Abstract
Structural variants (SVs) can affect protein-coding sequences as well as gene regulatory elements. However, SVs disrupting protein-coding sequences that also function as cis-regulatory elements remain largely uncharacterized. Here, we show that craniosynostosis patients with SVs containing the histone deacetylase 9 (HDAC9) protein-coding sequence are associated with disruption of TWIST1 regulatory elements that reside within the HDAC9 sequence. Based on SVs within the HDAC9‐TWIST1 locus, we defined the 3′-HDAC9 sequence as a critical TWIST1 regulatory region, encompassing craniofacial TWIST1 enhancers and CTCF sites. Deletions of either Twist1 enhancers (eTw5-7Δ/Δ) or CTCF site (CTCF-5Δ/Δ) within the Hdac9 protein-coding sequence led to decreased Twist1 expression and altered anterior/posterior limb expression patterns of SHH pathway genes. This decreased Twist1 expression results in a smaller sized and asymmetric skull and polydactyly that resembles Twist1+/− mouse phenotype. Chromatin conformation analysis revealed that the Twist1 promoter interacts with Hdac9 sequences that encompass Twist1 enhancers and a CTCF site, and that interactions depended on the presence of both regulatory regions. Finally, a large inversion of the entire Hdac9 sequence (Hdac9INV/+) in mice that does not disrupt Hdac9 expression but repositions Twist1 regulatory elements showed decreased Twist1 expression and led to a craniosynostosis-like phenotype and polydactyly. Thus, our study elucidates essential components of TWIST1 transcriptional machinery that reside within the HDAC9 sequence. It suggests that SVs encompassing protein-coding sequences could lead to a phenotype that is not attributed to its protein function but rather to a disruption of the transcriptional regulation of a nearby gene.
Structural variants (SVs) involve at least 50 nucleotides, rearranging large segments of DNA and often having profound consequences in evolution and human disease (Audano et al. 2019; Collins et al. 2020). Given their size and abundance, SVs represent an important mutational force that shapes genome function and contributes to germline and somatic diseases. A recent study analyzing ∼15,000 genomes discovered that SVs are responsible for ∼25% of all rare protein-truncating events per genome, indicating that SVs have a major effect on protein sequences (Collins et al. 2020). Moreover, an underscored modest selection was found against noncoding SVs in cis-regulatory elements that control spatiotemporal gene expression. The profound effect of SVs is also attributable to the numerous mechanisms by which they disrupt protein-coding genes and cis-regulatory architecture (Spielmann et al. 2018). SVs can alter the copy number of regulatory elements or 3D genome structure by disrupting higher-order chromatin organization such as topologically associating domains (TADs) (Lupiáñez et al. 2015). As a result of these position effects, SVs can also influence the expression of genes distant from the SV breakpoints, causing disease. However, the effect of SVs on gene regulatory mechanisms remains poorly understood.
We have previously demonstrated that protein-coding DNA sequences could also function as enhancers of nearby genes, indicating a dual function of DNA sequences (Birnbaum et al. 2012a,b, 2014; Hirsch and Birnbaum 2015; Hirsch et al. 2018). We have studied the TWIST1-HDAC9 locus as an example of this dual function of DNA sequences. TWIST1 is a transcription factor (TF) that plays a critical role in mesodermal development (Qin et al. 2012). TWIST1 regulates the expression of various other TFs and signaling pathways in the developing craniofacial and limb tissues and TWIST1 haploinsufficiency is associated with a range of craniofacial and limb malformations (Zhang et al. 2010). TWIST1 protein-coding mutations, including deletions, are associated with Saethre–Chotzen syndrome (OMIM #101400), an autosomal dominant craniosynostosis disorder (premature closure of the sutures) also associated with distal limb malformations (Cho et al. 2013; Miller et al. 2017). Mouse studies have shown that Twist1 homozygous null mice are lethal, but Twist1 heterozygous mice exhibit a craniosynostosis-like phenotype along with polydactyly (Zhang et al. 2010; Parsons et al. 2014). These findings indicate the existence of a Twist1 dosage threshold that is likely regulated by tissue-specific enhancers that are essential for correct craniofacial and limb formation. Recently, we identified eight tissue-specific enhancers located in the HDAC9-TWIST1 locus that recapitulate Twist1 expression (Hirsch et al. 2018). Two of these enhancers, Rr135 and Rr136 (also known as eTw6 and eTw7, respectively), reside in HDAC9 exons and have a dual function as protein-coding sequences and as limb/fin/branchial arch enhancers. These active enhancers comprise a spatiotemporal regulatory network of Twist1 transcription in the developing limbs/fins (Hirsch et al. 2018).
Histone deacetylase 9 (HDAC9) encodes an enzyme that modifies the N-terminal tail of histones and leads to compact chromatin structure and reduced gene expression (Zhou et al. 2001). The HDAC9 protein is highly expressed in mouse adult brain and heart tissues (Zhang et al. 2002; Sugo et al. 2010; Lang et al. 2012; Malhotra et al. 2019). In the brain, Hdac9 expression is limited to postmitotic neurons and highly expressed in the hippocampus and cerebral cortex (Lang et al. 2012). Copy number variants of HDAC9 are associated with schizophrenia in humans and with neuropathological changes in the hippocampus and cerebral cortex in mice (Lang et al. 2012). Hdac9 also inhibits skeletal myogenesis and is involved in heart development (Lu et al. 2000). Whereas Hdac9-null mice are fertile and have a normal life span, they develop cardiac hypertrophy with age and in response to pressure overload (Zhang et al. 2002). Moreover, a polydactyly phenotype has also been discovered in Hdac9-null mice (Morrison and D'Mello 2008). However, Hdac9 is not expressed in the developing limb, suggesting that the polydactyly phenotype in these mice is independent of HDAC9 protein function (Hirsch et al. 2018). Instead, the polydactyly phenotype could result from a disruption of Twist1 regulatory elements residing in the Hdac9 sequence, leading to haploinsufficiency of Twist1.
In this study, we aim to elucidate the effects of disruption of protein-coding sequences located in a critical regulatory region for a nearby gene. We identify and characterize craniofacial enhancers and bound CTCF regions in the HDAC9 sequence. These regulatory elements are part of a critical TWIST1 regulatory region that is disrupted by HDAC9-encompassing SVs in patients with craniosynostosis. Modeling these HDAC9 SVs in mice led to craniosynostosis-like phenotype and polydactyly. By combining 3D chromatin conformation and expression assays, we aim to demonstrate how these phenotypes correlate with reduced frequency of chromatin interactions between Twist1 regulatory elements and with reduced Twist1 expression. Overall, we explore how HDAC9-encompassing SVs can cause craniosynostosis through the disruption of a critical TWIST1 regulatory region that resides within the HDAC9 sequence.
Results
TWIST1 craniofacial enhancers reside within the HDAC9 sequence
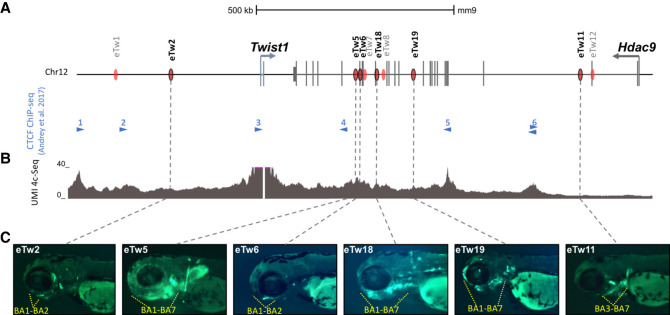
We previously described TWIST1 limb enhancers during embryonic development and characterized their function and activity (Hirsch et al. 2018). Nevertheless, the regulatory mechanisms controlling TWIST1 expression in craniofacial tissues are barely known. To explore the regulatory elements of TWIST1 during craniofacial development and skull formation, we focused on the TWIST1-HDAC9 locus (hg19: Chr 7: 18,050,988–19,741,484) and analyzed chromatin immunoprecipitation sequencing (ChIP-seq) of multiple histone modifications (H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K27ac, H3K27me3) from early human embryonic craniofacial tissues (stages CS13–CS14, corresponding to mouse embryonic day 10.5–11.5) (Wilderman et al. 2018). Twenty-five regions within the HDAC9-TWIST1 locus that were marked with active enhancer marks in both CS13-CS14 developmental stages were identified. Then, we further analyzed ATAC-seq and histone modification ChIP-seq of mouse E10.5 craniofacial tissues (maxilla, mandibula, pharyngeal arch 2, and frontal nasal plate) (Minoux et al. 2017), searching for sequences marked as active enhancers in mouse craniofacial developmental tissues. We defined sequences as enhancer candidates if they were necessarily marked in CS13–CS14 human data and in at least one of the craniofacial mouse data sets (H3K27ac/ATAC). These analyses identified 15 sequences, corresponding to five intergenic, six HDAC9 intronic, and four HDAC9 exonic sequences, that might regulate TWIST1 transcription during craniofacial development (Supplemental Fig. S1; Supplemental Table S1). Then, we used unique molecular identifier (UMI) circularized chromosome conformation capture sequencing (UMI-4C) on mouse E11.5 branchial arches (BA) 1–2 to determine the chromatin interaction profile of the Twist1-Hdac9 locus, using the Twist1 promoter region as a viewpoint. We found that the Twist1 promoter region frequently interacts with several CTCF sites and regions encompassing enhancer candidates such as Rr134 (also known as eTw5) and eTw6 (Fig. 1A,B), suggesting that these regulatory elements might play a role in Twist1 transcription regulation. Next, we tested these candidates for functional activity using a transgenic zebrafish enhancer assay. Of the 15 sequences, we previously reported on four sequences (eTw2, eTw5, eTw6, eTw11) as craniofacial and/or limb enhancers (Supplemental Table S2; Hirsch et al. 2018). The zebrafish enhancer assay showed that eTw2 and eTw6 drove specific GFP expression in branchial arches 1–2 (BA1–2) at 72 h postfertilization (72 hpf), which are homologous to mammalian mandibular arch, maxillar arch, and hyoid. eTw5 displayed a broad activity pattern in branchial arches and drove GFP in branchial arches 1–7 (BA1–7) at 72 hpf. eTw11 drove GFP expression in the posterior part of the branchial arches 3–7 (BA3–7) at 72 hpf (Fig. 1C). Two additional craniofacial enhancer candidates, eTw18 and eTw19, were shown to have functional activity in the craniofacial tissues of 72-hpf zebrafish embryos (Fig. 1C). eTw18 drove GFP expression in the branchial arches 1–7 with similarity to eTw5, and eTw19 drove GFP expression in branchial arches 1–7 and also in the front nasal and maxillary prominences at 72 hpf (Fig. 1C; Supplemental Fig. S2). Thus, each enhancer has a discrete activity pattern and together comprise a spatiotemporal regulatory network that likely controls Twist1 expression in the developing craniofacial tissues (Supplemental Table S2).
Figure 1.
Twist1 craniofacial enhancers in the Hdac9-Twist1 locus. (A) A scheme of the Hdac9-Twist1 locus. Blue lines represent Twist1 exons and black lines represent Hdac9 exons. Red ovals represent positive enhancer sequences in zebrafish enhancer assay. Blue arrowheads represent the directionality of CTCF-bound sites in E11.5 limb buds (Andrey et al. 2017). (B) UMI-4C interaction profile (based on two biological replicates) using the Twist1 promoter as a viewpoint in the branchial arches of mouse E11.5 embryos. (C) Activity pattern of Twist1 craniofacial enhancer in zebrafish. As zebrafish has seven branchial arches (BA1–7), yellow dashed lines show the GFP expression patterns in the branchial arches of zebrafish embryos at 72 hpf.
Structural variants compromise the HDAC9 coding sequence in patients with craniofacial malformations
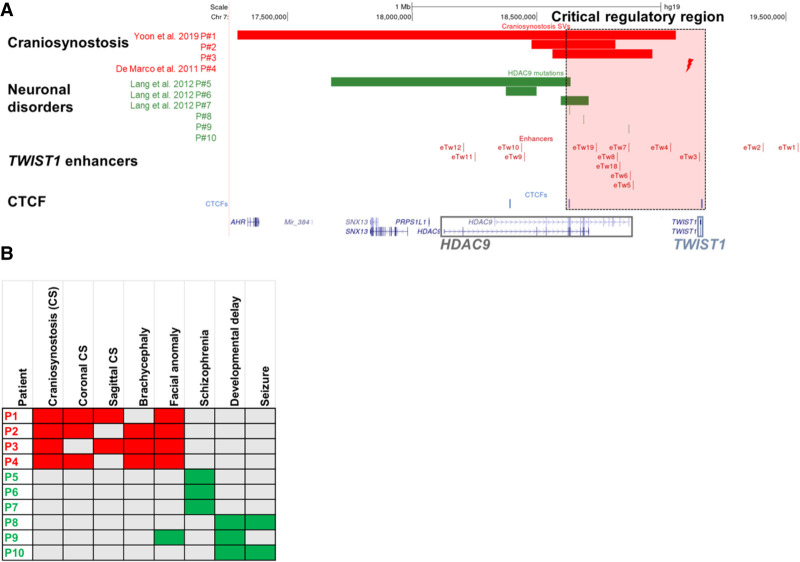
To demonstrate that disruption of protein-coding sequences can also affect regulatory elements of nearby genes, we collected patients with SVs in HDAC9. Through the international Matchmaker Exchange initiative (Philippakis et al. 2015), we found craniofacial malformation patients with deletions containing HDAC9 coding sequences that are also thought to function as regulatory elements of the neighboring TWIST1 gene (Supplemental Table S3; Fig. 2A,B). Two craniosynostosis patients were reported with an HDAC9 deletion (P1) and a translocation with an intergenic breakpoint between HDAC9 and TWIST1 (P4), respectively (De Marco et al. 2011; Yoon et al. 2020). We found two additional craniosynostosis patients with HDAC9 deletions, in which the TWIST1 protein-coding sequence is not disrupted (P2–P3) (Fig. 2A,B). Conversely, other HDAC9 deletions overlapping the 5′ region of HDAC9 are associated with neuronal disorders, such as schizophrenia (Lang et al. 2012), without reported craniofacial phenotype (P5–P7) (Fig. 2A,B). In addition, we identified HDAC9 single nucleotide variants (SNVs), including a splice site, frameshift, and missense variant in patients with global developmental delay, thin corpus callosum, and seizures, but without craniosynostosis (P8–P10) (Supplemental Table S3). The enrichment for loss-of-function variants in our cohort is consistent with the constraint data from the Exome Aggregation Consortium (ExAC) database, suggesting that HDAC9 is extremely intolerant to loss-of-function variations (probability of being loss-of-function intolerant [pLI] = 1) (Karczewski et al. 2020). Therefore, we concluded that SVs affecting the 3′ sequence of HDAC9, in the region delineated by two convergent CTCF sites (e.g., 3 and 5) and containing TWIST1 enhancers, are likely associated with craniofacial malformations, suggesting that the 3′-terminal part of HDAC9 is critical for TWIST1 transcriptional regulation (Fig. 2).
Figure 2.
Structural variants containing HDAC9 (but not TWIST1) in patients with craniosynostosis and neuronal disorders. (A) SVs encompassing HDAC9 but not TWIST1 indicate the potential location of a critical TWIST1 regulatory region (highlighted by the dashed light pink rectangle). Red bars represent three craniosynostosis patients with de novo HDAC9 deletions (P1–P3), and red lightning represents the translocation t(7;12)(p21.2;p12.3) breakpoint located between HDAC9 and TWIST1 in craniosynostosis patient (P4). Green bars represent three schizophrenia patients (P5–P7) with de novo HDAC9 deletions, and green lines represent three patients with neurological phenotypes (P8–P10) with SNVs in HDAC9. Blue lines represent CTCF sites involved in chromatin looping in the HDAC9-TWIST1 locus. Red lines represent TWIST1 enhancers located in introns or exons of the HDAC9 sequence and intergenic regions. (B) Human Phenotype Ontology heat map of patients’ common clinical features. Gray boxes represent either absent or unreported symptoms.
Alteration of Twist1 regulatory elements leads to a craniosynostosis-like phenotype
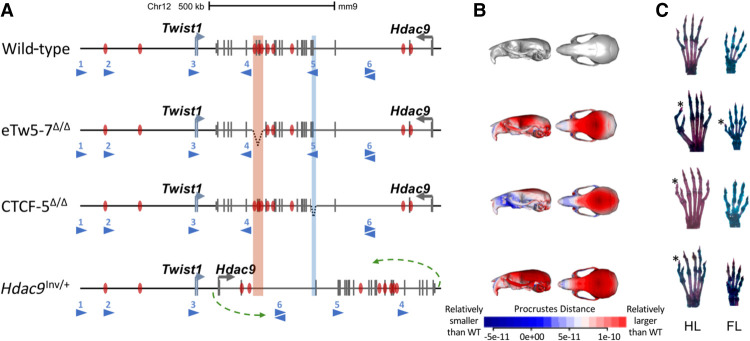
To evaluate the in vivo effect of aberrations within the TWIST1 critical regulatory region on craniofacial and limb development, we investigated three mouse models. In the first two models, Twist1 enhancers and a CTCF-5 site residing in the Hdac9 sequence were disrupted. Using the CRISPR-Cas9 genome editing system, we generated an eTw5-7Δ/Δ mouse model, homozygous for a deletion of three Twist1 enhancers (eTw5-7) and four Hdac9 exons (exons 20–23, NM_001271386.1) (Fig. 3A). In addition, we used a CTCF-5Δ/Δ model (Morrison and D'Mello 2008), in which the CTCF binding site #5 involving chromatin looping within the Twist1 locus and located in the Hdac9 sequence was deleted (Fig. 3A). However, in both models (eTw5-7Δ/Δ and CTCF-5Δ/Δ), the engineered deletions also encompass Hdac9 protein-coding sequences, giving rise to the possibility that the HDAC9 protein is also involved in the phenotypic outcomes. To address this question, we generated a third mouse model carrying a large inversion (∼1 Mb) of the entire Hdac9 sequence that does not disrupt the Hdac9 protein-coding sequence but repositions Twist1 enhancers and potentially interferes with promoter-enhancer looping (Fig. 3A).
Figure 3.
Alteration of Twist1 regulatory elements in mice. (A) Scheme of the aberrations within the Hdac9 sequence. In the eTw5-7Δ/Δ model, a 23-kb sequence, containing Hdac9 exons 20–23 and Twist1 enhancers (eTw5-7), was deleted (highlighted red rectangle). In the CTCF-5Δ/Δ model, a 1.5-kb sequence containing Hdac9 exons 6–7 was deleted (highlighted blue rectangle). In the Hdac9INV/+ model, the whole Hdac9 sequence, 890 kb long, was inverted. The inversion breakpoints are marked by green arrows (mm9, Chr 12: 34,721,220–35,613,000). (B) Heat maps showing anatomical distributions of shape change compared to the wild type for eTw5-7Δ/Δ, CTCF-5Δ/Δ, and Hdac9INV/+ mice, demonstrated by side view (right) and superior view (left) (red is larger and blue is smaller compared to the grand mean). (C) Polydactyly was found in both hindlimb (HL) (71%) and forelimb (FL) (50%) of eTw5-7Δ/Δ mice, whereas polydactyly was found only in the HL of CTCF-5Δ/Δ (32%) and Hdac9INV/+ (8%) mice. The polydactyly is marked by asterisks.
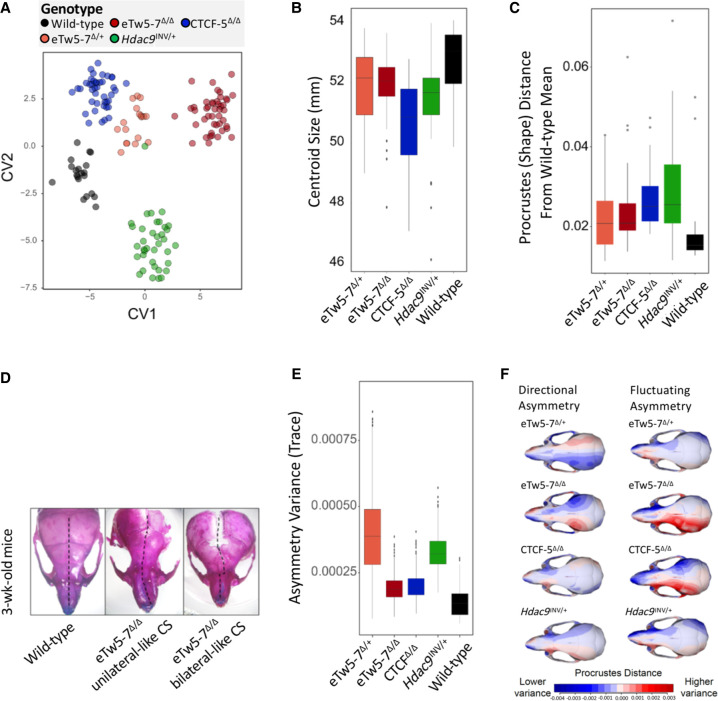
As TWIST1 haploinsufficiency leads to unilateral and bilateral coronal synostosis in both humans and Twist1−/+ mouse models, we tested whether deletions (eTw5-7Δ/+, eTw5-7Δ/Δ, and CTCF-5Δ/Δ) or relocation (Hdac9INV/+) of Twist1 regulatory elements have a significant impact on craniofacial development, including brachycephaly and craniosynostosis-like phenotype (Fig. 3B). We used skeletal staining and microcomputed tomography (micro-CT) to quantify morphological effects on the skull. We analyzed adult (∼80 d of age) mouse skulls from our mouse models for Twist1 regulatory elements (45 skulls of eTw5-7Δ/Δ, 18 skulls of eTw5-7Δ/+, 44 skulls of CTCF-5Δ/Δ, 38 skulls of Hdac9INV/+) and compared them with a cohort of 25 wild-type littermates. We quantified 3D craniofacial shape from micro-CT scan images using 68 standardized skeletal landmarks as in previous work (Supplemental Fig. S3; Percival et al. 2019) using geometric morphometric methods. From the visualization of canonical variate analysis (CVA) and by comparing mean shapes among groups, we determined that phenotypic effects are not confined to a single feature but involve multiple regions of the skull. Our results indicate that deletion of Twist1 enhancers (eTw5-7Δ/Δ) can cause a distinct set of alterations compared to wild-type morphology (Supplemental Movies S1, S2). The first canonical variate (CV1) most clearly separates wild-type mice from eTw5-7+/Δ and eTw5-7Δ/Δ (Fig. 4A), whereas the second canonical variate (CV2) demonstrates the morphological separation of wild-type from CTCF-5Δ/Δ and Hdac9INV/+ mice (Fig. 4A; Supplemental Movies S3–S6). The one-way analysis of variance (ANOVA) model showed that skulls of the mouse models (eTw5-7Δ/Δ, CTCF-5Δ/Δ, and Hdac9INV/+) significantly differ from wild-type skulls and their small skulls, recapitulating the skull phenotype of Twist1−/+ mice (Figs. 3B, 4B; Parsons et al. 2014). The multivariate analysis of variance (MANOVA) showed significant shape differences among the different groups (Supplemental Table S4). Analyzing procrustes (shape) distance for each individual from the wild-type mean shape showed a significant difference in the average shape of eTw5-7+/Δ, eTw5-7Δ/Δ, CTCF-5Δ/Δ, and Hdac9INV/+ mice from wild-type mice (Fig. 4C). Using alcian blue/alizarin red staining, we found notable asymmetry in the cranial morphology of eTw5-7Δ/Δ mice that resembles the unilateral and bilateral coronal synostosis found in humans with heterozygous TWIST1 mutations (Fig. 4D). To associate our mouse models to the uni- and bilateral craniosynostosis phenotype in humans, we quantified and compared asymmetry variances of the skulls using MANOVA for the asymmetric component of shape variation (the residuals of asymmetric procrustes superimposition). The skulls of both eTw5-7+/Δ and Hdac9INV/+ mice have significantly elevated asymmetry variances compared to wild type (Fig. 4E,F; Supplemental Tables S5, S6). However, no significant differences in cranial asymmetry were found between homozygous eTw5-7Δ/Δ and CTCF-5Δ/Δ and the wild type (Fig. 4E,F; Supplemental Tables S5, S6). Whereas all of our mouse models (e.g., both heterozygous and homozygous) showed significantly smaller skulls, only the heterozygous mice (e.g., eTw5-7+/Δ and Hdac9INV/+) showed significant asymmetry of the skull, linking uni- and bilateral craniosynostosis in humans to a partial functional dose of Twist1 during craniofacial development. Overall, our results show that the uni- or bilateral craniosynostosis phenotype caused by Twist1 haploinsufficiency is due to disrupted Twist1 regulatory elements and alteration of its spatiotemporal expression during development.
Figure 4.
Deletions of Twist1 regulatory elements lead to craniosynostosis-like morphology. (A) Canonical variate analysis (CVA) of micro-CT data from eTw5-7Δ/Δ, eTw5-7Δ/+, CTCF-5Δ/Δ, and Hdac9INV/+ mice compared to wild type. (B) Cranial size by genotype. (C) Box plot for procrustes (shape) distances for each individual from the wild-type mean shape. Note that because the procrustes distance is unsigned, the average and minimum procrustes distance for the wild type is positive and not 0. (D) Representative pictures of stained skulls of 3-wk-old wild-type and eTw5-7Δ/Δ mice, showing a range of variation in skull morphology. The dashed black line represents the asymmetric structure with unilateral (middle) and bilateral (right) craniosynostosis-like phenotypes. (E) Box plot of the asymmetry variance differences among the different genotypes and the wild type. It depicts group differences in the amount of variance explained by the asymmetric component of shape variation of the skull. (F) Heat map visualizations for each asymmetry component (directional asymmetry and fluctuating asymmetry), displaying the patterns of shape variation in each genotype compared to the wild type.
Deletions of Twist1 regulatory elements lead to preaxial polydactyly
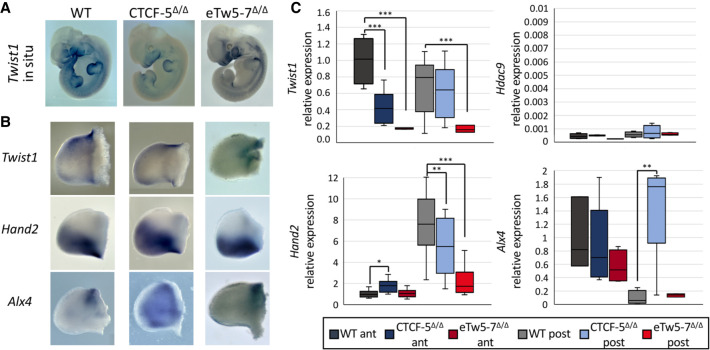
As Twist1 haploinsufficiency leads to limb malformations such as polydactyly, we evaluate the in vivo effect of disrupting Twist1 regulatory elements on limb development using our mouse models. Whereas homozygous Twist1-null mice (Twist1−/−) are lethal and heterozygous Twist1−/+ mice showed partial polydactyly penetrance (32%), the eTw5-7Δ/Δ mouse model showed high penetrance of polydactyly in the hindlimb (HL) (71%) and forelimb (FL) (50%) (Fig. 3C). This high penetrance of polydactyly showed variable expression, as only 15% of the mice showed bilateral polydactyly, and most of them showed right or left polydactyly in the HL and/or FL. The CTCF-5Δ/Δ model (Morrison and D'Mello 2008) showed HL polydactyly with a similar penetrance (32%) as Twist1−/+ mice (Fig. 3C). The polydactyly was observed in the right and/or left HL, with 20% of the mice showing bilateral polydactyly. Next, we evaluated whether Twist1 is differentially expressed in our deleted Hdac9 mouse embryos (eTw5-7Δ/Δ and CTCF-5Δ/Δ). Using qPCR and whole-mount in situ hybridization, we discovered a reduction of Twist1 expression in the branchial arch and in the anterior HL bud of eTw5-7Δ/Δ and CTCF-5Δ/Δ mice at E11.5 (Supplemental Fig. S4; Fig. 5A–C). These observations indicate that the dosage of Twist1 is critical and its haploinsufficiency can lead to partial penetrance and variable phenotypic expression. However, in these two models (eTw5-7Δ/Δ and CTCF-5Δ/Δ), the disrupted Twist1 regulatory elements encompass the Hdac9 protein-coding sequences, suggesting that the lack of HDAC9 protein could also contribute to the phenotype. We showed that Hdac9 is not expressed in either wild-type or eTw5-7Δ/Δ/CTCF-5Δ/Δ mice at the E11.5 limb bud indicating that HDAC9 does not contribute to the polydactyly phenotype (Fig. 5C; Hirsch et al. 2018). Moreover, we showed that, although homozygosity for the inversion (Hdac9INV/INV) is lethal as in Twist1-null mice, the heterozygous mice (Hdac9INV/+) are viable and showed HL polydactyly (8%) (Fig. 3C). We found that Twist1 levels were reduced in HL of Hdac9INV/+ embryos compared to wild types (Supplemental Fig. S5). Furthermore, no reduction in HDAC9 protein level was found in the cortical brain (where Hdac9 is highly expressed) compared to adult control mice (Supplemental Fig. S6). Our results imply that the polydactyly phenotype of our mouse models is due to the alteration of Twist1 regulation and not the HDAC9 protein.
Figure 5.
Deletions in Hdac9 that alter Twist1 regulatory elements lead to dysregulation of SHH pathway genes. (A) Whole-mount in situ hybridization of E11.5 mouse embryos showing reduced Twist1 expression pattern in eTw5-7Δ/Δ and CTCF-5Δ/Δ. (B) Whole-mount in situ of E11.5 hindlimb mouse embryos for Twist1 and its target SHH pathway genes, Hand2 and Alx4. Twist1 expression is reduced, especially in the anterior limb bud. Hand2 and Alx4 expression along the anterior/posterior axes is lost in the mutants. (C) Quantitative real-time PCR analyses in E11.5 HL buds show a significantly decreased Twist1 expression in anterior limb buds of CTCF-5Δ/Δ (P = 9 × 10−4) and in the entire limb bud of eTw5-7Δ/Δ (P = 5 × 10−4). No expression of Hdac9 in the limb buds of wild-type, CTCF-5Δ/Δ, or eTw5-7Δ/Δ embryos. Significant decrease of Hand2 expression in posterior limb buds of CTCF-5Δ/Δ (P = 3 × 10−2) and eTw5-7Δ/Δ (P = 1 × 10−5) embryos. Significant increase of Alx4 expression in posterior limb buds of CTCF-5Δ/Δ embryos (P = 8.8 × 10−3). Expression levels were normalized to actin, beta expression (Student's t-test, [*] P-value <0.05, [**] P-value <0.01, [***] P-value <0.001).
Deletion of Twist1 regulatory elements alters the expression of SHH pathway genes
To evaluate the effect of altered regulatory elements on TWIST1 target genes, we analyzed the expression of Hand2, Ptch1, Alx4, and Gli3, known TWIST1 target genes in the SHH pathway, both directly and indirectly. These transcription factors have unique and specific expression patterns along the anterior-posterior (A-P) axis of the limb bud, required for digit number and identity. Indeed, we showed that disrupted Twist1 regulatory elements not only reduced the expression of Twist1 but affected the spatiotemporal expression of its target genes along the A-P axis (Fig. 5B,C). Using qPCR, we observed that the expression pattern of Hand2 was reduced in the posterior limb bud of eTw5-7Δ/Δ and CTCF-5Δ/Δ (P < 0.05) (Fig. 5). Moreover, using in situ hybridization, Hand2 expression extended beyond the posterior boundary and diffused toward the anterior domain. This ectopic Hand2 expression led to a disproportion of the A-P pattern in the limb bud (Fig. 5B,C). Ptch1, a gene coding for a transmembrane SHH receptor that is differentially expressed in A-P axes, also lost its A-P limb bud expression in eTw5-7Δ/Δ (Supplemental Fig. S7). Furthermore, consistent with the negative feedback loop of SHH regulators, SHH antagonists Alx4 and Gli3 are both lacking precise anterior expression patterns and are extended toward posterior domains in CTCF-5Δ/Δ and eTw5-7Δ/Δ embryos (Fig. 5B,C; Supplemental Fig. S7). Alx4 expression levels are significantly increased in the posterior limb buds and Gli3 expression level is reduced in the anterior limb buds of CTCF-5Δ/Δ embryos (Fig. 5B,C; Supplemental Fig. S7). Therefore, deletions of the eTw5-7 enhancers or CTCF-5 site led to a significant reduction of Twist1 expression levels and ectopic expression of Hand2 and Alx4. These SHH pathway genes are no longer differentially expressed along the A-P axis and have a diffused pattern that likely led to inconsistency of SHH pathway signals and eventually resulted in high penetrance polydactyly.
Deletions of Twist1 enhancers and CTCF-5 site affect chromatin looping at the Twist1‐Hdac9 locus
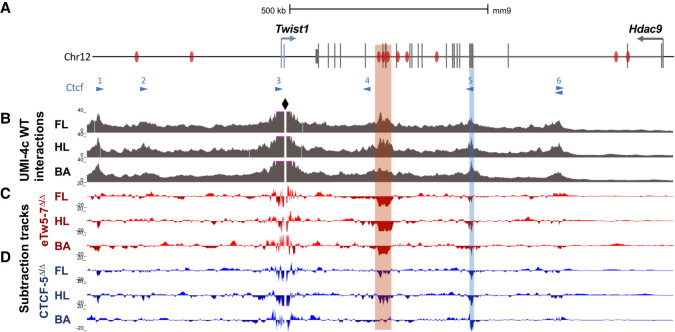
To elucidate the effect of Twist1 regulatory element deletions on chromatin organization within the Twist1-Hdac9 locus, we performed UMI-4C on E11.5 forelimbs, hindlimbs, and branchial arches. We compared the interaction maps of wild-type, eTw5-7Δ/Δ, and CTCF-5Δ/Δ embryos using the Twist1 promoter as a viewpoint (Fig. 6A; Supplemental Fig. S8). The Twist1 promoter displayed frequent interactions with a region that contains the cluster of three enhancers, eTw5, 6, and 7 in limb buds and the branchial arch, with the highest interaction frequency being observed in the limb buds (Fig. 6; Supplemental Fig. S8). These three enhancers were characterized with limb enhancer activity, and two of them (eTw5, 6) were also active in the branchial arches (Hirsch et al. 2018). In addition, the Twist1 promoter region was also involved in frequent interactions with two upstream CTCF sites (CTCF-1 and CTCF-2) and two downstream CTCF sites (CTCF-5 and CTCF-6), both in limb buds and branchial arch (Fig. 6B). Whereas the Twist1-Hdac9 locus contains several CTCF sites, these four CTCF-bound interacting regions are occupied by CTCF in the limb bud at E11.5 (Andrey et al. 2017), supporting their role in the 3D chromatin organization of the locus, facilitating Twist1 regulatory interactions (Fig. 6B). As expected, in both eTw5-7Δ/Δ and CTCF-5Δ/Δ embryos, interactions between the Twist1 promoter and the targeted region (eTw5-7 and CTCF-5, respectively) were severely reduced (Fig. 6B,C; Supplemental Fig. S9). In eTw5-7Δ/Δ embryos, the Twist1 promoter region showed a 5.9-fold reduction in interaction frequency with the eTw5-7 region in the HL and a 3.4-fold reduction in the FL (P = 1.55 × 10−62, 1.42 × 10−22, respectively). In CTCF-5Δ/Δ embryos, there was a 3.2-fold reduction in interaction frequency with the CTCF-5 region in the HL (P = 1.42 × 10−11) and a 2.8-fold reduction in the FL (P = 1.97 × 10−8). We also observed reduced interactions of the Twist1 promoter with the CTCF-3 site in eTw5-7Δ/Δ embryos and vice versa in CTCF-5Δ/Δ. In eTw5-7Δ/Δ embryos, the deletion of the eTw5-7 enhancer cluster led to a 1.7-fold reduction in interaction frequency with the CTCF-5 region in the HL (P = 0.05) and a 1.2-fold reduction in the FL (P > 0.05) (Fig. 6C). In CTCF-5Δ/Δ embryos, the deletion of the CTCF-5 site altered the interaction frequency with the eTw5-7 region, with a 1.4-fold reduction of interaction frequency detected in the HL (P = 5.42 × 10−8) and a 1.3-fold reduction in the FL (P = 6.39 × 10−5) (Fig. 6D). These interaction profiles imply that both the eTw5-7 enhancer cluster and the CTCF-5 site are partially dependent on each other for interaction with the Twist1 promoter. Similar interaction profiles were found in E11.5 BA (Fig. 6B). In eTw5-7Δ/Δ embryos, the Twist1 promoter region showed a 2.6-fold loss of interactions with the eTw5-7 region (P = 1.3 × 10−19), but no significant change was observed at the CTCF-5 site in the BA (Fig. 6C). Furthermore, in CTCF-5Δ/Δ embryos, the Twist1 promoter region showed a threefold loss of interactions with the CTCF-5 region (P = 1.4 × 10−6), but no significant change was observed at the eTw5-7 region (Fig. 6D). This suggests, that although the chromatin looping at the Hdac9-Twist1 locus is similar in both limb bud and BA, the interaction frequencies and the dependency of the regulatory elements involved in the Twist1 transcriptional mechanism are different. Overall, the Twist1 promoter region along with enhancers and CTCF binding sites compose a regulatory unit, likely functioning together to execute precise Twist1 expression during limb and craniofacial development.
Figure 6.
Deletions of Twist1 regulatory regions affect chromatin looping in the Hdac9-Twist1 locus. (A) A scheme of the Hdac9-Twist1 locus with Twist1 enhancers marked by black bars and CTCF-bound sites (Andrey et al. 2017) (and motif directionality) marked by blue triangles. (B) UMI-4C tracks (merged from two biological replicates) of WT E11.5 forelimb (FL), hindlimb (HL), and branchial arch (BA) demonstrate that Twist1 interaction patterns are largely conserved between limb buds and branchial arch. The Twist1 promoter serves as a viewpoint and is highlighted by the black diamond shape. Targeted eTw5-7 and CTCF regions are indicated by red and blue rectangles, respectively. (C,D) Subtraction tracks showing differential Twist1 promoter interactions in FL, HL, and BA of eTw5-7Δ/Δ and CTCF-5Δ/Δ embryos compared to WT (see also Supplemental Figs. S8, S9).
Discussion
In this study, we deciphered the role of SVs at the HDAC9-TWIST1 locus in craniosynostosis and craniofacial malformations. We identified several chromosomal aberrations affecting HDAC9 but not TWIST1 in patients with craniosynostosis. This suggests that the disruption of a critical TWIST1 regulatory region, within the HDAC9 sequence, leads to craniofacial malformations. We initially characterized six transcriptional enhancers located in the HDAC9-TWIST1 synteny block that regulates spatiotemporal TWIST1 activity during craniofacial development. These enhancers also reside in regions that likely interact with the Twist1 promoter in BA and limb bud of E11.5 mouse embryos. Each enhancer has a discrete activity pattern that recapitulated aspects of Twist1 expression during development. The partially overlapping activity pattern of the analyzed Twist1 enhancers might ensure the robustness of Twist1 expression during craniofacial and limb development. For example, eTw5, eTw18, and eTw19 are active enhancers in zebrafish BA1–7, whereas eTw2 and eTw6 enhancers are active in BA1–2, and eTw11 enhancer is active in BA3–7, indicating that the activity of each enhancer, along with the overlapping activity between enhancers, is important for proper spatiotemporal Twist1 expression.
SVs in this locus could disrupt the HDAC9 protein-coding sequence, TWIST1 regulatory elements, or both. Depending on the size and location of the SV, the effect of SV on the phenotype is based on the number and type of disrupted TWIST1 regulatory elements. Our mouse models emphasize that deletion of regulatory elements (i.e., enhancers and CTCF site) could lead to similar phenotypes with partial penetrance and variable expression. Indeed, in patients #1 and #3 with craniosynostosis, the deletions encompass the characterized regulatory elements including CTCF sites and eTw5-7, 18, 19 enhancers (Fig. 2). In patient #2, who also has craniosynostosis, the deletion encompasses CTCF binding sites and eTw19, which raises the question of whether disruption of eTw19 might be sufficient for craniosynostosis phenotype. On the contrary, deletion/disruption of enhancer/s could be compensated by the function of additional/shadow enhancers. In patient #5, in whom no craniosynostosis phenotype was reported, the deleted region encompasses eTw11 and eTw12 enhancers but not the active eTw5-7, 18, and 19 enhancers, suggesting that the remaining regulatory elements might compensate for the lack of eTw11 and eTw12 enhancers. Thus, SVs affecting the 3′ sequence of HDAC9, in the region delineated by two convergent CTCF sites (i.e., CTCF-3 and CTCF-5) and containing TWIST1 enhancers, are likely associated with craniofacial malformations, emphasizing the role of the number and type of regulatory elements in the TWIST1 transcriptional regulation.
Loss of function of craniofacial and limb Twist1 enhancers and CTCF sites emphasizes the crucial role of TWIST1 transcriptional regulation on the penetrance and expression level of the craniosynostosis and limb phenotype. All three models (eTw5-7Δ/Δ, CTCF-5Δ/Δ, and Hdac9INV/+) showed a skull phenotype, but the effect on the skull shape depended on the characteristics of the disrupted regulatory elements (Fig. 4E,F). For example, both eTw5-7Δ/Δ and CTCF-5Δ/Δ showed a small centroid size but with a different effect on skull shape (Fig. 4; Supplemental Movie S1). In addition, we found significant asymmetry in the skulls of both eTw5-7+/Δ and Hdac9INV/+ mice that resembles unilateral craniosynostosis, whereas homozygous mice show a small skull with no significant asymmetry that resembles bilateral craniosynostosis (Fig. 4E,F). The partial penetrance and variable expression of the phenotype are also emphasized in the developing limb. Whereas homozygous eTw5-7Δ/Δ mice showed both hindlimb and forelimb polydactyly with high penetrance (77%) (Fig. 3) and variable expression (right and/or left hind/forelimb), heterozygous eTw5-7Δ/+ mice have no limb phenotype. Similar observations were made for homozygous CTCF-5Δ/Δ mice, which showed hindlimb polydactyly with variable expression (right and/or left hindlimb) and partial penetrance (32%) as seen in Twist1−/+ mice (Fig. 3). The phenotypic similarity of the Twist1-null mouse was also observed for the homozygous Hdac9 inversion mice, which are lethal, whereas the heterozygous mice (Hdac9INV/+) are viable and show hindlimb polydactyly (8%) (Fig. 3). In addition, the deletions of eTw5-7 enhancers or the CTCF-5 site led to a significant reduction of Twist1, specifically in anterior limb buds, and ectopic expression of TWIST1 target genes, including Hand2, and Alx4 as well as Ptch1 and Gli3, respectively (Fig. 5; Supplemental Fig. S7). These SHH pathway genes are no longer differentially expressed along the A-P axis and have a diffused expression pattern without significant difference between anterior and posterior of the limb bud, which likely led to inconsistency of SHH pathway signals and eventually resulted in variable penetrance polydactyly. Indeed, each of our mouse models shows a specific disruption of Twist1 expression, emphasizing the role of Twist1 regulatory elements in fine-tuning spatiotemporal Twist1 expression. Overall, the type and number of disrupted regulatory elements can be associated with a uni- and bilateral (or none) craniosynostosis-like phenotype, as well as polydactyly based on a threshold dose of Twist1.
The tissue-specific activities of these craniofacial and limb enhancers are supported by specific chromatin interactions within the Twist1-Hdac9 locus in the limb bud and branchial arch. UMI-4C data demonstrated that the Twist1 promoter region that contains the CTCF-3 site interacts with four distal CTCF-bound sites, which together enable the 3D chromatin interactions required for Twist1 transcriptional regulation (Fig. 6). Moreover, the suggested 3D chromatin interactions enable the Twist1 promoter region to interact with regions that encompass eTw5, 6, 7, and 18 enhancers in the limb bud and branchial arch. Indeed, reduced chromatin looping was observed upon deletion of eTw5-7 and CTCF-5 (Fig. 6). In limb buds, Twist1 interaction frequency with both sites appeared to be codependent. This suggests that the Twist1 promoter region along with enhancer elements and architectural CTCF-bound sites compose a regulatory unit, functioning in unison to fine-tune precise Twist1 expression.
In summary, SVs represent an important mutational force that shapes genome function and contributes to germline and somatic diseases. As SVs are responsible for ∼25% of all rare protein-truncating events per genome, they have a major effect on protein sequences but also on cis-regulatory elements that control spatiotemporal gene expression. Here, we showed that SVs that affect the HDAC9 sequence can also modulate basic mechanisms of gene regulation controlling the expression of a nearby gene, TWIST1. These SVs affect TWIST1 regulatory elements and disrupt higher-order chromatin organization, leading to a phenotype that is not associated with the HDAC9 protein. Thus, careful interpretation is required when considering the molecular basis of SVs identified in human patients.
Methods
Ethics statement
DNA samples were obtained from all available samples following informed consent and approval of the Soroka Medical Center Internal Review Board (IRB). Clinical phenotyping was determined by an experienced pediatrician and geneticist.
All animal work was approved by the Ben-Gurion Institutional Animal Care and Use Committee, protocol number 52-09-2016.
Generating Hdac9/Twist1 mouse models
Two mouse strains, eTw5-7Δ/Δ and Hdac9 Inv/+, were created using a modified CRISPR-Cas9 protocol (Yang et al. 2014). In addition, we exploited a mouse model where exons 6 and 7 of the Hdac9 sequence, along with a CTCF site at the interionic sequence were deleted (Morrison and D'Mello 2008). Briefly, for eTw5-7Δ/Δ, two sgRNAs targeting a 23-kb sequence that encompasses the three enhancers and exons 17–20 of Hdac9 (23,137 bp; Chr 12: 34,883,772–34,906,909; mm9) were designed using CHOPCHOP (Montague et al. 2014). Similarly, for Hdac9 inversion, two gRNAs were designed, targeting regions delimiting the whole Hdac9 gene (Chr 12: 34,721,220–35,613,000; 891,781 bp). No potential off-targets were found when searching for matches in the mouse genome, when allowing for up to two mismatches in the 20-nt-long sequence preceding the PAM sequence. The T7 promoter was added to the sgRNA template, and the whole cassette was chemically synthesized by IDT. The PCR-amplified T7-sgRNA product was used as a template for in vitro transcription using the MEGAshortscript T7 kit (Thermo Fisher Scientific). Cas9 mRNA was transcribed in vitro using the mMESSAGE mMACHINE T7 kit (Thermo Fisher Scientific). The DNA template for in vitro transcription containing the humanized Streptococcus pyogenes Cas9 gene was PCR-amplified from the px330 plasmid. eTw5-7Δ/Δ and Hdac9INV/+ mice were generated by injecting a mix of Cas9 mRNA (final concentration of 100 ng/µL) and sgRNA (50 ng/µL) in injection buffer (10 mM Tris, pH 7.5; 0.1 mM EDTA) into the cytoplasm of C57BL/6J embryos in accordance with the standard procedure approved by Ben-Gurion University. Female mice of the ICR (CD-1) strain were used as foster mothers. F0 mice were genotyped using PCR to detect the deletion of enhancers from the mouse genome (Supplemental Table S7).
Genomic data analyses
Publicly available ChIP-seq and ATAC-seq data sets of E10.5/E11.5 mice embryos craniofacial tissues (i.e., Mx, Md, PA2, FNP) which used enhancer-associated marks (EP300, under the NCBI Gene Expression Omnibus [GEO; https://www.ncbi.nlm.nih.gov/geo] accession number GSE49413; H3K27ac, under GEO accession number GSE89435) (Attanasio et al. 2013; Minoux et al. 2017) as well as human embryonic ChIP-seq data for chromatin modifications (H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K27ac, H3K27me3, under GEO accession number GSE97752) from stages CS13–CS15 on craniofacial tissues (Wilderman et al. 2018) were obtained and analyzed for redundancy, as well as evolutionary conservation. Overlapping peaks were defined as when at least 1-bp regions overlapped. CTCF ChIP-seq data from the E11.5 limb bud (GEO: GSE84795) was also used in this study (Andrey et al. 2017). All the genomic data analyses presented are aligned to human (GRCh37/hg19) or mouse (NCBI37/mm9) genome assemblies. As this study is focused on a specific locus, HDAC9-TWIST1, which is a synteny block and evolutionary conserved, the use of hg19 and mm9 assemblies should not have any significant effect on the conclusions.
Transgenic zebrafish enhancer assay
Primers were designed to amplify the candidate enhancer sequences from human genomic DNA (Supplemental Table S7). PCR products were cloned into the E1b-GFP-Tol2 enhancer assay vector containing an E1b minimal promoter followed by the gene for GFP (Li et al. 2010). These constructs were injected into zebrafish embryos using standard procedures. For statistical significance, at least 100 embryos were injected per construct in at least two different injection experiments along with Tol2 mRNA to facilitate genomic integration (Fisher et al. 2006). GFP expression was observed and annotated 48 and 72 hpf. An enhancer was considered as a positive enhancer when at least 30% of the live embryos showed a consistent GFP expression pattern.
Craniofacial morphometric analysis and statistical analysis
Mice were scanned using a Scanco VivaCT scanner at 35-micron resolution. The total sample size for this analysis was 170 skulls (25 wild type, 38 HdacΔ/+, 44 CTCF-5Δ/Δ, 18 eTw5-7Δ/+, and 45 eTw5-7Δ/Δ). To quantify craniofacial shape, we used the standard set of 68 3D landmarks used in previous work (Percival et al. 2017; Katz et al. 2020). We used geometric morphometric methods to perform quantitative analyses to statistically evaluate and visualize patterns of variation in craniofacial shape and size, using R (R Core Team 2021). We performed a Generalized Procrustes Superimposition Analysis (GPA) to extract the aligned procrustes shape coordinates from the 3D landmark data, using the R package geomorph (https://www.rdocumentation.org/packages/geomorph/ versions/4.0.3). We quantified cranial size as the centroid of each landmark configuration, which is calculated as the square root of the sum of the squared distances from each landmark to the landmark set centroid for each specimen, using geomorph. We fitted a multivariate analysis of variance, implemented in the geomorph R package, to determine differences in craniofacial size and shape between the different genotypes and the wild-type group.
To quantify shape differences between each genotype and the wild type, we calculated the shape (procrustes) distance for each individual in the sample to the wild-type mean. This generates a distribution of distances for each genotype including the wild type. Because no individual is identical to its group genotype mean and because the procrustes distance is unsigned, the mean shape distance of wild-type individuals from the wild-type mean is positive and not zero. To visualize the shape effects that distinguish each genotype from the wild-type mean, we determined the vector of procrustes coordinate differences between each genotype mean and the wild-type mean. This vector was then used to generate a heat map showing the anatomical distribution of these distances and to generate 3D morphs. Because the mean differences themselves are fairly subtle and difficult to see on 3D morphs, we can exaggerate them by multiplying the shape difference vector by an arbitrary constant. This method was used to generate exaggerated versions of the genotype effects in Figure 3 and the 3D morph movies in the Supplemental Material.
To assess craniofacial asymmetry, we decomposed skull shape variation and extracted directional asymmetry (DA) and fluctuating asymmetry (FA) data. Directional asymmetry is the pattern of consistent shape variation between left and right sides, involving developmental mechanisms and/or differential gene expression (Klingenberg 2015), and the DA component for each individual is calculated as the shape difference between sides (left and right). Fluctuating asymmetry captures small asymmetric shape differences between left and right sides that, unlike DA, do not follow a pattern and is displayed as random or residual shape variation (Klingenberg 2015). A higher amount of shape variance explained by the FA component within the asymmetric shape component can be associated with developmental instability (Palmer and Strobeck 2003; Benítez et al. 2020). The FA component is calculated for each specimen and each side (left and right) as the adjusted deviation from the mean DA.
To visualize shape changes among groups and variation along the axes that distinguish groups, we performed canonical variate analysis as well as principal components analysis (PCA), using the R packages geomorph and morpho (Schlager 2017). The vector displacements from these analyses were used to visualize shape variation using morphs of 3D meshes or as heat maps, using a thin-plate spline method to calculate distances between the reference mesh and the target mesh. Negative values (inside the reference mesh) were visualized with red colors, whereas positive values (outside the reference mesh) were visualized with blue color values. Distances close to 0 (target mesh practically in the same position as the reference mesh) were visualized as white. Both the morphs and heat maps displaying differences between wild type (reference) and each genotype (target) were generated using the wild-type average mesh and wild-type landmark coordinates and the average shape coordinates for each genotype (Fig. 3B). Morphs and heat maps from the CVA analyses were generated from the average mesh and average landmark coordinates and the shape coordinates of the extreme ends of the CVA axes (CV1–2). Importantly, the determination of the statistical significance of differences among groups is based on the MANOVA and not on the CVA analyses, which were only performed to visualize the patterns of shape variation found in the MANOVA (Mitteroecker and Gunz 2009).
Gene expression analyses
Mouse E10.5 and E11.5 limb buds (whole or dissected for anterior/posterior), as well as adult brains, were dissected. Total RNA was isolated using a Total RNA purification micro kit (NORGEN, Cat. 35300) according to the manufacturer's protocol. Next, cDNA was synthesized using 5× All-In-One master mix (abm, Cat. G592). qPCR was performed using KAPA SYBR FAST qPCR master mix (Sigma-Aldrich) and run on the C1000 Touch thermal cycler (Bio-Rad). Samples were tested in duplicate. The specificity and absence of primer dimers were controlled by denaturation curves. Actin, beta (Actb) mRNA was used for normalization.
Whole-mount in situ hybridization
Mouse E10.5 and E11.5 embryos were fixed in 4% paraformaldehyde. Vectors containing mouse Twist1, Hand2, and Alx4 were used as templates for digoxygenin-labeled probes. Mouse whole-mount in situ hybridizations were performed according to standard procedures (Hargrave et al. 2006).
UMI 4C
UMI-4C was performed according to the protocol by Schwartzman et al. (2016). In brief, limb bud and branchial arch tissues of mouse E11.5 embryos were microdissected, dissociated into single cells, and crosslinked for 10 min using 2% formaldehyde. Crosslinked cell pellets were snap-frozen and stored at −80°C until further processing (Schwartzman et al. 2016). Frozen pellets were resuspended in 250 µL prediluted DpnII buffer and 7.5 µL preheated 10% SDS and incubated on a thermomixer for 1 h at 37°C, shaking at 900 RPM. After adding 75 µL 10% Triton X-100, the solution was incubated again (1 h, 37°C, 900 RPM). The chromatin was digested using 300 U DpnII (NEB, R0543L) in three stages (100 U for 2 h, 100 U overnight, 100 U for 2 h) at 37°C and 900 RPM. The solution was incubated at 65°C for 20 min to inactivate the restriction enzyme and put on ice. Next, the chromatin was ligated by adding 2000 U of T4 DNA ligase (NEB, M0202M) and 10× T4 DNA ligase buffer to a total volume of 650 µL, and the solution was incubated overnight at 16°C and 300 RPM. Ligated chromatin was de-crosslinked by incubation with 4 µL Proteinase K (20 mg/mL, Qiagen, Cat. 19131) overnight at 65°C and 300 RPM. A 3C template was then purified using 1× Ampure XP beads (Beckman Coulter, A63881). Up to 4 µg 3C template per sample were sheared using microTUBE snap-cap tubes (Covaris, 520045) in a Covaris M220 sonicator to an average fragment length of 300 bp. UMI-4C sequencing libraries were generated using the NEBNext Ultra II library prep kit (NEB, E7645L). For each sample, library prep was performed in four parallel reactions with a maximum input of 1000 ng per reaction, including a size selection targeting fragments with a length of 300–400 bp (unless the input was <100 ng) and 4–8 cycles of PCR enrichment depending on the input. Next, two nested PCR reactions were performed to enrich for fragments captured by the viewpoint of interest, both using 2 µL 10 mM Illumina enrichment primer 2 (CAAGCAGAAGACGGCATACGA) and either a viewpoint-specific “upstream” (reaction 1, 2 µL 10 mM, CTGTGACAGCAGTAGTGGCA) or “downstream” (reaction 2, 2 µL 10 mM, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTTCGACGCTCTGGGTGAT) primer. For each sample, we performed up to eight nested PCR reactions in parallel with an input of 100–200 ng per reaction using the KAPA2G Robust ready mix (Sigma-Aldrich, KK5702) in a volume of 50 µL. PCR program: 3 min at 95°C, 20 cycles (18 cycles for reaction 2) of 15 sec at 95°C, 15 sec at 55°C, 60 sec at 72°C, and final elongation of 5 min at 72°C. Between PCR reactions, the product was cleaned up using 1× AmpureXP beads and eluted in 21 µL. The final PCR product was cleaned up using 0.7× AmpureXP beads and eluted in 25 µL. Reactions per sample were pooled, and library concentration was quantified via qPCR using the KAPA SYBR FAST qPCR master mix (Sigma-Aldrich, KK4602). UMI-4C libraries were pooled and sequenced on an Illumina HiSeq or NovaSeq (paired-end, 2 × 150 cycles). Reads were first filtered based on the presence of the downstream primer sequence (20% mismatch allowed). The resulting FASTQ files were then used as input to the UMI-4C R package (https://github.com/tanaylab/umi4cpackage) (Schwartzman et al. 2016) to generate genomic interaction tracks, representing UMI counts (i.e., unique interactions) per genomic restriction fragment. The package was then used to generate smoothed, viewpoint-specific interaction profiles for the region of interest. For each profile, interaction counts were normalized to the total UMI count within the profile. Normalized profiles were subtracted to identify differentially interacting regions. Statistical significance of differential interactions was tested for two loci (“eTw5-7”: Chr 12: 34,883,878–34,906,858, “CTCF-5”: Chr 12: 35,116,684–35,117,991) using a Chi-square test within the UMI-4C R package (p4cIntervalsMean function) (Schwartzman et al. 2016).
Alcian blue/alizarin red staining
Skulls of 3-wk-old mice were processed and stained for bone and cartilage as previously described (Rigueur and Lyons 2014). Mice were euthanized and their skulls were dissected. The skin, eyes, organs, and adipose tissue were removed using forceps. Skulls were then fixed by two overnight changes of 95% ethanol, followed by 2 d in acetone. Next, the cartilage was stained using alcian blue staining solution (0.03% [w/v], 80% EtOH, 20% [glacial] acetic acid) for 2 d. Then, skulls were destained and postfixed by two overnight changes of 95% ethanol and then cleared in 1% KOH overnight at 4°C, followed by bone staining with alizarin red (0.005% [w/v] in 1% [w/v] KOH) for 3 d. Finally, specimens were cleared using 1% KOH and stored in 100% glycerol.
Supplementary Material
Acknowledgments
This research was supported by the Israel Science Foundation (https://www.isf.org.il). N.H. and R.Y.B. were partially supported by this grant (2334/21). This work was partly supported by the Research Foundation Flanders (FWO) under grants G044615N and 1520518N. In addition, E.D. and S.V. were, respectively, supported by a doctoral and postdoctoral fellowship of the FWO Research Fund.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276196.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Andrey G, Schöpflin R, Jerković I, Heinrich V, Ibrahim DM, Paliou C, Hochradel M, Timmermann B, Haas S, Vingron M, et al. 2017. Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res 27: 223–233. 10.1101/gr.213066.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, et al. 2013. Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342: 1241006. 10.1126/science.1241006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez HA, Lemic D, Villalobos-Leiva A, Bažok R, Órdenes-Claveria R, Pajač Živković I, Mikac KM. 2020. Breaking symmetry: fluctuating asymmetry and geometric morphometrics as tools for evaluating developmental instability under diverse agroecosystems. Symmetry (Basel) 12: 1789. 10.3390/sym12111789 [DOI] [Google Scholar]
- Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, Pappalardo Z, Clarke SL, Wenger AM, Nguyen L, et al. 2012a. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res 22: 1059–1068. 10.1101/gr.133546.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum RY, Everman DB, Murphy KK, Gurrieri F, Schwartz CE, Ahituv N. 2012b. Functional characterization of tissue-specific enhancers in the DLX5/6 locus. Hum Mol Genet 21: 4930–4938. 10.1093/hmg/dds336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum RY, Patwardhan RP, Kim MJ, Findlay GM, Martin B, Zhao J, Bell RJ, Smith RP, Ku AA, Shendure J, et al. 2014. Systematic dissection of coding exons at single nucleotide resolution supports an additional role in cell-specific transcriptional regulation. PLoS Genet 10: e1004592. 10.1371/journal.pgen.1004592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Yang TH, Shin ES, Byeon JH, Kim GH, Eun BL. 2013. Saethre–Chotzen syndrome with an atypical phenotype: identification of TWIST microdeletion by array CGH. Childs Nerv Syst 29: 2101–2104. 10.1007/s00381-013-2235-0 [DOI] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, et al. 2020. A structural variation reference for medical and population genetics. Nature 581: 444–451. 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco P, Raso A, Beri S, Gimelli S, Merello E, Mascelli S, Baldi M, Baffico AM, Pavanello M, Cama A, et al. 2011. A de novo balanced translocation t(7;12)(p21.2;p12.3) in a patient with Saethre–Chotzen-like phenotype downregulates TWIST and an osteoclastic protein-tyrosine phosphatase, PTP-oc. Eur J Med Genet 54: e478–e483. 10.1016/j.ejmg.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. 2006. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1: 1297–1305. 10.1038/nprot.2006.230 [DOI] [PubMed] [Google Scholar]
- Hargrave M, Bowles J, Koopman P. 2006. In situ hybridization of whole-mount embryos. Methods Mol Biol 326: 103–113. 10.1385/1-59745-007-3:103 [DOI] [PubMed] [Google Scholar]
- Hirsch N, Birnbaum RY. 2015. Dual function of DNA sequences: protein-coding sequences function as transcriptional enhancers. Perspect Biol Med 58: 182–195. 10.1353/pbm.2015.0026 [DOI] [PubMed] [Google Scholar]
- Hirsch N, Eshel R, Bar Yaacov R, Shahar T, Shmulevich F, Dahan I, Levaot N, Kaplan T, Lupiáñez DG, Birnbaum RY. 2018. Unraveling the transcriptional regulation of TWIST1 in limb development. PLoS Genet 14: e1007738. 10.1371/journal.pgen.1007738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DC, Aponte JD, Liu W, Green RM, Mayeux JM, Pollard KM, Pomp D, Munger SC, Murray SA, Roseman CC, et al. 2020. Facial shape and allometry quantitative trait locus intervals in the Diversity Outbred mouse are enriched for known skeletal and facial development genes. PLoS One 15: e0233377. 10.1371/journal.pone.0233377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. 2015. Analyzing fluctuating asymmetry with geometric morphometrics: concepts, methods, and applications. Symmetry (Basel) 7: 843–934. 10.3390/sym7020843 [DOI] [Google Scholar]
- Lang B, Alrahbeni TM, Clair DS, Blackwood DH, International Schizophrenia Consortium, McCaig CD, Shen S. 2012. HDAC9 is implicated in schizophrenia and expressed specifically in post-mitotic neurons but not in adult neural stem cells. Am J Stem Cells 1: 31–41. [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ritter D, Yang N, Dong Z, Li H, Chuang JH, Guo S. 2010. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev Biol 337: 484–495. 10.1016/j.ydbio.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Zhang CL, Olson EN. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell 6: 233–244. 10.1016/S1097-2765(00)00025-3 [DOI] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra R, Mauer AC, Lino Cardenas CL, Guo X, Yao J, Zhang X, Wunderer F, Smith AV, Wong Q, Pechlivanis S, et al. 2019. HDAC9 is implicated in atherosclerotic aortic calcification and affects vascular smooth muscle cell phenotype. Nat Genet 51: 1580–1587. 10.1038/s41588-019-0514-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Twigg SR, McGowan SJ, Phipps JM, Fenwick AL, Johnson D, Wall SA, Noons P, Rees KE, Tidey EA, et al. 2017. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J Med Genet 54: 260–268. 10.1136/jmedgenet-2016-104215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Holwerda S, Vitobello A, Kitazawa T, Kohler H, Stadler MB, Rijli FM. 2017. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science 355: eaal2913. 10.1126/science.aal2913 [DOI] [PubMed] [Google Scholar]
- Mitteroecker P, Gunz P. 2009. Advances in geometric morphometrics. Evol Biol 36: 235–247. 10.1007/s11692-009-9055-x [DOI] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42: W401–W407. 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison BE, D'Mello SR. 2008. Polydactyly in mice lacking HDAC9/HDRP. Exp Biol Med 233: 980–988. 10.3181/0802-RM-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AR, Strobeck C. 2003. Fluctuating asymmetry analysis revisited. In Developmental instability: causes and consequences (ed. Polak M), pp. 279–319. Oxford University Press, New York. [Google Scholar]
- Parsons TE, Weinberg SM, Khaksarfard K, Howie RN, Elsalanty M, Yu JC, Cray JJ Jr. 2014. Craniofacial shape variation in Twist1+/− mutant mice. Anat Rec (Hoboken) 297: 826–833. 10.1002/ar.22899 [DOI] [PubMed] [Google Scholar]
- Percival CJ, Marangoni P, Tapaltsyan V, Klein O, Hallgrimsson B. 2017. The interaction of genetic background and mutational effects in regulation of mouse craniofacial shape. G3 (Bethesda) 7: 1439–1450. 10.1534/g3.117.040659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival CJ, Devine J, Darwin BC, Liu W, van Eede M, Henkelman RM, Hallgrimsson B. 2019. The effect of automated landmark identification on morphometric analyses. J Anat 234: 917–935. 10.1111/joa.12973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, et al. 2015. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat 36: 915–921. 10.1002/humu.22858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Xu Y, He T, Qin C, Xu J. 2012. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res 22: 90–106. 10.1038/cr.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rigueur D, Lyons KM. 2014. Whole-mount skeletal staining. Methods Mol Biol 1130: 113–121. 10.1007/978-1-62703-989-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager S. 2017. Chapter 9. Morpho and Rvcg – Shape Analysis in R R-packages for geometric morphometrics, shape analysis and surface manipulations. In Statistical shape and deformation analysis (ed. Zheng G, et al. ). Academic Press, New York. [Google Scholar]
- Schwartzman O, Mukamel Z, Oded-Elkayam N, Olivares-Chauvet P, Lubling Y, Landan G, Izraeli S, Tanay A. 2016. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat Methods 13: 685–691. 10.1038/nmeth.3922 [DOI] [PubMed] [Google Scholar]
- Spielmann M, Lupiáñez DG, Mundlos S. 2018. Structural variation in the 3D genome. Nat Rev Genet 19: 453–467. 10.1038/s41576-018-0007-0 [DOI] [PubMed] [Google Scholar]
- Sugo N, Oshiro H, Takemura M, Kobayashi T, Kohno Y, Uesaka N, Song WJ, Yamamoto N. 2010. Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur J Neurosci 31: 1521–1532. 10.1111/j.1460-9568.2010.07218.x [DOI] [PubMed] [Google Scholar]
- Wilderman A, VanOudenhove J, Kron J, Noonan JP, Cotney J. 2018. High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep 23: 1581–1597. 10.1016/j.celrep.2018.03.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Jaenisch R. 2014. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9: 1956–1968. 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
- Yoon JG, Hahn HM, Choi S, Kim SJ, Aum S, Yu JW, Park EK, Shim KW, Lee MG, Kim YO. 2020. Molecular diagnosis of craniosynostosis using targeted next-generation sequencing. Neurosurgery 87: 294–302. 10.1093/neuros/nyz470 [DOI] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110: 479–488. 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sui P, Dong A, Hassell J, Cserjesi P, Chen YT, Behringer RR, Sun X. 2010. Preaxial polydactyly: interactions among ETV, TWIST1 and HAND2 control anterior-posterior patterning of the limb. Development 137: 3417–3426. 10.1242/dev.051789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Marks PA, Rifkind RA, Richon VM. 2001. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci 98: 10572–10577. 10.1073/pnas.191375098 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.