Abstract
Synthesis of the OmpF porin of Escherichia coli is regulated in response to environmental and growth phase signals. In order to identify constituents of the various regulatory pathways involved in modulating ompF transcriptional expression, transposon insertion mutagenesis was performed and mutations that increased ompF′-lacZ activity were identified as previously described. Mutations mapping to a previously identified gene of unknown function, lrhA, were obtained. We found that LrhA, a LysR homolog, functions as a regulatory component in the RpoS-dependent growth phase repression of ompF. In addition to altered growth phase regulation of ompF, these lrhA mutants have pleiotropic stationary-phase defects as a result of decreased RpoS levels. We provide evidence that LrhA promotes degradation of RpoS by functioning within a genetic pathway that includes the response regulator SprE and the ClpXP protease. LrhA functions upstream of the other components in the pathway and appears to modulate the activity of SprE.
Physiological adaptation to changing environmental conditions is essential to microbial viability. This is particularly important for bacteria that alternatively inhabit distinct niches differing in conditions of oxidative stress, temperature, pH, and osmolarity, as well as nutrient availability and exposure to various toxins. OmpF and OmpC, the major porins of the gram-negative bacterium Escherichia coli, allow nonspecific diffusion of solutes through the protective outer membrane. Because the abundant OmpF porin allows for the more rapid diffusion of solutes through the outer membrane, it is a major entry point for most molecules that E. coli encounters, contributing significantly to outer membrane permeability and thus susceptibility to environmental hazards (33). For this reason, regulation of OmpF synthesis is necessarily complex and responsive to nutrient limitation (27) and a variety of environmental conditions (34), the best understood being medium osmolarity (10, 11, 28, 35). Since OmpF synthesis is highly responsive to fluctuation in various environmental parameters, studying its regulation provides us with an opportunity to understand the sensing mechanisms and complex regulatory networks necessary for E. coli adaptive physiology.
Synthesis of OmpF is responsive to the bacterial growth rate such that during rapid logarithmic growth OmpF levels are high, but as bacterial growth begins to be limited by nutrient availability, OmpF synthesis decreases (27). This, and many other stationary-phase-dependent alterations in envelope physiology, presumably aids in survival under conditions of decreased metabolic capacity by encapsulating the bacterium within a protective, less permeable cell envelope (19). In addition to protective envelope adaptations, synthesis of a core set of cytoplasmic proteins is induced, independently of the nature of the limiting nutrient, as E. coli enters into stationary phase, resulting in a multiple-stress-resistant physiological state. For instance, stationary-phase E. coli is resistant to heat and osmotic and oxidative shock, as well as treatment with alkylating agents, ethanol, and acidic or basic pH (24, 27). These protective functions (22, 27), and in particular the stationary-phase-dependent decrease in OmpF synthesis (27, 36), were found to be mediated by the alternative primary sigma factor ςS (ς38), encoded by the rpoS gene. RpoS-dependent stationary-phase regulation of OmpF synthesis is mediated at the transcriptional level, as indicated by the approximately three- to fivefold derepression of ompF′-lacZ expression observed in an rpoS null mutant specifically during stationary-phase growth (36). However, the precise mechanism by which RpoS promotes transcriptional repression at ompF is unclear.
RpoS functions as a global regulator of many genes required for the physiological transition into stationary-phase growth, and as such, its levels are highly responsive to the bacterial growth rate. During rapid growth, RpoS is maintained at a low level. However, during the transition to stationary phase, RpoS levels rise dramatically. This strict growth phase regulation of RpoS accumulation is quite complex in that it has been proposed to be mediated at all levels of synthesis and stability and via multiple inputs (15). Expression of various rpoS′-lacZ operon fusions increases approximately four- to fivefold during entry into stationary phase in rich medium and is weakly induced in response to starvation for carbon, nitrogen, or phosphate in minimal medium (22). The weak starvation induction ratio seen with rpoS′-lacZ transcription fusions is in stark contrast to the strongly increased amount of RpoS observed under the same medium conditions (20), implying that there is significant posttranscriptional growth phase regulation of RpoS. In fact, a greater stationary-phase induction ratio of RpoS at the level of translation was demonstrated with various rpoS′-′lacZ protein fusions (20, 23, 26).
However, it has recently been argued that the stationary-phase-dependent induction of RpoS occurs exclusively through regulation of its susceptibility to proteolysis (46). Logarithmic-phase RpoS accumulation is minimal due to rapid degradation by the ClpXP protease, which requires amino acid residues 173 to 188 of RpoS for target recognition (40). ClpXP-mediated degradation of RpoS absolutely requires the two-component response regulator SprE (36), also called RssB (29) or, in Salmonella typhimurium, MviA (1). Constitutively active alleles of sprE prevent full induction of RpoS as bacteria enter stationary phase (36), while sprE null mutants allow logarithmic-phase RpoS accumulation to levels nearly equivalent to those observed in stationary phase (29, 36). SprE functions upstream of ClpXP to promote RpoS degradation in a substrate-specific manner which leaves ClpXP protease activity unaltered (36). Thus, SprE is the first response regulator to be implicated in regulating protein degradation, and its novel C-terminal domain reflects this unique function.
For all known two-component response regulators, activity is modulated through phosphorylation at a conserved aspartic acid residue (16, 43). The N-terminal receiver domain of SprE contains this conserved residue, and so SprE activity is also predicted to be modulated via phosphorylation. The phosphorylation of SprE at the conserved aspartic acid has been demonstrated in vitro with acetyl phosphate (5). Additionally, it has been shown that Δ(pta ackA) mutants, which can no longer synthesize acetyl phosphate, affect the in vivo function of SprE such that it no longer efficiently promotes RpoS degradation (5). This implies that it is the phosphorylated form of SprE that is functionally active. In the absence of acetyl phosphate, however, RpoS accumulation remains growth phase regulated, indicating that there must be an additional factor(s) that influences SprE activity. The additional growth phase signal(s) and effector molecule(s) that regulate SprE remain to be discovered. Here, we describe the identification and initial characterization of another component of the SprE/ClpXP pathway that functions to modulate SprE activity.
MATERIALS AND METHODS
Bacteria and bacteriophage.
All bacterial strains constructed for this study (Table 1) are derivatives of MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR] (7). Bacteria were grown at 37°C, and standard microbiological techniques were used for strain construction (41). Genetic map positions were determined by standard techniques, including Hfr mapping and P1vir transduction.
TABLE 1.
Bacterial strains
| Strain | Genotype | Referencea |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 ptsF25 deoC1 thiA1 | 7 |
| KEG400 | MC4100 ompR107 Φ(ompF′-lacZ+)16-13 | |
| KEG403 | KEG400 lrhA49::cam | |
| KEG404 | KEG400 rpoS::kan | |
| KEG405 | KEG403 rpoS::kan | |
| KEG408 | RO91 lrhA49::cam | |
| KEG409 | RO91 lrhA::spc | |
| KEG410 | KEG408 lrhA49::cam::spc | |
| KEG415 | LP801 lrhA49::cam | |
| KEG416 | LP801 lrhA::spc | |
| KEG418 | MC4100 lrhA49::cam | |
| KEG419 | MC4100 lrhA::spc | |
| KEG420 | LP810 lrhA::spc | |
| KEG421 | KEG408 clpX::kan | |
| KEG422 | KEG409 clpX::kan | |
| KEG423 | KEG412 rpoS::kan | |
| KEG429 | MC4100 clpX::kan clpP::cam | |
| KEG445 | ZK916 lrhA49::cam | |
| KEG446 | ZK916 lrhA::spc | |
| KEG448 | KEG445 lrhA49::cam::spc | |
| KEG449 | LP867 lrhA49::cam | |
| KEG447 | LP867 lrhA::spc | |
| LP801 | RO91 sprE::IS1 trpC::Tn10 | 36 |
| LP810 | RO91 sprE19::cam | 36 |
| LP855 | RO91 clpX::kan clpP::cam | 36 |
| LP867 | ZK916 cysC::Tn10 rpoS::kan | 37 |
| RO91 | MC4100 λΦ(rpoS742′-′lacZ) | 20 |
| ZK916 | MC4100 Φ(bolA′-lacZ) | 21 |
Strains for which no reference is given were created in this study.
Media, reagents, and enzymes.
Luria-Bertani (LB) growth medium was prepared as described previously (41). ONPG (ortho-nitrophenyl-β-d-galactoside) utilized for β-galactosidase assays was purchased from Sigma. β-Agarase (New England Biolabs), T4 DNA ligase (New England Biolabs), Taq polymerase (United States Biochemical Corp.), and reagents used for DNA sequence analysis (United States Biochemical Corp.) were used according to the recommendations of the respective manufacturers. For immunoblot analysis, proteins were transferred to a nitrocellulose membrane (pore size of 0.2 μm) purchased from Schleicher & Schuell. Monoclonal antibodies to RpoS were a gift of R. Burgess (32). Rabbit antibodies to ClpX and ClpP were a gift of S. Gottesman (12, 25). The ECL antibody detection kit and the anti-mouse and anti-rabbit immunoglobulin horseradish peroxidase-linked whole antibody (from sheep) were purchased from Amersham. Synthesis of oligonucleotide primers was provided by the Princeton University Department of Molecular Biology synthesis facility.
Biochemical analysis.
β-Galactosidase assays were performed as previously described (42). Activities are expressed as (units/A600) × 103, where 1 unit is 1 μmol of orthonitrophenol formed per minute. The data are averages of at least four independent assays, and the standard deviations are indicated as error bars.
For immunoblot analysis, whole-cell extracts were prepared by pelleting the bacterial culture and resuspending the pellet in a volume of loading buffer (41) that was determined to standardize for total protein based on culture optical density at 600 nm (OD600). The samples were boiled for 10 min, and an equal volume of sample was then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Immunoblot analysis was performed as described previously (32). The anti-RpoS monoclonal antibody was used at a dilution of 1:1,000, and the anti-ClpP and anti-ClpX rabbit polyclonal antibodies were used at a dilution of 1:5,000.
PCR amplification, DNA sequence analysis, and DNA cloning.
Chromosomal DNA adjacent to the lrhA49::cam insertion was amplified by a two-round PCR procedure (6). The first round of PCR (30 cycles) was performed at low stringency (annealing temperature of 45°C) with a primer recognizing the cat gene within mini-Tncam (out1-L, 5′CAGGCTCTCCCCGTGGAG) and a set of degenerate primers with a 5′ tag (ARB1, 5′GGCCACGCGTCGACTAGTACN10GATAT). The second round of PCR (30 cycles) was performed at a higher stringency (annealing temperature of 55°C) by using as the DNA template the PCR products obtained in the first round. The second round of PCR utilized a primer recognizing the insertion element of Tncam (1-L, 5′CTGCCTCCAGAGCCTG) and a primer annealing to the 5′ tag created in the first round (ARB2, 5′GGCCACGCGTCGACTAGTAC). The resulting PCR products were gel purified and then used as a template for DNA sequence analysis, as previously described (37). The lrhA and o405 (accession no. AE000318) genes were cloned into the high-copy pCR-Script vector (Stratagene). DNA encoding each gene was generated by PCR amplification from a chromosomal preparation of the Tncam49 mutant. We used a primer annealing within Tncam for lrhA (out1-L, CAGGCTCTCCCCGTGGAG) and o405 (out1-R, CTCCACGGGGAGAGCCTG) in combination with a primer overlapping the 3′ end of either gene (lrhA3′, CTATCGTCCGTCG; o4053′, GGCAGTGAAATTAAG). The PCR products were purified and used for subsequent ligation into the pCR-Script vector according to the manufacturer’s recommended procedure [Amp SK(+) cloning kit; Stratagene].
RESULTS
ompF growth phase regulation.
In order to identify loci involved in RpoS-dependent repression at ompF, a Tncam insertion mutagenesis screen was performed and mutations which increased expression of ompF′-lacZ in the presence of the constitutively repressing ompR107 allele were isolated (36). The expectation was that some of the Lac+ insertions would result from an alleviation of stationary-phase repression and thereby identify regulatory components functioning either upstream or downstream of RpoS. In order to distinguish the mutations that altered RpoS-dependent expression of ompF′-lacZ from those affecting other regulatory pathways, each was placed in double-mutant combination with an rpoS::kan null allele (36), with the expectation that mutations which specifically perturbed RpoS regulation would be nonadditive in comparison to the rpoS::kan single-mutant phenotype. There were two loci identified, sprE and crl, which met this specific requirement (36, 37). Here, we describe the characterization of a subset of the mutations which did not behave in clearly additive fashion with rpoS::kan in this simple double-mutant test. However, these mutations specifically perturbed growth phase regulation of ompF′-lacZ, and for this reason we continued to pursue the nature of the stationary-phase defect and its relation to RpoS regulation.
The 52-min linkage group.
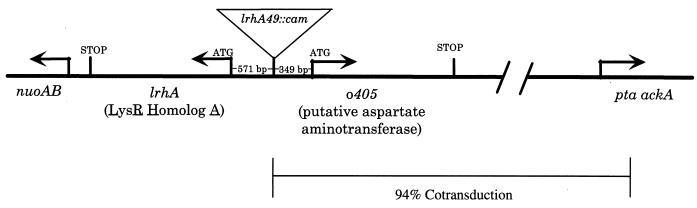
We found that eight independently isolated Tncam insertions obtained in the screen described above belong within a single linkage group. These mutations were mapped by Hfr crosses to approximately 52 min and were subsequently found to cotransduce with phage P1vir at 94% with Δ(ackA pta) at 51.8 min (45). In order to avoid confusion, we will refer to a representative mutant from this linkage group as lrhA49::cam and will justify this nomenclature below.
lrhA49::cam causes pleiotropic stationary-phase defects.
The transcriptional activity at ompF responds to two major signals: medium osmolarity (13) and stationary-phase growth (27, 36). As a first approximation in determining which, if either, regulatory pathway was perturbed, the ompF′-lacZ phenotype of each mutant was compared to that of the isogenic parent in logarithmic and stationary phase. We expected that altered osmotic regulation would be detected as a derepression of ompF′-lacZ under both growth conditions. In contrast, altered growth phase regulation would be detected as a specific defect in stationary-phase expression of ompF′-lacZ, as was previously observed in an rpoS null mutant (36).
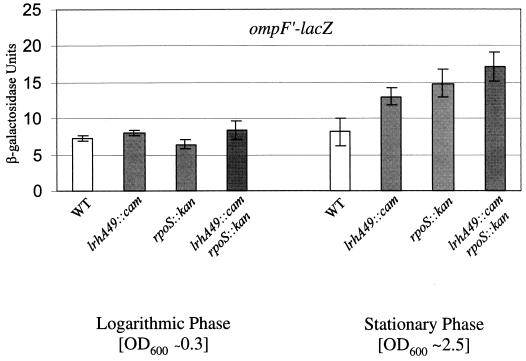
Each of the mutations in the 52-min linkage group conferred a similar growth phase defect in expression of ompF′-lacZ. A representative mutant, lrhA49::cam, resulted in an approximately twofold derepression of ompF′-lacZ expression specifically during stationary-phase growth, similar to what we observed with the rpoS::kan null allele (Fig. 1). Logarithmic-phase expression, and more specifically osmotic regulation (data not shown), of ompF′-lacZ remained unperturbed (Fig. 1). The lrhA49::cam mutation has no effect on ompF′-lacZ expression in an ompR+ background (data not shown). This is not surprising, since rpoS::kan results in only a modest increase in ompF′-lacZ in the presence of ompR+ (36), and it further supports the conclusion that lrhA49::cam plays no role in modulating the OmpR/EnvZ regulatory pathway. As was alluded to previously, ompF′-lacZ expression in the lrhA49::cam rpoS::kan double mutant may be subtly increased in comparison to the single mutants (Fig. 1); however, we suggest that the effects of lrhA49::cam are largely dependent upon RpoS (see Discussion).
FIG. 1.
Effect of lrhA49::cam on ompF′-lacZ expression. Each strain was grown in LB broth at 37°C with aeration overnight and then subcultured at 1:100 into LB broth. To determine logarithmic-phase phenotypes, β-galactosidase activity from the ompF′-lacZ gene fusion was assayed when cultures reached an OD600 of ∼0.3. The same strains were grown in LB broth at 37°C for 24 h to obtain analogous stationary-phase cultures. The wild-type (WT) parent is KEG400 (ompF′-lacZ), and the mutant derivatives are KEG403 (lrhA49::cam), KEG404 (rpoS::kan), and KEG405 (lrhA49::cam rpoS::kan).
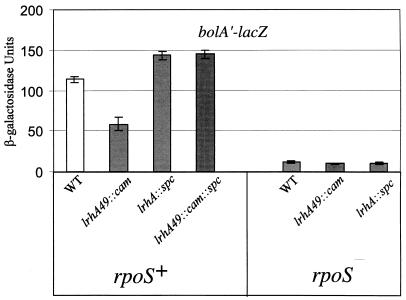
To determine if lrhA49::cam affected the stationary-phase response in general, we examined the expression of several other genes known to be regulated by RpoS. The lrhA49::cam mutant was unable to hydrolyze H2O2 to the same degree as the wild type when a 30% H2O2 solution was dropped onto patched bacteria (determined visually as a bubbling that reflected the amount of O2 generated). This “fizzing” phenotype depends upon synthesis of the KatE catalase, which is induced by RpoS as bacteria enter stationary phase (31). The stationary-phase expression of bolA′-lacZ (21) (Fig. 2) and to a lesser extent poxB′-lacZ (9) (data not shown), which are subject to RpoS-dependent stationary-phase induction, were also decreased in the presence of lrhA49::cam. These observations indicate that the lrhA49::cam mutation disrupts the activity of a factor involved globally in growth phase regulation, rather than specifically in ompF stationary-phase expression, such that proper development of multiple aspects of stationary-phase physiology is hindered.
FIG. 2.
Effect of lrhA49::cam on bolA′-lacZ expression. Each strain was grown in LB broth at 37°C with aeration, and cells were harvested at 24 h of growth to determine the β-galactosidase activity of the bolA′-lacZ strain during stationary phase. The wild-type (WT) parent is ZK916, and its derivatives are KEG445 (lrhA49::cam), KEG446 (lrhA::spc), LP867 (rpoS::kan), KEG448 (rpoS::kan lrhA49::cam), and KEG449 (rpoS::kan lrhA::spc).
lrhA49::cam results in overexpression of lrhA.
Through PCR amplification and DNA sequence analysis of the chromosomal DNA adjacent to the Tncam mutations, we were able to determine their precise insertion junctions. These mutations were found to be inserted at the same site within an intergenic region between two genes of unknown function, suggesting that the insertions most likely alter the expression of one or both of the adjacent genes (Fig. 3). The lrhA gene has significant homology to LysR (3), suggesting that it functions as a DNA-binding transcription factor, while the o405 gene was recognized to possess homology to aspartate aminotransferase (2).
FIG. 3.
Diagrammatic representation of lrhA49::cam physical location. lrhA49::cam (as well as seven other insertions) was mapped by Hfr and P1vir two-factor crosses to approximately 52 min on the E. coli chromosome. Sequencing of the lrhA49::cam insertion junction revealed its location between two open reading frames, lrhA and o405 (accession no. AE000318). Directly downstream of lrhA is the nuoAB operon encoding the NADH dehydrogenase I enzyme complex.
Both genes, lrhA and o405, were cloned following PCR amplification from the E. coli chromosome into the pCR-Script vector such that gene expression would be under the control of the native promoter. Each plasmid was then tested for regulatory effects on ompF in KEG400 (MC4100 ompF′-lacZ ompR107). The presence of lrhA in multicopy increased the expression of ompF′-lacZ twofold, phenocopying lrhA49::cam, while o405 in multicopy had no effect on ompF′-lacZ expression. This result suggests that these Tncam mutations result in either increased or constitutive expression of lrhA.
Consistent with the previous observation, the null allele lrhA::spc (3) was found to have a phenotype opposite that of lrhA49::cam and resulted in decreased ompF′-lacZ expression (data not shown), while expression of poxB′-lacZ (data not shown) and bolA′-lacZ (Fig. 2) was increased. Further, we determined the bolA′-lacZ phenotype of a double mutant in which the lrhA49::cam insertion was combined in cis with the lrhA::spc null allele. The phenotype of the double mutant was indistinguishable from that of the lrhA::spc mutant (Fig. 2). Since this lrhA::spc allele was previously shown to have no effect upon expression of the nuo locus (3), these data argue against the possibility that lrhA49::cam alters stationary-phase regulation through effects on these genes. We conclude from this series of observations that lrhA49::cam is a gain-of-function mutation that increases lrhA expression and that o405 plays no role in the altered stationary-phase regulation we observe. This justifies the designation lrhA49::cam for this insertion mutation.
LrhA functions within the RpoS pathway to alter stationary-phase gene expression.
As described above, lrhA mutations perturb stationary-phase expression of bolA′-lacZ (Fig. 2). We were thus able to utilize this transcription fusion to determine the epistatic relationship between lrhA and rpoS. The lrhA49::cam allele was introduced into an rpoS::kan mutant, and the stationary-phase phenotypes of the single and double mutants were determined. We reasoned that if LrhA functions in a growth phase regulatory pathway other than the RpoS pathway, we should observe an additive double-mutant phenotype such that bolA′-lacZ expression in the double mutant would be decreased in comparison to either single mutant. However, if LrhA functions within the RpoS pathway, the double mutant should have a phenotype similar to that of either the rpoS::kan null mutant or the lrhA49::cam overexpression mutant, depending upon where each gene functions within the RpoS pathway. If rpoS functions upstream of lrhA, then we should expect the double mutant to have a phenotype similar to lrhA49::cam. However, if lrhA functions upstream of rpoS, we should expect the double-mutant phenotype to be identical to that of the rpoS::kan mutant. What we observed in the double mutant was a nonadditive level of bolA′-lacZ expression equivalent to that of the rpoS::kan single mutant (Fig. 2). We found that the lrhA::spc rpoS::kan double mutant is also phenotypically equivalent to the rpoS::kan single mutant (Fig. 2), further supporting the conclusion that rpoS is epistatic to lrhA. From these results, we conclude that LrhA acts upstream of RpoS to down-regulate the RpoS pathway and thus indirectly perturbs stationary-phase expression of bolA, and probably katE, poxB, and ompF as well.
lrhA mutations perturb growth phase regulation of RpoS posttranslationally.
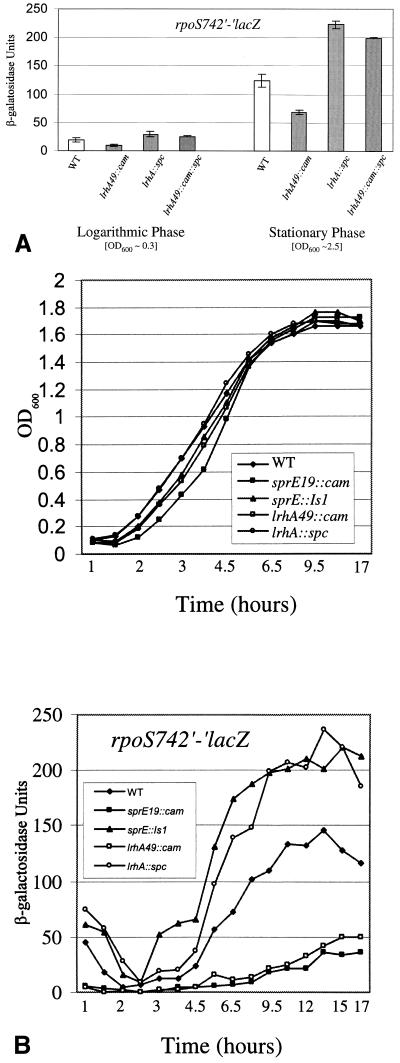
Since LrhA plays a role in modulating the activity of the RpoS pathway, we wanted to determine whether it functions to vary the levels of RpoS or whether LrhA acts in concert with RpoS to regulate stationary-phase development. Using the rpoS742′-lacZ operon fusion (20), we found that transcription of rpoS is unperturbed in the presence of lrhA49::cam (data not shown). In contrast, lrhA49::cam decreases expression of the rpoS742′-′lacZ protein fusion (30) by approximately twofold (Fig. 4), revealing that LrhA functions to regulate RpoS levels posttranscriptionally. The lrhA::spc mutant has the opposite phenotype, resulting in increased rpoS742′-′lacZ expression, and it overrides lrhA49::cam in the double mutant (Fig. 4A).
FIG. 4.
Effects of lrhA mutations on rpoS742′-′lacZ expression. (A) Strains were grown in LB broth at 37°C with aeration overnight and then subcultured at 1:100 into LB broth. To determine logarithmic-phase phenotypes, β-galactosidase activity from the rpoS742′-′lacZ gene fusion was assayed when cultures reached an OD600 of ∼0.3. The same strains were grown in LB broth at 37°C for 24 h to obtain analogous stationary-phase cultures. The wild-type (WT) parent is RO91 (rpoS742′-′lacZ), and its derivatives are KEG408 (lrhA49::cam), KEG409 (lrhA::spc), and KEG410 (lrhA49::cam::spc). (B) Strains were grown in LB broth at 37°C with aeration overnight and then subcultured at 1:100 into LB broth. Aliquots of each culture were taken at the indicated time points, and both the OD600 and β-galactosidase activity the samples were determined. The wild-type (WT) parent is RO91, and its mutant derivates are indicated in the figure.
Similar regulatory effects are observed when rpoS742′-′lacZ expression is assayed in logarithmic phase (Fig. 4A). When rpoS742′-′lacZ expression was assayed throughout the growth curve, we found that lrhA49::cam caused decreased expression at all time points and allowed only weak induction as bacteria entered stationary phase (Fig. 4B). We also observed greater induction of rpoS742′-′lacZ during mid-logarithmic-phase growth in the lrhA::spc mutant (Fig. 4B).
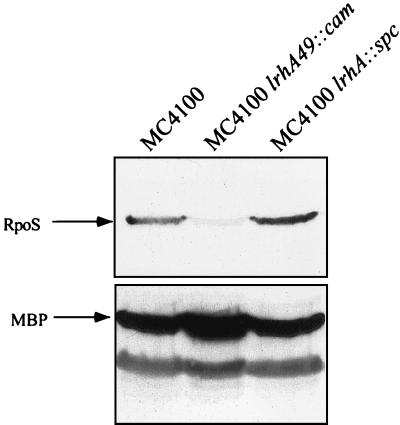
These lrhA growth curve phenotypes mimic what was observed with analogous overexpression and null alleles of sprE (36) (Fig. 4B) and suggested that LrhA could also be involved in growth phase regulation of RpoS accumulation. Therefore, we determined the steady-state level of RpoS during stationary phase by Western blot analysis in the lrhA mutant backgrounds (Fig. 5). In particular, the lrhA49::cam mutation results in dramatically decreased RpoS levels such that the protein is barely detectable. This inability to accumulate RpoS during the transition into stationary phase in the lrhA49::cam mutant has clear effects on the ability of these bacteria to induce genes such as bolA, katE, and poxB, which are important for viability during stationary phase (19, 24).
FIG. 5.
Immunoblot analysis of ςS. RpoS is indicated by an arrow in MC4100 and its lrhA mutant derivatives. Each strain was grown in LB broth at 37°C with aeration, and cells were harvested after 24 h of growth at an OD600 of ∼2.5 to obtain a whole-cell lysate, as described in Materials and Methods. After immunoblot analysis with anti-RpoS antibodies, the blot was stripped and reprobed with anti-MBP antibody to control for loading.
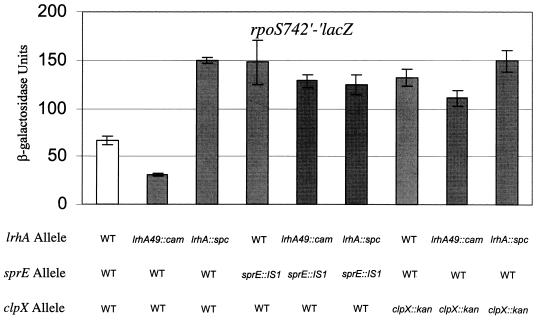
It has been proposed that modulation of the rate of RpoS degradation is the major control point for determining the amount of RpoS that accumulates throughout the growth curve (46). During logarithmic growth, RpoS is maintained at low levels due to degradation mediated by SprE and the ClpXP protease (29, 36, 40), and the rpoS742′-′lacZ protein fusion is susceptible to this regulated proteolysis (30). In order to determine whether LrhA acts within the SprE/ClpXP pathway, we performed tests of epistasis between alleles of lrhA and the null alleles sprE::IS1 and clpX::kan. We found that the presence of either the lrhA49::cam or the lrhA::spc allele had no phenotypic effect on rpoS742′-′lacZ in the absence of SprE or ClpX (Fig. 6). Therefore, lrhA functions to promote the degradation of RpoS through the genetically defined SprE/ClpXP pathway. Furthermore, LrhA must act upstream of the other known components of this pathway since the lrhA49::cam overexpression allele is phenotypically silent in the double mutants.
FIG. 6.
Epistasis test between lrhA and sprE::Is1 or clpX::kan. Each strain was grown in LB broth at 37°C with aeration, and cells were harvested after 24 h of growth at an OD600 of ∼2.5 to determine the stationary-phase rpoS742′-′lacZ phenotype. All strains are derived from RO91 (rpoS742′-′lacZ) and contain the indicated mutation(s). WT, wild type.
LrhA does not regulate synthesis of ClpX, ClpP, or SprE.
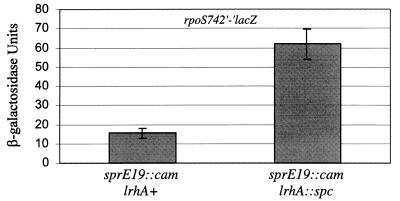
Since LrhA has homology to the LysR-like family of transcription factors (3), possessing a canonical helix-turn-helix DNA-binding domain, we wanted to determine whether LrhA functions to regulate the expression of sprE, clpX, or clpP and thereby to affect RpoS accumulation. We were able to genetically determine whether LrhA plays a regulatory role at the sprE promoter by utilizing the allele sprE19::cam (36). In sprE19::cam strains, sprE expression is uncoupled from its native promoter and is constitutively expressed independently of the native promoter’s activity. This Tncam is inserted 22 bp upstream of the start codon of sprE and has been found by Western blot analysis to result in constitutively increased levels of SprE (data not shown). There is a promoter within the transposon oriented towards sprE that is most likely responsible for stimulating this constitutive overexpression. We combined the sprE19::cam allele with lrhA::spc and determined the phenotype of the double mutant by assaying rpoS742′-′lacZ expression. Any effect of LrhA at the native sprE promoter should be blocked by the sprE19::cam mutation. However, we observed an increase in rpoS742′-′lacZ expression in the double mutant (Fig. 7), suggesting that LrhA does not function to regulate the activity of the native sprE promoter. Moreover, lrhA mutants have no effect upon stationary-phase SprE levels, as determined by Western blot analysis (data not shown). Thus, we conclude that LrhA does not regulate SprE synthesis.
FIG. 7.
Epistasis test between lrhA::spc and sprE19::cam. Each strain was grown in LB broth at 37°C with aeration, and cells were harvested after 24 h of growth at an OD600 of ∼2.5 to determine the stationary-phase rpoS742′-′lacZ phenotype. All strains are derived from LP801 (rpoS742′-′lacZ sprE19::cam) and contain the indicated lrhA allele.
We assayed for potential regulation of clpX and clpP by determining protein levels through Western blot analysis from both logarithmic- and stationary-phase cultures. We found no differences in the level of ClpX or ClpP in either stationary phase (Fig. 8) or logarithmic phase (data not shown) when comparing the lrhA mutants to the isogenic parental strain MC4100. The synthesis of neither SprE nor the ClpXP protease is altered in lrhA mutants. Since it was demonstrated previously that clpX and clpP are epistatic to sprE (36), and our epistasis data place lrhA function upstream of sprE; these final observations imply that LrhA acts to modulate the activity of the response regulator SprE.
FIG. 8.
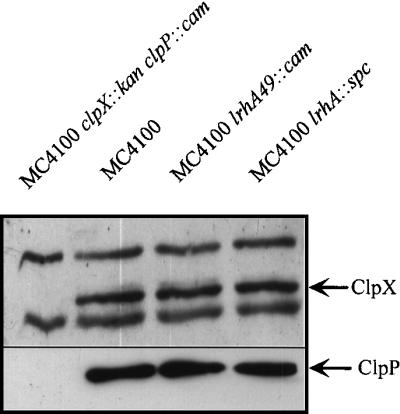
Immunoblot analysis of ClpX and ClpP. The ClpX and ClpP proteins in MC4100 and the indicated lrhA derivatives (the clpX::kan clpP::cam mutant is included as a control) are indicated by arrows. Each strain was grown in LB broth at 37°C with aeration, and cells were harvested after 24 h of growth at an OD600 of ∼2.5 to obtain a whole-cell lysate, as described in Materials and Methods. After immunoblot analysis with anti-ClpX antibodies, the blot was stripped and reprobed with anti-ClpP antibodies (the same result was obtained regardless of whether the blot had been previously probed and stripped [data not shown]).
DISCUSSION
To better understand the pathways which regulate ompF expression in response to environmental and growth phase signals, Tncam insertion mutagenesis was performed (36). This screen has successfully identified a number of novel components involved in stationary-phase-dependent RpoS repression at ompF. All of the components characterized thus far function to regulate RpoS accumulation and/or activity (36, 37). There are no known downstream effectors of RpoS regulation at ompF, suggesting that RpoS may repress expression at ompF directly. However, the mechanism of repression remains to be clarified.
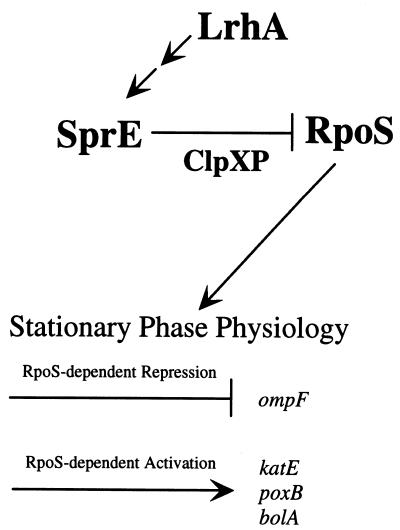
We have found that LrhA is another component of the ompF growth phase regulatory pathway and that it is involved in modulating stationary-phase-dependent RpoS accumulation. LrhA functions to modulate RpoS levels by promoting rapid degradation during logarithmic growth (Fig. 9). This conclusion is based partially on the observation that the lrhA49::cam overexpression allele allows very little stationary-phase accumulation of RpoS, while in contrast the lrhA::spc null allele allows greater RpoS accumulation during logarithmic growth. More telling is the observation that LrhA-dependent inhibition of RpoS accumulation absolutely requires the response regulator SprE and the ClpXP protease, placing LrhA upstream of SprE within this genetic pathway. Previous studies have determined that SprE acts upstream of ClpXP (36) in a substrate-specific manner to target RpoS for degradation (46). Since LrhA does not control the synthesis of SprE, we further suggest that LrhA promotes RpoS degradation by modulating the activity of SprE.
FIG. 9.
Genetic pathway required for growth phase-dependent regulation of RpoS degradation. LrhA is the most upstream component and somehow functions to modulate SprE activity. The phosphorylated form of SprE promotes degradation of RpoS by the ClpXP protease, probably in response to nutrient limitation. Then, RpoS, functioning as a sigma factor, is able to recruit RNAP to a subset of stationary-phase-inducible promoters and thereby acts as a global regulatory protein required for the physiological adaptations characteristic of stationary-phase bacteria.
SprE has been identified as a member of the family of two-component response regulators based on homology (4). As such, its activity is most likely modulated via phosphorylation at a conserved aspartic acid residue (43). However, a cognate sensor histidine kinase has yet to be identified for SprE, and so it remains unclear how SprE activity is growth phase modulated and to what specific signal(s) it responds. As has been observed with other response regulators, SprE can be phosphorylated in vitro by acetyl phosphate (5). This phosphorylation can be further detected in vivo as a SprE-dependent increase in rpoS742′-′lacZ expression in a Δ(pta ackA) mutant (5). However, as there is still stationary-phase induction of RpoS in the Δ(pta ackA) mutant, there must be an additional signal(s) which mediates growth phase regulation of SprE activity.
We propose that LrhA and acetyl phosphate modulate SprE activity independently. This is based upon initial experiments performed to elucidate LrhA function at ompF. In envZ::cam null strains, the orphaned response regulator OmpR is dependent upon phosphorylation by acetyl phosphate for DNA-binding activity at ompF (17), thereby allowing us to use OmpR-dependent activation at ompF to monitor acetyl phosphate levels. In an envZ::cam rpoS::kan background, we were able to determine whether LrhA had an effect upon ompF′-lacZ in the absence of SprE-dependent stationary-phase regulation. We observed that introduction of the lrhA::spc null mutant had no effect upon the phosphorylation level of OmpR; i.e., ompF′-lacZ expression remained unchanged (data not shown). If LrhA was involved in regulating acetyl phosphate levels, we would expect a decrease in ompF′-lacZ levels due to indirect effects upon OmpR activity. Instead, our observation suggests that LrhA regulates RpoS levels independently of acetyl phosphate and that there are probably multiple inputs which modulate SprE activity.
LrhA, as a putative DNA-binding transcription factor (3), is likely to regulate SprE activity indirectly. Perhaps LrhA regulates expression of an as-yet-unidentified cognate sensor histidine kinase. Alternatively, LrhA might regulate the expression of metabolic enzymes, as is observed with many LysR family members (39), the activity of which might in turn be sensed by SprE either indirectly, through a cognate histidine kinase sensor, or directly, through a small molecule phosphodonor or a phosphorylated metabolic enzyme. The direct regulatory targets of LrhA are unknown, but identification of these genes will provide an important link in our understanding of how SprE activity is modulated. Further characterization of LrhA function may also provide a means toward understanding how E. coli senses the growth rate in order to regulate RpoS appropriately.
LrhA was originally identified as a putative DNA-binding transcription factor based on homology to the LysR family of proteins. This LysR homology suggests that LrhA activity may be modulated via binding of a small molecule inducer (32). Typically, LysR homologs regulate expression of genes functioning within a metabolic pathway, and their activity is in turn modulated through binding of a metabolic intermediate, which is either utilized or produced by the regulated pathway. We do not yet understand how LrhA activity is regulated; i.e., the molecular nature of the coinducer and how its levels fluctuate are unknown. It may be that coinducer synthesis changes in response to the bacterial growth rate, and in this way LrhA would be able to modulate SprE activity, and thus RpoS accumulation, according to nutrient availability.
As noted above, additivity tests with lrhA49::cam and rpoS do not yield clear answers about the requirement for RpoS in the stationary-phase derepression of ompF. We suspect that this problem relates to nonspecific phenotypes caused by LrhA overexpression. The stress caused by excess LrhA probably affects ompF expression through an indirect mechanism that is unrelated to normal cellular function.
LrhA is most closely related to the PecT regulator of pectinolysis in Erwinia chrysanthemi (43) and the HexA regulator of pectinolysis and motility in Erwinia carotovora (14). E. chrysanthemi is a phytopathogenic enterobacterium that requires induction and secretion of a group of pectinolytic enzymes in order to cause soft rot disease in a variety of plants (18). The pectinase genes are regulated at the level of transcription in response to a variety of signals, including the presence of pectin and plant extracts, stationary-phase growth, osmolarity, low temperature, oxygen and nitrogen availability, and iron limitation. pecT was found in a screen designed to identify genes involved in regulating pectate lyase synthesis and was shown to function as a transcriptional repressor of five pectate lyase genes (43). As with LrhA, it is not known what environmental conditions regulate PecT activity.
In a subsequent study of pectinolysis regulation, a Tn5 insertion mutation which decreased the expression of a number of pectate lyases was isolated (8). This Tn5 was inserted within the intragenic region between pecT and the adjacent putative aspartate aminotransferase, 579 bp upstream of the pecT gene, in a manner highly analogous to the lrhA49::cam group of mutants at the lrhA locus described herein. The adjacent putative aspartate aminotransferase was shown to play no role in the Tn5 mutant phenotype or in pectate lyase regulation. Instead the Tn5 caused increased transcription at pecT by preventing PecT negative autoregulation. Since LrhA is 95% similar to PecT in the first 120 amino acids, which contain the putative DNA-binding domain, and since lrhA49::cam phenocopies lrhA overexpression, we infer that LrhA functions as a DNA-binding protein that negatively autoregulates its own expression. Furthermore, we would suggest, based on our observations, that perhaps PecT regulates the E. chrysanthemi pectate lyases in a growth phase-dependent manner through regulatory effects upon RpoS.
ACKNOWLEDGMENTS
We are thankful to L. Pratt for initial isolation of the lrhA49::cam mutant described in this paper and for helpful comments about this work. We are grateful to R. Burgess for the generous gift of RpoS monoclonal antibody and to S. Gottesman for generously providing us with both ClpX and ClpP polyclonal antibodies. We thank J. Bongaerts and G. Unden for providing us with the lrhA::spc allele. We thank B. Bassler, N. Hand, J. Huie, N. Ruiz, and especially T. Raivio for critical reading of the manuscript, and many thanks go to S. DiRenzo for help with preparation of the manuscript.
K.E.G. was supported by an NIGMS training grant (GM07388); T.J.S. was supported by NIGMS grant GM35791.
REFERENCES
- 1.Bearson S M, Benjamin W H J, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 4.Bosl M, Kersten H. Organization and functions of genes in the upstream region of tyrT of Escherichia coli: phenotypes of mutants with partial deletion of a new gene (tgs) J Bacteriol. 1994;176:221–231. doi: 10.1128/jb.176.1.221-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouche S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol. 1998;27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 6.Caetano-Annoles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–92. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 8.Castillo A, Reverchon S. Characterization of the pecT control region from Erwinia chrysanthemi 3937. J Bacteriol. 1997;179:4909–4918. doi: 10.1128/jb.179.15.4909-4918.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y Y, Wang A Y, Cronan J E J. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF) gene. Mol Microbiol. 1994;11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 10.Egger L A, Park H, Inouye M. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 11.Forst S A, Roberts D L. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol. 1994;145:363–373. doi: 10.1016/0923-2508(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 13.Hall M N, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K-12. J Mol Biol. 1981;46:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 14.Harris S J, Shih Y, Bentley S D, Salmond G P C. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence factors. Mol Microbiol. 1998;28:705–717. doi: 10.1046/j.1365-2958.1998.00825.x. [DOI] [PubMed] [Google Scholar]
- 15.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1995. pp. 1497–1512. [Google Scholar]
- 16.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 17.Hsing W, Silhavy T J. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 19.Huisman G W, Siegele D A, Zambrano M M, Kolter R. Morphological and physiological changes during stationary phase. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1672–1682. [Google Scholar]
- 20.Lange R, Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 21.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor ςS. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 23.Loewen P C, von Ossowski I, Switala J, Mulvey M R. KatF (ςS) synthesis in Escherichia coli is subject to posttranscriptional regulation. J Bacteriol. 1993;175:2150–2153. doi: 10.1128/jb.175.7.2150-2153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 25.Maurizi M R, Clark W P, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 26.McCann M P, Fraley C D, Matin A. The putative sigma factor KatF is regulated posttranscriptionally during carbon starvation. J Bacteriol. 1993;175:2143–2149. doi: 10.1128/jb.175.7.2143-2149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCann M P, Kidwell J P, Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 29.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 30.Muffler A, Traulsen D D, Lange R, Hengge-Aronis R. Posttranscriptional osmotic regulation of the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1996;178:1607–1613. doi: 10.1128/jb.178.6.1607-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen L H, Jenson D B, Thompson N E, Gentry D R, Burgess R R. In vitro functional characterization of overproduced Escherichia coli katF/rpoS gene product. Biochemistry. 1993;32:11112–11117. doi: 10.1021/bi00092a021. [DOI] [PubMed] [Google Scholar]
- 33.Nikaido H. Outer membrane. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 29–47. [Google Scholar]
- 34.Pratt L A, Hsing W, Gibson K E, Silhavy T J. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 35.Pratt L A, Silhavy T J. Porin regulation of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 105–127. [Google Scholar]
- 36.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt L A, Silhavy T J. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 38.Russo F, Slauch J M, Silhavy T J. Mutations that affect separate functions of OmpR, the phosphorylated regulator of porin transcription in Escherichia coli. J Mol Biol. 1993;231:261–273. doi: 10.1006/jmbi.1993.1281. [DOI] [PubMed] [Google Scholar]
- 39.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 40.Schweder T, Lee K-H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor (ςS) by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 42.Slauch J M, Silhavy T J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stock J B, Stock A M, Mottonen J M. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 44.Surgey N, Robert-Baudouy J, Condemine G. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J Bacteriol. 1996;178:1593–1599. doi: 10.1128/jb.178.6.1593-1599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanner B L, Wilmes-Riesenberg M R. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J Bacteriol. 1992;174:2124–2130. doi: 10.1128/jb.174.7.2124-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zgurskaya H I, Keyhan M, Matin A. The sigmaS level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol Microbiol. 1997;24:643–651. doi: 10.1046/j.1365-2958.1997.3961742.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Z, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]