Abstract
Microbial mass spectrometry imaging (MSI) is a powerful tool used to generate biological hypotheses about the roles of metabolites in microbial competition based upon their two-dimensional spatial distribution. The most commonly used ionization method for microbial MSI is matrix-assisted laser desorption ionization (MALDI). However, medium components and microbial metabolites influence the adhesion of agar samples to the MALDI target, limiting the applicability of MALDI MSI to microbes grown on specific media. Here, we describe a three-step process using a robotic sprayer for matrix application that improves adherence of agar samples to the MALDI target, enabling the use of different media for microbial growth and MSI analysis of larger sample surface areas.
Graphical Abstract
Mass spectrometry imaging (MSI) has been applied to microorganisms growing on agar to capture the spatial distribution of metabolic exchange between microbial competitors. This method of analysis of microbial cultures enables the generation of hypotheses regarding the biological function of a metabolites in inter-species interactions.1–3 The most commonly used ionization method for microbial MSI is matrix-assisted laser desorption ionization (MALDI).
In a classic microbial MALDI MSI experiment, a microbe is cultured on agar in a Petri dish, excised, transferred to a MALDI target, matrix is applied as a dry mixture of Universal Matrix (1:1 mixture of CHCA:DHB) using a sieve to hydrated agar cultures, and the sample is dehydrated prior to analysis.4 While applying matrix to hydrated samples via sieve is inexpensive and accessible, sample preparation is subjective, requires optimization for every medium and microbe, spatial resolution is limited due to matrix crystal size, and MALDI target surface area is constrained by the diameter of the sieve.5 In response to these limitations, multiple groups have developed spray-based approaches for matrix application to microbial cultures.6–9 Automated nebulizing sprayer methods for matrix application to agar samples have been shown to improve signal-to-noise and increase the number of metabolites detected compared to the sieve-based matrix application method.6–8
Regardless of the method used for matrix application, one of the major challenges of implementing microbial MALDI MSI more broadly to study microbial metabolite exchange is lack of adherence of agar samples to the MALDI target plate during sample preparation and/or data acquisition.5 Due to trapped air underneath the agar section, insufficient application of matrix, medium components, and/or metabolites produced by the microbes, the sample may lift or flake from the MALDI target plate preventing analysis or causing damage to the MALDI source. To circumvent challenges associated with sample adherence, specific media are used for microbial growth (such as ISP2 or MSgg5, 8), samples are inverted10, adhesion methods such as soft agar or soft landing are used9, 10, and DHB is included in the matrix or applied via sieve prior to function as a glue prior to application of the ionization matrix.5, 7, 11
Sieve-based matrix application for microbial MALDI MSI, either DHB for adherence or Universal Matrix for ionization, requires the use of a MALDI target that can fit within the sieve, which limits the surface area for analysis to a 96 spot MALDI target (~17.5 cm2 analysis area). Recent development of sprayer-based methods have enabled MSI imaging of microbial cultures on 384-spot target (~77 cm2 analysis area).11 However, in our hands, this method was not sufficient to prevent flaking of biosurfactant producing microbes, such as Bacillus subtilis, from the MALDI target.
Here, we describe the development of a three-step robotic sprayer-based method for matrix application to microbial agar samples for MALDI MSI that is both robust and reproducible. We followed the idea that DHB acts as a glue to enhance adherence of agar samples to MALDI targets and modified existing sprayer methods to incorporate applications of DHB to the MALDI target and samples prior to matrix coating for ionization.6, 7 We demonstrate the utility of this matrix application method on the well-characterized model bacterium B. subtilis strain 3610 grown on four different agar media.
MATERIALS AND METHODS
Microbial growth
B. subtilis 3610 was inoculated from glycerol stocks onto an LB plate and grown at 30 °C overnight. 4 mL LB broth was inoculated from overnight agar growth and incubated at 30 °C, overnight while shaking. 1.5 μL of B. subtilis 3610 broth culture was inoculated onto 12 mL of ISP2, Miller LB, BHI, or Tryptic Soy agar in 100 mm x 25 mm Petri dishes. B. subtilis 3610 monocultures were incubated for 72 hr at 30 °C prior to sample preparation for MALDI MSI. Expanded experimental details are in Supplemental Materials.
Sample preparation
Samples were prepared on a Bruker Daltonics ground steel MALDI 96 anchor target plate or 384 anchor target plate. The target plate was sparyed with DHB (‘plate coat’, 40 mg/mL in 70% ACN) using a TM Sprayer (HTX Technologies, Chapel Hill, NC). The following parameters were used: flow rate: 0.1 mL/min; passes: 4; nozzle temperature: 65°C; track spacing: 2 mm; nozzle velocity: 1200 mm/min; nozzle height: 65 mm (matrix density: 0.0067 mg/mm2). A region of agar, including the colony, was excised and laid upon the target. A ‘pre-coat’ of DHB (40 mg/mL in 90% ACN with 0.1% FA) was applied to the agar samples using the following sprayer parameters: flow rate 0.1 mL/min; passes: 6; nozzle temperature: 65°C; track spacing: 2 mm; nozzle velocity: 1200 mm/min; nozzle height: 65 mm (matrix density: 0.01 mg/mm2). Samples were dried at 37°C until dehydrated. After drying, a second matrix coat (‘final coat’) was applied to the samples for ionization of metabolites of interest. Sieved samples were prepared as previously described.4, 5 Additional experimental details are in Supplemental Materials.
MALDI MSI
MSI data was collected using an Autoflex Speed MALDI-TOF mass spectrometer equipped with a Nd:YAG smartbeam2 laser (Bruker Daltonics). The laser modulator was set to “small” mode. For each raster spot of 250 × 250 μm, 1000 shots (50 shots random walk) at a frequency of 2000 Hz were acquired in reflectron positive mode. All data were analyzed using FlexImaging v4.1 (Bruker Daltonics).
RESULTS AND DISCUSSION
Matrix Application on Agar.
The key principle underlying our matrix application method to improve adherence of agar samples to the MALDI target is the concept that DHB acts as a glue.5 In our hands, dehydration of agar samples containing microbial colonies directly onto the MALDI target led to flaking of the sample. Therefore, to improve agar sample adherence, DHB was sprayed onto the empty ground stainless steel MALDI target prior to mounting agar samples (Figure 1).11 While this ‘plate coat’ of matrix was sufficient to adhere ISP2 agar without microbial growth, it was not adequate to prevent agar containing B. subtilis colonies from flaking. Therefore, a second application of DHB was deposited onto the hydrated agar sample after mounting onto the MALDI target plate, aiming to replicate the effects of the sieve-based application of DHB.7 As matrix was applied to a hydrated sample, high organic solvent (90% ACN) was used to provide a drier spray for matrix application to minimize delocalization of metabolites. Spraying the hydrated sample with a ‘pre-coat’ of DHB enabled adherence of the sample to the MALDI target. After application of the ‘pre-coat’ of DHB, the agar sample was dehydrated and a ‘final coat’ of matrix was applied to enable ionization of metabolites.
Figure 1.
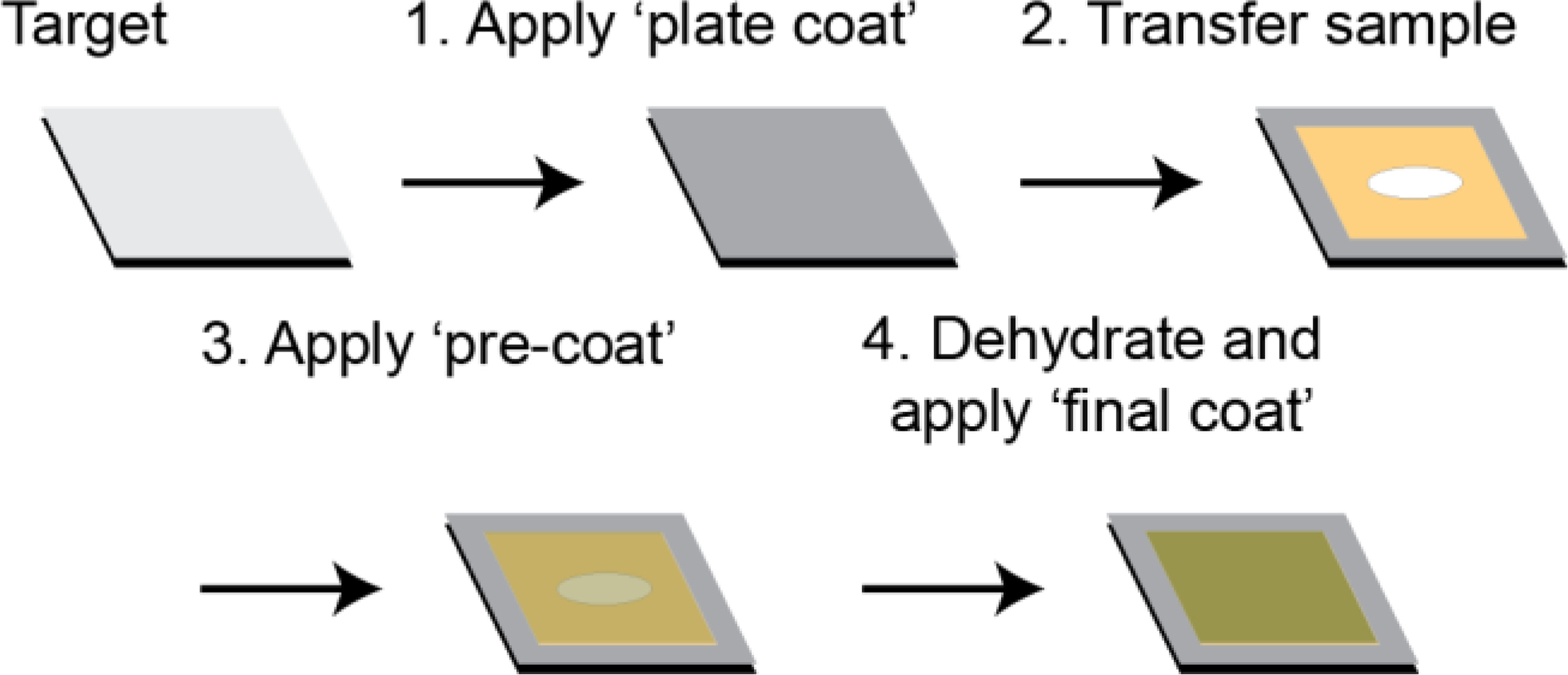
To adhere agar samples to MALDI target, the following steps were taken: (1) the target was coated with a solution of DHB, (2) the sample was mounted onto the target, (3) a ‘pre-coat’ of DHB was applied, and (4) the sample was dehydrated prior to application of the ‘final coat’ using the desired matrix for ionization.
MALDI MSI Analysis.
To verify that our method replicated reported increases in sensitivity using sprayer-based matrix application, MSI data was collected on B. subtilis colonies grown on ISP2 agar using the sieve method and the described three-step robotic sprayer method on the same target plate. As previously described, using an automated sprayer to apply matrix to the B. subtilis cultures resulted in better ionization of the surfactin and plipastatin molecular families and increased the total number of ions detected (Figure 2 (universal matrix), Figure S1 (DHB)).6, 7 Ionization of metabolites from the spray-based samples required lower laser power and detector gain likely due to enhanced co-crystallization of the metabolites with the matrix.
Figure 2.
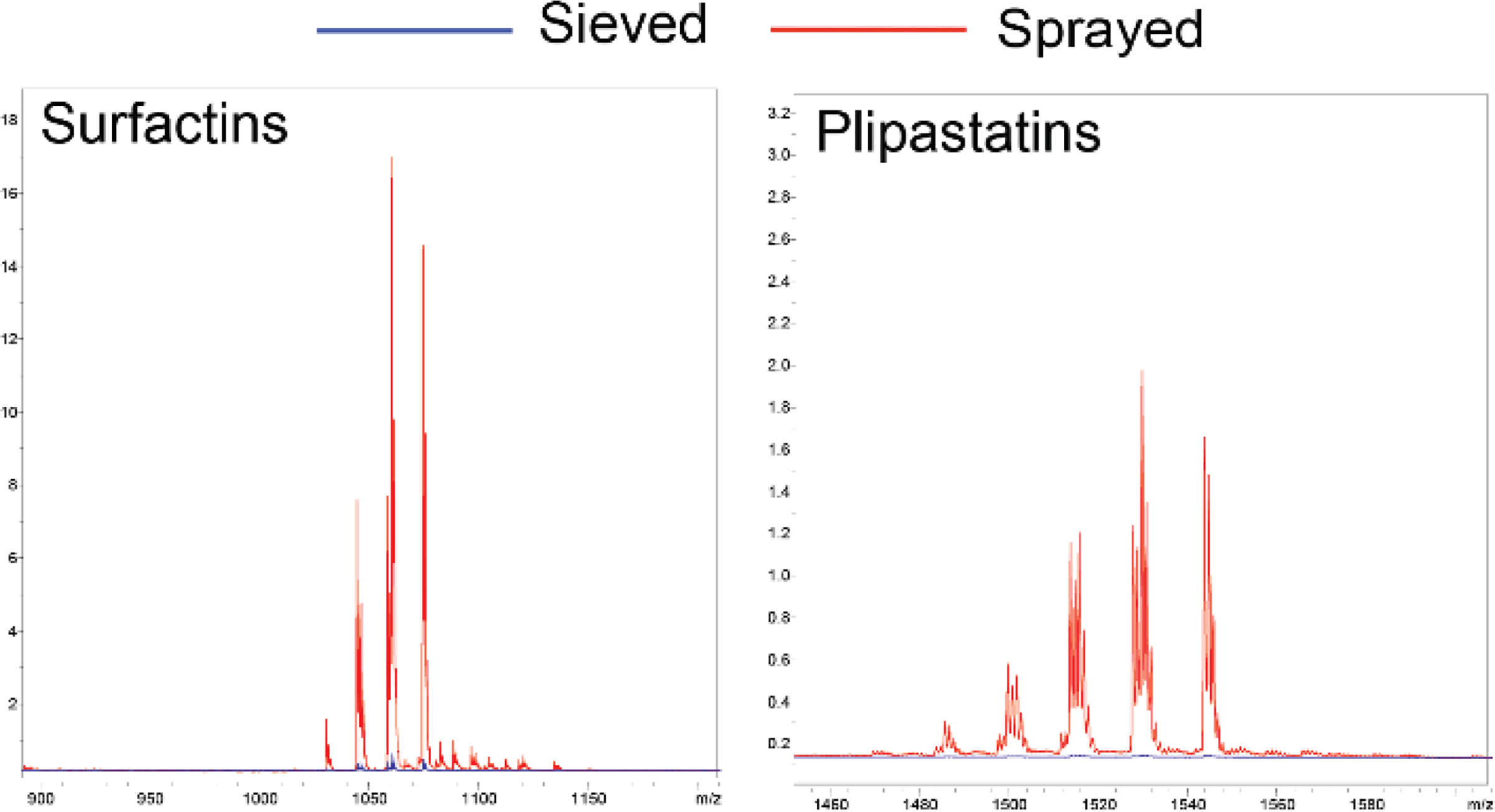
Average mass spectra comparison of MALDI MSI analysis of B. subtilis cultures grown on ISP2 agar prepared using the sieve (blue) or sprayer (red) method for matrix application using 1:1 CHCA:DHB. Data was acquired in a single acquisition. Annotation of surfactin and plipastatin molecular families are from ref.12
ISP2 agar has been predominantly used as the culture medium for microbial MALDI MSI due to its low background signal and demonstrated adherence to the target.5 However, variation in nutritional content of the cultivation environment influences microbial phenotypes, including morphological development and secondary metabolite production. Even media of the same type (e.g. LB) produced by different manufacturers can have nutrient differences. As a result, an inhibitory phenotype on one medium, may not be reproducible on another.
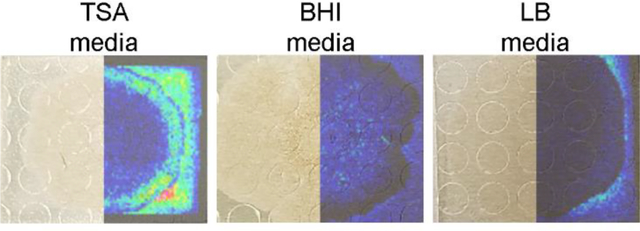
To test whether sample adhesion was improved for B. subtilis samples grown on different agar media, the three-step robotic sprayer method was used to apply DHB to B. subtilis colonies cultured on LB, BHI, and Tryptic Soy agar media placed on a 384-spot target and subjected to MALDI MSI. All media adhered to the target and differential metabolite ionization on different media was compared. Media specific metabolites were detected (Figure 3), although the lipopeptide surfactin and plipastatin molecular families were detected from cultures on all three media. While surfactin levels were similar across all B. subtilis cultures, plipastatin ionization was notably higher on TSA compared to BHI and LB.
Figure 3.
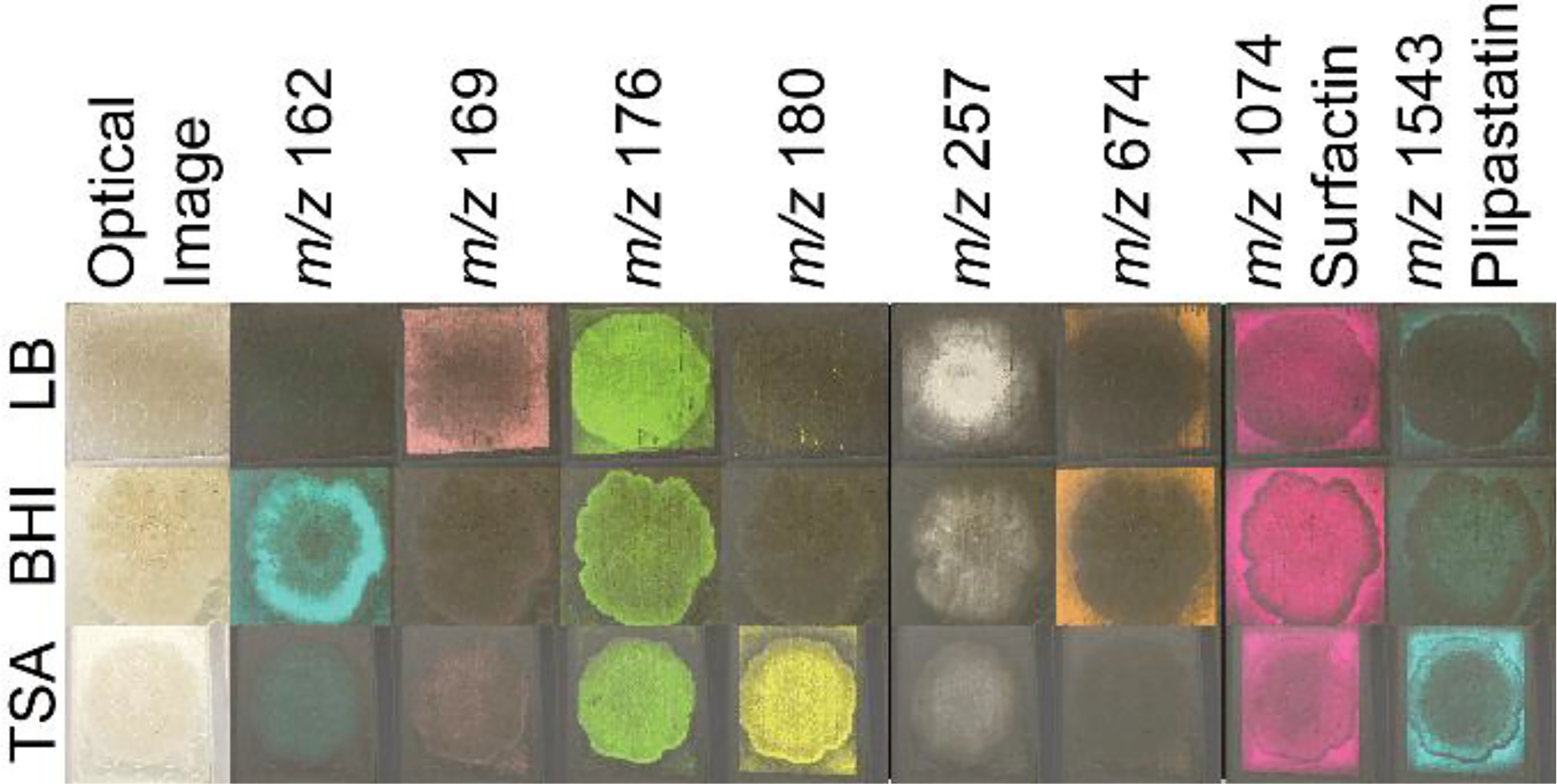
MALDI-IMS ion images of B. subtilis on TSA, BHI, and LB prepared using the three step sprayer method. DHB was used as the ionization matrix and data was acquired in a single acquisition. Putative annotations: m/z 162, 169, 176, 180, 257, and 674 are unknown metabolites; m/z 1074 is a surfactin; m/z 1543 is a plipastatin. All images were normalized to the total ion count and intensities were adjusted to improve visualization between samples. Annotation of compounds from the surfactin and plipastatin molecular families are from ref.12
In addition to widening the compatibility of microbial MALDI IMS to different types of agar media, improved sample adherence via this three-step spray-based matrix application enables the use for a 384-spot MALDI target for microbial MSI of biosurfactant producing microbes. Although we optimized the matrix application method to improve B. subtilis adherence to the MALDI target, we anticipate it being widely applicable to a broad variety of microbes on many different types of agar. The improved sample adherence via sprayer-based DHB application removes the sample size limitation of sieve-based matrix application, allows for true comparative analysis of metabolite levels and two-dimensional distribution between case and control samples, compatibility with agar-based high throughput screening, and rapid optimization of ionization conditions for metabolites of interest.
CONCLUSION
Challenges with sample adherence necessitated additional optimization of the spray-based matrix application for microbial MALDI MSI. Here, we demonstrated a three step robotic sprayer-based method for improved adherence to the MALDI target plate, which enabled comparative analysis of B. subtilis metabolites from colonies grown on three different agar media and enables the use of MALDI targets with larger analysis areas. While this matrix application method may need to be adjusted to increase the adherence of other microbes to MALDI targets, we anticipate that spray-based matrix applications will expand applicability of MALDI MSI to high throughput screening and rapid optimization of ionization conditions for metabolites of interest in agar based microbial cultures.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the members of the Phelan lab for their input and feedback on the manuscript. This work was supported by the National Institutes of Health R35 GM128690. V.V.P was partially supported by the L.S. Skaggs Professorship from the ALSAM Foundation.
Footnotes
Data files for all MSI analyses can be found in the MassIVE database under ID MSV000087768.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Figure S1 and full experimental details including microbial growth, matrix application, and data acquisition are in the Supplemental Material (PDF)
REFERENCES
- 1.Watrous JD; Dorrestein PC, Imaging mass spectrometry in microbiology. Nat Rev Microbiol 2011, 9 (9), 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunham SJB; Ellis JF; Li B; Sweedler JV, Mass Spectrometry Imaging of Complex Microbial Communities. Accounts Chem Res 2017, 50 (1), 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stasulli NM; Shank EA, Profiling the metabolic signals involved in chemical communication between microbes using imaging mass spectrometry. Fems Microbiol Rev 2016, 40 (6), 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang YL; Xu YQ; Straight P; Dorrestein PC, Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 2009, 5 (12), 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang JY; Phelan VV; Simkovsky R; Watrous JD; Trial RM; Fleming TC; Wenter R; Moore BS; Golden SS; Pogliano K; Dorrestein PC, Primer on Agar-Based Microbial Imaging Mass Spectrometry. J Bacteriol 2012, 194 (22), 6023–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann T; Dorrestein PC, Homogeneous Matrix Deposition on Dried Agar for MALDI Imaging Mass Spectrometry of Microbial Cultures. J Am Soc Mass Spectr 2015, 26 (11), 1959–1962. [DOI] [PubMed] [Google Scholar]
- 7.Anderton CR; Chu RK; Tolic N; Creissen A; Pasa-Tolic L, Utilizing a Robotic Sprayer for High Lateral and Mass Resolution MALDI FT-ICR MSI of Microbial Cultures. J Am Soc Mass Spectr 2016, 27 (3), 556–559. [DOI] [PubMed] [Google Scholar]
- 8.Li B; Comi TJ; Si T; Dunham SJB; Sweedler JV, A one-step matrix application method for MALDI mass spectrometry imaging of bacterial colony biofilms. J Mass Spectrom 2016, 51 (11), 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakeman CA; Moore JL; Noto MJ; Zhang YF; Singleton MD; Prentice BM; Gilston BA; Doster RS; Gaddy JA; Chazin WJ; Caprioli RM; Skaar EP, The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergeiner S; Schafferer L; Haas H; Muller T, Improved MALDI-TOF Microbial Mass Spectrometry Imaging by Application of a Dispersed Solid Matrix. J Am Soc Mass Spectr 2014, 25 (8), 1498–1501. [DOI] [PubMed] [Google Scholar]
- 11.Cleary J; Sanchez LM Large scale MALDI-TOF imaging of metabolites from filamentous fungi Bruker Application Note 2018, MSI08 [Google Scholar]
- 12.Watrous J; Roach P; Heath B; Alexandrov T; Laskin J; Dorrestein PC, Metabolic Profiling Directly from the Petri Dish Using Nanospray Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal Chem 2013, 85 (21), 10385–10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


