Abstract
Antimicrobial-resistant bacteria are a major public health problem. Of particular importance in the context of food safety is the prevalence of antimicrobial resistance (AMR) genes within nontyphoidal Salmonella, which is a leading bacterial cause of foodborne disease. We determined the prevalence of AMR genes across a very large number of Salmonella genomes (n = 25,647) collected from isolates from 16 common food sources. The average percentage of isolates from nonanimal foods, such as fruit, nuts and seeds, and vegetables, harboring at least one AMR gene was only marginally lower (72%) than that observed in isolates from animal foods such as beef, chicken, turkey, and pork (74%). This high prevalence of AMR genes was primarily driven by the high prevalence of aminoglycoside resistance genes in nearly all food isolates; genes for resistance to tetracycline and sulfonamide also were highly prevalent. However, evaluation of the number of genes per isolate revealed that the prevalence of AMR genes was higher in animal food isolates than in nonanimal food isolates (P = 0.018). A random forest analysis provided evidence that within a given serovar, resistance gene profiles differed according to isolate food source. AMR gene profiles could be used to correctly predict the food of origin for 71% of the isolates, but success differed according to serovar. This information can help inform AMR risk assessments of food commodities and refine processes for targeting interventions to limit the spread of AMR through the food supply.
Keywords: Antimicrobial resistance, Food, Public health, Salmonella, Whole genome sequencing
Nontyphoidal Salmonella is a leading bacterial cause of foodborne disease worldwide. In the United States, Salmonella annually causes approximately 1.2 million illnesses, 23,000 hospitalizations, and 450 deaths (28). From 2004 to 2012, the Centers for Disease Control and Prevention (CDC) estimated the number of culture-confirmed antimicrobial resistant nontyphoidal Salmonella infections at ~2 per 100,000 person years (21). Infants, elderly persons, and immunocompromised persons are at greatest risk of infection and severe outcomes from salmonellosis. Most people infected with Salmonella recover without treatment, but complicated infections require antibiotic therapy. The CDC (7) has estimated that 100,000 drug-resistant Salmonella infections occur each year in the United States. These drug-resistant infections can cause more severe clinical outcomes such as increased rates of hospitalization, sepsis, invasive disease, and death (10, 15, 32, 37).
Humans are exposed to antimicrobial-resistant Salmonella strains through food, animals, and the environment (8). Although antimicrobial resistance (AMR) in foodborne pathogens is a complex challenge, the use of antimicrobial agents in humans and food-producing animals is generally considered the main driver of resistance (12, 30). As a result, surveillance programs such as the National Antimicrobial Resistance Monitoring System (NARMS) have been established to track the emergence and spread of AMR that may result from agricultural livestock uses of antibiotics (34). Comparison of phenotypic and genotypic Salmonella data in the NARMS isolate collection revealed a high correlation (99.0%) between clinical resistance and the presence of known resistance genes and mutations (18). In a recent study of >5,000 Salmonella genomes, MICs were predicted with high confidence (~95%) from the genomic data alone (22). This high predictability of resistance and perhaps MICs from whole genome sequence (WGS) data in the absence of standard susceptibility testing results greatly expands the scope of surveillance to include any genomic sequence available for analysis.
The widespread use of antibiotics in individuals and groups affects the microbiomes of humans, animals, and the environment. As a result, AMR may be viewed within the One Health framework (19); it has been described as “the quintessential One Health issue” (27). An integral part of this One Health approach is understanding the distribution and prevalence of AMR determinants within animal and nonanimal agricultural products. This information can be used to reduce negative impacts on animal, human, and environmental health and for AMR risk assessments of various food commodities to identify the most likely sources of strains of resistant microorganisms that cause zoonotic and foodborne illnesses. This approach permits development of more precise intervention strategies based on the contribution of various sources to the burden of resistance in human infectious diseases.
In this study, we used WGS data to predict resistance and analyzed AMR gene profiles in a large collection of Salmonella isolates. The genomic sequences were from numerous food sources and have been deposited in the National Center for Biotechnology Information (NCBI) database by many agencies around the world (e.g., the U.S. Food and Drug Administration, U.S. Department of Agriculture, and Public Health England). We investigated the top 20 most abundant Salmonella serovars found in foods for the strength of association between specific AMR profiles and food categories. Our primary objective was to elucidate the patterns of AMR at the genomic level in Salmonella isolates from animal and nonanimal foods to more fully understand the nature and magnitude of AMR within the food supply.
MATERIALS AND METHODS
Data source, AMR gene detection, and serovar prediction.
The Salmonella database within the NCBI Pathogen Detection project (https://www.ncbi.nlm.nih.gov/pathogens/) was used as the source of information for this study. The WGS data for each isolate are also accompanied by metadata such as isolation source and collection date. Seventy-five percent of the isolates analyzed here that had an isolation date were collected after 2010. The NCBI also provides AMR gene profiles for each isolate based on the results of the AMRFinder tool (9), which identifies acquired resistance genes in protein, nucleotide, and genomic data. Individual AMR genes identified by AMRFinder were manually sorted into antimicrobial classes also according to AMRFinder (Table 1). The version of NCBI’s Salmonella Pathogen Detection database analyzed here was PDG000000002.1138.
TABLE 1.
Antimicrobials associated with AMR genes found in food isolates
| Antimicrobial type | No. of unique AMR genes | Total no. of AMR genes |
|---|---|---|
| Aminoglycosides | 44 | 20,325 |
| β-Lactams | 23 | 3,219 |
| Bleomycin | 2 | 201 |
| Fosfomycin | 3 | 2,293 |
| Macrolides-lincosamides-streptomycin | 11 | 74 |
| NoGenes | 1 | 3,219 |
| Phenicols | 8 | 1,035 |
| Quaternary ammonium compounds | 4 | 1,635 |
| Quinolones | 11 | 153 |
| Rifampin | 3 | 19 |
| Streptothricin | 2 | 4 |
| Sulfonamides | 4 | 3,726 |
| Tetracycline | 10 | 5,922 |
| Trimethoprim | 13 | 460 |
| Total | 139 | 42,285 |
Distribution of AMR genes differ by origin of isolates and serovar (6, 13); therefore, we stratified resistance gene comparisons across these categories. Although serovar is a field within the metadata, it is not standardized or required. Therefore, we used the SeqSero tool (41) to predict serovar from the WGS data for each isolate in the database. Isolates with ambiguous serovar predictions from SeqSero were removed. Supplemental Table S1 includes the predicted serovar, isolation sources sorted into the food categories, AMR gene, AMR gene class, and BioSample accession for the top 20 serovars.
Categorization of food sources.
Unique isolation sources were sorted into 16 food categories based on a modified version of a hierarchical isolation source categorization scheme (26); we removed data for isolates from nonfood or unknown sources. The 16 categories were beef, chicken, dairy, eggs, fish, fruit, game, grains, nuts and seeds, other meat, other poultry, pork, shellfish, turkey, vegetables, and multi-ingredient foods (Fig. 1). No distinction was made between isolates from beef or dairy cattle; samples from cattle were grouped under “beef.” Isolates in NCBI’s Pathogen Detection database do not represent a random sample; many isolates were collected as part of risk-based sampling strategies to focus on certain food types because of a long history of public health concerns regarding salmonellosis (e.g., in chicken).
FIGURE 1.
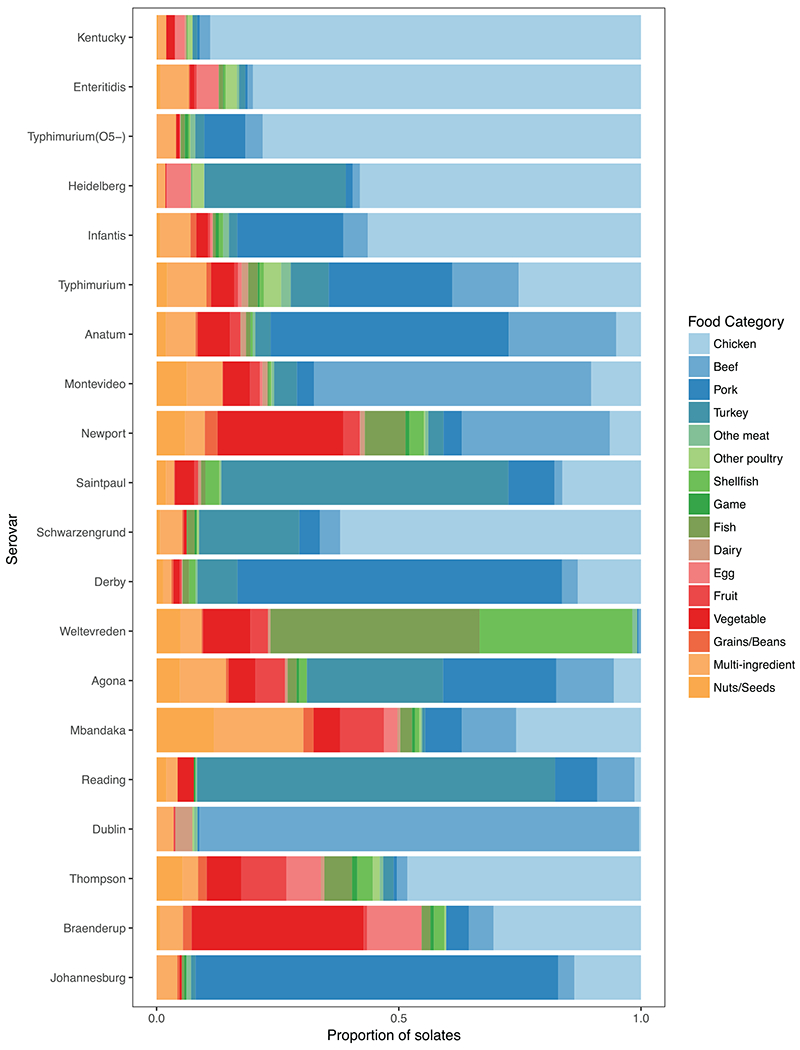
Proportion of Salmonella isolates of the top 20 most abundant serovars found in each of the food categories.
Data analysis.
A paired t test was performed in R v.3.3.3 (24) to determine whether there was a difference in the average number of AMR genes present in Salmonella isolates from nonanimal foods (fruit, vegetable, nuts and seeds, multi-ingredient, and grains and beans) compared with isolates from animal foods. We also analyzed the distribution of resistance genes by Salmonella serovar and food source for the top 20 most abundant serovars found within foods. We focused on the top 20 serovars because AMR genes are known to differ by serovar (6, 13). For each of the top 20 serovars, we also investigated how well AMR gene profiles of these isolates could be used to predict food category. We used a random forest approach within the R v.3.3.3 package randomForest v.4.6.12 (17). Within the randomForest analysis, the number of trees was 1,000, type was classification, and prediction accuracy was evaluated with type set to response using both out-of-bag error rates and via cross-validation (75% of data for training and 25% for testing); out-of-bag error rates are presented. Plots were generated with the R package ggplot2 v.2.2.1 (39).
RESULTS
Distribution of Salmonella isolates among food categories.
The 106,470 Salmonella isolates and associated metadata (e.g., AMR gene profiles and isolation source) represented 4,963 isolation sources and 552 predicted serovars. After removing isolates from nonfood or unknown sources, the data set was reduced to 25,647 isolates. To discern relationships between AMR and serovar, we focused on the top 20 most abundant serovars among isolates collected from a food.
Further subsetting the data to the top 20 serovars reduced the number of isolates by 37% to 16,095. Animal food categories (e.g., chicken, pork, turkey, and beef) had the greatest number of isolates in the full and the top 20 serovar data sets (Table 2), with the majority of isolates belonging to Salmonella serovars Kentucky, Enteritidis, and Typhimurium (O5–). Abundance of each of the top 20 serovars differed by food category, which is to be expected based on previous work (14); for example, the majority of Salmonella Saintpaul isolates were from turkey, the majority of Salmonella Enteritidis isolates were recovered from chicken, and Salmonella Newport isolates were found in nearly equal numbers in both beef and vegetables (Fig. 1 and Table 2).
TABLE 2.
Number of Salmonella isolates across various food categories for the 20 most abundant Salmonella serovarsa
| Salmonella serovar | No. of isolates |
Accuracyb | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken | Pork | Beef | Turkey | Multi-ingredient | Vegetable | Fish | Nuts, seeds | Egg | Shellfish | Fruit | Other poultry | Other meat | Grains, beans | Dairy | Game | Total | ||
| Kentucky | 2,142 | 12 | 53 | 23 | 40 | 42 | 4 | 8 | 47 | 3 | 3 | 24 | 0 | 0 | 8 | 0 | 2,409 | 0.92 |
| Enteritidis | 1,720 | 11 | 23 | 27 | 126 | 20 | 19 | 17 | 97 | 9 | 10 | 49 | 10 | 4 | 2 | 3 | 2,147 | 0.79 |
| Typhimurium (O5–) | 1,086 | 118 | 50 | 26 | 56 | 11 | 12 | 0 | 1 | 4 | 0 | 2 | 14 | 0 | 1 | 10 | 1,391 | 0.88 |
| Heidelberg | 735 | 18 | 19 | 370 | 19 | 2 | 2 | 4 | 63 | 1 | 1 | 30 | 1 | 0 | 1 | 1 | 1,267 | 0.74 |
| Infantis | 619 | 240 | 56 | 19 | 69 | 27 | 6 | 8 | 5 | 10 | 4 | 0 | 13 | 13 | 2 | 7 | 1,098 | 0.84 |
| Typhimurium | 267 | 270 | 145 | 83 | 86 | 51 | 21 | 23 | 8 | 9 | 8 | 38 | 21 | 10 | 14 | 4 | 1,058 | 0.60 |
| Anatum | 45 | 434 | 196 | 28 | 54 | 59 | 8 | 17 | 0 | 3 | 19 | 0 | 5 | 4 | 10 | 1 | 883 | 0.64 |
| Montevideo | 81 | 28 | 450 | 37 | 58 | 44 | 2 | 49 | 4 | 3 | 16 | 1 | 4 | 1 | 8 | 1 | 787 | 0.66 |
| Newport | 49 | 29 | 233 | 24 | 31 | 198 | 64 | 45 | 1 | 23 | 26 | 2 | 5 | 20 | 7 | 6 | 763 | 0.71 |
| Saintpaul | 108 | 63 | 11 | 394 | 11 | 27 | 6 | 13 | 0 | 19 | 5 | 1 | 2 | 1 | 4 | 0 | 665 | 0.76 |
| Schwarzengrund | 411 | 28 | 28 | 137 | 30 | 4 | 11 | 5 | 0 | 0 | 0 | 3 | 0 | 2 | 0 | 3 | 662 | 0.80 |
| Derby | 68 | 349 | 17 | 43 | 9 | 7 | 7 | 7 | 0 | 7 | 2 | 0 | 2 | 2 | 1 | 0 | 521 | 0.77 |
| Weltevreden | 0 | 1 | 2 | 0 | 17 | 38 | 167 | 19 | 0 | 122 | 14 | 0 | 4 | 1 | 2 | 0 | 387 | 0.47 |
| Agona | 21 | 88 | 45 | 106 | 36 | 21 | 7 | 18 | 0 | 6 | 23 | 0 | 0 | 2 | 2 | 2 | 377 | 0.57 |
| Mbandaka | 85 | 25 | 37 | 2 | 61 | 18 | 8 | 39 | 9 | 3 | 30 | 1 | 1 | 7 | 2 | 2 | 330 | 0.48 |
| Reading | 4 | 26 | 23 | 221 | 7 | 10 | 0 | 6 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 299 | 0.89 |
| Dublin | 1 | 1 | 258 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 10 | 0 | 284 | 0.97 |
| Thompson | 135 | 2 | 6 | 6 | 9 | 20 | 16 | 15 | 20 | 9 | 26 | 4 | 2 | 5 | 2 | 3 | 280 | 0.54 |
| Braenderup | 84 | 13 | 14 | 0 | 13 | 98 | 5 | 2 | 31 | 6 | 2 | 1 | 0 | 5 | 0 | 2 | 276 | 0.44 |
| Johannesburg | 29 | 158 | 7 | 2 | 9 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 211 | 0.80 |
| Total | 7,690 | 1,914 | 1,673 | 1,548 | 751 | 698 | 366 | 295 | 286 | 237 | 190 | 157 | 89 | 78 | 76 | 47 | 16,095 | 0.71c |
Rows are ordered by decreasing totals.
Accuracy refers to the classification success of a given isolate to food source based on its AMR gene profile within the random forest analysis.
Average accuracy of classification within the random forest analyses.
Distribution of AMR genes.
We found 279 unique AMR alleles across the entire data set, of which 139 were also found in isolates from foods; genes coding for sulfonamide, tetracycline, and aminoglycoside resistance were the most abundant (Table 1). The number of AMR genes within food isolates differed by serovar and food category (Table 2). Among the serovars, Salmonella Infantis appeared to have the broadest spectrum of resistance, with frequencies of >0.1 allele per isolate for eight antimicrobial classes. Other broad spectrum resistant Salmonella serovars were Agona, Derby, Dublin, Heidelberg, Newport, Saintpaul, Typhimurium, and Typhimurium (O5–), which all carried resistance genes found at >0.1 allele per isolate associated with five or more antimicrobial classes (Fig. 2). A significant portion of the isolates of these serovars came from terrestrial animal sources (Fig. 1). Among the least resistant Salmonella serovars were Enteritidis, Johannesburg, Montevideo, and Thompson; for all of these serovars, >40% of the isolates harbored no AMR genes (Fig. 2). Alleles conferring resistance to quinolones and the MLS (macrolides-lincosamides-streptomycin) class of antibiotics (considered highest priority within the critically important class of medically important antimicrobials (40)) were rarely present in any of the top 20 serovars (Fig. 2). However, alleles conferring resistance to another critically important class, aminoglycosides, were the most abundant in all serovars. Resistance to the β-lactams group, which includes alleles coding for both narrow and extended-spectrum β-lactamases, was present at variable levels across all serovars, with the highest levels observed in Salmonella Dublin (1.11 genes per isolate). The highest number of genes conferring phenicol resistance also were found in Salmonella Dublin (0.84 genes per isolate). Fosfomycin resistance was found at high frequencies within isolates of Salmonella serovars Agona, Derby, and Heidelberg (>0.95 genes per isolate; Fig. 2).
FIGURE 2.
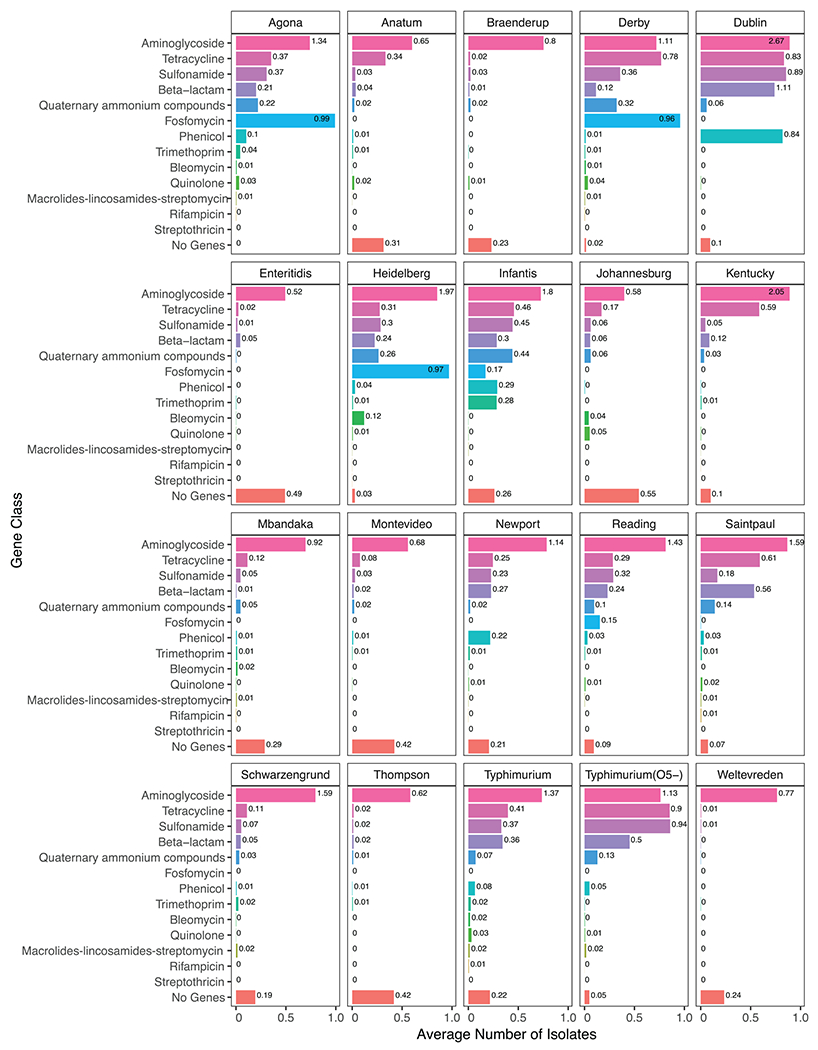
The proportion of isolates within each serovar whose genome included a given type of AMR gene or no AMR genes. Numbers at the ends of the bars represent the average number of AMR gene classes observed within isolates of that serovar (e.g., for Salmonella Agona, many isolates had multiple aminoglycoside AMR genes for an average of 1.34 aminoglycoside genes per isolate).
The average percentage of isolates from nonanimal foods (e.g., fruit, nuts and seeds, and vegetables) that harbored an AMR gene was 72% compared with 74% for isolates from animal foods (e.g., beef, chicken, turkey, and pork) (Fig. 3). This result was influenced in part by the presence of aminoglycoside resistance genes, which were found at roughly the same frequency (≥55% across all food types) in isolates from each food type (Fig. 3). However, when accounting for the number of AMR genes within each isolate, the average number of AMR genes per isolate was higher for isolates from animal foods than for those from nonanimal foods (t = 2.73, df = 12, P = 0.018). The chicken, beef, pork, and turkey categories had the highest percentages of isolates carrying genes conferring resistance to multiple antimicrobial classes (Fig. 3). These results were consistent even when considering all isolates, not just those from the top 20 serovars present in foods (Fig. S1). For example, regardless of serovar, within isolates from those four animal food categories, resistance genes for aminoglycosides, β-lactams, fosfomycin, sulfonamides, and tetracyclines were found in >10% of isolates. The abundance of resistance genes for fosfomycin, an antibiotic used to treat urinary tract infections in humans but not used in food animals in the United States, was highest in beef, pork, turkey, and eggs (Fig. 3). Salmonella serovars Agona, Derby, and Heidelberg had the highest percentage of isolates with fosfomycin resistance (Fig. 2). Although less abundant, resistance genes to the majority of antibiotic classes were also found in fish and nonanimal food products, including vegetables, fruits, and nuts and seeds (Fig. 3). Many of these isolates represented Salmonella serovars with a low prevalence of resistance genes (e.g., Weltevreden, Branderup, and Thompson; Fig. 2) regardless of isolate source.
FIGURE 3.
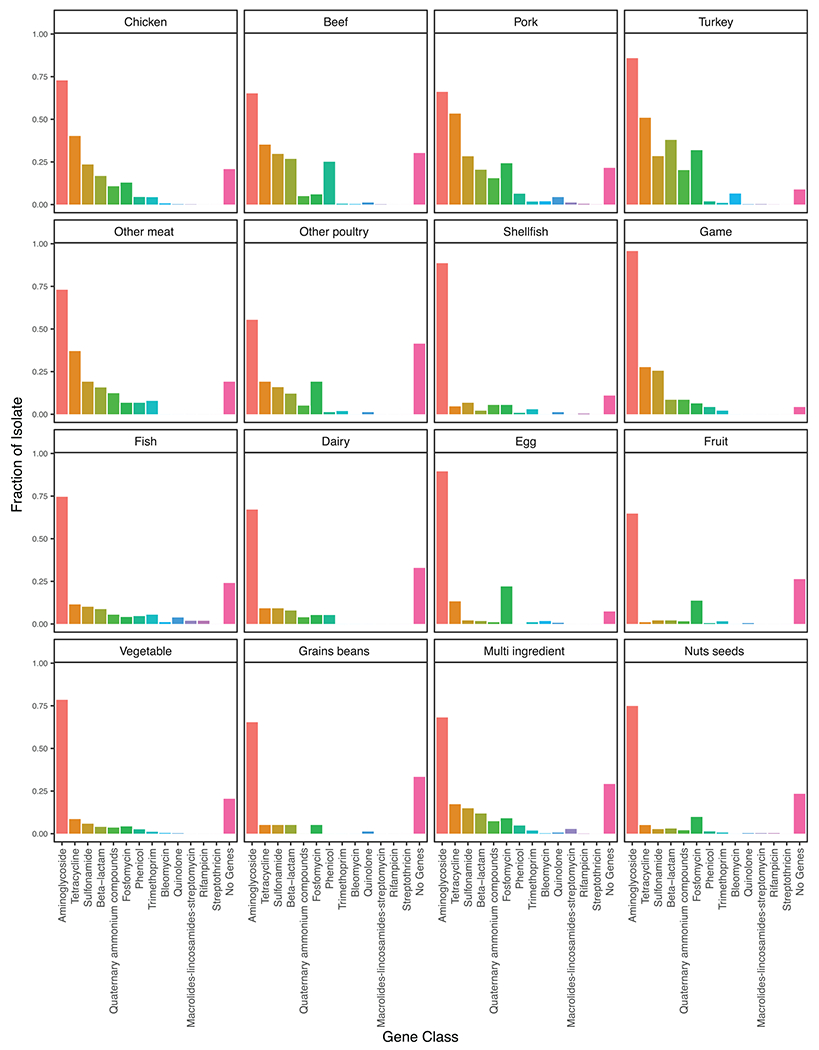
Fraction of isolates from the top 20 Salmonella serovars containing AMR gene classes separated by food category.
The hypothesis that AMR genes could be used to predict the food source of a specific Salmonella serovar was generally supported; however, the strength of that predictability varied depending on the serovar being considered (Table 2). For example, isolates of Salmonella serovars Dublin, Reading, Typhimurium (O5–), and Kentucky could be correctly classified to their known food source with >87% accuracy. In contrast Salmonella Agona isolates were classified to the correct food source with only 57% accuracy. The difference in predictability may be associated with the fact that serovars with the highest predictability were more prevalent in a single food category (e.g., Salmonella Dublin is predominantly found in beef; Table 2 and Fig. 1). In contrast, Salmonella Agona is more evenly distributed across several food (host) sources. However, Salmonella Derby is predominantly prevalent in pork but had a predictability of only 77%, suggesting that for certain food-serovar combinations an associated distinct signature of AMR genes may not be present.
To address the question of whether the AMR gene profile in isolates of particular Salmonella serovars was a better predictor of isolation source than was serovar alone, we also performed a random forest analysis using only serovar as a predictor of the food of origin. Under that scenario, we were able to correctly predict isolation source with 68% accuracy, which demonstrates that serovar is a fairly reliable predictor of food source. However, within each serovar, AMR gene profiles increase the accuracy of that prediction.
DISCUSSION
An understanding of the distribution of AMR genes across isolates from various food sources is important for developing effective methods for confronting the public health issues surrounding AMR. We determined the prevalence of resistance genes to various groups of antimicrobials across the top 20 Salmonella serovars isolated from foods. Because of the high prevalence of aminoglycoside resistance among Salmonella isolates from all food types, Salmonella isolates from terrestrial animals were not necessarily more likely to contain resistance genes than were Salmonella isolates from other food types (Fig. 3). However, when we considered the number of AMR genes per isolate we found that isolates from foods of animal origin, where various antimicrobials are more commonly used (16), had a higher prevalence of AMR genes than did isolates from nonanimal foods. This finding suggests that outbreaks of multidrug resistant (MDR) Salmonella would be more likely to involve animal-derived foods than nonanimal foods. This suggestion is concordant with the conclusions of Brown et al. (2), who found that strains causing MDR outbreaks were more likely to be linked to food from land animals (29%) than to plant-based foods (8%).
The use of antibiotics in animal agriculture is a longstanding practice, and many countries have implemented surveillance programs, such as NARMS, to track the development and spread of antibiotic resistance in animals, meat products, and humans. For major food animals (cattle, chickens, turkeys, and swine), our results mirror those of the NARMS data set, which comprises a significant portion of the isolates in our data set. Similar to the NARMS 2015 report (33), we found that the genes conferring resistance to critically important antimicrobials (macrolides and quinolones) were largely absent from food animal isolates. Likewise, nonanimal food isolates also bore few to none of these genes. We confirmed the NARMS finding that Salmonella Dublin isolates are highly resistant to multiple drug classes, including the broad β-lactams class (33). We also found high AMR among Salmonella Infantis isolates (Fig. 2), supporting growing evidence that a poultry-associated clonal MDR strain of Salmonella Infantis is currently in circulation around the globe (1, 31). We found a high prevalence of phenicol resistance genes in beef isolates, which supporting the hypothesis that antimicrobial use is the main driver of AMR. In animal agriculture, florfenicol is mainly used in beef and nonlactating dairy cattle, where it is approved for the treatment and control of bovine respiratory disease and foot rot (35). However, the finding of a high prevalence of fosfomycin resistance genes in poultry, swine, and egg isolates is ambiguous. Although fosfomycin is used mostly in South American countries (23) to treat infectious diseases of broilers and swine, it is not approved for use in the United States, where the majority of the swine, poultry, and egg isolates in our data set originated. However, the presence of fosA on a large MDR plasmid in Salmonella Infantis isolates from chicken indicates that these genes may be coselected through use of other unrelated antimicrobial agents (31, 35). The limited distribution of fosA may also be a consequence of biological phenomena rather than drug use. Other researchers have found that fosA genes are restricted to certain Salmonella serovars (25).
Isolates from game and other poultry and meat isolates had similar AMR gene classes as did isolates from other animal food sources, but the prevalence of those gene classes was reduced. Although these sources yielded Salmonella serovars with a high diversity of AMR gene classes, there was also a higher likelihood that these meats would be wild caught or raised organically. Our observation that dairy isolates contained fewer resistance genes supports those of other studies in which most Salmonella recovered from bulk tank milk were pansusceptible, although that finding was highly dependent on serovar distribution (29, 36). Egg isolates also had a lower prevalence of resistance genes than did chicken isolates. This difference could be due to differences in antibiotic use practices in hens laying eggs for human consumption, as has been suggested previously (1). A notable exception is the high frequency of fosfomycin resistance genes in isolates from eggs.
Because many Salmonella isolates from aquatic and nonanimal food sources also carried AMR genes, such foods should also be considered for possible AMR surveillance. For example, antimicrobials are used in the production of crops such as fruits, and AMR has been found in plant pathogens (38). Therefore, exposure to these antimicrobials in the environment by food species that carry Salmonella may also need to be considered (20, 38). Additional possible sources of AMR genes found in crops include wildlife, manure from treated animals used as fertilizer, and farm runoff into irrigation systems. In the United States, three antibiotic classes (tetracyclines, sulfonamides, and phenicols) are approved for use in fish (35), but unrestricted use of other antibiotic classes is permissible in some other countries (3, 11). In our study, fish and shellfish Salmonella isolates, which mostly belonged to serovar Weltevreden, were resistant to aminoglycosides, suggesting that the samples from which the isolates were recovered may not have been from the United States. Because many AMR genes (e.g., mcr-1 [colistin] and qnrB [quinolones]) are thought to have aquatic origins (4, 5), imported fish and shellfish certainly should be among those foods evaluated as potential AMR reservoirs.
Our ability to accurately predict the food source based on the type of AMR genes in recovered Salmonella isolates was highly dependent on the distribution of food sources in which each serovar was found and further illustrates the complexities of AMR bacteria within foods. For some Salmonella serovars, AMR genes could be used to predict with high accuracy the food source of an isolate, which suggests that different foods harbor isolates with distinct AMR profiles (Table 2). However, for other serovars prediction of food source was less certain; for example, there is little differentiation among foods in the prevalence of AMR gene classes in isolates of Salmonella Typhimurium and Salmonella Braenderup. Although the serovars with the highest prediction accuracy were those whose isolates were primarily from a single food category (e.g., 90% of isolates of Salmonella Kentucky were found in chicken; Table 2 and Fig. 1), that pattern does not hold for all serovars. For example, Salmonella Infantis has a predictability of 84%, but only 56% of Salmonella Infantis isolates came from chicken, which is the most frequently observed food source for this serovar.
Some limitations are evident in our study. The majority of the data were not obtained by random sampling but by risk-based sampling, where certain food sources were likely the focus of intense inspections and thus might be overrepresented. Some food categories and serovars also were selectively sequenced based on phenotypic results from traditional antimicrobial susceptibility tests, and our results may be biased toward higher levels of AMR. We also did not assess whether AMR changed over time or differed by geographic origin of the isolate. Industry- and regulation-driven changes in antibiotic administration impact AMR over time. For instance, NARMS data indicate a decrease in certain classes of AMR genes among Salmonella isolates from poultry, which could be the result of decreased use of third-generation cephalosporins in the poultry industry and greater emphasis on “no antibiotics ever” production (33). We also expect antibiotic use to vary within and among countries, so our findings may differ if we were to focus on only domestically produced foods. The degree to which AMR gene prevalence differs across food sources in relation to geography and time would be an appropriate topic for future research.
In this study, we characterized the prevalence of AMR genes among Salmonella isolates present in the NCBI Pathogen Detection database. We also assessed associations between specific AMR alleles and specific food categories. The results presented here may assist public health agencies develop illness prevention programs by contributing to a better understanding of the prevalence and distribution of antimicrobial resistant nontyphoidal Salmonella in contaminated food. Further utilization of this rich database by academia, industry food scientists, and other partners will help further define the increase in AMR among foodborne bacterial pathogens and inform efforts to address these issues and the clinical ramifications of increased AMR.
Supplementary Material
HIGHLIGHTS.
Aminoglycoside resistance was found in Salmonella isolates from nearly all food types.
The expected higher multiclass resistance was found among isolates from many animal foods.
AMR genes could be used to predict the isolate food type of origin with 71% accuracy.
ACKNOWLEDGMENTS
We thank the NCBI Pathogen Detection project for making the AMR predictions publicly available. We also thank the U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition and Center for Veterinary Medicine for support of this work.
Footnotes
SUPPLEMENTAL MATERIAL
Supplemental material associated with this article can be found online at: https://doi.org/10.4315/0362-028X.JFP-19-310.s1 and https://doi.org/10.4315/0362-028X.JFP-19-310.s2
REFERENCES
- 1.Brown AC, Chen JC, Watkins LKF, Campbell D, Folster JP, Tate H, Wasilenko J, Van Tubbergen C, and Friedman CR. 2018. CTX-M-65 extended-spectrum β-lactamase–producing Salmonella enterica serotype Infantis, United States. Emerg. Infect. Dis 24:2284–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown AC, Grass JE, Richardson LC, Nisler AL, Bicknese AS, and Gould LH. 2017. Antimicrobial resistance in Salmonella that caused foodborne disease outbreaks: United States, 2003–2012. Epidemiol. Infect 145:776–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabello FC 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol 8:1137–1144. [DOI] [PubMed] [Google Scholar]
- 4.Cabello FC, and Godfrey HP. 2018. Aquaculture, exaptation, and the origin of mcr-positive colistin resistance. Antimicrob. Agents Chemother 62:e01903–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabello FC, Tomova A, Ivanova L, and Godfrey HP. 2017. Aquaculture and mcr colistin resistance determinants. mBio 8:e01229–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll LM, Wiedmann M, den Bakker H, Siler J, Warchocki S, Kent D, Lyalina S, Davis M, Sischo W, Besser T, Warnick LD, and Pereira RV. 2017. Whole-genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington States reveals source and geographic associations. Appl. Environ. Microbiol 83:pii=e00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Available at: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. Accessed 17 June 2019.
- 8.Cohen ML, and Tauxe RV. 1986. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science 234:964–969. [DOI] [PubMed] [Google Scholar]
- 9.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu C-H, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, and Klimke W. 2019. Using the NCBI AMRFinder tool to determine antimicrobial resistance genotype-phenotype correlations within a collection of NARMS isolates. bioRxiv 550707. 10.1101/550707 [DOI] [Google Scholar]
- 10.Helms M, Simonson J, and Mølbak K. 2004. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J. Infect. Dis 190:1652–1654. [DOI] [PubMed] [Google Scholar]
- 11.Henriksson PJG, Rico A, Troell M, Klinger DH, Buschmann AH, Saksida S, Chadag MV, and Zhang W. 2018. Unpacking factors influencing antimicrobial use in global aquaculture and their implication for management: a review from a systems perspective. Sustain. Sci 13:1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes AH, Moore LS, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, Guerin PJ, and Piddock LJ. 2016. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387:176–187. [DOI] [PubMed] [Google Scholar]
- 13.Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, Olsen K, Bender JB, Wells S, Perez A, and Alvarez J. 2016. Serotypes and antimicrobial resistance in Salmonella enterica recovered from clinical samples from cattle and swine in Minnesota, 2006 to 2015. PLoS One 11:e0168016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson BR, Griffin PM, Cole D, Walsh KA, and Chai SJ. 2013. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerg. Infect. Dis 19:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krueger AL, Greene SA, Barzilay EJ, Henao O, Vugia D, Hanna S, Meyer S, Smith K, Pecic G, Hoefer D, and Griffin PM. 2014. Clinical outcomes of nalidixic acid, ceftriaxone, and multidrug-resistant nontyphoidal Salmonella infections compared with pansusceptible infections in FoodNet sites, 2006–2008. Foodborne Pathog. Dis 11:335–341. [DOI] [PubMed] [Google Scholar]
- 16.Landers TF, Cohen B, Wittum TE, and Larson EL. 2012. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 127:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liaw A, and Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
- 18.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, and Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother 60:5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwen SA, and Collignon PJ. 2018. Antimicrobial resistance: a One Health perspective. Microbiol. Spectr 6(2). 10.1128/microbiolspec.ARBA-0009-2017 [DOI] [PubMed] [Google Scholar]
- 20.McManus PS, Stockwell VO, Sundin GW, and Jones AL. 2002. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol 40:443–465. [DOI] [PubMed] [Google Scholar]
- 21.Medalla F, Gu W, Mahon BE, Judd M, Folster J, Griffin PM, and Hoekstra RM. 2016. Estimated incidence of antimicrobial drug–resistant nontyphoidal Salmonella infections, United States, 2004–2012. Emerg. Infect. Dis 23:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, and Davis JJ. 2018. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J. Clin. Microbiol 57(2):pii=e01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez DS, Tapia MO, and Soraci AL. 2014. Fosfomycin: uses and potentialities in veterinary medicine. Open Vet. J 4:26–43. [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- 25.Rehman MA, Yin X, Persaud-Lachhman MG, and Diarra MS. 2017. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob. Agents Chemother 61(8):pii=e00410–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson LC, Bazaco MC, Parker CC, Dewey-Mattia D, Golden N, Jones K, Klontz K, Travis C, Kufel JZ, and Cole D. 2017. An updated scheme for categorizing foods implicated in foodborne disease outbreaks: a tri-agency collaboration. Foodborne Pathog. Dis 14:701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson TP, Bu DP, Carrique-Mas J, Fevre EM, Gilbert M, Grace D, Hay SI, Jiwakanon J, Kakkar M, Kariuki S, Laxminarayan R, Lubroth J, Magnusson U, Thi Ngoc P, Van Boeckel TP, and Woolhouse ME. 2016. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg 110:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, and Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis 17:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnier JL, Karns JS, Lombard JE, Kopral CA, Haley BJ, Kim SW, and Van Kessel JAS. 2018. Prevalence of Salmonella enterica, Listeria monocytogenes, and pathogenic Escherichia coli in bulk tank milk and milk filters from US dairy operations in the National Animal Health Monitoring System Dairy 2014 study. J. Dairy Sci 101:1943–1956. [DOI] [PubMed] [Google Scholar]
- 30.Tadesse DA, Singh A, Zhao S, Bartholomew M, Womack N, Ayers S, Fields PI, and McDermott PF. 2016. Antimicrobial resistance in Salmonella in the United States from 1948 to 1995. Antimicrob. Agents Chemother 60:2567–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, Morales C, Tyson GH, Mukherjee S, Brown AC, Green A, Wilson W, Dessai U, Abbott J, Joseph L, Haro J, Ayers S, McDermott PF, and Zhao S. 2017. Comparative analysis of extended-spectrum–β-lactamase CTX-M-65–producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother 61(7). 10.1128/AAC.00488-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.U.S. Food and Drug Administration. 2012. Guidance for industry 209. The judicious use of medically important antimicrobial drugs in food-producing animals. Available at: https://www.fda.gov/downloads/AnimalVeterinary/GuidanceComplianceEnforcement/GuidanceforIndustry/UCM216936.pdf. Accessed 17 June 2019.
- 33.U.S. Food and Drug Administration. 2015. NARMS 2015 integrated report. U.S. Food and Drug Administration, Silver Spring, MD. [Google Scholar]
- 34.U.S. Food and Drug Administration. 2018. National Antimicrobial Resistance Monitoring System. Available at: https://www.fda.gov/animalveterinary/safetyhealth/antimicrobialresistance/nationalantimicrobialResistanceMonitoringSystem/default.htm. Accessed 17 June 2019.
- 35.U.S. Food and Drug Administration. 2019. FDA approved animal drug products. Available at: https://animaldrugsatfda.fda.gov/adafda/views/#/search. Accessed 17 June 2019.
- 36.Van Kessel JS, Sonnier J, Zhao S, and Karns JS. 2013. Antimicrobial resistance of Salmonella enterica isolates from bulk tank milk and milk filters in the United States. J. Food Prot 76:18–25. [DOI] [PubMed] [Google Scholar]
- 37.Varma JK, Mølbak K, Barrett TJ, Beebe JL, Jones TF, Rabatsky-Ehr T, Smith KE, Vugia DJ, Chang H-GH, and Angulo FJ. 2015. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J. Infect. Dis 191:554–561. [DOI] [PubMed] [Google Scholar]
- 38.Vidaver AK 2002.Uses of antimicrobials in plant agriculture. Clin. Infect. Dis 34(Suppl. 3):S107–S110. [DOI] [PubMed] [Google Scholar]
- 39.Wickham H 2009. ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- 40.World Health Organization. 2015. WHO model list of essential medicines. 19th list. World Health Organization, Geneva. [Google Scholar]
- 41.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, and Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J. Clin. Microbiol 53:1685–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


