Abstract
Accumulation of 16S rRNA and production of guanosine polyphosphates (pppGpp and ppGpp) were studied during amino acid starvation in three wild-type strains of Helicobacter pylori. All strains exhibit a relaxed phenotype with respect to accumulation of 16S rRNA. This constitutes the first example of a wild-type eubacterium showing a relaxed phenotype. The guanosine polyphosphate levels do not rise as a result of amino acid starvation, as expected for relaxed organisms. However, in both growing and starved cells, basal levels of the two polyphosphates appeared to be present, demonstrating that the enzymatic machinery for guanosine polyphosphate production is present in this organism. These findings are discussed within the framework of the hypothesis that stringent control is a physiological control mechanism more important for the fitness of prokaryotes growing in the general environment than for those that inhabit protected niches.
Stringent control (SC) was originally identified as a mechanism that enables wild-type bacterial cells to rapidly inhibit stable RNA (sRNA) synthesis during amino acid starvation (10, 31). Experimentally, this response can be provoked by amino acid starvation, although the signal for SC involves charging of tRNA rather than unavailability of the free amino acids themselves (26). Over the years, a number of rel mutations have been isolated in several loci, defined relaxed in contrast to the wild-type, stringent behavior (2). The first such mutation to be defined was the relA gene, the central gene for SC; mutations in this gene completely abolish the stringent response in the eubacteria. The mutant response consisted of continued sRNA accumulation during amino acid deprivation (7). It was later shown that many other aspects of cell physiology are positively or negatively regulated during the stringent response (7).
Most stringent eubacteria accumulate ppGpp and pppGpp during SC (5–7). Accumulation of (p)ppGpp can also be provoked by nutritional or other stress conditions (4, 13). The enzymes responsible for (p)ppGpp synthesis are the relA gene product, (p)ppGpp synthetase I, and the spoT gene product, (p)ppGpp synthetase II. The spoT gene product is a bifunctional enzyme possessing both (p)ppGpp synthetic activity as well as (p)ppGpp degrading activity and is responsible for (p)ppGpp production independently of amino acid starvation and SC (39).
SC over sRNA synthesis has been shown to be present in all eubacteria examined (1, 7, 8, 11, 21, 28, 30, 34, 38) but in only one of the six archaeal strains studied to date (3, 9). It is widely accepted that ppGpp has a role in effecting several aspects of the stringent response, including SC over sRNA accumulation (7). In fact, an increase in the levels of ppGpp and pppGpp during the stringent response occurs in most eubacteria, but there are notable exceptions in which the correlation between increase in the level of ppGpp and inhibition of sRNA accumulation is absent (1, 7, 12, 29, 33). It should be noted that in wild-type eubacteria, a basal level of (p)ppGpp is always present, presumably as a result of the activity of the spoT gene. In all archaea examined, (p)ppGpp production has been shown to be totally absent, both during amino acid starvation in the unique (p)ppGpp-independent stringent case (9), as well as under a number of other conditions (3, 32). Thus, in most eubacteria, ppGpp may be the effector for SC over sRNA, whereas in some eubacteria, SC operates in the absence of ppGpp. In the archaea, SC is mostly absent, and ppGpp is never produced.
Archaeal organisms are extremophilic, that is to say, organisms that have become adapted to harsh environmental conditions, such as high temperature, extreme pH, high salt, or to a combination of such conditions, while the eubacteria are mostly mesophilic. Extremophiles live in protected niches where competition with other organisms is scarce or absent, whereas mesophiles are generally in active competition with other organisms. This suggests two different explanations for the fact that all eubacterial strains are stringent and produce ppGpp, whereas the archaea never produce ppGpp and tend to be relaxed: (i) SC and ppGpp production arose as part of the evolutionary process that defined the eubacteria as distinct from the archaea and the eukaryotes, and (ii) SC is an important element of the set of functions that enhance the fitness of microorganisms that compete for survival with other organisms in an environment that often undergoes rapid changes in surrounding conditions. This is probably not the case for organisms adapted to an extreme but stable environment where interspecific competition is lower. In order to distinguish between the two hypotheses, it would be useful to determine the phenotypes of eubacteria living in protected niches, with respect both to SC and to the presence or absence of (p)ppGpp. Under the second hypothesis, one would expect that wild-type eubacteria belonging to the latter category could have the relaxed phenotype.
Helicobacter pylori, the organism that is strongly associated with some forms of human gastroduodenal disease (15, 37), is the only organism capable of colonizing the gastral antral mucosa (22). A number of adaptative mechanisms allows H. pylori to occupy a protected niche where significant competition with other microorganisms does not occur (20, 35). We have therefore set out to examine various clinical isolates and collection strains of H. pylori in order to determine both the presence of SC and production of (p)ppGpp in this organism.
MATERIALS AND METHODS
Bacteria and plasmids.
One strain (H. pylori NCTC11637) is a collection organism obtained from the National Collection of Type Cultures, London, United Kingdom. All other strains were supplied by Ida Luzzi (Istituto Superiore di Sanità, Rome, Italy). Three H. pylori strains, NCTC11637, C3, and D1, among a number of strains analyzed, were able to grow in the complex liquid medium used in this study. Two strains, NCTC11637 and C3, bearing an 8-kb plasmid, are cytotoxic. The D1 strain neither bears a plasmid nor is cytotoxic. Salmonella typhimurium TA997 (aroC5 purF145 hisD2655) was obtained from R. Cortese (IRBM, Pomezia, Italy). The Escherichia coli CF5746 and CF5969, bearing the plasmids pALS10 (relA) and pHX41 (spoT), respectively, were both furnished by M. Cashel (National Institute of Health, Bethesda, Md.).
Sources of reagents.
Dehydrated Bacto Brucella Broth, Bacto Yeast Extract, and Bacto Tryptone were furnished by Difco (Detroit, Mich.); Columbia agar base, laked horse blood, growth (Vitox) and selective (Dent) supplements were from Oxoid, Unipath Ltd., Basingstoke, Hampshire, England; fetal calf serum was from Biological Industries, Kibbutz Beit Haemek 25155, Israel; pseudomonic acid (PA) was furnished by SmithKline Beecham Pharmaceuticals (Worthing, United Kingdom); dl-serine hydroxamate, guanidine thiocyanate, betacyclodextrin, and 2-chloro-6-(trichloro-methyl)pyridine (nitrapyrin), were from Sigma, St. Louis, Mo. All other chemicals, unless otherwise noted, were obtained from Merck (Darmstad, Germany). 3H-labeled amino acids were from New England Nuclear (Du Pont de Nemours, Firenze, Italy); [14C]uridine and 32Pi were from Amersham (Amersham, United Kingdom). Type I DNase was obtained from Boehringer Mannheim Biochemicals (Indianapolis, Ind.).
Media and growth conditions.
Solid medium for H. pylori was composed as follows: Columbia agar base (39 g/liter), laked horse blood (5%), Vitox and Dent, diluted according to the suppliers’ instructions, i.e., one vial per 500 ml of medium. Liquid media were BBCD, consisting of 2.8% brucella broth supplemented with 0.1% cyclodextrin, pH 7 (19, 24, 27); BBSN, modified from that of Kangatharalingam and Amy (17) and containing 2.8% brucella broth, 15 mM K2HPO4, 15 mM KH2PO4, 10 μM NH4Cl in 90 ml of H2O. Ten milliliters of sterile fetal calf serum and 0.5 μg of filter-sterilized nitrapyrin per ml were added to BBSN at 40 to 50°C after being autoclaved. Liquid cultures were prepared as follows: sterile tubes containing 2 to 6 ml of liquid medium were incubated overnight at 37°C under microaerophilic conditions on a rotor rotating at 40 to 60 rpm, after which the tubes were inoculated. In particular, use of BBSN resulted in a considerable improvement over previous work (17, 24, 27), as yields greater than 3.5 optical density at 600 nm (OD600) units were routinely obtained. S. typhimurium, when used in control experiments, was grown in BBCD and BBSN supplemented with 0.5% NaCl.
Analysis of protein synthesis.
Bacteria from a 2-day-old plate were resuspended in 2 ml of BBCD or BBSN to an OD600 of approximately 2.5 and incubated for 10 to 30 min under microaerophilic conditions at 37°C; this culture was used as an inoculum for 3-ml cultures. The inoculum was added to an initial OD600 of approximately 0.25. After 30 min, [3H]serine or [3H]glutamic acid was added, both at 15 μCi/ml (final concentration). After at least one doubling, amino acid starvation was accomplished by adding either PA or serine hydroxamate to the cultures. The antibiotic PA produces cellular effects similar to those of isoleucine starvation by preventing the charging of tRNAIle due to inhibition of isoleucyl tRNA synthetase in prokaryotes (3, 8, 9, 14, 16, 21, 38), whereas serine hydroxamate is a competitive inhibitor of seryl-tRNA synthetase (36). The MICs of these drugs for the various strains used were determined, and inhibition of protein synthesis was achieved by using a concentration of either drug corresponding to four times the MIC. Samples were then placed in 5% trichloracetic acid at different time intervals. The bacterial precipitates were collected on Millipore filters (pore size, 0.45 μm), and the radioactivity was counted as previously described (11).
Analysis of rRNA synthesis.
Preliminary experiments showed that, unlike amino acids, uridine was not significantly taken up by H. pylori from the growth medium (unpublished data). To detect rRNA synthesis in H. pylori, we used a specific oligonucleotide-rRNA hybridization technique. Bacteria from a 2-day-old plate were resuspended in 2 ml of BBCD or BBSN to an OD600 of approximately 2.5 and incubated for 10 to 30 min under microaerophilic conditions at 37°C; this culture was used to inoculate 3-ml cultures at an initial OD600 of approximately 0.25. At the appropriate times, 100-μl samples were taken and lysed by dilution with 400 μl of GED solution (5 M guanidine thiocyanate, 0.1 M EDTA [pH 7], 10 mM dithiothreitol solution). After 4 to 6 h (depending on the doubling time of the strain used), the culture was split in two. Amino acid starvation was accomplished in one of the two cultures by adding PA or serine hydroxamate; the other culture served as a nonstarved control. Aliquots (50 to 200 μl) of the bacterial cell lysates were filtered onto a nylon transfer membrane (Hybond-N+; Amersham International plc, Amersham, United Kingdom) by vacuum aspiration with a microsample filtration manifold (Hybri-Dot Manifold; BRL Life Technologies Inc, Gaithersburg, Md.) after which the filters were air dried. The filters were then treated with DNase in a solution containing 46 U of type I DNase per ml and 50 mg of bovine serum albumin fraction V per ml at 30°C for 30 min, washed three times with 2× SSC (1× SSC is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) at room temperature for 5 min, and allowed to air dry before hybridization (23). The oligonucleotide probe sequence 5′d(GGACATAGGCTGATCTCTTAGC) used for hybridization is complementary to the 16S rRNA sequences of H. pylori reported by Morotomi et al. (23). This oligonucleotide (polyacrylamide gel electrophoresis grade) was synthesized and purified by Genenco (M-Medical srl, Florence, Italy).
Guanosine polyphosphate assay.
Cells were labeled with [32P]orthophosphate, and guanosine polyphosphate production was analyzed by one-dimensional chromatography of formic extracts (9). The locations of the polyphosphates were detected by autoradiography.
RESULTS
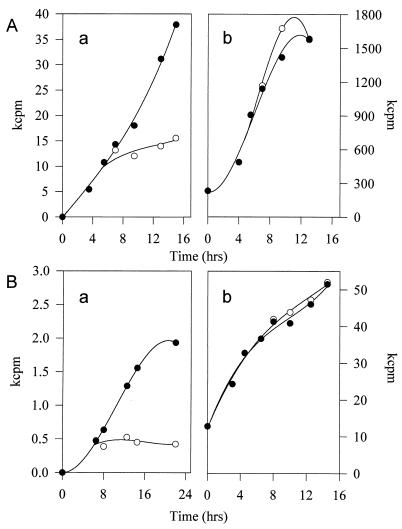
Accumulation of rRNA under amino acid starvation induced by the addition of PA was analyzed in three H. pylori strains, NCTC11637, C3, and D1. Analysis of protein synthesis showed that protein synthesis was shut down efficiently in all strains. Amino acid starvation was also provoked, in NCTC11637, by the addition of serine hydroxamate. The results of two typical experiments, displayed in Fig. 1, show that H. pylori NCTC11637 displays a relaxed phenotype. In fact, following the inhibition of protein synthesis, rRNA synthesis is either not affected or possibly enhanced, in the case of PA inhibition. A relaxed phenotype is exhibited by all three strains, as shown in Table 1, where rates of protein and rRNA synthesis under amino acid starvation are reported.
FIG. 1.
Protein synthesis and rRNA accumulation in H. pylori NCTC11637. Protein synthesis (panels a) was measured by the incorporation of 3H-labeled amino acids into acid-insoluble material; accumulation of 16S RNA (panels b) was measured by a specific oligonucleotide-rRNA hybridization technique, as described in Materials and Methods. (A) Experiments were performed during either exponential growth (solid circles) or starvation for isoleucine (open circles); PA was added at 5.5 h. (B) Same as for panel A, except that starvation was obtained by the addition of serine hydroxamate at 6.5 h. kcpm, 1,000 cpm.
TABLE 1.
Effect of amino acid starvation on RNA and protein synthesis
| Inhibitor | Inhibitor concn (μg/ml) | Bacterial strain | Rate of synthesisa of:
|
|
|---|---|---|---|---|
| RNA | Protein | |||
| PA | 0.06 | H. pylori NCTC11637 | 1.53 | 0.12 |
| 0.05 | H. pylori C3 | 1.07 | 0.11 | |
| 0.04 | H. pylori D1 | 0.69 | 0.04 | |
| Serine hydrox-amate | 4,800 | H. pylori NCTC11637 | 1.07 | 0.005 |
| Noneb | E. coli (relA+) | 0.06 | 0.08 | |
| E. coli (relA) | 0.60–1.00 | 0.05 | ||
The values reported are for treated cultures and are expressed as percentages of the rates of synthesis in the untreated cultures. Rates of synthesis were obtained by carrying out regression analyses on the portion of the incorporation curves after addition of the inhibitor.
Starvation was performed by removing the required amino acid from the growth medium of the auxotrophic strain; values reported are from Donini et al. (12).
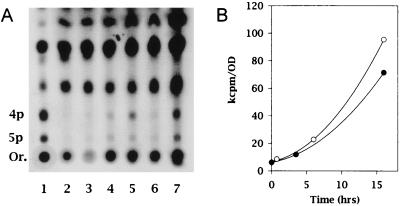
All three H. pylori strains were then analyzed for the production of guanosine polyphosphates. In a control experiment, it was shown that the relA+ strain S. typhimurium TA997, growing in a medium having the same organic composition as that of BBCD or BBSN and amino acid starved using PA, produced large amounts of (p)ppGpp, as expected. In contrast, during amino acid starvation, none of the three H. pylori strains accumulated either ppGpp or pppGpp. The results of a typical experiment carried out with H. pylori NCTC11637 is shown in Fig. 2A. The (p)ppGpp assay was performed over extended time periods: 9 h for the faster-growing C3 (doubling time, ∼7 h), 16 h for NCTC11637 (doubling time, ∼8 h), and 18.5 h for D1 (doubling time, ∼10 h). Figure 2A also shows the presence of faint spots with chromatographic mobilities identical to those of pppGpp and ppGpp. Such spots did not increase in intensity as a result of amino acid starvation and had similar intensities in both the treated and control samples. Figure 2B shows the results obtained by measuring the intensities of the spots corresponding to ppGpp. The level of the nucleotide increases with similar kinetics in the treated and untreated samples, confirming that amino acid starvation does not cause an immediate and significant increase in the level of ppGpp. Experiments carried out with C3 and D1 gave results very similar to those obtained with NCTC11637 (data not shown).
FIG. 2.
Guanosine polyphosphates in H. pylori NCTC11637. (A) (p)ppGpp in growing and amino acid-starved cells. Lane 1, control (S. typhimurium TA997), 10 min after addition of PA; lanes 2 to 5, H. pylori 0, 45 min, 6 h, and 16 h after addition of PA, respectively; lanes 6 and 7, H. pylori control samples 3.5 h and 16 h after time 0, Abbreviations: Or., origin; 5p, pppGpp; 4p, ppGpp. (B) Basic ppGpp levels in H. pylori NCTC11637. The spots corresponding to ppGpp on the polyethyleneimine-cellulose chromatograms from the experiment shown in panel A were counted with a PhosphorImager scan analysis apparatus. The counts were normalized to the corresponding optical densities at 600 nm (OD). Experiments were performed during either exponential growth (solid circles) or starvation for isoleucine (open circles). The computer-generated image shown in panel A was acquired with Adobe Photoshop 4.0 software on a Umax Astra 1200 scanner.
Hybridization experiments carried out using labeled relA and spoT E. coli gene fragments and NCTC11637 DNA gave negative results (data not shown).
DISCUSSION
It seems reasonable to hypothesize that stringency is a physiological mechanism primarily designed to prevent an imbalance of cellular macromolecules when bacteria growing in the wild are transferred from a nutritionally rich to a nutritionally poor environment (10). To date, relA-dependent SC over sRNA has been shown to be present in all wild-type eubacterial species examined and to be absent in the archaea (3, 9, 32).
The work reported here was carried out to test the hypothesis that SC is a physiological control mechanism more important for the fitness of prokaryotes living in the general environment than those living in protected niches. Our study shows that three wild-type strains of the eubacterium H. pylori exhibit a relaxed phenotype with respect to accumulation of 16S rRNA, which constitutes the first case of a wild-type eubacterium with a relaxed phenotype. Unlike the situation in the archaea, (p)ppGpp is present at low levels in all H. pylori strains examined, both in control samples and starved samples, showing that the enzymatic machinery for (p)ppGpp production exists in this organism. As expected for relaxed microorganisms, the (p)ppGpp levels do not rise as a result of amino acid starvation. These findings lend support to our proposition that SC is present in eubacterial mesophiles as a genetic control mechanism that increases the fitness of prokaryotes occupying the general environment. In protected niches, SC is either absent, such as in H. pylori and in most of the archaea examined, or present in a (p)ppGpp-independent form, such as in the stringent halobacterium Haloferax volcanii (9). The presence of a stringent response in the latter organism may be related to its inclusion among the “borderline” extreme halophiles (18, 25) that are capable of living in environments where competition with other microorganisms is more likely to occur.
Tomb and his colleagues have reported the complete genome sequence of H. pylori (35), and sequence analysis shows no sequence homology to the relA gene of E. coli, whereas the putative gene region HP0775 has a 36.7% base identity in common with the E. coli spoT gene. The lack of a relA gene in H. pylori provides a convincing explanation for our finding that this organism exhibits a relaxed phenotype. Moreover, the sequence data also account for our inability to detect sequence homology between the two E. coli genes and the H. pylori genome by hybridization. In fact, a 36.7% DNA homology is not easily detectable by standard hybridization techniques. The presence of a spoT-like gene in the H. pylori genome explains the existence of basal levels of (p)ppGpp, presumably a result of the activity of this gene. To this end, it would be useful to subject H. pylori to a set of physiological conditions other than amino acid starvation (temperature shifts, carbon or total starvation, cyanide treatment) that are known to cause production of guanosine polyphosphates in mesophilic eubacteria via the relA-independent (spoT) pathway (13).
The fact that the eubacterium H. pylori is a (p)ppGpp producer, despite having a relaxed phenotype, confirms the notion that (p)ppGpp production appears to be a feature that separates the eubacteria from the archaea and the eukaryotes. The evolution of (p)ppGpp production and its involvement in some aspects of SC may be a part of the process that has caused an efficient form of SC to evolve among the mostly mesophilic eubacteria.
ACKNOWLEDGMENT
This work was supported in part by a grant from the Italian Scientific and Technological Research Ministry (MURST).
REFERENCES
- 1.Acosta R, Lueking D R. Stringency in the absence of ppGpp accumulation in Rhodobacter sphaeroides. J Bacteriol. 1987;169:908–912. doi: 10.1128/jb.169.2.908-912.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alföldi L, Stent G S, Clowes R C. The chromosomal site of the RNA control (RC) locus in Escherichia coli. J Mol Biol. 1962;5:348–355. doi: 10.1016/s0022-2836(62)80077-1. [DOI] [PubMed] [Google Scholar]
- 3.Beauclerk A A D, Hummel H, Holmes D J, Böck A, Cundliffe E. Studies of the GTPase domain of archaebacterial ribosomes. Eur J Biochem. 1985;151:245–255. doi: 10.1111/j.1432-1033.1985.tb09095.x. [DOI] [PubMed] [Google Scholar]
- 4.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger A E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 5.Cashel M, Gallant J. Two compounds implicated in the function of RC gene of Escherichia coli. Nature (London) 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 6.Cashel M, Gallant J. Cellular regulation of guanosine tetraphosphate and guanosine pentaphosphate. In: Nomura M, Tisseries A, Lengyel P, editors. Ribosomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. pp. 733–745. [Google Scholar]
- 7.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger A E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 8.Cassels R, Oliva B, Knowles D. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J Bacteriol. 1995;177:5161–5165. doi: 10.1128/jb.177.17.5161-5165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimmino C, Scoarughi G L, Donini P. Stringency and relaxation among the halobacteria. J Bacteriol. 1993;175:6659–6662. doi: 10.1128/jb.175.20.6659-6662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donini P. Amino acid control over deoxyribonucleic acid synthesis in Escherichia coli infected with T-even bacteriophage. J Bacteriol. 1970;102:616–627. doi: 10.1128/jb.102.3.616-627.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donini P, Santonastaso V, Roche J, Cozzone A J. The relationship between guanosine tetraphosphate, polysomes and RNA synthesis in amino acid starved Escherichia coli. Mol Biol Rep. 1978;4:15–29. doi: 10.1007/BF00775174. [DOI] [PubMed] [Google Scholar]
- 13.Edlin G, Donini P. Synthesis of guanosine 5′ diphosphate, 2′-(or 3′) diphosphated and related nucleotides in a variety of physiological conditions. J Biol Chem. 1971;246:4371–4373. [PubMed] [Google Scholar]
- 14.Fuller A T, Banks G T, Mellows G, Barrow K D, Woolford M, Chain E B. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature (London) 1971;234:416–417. doi: 10.1038/234416a0. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin C S, Blinkow E D, Warren J R, Waters T E, Sanderson C R, Easton L. Evaluation of cultural techniques for isolating Campylobacter pyloridis from endoscopic biopsies of gastric mucosa. J Clin Pathol. 1986;39:353–365. doi: 10.1136/jcp.38.10.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes J, Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kangatharalingam N, Amy P S. Helicobacter pylori comb. nov. exhibits facultative acidophilism and obligate microaerophilism. Appl Environ Microbiol. 1994;60:2176–2179. doi: 10.1128/aem.60.6.2176-2179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kushner D J. The halobacteriaceae. In: Sokatch J R, Ornston L N, editors. The bacteria. Orlando, Fla: Academic Press; 1985. pp. 171–214. [Google Scholar]
- 19.Marchini A, Massari P, Manetti R, Olivieri R. Optimized conditions for the fermentation of Helicobacter pylori and production of vacuolating cytotoxin. FEMS Microbiol Lett. 1994;124:55–60. doi: 10.1111/j.1574-6968.1994.tb07261.x. [DOI] [PubMed] [Google Scholar]
- 20.McGowan C C, Cover T L, Blaser M. Helicobacter pylori and gastric acid: biological and therapeutic implications. Gastroenterology. 1996;110:926–938. doi: 10.1053/gast.1996.v110.pm8608904. [DOI] [PubMed] [Google Scholar]
- 21.Mechold U, Malke H. Characterization of the stringent and relaxed response of Streptococcus equisimilis. J Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morotomi M, Hoshina S, Green P, Neu H C, LoGerfo P, Watanabe I, Mutai M, Weinstein I B. Oligonucleotide probe for detection and identification of Campylobacter pylori. J Clin Microbiol. 1989;27:2652–2655. doi: 10.1128/jcm.27.12.2652-2655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morshed M G, Karita M, Konishi H, Okita K, Nakazawa T. Growth medium containing cyclodextrin and low concentration of horse serum for cultivation of Helicobacter pylori. Microbiol Immunol. 1994;38:897–900. doi: 10.1111/j.1348-0421.1994.tb02143.x. [DOI] [PubMed] [Google Scholar]
- 25.Mullakhanbhai M F, Larsen H. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch Microbiol. 1975;104:207–214. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- 26.Neidhardt F C. Properties of a bacterial mutant lacking amino acid control of RNA synthesis. Biochim Biophys Acta. 1963;68:365–379. doi: 10.1016/0006-3002(63)90158-6. [DOI] [PubMed] [Google Scholar]
- 27.Olivieri R, Bugnoli M, Armellini D, Biancardi S, Rappuoli R, Bayeli P F, Abate L, Esposito E, De Gregorio L, Aziz J, Basagni C, Figura N. Growth of Helicobacter pylori in media containing cyclodextrins. J Clin Microbiol. 1993;31:160–162. doi: 10.1128/jcm.31.1.160-162.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Östling J, Holmquist L, Kjelleberg S. Global analysis of the carbon starvation response of a marine Vibrio species with disruptions in genes homologous to relA and spoT. J Bacteriol. 1996;178:4901–4908. doi: 10.1128/jb.178.16.4901-4908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pao C C, Dyess B T. Stringent control of RNA synthesis in the absence of guanosine 5′-diphosphate-3′-diphosphate. J Biol Chem. 1981;256:2252–2257. [PubMed] [Google Scholar]
- 30.Price V L, Gallant J A. A new relaxed mutant of Bacillus subtilis. J Bacteriol. 1982;149:635–641. doi: 10.1128/jb.149.2.635-641.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sands M K, Roberts R B. The effects of a tryptophane-histidine deficiency in a mutant of Escherichia coli. J Bacteriol. 1952;63:505–511. doi: 10.1128/jb.63.4.505-511.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scoarughi G L, Cimmino C, Donini P. Lack of production of (p)ppGpp in Halobacterium volcanii under conditions that are effective in the eubacteria. J Bacteriol. 1995;177:82–85. doi: 10.1128/jb.177.1.82-85.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spadaro A, Spena A, Santonastaso V, Donini P. Stringency without ppGpp accumulation. Nature (London) 1981;291:256–258. doi: 10.1038/291256a0. [DOI] [PubMed] [Google Scholar]
- 34.Strauch E, Takano E, Baylis H A, Bibb M J. The stringent response in Streptomyces coelicolor A3(2) Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 35.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 36.Tosa T, Pizer L I. Biochemical basis for the antimetabolite action of l-serine hydroxamate. J Bacteriol. 1971;106:972–982. doi: 10.1128/jb.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warren J R, Marshall B J. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 38.Whitehead K E, Webber G M, England R R. Accumulation of ppGpp in Streptococcus pyogenes and Streptococcus rattus following amino acid starvation. FEMS Microbiol Lett. 1998;159:21–26. doi: 10.1111/j.1574-6968.1998.tb12836.x. [DOI] [PubMed] [Google Scholar]
- 39.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]