diagnosis of COVID-19. In first way, whole CXR image can be considered for analysis whereas in second way image segmentation is performed firstly to extract relevant lung regions for CXR analysis. To achieve segmentation task other annotated datasets are used containing corresponding output mask associated with each input image for pixel level classification by training various segmentation CNN models. These models are basic UNet, UNet++, UNet with ResNet block, UNet with DenseNet block and other variants of UNet model. Once the segmentation models are trained, then these models can be used to extract lung regions of existing dataset taken into consideration for diagnosis of COVID-19 pathology. After automated annotation of lung regions from CXR images both paths follow similar sequence of steps. Initially universal pre-processing operations like resizing, normalization and formatting is performed, followed by advanced image pre-processing operations like image denoising, image enhancement. After this, pre-processed images are fed into deep learning approaches for automatic feature extraction and classification of CXR images into one of pathology class. Finally, to make results interpretable, visualization of features learned by model can be seen by feature visualization approaches like CAM, Grad-CAM and Grad-CAM++.
Fig. 1.
Block diagram of two ways and sequence of basic elements for analysis of COVID-19 among other pathology classes
Datasets considered
Most of the studies performed for diagnosis of COVID-19 collects images from public datasets and make dataset by fusion of 3-classes or 4-classes. In 3-way classification these classes are COVID-19, normal and pneumonia whereas normal and pneumonia samples are taken form RSNA pneumonia detection challenge [65] and covid-samples collected from COVID-19 chest Xray-dataset [13]. In 4-way classification pneumonia class is further separated as virus and bacterial pneumonia taken from chest-Xray-pneumonia dataset [46]. Similarly, for the purpose of segmentation the available benchmark datasets are JSRT dataset [62], Montgomery and Shenzhen dataset [28].
Issues affecting results in literature
After analysing existing literature, we find that despite of having good results there exist several shortcomings affecting results of existing studies. Major drawbacks of existing studies are lack of proper pre-processing strategy, size of dataset and weak interpretation of models. In best of authors knowledge, none of public dataset comprises classes taken under similar conditions, as COVID-19 samples are taken for one dataset and normal, pneumonia from another dataset. Making of new dataset by fusion of dissimilar classes from different repositories may give biased results. Firstly, advanced pre-processing strategy is missing or not described explicitly in almost all the studies. Only few papers have randomly chosen advanced pre-processing strategy; among one of HE, gamma correction or Total variation filter. Secondly, majority of initial studies have chosen small subset of classes for exploring deep learning methodologies but their comparison is invalid. Another drawback regarding these studies is weak interpretation of models where models make correct classification of sample by focusing on irrelevant features for classification. This is due to lack of proper pre-processing and segmentation strategy [17] of models.
Several minor issues that affect results in literature and be advised for proper documentation to improve quality and clarity of publication. Firstly, specifications of hyperparameters like batch size, learning rate, optimization algorithm and proper training, testing approach is missing. In some of publications it was not clearly mentioned how their final model was selected. Also, COVID-19 is a minority class with a smaller number of samples. Most of publications used cross-entropy loss without handling class imbalance problem that in terms discriminate results of minority class. Further, proper validation set and class-wise sensitivity analysis of models is missing. It is of utmost important to resolve all these minor issues to add new knowledge in existing state-of-the-art studies.
In this study, exploration and making of advanced pre-processing pipeline by cumulative analysis of image denoising and image enhancement techniques based on deep learning architectures. The study focuses on performance analysis by implementation of advanced image pre-processing combinations for diagnosis of COVID-19 using whole image analysis. Proposed research work is advantageous as advanced pre-processing of images, in terms improve interpretability and performance of models. As per literature survey Histogram equalization, CLAHE and gamma correction [25] are best for enhancement of X-ray images are tested in combination with denoising approaches. In addition to this, we resolve class imbalance issue by weighted cross entropy loss function that improves sensitivity of COVID-19 diagnosis. All other minor issues regarding training and analysis have been addressed properly. Aim of this study is to find best individual enhancement technique as well as other combinations that further improves performance and interpretation of models by considering these datasets.
Materials
In this section, we discuss the making of two benchmark datasets to study the effects of image enhancement techniques by collecting data samples from different resources. We make two separate datasets COVIDx and COVIDz by utilizing public CXR datasets that are used in majority of research studies. The reason for making two separate datasets is to study the effect of image enhancement approaches on two different types of studies i.e., 3-class classification and 4-class classification carried out by researchers with dissimilar samples. However, both datasets use COVID-19 CXR samples from similar repository but by considering different views of images. The COVIDx dataset comprises four classes namely COVID-19, normal, virus pneumonia and bacterial pneumonia. In this samples of COVID-19 are collected from chest Xray-dataset [13] with PA, AP and AP supine view. Another chest-Xray-pneumonia dataset [46] is used that comprises 3 classes as normal, viral pneumonia and bacterial pneumonia labelled separately. Due to similarity in COVID-19 and virus pneumonia this dataset helps to understand the impact of enhancement techniques on COVID-19 pneumonia when bacterial pneumonia (BP) and viral pneumonia (VP) are included separately. Another COVIDz dataset make by fusion of three popular publicly available datasets i.e. COVID-19 chest Xray-dataset [13] with only PA view, RSNA (Radiological Society of North America [65]) and USNLM (U.S. national library of medicine [28]). Both datasets contain different view of COVID-19 samples. We consider PA view because majority of studies includes only PA view of samples. This helps to understand the changes in COVID-19 sensitivity by considering view as variability factor that may impact other classes results. Covid chest xray dataset is a publicly available database of CXR images collected by Cohen et al. [13] related to MERS, ARDS, SARS, COVID-19 pneumonia, viral pneumonia etc. from various resources accessible at different public domains. This dataset comprises findings such as COVID-19, SARSr-CoV-1 or SARS, ARDS, SARSr-CoV-2 or, Pneumocystis spp. And Streptococcus spp. with following features: patient ID, sex, age, offset, survival, modality, date, finding, view, location, filename, URL, doi, license and other notes [13]. RSNA [65] dataset contains 30,000 images, 7500 examinations labelled normal, 15,000 examinations labelled pneumonia and remaining 7500 examinations contain symptoms other than pneumonia. Also, NIH CXR14 [28] dataset is used for collecting normal CXR images. Table 3 shows complete description about making of datasets with sources used for taking CXR images, their respective class, number of samples present in class and total number of samples.
Table 3.
Class summary and data resources related to datasets
| Dataset used | class | # | Total | Dataset used | class | # | Total | ||
|---|---|---|---|---|---|---|---|---|---|
| COVIDx Data | Covid CXR [13] | COVID-19 | 476 | 476 | COVIDz Data | Covid CXR [13] | COVID-19 | 193 | 193 |
| CXR Pneumonia [46] | Normal | 1583 | 1663 | Pneumonia | 38 | 520 | |||
| NLM(MC) [28] | Normal | 80 | RSNA [65] | Pneumonia | 482 | ||||
| CXR Pneumonia [46] | VP | 1493 | 1493 | Normal | 443 | 523 | |||
| BP | 2780 | 2780 | NLM(MC) [28] | Normal | 80 | ||||
| Total COVIDx samples | 6412 | Total COVIDz samples | 1236 | ||||||
This study has been carried out under two experiments each using separate dataset COVIDx and COVIDz. The reason for making two datasets is to analyse the effect of various combinations of image enhancement techniques on two major types of studies (3-way classification and 4-way classification). COVIDz dataset helps to add more validity to study effective analysis of image enhancement pipeline selected and tested during experiment 1 for diagnosis of COVID-19 pathology containing different view of images.
Proposed research methodology
In following sections, we discuss the methodology adopted for better identification of COVID-19 CXR samples. In first subsection, we introduce an overview of image denoising and enhancement frameworks used. In second sub-section, we analyse various combinations of image enhancement and denoising approaches in search of better pre-processing pipeline for COVID-19 diagnosis. Following this, a brief discussion about class imbalance, classification process, classification network architecture and performance metrics is provided.
Background
In this section, we first produce an overview of how proposed approaches are working for the sake of better understanding of methodology. Firstly, we give brief description of image denoising approaches used in combination with image enhancement. Following this, we particularly give insightful overview of each image enhancement technique with their framework diagrams in Fig. 2.
Fig. 2.
Framework diagrams for image enhancement approaches; where (a) corresponds to Histogram equalization, (b) represents CLAHE flowchart, (c) represents Gamma correction flowchart
Image denoising approaches
Image denoising [47] is one of essential image pre-processing or post-processing step to eliminate noise or distortions in an image, helpful for further analysis. Typically, traditional denoising approaches focus on smoothing images by assigning equal weight to all the pixels in an image. However, a better denoising approach assigns unequal weight to the pixels, inversely proportional to distance from the central pixel I [Xc, Yc] in image. Specifically, gaussian filter [47] is a linear smoothing filter that reduces weights assigned to a pixel with increasing distance from central pixel based on Gaussian function. The input pixel in gaussian filters is weighted according to Eq. (1),
| 1 |
Where, , corresponds to distance of pixel [x, y] from central pixel [xc, yc].
On the other hand, Median filter [47] belongs to class of non-linear filter where pixels inside a selected window are ranked and filtering is performed based on pixel ordering [42]. This involves filtering parameter as size and shape of filtering window rather then weighted mask. It removes isolated impulsive noise from X-ray images by preserving the edges opposing to average filtering. Let A[i]i = 0…(n − 1) be ranked array of selected window, then median filter assigns new pixel value as median of ranked pixel values inside window computed as A[(n-1)/2].
Adaptive median filter is [47] modified version of median filter where window size is changed dynamically during operation. This is performed when neighborhood pixels has reached its maximum size but filtering operation is not applied because of lack in pixel values. Compared to median filter AMF has higher probability to handle impulse noise. Let Zmin, Zmax, Zmed represents the minimum, maximum and median gray-scale [42] value in Sxy. Also, Smax and Zxy represents maximum allowed size of Sxy and gray-scale value at pixel [x, y]. Then algorithm of AMF works in two levels as:
Algorithm 1: Two levels for adaptive median filter

Due to capturing of images under different conditions adds uncertainty to the pixel values present in an image. Image denoising with total variation filter [52] resolves such uncertainty present at pixel level. Let I be a grayscale image over given bounded set Ω subset of R2 then denoised image d approximately matches an observed image y = (y1, y2)∈Ω, is represented by Eq. (2),
| 2 |
Where , Gaussian Kernel (Gσ) with variance(σ), contrast parameter (K > 0) and a convolution operator (*).
Histogram equalization (HE)
The Histogram equalization [21] approach aims to enhance the quality of image by distributing gray levels in global information of an image [17]. It enhances quality by changing low contrast and brightness of dark images by ensuring each level has equal probability to occur. The information inside image is skewed towards lower grayscale end for a dark image. In order to make image clearer, it can re-distribute grey levels towards dark end of the histogram. The function representation for the histogram with intensity of image in range [0, L-1] is defined by Eq. (3), where nk represents the count of pixels having intensity kth intensity value (rk). Finally, generated histograms are normalized by total pixels in M by N image by Eq. (4), representing chance of occurrence of kth intensity level in an image.
| 3 |
| 4 |
Contrast limited adaptive histogram equalization (CLAHE)
An upgraded variant of HE was introduced as Adaptive Histogram Equalization (AHE) [47]. AHE is a local approach that divides entire image into small regions and thus apply HE to enhance contrast of these small patches separately. However, this local approach is better than global approach like HE but it considers noise component in an image too. To resolve this issue, CLAHE [35] was introduced to give more natural appearance to enhanced images. Also, threshold parameter is used to limit the contrast enhancement in selected regions respectively. To achieve this, RGB color space is converted to HSV (Hue, saturation and value) i.e., human sense color space. The value component is only considered by CLAHE without changing saturation and hue component. Then CLAHE is applied by re-distributing gray-levels to small patches by user provided threshold limit. Finally, processed images are converted to RGB color space.
Gamma correction
Typically, image normalization is based on application of linear operations such as addition, scalar multiplication and subtraction on specific pixel values. However, non-linear operations are supported by gamma correction [11, 18, 36, 49] on image pixels. GC enhances the image by changing the value of pixel by a projection relationship of input pixel value with gamma value as per internal map. If P corresponds to pixel value in range [0,255], Γ represents gamma mapping set, x belongs to P corresponds to grayscale pixel value and Ω is angle value. Let xm be midpoint in range [0,255]. Then, linear mapping from set P to set Ω and set Ω to set Γ is defined by Eqs. (5), (6) and (7) respectively.
| 5 |
| 6 |
| 7 |
Based on above equations, set P is mapped to set Γ values. Let γ (x) = h(x), then corresponding gamma correction function g(x) gives output pixel vector computed using Eq. (8). Figure 2 presents the flowchart for each enhancement approaches i.e., HE, CHALE and gamma correction.
| 8 |
Image pre-processing pipeline
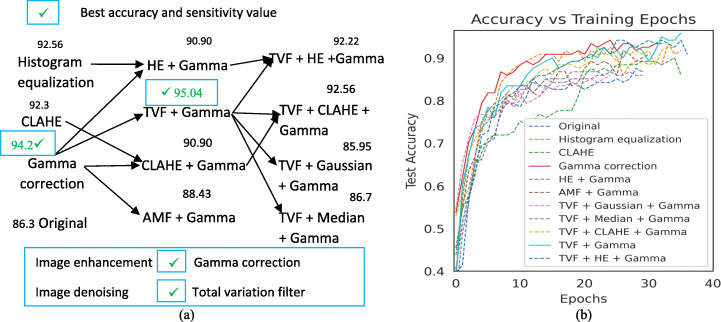
With advancement in application of deep learning approaches in computer aided diagnosis (CAD) [45] assists radiologists to diagnose diseases more accurately in early stages. After the acquisition of samples, image preprocessing is an essential phase that reduces training time of an algorithm as well as error rate. For image enhancement best suitable techniques for x-ray images, Histogram equalization, CLAHE and gamma correction have been tested with Adaptive median filter (AMF), Median filter (MF), total variation filter (TVF) and gaussian denoising filters. In this study, we analyse the cumulative effects of image denoising and image enhancement techniques on benchmark datasets used by researchers to study COVID-19. Initially model having more depth i.e., DenseNet201 is selected to make initial pipeline by training and testing model on COVIDx dataset containing COVID-19, normal, viral pneumonia and bacterial pneumonia classes. Due to large dataset size and complexity of processing huge data, initial testing has been carried out on subset of COVIDx dataset comprises of 193 COVID, 213 normal, 116 viral pneumonia and 116 bacterial pneumonia cases to get intuition of improvements if any. Figure 3a presents the greedy scenario in which eleven combinations are tested and their corresponding accuracy values obtained are written on each combination. Also, Fig. 3b presents the improvements in testing accuracy curve with respect to epochs. These combinations are Histogram equalization, CLAHE, gamma correction, HE + Gamma, CLAHE + Gamma, AMF + Gamma, TVF + Gamma, TVF + Gaussian + Gamma, TVF + Median + Gamma, TVF + CLAHE + Gamma and TVF + HE + Gamma. During initial testing on subset of dataset, gamma correction gain accuracy of 94.21%, TVF + Gamma gain accuracy score of 95.04% which is higher as compared to original images under similar learning rate 10−5, batch size as 32, optimizer as Adam and loss function as weighted cross-entropy loss. Based on results, tick at top head of some model represents best improvement in accuracy and COVID-19 sensitivity values as compared to others image enhancement combinations. Finally, we implemented these combinations on whole COVIDx dataset and came with improvements as compared to plane images. Gamma correction, TVF + Gamma and CLAHE achieve more improvements in sensitivity value as compared to other enhancement techniques. But image denoising with TVF before gamma correction helps to converge top results faster as compared to CLAHE and HE. Results of various proposed combinations on whole COVIDx dataset are shown in Table 7. The proposed pipeline is tested on ten CNN models under 3-way and 4-way classification scenario using two datasets and outperforms results achieved without image preprocessing pipeline. Finally, best combinations of image enhancement techniques are compared again with top performing models to come with robust comparison among various models and enhancement approaches effects. Individually gamma correction performs best as it improves accuracy score of all classes and application of total variation denoising approach before gamma correction improves performance and convergence time on benchmark datasets [63, 64]. Also, gamma correction is equally favourable to all the classes.
Fig. 3.
Sequence of image enhancement and denoising combinations tested for finding best enhancement technique
Table 7.
Comparison of various combinations of image enhancement techniques with DenseNet201 model
| Enhancement approach | Accuracy (%) | Sensitivity (%) | Precision (%) | F1-score | COVID-19 sensitivity (%) | COVID-19 PPV (%) |
|---|---|---|---|---|---|---|
| Original | 92.07 | 92.07 | 92.15 | 92.10 | 96.20 | 96.83 |
| HE | 91.04 | 91.04 | 91.21 | 91.01 | 96.20 | 96.20 |
| CLAHE | 92.80 | 92.80 | 92.92 | 92.76 | 97.47 | 96.25 |
| Gamma correction | 92.95 | 92.95 | 93.08 | 92.43 | 94.94 | 97.40 |
| HE + Gamma | 91.48 | 91.48 | 91.65 | 91.51 | 96.20 | 96.20 |
| CLAHE + Gamma | 92.36 | 92.36 | 92.45 | 92.34 | 100 | 96.34 |
| TVF + Gamma | 93.10 | 93.10 | 93.28 | 93.06 | 96.20 | 96.20 |
| AMF + Gamma | 88.11 | 88.11 | 88.81 | 88.24 | 100 | 90.80 |
| TVF + CLAHE +Gamma | 92.22 | 92.22 | 92.57 | 92.03 | 98.73 | 97.50 |
| TVF + HE + Gamma | 90.31 | 90.31 | 90.98 | 90.50 | 96.20 | 97.44 |
| TVF + gaussian + Gamma | 90.90 | 90.90 | 91.19 | 90.72 | 100 | 96.34 |
| TVF + median + Gamma | 89.72 | 89.72 | 89.88 | 89.64 | 92.48 | 98.65 |
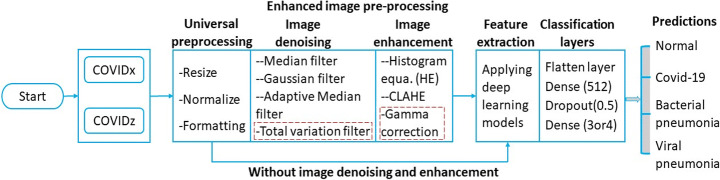
The steps involved in the classification process pipeline are shown in Fig. 4 are explained as: CXR images used in this study are collected from heterogeneous data sources. These sources may represent heterogeneity in size, acquisition condition, shape, datatype, range, scanning condition, and postprocessing [43] etc. CXR images from both datasets are reshaped to 320*320*3, normalized images to ensure sample mean value to zero and standard deviation value to one [57] and change datatypes from uint8/uint16 converted to uniform format float32. Following this image are denoised and enhanced with various enhancement approaches. Once pre-processing is completed, images are fed into deep learning models for feature extraction and classification as shown in Fig. 4. Also, images are resized as per requirement of DL networks. In Fig. 4 red dotted total variation filter and gamma correction achieves better interpretation and evaluation compared to universal preprocessing in this case.
Fig. 4.
Proposed image classification pipeline
After number of experiments on the combination of image denoising and image enhancement techniques [44] we found some useful preprocessing pipelines that improve accuracy as well as covid-19 sensitivity. Initially total variation filter approach [10] is used for denoising [30] uncertainty at pixel level. Then, image enhancement technique gamma correction (GC) is used to enhance contrast value by changing intensity. It helps to decode and encode luminance to improve accuracy value. The resulting enhanced image acts as input to the deep learning models for classification of given image one of the classes. Also, to implement HE and CLAHE, image from RGB color space are converted to HSV color space. V is value component that is taken into consideration by HE [2] and CLAHE for contrast enhancement and merged back with H and S channel. Figure 5 shows output preprocessed images and their corresponding distribution of pixel values after application of various preprocessing sequences as shown, Raw image, Histogram equalized, CLAHE, Gamma corrected, HE + Gamma, TVF + Gamma, TVF + HE +Gamma and finally normalized image of last combination. Even while performing experiments image normalization is performed initially but to show the effects of various enhancement approaches on real CXR images image normalization is shown last.
Fig. 5.
Visualization of effects on image and its pixel distribution after various preprocessing steps
Imbalanced learning approach
In medical image analysis key challenge is the presence of class imbalance. The CXR datasets COVIDx and COVIDz collected for this experimental research study are imbalanced. However, imbalanced dataset [4] does not ensure better learning of models. So, we used a class weighted loss-based approach to resolve class imbalance issue. Ideally, training of models occurs on balanced data that includes equal count of negative as well as positive training samples that would contribute equally to the loss. Usage of cross-entropy loss function on imbalanced data authorize models [20] to prioritize learning process of majority class only. Since, majority of studies on COVID-19 uses CXR dataset collected by Cohen et al. containing less examples as compared with other pathology datasets considered. Since, Simple cross-entropy loss for any ith input feature is given as Eq. (9):
| 9 |
where, yi is label (0 or 1), xi is input feature and f(xi) is output probability that feature is positive. For complete training set T average cross-entropy loss with N samples is given as Eq. (10). To balance the loss, first we calculate contribution frequency of positive and negative i.e., freqpos and freqneg by adding individual contribution of training examples for each class using Eqs. (11) and (12):
| 10 |
| 11 |
| 12 |
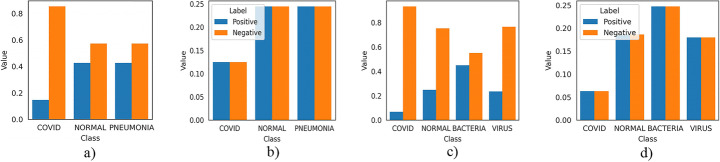
As shown in Fig. 6a and c that positive samples in both datasets corresponding each pathology contribute significantly lower to the loss than negative ones. However, balancing of data need equal contribution to loss for positive as well as negative cases of each class. For this we simply multiply each example in training set by a class specific weighted factor (Wpos and Wneg) corresponding to each class, so that each class has same positive and negative contribution. To achieve this by following Eqs. (13) and (14):
| 13 |
| 14 |
Fig. 6.
Contribution of each class to loss function with and without weighted factor
Using above weights, contribution of negative as well as positive examples within each class is balanced and Fig. 6b and 6d) shows equal contribution to loss function for both datasets. By using these weights, our improved weighted loss function for each training example will be given by Eq. (15) as:
| 15 |
Finally, to calculate multilabel loss, simply sum up average loss for each individual class as shown in Eq. (16) for COVIDx dataset and each one is calculated using Eq. (17). Also, before taking logs a small value ϵ is added to predicted values to avoid certain numerical error if predicted value probabilities to be zero. However, Fig. 6 shows the contribution of positive and negative samples of each class to loss function before and after application of weighted factor. Usage of this weighted loss function handles class imbalance and have shown improvements in sensitivity and positive predictive values for each pathology.
| 16 |
| 17 |
Classification process
Based on the number of classes present in COVIDx and COVIDz CXR image datasets we have 4-way classification as COVID-19 vs Normal vs Viral pneumonia vs Bacterial pneumonia and 3-way classification scenario as COVID-19 vs Normal vs Pneumonia. Independent test set is used for comparative analysis whereas Model development have been carried out using Training and validation sets. Tables 4 and 5 clearly depicted that how many CXR images are distributed into each training, testing and validation sets in COVIDx and COVIDz dataset. Since, COVID CXR dataset used for proposed study does not previously hold any data for training and testing purpose. So, we randomly distribute CXR fused datasets into training, testing and validation set. Data splitting has been performed on patient level to ensure no data leakage between the training, validation and testing dataset.
Table 4.
Distribution of samples for 4-way classification in all infection types
| set | Normal | COVID-19 | VP | BP | Total |
|---|---|---|---|---|---|
| Training | 1293 | 353 | 1228 | 2348 | 5222 |
| Validation | 150 | 44 | 125 | 190 | 509 |
| Test | 220 | 79 | 140 | 242 | 681 |
| Total | 1663 | 476 | 1493 | 2780 | 6412 |
Table 5.
Distribution of samples for 3-way classification in all infection types
| set | Normal | Pneumonia | COVID-19 | Total |
|---|---|---|---|---|
| Training | 426 | 426 | 146 | 998 |
| Validation | 39 | 38 | 16 | 93 |
| Test | 58 | 56 | 32 | 146 |
| Total | 523 | 520 | 194 | 1236 |
Classification network architecture
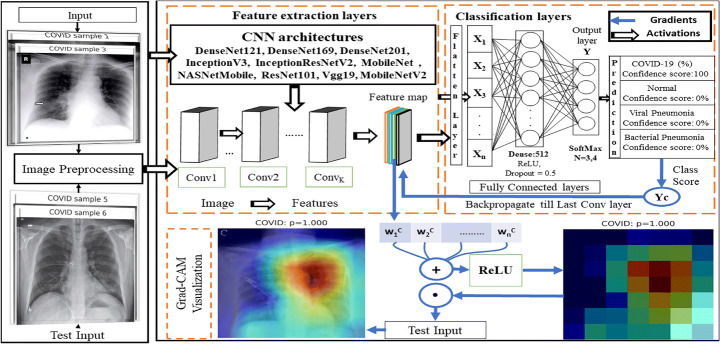
The primary aim of classification network is to categorize CXR images grounded on various features corresponding to different type of pathology. We implemented various state-of-the-art (SOTA) deep transfer learning models as a backbone for building new classification network. Deep learning models require huge volume of data for better training of models. To work with limited training CXR examinations we apply deep transfer learning-based approaches using pretrained ImageNet weights. For the purpose of classification we choose ten popular models, VGG19, InceptionV3, InceptionResNetV2,MobileNet, MobileNetV2, ResNet50, NASNetMobile, DenseNet121, DenseNet169 and DenseNet201 [10, 23, 32, 54] as baseline architectures. In Fig. 7 we have shown the classification network architecture that take input samples from two datasets COVIDx and COVIDz based on different pathological characteristics. For training the network, data samples from training and validation set are given as input to pretrained CNN architectures with and without image preprocessing pipeline. Feature extraction layers convert input images to feature maps after several convolutions, Batch normalization and pooling operations. On the top of feature extraction layers, we added new classification layers as flatten layer to convert feature maps into 1D vector followed by dense layer having 512 neurons, dropout layer with value of 0.5 to avoid overfitting accompanied with ReLU activation. Finally, dense inference layer with SoftMax activation transforms feature vectors into probability values between [0,1] corresponding to each class. In addition, output layer consists of 3 or 4 neurons based on the number of classes in the classification scenario. In the end we train and finetune the classification network. On given test CXR sample, these models classify input image into one pathology class with maximum confidence score. Finally, we visualize learning process by backtracking till last convolutional layer and extracting feature maps weights to interpret heatmap to know why given sample is classified to particular class. All these images are processed in 3 channel and resized before training according to the image size appropriate for various SOTA deep learning approaches. Minibatch gradient descent and Adam optimizer are used for training and testing. These baseline architectures use class weighted loss function to handle class imbalance over classification scenarios.
Fig. 7.
Classification network architecture
All these baseline models are variants of CNN architecture and were found extremely useful in the automated detection of COVID-19 pathology. MobileNet and MobileNetV2 are CNN architectures developed by Howard et al. in the year 2017 and 2019 for mobile vision applications. These are lightweight architecture with 17 Mb and 14 Mb of size and consisting of 80 layers. Both were trained on ImageNet dataset consisting of 1000 classes with suitable image size (224,224,3). VGG19 architecture was introduced in the 2014 consisting of 19 layers. It has 16 convolution layers, 5 MaxPool, 3 Fully connected and 1 SoftMax layer. It accepts image in 3 channels (RGB) with shape (224,224). NASNetMobile is another powerful architecture proven useful for image classification came in year 2018. Experiments are performed for finding best convolutional layer using CIFAR-10 dataset and then applied this on ImageNet dataset. It gives 2.4% error rate on CIFAR-10 and achieves 96.2% top-5 and 82.7% top-1 accuracy on ImageNet. Also, (224,224,3) is suitable image shape for this model. InceptionResNetV2 was developed in year 2016. It focuses more on residuals connection instead of merge and split approach. Like others it uses 3 channel images with appropriate size of (299, 299) and has 56 M parameters. In traditional CNN models with L layers, there were only one connection between two layers with L connections in total. But series of densely CNN models (DenseNet) connects each layer to all other layers in feed forward manner resulted in total L(L + 1)/2 connections in total. The network includes L layers, each layer implements convolution (Conv), ReLU (rectified linear units), pooling, batch normalization and non-linear transformation. These architectures proposed in the year 2017 applied to CIFAR-10+, CIFAR-10, CIFAR-100+ and CIFAR-100, SVHN and ImageNet. In Table 6 we have shown some basic features like depth, number of parameters, size of pretrained model and appropriate image size related to baseline architectures used. In this study, we build new model by instainting these models using pretrained weights of ImageNet by freezing all the layers of these base models in beginning and then perform finetuning by at the end. The entire process is accomplished by two experiments. In experiment 1, classification network is trained and tested using COVIDx dataset. In experiment 2, classification network is trained and tested using COVIDz dataset to make one of these possible predictions based on maximum probability value i.e., a) Normal, b) Pneumonia and c) COVID-19 infection. This entire work has been carried out using Tensorflow, Keras and scikit-image via NVIDIA Tesla P100 GPU.
Table 6.
Basic features about deep learning baseline architectures
| CNN model | Depth | Parameters | Size (MB) | Image size |
|---|---|---|---|---|
| VGG19 | 26 | 144 M | 549 | [224, 224] |
| MobileNet | 88 | 4.2 M | 16 | [224, 224] |
| MobileNetV2 | 88 | 3.5 M | 14 | [224, 224] |
| InceptionV3 | 159 | 23.8 M | 92 | [299, 299] |
| InceptionResNetV2 | 572 | 55.9 M | 215 | [299, 299] |
| ResNet50 | – | 25.7 M | 98 | [224, 224] |
| NesNetMobile | – | 5.3 M | 23 | [224, 224] |
| DenseNet121 | 121 | 8 M | 33 | [224, 224] |
| DenseNet169 | 169 | 14.3 M | 57 | [224, 224] |
| DenseNet201 | 201 | 20.2 M | 80 | [224, 224] |
Training and Finetuning
To understand the impact of image enhancement techniques on performance, we consider ten baseline pretrained models as feature extractor and build new classification architecture by adding few classification layers on the top of pretrained models as shown in Fig. 7. Two phase training process is explained as follows:
In first phase, above pretrained models are used as backbone with their weights are frozen by training only fully connected layers on our datasets. Adam is an optimizer used for training with an initial learning rate of 0.000001 using Minibatch gradient descent having batch size 32 and model trained for about 30 epochs. Training is carried out using early stopping strategy with patience value 5 whereas learning rate is reduced by factor 10 if training loss does not improve. Model having lowest validation loss with enhancement technique is taken for next stage.
In second phase, we perform fine-tuning which involves unfreezing some layers and retraining with low learning rate. While finetuning, similar Adam optimizer with low learning rate of 0.000001 is used by unfreezing top 20 layers except batch normalization layer and trained for about 5 epochs. All other similar hyperparameters of training phase are used. This considerably improved the accuracy of classifying COVID-19 cases against others
Performance metrics
Accuracy, sensitivity (Recall), positive predictive value (PPV) and F1-score are metrics used for measuring performance of classification models are calculated using Eqs. (18), (19), (20), (21), (22) and (23). Whereas true positive (TP), false positive (FP), false negative (FN) and true negative (TN) are calculated using confusion matrix [59, 61]. COVID-19 sensitivity and COVID-19 PPV are also taken into consideration. For any input image IX, class having maximum confidence score is considered as final prediction for calculating confusion matrix and various evaluation measures.
| 18 |
| 19 |
| 20 |
| 21 |
| 22 |
| 23 |
Results and analysis
In this section, we discuss results achieved after experimentally analysing CXR images using various image denoising and image enhancement techniques combined with transfer learning architectures. Most suitable image enhancement approaches Histogram equalization, CLAHE and gamma correction has been tested individually as well as in combination with image denoising approaches in a greedy manner. Based on initial processing on subset of COVIDx dataset, similar combinations of image enhancement and denoising techniques are tested on whole COVIDx dataset as listed in Table 7. Image enhancement approach having minimum loss and maximum accuracy is considered for comparison with other deep learning approaches. For fair and reasonable comparison, we compared performance of proposed pipeline with ten deep learning models and obtained result confirm improvements as shown in Tables 7, 8 and 9. In Table 7, we present the results obtained by best possible combinations of enhancement and denoising techniques. From these results, it is clear that individually gamma correction achieves better accuracy score and validation loss when compared to original, HE and CLAHE under similar conditions having initial learning rate as 0.000001, optimizer as Adam, loss function as class weighted loss function and batch size as 32. Further denoising of images with Total variation filter before gamma correction achieved higher accuracy, least loss value and improved COVID-19 sensitivity over gamma correction as shown in Table 7. Although, gamma correction, TVF + Gamma not only advantageous in accuracy score but also in time taken by model to converge the best possible results.
Table 8.
Models comparison of classification results with and without TVF + Gamma
| Classes | Process | Classification Model | Acc. (%) | Sen. (%) | PPV (%) | F1-score (%) | COVID-19 sensitivity | COVID-19 (PPV) (%) |
|---|---|---|---|---|---|---|---|---|
| COVID-19, Normal and Viral and Bacterial pneumonia 4-way classification (Experiment 1) | With original | MobileNet | 91.34 | 91.34 | 91.66 | 91.26 | 98.73 | 98.73 |
| MobileNetV2 | 89.87 | 89.87 | 90.24 | 89.85 | 96.20 | 98.70 | ||
| VGG19 | 92.66 | 92.66 | 92.86 | 92.61 | 97.47 | 96.25 | ||
| ResNet50 | 91.34 | 91.34 | 91.77 | 91.38 | 93.67 | 98.67 | ||
| NASNetMobile | 88.40 | 88.40 | 88.45 | 88.28 | 98.73 | 96.30 | ||
| InceptionV3 | 88.69 | 88.69 | 89.38 | 88.67 | 91.14 | 98.63 | ||
| InceptionResNetV2 | 90.16 | 90.16 | 90.33 | 90.11 | 97.47 | 97.47 | ||
| DenseNet121 | 92.66 | 92.66 | 92.78 | 92.68 | 97.20 | 97.44 | ||
| DenseNet169 | 92.22 | 92.22 | 92.45 | 92.16 | 98.73 | 96.30 | ||
| DenseNet201 | 92.07 | 92.07 | 92.15 | 92.10 | 96.20 | 96.83 | ||
| With TVF + Gamma | MobileNet | 93.25 | 93.25 | 93.50 | 92.14 | 98.72 | 100 | |
| MobileNetV2 | 90.31 | 90.31 | 90.52 | 90.21 | 97.47 | 93.90 | ||
| VGG19 | 92.80 | 92.80 | 93 | 92.76 | 97.47 | 96.25 | ||
| ResNet50 | 91.48 | 91.48 | 91.74 | 91.43 | 94.94 | 92.59 | ||
| NASNetMobile | 92.07 | 92.07 | 92.17 | 92.06 | 96.20 | 95 | ||
| InceptionV3 | 87.08 | 87.08 | 87.28 | 86.93 | 91.14 | 94.74 | ||
| InceptionResNetV2 | 89.13 | 89.13 | 89.44 | 89.12 | 97.47 | 98.72 | ||
| DenseNet121 | 92.80 | 92.80 | 93.01 | 92.86 | 96.20 | 96.20 | ||
| DenseNet169 | 92.36 | 92.36 | 92.54 | 92.44 | 100 | 96.34 | ||
| DenseNet201 | 93.10 | 93.10 | 93.28 | 93.06 | 96.20 | 96.20 | ||
| COVID-19, Normal and Pneumonia 3-way classification (Experiment 2) | With original | MobileNet | 86.99 | 86.99 | 87.11 | 86.98 | 90.62 | 93.55 |
| MobileNetV2 | 80.82 | 80.82 | 81.37 | 80.63 | 78.12 | 86.21 | ||
| VGG19 | 76.71 | 76.71 | 77.58 | 76.34 | 56.25 | 85.71 | ||
| ResNet50 | 85.62 | 85.62 | 86.73 | 85.81 | 84.38 | 100 | ||
| NASNetMobile | 78.77 | 78.77 | 79.89 | 78.70 | 65.62 | 91.30 | ||
| InceptionV3 | 87.67 | 87.67 | 88.08 | 87.71 | 87.50 | 90.32 | ||
| InceptionResNetV2 | 78.77 | 78.77 | 80.50 | 78.89 | 68.75 | 100 | ||
| DenseNet121 | 78.77 | 78.77 | 79.14 | 78.90 | 84.38 | 93.10 | ||
| DenseNet169 | 87.67 | 87.67 | 87.83 | 87.72 | 90.62 | 96.67 | ||
| DenseNet201 | 89.73 | 89.73 | 89.81 | 89.65 | 100 | 96.97 | ||
| With TVF + Gamma | MobileNet | 88.36 | 88.36 | 88.48 | 88.34 | 96.88 | 96.88 | |
| MobileNetV2 | 89.04 | 89.04 | 89.18 | 89.06 | 90.62 | 96.67 | ||
| VGG19 | 89.04 | 89.04 | 89.04 | 88.97 | 96.88 | 93.94 | ||
| ResNet50 | 86.30 | 86.30 | 87.07 | 86.35 | 93.75 | 100 | ||
| NASNetMobile | 80.82 | 80.82 | 81.68 | 80.85 | 84.38 | 84.38 | ||
| InceptionV3 | 77.40 | 77.40 | 78.34 | 77.43 | 68.75 | 91.67 | ||
| InceptionResNetV2 | 80.14 | 80.14 | 81.43 | 80.39 | 78.12 | 100 | ||
| DenseNet121 | 85.62 | 85.62 | 85.52 | 85.54 | 96.88 | 91.18 | ||
| DenseNet169 | 89.73 | 89.73 | 89.82 | 89.77 | 96.88 | 100 | ||
| DenseNet201 | 91.10 | 91.10 | 91.10 | 91.09 | 100 | 100 |
Table 9.
Comparison of image enhancement techniques for the classification of COVID-19 with top performing CNN architectures
| CNN model | Enhancement technique | Acc. (%) | Sen. (%) | PPV (%) | F1-score (%) | COVID-19 Sensitivity (%) | COVID-19 (PPV) (%) | |
|---|---|---|---|---|---|---|---|---|
| 4-way classification | MobileNet | CLAHE | 90.60 | 90.60 | 90.88 | 90.52 | 100 | 94.05 |
| Gamma | 92.51 | 92.51 | 92.72 | 92.51 | 98.73 | 96.30 | ||
| VGG19 | CLAHE | 91.91 | 91.91 | 92.06 | 91.90 | 97.47 | 96.25 | |
| Gamma | 92.95 | 92.95 | 93.09 | 92.94 | 96.20 | 97.44 | ||
| DenseNet169 | CLAHE | 92.22 | 92.22 | 92.40 | 92.10 | 100 | 96.34 | |
| Gamma | 91.48 | 91.48 | 91.94 | 91.30 | 100 | 96.34 | ||
| DenseNet201 | CLAHE | 92.80 | 92.80 | 92.92 | 92.76 | 97.47 | 96.25 | |
| Gamma | 92.95 | 92.95 | 93.08 | 92.43 | 94.94 | 97.40 | ||
| 3-way classification | MobileNet | CLAHE | 88.36 | 88.36 | 88.36 | 88.36 | 93.75 | 93.75 |
| Gamma | 89.73 | 89.73 | 89.93 | 89.74 | 87.50 | 96.55 | ||
| VGG19 | CLAHE | 80.82 | 80.82 | 83.57 | 80.94 | 100 | 65.31 | |
| Gamma | 89.04 | 89.04 | 89.29 | 89.08 | 93.75 | 93.75 | ||
| DenseNet169 | CLAHE | 88.36 | 88.36 | 88.86 | 88.32 | 96.88 | 100 | |
| Gamma | 88.36 | 88.36 | 88.53 | 88.37 | 96.88 | 100 | ||
| DenseNet201 | CLAHE | 89.73 | 89.73 | 89.81 | 89.65 | 100 | 96.97 | |
| Gamma | 91.10 | 91.10 | 91.10 | 91.09 | 100 | 100 |
In Fig. 8, we have shown graph presenting validation accuracy and validation loss achieved by DenseNet201 model vs epochs over best combinations considered in study for 35 epochs. It gives clear intuition that TVF + Gamma correction takes 24 epochs to reach top accuracy of 93.10% whereas Gamma achieve its higher accuracy score after 30 epochs to reach 92.95% respectively which is considerably better than original as well as other neighbour techniques in Table 7. However, gamma correction also shows advantage in training better and smother than HE and CLAHE. Due to more deep architecture of DenseNet201 with TVF + Gamma achieves higher accuracy and lower loss as compared to enhancement techniques. Also, gamma correction gives comparable accuracy score but denoising using TVF before gamma reaches top accuracy faster as compared to gamma. So, we consider it as preprocessing pipeline in next stage to perform model-based comparisons to cross check its effect using ten state-of-the-art CNN models performance.
Fig. 8.
Visualization of validation accuracy and validation loss vs epochs under various preprocessing combinations by DenseNet201 model
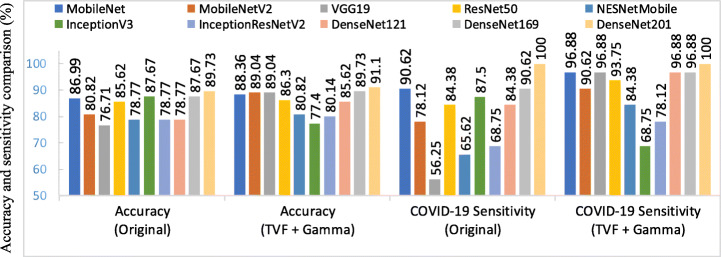
In Table 8, we present results that verify optimized image preprocessing pipeline TVF + Gamma comparing with original images without pre-processing pipeline in 3-way (COVID-19, normal and pneumonia) classification and 4-way (COVID-19, normal, viral pneumonia and bacterial pneumonia) classification. The finest results attained by ten CNN architectures each with technique referred as original and TVF + Gamma is Shown in Table 8. Image preprocessing pipeline TVF + Gamma used with CNN models outperforms the models experimented with original images in terms of accuracy as well as sensitivity. Figures 9 and 10, presents accuracy and COVID-19 sensitivity comparison of all the models with original and TVF + Gamma approach corresponding to COVIDx and COVIDz dataset. It clearly depicts improvements shown in accuracy and sensitivity by all models except inception model in experiment 1 and experiment 2. Although, no single model performs well on all datasets. However, MobileNet model outperforms all other models in experiment 1(4-way classification) using proposed preprocessing pipeline with accuracy increase of 1.94% in accuracy score followed by DenseNet201 with accuracy score of 93.10%. Similarly, in experiment 2 (3-way classification) DenseNet201 model achieved an accuracy score of 91.11% with 1.47% increase in accuracy score.
Fig. 9.
Models’ comparison of accuracy and COVID-19 sensitivity with and without TVF + Gamma corresponding to COVIDx dataset
Fig. 10.
Models’ comparison of accuracy and COVID-19 sensitivity with and without TVF + Gamma corresponding to COVIDz dataset
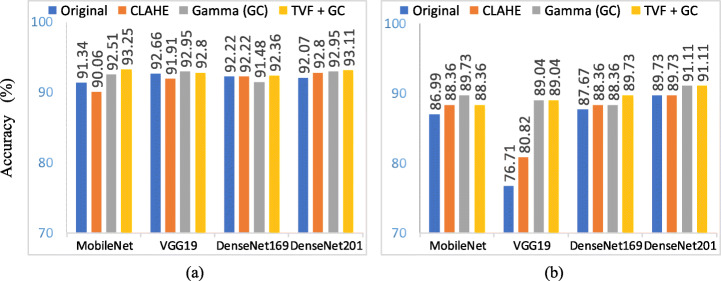
Further analysis of top performing models with top performing individual as well as hybrid enhancement techniques have been compared to know more about performance sequence and effects of these enhancement techniques on various models. Four CNN models, MobileNet, VGG19, DenseNet169 and DenseNet201 has been selected for comparison with enhancement techniques shown in Table 9. We selected top performing image preprocessing combinations as gamma correction, CLAHE and TVF+ Gamma. Each best selected model is tested with Gamma and CLAHE in both 3-way and 4-way classification and their results are shown in Table 9. Major focus of Table 9 study is to compare TVF + Gamma with other selected enhancement approaches with four best performing models. It helps to rank image enhancement techniques more accurately. Results of Tables 8 and 9 shows that among all combinations TVF + Gamma outperforms in all exception in inception-based model. In Fig. 11, we have shown the comparison of results over original vs CLAHE vs gamma correction and TVF + Gamma in both the experiments. Individually gamma correction achieves better overall sensitivity as compared to HE and CLAHE for classification of COVID-19 both in terms of time and accuracy. But in some cases, it gains less COVID-19 sensitivity as compared to HE and CLAHE. So, performing image denoising first using TVF then apply gamma correction performs better than adaptive median filter with Gamma, HE + Gamma, CLAHE + Gamma, AMF + Gamma, TVF + gaussian + Gamma and TVF + median + Gamma.
Fig. 11.
Accuracy comparison of models with various image enhancement combinations in experiment 1(COVIDx) and experiment 2 (COVIDz)
Among all ten deep CNN architecture MobileNet achieves best accuracy score and outperforms all other models when applied with TVF + Gamma in experiment 1 and DenseNet201 in experiment 2. In fact, deep CNN models like MobileNet and DenseNet201 are best suitable for classification of COVID-19 due to its similarity with other classes like viral pneumonia. But usage of image enhancement techniques shows improvements in results of shallow CNN models also. In Fig. 11 proposed TVF + Gamma has better or comparable accuracy in MobileNet, VGG19, DenseNet169 DenseNet201 in both 3-way and 4-way classification. Whereas application of gamma correction helps models to converge faster towards better results and combining it with TVF improves COVID-19 sensitivity that in improves overall performance. The finest accuracy is achieved by MobileNet model with accuracy score of 93.25% and COVID-19 sensitivity score of 98.75% when preprocessing pipeline used is TVF + Gamma. This is because gamma correction is more favourable to viral pneumonia and TVF is better for COVID-19 identification. So, combining these improves accuracy of CNN models. In ranking of enhancement techniques, TVF + Gamma is better than gamma correction which is again better than HE and CLAHE in accuracy as well as time. These combinations improve performance over original images and can be useful in CAD systems. Importance of image enhancement and image denoising not only improves performance of models but also reduces time required to converge best possible results.
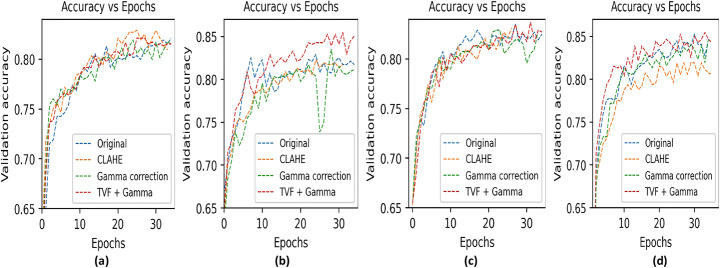
In Fig. 12 we have shown validation accuracy curve with respect to epochs corresponding to VGG19, MobileNet, DenseNet169 and DenseNet201 models with original and preprocessing pipeline as CLAHE, gamma correction and TVF + Gamma in 4-class classification. Also, based on principle of class having maximum confidence score as final predicted class, we have shown resulted confusion matrices of top image enhancement techniques based on DenseNet201 model and confusion matrix of best performing model MobileNet in 4-class classification in Fig. 13. In Fig. 13, subfigures a, b, c, d, e are confusion metrics obtained by DenseNet201 model with and without enhancement combination. We can clearly see improvements in confusion metrics with enhancement techniques. Without enhancement techniques a greater number of samples in other classes are confused with COVID-19 class, also there exists confusion between bacterial and virus classes. With gamma correction and TVF + Gamma such confusion is minimized. Moreover, further observations sight that in experiment 1 viral class miserably confused with bacterial and vice-versa. Subfigure (f) represent confusion matrix of MobileNet model with TVF + Gamma enhancement. MobileNet model improves COVID-19 sensitivity as well there is no overlapping of other classes to COVID-19 class but it has less sensitivity corresponding to virus pneumonia.
Fig. 12.
Visualization of validation accuracy vs epochs of CNN models with preprocessing pipeline as CLAHE, gamma correction and TVF + Gamma in 4-way classification using COVIDx dataset
Fig. 13.
Confusion matrices by DenseNet201 and MobileNet model corresponding to 4-class configurations with various enhancement techniques. Subplots a, b, c, d, e corresponds to DenseNet201 model for comparison of image enhancement combinations and subplot f), corresponds to best performing MobileNet model with TVF + Gamma
Class-wise sensitivity analysis
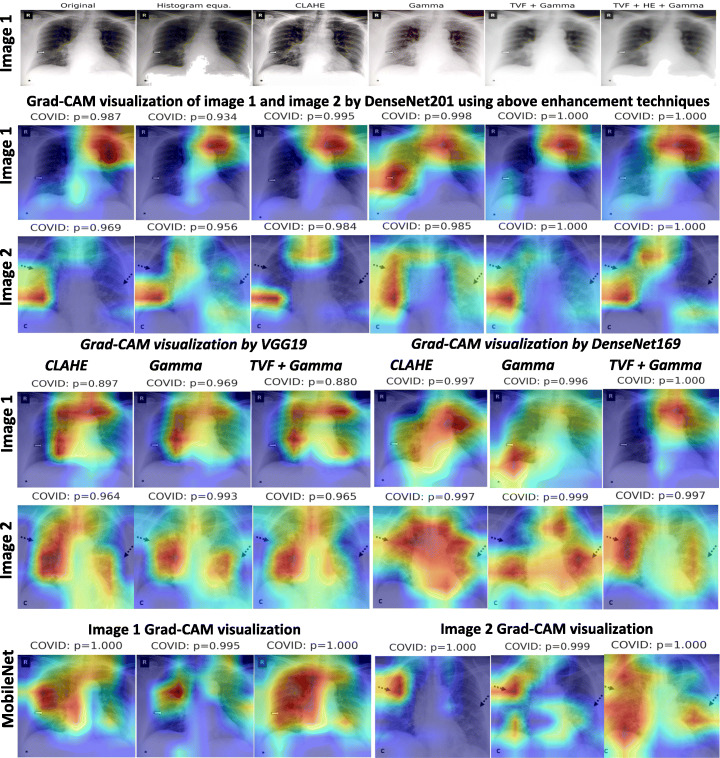
Figure 14 presents schematic representation of class-wise sensitivity [11] achieved by various models with and without image enhancement approaches. It helps in understanding the models and image enhancement approaches suitability for each of the class. As, none of the model and image enhancement approach appears equally suitable for all the classes. Although, simple conclusions can be drawn that can may help in making of models by consideration of all the classes. It is observed that usage of image enhancement techniques as gamma and TVF + Gamma improves sensitivity in almost all the cases. Precisely, CLAHE is better for COVID-19 features whereas gamma correction is better for normal, BP and VP classes. Since, application of denoising with TVF followed by gamma correction improves COVID-19 and BP sensitivity. However, average sensitivity of all the classes is better with TVF with gamma correction. Finally, MobileNet with TVF + Gamma achieves higher average sensitivity compared to others.
Fig. 14.
Class-wise sensitivity analysis of various models with respect to image enhancement techniques on COVIDx dataset
Confidence analysis
To evaluate effectiveness of enhancement techniques we compared enhancement techniques with our DenseNet201 model, in which the confidence threshold Thconf ranges from 0.5 to 0.95. The results obtained on COVIDx dataset as accuracy, recall and COVID-19 sensitivity based on different enhancement techniques are shown in Fig. 15. From analysis we perceive that usage of enhancement techniques improves confidence score [18] corresponding to each class. Figure 15a, b and c reveals the strength of proposed pipeline by achieving more accuracy and sensitivity with TVF + Gamma at various threshold values. Results shows that even at higher threshold values 0.95, TVF + Gamma achieves higher accuracy of 92%, recall of 70% and COVID-19 sensitivity of 92.4% as compared with other enhancement techniques. This ensures that proposed pre-processing pipeline is superior than others. Also, gamma correction achieves higher values but denoising with TVF seems more useful in correct identification of samples. So, it is strongly recommended to use image enhancement techniques for better pre-processing before application of deep learning approaches. In Table 10 we have shown some key points observed after analysing effects of various image enhancement techniques on performance of the models in terms of accuracy, sensitivity and time.
Fig. 15.
Comparison of various image enhancement techniques at different threshold values by considering DenseNet201 model with COVIDx dataset; where (a) corresponds to accuracy comparison, (b) represents Sensitivity comparison and (c) represents COVID-19 sensitivity comparison
Table 10.
Comparison of image enhancement methods based on results for effective diagnosis of COVID-19
| Technique | Accuracy | Sensitivity | Time |
|---|---|---|---|
| TVF + Gamma | Achieves accuracy better than HE, CLAHE and gamma correction even at large threshold values and having minimal overlapping with COVID-19 class. | Achieves more COVID-19 sensitivity and precision in deep CNN models. | Takes time comparable to gamma correction but less than HE and CLAHE. |
| Gamma correction | Achieves better accuracy then HE and CLAHE because of more improvement in true positives of virus class. | Sensitivity more than HE and CLAHE but Less than TVF + Gamma. | Takes less time to converge best possible results. |
| CLAHE | Accuracy more than to HE but less than Gamma, TVF + Gamma due to maximal overlapping of viral and bacterial pneumonia. | More COVID-19 sensitivity in both shallow and deep CNN models but less sensitivity of virus class. | Takes more time then TVF + Gamma and gamma correction to converge best possible result. |
| Histogram equalization | Accuracy less than CLAHE, Gamma, TVF + Gamma because of less accuracy of normal and virus class. | Least sensitive to covid class than CLAHE but more than gamma correction. | Takes more time than gamma correction but less than CLAHE. |
| Original | Accuracy is less. | Sensitivity is less at higher threshold values. | Takes more time to converge best possible results. |
From global perspective, this research study gives promising results that can improve performance and interpretability of existing state-of-the-art models. We ensure supremacy of our proposed pipeline on the basics of learning curves, confusion matrix, class-wise sensitivity, confidence score analysis and Gradient-CAM visualization maps. We tested our proposed pipeline with ten deep learning models on two different benchmark datasets. Out of total 20 combinations our proposed pre-processing pipeline wins for 17 times only except inception-based models. Confusion matrix obtained by our proposed model shows zero overlapping of other classes with COVID-19 class. Confidence score prediction ensures that proposed approach predicts classes with higher probability that in terms achieves higher accuracy, sensitivity and COVID-19 sensitivity as per Section 5.2. Regarding interpretation of CXR images, our proposed model classify image to COVID-19 class by looking at relevant lung regions as shown in Section 5.6. All these quantitative and qualitative outputs ensures that proposed model MobileNet with pre-processing pipeline TVF + Gamma is superior to others.
The reason for the success of MobileNet model is because of its structural characteristics of depthwise separation convolutions followed by point wise convolutions. It achieves better results because of better classification ability of COVID-19, normal and BP classes with TVF + Gamma whereas for VP it is comparable to others. Because of its higher depth its interpretation is also better as Grad-CAM attention maps shows model focus on more relevant lung regions. Also, interpretation of DenseNet201 model is comparable to MobileNet as models having more depth and comparable performance tries to capture more relevant features for classification. Finally, we compare the results obtained by our best model MobileNet in 4-way classification and DenseNet201 in 3-way classification using proposed preprocessing pipeline with various state-of-the-art approaches as shown in Tables 11, 12 and 13. In Table 11 we compared our proposed DenseNet201 with very first COVID CXR classification COVID-Net [71] model. In Table 12 we further compare proposed MobileNet model with very popular COVID detection model named COVID-AID [39] and In Table 13 we compared proposed models with various state-of-the-art approaches.
Table 11.
Class wise comparison of DenseNet201 with COVID-Net
| Pathology | COVID-Net (%) | Proposed (%) | ||
|---|---|---|---|---|
| Sensitivity | PPV | Sensitivity | PPV | |
| COVID-19 | 91.0 | 98.9 | 100 | 100 |
| Normal | 95.0 | 90.5 | 89.66 | 88.14 |
| Pneumonia | 94.0 | 91.3 | 89.09 | 87.50 |
Table 12.
Class wise comparison of DenseNet201 with Covid-AID
| Pathology | Covid-AID (%) | Proposed (%) | |||
|---|---|---|---|---|---|
| Sensitivity | PPV | Sensitivity | PPV | ||
| 4-class | COVID-19 | 100 | 93.8 | 98.73 | 100 |
| Normal | 76.1 | 98.9 | 93.64 | 98.56 | |
| VP | 87.2 | 72.1 | 78.57 | 90.91 | |
| BP | 96.1 | 88.1 | 99.59 | 88.28 | |
Table 13.
Comparison of proposed model with other state-of-the-art models
| Ref. | Best model | Classes | # Of COVID samples | Technique | Performance metrics (%) | ||
|---|---|---|---|---|---|---|---|
| Acc. | Sencov | Other | |||||
| [67] | CapsNet | 3 | 231 | – | 84 | 94.57 | F1–84.21 |
| [12] | VGG16 | 3 | 132 | Fine tuning | 85 | 100 | F1–85 |
| [51] | DarkNet | 3 | 127 | – | 87.02 | 97.9 | F1–87.37 |
| [14] | Xception | 3 | 127 | Fine tuning | – | 97 | F1cov - 96.9 |
| [33] | Xception | 3 | 290 | Fine tuning | 90.21 | 89 | F1–91 |
| [5] | MobileNetV2 | 3 | 224 | Fine tuning | 92.85 | 98.66 | Spcov − 96.46 |
| [40] | ResNet50 | 3 | 184 | Fine tuning | – | 98 | Spcov - 93 |
| [71] | COVID-Net | 3 | 268 | – | 93.3 | 91 | PPVcov-98.9 |
| [1] | DeTrac | 3 | 105 | Fine tuning | 93.1 | 100 | Sp - 85.1 |
| [49] | Shallow-CNN | 3 | 321 | – | – | 100 | PPVcov - 99.38 |
| [54] | DenseNet-201 | 3 | 683 | Fine tuning | – | 98.93 | Sp - 98.77 |
| DenseNet201 with TVF+ Gamma | 3 |
193 (PA view) |
Fine tuning | 91.10 | 100 |
F1–91.09 PPVcov − 100 |
|
| [37] | GoogleNet | 4 | 69 | Fine tuning | 80.6 | 100 | F1–82.32 |
| [38] | CovXNet | 4 | 305 | – | 90.2 | 95 | F1–90.4 |
| [50] | ResNet18 | 4 | 180 | Fine tuning | 88.9 | 92.5 | F1–84.4 |
| [24] | AlexNet | 4 | 371 | Fine tuning | 93.42 | 89.18 | Sp - 98.9 |
| [39] | DenseNet121 | 4 | 155 | Fine tuning | 90.5 | 100 | F1–92.30 |
| MobileNet with TVF + Gamma | 4 | 476 (PA, AP, AP Supine) | Fine tuning | 93.25 | 98.72 |
PPVcov - 100 F1–92.14 |
|
Bold signifies the results of proposed model
Comparison with Covid-net
COVID-Net [71] is one of the first popular and most effective SOTA deep CNN based learning approach for distinguishing COVID-19 examples from pneumonia and control cases. So, we compare results achieved by Densenet201 with COVID-Net as described in Table 11. We used identical dataset to study 3-way classification of COVID-19 vs Normal vs Pneumonia. With similar dataset, DenseNet201 with proposed preprocessing pipeline presented improved COVID-19 sensitivity and PPV related to COVID-Net with same covid dataset. In addition, this model uses only 20.2 M parameters and advantageous in terms of stability and performance whereas COVID-Net uses 116.6 M parameters.
Comparison with Covid-AID
Covid-AID [39] is another popular AI based detection approach for identification of COVID-19 in CXR samples. It is state-of-the-art deep CNN architecture that uses DenseNet121 as baseline model where model weights are initialized via CheXNet application. Covid-AID usages covid-chestxray-dataset [13] and accomplish classification of COVID-19 vs normal vs viral pneumonia vs bacterial pneumonia with 90.5% accuracy. With similar dataset, we meaningfully improved the results with accuracy score of 93.10% in 4-class classification. However, Covid-AID uses only PA view samples. Also, Covid-AID combines VP and BP classes to perform 3-way classification. So, our DenseNet-201 with TVF + Gamma achieved an accuracy score of 99% in similar 3-way classification. Also, class wise evaluation of DenseNet201 model and Covid-AID over COVID-19 sensitivity and PPV is shown in Table 12.
Comparison with the state-of-the-art models
To demonstrate robustness of CXR enhancement on performance of deep learning models, we make use of different combinations of image denoising and image enhancement techniques as preprocessing pipeline with ten deep CNN models to facilitate transfer learning. Further to confirm the effectiveness of our proposed model, we compare performance of model with several state-of-the-art existing models in similar 3-way and 4-way classification scenarios as shown in Table 13. Table 13 contains features as the reference considered for comparison, best architecture, number of classes, number of covid samples for analysis, technique used and performance measure in terms of overall Accuracy (A), COVID-19 sensitivity (Spcov), F1-score, COVID-19 positive predictive value (precision) as per metrics taken in reference. Proposed model outperform state-of-the art models in terms of COVID-19 sensitivity, F1-score and PPVcov (precision) in 3-way and 4-way classification because of image enhancement effects. It is advantageous because of better pre-processing of input samples with TVF + Gamma and gamma correction itself. Alone, gamma correction improves sensitivity of virus class and normal class and TVF improves COVID-19 sensitivity as well. Like major studies, our study considers only PA view images of CXR for COVID-19 analysis in 3-way classification whereas 4-way classification considers PA, AP and AP supine view of COVID-19 samples. We observed that with PA view sensitivity of proposed model is 100% but it reduces to 98.72% by considering AP and AP supine view samples. Thus, hybrid of both Gamma correction and total variation filter improves overall performance of models. In 3-way classification, proposed DenseNet201 with TVF + Gamma outperforms [12, 14, 33, 40, 48, 50, 53, 67] under similar scenario covid vs normal vs pneumonia in accuracy, sensitivity, F1-score and positive predictive value. Accuracy of these model is less because of overlapping between COVID-19 and Viral pneumonia due to their similar characteristics. Success of our proposed DenseNet201 model lies in image pre-processing pipeline as there is no overlapping between viral pneumonia and COVID-19 pneumonia. Also, proposed model is less accurate than [1, 5, 71]. As in [5], proposed model compared COVID-19 samples with normal and bacterial pneumonia samples with more training samples than this study. Accuracy improves because of less overlapping between bacterial, covid samples and more bacterial training samples than this study. In [71], study is carried out using large dataset of 13,975 CXR images comprises a corpus of 268 COVID samples, 5538 pneumonia and 8066 normal cases. Instead of having such large dataset for better training but study considers only 100 samples from each class for testing because of more focus on better classification of COVID cases. Accuracy of normal and pneumonia class this study is 95% and 94% but COVID accuracy is 91% which is less than our proposed model. We believe usage of image preprocessing will further improve accuracy of pneumonia and normal samples due to large training samples. In [1], study was carried out using corpus of 105 COVID, 11 SARS and 80 normal samples by using concept of class decomposition that improves accuracy of this model. Similarly in 4-way classification our proposed MobileNet with TVF + Gamma model outperforms state-of-the-art models in similar scenario of covid vs virus vs bacteria vs normal under similar evaluation measures considered in the study. This study enlightens notable accuracy, COVID-19 sensitivity and PPV boost in model performance after application of image enhancement and image denoising techniques.
In this study, we gain insight of few observations on the basics of literature review and performance analysis of various models. Usage of Deep CNN models i.e., DenseNet series and MobileNet series models give better performance as compared to other models because due to more depth it is better in distinguishing overlapped features of COVID-19 and virus pneumonia. DenseNet and MobileNet series models with TVF + Gamma models has higher confidence score as output probability and interpretation for COVID-19 pathology class than all other CNN models. Also, it is better to use weighted class loss function as compared to undersampling or oversampling of training samples [53]. Also, studies depict that classification accuracy is better in covid vs bacterial pneumonia than covid vs viral pneumonia due to similarity in covid and viral pneumonia. But usage of proposed image preprocessing pipeline with deep CNN models improves performance by distinguishing overlapping in viral pneumonia and covid pneumonia more accurately. With gamma correction models gains more advantage than CLAHE and HE as enhancement techniques and improves sensitivity value of all classes even at higher threshold level. Although, no single model performs well on all datasets but with image preprocessing pipeline achieves better performance than other state-of-the-art studies. However, results of this study can be generalizable to even more large datasets for diagnosis of COVID-19 in terms of time and performance as compared with plain image analysis.
INSPECTION OF MODEL’S DECISION by grad-CAM
To demonstrate output qualitatively, we visualize learning of proposed model with gradient weighted class activation mapping (Grad-CAM). Due to very complex architectures of deep learning models, it is harder to interpret why the model has classified given input to particular class. Grad-CAM aimed at increasing interpretability by looking at where model has focused while classifying given input image to particular class. In Fig. 16, we have shown the effects of different image enhancement techniques on Grad-CAM attention visualization of correctly classified samples by DenseNet201, VGG19, MobileNet and denseNet169 models. On given input test image, we use Grad-CAM technique to build heatmap by extracting class specific gradients flowing into last convolutional layer corresponding every class showing significant regions considered by model for predicting certain pathological condition. This is indeed important to understand whether our model focus on right regions while classifying given input image so that experts could validate the prediction process. Grad-CAM visualization in Fig. 16 is shown on two images. In Fig. 16, first row shows effect of various image enhancement techniques on plain input test image 1 corresponding to that Grad-CAM visualization has performed. Second and third row shows silancy map corresponding to image 1 and another considered image 2 by best performing DenseNet201 model trained using image enhancement techniques shown in first row. Fourth, fifth and sixth row shows heatmap generated by models VGG19, DenseNet169 and MobileNet with image enhancement techniques used among CLAHE, gamma correction and TVF + Gamma. In saliency map, Red signifies region of more importance and each Grad-CAM generated image has confidence score at the top. We observed from the study that application of enhancement techniques helps model to focus more on lung regions due to changes in the distribution of pixels. Moreover, we observe that with gamma correction models focus more on the lung regions as marked by radiologists better than HE and CLAHE. Also, DenseNet201 model gives higher confidence score to correctly classified images when preprocessing pipeline is Gamma and TVF + Gamma. Even disadvantage with plain CXR image is that models may consider sometimes other irrelevant features rather than lungs, so it is better to segment lung regions. Usage of Gamma and TVF + Gamma not only improves performance of plain images but also applicable to segmented lung images because it denoises and enhances lung regions for better analysis. Application of gamma correction improves performance and reduces time of convergence whereas hybrid of TVF + Gamma improves COVID-19 accuracy and confidence score for classification of given COVID infected image.
Fig. 16.
Effects of different image enhancement techniques on attention visualization by Grad-CAM of correctly classified COVID-19 CXR images using different models
Discussion
This experimental study discusses the importance of image denoising and image enhancement approaches on the performance of ten deep learning models using two CXR datasets. This study concludes that deep learning models extract more relevant features if better pre-processing pipeline is utilized before feature extraction. Proposed study discloses that the application of image enhancement approach as gamma correction and TVF + Gamma improves interpretability and performance of state-of-the-art approaches. Individually Gamma correction is better for BP, VP and normal classes whereas CLAHE is better for identification of COVID-19 features. Out of 20 trails with 10 CNN feature extractors, our proposed pre-processing pipeline wins for 17 trails. It minimizes the overlapping of other classes to COVID-19 class that improves precision and COVID-19 sensitivity. The results of proposed methodology indicate, when CXR images are pre-processed with TVF + Gamma and features extraction is performed using MobileNet model achieves higher accuracy score of 93.25% followed by DenseNet201 with accuracy score of 93.10%. Proposed model achieves zero overlapping with 100% precision for COVID-19 class. Also, we perceive that deep CNN models like MobileNet, DenseNet series have higher sensitivity, accuracy and interpretation with proposed pre-processing pipeline as compared to other baseline models. Similarly, in our case deep CNN models like MobileNet and DenseNet201 achieves highest accuracy and sensitivity value among other models.
Recommendations for authors
For authors, we recommend preparing their publication atleast by considering the mandatory suggestions with proper documentation so that their modelling approach results can be reproduced. Our suggestions are based on the limitations of existing state-of-the-art studies. Firstly, authors should properly specify their universal pre-processing approach like resizing, type of normalization, cropping and formatting in sufficient detail. As good quality of data leads to better interpretation and analysis of the model. Thus, authors need to focus more on advanced pre-processing techniques in addition to modelling. Regarding pre-processing in addition to image denoising and enhancement approaches, image segmentation can be implemented to extract relevant lung regions for better interpretation of models. Secondly, COVID-19 dataset constitutes of a smaller number of samples as compared to other classes of comparison. Thus, it is mandatory to handle class imbalance either by generating synthetic data or by weighted cross entropy loss for better training of minority class. Proper documentation of training and testing parameters like learning rate, batch size, number of epochs etc. is required to reproduce results and make models operational in future. Also, hyperparameter setting should be carefully done by considering validation dataset but test dataset is used for final evaluation only. Finally, authors should clearly mention how they selected their final model and how their model performs for each of the class by class-wise sensitivity analysis.
Conclusions
This article presents a comparative study of various combinations of image enhancement and image denoising approaches using ten different state-of-the-art transfer learning models for automated and early diagnosis of COVID-19. Due to exponential behaviour of COVID-19 epidemic, it completely shattered healthcare sector worldwide because of its unidentified nature and similarity with other pathology classes in early stages. We believe that timely inference and correct diagnosis of type of disease can save millions of lives every year. Thus, computer-aided diagnosis significantly assists radiologists to capture better images and Real time identification of COVID-19 or pneumonia just after acquisition. Image preprocessing and denoising play very important role in improving the quality of image that improves performance and reduces the time required for training models. Image preprocessing improves the diagnostics power of CAD tools to identify disease in early stages more accurately. It will be useful for screening process at airports for early diagnosis of COVID-19, Pneumonia and others type of disease. After the extensive number of trials, this study concludes that deep CNN models help in the identification of COVID-19 pathology more accurately as compared to shallow CNN models. Also, series of DenseNet and MobileNet models have higher potential in detecting abnormalities in chest x-ray images of COVID-19 among other pathology classes with higher confidence score. In this study, we conclude that individually gamma correction is best image enhancement technique in terms of performance as well as time of convergence. But application of TVF along with gamma further improves accuracy, COVID-19 sensitivity and confidence score of prediction as well. In this study, our proposed image preprocessing pipeline tested successfully on two datasets with improvements in accuracy, sensitivity and confidence score better then HE, CLAHE and gamma correction. This study concludes best model as MobileNet with TVF + Gamma outperforms all other models and enhancement combinations in terms of accuracy and class wise sensitivity. Our model improves COVID-19 sensitivity as well as there is no overlapping of other classes to COVID-19 class. The outcomes of this study can be generalizable with better improvements on large datasets and applicable to CAD systems. To interpret output qualitatively, we visualize learning of model with gradient weighted class activation mapping (Grad-CAM). Finally, in global perspectives after critically analysing the related literature and obtained results, we can say that application of image enhancement techniques with deep learning models has enormous improvements in classification of COVID-19 patients against others. In addition, series of deep CNN models has higher sensitivity to COVID-19 and likely to be more supportive in early classification of COVID-19 cases among others. In future, we hope to use segmentation models to segment lung regions from large CXR datasets and to train deep CNN models like DenseNet, GoogleNet, MobileNet and ResNet using proposed image preprocessing pipelines.
Acknowledgements
The authors are highly thankful to the Editor-in-Chief and anonymous reviewers, whose valuable comments helped us greatly to improve this article. The first author is grateful to the University Grants Commission (UGC), New Delhi, India, for the research fellowship provided through UGC-JRF Scheme. The second author greatly acknowledge the IoE grant of the Banaras Hindu University.
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ajay Sharma, Email: ajay.sharma17@bhu.ac.in.
Pramod Kumar Mishra, Email: mishra@bhu.ac.in.
References
- 1.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2021;51(2):854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdullah-Al-Wadud M, Kabir MH, Dewan MAA, Chae O. A dynamic histogram equalization for image contrast enhancement. IEEE Trans Consum Electron. 2007;53(2):593–600. doi: 10.1109/TCE.2007.381734. [DOI] [Google Scholar]
- 3.Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A. Covid-caps: a capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recogn Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand A, Pugalenthi G, Fogel GB, Suganthan PN. An approach for classification of highly imbalanced data using weighting and undersampling. Amino Acids. 2010;39(5):1385–1391. doi: 10.1007/s00726-010-0595-2. [DOI] [PubMed] [Google Scholar]
- 5.Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 43(2):635–640 [DOI] [PMC free article] [PubMed]
- 6.Arias-Londoño JD, Gomez-Garcia JA, Moro-Velázquez L, Godino-Llorente JI (2020) Artificial intelligence applied to chest X-ray images for the automatic detection of COVID-19. A thoughtful evaluation approach. IEEE Access [DOI] [PMC free article] [PubMed]
- 7.Ayaz M, Shaukat F, Raja G (2021) Ensemble learning based automatic detection of tuberculosis in chest X-ray images using hybrid feature descriptors. Phys Eng Sci Med, 1-12 [DOI] [PMC free article] [PubMed]
- 8.Calderon-Ramirez S, Yang S, Moemeni A, Colreavy-Donnelly S, Elizondo DA, Oala L, ..., Molina-Cabello MA (2021) Improving uncertainty estimation with semi-supervised deep learning for COVID-19 detection using chest X-ray images. IEEE Access [DOI] [PMC free article] [PubMed]
- 9.Chakraborty M, Dhavale SV, Ingole J (2021) Corona-Nidaan: lightweight deep convolutional neural network for chest X-Ray based COVID-19 infection detection. Appl Intell 51(5):3026–3043 [DOI] [PMC free article] [PubMed]
- 10.Chaturvedi SS, Tembhurne JV, Diwan T (2020) A multi-class skin cancer classification using deep convolutional neural networks. Multimed Tools Appl 79(39):28477–28498
- 11.Chen YH, Hong WC, Shen W, Huang NN (2016) Electric load forecasting based on a least squares support vector machine with fuzzy time series and global harmony search algorithm. Energies 9(2):70
- 12.Civit-Masot J, Luna-Perejón F, Domínguez Morales M, Civit A (2020) Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Appl Sci 10(13):4640
- 13.Cohen JP, Morrison P, Dao L (2020) Covid-19 image data collection. arXiv preprintarXiv:2003.11597
- 14.Das NN, Kumar N, Kaur M, Kumar V, Singh D (2020) Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. Irbm [DOI] [PMC free article] [PubMed]
- 15.Dhiman G, Chang V, Kant Singh K, Shankar A (2021) ADOPT: automatic deep learning and optimization-based approach for detection of novel coronavirus COVID-19 disease using X-ray images. J Biomol Struct Dyn, 1-13 [DOI] [PMC free article] [PubMed]
- 16.El-Kenawy ESM, Mirjalili S, Ibrahim A, Alrahmawy M, El-Said M, Zaki RM, Eid MM (2021) Advanced meta-heuristics, convolutional neural networks, and feature selectors for efficient COVID-19 X-ray chest image classification. IEEE Access 9:36019–36037 [DOI] [PMC free article] [PubMed]
- 17.Eslami M, Tabarestani S, Albarqouni S, Adeli E, Navab N, Adjouadi M. Image-to-images translation for multi-task organ segmentation and bone suppression in chest x-ray radiography. IEEE Trans Med Imaging. 2020;39(7):2553–2565. doi: 10.1109/TMI.2020.2974159. [DOI] [PubMed] [Google Scholar]
- 18.Fan GF, Qing S, Wang H, Hong WC, Li HJ (2013) Support vector regression model based on empirical mode decomposition and auto regression for electric load forecasting. Energies 6(4):1887–1901
- 19.Fang Z, Ren J, MacLellan C, Li H, Zhao H, Hussain A, Fortino G (2021) A novel multi-stage residual feature fusion network for detection of COVID-19 in chest X-ray images. IEEE Trans Mol Biol Multi-Scale Commun 8(1):17–27 [DOI] [PMC free article] [PubMed]
- 20.Gao L, Zhang L, Liu C, Wu S (2020) Handling imbalanced medical image data: A deep-learning-based one-class classification approach. Artif Intell Med 108:101935 [DOI] [PMC free article] [PubMed]
- 21.Goel T, Murugan R, Mirjalili S, Chakrabartty DK (2020) OptCoNet: an optimized convolutional neural network for an automatic diagnosis of COVID-19. Appl Intell, 1-16 [DOI] [PMC free article] [PubMed]
- 22.Hemdan EED, Shouman MA, Karar ME (2020) COVIDz-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055
- 23.Huan EY, Wen GH. Transfer learning with deep convolutional neural network for constitution classification with face image. Multimed Tools Appl. 2020;79:1–15. doi: 10.1007/s11042-019-08376-5. [DOI] [Google Scholar]
- 24.Ibrahim AU, Ozsoz M, Serte S, Al-Turjman F, Yakoi PS (2021) Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cogn Comput 1–13 [DOI] [PMC free article] [PubMed]
- 25.Ikhsan IAM, Hussain A, Zulkifley MA, Tahir NM, Mustapha A (2014) An analysis of x-ray image enhancement methods for vertebral bone segmentation. In 2014 IEEE 10th international colloquium on signal processing and its applications (pp. 208-211). IEEE
- 26.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, ..., Ng AY (2019) Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence (Vol. 33, No. 01, pp. 590–597)
- 27.Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Informatics in medicine unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quantitative imaging in medicine and surgery. 2014;4(6):475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ketu S, Mishra PK (2020) Enhanced Gaussian process regression-based forecasting model for COVID-19 outbreak and significance of IoT for its detection. Appl Intell, 1-21 [DOI] [PMC free article] [PubMed]
- 30.Ketu S, Mishra PK (2020) A hybrid deep learning model for COVID-19 prediction and current status of clinical trials worldwide. Computers, Materials and Continua 66(2)
- 31.Ketu S, Mishra PK (2021) Hybrid classification model for eye state detection using electroencephalogram signals. Cognitive Neurodynamics pp:1–18 [DOI] [PMC free article] [PubMed]
- 32.Khan S, Islam N, Jan Z, Din IU, Rodrigues JJC (2019) A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett 125:1–6
- 33.Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Prog Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khatami F, Saatchi M, Zadeh SST, Aghamir ZS, Shabestari AN, Reis LO, Aghamir SMK. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-80061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Wang Y, Wang S, Wang J, Liu J, Jin Q, Sun L. Multiscale attention guided network for COVID-19 diagnosis using chest X-ray images. IEEE Journal of Biomedical and Health Informatics. 2021;25(5):1336–1346. doi: 10.1109/JBHI.2021.3058293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li MW, Wang YT, Geng J, Hong WC. Chaos cloud quantum bat hybrid optimization algorithm. Nonlinear Dynamics. 2021;103(1):1167–1193. doi: 10.1007/s11071-020-06111-6. [DOI] [Google Scholar]
- 37.Loey M, Smarandache F, Khalifa MNE (2020) Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry 12(4):651
- 38.Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020;122:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangal A, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora C (2020) CovidAID: COVID-19 detection using chest X-ray. arXiv preprint arXiv:2004.09803
- 40.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ (2020) Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal 65:101794 [DOI] [PMC free article] [PubMed]
- 41.Mishra S. Financial management and forecasting using business intelligence and big data analytic tools. Int J Financ Eng. 2018;5(02):1850011. doi: 10.1142/S2424786318500111. [DOI] [Google Scholar]
- 42.Mishra S, Tripathi AR. IoT platform business model for innovative management systems. Int J Financ Eng (IJFE) 2020;7(03):1–31. [Google Scholar]
- 43.Mishra S, Tripathi AR. Literature review on business prototypes for digital platform. J Innov Entrepreneurship. 2020;9(1):1–19. doi: 10.1186/s13731-020-00126-4. [DOI] [Google Scholar]
- 44.Mishra S, Tripathi AR. AI business model: an integrative business approach. J Innov Entrepreneurship. 2021;10(1):1–21. doi: 10.1186/s13731-021-00157-5. [DOI] [Google Scholar]
- 45.Mishra S, Triptahi AR. Platforms oriented business and data analytics in digital ecosystem. Int J Financ Eng. 2019;6(04):1950036. doi: 10.1142/S2424786319500361. [DOI] [Google Scholar]
- 46.Mooney P (2018) Kaggle chest x-ray images (pneumonia) dataset. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. [Online; accessed Dec 5, 2020]
- 47.Motwani MC, Gadiya MC, Motwani RC, Harris FC (2004) Survey of image denoising techniques. In proceedings of GSPX (Vol. 27, pp. 27-30). Proceedings of GSPX
- 48.Mukherjee H, Ghosh S, Dhar A, Obaidullah SM, Santosh KC, Roy K (2021) Shallow convolutional neural network for COVID-19 outbreak screening using chest X-rays. Cogn Comput:1–14 [DOI] [PMC free article] [PubMed]
- 49.Oh Y, Park S, Ye JC (2020) Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imaging 39(8):2688–2700 [DOI] [PubMed]
- 50.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perumal V, Narayanan V, Rajasekar SJS (2021) Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl Intell 51(1):341–358 [DOI] [PMC free article] [PubMed]
- 52.Punn NS, Agarwal S (2021) Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. Appl Intell 51(5):2689–2702 [DOI] [PMC free article] [PubMed]
- 53.Rahman S, Sarker S, Miraj MAA, Nihal RA, Nadimul Haque AKM, Noman AA (2021) Deep learning–driven automated detection of Covid-19 from radiography images: A comparative analysis. Cogn Comput 1–30 [DOI] [PMC free article] [PubMed]
- 54.Rajinikanth V, Joseph Raj AN, Thanaraj KP, Naik GR. A customized VGG19 network with concatenation of deep and handcrafted features for brain tumor detection. Appl Sci. 2020;10(10):3429. doi: 10.3390/app10103429. [DOI] [Google Scholar]
- 55.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, ..., Ng AY (2017) Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225
- 56.Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz CP, Patel BN, Yeom KW, Shpanskaya K, Blankenberg FG, Seekins J, Amrhein TJ, Mong DA, Halabi SS, Zucker EJ, Ng AY, Lungren MP. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15(11):e1002686. doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rehman A, Saba T. Neural networks for document image preprocessing: state of the art. Artif Intell Rev. 2014;42(2):253–273. doi: 10.1007/s10462-012-9337-z. [DOI] [Google Scholar]
- 58.Sedik A, Hammad M, Abd El-Samie FE, Gupta BB, Abd El-Latif AA (2022) Efficient deep learning approach for augmented detection of coronavirus disease. Neural Comput Applic 34(14):11423–11440 [DOI] [PMC free article] [PubMed]
- 59.Sharma A, Mishra PK. State-of-the-art in performance metrics and future directions for data science algorithms. J Sci Res. 2020;64(2):221–238. [Google Scholar]
- 60.Sharma A, Mishra PK (2021) Deep learning approaches for automated diagnosis of Covid-19 using imbalanced training CXR data. In: International Conference on Advanced Network Technologies and Intelligent Computing. Springer, Cham, pp 453–472
- 61.Sharma A, Mishra PK (2022) Performance analysis of machine learning based optimized feature selection approaches for breast cancer diagnosis. Int J Inf Technol 14(4):1949–1960
- 62.Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu KI, Matsui M, Fujita H, Kodera Y, Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists' detection of pulmonary nodules. Am J Roentgenol. 2000;174(1):71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 63.Srivastava A, Mishra PK. A survey on WSN issues with its heuristics and Meta-heuristics solutions. Wirel Pers Commun. 2021;121:1–70. doi: 10.1007/s11277-021-08659-x. [DOI] [Google Scholar]
- 64.Srivastava A, Mishra PK (2022) Covid-MANet: Multi-task attention network for explainable diagnosis and severity assessment of COVID-19 from CXR images. Pattern Recognit 108826 [DOI] [PMC free article] [PubMed]
- 65.Stein A (2018) Pneumonia dataset annotation methods. rsna pneumonia detection challengediscussion. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/discussion/ (2020). [Online; accessed Dec 5, 2020]
- 66.Tang S, Wang C, Nie J, Kumar N, Zhang Y, Xiong Z, Barnawi A (2021) EDL-COVID: Ensemble deep learning for COVID-19 case detection from chest X-ray images. IEEE Trans Industr Inform 17(9):6539–6549 [DOI] [PMC free article] [PubMed]
- 67.Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos, Solitons Fractals. 2020;140:110122. doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. Covidgan: data augmentation using auxiliary classifier Gan for improved covid-19 detection. Ieee Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waller JV, Kaur P, Tucker A, Lin KK, Diaz MJ, Henry TS, Hope M. Diagnostic tools for coronavirus disease (COVID-19): comparing CT and RT-PCR viral nucleic acid testing. Am J Roentgenol. 2020;215(4):834–838. doi: 10.2214/AJR.20.23418. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2097-2106)
- 71.Wang L, Lin ZQ, Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.WHO: Coronavirus disease 2019 (covid-19) Dashboard (2021) https://covid19.who.int/. [Online; accessed Feb 7, 2021]
- 73.Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) Covid-19 screening on chest x-ray images using deep learning-based anomaly detection. arXiv preprint arXiv:2003.12338
- 74.Zhou T, Lu H, Yang Z, Qiu S, Huo B, Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl Soft Comput. 2021;98:106885. doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zreik M, Van Hamersvelt RW, Wolterink JM, Leiner T, Viergever MA, Išgum I (2018) A recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Trans Med Imaging 38(7):1588–1598 [DOI] [PubMed]