Abstract
Understanding the pharmacogenomics of opioid metabolism and behavior is vital to therapeutic success, as mutations can dramatically alter therapeutic efficacy and addiction liability. We found robust, sex-dependent BALB/c substrain differences in oxycodone behaviors and whole brain concentration of oxycodone metabolites. BALB/cJ females showed robust state-dependent oxycodone reward learning as measured via conditioned place preference when compared with the closely related BALB/cByJ substrain. Accordingly, BALB/cJ females also showed a robust increase in brain concentration of the inactive metabolite noroxycodone and the active metabolite oxymorphone compared with BALB/cByJ mice. Oxymorphone is a highly potent, full agonist at the mu opioid receptor that could enhance drug-induced interoception and state-dependent oxycodone reward learning. Quantitative trait locus (QTL) mapping in a BALB/c F2 reduced complexity cross revealed one major QTL on chromosome 15 underlying brain oxymorphone concentration that explained 32% of the female variance. BALB/cJ and BALB/cByJ differ by fewer than 10,000 variants, which can greatly facilitate candidate gene/variant identification. Hippocampal and striatal cis-expression QTL (eQTL) and exon-level eQTL analysis identified Zhx2, a candidate gene coding for a transcriptional repressor with a private BALB/cJ retroviral insertion that reduces Zhx2 expression and sex-dependent dysregulation of cytochrome P450 enzymes. Whole brain proteomics corroborated the Zhx2 eQTL and identified upregulated CYP2D11 that could increase brain oxymorphone in BALB/cJ females. To summarize, Zhx2 is a highly promising candidate gene underlying brain oxycodone metabolite levels. Future studies will validate Zhx2 and its site of action using reciprocal gene editing and tissue-specific viral manipulations in BALB/c substrains.
SIGNIFICANCE STATEMENT
Our findings show that genetic variation can result in sex-specific alterations in whole brain concentration of a bioactive opioid metabolite after oxycodone administration, reinforcing the need for sex as a biological factor in pharmacogenomic studies. The cooccurrence of female-specific increased oxymorphone and state-dependent reward learning suggests that this minor yet potent and efficacious metabolite of oxycodone could increase opioid interoception and drug-cue associative learning of opioid reward, which has implications for cue-induced relapse of drug-seeking behavior and for precision pharmacogenetics.
Introduction
Opioids are analgesics prescribed for severe pain and have a high addiction liability. The heritability of opioid use disorder is estimated to be between ∼25% and 70% (Tsuang et al., 1996, 1998; Kendler et al., 2000, 2003; Goldman et al., 2005; Chan et al., 2011). Genome-wide association studies have identified candidate genes contributing to the heritability of opioid use disorder in humans (Crist et al., 2019), but we still know very little regarding the genetic contributions to opioid use disorder.
Genetic variation can influence the pharmacokinetic properties of xenobiotics such as opioids, and in turn, the therapeutic potency, efficacy, and addiction liability. Oxycodone (OXY) is a commonly prescribed semisynthetic opioid that is metabolized via phase I metabolism in humans into biologically inactive noroxycodone (NOR; ∼45% via CYP3A4) and the biologically active oxymorphone (OMOR; ∼19% via CP2D6) (Huddart et al., 2018). Oxymorphone exhibits greater potency and efficacy at activating the mu opioid receptor than oxycodone (Thompson et al., 2004; Lalovic et al., 2006). An unresolved question is whether oxymorphone contributes to individual differences in the analgesic and addictive properties of oxycodone (Candiotti et al., 2009; Zwisler et al., 2009, 2010; Lemberg et al., 2010; Andreassen et al., 2012; McMillan et al., 2019). Given the widespread use and misuse of oxycodone, the contribution of oxymorphone to oxycodone’s motivational and therapeutic properties is an important question.
The use of mice in unbiased, discovery-based genetic analysis of opioid traits provides complimentary advantages to human genetic studies by allowing control of genetic background, precise dosing, collection of endpoint tissues at appropriate time points, and the ability to validate causal variants through gene editing. Inbred mouse strains show large variation in opioid behaviors (Belknap et al., 1993, 1995; Bergeson et al., 2001; Kest et al., 2002; Wilson et al., 2003; Solecki et al., 2009; Bubier et al., 2020), and forward genetic studies have identified candidate genes influencing such traits as opioid-induced respiratory depression (Galnt11; Bubier et al., 2020); locomotor stimulation (Csnk1e; Bryant et al., 2009, 2012); antinociception (Kcnj9; Smith et al., 2008 and Car8; Levitt et al., 2017); physiologic withdrawal (Gnao1; Kest et al., 2009 and Cnih3; Nelson et al., 2016); tolerance; and hyperalgesia (Mpdz; Donaldson et al., 2016).
More recently, the use of near-isogenic substrains, distinguished primarily by variants that have arisen from genetic drift, have proven to be useful for genetic mapping of complex traits in “reduced complexity crosses” (Bryant et al., 2018, 2020). Large phenotypic variance combined with a drastic reduction in the density of genetic polymorphisms facilitates the identification of causal genes/variants that can be readily validated via gene editing (Mulligan et al., 2019). Reduced complexity crosses have been used between multiple mouse substrains (e.g., C57BL/6, DBA/2, BALB/c) to map the genetic basis of complex phenotypes such as psychostimulant sensitivity, binge-like eating, and thermal nociception (Kumar et al., 2013; Harkness et al., 2015; Shi et al., 2016; Kirkpatrick et al., 2017; Miner et al., 2017; Reed et al., 2018; Goldberg et al., 2021; Beierle et al., 2022).
Regarding this study, BALB/cJ (J) and BALB/cByJ (By) substrains were separated in 1935 after the F37 generation of inbreeding and have since been maintained as separate substrains. Fewer than 10,000 single nucleotide polymorphisms (SNPs) and indels are estimated to distinguish J from By, comprising a 500-fold reduction in genetic complexity compared with C57BL/6J versus most classic inbred strains (Keane et al., 2011; Yalcin et al., 2011). The J and By substrains differ in several interesting neurobehavioral phenotypes (Hilakivi and Lister, 1989; Perincheri et al., 2005; Turner et al., 2008; Velez et al., 2010; Sittig et al., 2014; Dam et al., 2019; Poyntz et al., 2019; Jager et al., 2020). Given this phenotypic variation, BALB/c substrains likely harbor readily identifiable causal genetic variants mediating important behavioral traits, and we have previously employed a reduced complexity cross in BALB/c substrains to map quantitative trait loci (QTL), RNA expression QTL (eQTL), and candidate genes underlying thermal nociception and brain weight (Beierle et al., 2022).
In this study, we observed robust sex-dependent BALB/c substrain differences in state-dependent expression of oxycodone reward learning in a conditioned place preference paradigm. We also observed robust, sex-dependent strain differences in brain concentrations of oxycodone (OXY) and its metabolites noroxycodone (NOR) and oxymorphone (OMOR). To identify candidate genes underlying brain OXY and metabolite concentration, we conducted metabolite QTL mapping and gene-level and exon-level eQTL mapping in a BALB/c reduced complexity cross as well as complementary whole brain proteomic and liver transcriptomic analysis. The combined results strongly implicate Zhx2 as a candidate gene underlying brain concentration of OMOR.
Materials and Methods
Mice
All experiments were conducted in accordance with the National Institutes of Health Guidelines for the Use of Laboratory Animals, 8th ed (National Research Council, 2011) and were approved by the Institutional Animal Care and Use Committee at Boston University School of Medicine. BALB/cJ (J) and BALB/cByJ (By) mice (7 weeks old) were purchased from The Jackson Laboratory (#000651, #001026; Bar Harbor, ME), housed four per cage, and allowed 6 days to acclimate before testing. BALB/cJ and BALB/cByJ mice were also bred in-house for select parental strain experiments and were always conducted with mice ordered from Jackson Laboratory to avoid the fixation of genetic drift in our breeding population. These mice were tested between the ages of 47 and 83 days old. BALB/cJ x BALB/cByJ-F1 and -F2 mice were bred in-house as described below and were tested between the ages of 56 and 134 days old. The age range was larger than our typical range of 50–100 days old because of the COVID-19 pandemic. All mice were maintained on Teklad 18% protein diet (#2018; Envigo, IN) and were tested from 10:30 to 13:00 in the light phase of a 12-hour light/dark cycle (lights on at 0630).
Drugs
Oxycodone hydrochloride was purchased from Sigma-Aldrich (St. Louis, MO), was dissolved in sterilized saline (0.9% NaCl), and was administered in an injection volume of 10 μl/g.
Conditioned Place Preference in BALB/c Substrains
BALB/cJ and BALB/cByJ substrains for parental strain experiments were trained and tested for conditioned place preference (CPP) using a 9-day protocol in a two-chamber apparatus (20 cm × 40 cm) as previously described (Kirkpatrick and Bryant, 2015). On day 1, mice were administered saline (SAL), placed into the left side, and provided open access to both sides to assess initial preference. On days 2 and 4, mice were confined to the right chamber and administered 1.25 mg/kg OXY (i.p.) or volume-matched SAL (10 μl/g, i.p.). The left and right sides were distinguished by different plastic floor texture inserts. On day 3 and day 5, mice were confined to the left chamber and administered SAL (i.p.). Drug-free preference was measured on day 8 after a SAL injection (i.p.), placement into the left side and providing open access to both sides for 30 minutes. State-dependent preference was measured on day 9, which was procedurally identical to day 8 except that OXY (1.25 mg/kg, i.p.) was injected into OXY-trained mice and SAL (i.p.) into SAL-trained mice. Preference was defined as the difference in time spent on the drug-paired side between day 1 and either day 8 or day 9. All testing started at 1100, all training and testing sessions lasted 30 minutes, and all testing apparatuses were housed in unlit sound attenuating chambers. Video was captured with overhead cameras (Swann Security Inc, Santa Fe Springs, CA) and graded using ANYMAZE tracking software (Wood Dale, IL). These experiments were conducted in nine cohorts and with separate distinct aims.
To establish behavioral differences between substrains, three cohorts totaling 88 mice were ordered from The Jackson Laboratory and used to assess several OXY related phenotypes over 7 weeks. The first 9 days of this testing was CPP as described above, and for brevity and specificity we have omitted subsequent testing. Three subsequent cohorts of inbred BALB/cJ and BALB/cByJ substrains were bred in-house using mice ordered from The Jackson Laboratory to ensure that observed differences in behaviors could be replicated in mice bred at Boston University, as all F2 mice would be generated in our facilities. This group constituted 77 mice from 47 to 83 days old at day 1 of testing. Finally, three more cohorts totaling 120 mice, ordered at 7 weeks old from The Jackson Laboratory, were tested for CPP, and tissues were subsequently taken for scRNA-seq (single-cell RNA sequencing (data not included), whole brain proteomics, and liver RNA-seq (RNA sequencing) and proteomic testing.
Samples Generated for Assessment of Whole Brain OXY, NOR, and OMOR Concentrations
Whole brain concentrations were assayed in BALB/cJ and BALB/cByJ substrains (20 J, 20 By) from two different cohorts with slightly different protocols. The first cohort consisted of 16 mice [seven J (four females, three males) and nine By (six females, three males)], bred in-house and 56–82 days old, that were tested using a 3-day protocol. On day 1 and day 2, mice were administered saline (10 μl/g, i.p.) and placed in a 20 cm × 40 cm open arena (the CPP chambers minus the floor textures and divider) to allow for habituation to injection and context. On day 3, mice were administered 1.25 mg/kg OXY (i.p.) and recorded for 30 minutes within the apparatus. Immediately after testing, mice were sacrificed by rapid decapitation, brains were dissected from the skull, olfactory bulbs trimmed, and brainstem trimmed at the pons-medulla boundary. Brains were then flash frozen in a bath of alcohol and dry ice, weighed, and stored at −80°C until processed. All testing started at 1100, all testing sessions lasted 30 minutes, and all testing apparatuses were housed in sound attenuating chambers. Video was captured with overhead cameras and graded using ANYMAZE tracking software.
The second cohort of mice comprised 25 OXY-treated mice [13 J (six females, seven males) and 12 By (six females, six males)] that were trained for state-dependent OXY-CPP (the same samples were used for the liver RNA-seq experiment that is described below). Immediately after day 9 of CPP (30 minutes after the third dose of OXY; 1.25 mg/kg, i.p.), brains were dissected as described above and stored at −80°C before analysis. Thus, the two cohorts used for metabolite analysis differed with regard to the number of OXY injections (one vs. three) and the size of the arena in which they were placed (20 cm × 40 cm or 20 cm × 20 cm).
Oxycodone and Metabolite Sample Preparation
Frozen and preweighed mouse brains were shipped from Boston University School of Medicine to the University of Utah Center for Human Toxicology on dry ice. The samples were stored at −30°C until processed. Briefly, the samples were removed from the freezer in groups of 10 and kept on ice. The brain sample was then transferred to a 15-ml polypropylene centrifuge tube and 4 ml of Type 1 water was added to the tube. The samples were then homogenized using the Sonics Vibra-Cell sonicator fitted with the microtip at 30% power. Homogenization was done in three 15-second cycles with approximately 2 seconds between the cycles, resulting in ∼4.5 ml of brain homogenate. After all 10 samples were sonicated, 1 ml of each homogenate was aliquoted into an individually marked 16 × 100 silanized glass tube. After these operations were completed, all test tubes were placed back into a freezer at −30°C until further processing and analysis.
Quantification of Oxycodone and Metabolites by Liquid Chromatography-Mass Spectrometry
OXY, NOR, and OMOR concentrations were measured using a validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) method (Fang et al., 2013). Deuterated OXY-d6, NOR-d3, and OMOR-d3 were added as internal standards to each homogenate sample and mixed. The samples were then extracted under basic conditions (100 μl of concentrated ammonium hydroxide) with 4 ml of freshly prepared n-butyl chloride:acetonitrile (4:1 v/v). Briefly, the samples were vortex mixed for ∼30 seconds and then centrifuged at 1200 g for 10 minutes. The upper organic layer of each sample was then transferred into a clean silanized glass tube and evaporated to dryness under a stream of filtered air at 40°C. The extracts were reconstituted in 75 μl of 0.1% formic acid in water and centrifuged to clarify, and the supernatants were transferred into autosampler vials. The LC-MS/MS system was an Agilent 1100 Series high-performance liquid chromatography (HPLC) coupled to a Thermo Scientific TSQ Quantum Access Triple Stage Quadruple mass spectrometer. A YMC-Pack ODS-AQ 5μm 2.0 × 100 mm column (Waters, Milford, MA) was used for analysis, and the mass spectrometer was run in positive electrospray mode. The concentrations of OXY, NOR, and OMOR were determined from the ratio of the peak area of each drug to the peak area of its internal standard and comparison with the calibration curve that was generated from the analysis of human plasma fortified with known concentrations of OXY, NOR, OMOR, and their internal standards. The lower limit of quantitation of the assay for all analytes was 0.2 ng/ml (dynamic range of 0.2–250 ng/ml).
F2 Breeding and Genotyping
BALB/c F2 mice were generated as described (Beierle et al., 2022). Testing began between 56 and 134 days old, a range increased beyond our normal testing window of 50–100 days because of the COVID-19 shutdown. Genotypes were determined using the miniMUGA microarray (Sigmon et al., 2020), and we refined these results to 304 that were polymorphic and reliable between our BALB/c substrains.
Whole Genome Sequencing and Genotype Calling of BALB/c Substrains
We used existing whole genome sequences for BALB/c substrains for variant identification as we previously reported (Beierle et al., 2022).
F2 Mice for Metabolite Phenotyping
F2 mice were phenotyped for CPP as described above. Three days after the completion of testing, mice were tested for baseline hot plate latencies as previously described (Beierle et al., 2022) and were subsequently injected with OXY so that the samples could be assayed for metabolism. At 12:00, after hot plate testing, saline control (OXY naïve) F2 mice who had previously received three SAL injections during the CPP experiment were administered an acute OXY injection (1.25 mg/kg i.p.) and placed in a clean mouse cage for 30 minutes, and then brains were collected and flash frozen as described above. Sample processing and analysis were conducted as previously described.
QTL Mapping of Brain OXY and Metabolite Concentrations
QTL mapping in the F2 cross was conducted as reported (Beierle et al., 2022). Briefly, poor quality markers and animals were excluded based on missing calls, non-Mendelian inheritance, and inappropriate crossover counts within a mouse. After quality control, there were 133 F2 mice (68 females, 65 males) and 218 polymorphic markers from the miniMUGA with which to conduct QTL mapping for whole brain oxycodone and metabolites. To determine the QTL model, a three-way ANOVA considering sex, cohort, and age on brain [OMOR] revealed a significant effect of sex but no other main effects or interactions. We therefore included sex as an additive covariate in the model. Calculating QTLs using Haley-Knott regression was conducted as described (Beierle et al., 2022). Given the sparsity of miniMUGA markers within our chromosome 15 QTL interval, we designed an additional marker using the TaqMan fluorescent genotyping assay (Thermo Fisher Scientific). We targeted rs264203947 (chr15:57104609 bp). The assay was conducted using a 10-μl reaction consisting of 5 ng of DNA and 0.5 μl of the TaqMan assay in 1× TaqMan Genotyping Master Mix (Thermo Fisher Scientific, 4371355) and was run using the following cycle in a 12K QUANT STUDIO 12K FLEX (Thermo Fisher Scientific): 1 × 95°C for 10 minutes, 40 × 95°C for 15 seconds, and 60°C for 1 minute. Power analysis of F2 whole brain [OMOR] was conducted using the package R/qtlDesign (Sen et al., 2007). Power analysis illustrating effect sizes versus sample sizes required was computed for day 9 preference in R/qtlDesign (Sen et al., 2007) using the effect size derived from the parental strain differences.
Gene-Level, Exon-Level Expression QTL Mapping in F2 Mice
A subset of 64 F2 mice were sacrificed for eQTL analysis after D9 CPP testing as described in Beierle et al., 2022. Briefly, we analyzed hippocampal and striatal RNA-seq counts using R/MatrixEQTL (Shabalin, 2012) using the ‘linear cross’ model and included Sex, RNA extraction Batch (RNA extraction), and Prior Treatment (SAL, OXY) as covariates. The same model was used to analyze intron and exon feature counts from the R/ASpli (Mancini et al., 2021) as described (Beierle et al., 2022).
Whole Brain Peptide Proteomics in BALB/c Parental Substrains
Twenty-four BALB/cJ (J) and 24 BALB/cByJ (By) mice (12 per sex per substrain) were trained and tested for drug-free and state-dependent CPP using 1.25 mg/kg OXY (i.p.) and published procedures (Kirkpatrick and Bryant, 2015) that are summarized above. Immediately after testing for state-dependent CPP on day 9 (30 minutes post-OXY), mice were sacrificed and 16 brains were harvested for proteomic analysis as described in Beierle et al. (2022). Briefly, eight BALB/cJ (two per sex per treatment) and eight BALB/cByJ (two per sex per treatment) whole brains were collected, homogenized, and fractionated and MS/MS spectra was acquired. Spectra were searched against the complete Swiss-Prot mouse proteome, MaxQuant was used for data normalization, and in-house scripts were used for statistical analysis. A complete list of differentially expressed proteins can be found in Beierle et al. (2022).
Parental Strain Liver Dissection, Extraction, RNA-Seq, and Differenitally Expressed Gene Analysis
Sixteen BALB/cJ (eight females, eight males) and 16 BALB/cByJ mice (eight females, eight males) were trained and tested for state-dependent OXY-CPP using 1.25 mg/kg OXY (i.p.) as described above and sacrificed immediately after day 9 testing (30 minutes post-OXY) by rapid decapitation. The left lobe of the liver was harvested and submerged in RNA Stabilization Solution RNAlater Solution (Thermo Fisher) and stored at 4°C. Briefly, RNA was extracted using TRIzol and spin columns, sequenced on an Illumina NovaSEQ6000, then FASTQ files demultiplexed, trimmed, and aligned to the mm10 mouse reference genome (Ensembl). We assessed the effect of substrain on differential gene expression in the sex-collapsed dataset while controlling for the effects of prior treatment. We also conducted separate analyses in females and males while controlling for the effect of treatment. Full details describing RNA extraction and data processing can be found in Beierle et al. (2022).
Liver Proteomics in BALB/c Parental Substrains
The same 16 SAL-treated mice used for liver RNA-seq were also used for liver proteomics—eight BALB/cJ (four males, four females) and eight BALB/cByJ mice (four males, four females). The left lateral lobes of the liver were homogenized and fractionated, and MS/MS spectra was acquired. Livers were processed and analyzed in the same manner as the whole brain samples above Notable, and one mouse (female By) was excluded from analysis because the tandem mass tag multiplexing channel corresponding to this sample was empty.
Results
BALB/cJ Mice Show Increased State-Dependent OXY-CPP Compared with BALB/cByJ Mice
The CPP protocol and apparatus are pictured in Fig. 1A. In examining drug-free OXY-CPP on day 8 using a three-way ANOVA, there was a significant main effect of treatment [F(1, 276) = 5.8; #P = 0.019], indicating a significant preference for OXY. However, there was no effect of substrain [F(1, 276) = 1.9; P = 0.17], sex [F(1, 276) = 1.92; P = 0.16], or any interactions (P = 0.12–0.74; Supplemental Fig. 1). One video recording of the 285 recordings was lost for day 8 and was therefore not in the present analysis.
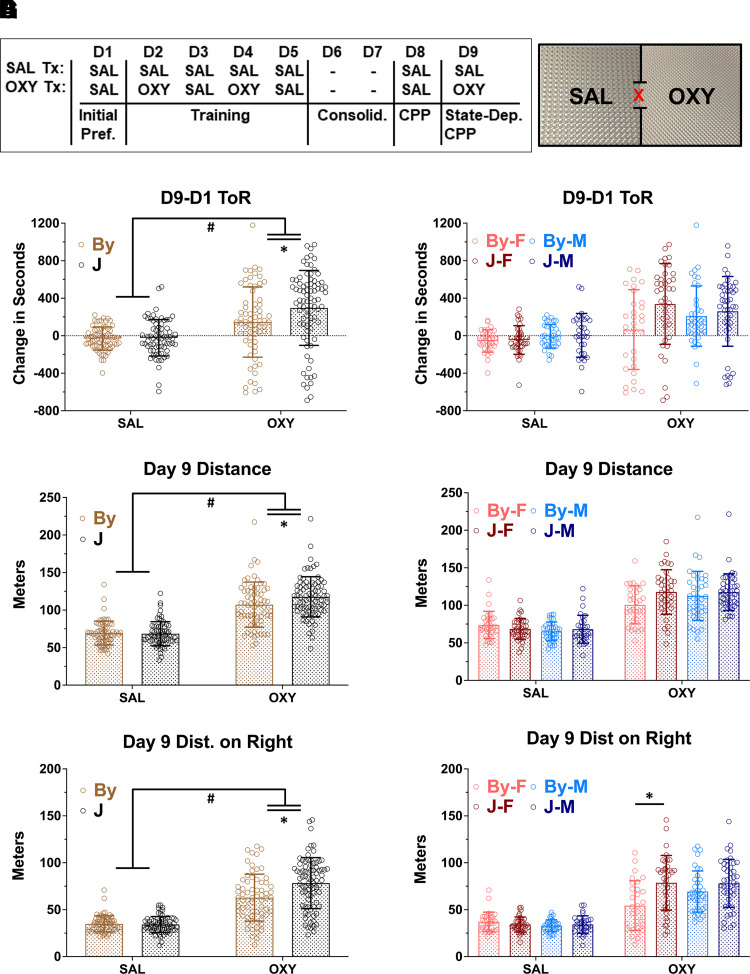
Fig. 1.
BALB/cJ (J) mice show enhanced state-dependent learning of OXY-CPP (1.25 mg/kg, i.p.) and concomitant locomotion compared with BALB/cByJ (By) mice. (A) Schematic of the CPP regimen and apparatus. (B) Results were analyzed using a three-way ANOVA considering substrain, treatment, and sex; error bars represent standard deviation. There was a treatment × substrain interaction (P = 0.048) that was explained by J mice showing greater OXY-CPP than By mice (Tukey’s post hoc *P = 0.015). (C) Female mice accounted for a majority of the substrain difference in OXY-CPP. (D) For total OXY-induced locomotor activity, there was a significant treatment × substrain interaction (P = 0.043) that was explained by J mice showing greater OXY-induced locomotion than By mice (Tukey’s post hoc *P = 0.037). (E) Females accounted for a majority of the substrain difference in total OXY-induced locomotor activity. (F) For OXY-induced locomotor activity specifically on the OXY-paired side (right side), there was a significant treatment × substrain interaction (P = 0.043) that was explained by J mice showing greater OXY-induced locomotor activity than By mice (Tukey’s post hoc *P = 0.037). (G) When sex was included in the ANOVA model, there was a significant treatment × substrain × sex interaction (P = 0.035) that was driven by increased OXY-induced locomotor activity in J females compared with By females (Tukey’s post hoc *P = 2.3e-5). Sample sizes from left to right (panel B) were 32, 37, 36, 32, 29, 38, 37, and 43.
# = main effect of treatment p<0.05, * = Tukey's Post Hoc T-test p < 0.05.
In examining state-dependent OXY-CPP on day 9 using a three-way ANOVA considering substrain, treatment, and sex, there was a main effect of treatment [F(1, 276) = 51.06; #P = 8.01e-12], indicating the presence of overall state-dependent CPP while under the influence of OXY (1.25 mg/kg, i.p.; Fig. 1B). There was also a main effect of substrain [F(1, 276) = 5.13; P = 0.024; Fig. 1B], and an interaction between treatment and substrain [F(1, 276) = 3.92; P = 0.048]. Tukey post hoc test revealed a significant increase in state-dependent OXY-CPP in the OXY J versus the OXY By group (*adjusted P = 0.015). Adding sex to the model did not reveal any significant main effect of sex [F(1, 276) = 1.03; P = 0.31] or interactions between sex and strain [F(1, 276) = 2.53; P = 0.11], sex and treatment [F(1, 276) = 0.17; P = 0.68] or three-way interaction [F(1, 276) = 2.37; P = 0.12]. Nevertheless, to facilitate later comparison with the metabolite results, we also present the same data stratified by sex (Fig. 1C). One video recording of the 285 recordings was lost for day 9 and is therefore not in the present analysis.
Analysis of OXY-induced locomotor activity (m) during state-dependent CPP assessment on day 9 revealed a main effect of treatment [F(1, 276) = 258.52; #P = 2e-16], a trending effect of strain [F(1, 276) = 3.2; P = 0.075] and an interaction between treatment and substrain [F(1, 276) = 4.78; P = 0.042]. Tukey’s post hoc test revealed a significant increase in J versus By OXY groups (*adjusted P = 0.037; Fig. 1D). When considering sex, we observed no main effect [F(1, 276) = 0.05; P = 0.82] nor a treatment × sex interaction [F(1, 276) = 3.08; P = 0.08] or treatment × substrain × sex interaction [F(1, 276) = 3.356; P = 0.068; Fig. 1E].
Analysis of day 9 locomotion on the right side revealed a main effect of treatment [F(1, 276) = 251.04, #P = 2e-16], a main effect of substrain [F(1, 276) = 10.75; P = 0.001] and interaction between treatment and substrain [F(1, 276) = 10.78; P = 6.3e-4; Fig. 1F]. Concerning sex, we observed no main effects [F(1, 276) = 1.12; P = 0.29] but did observe a significant treatment × substrain × sex interaction [F(1, 276) = 4.485; P = 0.035]. Tukey’s post hoc test revealed that the three-way interaction was driven by a significant increase in OXY-induced locomotor activity on the right side in OXY J females versus OXY By females (*adjusted P = 2.3e-5; Fig. 1G).
Increased Whole Brain [OXY], [NOR], and [OMOR] in BALB/c J versus BALB/cByJ Mice
Given the differences in testing protocol between the two cohorts used for parental strain whole brain concentrations, we analyzed the data separately and we observed the same overall pattern of results. For cohort 1, analysis of whole brain [OXY] revealed a significant effect of substrain [F(1, 12) = 8.3; P = 0.014] and interaction between substrain and sex [F(1, 12) = 5.68; P = 0.035; Fig. 2A] but no effect of sex [F(1, 12) = 0.16; P = 0.7]. Tukey’s post hoc test revealed this interaction to be mediated by a significant increase in whole brain [OXY] between the males (*P = 0.015) not present in females (P = 0.85; Fig. 2A). For whole brain [NOR], there was a significant effect of substrain [F(1, 12) = 7.88; P = 0.016], sex [F(1, 12) = 14.72; P = 0.002], and a trending interaction [F(1, 12) = 3.3; P = 0.094; Fig. 2B]. Tukey’s post hoc test revealed a significant increase in whole brain [NOR] in the J females compared with all other groups (*P = 0.016–0.003; Fig. 2B).
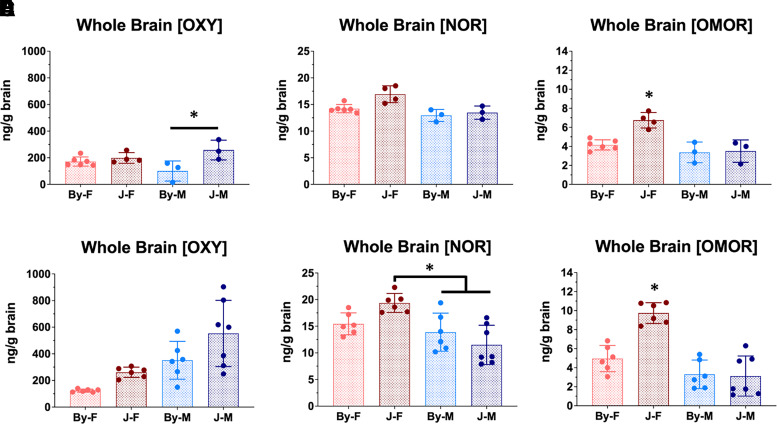
Fig. 2.
BALB/cJ mice show elevated whole brain concentrations of OXY, NOR, and OMOR compared with BALB/cByJ mice at 30 minutes postinjection of OXY (1.25 mg/kg, i.p.). (A–C) All analysis was conducted using a two-way ANOVA considering substrain and sex; error bars represent standard deviation. For the first cohort of mice, we observed an effect of substrain on brain concentrations of all compounds. We also detected significant main effects of sex and interactions of sex with substrain for [NOR] (P = 0.048) and [OMOR] (P = 0.005). In both cases, this effect was driven by increased drug concentrations in J females (Tukey’s post hoc *P = 0.022–9.9e-4). Sample sizes from left to right in panel A were6, 4, 3, and 3. (D) For the second cohort of mice in which there were slightly different procedures (see Materials and Methods), for whole brain [OXY] we observed an effect of substrain (P = 0.007) and sex (P = 3.1e-4) but no interaction. (E) For whole brain [NOR], there was no effect of substrain, but there was a significant effect of sex (P = 5.2e-4) and a sex × substrain interaction (P = 0.014) that was driven by a significant increase in J females vs. J males (Tukey’s post hoc *P = 5.2e-4). (F) Finally, for whole brain [OMOR], there were main effects of substrain (P = 0.004) and sex (P = 1.5e-6) and a substrain × sex interaction (P = 8.7e-4) that was driven by the increase in [OMOR] in J females compared with all three other groups (Tukey’s post hoc *P < 2.1e-4). Sample sizes from left to right (panel A) were 6, 6, 6, and 7.
* = Tukey's Post Hoc T-test p < 0.05.
Analysis of whole brain [OMOR] revealed a main effect of substrain [F(1, 12) = 11.48; P = 0.005], sex [F(1, 12) = 19.18; P = 7e-4], and an interaction [F(1, 12) = 7.86; P = 0.016] that was also driven by an increase in brain [OMOR] in J females compared with the three other groups (*P = 0.002–0.001; Fig. 2C).
For whole brain [OXY] in cohort 2, we observed a main effect of substrain [F(1, 21) = 8.9; P = 0.007] and sex [F(1, 21) = 18.6; P = 3.1e-4] but no interaction [F(1, 21) = 0.28; P = 0.6, Fig. 2D]. Analysis of whole brain [NOR] revealed a main effect of sex [F(1, 21) = 16.8; P = 5.2e-4], and a substrain × sex interaction [F(1, 21) = 7.13; P = 0.014] but no main effect of substrain [F(1, 21) = 0.151; P = 0.7]. Tukey’s post hoc test revealed that the substrain × sex interaction was driven by an increase in [NOR] in J females compared with J males (*P = 0.02; Fig. 2E) and By males (*P = 5.1e-4) but not By females (P = 0.13). Finally, analysis of whole brain [OMOR] revealed a significant effect of substrain [F(1, 21) = 10.24; P = 0.01, sex [F(1, 21) = 43.7; P = 1.5e-6], and a substrain × sex interaction [F(1, 21) = 15.04; P = 8.7e-4] that was driven by an increase in J females compared with the three other groups (*P = 2e-4–1.4e-5; Fig. 2F).
QTL Model Selection for Whole Brain [OMOR] in F2 Mice
A genetic map of the 219 markers that passed quality control measures is shown in Supplemental Fig. 2A. Details concerning the TaqMan fluorescent genotyping assay are provided in Supplemental Table 1. To derive an appropriate model for QTL mapping, we considered the effects of sex, cohort, and age on whole brain [OMOR] via ANOVA in our F2 mice and observed a main effect of sex [F(1, 125) = 17.49; P = 5.4e-5] but no effect of cohort [F(1, 125) = 0.62; P = 0.43], age [F(1, 125)= 0.064, P = 0.8], or any interactions (p values > 0.49; Supplemental Fig. 2B). Therefore, we included sex as an additive covariate in our QTL analysis.
A Major QTL on Chromosome 15 Underlying Whole Brain [OMOR]
We did not identify any genome-wide significant QTLs underlying BALB/c substrain differences in state-dependent OXY-CPP when considering treatment as an interactive covariate and sex as an additive covariate (Supplemental Fig. 3, A and B). This null result supports previous observations from our group and others that CPP is not a highly heritable trait (Cunningham et al., 1991; Philip et al., 2010; Bryant et al., 2014; Kirkpatrick et al., 2017; Gonzales et al., 2018; Ruan et al., 2020). Nevertheless, CPP is clearly a useful phenotype as it led us to measure a more heritable trait, brain concentration of OXY and its metabolites, and to ultimately identify a plausible candidate gene that could influence both traits.
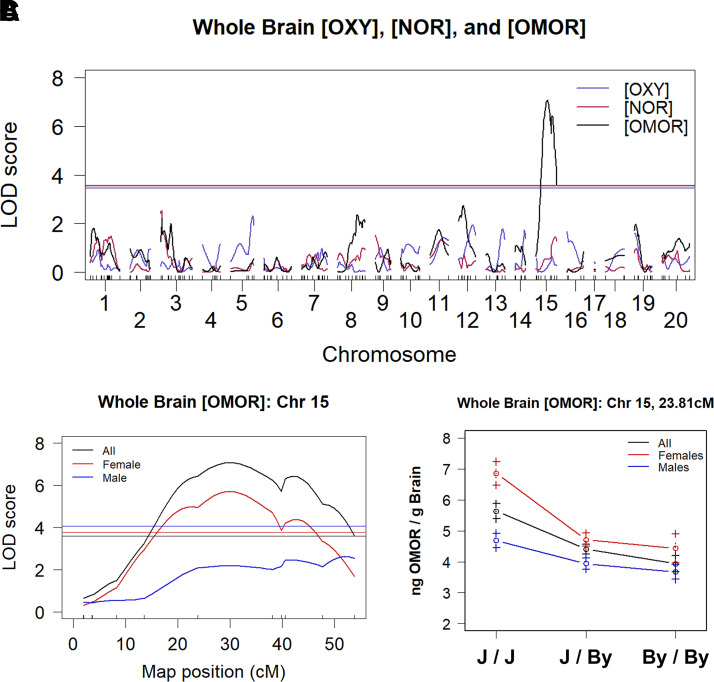
We identified a genome-wide significant QTL on chromosome 15 [Logarithm of the Odds (LOD) = 7.07; P < 0.001] for whole brain [OMOR] that peaked at 30 centimorgans (cM) [67 megabases (67 Mb)] and explained 19% of the variance in all mice and 32% in females (Fig. 3, A and B; Table 1). Notably, we did not observe significant QTLs for whole brain [OXY] or [NOR]. At the peak associated marker for whole brain [OMOR] (rs264203947), the effect of genotype and sex recapitulated the parental substrain difference, in which females homozygous for the J allele showed an increase in whole brain [OMOR] (Fig. 3C). Table 1 summarizes the QTL mapping results. There are 55 polymorphic protein coding genes within the Bayes confidence interval (Supplemental Table 2).
Fig. 3.
A major QTL on chromosome 15 underlies variation in whole brain OMOR concentration after OXY administration (1.25 mg/kg, i.p.). (A) In 133 F2 mice and considering sex and additive covariate, a single genome-wide significant QTL peaked on medial chromosome 15 (LOD = 7.07; P < 0.001) that explained 19% of the sex-combined phenotypic variance. (B) Chromosome 15 QTL plot shows a peak association at 30 cM (68 Mb). (C) At the peak-associated marker, F2 mice sorted by BALB/c genotype precisely recapitulated the result from the parental substrains (J > By) of increased whole brain [OMOR] in females with the homozygous J/J genotype vs. the homozygous By/By genotype.
TABLE 1.
Summary of the chromosome (chr) 15 QTL for whole brain [OMOR]
Summary statistics are provided for the sex-combined and females-only analysis.
| N | Mice | Phenotype | Model | Chr | Peak (cM) | Peak (Mb) | LOD | P Value | Bayesian Interval (cM) | Bayesian Interval (Mb) | 1.5 LOD Drop (cM) | 1.5 LOD Drop (Mb) | % Variance Explained |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 133 | All | Brain [OMOR] | Sex Additive | 15 | 29.95 | 68.64 | 7.07 | <0.001 | 20.95–43.95 | 53.79–88.49 | 18.95–46.95 | 50.39–95.94 | 19.10 |
| 68 | Female | Brain [OMOR] | No Covariates | 15 | 29.95 | 68.64 | 5.71 | <0.001 | 16.95–43.95 | 44.63–88.49 | 12.95–47.8 | 31.48–93.65 | 32.05 |
Power analysis using qtlDesign (Sen et al., 2007) indicated that using whole brain [OMOR] from 133 F2 mice, we had 80% power to detect additively inherited QTLs that explained 14% of phenotypic variance (Supplemental Fig. 4A). Analysis of D9 preference revealed that we were underpowered to map our observed P0 differences in preference, needing to test 311 OXY-treated F2 mice compared with our 200 (Supplemental Fig. 4B). Our current sample size could only successfully map QTLs explaining ∼12% of the observed variance (Supplemental Fig. 4C).
Transcript-Level eQTL and Exon-Level eQTL Analysis of Chromosome 15 Identifies Zhx2 As a Candidate Gene Underlying Whole Brain [OMOR]
We obtained an average of 33.9 million paired-end reads across all 128 RNA-seq samples from the 64 F2 mice (16 per sex per treatment). Table 3 shows a list of transcripts with significant eQTLs on chromosome 15 within the striatum and hippocampus [false discovery rate (FDR) < 0.05]. In considering both striatal and hippocampal tissue, Zhx2 was the only transcript containing an eQTL (FDR < 0.05) that peaked with the same marker (24 cM) that was identified for whole brain [OMOR] (striatum FDR = 1.4e-20; hippocampus FDR = 7.2e-8). A complete list of eQTL genes with unadjusted P values can be found in Beierle et al. (2022). QTL analysis at the exon/intron level identified 39 differentially used, non-binned features within the striatum and 55 in the hippocampus associated with the peak marker for whole brain [OMOR] (FDR < 0.05). In both tissues Zhx2 features comprised the most significant cis-exon-level eQTL, and in both tissues introns 1 and 3 and exons 3 and 4 of Zhx2 were differentially regulated as a function of genotype (Table 3). Interestingly, although the use of short-read DNA sequencing failed to identify any variants assigned to Zhx2, there is a well documented structural variant within intron 1 of this gene that decreases Zhx2 expression (Perincheri et al., 2005). Indeed, closer examination of short-read alignment showed a lack of reads aligning to the start site of the structural variant in BALB/cJ (Supplemental Fig. 5), thus corroborating the presence of this historical structural variant that is private to BALB/cJ. Raw data can be found at the NCBI Gene Expression Omnibus ascension GSE196352 (striatum) and GSE196334 (hippocampus).
TABLE 3.
Exon-level cis-eQTLs on chromosome 15 for hippocampus and striatum
| Region | SNP | Name | Feature | Start (bp) | Length | P Value | FDR |
|---|---|---|---|---|---|---|---|
| Striatum | rs264203947 | Zhx2 | Exon 3 | 57821111 | 2641 | 3.00E-26 | 3.44E-20 |
| rs264203947 | Zhx2 | Intron 1 | 57695787 | 66859 | 2.73E-24 | 3.10E-18 | |
| rs264203947 | Zhx2 | Exon 4 | 57838591 | 1242 | 1.00E-22 | 9.07E-17 | |
| rs264203947 | Zhx2 | Intron 3 | 57823752 | 14839 | 7.76E-16 | 3.53E-10 | |
| Hippocampus | rs264203947 | Zhx2 | Intron 1 | 57695787 | 66859 | 1.71E-14 | 2.92E-08 |
| rs264203947 | Zhx2 | Exon 3 | 57821111 | 2641 | 3.95E-14 | 4.08E-08 | |
| rs264203947 | Zhx2 | Exon 4 | 57838591 | 1242 | 2.04E-11 | 8.70E-06 | |
| rs264203947 | Zhx2 | Intron 3 | 57823752 | 14839 | 3.10E-08 | 0.004925 |
bp, base pair; SNP, single nucleotide polymorphism.
Exon-level cis-eQTLs for Zhx2 on (FDR < 0.05) were conducted in 64 F2 mice (16 per sex per treatment), and analysis included sex, RNA extraction batch, and treatment as covariates.
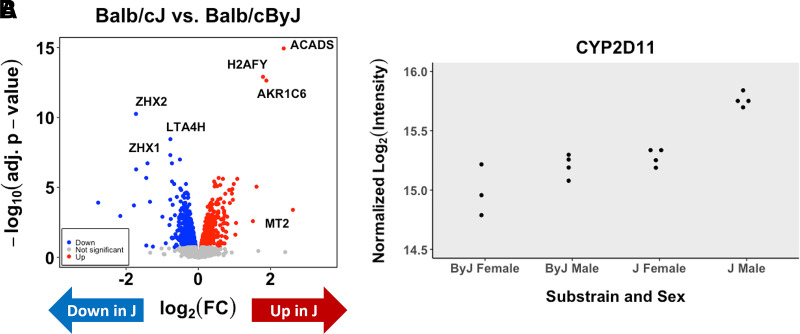
Proteomic Analysis of Whole Brain Tissue from the BALB/c Parental Substrains Corroborates ZHX2 As a Functional Candidate Protein Underlying Whole Brain [OMOR]
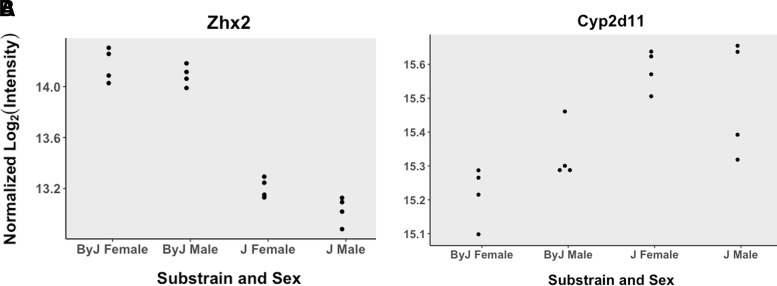
Analysis of parental substrain whole brain homogenate protein levels through mass spectrometry revealed 386 differentially expressed proteins with an adjusted P value < 0.05 and 1377 genes with an unadjusted P value < 0.05. Of the top 386 proteins (adjusted P < 0.05), only six were contained within the Bayes confidence interval of the chromosome 15 QTL for whole brain [OMOR] (MRPL13, ZHX2, LY6A, BOP1, GCAT, and CYP2D11). Only one of these genes, MRPL13, contains an annotated mutation based on short read sequencing data. Individual expression plots based on substrain and sex are shown for ZHX2 (Fig. 4A) and CYP2D11 (Fig. 4B). Interestingly, when analyzed separately by sex, we observe that female mice show a significant difference and are driving the observed difference [log fold change (logFC) = 0.37; logOdds = 0.97] as opposed to males who did not reach significance (logFC = 0.17; logOdds = −4.37). A complete data set can be found in Beierle et al. (2022).
Fig. 4.
Whole brain proteomics of BALB/c substrains corroborates the Zhx2 eQTL and identifies CYP2D11 as an upregulated protein in J mice. (A) Data generated from eight BALB/cJ (two per sex per treatment) and eight BALB/cByJ (two per sex per treatment) mice. Data from normalized intensity for Zhx2 in J and By mice (adjusted P = 2.79e-9). (C) Normalized intensity for CYP2D11 in J and By mice (adjusted P = 0.0066).
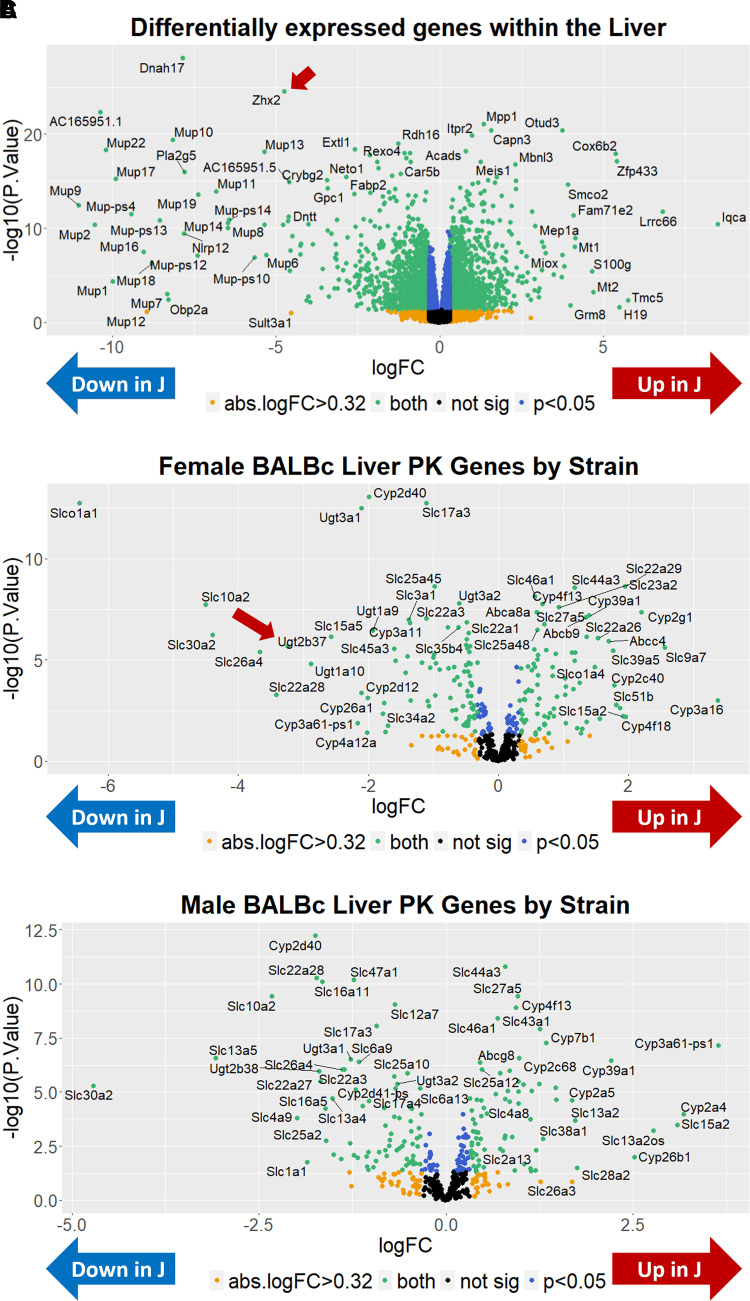
Transcriptome Analysis of Liver between BALB/c Substrains
Given the identification of a QTL and candidate gene underlying OXY metabolite concentration in the brain, to gain insight into the downstream mechanism, in the absence of having harvested F2 livers, we turned to analysis of the liver in the parental BALB/c substrains, which is the primary site of OXY metabolite production. The results of differential gene expression across substrains are shown as a volcano plot in Fig. 5A. In considering genes with an absolute log2 fold change greater than 0.32 (absolute fold change > 1.25), we observed a total of 2465 differentially expressed transcripts with an adjusted P < 0.05 (Supplemental Table 3). Sixty-one of these transcripts were located within the Bayes QTL interval for brain [OMOR] levels. These genes include Zhx2 but also four cytochrome P450 (Cyp) genes, including Cyp2d12, Cyp2d37-ps, Cyp2d40, and CYP2d41-ps, which were all downregulated in J mice. Because both the increase in state-dependent OXY-CPP and increase in oxymorphone brain concentration were driven by J females, we also investigated potential sex-specific differential gene expression within the liver (Supplemental Tables 4 and 5). To facilitate legibility, we also plotted female- and male-specific alterations in pharmacokinetics (PK)-related gene expression (defined for this purpose as any Cyp, UDP-glucuronosyltransferase (Ugt), ATP-binding cassette (Abc), or solute carrier (Slc) gene and any gene found within the Gene Ontology (GO) terms xenobiotic metabolic process or xenobiotic transport (GO:0006805, 0042908; Fig. 5, B and C). Sex-stratified analysis identified Ugt2b37 as a robustly differentially expressed transcript in females (logFC = −3.24; adjusted P = 6.96e-05) but not in males (logFC = −0.68; adjusted P = 0.16). Analysis of Ugt2b37 transcript levels revealed a significant main effect of substrain [F(1, 28) = 20.51; P = 1e-4], sex [F(1, 28) = 15.82; P = 4.4e-4], and an interaction [F(1, 28) = 18.07; P = 2.1e-4]. Tukey’s post hoc test revealed that the interaction was driven by a significant increase in Ugt2b37 in By females versus all other groups (adjusted P = 1.71e-5–6.0e-6), raising the interesting hypothesis that a decrease in Ugt2b37 expression in J females could increase plasma [OMOR] and, in turn, increase brain [OMOR]. Raw data can be found at the NCBI Gene Expression Omnibus ascension GSE198375.
Fig. 5.
Transcriptome analysis of the liver in BALB/c substrains. (A) Sixteen BALB/cJ (four per sex per treatment) and 16 BALB/cByJ mice (four per sex per treatment) data were analyzed blocking the effects of sex and treatment. Sex-combined: a volcano plot shows the distribution of differentially expressed genes in liver homogenate of all mice (J-By expression). Zhx2 is denoted with an arrow. (B) Females: Using only females, data were analyzed blocking the effect of treatment. A volcano plot shows the distribution of differentially expressed PK genes in liver homogenate. For this figure, PK genes include Cyp, Ugt, Abc, and Slc genes. Note the robust downregulation of Ugtb37 in J females, denoted with an arrow. (C) Males: Using only males, data were analyzed blocking the effect of treatment. A volcano plot shows the distribution of differentially expressed PK genes in liver homogenate of male mice.
Proteomic Analysis of Liver between BALB/c Substrains
Analysis of gene expression differences in the liver homogenate protein levels between parental substrains through mass spectrometry revealed 693 differentially expressed proteins with an adjusted P value < 0.05 and 1667 proteins with an unadjusted P value < 0.05 (Fig. 6A). Of the top 693 proteins (adjusted P < 0.05), only 14 were contained within the Bayes confidence interval of the chromosome 15 QTL for whole brain [OMOR] (ZHX2, ZHX1, GM29394, SQLE, NDRG1, ST3GAL1, KHDRBS3, PLEC, LGALS1, PLA2G6, CYP2D11, SERHL, ARFGAP3, and FBLN1). Only one of these genes, KHDRBS3, contains an annotated mutation based on short-read sequencing data. Notably, we do observe higher expression of CYP2D11 in J mice (Fig. 6B). A complete list of differentially expressed liver proteins can be found in Supplemental Table 6.
Fig. 6.
Proteomic analysis of the liver in BALB/c substrains. (A) Eight BALB/cJ (four females, four males) and seven BALB/cByJ saline-treated mice (three females, four males) were analyzed. A volcano plot shows the distribution of differentially expressed proteins in liver homogenate of all mice (J-By expression). (B) Overlap between differentially expressed RNA transcripts and proteins in the liver. (C) Normalized intensity of CYP2D11 across strain and sex.
Discussion
We identified robust state-dependent CPP in J females and an absence in By females, whereas all male mice showed a comparable phenotype (Fig. 1). A similar trend was observed for OXY-induced locomotion during state-dependent CPP (Fig. 1, E–G). Because this substrain difference was only observed after OXY administration, we considered whether brain concentration of OXY and/or its metabolites could explain the state-dependent recall of OXY-CPP. We found a robust 1.5- to 2-fold increase in brain [OMOR] in J females at 30 minutes post-OXY in two independent experiments (Fig. 2, C and F), implicating increased brain [OMOR] as a potential mechanism underlying increased state-dependent OXY-CPP.
Although we have not yet made the link between increased brain [OMOR] and increased OXY-induced behavior in J females, it is worthy of discussion. Because OMOR has higher affinity and efficacy at the mu opioid receptor compared with OXY (Thompson et al., 2004; Peckham and Traynor, 2006; Volpe et al., 2011), increased whole brain [OMOR] could facilitate and enhance drug-induced interoception. We hypothesize that increased brain [OMOR] enhances the speed and intensity of the opioid interoceptive effect, which serves as a more effective unconditioned stimulus, leads to stronger conditioned stimulus–unconditioned stimulus association with the drug-paired context, and increases state-dependent recall of OXY-CPP.
QTL mapping identified a single genome–wide QTL on chromosome 15 for whole brain [OMOR] (Fig. 3, A and B). Despite parental substrain differences, we did not detect significant QTLs for brain [OXY] or [NOR]. The large genetic effect of the chromosome 15 QTL (Fig. 3C) mirrors parental substrain data (Fig. 2D) and corresponds to a larger portion of the observed female variance compared with the sex-combined analysis (32% vs. 19%; Table 1). eQTL analysis (Table 2) combined with proteomics (Fig. 4) revealed Zhx2 as a candidate gene. Zhx2 was the only transcript with a cis-eQTL that peaked in the same location as the chromosome 15 QTL for brain [OMOR]. Exon-level eQTL analysis revealed that the only coding exon (3) within Zhx2 was differentially used between genotypes at this locus, as were three other introns/noncoding exons (Table 3). Both whole brain and liver proteomics showed ZHX2 downregulation in J mice, supporting prior studies (Perincheri et al., 2005). Short-read sequencing did not detect variants within or near Zhx2; however, there is a known 6.2-kilobase (kb) mouse endogenous retroviral element (MERV) within Zhx2 (Perincheri et al., 2005). Insertion of this MERV leads to inappropriate splicing of Zhx2 mRNA and decreased protein expression (Perincheri et al., 2008). Lack of alignment in our short-read whole genome sequencing (WGS) data corroborates the private MERV insertion in the BALB/cJ substrain (Supplemental Fig. 5). There is also a cluster of Cyp2d genes at the distal end of the QTL interval for brain [OMOR], including Cyp2d11, Cyp2d10, Cypd29, Cyp2d12, Cyp2d34, Cyp2d26, and a polymorphic Cyp2d40 gene and pseudogene Cyp2d37-ps that is located 15 Mb distally from the QTL peak (Table 1). We did not identify eQTLs for these transcripts, largely due to low levels of Cyp2d transcripts in the eQTL datasets. Given the linkage disequilibrium between Zhx2 and the Cyp2d locus, some degree of linkage between the Zhx2 marker and Cyp2d11 expression would have been expected if the transcript had been detectable.
TABLE 2.
Transcript-level cis-eQTLs on chromosome 15 for striatum and hippocampus
| Region | Gene | Chr | Location (Mb) | Location (cM) | P Value | FDR |
|---|---|---|---|---|---|---|
| Striatum | Zhx2 | chr15 | 4.31 | 1.95 | 3.1E-07 | 5.7E-03 |
| Zhx2 | chr15 | 4.99 | 1.983 | 3.1E-07 | 5.7E-03 | |
| Zhx2 | chr15 | 7.83 | 3.603 | 2.6E-07 | 4.8E-03 | |
| Zhx2 | chr15 | 8.12 | 3.702 | 2.6E-07 | 4.8E-03 | |
| Zhx2 | chr15 | 17.35 | 7.918 | 1.7E-07 | 3.4E-03 | |
| Zhx2 | chr15 | 21.04 | 8.263 | 6.5E-08 | 1.3E-03 | |
| Zhx2 | chr15 | 32.32 | 13.546 | 2.5E-13 | 7.3E-09 | |
| Zhx2 | chr15 | 57.11 | 23.8 | 6.6E-26 | 1.4E-20 | |
| Zhx2 | chr15 | 81.39 | 38.094 | 1.9E-09 | 4.7E-05 | |
| Zhx2 | chr15 | 84.44 | 39.864 | 3.9E-08 | 8.0E-04 | |
| Zhx2 | chr15 | 86.26 | 40.584 | 3.9E-08 | 8.0E-04 | |
| Hippocampus | Zhx2 | chr15 | 32.32 | 13.546 | 6.2E-09 | 2.7E-04 |
| Zhx2 | chr15 | 57.11 | 23.8 | 1.8E-13 | 7.2E-08 | |
| Zhx2 | chr15 | 81.39 | 38.094 | 5.4E-09 | 2.4E-04 | |
| Zhx2 | chr15 | 84.44 | 39.864 | 1.2E-07 | 4.5E-03 | |
| Zhx2 | chr15 | 86.26 | 40.584 | 1.2E-07 | 4.5E-03 |
Transcripts possessing significant eQTLs on chromosome 15 (FDR < 0.05) in the striatum and hippocampus. Bold rows indicate the peak location of each cis-eQTL, which in both cases is the same location as the QTL for brain [OMOR]. Analysis was conducted in 64 F2 mice (16 per sex per treatment), and analysis included sex, RNA extraction batch, and treatment as covariates.
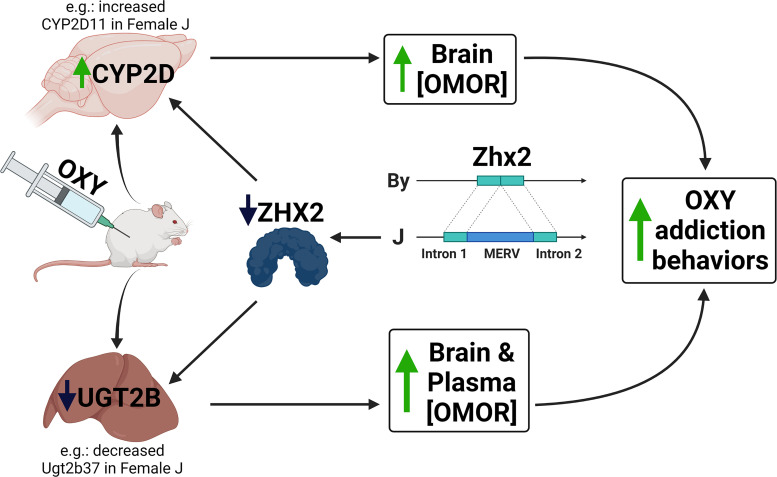
Zhx2 (zinc fingers and homeoboxes 2) codes for a globally expressed transcriptional repressor that homo- or heterodimerizes with ZHX-1 and ZHX-3, binds to DNA using zinc finger and homeobox domains, and silences gene expression (Kawata et al., 2003a,b). Decreased ZHX2 protein caused by the MERV alters the liver transcriptome (Perincheri et al., 2005; Jiang et al., 2017), including sex-specific alterations to expression of Cyp genes (Creasy et al., 2016). The Zhx2 MERV could also impact P450 expression via incomplete silencing of fetal liver proteins during development, as observed in BALB/cJ mice (Perincheri et al., 2005). We hypothesize that the constitutive decrease in Zhx2 expression in J mice leads to female-specific alterations in the expression of one or more pharmacokinetic transcripts such as Cyp2d11, culminating in increased brain concentrations of OMOR (Fig. 7).
Fig. 7.
Proposed quantitative trait mechanism linking decreased Zhx2 expression with increased brain concentration of OMOR and increased OXY-induced behaviors. We hypothesize that the insertion of a MERV into Zhx2 leads to decreased protein expression and, in turn, changes in expression of liver enzymes that metabolize OXY. The functional consequence is an increase in brain concentration of the highly potent metabolite OMOR, which serves as both a potent unconditioned and conditioned stimulus that facilitates both state-dependent learning and state-dependent memory of OXY reward and possibly other OXY-induced behaviors. Created with BioRender.com.
A critical unanswered question is whether increased whole brain [OMOR] originates from the liver, the brain, or both. There is growing appreciation for drug metabolism within the brain, including CYP2Ds (Miksys and Tyndale, 2006; Ferguson and Tyndale, 2011; Ravindranath and Strobel, 2013), and manipulation of central CYP2Ds has been shown to modulate OXY, tramadol, and codeine analgesia (McMillan and Tyndale, 2015; McMillan et al., 2019). Although liver transcriptomics revealed a decrease in several Cyp2d transcripts in the J substrain, both liver and brain proteomics revealed an increase in CYP2D11 protein in J mice. In the brain, this increase is most robust in J females. Thus, proteomics data supports one hypothesis that increased CYP2D11 leads to increased phase I metabolism of OXY in the J female brain, ultimately explaining increased brain [OMOR].
A second hypothesis explaining increased brain [OMOR] is reduced glucuronidation of oxymorphone by one or more UGT enzymes, resulting in reduced clearance and increasing plasma and brain [OMOR] in J females. In support, we observed a downregulation of Ugt2b37 liver RNA in J mice that was much more robust in J females compared with J males (female logFC = −3.24; male logFC = −0.68). Ugt2b37 codes for a UGT enzyme that mediates phase 2 glucuronidation of morphine in mice (Kurita et al., 2017). Because ZHX2 can also act as a transcriptional activator (Zhao et al., 2022), reduced ZHX2 could lead to decreased Ugt2b37 liver transcription or could influence Ugt2b37 transcript levels through indirect mechanisms (e.g., developmental) or Zhx2 expression may be completely unrelated to Ugt2b37 transcript levels. Importantly, we were unable to directly confirm differential liver UGT2B37 expression at the protein level, as it was not detectable under our assay conditions.
Isolating the effects of Zhx2 genetic variation via gene editing will provide additional support for one or more hypothesized DNA targets of ZHX2 (e.g., Cyp2d11 and Ugt2b37) that we can then assess directly. We favor the Cyp hypothesis over the Ugt hypothesis as the most direct explanation for increased brain [OMOR]. Ultimately, the mechanism could be more complicated than the regulation of one PK-related gene in one tissue given the ubiquitous expression of Zhx2 across tissues and its functional role in development (Perincheri et al., 2005; Wu et al., 2009; Kawamura et al., 2018).
There are limitations to this study. First, our metabolite profiling relies on a single dose, time point, route of administration, and tissue. Full tissue-concentration curves (e.g., brain vs. plasma) are necessary to inform the underlying pharmacokinetic mechanism. Second, we were unable to identify QTLs underlying state-dependent learning of OXY reward. The heritability of CPP is notoriously low (Cunningham et al., 1991; Philip et al., 2010; Bryant et al., 2014) due to the large environmental variance, necessitating very large sample sizes for detecting QTLs. Based on parental strain effect sizes, we would have needed 311 OXY-treated F2 mice to detect QTLs with 80% power (622 with saline controls). Rather than test more F2 mice, we will test the effect of Zhx2 on OXY behavior in future gene editing studies focused on the quantitative trait mechanism linking Zhx2 to brain [OMOR].
This study highlights the importance of assessing pharmacokinetic measures in genetic analysis of drug-induced behaviors. It also highlights how sex can dramatically alter brain metabolite concentration in a genotype-dependent manner, which is important given sex differences in substance use disorders (Cornish and Prasad, 2021), including opioid use disorder (Chartoff and McHugh, 2016). This study also highlights the limitations of relying solely on short-read DNA sequencing to annotate genetic variants and the utility of including proteomics in genetic regulation of gene expression associated with complex traits. Finally, this study highlights the power of reduced complexity crosses for rapidly identifying functional candidate genes underlying complex traits (Bryant et al., 2018, 2020).
In summary, we identified a QTL containing a highly plausible candidate gene underlying whole brain [OMOR]. We hypothesize that increased brain [OMOR] in females is mediated by a MERV-mediated loss of Zhx2 function, supported by QTL mapping, eQTL analysis, proteomics, and published literature (Fig. 7). Furthermore, we hypothesize that dysregulation of one or more proteins involved in OXY pharmacokinetics increases whole brain [OMOR] and state-dependent OXY reward learning (Fig. 7). Although we have speculated on candidate mechanisms downstream of Zhx2, future studies will be needed to test these hypotheses. We plan to use reciprocal gene editing on the two BALB/c genetic backgrounds and generate full tissue-concentration profiles of OXY and metabolites in these lines, allowing for the direct validation of Zhx2 involvement in brain OMOR and its downstream effects, including validating candidate pharmacokinetic mechanisms. Furthermore, it will be important to test the role of Zhx2 in the pharmacokinetic profiles of other opioid drugs, addiction-relevant model behaviors, and antinociception. Finally, given the nominal association of the Zhx2 locus with the quantity and frequency of nicotine use in humans (P = 7 × 10−7) (McGue et al., 2013), we will test the influence of Zhx2 in the pharmacokinetic profile and addiction liability to other drugs of abuse.
Acknowledgments
This research was supported by generous ongoing support from BU to AE and the CNSB.
Abbreviations
- By
BALB/cByJ
- chr
chromosome
- cM
centimorgan
- CPP
conditioned place preference
- Cyp
cytochrome P450
- eQTL
RNA expression QTL
- FDR
false discovery rate
- J
BALB/cJ
- logFC
log fold change
- Mb
megabase
- MERV
mouse endogenous retroviral element
- MS/MS
tandem mass spectrometry
- NOR
noroxycodone
- OMOR
oxymorphone
- OXY
oxycodone
- PK
pharmacokinetics
- QTL
quantitative trait locus
- RNA-seq
RNA sequencing
- SAL
saline
- Ugt
UDP-glucuronosyltransferase
Authorship Contributions
Participated in research design: Beierle, Ferris, Bryant.
Conducted experiments: Beierle, Yao, Goldstein, Lynch, Scotellaro, Shah, Sena, Averin, Moody, Reilly, Emili.
Contributed new reagents or analytic tools: Linnertz, Ferris.
Performed data analysis: Beierle, Yao, Goldstein, Lynch, Scotellaro, Shah, Sena, Wong, Averin, Moody, Reilly, Emili.
Wrote or contributed to the writing of the manuscript: Beierle JA, Reilly, Peltz, Emili, Ferris, Bryant.
Footnotes
This research was funded by National Institutes of Health National Institute on Drug Abuse [Grant U01-DA050243] (C.D.B.), [Grant R01-DA039168] (C.D.B.), [Grant 5U01-DA04439902] (G.P.), and [Grant N01-DA198951] (D.E.M.); National Institute of Allergy and Infectious Diseases [Grant U19-AI100625] (Dr. Fernando Pardo-Manuel De Villena) and [Grant P01-AI132130] (Dr. Fernando Pardo-Manuel De Villena); National Institute of General Medical Sciences [Grant T32-GM008541] (Dr. David Farb); and the Burroughs Welcome Fund Transformative Training Program in Addiction Science [Grant 1011479] (Dr. Lindsay Farrer).
No author has an actual or perceived conflict of interest with the contents of this article.
Primary Laboratory of Origin: Boston University School of Medicine, Laboratory of Addiction Genetics, Department of Pharmacology and Experimental Therapeutics and Psychiatry (Boston, MA).
A preprint of this article was deposited in bioRxiv [https://doi.org/10.1101/2022.03.18.484877v1].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Andreassen TN, Eftedal I, Klepstad P, Davies A, Bjordal K, Lundström S, Kaasa S, Dale O (2012) Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur J Clin Pharmacol 68:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beierle JAYao EJGoldstein SIScotellaro JLSena KDLinnertz CAWillits ABKader LYoung EEPeltz G, et al. (2022) Genetic basis of thermal nociceptive sensitivity and brain weight in a BALB/c reduced complexity cross. Mol Pain 18:17448069221079540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Riggan J, O’Toole LA (1993) Voluntary consumption of morphine in 15 inbred mouse strains. Psychopharmacology (Berl) 112:352–358. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Mogil JS, Helms ML, Richards SP, O’Toole LA, Bergeson SE, Buck KJ (1995) Localization to chromosome 10 of a locus influencing morphine analgesia in crosses derived from C57BL/6 and DBA/2 strains. Life Sci 57:PL117–PL124. [DOI] [PubMed] [Google Scholar]
- Bergeson SE, Helms ML, O’Toole LA, Jarvis MW, Hain HS, Mogil JS, Belknap JK (2001) Quantitative trait loci influencing morphine antinociception in four mapping populations. Mamm Genome 12:546–553. [DOI] [PubMed] [Google Scholar]
- Bryant CD, Ferris MT, De Villena FPM, Damaj MI, Kumar V, Mulligan MK (2018) Reduced complexity cross design for behavioral genetics, in Molecular-Genetic and Statistical Techniques for Behavioral and Neural Research (Gerlai RT, ed) pp 165–190, Academic Press, Cambridge, MA. [Google Scholar]
- Bryant CD, Graham ME, Distler MG, Munoz MB, Li D, Vezina P, Sokoloff G, Palmer AA (2009) A role for casein kinase 1 epsilon in the locomotor stimulant response to methamphetamine. Psychopharmacology (Berl) 203:703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CD, Guido MA, Kole LA, Cheng R (2014) The heritability of oxycodone reward and concomitant phenotypes in a LG/J × SM/J mouse advanced intercross line. Addict Biol 19:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CDParker CCZhou LOlker CChandrasekaran RYWager TTBolivar VJLoudon ASVitaterna MHTurek FW, et al. (2012) Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology 37:1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CD, Smith DJ, Kantak KM, Nowak TS Jr, Williams RW, Damaj MI, Redei EE, Chen H, Mulligan MK (2020) Facilitating complex trait analysis via reduced complexity crosses. Trends Genet 36:549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier JA, He H, Philip VM, Roy T, Hernandez CM, Bernat R, Donohue KD, O’Hara BF, Chesler EJ (2020) Genetic variation regulates opioid-induced respiratory depression in mice. Sci Rep 10:14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiotti KA, Yang Z, Rodriguez Y, Crescimone A, Sanchez GC, Takacs P, Medina C, Zhang Y, Liu H, Gitlin MC (2009) The impact of CYP2D6 genetic polymorphisms on postoperative morphine consumption. Pain Med 10:799–805. [DOI] [PubMed] [Google Scholar]
- Chan G, Gelernter J, Oslin D, Farrer L, Kranzler HR (2011) Empirically derived subtypes of opioid use and related behaviors. Addiction 106:1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartoff EH, McHugh RK (2016) Translational studies of sex differences in sensitivity to opioid addiction. Neuropsychopharmacology 41:383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish JL, Prasad AA (2021) Sex differences in substance use disorders: a neurobiological perspective. Front Glob Womens Health 2:778514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy KT, Jiang J, Ren H, Peterson ML, Spear BT (2016) Zinc fingers and homeoboxes 2 (Zhx2) regulates sexually dimorphic Cyp gene expression in the adult mouse liver. Gene Expr 17:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist RC, Reiner BC, Berrettini WH (2019) A review of opioid addiction genetics. Curr Opin Psychol 27:31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Hallett CL, Niehus DR, Hunter JS, Nouth L, Risinger FO (1991) Assessment of ethanol’s hedonic effects in mice selectively bred for sensitivity to ethanol-induced hypothermia. Psychopharmacology (Berl) 105:84–92. [DOI] [PubMed] [Google Scholar]
- Dam SA, Jager A, Oomen CA, Buitelaar JK, Arias-Vasquez A, Glennon JC (2019) Inhibitory control in BALB/c mice sub-strains during extinction learning. Eur Neuropsychopharmacol 29:509–518. [DOI] [PubMed] [Google Scholar]
- Donaldson R, Sun Y, Liang D-Y, Zheng M, Sahbaie P, Dill DL, Peltz G, Buck KJ, Clark JD (2016) The multiple PDZ domain protein Mpdz/MUPP1 regulates opioid tolerance and opioid-induced hyperalgesia. BMC Genomics 17:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang WB, Lofwall MR, Walsh SL, Moody DE (2013) Determination of oxycodone, noroxycodone and oxymorphone by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry in human matrices: in vivo and in vitro applications. J Anal Toxicol 37:337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CS, Tyndale RF (2011) Cytochrome P450 enzymes in the brain: emerging evidence of biological significance. Trends Pharmacol Sci 32:708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg LRYao EJKelliher JCReed ERWu Cox JParks CKirkpatrick SLBeierle JAChen MMJohnson WE, et al. (2021) A quantitative trait variant in Gabra2 underlies increased methamphetamine stimulant sensitivity. Genes Brain Behav 20:e12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F (2005) The genetics of addictions: uncovering the genes. Nat Rev Genet 6:521–532. [DOI] [PubMed] [Google Scholar]
- Gonzales NM, Seo J, Hernandez Cordero AI, St Pierre CL, Gregory JS, Distler MG, Abney M, Canzar S, Lionikas A, Palmer AA (2018) Genome wide association analysis in a mouse advanced intercross line. Nat Commun 9:5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness JH, Shi X, Janowsky A, Phillips TJ (2015) Trace amine-associated receptor 1 regulation of methamphetamine intake and related traits. Neuropsychopharmacology 40:2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilakivi LA, Lister RG (1989) Comparison between BALB/cJ and BALB/cByJ mice in tests of social behavior and resident-intruder aggression. Aggress Behav 15:273–280 DOI: 10.1002/ab.2480150402. [Google Scholar]
- Huddart R, Clarke M, Altman RB, Klein TE (2018) PharmGKB summary: oxycodone pathway, pharmacokinetics. Pharmacogenet Genomics 28:230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager A, Dam SA, Van Der Mierden S, Oomen CA, Arias-Vasquez A, Buitelaar JK, Kozicz T, Glennon JC (2020) Modulation of cognitive flexibility by reward and punishment in BALB/cJ and BALB/cByJ mice. Behav Brain Res 378:112294. [DOI] [PubMed] [Google Scholar]
- Jiang J, Creasy KT, Purnell J, Peterson ML, Spear BT (2017) Zhx2 (zinc fingers and homeoboxes 2) regulates major urinary protein gene expression in the mouse liver. J Biol Chem 292:6765–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Yamanaka K, Poh B, Kuribayashi H, Koso H, Watanabe S (2018) The role of Zhx2 transcription factor in bipolar cell differentiation during mouse retinal development. Biochem Biophys Res Commun 503:3023–3030. [DOI] [PubMed] [Google Scholar]
- Kawata H, Yamada K, Shou Z, Mizutani T, Miyamoto K (2003a) The mouse zinc-fingers and homeoboxes (ZHX) family; ZHX2 forms a heterodimer with ZHX3. Gene 323:133–140. [DOI] [PubMed] [Google Scholar]
- Kawata H, Yamada K, Shou Z, Mizutani T, Yazawa T, Yoshino M, Sekiguchi T, Kajitani T, Miyamoto K (2003b) Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem J 373:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TMGoodstadt LDanecek PWhite MAWong KYalcin BHeger AAgam ASlater GGoodson M, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC (2003) Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry 160:687–695. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Neale MC, Prescott CA (2000) Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch Gen Psychiatry 57:261–269. [DOI] [PubMed] [Google Scholar]
- Kest B, Palmese CA, Hopkins E, Adler M, Juni A, Mogil JS (2002) Naloxone-precipitated withdrawal jumping in 11 inbred mouse strains: evidence for common genetic mechanisms in acute and chronic morphine physical dependence. Neuroscience 115:463–469. [DOI] [PubMed] [Google Scholar]
- Kest B, Smith SB, Schorscher-Petcu A, Austin J-S, Ritchie J, Klein G, Rossi GC, Fortin A, Mogil JS (2009) Gnao1 (G alphaO protein) is a likely genetic contributor to variation in physical dependence on opioids in mice. Neuroscience 162:1255–1264. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick SL, Bryant CD (2015) Behavioral architecture of opioid reward and aversion in C57BL/6 substrains. Front Behav Neurosci 8:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick SLGoldberg LRYazdani NBabbs RKWu JReed ERJenkins DFBolgioni AFLandaverde KILuttik KP, et al. (2017) Cytoplasmic FMR1-interacting protein 2 is a major genetic factor underlying binge eating. Biol Psychiatry 81:757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar VKim KJoseph CKourrich SYoo S-HHuang HCVitaterna MHde Villena FP-MChurchill GBonci A, et al. (2013) C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342:1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita A, Miyauchi Y, Ikushiro S, Mackenzie PI, Yamada H, Ishii Y (2017) Comprehensive characterization of mouse UDP-glucuronosyltransferase (Ugt) belonging to the Ugt2b subfamily: identification of Ugt2b36 as the predominant isoform involved in morphine glucuronidation. J Pharmacol Exp Ther 361:199–208. [DOI] [PubMed] [Google Scholar]
- Lalovic B, Kharasch E, Hoffer C, Risler L, Liu-Chen L-Y, Shen DD (2006) Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin Pharmacol Ther 79:461–479. [DOI] [PubMed] [Google Scholar]
- Lemberg KK, Heiskanen TE, Neuvonen M, Kontinen VK, Neuvonen PJ, Dahl M-L, Kalso EA (2010) Does co-administration of paroxetine change oxycodone analgesia: an interaction study in chronic pain patients. Scand J Pain 1:24–33. [DOI] [PubMed] [Google Scholar]
- Levitt RCZhuang GYKang YErasso DMUpadhyay UOzdemir MFu ESSarantopoulos KDSmith SBMaixner W, et al. (2017) Car8 dorsal root ganglion expression and genetic regulation of analgesic responses are associated with a cis-eQTL in mice. Mamm Genome 28:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E, Rabinovich A, Iserte J, Yanovsky M, Chernomoretz A (2021) Corrigendum to: ASpli: integrative analysis of splicing landscapes through RNA-Seq assays. Bioinformatics 37:2609–2616. [DOI] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Malone S, Oetting WS, Iacono WG (2013) A genome-wide association study of behavioral disinhibition. Behav Genet 43:363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DM, Miksys S, Tyndale RF (2019) Rat brain CYP2D activity alters in vivo central oxycodone metabolism, levels and resulting analgesia. Addict Biol 24:228–238. [DOI] [PubMed] [Google Scholar]
- McMillan DM, Tyndale RF (2015) Nicotine increases codeine analgesia through the induction of brain CYP2D and central activation of codeine to morphine. Neuropsychopharmacology 40:1804–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksys S, Tyndale RF (2006) Nicotine induces brain CYP enzymes: relevance to Parkinson’s disease, in Parkinson’s Disease and Related Disorders (Riederer P, Reichmann H, Youdim MBH, Gerlach M, eds) pp 177–180, Springer, New York. [DOI] [PubMed] [Google Scholar]
- Miner NB, Elmore JS, Baumann MH, Phillips TJ, Janowsky A (2017) Trace amine-associated receptor 1 regulation of methamphetamine-induced neurotoxicity. Neurotoxicology 63:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Abreo T, Neuner SM, Parks C, Watkins CE, Houseal MT, Shapaker TM, Hook M, Tan H, et al. (2019) Identification of a functional non-coding variant in the GABA A receptor α2 Subunit of the C57BL/6J mouse reference genome: Major implications for neuroscience research. Frontiers in Genetics 10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (2011) Guide for the Care and Use of Laboratory Animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- Nelson ECAgrawal AHeath ACBogdan RSherva RZhang BAl-Hasani RBruchas MRChou Y-LDemers CH, et al. (2016) Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry 21:608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham EM, Traynor JR (2006) Comparison of the antinociceptive response to morphine and morphine-like compounds in male and female Sprague-Dawley rats. J Pharmacol Exp Ther 316:1195–1201. [DOI] [PubMed] [Google Scholar]
- Perincheri S, Dingle RWC, Peterson ML, Spear BT (2005) Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc Natl Acad Sci USA 102:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perincheri S, Peyton DK, Glenn M, Peterson ML, Spear BT (2008) Characterization of the ETnII-α endogenous retroviral element in the BALB/cJ Zhx2 ( Afr1 ) allele. Mamm Genome 19:26–31. [DOI] [PubMed] [Google Scholar]
- Philip VMDuvvuru SGomero BAnsah TABlaha CDCook MNHamre KMLariviere WRMatthews DBMittleman G, et al. (2010) High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav 9:129–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyntz HCJones AJauregui RYoung WGestin AMooney ALamiable OAltermann ESchmidt AGasser O, et al. (2019) Genetic regulation of antibody responsiveness to immunization in substrains of BALB/c mice. Immunol Cell Biol 97:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath V, Strobel HW (2013) Cytochrome P450-mediated metabolism in brain: functional roles and their implications. Expert Opin Drug Metab Toxicol 9:551–558. [DOI] [PubMed] [Google Scholar]
- Reed C, Baba H, Zhu Z, Erk J, Mootz JR, Varra NM, Williams RW, Phillips TJ (2018) A spontaneous mutation in Taar1 impacts methamphetamine-related traits exclusively in DBA/2 mice from a single vendor. Front Pharmacol 8:993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan QTYazdani NBlum BCBeierle JALin WCoelho MAFultz EKHealy AFShahin JRKandola AK, et al. (2020) A mutation in Hnrnph1 that decreases methamphetamine-induced reinforcement, reward, and dopamine release and increases synaptosomal hnRNP H and mitochondrial proteins. J Neurosci 40:107–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Satagopan JM, Broman KW, Churchill GA (2007) R/qtlDesign: inbred line cross experimental design. Mamm Genome 18:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA (2012) Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28:1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Walter NAR, Harkness JH, Neve KA, Williams RW, Lu L, Belknap JK, Eshleman AJ, Phillips TJ, Janowsky A (2016) Genetic polymorphisms affect mouse and human trace amine-associated receptor 1 function. PLoS One 11:e0152581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmon JSBlanchard MWBaric RSBell TABrennan JBrockmann GABurks AWCalabrese JMCaron KMCheney RE, et al. (2020) Content and performance of the MiniMUGA genotyping array: a new tool to improve rigor and reproducibility in mouse research. Genetics 216:905–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittig LJ, Jeong C, Tixier E, Davis J, Barrios-Camacho CM, Palmer AA (2014) Phenotypic instability between the near isogenic substrains BALB/cJ and BALB/cByJ. Mamm Genome 25:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SBMarker CLPerry CLiao GSotocinal SGAustin J-SMelmed KClark JDPeltz GWickman K, et al. (2008) Quantitative trait locus and computational mapping identifies Kcnj9 (GIRK3) as a candidate gene affecting analgesia from multiple drug classes. Pharmacogenet Genomics 18:231–241. [DOI] [PubMed] [Google Scholar]
- Solecki W, Turek A, Kubik J, Przewlocki R (2009) Motivational effects of opiates in conditioned place preference and aversion paradigm--a study in three inbred strains of mice. Psychopharmacology (Berl) 207:245–255. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Wojno H, Greiner E, May EL, Rice KC, Selley DE (2004) Activation of G-proteins by morphine and codeine congeners: insights to the relevance of O- and N-demethylated metabolites at μ- and δ-opioid receptors. J Pharmacol Exp Ther 308:547–554. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, Meyer JM, Toomey R, Faraone SV, Eaves L (1996) Genetic influences on DSM-III-R drug abuse and dependence: a study of 3,372 twin pairs. Am J Med Genet 67:473–477. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L (1998) Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry 55:967–972. [DOI] [PubMed] [Google Scholar]
- Turner JK, McAllister MM, Xu JL, Tapping RI (2008) The resistance of BALB/cJ mice to Yersinia pestis maps to the major histocompatibility complex of chromosome 17. Infect Immun 76:4092–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez L, Sokoloff G, Miczek KA, Palmer AA, Dulawa SC (2010) Differences in aggressive behavior and DNA copy number variants between BALB/cJ and BALB/cByJ substrains. Behav Genet 40:201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe DA, McMahon Tobin GA, Mellon RD, Katki AG, Parker RJ, Colatsky T, Kropp TJ, Verbois SL (2011) Uniform assessment and ranking of opioid μ receptor binding constants for selected opioid drugs. Regul Toxicol Pharmacol 59:385–390. [DOI] [PubMed] [Google Scholar]
- Wilson SG, Smith SB, Chesler EJ, Melton KA, Haas JJ, Mitton B, Strasburg K, Hubert L, Rodriguez-Zas SL, Mogil JS (2003) The heritability of antinociception: common pharmacogenetic mediation of five neurochemically distinct analgesics. J Pharmacol Exp Ther 304:547–559. [DOI] [PubMed] [Google Scholar]
- Wu C, Qiu R, Wang J, Zhang H, Murai K, Lu Q (2009) ZHX2 interacts with ephrin-B and regulates neural progenitor maintenance in the developing cerebral cortex. J Neurosci 29:7404–7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin BWong KAgam AGoodson MKeane TMGan XNellåker CGoodstadt LNicod JBhomra A, et al. (2011) Sequence-based characterization of structural variation in the mouse genome. Nature 477:326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YGao LJiang CChen JQin ZZhong FYan YTong RZhou MYuan A, et al. (2022) The transcription factor zinc fingers and homeoboxes 2 alleviates NASH by transcriptional activation of phosphatase and tensin homolog. Hepatology 75:939–954. [DOI] [PubMed] [Google Scholar]
- Zwisler ST, Enggaard TP, Mikkelsen S, Brosen K, Sindrup SH (2010) Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand 54:232–240. [DOI] [PubMed] [Google Scholar]
- Zwisler ST, Enggaard TP, Noehr-Jensen L, Pedersen RS, Mikkelsen S, Nielsen F, Brosen K, Sindrup SH (2009) The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharmacol Toxicol 104:335–344. [DOI] [PubMed] [Google Scholar]