Abstract
ς54 is the subunit of bacterial RNA polymerase that transcribes from promoters with enhancer elements bound by enhancer-binding proteins. By computer searches of Helicobacter pylori genomic sequences, chromosomal gene disruption, and RNA analyses, we have identified ς54-recognized promoters that regulate transcription of flagellar basal body and hook genes, as well as the enhancer-binding protein FlgR (flagellum regulator), a transactivating protein of the NtrC family. We demonstrate that FlgR is required for bacterial motility and transcription of five promoters for seven basal body and hook genes. In addition, FlgR acts as a repressor of transcription of the ς28-regulated flaA flagellin gene promoter, while changes in DNA topology repress transcription of the ς54-regulated flaB flagellin gene promoter. Our data indicate that regulation of flagellar gene expression in H. pylori shows similarities with that in enterobacteriaceae and Caulobacter.
Helicobacter pylori is a curved, microaerophilic, gram-negative bacterium which was first described by Warren and Marshall (35). This organism is a human gastric pathogen associated with peptic ulcer disease as well as chronic gastritis (5, 10), which may predispose to gastric cancer (25, 27). Several bacterial factors, including urease (9), the cytotoxin-associated protein (7, 34), the vacuolating toxin (8, 32), and flagellins (23, 14, 30), have been suggested to play a role in pathogenesis. In addition, motility has been shown to constitute an essential colonization factor for H. pylori, in that nonmotile mutants are unable to establish a persistent infection in the gnotobiotic piglet model (11).
H. pylori cells normally possess from two to six polar sheathed flagella, which enable the bacterium to move in the highly viscous mucous layer of the gastric epithelium. As with enteric bacteria, the H. pylori flagella are composed of three structural elements: a basal body which serves as a cell anchor and contains the proteins required for rotation and chemotaxis, a curved hook structure, and the helically shaped flagellar filament. Thus far, only a few of the numerous proteins involved in the formation of this complex structure have been characterized in some detail; these include the components of the flagellar filament FlaA (23) and FlaB (30), which are expressed by cultured cells to very different levels, FlaA being the predominant subtype. In contrast to the situation in the closely related organism Campylobacter coli, where both flagellin genes are contiguous and encode proteins of almost identical amino acid sequence, the H. pylori genes are unlinked on the chromosome and share only limited acid sequence similarity (33). Studies with isogenic H. pylori mutants defective in either FlaA or FlaB have demonstrated that both flagellins are necessary for full motility (17). Furthermore, the presence of both flagellins is required for the establishment of a persistent infection in the gnotobiotic piglet model (11).
Available data suggest that expression of the two flagellins in H. pylori is regulated by different sigma factors (ς54 in the case of flaB and ς28 in the case of flaA). Although only a few experimental data are available, it seems likely that both genes can be regulated differentially by environmental conditions. In support of this hypothesis is the finding that flaB expression in the related organism C. coli can be modulated by certain growth conditions (1). A surprising peculiarity of the H. pylori system is the fact that mutations in the flagellar hook protein FlgE do not prevent the synthesis of either FlaA or FlaB (26). This finding is in sharp contrast with the situation in enteric bacteria, where the flagellin can be synthesized only when the assembly of the basal body-hook complex is completed. Interestingly, like flaB, the flgE gene is preceded by a consensus sequence similar to a ς54-recognized promoter. This finding may suggest that both genes are regulated coordinately by the same factors and may constitute members of a yet unknown larger regulon of flagellar structural and biosynthetic genes.
In this work, we identified a series of basal body and hook genes as members of a regulon and provide evidence that these genes are all regulated by the same master transcriptional factor, the NtrC homolog FlgR. This is the first direct demonstration of transcriptional control in H. pylori flagellar gene expression and is believed to be a basis for further investigations, which should lead to an elucidation of the regulatory hierarchies that govern flagellum synthesis in H. pylori.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli DH5α was used for cloning and plasmid preparations. H. pylori G27 cells were recovered from frozen stocks on Columbia agar plates containing 5% horse blood, 0.2% cyclodextrin, and Dent’s or Skirrow’s antibiotic supplement under microaerophilic conditions (Oxoid) for 2 to 3 days. After passage on fresh plates, bacteria were cultured in a 5% CO2–95% air atmosphere. Liquid cultures of H. pylori were grown in modified brucella broth containing Dent’s or Skirrow’s antibiotic supplement and 5% fetal calf serum. When required, kanamycin or novobiocin was added to a final concentration of 25 or 100 μg/ml, respectively. E. coli DH5α was cultured in Luria-Bertani medium. Natural transformation of H. pylori G27 was carried out by adding 1 to 5 μg of plasmid DNA to a spot of fresh bacteria which had been incubated for 5 h at 37°C. After overnight incubation at 37°C, bacteria were collected and streaked on selective agar plates. Single colonies were then selected for further analysis. Motility of H. pylori strains was assayed by stab-inoculating bacteria with a pipette tip into 0.3% agar plates containing brucella broth supplemented with 10% fetal calf serum and Dent’s or Skirrow’s antibiotic supplement.
DNA techniques.
DNA manipulations were carried out by general techniques as described by Sambrook et al. (28). Mid-scale plasmid preparations were carried out with a Qiagen Midi column plasmid purification kit (Qiagen Inc.). DNA fragments or PCR amplification products for cloning purposes were purified from agarose gels with a QiaEX DNA purification kit (Qiagen Inc.). For Southern blot analyses, the Amersham ECL direct nucleic acid labeling and detection system was used. PCRs were performed in a Perkin-Elmer 2400 thermal cycler with Taq DNA polymerase (Boehringer Mannheim). In each reaction, 100 ng of H. pylori G27 chromosomal DNA was mixed with 100 pmol of each specific primer in a 100-μl sample containing standard concentrations of deoxynucleotides and MgCl2 (Boehringer Mannheim). Reactions were performed by denaturing DNA at 94°C for 5 min, annealing at appropriate temperatures calculated on the basis of the melting temperature of the oligonucleotides used for 1 min, and extending at 72°C for 1 min (30 cycles). PCR-amplified promoter fragments were sequenced according to the dideoxy-chain termination method using either [α-33P]dATP (Amersham) and a T7 sequencing kit (Pharmacia) or an Applied Biosystems 373 automated DNA sequencer.
Construction of an isogenic flgR mutant of H. pylori G27 by allelic exchange mutagenesis.
An isogenic flgR mutant was obtained by transforming H. pylori G27 with a pGEM3 vector (Promega) carrying the C. coli kanamycin cassette (22) flanked by a 403-bp and a 412-bp fragment obtained by PCR performed on H. pylori chromosomal DNA with primer pairs ntr1-ntr2 and ntr8-ntr9, respectively. The resulting mutant had bp 49 to 585 of the flgR coding sequence replaced by the kanamycin cassette.
Cloning and sequencing of promoter fragments.
Promoter sequences were PCR amplified from chromosomal DNA of H. pylori G27 by using primer pairs gyr1-gyr2 (orf698-orf699-dgkA-gyrA-orf702-flgR), fla1-fla2 (flaB-topA), flgE3-flgE4 (orf906-flgD-flgE′), flgE5-flgE6 (flgE), flgB1-flgB2 (flgB-flgC), and flgK1-flgK2 (orf1120-flgK). PCR fragments were then cloned into pGEM3 (Promega) and sequenced.
RNA preparation.
Total H. pylori RNA was extracted as described previously (29). Briefly, 25 ml of H. pylori grown in modified brucella broth at 37°C was harvested at an optical density at 590 nm of 1.0 and stored at −20°C. Cells were lysed in 3.7 ml of 100 mM Tris-HCl (pH 7.5)–2 mM Na2EDTA–1% sodium dodecyl sulfate for 5 min at 95°C. After 10 min of incubation on ice in the presence of 80 mM KCl, cellular debris was removed by centrifugation at 8,000 rpm for 10 min in a JA20 rotor. Then 4.56 g of CsCl was added to 3.5 ml of supernatant, and the RNA was sedimented by centrifugation in an SW65 rotor for 15 to 20 h at 35,000 rpm. The RNA pellet was then resuspended in 500 μl of TE (10 mM Tris-HCl [pH 8], 1 mM Na2EDTA), extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, resuspended in 200 μl of TE, reprecipitated, and stored at −20°C.
Primer extension analysis.
Three picomoles of oligonucleotide was 5′ end labeled in the presence of [γ-32P]ATP (5,000 Ci/mmol; Amersham) and T4 polynucleotide kinase (New England Biolabs). Labeled oligonucleotide (0.4 to 1.0 pmol) was then coprecipitated with 15 μg of H. pylori total RNA and resuspended in a mixture of 5 μl of H2O, 2 μl of 2 mM deoxynucleoside triphosphates, and 2 μl of 5× reverse transcription buffer (cDNA synthesis kit; Boehringer Mannheim). The reaction mixture was incubated for 1 min at 95°C, 1 μl of reverse transcriptase (20 U/μl; Boehringer Mannheim) was added, and reverse transcription was carried out by incubation at 45°C for 45 min. The sample was then incubated for 10 min at room temperature with 1 μl of RNase A (1 mg/ml) for RNA digestion, extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, and resuspended in 6 μl of sequencing loading buffer. After denaturation at 95°C for 2 min, samples were subjected to 6 or 10% urea-polyacrylamide gel electrophoresis and autoradiographed. For quantitative analysis of the bands, the same gels were then exposed to a Multiscan PhosphorImager (Molecular Dynamics).
RESULTS
Identification of flagellar genes transcribed by ς54 promoters.
Analysis of the complete genome sequence of H. pylori has revealed the presence of at least 20 flagellar structural genes (33). These genes are distributed throughout the genome, and some are organized in operon-like structures. In an attempt to identify ς54-regulated loci among the 20 flagellar genes, we searched for the presence of a ς54 core consensus sequence (GGN10GC) within their 5′ untranslated regions. The analysis showed the presence of this consensus sequence upstream of five operons encoding seven flagellar genes. Figure 1A shows the structural organization of these operons.
FIG. 1.
Structural organization of the indicated genes and operons. Dark gray bars indicate flagellar genes; open boxes indicate ORFs of unknown functions and are marked as reported by Tomb and coworkers (33); light gray boxes represent regulatory genes. The kanamycin cassette is indicated by boxes with vertical bars. (A) Genes transcribed by ς54-regulated promoters. flgD and flgE′ encode proteins with 25.5 and 30.5% amino acid identity to the flagellar hook assembly protein FlgD of Salmonella choleraesuis (18) and the structural hook protein FlgE of H. pylori (26), respectively. flgB and flgC constitute an operon encoding two proteins with 31 and 46% amino acid identity to the proximal flagellar rod proteins FlgB of S. choleraesuis (16) and FlgC of Borrelia burgdorferi (13), respectively. flgK encodes for a protein with 27.6% amino acid identity to the flagellar hook-associated protein FlgK (HAP1) of S. choleraesuis (15). (B) Construction of the G27[flgR] mutant strain. To study the regulatory protein involved in transcriptional activation of the putative ς54-dependent genes, we searched for NtrC homologs in the genome sequence of H. pylori (33). One protein (HP703) with significant amino acid sequence homology to a series of NtrC-like regulators was detected. The highest degree of amino acid similarity (44.2% identity, 69.1% similarity) was with FleQ, an activator of mucin adhesion and flagellum expression in Pseudomonas aeruginosa (3).
Besides flaB, for which an RNA 5′ end has been mapped downstream from a ς54 consensus promoter sequence (30), and flgE, for which a ς54 consensus sequence has been noted (26), we identified three additional putative operons. The first operon consists of an open reading frame (ORF) of unknown function (orf906), one gene, flgE, encoding the flagellar hook assembly protein FlgD, and one gene encoding a homolog of the structural hook protein FlgE (33). The second operon is constituted of two genes encoding the proximal flagellar rod proteins FlgB and FlgC. The third operon contains one ORF of unknown function (orf1120) and the gene encoding the flagellar hook-associated protein FlgK. It is likely that five putative ς54-recognized promoters regulate expression of seven basal body and hook genes of H. pylori.
FlgR, an NtrC homolog, is required for bacterial motility.
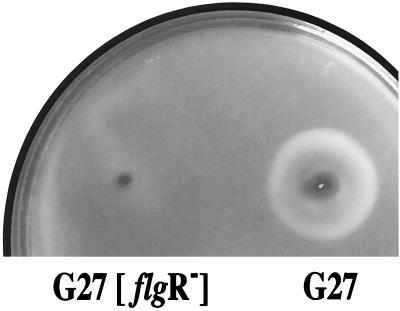
In all cases described thus far, transcription from ς54-recognized promoters is dependent on distant upstream (>100 bp) enhancer-like sequences which are bound by a class of transactivating proteins, usually referred to as the NtrC (NR-I) family (20). Therefore, we selected from the putative transcriptional regulators of H. pylori that one with the highest amino acid homology to this family of proteins to study transcription from the putative ς54 promoters. Interestingly, the selected protein (named HP703 in reference 33) shows also a high degree of similarity to proteins involved in regulation of flagellar biosynthesis. These include FlrC (43.0% identity in a 386-amino-acid overlap) of Vibrio cholerae and FlbD (35.3% identity in a 377-amino-acid overlap) of Caulobacter crescentus. As a preliminary test for the role of this putative regulator in transcriptional activation of flagellar genes, we constructed an HP703 isogenic mutant of H. pylori G27 and assayed for bacterial motility. The DNA sequence corresponding to the N-terminal part of the HP703 proteins was substituted with a kanamycin resistance gene (Fig. 1B). Correct replacement of the wild-type sequence with the kanamycin cassette was assessed by means of PCR, using oligonucleotide pair ntr1-ntr9 (Table 1), complementary to regions flanking the insertion site, and by Southern blot analysis using HaeIII-digested chromosomal DNAs of the mutant and wild-type strains, respectively (data not shown). Bacterial motility was investigated by assaying the ability of the cells to spread on soft agar plates. Figure 2 shows that the area of spreading of the HP703 knockout (FlgR−) mutant strain (see below) is much less than the area covered by the wild-type strain. In contrast, this mutant strain grew similarly to the previously characterized nonmotile CheY mutant strain (reference 4 and data not shown), thus showing a loss of motility functions.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′ to 3′)a | Restriction recognition site | Strand | Used to map transcription start upstream of: | Nucleotide positionb |
|---|---|---|---|---|---|
| gyrN | CCAATAACCACCATCCAAG | − | orf698 | 751122–751104 | |
| flaB | GCATGAGAAGTTAAAGCGGC | + | flaB | 124446–124465 | |
| flgE | GACACCAGACCATAAAGACC | + | flgE | 922547–922566 | |
| flgE2 | GGATTAATGGGAGATGGCATG | − | orf906 | 956504–956484 | |
| flgB | AAGACCGATAATCCAACGCC | + | flgB | 1641322–1641341 | |
| flgK | GATGTCTCTTATATCGCGCTCGG | + | orf1120 | 1186790–1186812 | |
| flaA | CGCATTGATATTTGTATTGACCTG | + | flaA | 637268–637291 | |
| cagN | GTCAATGGTTTCGTTAGTC | − | cagA | 579941–579923 | |
| ureA | CATAGTGGAGCATCAAC | + | ureA | 77911–77927 | |
| ntrY | GAATGAAAAGAAACGCATCACTC | − | orf702 | 755012–754990 | |
| gyrA | CTGAATTATCTTGCATGTGTC | − | gyrA | 752527–752507 | |
| ntrN | ATCATCTTCTACAATGGCG | − | flgR | 755491–755473 | |
| ntr1 | aatcgtgaattcTAGAGTGATGGGCGAAG | EcoRI | + | 755102–755130 | |
| ntr2 | aaaaaaggatccAGGCTTTTACGCATG | BamHI | − | 755523–755497 | |
| ntr8 | tgtccgggatccCAGAGCATCTATTAGAAAAGCGAG | BamHI | + | 756042–756076 | |
| ntr9 | tatgcactgcagCCCAAATGATACGCATCGCACAC | PstI | − | 756437–756403 | |
| flgE3 | attataggatccGCTATTCAAAGCGTTTGCGTTGG | BamHI | − | 956559–956525 | |
| flgE4 | attattgaattcTGTTCTCATTAAGCGCGAATAACG | EcoRI | + | 956298–956333 | |
| flgE5 | GCGATTTGGTGGGCTTggatccATTGACACCAGAC | BamHI | + | 922521–922556 | |
| flgE6 | CTAAAGCGAGTTgaattcTTAAGCTTGAGCGATAAC | EcoRI | − | 922784–922749 | |
| flgB1 | AAAGGGggatccACATTAGCGATGTTAGAAG | BamHI | + | 1641273–1641303 | |
| flgB2 | AATGGCTCTgaattcGCTTATCGCTCAAGC | EcoRI | − | 1641514–1641485 | |
| flgK1 | attattgaatcAAAAGCTTGAATCGCTAGCTG | EcoRI | + | 1186713–1186745 | |
| flgK2 | attattggatccCAAGCGGGGAATGCGATGAGC | BamHI | − | 1187086–1187054 | |
| fla1 | attataggatccGCATGAGAAGTTAAAGCGGCG | BamHI | + | 124434–122466 | |
| fla2 | attatagaattcCCTAACATGCCCTTTAGAGGC | EcoRI | − | 124771–124739 | |
| gyr1 | tatttaggatccCCAATAACCACCATCCAAGACATG | BamHI | − | 751134–751099 | |
| gyr2 | attattgaattccGATTGGCTAGGCATACAGCCCCCAG | EcoRI | + | 750711–750746 |
Capital letters, H. pylori-derived sequences; lowercase letters, sequences added for cloning purposes; underlined letters, restriction recognition sites.
According to the genome sequence published by Tomb et al. (33).
FIG. 2.
Bacterial motility assay. The indicated strains were stabbed into semisolid agar medium and incubated at 37°C for 72 h.
It is possible that the NtrC-homologous protein is the specific master activator of the hypothetical ς54-regulated flagellar genes. Therefore, we propose to name this protein FlgR (for flagellum regulatory protein).
Transcriptional analysis of the flgR gene.
Figure 1B shows that the flgR gene is located at the 3′ end of a putative operon which contains the gene encoding diacylglycerol kinase (dgkA), the gene encoding the subunit A of DNA gyrase (gyrA), and three ORFs of unknown function (orf698, orf699, and orf702). Unexpectedly, no cognate sensor kinase could be detected in this operon. To assess the transcriptional regulation of FlgR expression, we designed a series of oligonucleotides complementary to the 5′ ends of the coding sequences of flgR, orf702, gyrA, and orf699 (Table 1) and performed primer extension analysis of total RNA extracted from H. pylori G27. While reverse transcription of primers complementary to flgR, orf702, and gyrA yielded numerous bands migrating to different positions in the gel (data not shown), extension of the orf699-specific primer revealed one major band of high molecular weight, indicating the presence of a promoter upstream of orf698 (Fig. 1B). Cloning and sequencing of the 5′ region of orf698 from strain G27 allowed us to map the transcription start site of the operon at 18 nucleotides (nt) upstream of the ATG start codon of translation. A similar primer extension experiment using total RNA extracted from the FlgR− mutant strain yielded the same extension product in comparable amounts. Nucleotide sequence analysis upstream of the transcription start site revealed the presence of a −10 region with a perfect match to the E. coli ς70 consensus sequence (TATAAT), an AT-rich region (86% AT) spanning positions −18 to −60, and no consensus for a −35 region.
Our data suggest that the flgR gene is included in an operon with five other genes and that it is regulated by a ς70-like promoter in both wild-type and FlgR− mutant strains.
FlgR is the master regulator of basal body and hook genes transcribed by ς54 promoters.
To define the start point of transcription of the hypothetical ς54-regulated flaB, flgE, orf906-flgDE′, flgBC, and orf1120-flgK genes, we carried out primer extension assays of total H. pylori RNA. The oligonucleotides complementary to these coding sequences are shown in Table 1. To ensure correct mapping of the extension products, we cloned and sequenced in parallel the respective promoter regions of these loci from the chromosomal DNA of H. pylori G27.
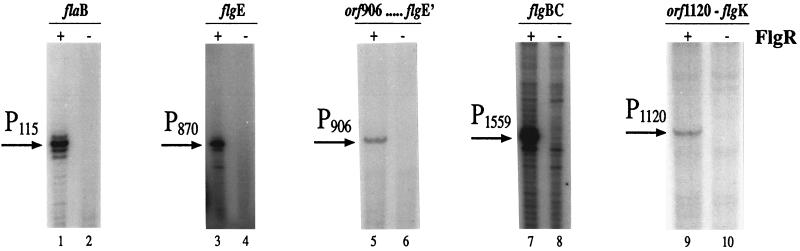
The urea-acrylamide gels in Fig. 3 show the results of primer extension experiments carried out by hybridizing each oligonucleotide to total RNA extracted from H. pylori. Each reverse transcription of RNA shows a major band, P, which defines the indicated start site of RNA transcription (lanes 1, 3, 5, 7, and 9). In the case of flaB, the experiment located the start site of RNA transcription at 25 nt upstream from the ATG start codon, which defines the P115 promoter. Transcription of flgE, directed by promoter P870, started at 30 nt upstream of the ATG start codon, while orf906-flgDE′, flgBC, and orf1120-flgK transcripts mapped at 33, 22, and 24 nt upstream of the respective ATG start codons, defining promoters P906, P1559, and P1120, respectively.
FIG. 3.
Primer extension analyses of H. pylori RNAs. The elongated primers are indicated by arrows and labeled “P.” Subscript numbers refer to the downstream ORF as designated by Tomb et al. (33). Total RNA was extracted from wild-type H. pylori G27 (lanes +) or from the flgR isogenic mutant (lanes −), annealed to specific primers, and elongated with reverse transcriptase as detailed in Materials and Methods.
To demonstrate that FlgR is the transcriptional regulator of the basal body and hook genes, we extracted total H. pylori RNA from liquid cultures of the G27[flgR] mutant and carried out primer extension analyses. As shown in Fig. 3 (lanes 2, 4, 6, 8, and 10), a primer extension product could not be detected for either of the ς54-dependent flagellar genes, thus demonstrating the positive role of FlgR on transcription of these genes.
Nucleotide sequences of these promoters were aligned with respect to the initiation site of transcription (Fig. 4). As expected, conserved sequences at positions −12 (GC) and −24 (GG) of the core consensus of ς54-regulated promoters are found. In addition, this core consensus could be extended to conserved nucleotides both upstream and downstream from the core consensus.
FIG. 4.
Nucleotide sequences of H. pylori ς54 promoters. Shown is alignment of the sequence from +1 to −60 of the indicated promoter genes and a derived consensus sequence. The promoter region is shaded; identical bases are shown in boldface.
These data suggest that each of the analyzed genes or operons is transcribed from a single promoter recognized by the sigma factor ς54.
FlgR represses transcription of flaA.
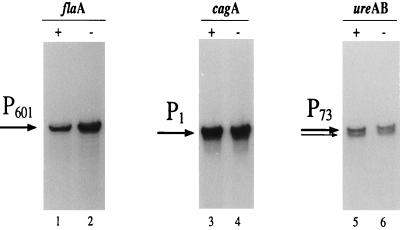
To assess the possible influence of FlgR on flaA transcription, which is regulated by the alternative signal factor ς28 (23), we hybridized total RNA extracted from H. pylori G27 and from the isogenic FlgR− mutant strain to an oligonucleotide specific for flaA (Table 1). The primer extension experiment shown in Fig. 5 (lanes 1 and 2) shows a typical result. In both cases, one major band (P601) which corresponds to the ς28-dependent flaA transcription start site at 50 nt upstream of the flaA ATG start codon reported by Leying et al. (23) was detected. Surprisingly, the intensity of the P601 band was higher in the mutant strain (lane 2) than in the wild-type strain (lane 1), suggesting that the P601 RNA is increased in the FlgR− mutant strain. To assess the difference in the amount of RNA, we evaluated the relative intensities of the bands by exposure of the same gel to a PhosphorImager. The evaluation revealed a twofold increase in the amount of transcript in the mutant strain, suggesting that FlgR exerts a negative feedback on FlaA expression by specifically repressing transcription of the flaA gene. In contrast, primer extension analysis of the cagA gene, which was demonstrated to be regulated by the ς70-dependent P1 promoter (29), showed no difference in transcript size and/or amount between the wild-type and mutant strains (Fig. 5, lanes 3 and 4). Similarly, expression of the ureAB operon encoding the two subunits of the H. pylori urease enzyme was not altered in the mutant strain (Fig. 5, lanes 5 and 6). Transcription of this operon was initiated at the P73 promoter, starting at two C nucleotides mapping 56 and 57 nt upstream of the ureA ATG start codon. Seven nucleotides upstream of the transcription start, a ς70 −10 extended-like promoter (TGCTACAAT) with only one mismatch with respect to the E. coli consensus sequence (TGNTATAAT) was detected (19), indicating ς70-regulated transcription of ureAB rather than ς54-dependent expression as proposed previously (21).
FIG. 5.
Transcriptional regulation of flaA by FlgR. The RNA used was from the same preparation as that used in the experiment shown in Fig. 3. Symbols are as in Fig. 3. Control RNAs, cagA and ureAB.
Transcription of the flaB gene is sensitive to DNA topology.
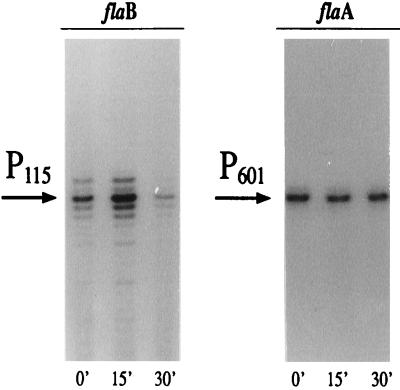
Since the direction of transcription of the flaB gene is opposite that of the topA gene (Fig. 1A), the two promoters result in overlapping and divergent configurations (31). To assess whether the flaB promoter is sensitive to DNA topology, we incubated liquid cultures of H. pylori G27 with the DNA gyrase inhibitor novobiocin and assayed the accumulation of transcripts at this promoter by primer extension analyses. Figure 6 shows a primer extension experiment carried out with specific oligonucleotides on RNA extracted before and after addition of novobiocin to the culture medium. Following addition of novobiocin, the amount of flaB transcript (P115) changed with time (lanes 0’, 15’, and 30’). The P115 transcript increased after 15 min of treatment and decreased after 30 min of treatment. This suggests an immediate positive response to the perturbation followed by a negative response. In contrast, the amount of flaA transcript (P601) was not changed by the addition of novobiocin to the culture medium (Fig. 5), thus confirming a specific response of promoter P115 to novobiocin treatment.
FIG. 6.
Novobiocin affects transcription of flaB but not flaA. Total RNA extracted from H. pylori G27 before addition of novobiocin (time zero [lane 0’]) and after 15 and 30 min (lanes 15’ and 30’).
It is likely that transcription from the P115 (flaB) promoter is sensitive to changes in DNA topology, suggesting a complex mechanism of regulation of the flaB and topA genes.
DISCUSSION
Motility has been shown to be a key factor for the ability of H. pylori to colonize the gastric mucosa (11). While a few structural components of the flagella (23, 30) and the flagellar motor switch protein CheY (4) have been characterized in some detail, little is known about factors that regulate expression of genes involved in motility and chemotaxis.
We have identified the flagellar regulon of H. pylori, whose transcription is under the positive control of the alternative sigma factor ς54 and the transcriptional activator FlgR. A line of evidence demonstrating that FlgR and ς54 regulate the basal body and hook genes is based on the finding that ς54 is unable to activate transcription of the regulon in a FlgR− background. Conversely, flaA, the gene encoding the major flagellin, is regulated by the alternative sigma factor ς28 (23). Interestingly, transcription from the flaA promoter (P601) is enhanced in the FlgR− background, suggesting a negative feedback exerted by FlgR on transcription of this ς28-regulated promoter (Fig. 5). This, in turn, implies that in the context of flagellar gene expression, the efficiency of ς28-dependent transcription is dependent on ς54 holoenzyme transcription. Since transcription of the two flagellin genes flaA and flaB is regulated by two different ς factors (ς28 and ς54, respectively), these genes may be differentially expressed, depending on environmental conditions. This regulation may enable H. pylori to produce flagella which are particularly suited for motility within a given environment (high-viscosity mucus, low or high osmolarity or pH, etc.) by changing flagellar parameters.
Transcription from the flaA promoter was unchanged by treatment of the cells with novobiocin, an inhibitor of bacterial gyrases, while transcription from the flaB promoter changed with time after drug treatment (Fig. 6). Following addition of novobiocin to the culture medium, transcription of the flaB gene was first enhanced and subsequently reduced, thus suggesting that a perturbation of the local density of DNA supercoil may modulate transcription of the flaB gene. This is in agreement with the features highlighted by studies of the NtrC protein from E. coli (6). In those studies, it was reported that the binding site of NtrC can be substituted by a DNA element possessing an intrinsic supercoil structure. Consequently, expression of flagellar genes may depend on the supercoil state of some promoters, which in turn may depend on environmental conditions. In this context, it should be noted that flgR is cotranscribed with gyrA, the gene encoding subunit A of DNA gyrase, and that both genes are coregulated by a single promoter (Fig. 1B). Therefore, it is tempting to speculate that changes in the expression of genes involved in regulation of DNA supercoiling may coordinately change expression of the flagellar genes, and vice versa. In support of such a hypothesis is also the finding that the FlgR-regulated flaB promoter overlaps the divergently transcribing promoter of topA (31), the gene encoding topoisomerase I. This may suggest a coordinate regulation of both genes in response to the same environmental stimuli.
Regulation of motility and chemotaxis has been extensively studied in E. coli, Salmonella spp., and Caulobacter crescentus (reviewed in references 2, 12, 24, and 36). In these organisms, the genes required for flagellar biosynthesis are sequentially expressed according to a hierarchical pathway. In enterobacteriaceae, three classes of flagellar biosynthetic genes have been identified. Environmental signals trigger expression of class I (early) genes, which encode two master regulatory proteins. These regulatory proteins are required for Eς70-dependent expression of class II (middle) genes, which encode the components of the basal body and the hook. Assembly of the basal body-hook complex in turn acts as signal for Eς28-regulated expression of class III (late) genes, including the genes for chemoreception and the flagellin gene, by allowing the release of an anti-sigma factor protein (36). In contrast, four classes of flagellar biosynthetic genes have been identified in Caulobacter (36). An early signal in the cell cycle is assumed to trigger activation of the class I gene product, a transcriptional regulator, which in turn activates Eς70-dependent transcription of class II genes, encoding structural components of the MS ring-switch complex. Among class II genes are also the alternative ς factor ς54 and its cognate transcriptional regulator (FlbD) which activate expression of class III and class IV genes, encoding structural proteins of the basal body-hook complex and flagellar filament, respectively. In contrast to the situation in enterobacteriaceae, two different checkpoints in the assembly of the flagellar structure control activation of these late flagellar genes. Completion of the MS ring-switch complex acts as a signal for transcription of class III genes, while completion of the basal body-hook complex is required for activation of class IV genes.
Our data suggest that the transcriptional hierarchy that governs flagellar synthesis in H. pylori has similarities to both systems. As in Caulobacter, Eς54 and a cognate transcriptional activator (FlgR) are required for transcription of genes coding for structural proteins of the basal body and hook. In contrast, expression of the major flagellin (FlaA) appears to be regulated by Eς28, thus reflecting the situation observed in enterobacteriaceae. In contrast to both systems, no checkpoints in flagellar assembly that regulate transcription of flagellin genes are obvious in H. pylori. In fact, disruption of the flgR gene, and the resulting defect in expression of basal body and hook genes, does not prevent FlaA synthesis. Thus, it seems unlikely that an anti-sigma factor protein blocks transcription of ς28-dependent genes in the absence of a functional basal body-hook complex in H. pylori. In support of this hypothesis, analysis of the complete genome sequence has revealed no genes coding for proteins with significant homology to an anti-sigma factor protein (33). Nevertheless, FlgR exerts a slightly similar effect by down-modulating transcription of the flaA gene (Fig. 5).
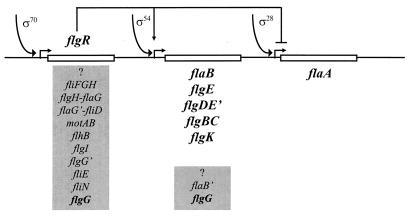
Transcription of the flgR gene is under the control of a ς70-dependent promoter (Fig. 1B). In fact, the structure of this promoter resembles the structure of the P1 promoter of cagA (29), which is recognized by ς70 and regulated through the interaction of the AT-rich upstream element with the α subunit of RNA polymerase. A search for such promoters upstream of the H. pylori flagellar genes revealed the presence of ς70 consensus sequences upstream of nine operons (Fig. 7). For one of these, the flgG homolog gene (HP1585 in reference 33), a putative ς54 promoter was also detected, suggesting a possible coordinate regulation of this gene by means of two alternative overlapping promoters. A putative ς54 promoter was also detected upstream of the flaB′ gene encoding a homolog of the FlaB flagellin (HP295 in reference 33). Based on these observations and taking into account our data, we propose a model for the regulation of flagellar gene expression in H. pylori (Fig. 7). In this model, Eς70 directs transcription of the master regulator FlgR and genes coding for structural components of the flagellar export apparatus, motor, and basal body. FlgR in turn activates transcription of ς54-dependent genes encoding structural components of the basal body-hook complex and represses transcription of the ς28-dependent gene encoding the major flagellin subunit FlaA.
FIG. 7.
Proposed model for flagellar gene expression in H. pylori. Dependence of transcription on the specified ς factors is shown for genes and operons indicated in boldface. Genes and operons whose promoters have been deduced on the basis of nucleotide sequence analysis are enclosed by gray boxes.
ACKNOWLEDGMENTS
We thank Gary Jennings and Isabel Delany for critical reading of the manuscript, C. Mallia for editing, and G. Corsi for the figures.
G.S. was partially supported by a fellowship of the Gottlieb-Daimler und Karl-Benz-Stiftung. This work was supported in part by grant FMRX-CT98-0164 from the European Union.
REFERENCES
- 1.Alm R A, Guerry P, Trust T J. The Campylobacter ς54flaB flagellin promoter is subject to environmental regulation. J Bacteriol. 1993;175:4448–4455. doi: 10.1128/jb.175.14.4448-4455.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsler C D, Matsumura P. Chemotactic signal transduction in Escherichia coli and Salmonella typhimurium. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 89–103. [Google Scholar]
- 3.Arora S K, Ritchings B W, Almira E A, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier D, Spohn G, Rappuoli R, Scarlato V. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol. 1997;179:4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser M J. Gastric Campylobacter-like organisms, gastritis and peptic ulcer disease. Gastroenterology. 1987;93:371–382. doi: 10.1016/0016-5085(87)91028-6. [DOI] [PubMed] [Google Scholar]
- 6.Brahms G, Brahms S, Magasanik B. A sequence-induced superhelical DNA segment serves as transcriptional enhancer. J Mol Biol. 1995;246:35–42. doi: 10.1016/s0022-2836(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 7.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10556–10573. [PubMed] [Google Scholar]
- 9.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick J D. Helicobacter (Campylobacter) pylori: a twist on an old disease. Annu Rev Microbiol. 1990;108:70–90. doi: 10.1146/annurev.mi.44.100190.001341. [DOI] [PubMed] [Google Scholar]
- 11.Eaton K A, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbach M. Control of chemotaxis. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 13.Ge Y, Old I G, Saint Girons I, Charon N W. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus ς70 promoter. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas R, Meyer T F, van Putten J P M. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using shuttle mutagenesis. Mol Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 15.Homma M, DeRosier D J, Macnab R M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 16.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab R M. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 17.Josenhans C, Labigne A, Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J Bacteriol. 1995;177:3010–3020. doi: 10.1128/jb.177.11.3010-3020.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsukake K, Doi H. Nucleotide sequence of the flgD gene of Salmonella typhimurium which is essential for flagellar hook formation. Biochim Biophys Acta. 1994;1218:443–446. doi: 10.1016/0167-4781(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35 recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 20.Kustu S, North K, Weiss D. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 21.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leying H, Suerbaum S, Geis G, Haas R. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol Microbiol. 1992;6:2863–2874. doi: 10.1111/j.1365-2958.1992.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 24.Macnab R B. Flagellar switch. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 181–199. [Google Scholar]
- 25.Nomura A, Stemmermann G N, Chyou P-H, Kato I, Perez-Perez G I, Blaser M J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 26.O’Toole P W, Kostrzynska M, Trust T J. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol Microbiol. 1994;14:691–703. doi: 10.1111/j.1365-2958.1994.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 27.Parsonnet J, Friedman G D, Vandersteen D P, Cgang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and risk of gastric carcinoma. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Spohn G, Beier D, Rappuoli R, Scarlato V. Transcriptional analysis of the divergent cagAB genes encoded by the pathogenicity island of Helicobacter pylori. Mol Microbiol. 1997;26:361–372. doi: 10.1046/j.1365-2958.1997.5831949.x. [DOI] [PubMed] [Google Scholar]
- 30.Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J Bacteriol. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suerbaum S, Brauer-Steppkes T, Labigne A, Cameron B, Drilica K. Topoisomerase I of Helicobacter pylori: juxtaposition with a flagellin gene and functional requirement of a zinc finger motif. Gene. 1998;210:151–161. doi: 10.1016/s0378-1119(98)00065-1. [DOI] [PubMed] [Google Scholar]
- 32.Telford J L, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Censini S, Covacci A, Xiang Z, Papini E, Montecucco C, Parente L, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerland L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.Tummuru M K R, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren J R, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;i:1273–1275. [PubMed] [Google Scholar]
- 36.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy: not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]