Abstract
Deinococcus radiodurans R1 is extremely resistant to both oxidative stress and ionizing radiation. A simple and general targeted mutagenesis method was developed to generate catalase (katA) and superoxide dismutase (sodA) mutants. Both mutants were shown to be more sensitive to ionizing radiation than the wild type.
Deinococcus (Micrococcus) radiodurans R1 has extreme resistance to genotoxic chemicals, oxidative damage, high levels of ionizing and UV radiation, and desiccation (4, 7, 12, 19). It has been suggested that the extreme resistance to ionizing radiation is attributable to an effective DNA repair system and a special chromosome structure (8, 13, 14, 16–18). However, protective mechanisms against oxidative damage may also be involved in this extreme radiation resistance. The lethal effect of ionizing radiation is known to be oxygen enhanced (15) by generating hydrogen peroxide and oxygen free radicals, which damage cell membranes, proteins, and nucleic acids. It has been shown that a pretreatment of D. radiodurans at early exponential-growth phase with hydrogen peroxide enhances its resistance to radiation (22). Moreover, D. radiodurans expresses relatively high levels of catalase and superoxide dismutase activities (5). However, the importance of the catalase and superoxide dismutase activities in the extreme radiation resistance of D. radiodurans has never been tested.
In this study, we have developed a general and simple method to inactivate any targeted gene in D. radiodurans. We have successfully constructed both catalase (katA) and superoxide dismutase (sodA) mutants for investigating their roles in the extreme resistance to ionizing radiation.
Construction of katA and sodA mutants.
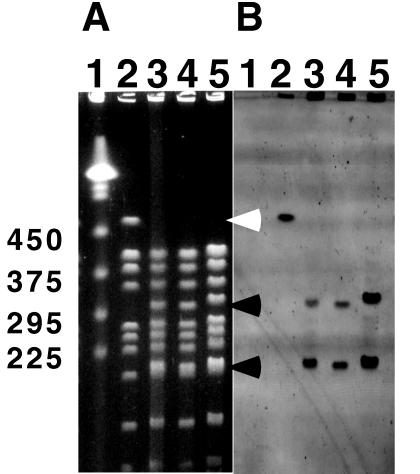
The plasmids and strains used in this study are listed in Table 1. Based on the available sequence of the katA gene from D. radiodurans, three different plasmids were constructed carrying different parts of the gene. A 705-bp DNA fragment from codon 154 to 386 of the deinococcal katA gene (accession no. D63898) was PCR amplified with katA-F (5′-GGACTTCGTCGTCAACAACCTC-3′) and katA-B (5′-ATCGGCAGTTGCAGGTAGTTGG-3′) primers. A 2,970-bp PCR fragment containing the complete regulatory and coding regions of katA was PCR amplified with katA-F1 (5′-GCTCTTCCATCCCGATCAC-3′) and katA-B6 (5′-CCAGAAAAGCACCGTACTGG-3′) primers. Amplified PCR fragments were blunt end cloned into a pCR-Blunt cloning vector (Kmr; Invitrogen, Carlsbad, Calif.). D. radiodurans was transformed with the plasmid constructs, selecting for kanamycin resistance at 25 μg/ml. The transformation protocol was described previously (20). Since pCR-Blunt is a ColE1 plasmid derivative and does not replicate as a plasmid in D. radiodurans, kanamycin resistance in D. radiodurans was produced by duplication insertion of the plasmid into the chromosome and subsequent amplification of the plasmid (20). To confirm that the plasmid constructs had integrated into the chromosome, high-molecular-weight DNA plugs were prepared and analyzed by pulsed-field gel electrophoresis (PFGE) as described previously (23). DNA plugs were digested with NotI restriction enzyme and resolved by PFGE (Fig. 1). Because the pCR-Blunt vector has a unique NotI restriction site, its integration into the chromosome creates an extra NotI site in the 485-kb NotI fragment (Fig. 1A, lane 2) of wild-type R1, yielding 290- and 195-kb fragments (Fig. 1A, lanes 3 to 5). The integration of the plasmid constructs was further confirmed by Southern blot analysis (Fig. 1B) with a nonradioactive probe generated from pKKW1 plasmid DNA by a method described previously (24).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Source |
|---|---|---|
| Strain | ||
| D. radiodurans | ||
| R1 | Wild type, catalase A and superoxide dismutase positive | ATCCa |
| KKW7001 | R1 transformed with pKKW1, catalase A positive | This study |
| KKW7002 | R1 transformed with pKKW2, catalase A positive | This study |
| KKW7003 | R1 transformed with pKKW3, catalase A negative | This study |
| KKW7004 | R1 transformed with pKKW4, superoxide dismutase negative | This study |
| E. coli Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL(StrR) endA1 nupG | Invitrogen |
| Plasmids | ||
| pCR-Blunt | pUC derived (Kanr) | Invitrogen |
| pKKW1 | A 2,970-bp PCR fragment containing the complete regulatory and coding regions of katA cloned into pCR-Blunt | This study |
| pKKW2 | A 547-bp SacII fragment (the promoter region of katA and part of the coding region) deleted from pKKW1 | This study |
| pKKW3 | A 705-bp PCR fragment from codon 154 to 386 of katA cloned into pCR-Blunt | This study |
| pKKW4 | A 475-bp PCR fragment from codon 24 to 182 of sodA cloned into pCR-Blunt | This study |
ATCC, American Type Culture Collection.
FIG. 1.
(A) PFGE analysis of wild type and transformants. DNA agarose plugs were digested with NotI restriction enzyme, and digested fragments were resolved with a contour-clamped homogeneous electric field mapper. The gel was stained with ethidium bromide. Lane 1, yeast markers; lane 2, R1 (wild type); lane 3, KKW7001 (complete katA); lane 4, KKW7002 (katA with its promoter region deleted); lane 5, KKW7003 (partial internal coding region of katA). The 485-kb NotI fragment (which disappears) is indicated by the white arrowhead. The two new NotI fragments are indicated by black arrowheads. The molecular sizes in kilobases are shown on the left. (B) Southern blot from gel shown in panel A and hybridized with fluorecein-labelled probe generated from pKKW1 plasmid DNA. The hybridization signal was detected with a FluorImager SI as described previously (24).
To construct the sodA mutant, sequence data of D. radiodurans was obtained through early release from The Institute for Genomic Research at www.tigr.org and searched for putative sodA sequence. A putative deinococcal sodA gene, which shares 57% identity in deduced amino acid sequence with the Escherichia coli sodA gene, was identified (data not shown). A 475-bp DNA fragment from codon 24 to 182 of the putative deinococcal sodA gene was PCR amplified with SodA-F1 (5′-AGTTGAGGTAGTAGGCGTGTTCCC-3′) and SodA-B3 (5′-GGAAATTCACCACACCAAGCATC-3′) primers. Amplified PCR fragments were blunt end cloned into pCR-Blunt cloning vector. D. radiodurans was transformed with the plasmid construct, selecting for kanamycin resistance at 25 μg/ml by using a previously described protocol (20).
Enzymatic activities of katA and sodA mutants.
Transformants with katA plasmid constructs (pKKW1, pKKW2, and pKKW3) were tested for catalase activities by adding 10 μl of 3% hydrogen peroxide to 200 μl of overnight culture in a microtiter plate. The robust bubbling due to the formation of oxygen from hydrogen peroxide was an indication of catalase activity. This test indicated that only transformants from plasmid pKKW3 had lost catalase activity. Transformants with a sodA plasmid construct (pKKW4) were tested for loss of superoxide dismutase activity by a paraquat sensitivity test with a standardized single filter paper disk method (1). Briefly, 10 μl of 13 mM paraquat was spotted on a 5-mm-diameter sterilized filter paper disk which was placed on a lawn of tested bacteria. Wild-type R1 strain was resistant to paraquat and no zone of growth inhibition was observed, but sodA mutants were sensitive to paraquat and a zone of growth inhibition about 25 mm in diameter around the filter disk was observed.
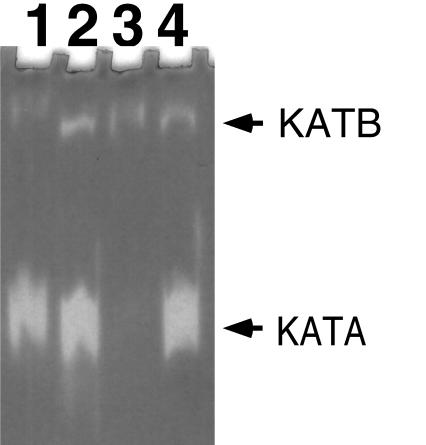
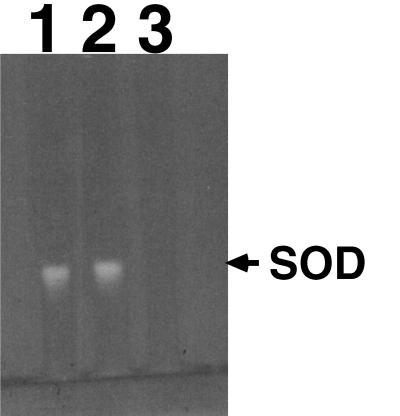
A few selected mutant strains were further confirmed for the loss of catalase or superoxide dismutase activities. Enzymatic activities of the transformants were analyzed by separating total soluble protein extract in a 7.5% nondenaturing gel and staining for catalase activity (Fig. 2) or superoxide dismutase activity (Fig. 3). To prepare the protein extract, D. radiodurans cultures were pelleted and resuspended in butanol-saturated phosphate buffer to remove the outer membrane. Cells were washed in 50 mM phosphate buffer (pH 7) and then resuspended again in the same buffer. Cell suspensions were sonicated at 4°C (15 s pulse on, 10 s pulse off) for a total of 5 min by using a Model 60 Sonic Dismembrator set at an intensity of 8 (Fisher Scientific, Pittsburgh, Pa.). Cell debris was removed by centrifugation (13,000 × g, 5 min, 4°C), and the cell extracts were either stored on ice or quick frozen at −70°C until subsequent analysis. Protein concentration was determined by using the Bradford protein dye assay (Bio-Rad, Hercules, Calif.) (3). Samples of cell extracts (5 or 10 μg per lane) were separated electrophoretically on 7.5% nondenaturing polyacrylamide gels (22). Gels were stained for catalase activity by the horseradish peroxidase-diaminobenzidine method (6) or stained for superoxide dismutase activity by the nitroblue tetrazolium-riboflavin method (2). As predicted, only transformants derived from the plasmid, which carries part of the internal coding region of the katA gene (pKKW3), lost the major catalase activity (Fig. 2). Strain KKW7003 (katA mutant) had lost the major catalase, but a minor catalase was still present (Fig. 2). Strain KKW7004 was also confirmed to have lost superoxide dismutase activity (Fig. 3). These results confirmed the validity of the developed targeted mutagenesis method.
FIG. 2.
Analysis of catalase activity. Lane 1, R1 (wild type); lane 2, KKW7001 (complete katA); lane 3, KKW7003 (internal coding region of katA); lane 4, KKW7002 (katA with promoter region deleted). The two catalase bands shown are labelled KATA (catalase A) and KATB (catalase B).
FIG. 3.
Analysis of superoxide dismutase activity. Lane 1, R1 (wild type); lane 2, KKW7003 (katA mutant); lane 3, KKW7004 (sodA mutant). SOD, superoxide dismutase.
This is the first report to demonstrate that duplication insertion (20) can be exploited to create insertional mutations in D. radiodurans, although a similar strategy has been used with budding yeast and Listeria spp. (9, 25). Unlike the previous direct insertion technique used with D. radiodurans (10, 11), which involves complex in vitro DNA manipulation and numerous cloning steps, the current technique is simpler, calling for (i) PCR of the internal fragment of the gene, (ii) a single cloning step to put the PCR fragment into the plasmid, and (iii) transformation of the host. The developed targeted mutagenesis method should be generally applicable for inactivating other genes, thereby facilitating functional studies of targeted genes with unknown function in D. radiodurans, whose genome is almost completely sequenced.
Testing ionizing radiation resistance of katA and sodA mutants.
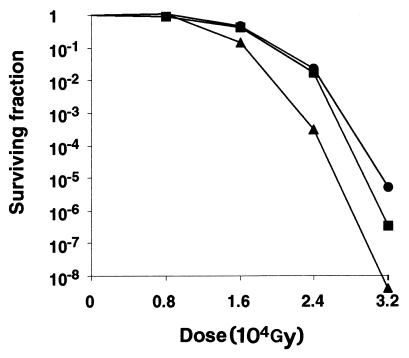
Five-milliliter cultures of wild-type (R1), katA mutant (KKW7003), and sodA mutant (KKW7004) strains, grown in Falcon 2097 culture tubes to exponential phase at 32°C, were placed on ice inside a 250-ml beaker. All bacterial cultures were simultaneously exposed to ionizing radiation with a dose of 8,000 to 32,000 Gy from a 60Co source irradiator (GammaBeam 650). The katA mutant and wild type have similar sensitivities to ionizing radiation when exposed to 8,000 Gy, but the katA mutant is reproducibly more sensitive (2- to 15-fold) than the wild-type strain to gamma radiation at doses of 16,000 Gy or higher (Fig. 4). A previous analysis of changes in cellular proteins of D. radiodurans after gamma irradiation by two-dimensional gel electrophoresis has identified a number of proteins that increase in amount after irradiation, including a 60-kDa protein, RIP60 (21). The N-terminal end of RIP60, which has been partially determined, has an amino acid sequence of DENNKGV (21), which we were able to match perfectly with the katA gene when the amino acid sequence was reverse transcribed into DNA sequence, and the calculated size of deduced katA gene product is also 60 kDa (data not shown). Thus, the katA gene product is likely the unidentified RIP60 protein that increases in amount after gamma radiation, which is consistent with our finding that katA is required for extreme radiation.
FIG. 4.
Cell survival after gamma ray exposure. Gamma radiation was from a 60Co source (GammaBeam 650). Circles, strain R1 (wild type); squares, strain KKW7003 (katA mutant); triangles, strain KKW7004 (sodA mutant). The exposure rate was 368 Gy per min.
On the other hand, the sodA mutant is reproducibly more sensitive (3- to 90-fold) than the wild-type strain and the katA mutant to ionizing radiation at a dose of 16,000 Gy or higher (Fig. 4). This result suggested that superoxide dismutase encoded by sodA is more important than the catalase encoded by katA in protecting the bacteria from high doses of ionizing radiation. It is possible that the existence of a secondary catalase B in the katA mutant (Fig. 2) is enough to handle most of the hydrogen peroxide generated during ionizing radiation. However, apparently, there does not exist a secondary superoxide dismutase in the sodA mutant (Fig. 3) to handle the superoxide anions generated during ionizing radiation. Through analysis of sequence data of D. radiodurans obtained through early release from The Institute for Genomic Research, at www.tigr.org, we have identified other genes that encode putative oxidative defense enzymes such as glutathione reductase (gor) and alkyl hydroperoxide reductase (ahpA). We are currently creating mutations in these genes and constructing mutants with double mutations in order to decipher the oxidative defense systems in D. radiodurans.
Acknowledgments
We thank Ken Minton and Rita Cheng for their comments on the manuscript. We are also grateful to Herb Schellhorn for the protocol of catalase activity staining in a nondenaturing polyacrylamide gel and especially to Kevin Groch for operating the 60Co gamma radiation source.
This work was supported by the Department of Energy’s Environmental Management Science Program and Microbial Genome Program through a DOE contract, DE-AC06-76RLO 1830, to K.K.W.
REFERENCES
- 1.Bauer A W, Kirby W M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Patrol. 1966;45:493–496. [PubMed] [Google Scholar]
- 2.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Carbonneau M A, Melin A M, Perromat A, Clerc M. The action of free radicals on Deinococcus radiodurans carotenoids. Arch Biochem Biophys. 1989;275:244–251. doi: 10.1016/0003-9861(89)90370-6. [DOI] [PubMed] [Google Scholar]
- 5.Chou F I, Tan S T. Manganese(II) induces cell division and increases in superoxide dismutase and catalase activities in an aging deinococcal culture. J Bacteriol. 1990;172:2029–2035. doi: 10.1128/jb.172.4.2029-2035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clare D A, Duong M N, Darr D, Archibald F, Fridovich I. Effects of molecular oxygen on the detection of superoxide radical with nitroblue tetrazolium and an activity stain for catalase. Anal Biochem. 1984;140:532–537. doi: 10.1016/0003-2697(84)90204-5. [DOI] [PubMed] [Google Scholar]
- 7.Dean C J, Feldschreiber P, Lett J T. Repair of x-ray damage to the deoxyribonucleic acid in Micrococcus radiodurans. Nature. 1966;209:49–52. doi: 10.1038/209049a0. [DOI] [PubMed] [Google Scholar]
- 8.Evans D M, Moseley B E. Roles of the uvsC, uvsD, uvsE, and mtcA genes in the two pyrimidine dimer excision repair pathways of Deinococcus radiodurans. J Bacteriol. 1983;156:576–583. doi: 10.1128/jb.156.2.576-583.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutman P D, Carroll J D, Masters C I, Minton K W. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene. 1994;141:31–37. doi: 10.1016/0378-1119(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 11.Gutman P D, Fuchs P, Ouyang L, Minton K W. Identification, sequencing, and targeted mutagenesis of a DNA polymerase gene required for the extreme radioresistance of Deinococcus radiodurans. J Bacteriol. 1993;175:3581–3590. doi: 10.1128/jb.175.11.3581-3590.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattimore V, Battista J R. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minton K W. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat Res. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 14.Minton K W, Daly M J. A model for repair of radiation-induced DNA double-strand breaks in the extreme radiophile Deinococcus radiodurans. Bioessays. 1995;17:457–464. doi: 10.1002/bies.950170514. [DOI] [PubMed] [Google Scholar]
- 15.Mirsa H P, Fridovich I. Superoxide dismutase and the oxygen enhancement of radiation lethality. Arch Biochem Biophys. 1976;176:577–581. doi: 10.1016/0003-9861(76)90201-0. [DOI] [PubMed] [Google Scholar]
- 16.Moseley B E, Copland H J. Isolation and properties of a recombination-deficient mutant of Micrococcus radiodurans. J Bacteriol. 1975;121:422–428. doi: 10.1128/jb.121.2.422-428.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moseley B E, Evans D M. Isolation and properties of strains of Micrococcus (Deinococcus) radiodurans unable to excise ultraviolet light-induced pyrimidine dimers from DNA: evidence for two excision pathways. J Gen Microbiol. 1983;129:2437–2445. doi: 10.1099/00221287-129-8-2437. [DOI] [PubMed] [Google Scholar]
- 18.Moseley B E, Mattingly A, Shimmin M. Isolation and some properties of temperature-sensitive mutants of Micrococcus radiodurans defective in DNA synthesis. J Gen Microbiol. 1972;70:399–409. doi: 10.1099/00221287-70-3-399. [DOI] [PubMed] [Google Scholar]
- 19.Setlow J K, Boling M E. The resistance of Micrococcus radiodurans to ultraviolet radiation. II. Action spectra for killing, delay in DNA synthesis, and thymine dimerization. Biochim Biophys Acta. 1965;108:259–265. doi: 10.1016/0005-2787(65)90010-9. [DOI] [PubMed] [Google Scholar]
- 20.Smith M D, Lennon E, McNeil L B, Minton K W. Duplication insertion of drug resistance determinants in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 1988;170:2126–2135. doi: 10.1128/jb.170.5.2126-2135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A, Hirano H, Kikuchi M, Kitayama S, Watanabe H. Changes in cellular proteins of Deinococcus radiodurans following gamma-irradiation. Radiat Environ Biophys. 1996;35:95–99. doi: 10.1007/BF02434031. [DOI] [PubMed] [Google Scholar]
- 22.Wang P, Schellhorn H E. Induction of resistance to hydrogen peroxide and radiation in Deinococcus radiodurans. Can J Microbiol. 1995;41:170–176. doi: 10.1139/m95-023. [DOI] [PubMed] [Google Scholar]
- 23.Wong K K, McClelland M. A BlnI restriction map of the Salmonella typhimurium LT2 genome. J Bacteriol. 1992;174:1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong K K, Stillwell L C, Dockery C A, Saffer J D. Use of tagged random hexamer amplification (TRHA) to clone and sequence minute quantities of DNA—application to a 180 kb plasmid isolated from Sphingomonas F199. Nucleic Acids Res. 1996;24:3778–3783. doi: 10.1093/nar/24.19.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuenscher M D, Kohler S, Goebel W, Chakraborty T. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol Gen Genet. 1991;228:177–182. doi: 10.1007/BF00282463. [DOI] [PubMed] [Google Scholar]