Abstract
AniA (formerly Pan1) is the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. AniA has been shown to be a major antigen in patients with gonococcal disease, and we have been studying its regulation in order to understand the gonococcal response to anaerobiosis and its potential role in virulence. This study presents a genetic analysis of aniA regulation. Through deletion analysis of the upstream region, we have determined the minimal promoter region necessary for aniA expression. This 130-bp region contains a sigma 70-type promoter and an FNR (fumarate and nitrate reductase regulator protein) binding site, both of which are absolutely required for anaerobic expression. Also located in the minimal promoter region are three T-rich direct repeats and several potential NarP binding sites. This 80-bp region is required for induction by nitrite. By site-directed mutagenesis of promoter sequences, we have determined that the transcription of aniA is initiated only from the sigma 70-type promoter. The gearbox promoter, previously believed to be the major promoter, does not appear to be active during anaerobiosis. The gonococcal FNR and NarP homologs are involved in the regulation of aniA, and we demonstrate that placing aniA under the control of the tac promoter compensates for the inability of a gonococcal fnr mutant to grow anaerobically.
Neisseria gonorrhoeae, like other pathogenic bacteria, regulates the expression of proteins in response to environmental stimuli. While previously considered to be an obligate aerobe (39), N. gonorrhoeae has been shown to grow anaerobically in the laboratory when provided with nitrite as a terminal electron acceptor for anaerobic respiration (24). We are particularly interested in how the gonococcus alters protein expression in response to anaerobiosis. It has been found that at least three gonococcal outer membrane proteins (OMPs) are induced and that at least five OMPs are repressed by anaerobic growth in gonococcal strain F62. AniA (formerly Pan1), the major anaerobically induced OMP, is tightly regulated, and its expression is restricted to anaerobically grown cells (7).
Western blot analyses with sera from patients with gonococcal disease indicated that AniA was a major antigen in patients with both complicated and uncomplicated diseases (8). These results suggested that AniA is expressed in the host and that the gonococcus encounters an anaerobic environment during infection. An antigenically related anaerobically induced OMP was detected in all strains of gonococci tested and in a number of commensal Neisseria strains but was poorly expressed in N. meningitidis strains (19).
The initial cloning and characterization of the aniA gene have been reported (20). In that study, Northern analysis demonstrated the lack of an aniA message in aerobically grown cells. The primer extension data from anaerobically grown cells suggested the presence of two RNA transcripts differing in length by only 9 bp. Consistent with this finding, two overlapping corresponding promoter sequences were proposed. The −10 sequence of the promoter for the longer, less abundant message was homologous to the sequence of Escherichia coli ς70 promoters, while the sequence of the promoter for the shorter, more abundant message shared 11 of 14 bases with the E. coli gearbox promoter consensus sequence (20). Gearbox promoters were named for their characteristic of producing a gene product at a rate inversely proportional to the growth rate of the cell. These promoters are induced during the stationary phase in E. coli, and some are dependent on ςs, a stationary-phase sigma factor (1, 2, 25, 40). When aniA was initially sequenced, there were no homologous proteins for AniA in the databases; it has since been reported that AniA shares significant identity with copper-containing nitrite reductases (6, 29).
In this paper, we present the nucleotide sequence of the region upstream of the aniA gene and an initial characterization of the elements involved in the regulation of aniA in strain F62.
(This work was presented in part at the 97th General Meeting of the American Society for Microbiology, Miami Beach, Fla., 4 to 8 May 1997 [33a].)
MATERIALS AND METHODS
Growth of gonococcal strains.
All gonococcal strains were derived from strain F62 and were grown on plates containing GC medium base (Difco Laboratories, Detroit, Mich.) with 1% Kellogg’s supplement (GCK) (23). When necessary, chloramphenicol was added at 1 μg ml−1, erythromycin was added at 2 μg ml−1, or kanamycin was added at 40 μg ml−1. Aerobic cultures were grown at 37°C in a 5% CO2 incubator. Anaerobic cultures were incubated in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.) at 37°C for 20 h in an atmosphere of 85% N2–10% H2–5% CO2. Nitrite was provided for anaerobically grown cultures by placing 40 μl of a 20% (wt/vol) NaNO2 solution on a sterile cellulose disk in the center of a plate. Cultures with nitrite grow in a characteristic halo around the nitrite disk, while cultures without nitrite remain viable but do not grow (35).
Gonococcal transformation.
A light suspension of type 1 cells (23) was made with 2 ml of GCK broth containing 0.042% NaHCO3 and 10 mM MgCl2. Purified DNA or a ligation mixture was added, and cultures were grown for 5 to 6 h with shaking at 37°C. Cultures were then plated on GCK plates containing the appropriate antibiotic.
Extraction of gonococcal chromosomal DNA.
Gonococci were harvested from plates, suspended in 0.5 ml of 50 mM Tris-HCl (pH 8.5)–50 mM EDTA–15% sucrose–1 mg of lysozyme ml−1, and incubated at room temperature for 10 min. Sodium dodecyl sulfate was added to 0.4% to lyse the cells, and the solution was mixed by inversion of the tube. After incubation for 5 min at 70°C, 100 μl of 5 M potassium acetate was added, and the mixture was chilled on ice for 30 min. The precipitated proteins were pelleted by centrifugation at 12,000 × g for 15 min. The supernatant containing the DNA was removed to a clean tube, to which 2 volumes of cold 95% ethanol was added. This mixture was centrifuged for 5 min at maximum speed in an SS-34 rotor of an RC-5B centrifuge (Sorvall, Newtown, Conn.). The supernatant was decanted, and the pellet was allowed to dry. The pellet was then resuspended in 50 μl of TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA [pH 8.0]) containing 10 μg of RNase A ml−1.
PCR.
The primers used for the creation of reporter constructs by PCR are listed in Table 1.
TABLE 1.
Primer sequences used in the construction of deletions and mutations
| Primer | Sequence (5′ to 3′)a | Locationb |
|---|---|---|
| P1 | CCTTTTAAGGATCCGCCGTCTGAAAATTCACAAAATATGAATGTTA | −364 |
| P2 | GGCTAATGGATCCCGTTTCATAATGTTTTCCTTTTG | +31 |
| P3 | GATGAAACGCCGAGTTAACGCC | +470c |
| P4 | ACTTAAGGATCCATGCCCGCAATGGGACAACC | −15 |
| P5 | ATGAATGGATCCGATTGTAGCATGGTTTACCG | −250 |
| P6 | ACCGCAATTACACACCAAGTTCTTAACTAATC | −7 |
| P7 | TGTGTAATTGCGGTTGTCCCATTGCGGGC | +15 |
| P8 | TCACACGTTCATCTTAACTAATCC | +15 |
| P9 | AAGATGAACGTGTGATTATGCGGTTG | −19 |
| P10 | CAGGTTATTTGACGTAAATTAAAATGC | −31 |
| P11 | CGTCAAATAACCTGTAAAATATGAAC | −69 |
| P12 | AATCTCTAGACACCAAGTTCTTAACTAATC | +14 |
| P13 | TGGTGTCTAGAGATTATGCGGTTGTCCCATTG | −27 |
| P14 | AAAGGATCAAATAATGAAACGGGATCCCGTCG | +65C |
| P15 | GATCCTTTGTATAGAAAAGTAGGGGGGATTAG | +9 |
| P16 | CTTAAAATTTTATGCCCGCAATGGGAC | −19 |
| P17 | GGGCATAAAATTTTAAGTCAAATAATTC | −46 |
| P18 | CCGGAGGGATCCGCAAATCAGCCTATTCATTG | −111 |
| P19 | ATGAATTATCTAACTTAAATTAAAATGC | −31 |
| P20 | TAAGTTAGATAATTCATAAAATATGAAC | −69 |
| P21 | TTTTATGGATCCTTTGACTTAAATTAAAATGC | −31 |
| P22 | GTCATTTTGGATCCATATTTTATGAATTATTTGAC | −45 |
| P23 | ATCGTACTCGAGCCAGTAGTTCGGGCGGCCTTT | +2650 |
| P24 | TAATGTCTCGAGTTTGTAAGAAAAGTAGGGGGGAT | +12 |
| P25 | ACAAAAGGATCCCATTATGAAACGCCAAGCAT | +62 |
| P26 | TTATAAGGATCCAGAAGCGTCATTTTAAGTTC | −65 |
Changes in the sequence from the wild-type sequence are indicated by bold type.
Location of the 3′ end of the primer relative to the sigma 70-type promoter transcription start site.
Relative to the sigma 70-type promoter transcription start site in the lacZ fusion.
Nuclease protection assay.
A PCR product amplified from pLES940 (36) and containing approximately 350 bp of the aniA upstream region, the junction, and about 20 bp of lacZ was cloned into the Bluescript II SK(+) phagemid (Stratagene, LaJolla, Calif.), which had been digested with XbaI and EcoRI. The phagemid containing the insert was linearized by restriction digestion with EcoRI. An [α-32P]UTP-labeled antisense RNA probe was generated by in vitro transcription with T3 RNA polymerase and a MAXIscript kit (Ambion Inc., Austin, Tex.) in accordance with the manufacturer’s instructions. An RNeasy total RNA kit (Qiagen, Chatsworth, Calif.) was used to prepare total RNA from RUG7001 (wild-type aniA′-′lacZ fusion in a wild-type background; see below) harvested from anaerobic plates with nitrite disks.
Nuclease protection was performed with an RPA II kit (Ambion), substituting a 1:50 dilution of S1 nuclease for the RNase and 10× S1 nuclease buffer (300 mM Na acetate [pH 4.6], 10 mM Zn acetate, 500 mM NaCl, 50% glycerol) for buffer Bx. Hybridization was performed overnight at 42°C, and the nuclease reaction was carried out at 37°C for 45 min. The reaction products were electrophoresed along with a sequence of the probe made with a T3 primer and the dsDNA cycle sequencing system (Life Technologies Inc., Gaithersburg, Md.).
Construction of lacZ fusions.
Deletions and mutations of the aniA upstream region were created by PCR with the primer pairs listed in Table 2. Translational lacZ fusions were made with pLES94 (36). Gonococcal strain F62 chromosomal DNA was used as the template for PCR. PCR products and pLES94 were cut with BamHI. Digested insert and plasmid were ligated and transformed into E. coli MC1061. Transformants were selected on Luria-Bertani medium plates containing ampicillin at 100 μg ml−1 and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 40 μg ml−1. After overnight incubation at 37°C, blue colonies were identified, and their plasmids were extracted. Plasmids were checked for the presence and orientation of the insert by PCR. Plasmids containing an insert in the correct orientation were used to transform gonococcal strain F62. Transformants were plated on GCK plates containing chloramphenicol. Chromosomal DNA was prepared from chloramphenicol-resistant colonies, and PCR was used to confirm the presence of the reporter construct. The PCR product was sequenced to ensure that the desired alteration had been made.
TABLE 2.
Primer pairs and plasmids used in the construction of reporter strains
| Strain | Plasmid | Primer pair(s) |
|---|---|---|
| RUG7001 | pLES940 | P1, P2 |
| RUG7004 | pTCH1 | P4, P2 |
| RUG7005 | pTCH2 | P5, P2 |
| RUG7006 | pTCH3 | P1, P7; P6, P2 |
| RUG7007 | pTCH4 | P1, P9; P8, P2 |
| RUG7008 | pTCH5 | P1, P11; P10, P2 |
| RUG7009 | pTCH6 | P1, P13; P12, P2 |
| RUG7012 | pLES943 | P22, P2 |
| RUG7014 | pLES945 | P1, P2 |
| RUG7015 | pLES946 | P1, P2 |
| RUG7020 | pTCH8 | P1, P17; P16, P2 |
| RUG7023 | pTCH9 | P1, P15; P14, P3 |
| RUG7024 | pTCH10 | P18, P2 |
| RUG7040 | pTCH11 | P1, P20; P19, P2 |
| RUG7041 | pTCH12 | P21, P2 |
| RUG7045 | pTCH13 | P26, P2 |
Construction of deletions.
Deletions were created by PCR with primers containing BamHI sites at the desired deletion sites in the aniA upstream region. The inserts were amplified with these upstream primers and primer P2 (Table 2).
Site-directed mutagenesis.
All mutations in the aniA upstream region were created by PCR overlap extension (18, 32). Briefly, two PCRs were used to create 5′ and 3′ fragments whose sequences overlapped by several base pairs at the mutation. Both fragments were used as the template for a third PCR. The plus strand from one fragment and the minus strand from the other fragment annealed, acting as both a primer and a template. When the product was extended, it created the full-length insert; this insert was then amplified with primers P1 and P2, which were included in the PCR. Due to the proximity of the symmetric repeat to the P2 primer site, the pTCH9 insert, which contains a mutation of the symmetric repeat, was created with a plasmid pLES940 preparation as a template and the P3 primer instead of P2.
The reporter constructs in strains RUG7014 and RUG7015 contained the aniA upstream regions amplified from N. meningitidis RUN5645 and RUN5646, respectively (19).
Construction of an aniA null mutant.
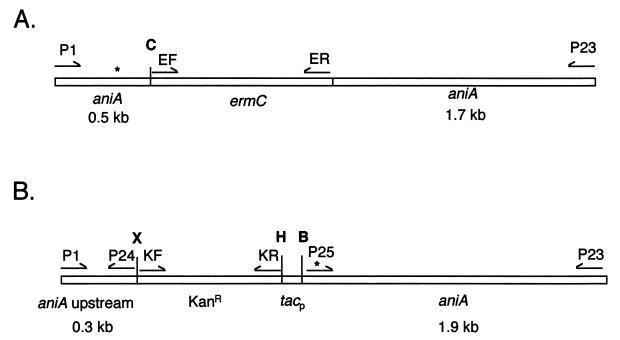
With F62 chromosomal DNA as a template, aniA was amplified by PCR with primers P1 and P23. The PCR product was digested with ClaI, creating two fragments 0.5 and 1.7 kb long. The ends of the 1.7-kb fragment were made flush with the Klenow fragment of E. coli DNA polymerase I. The ermC gene, which confers erythromycin resistance, was amplified from pHSS23 (41) (gift from Joe Dillard) with primers EF (5′-ATGTTTGCGCCGGTATCATCGATAAGCTTTGGC-3′) and ER (5′-CTGGATGATCCTCCAGCGCG-5′) and digested with ClaI and SmaI. The ermC fragment was ligated to the two aniA fragments, a step which resulted in ermC interrupting aniA. The ligation mixture was transformed into strain RUG7001. Transformants were plated on GCK plates containing erythromycin, and the insertion was confirmed by PCR. This process created strain RUG7011 (Fig. 1A).
FIG. 1.
Maps of the aniA null (A) and constitutive (B) mutants. Primers used to amplify the fragments are indicated above the genes. An asterisk denotes the location of the ATG start codon in each construct. Maps are not drawn to scale. C, ClaI; X, XhoI; H, HindIII; B, BamHI.
Construction of a constitutive aniA mutant.
With F62 chromosomal DNA as a template, the upstream region and the coding region of aniA were amplified by PCR into two separate fragments. The upstream region (335 bp) was amplified with primers P1 and P24 and then digested with XhoI. The coding region (1,894 bp), including the ATG start codon, was amplified with primers P25 and P23 and then digested with BamHI. The kanamycin resistance gene was amplified from pHSS23 (41) with primers KF (5′-TGAGCGAAGCTTCGGAAGAGCGCCTGATGCGG-3′) and KR (5′-AGAACTCTCGAGTGAGATCCCCGCGCTGGAGG-3′) and digested with XhoI and HindIII. The tac promoter (Pharmacia Biotech Inc., Piscataway, N.J.) was cloned into the HindIII and BamHI sites of pBluescript II SK(+) (Stratagene). The kanamycin resistance gene was then cloned into the XhoI and HindIII sites, placing it upstream of the tac promoter. The kanamycin resistance gene-tac promoter fragment was amplified by PCR from this construct and digested with XhoI and BamHI. The fragment was ligated to the two aniA fragments, and the ligation mixture was transformed into F62. This step inserted the kanamycin resistance gene-tac promoter fragment between the upstream region and the coding region of aniA and placed the tac promoter and the ribosome binding site in the proper orientation relative to the ATG start codon of the aniA coding region (Fig. 1B).
To transfer constitutive aniA to strain RUG7001, the entire construct was amplified with primers P1 and P23. The PCR product was used to transform RUG7001, creating RUG7035. This PCR product was also transformed into fnr and narP mutants (27) containing the aniA′-′lacZ fusion (creating strains RUG7025 and RUG7039, respectively). For all strains containing the constitutive aniA mutation, transformants were selected on GCK plates containing kanamycin, and the presence of the kanamycin resistance gene and the tac promoter was confirmed by PCR.
β-gal assays.
Anaerobically grown and anaerobically incubated cultures were assayed for β-galactosidase (β-gal) activity by the method of Miller (30). For β-gal assays with plate cultures, the plates were inoculated with 100 μl of a cell suspension with an A600 of approximately 10, and the incubation time was standardized to 20 h. These cultures were harvested with sterile swabs and suspended in Z buffer (30). When nitrite disks were used, only the halo of growth around the nitrite disks was harvested; the full plate was harvested for cultures incubated in the absence of nitrite. Samples containing broth were centrifuged to remove the medium, and the cell pellets were resuspended in Z buffer. Cells were lysed with toluene–0.1% sodium dodecyl sulfate and assayed as described (30). Activity is reported in Miller units. Results reported are the averages of at least three independent assays performed in triplicate on each day that the cultures were grown for each strain.
Time course for aniA induction.
Gonococcal cultures were grown in GCK broth containing 0.042% NaHCO3 in a Gyrotory water bath shaker (New Brunswick Scientific Co., Edison, N.J.) at 37°C to an A600 of approximately 1.0. A sample of each culture was taken for initial measurements of β-gal activity. The remaining culture was transferred into the anaerobic chamber, where 3.3 ml was added to 6.6 ml of prereduced supplemented GCK broth in 12-ml serum vials containing stir bars. The cultures were allowed to equilibrate for 15 min on a stir plate at 37°C. NaNO2 (5 mM, final concentration) was added to appropriate vials. All vials were stirred for 5 min, removed from the anaerobic chamber, and placed on a stir plate in a 37°C room. Vial caps were loosened to create an environment with reduced oxygen tension. Samples were removed each hour for 4 h with a needle and syringe through the septa in the caps and assayed for β-gal activity. RUG7001 served as the wild-type control.
Oligonucleotides and DNA sequencing.
All oligonucleotide syntheses and confirmatory DNA sequencing were performed at the University of Rochester Core Nucleic Acid Laboratory.
Molecular biology techniques.
General techniques were performed in accordance with standard protocols (3, 4, 33). Plasmid preparations were obtained with Wizard Plus Minipreps or Wizard Plus SV Minipreps kits (Promega Corp., Madison, Wis.). DNA fragments were purified with a Wizard PCR Preps kit (Promega), by the freeze-squeeze technique, or with a MERmaid kit (Bio 101, Inc., Vista, Calif.).
Nucleotide sequence accession number.
The sequence upstream of aniA has been deposited in GenBank under accession no. AF082184.
RESULTS
Anaerobic growth of N. gonorrhoeae.
Unlike organisms such as E. coli, most strains of gonococci, including F62, do not grow anaerobically in broth cultures. It is possible to grow F62 in broth under oxygen-limiting conditions and to obtain the induction of aniA (21), but in order to study the regulation of aniA under completely anaerobic conditions, it was necessary to utilize plate cultures. It has been reported that gonococci require a continuous supply of low levels of nitrite to support anaerobic growth, and this was obtained by placing a nitrite disk on the plate (24). While not completely ideal conditions, this method produced visible growth, and β-gal assays performed on these cultures were reproducible between assays and laboratory personnel. This method was used in this study as well as those previously reported from our laboratory (7, 8, 19–21, 36).
Sequence upstream of aniA.
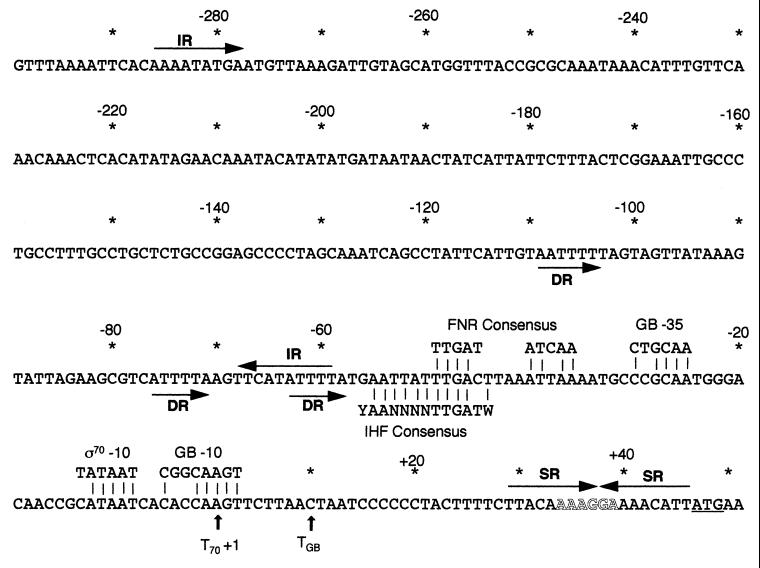
An approximately 350-bp sequence upstream of the aniA start codon was cloned, and analysis of the sequence (Fig. 2) revealed the presence of several interesting motifs. One base upstream of the ATG is a 9-bp symmetric repeat which contains the Shine-Dalgarno sequence (centered at +37.5 from the ς70 transcription start site). There are two overlapping putative promoters; the sequence homologous to E. coli gearbox promoters contains both −10 and −35 consensus sequences, while the sigma 70-type promoter has a potential FNR (fumarate and nitrate reductase regulator protein) consensus binding site (14, 37) in lieu of a −35 sequence (centered at −42.5). An IHF (integration host factor) consensus binding site (10) overlaps the FNR consensus. A T-rich region is located just upstream of the IHF consensus and contains three direct repeats (at −105, −73.5, and −60.5). Finally, there is a 10-bp inverted repeat with approximately 200 bp separating the two halves (at −280.5 and −63.5).
FIG. 2.
Nucleotide sequence of the aniA upstream region. The sequence is numbered in relation to the sigma 70-type promoter transcription start site. The ATG start codon is indicated by underlining, and the ribosome binding site is indicated by outlined letters. The previously reported transcription start sites are indicated by vertical arrows beneath the sequence: T70, start of transcription from the putative ς70 promoter; TGB, start of transcription from the putative gearbox (GB) promoter. The inverted and symmetric repeats (IR and SR, respectively) are labeled with arrows above the sequence, and the direct repeats (DR) are indicated by arrows beneath the sequence. Sequences homologous to known promoters and consensus binding sites are indicated; matching bases within these sequences are indicated with vertical lines.
Nuclease protection assay of aniA transcripts.
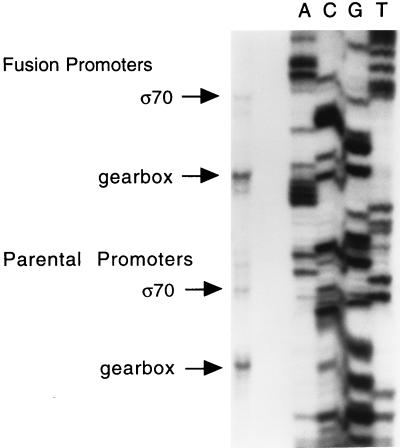
Previous primer extension data suggested the presence of two aniA transcripts (20). To confirm these results, nuclease protection was performed on strain RUG7001, which contains both the parental aniA and the aniA′-′lacZ fusion. The RNA probe was designed to contain the aniA upstream region fused to a portion of lacZ to enable us to detect transcripts from both the parental and the fusion promoters (expected sizes of 46/55 bp and 68/77 bp [gearbox promoter/ς70 promoter], respectively). The results from this assay indicated the presence of two RNA transcripts, one corresponding to each of the putative promoters, and that the shorter fragment accounted for the majority of the aniA message (Fig. 3). The lengths of the protected fragments (46/54 bp and 69/80 bp) correlated well with the primer extension data. In addition, it was evident from these data that the levels of message from the fusion promoter were similar to those from the parental promoter, thus validating our use of the lacZ fusion to measure the induction of aniA. The minor bands present in Fig. 3 can be attributed to the difficulty in obtaining clean results when one is working with very short protected fragments in a nuclease protection assay.
FIG. 3.
Nuclease protection. Autoradiogram of S1 nuclease protection products from both fusion and parental promoters. The larger products of each set correspond to the sigma 70-type promoter, while the smaller products correspond to the gearbox promoter. A sequence generated by the same primer as that used to produce the probe is used as a size marker (lanes A, C, G, and T).
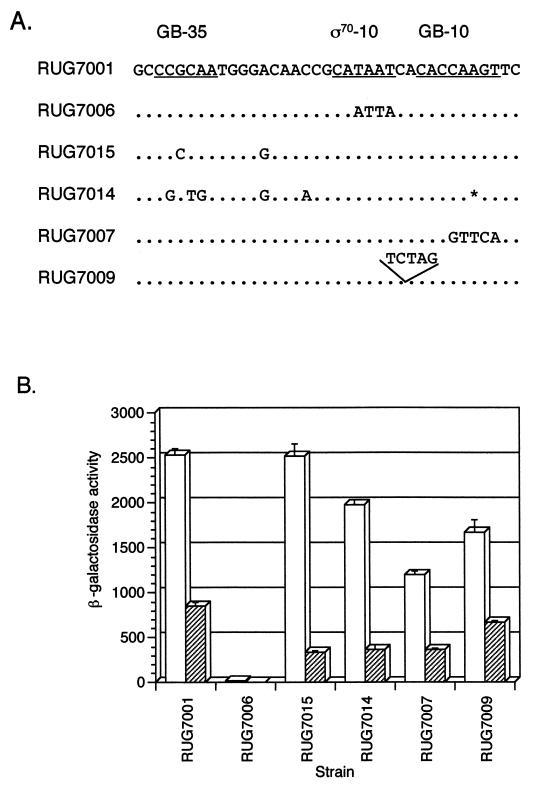
Deletion analysis of the aniA upstream region.
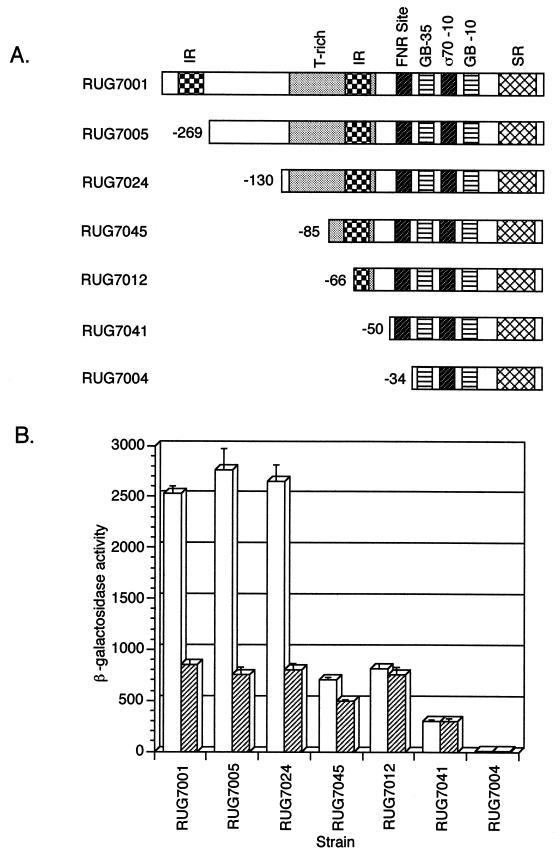
To determine which cis elements are important in the regulation of aniA, we began with a deletion analysis of the upstream region. We had previously cloned 360 bp of the aniA upstream region, including the ATG start codon and the codons for the first 3 amino acids, into vector pLES94, forming a translational lacZ fusion (36). The resulting plasmid, pLES940, was transformed into gonococcal strain F62. The lacZ fusion was integrated into the chromosome by homologous recombination into the proAB genes, creating a single-copy reporter system for the expression of aniA. The resulting strain, RUG7001, contains what is designated the “wild-type” lacZ fusion in a wild-type background. While β-gal activity was negligible in aerobic cultures (<10 Miller units for all strains reported in this study), when cultures of RUG7001 were incubated anaerobically, β-gal activity increased to approximately 900 Miller units. When nitrite was provided to the cultures to allow anaerobic growth, β-gal activity increased about threefold to yield 2,500 Miller units (Fig. 4).
FIG. 4.
Deletion results. (A) Schematic of the deletions constructed in the lacZ reporter fusions (not drawn to scale). The 5′ ends of the deletions are numbered in relation to the sigma 70-type promoter transcription start site. IR, inverted repeats; GB, gearbox promoter sequences; SR, symmetric repeat. (B) Anaerobic β-gal assay results (mean ± standard error of the mean Miller units). Cultures grown with nitrite are indicated by open bars; cultures incubated without nitrite are indicated by hatched bars.
Six lacZ fusions containing aniA deletion mutations were constructed and placed into a wild-type F62 background. These are depicted schematically in Fig. 4A; the β-gal assay results are shown in Fig. 4B. The fusion in RUG7005 contains 334 bp and eliminates the 5′ half of the inverted repeat; β-gal activity was not changed from that of the wild type. The fusion in RUG7024 contains 195 bp of the aniA upstream region and eliminates everything upstream of the T-rich region. As shown in Fig. 4B, β-gal activity was the same as that in the wild type. The fusion in RUG7045 contains 150 bp and deletes the 5′ half of the inverted repeat and the T-rich region located at −102 to −108, while the fusion in RUG7012 contains 131 bp and deletes the 5′ half of the inverted repeat, most of the T-rich region, and a portion of the 3′ half of the inverted repeat; β-gal activity in these two strains grown in the presence of nitrite decreased to approximately the levels seen in the absence of nitrite. This result suggests that an element present in the RUG7024 fusion but not in the RUG7045 fusion is involved in responding to a second induction signal. This signal is most likely nitrite. The RUG7041 fusion contains 115 bp of the aniA upstream region, and the deletion was made just upstream of the FNR consensus binding site, eliminating the inverted repeat and the T-rich region; β-gal activity in this strain was essentially the same in the presence or absence of nitrite but was lower than has been seen previously (300 Miller units). The RUG7004 fusion contains only 99 bp of the aniA upstream region, and the deletion eliminated a required element, as evidenced by the absence of β-gal activity in cultures grown either with or without nitrite.
Mutation of the aniA promoter.
Data from a previous primer extension experiment (20) and the nuclease protection assay (Fig. 3) suggested the presence of two transcripts corresponding to two putative promoters. The shorter, more abundant message corresponded to sequences homologous to E. coli gearbox promoters, and the longer, less abundant message corresponded to a sigma 70-type promoter. We were particularly interested in confirming the presence of two active promoters. To evaluate the roles of the two putative promoters, mutations were made in each promoter separately and were cloned into the lacZ reporter system. To determine the role of the sigma 70-type promoter, the −10 sequence was changed from CATAAT to CAATTA (RUG7006). This change eliminated virtually all activity from the lacZ fusion, indicating an absolute requirement for this promoter (Fig. 5B).
FIG. 5.
Mutation analysis of promoter sequences. (A) Putative −10 and −35 promoter sequences are underlined. Base changes in each strain are indicated. An asterisk represents a base deletion. GB, gearbox. (B) Anaerobic β-gal assay results (mean ± standard error of the mean Miller units). Cultures grown with nitrite are indicated by open bars; cultures incubated without nitrite are indicated by hatched bars.
The mutation analysis of the sigma 70-type promoter suggested that there was a single active promoter, while the primer extension analysis and nuclease protection assay suggested that the gearbox was the major promoter. Although it was impossible to evaluate the gearbox promoter without an active sigma 70-type promoter, we performed mutation analysis on the gearbox sequences to resolve the conflict of data that we had obtained. We initially used two aniA promoters from strains of N. meningitidis. The fusions in RUG7015 and RUG7014 contain sequences amplified from meningococcal strains RUN5646 and RUN5645, respectively. These are N. meningitidis strains which possess the aniA gene (6a) but in which the AniA protein is undetectable by Western blotting (19). When compared to that of F62, the aniA upstream region of these meningococcal strains contained nucleotide differences in the gearbox promoter sequences (Fig. 5A). We hypothesized that these differences may be responsible for the lack of AniA in these strains.
The single-base-pair change in the gearbox −35 sequence in RUG7015 resulted in no decrease in β-gal activity when cultures were grown in the presence of nitrite (Fig. 5B). Cultures incubated anaerobically without nitrite produced about one-third the β-gal activity of the wild type. Similarly, the 3-bp change in the −35 sequence and the single-base-pair deletion in the −10 sequence in RUG7014 did not produce a dramatic decrease in β-gal activity. Site-directed mutagenesis was used to change the gearbox −10 sequence from CACCAAGT to CACGTTCA (RUG7007). This mutation decreased β-gal activity by only one-half. Altering the number of bases between the −10 and −35 sequences is known to significantly affect expression from E. coli promoters (31 and references therein). We therefore introduced an additional 5 bp between the gearbox −10 and −35 sequences without changing the sigma 70-type promoter spacing (RUG7009). This mutation decreased β-gal activity in anaerobically grown cultures by only one-third. All of these results indicate that the gearbox promoter is not an active promoter during anaerobic growth.
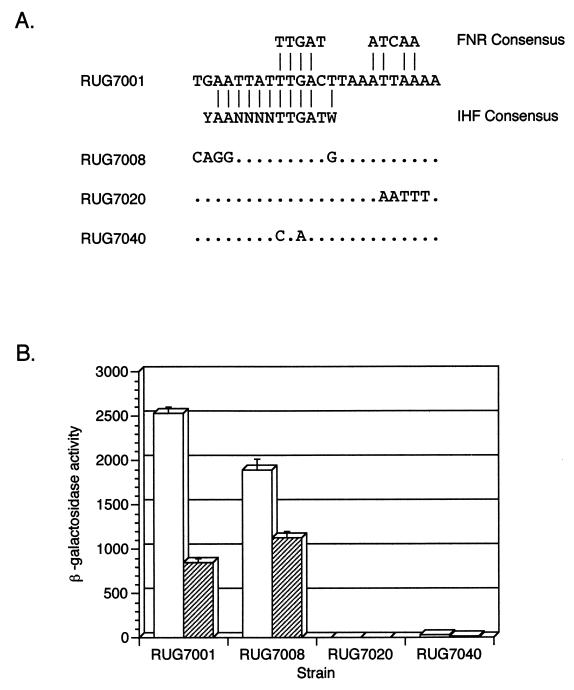
Mutation of the IHF and FNR consensus binding sites.
As the putative IHF and FNR binding sites overlap, it was impossible to completely mutate one site without potentially affecting the other. Bases which matched one consensus and were not a part of the other consensus were mutated as shown in Fig. 6. Mutation of the IHF consensus only slightly affected β-gal activity (RUG7008). In contrast, mutation of the 3′ half of the FNR consensus decreased β-gal activity to almost zero (RUG7020). To further investigate the putative FNR binding site, the first and third bases of the consensus, which are known to be required for the E. coli nirB promoter (5), were mutated. This mutation (RUG7040) also dramatically decreased β-gal activity.
FIG. 6.
Mutation of FNR and IHF binding sites. (A) Base changes in each strain are indicated. (B) Anaerobic β-gal assay results (mean ± standard error of the mean Miller units). Cultures grown with nitrite are indicated by open bars; cultures incubated without nitrite are indicated by hatched bars.
Evaluating the role of the symmetric repeat.
Positioned around the ribosome binding site, the symmetric repeat was a possible repressor binding site (reviewed in references 9 and 16). When bound to the DNA, a protein at that position could prevent procession of the RNA polymerase or could prevent the binding of the ribosome by binding to the message. To investigate this possibility, the sequence of the repeat was scrambled, except for the 6-bp Shine-Dalgarno sequence, changing wild-type TTACAAAAGGAAAACATT to TATACAAAGGATCAAATA (RUG7023). This mutation had little effect on β-gal activity. The β-gal activity of cultures grown in the presence of nitrite was 1,930 ± 65 Miller units, while the β-gal activity of cultures incubated without nitrite was 511 ± 19 Miller units.
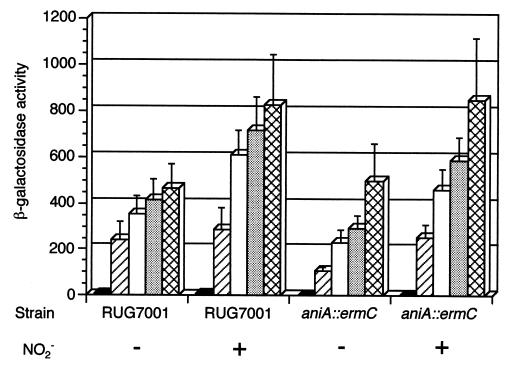
Requirement for an FNR homolog.
Mutation analysis of the aniA promoter indicated that there is an absolute requirement for the FNR binding site (Fig. 6). The gonococcal FNR homolog was recently identified and will be reported elsewhere (27). When the gonococcal fnr gene was insertionally inactivated, strain F62 could no longer grow anaerobically. A wild-type promoter fusion in this strain (RUG7022) resulted in negligible β-gal activity (Table 3). Placing the aniA gene under the control of the tac promoter in this background (RUG7025) restored the ability to grow anaerobically but had no effect on β-gal activity.
TABLE 3.
Anaerobic β-gal activity and growth characteristics of aniA, fnr, and narP mutants
| Strain | Relevant genotype | β-gal activity in the presence ofa:
|
Anaerobic growthb | |
|---|---|---|---|---|
| −O2 +NO2 | −O2 −NO2 | |||
| RUG7001 | Wild type | 2,530 ± 70 | 853 ± 46 | + |
| RUG7011 | aniA::ermC | 493 ± 30 | 1,003 ± 102 | − |
| RUG7035 | tacp::aniA | 3,182 ± 109 | 740 ± 64 | + |
| RUG7022 | fnr::ermC | 6 ± 0.6 | 5 ± 0.2 | − |
| RUG7025 | fnr::ermC tacp::aniA | 12 ± 0.3 | 7 ± 0.5 | + |
| RUG7036 | narP::ermC | 669 ± 96 | 101 ± 8 | + |
| RUG7039 | narP::ermC tacp::aniA | 472 ± 19 | 72 ± 7 | + |
Expressed in Miller units as mean ± standard error of the mean. −O2, no oxygen; +NO2, nitrite; −NO2, no nitrite.
Presence (+) or absence (−) of growth around the nitrite disk on GCK plates.
Full induction requires a NarP homolog.
Nitrate and nitrite regulation of genes in E. coli is mediated by dual two-component systems. The NarX-NarL and NarQ-NarP systems respond differentially to nitrate and nitrite (reviewed in reference 11). Since aniA seems to respond to a nitrite signal, we were interested in these regulatory systems. The gonococcal NarP homolog has been identified (27). Insertionally inactivating the narP gene resulted in a strain (RUG7036) that would grow anaerobically but that produced decreased β-gal activity from the aniA′-′lacZ fusion. As shown in Table 3, β-gal activity was diminished in cultures both with and without nitrite. Placing aniA under the control of the tac promoter in this strain (RUG7039) had no significant effect on β-gal activity.
Autoregulation.
As reported for gonococcal strain MS11 (29), an aniA null mutant was unable to grow anaerobically. This F62 derivative (RUG7011) had one-fifth the wild-type β-gal activity in the presence of nitrite (Table 3). A strain with aniA constitutively expressed from the tac promoter was also created (RUG7035). This strain grew very well anaerobically; β-gal activity in this strain was increased over that in the wild-type strain when cultures were grown in the presence of nitrite but were unchanged when cultures were incubated without nitrite (Table 3). These results can be attributed either to autogenous regulation or to the growth characteristics of these strains.
While we have reported β-gal activity after 20 h of incubation, it was desirable to look at the initial induction of the aniA promoter under conditions in which the cultures could grow. A 4-h induction assay was performed on the aniA null mutant (RUG7011), and the results were compared to those for wild-type strain RUG7001 in the same assay. The A600 of cultures of RUG7001 without nitrite doubled on average during this time period, while the A600 of cultures of RUG7001 with nitrite increased by 2.7-fold. The increases in the A600 for strain RUG7011 without and with nitrite were 1.6- and 1.4-fold, respectively. The β-gal data (Fig. 7) indicated that the aniA null mutant responded to the presence of nitrite in the same manner as the wild-type strain and that its decreased 20-h β-gal levels were due to its inability to grow anaerobically.
FIG. 7.
Time course for aniA induction. Aerobic cultures of RUG7001 and RUG7011 were shifted to oxygen-limited conditions in the absence (−) or presence (+) of nitrite as indicated. Samples were removed before the shift (solid bar), at 1 h (hatched bar), at 2 h (open bar), at 3 h (stippled bar), and at 4 h (cross-hatched bar) and assayed for β-gal activity (expressed as mean ± standard error of the mean Miller units).
DISCUSSION
Pathogenic bacteria often coordinately regulate virulence genes in response to environmental stimuli (recently reviewed in reference 28). Previous studies on N. gonorrhoeae indicated that anaerobiosis is likely a physiologically relevant state in human infection (8) and that the organism alters outer membrane protein expression in response to oxygen levels (7). We have undertaken the study of how aniA is regulated to better understand the gonococcal response to anaerobiosis and to evaluate its potential role in virulence.
An examination of the sequence upstream of aniA revealed several interesting motifs which have been the main focus of this report. We were most interested in the status of the two putative promoters. The mutations that we constructed indicated that the sigma 70-type promoter is the only active promoter during anaerobiosis. Mutating the sigma 70-type promoter −10 sequence eliminated expression, while mutations in the gearbox sequences decreased expression relatively slightly. These results were surprising considering the primer extension data (20) and nuclease protection assay data (Fig. 3), which indicated that the gearbox was the major promoter. This conflict in the data can be resolved when one takes into account that both primer extension and nuclease protection detect not only initiated transcripts but also degraded and processed RNA species. Given the distinct products seen with both of these biochemical techniques, it is possible that specific cleavage of 9 bp from the 5′ end of the message occurs, although the purpose of such processing is unknown at this time. The fact that changing the spacing in the gearbox promoter (RUG7009) does not cause a significant decrease in β-gal activity makes it clear that the gearbox sequences do not act as a promoter.
In conjunction with the requirement for the sigma 70-type promoter is the requirement for the FNR binding site. It is common for anaerobically induced genes to possess an FNR binding site in place of the traditional −35 sequence, so it is not at all surprising that this motif is present in aniA. The FNR binding site is centered at −42.5 and is therefore in position to act as a conventional class II transcription activator (22). It is still unclear if there is a role for the IHF binding site which overlaps the FNR binding site. IHF is known to coordinate with FNR and NarL in the regulation of the nitrate reductase operon in E. coli (34). Gonococcal IHF has been shown to bind to the pilE promoter in gel mobility shift assays (17), and mutation of the IHF binding site results in a large decrease in expression from the pilEp1 promoter (15). However, mutation of the consensus sequence in aniA resulted in little change in β-gal activity (Fig. 6). The decrease seen may have been due to an alteration of the context of the FNR binding site. It is therefore unlikely that IHF functions in the regulation of aniA, but it remains to be determined if IHF actually binds to the aniA promoter.
The deletion studies reported here indicated that the length of the upstream region required for full induction is approximately 170 bp. Although repeated sequences within promoter regions have often proved to be binding sites for either activators or repressors, our deletion and mutation analyses of the inverted and symmetric repeats in the aniA promoter region failed to reveal any function for these two motifs. The direct repeats, however, are part of the region which seems to respond to a second induction signal and is required for full expression during anaerobic growth. As with other nitrite reductases, it is likely that this second signal is nitrite.
The requirement for the gonococcal homologs of FNR and NarP was not unexpected. We had an indication of the necessity for FNR through the mutation studies of the putative FNR binding site, and NarP was a likely candidate for an activator, considering its role in the regulation of other genes induced by nitrite (recently reviewed in reference 11). By placing aniA under the control of the tac promoter, we restored the ability of the fnr mutant to grow anaerobically. This result indicates that aniA is the only gene regulated by FNR that is essential for anaerobic growth. The fact that the narP mutant had decreased β-gal activity due to anaerobiosis alone suggests that NarP and FNR may act in synergy to regulate aniA, as has been noted for E. coli nirB (38).
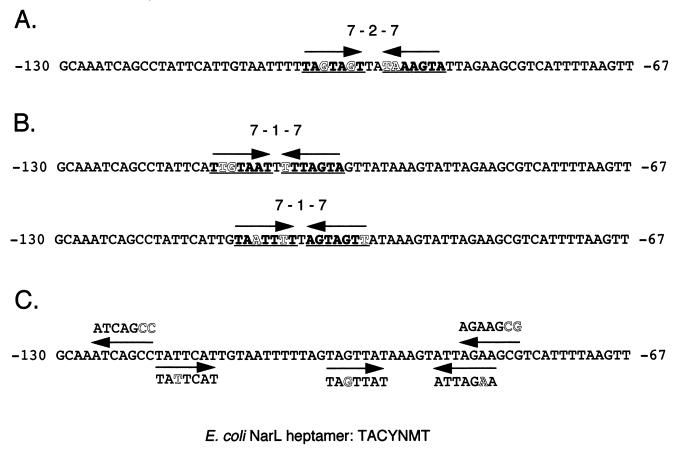
The data that we obtained from the deletion analysis and the narP mutant analysis suggest that NarP interacts directly with the aniA promoter at the T-rich region. We closely scrutinized this region for the presence of potential NarP binding sites. In E. coli, NarL and NarP bind to a consensus sequence, TACYNMT (Y, C or T; M, A or C), which has been called the NarL heptamer. Such heptamers can be found as direct repeats, inverted repeats of the form 7-2-7 (heptamers separated by 2 bp), or individually as half sites (11, 13, 26, 38). It was recently reported that NarP binds only to the 7-2-7 motif, while NarL can bind to any heptamer arrangement (12). We found no less than 11 heptamers in the region from −67 to −130. As shown in Fig. 8, these heptamers deviated from the E. coli consensus by only 1 or 2 bases each and were found as a single 7-2-7 motif, as two 7-1-7 motifs (heptamers separated by 1 bp), and as several half sites. aniA is the first gonococcal gene found to be regulated by NarP; therefore, the binding site sequence is unknown. However, the deletion in RUG7045 pinpoints the region responsible for nitrite induction to between −85 and −130, a region which contains the majority of the NarL heptamers. This finding suggests that gonococcal NarP may have a binding site similar to that of NarL or NarP in E. coli. It will be necessary to perform further mutagenesis studies as well as gel mobility shift assays to determine to which, if any, of these sites NarP can bind. These studies, as well as those necessary to confirm that FNR interacts directly with the FNR consensus binding site in the aniA promoter, are in progress.
FIG. 8.
Potential NarP binding sites in the aniA upstream region. The plus-strand sequence of the region from −130 to −67 from the sigma 70-type promoter transcription start site is presented. NarL heptamers and their direction are indicated by arrows. Nucleotides which do not match the E. coli consensus are indicated by outlined letters. The E. coli consensus is indicated at the bottom (Y, C or T; M, C or A). As the binding site sequence for gonococcal NarP is unknown, any of these sites is a candidate. (A) A single 7-2-7 motif consisting of an inverted repeat of the heptamer separated by 2 bp was found. Heptamers are also indicated by bold type and underlining. (B) Two similar heptamer inverted repeats in a 7-1-7 arrangement (heptamers separated by 1 bp). Heptamers are also indicated by bold type and underlining. (C) Remaining unpaired half sites.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant RO1 AI11709 from the National Institutes of Health to V.L.C. and by project grant G9603098 from the United Kingdom Medical Research Council to J.A.C. T.C.H. was supported by National Institutes of Health training grant T32 AI07363.
We thank Lin Silver for constructing the constitutive aniA mutant and several of the reporter constructs, Joe Dillard for the kind gift of pHSS23, Lou Passador for critical comments on the manuscript, and Doug Browning for assistance in identifying NarL heptamers.
REFERENCES
- 1.Aldea M, Garrido T, Hernandez-Chico C, Vicente M, Kushner S R. Induction of a growth-phase-dependent promoter triggers transcription of bolA, an Escherichia coli morphogene. EMBO J. 1989;8:3923–3931. doi: 10.1002/j.1460-2075.1989.tb08573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 5.Bell A I, Cole J A, Busby S J W. Molecular genetic analysis of an FNR-dependent anaerobically inducible Escherichia coli promoter. Mol Microbiol. 1993;4:1753–1763. doi: 10.1111/j.1365-2958.1990.tb00553.x. [DOI] [PubMed] [Google Scholar]
- 6.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 6a.Clark, V. L. Unpublished results.
- 7.Clark V L, Campbell L A, Palermo D A, Evans T M, Klimpel K W. Induction and repression of outer membrane proteins by anaerobic growth of Neisseria gonorrhoeae. Infect Immun. 1987;55:1359–1364. doi: 10.1128/iai.55.6.1359-1364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark V L, Knapp J S, Thompson S, Klimpel K W. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb Pathog. 1988;5:381–390. doi: 10.1016/0882-4010(88)90038-1. [DOI] [PubMed] [Google Scholar]
- 9.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig N L, Nash H A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 11.Darwin A J, Stewart V. The NAR modulon systems: nitrate and nitrite regulation of anaerobic gene expression. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 343–359. [Google Scholar]
- 12.Darwin A J, Tyson K L, Busby S J W, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 13.Dong X-R, Li S F, DeMoss J A. Upstream sequence elements required for NarL-mediated activation of transcription from the narGHJI promoter of Escherichia coli. J Biol Chem. 1992;267:14122–14128. [PubMed] [Google Scholar]
- 14.Eiglmeier K, Honore N, Iuchi S, Lin E C C, Cole S T. Molecular genetic analysis of FNR-dependent promoters. Mol Microbiol. 1989;3:869–878. doi: 10.1111/j.1365-2958.1989.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 15.Fyfe J A M, Davies J K. An AT-rich tract containing an integration host factor-binding domain and two UP-like elements enhances transcription from the pilEp1 promoter of Neisseria gonorrhoeae. J Bacteriol. 1998;180:2152–2159. doi: 10.1128/jb.180.8.2152-2159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gralla J D. Transcription control—lessons from an E. coli promoter data base. Cell. 1991;66:415–418. doi: 10.1016/0092-8674(81)90001-5. [DOI] [PubMed] [Google Scholar]
- 17.Hill S A, Samuels D S, Carlson J H, Wilson J, Hogan D, Lubke L, Belland R J. Integration host factor is a transcriptional cofactor of pilE in Neisseria gonorrhoeae. Mol Microbiol. 1997;23:649–656. doi: 10.1046/j.1365-2958.1997.2321612.x. [DOI] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Hoehn G T, Clark V L. Distribution of a protein antigenically related to the major anaerobically induced gonococcal outer membrane protein among other Neisseria species. Infect Immun. 1990;58:3929–3933. doi: 10.1128/iai.58.12.3929-3933.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoehn G T, Clark V L. Isolation and nucleotide sequence of the gene (aniA) encoding the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae. Infect Immun. 1992;60:4695–4703. doi: 10.1128/iai.60.11.4695-4703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoehn G T, Clark V L. The major anaerobically induced outer membrane protein of Neisseria gonorrhoeae, Pan 1, is a lipoprotein. Infect Immun. 1992;60:4704–4708. doi: 10.1128/iai.60.11.4704-4708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellogg D S, Jr, Peacock W L, Jr, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knapp J S, Clark V L. Anaerobic growth of Neisseria gonorrhoeae coupled to nitrite reduction. Infect Immun. 1984;46:176–181. doi: 10.1128/iai.46.1.176-181.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor ςs. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Stewart V. Localization of upstream sequence elements required for nitrate and anaerobic induction of fdn (formate dehydrogenase-N) operon expression in Escherichia coli K-12. J Bacteriol. 1992;174:4935–4942. doi: 10.1128/jb.174.15.4935-4942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lissenden, S., T. Regan, H. Crooke, J. Cardinale, T. C. Householder, V. L. Clark, H. Smith, and J. A. Cole. Submitted for publication.
- 28.Mahan M J, Slauch J M, Mekalanos J J. Environmental regulation of virulence gene expression in Escherichia, Salmonella, and Shigella spp. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2803–2815. [Google Scholar]
- 29.Mellies J, Jose J, Meyer T F. The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol Gen Genet. 1997;256:525–532. doi: 10.1007/s004380050597. [DOI] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 31.Mulligan M E, Brosius J, McClure W R. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985;260:3529–3538. [PubMed] [Google Scholar]
- 32.Pogulis R J, Vallejo A N, Pease L R. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol Biol. 1996;57:167–176. doi: 10.1385/0-89603-332-5:167. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33a.Schipp T C, Clark V L. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. 1997. p. 61. , abstr. B-188. [Google Scholar]
- 34.Schroder I, Darie S, Gunsalus R P. Activation of the Escherichia coli nitrate reductase (narGHJI) operon by NarL and Fnr requires integration host factor. J Biol Chem. 1993;268:771–774. [PubMed] [Google Scholar]
- 35.Short H B, Clark V L, Kellogg D S, Jr, Young F E. Anaerobic survival of clinical isolates and laboratory strains of Neisseria gonorrhoeae: use in transfer and storage. J Clin Microbiol. 1982;15:915–919. doi: 10.1128/jcm.15.5.915-919.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silver L E, Clark V L. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene. 1995;166:101–104. doi: 10.1016/0378-1119(95)00605-6. [DOI] [PubMed] [Google Scholar]
- 37.Spiro S, Guest J R. Regulation and over-expression of the fnr gene of Escherichia coli. J Gen Microbiol. 1987;133:3279–3288. doi: 10.1099/00221287-133-12-3279. [DOI] [PubMed] [Google Scholar]
- 38.Tyson K L, Bell A I, Cole J A, Busby S J W. Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol Microbiol. 1993;7:151–157. doi: 10.1111/j.1365-2958.1993.tb01106.x. [DOI] [PubMed] [Google Scholar]
- 39.Vedros N A. Genus I. Neisseria Trevisan. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 290–296. [Google Scholar]
- 40.Vicente M, Kushner S R, Garrido T, Aldea M. The role of the ‘gearbox’ in the transcription of essential genes. Mol Microbiol. 1991;5:2085–2091. doi: 10.1111/j.1365-2958.1991.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 41.Wainwright L A, Pritchard K H, Seifert H S. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol. 1994;13:75–87. doi: 10.1111/j.1365-2958.1994.tb00403.x. [DOI] [PubMed] [Google Scholar]