Summary
Adventitious rooting is an essential biological process in the vegetative propagation of economically important horticultural and forest tree species. It enables utilization of the elite genotypes in breeding programmes and production. Promotion of adventitious root (AR) formation has been associated with starvation of inorganic phosphate and some factors involved in low phosphorus (LP) signalling. However, the regulatory mechanism underlying LP‐mediated AR formation remains largely elusive. We established an efficient experimental system that guaranteed AR formation through short‐term LP treatment in Populus ussuriensis. We then generated a time‐course RNA‐seq data set to recognize key regulatory genes and regulatory cascades positively regulating AR formation through data analysis and gene network construction, which were followed by experimental validation and characterization. We constructed a multilayered hierarchical gene regulatory network, from which PuMYB40, a typical R2R3‐type MYB transcription factor (TF), and its interactive partner, PuWRKY75, as well as their direct targets, PuLRP1 and PuERF003, were identified to function upstream of the known adventitious rooting genes. These regulatory genes were functionally characterized and proved their roles in promoting AR formation in P. ussuriensis. In conclusion, our study unveiled a new hierarchical regulatory network that promoted AR formation in P. ussuriensis, which was activated by short‐term LP stimulus and primarily governed by PuMYB40 and PuWRKY75.
Keywords: adventitious root, low phosphorus signalling, poplar, PuMYB40, PuWRKY75, gene regulatory network, PuLRP1, PuERF003
Introduction
As inorganic phosphate (Pi) is a basal component for membrane (phospholipid), molecular energy battery (e.g., ATP) and nucleic acids (DNA and RNA), its shortage and low mobility in soil can threaten plant growth and development. To cope with low phosphate (LP), plants have evolved various mechanisms to enhance Pi acquisition (Chiou and Lin, 2011). One of these mechanisms is to alter root system architecture (RSA) for enlarging the root surface area to efficiently uptake Pi from the soil and enhance Pi transport and absorption (Jain et al., 2007; Wen et al., 2019). Previous studies in Arabidopsis have shown that Pi deficiency has a strong effect on modulating of RSA, such as inhibition of the primary root (PR) growth, enhancement of the density, and length of lateral roots (LRs) as well as the number of root hairs (Lopez‐Bucio et al., 2005; Péret et al., 2011; Williamson et al., 2001). These modifications of RSA increase the overall root system surface significantly (Huang and Zhang, 2020). Adventitious roots (AR) are known to originate from non‐rooted and non‐meristematic tissues (Legué et al., 2014); its initiation and elongation have been proven to be promoted by Pi starvation in several plant species including Hordeum vulgare (Guo et al., 2018), Oryza sativa (Dai et al., 2012; Hu et al., 2011; Negi et al., 2016; Zhou et al., 2008), Phaseolus vulgaris (Lynch, 2011; Miller et al., 2003; Ochoa et al., 2006; Walk et al., 2006), and Solanum lycopersicum (Kim et al., 2008). ARs play an important role in Pi acquisition because they typically have shallow root growth angles and enhance plant foraging in phosphorus‐rich topsoil (Kim et al., 2008; Lynch, 2011).
A complex and extensive signalling network has been uncovered to underlie the plants responses to Pi starvation (Crombez et al., 2019). Among the plethora of factors involved in the network, the phytohormones, which include auxin, cytokinin, ethylene, abscisic acid, gibberellins, jasmonate, brassinosteroids, strigolactones, and peptide hormones, are the very important modulators of LR and AR development (Abarca, 2021; Bellini et al., 2014; Crombez et al., 2019). Phytohormone signallings of LR and AR formation have been evidenced to be subject to the regulatory mechanisms in which TFs are the major players (Bellini et al., 2014; Crombez et al., 2019). Among Pi starvation‐related TFs, PHOSPHATE RESPONSE 1 (PHR1), a MYB coiled‐coil domain TF (Dubos et al., 2010; Sega and Pacak, 2019), is known as the master regulator that mediates phosphate starvation response in Arabidopsis (Devaiah et al., 2009), bread wheat (Zheng et al., 2020), rice (Dai et al., 2012; Rubio et al., 2001), and soybean (Xue et al., 2017); thousands of phytohormone signalling factors and many other LR‐related TFs have been identified to be the direct targets of PHR1 (Castrillo et al., 2017). PHR1 and related TFs have been suggested to be the major players in Pi‐starvation‐induced LR formation (Crombez et al., 2019). Also, the PHR1‐independent TFs such as WRKYs (Bakshi et al., 2015; Dai et al., 2016; Devaiah et al., 2007a) and ZAT6 (Devaiah et al., 2007b) in Arabidopsis and rice have been reported to be involved in Pi‐starvation‐induced LR formation. However, the regulatory mechanisms involving the TFs mentioned above remains largely unknown.
Adventitious rooting is important in vegetative propagation of horticultural and woody plant species that have high economical values (Rigal et al., 2012). Although AR and LR originate from different organs, they resemble each other in some aspect; (a) their development is evidenced to share key components of the genetic and hormonal regulatory networks; (b) both are promoted by Pi starvation (Bellini et al., 2014). These findings advance our understanding of molecular mechanisms that control AR formation. Since then, most of previous works on adventitious rooting have been focused on herbaceous plants, it is unclear whether the data from those studies can be translated to woody plants.
In this study, we identified an R2R3‐type MYB TF, PuMYB40, in Populus ussuriensis. We also identified its regulatory targets (PuLRP1 and PuERF003) and a co‐factor (PuWRKY75) during short‐term LP‐driven AR formation through constructing a multilayered hierarchical gene regulatory network (ML‐hGRN). In this ML‐hGRN, PuMYB40 functioned as a high hierarchical regulator to modulate adventitious rooting. Short‐term LP treatment of P. ussuriensis stem cuttings significantly increased the expression of PuMYB40, PuWRKY75, PuLRP1, and PuERF003 which promoted AR formation at the basal ends of stem cuttings. We experimentally validated the interaction between PuMYB40 and PuWRKY75, which activated PuLRP1 and PuERF003 to promote AR formation in P. ussuriensis. In summary, these results provide a new insight into molecular mechanism of short‐term LP‐driven AR formation in P. ussuriensis, thereby advancing our understanding of the regulatory mechanisms of AR formation in woody plants.
Results
LP stimulates AR formation in P. ussuriensis
To exert LP treatment and examine if it can promote the AR formation, we treated in vitro stem cuttings of P. ussuriensis with two conditioned‐media, the low phosphorus (LP) (10 μm) and the normal phosphorus (NP) (200 μm). After five‐day culture on NP solid medium, the root primordia emerged at the basal ends of stem cuttings, which were then split and transferred to LP and NP liquid medium (Figure S1). A significant increase in the number of ARs for the cuttings transferred to LP medium was observed within 24 h (Figure 1a,b) as compared with that of cuttings in NP medium. In addition, the average root length was shorter in LP media compared to that in NP media as measured at 48 h after the transfer (Figure S2). Paraffin sectioning of the basal ends of stem cuttings on two media revealed that more new AR primordia were stimulated at the basal ends of the stem cuttings on LP than NP at 12 h (Figure 1c). Therefore, it can be speculated that LP treatment promotes AR formation by triggering more non‐root cells to become primordia. In addition, no significant change in AR number was observed for the modified LP (10 μm) liquid culture in which 5‐day pre‐culture on NP solid medium was omitted (Figure S3), suggesting an important role of 5‐day pre‐culture on NP solid medium in LP‐stimulated AR formation in our study.
Figure 1.
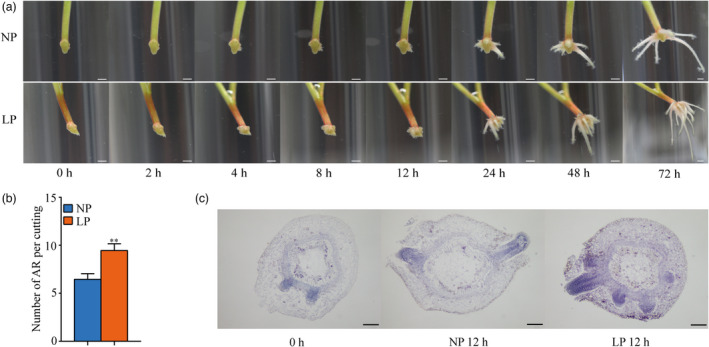
Short‐term low phosphorus (LP)‐accelerated AR formation in P. ussuriensis. (a) Comparison of various rooting stages of cuttings in WT under LP condition and normal phosphorus (NP). Bar: 5 mm. NP represents liquid WPM supplied with 200 μm of KH2PO4, LP represents liquid WPM supplied with 10 μm of KH2PO4. (b) Comparison of AR numbers in WT under LP and NP conditions. Means and standard deviations (SD) were calculated from 50 cuttings. The asterisks indicate significant (**P < 0.01) differences of AR numbers in WT between LP and NP conditions, based on one‐way ANOVA and Duncan’s multiple range test. (c) The paraffin sections in the stem segments of cuttings under LP and NP conditions for 0 and 12 h. Bar: 200 μm.
Construction of LP‐driven AR formation ML‐hGRN
We designed a time series (2, 4, 8, 12, and 24 h) RNA‐seq experiment to obtain the gene expression profiles of LP‐driven AR formation in primordium‐formed cuttings of P. ussuriensis. We used a scalpel to precisely harvest the sites of AR primordium induction (<2 mm in length) at the basal ends of the stem cuttings for RNA extraction and sequencing library construction. Analysis of RNA‐seq led to the identification of a total of 1706 significantly differentially expressed genes at five time points, including 817 up‐regulated and 889 down‐regulated DEGs (Tables 1 and S1). We selected nine up‐regulated DEGs which are homologous to the well‐known Pi starvation induction‐related genes in Arabidopsis thaliana including AtSPX2 (Duan et al., 2008), AtPS3 (Miura et al., 2005), AtACP5 (Pozo et al., 2002), AtGDPD1 (Cheng et al., 2011), AtPAP12 (Tran et al., 2010), AtPAP17 (Miura et al., 2005), AtPECP1 (Angkawijaya and Nakamura, 2017), AtRNS1 (Bariola et al., 1994), and AtSPX2.1 (Duan et al., 2008), to perform RT‐qPCR. The result showed that all tested DEGs were significantly up‐regulated by short‐term LP treatment (Figure S4), suggesting that the induction of AR formation under short‐term LP treatment is due to the LP stress. We selected 29 root development‐, auxin‐, and low phosphate starvation‐related DEGs (Table S2) and used them as bottom‐layered genes to construct upstream ML‐hGRN (Kumari et al., 2016; Wei et al., 2013) for identifying TFs that potentially regulate them. Since the root induction by LP was a rapid process, the differentially expressed TFs at the five time points might not cover some important regulators. In addition, the MYB40 was probably expressed in a small number of pericycle cells from which the ARs were originated. Therefore, we used the whole TFs as the inputs for construction upper layers of ML‐hGRN on the competitive basis. The interference frequency of each TF represents its regulation on the layer immediately below it; a larger interference frequency represents a tighter regulation during AR formation (Wei, 2019). From the ML‐hGRN we built, PuMYB40 possesses the highest interference frequency, which is the times an edge (or a regulatory relationship) between a TF and a target gene was discovered in all combined triple‐gene blocks evaluated during ML‐hGRN construction. The ML‐hGRN was plotted in such a way that the width of each edge was proportional to its interference frequency. The results showed that PuMYB40 had the highest sum of interference frequencies to the genes in the next layer below it (Figure S5, Table S3), indicating its regulatory strength in governing the LP‐driven AR formation in P. ussuriensis.
Table 1.
The numbers of differentially expressed genes (DEGs) in the newly formed root primordia at the basal ends of Populus ussuriensis stem cuttings at different time points after low phosphorus (LP) treatment in comparison with normal phosphorus (NP) treatment
| 2 h | 4 h | 8 h | 12 h | 24 h | |
|---|---|---|---|---|---|
| Up‐regulated DEGs | 71 | 166 | 158 | 269 | 153 |
| Down‐regulated DEGs | 66 | 177 | 180 | 232 | 234 |
| Total | 137 | 343 | 338 | 501 | 387 |
Expression profiles and transcriptional activation of PuMYB40
Since the PuMYB40 was not among the DEGs in the AR primordium induction sites which were obtained from 2‐mm long basal ends of stem cuttings, we extended the sample size from 2 to 5 mm and then, performed RT‐qPCR and consequently, validated up‐regulation of the PuMYB40 at all the time points under LP conditions compared with that under NP conditions in the vicinity of the AR primordium induction (Figure 2a). This result was consistent with the predicted by the ML‐hGRN.
Figure 2.
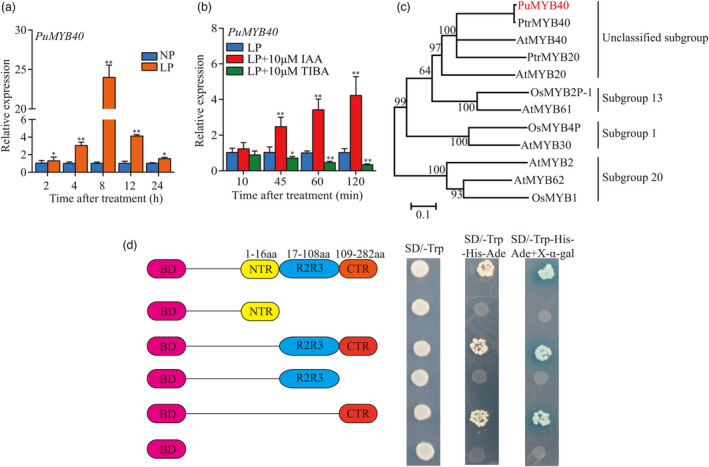
The transcriptional activation activity of PuMYB40 and its expression pattern during short‐term low phosphorus (LP)‐accelerated AR formation. (a) RT‐qPCR analysis of PuMYB40 expression during short term LP‐accelerated AR formation. The asterisks indicate significant differences of PuMYB40 expression level between LP and normal phosphorus (NP) conditions, based on Student’s t‐test: *P < 0.05, **P < 0.01. (b) RT‐qPCR analysis of PuMYB40 expression levels under LP condition and LP with a supply of 10 μm IAA or 10 μm TIBA treatment. The asterisks indicate significant differences of PuMYB40 expression level between LP and LP with a supply of 10 μm IAA or 10 μm TIBA treatment, based on Student’s t‐test: *P < 0.05, **P < 0.01. In (a) and (b), the basal ends of stem cuttings (5 mm) were harvested at different time points under LP and NP conditions. Each value represents the mean SD of three biological replicates. (c) The phylogenetic analysis of PuMYB40 and some other MYB genes with the high similarity. The MEGA 7.0. software (Kumar et al., 2016) was used to construct the phylogenetic tree by the neighbour‐joining (NJ) method. The numbers above or below the branches are the bootstrap values from 1000 replicates. Pu: P. ussuriensis, At: A. thaliana, Ptr: P. trichocarpa, Os: O. sativa. (d) The transcription activation of PuMYB40 in yeast. N‐terminal region (NTR), R2R3‐C, R2R3 and C terminal region (CTR) of PuMYB40 were cloned into pGBKT7 vector to examine each fragment’s role in gene activation. Magenta rectangles represent BD, yellow rectangles represent NTR, blue rectangles represent R2R3, and red rectangles represent CTR.
We examined the temporal expression pattern of PuMYB40 from the 5‐mm long basal ends of primordium‐formed cuttings under LP supplemented with indole‐3‐acetic acid (IAA) and an auxin biosynthesis inhibitor, TIBA (Ludwig‐Muller et al., 2005). The results showed 10 μm IAA increased, and 10 μm TIBA decreased PuMYB40 expression under LP conditions (Figure 2b). This indicates that LP induced up‐regulation of PuMYB40 expression might be partially controlled by auxin.
Phylogenetic analysis for some MYB genes of rice, A. thaliana, and P. trichocarpa showed that PuMYB40 is homologous to AtMYB40 and was classified into unclassified subfamily (Figure 2c). Transcriptional activation assay showed that PuMYB40 possessed transcriptional activity, and the transactivation site was within the C‐terminal region (CTR) (109‐282 aa) (Figure 2d).
PuMYB40 promoted the AR formation
We generated 11 PuMYB40 overexpression (PuMYB40‐OE), and 13 dominant repression lines by fusing a 27‐bp SUPERMAN repression domain X (SRDX) motif, which inhibits the transcription of target genes without affecting its own transcription (Hiratsu et al., 2003), to PuMYB40 (PuMYB40‐SRDX). We used PCR and RT‐qPCR to screen the transgenic lines, and then selected the two lines with the highest expression levels for each construct for further phenotype analysis (Figure S6a–d). Under NP condition, the average number of AR in PuMYB40‐OE lines was significantly higher than that in WT, whereas the average number of AR in PuMYB40‐SRDX lines was significantly lower than that in WT (Figure 3a,b). Also, the adventitious rooting process in PuMYB40‐OE lines was significantly expedited; the adventitious roots emerged 1 day earlier than WT. On the contrary, the adventitious rooting for PuMYB40‐SRDX lines emerged 1 day later than WT (Figure 3c). The biomass analysis showed that the root dry weight of PuMYB40‐OE lines increased 37.23–40.24% compared to WT, whereas the root dry weight decreased 49.89–51.77% in PuMYB40‐SRDX lines compared to WT (Figure 3d). However, PuMYB40‐SRDX lines did not display significant increase in AR number under LP conditions (Figure 3e) as compared to WT, reflecting the indispensable role of PuMYB40 in LP‐driven AR formation in P. ussuriensis.
Figure 3.
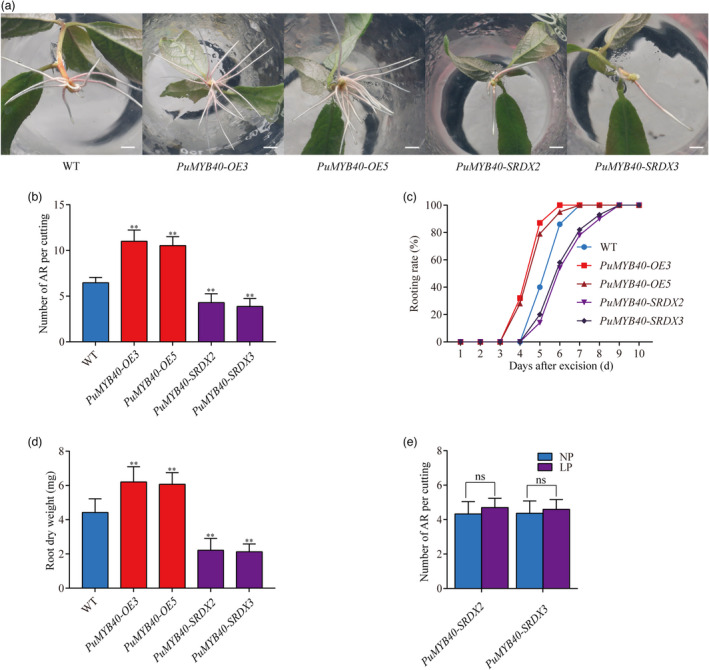
PuMYB40 accelerates adventitious root (AR) formation in P. ussuriensis. (a) Comparison of root growth of PuMYB40‐OE, PuMYB40‐SRDX lines, and WT on the tenth day after cutting. Bar: 1cm. (b) Number of ARs per cutting after cutting for 10 days. (c) Rooting rate (%) of 30 stem segments in the 10‐day period following cutting. (d) Comparison of AR dry weight 3 week after cutting. (f) Number of ARs per cutting of PuMYB40‐SRDX lines under LP and NP conditions after cutting for 10 days. In (b), (d), and (e), values represent the mean ± SD of 30 plants. Significant differences compared based on one‐way ANOVA and Duncan’s multiple range test: **P < 0.01.
Identification of PuWRKY75 as PuMYB40‐interacting protein
In the ML‐hGRN for short‐term LP‐driven AR formation, PuWRKY75 was located at the top layer (or the third layer from the bottom) and possessed the second highest interference frequency (Table S3). The phylogenetic tree shows that PuWRKY75 shares the closest similarity to Populus trichocarpa WRKY75 (PtrWRKY75) (99.99%) and then Arabidopsis thaliana WRKY75 (AtWRKY75) (76.09%) (Figure S7a). Multiple sequence alignment analysis showed that PuWRKY75 contains one classical WRKYGQK conserved domain and a CX4C23HXH zinc‐finger structure and has a high protein sequence similarity to the Group Ⅱc (Figure S7a). We performed RT‐qPCR on the 5‐mm basal ends of primordium‐formed cuttings cultured under two different conditions of LP and LP plus10 μM TIBA. The result showed that PuWRKY75 was significantly up‐regulated by LP (Figure S7b) and 10 μm TIBA was sufficient to suppress LP‐induced PuWRKY75 up‐regulation (Figure S7c).
We then generated 11 PuWRKY75‐OE and 13 PuWRKY75‐SRDX lines and selected the two lines with the highest expression levels for each construct for further phenotype analysis (Figure S8a–d). As shown in Figure 4a,b, the AR numbers of PuWRKY75‐OE lines were higher than that of WT under NP condition, whereas PuWRKY75‐SRDX lines had the reduced AR numbers than WT. The dry weights of both PuWRKY75‐OE lines increased 13.71%–27.58% compared to that of WT, whereas the dry weights of both PuWRKY75‐SRDX lines decreased 40.69%–42.50% compared to that of WT (Figure 4c). Notably, the AR numbers of PuWRKY75‐SRDX8 and PuWRKY75‐SRDX13 showed no significant difference between LP and NP conditions (Figure 4d).
Figure 4.
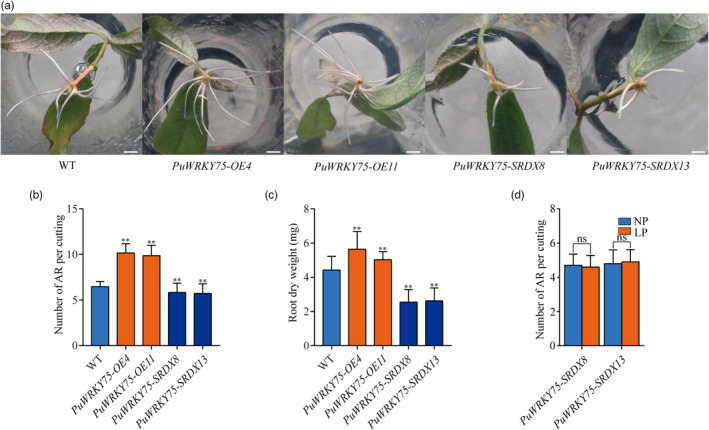
PuWRKY75 accelerates adventitious root (AR) formation in P. ussuriensis. (a) Comparison of root growth of PuWRKY75 transgenic lines and WT on the tenth day after cutting. Bar: 1 cm. (b) Number of ARs per cutting 10 day after cutting. (c) Comparison of AR dry weight after cutting for 3 weeks. (d) Number of ARs per cutting of PuWRKY75‐SRDX lines under LP and NP conditions on the tenth day after cutting. In (b), (c), and (d), values represent the mean ± SD of 30 plants. Significant differences compared based on one‐way ANOVA and Duncan’s multiple range test: **P < 0.01.
In the ML‐hGRN for short‐term LP‐driven AR formation, some of predicted target genes were both the targets of PuMYB40 and PuWRKY75. We hypothesized that PuMYB40 interacts with PuWRKY75 in promoting AR formation in P. ussuriensis. To verify this hypothesis, yeast two‐hybrid (Y2H), Bimolecular Fluorescent Complementary (BiFC), and Co‐immunoprecipitation (Co‐IP) assay were performed. Transcriptional activation activity assay performed above proved that PuMYB40 CTR (109–282 aa) contains transcriptional activation site (Figure 2d). Later, we proved that transcriptional activation site of PuMYB40 was within 252–282 aa. Since it might influence the result of Y2H, we removed the transcriptional activation site (252–282 aa) through PCR‐based technology. As shown in Figure 5a, Y2H results suggest that PuMYB40 interacts with PuWRKY75 in vitro and the PuMYB40▵109–252 and PuWRKY75▵1–100 functioned as the interaction domains. The BiFC results showed that the yellow fluorescent protein (YFP) signal was only detected in the nucleus containing MYB40:YFPN/WRKY75:YFPC (Figure 5b), indicating that PuMYB40 interacts with PuWRKY75 in nuclei. The interaction was further confirmed by Co‐IP assay as MYB40‐FLAG co‐precipitated with WRKY75‐GFP (Figure 5c). These results indicate that the PuMYB40 interacts with PuWRKY75 to mediate short‐term LP‐driven AR formation in P. ussuriensis.
Figure 5.
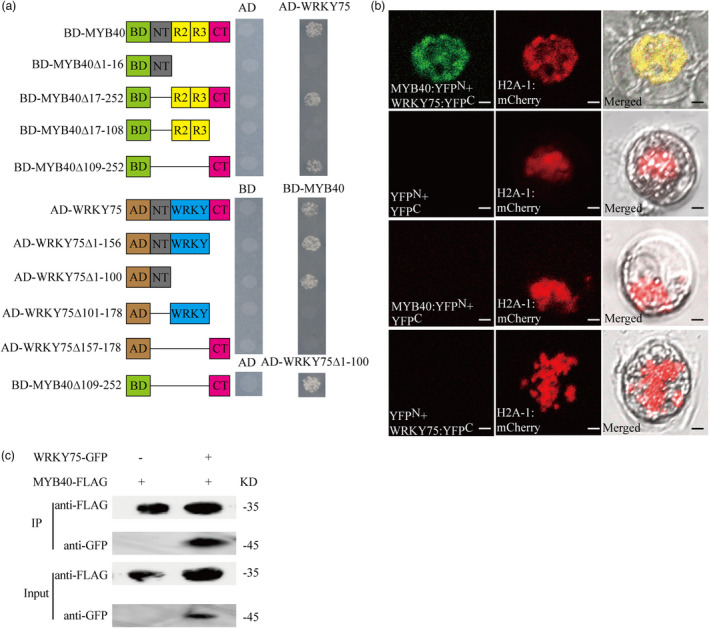
PuMYB40 interacted with PuWRKY75. (a) Yeast two‐hybrid (Y2H) assay was used to identify the interaction of PuMYB40 with PuWRKY75 in yeast. The green rectangles represent GAL4 BD; the brown rectangles represent GAL4 AD; the grey rectangles represent N‐terminal region; the yellow rectangles represent R2R3 domain; the blue rectangles represent WRKY domain, and the magenta rectangles represent C‐terminal region. (b) Bimolecular Fluorescent Complimentary assay demonstrating PuMYB40 interacted with PuWRKY75. Plasmids were co‐transformed into “Yinzhong” Qu 2 protoplasts using PEG‐mediated method. H2A‐1:mCherry served as nuclear marker. All experiments were performed with at least three independent biological repeats. (c) Co‐Immunoprecipitation assay showed that PuMYB40 interacted with PuWRKY75. The indicated Flag‐tagged PuMYB40 was co‐expressed with GFP‐tagged PuWRKY75 in tobacco leaves, while single flag‐tagged PuMYB40 served as control. Both anti‐GFP antibody and anti‐FLAG antibody were diluted 1000‐fold.
PuLRP1 and PuERF003 are direct targets of PuMYB40 and PuWRKY75 as evidenced Yeast one‐hybrid, ChIP‐qPCR, and electrophoresis mobility shift assay
The ML‐hGRN built with the RNA‐seq data from the short‐term LP‐driven AR formation process revealed that PuLRP1 and PuERF003 were located at the layer immediately below the layer that comprised PuMYB40 and PuWRKY75 and were directly regulated by PuMYB40 and PuWRKY75. The sum of the interference frequencies of all outgoing edges of PuWRKY75 was second to that of PuMYB40 (Table S3, Figure S5). We performed yeast one‐hybrid (Y1H), chromatin‐immunoprecipitation quantitative PCR (ChIP‐qPCR) and electrophoresis mobility shift assay (EMSA), which demonstrated that PuMYB40 was capable of binding to several specific MYB‐binding site (MBS) motifs, including MYB core‐binding site CNGTT(A/G) (Romero et al., 1998) and (T/C)AAC(T/G)G (Romero et al., 1998) in promoter sequences of the PuLRP1 and PuERF003, respectively (Figure 6a–h). The promoter sequences of PuLRP1 and PuERF003 were listed in (Figure S9).
Figure 6.
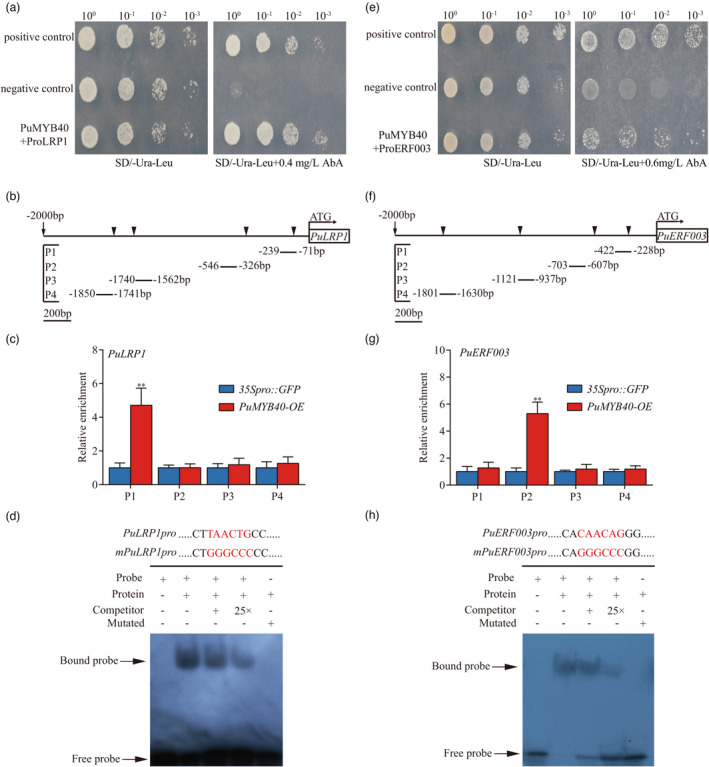
PuMYB40 binding to PuLRP1 and PuERF003 promoters in vitro and in vivo in P. ussuriensis, respectively. (a) Yeast one‐hybrid (Y1H) assay was used to verify PuMYB40 bound to the PuLRP1 promoter region. The pGAD‐Rec‐p53/P53‐pAbAi and pGADT7‐Rec‐PuMYB40/p53‐pAbAi served as positive and negative controls, respectively. The AbA concentration of 0.4 mg/L was the one beyond which yeast growth was completely suppressed. (b) The MYB binding sites of PuMYB40 in PuLRP1 promoter regions. (c) ChIP‐qPCR analysis of the MYB binding sites in PuLRP1 promoter region. The relative abundances of PuLRP1 promoter fragments in the chromatin isolated from PuMYB40‐OE and 35Spro::GFP lines were determined. Each value represents the mean SD of three biological replicates. The asterisks indicate significant (**P < 0.01) difference in enrichment fold between PuMYB40‐OE and 35Spro::GFP lines, based on Student’s t‐test. (d) The use of electrophoretic mobility shift assay (EMSA) to verify the binding of PuMYB40 the MBS cis‐elements in PuLRP1 promoters. The biotin‐labelled probe was used as negative control (Lane 1); the biotin‐labelled probe incubated with PuMYB40 protein was tested (Lane 2); the competitive probes of 1× and 25× (lack of biotin label) were used (Lanes 3 and 4, respectively), the mutant probe incubated with PuMYB40 protein was employed to test the authenticity of PuMYB40’s binding to MBS cis‐element in PuLRP1 promoters (Lane 5). (e) Y1H assay was used to verify PuMYB40 bound to the PuERF003 promoter region. The pGAD‐Rec‐p53/P53‐pAbAi and pGADT7‐Rec‐PuMYB40/p53‐pAbAi served as positive and negative controls, respectively. The AbA concentration of 0.6 mg/L was the one beyond which yeast growth was completely suppressed. (f) The MYB binding sites of PuMYB40 in PuERF003 promoter regions. (g) Chromatin‐immunoprecipitation quantitative PCR (ChIP‐qPCR) analysis of the MYB‐binding sites in PuERF003 promoter region. The relative abundances of PuERF003 promoter fragments in chromatin isolated from PuMYB40‐OE and 35Spro::GFP lines were determined. Each value represents the mean SD of three biological replicates. The asterisks indicate significant (**P < 0.01) difference of enrichment fold between PuMYB40‐OE and 35Spro::GFP lines, based on Student’s t‐test. (h) The use of EMSA to verify the binding of PuMYB40 to the MBS cis‐elements in PuERF003 promoters. The biotin‐labelled probe was used as negative control (Lane 1); the biotin‐labelled probe incubated with PuMYB40 protein was tested (Lane 2); the competitive probes of 1× and 25× (lack of biotin label) were used (Lanes 3 and 4, respectively); the mutant probe incubated with PuMYB40 protein was employed to test the authenticity of PuMYB40’s binding to MBS cis‐elements in PuERF003 promoters (Lane 5).
In addition, we performed Y1H, ChIP‐qPCR, and EMSA and demonstrated the binding of PuWRKY75 to the specific W‐box motif (C/T)TGAC(C/T) in promoter sequences of PuLRP1 and PuERF003 (Figure 7a–h). This is in an agreement with the previous study where WRKY75 was shown to bind to W‐box in A. thaliana (Rushton et al., 2010).
Figure 7.
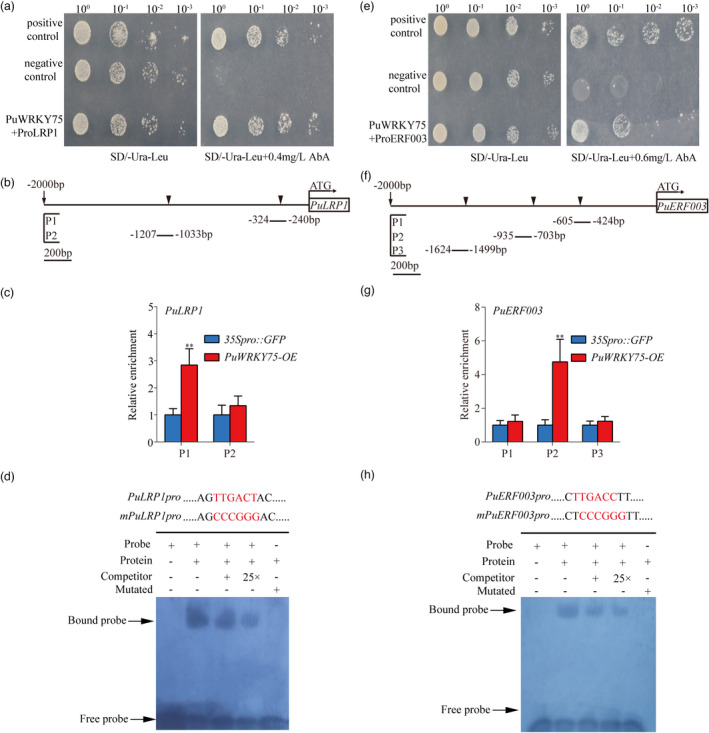
PuWRKY75 binding to PuLRP1 and PuERF003 promoters in vitro and in vivo in P. ussuriensis. (a) Yeast one hybrid (Y1H) assay was used to verify the binding of PuWRKY75 to the PuLRP1 promoter region. The pGAD‐Rec‐p53/P53‐pAbAi and pGADT7‐Rec‐PuWRKY75/p53‐pAbAi served as positive and negative controls, respectively. The AbA concentrations of 0.4 mg/L were one beyond which yeast growth was completely suppressed. (b) The W‐box binding sites of PuWRKY75 in PuLRP1 promoter regions. (c) ChIP‐qPCR analysis of the W‐box cis‐element in PuLRP1 promoter region. The relative abundances of PuLRP1 promoter fragments in the chromatin isolated from PuWRKY75‐OE and 35Spro::GFP lines were determined. Each value represents the mean SD of three biological replicates. The asterisks indicate significant (**P < 0.01) differences of enrichment fold between PuWRKY75‐OE and 35Spro::GFP lines, based on Student’s t‐test. (d) The use of electrophoretic mobility shift assay (EMSA) to verify the binding of PuWRKY75 to the W‐box cis‐elements in PuLRP1 promoters. The biotin‐labelled probe was used as negative control (Lane 1); the biotin‐labelled probe incubated with PuWRKY75 protein was tested (Lane 2); the competitive probes of 1× and 25× (lack of biotin label) were used (Lanes 3 and 4, respectively); the mutant probe incubated with PuWRKY75 protein was employed to test the authenticity of PuWRKY75’s binding to W‐box cis‐element in PuLRP1 promoter (Lane 5). (e) Y1H assay was used to verify the binding of PuWRKY75 to the PuERF003 promoter region. The pGAD‐Rec‐p53/P53‐pAbAi and pGADT7‐Rec‐PuWRKY75/p53‐pAbAi served as positive and negative controls, respectively. The AbA concentrations of 0.6 mg/L were the one beyond which yeast growth was completely suppressed. (f) The W‐box binding sites of PuWRKY75 in PuERF003 promoter region. (g) ChIP‐qPCR analysis of the W‐box cis‐element in PuERF003 promoter region. The relative abundances of PuERF003 promoter fragments in the chromatin isolated from PuWRKY75‐OE and 35Spro::GFP lines were determined. Each value represents the mean SD of three biological replicates. The asterisks indicate significant (**P < 0.01) differences of enrichment fold between PuWRKY75‐OE and 35Spro::GFP lines, based on Student’s t‐test. (h) The use of EMSA to verify the binding of PuWRKY75 to the W‐box cis‐elements in PuERF003 promoters. The biotin‐labelled probe was used as negative control (Lane 1); the biotin‐labelled probe incubated with PuWRKY75 protein was tested (Lane 2); the competitive probes of 1× and 25× (lack of biotin label) were used (Lane 3 and 4, respectively); the mutant probe incubated with PuWRKY75 protein was employed to test the authenticity of PuWRKY75’s binding to W‐box cis‐element in PuERF003 promoter (Lane 5).
Upregulation of PuLRP1 and PuERF003 promoted AR formation in transgenics
We generated 10 PuLRP1‐OE, 14 PuLRP1‐SRDX, 14 PuERF003‐OE, and 12 PuERF003‐SRDX lines and selected the two lines with the highest expression levels for each construct for further phenotype analysis (Figure S10a–h). The basal ends of stem cuttings from PuLRP1‐OE, PuLRP1‐SRDX, PuERF003‐OE, and PuERF003‐SRDX lines were cultured on NP media to observe the AR phenotype compared to that of WT. As expected, the numbers of ARs of both PuLRP1‐OE and PuERF003‐OE observed were much higher than that in WT. In contrast, PuLRP1‐SRDX and PuERF003‐SRDX lines produced less ARs compared with WT (Figure 8a–d). The biomass analysis showed that the root dry weights of PuLRP1‐OE and PuERF003‐OE lines increased 30.97%–43.10% and 42.73%–44.08%, respectively, compared to WT, whereas the root dry weights of PuLRP1‐SRDX and PuERF003‐SRDX lines decreased 34.82%–37.68% and 39.49%–40.32%, respectively (Figure 8e,f). Besides, the AR numbers of PuLRP1‐SRDX and PuERF003‐SRDX lines had no significant difference between LP and NP conditions (Figure 8g,h), substantiating the involvement of PuLRP1 and PuERF003 in LP‐driven AR formation again. These results demonstrated that PuLRP1 and PuERF003 had similar effects on AR formation as PuMYB40 and PuWRKY75 did, and thus are likely to be involved in modulating short‐term LP‐driven AR formation as the target genes of PuMYB40.
Figure 8.
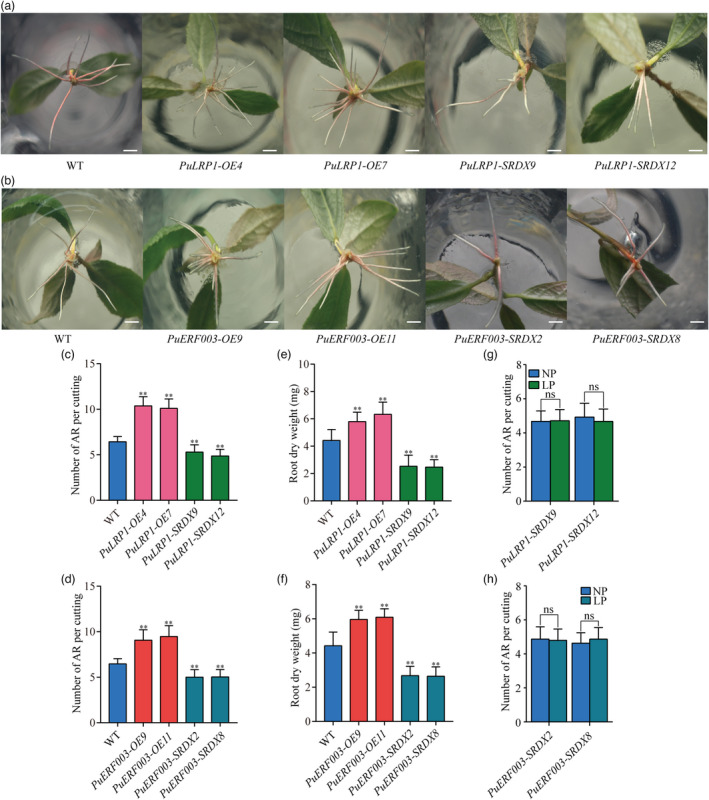
PuLRP1 and PuERF003 accelerated AR formation in P. ussuriensis. (a) Comparison of root growth of PuLRP1 transgenic lines and WT after cutting for 10 days. Bar: 1 cm. (b) Comparison of root growth of PuERF003 transgenic lines and WT after cutting for 10 days. Bar: 1 cm. (c) Number of ARs per cutting of PuLRP1 transgenic lines and WT after cutting for 10 days. (d) Number of ARs per cutting of PuERF003 transgenic lines and WT after cutting for 10 days. (e) Comparison of AR dry weight of PuLRP1 transgenic lines and WT after cutting for 3 weeks. (f) Comparison of AR dry weight of PuERF003 transgenic lines and WT after cutting for 3 weeks. (g) Number of ARs per cutting of PuLRP1‐SRDX lines under LP and NP conditions after cutting for 10 days. (h) Number of ARs per cutting of PuERF003‐SRDX lines under LP and NP after cutting for 10 days. In (c–h), values represent the mean ± SD of 30 plants. Significant differences compared based on one‐way ANOVA and Duncan’s multiple range test: **P < 0.01.
Upregulation of PuLRP1 and PuERF003 in WT under LP condition and in PuMYB40 and PuWRKY75 overexpression lines
Then, we examined the expression patterns of PuLRP1 and PuERF003 during short‐term LP treatment using 5‐mm long basal ends of primordium‐formed cuttings. In WT, PuLRP1 and PuERF003 were highly expressed under LP conditions (Figure 9a,b); a treatment with TIBA (10 μM), an auxin transport inhibitor, decreased the expression levels of PuLRP1 and PuERF003 compared with LP treatment (Figure 9c,d). In PuMYB40‐OE and PuMYB40‐SRDX lines, the expression levels of PuLRP1 and PuERF003 were highly up‐ and down‐regulated, respectively, in 5‐mm long basal ends of primordium‐formed cuttings under LP treatment (Figure 9e,f). Taken together, these results manifested that PuLRP1 and PuERF003 were the direct targets of PuMYB40, and both were up‐regulated by PuMYB40 under LP condition. The down‐regulation of PuLRP1 and PuERF003 caused by TIBA resembled that of PuMYB40 downregulation, implying the involvement of auxin signalling in the LP induced up‐regulation of both genes.
Figure 9.
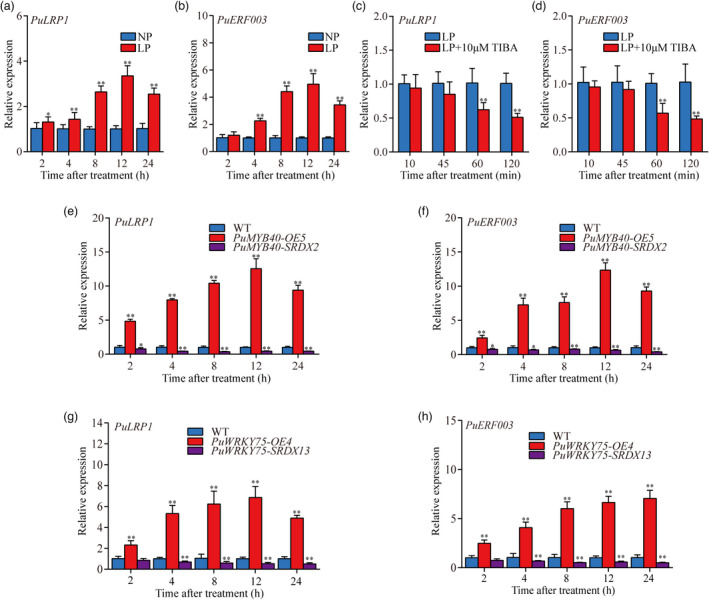
The expression patterns of PuLRP1 and PuERF003. (a) and (b) RT‐qPCR analysis of PuLRP1 and PuERF003 expression patterns during short‐term LP‐accelerated AR formation. (c) and (d) RT‐qPCR analysis of PuLRP1 and PuERF003 expression patterns during LP treatment and LP supplied with 10 μm TIBA treatment. (e) and (f) RT‐qPCR analysis of PuLRP1 and PuERF003 expression patterns in PuMYB40 transgenic lines under LP condition. (g) and (h) RT‐qPCR analysis of PuLRP1 and PuERF003 expression patterns in PuWRKY75 transgenic lines under LP condition. The basal ends of stem cuttings (5 mm) were harvested at different time points. The basal ends of stem cuttings (5 mm) were harvested at different time points under LP or NP condition. Each value represents the mean SD of three biological replicates. The asterisks indicate significant difference in PuLRP1 and PuERF003 expression level, based on Student’s t‐test: *P < 0.05, **P < 0.01.
We also investigated the expression patterns of PuLRP1 and PuERF003 in PuWRKY75 transgenic lines. Both PuLRP1 and PuERF003 were significantly up‐regulated in PuWRKY75‐OE lines and significantly down‐regulated in PuWRKY75‐SRDX lines (Figure 9g,h) in 5‐mm long basal ends of primordium‐formed cuttings under LP conditions, indicating that PuWRKY75 promotes AR formation through up‐regulating PuLRP1 and PuERF003 transcription.
PuMYB40 and PuWRKY75 co‐regulate PuLRP1 and PuERF003 using dual‐luciferase assay
In LUC assay, coexpression of PuMYB40 with ProPuLRP1:LUC and ProPuERF003:LUC in Nicotiana benthamiana leaves led to significant increase in LUC activity, respectively (Figure 10a,b). Also, coexpression of PuWRKY75 with ProPuLRP1:LUC and ProPuERF003:LUC significantly increased LUC activity in N. benthamiana. When PuMYB40 was coexpressed with PuWRKY75, ProPuLRP1:LUC, and ProPuERF003:LUC constructs, the LUC activity was further increased as compared with the above circumstances where only PuMYB40 or PuWRKY75 was used (Figure 10b). The PuMYB40‐binding site (TAACTG) and the PuWRKY75‐binding site (TTGACT) are both present in close proximity in the promoter regions of both PuLRP1 and PuERF003 (Figure S9). Taken together, these results provided direct evidence to substantiate that PuMYB40 and PuWRKY75 co‐regulate the same target genes, PuLRP1 and PuERF003, and the presence of both PuMYB40 and PuWRKY75 enhanced PuLRP1 and PuERF003 to higher expression levels additively. As to whether PuMYB40 and PuWRKY75 acted independently or in a heterodimer, additional evidence is needed.
Figure 10.
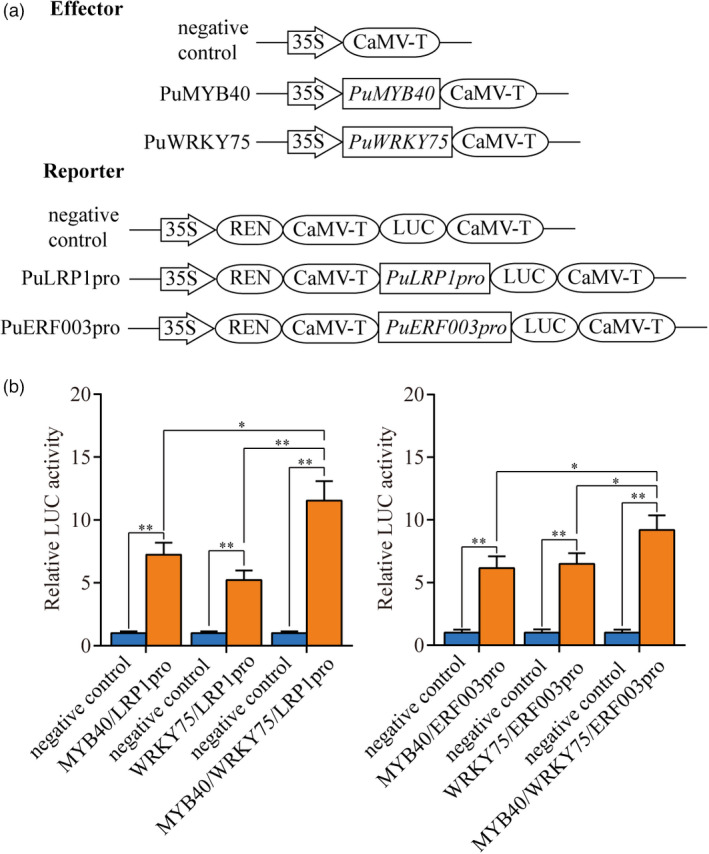
PuMYB40 and PuWRKY75 controls PuLRP1 and PuERF003. (a) Schematic diagrams of effector and reporter constructs used for the dual‐luciferase assay. (b) Dual‐luciferase reporter assay showed that PuMYB40 and PuWRKY75 positively regulated the expression of PuLRP1 and PuERF003, respectively. The pGreen 62SK/pGreen 0800‐LUC served as control and the LUC/REN ratios of control were set as 1. Each value represents the mean SD of three biological replicates. The asterisks indicate significant difference based on Student’s t‐test: *P < 0.05, **P < 0.01.
Discussion
In this study, the LP‐driven AR formation in P. ussuriensis was demonstrated through transfer of root primordium‐formed cuttings to LP conditioned medium after a five‐day culture on WPM medium (NP condition), which is perceived as the deprivation of phosphate for stimulating AR initiation and development. Following that, we dissected the major underlying molecular mechanisms that governed this process via computational and experimental combined approaches and revealed that PuMYB40 and PuWRKY75 were the major high hierarchical regulators that administrated this process through regulating PuLRP1 and PuERF003.
Wounding stress initiated the AR formation, but LP treatment augmented it
It is intriguing that within short‐term (12 h) culture after transferring the root primordium‐formed cuttings to LP media, LP‐treated cuttings produced two times more ARs than in NP condition (Figure 1c). It can be questioned how this rapid response was perceivable within 12 h.
This rapid response did not start from ground up. First, the LP‐driven AR formation was preceded by a five‐day culture in NP condition, where the cuttings already came into the stage of primordium formation after five days (Figure 1c). This could lead to a misconception that NP condition caused AR formation too. However, the real player is the wounding stress; present knowledge has shown that wounding induces early transient accumulation of jasmonate and auxin, initiating AR formation (Druege et al., 2016). Wounding induces WIND TFs to initiate cellular reprogramming that eventually confers AR competence on cut site cells (Iwase et al., 2011). Secondly, transient accumulation of auxin at rooting zone activates WOX11/12 to convert non‐root cells into root founder cells. Finally, WOX11/12 up‐regulates LBD16/29 and WOX5/7 to initiate root meristem formation (Ikeuchi et al., 2016; Jing et al., 2020; Yu et al., 2017).
It should be noted here that not all the AR competent cells are converted into root founder cells. Given that auxin accumulation is ‘transient’ event, not all WOX11/12 gene in all the cells can be activated (Jing et al., 2020). As demonstrated earlier, oscillatory auxin responses have been demonstrated to periodically convert some (not all) of the AR competent cells into root founder cells (Santos and Ten, 2019). Therefore, it can be hypothesized that the unconverted AR competent cells wait for additional signal impulse to become founder cells. LP stress is known to significantly enhance the auxin sensitivity in LR initiation and development (Crombez et al., 2019; Santos and Ten, 2019). It can be inferred that LP treatment activates the dormant AR competent cells by increasing local auxin level, rendering them to develop into root meristems. In addition, ARs formed after LP treatment were shorter than those in NP media in our study, which appears to be in agreement with the well‐known effect of auxin as a root elongation inhibitor in most species (Costa et al., 2013).
Construction of ML‐hGRN that encompassed key regulators controlling LP‐stimulated AR formation
One of primary goals of this study was to delve into gene regulatory networks for identifying TFs involved in short‐term LP‐driven AR formation. Therefore, we employed the Bottom‐up GGM algorithm (Kumari et al., 2016; Wei, 2019) to construct the ML‐hGRN that operated above root formation biological processes. As the effort to identify DEGs from the time‐course RNA‐seq data with large‐time intervals upon LP treatment led to the small number of TFs that were not sufficient for inferring comprehensive ML‐hGRN relationships, we selected whole genome‐wide TFs as candidate input genes to construct the network. As a result, PuMYB40 emerged at the top layer of ML‐hGRN after a competitive competing process. To our surprise, the PuMYB40 was not among the DEGs identified at time‐course LP RNA‐seq data, where the samples were collected at the primordium induction sites (<2 mm in length) at the bases of the stem cuttings, presumably the time points or the sites at which we collected samples did not cover MYB40 up‐regulation (Table S1) or MYB40 was expressed in only a very small fraction of pericycle cells under the primordia. We extended the sample size to the 5‐mm basal ends of stem cuttings and performed RT‐qPCR to determine expression pattern of PuMYB40, and the result showed that PuMYB40 was significantly up‐regulated under LP treatment compared with that under NP conditions. It might be due to the ectopic regulation of target genes by PuMYB40 through intercellular trafficking. Intercellular trafficking of RNAs, proteins, hormones, and metabolites via plasmodesmata has been recently reviewed for playing important roles in local signalling events (Li et al., 2021; Sorkin and Nusinow, 2021). It requires more future endeavours to uncover intercellular trafficking of PuMYB40 during short‐term LP‐driven AR formation.
PuMYB40‐mediated ML‐hGRN not only accelerated wounding‐induced AR formation but also served as a major player in the LP‐stimulated AR formation
The ML‐hGRN that we constructed consists of two cores collaborative and interactive TFs, PuMYB40 and PuWRKY75, and their target genes, PuLRP1 and PuERF003, which were experimentally validated (Figures 6a–h, 7a–h and 9a–h). The counterparts of PuLRP1 and PuERF003 promote AR formation in A. thaliana (Smith and Fedoroff, 1995) and poplar (Trupiano et al., 2013). In A. thaliana, LRP1, which encodes a plant‐specific SHORT INTERNODES/STYLISH (SHI/STY) protein, is up‐regulated during LR and AR formation (Singh et al., 2020; Smith and Fedoroff, 1995). More importantly, LRP1 was found to activate downstream of several auxin signalling modules such as SLR/IAA14‐ARF7‐ARF19, IAA28‐ARF7‐ARF19‐GATA23, BDL/IAA12 MP/ARF5, and IAA3/SHY2‐ARFs and its overexpression leads to more LR formation loci and primordia. In addition, LRP1 and the SHI/STY‐related proteins have been found to induce auxin biosynthesis‐related gene, YUC4, which increases endogenous auxin level. Thus, enhancement of auxin signalling by LRP1 overexpression may result in more LR initiation sites (Singh et al., 2020). The evidence suggests that LRP1 and auxin modulate each other during development of LR. In hybrid poplar (P. tremula × P. alba), PtaERF003, which belongs to Va group of the AP2/ERF family, was identified, and characterized to promote LR and AR formation, however, its exact function remains unknown (Trupiano et al., 2013). It has been shown that the AP2/ERF TFs are also responsible for endogenous auxin biosynthesis as well as cell fate transition from non‐root cells to root meristem cells during AR formation (Bellini et al., 2014; Druege et al., 2016). Based on the evidence above, the PuMYB40‐targeted genes, PuLRP1 and PuERF003, are suggested to enhance auxin signalling. In addition to PuMYB40, PuWRKY75, a WRKY family TF whose counterpart is known to control AR primordium formation (Devaiah et al., 2007a), played a role in AR formation, but the regulatory role of AtWRKY75 was not characterized before. In this study, we demonstrated that PuWRKY75 and PuMYB40 interacted with each other (Figure 5b) and directly co‐regulate PuLRP1 and PuERF003 (Figures 6a–h and 10a,b) to promote AR formation. Since wounding stress‐induced AR formation was in progress (Figure 1c) in the stem cuttings prior to LP treatment, LP stimulus activated PuMYB40/PuWRKY75‐mediated ML‐hGRN, leading to significant increase in auxin responses that in turn accelerated AR formation by enhancing pre‐established auxin signalling. Conversely, the PuMYB40‐centred network genes seem to be positively regulated by auxin. Blocking auxin transport by TIBA treatment dramatically declined the expression levels of PuMYB40, PuLRP1, and PuERF003 even under LP condition (Figures 2b and 9c,d). This provided the evidence for reciprocal interaction between the PuMYB40‐centred network and auxin signalling. However, specific mechanism underlying the cross‐talk between PuMYB40 network and auxin signalling needs to be further demonstrated.
LP treatment initiation time rather than duration played a more important role in promoting AR number
We placed the stem cuttings under LP condition immediately after excising, and cultured for seven days, which can be regarded as long‐term LP treatment. Interestingly, it did not show significant increase in AR phenotypes compared to NP (Figure S3). This result showed that the initiation of LP treatment immediately after cutting did not necessarily augment AR formation and numbers, which implies that the time of LP treatment initiation is more important than the LP treatment duration, albeit underlying molecular mechanisms remain unclear and need further investigations. In this study, 12–24 h LP treatment at five days after wounding was demonstrated as an appropriate time to escalate AR formation in P. ussuriensis. It is of great significance that we established for short term LP‐driven AR formation, since it could be a time‐saving strategy in vegetative propagation of P. ussuriensis.
In summary, LP treatment up‐regulated PuMYB40 and PuWRKY75 in the root primordia emerged from the basal ends of stem cuttings in P. ussuriensis. The activated PuMYB40 and PuWRKY75 directly targeted PuLRP1 and PuERF003, both of which are known to enhance AR‐associated auxin signalling and enhance the number of ARs. Thus, our new results and data unveiled the major high hierarchical regulators and their top‐down regulatory “command chains” that augment AR formation and subsequent whole plant growth upon short‐term LP treatment in P. ussuriensis. The results advanced our understanding the regulatory mechanisms underlying AR formation in woody species.
Experimental procedures
Plant materials and growth conditions
The plants of P. ussuriensis clone termed “Donglin” were maintained in vitro at 25 °C under 46 μmol photons m−2 s−1 cool white fluorescent lights illumination with 16 h‐light/8 h‐darkness photoperiod. New cuttings (c. 3 cm) from 3‐week‐old in vitro plants were cultured on the Woody Plant Medium (WPM) (Lloyd, 1980) supplemented with 20 g/L sucrose and 3 g/L Gelrite (Duchefa, Haarlem, The Netherlands), in which Pi concentration was conditioned as 200 μm KH2PO4. After 5 days of culture, the root primordium‐formed cuttings were transferred to two types of media: (i) liquid WPM with 200 μm KH2PO4 (NP); (ii) For the low phosphate WPM medium, the Pi concentration was 10 μm and the amount of reduced potassium ions were supplied by addition of 95 μm K2SO4 to make the final concentration of potassium the same as the normal WPM medium.
Paraffin section assay
12 h after being cultured in the above liquid media, the basal ends of root primordium‐formed cuttings were fixed in FAA solution containing 5:90:5 (v:v:v) of 37% formaldehyde, 70% alcohol, and glacial acetic acid for 24 h. Then, the cuttings were stained with haematoxylin for 24 h and washed with cold running water overnight. After dehydration using graded ethanol and vitrification by dimethylbenzene, the cuttings were embedded in paraffin. Following that, 8‐μm sections were cut and dewaxed by dimethylbenzene and rehydrated by graded ethanol. Finally, the sections were observed and imaged using a stereo microscope (Olympus SZX7, Tokyo, Japan).
RNA extraction and RT‐qPCR analysis
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Duesseldorf, Germany, Cat. No. 74904). First‐strand cDNA synthesis was carried out with HIScriptⅡ QRT SuperMix for qPCR gDNA wiper Kit (Vazyme, Nanjing, China) following the protocol provided by the manufacturer. RT‐qPCR was performed using TransStart® Top Green qPCR SuperMix (TransGen Biotech, Beijing, China). qTOWER 3G Cycler was used to work program. The relative expression levels of genes were calculated using the method (Arocho et al., 2006). The PuActin7 and PuUBQ10 served as the internal control (Wei et al., 2020a,2020b).
RNA‐seq library construction and sequencing
A scalpel was used to precisely harvest the site of AR primordium induction (<2 mm in length) at the basal ends of the stem cuttings at 2, 4, 8, 12, and 24 h after the root primordium‐formed cuttings were transferred into liquid WPM containing 10 or 200 μm KH2PO4. Samples were collected with two biological replicates (50 cuttings each). Total RNA isolation, library construction, and sequencing were performed by Annoroad Gene Technology Corporation (Beijing, China). Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (#E7530L; NEB, Beverly, MA, USA) following the manufacturer’s procedures. The libraries were sequenced on an Illumina Hiseq 2500 platform to generate paired‐end reads of 150 nt in length. NCBI Gene Expression Omnibus (GEO) database accession number of RNA‐seq samples is GSE174726.
Differential expression analysis and construction of ML‐hGRN
Based on the read counts, differentially expressed genes (DEGs) for each time point were identified using the edgeR method contained in the Pop’s pipe package (Li et al., 2014b), with the cut‐off thresholds of log2 (Fold change) >1 and P‐value < 0.05. Then, DEGs for each time point were used as an input for GO enrichment analysis using the same package. After that, all genes involved in biological processes of root development (GO: 0048364), low phosphate (GO: 0016036), and auxin signalling (GO: 0009733) were used as the bottom layer to construct a multi‐layered hierarchical gene regulatory network (ML‐hGRN), with all TFs being used as the input for constructing upper layers. The algorithm used is Bottom‐up GGM algorithm that has been described early (Kumari et al., 2016; Lu et al., 2013; Wei, 2019). Finally, three‐layered gene regulatory network of LP‐driven AR formation in P. ussuriensis was obtained.
Auxin inhibitor treatment
Root primordium‐formed cuttings were cultured in liquid LP WPM with or without 10 μm IAA or 10 μm TIBA (auxin transport inhibitor). After 10‐, 45‐, 60‐, and 120‐min culture, 5‐mm long basal ends of the root primordium‐formed cuttings were harvested to analyse impact of TIBA on temporal expression level of PuMYB40, PuWRKY75, PuLRP1, and PuERF003 by RT‐qPCR.
Gene cloning, vector constructs, and poplar transformation
We synthesized the first‐stranded complementary DNA (cDNA) using PrimeScript RT First Strand cDNA Synthesis kit (Takara, Dalian, China) and mRNA from all the time points. Following that, the PuMYB40, PuWRKY75, PuLRP1, and PuERF003 were amplified by PCR with KOD DNA Polymerase (TOYOBO, Osaka, Japan), the gene‐specific primers, and P. ussuriensis cDNAs. The full‐length coding sequences of PuMYB40, PuWRKY75, PuLRP1, and PuERF003 were cloned into pBI121 vector under the control of cauliflower mosaic virus 35S promoter (35S) for overexpression. A 27‐bp DNA sequence encoding the SUPERMAN repression domain X (Hiratsu et al., 2003) was fused in‐frame to the 3’ ends of the PuMYB40, PuWRKY75, PuLRP1, and PuERF003, which were then cloned into the pBI121‐GFP to construct repression vector. Transgenic lines were generated using leaf disc method as described earlier (Maheshwari and Kovalchuk, 2016). All transgenic lines were verified by PCR and RT‐qPCR analysis. The primers used in vector construction were listed in Table S4.
Transcriptional activation activity assay and yeast two‐hybrid assay
The full‐length and different domain segments of coding region of PuMYB40 were fused into pGBDT7‐BD vector to identify the activation domain. The resulting recombinant plasmids were transformed into Y2HGold yeast competent using YeastmakerTM Yeast Transformation System 2 (Takara, Beijing, China), and the transformed yeast competent cells were then placed on SD/‐Trp, SD/‐Trp‐His‐Ade and SD/‐Trp‐His‐Ade containing 0.1 m X‐α‐Gal yeast deficient medium at 30 °C for three days.
The Y2H experiment was conducted following the manual of Matchmaker Gold Yeast Two‐Hybrid System Kit (Clontech, Mountain View, CA, USA). In brief, full‐length, and segmental coding sequences of PuMYB40 and PuWRKY75 were inserted into pGBKT7‐BD and pGADT7‐AD vector, respectively. Recombinant plasmids were co‐transformed into Y2HGold yeast competent using YeastmakerTM Yeast Transformation System 2 (Takara). The transformed yeast cells were spread on SD/‐Leu‐Trp‐His‐Ade yeast deficient mediums. The full‐length, and segmental coding sequences of PuMYB40 inserted into pGBKT7‐BD, which were co‐transformed with pGADT7‐AD vector into Y2HGold yeast competent, served as a negative control; additionally, the full‐length and segmental coding sequences of PuWRKY75 inserted into pGADT7‐AD that was co‐transformed with pGBKT7‐BD vector into Y2HGold yeast competent served as the other negative control. The primers used were listed in Table S4.
Chromatin immunoprecipitation assay
Mature roots of three‐week‐old PuMYB40‐OE, PuWRKY75‐OE, and 35Spro::GFP lines were used to perform ChIP assay following previous protocol with minor modification (Li et al., 2014a). Three grams of mature roots were collected, and cross‐linking was done using the 1% (v/v) formaldehyde under vacuum for 15 min at room temperature. Then, 0.125 m glycine was added to quench the cross‐linking. Samples were ground to fine powder with liquid nitrogen to isolate cell nuclei. Chromatin was dissolved and sheared into fragments of 200–500 bp by sonication. All experiments were performed with three independent biological replicates. Primers used for ChIP‐qPCR were listed in Table S4.
Yeast One‐Hybrid assay
Y1H assays were performed using the Matchmaker yeast one‐hybrid system (Clontech). Briefly, PuMYB40 and PuWRKY75 coding sequences were fused into pGADT7‐AD vector. PuLRP1 and PuERF003 promoter sequences were cloned and inserted into pAbAi vector; the resulting constructs were co‐transformed into Y1HGold yeast strain using YeastmakerTM Yeast Transformation System 2 (Takara). The transformed yeast cells were placed on SD/‐Ura‐Leu and SD/‐Ura‐Leu with 0.4 mg/mL Aureobasidin A (AbA) and 0.6 mg/L AbA yeast deficient medium for three days at 30 °C. All primers used in Y1H assay were listed in Table S4.
Electrophoretic mobility shift assay
The full‐length coding regions of PuMYB40 and PuWRKY75 were fused into pMAL‐c5X vector and then transformed into ER2523 competence cells (NEB, UK). Recombination proteins were expressed and purified using Amylose Resin (NEB, Beverly, MA, USA) following the manufacturer’s instructions. Oligonucleotide probes containing the MBS and W‐box motifs, which are specific for promoters of PuLRP1 and PuERF003, both were labelled with biotin by using EMSA Probe Biotin Labeling Kit (Beyotime, Jiangsu, China) following the manufacturer’s instructions. EMSA was performed following the procedure described early (Wei et al., 2020). The construct‐based primer and probe sequences used for EMSA were listed in Table S4.
Dual‐Luciferase assay
The full‐length coding regions of PuMYB40 and PuWRKY75 were fused into pGreenII 62‐SK vector. The promoter sequences of PuLRP1 and PuERF003 were fused into pGreenⅡ0800‐LUC vector. All plasmid vectors were separately transformed into Agrobacterium EHA105. Then mixed the Agrobacteria of PuMYB40‐62SK/PuLRP1‐0800LUC, PuMYB40‐62SK/PuERF003‐0800LUC, PuWRKY75‐62SK/PuLRP1‐0800LUC, and PuWRKY75‐62SK/PuERF003‐0800LUC were co‐infiltrated in leaves of 6‐week‐old Nicotiana benthamiana. With the PGreenII‐62SK/pGreenⅡ0800‐LUC served as the control. Dual‐Luciferase® Reporter Assay System (Promega, E1910, Madison, WI, USA) was used to perform LUC assay; the firefly luciferase activity and Renillia luciferase (REN) activity were measured using Synergy HI Microplate Reader (Bio‐Tek, Biotek Winooski, Vermont, USA) following the manufacturer’s instructions. The primers used for LUC assays were listed in Table S4.
Co‐immunoprecipitation assay
Full‐length PuMYB40 and PuWRKY75 coding sequences were inserted into the pMpGWB111 vector with the 3× Flag tag and pBI121‐GFP vector with the GFP tag, respectively. The bacteria carrying above constructs were infiltrated into the leaves of six‐week‐old N. benthamiana. Co‐IP assay was performed as previously described (Li et al., 2020; Liu et al., 2020). In brief, the infiltrated N. benthamiana leaves were grounded in liquid N and resuspended the powder in 1 mL protein extraction buffer (50 mm of Tris‐HCL at pH 7.4, 2.5 mm of EDTA, 150 mm of NaCl, 0.2% [w/v] NP‐40, 20% [v/v] glycerol, 1 mm of PMSF and 1% plant cocktail [v/v]; Sigma, St. Louis, MO, USA). The lysate was then centrifuged at 12 000 g for 5 min at 4 °C, and the supernatant was centrifuged one more time at 12 000 g for 5 min at 4 °C. The supernatant was incubated with 5 μL Anti‐FLAG antibody (Abcam) to immunoprecipitated the protein complex by shaking at 40 rpm, 4 °C overnight. Protein A/G beads (Thermo Fisher Scientific, Waltham, MA, USA) were washed with 1 mL protein extraction buffer 4 times. One hundred microlitres washed Protein A/G beads were added to protein‐antibody complexes and shaking at 40 rpm, 4 °C for 2 h. The beads were collected and washed two times with 300 μL of Elution Buffer (10 µL 20% SDS, 20 µL 1 m NaHCO3 and 170 µL ddH2O). Western blot was performed using anti‐GFP (Abcam, Cambridge, MA, USA) and anti‐FLAG (Abcam, Cambridge, MA, USA) antibody to detect the target proteins. The imaging was performed using GE Fujifilm Image Quant LAS4000. The primers used for Co‐IP assay were listed in Table S4.
Bimolecular fluorescent complementary assay
Populus alba × P. berolinensis protoplasts were isolated and transformed following the procedures as described early (Lin et al., 2014; Liu et al., 2021). In brief, PuMYB40 and PuWRKY75 coding regions were infused into the pENTER/D‐TOPO vector, and then recombined with the nEYFP/pUGW2 and cEYFP/pUGW2 using Gateway recombinant technology, respectively. High‐purity plasmids were extracted using the CsCl gradient ultracentrifugation method as described in early (Lin et al., 2014). MYB40:YFPN/WRKY75:YFPC, MYB40:YFPN/YFPC, YFPN/WRKY75:YFPC and YFPN/YFPC were co‐transformed with H2A‐1:mCherry into poplar protoplasts through PEG‐mediated method (Lin et al., 2014). After 14 h co‐culture, protoplasts were collected and then, the fluorescence signals were observed using a laser focusing microscope (Synergy HI Microplate Reader (Bio‐Tek). The primers used were listed in Table S4.
Author contributions
CL, HRW, and HZW designed the study; HZW, SP, JY, YW, WL, and HF performed the research; HZW, HRW, and WL analysed the data; HZW, SP, HRW, and CL wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Figure S1 A scheme showing the experimental steps of low phosphate treatment experimental steps.
Figure S2 Comparison of root length in WT treated with low phosphorus (LP) and normal phosphorus (NP) conditions.
Figure S3 Comparison of AR number in cuttings directly treated with low phosphorus (LP) and normal phosphorus (NP) conditions.
Figure S4 RT‐qPCR analysis of Pi starvation induction related DEGs from RNA‐seq experiment.
Figure S5 The multilayered hierarchical gene regulatory network (ML‐hGRN) built with Bottom‐up GGM algorithm where PuMYB40 and PuWRKY75 were located at the third layer and directly regulated LPR1 and ERF003.
Figure S6 PCR and RT‐qPCR verified the expression level of PuMYB40 in PuMYB40‐OE and PuMYB40‐SRDX lines.
Figure S7 Phylogenetic analysis and expression pattern of PuWRKY75.
Figure S8 PCR and RT‐qPCR verified the expression level of PuWRKY75 in PuWRKY75‐overexpression (OE) lines and PuWRKY75‐ SUPERMAN repression domain X (SRDX) lines.
Figure S9 The promoter sequence of PuLRP1 and PuERF003.
Figure S10 PCR and RT‐qPCR verified the expression level of PuLRP1 and PuERF003 in PuLRP1 and PuERF003 transgenic lines, respectively.
Table S1 Total differential expression genes (DEGs) of low phosphorus (LP) time‐course RNA‐seq data
Table S2 The GO annotation of 29 root development‐, auxin‐ and low phosphate starvation‐ related DEGs in low phosphorus (LP) time‐course RNA‐seq data
Table S3 The short‐term low phosphorus (LP)‐driven AR formation ML‐hGRN
Table S4 Primers used in this study
Table S5 Statistical report of t tests and ANOVA results for the data presented in each figure
Acknowledgement
This work was supported by the NSFC (31971671), the Fundamental Research Funds for the Central Universities of China (2572018CL04), the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (2020A02) and the Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Wang, H. , Pak, S. , Yang, J. , Wu, Y. , Li, W. , Feng, H. , Yang, J. , Wei, H. and Li, C. (2022) Two high hierarchical regulators, PuMYB40 and PuWRKY75, control the low phosphorus driven adventitious root formation in Populus ussuriensis . Plant Biotechnol. J., 10.1111/pbi.13833
Contributor Information
Hairong Wei, Email: hairong@mtu.edu.
Chenghao Li, Email: chli@nefu.edu.cn.
Data availability
The sequence data of genes in this article have been deposited into GenBank under the following accession numbers: PuMYB40 (MZ687801), PuWRKY75 (MZ687802), PuLRP1 (MZ687803), PuERF003 (MZ687804), PuActin7 (MH644084), and PuUBQ10 (MN872590). The raw data of RNA‐seq experiments reported in this study have been deposited in the Gene Expression Omnibus with accession numbers of GSE174726.
References
- Abarca, D. (2021) Identifying molecular checkpoints for adventitious root induction: are we ready to fill the gaps? Front. Plant Sci. 12, 621032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angkawijaya, A.E. and Nakamura, Y. (2017) Arabidopsis PECP1 and PS2 are phosphate starvation‐inducible phosphocholine phosphatases. Biochem. Biophys. Res. Commun. 494, 397–401. [DOI] [PubMed] [Google Scholar]
- Arocho, A. , Chen, B. , Ladanyi, M. and Pan, Q. (2006) Validation of the 2‐DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR‐ABL P210 transcripts. Diagn. Mol. Pathol. 15, 56–61. [DOI] [PubMed] [Google Scholar]
- Bakshi, M. , Vahabi, K. , Bhattacharya, S. , Sherameti, I. , Varma, A. , Yeh, K.W. , Baldwin, I. et al. (2015) WRKY6 restricts Piriformospora indica‐stimulated and phosphate‐induced root development in Arabidopsis . BMC Plant Biol. 15, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola, P.A. , Howard, C.J. , Taylor, C.B. , Verburg, M.T. , Jaglan, V.D. and Green, P.J. (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6, 673–685. [DOI] [PubMed] [Google Scholar]
- Bellini, C. , Pacurar, D.I. and Perrone, I. (2014) Adventitious roots and lateral roots: similarities and differences. Annu. Rev. Plant Biol. 65, 639–666. [DOI] [PubMed] [Google Scholar]
- Castrillo, G. , Teixeira, P.J. , Paredes, S.H. , Law, T.F. , de Lorenzo, L. , Feltcher, M.E. , Finkel, O.M. et al. (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature, 543, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Zhou, W. , Sheery, N.I. , Peters, C. , Li, M. , Wang, X. and Huang, J. (2011) Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid‐localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 66, 781–795. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J. and Lin, S.I. (2011) Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Costa, C.T. , Almeida, M.R. , Ruedell, C.M. , Schwambach, J. , Maraschin, F.S. and Fettneto, A.G. (2013) When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 4, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombez, H. , Motte, H. and Beeckman, T. (2019) Tackling plant phosphate starvation by the roots. Dev. Cell, 48, 599–615. [DOI] [PubMed] [Google Scholar]
- Dai, X. , Wang, Y. , Yang, A. and Zhang, W.H. (2012) OsMYB2P‐1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate‐starvation responses and root architecture in rice. Plant Physiol. 159, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , Wang, Y. and Zhang, W.H. (2016) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah, B.N. , Karthikeyan, A.S. and Raghothama, K.G. (2007a) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis . Plant Physiol. 143, 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah, B.N. , Madhuvanthi, R. , Karthikeyan, A.S. and Raghothama, K.G. (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis . Mol. Plant, 2, 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah, B.N. , Nagarajan, V.K. and Raghothama, K.G. (2007b) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 145, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druege, U. , Franken, P. and Hajirezaei, M.R. (2016) Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 7, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, K.E. , Yi, K. , Dang, L. , Huang, H. , Wu, W. and Wu, P. (2008) Characterization of a sub‐family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54, 965–975. [DOI] [PubMed] [Google Scholar]
- Dubos, C. , Stracke, R. , Grotewold, E. , Weisshaar, B. , Martin, C. and Lepiniec, L. (2010) MYB transcription factors in Arabidopsis . Trends Plant Sci. 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Chen, G. , Zhang, X. , Li, T. , Yu, H. and Liu, C. (2018) Quantitative trait locus analysis of adventitious and lateral root morphology of barley grown at low and high P. Funct. Plant Biol. 45, 957–967. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K. , Matsui, K. , Koyama, T. and Ohme‐Takagi, M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis . Plant J. 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hu, B. , Zhu, C. , Li, F. , Tang, J. , Wang, Y. , Lin, A. , Liu, L. et al. (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156, 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G. and Zhang, D. (2020) The plasticity of root systems in response to external phosphate. Int. J. Mol. Sci. 21, 5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi, M. , Ogawa, Y. , Iwase, A. and Sugimoto, K. (2016) Plant regeneration: cellular origins and molecular mechanisms. Development, 143, 1442–1451. [DOI] [PubMed] [Google Scholar]
- Iwase, A. , Mitsuda, N. , Koyama, T. , Hiratsu, K. , Kojima, M. , Arai, T. , Inoue, Y. et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis . Curr. Biol. 21, 508–514. [DOI] [PubMed] [Google Scholar]
- Jain, A. , Poling, M.D. , Karthikeyan, A.S. , Blakeslee, J.J. , Peer, W.A. , Titapiwatanakun, B. , Murphy, A.S. et al. (2007) Differential effects of sucrose and auxin on localized phosphate deficiency‐induced modulation of different traits of root system architecture in Arabidopsis . Plant Physiol. 144, 232–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, T. , Ardiansyah, R. , Xu, Q. , Xing, Q. and Muller‐Xing, R. (2020) Reprogramming of cell fate during root regeneration by transcriptional and epigenetic networks. Front. Plant Sci. 11, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J. , Lynch, J.P. and Brown, K.M. (2008) Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant Cell Environ. 31, 1744–1755. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. and Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, S. , Deng, W. , Gunasekara, C. , Chiang, V. , Chen, H.S. , Ma, H. , Davis, X. et al. (2016) Bottom‐up GGM algorithm for constructing multilayered hierarchical gene regulatory networks that govern biological pathways or processes. BMC Bioinformatics, 17, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legué, V. , Rigal, A. and Bhalerao, R.P. (2014) Adventitious root formation in tree species: involvement of transcription factors. Physiol Plant. 151, 192–198. [DOI] [PubMed] [Google Scholar]
- Li, C. , Qi, W. , Liang, Z. , Yang, X. , Ma, Z. and Song, R. (2020) A SnRK1‐ZmRFWD3‐Opaque2 signaling axis regulates diurnal nitrogen accumulation in maize seeds. Plant Cell, 32, 2823–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Lin, Y.C. , Li, Q. , Shi, R. , Lin, C.Y. , Chen, H. , Chuang, L. et al. (2014a) A robust chromatin immunoprecipitation protocol for studying transcription factor‐DNA interactions and histone modifications in wood‐forming tissue. Nat. Protoc. 9, 2180–2193. [DOI] [PubMed] [Google Scholar]
- Li, X. , Gunasekara, C. , Guo, Y. , Zhang, H. , Lei, L. , Tunlaya‐Anukit, S. , Busov, V. et al. (2014b) Pop's Pipes: poplar gene expression data analysis pipelines. Tree Genet. Genomes, 10, 1093–1101. [Google Scholar]
- Li, Z.P. , Paterlini, A. , Glavier, M. and Bayer, E.M. (2021) Intercellular trafficking via plasmodesmata: molecular layers of complexity. Cell. Mol. Life Sci. 78, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.C. , Li, W. , Chen, H. , Li, Q. , Sun, Y.H. , Shi, R. , Lin, C.Y. et al. (2014) A simple improved‐throughput xylem protoplast system for studying wood formation. Nat. Protoc. 9, 2194–2205. [DOI] [PubMed] [Google Scholar]
- Liu, C. , Li, K. , Wang, M. , Fan, E. , Yang, C. , Wang, J. , Fu, P. et al. (2021) Qu‐2, a robust poplar suspension cell line for molecular biology. J. For. Res. 32, 733–740. [Google Scholar]
- Liu, Y. , Ma, M. , Li, G. , Yuan, L. , Xie, Y. , Wei, H. , Ma, X. et al. (2020) Transcription factors FHY3 and FAR1 regulate light‐Induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis . Plant Cell, 32, 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, G. and McCown, B. (1980) Commercially feasible micropropagation of mountain laurel, (Kalmia latifolia) by use of shoot tip culture. Comb. Proc. Int. Plant Propag. Soc. 30, 421–427. [Google Scholar]
- Lopez‐Bucio, J. , Hernandez‐Abreu, E. , Sanchez‐Calderon, L. , Perez‐Torres, A. , Rampey, R.A. , Bartel, B. and Herrera‐Estrella, L. (2005) An auxin transport independent pathway is involved in phosphate stress‐induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 137, 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S. , Li, Q. , Wei, H. , Chang, M.J. , Tunlaya‐Anukit, S. , Kim, H. , Liu, J. et al. (2013) Ptr‐miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa . Proc. Natl Acad. Sci. USA, 110, 10848–10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig‐Muller, J. , Vertocnik, A. and Town, C.D. (2005) Analysis of indole‐3‐butyric acid‐induced adventitious root formation on Arabidopsis stem segments. J. Exp. Bot. 56, 2095–2105. [DOI] [PubMed] [Google Scholar]
- Lynch, J.P. (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 156, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari, P. and Kovalchuk, I. (2016) Agrobacterium‐mediated stable genetic transformation of Populus angustifolia and Populus balsamifera . Front. Plant Sci. 7, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C.R. , Ochoa, I. , Nielsen, K.L. , Beck, D. and Lynch, J.P. (2003) Genetic variation for adventitious rooting in response to low phosphorus availability: potential utility for phosphorus acquisition from stratified soils. Funct. Plant Biol. 30, 973–985. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Rus, A. , Sharkhuu, A. , Yokoi, S. , Karthikeyan, A.S. , Raghothama, K.G. , Baek, D. et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl Acad. Sci. USA, 102, 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi, M. , Sanagala, R. , Rai, V. and Jain, A. (2016) Deciphering phosphate deficiency‐mediated temporal effects on different root traits in rice grown in a modified hydroponic system. Front. Plant Sci. 7, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa, I.E. , Blair, M.W. and Lynch, J.P. (2006) QTL analysis of adventitious root formation in common bean under contrasting phosphorus availability. Crop Sci. 46, 1609–1621. [Google Scholar]
- Péret, B. , Clement, M. , Nussaume, L. and Desnos, T. (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 16, 442–450. [DOI] [PubMed] [Google Scholar]
- Pozo, J.C. , Allona, I. , Rubio, V. , Leyva, A. , Pena, A.D. , Aragoncillo, C. and Paz‐Ares, J. (2002) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 19, 579–589. [DOI] [PubMed] [Google Scholar]
- Rigal, A. , Yordanov, Y.S. , Perrone, I. , Karlberg, A. , Tisserant, E. , Bellini, C. , Busov, V.B. et al. (2012) The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 160, 1996–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, I. , Fuertes, A. , Benito, M.J. , Malpica, J.M. , Leyva, A. and Paz‐Ares, J. (1998) More than 80 R2R3‐MYB regulatory genes in the genome of Arabidopsis thaliana . Plant J. 14, 273–284. [DOI] [PubMed] [Google Scholar]
- Rubio, V. , Linhares, F. , Solano, R. , Martin, A.C. , Iglesias, J. , Leyva, A. and Paz‐Ares, J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Santos, T.J. and Ten, T.K. (2019) The systems biology of lateral root formation: connecting the dots. Mol. Plant, 12, 784–803. [DOI] [PubMed] [Google Scholar]
- Sega, P. and Pacak, A. (2019) Plant PHR transcription factors: put on a map. Genes, 10, 1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Yadav, S. , Singh, A. , Mahima, M. , Singh, A. , Gautam, V. and Sarkar, A.K. (2020) Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis . Plant J. 101, 87–100. [DOI] [PubMed] [Google Scholar]
- Smith, D.L. and Fedoroff, N.V. (1995) LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis . Plant Cell, 7, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin, M.L. and Nusinow, D.A. (2021) Time will tell: intercellular communication in the plant clock. Trends Plant Sci. 26, 706–719. [DOI] [PubMed] [Google Scholar]
- Tran, H.T. , Qian, W. , Hurley, B.A. , She, Y.‐M. , Wang, D. and Plaxton, W.C. (2010) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate‐starved Arabidopsis thaliana . Plant Cell Environ. 33, 1789–1803. [DOI] [PubMed] [Google Scholar]
- Trupiano, D. , Yordanov, Y. , Regan, S. , Meilan, R. , Tschaplinski, T. , Scippa, G.S. and Busov, V. (2013) Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus . Planta, 238, 271–282. [DOI] [PubMed] [Google Scholar]
- Walk, T.C. , Jaramillo, R. and Lynch, J.P. (2006) Architectural tradeoffs between adventitious and basal roots for phosphorus acquisition. Plant Soil, 279, 347–366. [Google Scholar]
- Wei, H. (2019) Construction of a hierarchical gene regulatory network centered around a transcription factor. Brief. Bioinform. 20, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Wei, H. , Yordanov, Y.S. , Georgieva, T. , Li, X. and Busov, V. (2013) Nitrogen deprivation promotes Populus root growth through global transcriptome reprogramming and activation of hierarchical genetic networks. New Phytol. 200, 483–497. [DOI] [PubMed] [Google Scholar]
- Wei, M. , Chen, Y. , Zhang, M. , Yang, J. , Lu, H. , Zhang, X. and Li, C. (2020) Selection and validation of reference genes for the qRT‐PCR assays of Populus ussuriensis gene expression under abiotic stresses and related ABA treatment. Forests, 11, 476. [Google Scholar]
- Wei, M. , Liu, Q. , Wang, Z. , Yang, J. , Li, W. , Chen, Y. , Lu, H. et al. (2020) PuHox52‐mediated hierarchical multilayered gene regulatory network promotes adventitious root formation in Populus ussuriensis . New Phytol. 228, 1369–1385. [DOI] [PubMed] [Google Scholar]
- Wen, Z. , Li, H. , Shen, Q. , Tang, X. , Xiong, C. , Li, H. , Pang, J. et al. (2019) Tradeoffs among root morphology, exudation and mycorrhizal symbioses for phosphorus‐acquisition strategies of 16 crop species. New Phytol. 223, 882–895. [DOI] [PubMed] [Google Scholar]
- Williamson, L.C. , Ribrioux, S.P. , Fitter, A.H. and Leyser, H.M. (2001) Phosphate availability regulates root system architecture in Arabidopsis . Plant Physiol. 126, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y.B. , Xiao, B.X. , Zhu, S.N. , Mo, X.H. , Liang, C.Y. , Tian, J. , Liao, H. et al. (2017) GmPHR25, a GmPHR member up‐regulated by phosphate starvation, controls phosphate homeostasis in soybean. J. Exp. Bot. 68, 4951–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Liu, W. , Liu, J. , Qin, P. and Xu, L. (2017) Auxin control of root organogenesis from callus in tissue culture. Front. Plant Sci. 8, 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. , Liu, C. , Qiao, L. , Zhao, J. , Han, R. , Wang, X. , Ge, C. et al. (2020) The MYB transcription factor TaPHR3‐A1 is involved in phosphate signaling and governs yield‐related traits in bread wheat. J. Exp. Bot. 71, 5808–5822. [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Jiao, F. , Wu, Z. , Li, Y. , Wang, X. , He, X. , Zhong, W. et al. (2008) OsPHR2 is involved in phosphate‐starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 A scheme showing the experimental steps of low phosphate treatment experimental steps.
Figure S2 Comparison of root length in WT treated with low phosphorus (LP) and normal phosphorus (NP) conditions.
Figure S3 Comparison of AR number in cuttings directly treated with low phosphorus (LP) and normal phosphorus (NP) conditions.
Figure S4 RT‐qPCR analysis of Pi starvation induction related DEGs from RNA‐seq experiment.
Figure S5 The multilayered hierarchical gene regulatory network (ML‐hGRN) built with Bottom‐up GGM algorithm where PuMYB40 and PuWRKY75 were located at the third layer and directly regulated LPR1 and ERF003.
Figure S6 PCR and RT‐qPCR verified the expression level of PuMYB40 in PuMYB40‐OE and PuMYB40‐SRDX lines.
Figure S7 Phylogenetic analysis and expression pattern of PuWRKY75.
Figure S8 PCR and RT‐qPCR verified the expression level of PuWRKY75 in PuWRKY75‐overexpression (OE) lines and PuWRKY75‐ SUPERMAN repression domain X (SRDX) lines.
Figure S9 The promoter sequence of PuLRP1 and PuERF003.
Figure S10 PCR and RT‐qPCR verified the expression level of PuLRP1 and PuERF003 in PuLRP1 and PuERF003 transgenic lines, respectively.
Table S1 Total differential expression genes (DEGs) of low phosphorus (LP) time‐course RNA‐seq data
Table S2 The GO annotation of 29 root development‐, auxin‐ and low phosphate starvation‐ related DEGs in low phosphorus (LP) time‐course RNA‐seq data
Table S3 The short‐term low phosphorus (LP)‐driven AR formation ML‐hGRN
Table S4 Primers used in this study
Table S5 Statistical report of t tests and ANOVA results for the data presented in each figure
Data Availability Statement
The sequence data of genes in this article have been deposited into GenBank under the following accession numbers: PuMYB40 (MZ687801), PuWRKY75 (MZ687802), PuLRP1 (MZ687803), PuERF003 (MZ687804), PuActin7 (MH644084), and PuUBQ10 (MN872590). The raw data of RNA‐seq experiments reported in this study have been deposited in the Gene Expression Omnibus with accession numbers of GSE174726.


