Abstract
BRDT, a member of the BET family of double bromodomain-containing proteins, is essential for spermatogenesis in the mouse and has been postulated to be a key regulator of transcription in meiotic and post-meiotic cells. To understand the function of BRDT in these processes, we first characterized the genome-wide distribution of the BRDT binding sites, in particular within gene units, by ChIP-Seq analysis of enriched fractions of pachytene spermatocytes and round spermatids. In both cell types, BRDT binding sites were mainly located in promoters, first exons, and introns of genes. BRDT binding sites in promoters overlapped with several histone modifications and histone variants associated with active transcription, and were enriched for consensus sequences for specific transcription factors, including MYB, RFX, ETS and ELF1 in pachytene spermatocytes, and JunD, c-Jun, CRE and RFX in round spermatids. Subsequent integration of the ChIP-seq data with available transcriptome data revealed that stage-specific gene expression programs are associated with BRDT binding to their gene promoters, with most of the BDRT-bound genes being up-regulated. Gene Ontology analysis further identified unique sets of genes enriched in diverse biological processes essential for meiosis and spermiogenesis between the two cell types, suggesting distinct developmentally stage-specific functions for BRDT. Taken together, our data suggest that BRDT cooperates with different transcription factors at distinctive chromatin regions within gene units to regulate diverse downstream target genes that function in male meiosis and spermiogenesis.
Keywords: BRDT, male meiosis, spermiogenesis, transcription
Graphical Abstract
We identified and characterized that BRDT binds preferentially at highly transcribed genes which have known functions during spermatogenesis in pachytene spermatocytes and round spermatids. BRDT-bound regions are enriched for consensus sequences for the transcription factors MYB, RFX, ETS and ELF1 in PS, and JunD, c-Jun, CRE and RFX in RS.
Introduction
The bromodomain is a highly conserved eukaryotic 110-amino acid long motif which binds acetylated lysine residues (Mujtaba, Zeng, & Zhou, 2007) in histones (Kanno et al., 2004) and non-histone proteins as well (Barlev et al., 2001; Gamsjaeger et al., 2011; LeRoy, Rickards, & Flint, 2008; Sinha, Faller, & Denis, 2005). Bromodomains are found in chromatin-associated factors including histone modifying enzymes (Agricola, Randall, Gaarenstroom, Dupont, & Hill, 2011; Gregory et al., 2007; Jacobson, Ladurner, King, & Tjian, 2000; Wang et al., 1997), ATP-dependent chromatin-remodeling factors (Liu, Mulholland, Fu, & Zhao, 2006; Racki et al., 2009; Thompson, 2009), and transcriptional regulators (Denis, 2010; Sankar et al., 2008; Tae et al., 2011; Tamkun, 1995; Zhou & Grummt, 2005). The BET family of bromodomain proteins are distinct in that they possess two bromodomains and an extra-terminal (ET) domain (Taniguchi, 2016), which is believed to be involved in protein-protein interactions (Crowe et al., 2016; Matangkasombut, Buratowski, Swilling, & Buratowski, 2000).
The founding member of the BET family is the Drosophila female sterile (1) homeotic (fs(1)h) gene and is the only BET gene in flies. It has been shown to regulate the transcription of the segmentation genes Ultrabithorax (Ubx) and trithorax (trx) (D. H. Huang & Dawid, 1990), as well as the genes tailless (tll) and hückebein (hkb), which govern head and gut formation (B. L. Florence & Faller, 2008). The yeast S. cerevisiae has two BET family proteins, BDF1 and BDF2, and both are components of the basal transcriptional machinery (Durant & Pugh, 2007; Matangkasombut et al., 2000). BDF1 has been shown to be involved in depositing histone variants in chromatin (Krogan et al., 2003; Sawa et al., 2004) and to control the transcription of several members of the U family of small nuclear RNAs (Lygerou et al., 1994).
There are four mammalian BET proteins, BRD2, BRD3, BRD4 and BRDT, all of which have been implicated in transcription regulation (B. Florence & Faller, 2001) including binding to the hyperacetylated body region of actively transcribing genes and facilitating transcriptional elongation by RNA polymerase II (Pol II) (Dhar, Thota, & Rao, 2012; Gaucher et al., 2012; Jang et al., 2005; LeRoy et al., 2008). BRD2 interacts with TATA binding protein (TBP) through its first bromodomain (BD1) (Peng et al., 2007) and with E2F through its ET domain and plays a role in modulating E2F-regulated transcription (Denis, Vaziri, Guo, & Faller, 2000; Guo, Faller, & Denis, 2000). Similarly, BRD3 binds via its BD1 to multi-acetylated GATA1, and modulates GATA1-regulated transcription (Lamonica et al., 2011). BRD4 is a regulatory component of the positive transcriptional elongation factor beta (P-TEFb) complex, and is required for P-TEFb-dependent transcription (Jang et al., 2005; Z. Yang et al., 2005). BRD4 binding via its C-terminal domain (CTD) frees cyclin T1 (CCNT1) and cyclin dependent kinase 9 (CDK9), the major components of P-TEFb, from inhibition by the 7SK small nuclear RNA and HEXIM1 (Bisgrove, Mahmoudi, Henklein, & Verdin, 2007; Schroder et al., 2012; Yik et al., 2003). The CTDs of Fs(1)h and BRDT were also found to be capable of this interaction (Bisgrove et al., 2007).
BRDT is unique among the mammalian BET family proteins in that it is most abundantly if not exclusively expressed in the testis (Jones, Numata, & Shimane, 1997), specifically in pachytene spermatocytes (PS) through to post-meiotic round spermatids (RS) in the mouse (Shang, Nickerson, Wen, Wang, & Wolgemuth, 2007; Shang et al., 2004). It was shown to be involved in the expression of over 3000 genes (Gaucher et al., 2012), two-thirds of which correspond to genes activated by BRDT and one-third of which are repressed by BRDT. In certain genes, BRDT mediates transcription by recruiting the P-TEFb complex to the TSS (Gaucher et al., 2012). Along with modulating transcription per se, BRDT also affects gene expression as part of splicing machinery and determining the length of the 3’UTR of mRNA transcribed in RS (Berkovits, Wang, Guarnieri, & Wolgemuth, 2012).
To further understand the role of BRDT in regulating gene expression during spermatogenesis, we identified and characterized in detail the distribution and features of BRDT binding sites within gene units in the genome by performing genome-wide occupancy studies of BRDT complexes in enriched populations of PS and RS. We found that BRDT preferentially binds to gene units of highly transcribed genes in PS and RS. Most of the BRDT binding sites co-localized with known histone modifications and histone variants that correlate with active transcription. Further, BRDT binding sites in promoters were enriched for the binding motifs of a several different transcription factors, including MYB, RFX, ETS and ELF1 in PS and JunD, c-Jun, CRE and RFX in RS. This suggests that BRDT has multiple binding partners, and that these different BRDT-containing complexes may have distinct functions that modulate the unique spermatogenic and spermiogenic transcriptional programs. Integration of the ChIP-seq data with available transcriptomic data corresponding to specific stages of differentiating mouse spermatogenic cells highlighted a particular relevance of BRDT binding to promoters to gene expression patterns, revealing that stage-specific gene expression programs during meiosis and post-meiosis are associated with BRDT binding to their gene promoters. Gene ontology analyses showed that the BRDT-bound genes corresponded to genes with essential roles in diverse cellular processes and in organelle functions that are essential for proper spermatogenesis and male fertility. Overall, these results define genomic features and signatures that characterize the BRDT binding sites in genes transcriptionally active in meiotic and post-meiotic cells, and strongly suggest transcriptional partners that may cooperate with BRDT to regulate transcriptional activity during spermatogenesis.
Results
Genome-wide analysis of BRDT binding in PS and RS
As noted above, BRDT is expressed uniquely in the testis from early PS through RS (Shang et al., 2004) and has been shown to be essential for spermatogenesis (Gaucher et al., 2012; Shang et al., 2007). Previous ChIP-Seq analysis showed that while most of the BRDT binding sites are intergenic, a significant fraction of BRDT binds to acetylated TSS of genes activated by the end of meiosis (Gaucher et al., 2012). These observations led us to explore in more detail the genomic binding sites of BRDT in enriched fractions of PS and RS, with a particular focus on gene units, using ChIP-Seq of formaldehyde-fixed chromatin and a C-terminal anti-BRDT antibody (Berkovits & Wolgemuth, 2011).
We identified 5452 BRDT-bound genomic loci in PS chromatin, of which 26% were located in promoters, 12% in exons, and 37% in introns, as designated in the annotated RefSeq genes (Figure 1A, B). The remaining 25% were in intergenic regions, defined as not in promoters, exons or introns. In RS, there were 5618 BRDT-bound genomic loci, of which 16% were located in promoters, 8% in exons, 43% in introns and 33% in intergenic regions (Figure 1A, B). These results differ somewhat from the BRDT distribution reported by Gaucher et al. (Gaucher et al., 2012), in that we found that 25% (PS) and 33% (RS) of BRDT binding sites are intergenic, compared to their report of 66% and 60%, respectively. A possible explanation for the differences may be the criteria used to define and classify intergenic regions, which is key to avoid overlap between distinct regions, even more so when gene units are adjacent to one another. It should further be noted that different BRDT antibodies were used in the studies. Previous analyses on ChIP-Seq binding to intergenic regions of the other three BET proteins (BRD2, BRD3 and BRD4) in HEK293 cell lines revealed a distribution similar to our findings (LeRoy et al., 2012). That is, ~65% of BRD-binding sites were located in gene units (defined as promoter and gene body) and ~35% of binding sites were intergenic. Although BRDT is the most divergent of the BET family proteins, and testicular cells are very different from HEK293 cells, the ratio of 2:1 gene binding to intergenic binding found by Leroy and colleagues more closely matches the ratios we observed for BRDT binding (LeRoy et al., 2012).
Figure 1.
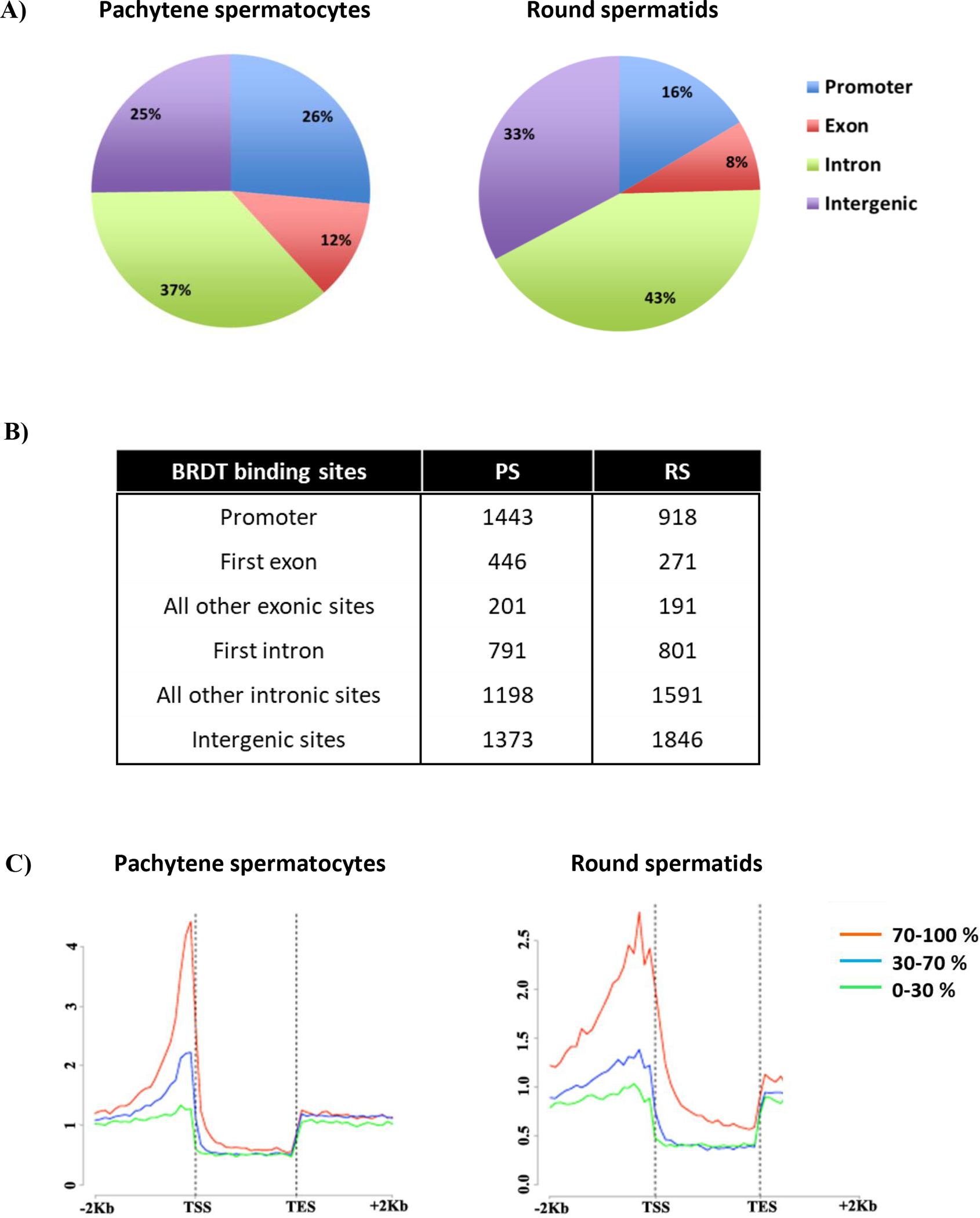
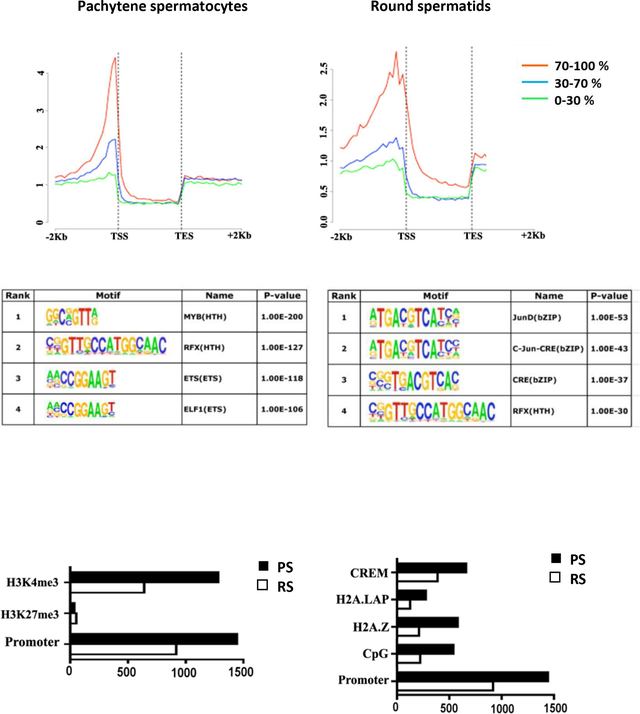
High–resolution genome-wide analysis of BRDT-containing complexes occupancy in pachytene spermatocytes (PS) and round spermatids (RS) of wild type mice. (A-B) Pie chart shows genomic annotation of location and number of BRDT binding sites across the genome in wild type PS and RS. (C) High-resolution analysis of average tag density across a gene body in wild type mouse testicular cells. In each plot, genes were classified into 3 groups based on expression levels in PS and RS: highly expressed (the top 30% of genes indexed by expression levels, red lines), medium expression (the middle 40% of genes, blue lines) and lowly expressed (the lowest 40% of genes, green lines). Average tag density across the gene unit was plotted for each group. TSS, Transcription Start Sites; TES, Transcription End Sites
Considering that one of the most important proposed functions of BRDT in spermatogenesis is to drive a developmentally regulated testis-specific gene expression program (Gaucher et al., 2012), we then focused our analysis on BRDT’s role in transcription. We first assessed the number of BRDT binding sites in promoter regions in PS and RS. In PS, 1443 BRDT binding sites were located in the promoters of 1523 genes (Supplemental Table S1). This excess of genes above the number of binding sites is caused by the presence of a binding site between two genes that transcribe in opposite directions. That is, one binding site can be in the promoter of two different genes. In RS, 918 BRDT binding sites were located in the promoters of 946 genes (Supplemental Table S2). Comparison of the two lists of putative BRDT target genes revealed that 233 genes bound BRDT in the promoter region in both cell types, which corresponded to 15% and 25% of the promoter-bound genes in PS and RS, respectively.
We next investigated the distribution of BRDT binding at 5’ introns and exons. Introns in the 5′ proximal region of a gene (first or ‘early’ introns) have often been shown to be involved in regulation of gene expression (Bradnam & Korf, 2008). Interestingly, 791 of the 1989 (40%) intronic sites of BRDT binding in PS and 801 of the 2392 (33%) intronic sites of BRDT binding in RS occurred in the first intron (Figure 1B), the most common location for intron-mediated enhancement (IME) (Mascarenhas, Mettler, Pierce, & Lowe, 1990). Of the 647 exonic sites of BRDT binding in PS, 446 (69%) and 271 of the 462 (59%) exonic sites of BRDT binding in RS occur in the first exon (Figure 1B).
As three-quarters of the binding sites in PS and two-thirds of the sites in RS were within gene units, we next analyzed the average BRDT tag density distribution across a gene unit. In both cell types, a major peak of BRDT occupancy was observed just 5’ to the TSS (Figure 1C). Although a greater number of binding sites was present in the gene body as compared to the promoter, the promoter binding clustered one to two nucleosomes before the TSS, whereas the intronic and exonic binding sites were spread relatively evenly over the whole gene, albeit with a slight bias toward the 5’ end of genes (Figure 1C).
Enriched DNA binding sites in BRDT-bound regions suggest possible interaction with specific transcription factors
We next asked whether the promoter regions bound by BRDT would be enriched with distinct transcription factors (TFs) binding motifs, which would suggest a possible interaction between BRDT and specific TFs. Consensus and optimal in vitro DNA binding motifs in BRDT-bound regions were therefore ascertained from JASPAR, UniPROBE and Transfac. Multiple Expression motifs for Motif Elicitation (MEME) was used to find motifs enriched in the 500 BRDT promoter-binding peaks with the highest number of reads in each cell type. Each motif was scored for significantly increased binding frequency at these peaks as compared to background. In PS, the four most highly enriched motifs were for binding of the TFs MYB, RFX, ETS and ELF1 (Figure 2A) and in RS, the four highest peaks were for JunD, c-Jun, CRE and RFX (Figure 2A). This is of particular interest as it suggests a dynamic interaction between TFs and epigenetic readers such as BRDT, which could be recruited to promoters to co-regulate a subset of genes during spermatogenesis.
Figure 2.
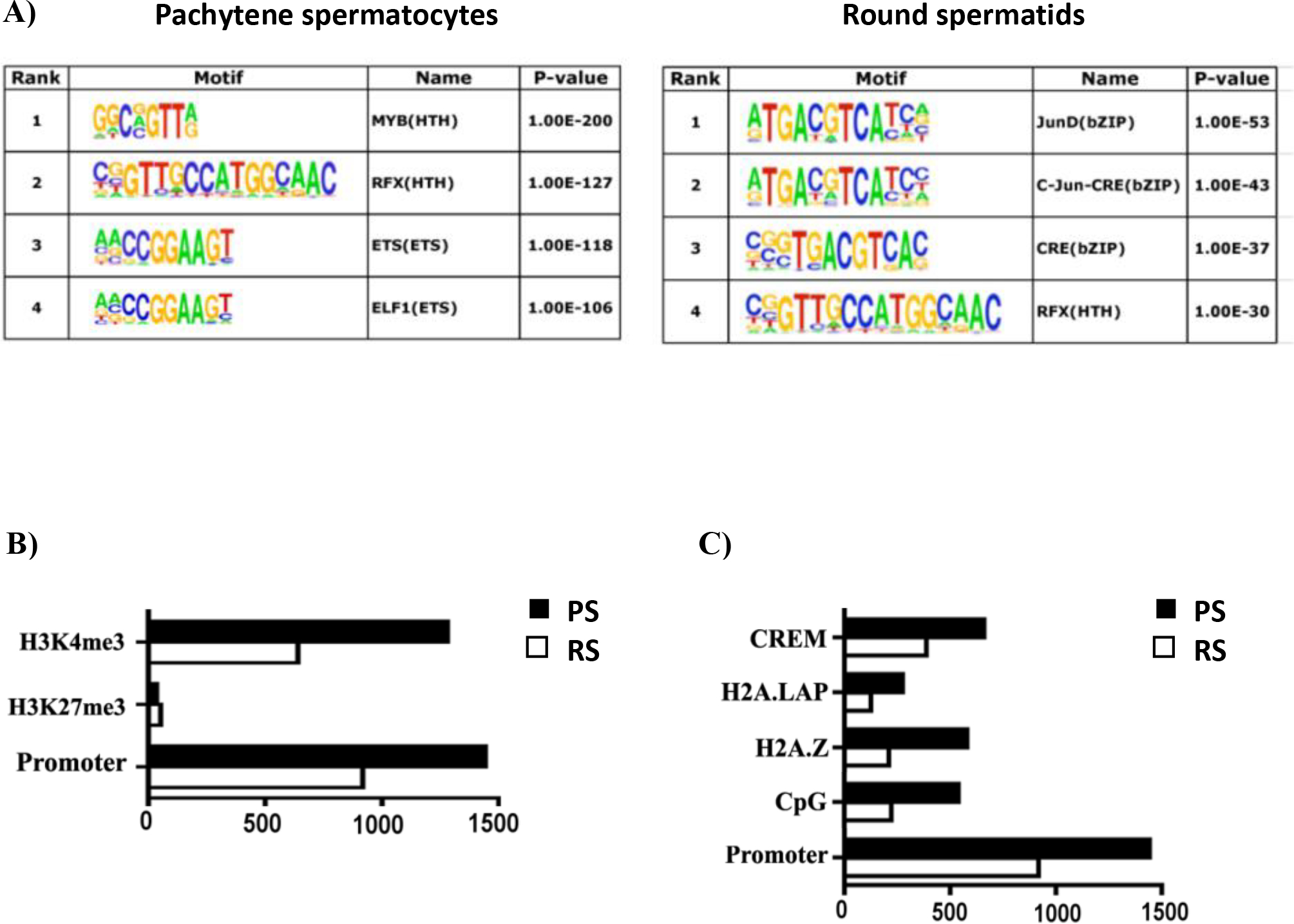
Characterization of BRDT binding site occupancy in pachytene spermatocytes (PS) and round spermatids (RS). (A) BRDT in vivo consensus binding site predicted by MEME and presented as a sequence LOGO. A total of 500 peak regions were analyzed for overrepresented motifs. Enriched transcription factor motifs lying within BRDT binding sites in PS and RS. (B) Correlative analysis of H3K4me3, H3K27me3 with BRDT binding site occupancy. (C) Correlative analysis of CpG, H2AZ, H2A.LAP, CREM with BRDT binding site occupancy. The x-axis in (B) and (C) refers to the number of associations with BRDT binding sites.
The majority of BRDT binding sites in gene units co-localize with features of active chromatin
To assess the association of BRDT promoter binding with active transcription and the epigenetic features of the BRDT binding sites at promoter regions, we next compared our BRDT ChIP-Seq data with published ChIP-seq data sets for features and markers of transcription. The active chromatin histone mark H3K4me3 and the repressive histone mark H3K27me3 have been examined in extracts obtained from total testicular cells (Cui et al., 2012). Comparison of these data with our results for BRDT binding revealed that ~89% (1279) and ~70% (639) of BRDT promoter binding sites are associated with H3K4me3 in PS and RS, respectively, and only 2.4% (52) and 5.7% (34) of BRDT promoter binding sites associated with H3K27me3 in the two populations (Figure 2B).
We further extended this analysis of BRDT binding and features of active transcription by comparing our data with published ChIP-Seq data sets for CpG islands and the histone H2A variants H2A.Z and H2A.LAP1. Interestingly, we observed that 37.4% (540) and 24.3% (223) of BRDT promoter binding sites in PS and RS, respectively, overlapped CpG islands (Figure 2C). Both H2A.Z and H2A.LAP1 have been reported to be components of the chromatin of active promoters (Soboleva, Nekrasov, Ryan, & Tremethick, 2014) and had been examined in extracts obtained from total testicular cells (Cui et al., 2012; Martianov et al., 2010; Soboleva et al., 2011).. It has been reported that BRD2 is recruited to androgen receptor (AR)–regulated genes in an H2A.Z-dependent manner and that chemical inhibition of BRD2 recruitment greatly inhibited AR–regulated gene expression (Draker et al., 2012). We found that 40% (580) and 23% (212) of BRDT promoter binding sites associated with H2A.Z in PS and RS, respectively (Figure 2C). The association of BRDT promoter binding sites with H2A.LAP1, however, revealed overlap for only 19.2% (276) and 13.6% (125) of binding sites in PS and RS, respectively (Figure 2C).
Our MEME analysis mentioned above identified CRE motif binding sites in RS (Figure 2A) and it is known that the CREMτ isoform of the TF CREM is strongly and selectively expressed in RS (Blendy, Kaestner, Weinbauer, Nieschlag, & Schutz, 1996; Foulkes, Mellstrom, Benusiglio, & Sassone-Corsi, 1992), suggesting that BRDT is associated with CREMτ to potentially regulate gene expression in these cells. To this end, we next compared our BRDT ChIP-Seq data with CREMτ ChIP-Seq data (Martianov et al., 2010). Our analysis revealed a notable overlap of BRDT and CREMτ binding in promoters, with 42% (389) of all BRDT binding sites shared with reported CREMτ binding sites (Figure 2C). This overlap indicates that a possible collaboration between these two factors during spermiogenesis. For example, CREB binding protein (CBP), which binds to CRE elements, can catalyze lysine butyrylation in histones (Chen et al., 2007), and butyrylated H4K5 is sufficient to abolish the interaction between BRDT and histone H4 (Goudarzi et al., 2016).
ChIP-seq and transcriptome analysis reveals that stage-specific gene expression is associated with BRDT binding to their gene promoters
We then asked whether BRDT binding to genes in PS and RS correlated with gene expression in these cells, using available data sets of RNA-seq analysis of spermatogonia (Sg), PS, and RS (GSE100964). The expression levels of genes in the different cell types were analyzed based on the probe name suffix and signal intensity as described in Wichman et al.(Wichman et al., 2017). We then interrogated the list of BRDT-bound genes in PS and RS (Supplemental Table S1 and S2) as related to the transcriptomic data. In total, 1,407 and 829 genes among the BRDT-bound genes in PS and RS, respectively, were represented in the transcriptomic data. Of these, 1,295 (92%) and 744 genes (89%) were expressed in PS and RS, respectively, while 112 genes (8%) and 85 genes (11%) were not expressed (Figure 3A; Supplemental Table S3 and S4), indicating that most BRDT-bound genes in PS and RS were expressed in the corresponding cell types. The observation of a large portion of expressed genes among the BRDT-bound genes in PS and RS was consistent with our concomitant analyses which utilized microarray datasets, GSE4193 (data not shown). That is, 83% and 85% of BRDT-bound genes were expressed in PS and RS, respectively, while 17% and 15% were not expressed.
Figure 3.
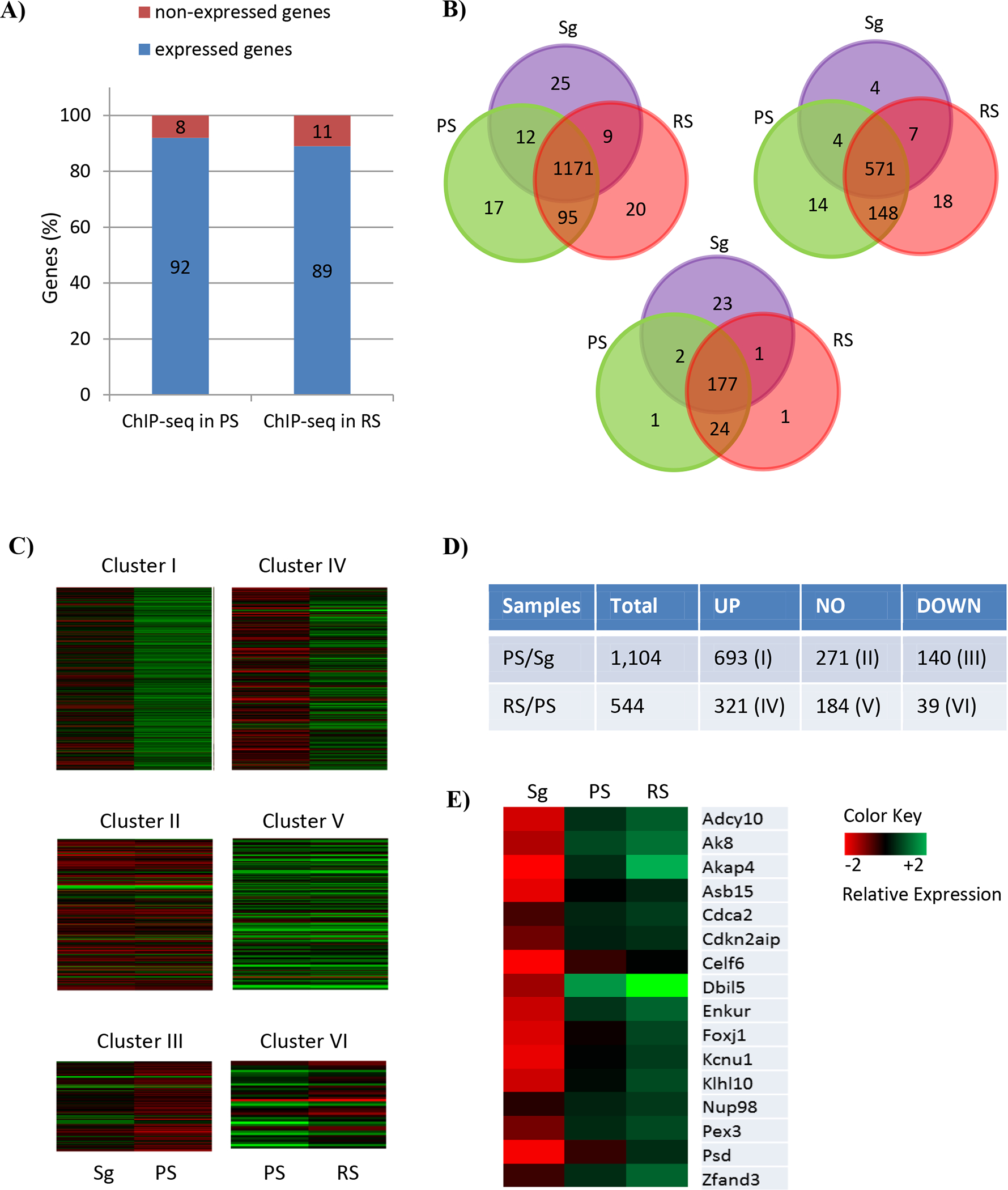
Comparative BRDT ChIP-seq and transcriptome analysis corresponding to the staged mouse spermatogenic cells. (A) Respective proportions of BRDT-bound genes in PS and RS, which are expressed and non-expressed in PS and RS, respectively. A total of 1,444 and 888 BRDT-bound genes in PS and RS, respectively were included. (B) Venn diagram of the genes expressed in three cell types, Sg, PS and RS, within the BRDT-bound genes either in PS (left) or RS (right) and both PS and RS (bottom). Distinct and overlapping expression between and among the cell types is shown. Purple, green and red cycles represent Sg, PS, and RS, respectively. (C) Heat maps showing the expression of 6 clusters of BRDT-bound genes that are expressed in PS and RS. The following comparisons were performed: PS vs. Sg; RS vs. PS (Log2 Fold Change (|FC|) ≥ 2; p < 0.01). Columns indicate specific cell types and rows indicate genes. The low versus high expression levels are represented on a red to green scale. (D) Total number of up-, down-, and non-differentially regulated genes belonging to each of the Clusters (|FC| ≥ 2; p < 0.01 (E) Heat map showing relative expression levels of the 16 genes overlapping between Cluster I and Cluster IV. The low versus high expression levels are represented on a red to green scale.
We next examined whether the 1,295 and 744 genes, which were identified to be expressed in PS and RS, overlapped in expression or were unique to the particular cell type. Fig. 3B shows that of the 1,295 genes which are expressed in PS, 1,183 (91%) and 1,266 (97%) were also expressed in Sg and RS, respectively. Only 17 genes of the 1,295 (1.3%) were expressed only in PS. (Figure 3B, left). Of the 744 genes which are expressed in RS, 719 (96%) and 578 (78%) were also expressed in PS and Sg, respectively, and 18 (2.4%) were expressed only in RS (Figure 3B, right).
As mentioned before, our ChIP-seq analysis identified a total of 233 genes that overlapped in their binding to BRDT between PS and RS. We therefore interrogated the data set to compare the expression patterns for these genes. Of the 233, a total of 208 genes were represented in the data set, of which 204 (98%) and 203 (97.5%) genes were expressed in PS and RS, respectively, and 201 (96%) were expressed in both PS and RS. Of these, 177 (88%) were also expressed in Sg, representing the proportion of genes expressed in all three cell types; Sg, PS and RS. (Figure 3B, bottom).
Cluster analysis of genes bound by BRDT reveals that BRDT is mainly involved in up-regulation of gene expression
We first defined significantly differentially expressed genes (DEGs) in spermatogenic cells as those which were more than 2-fold increased or decreased with a p adjusted value p < 0.05. Among all the BRDT-bound genes in PS and RS, 833 (76%) and 360 (66%) were significantly changed in expression levels in PS compared to Sg, and in RS compared to PS, respectively (Figure 3C and 3D). These observations were also consistent with our other studies which utilized GSE4193, showing DEGs among all the BRDT-bound genes in 67% and 66% in PS compared to Sg, and in RS compared to PS, respectively (data not shown). We then classified BRDT-bound genes into 6 clusters according to their expression patterns. Cluster I represents the genes whose expression significantly increased in PS compared to Sg. Cluster II represents the genes whose expression is unchanged between PS and Sg. Cluster III represents the genes whose expression significantly decreased in PS compared to Sg. Cluster IV represents the genes whose expression significantly increased in RS compared to PS. Cluster V represents the genes whose expression is similar between RS and PS. Finally, Cluster VI represents the genes whose expression significantly decreased in RS compared to PS.
Cluster analysis of BRDT-bound DEGs revealed that Cluster I and IV, which represent the up-regulated genes in PS and RS, were the most abundant (63% in PS and 59% in RS), while Cluster III and VI, which represent the down-regulated genes in PS and RS, were the lowest (13% in PS and 7% in RS) (Figure 3C, D). As can be seen from the heat maps in Figure 3C and the fold-changes in the genes listed in Supplemental Table S5, the majority of Cluster I genes exhibited strong increases in gene expression, for example Ldhc, Mroh2b, Fbp1, Aqp9 and Ccdc60. While there were fewer down-regulated genes in Cluster III, in those that were changed, the decreases were quite significant, for example Hist1h4m, Fbln2, Crispld2, Junb and Hnrnpa1 (Supplemental Table S5). Similarly, the heat maps in Figure 3C and fold- changes in Supplemental Table S6, the majority of Cluster IV genes were strongly up-regulated, for example, Golga7b, Srxn1, Agpat9, Prss54 and Chd5 (Supplemental Table S6). And again, while a lower proportion of genes were found in Cluster VI genes, those that were decreased in RS exhibited strong changes, including Arf5, Calm2, Cby1, Coa5 and Ddx19b (Supplemental Table S6).
Comparison of the genes from Clusters I and IV identified a total of 16 genes that overlapped between the two clusters (Figure 3E). As can be seen from the heat maps in Fig. 3E, this subgroup of genes showed very low or undetectable levels of expression in Sg, but dramatically increased in PS and RS. These genes include Adcy10, Ak8, Akap4, Asb15, Dbil5, Enkur, Foxj1 and Klhl10 (Supplemental Table S7).
GO enrichment analysis of BRDT-bound genes whose expression significantly increased in PS and RS
Given that Cluster I and IV genes were most enriched in the corresponding cell types, we next obtained a global overview of the biological functions of proteins encoded by the genes in these two clusters. Specifically, we performed a functional annotation using DAVID (Database for Annotation, Visualization, and Integrated Discovery) analysis for biological processes Gene Ontology (GO) terms (Huang da, Sherman, & Lempicki, 2009a, 2009b) (Supplemental Table S8 and S9). GO enrichment analysis revealed that in PS, the Cluster I gene set was most significantly enriched for protein modification by small protein conjugation or removal (GO: 0070647; P-value = 9.79E-12), followed by cell cycle (GO:0007049; P-value = 1.16E-11), and cilium organization (GO: 0044782; P-value = 2.12E-8) (Figure 4A; Supplemental Table S10). We also observed enrichment for genes involved in ubiquitin-dependent protein catabolic process, microtubule cytoskeleton organization, protein localization to organelle, spermatogenesis, DNA repair, response to endoplasmic reticulum stress and regulation of response to DNA damage stimulus. (Figure 4A; Supplemental Table S10). In RS, the Cluster IV gene set was most significantly enriched for spermatogenesis (GO:0007283; P-value = 2.24E-19), followed by sperm capacitation (GO:0048240; P-value = 3.56E-05), and protein ubiquitination (GO:0016567; P-value = 5.86E-04) (Figure 4B; Supplemental Table S10). We also observed enrichment for genes involved in single fertilization, microtubule-based process, regulation of mRNA 3’-end processing, glycerolipid catabolic process, negative regulation of cell cycle, epithelial cilium movement and multicellular organismal homeostasis. (Figure 4A; Supplemental Table S10). It is of note that protein ubiquitination (GO: 0016567) and microtubule-based process (GO:0007017), which are third and fifth among the GO enriched terms for Cluster IV, are derivative functions of protein modification by small protein conjugation or removal (GO: 0070647) and microtubule cytoskeleton organization (GO:0000226), which are first and fifth enriched GO-terms for Cluster I (according to the GO annotation).
Figure 4.
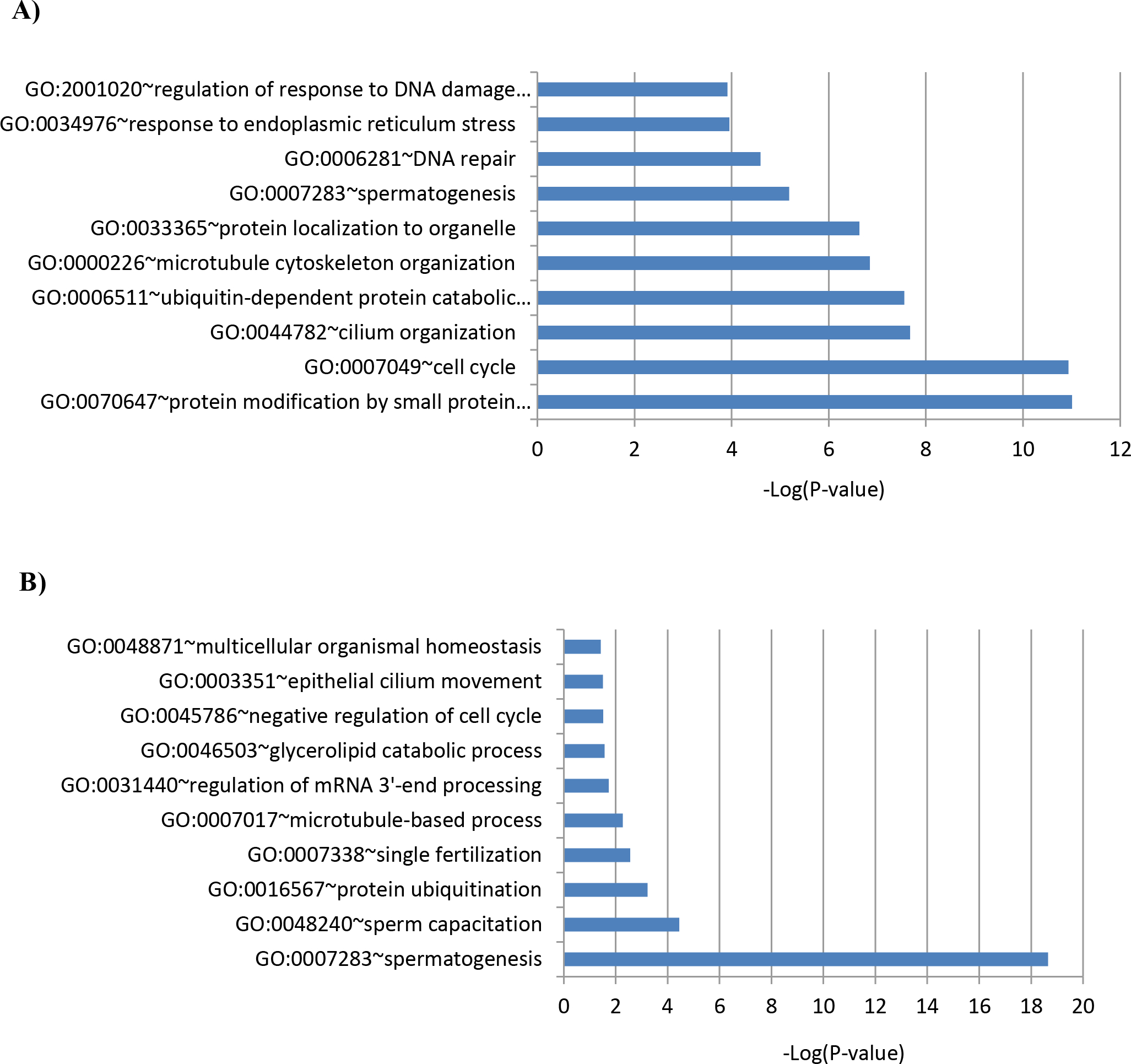
Enriched biological process GO terms of the genes belonging to (A) Cluster I (up-regulated genes in PS vs. Sg) and (B) Cluster IV (up-regulated genes in RS vs. PS).
Among the 16 genes which overlapped between Cluster I and IV, 8 were detected by our GO analysis; Adcy10, Akap4, Asb15, Cdca2, Dbil5, Foxj1, Klhl10 and Pex3. Seven genes belong to the top 3 biological processes enriched for both Cluster I and Cluster IV. Pex3 belongs to the fifth among the GO enriched terms for both Cluster I and Cluster IV: microtubule cytoskeleton organization (GO:0000226) and microtubule-based process (GO:0007017), which is a derivative function of microtubule cytoskeleton organization (Supplemental Table S10). Adcy10 and Dbil5 were listed under spermatogenesis (GO:0007283), and Akap4 and Foxj1 are listed under cilium organization (GO:0044782). Cdca2 is listed under cell cycle (GO:0007049). Asb15 and Klhl10 are part of protein modification by small protein conjugation or removal (GO: 0070647) and protein ubiquitination (GO:0016567). Klhl10 is also listed under spermatogenesis (GO:0007283), suggesting its crucial functions in both meiotic and post-meiotic stages of spermatogenesis.
Visualization of the localization of BRDT-binding sites at promoters of the up-regulated genes in PS and RS
Based on the enriched biological processes identified in our GO analysis and potential relevance to BRDT’s functions as shown in previous studies from our laboratory (Berkovits et al., 2012; Berkovits & Wolgemuth, 2011; Manterola et al., 2018; Wang & Wolgemuth, 2016), we selected three critical processes and specific examples therein with which to localize in detail the sites of BRDT binding, namely spermatogenesis, regulation of response to DNA damage stimulus and regulation of mRNA 3’-end processing (Figure 5A, B and C). Genome browser visualization of the BRDT-bound at the promoter of gene loci in both cell types showed a distinct pattern of BRDT binding to promoters between the cell types: both PS and RS, only PS and only RS.
Figure 5.
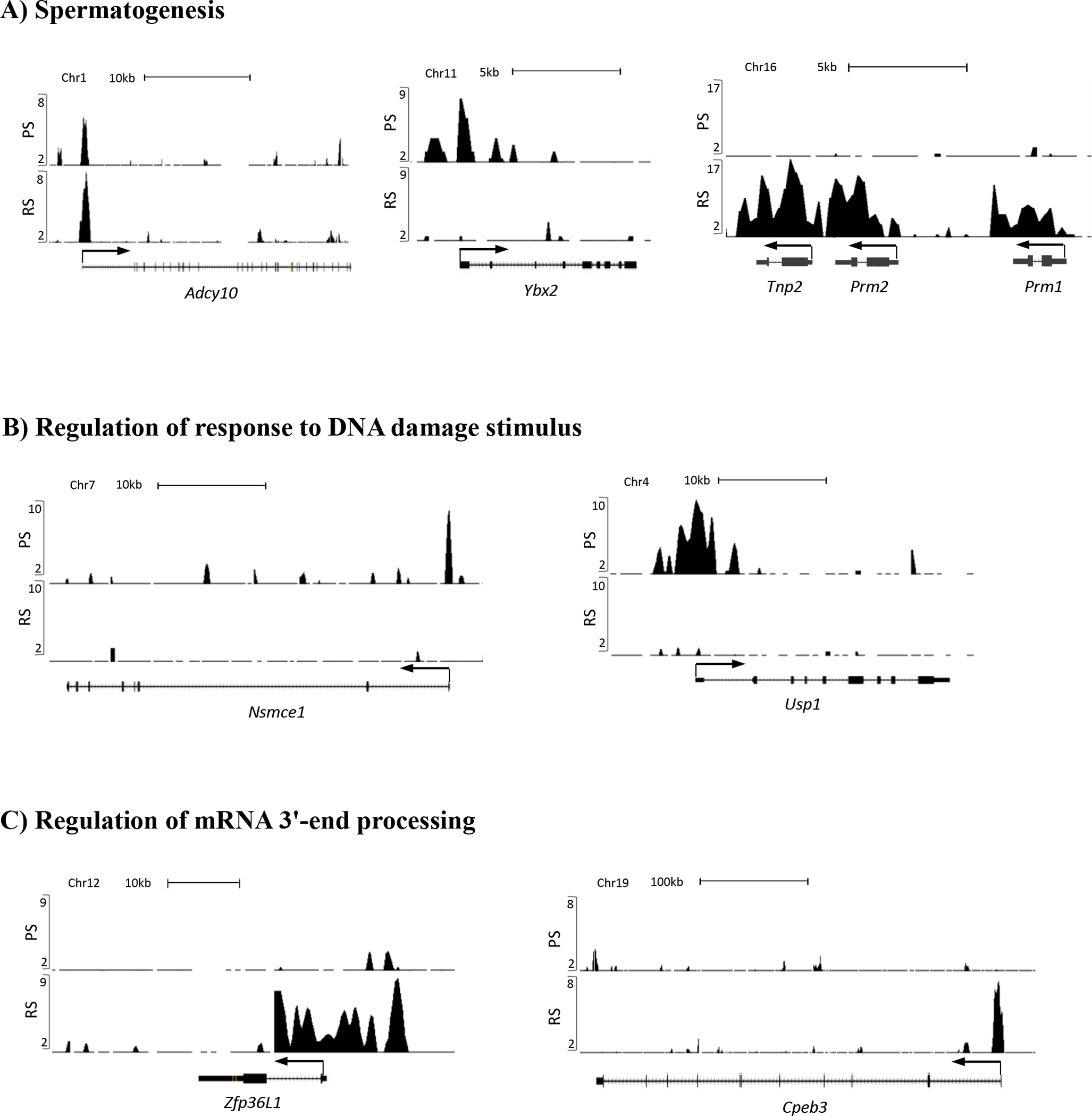
UCSC Genome Browser screenshot localization of BRDT ChIP-seq signals at the gene promoters implicated in (A) spermatogenesis, (B) regulation of response to DNA damage stimulus, and (C) regulation of mRNA 3’-end processing. Graphic representation of ChIP-Seq reads derived from pachytene spermatocytes (PS) and round spermatids (RS) are shown on the vertical axis on all panels. ChIP-seq signals are displayed as normalized RPM (reads per million reads) values.
In both Cluster I and IV, spermatogenesis was a significantly enriched GO term and included several genes known to be essential for male fertility. For example, Adcy10, which encodes Testicular Soluble Adenylyl Cyclase, has a critical role in mammalian spermatogenesis by producing the cAMP which regulates cAMP-responsive nuclear factors (Schmid et al., 2007). Adcy10-null mice are sterile and produce spermatozoa with deficits in progressive motility and are unable to fertilize zona-intact eggs (Esposito et al., 2004; Xie et al., 2006). Our ChIP-seq analysis showed that BRDT strongly bound its promoter in both cell types (Figure 5A, left). Another example of genes involved in spermatogenesis, Ybx2, is involved in the regulation of the stability and/or translation of germ cell mRNAs via direct interaction with target RNAs such as Prm1(Chowdhury & Kleene, 2012). Loss of Ybx2 (Ybx2−/− mice) results in male infertility due to a spermatogenic arrest in post-meiotic cells with many misshapen and multinucleated spermatids (J. X. Yang et al., 2005). Ybx2 is bound by BRDT extensively in only PS (Figure 5A, middle). Also in the GO biological process of spermatogenesis are the sperm-specific nuclear protein genes protamine 1 and 2 (Prm1, Prm2) and transition protein 2 (Tnp2), which are essential for generating elongating spermatids and mature spermatozoa (Brewer, Corzett, & Balhorn, 1999) and are involved in intermediate stages in the replacement of histones by protamines (Meistrich, Mohapatra, Shirley, & Zhao, 2003; O’Carroll et al., 2000; Yu et al., 2000). Strong BRDT binding was also observed in only RS not only at the promoter but also throughout the genomic region (Figure 5A, right).
In PS, Cluster I genes encoding proteins of regulation of response to DNA damage stimulus, for example, Nsmce1 and Usp1 are both showed a strong signal peak of BRDT in PS. NSMCE1 is a component of SMC5-SMC6 complex which is highly expressed in mouse testis and associate with sex chromosomes during the pachytene stage (Taylor et al., 2001). NSMCE1 is involved in maintenance of genome integrity and DNA damage response and regulation of homologous recombination-mediated DNA repair (Fujioka, Kimata, Nomaguchi, Watanabe, & Kohno, 2002; Harvey, Sheedy, Cuddihy, & O’Connell, 2004). Our ChIP-seq analysis showed that BRDT strongly bound its promoter in PS (Figure 5B, left). As another example of genes involved in DNA damage, Usp1 is a key modulator of DNA repair, partly through de-ubiquitination of its known targets such as FANCD2 (Oestergaard et al., 2007) and PCNA (T. T. Huang et al., 2006). Deletion of mouse Usp1 (Usp1−/− mice) results in male infertility due to chromosome instability and extensive loss of spermatocytes and spermatids (Kim et al., 2009). Our ChIP-seq analysis revealed that the Usp1 promoter is bound by BRDT exclusively in PS (Figure 5B, right).
In RS, Cluster IV genes encoding proteins of regulation of mRNA 3’-end processing, for example, Zfp36l1 and Cpeb3, are bound by BRDT only in RS. BRDT binding at this class of genes is not surprising as we have shown that partial loss of BRDT function has an effect on mRNA splicing in RS (Berkovits et al., 2012). ZFP36L1 promotes cytoplasmic AU-rich element (ARE)-mediated mRNA deadenylation and decay processes (Son, Kim, Choi, Chun, & Chun, 2019) and also plays a role in the regulation of nuclear mRNA 3’-end processing and modulation of mRNA 3’-end maturation efficiency (Desroches-Castan, Cherradi, Feige, & Ciais, 2011). Zfp36l1 is bound by BRDT only in RS (Figure 5C, left). Cytoplasmic polyadenylation element-binding protein 3 (CPEB3), binds to the mRNA 3’-UTR of mRNA and negatively regulates their translation (Ford, Ling, Kandel, & Fioriti, 2019; Y. S. Huang, Kan, Lin, & Richter, 2006) and the CPEB3 ortholog ORB2 has been known to be required for the orderly differentiation of the spermatids after meiosis is complete (Xu, Hafer, Agunwamba, & Schedl, 2012). Our ChIP-seq analysis revealed that the Cpeb3 promoter is bound by BRDT only in RS (Figure 5C, right).
To validate the ChIP-seq data on BRDT-binding sites at promoters of specific genes, we performed chromatin immunoprecipitation (ChIP) analysis followed by qPCR in PS and RS of wild type mice (Supplemental Figure S1). We selected four genes analyzed in Figure 5: Ybx2 and Usp1, whose promoters are bound by BRDT in PS but not RS, and Prm1 and Zfp36L1, whose promoters are bound by BRDT in RS but not PS. Consistent with the ChIP-seq data shown in Figure 5, ChIP-qPCR confirmed BRDT occupancy at the promoters of Ybx2 and Usp1 only in PS with a greater than 7-fold enrichment and at the promoters of Prm1 and Zfp36L1 only in RS with a greater than 8-fold enrichment (Supplemental Figure S1). These data, together with the ChIP-seq analysis, further strongly supported a distinct pattern of BRDT binding to promoters correlated with their expression in specific cell types.
Discussion
The data in the present study significantly extend our understanding of the function of BRDT in regulating gene expression during spermatogenesis and hence, male germ cell differentiation. Our detailed ChIP-Seq analysis revealed genomic features of BRDT bound chromatin in PS and RS that showed that while BRDT binding was widely spread throughout the mouse genome, the majority of BRDT binding sites were located in gene units (the promoter, introns and exons), which suggested essential roles for BRDT in regulation of gene expression. For example, IMEs (which are mostly located in the first intron region) have been known to act as an enhancer of gene expression (Mascarenhas et al., 1990) and our ChIP-seq analysis revealed that IMEs are enriched for sites bound by BRDT. Further, the exons coding for 5’ UTRs can also be targets for transcriptional regulation (Al-Harthi & Roebuck, 1998; Turner et al., 2010), and these regions were also identified to be enriched sites bound by BRDT. These results suggest a possible function of BRDT in gene regulation via direct binding to both introns and exons in addition to the promoters.
Some of the early intron/exon binding by BRDT could be also involved in the selection of an alternative promoter, a process that is prevalent in the testis (DeJong, 2006; Freiman, 2009; Liu, Zhang, Li, Yan, & Li, 2010). The utilization of gene isoform expression has been known to be regulated by alternative promoter usage (Ayoubi & VanDeVen, 1996) and interestingly, we have previously identified a function of BRDT in modulating gene isoforms expression as part of the splicing machinery (Berkovits et al., 2012), suggesting a possible function for BRDT in alternative promoter usage. However, since the BRDT binding peaks were broad (usually over two nucleosomes), we cannot exclude the possibility that some of the peaks might actually straddle the TSS. As we designated the midpoint of the peak as the BRDT binding site, some peaks might have been counted as being present in the first exon even though they partially overlap the promoter as well.
Our ChIP-seq analysis also identified numerous BRDT-binding sites located in intergenic regions (25% in PS and 33% in RS; Figure 1A, B). Intergenic regions are known to contain potentially important DNA regions that generate non-coding RNAs such as microRNAs (miRNA) (Bartel, 2004), PIWI interacting RNAs (piRNA) (Siomi, Sato, Pezic, & Aravin, 2011), and enhancer RNA (eRNA) (Li, Notani, & Rosenfeld, 2016). These RNAs have in turn been shown to function in the regulation of tissue-, cell- or state-specific gene expression and in the control of epigenetic regulation and chromosomal dynamics (Bernstein & Allis, 2005; Khalil et al., 2009). The most closely BRDT-related BET protein, BRD4, has been shown to bind to active enhancers and control cell identity gene induction in adipogenesis and myogenesis (Lee et al., 2017). Further, recent studies have shown that BRD4 interacts with eRNA that were produced from BRD4-bound enhancers in response to chronic immune signaling (Rahnamoun et al., 2018). While out of the scope of the present studies it will thus be of great interest to identify the specific BRDT binding regions in intergenic regions.
Another important finding of our studies was the identification of an enrichment for specific TF motifs, including MYB, RFX, ETS and ELF1 motifs in PS and JunD, c-Jun, CRE and RFX motifs in RS. In PS, MYB motifs were the most prevalent in PS and interestingly, MYB proteins are known transcriptional regulators in spermatocytes, and A-MYB in particular has been called a master regulator of meiotic genes in spermatocytes (Bolcun-Filas et al., 2011). Of note, A-MYB has an overlapping expression pattern in spermatocytes with BRDT and depletion of either protein results in arrest at the pachytene stage (Gaucher et al., 2012; Latham et al., 1996; Shang et al., 2007; Toscani et al., 1997). A-MYB has been shown to bind and up-regulate the Rfx2 promoter and conversely, Rfx2 has been suggested to be a candidate downstream amplifier of A-MYB regulation (Horvath, Kistler, & Kistler, 2009; Toscani et al., 1997). RFX motifs were also enriched at BRDT binding sites in promoters in both cell types examined (Figure 2A). This is of interest in light of previous observations that several RFX family TFs are up-regulated in post-meiotic cells and RFX2 in particular is expressed at high levels in pachytene spermatocytes (Horvath et al., 2009). Of note, the promoter of the testis-specific histone 1 variant H1t, which we had previously shown to be a direct target of BRDT-containing complexes (Shang et al., 2007), contains two imperfect inverted repeats which together comprise the X-box motif of RFX binding (VanWert, Wolfe, & Grimes, 2008). Of the 233 genes that were promoter-bound by BRDT in both PS and RS, 75% of them contained the RFX motif in their promoters.
In RS, the highest scores for an enrichment for specific TF motifs were observed for JunD, C-Jun, and CRE (Figure 2A). High levels of expression of JunD mRNA in post-meiotic RS (relative to meiotic PS) (Alcivar, Hake, Kwon, & Hecht, 1991). JunD-deficient males exhibited multiple age-dependent defects in spermatogenesis, with abnormalities in sperm head and flagellum structures (Thepot et al., 2000), defects reminiscent of those seen in BrdtΔbd1/Δbd1 mutant sperm (Shang et al., 2007). Our lab and others have identified that BRDT interacts with several transcription regulators such as SMARCE1, HDAC1, PRMT5 and TRIM28 (Dhar et al., 2012; Wang & Wolgemuth, 2016); whether this could be true for BRDT and JunD remains to be determined. Overall, these findings strongly suggest that BRDT may cooperate with specific TFs in distinct cellular contexts, and diverse BRDT containing-complexes may be recruited to different promoters to regulate specific genes in meiotic and post-meiotic cells.
It is known that transcriptional regulatory networks function within different epigenetic landscapes which in turn could play pivotal roles in modulating the expression of genes involved in the progression of meiosis and spermiogenesis (Crichton, Playfoot, & Adams, 2014; Kota & Feil, 2010). We have recently shown that in the absence of BRDT, the sex chromosomes in PS had reduced levels of H3K9me3 and H3K4me1, and increased levels of H3K9ac and RNA POL II (Manterola et al., 2018). Depletion of BRDT also led to more condensed and less accessible chromatin status and resulted in abnormal chromatin organization (Manterola et al., 2018). Furthermore, it has been known that BRDT may function in reorganizing haploid round spermatid chromatin (Shang et al., 2007), possibly in a histone H4 acetylation-dependent manner by cooperating with chromatin remodeling factors (Dhar et al., 2012). In this study, our ChIP-seq data of BRDT binding and marks of active transcription at the promoter regions showed a high overlap with CpG islands and the active histone modifications; H3K4me3 and H3K27ac, and the active histone variants; H2A.Z and H2A.LAP1 (Figure 2B and 2C). The location of CpG islands and GC-enriched regions to transcriptionally permissive chromatin is well-documented (R. Illingworth et al., 2008; R. S. Illingworth & Bird, 2009). Conversely, during spermiogenesis it is known that transition protein 2 (TP2) can use CpG island sequences to reorganize spermatid chromatin to an inactive configuration (Kundu & Rao, 1996). Together, these findings strongly suggest that BRDT plays a crucial role in regulation of transcriptional programs and chromatin organization by not only a selective binding of BRDT to its target promoters but also cooperating with its multiple binding partners.
Integration of our ChIP-seq data with available transcriptomic data revealed that the majority of BRDT-bound genes in PS and RS were enriched mRNA transcripts in the corresponding cell types (Figure 3A). Cluster analysis revealed that most of the BRDT-bound genes were found to be differentially expressed in PS and RS (76% and 66%, respectively), suggesting that the BRDT-bound DEGs in PS and RS are highly correlated with BRDT binding to their promoters. Of these, most of the genes were found to be significantly up-regulated in PS and RS (83% and 89%, respectively), indicating their potential roles in meiosis and post-meiotic meiosis, respectively (Clusters I and IV, Figure 3C, D). Others (17% in PS and 11% in RS) were found to be significantly down-regulated in PS and RS, indicating a possible role of BRDT in repressing gene expression at PS and RS stages (Clusters III and VI, Figure 3C, D).
Our cluster analysis also identified numerous genes, in Cluster II and V, whose expression is similar between Sg and PS, and between PS and RS, respectively (Figure 3C, D). This observation suggests that the binding of BRDT to the promoters in the corresponding to the genes may not simply act in regulation of gene transcription but rather, could modulate other molecular machinery involved in such processes as splicing, alternative 3’ end selection, etc. Alternatively, it may reflect a more transient binding of BRDT to the promoters in the corresponding genes. Transient binding of transcriptional regulators to the promoters is known to be insufficient to maintain the activation of transcription and rather, requires the constant presence of an activator to ensure open chromatin structure and accessibility for transcriptional machinery (Ptashne, 2011). In this regard, further analyses of BRDT binding patterns in parallel with the patterns for different aspects of epigenetic regulation including DNA methylation, chromatin accessibility and histone modifications, will help to understand the functionality of BRDT binding in the non-regulated its target genes.
Our GO enrichment analysis of the up-regulated genes in PS and RS revealed that the most significantly enriched biological processes are those essential for meiosis and spermiogenesis, respectively, and that there are unique sets of enriched genes in Clusters I and IV, suggesting distinct developmentally stage-specific functions for BRDT in PS and RS (Figure 4A, B; Supplemental Table S10). The observed overlap of 3 biological processes in both PS and RS (protein modification by small protein conjugation or removal/protein ubiquitination, microtubule cytoskeleton organization/microtubule-based process, and spermatogenesis), is highly statistically significant (Figure 4A, B). Conversely, there are clear stage-specific processes: ER stress response, DNA damage response, and protein localization were enriched only in PS while genes encoding sperm capacitation, fertilization and mRNA processing were enriched only in RS (Figure 4A, B). While the biological processes such as spermatogenesis, cell cycle process, and DNA damage response have previously been associated to BRDT (Gaucher et al., 2012; Manterola et al., 2018; Shang et al., 2007), we also observed enrichment in PS of biological processes not yet associated to BRDT function, including protein ubiquitination, ER stress response and protein localization (Figure 4A). Further, in RS, beyond the biological processes previously associated with BRDT, such as spermatogenesis, mRNA metabolic process and microtubule-based process (Berkovits et al., 2012; Berkovits & Wolgemuth, 2011; Dhar et al., 2012; Gaucher et al., 2012; Shang et al., 2007), we observed enrichment for genes involved in biological processes not yet associated to BRDT function, including glycerolipid catabolic process, negative regulation of cell cycle and epithelial cilium movement (Figure 4B). Genome browser visualization of the localization and ChIP-qPCR of BRDT-binding sites at promoters of specific examples of genes in the three selected critical processes further strongly supported distinct patterns of BRDT binding to promoters between the cell types (Figure 5 and Supplemental Figure S1).
In summary, the detailed genome-wide binding patterns of BRDT and their integration with gene expression patterns in the present study, provide comprehensive insight into BRDT function in mechanisms of gene regulation by BRDT during spermatogenesis. Further, the identification of overlap of BRDT binding with known TF binding motifs and determination of features of the epigenetic landscapes correlated with BRDT-binding revealed new correlations heretofore not appreciated.
Materials and Methods
Mice
The mice used in the present analysis were maintained in a National Institute of Health Research Animal Accredited Facility in accordance with the specifications of the Association for Assessment and Accreditation of Laboratory Animal Care. Mouse protocols were approved by the Institutional Animal Care and Use Committee of Columbia University.
Generation of populations of pachytene spermatocytes and round spermatids
The pachytene spermatocyte (PS) sample consisted of cellular suspensions obtained from testicular tubules isolated from 18 day-old male 129 Sv/Ev mice following our laboratory’s standard protocols as previously published (Wolgemuth, Gizang-Ginsberg, Engelmyer, Gavin, & Ponzetto, 1985). Briefly, testes were decapsulated and transferred to cold PBS, the seminiferous tubules were manually sheared with scissors and by pipetting, and then passed through a 40 μm filter. Although this population also includes spermatogonia (Sg), leptotene/zygotene spermatocytes, and some Sertoli cells, these cells do not express BRDT. Round spermatids (RS) were purified as previously described (Wolgemuth et al., 1985). Briefly, testes were dissected from seven adult mice and decapsulated. The cellular suspension was obtained from the tubular mass by digestion with 1.0 mg/ml collagenase (Sigma Cat# C0130) in RPMI buffer followed by digestion with 0.25 mg/ml trypsin (Sigma cat#T8003) in RPMI (Wolgemuth et al., 1985). Cell aggregates and connective tissue were removed by filtration through 74-pm nylon mesh (BD Falcon). The single cell suspension of germ cells was separated using gravity sedimentation on a 2–4% BSA in DPBS gradient. Fractions containing RS were pooled to yield samples containing ≥90% RS, as assessed by flow cytometric analysis (Golan et al., 2003) on a BD FACSCalibur Cell Analyzer. All steps of cell separation, fractionation and pooling were performed at 4oC in order to maintain cell viability and maximize nucleic acid integrity.
Chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-Seq)
To identify BRDT-containing complex binding sites, we prepared cross-linked chromatin samples for ChIP-Seq analysis. Approximately 107 cells from the two cellular preparations were fixed with 1% formaldehyde for 10 min at room temperature. The chromatin template was then fragmented to a size of 150 to 200 bp by sonication (Cuddapah et al., 2009). Aliquots of the chromatin samples were subjected to chromatin immunoprecipitation using a C-terminal anti-BRDT antibody generated by our laboratory (Shang et al., 2007) or anti-IgG as a control. This was followed by performing end repair and addition of “A” bases to the 3’ end of the DNA fragments, and ligation of adapters to the DNA fragments (Cuddapah et al., 2009). The resulting fragments were then size-selected for 150–350bp regions before generating the library, which was enriched for the adapter-modified DNA fragments by PCR. The purified DNA samples were subjected to analysis on the Ilumina Cluster Station and Genome Analyzer. In samples using the BRDT antibody, 17.5 and 18 million uniquely aligned short sequence reads in PS and RS, respectively, were obtained. Using the IgG antibody, 13 and 8.3 million reads in the two populations were obtained.
Data preprocessing
Sequenced tags were mapped to the mouse genome (mm8) using Bowtie algorithm (Langmead, Trapnell, Pop, & Salzberg, 2009) with the option ‘-m 1’. In order to reduce PCR amplification bias, at most one read was kept per genomic location. The resulting lists of uniquely mapped non-redundant reads were used for all downstream analyses. UCSC genome browser displayable BedGraph files for ChIP-Seq data were generated by counting tags in 200-bp windows across the genome and normalizing window tag counts by 10 million total sequenced reads to make different libraries directly comparable.
Genome-wide distribution of binding sites
BRDT binding sites from the ChIP-Seq data were identified using SICER software (Zang et al., 2009) using IgG data as control and a False Discovery Rate (FDR) threshold of 10%. The midpoint of each identified BRDT binding region (‘island’) is regarded as a BRDT binding site. Gene and exon location information was obtained from the Ensembl database (www.ensembl.org) using the BioMart tool. Based on this information, promoter, exonic, intronic, and intergenic regions were defined as follows. A promoter was defined as being 2 kb upstream of the TSS. To avoid double counting at the promoter overlaps, coordinates of all promoters were merged to define a non-redundant set of promoter regions. Similarly, other classes of regions (exons and introns) were merged to avoid double counting. Some BRDT binding sites are located in multiple classes of region, e.g. in the exon of one transcript isoform and in the intron of another transcript isoform of the same gene. In order to deal with such ambiguities in the assignment of BRDT binding sites to different classes, we used the following prioritization: promoter took precedence over an exon, followed by exon > intron > intergenic. Thus, for example, if a BRDT binding site is located in both an exon and an intron, then it is assigned as being located in the exon. The number of BRDT binding sites in the resulting non-redundant and non-overlapping classes of regions were counted and used to create pie charts to illustrate the distribution of binding sites.
Gene body assay
The sequence read density (per 100-bp window and 30 million non-redundant uniquely mapped reads) was determined along the gene-body and in the upstream/downstream 2 kb regions in the sample and control IgG libraries. The signal level in each window was computed as the difference between the read densities in the sample and control libraries, with the negative values set to zero. Genes were ranked based on their expression levels and stratified into three expression groups: the ‘high’ group (the top 30% genes in the ranked list), the ‘silent’ group (the bottom 30% genes), and the ‘medium’ group (the remaining genes). The mean signal levels over all genes in each group were determined and displayed as gene-body plots.
Motif analysis
MEME (Multiple Expectation maximization for Motif Elicitation) (Bailey et al., 2009) with default parameters was used to identify statistically overrepresented motifs within the inferred binding sites. In order to reduce noise for the motif identification, we used only DNA sequences at the top 500 BRDT binding sites in terms of ChIP-Seq tag density.
Comparison of our ChIP-seq data with the published ChIP-seq data on CREM, and H2A-variant, H3K4me3 and H3K27me3
As defined above, BRDT binding sites are midpoints of BRDT binding regions identified by the SICER algorithm. To assess the overlap of BRDT binding with regions previously reported to be bound by selected nuclear proteins, including CREM (Martianov et al., 2010), the H2A variants H2A.Lap1 and H2A.Z (Nekrasov et al., 2012), and H3K4me3 and H3K27me3 (Cui et al., 2012), the chromosomal coordinate of the published binding regions of these proteins was designated as (x1, x2). A BRDT binding site x is assumed to overlap this region if x1 ≤ x ≤ x2.
Functional enrichment analysis
DAVID Bioinformatics Resources 6.8 was used to analyze the gene lists derived from ChIP-seq data to find enriched biological functions (Huang da et al., 2009a, 2009b). We uploaded the significant BRDT-bound at the promoters of genes to investigate the enriched potential functions. The P-values were determined by Fisher’s Exact test with FDR multiple test correction. P-value < 0.1 and false discovery rate (FDR) < 0.05 were used as the cut-off criteria for the analysis.
Analyses of expression patterns using online expression data sets
The patterns of expression of the genes from staged spermatogenic cells were analyzed using transcriptomic data available from the GEO website: http://www.ncbi.nlm.nih.gov/geo/; GSE100964 and GSE4193. The raw expression data of the staged spermatogenic cells were downloaded from the GEO website and analyzed with the GeneSpring GX software (version 11, Agilent Technologies) using the RMA algorithm for summarization and normalization. The analysis was carried out as described in Wichman et al (Wichman et al., 2017) and Namekawa et al (Namekawa et al., 2006). Significantly differentially expressed genes (DEGs) were defined after comparing transcript abundance in Sg and PS as well as PS and RS, as those which were more than 2-fold increased or decreased with a p adjusted value p < 0.05. The heatmaps were obtained by representation of the normalized values of expression of these genes in the indicated color scale (red: low expression; green: high expression).
ChIP and real-time PCR
ChIP was performed as described in the Materials and Methods for the ChIP-Seq above. Promoter region primers spanning from −700 to +300 bp relative to the gene transcription start sites were used, and DNA binding was quantified using the Applied Biosystems 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA) containing FastStart Universal SYBR Green Master (Roche, Mannheim, Germany), cDNA and primers. PCR primers used are listed in Supplemental Tables S11. The data were normalized with β-actin according to the 2–ΔΔCt relative quantitation method in the manufacturer’s manual.
Supplementary Material
Supplemental Figure S1. ChIP-qPCR of BRDT binding to the promoters of Ybx2, Usp1, Prm1 and Zfp36L1 in PS and RS. Relative DNA binding is the percentage of input (normalized to IgG). Error bars indicate standard deviation of triplicate measurements. *P < 0.05, **P < 0.01, t-test.
Supplemental Tables S1. A list of genes with BRDT binding in the promoter in PS.
Supplemental Tables S2. A list of genes with BRDT binding in the promoter in RS.
Supplemental Table S3. A list of genes expressed in PS whose promoters are bound by BRDT in PS
Supplemental Table S4. A list of genes expressed in RS whose promoters are bound by BRDT in RS
Supplemental Table S5. A list of genes belonging to Cluster I, II and III in PS
Supplemental Table S6. A list of genes belonging to Cluster IV, V and VI in RS
Supplemental Table S7. 16 genes overlapping between Cluster I and Cluster IV
Supplemental Table S8. Enriched biological process GO-terms of the up-regulated genes in PS compared to Sg (Cluster I)
Supplemental Table S9. Enriched biological process GO-terms of the up-regulated genes in RS compared to PS (Cluster IV)
Supplemental Table S10. Top 10 Enriched biological process GO-terms in Clusters I and IV.
Supplemental Table S11. Real-time PCR primers used in the qPCR analyses.
Acknowledgments
This work was supported in part by a grant from the NIH, GM081767 (D.J.W.), by the Division of Intramural Research, NHLBI, NIH (K.Z.), and an “Estadías Cortas de Investigación Internacionales”- Internationalization Grant UCH-1566 (M.M.).
Funding information:
This work was supported in part by a grant from the NIH, GM081767 (D.J.W), by the Division of Intramural Research at the National Heart, Lung, and Blood Institute (K.Z.), and an “Estadías Cortas de Investigación Internacionales”- Internationalization Grant UCH-1566 (M.M).
Footnotes
Conflict of Interest Statement
The authors declare they have no competing interests.
Availability of data and materials
The GEO accession number for the raw and processed ChIP-Seq data described in this article is GSE98489.
References
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, & Hill CS (2011). Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell, 43(1), 85–96. doi: 10.1016/j.molcel.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Al-Harthi L, & Roebuck KA (1998). Human immunodeficiency virus type-1 transcription: role of the 5’-untranslated leader region (review). Int J Mol Med, 1(5), 875–881. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9852310 [DOI] [PubMed] [Google Scholar]
- Alcivar AA, Hake LE, Kwon YK, & Hecht NB (1991). junD mRNA expression differs from c-jun and junB mRNA expression during male germinal cell differentiation. Mol Reprod Dev, 30(3), 187–193. doi: 10.1002/mrd.1080300304 [DOI] [PubMed] [Google Scholar]
- Ayoubi TAY, & VanDeVen WJM (1996). Regulation of gene expression by alternative promoters. Faseb Journal, 10(4), 453–460. Retrieved from <Go to ISI>://WOS:A1996UE55700009 [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, . . . Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res, 37(Web Server issue), W202–208. doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, & Berger SL (2001). Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell, 8(6), 1243–1254. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11779500 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Berkovits BD, Wang L, Guarnieri P, & Wolgemuth DJ (2012). The testis-specific double bromodomain-containing protein BRDT forms a complex with multiple spliceosome components and is required for mRNA splicing and 3’-UTR truncation in round spermatids. Nucleic Acids Res, 40(15), 7162–7175. doi: 10.1093/nar/gks342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, & Wolgemuth DJ (2011). The first bromodomain of the testis-specific double bromodomain protein Brdt is required for chromocenter organization that is modulated by genetic background. Dev Biol, 360(2), 358–368. doi: 10.1016/j.ydbio.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, & Allis CD (2005). RNA meets chromatin. Genes Dev, 19(14), 1635–1655. doi: 10.1101/gad.1324305 [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, & Verdin E (2007). Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A, 104(34), 13690–13695. doi: 10.1073/pnas.0705053104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, & Schutz G (1996). Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature, 380(6570), 162–165. doi: 10.1038/380162a0 [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E, Bannister LA, Barash A, Schimenti KJ, Hartford SA, Eppig JJ, . . . Schimenti JC (2011). A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development, 138(15), 3319–3330. doi: 10.1242/dev.067645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, & Korf I (2008). Longer first introns are a general property of eukaryotic gene structure. PLoS One, 3(8), e3093. doi: 10.1371/journal.pone.0003093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer LR, Corzett M, & Balhorn R (1999). Protamine-induced condensation and decondensation of the same DNA molecule. Science, 286(5437), 120–123. doi: 10.1126/science.286.5437.120 [DOI] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, . . . Zhao Y (2007). Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics, 6(5), 812–819. doi: 10.1074/mcp.M700021-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury TA, & Kleene KC (2012). Identification of Potential Regulatory Elements in the 5 ‘ and 3 ‘ UTRs of 12 Translationally Regulated mRNAs in Mammalian Spermatids by Comparative Genomics. J Androl, 33(2), 244–256. doi: 10.2164/jandrol.110.012492 [DOI] [PubMed] [Google Scholar]
- Crichton JH, Playfoot CJ, & Adams IR (2014). The role of chromatin modifications in progression through mouse meiotic prophase. J Genet Genomics, 41(3), 97–106. doi: 10.1016/j.jgg.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Crowe BL, Larue RC, Yuan C, Hess S, Kvaratskhelia M, & Foster MP (2016). Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction. Proc Natl Acad Sci U S A, 113(8), 2086–2091. doi: 10.1073/pnas.1516813113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, & Zhao K (2009). Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res, 19(1), 24–32. doi: 10.1101/gr.082800.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P, Liu W, Zhao Y, Lin Q, Zhang D, Ding F, . . . Hu, S. (2012). Comparative analyses of H3K4 and H3K27 trimethylations between the mouse cerebrum and testis. Genomics Proteomics Bioinformatics, 10(2), 82–93. doi: 10.1016/j.gpb.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong J (2006). Basic mechanisms for the control of germ cell gene expression. Gene, 366(1), 39–50. doi: 10.1016/j.gene.2005.10.012 [DOI] [PubMed] [Google Scholar]
- Denis GV (2010). Bromodomain coactivators in cancer, obesity, type 2 diabetes, and inflammation. Discov Med, 10(55), 489–499. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/21189220 [PMC free article] [PubMed] [Google Scholar]
- Denis GV, Vaziri C, Guo N, & Faller DV (2000). RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ, 11(8), 417–424. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10965846 [PMC free article] [PubMed] [Google Scholar]
- Desroches-Castan A, Cherradi N, Feige JJ, & Ciais D (2011). A novel function of Tis11b/BRF1 as a regulator of Dll4 mRNA 3 ‘-end processing. Mol Biol Cell, 22(19), 3625–3633. doi: 10.1091/mbc.E11-02-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Thota A, & Rao MR (2012). Insights into role of bromodomain, testis-specific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. J Biol Chem, 287(9), 6387–6405. doi: 10.1074/jbc.M111.288167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, & Cheung P (2012). A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet, 8(11), e1003047. doi: 10.1371/journal.pgen.1003047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant M, & Pugh BF (2007). NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol Cell Biol, 27(15), 5327–5335. doi: 10.1128/MCB.00468-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Jaiswal BS, Xie F, Krajnc-Franken MAM, Robben TJAA, Strik AM, . . . Gossen JA (2004). Mice deficient for soluble adenylyl cyclase are infertile because of a severe sperm-motility defect (vol 101, pg 2993, 2004). Proc Natl Acad Sci U S A, 101(14), 5180–5180. doi: 10.1073/pnas.0401409101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B, & Faller DV (2001). You bet-cha: a novel family of transcriptional regulators. Front Biosci, 6, D1008–1018. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11487468 [DOI] [PubMed] [Google Scholar]
- Florence BL, & Faller DV (2008). Drosophila female sterile (1) homeotic is a multifunctional transcriptional regulator that is modulated by Ras signaling. Dev Dyn, 237(3), 554–564. doi: 10.1002/dvdy.21432 [DOI] [PubMed] [Google Scholar]
- Ford L, Ling E, Kandel ER, & Fioriti L (2019). CPEB3 inhibits translation of mRNA targets by localizing them to P bodies. Proc Natl Acad Sci U S A, 116(36), 18078–18087. doi: 10.1073/pnas.1815275116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes NS, Mellstrom B, Benusiglio E, & Sassone-Corsi P (1992). Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature, 355(6355), 80–84. doi: 10.1038/355080a0 [DOI] [PubMed] [Google Scholar]
- Freiman RN (2009). Specific variants of general transcription factors regulate germ cell development in diverse organisms. Biochim Biophys Acta, 1789(3), 161–166. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/19437618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Kimata Y, Nomaguchi K, Watanabe K, & Kohno K (2002). Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. Journal of Biological Chemistry, 277(24), 21585–21591. doi: 10.1074/jbc.M201523200 [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, & Mackay JP (2011). Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol, 31(13), 2632–2640. doi: 10.1128/MCB.05413-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, . . . Khochbin S (2012). Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J, 31(19), 3809–3820. doi: 10.1038/emboj.2012.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan R, Lewin LM, Soffer Y, Lotan G, Shochat L, & Vigodner M (2003). Evaluation of spermatogenesis using flow cytometry and confocal microscopy. Isr Med Assoc J, 5(7), 536. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12901259 [PubMed] [Google Scholar]
- Goudarzi A, Zhang D, Huang H, Barral S, Kwon OK, Qi S, . . . Khochbin S (2016). Dynamic Competing Histone H4 K5K8 Acetylation and Butyrylation Are Hallmarks of Highly Active Gene Promoters. Mol Cell, 62(2), 169–180. doi: 10.1016/j.molcel.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, . . . Blobel GA (2007). Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol, 27(24), 8466–8479. doi: 10.1128/MCB.00993-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Faller DV, & Denis GV (2000). Activation-induced nuclear translocation of RING3. J Cell Sci, 113 (Pt 17), 3085–3091. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10934046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SH, Sheedy DM, Cuddihy AR, & O’Connell MJ (2004). Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol Cell Biol, 24(2), 662–674. doi: 10.1128/Mcb.24.2.662-674.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath GC, Kistler MK, & Kistler WS (2009). RFX2 is a candidate downstream amplifier of A-MYB regulation in mouse spermatogenesis. BMC Dev Biol, 9, 63. doi: 10.1186/1471-213X-9-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, & Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 37(1), 1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, & Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 4(1), 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang DH, & Dawid IB (1990). The maternal-effect gene fsh is essential for the specification of the central region of the Drosophila embryo. New Biol, 2(2), 163–170. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1982070 [PubMed] [Google Scholar]
- Huang TT, Nijman SMB, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, . . . D’Andrea AD (2006). Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol, 8(4), 339–U313. doi: 10.1038/ncb1378 [DOI] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, & Richter JD (2006). CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. Embo Journal, 25(20), 4865–4876. doi: 10.1038/sj.emboj.7601322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, . . . Bird A (2008). A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol, 6(1), e22. doi: 10.1371/journal.pbio.0060022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, & Bird AP (2009). CpG islands--’a rough guide’. FEBS Lett, 583(11), 1713–1720. doi: 10.1016/j.febslet.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, & Tjian R (2000). Structure and function of a human TAFII250 double bromodomain module. Science, 288(5470), 1422–1425. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10827952 [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, & Ozato K (2005). The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell, 19(4), 523–534. doi: 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Jones MH, Numata M, & Shimane M (1997). Identification and characterization of BRDT: A testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics, 45(3), 529–534. doi: 10.1006/geno.1997.5000 [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, & Ozato K (2004). Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell, 13(1), 33–43. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14731392 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, . . . Rinn JL (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A, 106(28), 11667–11672. doi: 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, & D’Andrea AD (2009). Inactivation of Murine Usp1 Results in Genomic Instability and a Fanconi Anemia Phenotype. Dev Cell, 16(2), 314–320. doi: 10.1016/j.devcel.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota SK, & Feil R (2010). Epigenetic transitions in germ cell development and meiosis. Dev Cell, 19(5), 675–686. doi: 10.1016/j.devcel.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, . . . Greenblatt JF (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell, 12(6), 1565–1576. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14690608 [DOI] [PubMed] [Google Scholar]
- Kundu TK, & Rao MR (1996). Zinc dependent recognition of a human CpG island sequence by the mammalian spermatidal protein TP2. Biochemistry, 35(49), 15626–15632. doi: 10.1021/bi961271i [DOI] [PubMed] [Google Scholar]
- Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, . . . Blobel GA (2011). Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci U S A, 108(22), E159–168. doi: 10.1073/pnas.1102140108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, & Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol, 10(3), R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Litvin J, Orth JM, Patel B, Mettus R, & Reddy EP (1996). Temporal patterns of A-myb and B-myb gene expression during testis development. Oncogene, 13(6), 1161–1168. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8808690 [PubMed] [Google Scholar]
- Lee JE, Park YK, Park S, Jang Y, Waring N, Dey A, . . . Ge K (2017). Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat Commun, 8(1), 2217. doi: 10.1038/s41467-017-02403-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Chepelev I, DiMaggio PA, Blanco MA, Zee BM, Zhao K, & Garcia BA (2012). Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol, 13(8), R68. doi: 10.1186/gb-2012-13-8-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Rickards B, & Flint SJ (2008). The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell, 30(1), 51–60. doi: 10.1016/j.molcel.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, & Rosenfeld MG (2016). Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet, 17(4), 207–223. doi: 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- Liu H, Mulholland N, Fu H, & Zhao K (2006). Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol Cell Biol, 26(7), 2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang Y, Li S, Yan Y, & Li Y (2010). Dynamic regulation of glutamate decarboxylase 67 gene expression by alternative promoters and splicing during rat testis maturation. Mol Biol Rep, 37(7), 3111–3119. doi: 10.1007/s11033-009-9889-4 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, . . . Seraphin B (1994). The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res, 22(24), 5332–5340. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7816623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola M, Brown TM, Oh MY, Garyn C, Gonzalez BJ, & Wolgemuth DJ (2018). BRDT is an essential epigenetic regulator for proper chromatin organization, silencing of sex chromosomes and crossover formation in male meiosis. PLoS Genet, 14(3), e1007209. doi: 10.1371/journal.pgen.1007209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Choukrallah MA, Krebs A, Ye T, Legras S, Rijkers E, . . . Davidson I (2010). Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics, 11, 530. doi: 10.1186/1471-2164-11-530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas D, Mettler IJ, Pierce DA, & Lowe HW (1990). Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol Biol, 15(6), 913–920. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/2103480 [DOI] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, & Buratowski S (2000). Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev, 14(8), 951–962. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10783167 [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, & Zhao M (2003). Roles of transition nuclear proteins in spermiogenesis. Chromosoma, 111(8), 483–488. doi: 10.1007/s00412-002-0227-z [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, & Zhou MM (2007). Structure and acetyl-lysine recognition of the bromodomain. Oncogene, 26(37), 5521–5527. doi: 10.1038/sj.onc.1210618 [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, & Lee JT (2006). Postmeiotic sex chromatin in the male germline of mice. Curr Biol, 16(7), 660–667. doi: 10.1016/j.cub.2006.01.066 [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, . . . Tremethick DJ (2012). Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol, 19(11), 1076–1083. doi: 10.1038/nsmb.2424 [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, . . . Jenuwein T (2000). Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol, 20(24), 9423–9433. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11094092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, . . . Patel KJ (2007). Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell, 28(5), 798–809. doi: 10.1016/j.molcel.2007.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Dong W, Chen L, Zou T, Qi Y, & Liu Y (2007). Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol Cell Biochem, 294(1–2), 45–54. doi: 10.1007/s11010-006-9223-6 [DOI] [PubMed] [Google Scholar]
- Ptashne M (2011). Principles of a switch. Nat Chem Biol, 7(8), 484–487. doi: 10.1038/nchembio.611 [DOI] [PubMed] [Google Scholar]
- Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, . . . Narlikar GJ (2009). The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature, 462(7276), 1016–1021. doi: 10.1038/nature08621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, & Lauberth SM (2018). RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol, 25(8), 687–697. doi: 10.1038/s41594-018-0102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar N, Baluchamy S, Kadeppagari RK, Singhal G, Weitzman S, & Thimmapaya B (2008). p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene, 27(43), 5717–5728. doi: 10.1038/onc.2008.181 [DOI] [PubMed] [Google Scholar]
- Sawa C, Nedea E, Krogan N, Wada T, Handa H, Greenblatt J, & Buratowski S (2004). Bromodomain factor 1 (Bdf1) is phosphorylated by protein kinase CK2. Mol Cell Biol, 24(11), 4734–4742. doi: 10.1128/MCB.24.11.4734-4742.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Sutto Z, Nlend MC, Horvath G, Schmid N, Buck J, . . . Salathe M (2007). Soluble adenylyl cyclase is localized to cilia and contributes to ciliary beat frequency regulation via production of cAMP. Journal of General Physiology, 130(1), 99–109. doi: 10.1085/jgp.200709784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, . . . Ott M (2012). Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem, 287(2), 1090–1099. doi: 10.1074/jbc.M111.282855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E, Nickerson HD, Wen D, Wang X, & Wolgemuth DJ (2007). The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development, 134(19), 3507–3515. doi: 10.1242/dev.004481 [DOI] [PubMed] [Google Scholar]
- Shang E, Salazar G, Crowley TE, Wang X, Lopez RA, Wang X, & Wolgemuth DJ (2004). Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr Patterns, 4(5), 513–519. doi: 10.1016/j.modgep.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Sinha A, Faller DV, & Denis GV (2005). Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem J, 387(Pt 1), 257–269. doi: 10.1042/BJ20041793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, & Aravin AA (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol, 12(4), 246–258. doi: 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, & Tremethick DJ (2011). A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol, 19(1), 25–30. doi: 10.1038/nsmb.2161 [DOI] [PubMed] [Google Scholar]
- Soboleva TA, Nekrasov M, Ryan DP, & Tremethick DJ (2014). Histone variants at the transcription start-site. Trends Genet, 30(5), 199–209. doi: 10.1016/j.tig.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Son YO, Kim HE, Choi WS, Chun CH, & Chun JS (2019). RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. Nat Commun, 10. doi:ARTN 77 10.1038/s41467-018-08035-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tae S, Karkhanis V, Velasco K, Yaneva M, Erdjument-Bromage H, Tempst P, & Sif S (2011). Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res, 39(13), 5424–5438. doi: 10.1093/nar/gkr170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW (1995). The role of brahma and related proteins in transcription and development. Curr Opin Genet Dev, 5(4), 473–477. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7580139 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y (2016). The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. Int J Mol Sci, 17(11). doi: 10.3390/ijms17111849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EM, Moghraby JS, Lees JH, Smit B, Moens PB, & Lehmann AR (2001). Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Apr18 complex. Mol Biol Cell, 12(6), 1583–1594. doi:DOI 10.1091/mbc.12.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thepot D, Weitzman JB, Barra J, Segretain D, Stinnakre MG, Babinet C, & Yaniv M (2000). Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development, 127(1), 143–153. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10654608 [DOI] [PubMed] [Google Scholar]
- Thompson M (2009). Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie, 91(3), 309–319. doi: 10.1016/j.biochi.2008.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J, . . . Reddy EP (1997). Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature, 386(6626), 713–717. doi: 10.1038/386713a0 [DOI] [PubMed] [Google Scholar]
- Turner JD, Alt SR, Cao L, Vernocchi S, Trifonova S, Battello N, & Muller CP (2010). Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem Pharmacol, 80(12), 1860–1868. doi: 10.1016/j.bcp.2010.06.037 [DOI] [PubMed] [Google Scholar]
- VanWert JM, Wolfe SA, & Grimes SR (2008). Binding of RFX2 and NF-Y to the testis-specific histone H1t promoter may be required for transcriptional activation in primary spermatocytes. J Cell Biochem, 104(3), 1087–1101. doi: 10.1002/jcb.21694 [DOI] [PubMed] [Google Scholar]
- Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, . . . Berger SL (1997). Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol, 17(1), 519–527. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8972232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, & Wolgemuth DJ (2016). BET Protein BRDT Complexes With HDAC1, PRMT5, and TRIM28 and Functions in Transcriptional Repression During Spermatogenesis. J Cell Biochem, 117(6), 1429–1438. doi: 10.1002/jcb.25433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichman L, Somasundaram S, Breindel C, Valerio DM, McCarrey JR, Hodges CA, & Khalil AM (2017). Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility. Biol Reprod, 97(2), 313–323. doi: 10.1093/biolre/iox084 [DOI] [PubMed] [Google Scholar]
- Wolgemuth DJ, Gizang-Ginsberg E, Engelmyer E, Gavin BJ, & Ponzetto C (1985). Separation of mouse testis cells on a Celsep (TM) apparatus and their usefulness as a source of high molecular weight DNA or RNA. Gamete Res, 12, 1–10. doi: 10.1002/mrd.1120120102 [DOI] [PubMed] [Google Scholar]
- Xie F, Garcia MA, Carlson AE, Schuh SM, Babcock DF, Jaiswal BS, . . . Conti M (2006). Soluble adenylyl cyclase (sAC) is indispensable for sperm function and fertilization. Dev Biol, 296(2), 353–362. doi: 10.1016/j.ydbio.2006.05.038 [DOI] [PubMed] [Google Scholar]
- Xu SW, Hafer N, Agunwamba B, & Schedl P (2012). The CPEB Protein Orb2 Has Multiple Functions during Spermatogenesis in Drosophila melanogaster. PLoS Genet, 8(11). doi:ARTN e1003079 10.1371/journal.pgen.1003079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JX, Medvedev S, Yu JY, Tang LC, Agno JE, Matzuk MM, . . . Hecht NB (2005). Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci U S A, 102(16), 5755–5760. doi: 10.1073/pnas.0408718102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, & Zhou Q (2005). Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell, 19(4), 535–545. doi: 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, & Zhou Q (2003). Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell, 12(4), 971–982. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14580347 [DOI] [PubMed] [Google Scholar]
- Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, . . . Meistrich ML (2000). Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci U S A, 97(9), 4683–4688. doi: 10.1073/pnas.97.9.4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, & Peng W (2009). A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics, 25(15), 1952–1958. doi: 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, & Grummt I (2005). The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol, 15(15), 1434–1438. doi: 10.1016/j.cub.2005.06.057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. ChIP-qPCR of BRDT binding to the promoters of Ybx2, Usp1, Prm1 and Zfp36L1 in PS and RS. Relative DNA binding is the percentage of input (normalized to IgG). Error bars indicate standard deviation of triplicate measurements. *P < 0.05, **P < 0.01, t-test.
Supplemental Tables S1. A list of genes with BRDT binding in the promoter in PS.
Supplemental Tables S2. A list of genes with BRDT binding in the promoter in RS.
Supplemental Table S3. A list of genes expressed in PS whose promoters are bound by BRDT in PS
Supplemental Table S4. A list of genes expressed in RS whose promoters are bound by BRDT in RS
Supplemental Table S5. A list of genes belonging to Cluster I, II and III in PS
Supplemental Table S6. A list of genes belonging to Cluster IV, V and VI in RS
Supplemental Table S7. 16 genes overlapping between Cluster I and Cluster IV
Supplemental Table S8. Enriched biological process GO-terms of the up-regulated genes in PS compared to Sg (Cluster I)
Supplemental Table S9. Enriched biological process GO-terms of the up-regulated genes in RS compared to PS (Cluster IV)
Supplemental Table S10. Top 10 Enriched biological process GO-terms in Clusters I and IV.
Supplemental Table S11. Real-time PCR primers used in the qPCR analyses.
Data Availability Statement
The GEO accession number for the raw and processed ChIP-Seq data described in this article is GSE98489.


