Abstract
To optimize the utilization of photosynthate and avoid damage that can result from the absorption of excess excitation energy, photosynthetic organisms must rapidly modify the synthesis and activities of components of the photosynthetic apparatus in response to environmental cues. During nutrient-limited growth, cyanobacteria degrade their light-harvesting complex, the phycobilisome, and dramatically reduce the rate of photosynthetic electron transport. In this report, we describe the isolation and characterization of a cyanobacterial mutant that does not degrade its phycobilisomes during either sulfur or nitrogen limitation and exhibits an increased ratio of phycocyanin to chlorophyll during nutrient-replete growth. The mutant phenotype was complemented by a gene encoding a polypeptide with similarities to polypeptides that catalyze covalent bond formation between linear tetrapyrrole chromophores and subunits of apophycobiliproteins. The complementing gene, designated nblB, is expressed at approximately the same level in cells grown in nutrient-replete medium and medium devoid of either sulfur or nitrogen. These results suggest that the NblB polypeptide may be a constitutive part of the machinery that coordinates phycobilisome degradation with environmental conditions.
Microorganisms exhibit numerous mechanisms for sensing and responding to changes in their environment. During nutrient-limited growth, many microbes exhibit responses that are specific to the nutrient which is limiting (23, 25, 26, 29, 34, 35, 38, 44, 47, 49, 54, 56). Other responses are general and occur during any of a number of different nutrient limitation conditions (9, 12–14, 45). Specific nutrient limitation responses involve changes in the metabolism of the cell that facilitate the uptake and assimilation of the limiting nutrient. These changes may include increased synthesis of high-affinity transport systems (25, 26, 36, 38) and the production of hydrolytic enzymes (17, 40, 41) that enable organisms to assimilate alternate forms of the limiting nutrient. For example, extracellular phosphatases and sulfatases that catalyze the release of inorganic phosphate and sulfate from organic esters in the environment may be synthesized in response to phosphorus and sulfur deprivation, respectively (17, 40, 41). The general responses observed during starvation include the cessation of cell division and alterations in both the structure and metabolism of the cell (12, 14, 16, 46, 59). One of the general responses to nutrient limitation that is specific for photosynthetic organisms involves modifications of the photosynthetic apparatus (10, 11, 28, 30, 51, 53). For example, in cyanobacteria both the level and composition of the major light-harvesting complex, designated phycobilisomes (PBS), are controlled by changes in the light conditions (11, 28, 51, 53) and nutrient availability (1, 12, 60).
We have been analyzing the ways in which the cyanobacterium Synechococcus sp. strain PCC 7942 modulates PBS levels during nutrient limitation. Starvation for sulfur or nitrogen triggers the rapid and complete degradation of the PBS (1, 12, 13, 27, 60). This process occurs in an ordered fashion in which phycocyanin (PC) hexamers distal in the PBS rods are degraded, followed by the loss of whole rods and then the complete degradation of the light-harvesting complex. Proteolysis of the PBS provides the cells with some of the limiting nutrient (e.g., amino acids containing nitrogen and sulfur) but also helps minimize the absorption of excess excitation energy, which can lead to the production of reactive, harmful oxygen species (6) and overreduction of the photosynthetic electron transport chain.
The PBS gives cyanobacterial cells their typical blue-green color. Starvation for either nitrogen or sulfur triggers rapid PBS degradation in Synechococcus sp. strain PCC 7942, and the cells become a chlorotic yellow-green (1, 12, 13, 60). The loss of PBS during nutrient limitation provides a visual screen for isolating mutants that are aberrant for acclimation processes; this visual screen has led to the identification of factors that are directly involved in PBS degradation or that control the general acclimation responses (13, 45). Some of the mutants of Synechococcus sp. strain PCC 7942 that were unable to degrade their PBS during sulfur- or nitrogen-limited growth were complemented by the nblA (nonbleaching) gene, which encodes a polypeptide of 59 amino acids. Sequences similar to nblA are present on the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 (CyanoBase) and on the plastid genomes of red algae (2, 42, 55). The nblA transcript accumulates to high levels during sulfur and nitrogen starvation of Synechococcus sp. strain PCC 7942 and, to a lesser extent, during phosphorus starvation (13). The transcript is barely detectable during nutrient-replete growth. Furthermore, a strain induced for high level expression of nblA from the alkaline phosphatase promoter during phosphorus limitation becomes chlorotic as a result of the rapid and complete loss of the PBS, while a strain in which the nblA gene was inactivated exhibits a strong nonbleaching phenotype (13). We still do not know the mechanism by which NblA triggers PBS degradation and are only just beginning to learn about the ways in which the nblA gene is controlled (45).
Recently, we have focused on the control of the general nutrient stress responses in cyanobacteria, the role that these responses play in allowing survival during nutrient deprivation, and the ways in which multiple environmental signals affect these responses. We have isolated additional nonbleaching mutants of Synechococcus sp. strain PCC 7942 and identified a gene, designated nblR, encoding a transcriptional regulator that appears to control the activity of the nblA promoter. In addition to governing accumulation of the nblA transcript, NblR integrates the responses of cells to a number of different environmental cues, including high light intensity, and is absolutely essential for extended cell survival during stress conditions (45). In this report, we describe the isolation and characterization of another nonbleaching mutant that is complemented by a gene designated nblB. The role of this gene in PBS degradation is discussed.
MATERIALS AND METHODS
Strains, culture conditions and pigment analysis.
Synechococcus sp. strain PCC 7942 was grown in BG-11 medium in low light (LL) (50 μmol of photons m−2 s−1) or high light (HL) (500 μmol of photons m−2 s−1). For HL treatment, cultures were diluted to 5 × 107 cells per ml. When appropriate, the cells were starved for sulfur or nitrogen as described previously (12). For low-sulfur, solid medium, we used BG-11 medium containing 0.7% agarose (American Biorganics Inc., Niagara Falls, N.Y.) and lacking sulfur. When appropriate, antibiotics were included in the solid medium at final concentrations of 2 (ampicillin), 25 (spectinomycin), and 25 kanamycin (μg/ml). For experiments in which β-glucuronidase (GUS) activity was measured, cultures were preincubated for 18 h at a fluence of 10 μmol of photons m−2 s−1 before transfer to HL. The level of chlorophyll and PC were quantified by spectrophotometry as previously described (12).
Construction of the parental strain hliNG.
The chimeric gene hliA-GUS, in which the hliA promoter (a promoter regulated by high light intensity [see Results]) was fused to a promoterless uidA gene (which encodes GUS), was inserted into the genome of wild-type Synechococcus sp. strain PCC 7942 at a neutral site. The neutral site vector (pUC19 derivative) contained a 2.3-kbp HindIII/SalI fragment from wild-type genomic DNA with unique HpaI and NcoI sites located about 100 bp apart and 700 bp from the HindIII site in the polylinker. The spectinomycin resistance (Ω) cassette (carrying the 2.0-kbp aadA gene) (39) was inserted into the HpaI site, while a 2.9-kbp SalI/XbaI fragment containing the translational hliA-GUS fusion (18) was inserted into the NcoI site. The resulting plasmid, pND85, was used for transformation of the wild-type cells. As a result of a double homologous recombination, the hliA-GUS fusion and Ω cassette were integrated into the neutral site on the cyanobacterial genome to give strain hliGUS. The hliA promoter was also fused to the nblA coding region. This chimeric gene contained 494 bp upstream of the AUG initiator codon of hliA fused in frame at the AUG start codon of nblA. The translational fusion of hliA-nblA was cloned as a 680-bp SalI/SacI fragment into the autonomously replicating vector pCB4′(kan), which was derived from pCB4′ (24) by excising the ampicillin resistance gene from the vector with DraI and replacing it with a kanamycin resistance cassette by blunt-end ligation. The resulting shuttle vector, pND89, was transformed into hliGUS to create the parental strain hliNG.
Mutagenesis and screening.
Strain hliNG was mutagenized by N-methyl-N′-nitrosoguanidine (NTG) as described earlier (13), with some modifications. A 50-ml culture of mid-logarithmic-phase cells was pelleted by centrifugation (5,000 × g, 5 min), resuspended in 1 ml of BG-11 medium, and adjusted to an NTG concentration of 0.37 μg/ml (by addition of 80 μl of a 5-mg/ml solution). Following a 30-min incubation, the cells were washed three times with BG-11, resuspended in 50 ml of BG-11, and allowed to recover overnight under LL conditions. The cells were then concentrated by centrifugation and resuspended in 1 ml of BG-11. Aliquots of 20 μl (103 to 104 CFU) were spread onto BG-11 solid medium containing spectinomycin (25 μg/ml) and kanamycin (25 μg/ml) and incubated at 20 μmol of photons m−2s−1 for 10 to 14 days. The plates, containing the mutagenized colonies, were exposed to 500 μmol of photons m−2s−1 for 12 h, and bleaching was visually monitored. Colonies that remained green were selected and restreaked to single colonies on BG-11 solid medium containing antibiotics. HL treatment and restreaking of the cells were repeated until single-cell-derived colonies that were homogeneous for the nonbleaching phenotype were isolated. All mutants were also tested for bleaching on low-sulfur, BG-11 solid medium.
Complementation of cyanobacterial mutant and sequence analysis.
Standard molecular techniques were performed as described by Sambrook et al. (43). One of the mutants that exhibited a nonbleaching phenotype both in HL and during sulfur deprivation was complemented by transformation with a genomic library. The library, in the vector pUC118, was constructed from Synechococcus sp. strain PCC 7942 genomic DNA that was partially digested with Sau3AI. The library contains inserts with an average size of 7 kbp (45). This plasmid library cannot replicate autonomously in, but can confer ampicillin resistance to, Synechococcus upon integration into the genome by a single homologous recombination event. Transformants were screened in HL for the bleaching phenotype characteristic of the original parental strain (hliNG). Colonies selected for bleaching during HL exposure were also tested for bleaching on solid, low-sulfur BG-11 medium. After several rounds of rescreening to ensure genetic homogeneity, the genomic DNA was isolated from the complemented strain (52). pUC118 with the Synechococcus sp. strain PCC 7942 flanking DNA was recovered by digesting purified genomic DNA with BamHI (which does not cut within the plasmid), diluting and ligating the BamHI fragments, and then transforming the ligated material into Escherichia coli DH5α. The rescued plasmid contained a 9.7-kbp insert of Synechococcus sp. strain PCC 7942 genomic DNA. This plasmid, as well as subclones of the 9.7-kbp insert, were tested for the ability to complement the original nonbleaching mutant. A 2.7-kbp ClaI-SacII fragment that was able to complement the nonbleaching phenotype was sequenced by using the ABI PRISM system (Perkin-Elmer Corporation, Foster City, Calif.).
Interposon inactivation.
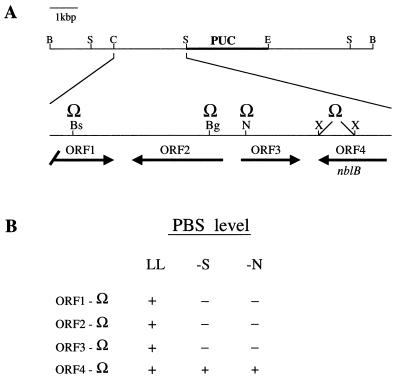
Four open reading frames (ORFs) identified by sequence analysis of the 2.7-kbp ClaI-SacII fragment (see Fig. 4) were each interrupted with the Ω cassette (34, 39). This cassette was inserted into the BsrGI, BglII, and NheI sites of ORF1, ORF2, and ORF3, respectively. ORF4 (also designated nblB) was interrupted by replacing a 400-bp XhoI fragment with the Ω cassette (see Fig. 4B). Spectinomycin-resistant transformants were screened for ampicillin sensitivity to ensure the occurrence of a double homologous recombination event (replacement of wild-type ORFs with sequences interrupted by the Ω cassette).
FIG. 4.
The region of cyanobacterial DNA that complements the nblB mutant phenotype. (A) The complementing plasmid linearized with BamHI (B). The plasmid contains a 9.7-kbp insert of which a 2.7-kbp ClaI (C)/SacII (S) fragment, enlarged and to the left of the vector sequence, is capable of complementing the mutant phenotype. The positions of ORF1, ORF2, ORF3, and ORF4 (or nblB) on the fragment are indicated. ORF1 is truncated at the 5′ end, while the remaining ORFs are complete. A restriction site for EcoRI (E) is also shown. The ORFs were individually inactivated by insertion of the Ω cassette at the BsrGI site (Bs) of ORF1, the BglI site (Bg) of ORF2, and the NheI site (N) of ORF3. ORF4 or nblB was interrupted by deletion of an XhoII fragment followed by insertion of the Ω cassette. (B) Effect of inactivation of each of the four ORFs on bleaching during sulfur (−S) and nitrogen (−N) limitation.
Quantitation of GUS activity.
GUS activity was measured by the protocol of Wilson et al. (58), and the protein concentrations of cell lysates were determined by using a Micro BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, Ill.).
RNA isolation and Northern analysis.
RNA was isolated as described previously (13). A DNA fragment extending from 119 bp upstream of the AUG start codon of the hliA gene to 97 bp downstream of the translation termination codon (18) and an nblA antisense transcript synthesized from KpnI-digested plasmid pJC40 (13) by using T7 RNA polymerase (Stratagene, La Jolla, Calif.) were used as gene-specific probes. An 800-bp SacII/PstI fragment containing the nblB gene was cloned into pBluescript SK+ to create pND20. Labeled antisense transcript from SacII-digested pND20 was synthesized with T7 RNA polymerase and used as a nblB gene-specific probe.
Nucleotide sequence accession number.
The sequence of the nblB gene has been deposited in the GenBank database and assigned accession no. AF079532.
RESULTS
Rationale of mutant screen and possible phenotypes.
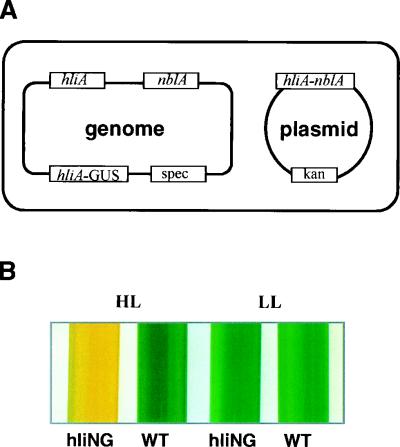
We developed a procedure for screening for mutants in two distinct regulatory pathways, one involving the induction of genes by blue UV-A light and the other involving the degradation of the PBS during nutrient-limited growth. Elements of the mutant screen are shown in Fig. 1. Synechococcus strain hliNG (Fig. 1A), which harbors two chimeric genes, was used as the parental strain for mutagenesis. Both of the chimeric genes have the promoter for hliA, but in one case it is fused to nblA (placed in a plasmid that replicates in both E. coli and Synechococcus sp. strain PCC 7942), and in the other it is fused to the uidA or GUS-encoding gene (integrated into the cyanobacterial genome). It should also be noted that strain hliNG has endogenous genomic copies of both hliA and nblA.
FIG. 1.
(A) Genetic background of the parental strain hliNG. This strain contains the native hliA and nblA genes, an autonomously replicating plasmid derived from pC4B (39) that confers kanamycin (kan) resistance to the cyanobacterium, the nblA gene fused to the hliA promoter and 5′ untranslated region at the initiator AUG codon (on the plasmid), and an hliA-GUS chimeric gene (see Materials and Methods), contiguous to the gene for spectinomycin (spec) and integrated into the cyanobacterial genome. (B) Pigmentation of wild-type (WT) cells and the hliNG strain in liquid medium in LL and HL.
The hliA gene encodes a polypeptide that resembles the Lhc polypeptides of vascular plants but appears to be part of a carotenoid-chlorophyll complex that increases when the cyanobacterium is exposed to HL (18). The hliA promoter is activated by HL, by blue /UV-A light, and to some extent by nutrient limitation. The nblA gene encodes a protein that is involved in the degradation of the light-harvesting PBS in Synechococcus sp. strain PCC 7942 (13). As shown in Fig. 1B, wild-type cells grown in liquid medium appear blue-green when exposed to either HL or LL. In contrast, when cultures of hliNG are exposed to HL, the PBS are degraded and the cells appear bleached. The bleaching is a consequence of elevated NblA levels reflecting light-activated transcription from the hliA-nblA reporter gene on the shuttle vector. Furthermore, GUS activity in this strain also increases in response to HL exposure (Fig. 2B).
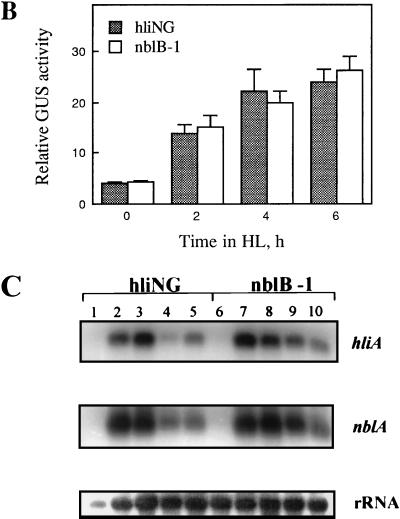
FIG. 2.
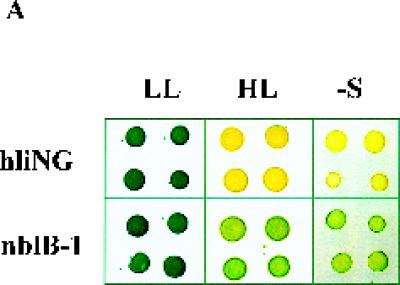
Characteristics of the nblB mutant. (A) Pigmentation phenotype of hliNG and the nblB mutant in LL, in HL, and during sulfur starvation (−S). Each grouping has four colonies derived from four individual cell aliquots. (B) GUS activity in strain hliNG and the nblB-1 mutant following exposure to HL. (C) Hybridization of labeled probes specific for hliA and nblA to RNA isolated from strain hliNG (lanes 1 to 5) or the nblB-1 strain (lanes 6 to 10) after growth at 10 μmol of photons m−2 s−1 (lane 1 and 6) or following exposure to HL for 30 min (lanes 2 and 7), 1 h (lanes 3 and 8), 2 h (lanes 4 and 9), and 4 h (lanes 5 and 10). rRNA was used as a loading control.
Following treatment of the hliNG strain with NTG, we screened for cells that could not degrade their PBS and remained blue-green during exposure to HL. Such mutants could have a lesion in the introduced hliA-nblA fusion gene, in the pathway that leads to the degradation of the PBS, or in the activation of the hliA promoter. These three different possibilities could be distinguished based on evaluation of the bleaching phenotype and measurement of GUS activity upon exposure of the potential mutants to HL, LL, and nutrient limitation. If the lesion was in the hliA-nblA reporter gene construct (Table 1, class 1), the mutant would not bleach but would show elevated GUS activity upon exposure to HL and would bleach normally during nutrient-limited (e.g., sulfur-deficient) growth. If the mutant was defective in expression from the hliA promoter (Table I, class 2), it would exhibit reduced bleaching and reduced GUS activity in HL but normal degradation of the PBS during nutrient deprivation. Finally, if the mutant was abnormal for the bleaching process (Table I, class 3), it would exhibit reduced bleaching upon exposure to HL or nutrient deprivation conditions and elevated GUS activity in HL.
TABLE 1.
Possible mutant phenotypes
| Phenotype | Phenotypea
|
|||
|---|---|---|---|---|
| Phycobilisome level
|
GUS activity in HL | |||
| LL | HL | −Sb | ||
| Parental strain hliNG | + | − | − | + |
| Class 1 (defect in hliA-nblA) | + | + | − | + |
| Class 2 (defect in expression from hliA promoter) | + | + | − | − |
| Class 3 (defect in PBS degradation) | + | + | + | + |
Visually monitored phenotype: +, high PBS or GUS level; −, low PBS or GUS level.
For −S conditions, the cells were exposed to sulfur starvation for 48 h.
Phenotype of the nblB-1 strain.
One of the strains obtained from the screen exhibited a phenotype expected of the class 3 mutants. This strain, with a mutation designated nblB-1 (nonbleaching mutant B, allele 1), does not degrade its PBS when exposed to either HL or sulfur deprivation (Fig. 2A). The pigmentation (both chlorophyll and phycobiliproteins) in the mutant does decline to some extent during sulfur starvation because the cells undergo a number of cell divisions and do not synthesize new PBS during growth. Furthermore, the GUS activity in nblB-1 is increased to a similar extent as in the parental strain hliNG upon exposure to HL (Fig. 2B). Finally, the endogenous hliA and nblA transcripts accumulate to normal levels when the mutant is exposed to HL (Fig. 2C) or to sulfur and nitrogen limitation (data not shown). These data suggest that nblB-1 can sense both HL and nutrient limitation and that it responds by synthesizing normal levels of the transcripts for hliA and nblA. However, despite the accumulation of the nblA transcript (and probably the NblA protein) under both HL and nutrient limitation conditions, the mutant is unable to degrade its PBS. Therefore, this mutant is likely to be defective in a process critical to the degradation of the PBS.
Complementation of the nblB1 mutant.
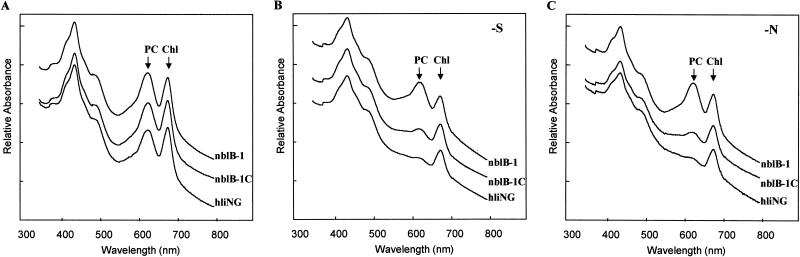
To identify the gene altered in the nblB-1 mutant strain we complemented the mutant with a wild-type recombinant DNA library in the vector pUC118 (45). As shown in the spectra in Fig. 3A, strain hliNG, the nblB-1 mutant, and the nblB-1C complemented strain all have high PC absorbance (peak at 620 nm) during nutrient-replete growth. The nblB-1 mutant consistently has higher levels of PC relative to chlorophyll than strain hliNG or the complemented strain. There is essentially no loss of PC when the nblB-1 mutant is starved for 24 h for either sulfur or nitrogen, while strain hliNG exhibits a dramatic reduction in PC (Fig. 3B and C). The nblB-1C strain is nearly indistinguishable from the parental strain hliNG, and its PBS is rapidly degraded during either sulfur or nitrogen limitation. Generally, the degradation of the PBS in the nblB-1C strain is slightly less complete than observed for hliNG. This could be a consequence of having both a mutant and a wild-type copy of the nblB gene present in a single cell.
FIG. 3.
Complementation of the nblB-1 mutant phenotype. Absorbance spectra from 300 to 800 nm of strain hliNG and the nblB-1 mutant, and nblB-1C complemented strain grown in nutrient-replete medium (A) or exposed to either sulfur (B) or nitrogen (C) limitation for 48 h are shown. Peaks for phycocyanin (PC) and chlorophyll (Chl) absorbance are indicated with arrows. The curves were offset from each other to make it easier to identify the absorbance differences.
Identification of the complementing gene.
The plasmid rescued from genomic DNA of nblB-1C digested with BamHI is shown in Fig. 4A. The smallest subclone from the rescued plasmid that was able to complement the mutant phenotype contained a 2.7-kbp ClaI-SacII fragment which in Fig. 4A is to the left of the plasmid sequence and expanded. This DNA fragment contains an ORF that is truncated at its 5′ end (ORF1) and three other complete ORFs (ORF2, ORF3, and ORF4). Inactivation of ORF1, ORF2, and ORF3 in wild-type cells had no effect on bleaching during sulfur deprivation. In contrast, inactivation of ORF4 resulted in a strain that did not bleach during sulfur or nitrogen deprivation (Fig. 4B) and contained an increased ratio of PC to chlorophyll in nutrient-replete cells (data not shown); this phenotype is similar to that of the original nblB-1 mutant. Hence, the gene containing ORF4 was named nblB. Sequence analyses of nblB in nblB-1, and of a second class 3 mutant (nblB-2) that was complemented by the 2.7-kbp ClaI-SacII fragment, localized a point mutation (see below) in the nblB genes of both strains.
Sequence of NblB.
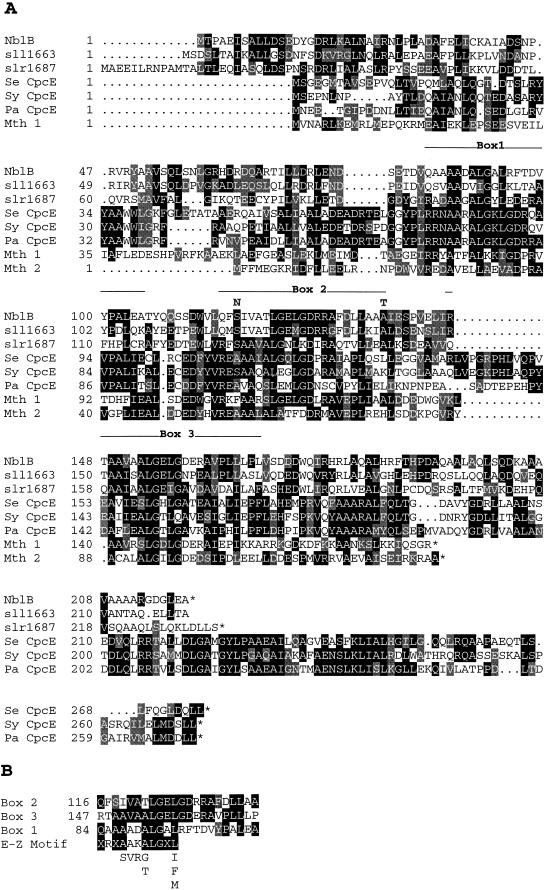
The deduced NblB protein (Fig. 5A) contains 219 amino acids, has a molecular mass of 24.3 kDa, and has an isoelectric point of 4.43. The deduced protein sequence is similar to sequences of two hypothetical proteins encoded on the genome of Synechocystis sp. strain PCC 6803 (sll1663 and slr1687) and the CpcE polypeptides from a number of cyanobacteria; the latter polypeptides are subunits of lyases (the other subunit is CpcF) that ligate linear tetrapyrrole chromophores to apophycocyanin α subunits (21, 22, 31, 32, 50, 61). Considerable blocks of similarity are present, especially within the second half of the NblB protein. There are two regions in NblB, amino acids 116 to 137 (box 2) and 147 to 168 (box 3), that have a high degree of similarity to sequences of the CpcE polypeptides. Interestingly, as shown in Fig. 5B, these two regions are similar to each other and to a less conserved region of the NblB protein from amino acids 84 to 105 (box 1). These repeat motifs are also present in the CpeZ polypeptides of Fremyella diplosiphon (32) and Pseudanabaena sp. strain PCC 7409 (19) and the PecE polypeptide of Anabaena sp. PCC 7120 (31); these polypeptides appear to be involved in chromophore attachment to phycoerythrin and phycoerythrocyanin subunits. In CpcE, the first of the three repeat motifs contains a sequence, designated the E-Z sequence (Fig. 5B), that is a signature for bilin lyase subunits (32, 61). The repeat sequences in NblB are similar but not identical to the E-Z sequence (both of the positions for positively charged amino acids in the E-Z motif are not present in the repeats of NblB).
FIG. 5.
(A) Amino acid sequence comparisons of NblB with two hypothetical proteins of Synechocystis sp. strain PCC 6803 (sll1663 and slr1687) (33), CpcE from S. elongatus (Se CpcE) (accession no. p50037), CpcE from Pseudanabaena (Pa CpcE) (20), CpcE from Synechocystis sp. strain 6803 (Sy CpcE) (slr1878) (33), and two hypothetical proteins from the methanobacteria (Mth 1 and Mth 2) (48). Amino acids identical between the NblB sequence and 30% or more of the other sequences presented are in a black background, while conserved amino acids are in a gray background. (B) Homology among the E-Z motif and the three repeat units, designated boxes 1, 2, and 3, of NblB. Identical residues are in a black background, while conserved residues are in a gray background. The position of the first amino acid of each repeat sequence is indicated. Alignments were performed with the Genetics Computer Group (Madison, Wis.) sequence analysis package.
Sequence analysis of the mutant strains demonstrated that the lesions in nblB-1 and nblB-2 would result in changes in amino acids at the borders of the repeat motif of box 2; a serine-to-asparagine change and an alanine-to-threonine change would occur at positions 118 and 138, respectively. These alterations in the mutant strains are shown above the NblB amino acid sequence presented in Fig. 5A.
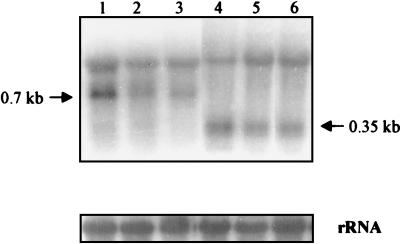
During sulfur and nitrogen limitation, the transcription of nblA dramatically increases; this increase is controlled by the nblR response regulator (45). To determine if the level of the nblB transcript is also regulated by the nutrient status of the environment, we isolated RNA from wild-type cells and the nblBΩ mutant, grown under nutrient-replete conditions or exposed to nitrogen or sulfur starvation for 4 h, and hybridized the RNA to a gene-specific nblB probe. As shown in Fig. 6, two hybridization signals were observed (e.g., lane 1). The upper band corresponds to the 16S rRNA and appears to represent nonspecific hybridization, an inference supported by the finding that this signal is still present in the nblBΩ strain (compare lanes 1 and 4). In contrast, the lower hybridization signal, approximately 700 nucleotides in wild-type cells (lanes 1 to 3), is truncated to 350 nucleotides in the nblBΩ strain (lanes 4 to 6); the smaller transcript in nblBΩ is of the size that would be predicted from the position at which the drug resistance cassette was inserted into the nblB gene. Furthermore, the nblB transcript is present at approximately the same level in cells grown in complete medium and cells starved for sulfur or nitrogen (compare lane 1 with lanes 2 and 3). Finally, the nblR mutant has normal levels of the nblB transcript under both nutrient-replete and starvation conditions (unpublished data). Hence, nblB is constitutively expressed and not under the control of NblR.
FIG. 6.
Northern blot analysis of nblB expression. RNA was isolated from both the wild type (lanes 1 to 3) and the nblBΩ mutant (lanes 4 to 6) after nutrient-replete growth (lanes 1 and 4) or growth under sulfur-deficient (lanes 2 and 5) and nitrogen-deficient (lanes 3 and 6) conditions. The RNA was hybridized to a nblB gene-specific probe described in Materials and Methods. The rRNA loading controls are shown at the bottom.
DISCUSSION
The light-harvesting PBS of cyanobacteria can comprise 30% of the total protein in the cyanobacterial cell. When cyanobacteria are starved for either nitrogen or sulfur, they degrade the PBS in an ordered fashion. We have isolated a gene, nblB, encoding a polypeptide that appears to be required for the degradation of the PBS during nutrient deprivation. The ratios of the different phycobiliprotein subunits in the nblB mutant remain the same in starved and unstarved cells (unpublished data), although the absolute level of PBS per cell declines in starved cells because the cells can undergo a few rounds of cell division without synthesizing additional PBS. Furthermore, nblB appears to be important for maintaining the ratio of PBS to chlorophyll during nutrient-replete growth. These results suggest that NblB is part of a pathway that coordinates PBS levels with ambient environmental conditions. In addition to NblB, the NblA polypeptide, which comprise only 59 amino acids, is also absolutely essential for the degradation of PBS during nutrient limitation (13).
Interestingly, the NblB polypeptide has similarity to the CpcE subunit of the PC α-subunit phycyanobilin lyases, for which genes have been characterized from a number of organisms (8, 21, 22, 32, 50, 61). These lyases are composed of two subunits, CpcE and CpcF. Mutants of the marine cyanobacterium Synechococcus sp. strain PCC 7002 that were mutated in either cpcE or cpcF accumulated very low levels of PC. Furthermore, while the β subunit of PC in these strains appeared to be normal, approximately 80% of the α subunits that were present were not associated with a chromophore (61). Instead, they were associated with a mixture of different bilin adducts (50); one of the major adducts was mesobiliverdin, which can be attached to apophycocyanin subunits in a nonenzymatic in vitro reaction (3–5). Inactivation of cpcE or cpcF in Synechococcus sp. strain PCC 6301 also resulted in reduced accumulation of PC (8). Furthermore, in vitro experiments using the CpcE and CpcF polypeptides overexpressed in E. coli demonstrated that these polypeptides formed heterodimers that were capable of catalyzing the addition of phycocyanobilin to the cysteine (Cys-84) of the chromophore binding site of the α subunits of PC (21, 22). Together, these results strongly suggest that both CpcE and CpcF are required for the attachment of the phycocyanobilin chromophore to the α subunits of PC.
Since the initial discovery of the PC α-subunit phycocyanobilin lyase, some of the genes encoding analogous lyases thought to play a role in attachment of bilin tetrapyrroles to phycoerythrin (32, 57) and phycoerythrocyanin (31) subunits have been identified. In all cases, the lyase genes are part of operons encoding phycobiliproteins. In Synechococcus sp. strain PCC 6301, cpcE and cpcF are located downstream of the cpcB2A2 gene cluster. The cpcE and cpcF genes are also located downstream of the cpcBA genes in Pseudanabaena (20), Anabaena sp. strain PCC 7120 (7), F. diplosiphon (15), and Calothrix sp. strain PCC 7601 (37).
While there is substantial similarity between NblB and CpcE, the latter polypeptides of Synechococcus elongatus (accession no. P50037), Pseudanabaena (20), and Synechocystis sp. strain PCC 6803 (slr1878) are longer at the C terminus by approximately 55 amino acids. There are two hypothetical proteins in Synechocystis sp. strain PCC 6803 (sll1663 and slr1687) that also show considerable similarity to NblB but are the same size, suggesting that they are functional analogues. Furthermore, there are two ORFs in the methanobacteria with considerable similarity to both NblB and CpcE (48). The methanobacterial ORFs, the phycobiliprotein lyases, and the putative NblB analogues in Synechocystis sp. strain PCC 6803 all have the repeated motif presented in Fig. 5B; these repeats may serve equivalent functions among all of these polypeptides.
The phenotype of the strain in which nblB is inactivated is dramatically different from that of a strain in which the cpcE gene is disrupted. Mutants of cpcE are not able to efficiently attach a chromophore to cysteine 84 of the α subunit of PC and appear chlorotic since most the PC that is synthesized cannot achieve stable assembly into PBS (and the α subunit of PC that is present is missing the chromophore or exhibits attachment of an atypical chromophore). In contrast, mutants of nblB are unable to degrade their PBS under conditions that normally favor PBS degradation, such as sulfur or nitrogen deprivation. Furthermore, the nblB mutants exhibit higher PC levels than wild-type cells grown in nutrient-replete conditions. Hence, it is likely that NblB is part of the machinery that coordinates the degradation of the PBS with changes in ambient environmental conditions. The similarity between NblB and CpcE (and CpeZ and PecE) may reflect the ability of both polypeptides to bind tetrapyrrole chromophores of phycobiliprotein subunits via the common repeated motif. However, the former may also facilitate cleavage of the tetrapyrrole from PBS subunits. A similar motif in the hypothetical methanobacterial polypeptides suggests that they also may function via an association with linear tetrapyrroles.
While NblB and NblA are both necessary to elicit PBS degradation during nutrient limitation, the nblB transcript, unlike the nblA transcript, does not accumulate during nutrient limitation. Instead, the nblB gene is constitutively expressed. This is consistent with the finding that engineered expression of nblA, under conditions that normally do not result in the loss of PBS, elicits the degradation of PBS. Hence, to initiate bleaching during nutrient limitation the only gene that has to be activated is nblA.
ACKNOWLEDGMENTS
We thank Rakefet Schwarz and Devaki Bhaya for discussion of the data and critical reading of the manuscript.
This work was supported by the Carnegie Institution of Washington and by USDA grant 94-37306-0344 and NSF grant MCB 9727836 awarded to A.R.G.
Footnotes
We dedicate this paper to Wolfhart Ruediger in celebration of his 65th birthday. Wolfhart has devoted much of his scientific career to analyzing the structure and function of tetrapyrroles in photosynthetic organisms.
Carnegie Institution of Washington publication no. 1377.
REFERENCES
- 1.Allen M M, Smith A J. Nitrogen chlorosis in blue-green algae. Arch Microbiol. 1969;69:114–120. doi: 10.1007/BF00409755. [DOI] [PubMed] [Google Scholar]
- 2.Apt K E, Grossman A R. Characterization and transcript analysis of the major phycobiliprotein subunit genes from Aglaothamnion neglectum (rhodophyta) Plant Mol Biol. 1993;21:27–38. doi: 10.1007/BF00039615. [DOI] [PubMed] [Google Scholar]
- 3.Arciero D M, Bryant D A, Glazer A N. In vitro attachment of bilins to apophycocyanin. I. Specific covalent adduct formation at cysteinyl residues involved in phycocyanobilin binding in C-phycocyanin. J Biol Chem. 1988;34:18343–18349. [PubMed] [Google Scholar]
- 4.Arciero D M, Dallas J L, Glazer A N. In vitro attachment of bilins to apophycocyanin. II. Determination of the structure of tryptic bilin peptides derived from the phycocyanobilin adduct. J Biol Chem. 1988;34:18350–18357. [PubMed] [Google Scholar]
- 5.Arciero D M, Dallas J L, Glazer A N. In vitro attachment of bilins to apophycocyanin. III. Properties of the phycoerythrobilin adduct. J Biol Chem. 1988;34:18358–18363. [PubMed] [Google Scholar]
- 6.Asada K. Production and action of active oxygen species in photosynthetic tissues. In: Foyer C H, Mullineaux P M, editors. Causes of photooxidative stress and amelioration of defense systems in plants. Boca Raton, Fla: CRC Press; 1994. pp. 77–104. [Google Scholar]
- 7.Belknap W R, Haselkorn R. Cloning and light regulation of expression of the phycocyanin operon of the cyanobacterium Anabaena. EMBO J. 1987;6:871–884. doi: 10.1002/j.1460-2075.1987.tb04833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhalerao R P, Lind L K, Gustafsson P. Cloning of the cpcE and cpcF genes from Synechococcus sp. PCC 6301 and their inactivation in Synechococcus sp. PCC 7942. Plant Mol Biol. 1994;26:313–326. doi: 10.1007/BF00039542. [DOI] [PubMed] [Google Scholar]
- 9.Bhaya, D., R. Schwarz, and A. R. Grossman. Molecular responses to environmental stress. In M. Potts and B. Whitton (ed.), Ecology of cyanobacteria: diversity in time and space, in press. Kluwer Publishers, Dortrecht, The Netherlands.
- 10.Bryant D A. The photoregulated expression of multiple phycocyanin species: general mechanism for control of phycocyanin synthesis in chromatically adapting cyanobacteria. Eur J Biochem. 1981;119:425–429. doi: 10.1111/j.1432-1033.1981.tb05625.x. [DOI] [PubMed] [Google Scholar]
- 11.Bryant D A, Cohen-Bazire G. Effects of chromatic illumination on cyanobacterial phycobilisomes: evidence for the specific induction of a second pair of phycocyanin subunits in Pseudanabaena 7409 grown in red light. Eur J Biochem. 1981;119:415–424. doi: 10.1111/j.1432-1033.1981.tb05624.x. [DOI] [PubMed] [Google Scholar]
- 12.Collier J L, Grossman A R. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol. 1992;174:4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier J L, Grossman A R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994;13:1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier J L, Herbert S K, Fork D C, Grossman A R. Changes in the cyanobacterial photosynthetic apparatus in response to macronutrient deprivation. Photosyn Res. 1994;42:173–183. doi: 10.1007/BF00018260. [DOI] [PubMed] [Google Scholar]
- 15.Conley P B, Lemaux P G, Grossman A R. Molecular characterization and evolution of sequences encoding light harvesting components in the chromatically adapting cyanobacterium Fremyella diplosiphon. J Mol Biol. 1988;199:447–465. doi: 10.1016/0022-2836(88)90617-1. [DOI] [PubMed] [Google Scholar]
- 16.Davies J, Yildiz F, Grossman A R. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 1996;15:2150–2159. [PMC free article] [PubMed] [Google Scholar]
- 17.de Hostos E L, Togasaki R K, Grossman A R. Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J Cell Biol. 1988;106:29–37. doi: 10.1083/jcb.106.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolganov N A M, Bhaya D, Grossman A R. Cyanobacterial protein with similarity to the chlorophyll a/b-binding proteins of higher plants: evolution and regulation. Proc Natl Acad Sci USA. 1995;92:636–640. doi: 10.1073/pnas.92.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubbs J M, Bryant D A. Molecular cloning and transcriptional analysis of the cpeBA operon from the cyanobacterium Pseudanabaena species PCC 7409. Mol Microbiol. 1991;5:3073–3085. doi: 10.1111/j.1365-2958.1991.tb01867.x. [DOI] [PubMed] [Google Scholar]
- 20.Dubbs J M, Bryant D A. Organization and transcription of the genes encoding two differentially expressed phycocyanins in the cyanobacterium Pseudoanabaena sp. PCC 7409. Photosyn Res. 1993;36:169–183. doi: 10.1007/BF00033036. [DOI] [PubMed] [Google Scholar]
- 21.Fairchild C D, Glazer A N. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin α subunit phycocyanobilin lyase. J Biol Chem. 1994;269:8686–8694. [PubMed] [Google Scholar]
- 22.Fairchild C D, Zhao J, Zhou J, Colson S E, Bryant D A, Glazer A N. Phycocyanin α-subunit phycocyanobilin lyase. Proc Natl Acad Sci USA. 1992;89:7017–7021. doi: 10.1073/pnas.89.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flores E, Guerrero M G, Losada M. Short-term ammonium inhibition of nitrate utilization by Anacystis nidulans and other cyanobacteria. Arch Microbiol. 1980;128:137–144. [Google Scholar]
- 24.Gendel S, Straus N, Pulleybank D, Williams J. Shuttle cloning vectors for the cyanobacterium Anacystis nidulans. J Bacteriol. 1983;156:148–154. doi: 10.1128/jb.156.1.148-154.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green L S, Grossman A R. Changes in sulfate transport characteristics and protein composition of Anacystis nidulans R2 during sulfur deprivation. J Bacteriol. 1988;170:583–587. doi: 10.1128/jb.170.2.583-587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grillo J F, Gibson J. Regulation of phosphate accumulation in the cyanobacterium Synechococcus. J Bacteriol. 1979;140:508–517. doi: 10.1128/jb.140.2.508-517.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossman A R, Bhaya D, Apt K E, Kehoe D M. Light-harvesting complexes in oxygenic photosynthesis: diversity, control and evolution. Annu Rev Genet. 1995;29:231–287. doi: 10.1146/annurev.ge.29.120195.001311. [DOI] [PubMed] [Google Scholar]
- 28.Grossman A R, Kehoe D M. Phosphorelay control of phycobilisome biogenesis during complementary chromatic adaptation. Photosyn Res. 1997;53:95–108. [Google Scholar]
- 29.Herrero A, Guerrero M G. Regulation of nitrate reductase in the cyanobacterium Anacystis nidulans. J Gen Microbiol. 1986;132:2463–2468. [Google Scholar]
- 30.Horton P, Ruban A B, Walters R G. Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:655–684. doi: 10.1146/annurev.arplant.47.1.655. [DOI] [PubMed] [Google Scholar]
- 31.Jung L J, Chan C F, Glazer A N. Candidate genes for the phycoerythrocyanin α subunit lyase. J Biol Chem. 1995;270:12877–12884. doi: 10.1074/jbc.270.21.12877. [DOI] [PubMed] [Google Scholar]
- 32.Kahn K, Mazel D, Houmard J, Tandeau de Marsac N, Schaefer M R. A role for CpeYZ in cyanobacterial phycoerythrin biosynthesis. J Bacteriol. 1997;179:998–1006. doi: 10.1128/jb.179.4.998-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa H, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein coding regions. DNA Res. 1996;2:153–166. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 34.Laudenbach D E, Grossman A R. Characterization and mutagenesis of sulfur-regulated genes in a cyanobacterium: evidence for function in sulfate transport. J Bacteriol. 1991;173:2739–2750. doi: 10.1128/jb.173.9.2739-2750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee T-Y, Makino K, Shinagawa H, Amemura M, Nakata A. Phosphate regulon in members of the family Enterobacteriaceae: comparison of the phoB-phoR operons of Escherichia coli, Shigella dysenteriae, and Klebsiella pneumoniae. J Bacteriol. 1989;171:6593–6599. doi: 10.1128/jb.171.12.6593-6599.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandueño F, Vega-Palas M A, Flores E, Herrero A. A cytoplasmic-membrane protein repressible by ammonium in Synechococcus R2 altered expression in nitrate-assimilation mutants. FEBS Lett. 1988;239:289–291. [Google Scholar]
- 37.Mazel D, Houmard J, Tandeau de Marsac N. A multigene family in Calothrix sp. PCC 7601 encodes phycocyanin, the major component of the cyanobacterial light-harvesting antenna. Mol Gen Genet. 1988;211:296–304. doi: 10.1007/BF00330607. [DOI] [PubMed] [Google Scholar]
- 38.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport of the cyanobacterium Synechococcus sp. PCC 7942. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 39.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 40.Quisel J, Wykoff D, Grossman A R. Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 1996;111:839–848. doi: 10.1104/pp.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray J M, Bhaya D, Block M A, Grossman A R. Isolation, transcription, and inactivation of the gene for an atypical alkaline phosphatase of Synechococcus sp. strain PCC 7942. J Bacteriol. 1991;173:4297–4309. doi: 10.1128/jb.173.14.4297-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reith M E, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Scanlan D J, Mann N H, Carr N G. The response of the picoplanktonic marine cyanobacterium Synechococcus sp. WH7803 to phosphate starvation involves a protein homologous to the periplasmic phosphate-binding protein of Escherichia coli. Mol Microbiol. 1993;10:181–191. doi: 10.1111/j.1365-2958.1993.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 45.Schwarz R, Grossman A R. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc Natl Acad Sci USA. 1998;95:11008–11013. doi: 10.1073/pnas.95.18.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman D M, Sherman L A. Effect of iron deficiency and iron restoration on ultrastructure of Anacystis nidulans. J Bacteriol. 1983;156:393–401. doi: 10.1128/jb.156.1.393-401.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirko A, Hryniewicz M, Hulanicka D, Bück A. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J Bacteriol. 1990;172:3351–3357. doi: 10.1128/jb.172.6.3351-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qui D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum DH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki I, Horie N, Sugiyama T, Omata T. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC 7942 required for maximum efficiency of nitrogen assimilation. J Bacteriol. 1995;177:290–296. doi: 10.1128/jb.177.2.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson R V, Zhou J, Leary J A, Williams T, de Lorimier R, Bryant D A, Glazer A N. Characterization of phycocyanin produced by cpcE and cpcF mutants and identification of an intergenic suppressor of the defect in bilin attachment. J Biol Chem. 1992;267:16146–16154. [PubMed] [Google Scholar]
- 51.Tandeau de Marsac N. Occurrence and nature of chromatic adaptation in cyanobacteria. J Bacteriol. 1977;130:82–91. doi: 10.1128/jb.130.1.82-91.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tandeau de Marsac N, Borrias W E, Kuhlemeier C J, Castets A M, van Arkel G A, van den Hondel C A M J J. A new approach for molecular cloning in cyanobacteria: cloning of an Anacystis nidulans met gene using a Tn901-induced mutant. Gene. 1982;20:111–119. doi: 10.1016/0378-1119(82)90092-0. [DOI] [PubMed] [Google Scholar]
- 53.Tandeau de Marsac N, Houmard J. Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms. FEMS Microbiol Rev. 1993;104:119–190. [Google Scholar]
- 54.Utkilen H C, Heldal M, Knutsen G. Characterization of sulfate uptake in Anacystis nidulans. Physiol Plant. 1976;38:217–220. [Google Scholar]
- 55.Valentin K, Maid U, Emich A, Zetsche K. Organization and expression of a phycobiliprotein gene cluster from the unicellular red alga Cyanidium caldarium. Plant Mol Biol. 1992;20:267–276. doi: 10.1007/BF00014494. [DOI] [PubMed] [Google Scholar]
- 56.Wanner B L. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- 57.Wilbanks S M, Glazer A N. Rod structure of a phycoerythrin II-containing phycobilisome. I. Organization and sequence of the gene cluster encoding the major phycobiliprotein rod components in the genome of marine Synechococcus sp. WH8020. J Biol Chem. 1993;268:1226–1235. [PubMed] [Google Scholar]
- 58.Wilson K J, Jefferson R A, Hughes S G. The Escherichia coli gus operon: induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria. In: Gallagher S R, editor. GUS protocols. Using the GUS gene as a reporter of gene expression. San Diego, Calif: Academic Press, Inc.; 1992. pp. 7–22. [Google Scholar]
- 59.Wykoff D D, Davies J P, Melis A, Grossman A R. The regulation of photosynthetic electron transport during nutrient deprivation of Chlamydomonas reinhardtii. Plant Physiol. 1998;117:129–139. doi: 10.1104/pp.117.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamanaka G, Glazer A N. Dynamic aspects of phycobilisome structure. Phycobilisome turnover during nitrogen starvation in Synechococcus sp. Arch Microbiol. 1980;124:39–47. [Google Scholar]
- 61.Zhou J, Gasparich G E, Stirewalt V L, de Lorimier R, Bryant D A. The cpcE and cpcF genes of Synechococcus sp. PCC 7002. Construction and phenotypic characterization of interposon mutants. J Biol Chem. 1992;267:16138–16145. [PubMed] [Google Scholar]