Abstract
Agrobacterium tumefaciens C58 and its derivatives give rise to spontaneous mutants resistant to tetracycline at a high frequency. We observed that a mutation affecting a tRNA processing function significantly affected the emergence of such mutants, suggesting that C58 contained a positively acting gene conferring resistance to tetracycline. A cosmid clone conferring resistance to tetracycline in Escherichia coli and Agrobacterium was isolated from a genomic bank of one such mutant. Subcloning, transposon mutagenesis, and DNA sequence analysis revealed that this DNA fragment contained two divergently transcribed genes, tetA and tetR, encoding products that were very similar to proteins of the Tet(A) class of tetracycline resistance systems. In the clone from this mutant, tetR was disrupted by an IS426. The homologous region from wild-type NT1 contained an intact tetR gene and did not confer resistance to tetracycline. Hybridization analysis showed that of 22 members of the genus Agrobacterium surveyed, only strains C58 and T37 contained the tet determinant. Moreover, only these two strains mutated to resistance to this antibiotic. Unlike other Tet(A) systems, neither tetracycline nor a series of its derivatives induced the expression of this tet gene unit. Other polycyclic compounds, including many of plant origin, also did not induce this tet gene system. The divergent promoter region of this tet system contained a single inverted repeat element identical to one such operator repeat in the promoter region of the tet determinant from the IncP1α R plasmid RP4. TetR repressor proteins from the Agrobacterium tet system and from RP4 interacted with the heterologous operators. While the repressive effect of the TetR protein from strain C58 (TetRC58) on the tetA gene from strain RP4 (tetARP4) was not relieved by tetracycline, repression of tetAC58 by TetRRP4 was lifted by this antibiotic.
Genes encoding resistance to antibiotics have been identified in a wide spectrum of prokaryotic organisms, including gram-negative and gram-positive bacteria. Although most of the genes characterized to date have been isolated from clinical isolates, these determinants are believed to have originated in soil bacteria (38). More recently, genes encoding proteins that confer resistance to several different drugs, the multidrug systems, have been identified and characterized (37). Horizontal transfer of these determinants among diverse bacteria are challenging the effectiveness of antibiotics in infectious disease control (39, 48).
Genes conferring resistance to tetracycline are among the most abundant of the identified drug resistance elements (29). Although mechanisms such as target site protection and drug inactivation are known (7, 44), most tetracycline resistance determinants function through the efflux mechanism, pumping a drug-metal ion complex out of the bacterial cell by a process energized by the membrane proton gradient (37, 45).
It is a well-recognized but unpublished observation that Agrobacterium tumefaciens C58 and its derivatives give rise at a high frequency to spontaneous mutants resistant to tetracycline. Most genetic and molecular analyses concerning A. tumefaciens and its plasmids are conducted in the C58 chromosomal background. Since many broad-host-range cloning vectors encode resistance to tetracycline as the selectable marker, the propensity of this strain to mutate to tetracycline resistance poses problems to the genetic analysis of this bacterium and to its use as a plant transformation agent.
During a study of mutants affecting the induction of tra genes of pTiC58 in NT1 (51), a Ti plasmid-cured derivative of C58 (53), we observed that one mutant, NTM7, while remaining able to mutate to tetracycline resistance, expressed the phenotype at a much lower level than did its wild-type parent. NTM7 later was found to carry a Tn5 insertion in a locus encoding a protein homologous to the product of the Escherichia coli rnd gene (30). In E. coli, a miaA mutant is unable to express Tet(M)-mediated tetracycline resistance (8). Since both the rnd and miaA genes are involved in tRNA modification (12, 52), we reasoned that there may be a positively acting tetracycline resistance element in the genome of strain C58 which can function normally in the wild-type bacterium but not in the rnd mutant. A study was initiated to investigate the nature of such tetracycline-resistant mutants. We confirmed our hypothesis and report here the molecular cloning, nucleotide sequence, and genetic characterization of a tetracycline resistance gene unit from this A. tumefaciens strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains of E. coli and A. tumefaciens and the plasmids used in this study are listed in Table 1. E. coli strains were grown at 37°C in L broth (LB), on L agar plates (Difco Laboratories, Detroit, Mich.), or in A medium (33). Agrobacterium strains were grown at 28°C in LB or on nutrient agar (NA) (Difco Laboratories). AB medium (11) supplemented with 0.2% mannitol as the sole carbon source was used as the defined minimal medium for Agrobacterium strains. Biotin was added to a final concentration of 2 μg/ml for biovar 2 strains. Agar (Difco Laboratories) was added at 1.5% to make solid medium. Except when specified in the text, antibiotics were used at the following concentrations: for E. coli, kanamycin at 50 μg/ml, tetracycline at 10 μg/ml, and ampicillin at 100 μg/ml; for Agrobacterium spp., kanamycin at 50 μg/ml, carbenicillin at 50 or 100 μg/ml, and tetracycline at 2 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma Chemical Co., St. Louis, Mo.) was included in the medium at 40 μg/ml to monitor the production of β-galactosidase.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 Δ(lacZYA-argF)U169 | 40 |
| S17-1 | Pro− Res− Mod+recA; integrated RP4-Tet::Mu-Kan::Tn7, Mob+ | 43 |
| S17-1(pHoHo1, pSShe) | Ampr Cmr; source for Tn3HoHo1 | 46 |
| LE392 | F−hsdR514(rK− mK+) supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 | Promega |
| A. tumefaciens | ||
| C58 | Wild-type A. tumefaciens strain containing the nopaline-type Ti plasmid pTiC58 | 53 |
| NT1 | pTiC58-cured derivative of strain C58 | 51 |
| NT1TcR1 | Randomly picked tetracycline-resistant mutant of NT1 | This study |
| Plasmids | ||
| pBluescriptSK(+) | Ampr; cloning vector | Stratagene |
| pCP13/B | Tcr; IncP1α; broad-host-range cosmid vector | 15 |
| pRK415K | Tcr Kmr; IncP1α; broad-host-range cloning vector | 14 |
| pDSK519 | Kmr; IncQ; broad-host-range cloning vector | 25 |
| pBAD22 | Ampr; expression vector controlled by PBAD | 19 |
| pRG970b | Spr/Strr Ampr; promoter selection vector | 12 |
| pJB3 | Ampr; broad-host-range cloning vector | 3 |
| pZL1 | 1.7-kb BglII cos fragment from pCP13/B cloned into pJB3 | This study |
| pBBRMCS-2 | Kmr; broad-host-range cloning vector of unknown Inc group | 27 |
| pDLB4 | Kmr; PBAD from pBAD22 cloned into pBBRMCS-2 | This study |
| pZLT1 | Cosmid clone containing Tcr trait from NT1TcR1 in pZL1 | This study |
| pZLE4.5 | 4.5-kb EcoRI fragment containing the Tcr trait in pZL1 | This study |
| pDLE4.5 | 4.5-kb EcoRI fragment containing the Tcr trait in pDSK519 | This study |
| pZLT2 | Cosmid clone containing the wild-type tetC58 locus from NT1 | This study |
| pZLHE1.6 | 1.6-kb HindIII/EcoRI fragment containing IS426-disrupted tetRC58 from pZLE4.5 cloned into pBluescript SK(+) | This study |
| pZLHS2.9 | 2.9-kb HindIII/SmaI fragment containing tetAC58 from pZLE4.5 cloned into pBluescript SK(+) | This study |
| pZLE4.8 | 4.8-kb EcoRI fragment containing portions of IS426 and tetRC58 from pZLT1 cloned into pBluescript SK(+) | This study |
| pZLE8.5 | 8.5-kb EcoRI fragment containing the wild-type tetC58 locus from pZLT2 cloned into pBluescript SK(+) | This study |
| pZLOP1 | 66-bp tetC58 intergenic region cloned into promoter selection vector pRG970b | This study |
| pBetR | tetRC58 cloned as a 2.7-kb NcoI fragment into pBAD22 | This study |
| pDLB4-tetR | tetRC58 cloned as a 2.7-kb EcoRI/BamHI fragment from pBetR into pDLB4 | This study |
Ampr, ampicillin resistant; Cmr, chloramphenical resistant; Kmr, kanamycin resistant; Spr, spectinomycin resistant; Tetr or Tcr, tetracycline resistant; Strr, streptomycin resistant.
DNA manipulations.
Plasmids were isolated by an alkaline lysis method (40). Cloning followed standard recombinant DNA techniques (40). Restriction digestions were carried out in accordance with the instructions of the manufacturers, and digestion products were separated by electrophoresis in agarose gels with Tris-borate-EDTA buffer (40). DNA fragments were recovered from agarose gels by using GenElute spin columns (Supelco Inc., Bellefonte, Pa.). Plasmids were introduced into E. coli by CaCl2-mediated transformation (40) and into A. tumefaciens by S17-1-mediated mating (43) or by electroporation (9).
Genomic bank construction.
Cosmid pZL1 was constructed by cloning the cos site from pCP13 (15) as a 1.7-kb BglII fragment into pJB3 (6). Genomic DNA was digested with Sau3AI to give an average fragment size of 40 kb and ligated to pZL1 that had been digested with BamHI and treated with alkaline phosphatase. The ligation mixture was packaged into phage λ by using the Packagene system (Promega Corporation, Madison, Wis.) and transduced into E. coli LE392.
Southern hybridization.
Genomic DNA was prepared from overnight cultures of Agrobacterium spp. as described previously (18). After digestion with restriction endonuclease, DNA fragments were separated on 0.7% agarose gels and transferred to a nylon membrane. DNA probes were randomly labeled by using the Genius digoxigenin kit (Boehringer GmbH, Mannheim, Germany). Hybridization, washing, and detection followed protocols provided by the manufacturer. Hybridization and washing were performed under conditions of medium stringency.
Tn3HoHo1 mutagenesis.
The insert in clone pDLE4.5 (Table 1) was mutagenized with Tn3HoHo1 as described previously (46). Sites and orientations of the insertions were determined by restriction mapping. Each mutant was tested for tetracycline resistance on LB medium containing this antibiotic.
DNA sequence analysis.
DNA fragments containing the tet region were sequenced by use of the chain termination method (41) by the Genetic Engineering Facility at the University of Illinois. DNA sequences were analyzed by using DNA Strider (32). The BLAST (4) protocols were used to search the GenBank database for related sequences. The GAP subroutine of the GCG program (Genetics Computer Group, Madison, Wis.) was used to compare sequences for similarity.
Isolation of tetracycline-resistant mutants.
Agrobacterium spp. were grown in LB liquid medium to saturation. A 200-μl volume of culture from each strain was spread onto NA medium containing tetracycline at 5 or 10 μg/ml. The capacity to give tetracycline-resistant mutants was assessed by the appearance of colonies on the plates after 72 h of incubation at 28°C.
Gene expression assays.
E. coli and A. tumefaciens strains were grown in A and AB-mannitol media, respectively. Overnight cultures were washed and diluted 40-fold in the same respective media, and incubation was continued until the optical density at 600 nm (OD600) reached about 0.7 for E. coli and 0.3 for A. tumefaciens. When needed, cultures were divided, and inducers (arabinose or tetracycline) were added to one subculture at the concentrations specified in Results. β-Galactosidase activity was determined quantitatively as described previously (33). β-Glucuronidase activity was measured as described previously (13, 24, 49). The activity of this enzyme was calculated by using the Miller formula (33), with OD420 replaced by OD415. All enzymatic activities were expressed as units per 109 CFU.
Induction assays.
The ability of tetracycline, its derivatives, and related compounds to induce the tet system of strain C58 (tetC58 system) was assessed two ways. In the first, reporter strains were inoculated onto medium containing X-Gal and the compound being tested. The appearance of blue colonies signalled the induction of the reporter gene. In the second, reporter strains were grown in 30 ml of liquid medium with appropriate antibiotics to saturation and mixed with 50 ml of warm 0.8% agar with X-Gal, and 6-ml volumes were layered over a base of LB or AB medium in 100-mm-diameter petri dishes. Discs of Whatman 3MM paper (0.5-cm diameter), onto which a few crystals of the compound to be tested were sprinkled, were laid on the surface of the soft agar. Dissolution and diffusion of the chemical give a concentration gradient around the disc. The plates were incubated at 28°C for A. tumefaciens reporter strains and at 37°C for E. coli reporter strains and examined at intervals over a 3-day period. Susceptibility to a given compound was indicated by a zone of growth inhibition around the disc. Induction of the reporter by the compound was indicated by a blue zone around the disc or, in cases where the compound also was inhibitory, by a blue zone in the area of growth at the edge of the zone of inhibition.
Nucleotide sequence accession number.
DNA sequences of wild-type tetC58 were deposited in the GenBank database under accession no. AF090987.
RESULTS
Cloning a tetracycline resistance determinant from A. tumefaciens C58.
Plating strain C58 onto medium containing tetracycline at concentrations ranging from 0.5 to 20 μg/ml results in the appearance at a high frequency of mutants resistant to this antibiotic (Fig. 1A and data not shown). However, we observed that a C58 mutant with a Tn5 insertion in a gene homologous to rnd (30) yielded tetracycline-resistant mutants that grew much more slowly on medium containing the antibiotic (Fig. 1B). Since a miaA mutant of E. coli does not express Tet(M)-mediated tetracycline resistance (8) and both rnd and miaA genes encode products involved in tRNA modification, we reasoned that strain C58 contains an active tetracycline resistance element that functions in the rnd+ but not in the rnd mutant background. To test this, a genomic library of NT1TcR1, a tetracycline-resistant mutant of the otherwise-wild-type strain NT1, was constructed in the broad-host-range cosmid vector pZL1 (Table 1). This bank, in E. coli LE392, consists of 856 clones containing cosmids with an average insert size of 42 kb. The bank was screened on LB medium containing tetracycline, and one cosmid that confers resistance to this antibiotic on its E. coli host was isolated. This clone, named pZLT1, also conferred resistance to high levels of this antibiotic on several Agrobacterium strains, including 15955, R10, and K84 (data not shown).
FIG. 1.
A mutation in a tRNA processing gene in NTM7 (29) affects expression of resistance in spontaneous tetracycline-resistant mutants of NT1. Approximately 108 cells of strains NT1 (A) and NTM7 (B) were spread on NA plates supplemented with tetracycline at 5 μg/ml. The plates were incubated for 3 (A) or 6 (B) days at 28°C. Note that the tetracycline-resistant mutants of NTM7 arising on the plate grow considerably more slowly than do those of strain NT1 on medium containing the antibiotic.
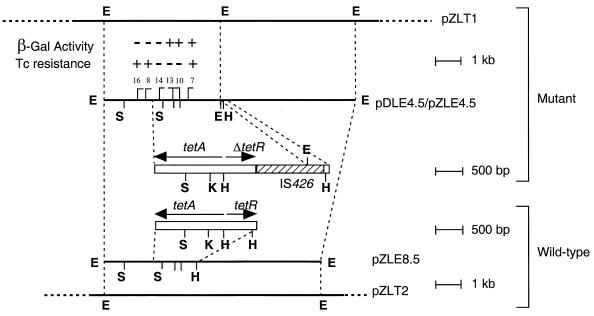
The tetracycline resistance trait was subcloned from pZLT1 as a 4.5-kb EcoRI fragment into pZL1 to generate pZLE4.5 (Fig. 2). To localize the tet determinant, this DNA fragment was recloned into pDSK519 (25) to give pDLE4.5, and this plasmid was mutagenized with Tn3HoHo1 (46). The location and orientation of the insertion in each mutant plasmid were determined, and each was tested for its ability to confer resistance to tetracycline. Of six mutations mapped, the three contiguous insertions that are located in the middle of the fragment abolished resistance to this antibiotic (Fig. 2).
FIG. 2.
Genetic organization of the tet locus from the tetracycline-resistant and wild-type genomes of A. tumefaciens C58. A restriction map and the locations of the independent Tn3HoHo1 insertions, identified in clone pDLE4.5, are shown. The response of each mutant to tetracycline was assessed as described in Materials and Methods and is expressed as resistant to the antibiotic (+) or susceptible to the antibiotic (−). The horizontal crossbar capping each insertion site indicates the direction of transcription of the lacZ reporter of Tn3HoHo1 as determined by restriction mapping. β-Galactosidase activity was assessed on medium containing X-Gal: the presence of blue colonies (+) or white colonies (−) is indicated. The IS426 element is indicated by the hatched portion of the bar. S, SmaI; E, EcoRI; K, KpnI; H, HindIII; Tc, tetracycline.
DNA sequence analysis.
DNA fragments corresponding to the region defined by the Tn3HoHo1 insertion were subcloned and subjected to sequence analysis. BLAST searches revealed that one such EcoRI-HindIII fragment from plasmid pZLHE1.6 (Table 1) contained a portion of a tetR(A) homologue. As sequencing continued, however, this gene was abruptly disrupted by a totally different DNA sequence identical to the sequence of IS426, an IS element identified from A. tumefaciens T37 (50). Apparently, the 4.5-kb EcoRI fragment from the mutant contains an incomplete tet locus; insertion of IS426 introduced an EcoRI site which divided this region into two fragments when this enzyme was used for cloning. To obtain the complete sequence of the mutant version, the adjacent 4.9-kb EcoRI fragment was cloned into pBluescript SK(+) from pZLT1 to generate pZLE4.8 (Table 1). Analysis of the insert identified sequences corresponding to the remainder of IS426 followed by the 3′ end of the tetR gene of strain C58 (tetRC58).
Analysis of sequence data from pZLHS2.9 (Table 1), a plasmid harboring an insert from the opposite end of the region encoding the tetR(A) homologue, identified a single open reading frame of 1183 bp (Fig. 2 and data not shown). BLAST analysis indicated that this open reading frame encodes a protein very similar to the tetA(A) gene product from other tet systems.
To obtain the wild-type version of the tet locus from C58, a portion of the tetA(A) homologue was used as a probe to screen a genomic bank of the wild-type, tetracycline-sensitive strain NT1 (16). A cosmid hybridizing strongly to the probe was identified and designated pZLT2 (Fig. 2). The hybridizing region was subcloned as an 8.5-kb EcoRI fragment into pBluescript SK(+) to generate pZLE8.5 (Fig. 2), and the insert in this plasmid was used as the template to sequence a 2.6-kb region containing the wild-type tet locus.
Examination of the sequence indicated that this region contains two divergently transcribed open reading frames (Fig. 2 and data not shown). Database searches revealed that the two open reading frames can code for products that are very similar to the Tet(A) class of tetracycline resistance genes from E. coli transposons, with the highest similarity to that of the Tn1721 tet system (2). One we call tetAC58 could code for a protein with an Mr of 41,458 and having 46% identity (55% similarity) with TetA from the IncP1α R plasmid RP4 (35) (Fig. 3A). The second, which could code for a protein with an Mr of 24,389 and having 46% identity (55% similarity) with TetR from RP4 (35), was named tetRC58 (Fig. 3B). The nucleotide sequences of tetAC58 and tetRC58 from the mutant and wild-type strains are identical. However, in the mutant, the sequence is interrupted by the IS426 insertion at bp 520 of tetRC58 (data not shown). This insertion generated a 5-bp duplication of the sequence 5′ GGGGG 3′ at the target site.
FIG. 3.
Relatedness of TetA and TetR from strain C58 to the gene products of tetA and tetR from RP4. Alignments between the predicted amino acid sequence of TetAC58 and TetARP4 (A) and of TetRC58 and TetRRP4 (B) were performed by using the GAP algorithm from the GCG package as described in Materials and Methods. Identical amino acids are shown as white letters on a black background, and conserved amino acids are shown as black letters on a shaded background.
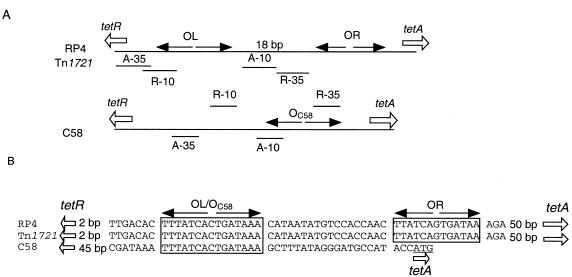
A hydropathy plot generated by the Kyte-Doolittle algorithm (28) revealed that TetAC58 contains 12 putative hydrophobic segments (data not shown). Such domains, which could represent transmembrane segments, are a typical feature of the major facilitator superfamily of proteins (31, 36). Sequence analysis indicated that, like other class A and class B TetR proteins, TetRC58 contains a typical helix-turn-helix domain in its N-terminal portion (data not shown). The tetAC58 and tetRC58 genes are separated by an 88-bp intergenic region with characteristics of regulated divergent promoters (Fig. 4A). A 15-bp inverted repeat (IR) element overlaps the putative −10 region of the tetAC58 promoter. This IR also overlaps the potential −35 site of the tetRC58 promoter. As shown in Fig. 4B, the sequence of this IR is identical to those of the operators found between the tetA and tetR genes of other class A and class B tet systems, including those of RP4 and Tn1721 (2, 35). However, the promoter region of tetC58 lacks the second IR element present in the corresponding region of tetRP4 and of tetTn1721. Furthermore, the intergenic region of tetC58 is considerably shorter than those of the tet elements of RP4 and Tn1721 (Fig. 4B).
FIG. 4.
Organization of the intergenic regulatory regions from several related tet systems. (A) Locations of putative promoter and operator sequences. (B) Comparison of the DNA sequences of operators from the tet systems of C58, RP4, and Tn1721. Inverted repeat sequences are boxed. The transcriptional directions of the tetA and tetR genes are indicated by arrows. A-10 and A-35, the −10 and −35 sites for the tetA promoters; R-10 and R-35, the −10 and −35 sites for the tetR promoters; OC58, putative operator of tetC58; OL and OR, the left and right operators, respectively, of the tet systems of Tn1721 and RP4.
Expression of tetC58 is not induced by tetracycline or related compounds.
In class A and class B tet systems, TetR represses expression of tetA and tetR, and this repression is relieved by subinhibitory levels of the antibiotic (3, 21, 23). Tetracycline does not induce the Agrobacterium tet system; as noted above, when a large number cells are spread onto medium containing this antibiotic, only a very small fraction of the cells grow to produce colonies. Moreover, of those such colonies tested, all now are constitutively resistant to tetracycline (data not shown). To study the regulation of the tetA and tetR genes of Agrobacterium spp., a 66-bp fragment of the intergenic region, which comprises sequence between the putative ribosome binding sites for the tetAC58 and tetRC58 genes, was cloned into the divergent promoter detection vector pRG970b (13) to generate pZLOP1. In this construct, tetAC58 and tetRC58 are transcriptionally fused to the lacZ and uidA genes, respectively, of the vector. In addition, the tetRC58 gene was cloned as a NcoI fragment from pZLT2 into the expression vector pBAD22 (19) to generate pBetR. This construct allows us to regulate the expression of tetRC58 with arabinose. To express tetRC58 in Agrobacterium spp., this gene was excised from pBetR as an EcoRI-BamHI fragment and cloned into pDBL4 (Table 1), a broad-host-range expression vector containing the arabinose promoter and downstream transcriptional terminator from pBAD22 cloned into pBBRMCS-2 (27), to construct pDBL4-tetR. Expression of tetRC58 in these two constructs is inducible in A. tumefaciens and E. coli by addition of arabinose to the culture medium at a final concentration of 0.4%.
Under conditions in which tetRC58 is repressed, expression levels of the tetAC58::lacZ and tetRC58::uidA fusions were lower in strain NT1(pZLOP1, pDLB4-tetR) than in strain NT1TcR1(pZLOP1, pDLB4-tetR). We attribute the lower expression levels of these fusions in NT1 to the chromosomal copy of tetRC58. However, when the cloned copy of tetRC58 was induced by adding arabinose, expression of the reporter fusions in both strains was inhibited to similar levels (Table 2). Similarly, expression of the tetAC58::lacZ reporter on pZLOP1 was repressed by TetRC58 in an E. coli host but only upon addition of arabinose (Table 2).
TABLE 2.
TetRC58 controls expression of tetAC58 and tetRC58
| Strain | tetRC58 source | β-Galactosidase activitya (tetAC58::lacZ)
|
β-Glucuronidase activitya (tetRC58::uidA)
|
||
|---|---|---|---|---|---|
| No ara | With ara | No ara | With ara | ||
| NT1TcR1(pZLOP1, pDLB4) | None | 77 | 85 | 53 | 45 |
| NT1TcR1(pZLOP1, pDLB4-tetR) | pDLB4-tetR | 65 | 2 | 18 | 6 |
| NT1(pZLOP1, pDLB4) | Chromosome | 19 | 14 | 14 | 20 |
| NT1(pZLOP1, pDLB4-tetR) | Chromosome/pDLB4-tetR | 25 | 2 | 14 | 7 |
| DH5α(pZLOP1, pDLB4) | None | 2,213 | 2,243 | NTb | NT |
| DH5α(pZLOP1, pDLB4-tetR) | pDLB4-tetR | 1,678 | 29 | NT | NT |
β-Galactosidase and β-glucuronidase activities are expressed as units per 109 CFU as described in Materials and Methods. Activities were determined in cultures with or without 0.4% arabinose (ara) added as an inducer.
NT, not tested (E. coli DH5α has an endogenous β-glucuronidase activity).
Tetracycline, when added at final concentrations ranging in 10-fold increments from 0.0001 to 1 μg/ml, had no effect on the expression of tetAC58 or tetRC58 in either host (data not shown). Although tetracycline does not induce the expression of tetAC58, we consider the possibility that compounds similar to this antibiotic could be the ligands for TetRC58. To test this, a total of 25 polycyclic compounds with structures similar to tetracycline were tested for their ability to induce the expression of the tetAC58::lacZ fusion. These included active derivatives of tetracycline as well as various nontoxic compounds, most of plant origin. None of these agents induced the expression of the tetAC58::lacZ fusion at a detectable level (data not shown). When the tetracycline-sensitive strains NT1(pZLOP1) and NT1(pZLOP1, pDLB4-TetR) were used as reporters, resistant mutants formed in the zones of growth inhibition caused by chlortetracycline and oxytetracycline. However, no such mutants appeared in the zone of growth inhibition caused by minocycline (data not shown).
Cross-operator recognition by TetRC58 and TetRRP4.
By sequence analysis, the putative 15-bp operator of tetC58 is identical to those of other tet systems (Fig. 4B). Furthermore, the sequence from residues 26 to 47 of TetRC58 is virtually identical to that of the N-terminal helix-turn-helix domain of several related TetR proteins, including TetRRP4, TetRTn10, and TetRTn1721 (Fig. 3B and data not shown). Given these similarities, we determined whether TetR from A. tumefaciens and from RP4 will recognize the operators of the noncognate homologues. Strains DH5α(pRK415K, pBetR) and DH5α(pRK415K, pZLOP1) were constructed for this study. Plasmid pRK415K carries the RP4 tetracycline resistance gene unit, and expression of this determinant is inducible by tetracycline (14, 25). If TetRC58 can repress the expression of tetARP4, upon induction of tetRC58, strain DH5α(pRK415K, pBetR) should fail to grow on medium containing tetracycline. As shown in Table 3, this strain grows well on LB medium with tetracycline at a concentration of 10 μg/ml. However, when the expression of tetRC58 on pBetR was induced by arabinose, this strain was unable to grow on the same medium. These results indicate that TetRC58 can repress at the tetARP4 promoter and that this repression is not responsive to tetracycline. To determine if TetRRP4 can repress tetAC58, the expression of the tetAC58::lacZ reporter was examined in strain DH5α(pRK415K, pZLOP1). Results from this test indicated that TetRRP4 from pRK415K indeed repressed tetAC58 expression, and like the repressive effect on its cognate tetA, the repression was responsive to tetracycline (Table 3). Thus, TetRRP4 can recognize and repress expression from the promoter of the A. tumefaciens tet system. Furthermore, unlike the tetC58 system, this repression of tetAC58 by TetRRP4 is relieved by tetracycline.
TABLE 3.
TetRC58 and TetRRP4 recognize noncognate tet operators
| Strain | β-Galactosidase activitya
|
Growth on TC mediumb
|
||
|---|---|---|---|---|
| No TC | With TC | No ara | With ara | |
| DH5α(pRK415K, pBetR) | NTc | NT | ++ | − |
| DH5α(pRK415K, pBAD22) | NT | NT | ++ | ++ |
| DH5α(pZLOP1) | 2,311 | 2,211 | NT | NT |
| DH5α(pZLOP1, pRK415K) | 387 | 2,154 | NT | NT |
β-Galactosidase activity is expressed as units per 109 CFU as described in Materials and Methods. When needed, tetracycline (TC) was added to the culture at 10 μg/ml as an inducer.
Growth was assessed on LB medium supplemented with tetracycline at 5 and 10 μg/ml (TC medium) and with or without 0.4% arabinose (ara) added as an inducer. ++, good growth; −, no growth.
NT, not tested.
Nature of the tetracycline-resistant mutants.
Our data indicate that a tet unit in the genome of strain C58 accounts for the appearance of tetracycline-resistant mutants when this strain is grown in the presence of this antibiotic. In the particular mutant used in this study, the tetracycline resistance phenotype arose from the transposition of IS426 into tetRC58. Since tetracycline does not induce the expression of tetAC58, we were interested in determining whether all tetracycline-resistant mutants were caused by the same type of mutation. To examine this, genomic DNA from eight independent tetracycline-resistant mutants was probed with a DNA fragment containing the tetC58 genes. These eight mutants grouped into four types based on the size of the hybridizing fragment (data not shown). One class, represented by three mutants, contained a fragment indistinguishable in size from that of the wild type. In the second, two of the eight mutants gave a fragment indistinguishable in size from that of NT1TcR1, the original mutant from which the tet locus was cloned. The remaining three mutants contained two restriction fragment length polymorphism (RFLP) patterns different from that of the wild type and of NT1TcR1 (data not shown).
Surveying for the presence of the tet locus in the genomes of Agrobacterium strains.
We examined a collection of wild-type isolates of Agrobacterium spp. for the tet locus by Southern analysis using a probe containing part of tetA and most of tetR. We also examined some commonly used derivatives of strain C58. Of those not derived from C58, only strain T37 contains DNA with detectable relatedness to the tet locus from strain C58 (Table 4). Moreover, the probe hybridized to an EcoRI fragment of the same size in the two strains (data not shown). Consistent with these results, of the 26 strains tested, only T37 and those derived from C58 gave rise to tetracycline-resistant mutants (Table 4). The probe hybridized with all derivatives of strain C58 tested, including strain UIA5, which lacks both pTiC58 and the 450-kb catabolic plasmid pAtC58 (Table 4). Consistent with this, UIA5 gives rise to mutants resistant to tetracycline.
TABLE 4.
Tetracycline resistance characteristics of Agrobacterium spp.
| Agrobacterium species | Strainc | Appearance of TC-resistant mutants on medium with TC ata:
|
Homology with tetC58b | |
|---|---|---|---|---|
| 5 μg/ml | 10 μg/ml | |||
| A. tumefaciens | C58 (NT1, UIA5, A281) | ++ | ++ | ++ |
| T37 | ++ | ++ | ++ | |
| K108 | − | − | − | |
| Ach5 (LBA4404) | − | − | − | |
| A6NC | − | − | − | |
| R10 | − | − | − | |
| 15955 | − | − | − | |
| B6 | − | − | − | |
| Bo542 | − | − | − | |
| Chry5 | − | − | − | |
| ANT4 | − | − | − | |
| A. rhizogenes | 2655 | − | − | − |
| 2657 | − | − | − | |
| K599 | − | − | − | |
| 8196 | − | − | − | |
| A4 | − | − | − | |
| 15834 | − | − | − | |
| A. vitis | K308 | − | − | − |
| Hm1 | − | − | − | |
| Ag57 | − | − | − | |
| Tm4 | − | − | − | |
| A. radiobacter | K84 | − | − | − |
Tetracycline (TC)-resistant mutants either appeared (++) or did not appear (−).
Determined by Southern analysis as described in Materials and Methods. Homology was (++) or was not (−) found.
Strains shown in parentheses are derivatives of the type strain that also were tested.
DISCUSSION
We report here the characterization of a tet system from A. tumefaciens C58. This active resistance system accounts for the long-known observation that C58 and its derivatives give rise to tetracycline-resistant mutants at a high frequency.
This resistance determinant is structurally similar to that of tet systems found in plasmids and transposons from E. coli and other gram-negative bacteria, with the highest relatedness being to the class A tetracycline resistance gene system from Tn1721. The tetC58 system confers resistance to tetracycline and derivatives such as chlortetracycline and oxytetracycline but not to minocycline. This pattern of resistance suggests that tetC58 does not belong to the Tet(S) class of tetracycline resistance genes, which confers resistance to minocycline as well as to tetracycline, chlortetracycline, and oxytetracycline (10).
Like other class A systems, tetC58 is organized as two genes, a regulatory gene and a gene coding for an efflux pump, transcribed divergently from an intergenic region. Potential promoter elements for both genes are overlapped by a single putative operator composed of a 15-bp perfect IR element. Our genetic analysis indicates that this intergenic region contains all of the cis-acting information required for the regulated expression of these two genes. The DNA sequence of the putative operator is identical to that of operators from other tet systems. However, the promoter region of tetC58 contains only one repeat while the promoter regions of the tet determinants of RP4 and Tn1721 each contain two copies of the operators (Fig. 4B).
Heterologous repressor-operator recognition occurs between different classes of tet systems, even those with dissimilar operators (26). The high degree of sequence similarity between operators from the tetC58 and tetRP4 systems suggested that TetR proteins from these two systems could recognize the noncognate operator. This proved to be the case: TetR of RP4 repressed the tet system of C58 and TetR of C58 repressed tetracycline resistance conferred by the RP4 tet system. The capacity of TetRRP4 to repress expression of the tetAC58::lacZ fusion suggests that the 15-bp IR is the operator and indicates that a single copy of this element is sufficient for regulation of both genes.
The expression of almost all tetracycline resistance gene systems examined to date is under strict control (29, 45). Among these, repression of the Tet(B) class by TetR is the best understood (21, 45). In the absence of tetracycline, the repressors bind to operators overlapping the promoters of both the tetA and tetR genes (21). Binding of tetracycline to TetR(B) induces a conformational change that leads to release from the operators and concomitant expression of tetA (22, 34). Tetracycline-inducible TetR-mediated regulation of other tet systems is believed to function by a similar mechanism (21). Two lines of evidence indicate that tetracycline does not induce the Agrobacterium tet system. First, when cells were spread onto medium containing tetracycline at concentrations ranging from 10 to 1 μg/ml, only a small portion of the cells grew into colonies (data not shown). Moreover, those that did grow now expressed constitutive resistance to tetracycline. If tetracycline functioned as the inducer for tetC58, virtually all of the cells should be resistant to this antibiotic. Furthermore, E. coli and Agrobacterium strains harboring pZLE8.5 (Table 1), which carries the wild-type tetC58 locus, are sensitive to tetracycline, even at concentrations to which tetRP4 confers resistance (data not shown). Second, while the tetAC58::lacZ and the tetRC58::uidA reporter fusions were expressed constitutively in cells lacking TetRC58, the expression of both fusions was depressed considerably in cells harboring tetRC58. Furthermore, neither reporter was inducible by tetracycline. Repression of tetARP4 by TetRC58 is not relieved by tetracycline. On the other hand, repression of tetC58 by TetRRP4 does respond to tetracycline. These two sets of results indicate that the failure of the tet system of C58 to respond to tetracycline is a function of TetR and not of the tet operator. Mutants of the related repressor from Tn10 that do not respond to tetracycline have been isolated and characterized. These noninducible mutants repress the expression of tetATn10::lacZ fusions by binding to the operators even in the presence of tetracycline (5, 20, 34). We propose that TetRC58 represents such a noninducible form of TetR. Clearly, this repressor can bind DNA. Why it no longer reacts to tetracycline remains to be determined. We considered the possibility that tetracycline is not the true ligand for TetRC58. However, none of the 25 compounds we tested induced the system. Certainly, this was not an exhaustive survey. But on balance, that the locus of strain C58 can be regulated by TetRRP4 in response to tetracycline strongly suggests that this gene unit once was responsive to this antibiotic.
The interchangeability with respect to operator recognition exhibited by the two TetR proteins could explain another observation concerning tetracycline resistance in strain C58. Derivatives of this A. tumefaciens strain harboring RP4 and vectors carrying tetRP4 express only low level resistance to tetracycline. We propose that TetRC58 is partially dominant over TetRRP4. Thus, since TetRC58 does not respond to tetracycline, the tetRP4 determinant is not fully expressed in its A. tumefaciens host.
Expression of tetracycline resistance in C58 apparently occurs only following a mutation affecting regulation. These mutations can be of several types; in the particular mutant used in this study, the tetracycline resistance phenotype resulted from the insertion of IS426 in tetRC58. In genetic analysis carried out with C58 and its derivatives, this element is commonly associated with phenotypes in which the selection gives derepressed mutants (17). However, diverse RFLP patterns in Southern analysis of eight independent mutants indicated that other types of mutations can result in the tetracycline resistance phenotype. While mutants with a pattern indistinguishable from that of NT1TcR1 may have arisen from IS426 transposition, those with a pattern similar to that of the wild type could represent point mutations or very small deletions or insertions in the tetRC58 gene. They also could represent operator-constitutive mutations that no longer allow TetRC58 to bind at the promoter.
Among the agrobacteria we examined, only strain T37 contains DNA with detectable homology to the tet locus from C58. Furthermore, only C58 and T37 give rise to tetracycline-resistant mutants (Table 4). These two strains were isolated from different parts of the United States (42). Moreover, although strain C58 induces unorganized tumors, strain T37 causes teratomas. Both strains contain a nopaline/agrocinopine-type Ti plasmid, but RFLP analysis indicates that these two elements have diverged considerably from each other (42). Our survey also shows that generation of tetracycline-resistant mutants is associated with the presence of a tetC58 homologue in the genome of the particular isolate; those lacking this element do not yield mutants at a detectable level. The presence of the tet locus in strain UIA5, a derivative of C58 lacking both pTiC58 and pAtC58, indicates that this resistance determinant is associated with one of the two chromosomes of strain C58 (1). Interestingly, this is not the only antibiotic resistance determinant in strain C58. This strain, which also mutates to resistance to chloramphenicol at a high frequency, contains a gene coding for an atypical chloramphenicol acetyltransferase (47). Whether this cat gene is present in strain T37 or any other isolate of Agrobacterium spp. is not known.
ACKNOWLEDGMENTS
We thank Abigail Salyers for supplying the tetracyclines and related chemicals and Stanton Gelvin and Ramon Peñalver for helpful discussions. We also thank Malcolm Winkler, whose seminar on tRNA processing got us started on the problem.
Portions of this research were supported by grants R01 GM52465 from the NIH and NC CNTRL SOY SKF ANTC from the North Central Soybean Association to S.K.F.
REFERENCES
- 1.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmeier H, Cresnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene. 1992;111:11–20. doi: 10.1016/0378-1119(92)90597-i. [DOI] [PubMed] [Google Scholar]
- 3.Altenbuchner J, Schmid K, Schmitt R. Tn1721-encoded tetracycline resistance: mapping of structural and regulatory genes mediating resistance. J Bacteriol. 1983;153:116–123. doi: 10.1128/jb.153.1.116-123.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Biburger M, Berens C, Lederer T, Crec T, Hillen W. Intragenic suppressors of induction-deficient TetR mutants: localization and potential mechanism of action. J Bacteriol. 1998;180:737–741. doi: 10.1128/jb.180.3.737-741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanty J M, Brautaset T, Winther-Larsen H C, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J Biol Chem. 1991;266:2872–2877. [PubMed] [Google Scholar]
- 8.Burdett V. tRNA modification activity is necessary for Tet(M)-mediated tetracycline resistance. J Bacteriol. 1993;175:7209–7215. doi: 10.1128/jb.175.22.7209-7215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cangelosi G A, Best E A, Marinetti G, Nester E W. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 10.Charpentier E, Gerbaud G, Courvalin P. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene. 1993;131:27–34. doi: 10.1016/0378-1119(93)90665-p. [DOI] [PubMed] [Google Scholar]
- 11.Chilton M D, Currier T C, Farrand S K, Bendich A J, Gordon M P, Nester E W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly D M, Winkler M E. Structure of Escherichia coli K-12 miaA and characterization of the mutator phenotype caused by miaA insertion mutations. J Bacteriol. 1989;171:3233–3246. doi: 10.1128/jb.173.5.1711-1721.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook D M. Ph.D. dissertation. Urbana: University of Illinois at Urbana-Champaign; 1995. [Google Scholar]
- 14.Cook D M, Farrand S K. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J Bacteriol. 1992;174:6238–6246. doi: 10.1128/jb.174.19.6238-6246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dazins A, Chakrabarty A M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984;159:9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrand S K, O’Morchoe S P, McCutchan J. Construction of an Agrobacterium tumefaciens C58 recA mutant. J Bacteriol. 1989;171:5314–5321. doi: 10.1128/jb.171.10.5314-5321.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrand S K, Tempé J, Dessaux Y. Localization and characterization of the region encoding catabolism of mannopinic acid from the octopine-type Ti plasmid pTi15955. Mol Plant Microbe Interact. 1990;3:259–267. doi: 10.1094/mpmi-3-259. [DOI] [PubMed] [Google Scholar]
- 18.Glickmann E, Gardan L, Jacquet S, Hussain S, Elasri M, Petit A, Dessaux Y. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact. 1998;11:156–162. doi: 10.1094/MPMI.1998.11.2.156. [DOI] [PubMed] [Google Scholar]
- 19.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hecht B, Muller G, Hillen W. Noninducible Tet repressor mutations map from the operator binding motif to the C terminus. J Bacteriol. 1993;175:1206–1210. doi: 10.1128/jb.175.4.1206-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillen W, Berens C. Mechanisms underlying expression of the Tn10 encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 22.Hinrichs W, Kisker C, Duvel M, Muller A, Tovar K, Hillen W, Saenger W. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- 23.Izaki K, Kiuchi K, Arima K. Specificity and mechanism of tetracycline resistance in a multiple-drug-resistant strain of Escherichia coli. J Bacteriol. 1966;91:628–633. doi: 10.1128/jb.91.2.628-633.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferson R A. Assaying chemeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 25.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 26.Klock G, Unger B, Gatz C, Hillen W, Altenbuchner J, Schmid K, Schmitt R. Heterologous repressor-operator recognition among four classes of tetracycline resistance determinants. J Bacteriol. 1985;161:326–332. doi: 10.1128/jb.161.1.326-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M I, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1992;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Levy S B. Tetracycline resistance determinants are widespread. ASM News. 1988;54:418–421. [Google Scholar]
- 30.Luo, Z.-Q., and S. K. Farrand. Unpublished data.
- 31.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 32.Mark C. “DNA Strider”: a “C” program for fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 34.Muller G, Hecht B, Helbl V, Saenger W, Hillen W. Characterization of non-inducible Tet repressor mutants suggests conformational changes necessary for induction. Nat Struct Biol. 1995;2:693–703. doi: 10.1038/nsb0895-693. [DOI] [PubMed] [Google Scholar]
- 35.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncP alpha plasmids, compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 36.Pao S S, Paulsen I, Saier J M H. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;6:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert M C. Epidemiology of tetracycline resistance determinants. Trends Microbiol. 1994;2:353–357. doi: 10.1016/0966-842x(94)90610-6. [DOI] [PubMed] [Google Scholar]
- 39.Salyers A A, Speer B S, Shoemaker N B. New perspectives on tetracycline resistance. Mol Microbiol. 1990;4:151–156. doi: 10.1111/j.1365-2958.1990.tb02025.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sciaky D, Montoya A L, Chilton M-D. Fingerprints of Agrobacterium Ti plasmids. Plasmid. 1977;1:238–253. doi: 10.1016/0147-619x(78)90042-2. [DOI] [PubMed] [Google Scholar]
- 43.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 44.Speer B S, Salyers A A. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J Bacteriol. 1989;171:148–153. doi: 10.1128/jb.171.1.148-153.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speer B S, Shoemaker N B, Salyers A A. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;5:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stachel S E, An G, Flores C, Nester E W. A Tn3lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tennigkeit J, Matzura H. Nucleotide sequence analysis of a chloramphenicol-resistance determinant from Agrobacterium tumefaciens and identification of its gene product. Gene. 1991;98:113–116. doi: 10.1016/0378-1119(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 48.Travis J. Reviving the antibiotic miracle. Science. 1994;264:360–362. doi: 10.1126/science.8153615. [DOI] [PubMed] [Google Scholar]
- 49.Van den Eede G, Deblaere R, Goethals K, Van Montagu M, Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- 50.Vanderleyden J, Desair J, De Meirsman C, Michiels K, Van Gool A P, Chilton M-D, Jen G C. Nucleotide sequence of an insertion sequence (IS) element identified in the T-DNA region of a spontaneous variant of the Ti-plasmid pTiT37. Nucleic Acids Res. 1986;14:6699–6709. doi: 10.1093/nar/14.16.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waston B, Currier T C, Gordon M P, Chilton M-D, Nester E W. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975;123:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Deutscher M P. Cloning, characterization, and overexpression of the Escherichia coli rnd gene encoding RNase D. J Bacteriol. 1988;170:522–527. doi: 10.1128/jb.170.2.522-527.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zimmerer R P, Hamilton R H, Pootjes C. Isolation and morphology of temperate Agrobacterium tumefaciens bacteriophage. J Bacteriol. 1966;92:746–750. doi: 10.1128/jb.92.3.746-750.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]