Supplemental Digital Content is Available in the Text.
Adult-onset ablation of Prdm12 using inducible conditional knockout mice deregulates gene expression and excitability of nociceptors and alters sensitivity to capsaicin and formalin.
Keywords: Sensory neuron, Nociception, Transcription factor, Prdm12, Formalin, Capsaicin
Abstract
Prdm12 is a conserved epigenetic transcriptional regulator that displays restricted expression in nociceptors of the developing peripheral nervous system. In mice, Prdm12 is required for the development of the entire nociceptive lineage. In humans, PRDM12 mutations cause congenital insensitivity to pain, likely because of the loss of nociceptors. Prdm12 expression is maintained in mature nociceptors suggesting a yet-to-be explored functional role in adults. Using Prdm12 inducible conditional knockout mouse models, we report that in adult nociceptors Prdm12 is no longer required for cell survival but continues to play a role in the transcriptional control of a network of genes, many of them encoding ion channels and receptors. We found that disruption of Prdm12 alters the excitability of dorsal root ganglion neurons in culture. Phenotypically, we observed that mice lacking Prdm12 exhibit normal responses to thermal and mechanical nociceptive stimuli but a reduced response to capsaicin and hypersensitivity to formalin-induced inflammatory pain. Together, our data indicate that Prdm12 regulates pain-related behavior in a complex way by modulating gene expression in adult nociceptors and controlling their excitability. The results encourage further studies to assess the potential of Prdm12 as a target for analgesic development.
1. Introduction
Nociception is an evolutionarily conserved mechanism crucial for survival and well-being.45 Nociceptors, sensory neurons specialized to detect noxious stimuli, are highly diverse and can be subdivided according to several features such as the stimuli they detect, their underlying molecular characteristics, and innervation patterns. The ability of nociceptors to detect various specific noxious stimuli is conferred by the combinatorial expression of specific ligand and voltage-gated ion channel receptors. An important property of nociceptors is that they can be sensitized after tissue injury or inflammation, which contributes to chronic pain,18,39 or desensitized after transient receptor potential vanilloid type 1 (TRPV1) activation.48 Sensitization of nociceptors involves dysregulation of receptor and ion channel expression and/or modulation of their activity.18 However, the mechanisms controlling changes in expression and function of ion channels and receptors in nociceptors are still largely unknown.
Prdm12 is an evolutionarily conserved epigenetic regulator of the PRDM (PRDI-BF1 & Riz1 homology domain) family of putative histone methyltransferases. In frog, chicken, and mouse embryos, Prdm12 expression is restricted to the developing nervous system.26,52,64 In the developing mouse peripheral nervous system, within the trigeminal ganglia (TG), superior-jugular ganglia (SJG) and accessory ganglia in the head region, and the dorsal root ganglia (DRG) in the body region, Prdm12 is restricted to the nociceptive lineage, where it is expressed from neural crest or placodal progenitors to differentiating postmitotic neurons.3,14 This high degree of conservation and restricted expression pattern suggest that Prdm12 may play important function(s) in nociceptors. Indeed, in the frog, Prdm12 knockdown and gain of function experiments lead to altered sensory neurogenesis.9,41,42 In mice, constitutive deletion of Prdm12 or its conditional deletion during neurogenesis (from E13.5, in differentiating post-mitotic cells) results in the selective loss of the entire nociceptive lineage.3,14 Such neuronal loss is most likely because of increased cell death linked to downregulation of the expression of neurotrophic receptor tyrosine kinase 1 (TrkA, encoded by Ntrk1) that binds nerve growth factor (NGF) and is required for nociceptor survival.14 In humans, mutations in PRDM12 have been found in patients with congenital insensitivity to pain (CIP). These patients with CIP are unable to feel pain since birth, although their mechanosensation and proprioception are unaffected. Consequently, PRDM12 CIP patients all present severe injuries, mainly because of automutilation and unnoticed trauma. In these CIP patients, skin biopsies have revealed a loss of nociceptive nerve endings in the epidermis, suggesting that, like in mouse embryos, PRDM12 is required for nociceptor development in humans.9 Single-cell RNA-seq studies and immunostaining we performed on mouse DRG neurons have shown that Prdm12 remains expressed at adulthood,9,14,55,65 but its role in mature nociceptors has not yet been characterized.
In this study, using Prdm12 inducible conditional knockout (icKO) mouse models, we demonstrate that in adults, Prdm12 plays a role in the control of the expression of a set of genes encoding receptors, channels, and neurotransmitters in nociceptors and therefore modulates their functionality.
2. Materials and methods
2.1. Mice
All mice were maintained on a C57BL/6J background and mice of both sexes were used. Mice were housed at room temperature with a 12-hour light/dark cycle in standard cages with litter, water, and food ad libitum. Air circulation in the facility was filtered and temperature monitored at a steady 20°C. Cages were also provided with cottons and cardboard rolls for enrichment. The experimental protocols were approved by the CEBEA (Comité d'éthique et du bien-être animal) of the IBMM-ULB and are conform to the European guidelines on the ethical care and use of animals; experiments conducted in Cambridge were regulated under the Animals (Scientific Procedure) Act 1986, Amendment Regulations 2012, and all protocols were approved by a U.K. Home Office project license (P7EBFC1B1) and reviewed by the University of Cambridge Animal Welfare and Ethical Review Body. The following mouse strains were used: Prdm12LacZ/+ and Prdm12fl/fl,14 Rosa26 LSL-tdTomato (Ai14, JAX#007908),40 AdvillinCreERT2 (JAX#026516),31 and Rosa26CreERT2 (JAX# 008463).56 Polymerase chain reaction (PCR) genotyping was done as follows: for Prdm12 floxed allele, using primers forward 5′-GCTGATCGAGTCCAGGAGAC-3′ and reverse 5′-CCAAACATCCACAACCTTCA-3′; for AdvillinCreERT2, using primers forward 5′-GACAGATTATCTGCAATCTCTCTAAG-3′and reverse 5′-GTTCCTGATGTTCCTGGCATCTGTC-3′; for Rosa26 LSL-tdTomato, using primers forward 5′-AAGGGAGCTGCAGTGGAGTA-3′ and reverse 5′-CCGAAAATCTGTGGGAAGTC-3′; and for Rosa26CreERT2, using primers forward 5′-CGTGATCTGCAACTCCAGTC-3′ and reverse 5′-AGGCAAATTTTGGTGTACGG-3′ to detect the Cre and forward 5′-CTGGCTTCTGAGGACCG-3′ and reverse 5′-CCGAAAATCTGTGGGAAGTC-3′ to detect the wild-type allele.
Tamoxifen (TAM) dissolved in corn oil has been administrated (1.9 mg/25 g body weight) intraperitoneally (i.p.) in more than 56-day-old AdvillinCreERT2;Prdm12fl/fl mice 2 times in a week, and 4 times in a week in the case of Rosa26CreERT2;Prdm12fl/fl mice. Injected animals were killed 28 to 35 days after the last TAM injection for analysis. For tissue recovery, mice were anesthetized with Domitor (1 mg/kg, i.p.) and Ketamine (75 mg/kg, i.p.) or with Ketamine (75 mg/kg, i.p.) and Rompun (1 mg/kg, i.p.). Whole-mount X-gal staining of dissected brain and cranial ganglia of Prdm12LacZ/+ mice was performed as described.14 For retrograde labelling studies, intra-articular injections of Fast Blue (1.5 µL, 2% in 0.9% saline; Polysciences) and complete Freund adjuvant (CFA) (7.5 µL, 10 mg/mL; Chondrex) were conducted in mice under anesthesia (ketamine, 100 mg/kg and xylazine, 10 mg/kg, i.p.).
2.2. Immunohistochemistry
For immunostainings, after intracardiac perfusion with ice cold phosphate-buffered saline (PBS) (1×), dissected DRG, TG, or SJG were fixed in 4% paraformaldehyde for 15 minutes, rinsed several times in cold PBS, then cryoprotected overnight in sucrose 30% (dissolved in PBS) at 4°C before embedding in freshly prepared gelatin 7.5%–sucrose 15% (dissolved in PBS at 40°C), and stored at −80°. The frozen blocks were then cryostat sectioned to obtain 16-µm sections and stored at −20°C until use for immunostainings. Primary antibodies used were as follows: homemade rabbit and guinea pig anti-PRDM12 (1:5000 and 1:2000, respectively), chicken anti-peripherin (1:1000, Abcam, Cambridge, United Kingdom, ab106276), goat anti-calcitonin gene related peptide (CGRP) (1:200, Abcam ab36001), rabbit anti-Nav1.8 (1:400, Abcam ab63331), rabbit anti-Trpv1 (1:1000, Abcam ab31895), and mouse anti-βIII-Tubulin (1:200, Covance MMS-435P). Isolectin B4 conjugated to Alexa Fluor 594 (Invitrogen, Waltham, MA; I21413) was used at 1:1000 dilution. Secondary antibodies used were as follows: goat anti-rabbit Alexa Fluor 594 (1:800, Invitrogen A11012), donkey anti-rabbit Alexa Fluor 488 (1:1000, Invitrogen A21206), donkey anti-goat Alexa Fluor 594 (1:2000, Invitrogen A11058), goat anti-guinea pig Alexa Fluor 488 (1:2000, Invitrogen A11073), donkey anti-guinea pig Alexa Fluor 488 (1:800, Bio connect), donkey anti-chicken Alexa Fluor 488 (1:1000, Bio connect–Jackson), and goat anti-mouse Alexa Fluor 594 (1:1000, Invitrogen A11017). Immunostainings were performed as described.52 Images collection and analysis were performed using a wide-field fluorescence microscope Zeiss Axio Observer Z1, a laser-scanning confocal microscope Zeiss LSM 710 using the Zeiss Zen microscopy software or a stereo microscope Olympus SZX16/Olympus BX51 and the softwares Cell^F or CellSens Entry V2.1. Image analysis was performed using ImageJ/Fiji and Microsoft Excel 2020 and GraphPad version 9. Cell counts were conducted with at least 8 DRG sections analysed per animal with an N ≥ 3.
2.3. RNA-seq and next-generation sequencing data analysis
Before tissue collection, mice were deeply anesthetized with Ketamine (75 mg/kg, i.p.) and Rompun (1 mg/kg, i.p.) and intracardiacally perfused with ice-cold RNAse-free (RF) PBS. Dorsal root ganglia or TG from inducible knockout animals were harvested 4 weeks after the last TAM injection. Controls were TAM injected Prdm12fl/fl or Prdm12fl/+ mice with no Cre transgene; corn oil–injected, Cre-expressing Prdm12fl/fl mice; or TAM-injected, Cre-expressing Prdm12+/+. Tissues were microdissected in ice-cold RF PBS and stored at −80°C in 1 mL TRIZOL (Thermofisher; ref 15596026). RNA was extracted using the illustra RNAspin Mini RNA isolation kit from GE Healthcare (25-0500-70). Quality and integrity of RNA was assessed with the Fragment Analyzer from Advanced Analytical by using the standard sensitivity RNA Analysis Kit (DNF-471). RNA-seq libraries were performed using 100 ng total RNA of a nonstranded RNA Seq, massively parallel mRNA sequencing approach from Illumina (TruSeq RNA Library Preparation Kit v2, Set A; 48 samples, 12 indexes, Cat. N°RS-122-2001). Libraries were prepared on the automation (Beckman Coulter's Biomek FXP workstation). For accurate quantitation of cDNA libraries, a fluorometric-based system, the QuantiFluordsDNA System from Promega (Madison, WI) was used. The size of final cDNA libraries was determined by using the dsDNA 905 Reagent Kit (Fragment Analyzer from Advanced Bioanalytical) exhibiting a sizing of 280 bp in average. Libraries were pooled and sequenced on the Illumina HiSeq 4000 (SE; 50 bp; 30 Mio reads/sample). Sequence images were transformed to BCL files with the Illumina software BaseCaller software, which were demultiplexed to fastq files using bcl2fastq v2.20.0.422. Sequencing quality was asserted using FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/)(version 0.11.5). Sequences were aligned to the reference genome Mus musculus (mm10 version 9104, https://www.ensembl.org/Mus_musculus/Info/Index) using the STAR aligner software version 2.7.8a,16 allowing for 2 mismatches within 50 bases. Subsequently, read counting was performed using featureCounts version 2.0.1.34 Read counts were analyzed in the R/Bioconductor environment version 3.6.1 (www.bioconductor.org) using the DESeq2 package version 1.32.0.37 Candidate genes were selected as those with an FDR-corrected P-value ≤0.05. Genes were annotated using the M musculus GTF file mm10 version 104 used to quantify the reads within genes. RNA-seq data have been deposited at Gene Expression Omnibus (GEO) under accession GSE166062 and GSE182904.
Gene ontology (GO) and pathway analysis was performed in RStudio using Bioconductor version 3.12 selecting all differentially expressed gene (DEG) with an FDR-adjusted p value <0.05 or filtering them by log2 fold-change, using as cut-off the fold-changes associated to some identified DEG considered as likely relevant Prdm12 targets. ClusterProfiler and enrichGO were used for GO using enrichKEGG.23–25 Analysis and sensory neuron classification of deregulated genes was performed using the gene expression visualization tools developed by Usoskin et al., 2015 (linnarssonlab.org) and Zeisel et al., 2018 (mousebrain.org). Deregulated genes were manually classified as “Neuronal” if they had significant expression in at least one neuronal cell type. Deregulated genes were manually classified as “Not neuronal” if they had unknown expression pattern, no significant expression at least in one neuronal cell type, or significant expression at least in one nonneuronal cell type. Volcano plots were created using ggplot2 function of the tidyverse package.59
2.4. Real-time quantitative polymerase chain reaction
Dorsal root ganglia were dissected individually in cold RF PBS and kept at −80°C until RNA extraction. RNA extraction and purification were performed using the Monarch Total RNA Miniprep kit (Ref: T2010S) following the manufacturer's instructions. RNA (400 ng) was transcribed to cDNA using the iScript cDNA Synthesis kit (Cat #1708891; BioRad). The obtained cDNA (40-50 ng) was then amplified using the GoTaq qPCR Master Mix (Cat #A6001; Promega) following the manufacturer's instructions. All qPCR lectures were performed using the CFX96 Dx In Vitro Diagnostics real-time PCR systems (Bio Rad, Hercules, CA). The comparative 2−ΔΔCT method36 was used to determine relative gene expression, comparing with the level observed in control tissue, normalizing to GADPH. The tailed Student T test (α = 0.05) was used to define statistical significance. Error bars show SD of between 4 and 9 biological replicates. The sequences of the primers are listed in the Appendix (available as supplementary digital content at http://links.lww.com/PAIN/B535).
2.5. In situ hybridization
For in situ hybridization (ISH), dissected DRG attached to the spinal cord were fixed in 4% PFA (dissolved in RF PBS) overnight and cryopreserved in 30% sucrose (in RF PBS) overnight. Afterward, the tissue was embedded in 15% sucrose/7.5% gelatine (dissolved in RF PBS at 40°C), put at 4°C for >3 hours and stored at −80°C. In situ hybridization was then performed on 20-µm cryosections. Plasmids used for generating probes for Prdm12, Ntrk1, and Trpm8 were described elsewhere.14 Chrna6, Grik1, Mrgprb5, and Cysltr2 cDNA used for probe generation were obtained by RT-PCR from RNA extracted from mouse embryonic DRG using the following primers: Chrna6, forward 5′-TTCCAGGTCGAAGGCAAGAC-3′ and reverse 5′-TTGGCAGGCCTCTTGGTATG-3′; Grik1, forward 5′-GAATGACAAAGGGGAGTGGA-3′ and reverse 5′-AAGGTCATTGTCGAGCCA-3′; Mrgprb5, forward 5′-CCATCAGTGTTGAGCGCTCT-3′ and reverse 5′-GTCCTGCATGGCTCTCTGAA-3′; Cysltr2, forward 5′-GATATTTGGGGACTTGGCCTG-3′ and reverse 5′-GCTTTGAAATTCTCCCCAGCA-3′. The obtained PCR products were cloned into the pCR4-Topo vector and the riboprobe synthetized (NotI, T3). In situ hybridization experiments were performed as previously described using antisense digoxygenin-labeled riboprobes.52
2.6. Pain behavior assays
For all behavioral tests, Cre-negative Prdm12fl/fl mice or Cre-expressing mice, either Prdm12+/+ (WT) or Prdm12fl/+, all injected with TAM, were used as controls. Experimenter was blind to the genotype of the mice. For the formalin test, mice were habituated for 5 minutes the day before the test in a plexiglass cage with 3 mirrors on the walls and litter covering the floor. The day after, the mouse was injected with 25 µL of a 3% formalin solution in the right hind paw and directly after observed for 30 minutes. From the time of injection, licking time of the injected paw was recorded during 5-minute periods up to 30 minutes after formalin injection.
For the thermal place preference test, we used the BIOSEB BIO-T2CT device that is composed of 2 joined thermal plates and is equipped with a camera to follow the transitions of the mouse from one plate to another and monitor the time spent on each plate. The day before the test, mice were habituated for 180 seconds on the device and both plates were set at 30°C, the reference temperature for all tests. Mice were recorded for 180 seconds for each temperature tested. The temperatures of 22, 10, 43, and 52°C have been tested sequentially, with a day off in between hot and cold temperatures. Each day, the reference and the test plates were reversed.
Regarding the capsaicin test, mice were habituated for 5 minutes the day before the test in a plexiglass cage such as for the formalin test. On the day of the test, mice were injected with 10 µL of a 0.35 µg/µL capsaicin solution (0.9% saline, 10% ethanol, 10% Tween-20). Mice were recorded for 15 minutes after injection and the licking time of the injected paw was measured.
Tail flick response to heat was also tested in mice using the BIOSEB device BX-TF. Mice were restrained in a cylinder with the tail outside of the cylinder in a notch and the tail was subjected to a radiant heat source. The focus 30 was selected for this test meaning that the temperature applied on the tail was at 50°C after 5 seconds and at 60°C after 10 seconds. The withdrawal latency to the heat stimulus applied to the tail was therefore measured. In total, 3 measurements were carried out with a rest time of 15 seconds between measurements.
A cold plantar assay was used to evaluate the cold sensitivity in mice.4 Mice were habituated in small cages on a 3-mm-thick glass plate for 20 minutes. A 5-mL syringe cut at the end and filled with powdered dry ice was applied under the hind paw against the glass plate and the withdrawal latency was measured. Three measurements were taken per paw and both paws were tested with a time interval of 5 minutes between measurements.
The mechanical conflict avoidance test was used to evaluate the response to mechanical stimuli.20 For the habituation, mice were placed on the device for 5 minutes and were free to move around the different parts of the box. For the test, mice were placed in the first part which had a separation with the second part, the light was turned on after 10 seconds, and the separation was removed after 15 seconds. Mice went through the second part of this box, which is covered with nails, to get to the last part, which was dark. The time took for each mouse to cross half of the part with nails was measured. Two heights of nails were tested: 2 and 5 mm on 2 different days and 3 measurements were taken per height with a rest time of 30 minutes between each measurement.
For the LTC4-induced itch assay, mice were habituated in the same way as for the formalin and the capsaicin tests. On the day of the test, mice were injected in the right cheek with 0.75 µg of LTC4 solution (Santa Cruz, sc-205407). Mice were placed in the cage and recorded for 40 minutes after injection. Number of scratchings was then measured.50
An open-field assay was used to analyse the activity of mice and their exploratory behaviour. Mice were placed for 10 minutes in a plexiglass cage (44 × 44 cm) with a camera placed above the cage and the behaviour was analysed with a Noldus software. Several parameters have been measured, such as the distance travelled, the velocity, the time moving, and the time spent in the centre and on the borders.
For motor coordination in mice, we used the beam walking test. Mice were habituated the day before to walk across a narrow beam from one side to the other. On the day of the test, the time to cross half of the beam was measured and 3 measurements were done per animal. Statistical analysis was conducted with the Student t test. All graphs were processed with GraphPad.
2.7. Dorsal root ganglia neuron culture
Mice (3- to 7-month-old) were deeply anesthetized with a mixture of Domitor (1 mg/kg, i.p.) and Ketamine (75 mg/kg, i.p.) and an intracardiac perfusion with PBS was performed. Dorsal root ganglia from thoracic and lumbar levels were collected and then digested for 35 minutes at 37°C in an enzyme mixture containing Collagenase (12.5 mg/mL) and Dispase (3 mg/mL). Dorsal root ganglia were quickly centrifuged at 150g after digestion and then the enzyme mix was removed and replaced by DMEM/F12. Dissociation using a P1000 was carried out and the cell suspension was then filtered through a 0.2-µm filter followed by a 7-minute centrifugation at 150g. Cells were then resuspended in complete medium containing DMEM/F12, 10% of fetal bovine serum, and 1% of penicillin-streptomycin. Cells were plated in 4-well plates on glass coverslips coated with 0.1 mg/mL of poly-l-lysine and 10 µg/mL of laminin and kept in complete medium at 37°C.
2.8. Electrophysiology
Patch-clamp recordings were conducted at room temperature using an amplifier A-M system, INC Model 2400 controlled by WIN WCP software V5.52. Small DRG neurons (diameter <30 μm) were selected for current-clamp recording. In TAM-injected AdvillinCreERT2;Rosa26Ai14;Prdm12fl/fl mice, icKO neurons were identified by the presence of red Tomato signal (Olympus IX70 Inverted microscope equipped with Led fluorescence system). Electrodes were fabricated from 1.6 mm outer diameter borosilicate glass micropipettes (G86150T-4; Warner Instruments) using a Sutter Instruments P-97. Piezoelectric Burleigh micromanipulator was used to lower the electrode on cells. Liquid junction potential was not corrected. Cells were excluded from analysis if the resting membrane potentials were more positive than −40 mV. Neurons were recorded within 24 hours of culture to prevent neurite outgrowth that degrades space clamp.
For current-clamp recordings, electrodes had a resistance of 2.5 to 3.5 MΩ when filled with the pipette solution of the following composition (in millimolor): 140 K-Aspartate, 10 NaCl, 10 EGTA, 1 MgCl2, 10 HEPES, pH 7.20, with KOH (290 ± 5 MΩ). The extracellular solution was composed of (in millimolor): 150 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 10 HEPES, 10 glucose, pH 7.40, with NaOH (290 ± 5 MΩ). Whole-cell configuration was obtained in voltage-clamp mode before proceeding to the current-clamp recording mode.
Three variables were assessed in each neuron: resting membrane potential, threshold current, and action potential frequency. Threshold current was determined by the first action potential elicited by a series of depolarizing current injections (300 ms) that increased in 20 pA increments. Action potential frequency was determined by quantifying the number of action potentials elicited in response to depolarizing current injections (2 seconds, 1500 pA). Any modification in basal excitability will be evidenced by change in the threshold current and/or action potential frequency.
3. Results
3.1. Prdm12 expression in mature dorsal root ganglia nociceptors is dispensable for their survival
To visualize Prdm12 expression in adult mouse CNS, we performed X-gal staining on dissected neural tissues from Prdm12LacZ/+ mice. Figure 1A shows that Prdm12 remains expressed in all trunk and cranial ganglia containing somatosensory nociceptors. Sections of the brains reveal that Prdm12 is only detected in a few specific cells in the olfactory region and in some brain nuclei, such as the lateral septal nucleus, lateral hypothalamic area, and in the A13 dopaminergic nucleus of the incerto-hypothalamic area (Fig. 1B). Double immunostainings on transverse sections of TG, SJG, and DRG showed that in these distinct ganglia, Prdm12 is absent from mechanoreceptors and proprioceptors (TrkB+, TrkC+) and is expressed in both peptidergic (TrkA+, CGRP+) and nonpeptidergic (Ret+) nociceptors (NaV1.8+, TrpV1+), as well as in some itch-mediating somatostatin (Sst+) neurons and c-fiber low-threshold mechanoreceptors (C-LTMRs) expressing tyrosine hydroxylase (TH+) (Fig. 1C).
Figure 1.
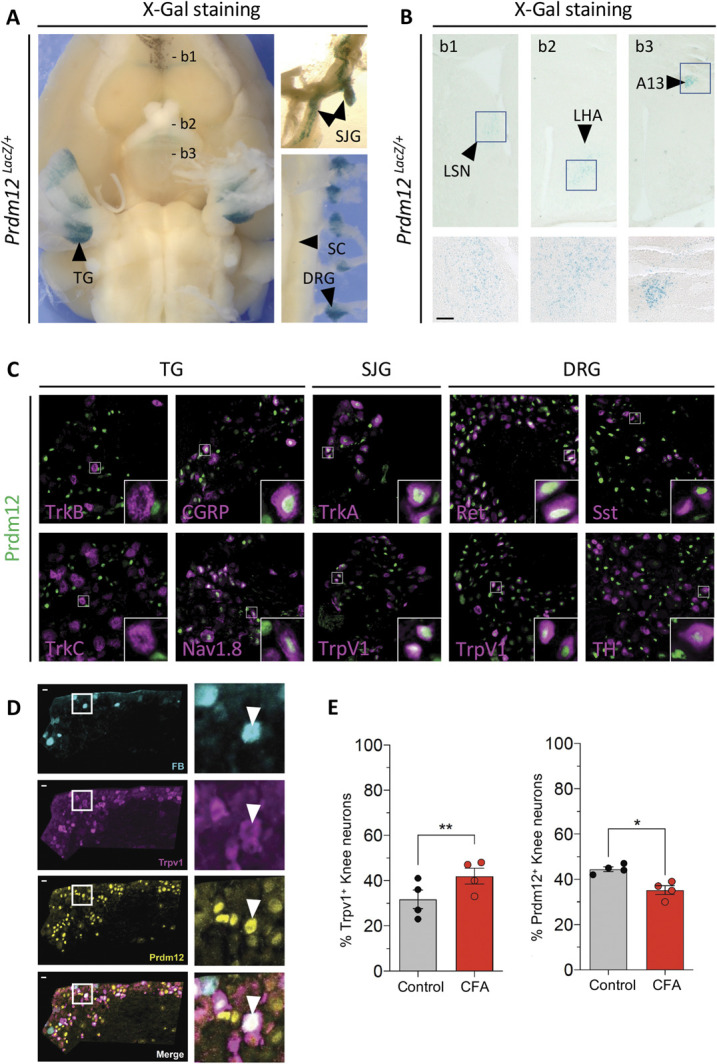
Prdm12 is expressed in nociceptors of adult mice and is downregulated upon CFA-induced inflammation. (A) X-Gal staining of the brain (ventral view) with attached trigeminal ganglia (TG) of an adult Prdm12LacZ/+ mouse. Superior jugular ganglia (SJG) and spinal cord (SC) with attached dorsal root ganglia (DRG) are shown on the right. (B) Coronal sections at levels indicated in (A) showing that Prdm12 expression in the brain is restricted to specific nuclei such as the lateral septal nucleus (LSN), lateral hypothalamic area, and the diencephalic A13 nucleus. Scale bar in insets = 100 µm. (C) Double immunostainings on transverse sections of TG, SJG, and DRG of adult mice with the indicated markers. (D) Triple staining showing Prdm12+ neurons, TrpV1+ neurons, and FastBlue (FB) retrolabelled knee-innervating neurons in L3 to L4 DRG of CFA-injected mice. Inset shows a knee-innervating DRG neuron coexpressing Prdm12 and TrpV1. (E) Percentage of Prdm12- and TrpV1-positive cells counted from the contralateral (Ctrl) and CFA-injected sides (n = 4, >300 neurons counted in each condition, scale bar = 50 μm). *P < 0.05, **P < 0.01, paired t test. CFA, complete Freund adjuvant; SJG, superior jugular ganglia.
Altered epigenetic mechanisms are known to contribute to inflammation-induced pain hypersensitivity.13 Injection of CFA into the knee joint produces acute and inflammatory pain in mice that models human arthritis.10 To determine whether Prdm12 expression is modulated in such an inflammatory condition, we performed unilateral knee injections of CFA, 7 days after retrograde labeling of knee-innervating neurons with Fast Blue. Twenty-four hours after CFA injection, immunostaining was performed using anti-Prdm12 and anti-Trpv1 antibodies on whole DRG sections (Fig. 1D). Comparing the ipsilateral (CFA injected) and contralateral (control) sides of the injected mice, we observed a statistically significant decrease in the number of knee-innervating neurons that express Prdm12 while the number of Trpv1+ neurons was increased as expected6 (Fig. 1E).
To gain insight into the role of Prdm12 in mature nociceptors in adult mice, females carrying Prdm12 floxed alleles were crossed with AdvillinCreERT2 TAM-inducible Cre driver males that express Cre specifically in somatosensory neurons.31 Rosa26CreERT2 mice that express Cre ubiquitously after treatment with TAM were also used given the breeding issues we have had with the AdvillinCreERT2 line, the inheritance of the Cre being no longer Mendelian. To ensure efficient excision of the floxed alleles, mice with 2 Rosa26CreERT2 alleles were used.47 The resulting conditional knockout offspring AdvillinCreERT2;Prdm12 fl/fl mice and Rosa26CreERT2;Prdm12 fl/fl mice are here designated Avil icKO and Rosa26 icKO. By immunostaining, we validated the TAM-induced loss of Prdm12 in the DRG of both Avil icKO and Rosa26 icKO mice (Sup. Fig. 1, http://links.lww.com/PAIN/B535).
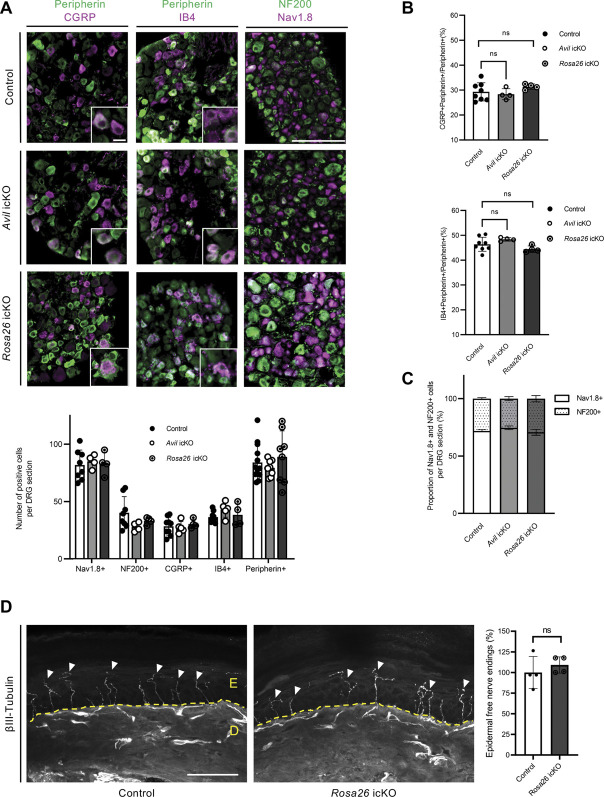
In contrast to Prdm12 ablation during development, we found no evidence for neuronal loss in DRG of Avil icKO and Rosa26 icKO mice. Using antibodies against peripherin as a general marker of peripheral neurons, NF200 as a mechano-proprioceptive marker, NaV1.8 marking all nociceptors, CGRP labeling peptidergic nociceptors, and performing isolectin B4 (IB4) staining to visualize nonpeptidergic nociceptors, no significant difference in the absolute number of cells positive with these markers was observed in DRG between mutant and control mice (Fig. 2A). No difference was also detected in the proportion of CGRP+ or IB4+ neurons among peripherin+ cells (Fig. 2B) nor in the proportion of DRG neurons positive for NaV1.8+ and NF200+ (Fig. 2C). Moreover, in Avil icKO mice, in which a Cre-dependent tdTomato reporter was used to mark Advillin-expressing cells, counting analyses revealed a similar number (∼50%) of tdTomato+ cells in DRG of both icKO and control mice (Sup. Fig. 1, http://links.lww.com/PAIN/B535).
Figure 2.
Loss of Prdm12 in nociceptor of adult mice does not alter diversity of DRG neuronal subpopulations. (A) Representative images of DRG sections of Avil icKO and Rosa26 iKO mice injected with tamoxifen or with corn oil as control immunolabelled for Peripherin, CGRP, Nav1.8, and NF-200 or stained with IB4-Alexa594, with quantification showing that the number of cells positive for the different markers is similar in DRG sections of icKO and control mice. Scale bar: 100 µm, inset scale bar: 25 µm. (B) Proportion of CGRP+ neurons and of IB4+ neurons among Peripherin+ neurons in DRG are similar in both icKO and control mice. (C) Proportions of DRG neurons positive for Nav1.8 and NF200 are unchanged in DRG sections of both icKO. (D) Left panel, βIII-Tubulin immunostainings on skin tissue of TAM-injected Rosa26 icKO and control mice. Arrows point skin epidermis innervation (E = epidermis; D = dermis). Scale bar: 50 µm. Right panel, Quantification showing the percentage of labelled sensory terminals invading the epidermis in TAM-injected Rosa26 icKO mice compared to controls. In all experiments, n ≥ 3 for both genotypes. All quantifications were submitted to the 2 tailed student's t test or two-way ANOVA test. Values are represented as mean ± SD. DRG, dorsal root ganglia; icKO, inducible conditional knockout.
Given that CIP patients show a loss of intraepidermal nerve endings, we also performed immunohistochemical analysis of hind paw skin sections from both Avil icKO and Rosa26 icKO mice using βIII-Tubulin, a marker of intraepidermal nerve fibers.32 In both mutant mice, normal epidermal βIII-Tubulin+ innervation was observed (Fig. 2D; data not shown). Thus, loss of Prdm12 appears dispensable for nociceptor survival in adult mice.
3.2. Loss of Prdm12 affects gene expression in nociceptors of adult mice
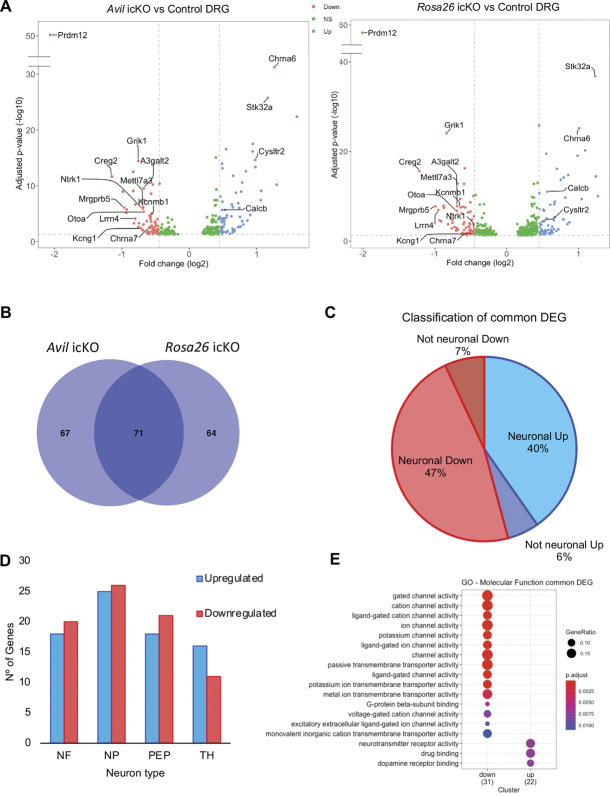
To determine whether Prdm12 plays a role in the control of gene expression in mature nociceptors, we performed a transcriptomic analysis by bulk RNA-seq on dissected lumbar DRG of both Avil and Rosa26 icKO models and control mice 1 month after TAM injection. Using an FDR-adjusted P-value ≤0.05 and an absolute log2 fold-change cut-off of ≥0.449 (based on the fold-change of Ntrk1, recently reported as reduced in adult Prdm12 knockout mice30) we obtained a list of 140 candidate genes in Avil icKO mice and 134 in Rosa26 icKO mice that are dysregulated in the absence of Prdm12 (Fig. 3A and Sup. Tables 1 and 2, http://links.lww.com/PAIN/B535).
Figure 3.
Transcriptomic analysis of the consequences of Prdm12 depletion in DRG of adult mice. (A) Volcano plots showing the –log10 adjusted P-value as a function of the log2 fold-changes of deregulated genes in DRG of Avil icKO mice (icKO mice, n = 5 vs control mice, n = 4) and Rosa26 icKO mice (icKO mice, n = 6 vs control mice, n = 6) 1 month after tamoxifen injection. The genes were identified by bulk RNA-seq and selected based on adjusted P-value <0.05. Genes with a log2 fold-change >0.449 are indicated in red, genes below the threshold are in light grey. (B) Venn diagram showing the overlap between the DEGs identified in Avil icKO and Rosa26 icKO mice. (C) Proportion of neuronal genes among the common DEGs. Neuronal genes upregulated and downregulated by the loss of Prdm12 are indicated in blue and red, respectively. (D) Classification of the common DEGs based on their expression in the major different subtypes of DRG neurons, with downregulated genes in red and upregulated genes in blue. (E) Gene ontology classification of the DEGs. The graph shows the enriched biological molecular function associated with downregulated and/or upregulated genes. DRG, dorsal root ganglia; icKO, inducible conditional knockout.
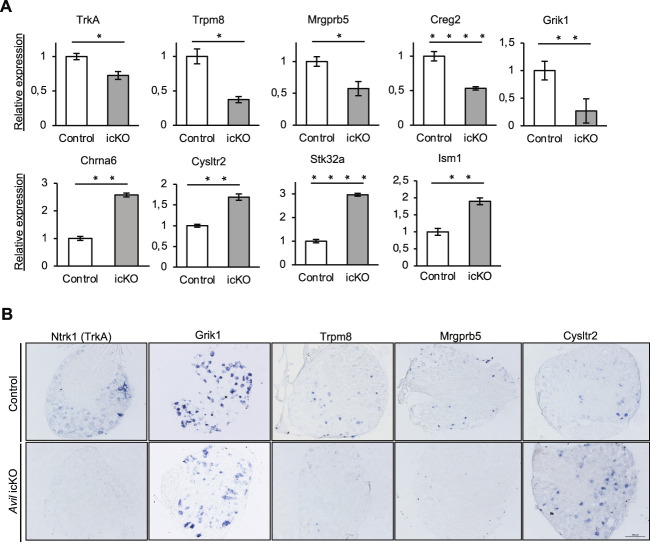
Comparing the 2 datasets, we identified 71 transcripts common to both models, suggesting that despite the rather small log2 fold-changes observed, they are biologically relevant DEGs. Among them, 39 were downregulated and 33 upregulated by the loss of Prdm12 (Fig. 3B and Sup. Data Table 1 and 2, http://links.lww.com/PAIN/B535). Most (87%) of these common DEGs are neuronal genes (Fig. 3C). Gene expression distribution analysis of these common DEGs in the major different DRG nociceptors revealed that many of them are characteristic nociceptive genes or genes expressed in TH+ C-LTMRs but that some genes expressed in neurofilament neurons are also found (Fig. 3D). Gene ontology analysis based on molecular function of these common DEGs revealed that many downregulated DEGs encode channel proteins, transmembrane receptors, and other membrane proteins, whereas some of the upregulated genes encode neurotransmitter receptors (Fig. 3E). Further GO analysis based on all categories indicates enrichment in genes involved in catecholamine transport, detection of stimulus, detection/sensory perception of temperature stimulus, and positive regulation of ion transport and blood vessel diameter (Sup. Fig. 2A, http://links.lww.com/PAIN/B535). KEGG pathway analysis revealed that the 3 most significantly deregulated pathways are neuroactive ligand–receptor interaction, Ca2+ signaling and cholinergic synapse (Sup data Fig. 2B, http://links.lww.com/PAIN/B535). Focusing on the neuroactive ligand–receptor interaction category, 6 genes appear upregulated while 3 appear downregulated (Sup. Fig. 2C, http://links.lww.com/PAIN/B535). Besides Prdm12, among the common DEGs are genes encoding membrane proteins, such as the NGF receptor TrkA,12 the sensory MAS-related specific G protein-coupled receptor, member 5 (Mrgprb5),35 the α 6 and α 7 subunits of the nicotinic acetylcholine receptor (Chrna6 and Chrna7),1,44,51,60 the ionotropic glutamate receptor kainate 1 (Grik1/Glur5),27 the cysteinyl-leukotriene receptor 2 (Cysltr2),50 the neuronal-specific leucine-rich protein 4 (Lrrn4), and the glycosylphosphatidylinositol-anchored protein otoancorin.68 Other DEGs encode ion channel proteins such as the cold-activated transient receptor potential cation channel, subfamily M, member 8 (Trpm8),15 the K+ large conductance calcium-activated channel, subfamily M, beta member 1 (Kcnmb1) and voltage-gated channel, subfamily G, member 1 (Kcng1),49 extracellular proteins such as the secreted glycoprotein cellular repressor of E1A-stimulated genes 229 and the secretory neuropeptide calcitonin-related polypeptide beta (Calcb),61 and enzymes such as alpha 1,3-galactosyltransferase 2 (A3galt2) and the methyltransferase like 7A3 (Mettl7a3) that are strongly expressed in sensory neurons and serine/threonine kinase 32A (Stk32a) that is highly expressed in sympathetic neurons (mousebrain.org). The differential expression of some of these genes was confirmed by RT-qPCR and ISH on DRG sections from Avil icKO mice (Fig. 4) and by RT-qPCR alone in Rosa26 icKO mice (Sup. Fig. 3, http://links.lww.com/PAIN/B535).
Figure 4.
Validation of some of the identified DEGs as Prdm12 targets in DRG of Avil icKO mice. (A) RT-qPCR analysis of the expression of the indicated DEGs. (B) ISH analysis of the expression of the indicated genes. *, **, ***, and **** indicate respective P values <0.05, <0.01, <0.001, and <0.0001 (Student t test, α = 0.05). DRG, dorsal root ganglia; icKO, inducible conditional knockout; ISH, in situ hybridization.
As TG have a more complex embryonic origin than DRG, being derived from both neural crest and placodal cells, we also performed bulk RNA-seq analysis on TG samples from TAM-injected Rosa26 icKO and control mice. Using an FDR-adjusted P-value ≤0.05 and an absolute log2 fold-change cut-off of ≥0.31 (based on the fold-change of Mrgprb5 that appears as true DEG in DRG), we identified 125 DEGs, with 35 upregulated and 95 downregulated genes (Fig. 5A and Sup. Table 3, http://links.lww.com/PAIN/B535). As in DRG, most of these DEGs are nociceptive neuronal genes, with many of them encoding proteins with channel activity (Fig. 5B). Comparing the 2 list of DEG identified in DRG and TG of Rosa26 icKO mice, we found 41 common dysregulated transcripts (Fig. 5C). These transcripts common to both ganglia, designed here as Prdm12 core DEGs, are listed in Figure 5D. Together, these results strongly support a role for Prdm12 in mature nociceptors in the transcriptional regulation of a battery of genes essential for their function.
Figure 5.
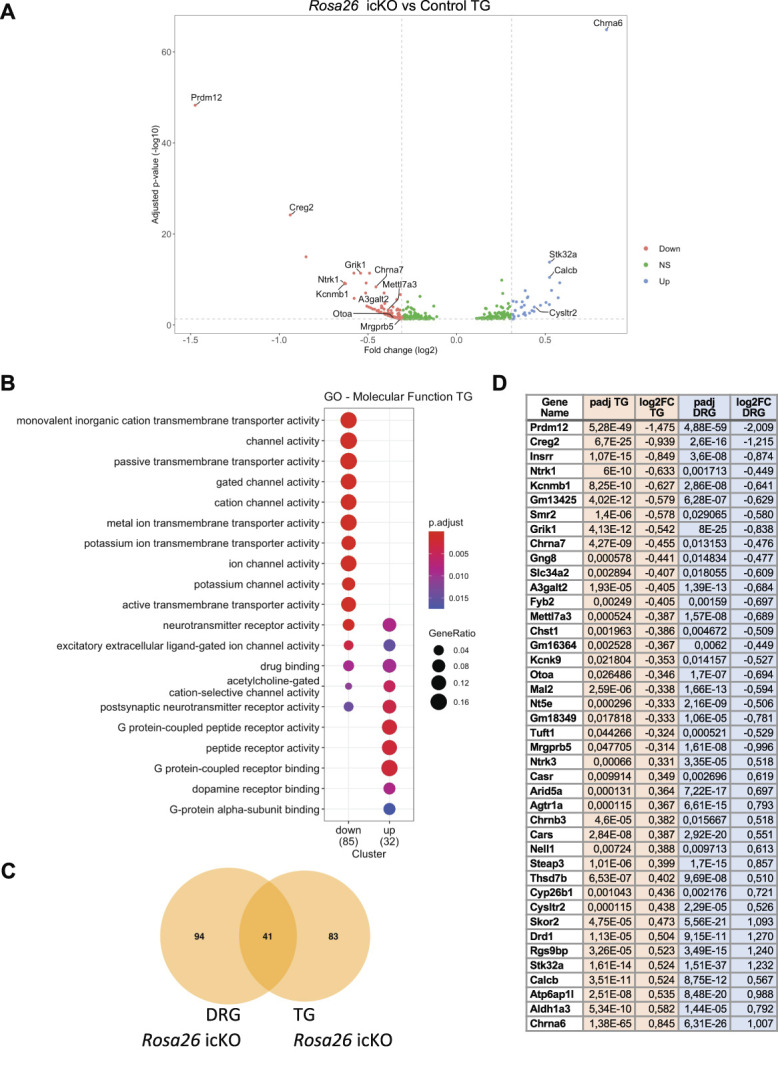
Transcriptomic analysis of the consequences of the loss of Prdm12 in TG of adult mice. (A) Volcano plots showing the –log10 adjusted P-value as a function of the log2 fold-changes of deregulated genes in TG of Rosa26 icKO mice (icKO mice, n = 6 vs control mice, n = 6) 1 month after tamoxifen injection. The genes were identified by bulk RNA-seq and selected based on adjusted P-value <0.05. Genes with a log2 fold-change >0.31 are indicated in red, genes below the threshold are in light grey. (B) Gene ontology classification of the DEGs. The graph shows the enriched biological molecular function associated with downregulated and/or upregulated genes. (C) Venn diagram showing the overlap between the DEGs identified in TG and DRG of Rosa26 icKO mice. (D) List of common DEGs in TG and DRG of Rosa26 icKO. DRG, dorsal root ganglia; TG, trigeminal ganglia; icKO, inducible conditional knockout.
3.3. Loss of Prdm12 affects dorsal root ganglia neuron excitability
To determine whether the molecular defects observed in sensory neurons of icKO lead to changes in neuronal excitability, current-clamp recordings were performed on cultured small diameter DRG neurons (ie, putative nociceptors) from Avil icKO (marked by tdTomato+ fluorescence) and control mice. Measurements were done 24 hours after mechanical dissociation, which is known to induce hyperexcitability in DRG neurons67 by mechanisms that may present common points with in vivo inflammation-induced hyperexcitability.33 Representative action potential traces from control and Avil icKO DRG neurons in response to depolarizing current steps showed that an action potential was elicited at a threshold of 1310 and 750 pA, respectively (Fig. 6A). The mean current threshold for action potential firing in Avil icKO neurons was significantly lower compared with control DRG neurons (control, 1420 ± 161 pA, n = 19; Avil icKO, 850 ± 109 pA, n = 30; Fig. 6B). Moreover, the absence of Prdm12 doubled the maximal number of action potentials evoked by a 1-sec-current step in small DRG neurons (Fig. 6C). There was no significant difference in resting membrane potential for DRG neurons isolated from control or Avil icKO mice (Fig. 6D). Thus, loss of Prdm12 alters DRG neuronal excitability and, in the experimental conditions used, results in hyperexcitability of DRG neurons.
Figure 6.
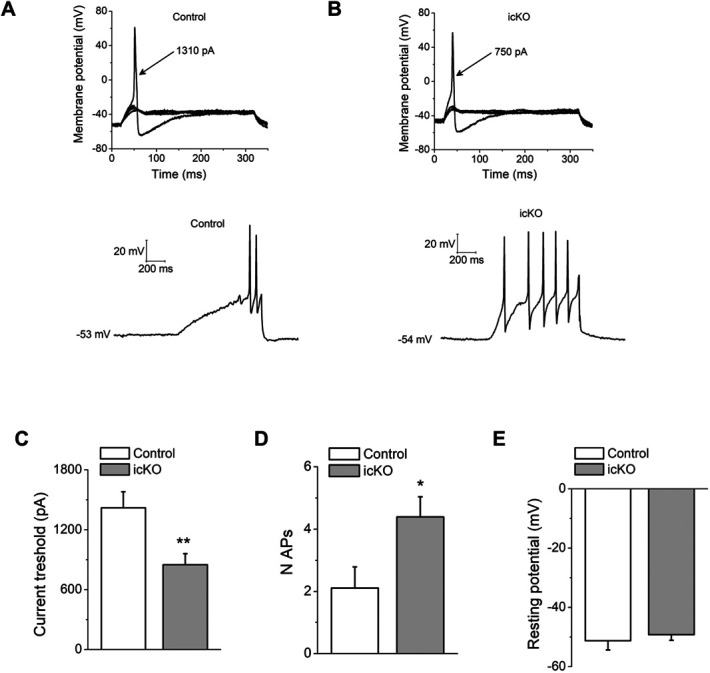
Loss of Prdm12 leads to increased DRG neuron excitability. (A) Representative traces from a control DRG neuron, showing subthreshold responses and subsequent action potential induced by injection of 1310 pA (top), and action potential frequency induced by a 1-second 1500 pA step (bottom). (B) The same protocol was applied to an Avil icKO DRG neuron. The current threshold was 750 pA for this neuron. Arrows indicate the current amplitude used to elicit the labeled response. (C) Comparison of current threshold in control (n = 19) and Avil icKO (n = 30) DRG neurons. (D) Maximal number of action potentials evoked in response to external current stimuli of 1 second, up to 1500 pA in control (n = 19) and Avil icKO (n = 30) DRG neurons. (E) Resting membrane potential in control (n = 19) and Avil icKO (n = 30) DRG neurons. Each column represents mean ± SEM. *P < 0.05, **P < 0.01 (Student t test). DRG, dorsal root ganglia; icKO, inducible conditional knockout.
3.4. Conditional loss of Prdm12 alters sensitivity to capsaicin and formalin
To determine whether loss of Prdm12 in mature nociceptors of adult mice alters pain sensation, pain behavior assays were performed on icKO and control mice 1 month after TAM injection. Given the difficulties encountered to generate Avil icKO mice because of non-Mendelian transmission of the Cre and based on the consistency of the transcriptional changes observed in DRG using the 2 mouse models, both Avil icKO and Rosa26 icKO mice were used. All behavior tests were performed using both male and female mice. To evaluate thermal nociception, we performed a thermal place preference test in Avil icKO mice and tail flick and cold plantar tests in Rosa26 icKO mice. Mechanical nociception was evaluated in Rosa26 icKO mice using the mechanical conflict avoidance test. Using these tests, no difference between mutant and control mice was observed in mechanosensation and thermosensation. However, mutant mice did show a trend not reaching significance (P = 0.07), of greater heat sensitivity in the temperature place preference assay, in the 52°C vs 30°C condition (Figs. 7A–D). In addition, as a control for motor behavior, we also tested Rosa26 icKO and control mice in the open-field and beam walking tests (Sup. Fig. 4A,B, http://links.lww.com/PAIN/B535). Similarly, no difference was found between the 2 groups.
Figure 7.
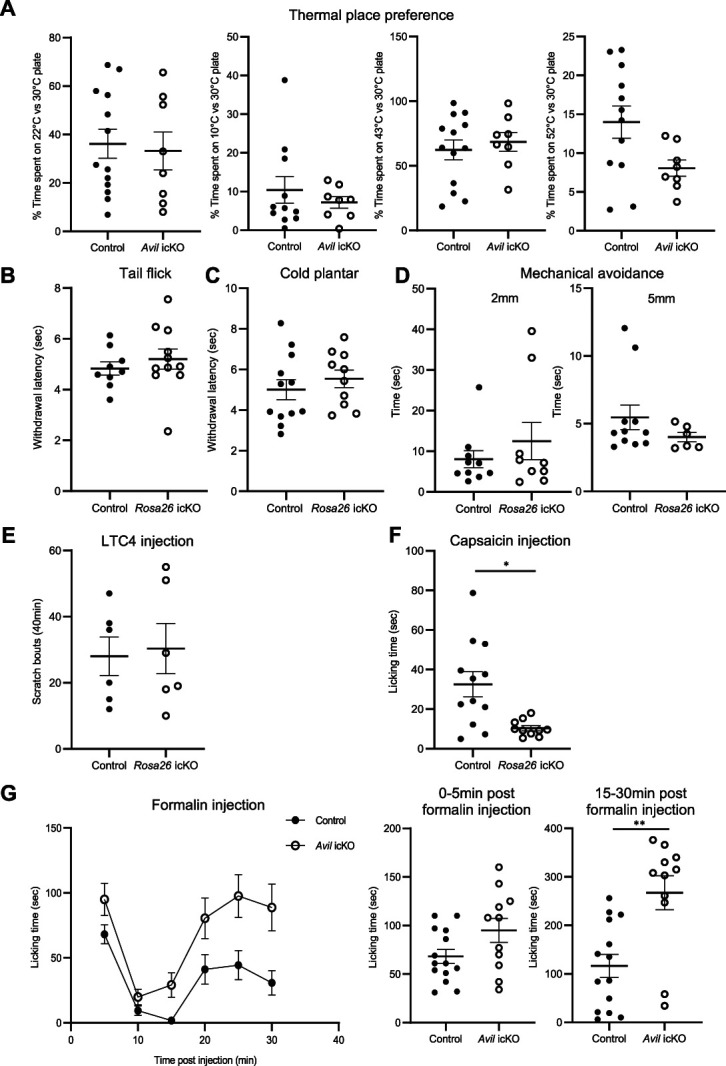
Conditional knockout of Prdm12 in mature nociceptors does not alter thermal and mechanical nociception but affects responses to formalin and capsaicin. (A) Avil icKO and control mice spent similar time on the test side in a 2-temperature choice assay. (B) Rosa26 icKO (n = 11) and control mice (n = 9) show similar withdrawal latency in the tail flick test using a focus 30. (C) Rosa26 icKO (n = 10) and control mice (n = 12) show no difference in withdrawal latency in the cold plantar assay. (D) Rosa26 icKO and control mice show the same behavior in the mechanical conflict avoidance test. They spent the same time to cross half of the second chamber with nails on the floor. Results obtained with 2 different nail heights (2 and 5 mm) are shown. (E) Intradermal cheek injection of N-met LTC4 elicits scratching bouts that are not significantly different between Rosa26 icKO and control mice. (F) Rosa26 icKO mice (n = 10) spent less time licking their paw than control mice (n = 12) after capsaicin injection (P = 0.011). (G) Time course of the nocifensive response (licking time in seconds) of Avil icKO (n = 11) and control mice (n = 14) until 30 minutes after formalin injection. Response of individual Avil icKO and control mice in the first (0-5 minutes after formalin injection) and second phase (15-30 minutes after formalin injection) is shown on the right. Note that Avil icKO mice spent more time licking their paw when compared with control mice during the second phase (P = 0.001). Values are represented as mean ± SEM. *P < 0.05, **P < 0.01 (Student t test, α = 0.05). icKO, inducible conditional knockout.
Based on the upregulation of the Cysltr2 receptor that mediates cysteinyl leukotriene C4-driven itch,57 we investigated itch behavior of Rosa26 icKO mice. Therefore, we injected LTC4 into the cheek of control and mutant mice and measured their scratching behavior as described. Unexpectedly, no differences were observed between control and icKO mice (Fig. 7E).
We next tested the response of Rosa26 icKO mice to capsaicin, the active component of chili that, by binding to its receptor TRPV1, leads to nociceptor activation. Capsaicin was injected subcutaneously in the hind paw and the time mice spent licking the injection site was recorded for 15 minutes after injection. We observed that the response of icKO mice was significantly reduced in comparison with control mice (Fig. 7F), suggesting a downregulation of TRPV1-related pain transmission in adult nociceptors lacking Prdm12.
Given our observation that Prdm12 expression is decreased in a model of joint inflammation, we hypothesized that loss of Prdm12 might lead to changes in inflammation-related nociceptive behavior of adult mice. We thus examined behavior responses of Avil icKO mice in the formalin test, a common model for tissue injury-induced pain, which produces paw swelling and an inflammatory response.11 This acute inflammation is caused by cell damages that induce the production of endogenous mediators and subsequently the release of inflammatory mediators in the paw. In this test, 2 distinct periods of high licking activity of the injected paw can be identified: an early, short-lasting phase of intense licking caused by the direct chemonociceptive effect of formalin on nociceptors followed by a short pause and a second prolonged phase of continuous tonic licking induced, at least in part, by inflammatory responses caused by formalin-induced cellular damages.53 As shown in Figure 7G, Avil icKO mice showed similar responses to control animals during the first phase. However, they spent more time licking when compared with control mice during the second phase. Behavioral responses of Rosa26 icKO mice have been also examined in the formalin test. An increase in sensitivity in the second phase of the phenotype was also observed (Sup data Fig. 4C, http://links.lww.com/PAIN/B535). These results suggest a role for Prdm12 in inflammatory pain. Thus, adult-onset ablation of Prdm12 modulates in a complex way pain-related behaviors.
4. Discussion
In this study, we show that in adult mice, Prdm12 remains strongly expressed in mature nociceptors and that in the CNS, it is only found in a few specific brain nuclei. In a recent report, Prdm12 has been shown to be enriched in hypothalamic POMC neurons where it plays a role in their development.8,19 In addition, our data suggest that CFA-induced peripheral inflammation decreases Prdm12 expression in mature nociceptors. Deregulated expression of Prdm12 has also been observed in response to nerve injury30 and in itch-sensitive neurons in response to dermatitis.63 Whether this deregulation of Prdm12 expression plays a causative role in the transcriptional changes that occurs in these conditions remains unknown.
The consistency of the RNA-seq data obtained analysing DRG of both Avil icKO and Rosa26 icKO mice and TG of Rosa26 icKO mice strongly support a role for Prdm12 in mature nociceptors in the transcriptional regulation of a battery of genes important for their functional properties. We found both upregulated and downregulated DEGs in DRG and TG of Prdm12 mutant mice. Landy and colleagues recently published the results of bulk RNA-seq analysis of DRGs of adult Prdm12 knockout mice, generated as we have done by crossing AvilCreERT2 mice to Prdm12 fl/fl mice.30 In contrast to our results, they found that almost all DEGs were decreased in mutant mice. Comparing their transcriptomic data to our list of DEGs identified in DRG, surprisingly, only 1 common gene was found, Chrna6. Besides differences in the RNA-seq approach used (ie, choice of fold-change cut-off), one possible explanation for these discrepancies is that in the conditional knockout mouse model they used, only the last exon of Prdm12 (exon V) is deleted. In this knockout, a transcript composed of exons I to IV is still detected. This transcript encodes a truncated protein with the conserved PR domain but lacking the zinc fingers. As it is thought that Prdm12 does not bind directly to DNA, this truncated protein may thus potentially retain some activity. Whether Prdm12 acts as an activator, a repressor, or both in mature nociceptors remains unknown. The identification of its direct targets will be a crucial step towards a better understanding of its mechanism of action.
Despite the effects of Prdm12 deficiency on transcription, we found that mice lacking Prdm12 exhibit normal thermosensation and itch response. Unaltered response to noxious cold is somehow surprising given the fact that we found that Trpm8 expression is reduced in Prdm12 icKO mice, as observed by Landy et al. (2021). This unaltered response to acute noxious cold may be because of the fact that Trpm8 expression is reduced but not abolished in DRG neurons of icKO mice. It also likely reflects the complexity of the cold-sensing mechanisms in mouse DRG that involve other molecular candidates apart from Trpm8,39 such as TrpA1,62 that is not deregulated in icKO DRG. Unchanged itch behavior was also unexpected, given the increased expression of Cysltr2, a sensor of mast cell–induced itch,50 although it should be noted that the increased expression was measured in lumbar DRG and the behavioral paradigm used activates TG neurons. Mechanical nociception was also unaltered in icKO mice, a result in accordance with the absence of modification of the expression of the TRPA1, TRPV4, and Piezo2 channels involved in mechanical pain.21
In contrast, we found that icKO mice spent less time licking their paw than control mice after capsaicin injection. This reduced sensitivity is at first glance surprising because TRPV1 expression is not modified. This could, however, be because of differences between icKO and controls in the localization of TRPV1 receptors that is rapidly internalized after stimulation. Indeed, capsaicin can be used as analgesic and has been shown to reduce lesion or inflammation-induced pain by triggering receptor endocytosis48,58 or nerve retraction.17,22,43 It could also be because of posttranscriptional modifications of TRPV1, its activity being modulated by kinases such as calmodulin-dependent protein kinase (CaMKII), protein kinase C (PKC) and protein kinase A (PKA), and phosphatases (calcineurin) as well as lipids such as PIP2,46 themselves regulated by G protein-coupled membrane receptors. Nerve growth factor plays a role in the sensitization of nociceptors via different mechanisms, including through the control of trafficking of TRPV1 channels to the plasma membrane.66 The downregulation of Ntrk1 encoding the TrkA receptor may thus play a role in the decreased response to capsaicin of icKO mice.
In addition, we also found that icKO mice develop increased sensitivity to formalin-induced inflammatory pain. Our gene expression analysis of DRG of mutant mice revealed that the loss of Prdm12 dysregulates a set of genes known to contribute to formalin-induced nociceptor sensitization. Among them Trpm8, Ntrk1, and Grik1 are downregulated and Chrna6 and Cysltr2 are upregulated in the absence of Prdm12 in basal conditions. The upregulation of Chrna6 is unexpected because its overexpression is protective against tactile allodynia associated with inflammatory injuries.60 The downregulation of Grik1 and Ntrk1 was also unpredicted because Grik1 deletion reduces inflammatory pain27 and NGF acts as a mediator of inflammatory pain.2,12,54 However, the deregulation of Trpm8 may play a role in the observed hypersensitivity of icKO mice. Indeed, previous studies have shown that formalin-evoked pain can be alleviated by cooling analgesia through Trpm8-dependent receptor activation.15 Moreover, recent studies indicate that activation of Trpm8 primarily affects late, but not early, phases of DRG neuron activity after formalin injection.7 The upregulation of Cysltr2 may also contribute to the hyperalgesia. Indeed, Cysltr2 is a known mediator of inflammatory reactions.50 As this gene expression analysis was done on noninflamed DRGs and the increase of sensitivity was observed upon inflammation, we wanted to assess changes in gene expression because of the loss of Prdm12 in inflammatory conditions. Therefore, we performed unilateral injections of formalin in the hind paw of TAM-injected Avil icKO and control mice, dissected L4 to L6 DRG of the injected and uninjected sides of mutant and control mice 24 hours after formalin injection, and analysed their transcriptome by bulk RNA-seq. A highly overlapping list of DEGs was identified in basal (comparing uninjected sides of icKO and controls) and inflammatory (comparing formalin-injected sides of icKO and controls) conditions (Sup data Fig. 5). However, comparing injected vs noninjected sides of mice and performing the analysis without a cut-off on the log2 fold-change, a number of significant DEGs were observed in icKO mice but not in control mice. Gene ontology analysis revealed an enrichment in genes encoding proteins involved in transport, regulation, and secretion of neurotransmitters and synapse organization and maturation (Sup data Fig. 6). These results further suggest a role for Prdm12 in inflammatory pain. Although our RNA-seq work does not provide mechanical insights into the formalin phenotype, the increased excitability of cultured DRG neurons from icKO mice may provide an initial explanation for their hypersensitivity in the formalin test. The downregulation of some K+ channel genes, such as Kcnmb1, Kcnj11, and Kcng1, could be involved in this increased excitability, as it was shown for Kcnn4.38
Taken together, our results show that Prdm12 remains closely linked to NGF at adulthood, as observed during embryogenesis. Like NGF, unlike in development, Prdm12 is not required anymore for the maintenance of adult nociceptors but plays a role in the excitability properties of nociceptors and affects pain behavior in adulthood. In their recent report, Landy and colleagues found that the knockout of Prdm12 exon V does not substantially modify pain sensitivity. A tendency not reaching significance to a reduction of the licking time is, however, apparent in their exon V Prdm12 knockout model after capsaicin injection. In the formalin test, although they conclude that Prdm12 exon V icKO mice show no differences in their response to formalin,27 a transient increase of licking time of the icKO mice is, however, observed 20 minutes after injection. We posit that the retention of exon II to IV in their knockout models, absent in our exon II Prdm12 knockout model,14 may be the cause of this milder phenotype. Although this article was under revision, Kokotovic and colleagues also reported that Prdm12 is required for nociceptor function in adult mice.5,28 Further studies are obviously needed to better understand the complex mechanisms by which Prdm12 controls nociceptive neuron properties.
Conflict of interest statement
All or part of the results herein are the subject of a pending patent, the applicants of which being Université Libre de Bruxelles et Université de Mons and the inventors being A. Latragna, A. Sabaté San José, S. Vermeiren, R. Gualdani, L. Ris, and E.J. Bellefroid. The remaining authors have no conflicts of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B535.
Supplementary Material
Acknowledgments
The authors thank members of the Centre for Microscopy and Molecular Imaging (CMMI) for help with microscopy and Louis Delhaye for technical help. The authors are also grateful to Patrick Di Stefano (ULB Knowledge Transfer Office) for helpful discussions during the project and Franziska Denk and Alexandre Pattyn for their valuable scientific input. This work was supported by grants from the Walloon Region (Win2Wal project PANOPP 1810123, to E.J.B. and L.R.), the FNRS (PDR T002020F) and the Fonds Defay (to E.J.B.), and vs Arthritis RG21973 (to E.St.J.S.). A.L., A.S. and P.T. are doctoral fellows from the FNRS (supported by Télévie, Aspirant, and FRIA fellowships, respectively). S. Vermeiren was a FRIA doctoral fellow and, together with R. Gualdani and S. Desiderio, a postdoctoral fellow, supported by the Walloon Region Win2Wal grant PANOPP.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
A. Latragna and A. Sabaté contributed equally to this work.
L. Ris and E. J. Bellefroid also contributed equally to this work.
Contributor Information
Aurore Latragna, Email: Aurore.LATRAGNA@umons.ac.be.
Alba Sabaté San José, Email: alsasajo95@gmail.com.
Panagiotis Tsimpos, Email: ptsibos@gmail.com.
Simon Vermeiren, Email: svermeir@ulb.ac.be.
Roberta Gualdani, Email: roberta.gualdani@umons.ac.be.
Sampurna Chakrabarti, Email: sc968@cantab.ac.uk.
Gerard Callejo, Email: gerard.callejo@ub.edu.
Simon Desiderio, Email: simon.desiderio@ulb.ac.be.
Orr Shomroni, Email: orr.shomroni@med.uni-goettingen.de.
Maren Sitte, Email: Maren.Sitte@med.uni-goettingen.de.
Sadia Kricha, Email: Sadia.Kricha@ulb.ac.be.
Maëlle Luypaert, Email: maelle.luypaert@ulb.be.
Benoit Vanhollebeke, Email: Benoit.Vanhollebeke@ulb.be.
Geoffroy Laumet, Email: GOLaumet@mdanderson.org.
Gabriela Salinas, Email: gabriela.salinas-riester@medizin.uni-goettingen.de.
Ewan St. John Smith, Email: es336@cam.ac.uk.
Laurence Ris, Email: Laurence.RIS@umons.ac.be.
References
- [1].Abdrakhmanova GR, AlSharari S, Kang M, Damaj MI, Akbarali HI. α7-nAChR-mediated suppression of hyperexcitability of colonic dorsal root ganglia neurons in experimental colitis. Am J Physiol 2010;299:G761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Barker PA, Mantyh P, Arendt-Nielsen L, Viktrup L, Tive L. Nerve growth factor signaling and its contribution to pain. J Pain Res 2020;13:1223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bartesaghi L, Wang Y, Fontanet P, Wanderoy S, Berger F, Wu H, Akkuratova N, Bouçanova F, Médard JJ, Petitpré C, Landy MA, Zhang MD, Harrer P, Stendel C, Stucka R, Dusl M, Kastriti ME, Croci L, Lai HC, Consalez GG, Pattyn A, Ernfors P, Senderek J, Adameyko I, Lallemend F, Hadjab S, Chrast R. PRDM12 is required for initiation of the nociceptive neuron lineage during neurogenesis. Cell Rep 2019;26:3484–92.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brenner DS, Golden JP, Gereau RW, IV. A novel behavioral assay for measuring cold sensation in mice. PLoS One 2012;7:39765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 2000;288:306–13. [DOI] [PubMed] [Google Scholar]
- [6].Chakrabarti S, Pattison LA, Singhal K, Hockley JRF, Callejo G, Smith ESJ. Acute inflammation sensitizes knee-innervating sensory neurons and decreases mouse digging behavior in a TRPV1-dependent manner. Neuropharmacology 2018;143:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen C, Zhang J, Sun L, Zhang Y, Gan WB, Tang P, Yang G. Long-term imaging of dorsal root ganglia in awake behaving mice. Nat Commun 2019;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen X, Wyler SC, Li L, Arnold AG, Wan R, Jia L, Landy MA, Lai HC, Xu P, Liu C. Comparative transcriptomic analyses of developing melanocortin neurons reveal new regulators for the anorexigenic neuron identity. J Neurosci 2020;40:3165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen YC, Auer-Grumbach M, Matsukawa S, Zitzelsberger M, Themistocleous AC, Strom TM, Samara C, Moore AW, Cho LTY, Young GT, Weiss C, Schabhüttl M, Stucka R, Schmid AB, Parman Y, Graul-Neumann L, Heinritz W, Passarge E, Watson RM, Hertz JM, Moog U, Baumgartner M, Valente EM, Pereira D, Restrepo CM, Katona I, Dusl M, Stendel C, Wieland T, Stafford F, Reimann F, Von Au K, Finke C, Willems PJ, Nahorski MS, Shaikh SS, Carvalho OP, Nicholas AK, Karbani G, McAleer MA, Cilio MR, McHugh JC, Murphy SM, Irvine AD, Jensen UB, Windhager R, Weis J, Bergmann C, Rautenstrauss B, Baets J, De Jonghe P, Reilly MM, Kropatsch R, Kurth I, Chrast R, Michiue T, Bennett DLH, Woods CG, Senderek J. Transcriptional regulator PRDM12 is essential for human pain perception. Nat Genet 2015;47:803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chillingworth NL, Donaldson LF. Characterisation of a Freund's complete adjuvant-induced model of chronic arthritis in mice. J Neurosci Methods 2003;128:45–52. [DOI] [PubMed] [Google Scholar]
- [11].Damas J, Liégeois JF. The inflammatory reaction induced by formalin in the rat paw. Naunyn Schmiedebergs Arch Pharmacol 1999;359:220–7. [DOI] [PubMed] [Google Scholar]
- [12].Denk F, Bennett DL, McMahon SB. Nerve growth factor and pain mechanisms. Annu Rev Neurosci 2017;40:307–25. [DOI] [PubMed] [Google Scholar]
- [13].Denk F, McMahon SB. Chronic pain: emerging evidence for the involvement of epigenetics. Neuron 2012;73:435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Desiderio S, Vermeiren S, Van Campenhout C, Kricha S, Malki E, Richts S, Fletcher EV, Vanwelden T, Schmidt BZ, Henningfeld KA, Pieler T, Woods CG, Nagy V, Verfaillie C, Bellefroid EJ. Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF receptor TrkA. Cell Rep 2019;26:3522–36.e5. [DOI] [PubMed] [Google Scholar]
- [15].Dhaka A, Murray AN, Mathur J, Earley TJ, Petrus MJ, Patapoutian A. TRPM8 is required for cold sensation in mice. Neuron 2007;54:371–8. [DOI] [PubMed] [Google Scholar]
- [16].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Frias B, Merighi A. Capsaicin, nociception and pain. Molecules 2016;21:797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gold MS, Gebhart GF. Nociceptor sensitization in pain pathogenesis. Nat Med 2010;16:1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hael CE, Rojo D, Orquera DP, Low MJ, Rubinstein M. The transcriptional regulator PRDM12 is critical for Pomc expression in the mouse hypothalamus and controlling food intake, adiposity, and body weight. Mol Metab 2020;34:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Harte SE, Meyers JB, Donahue RR, Taylor BK, Morrow TJ. Mechanical conflict system: a novel operant method for the assessment of nociceptive behavior. PLoS One 2016;11:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hill RZ, Bautista DM. Getting in touch with mechanical pain mechanisms. Trends Neurosci 2020;43:311–25. [DOI] [PubMed] [Google Scholar]
- [22].Ishida H, Zhang Y, Gomez R, Shannonhouse J, Son H, Kim YS. Capsaicin pretreatment alleviates postoperative pain and reduces primary sensory neuron Ca2+ activity. bioRxiv 2021:2021.05.21.445191. doi: 10.1101/2021.05.21.445191. [DOI] [Google Scholar]
- [23].Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci 2019;28:1947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 2021;49:D545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kinameri E, Inoue T, Aruga J, Imayoshi I, Kageyama R, Shimogori T, Moore AW. Prdm proto-oncogene transcription factor family expression and interaction with the notch-hes pathway in mouse neurogenesis. PLoS One 2008;3:e3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ko S, Zhao MG, Toyoda H, Qiu CS, Zhuo M. Altered behavioral responses to noxious stimuli and fear in glutamate receptor 5 (GluR5)- or GluR6-deficient mice. J Neurosci 2005;25:977–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kokotović T, Langeslag M, Lentartowicz EM, Manion J, Fell C, Alehabib E, Tafakhori A, Darvish H, Bellefroid EJ, Neely G, Kress M, Penninger JM, Nagy V. PRDM12 is required for pain sensation throughout life. Front Mol Neurosci 2021. Submitted. doi: 10.3389/fnmol.2021.720973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kunita R, Otomo A, Ikeda JE. Identification and characterization of novel members of the CREG family, putative secreted glycoproteins expressed specifically in brain. Genomics 2002;80:456–60. [PubMed] [Google Scholar]
- [30].Landy MA, Goyal M, Casey KM, Liu C, Lai HC. Loss of Prdm12 during development, but not in mature nociceptors, causes defects in pain sensation. Cell Rep 2021;34:108913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lau J, Minett MS, Zhao J, Dennehy U, Wang F, Wood JN, Bogdanov YD. Temporal control of gene deletion in sensory ganglia using a tamoxifen-inducible Advillin-Cre-ERT2 recombinase mouse. Mol Pain 2011;7:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lauria G, Borgna M, Morbin M, Lombardi R, Mazzoleni G, Sghirlanzoni A, Pareyson D. Tubule and neurofilament immunoreactivity in human hairy skin: markers for intraepidermal nerve fibers. Muscle and Nerve 2004;30:310–16. [DOI] [PubMed] [Google Scholar]
- [33].Li ZH, Cui D, Qiu CJ, Song XJ. Cyclic nucleotide signaling in sensory neuron hyperexcitability and chronic pain after nerve injury. Neurobiol Pain 2019;6:100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- [35].Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng HJ, Geng Y, Undem BJ, Kollarik M, Chen ZF, Anderson DJ, Dong X. Sensory neuron-specific GPCR mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 2009;139:1353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [37].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lu R, Flauaus C, Kennel L, Petersen J, Drees O, Kallenborn-Gerhardt W, Ruth P, Lukowski R, Schmidtko A. KCa3.1 channels modulate the processing of noxious chemical stimuli in mice. Neuropharmacology 2017;125:386–95. [DOI] [PubMed] [Google Scholar]
- [39].Luiz AP, MacDonald DI, Santana-Varela S, Millet Q, Sikandar S, Wood JN, Emery EC. Cold sensing by Na V 1.8-positive and Na V 1.8-negative sensory neurons. Proc Natl Acad Sci U S A 2019;116:3811–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matsukawa S, Miwata K, Asashima M, Michiue T. The requirement of histone modification by PRDM12 and Kdm4a for the development of pre-placodal ectoderm and neural crest in Xenopus. Dev Biol 2015;399:164–76. [DOI] [PubMed] [Google Scholar]
- [42].Nagy V, Cole T, Van Campenhout C, Khoung TM, Leung C, Vermeiren S, Novatchkova M, Wenzel D, Cikes D, Polyansky AA, Kozieradzki I, Meixner A, Bellefroid EJ, Neely GG, Penninger JM. The evolutionarily conserved transcription factor PRDM12 controls sensory neuron development and pain perception. Cell Cycle 2015;14:1799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].O'Neill J, Brock C, Olesen AE, Andresen T, Nilsson M, Dickenson AH. Unravelling the mystery of capsaicin: a tool to understand and treat pain. Pharmacol Rev 2012;64:939–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pan AH, Lu DH, Luo XG, Chen L, Li ZY. Formalin-induced increase in p2x3 receptor expression in dorsal root ganglia: implications for nociception. Clin Exp Pharmacol Physiol 2009;36:e6–11. [DOI] [PubMed] [Google Scholar]
- [45].Pattison LA, Callejo G, Smith ESJ. Evolution of acid nociception: ion channels and receptors for detecting acid. Philos Trans R Soc B Biol Sci 2019;374:20190291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rosenbaum T, Simon SA, Liedtke WB, Heller S. TRPV1 receptors and signal transduction. In: TRP ion channel function in sensory transduction and cellular signaling. Chapter 5. Frontiers in Neuroscience. Boca Raton,FL: CRC Press/Taylor & Francis, 2007. [Google Scholar]
- [47].Sandlesh P, Juang T, Safina A, Higgins MJ, Gurova KV. Uncovering the fine print of the CreERT2-LoxP system while generating a conditional knockout mouse model of Ssrp1 gene. PLoS One 2018;13:e0199785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sanz-Salvador L, Andrés-Borderia A, Ferrer-Montiel A, Planells-Cases R. Agonist- and Ca2+-dependent desensitization of TRPV1 channel targets the receptor to lysosomes for degradation. J Biol Chem 2012;287:19462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Smith PA. K+ channels in primary afferents and their role in nerve injury-induced pain. Front Cell Neurosci 2020;14:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Solinski HJ, Kriegbaum MC, Tseng PY, Earnest TW, Gu X, Barik A, Chesler AT, Hoon MA. Nppb neurons are sensors of mast cell-induced itch. Cell Rep 2019;26:3561–73.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Souslova V, Cesare P, Ding Y, Akopian AN, Stanfa L, Suzuki R, Carpenter K, Dickenson A, Boyce S, Hill R, Nebenius-Oosthuizen D, Smith AJH, Kidd EJ, Wood JN. Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature 2000;407:1015–17. [DOI] [PubMed] [Google Scholar]
- [52].Thélie A, Desiderio S, Hanotel J, Quigley I, Van Driessche B, Rodari A, Borromeo MD, Kricha S, Lahaye F, Croce J, Cerda-Moya G, Fernandez JO, Bolle B, Lewis KE, Sander M, Pierani A, Schubert M, Johnson JE, Kintner CR, Pieler T, Van Lint C, Henningfeld KA, Bellefroid EJ, Van Campenhout C. Prdm12 specifies V1 interneurons through cross-repressive interactions with Dbx1 and Nkx6 genes in Xenopus. Dev 2015;142:3416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tjølsen A, Berge OG, Hunskaar S, Rosland JH, Hole K. The formalin test: an evaluation of the method. PAIN 1992;51:5–17. [DOI] [PubMed] [Google Scholar]
- [54].Ugolini G, Marinelli S, Covaceuszach S, Cattaneo A, Pavone F. The function neutralizing anti-TrkA antibody MNAC13 reduces inflammatory and neuropathic pain. Proc Natl Acad Sci U S A 2007;104:2985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015;18:145–53. [DOI] [PubMed] [Google Scholar]
- [56].Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661–5. [DOI] [PubMed] [Google Scholar]
- [57].Voisin T, Perner C, Messou MA, Shiers S, Ualiyeva S, Kanaoka Y, Price TJ, Sokol CL, Bankova LG, Frank Austen K, Chiu IM. The CysLT2R receptor mediates leukotriene C4-driven acute and chronic itch. Proc Natl Acad Sci U S A 2021;118:e2022087118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Vyklický L, Nováková-Toušová K, Benedikt J, Samad A, Touška F, Vlachova V. Calcium-dependent desensitization of vanilloid receptor TRPV1: a mechanism possibly involved in analgesia induced by topical application of capsaicin. Physiol Res 2008;57:S59–68. [DOI] [PubMed] [Google Scholar]
- [59].Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T, Miller E, Bache S, Müller K, Ooms J, Robinson D, Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. Welcome to the tidyverse. J Open Source Softw 2019;4:1686. [Google Scholar]
- [60].Wieskopf JS, Mathur J, Limapichat W, Post MR, Al-Qazzaz M, Sorge RE, Martin LJ, Zaykin DV, Smith SB, Freitas K, Austin JS, Dai F, Zhang J, Marcovitz J, Tuttle AH, Slepian PM, Clarke S, Drenan RM, Janes J, Al Sharari S, Segall SK, Aasvang EK, Lai W, Bittner R, Richards CI, Slade GD, Kehlet H, Walker J, Maskos U, Changeux JP, Devor M, Maixner W, Diatchenko L, Belfer I, Dougherty DA, Su AI, Lummis SCR, Damaj MI, Lester HA, Patapoutian A, Mogil JS. The nicotinic α6 subunit gene determines variability in chronic pain sensitivity via cross-inhibition of P2X2/3 receptors. Sci Transl Med 2015;7:287ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wimalawansa SJ. Calcitonin gene-related peptide and its receptors: molecular genetics, physiology, pathophysiology, and therapeutic potentials. Endocr Rev 1996;17:533–85. [DOI] [PubMed] [Google Scholar]
- [62].Winter Z, Gruschwitz P, Eger S, Touska F, Zimmermann K. Cold temperature encoding by cutaneous TRPA1 and TRPM8-carrying fibers in the mouse. Front Mol Neurosci 2017;10:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Xing Y, Chen J, Hilley H, Steele H, Yang J, Han L. Molecular signature of pruriceptive MrgprA3+ neurons. J Invest Dermatol 2020;140:2041–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zannino DA, Downes GB, Sagerström CG. Prdm12b specifies the p1 progenitor domain and reveals a role for V1 interneurons in swim movements. Dev Biol 2014;390:247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S. Molecular architecture of the mouse nervous system. Cell 2018;174:999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang X, Huang J, McNaughton PA. NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. EMBO J 2005;24:4211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zheng JH, Walters ET, Song XJ. Dissociation of dorsal root ganglion neurons induces hyperexcitability that is maintained by increased responsiveness to cAMP and cGMP. J Neurophysiol 2007;97:15–25. [DOI] [PubMed] [Google Scholar]
- [68].Zwaenepoel I, Mustapha M, Leibovici M, Verpy E, Goodyear R, Liu XZ, Nouaille S, Nance WE, Kanaan M, Avraham KB, Tekaia F, Loiselet J, Lathrop M, Richardson G, Petit C. Otoancorin, an inner ear protein restricted to the interface between the apical surface of sensory epithelia and their overlying acellular gels, is defective in autosomal recessive deafness DFNB22. Proc Natl Acad Sci U S A 2002;99:6240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B535.