ABSTRACT
Drug resistance derived from extracellular vesicles (EVs) is an increasingly important research area but has seldom been described regarding fungal pathogens. Here, we characterized EVs derived from a triazole-resistant but amphotericin B-susceptible strain of Candida auris. Nano- to microgram concentrations of C. auris EVs prepared from both broth and solid agar cultures could robustly increase the yeast’s survival against both pure and clinical amphotericin B formulations in a dose-dependent manner, resulting in up to 16-fold changes of minimum inhibitory concentration. Meanwhile, this effect was not observed upon addition of these EVs to C. albicans, nor upon addition of C. albicans EVs to C. auris. No change in susceptibilities was observed upon EV treatment for fluconazole, voriconazole, micafungin, and flucytosine. Mass spectrometry indicated the presence of immunogenic-/drug resistance-implicated proteins in C. auris EVs, including alcohol dehydrogenase 1 as well as C. albicans Mp65-like and Xog1-like proteins in high quantities. Based on these observations, we propose a potential species-specific role for EVs in amphotericin B resistance in C. auris. These observations may provide critical insights into treatment of multidrug-resistant C. auris.
KEYWORDS: Candida auris, extracellular vesicles, drug resistance, amphotericin B, solid media EV purification
Introduction
Candida auris is a recently emerged pathogenic fungus described as a serious threat to global health by the US Centers for Disease Control and Prevention (CDC) due to its propensity for nosocomial outbreaks and multi-drug resistance [1]. The yeast was first discovered and described in 2009 from the external ear canal discharge of a patient in Japan [2]. Since then, infections caused by C. auris have been reported in six continents, with a crude mortality rate as high as 78%, depending on the geographic clades to which the aetiological C. auris isolates belong [3]. This novel fungal pathogen is also notorious for its persistence in the environment, especially in healthcare facilities due to its resistance to common disinfectants [4]. In Hong Kong, C. auris was first reported in June 2019 [5]. Identification of C. auris colonization in the city has since increased exponentially to over 200 incidents across four different local hospitals, especially during July–December, 2020 when 85% of the positive isolations occurred; and C. auris detection continues to be reported from even the initial outbreak ward [5]. The isolate recovered from the index patient in Hong Kong belonged to the South Asian clade and was multi-drug resistant. In particular, it was resistant to fluconazole and was non-wild-type to all other triazole agents as well as the echinocandins caspofungin and anidulafungin; while it was only susceptible to amphotericin B and was wild-type to micafungin [5]. Recent reports have suggested that despite performing appropriate antifungal susceptibility testing, usage of corresponding clinical dosages can still result in treatment failure [6,7]. Therefore, further research into molecular and phenotypic mechanisms of drug resistance in C. auris is required.
Extracellular vesicles (EVs) collectively describe lipid bilayer-delimited secreted membrane vesicles [8]. They contain an assortment of cargo that vary according to the nutritional, metabolic, and environmental conditions of the cell. In mammalian and bacterial cells, EVs have been implicated in cell–cell communication [8–11], immunomodulation [12], and drug resistance [13,14]. For example, bacterial outer membrane vesicles (OMVs) can horizontally transfer carbapenemases and their corresponding genes [15,16], sequester antimicrobial peptides [17,18], and act as bacteriophage decoys [17]. Yet, the roles of EVs in fungi have not been as well-defined [8]. Thus far, drug resistance derived from fungal EVs has only been described in two studies; where EVs produced by Candida albicans and Saccharomyces cerevisiae were demonstrated to provide cellular protection against the antifungal drugs fluconazole and caspofungin, respectively [19, 20]. Notably, it was shown that the cargo of S. cerevisiae EVs included a number of cell wall modification enzymes; and therefore may have helped compensate for the inhibition of (1,3)-β-d-glucan synthase upon caspofungin treatment [20]. Here, we show that EVs isolated from an amphotericin B-sensitive strain of C. auris can be back-added to increase the minimum inhibitory concentration (MIC) of amphotericin B by at least up to 16-fold in a dose-dependent manner. These findings may help explain the frequent failure to treat supposedly amphotericin B-susceptible C. auris with amphotericin B and may constitute a novel mechanism of resistance against the third class of effective antifungal drugs: the polyenes.
Materials and methods
Fungal strains. C. auris strain Cau1901 was isolated from the index patient in Hong Kong from Princess Margaret Hospital in June 2019 [5]. The isolate was cryopreserved at −80°C or maintained on Sabouraud dextrose agar (SDA; Difco, BD Diagnostics Systems, USA) supplemented with chloramphenicol (50 µg/mL; Calbiochem, USA) at 37°C. The reference strain C. albicans ATCC 90028 was obtained from the American Type Culture Collection (ATCC), USA. The quality control strain for susceptibility testing C. parapsilosis ATCC 22019T was obtained from the ATCC; strains C. albicans CNM-CL F8555 and Pichia kudriavzevii (synonym: C. krusei) CNM-CL-3403 were obtained from Statens Serum Institut (SSI), Denmark; and strain P. kudriavzevii NRRL Y-413 ( = ATCC 6258) was obtained from the Agricultural Research Service (ARS) Culture Collection (NRRL), Department of Agriculture, USA.
Isolation of fungal EVs. (i) Broth culture. For both C. auris Cau1901 and C. albicans ATCC 90028, a single colony (≥1 mm in diameter) on SDA was collected and transferred to 10 mL of Sabouraud dextrose broth (SDB, Sigma-Aldrich, USA) for growth at 37°C in a shaking incubator at 250 rpm. After 24 h of incubation, 1 mL of the liquid culture was transferred to 1.25 L of fresh SDB in a 2 L conical flask for further growth at 37°C in a shaking incubator at 250 rpm. After a further 48 h of incubation, the liquid cultures were centrifuged at 15,000 ×g for 15 min at 4°C, and the supernatant collected was vacuum filtered through 0.45 μm-polyethersulphone (PES) membranes (Nalgene, USA). Every 1.25 L of filtered supernatant was concentrated via centrifugation at 3000 ×g for 10 min at 4°C through a 100 kDa cellulose membrane concentrator (Merck, Germany) to a final volume of 25 mL. The concentrate was then collected for subsequent ultracentrifugation. (ii) Agar plate culture. For both C. auris Cau1901 and C. albicans ATCC 90028, fungal material was collected using a 10 μL inoculation loop and resuspended in 30 mL of phosphate-buffered saline (PBS; Oxoid, UK). After centrifugation at 3000 ×g for 5 min, the supernatant was collected and vacuum filtered through 0.45 μm-PES membrane. The filtrate was then collected for subsequent ultracentrifugation. (iii) EV isolation and purification. For both supernatant concentrate collected from broth cultures and filtrate collected from agar plate cultures, ultracentrifugation was performed at 100,000 ×g for 1.5 h at 4°C using the Optima XE (Beckman Coulter, USA) ultracentrifuge equipped with a SW32-Ti rotor (Beckman Coulter). Next, the pellets were resuspended in PBS and then subjected to particle separation via layered iodixanol (OptiPrep, Stemcell Technologies, Canada) density gradient ultracentrifugation at 100,000 ×g for 16 h at 4°C. The separated layers of different densities were then collected sequentially, and each fraction was washed with PBS by ultracentrifugation at 100,000 ×g for 1.5 h at 4°C twice. The density of each fraction was calculated from the measurement using a refractometer.
Isolation of human reticulocyte EVs. Human red blood cell EVs were isolated according to Usman and colleagues’ protocol [21].
EV quantification and measurements. Nanoparticle tracking analysis (NTA) was performed for each iodixanol fraction obtained using the ZetaView PMX-220 TWIN Laser system (Particle Metrix, Germany) for size and particle number quantification. Briefly, for each iodixanol fraction 1 µL of the solution was diluted in 10 mL of PBS prior to NTA for size and concentration and 1 mL of each diluted sample was used for the analysis. Each experiment was recorded at 11 random positions capturing 100–1000 particles (the “green” range) with 75 arbitrary units of sensitivity and a shutter speed of 0.01 s. All other parameters were kept at the manufacturer’s default settings. The protein content of each iodixanol fraction was also measured using the Pierce BCA Protein Assay Kit (Thermo Scientific, USA) according to the manufacturer’s protocol. Briefly, 10 μL of each undiluted iodixanol fraction was added to an alkaline reagent mix composed of reagent A and reagent B and then incubated for 30 min at 37°C. The resulting colour change was measured using the VICTOR Multilabel Plate Reader (Perkin Elmer, USA) and quantified against a standard bovine serum albumin (Thermo Scientific) protein curve. Measurements were performed in duplicate and averaged.
Transmission electron microscopy (TEM). Each iodixanol fraction was examined under TEM for particle visualization. Briefly, 1 µL of each iodixanol fraction was diluted in 5 μL of autoclaved Milli-Q water. Samples were negatively stained with 2.5% uranyl acetate and analysed using the transmission electron microscope Phillips CM100 (Philips Electron Optics, the Netherlands) equipped with the Tengra charge-coupled device (CCD) camera (Olympus Soft Imaging Solutions, Germany). At least three grid positions were interrogated per sample, and two independent experiments were performed.
Protein profile analysis. For each iodixanol fraction, 20 μL of the solution was subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) under a reducing condition at 120 V using 12% Mini-PROTEAN TGX precast gels (Bio-Rad, USA) for 30 min. The separated proteins were then stained using the Pierce Silver Stain for Mass Spectrometry (Thermo Scientific) kit according to the manufacturer’s instructions. The 25–35 kDa band from Fraction F6 (1.087 g/L iodixanol) from the gel was excised, purified, and trypsin-digested at the Centre for PanorOmic Sciences (CPOS) Proteomics and Metabolomics Core at the Li Ka Shing Faculty of Medicine of The University of Hong Kong (HKU). The peptides were identified via liquid chromatography–mass spectrometry (LC–MS) using the timsTOF Pro mass spectrometer (Bruker Daltonics, Germany) and results were analysed using MaxQuant v1.16.17 (Max-Planck-Institute of Biochemistry, Germany). The resulting protein profile was subject to Gene Ontology (GO) analysis using the Candida Genome Database Gene Ontology Slim Mapper [22].
Antifungal susceptibility testing. Susceptibility testing experiments were conducted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines for yeast using the broth microdilution method [23]. Briefly, 100 μL of 2× RPMI 1640 medium (Gibco, USA) supplemented with 2% glucose (VWR Chemicals, United Kingdom), buffered with 3-(N-morpholino)propanesulphonic acid (MOPS; Merck KGaA, Germany), and adjusted to pH 7.0 using NaOH (Sigma Aldrich) with various drug dilutions (all drugs were obtained from TargetMol [USA] except that pure amphotericin B powder was obtained from Cayman Chemical [USA], liposomal amphotericin B [AmBisome] from Gilead Sciences [USA] and amphotericin B deoxycholate from Bristol-Myers Squibb [USA]) was added to each well of a flat-bottom tissue-treated 96 well plate (Eppendorf, Germany). Next, EVs (0, 1 or 5 μg in 10 μL of PBS) were added to each well, followed by inoculation with 100 μL of C. auris Cau1901 or C. albicans ATCC 90028 adjusted to 0.5 McFarland standard (1–5 × 105 cells/mL). 10 μL of PBS was added to each positive control well to ensure that the observed effects were not due to the dilution of growth media. Measurements of growth were read using the VICTOR Multilabel Plate Reader at 405 nm 24 h post-inoculation. The MIC was defined as 50% inhibition of growth for the triazoles (fluconazole and voriconazole), echinocandins (anidulafungin and micafungin), and flucytosine, while the MIC for amphotericin B was defined as 90% inhibition of growth. Apart from the addition of EVs into susceptibility testing, yeast cell lysates were also used to serve as controls. Briefly, a loopful of cells were resuspended in 500 μL of PBS supplemented with the cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, Switzerland) containing 100 μL of 425–600 μm unwashed glass beads (Sigma-Aldrich). The cells were then mechanically disrupted using the TissueLyser II (Qiagen, Germany) for 1 min at 30 Hz. The lysed cell suspensions were then centrifuged at 3000 ×g for 10 min at 4°C; and the supernatant was collected and their protein content was quantified using the Pierce BCA Protein Assay Kit.
Results
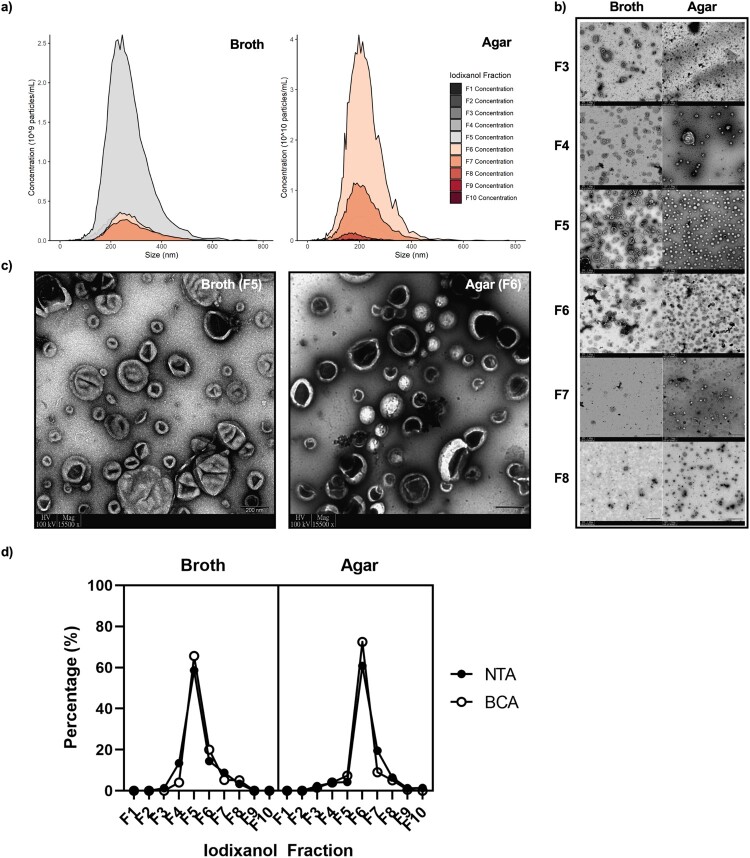
Morphology and characterization of C. auris EVs. C. auris EVs could be isolated from both broth and solid agar cultures and were similarly sized, though mainly localized in separate fractions upon density ultracentrifugation (Figure 1). TEM, NTA, and BCA all confirmed that broth culture EVs were found in F4–F7, while agar culture EVs were found in F3–F8 (Figure 1). The median sizes for broth culture- and agar culture-derived EVs were 215 and 207 nm, respectively, according to NTA (Figure 1(a)) and were in accordance with the sizes of various fungal EVs reported thus far [8, 24–26]. The purity of the preparation was confirmed through negative staining TEM using uranyl acetate, which showed a lack of protein aggregates and contaminating material (Figure 1(c)). Here, the classically described concave-cupped EVs [27] were successfully isolated from broth and agar cultures of C. auris.
Figure 1.
Differences and similarities of extracellular vesicles (EVs) derived from Candida auris using two culture methods. F3–F8 fractions correspond to densities of 1.044, 1.061, 1.065, 1.087, 1.101, and 1.127 ± 0.01 g/L, respectively. (a) Nanoparticle tracking analysis (NTA) of all separated iodixanol fractions from broth and agar cultures, n = 3. (b) Transmission electron microscopy (TEM) of separated iodixanol fractions from broth and agar cultures. Three grid positions were interrogated per sample, n = 3. Scale bars = 200 nm. (c) Representative TEM micrographs of the fractions containing the highest concentration of EVs. (d) Pooled comparison of the relative amount of EVs found in each fraction measured through NTA and bicinchoninic acid protein measurement assay, n = 3.
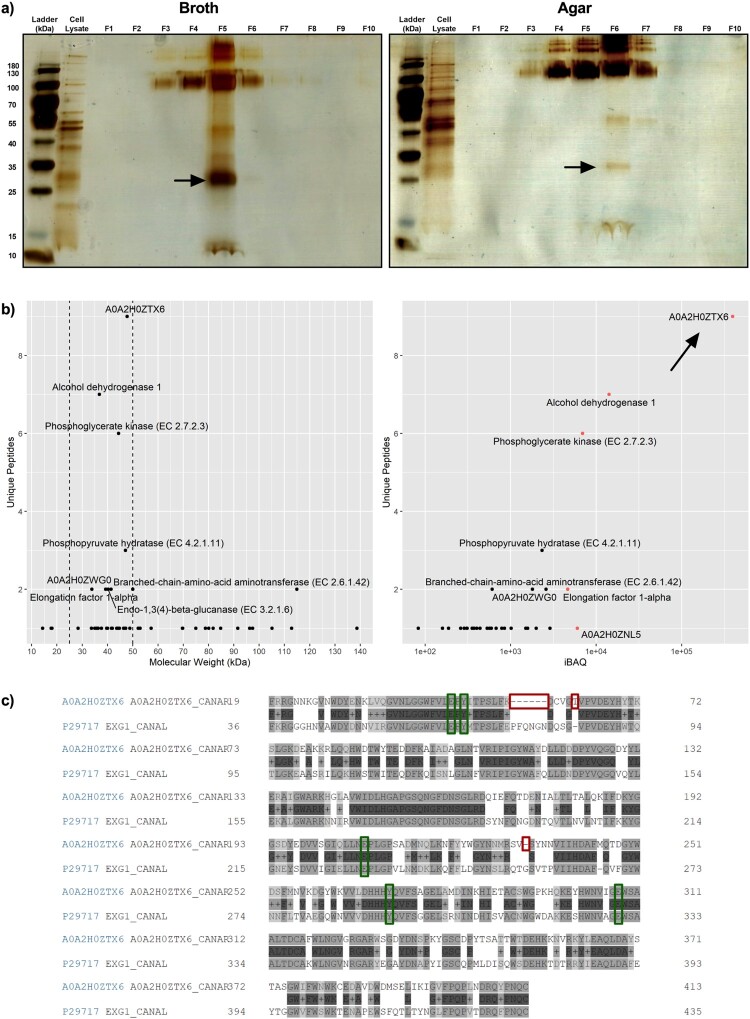
An SDS–PAGE for the iodixanol-separated fractions was performed to separate C. auris EV proteins by mass. The protein amounts as measured by BCA correlated with the intensity of the bands on the silver-stained gel (Figure 2(a)). Additionally, a distinct band was found around ∼25–35 kDa in the fractions with the highest concentration of EVs in both broth (F5) and agar culture (F6) preparations. This band from F6 of agar culture preparation was excised and subjected to LC–MS analysis. The resulting protein profile returned 49 proteins with at least one unique peptide detected (Supplementary Data File 1). To infer whether patterns of biological significance exist, the list underwent GO analysis for cellular location, biological processes, and molecular function (Figure S1). The GO analysis was generally nondescript and indicated most proteins were of cytoplasmic and mitochondrial origins (25/49 and 13/49, respectively). These proteins were usually associated with enzymatic function and not structural (Figure S1). Because GO analysis did not reveal obvious clues to C. auris EV function, a manual approach to investigating the protein list was taken. The list was first narrowed to appropriate sizes (25–50 kDa) and five proteins with high protein intensities (iBAQ > 5,000, Figure 2(b)) were identified. These included alcohol dehydrogenase 1, elongation factor 1α, phosphoglycerate kinase, and two uncharacterized proteins (A0A2H0ZTX6 and A0A2H0ZNI5). A0A2H0ZTX6 and A0A2H0ZNI5 shared 61.4% and 60.9% amino acid similarity to C. albicans glycosidase Xog1 and a major antigen mannoprotein Mp65, respectively. Amongst the five proteins identified, A0A2H0ZTX6 had both the highest protein intensity and number of unique peptides found. All described active sites and substrate binding domains in C. albicans Xog1 were found to be conserved in C. auris Xog1 (Figure 2(c)).
Figure 2.
Proteomic analysis of abundant small proteins in Candida auris extracellular vesicles reveals immunogenic and drug-related cargo. (a) Representative silver-stained SDS–PAGE of broth and agar-derived density gradient-separated fractions, n = 2. (b) Detailed analysis of protein list obtained using tandem liquid chromatography mass spectrometry of excised 25–35 kDa band. Proteins were first separated by molecular weight (25–50 kDa, left), then plotted against protein intensity (right). The top 5 proteins in iBAQ are highlighted red. (c) Aligned protein sequences of C. auris A0A2H0ZTX6 (above) and C. albicans Xog1 (below, P29717; alternative name: EXG1). Green boxes indicate conserved glycosylation sites and/or active sites, red boxes indicate insertions or deletions for C. auris A0A2H0ZTX6 when compared with C. albicans Xog1.
Effect of C. auris EV treatment on antifungal susceptibility. Drug susceptibility testing was performed for C. auris Cau1901 and C. albicans ATCC 90028 against a panel of antifungal drugs to establish baseline susceptibility prior to EV addition. Similar to most isolates in C. auris clade I [28], C. auris Cau1901 exhibited significant levels of resistance towards the triazole class of antifungals (MIC >128 μg/mL for fluconazole and MIC >16 μg/mL for voriconazole; Figures S2a–b). On the other hand, MICs for amphotericin B, flucytosine, and micafungin were 1, 0.25, and ≤0.03 μg/mL, respectively (Figure 3(a) and Figures S2c–d,).
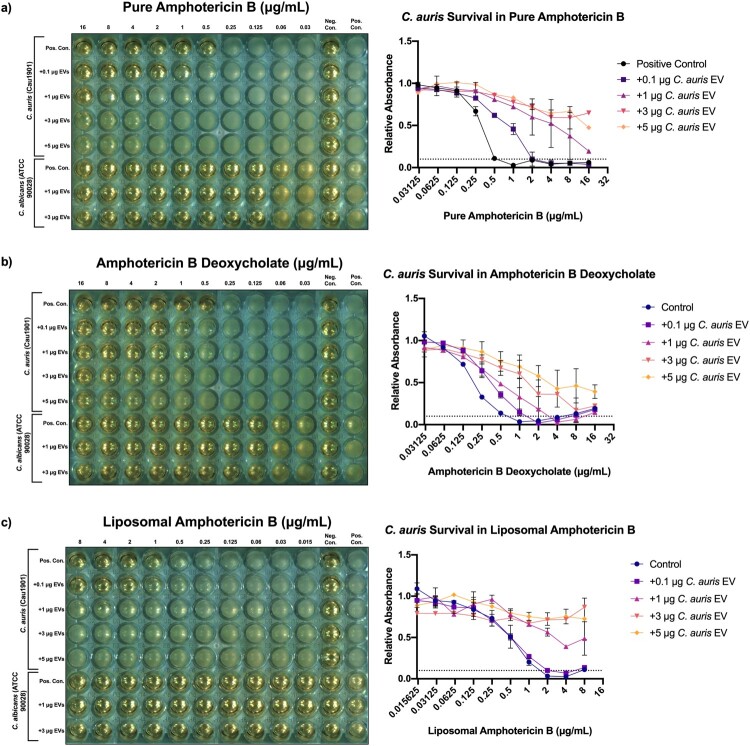
Figure 3.
The addition of Candida auris extracellular vesicles to in vitro C. auris cultures increases their resistance to amphotericin B in a dose-dependent manner, but not C. albicans. Left: Representative European Committee on Antimicrobial Susceptibility Testing (EUCAST) antifungal susceptibility testing (AFST) broth microdilution plate incubated with treatment groups, n = 3. Right: mean relative absorbance of AFST plates inoculated with C. auris Cau1901 measured at 450 nm after 24 h, n = 3. (a) Pure amphotericin B. (b) Water soluble amphotericin B deoxycholate. (c) Liposomal amphotericin B.
Given the antifungal resistance profile of C. auris Cau1901, we asked whether EVs derived from this multidrug-resistant organism could be a mechanism of drug resistance. Because a close correlation between the relative protein quantity and EV concentration was found (Figure 1(d)), protein quantity was used as a surrogate for EV concentration. C. auris EVs derived from agar and broth cultures were added to microdilution in vitro cultures of C. auris using human reticulocyte EVs as a control for fluconazole and voriconazole first. No difference in MIC was observed for the triazoles after the back-addition of EVs (Figure S2a–b). As flucytosine and micafungin are used in clinical practice against C. auris infection, an increased concentration of agar-derived EVs was added to observe if any effect was present. Still, no difference from the control was observed (Figure S2c–d).
On the other hand, the MICs for the pure pharmaceutical formulation and both of the clinical formulations of amphotericin B were increased upon EV addition in C. auris in a dose dependent manner (Figure 3). As little as 0.1 μg of EVs was able to quadruple the MIC and allowed for C. auris survival at 2 μg/mL of pure amphotericin B. Meanwhile, the addition of ≥1 μg of EVs allowed for robust survival of C. auris at 16 μg/mL of pure amphotericin B, suggesting a ≥16-fold increase in MIC (Figure 3(a)). To expand the significance of our findings, we performed the same experiments on two clinical formulations of amphotericin B. Similar potency was observed when adding C. auris EVs to plates containing amphotericin B deoxycholate, a formulation with increased solubility in water (Figure 3(b)). Most surprisingly, this resistance effect persisted when C. auris supplemented with EVs was treated against amphotericin B encapsulated in hydrogenated soy phosphatidylcholine bilayer liposomes (liposomal amphotericin B, Figure 3(c)), though no difference in MICs was observed between the control and the lowest concentration treatment group (addition of 0.1 μg EVs). Although official breakpoints have not yet been established for C. auris, an MIC ≥2 μg/mL alters the classification of C. auris Cau1901 from “wild-type” to “non-wild-type” against amphotericin B based on the tentative EUCAST epidemiological cut-off value (ECOFF) for C. auris [29] or from sensitive to resistant against this drug according to the tentative breakpoint published by the CDC [30]. On the other hand, when an equivalent protein amount of EVs derived from a commonly studied, amphotericin B-susceptible strain of C. albicans was added to wells containing C. auris and amphotericin B, a drastically reduced increase in MICs was observed (Figure S3). Moreover, the addition of 3 μg of C. auris EVs to C. albicans also did not increase its MICs in any of the formulations of amphotericin B (Figure 3).
Discussion
EVs were isolated and purified from both broth and agar cultures of the newly emerged fungal pathogen C. auris in this study. Given the relative novelty of the agar culture method compared to broth culture, we first assessed whether differences exist in the EVs produced by both methods. In addition to producing comparatively greater yield, decreased labour and material cost, as well as reduced biosafety risk [26], EV collection from colonies grown on a solid matrix may constitute a better physiological model for C. auris as a skin colonizer/pathogen. C. auris EVs were found to be slightly larger than previously reported EVs from other Candida species (100–200 nm) [24] (Figure 1(a)), though differences in preparation, strains, and measuring methods (dynamic light scattering vs NTA) may contribute to this phenomenon. Furthermore, NTA has been found to favour larger sized particles in a heterogenous population [31]. Thus, C. auris EVs fall within expected deviation for typical fungal EVs.
Proteins in C. auris EVs may provide insight on drug resistance-related EV function and possible avenues for immunotherapy. Five proteins were found in abundant quantities in C. auris EVs (Figure 2(b)). Three of them (alcohol dehydrogenase 1, A0A2H0ZTX6, and A0A2H0ZNI5) are suspected to be immunogenic [32, 33]. The two uncharacterized proteins were also identified to be homologues of C. albicans β−1,3-exoglucanase Xog1 and β−1,3-endoglucanase mannoprotein Mp65, respectively, which are major components of the C. albicans secretome. Xog1 is a hyphal-specific virulence factor which has been implicated in transporting glucan across membranes for biofilm formation, and thus may play some role in biofilm-derived drug resistance [33]. In a brief analysis, all of the glycosylation sites and substrate binding domains were found to be conserved among the two proteins. Importantly, C. albicans Xog1 is also the target for host antimicrobial peptide cathelicidin LL-37 which plays a key role in the antimicrobial barrier function in skin and epithelial surfaces [32]. Therefore, the presence of a Xog1-like protein in C. auris EVs may indicate a role for innate immune evasion through the release of EVs and may contribute to the success of C. auris as a skin colonizer. Despite abundant secretion in a soluble protein form and presence in biofilm EVs, however, Xog1 is absent in C. albicans yeast EVs and biofilm cell lysate [25]. In addition, certain deletions and an insertion were detected in C. auris’ A0A2H0ZTX6 when compared with C. albicans’ Xog1. These differentiations may account for the species-specific differences we observed when performing our susceptibility testing. Meanwhile, the presence of a homologue of an immunodominant mannoprotein Mp65 may also indicate the potential for C. auris EVs in immunotherapy. In C. albicans, both the mannosylated Mp65 and recombinant Mp65 without a mannan moiety induce a robust Th1 cytokine pattern in antigen-presenting cells and led to T cell activation [34]. This, combined with observations that mannoproteins and mannans primarily drive the host innate immune response to C. auris, suggested that C. auris EVs could be further investigated for their immunogenicity and vaccine potential.
C. auris EVs modulate amphotericin B susceptibilities in a species-specific manner. In this study, it was demonstrated that EVs from C. auris were able to decrease C. auris sensitivities towards amphotericin B, but not C. albicans (Figure 3). Conversely, C. albicans EVs were unable to affect C. auris MICs to amphotericin B (Figure S3). We hypothesize that this phenomenon may individually or synergistically work in three possible ways. First, C. auris EVs may modulate susceptibility to amphotericin B by drug sequestration or competitive binding. Amphotericin B demonstrates high binding affinity to the cell wall sterol ergosterol. As fungal EV membranes are largely composed of ergosterol [8, 35], it stands to reason that C. auris EVs may competitively bind amphotericin B molecules and reduce bioavailability of the drug, leading to greater fungal survival. However, the same effect of increased amphotericin B resistance was not observed for the addition of C. albicans EVs to C. auris (Figure S3), which was puzzling since membranes of C. albicans EVs are also largely composed of ergosterol [35]. Such an observation suggests that an alternative or supplementary model to this bioavailability theory is needed to explain how C. auris EVs modulate amphotericin B resistance. The second possible way in which C. auris EVs modulate amphotericin B resistance may be their role as a supplementary source of ergosterol to the cell. As suggested by Zarnowski et al., a similar composition of membrane proteins, lipids, and polysaccharides between the source cell and EVs combined with knowledge that EVs can bind cell walls and be internalized in C. albicans suggest that C. auris EVs can act as supplementary membrane material through membrane fusion [19]. The addition of a supplementary source of ergosterol from C. auris EVs may aid to stabilize the membrane by maintaining fluidity and help the fungus survive in the presence of amphotericin B. Indeed, this may also explain why C. auris EVs increased MIC to amphotericin B in C. auris cultures only but not C. albicans as the inter-species incorporation of a different membrane composition is less likely to occur. The presence of structural synthesis enzymes in C. auris EVs may represent the third possible way for how C. auris EVs modulate resistance to amphotericin B. The enrichment of compensatory cell wall remodelling enzymes in EVs from the distantly related model yeast S. cerevisiae such as glucan synthase subunit Fks1 and chitin synthase Chs3 suggests that a similar mechanism may exist for C. auris EVs. Indeed, one of the most highly expressed proteins in our EV samples was a Xog1 homologue (Figure 2(b)), which in C. albicans helps maintain cell wall integrity through β-glucan modifications [33]. Notably, thicker cell walls due to increased β-glucan composition in C. tropicalis have been associated with amphotericin B resistance [36], so structural enzymes present in EVs may act to repair or prevent membrane damage. Further work such as quantitative proteomic analysis on whole EV samples will help elucidate this answer.
Our research suggests that the discrepancies observed between in vitro antifungal susceptibility testing dosages and clinical dosages in C. auris infections can be partly attributed to the release of EVs. By extension, novel drugs targeting EV trafficking such as turbinmicin [37] may find synergistic efficacy when combined with existing antifungal drugs. Further research will be required to explore the potential clinical impact of our findings and whether other antimicrobial threats utilize similar mechanisms to evade drugs.
Supplementary Material
Acknowledgements
We thank the Department of Pathology, HKU for the use of their nanoparticle tracking analysis machine and technical expertise; the Electron Microscope Unit, HKU for their technical support and assistance in sample preparation for TEM; and the Proteomics and Metabolomics Unit, HKU for their technical support and assistance in performing LC–MS.
Funding Statement
This work was partly supported by the General Research Fund, Research Grants Council, University Grants Committee; as well as the framework of the Higher Education Sprout Project by the Ministry of Education (MOE-111-S-023-A) in Taiwan.
Disclosure statement
Patrick Chiu-Yat Woo has provided scientific advisory/laboratory services for Gilead Sciences, Incorporated; International Health Management Associates, Incorporated; Merck & Corporation, Incorporated; Micología Molecular S.L. and Pfizer, Incorporated. The other authors report no conflict of interest. The funding sources had no role in study design, data collection, analysis, interpretation, or writing of the report. The authors alone are responsible for the content and the writing of the manuscript.
Ethics statement
The use of clinical C. auris isolate in this study and leftover human blood specimens was approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW 16-365).
References
- 1.Chaabane F, Graf A, Jequier L, et al. Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front Microbiol. 2019;10:2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satoh K, Makimura K, Hasumi Y, et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Tian S, Han X, et al. Is the superbug fungus really so scary? A systematic review and meta-analysis of global epidemiology and mortality of Candida auris. BMC Infect Dis. 2020;20(1):827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ku TSN, Walraven CJ, Lee SA.. Candida auris: disinfectants and implications for infection control. Front Microbiol. 2018;9:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo PCY, Tsang C-C, Lau SKP.. Antifungal resistance: an emerging battlefield. Future Microbiol. 2020;15(8):571–574. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez JA, Arganoza MT, Boikov D, et al. Stable phenotypic resistance of Candida species to amphotericin B conferred by preexposure to subinhibitory levels of azoles. J Clin Microbiol. 1998;36(9):2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alatoom A, Sartawi M, Lawlor K, et al. Persistent candidemia despite appropriate fungal therapy: first case of Candida auris from the United Arab Emirates. Int J Infect Dis. 2021;70:36–37. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo J, Rodrigues ML, Janbon G.. Extracellular vesicles in fungi: past, present, and future perspectives. Front Cell Infect Microbiol. 2020;10:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paolicelli RC, Bergamini G, Rajendran L.. Cell-to-cell communication by extracellular vesicles: focus on microglia. Neuroscience. 2019;405:148–157. [DOI] [PubMed] [Google Scholar]
- 10.Crewe C, Joffin N, Rutkowski JM, et al. An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell. 2018;175(3):695–708.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow FW-N, Koutsovoulos G, Ovando-Vázquez C, et al. Secretion of an argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res. 2019;47(7):3594–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuipers ME, Hokke CH, Smits HH, et al. Pathogen-derived extracellular vesicle-associated molecules that affect the host immune system: an overview. Front Microbiol. 2018;9:2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Namee NM, O'Driscoll L.. Extracellular vesicles and anti-cancer drug resistance. Biochim Biophys Acta Rev Cancer. 2018;1870(2):123–136. [DOI] [PubMed] [Google Scholar]
- 14.Xavier CPR, Caires HR, Barbosa MAG, et al. The role of extracellular vesicles in the hallmarks of cancer and drug resistance. Cells. 2020;9(5):1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin JS, Kwon SO, Moon DC, et al. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS ONE. 2011;6(2):e17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rumbo C, Fernández-Moreira E, Merino M, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(7):3084–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning AJ, Kuehn MJ.. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni HM, Swamy Ch V, Jagannadham MV.. Molecular characterization and functional analysis of outer membrane vesicles from the antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J Proteome Res. 2014;13(3):1345–1358. [DOI] [PubMed] [Google Scholar]
- 19.Zarnowski R, Sanchez H, Covelli AS, et al. Candida albicans biofilm-induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018;16(10):e2006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao K, Bleackley M, Chisanga D, et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun Biol. 2019;2(1):305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usman WM, Pham TC, Kwok YY, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9(1):2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costanzo MC, Arnaud MB, Skrzypek MS, et al. The Candida Genome Database: facilitating research on Candida albicans molecular biology. FEMS Yeast Res. 2006;6(5):671–684. [DOI] [PubMed] [Google Scholar]
- 23.Arendrup MC, Meletiadis J, Mouton JW, et al. EUCAST Definitive Document E.Def 7.3.2 Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts 2020. Available from: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7.3.2_Yeast_testing_definitive_revised_2020.pdf. [DOI] [PubMed]
- 24.Karkowska-Kuleta J, Kulig K, Karnas E, et al. Characteristics of extracellular vesicles released by the pathogenic yeast-like fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells. 2020;9(7):1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson CS, Garcia-Ceron D, Rajapaksha H, et al. Protein markers for Candida albicans EVs include claudin-like Sur7 family proteins. J Extracell Vesicles. 2020;9(1):1750810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reis FCG, Borges BS, Jozefowicz LJ, et al. A novel protocol for the isolation of fungal extracellular vesicles reveals the participation of a putative scramblase in polysaccharide export and capsule construction in Cryptococcus gattii. mSphere. 2019;4(2):e00080–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1):3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- 28.Muñoz JF, Gade L, Chow NA, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9(1):5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arendrup MC, Prakash A, Meletiadis J, et al. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother. 2017;61(6):e00485–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Centers for Disease Control and Prevention . (2020). Candida auris. Laboratorians and Health Professionals. Antifungal Susceptibility Testing and Interpretation. [2021-10-03]. Available from: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html.
- 31.Krueger AB, Carnell P, Carpenter JF.. Characterization of factors affecting nanoparticle tracking analysis results with synthetic and protein nanoparticles. J Pharm Sci. 2016;105(4):1434–1443. [DOI] [PubMed] [Google Scholar]
- 32.Tsai P-W, Yang C-Y, Chang H-T, et al. Characterizing the role of cell-wall β−1,3-exoglucanase Xog1p in Candida albicans adhesion by the human antimicrobial peptide LL-37. PLoS ONE. 2011;6(6):e21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorgo AG, Heilmann CJ, Brul S, et al. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol Lett. 2013;338(1):10–17. [DOI] [PubMed] [Google Scholar]
- 34.Pietrella D, Lupo P, Rachini A, et al. A Candida albicans mannoprotein deprived of its mannan moiety is efficiently taken up and processed by human dendritic cells and induces T-cell activation without stimulating proinflammatory cytokine production. Infect Immun. 2008;76(9):4359–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas G, Honorato L, Guimarães AJ, et al. Protective effect of fungal extracellular vesicles against murine candidiasis. Cell Microbiol. 2020;22(10):e13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesa-Arango AC, Rueda C, Román E, et al. Cell wall changes in amphotericin B-resistant strains from Candida tropicalis and relationship with the immune responses elicited by the host. Antimicrob Agents Chemother. 2016;60(4):2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao M, Zhang F, Zarnowski R, et al. Turbinmicin inhibits Candida biofilm growth by disrupting fungal vesicle–mediated trafficking. J Clin Invest. 2021;131(5):e145123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.