Abstract
Brain maps, or atlases, are essential tools for studying brain function and organization. The abundance of available atlases used across the neuroscience literature, however, creates an implicit challenge that may alter the hypotheses and predictions we make about neurological function and pathophysiology. Here, we demonstrate how parcellation scale, shape, anatomical coverage, and other atlas features may impact our prediction of the brain’s function from its underlying structure. We show how network topology, structure–function correlation (SFC), and the power to test specific hypotheses about epilepsy pathophysiology may change as a result of atlas choice and atlas features. Through the lens of our disease system, we propose a general framework and algorithm for atlas selection. This framework aims to maximize the descriptive, explanatory, and predictive validity of an atlas. Broadly, our framework strives to provide empirical guidance to neuroscience research utilizing the various atlases published over the last century.
Keywords: Brain atlas, Networks, Epilepsy, Structure–function
1. Introduction
How we define anatomical brain structures and relate those structures to the brain’s function can either constrain or enhance our understanding of behavior and neurological diseases (Bohland et al., 2009; Dickie et al., 2017; Klein and Tourville, 2012; Mandal et al., 2012). Discoveries by scientists like Carl Wernicke and Pierre Paul Broca, who mapped specific brain regions to speech function, in addition to case studies from Phineas Gage and H.M., who lost specific brain regions with resultant changes in brain function and behavior, exemplify how brain structure and function are fundamentally linked (Barker, 1995; Beal et al., 2015; Van Horn et al., 2012). Properly labeling brain structures is paramount for enabling scientists to effectively communicate about the variability between healthy individuals and about the regions involved in neurological disorders (Mazziotta et al., 2001). Yet, no consensus has been reached on the most appropriate ways to label and delineate these regions, as evident by the wide variety of brain maps, or atlases, defining neuroanatomical structures (Evans et al., 2012).
In common usage, an atlas refers to a “collection of maps” National (2022) that typically defines geo-political boundaries and may include coarse borders (continental), fine borders (city), and anything in between (country; Fig. 1 a, left). Borders (National, 2022) are usually consistent across atlases of the world. In contrast, atlases of the brain are not consistent. Four separate atlases (Fig. 1 a, right) may define the superior temporal gyrus differently. For example, approximately ninety percent of the anterior superior temporal gyrus in the Harvard-Oxford atlas (Makris et al., 2006) overlaps with the posterior superior temporal gyrus in the Hammersmith atlas(Hammers et al., 2003). Atlases may also differ in other ways, including parcellation size, neuroanatomical coverage, and complexity of brain region shapes. For instance, the Yeo atlas (Thomas Yeo et al., 2011) contains 7 or 17 parcels while the Schaefer atlases (Schaefer et al., 2018) may have between 100 and 1000 parcels. Complicating matters further, atlases can differ in their intended use. The MMP atlas (Glasser et al., 2016) was intended for surface-based analyses (Coalson et al., 2018), yet a volumetric version (without sub-cortical structures) was independently created and used in connectivity studies (Wu et al., 2019). The plethora of available atlases poses a problem for reproducibility in studying healthy and diseased populations and for metanalyses describing the involvement of different regions of the brain in various diseases. This has been termed the Atlas Concordance Problem (Bohland et al., 2009).
Fig. 1. Many brain atlases are available in the neuroscience literature.
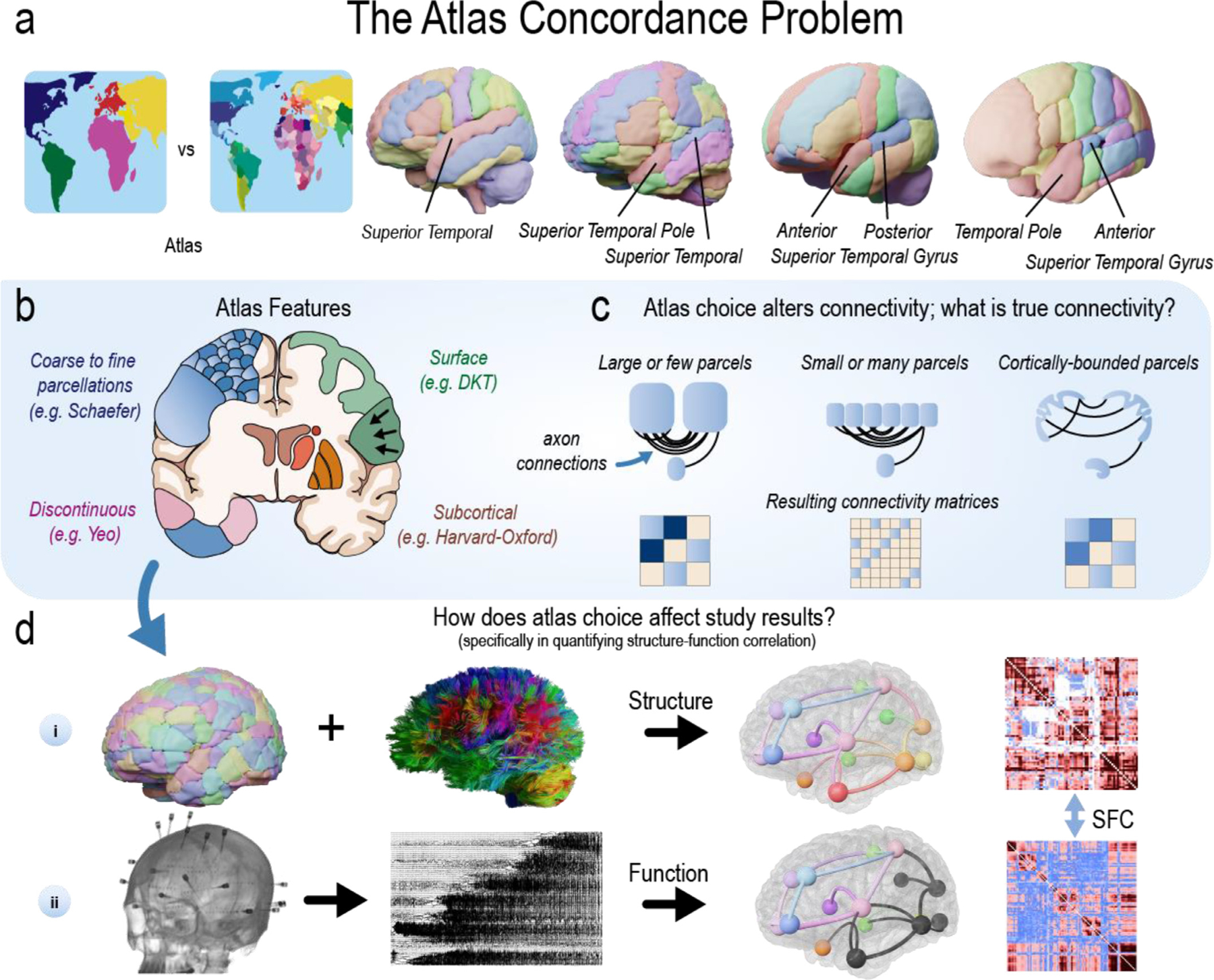
| a, In common usage, an atlas refers to a “collection of maps”National (2022) that defines geo-political borders at different scales. Although borders(National, 2022) are usually consistent across atlases of the world, they are typically not consistent across atlases of the brain. Four separate atlases (left-to-right: CerebrA, AAL, Hammersmith, Harvard-Oxford) may define the superior temporal gyrus differently. The lack of consistency across these labels poses a problem for reproducibility in cognitive, systems, developmental, and clinical studies, as well as metanalyses describing the involvement of different regions of the brain in various diseases (Bohland et al., 2009). This challenge has been previously referred to as the Atlas Concordance Problem. b, Atlases can have varying features (see also Table 1). c, Thus, all current connectivity studies in neuroscience may not accurately reflect some fundamentally “true” architecture. For example, atlases with either large or small parcels may affect the structural connectivity matrices that are used to define the ”true” network architecture of the brain, and subsequently that are used to test hypotheses or make predictions about the brain. d, When combined with white matter tracts reconstructed from diffusion MRI, atlases can be used to measure how different regions of the brain are structurally connected (i). Similarly, intracranial EEG (iEEG) implants can record neural activity to measure how different regions of the brain are functionally connected (ii). Technologies such as fMRI, MEG, and many others can also measure functional connectivity. The statistical similarity between structural and functional connectivity measurements can be calculated (e.g., structure–function correlation; SFC). Such estimates have been used to better understand the pathophysiology of disease. In this study, we evaluate how the varying atlases may alter the power to test a specific hypothesis about the brain’s structure–function relationship in epilepsy.
In the present study, we perform an extensive evaluation of the available atlases in the neuroscience literature (Table 1) by examining the effect of varying features such as parcellation size, coverage, and shape (Fig. 1 b) on structural connectivity (Fig. 1 c). We also examine how atlas choice changes structural network topology by measuring structure–function correlation (SFC) using an atlas-independent measure of functional connectivity (Fig. 1 d). We utilize a total of 55 brain atlases, including many routinely used in common neuroimaging software. Note the important distinction between the terms atlas, template, and stereotactic space (Evans et al., 2012) (see Fig. S1). We found that different atlases may alter the power to test a hypothesis about epilepsy pathophysiology that seizures propagate through the underlying structural connections of the brain. This hypothesis has been previously supported in prior research(Ashourvan et al., 2021; Proix et al., 2017; Shah et al., 2019; Wirsich et al., 2016).
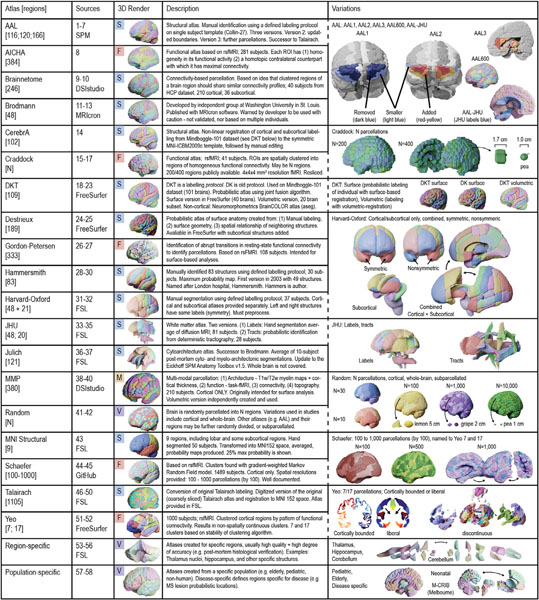
Table 1. Atlases.
|Atlas sources are detailed in Table S1 and abbreviations are in the glossary. S: Structurally defined atlas; F: Functionally defined atlas; M: Multi-modally defined atlas; V: A variably defined atlas that may be structurally, functionally, or multi-modally defined; ROI: region of interest; HCP: Human connectome project dataset (Van Essen et al., 2013); MS: multiple sclerosis.
|
In the context of our experimental design, we propose a new framework outlining how to appropriately choose an atlas when designing a neuroscience experiment. This framework is derived from historical foundations for assessing the validity and effectiveness of animal models (Willner, 1984), network models (Bassett et al., 2018), and psychometric tests (Association, 1954), which try to maximize the (1) descriptive, explanatory, and (3) predictive validity (Bassett et al., 2018) of a model. Atlases are a tool for investigators to test for causality and to make predictions about the brain. Thus, this framework incorporates a short discussion on explanatory modeling and predictive modeling, each with different goals (”To Explain or to Predict?”(Shmueli, 2010)). A one-size-fits-all approach may not exist for selecting an atlas, nor should it (Salehi et al., 2020); while there is one Planet Earth with a single atlas for a particular use (e.g., an atlas of the geo-political borders for a given point in time), there are many brains, with anatomical and functional variability across populations and species(Salehi et al., 2020). We hope our framework provides empirical guidance to neuroscience research utilizing the various atlases published over the last century.
2. Results
2.1. Clinical data
Forty-one individuals (mean age 34 ± 11; 16 female) underwent High Angular Resolution Diffusion Imaging (HARDI), composed of thirteen controls (mean age 35 ± 13; 6 female) and twenty-eight drug-resistant epilepsy patients (mean age 34 ±11; 12 female) evaluated for surgical treatment. Of the twenty-eight patients, twenty-four were implanted with stereoelectroencephalography (SEEG) and four with electrocorticography (ECoG). Ten SEEG patients (mean age 34 ±8; 4 female) had clinical seizure annotations, and the first seizure from each patient (mean duration 81 s) without artifacts was selected for SFC analyses. Patient and control demographics are included in Table S2.
2.2. Atlas morphology: sizes and shapes
We hypothesized that atlas morphological properties, including size and shape (Fig 2), affect SFC. To test this hypothesis, we first quantified the distributions of parcellation sizes (Fig 2 a) and shapes (Fig 2 b) in various atlases. These results exemplify the diversity of atlas parcellation morphology. Fig 2 c shows a comparison of individual parcellation volumes and sphericities. The remaining atlases are shown in Fig. S2. In contrast to standard atlases, random atlases have constant sphericity with respect to volume size. Note that the distribution of parcellation shapes (i.e. sphericity) is similar across parcellation sizes in random atlases and their parcellations may not represent true anatomical or functional boundaries. Thus, random atlases allow us to study how parcellation scale affects network structure and SFC while keeping the effect of shape constant. Crucially, random atlases also allow us to explore if accurate and precise anatomical boundaries are essential in some experimental designs(Albers et al., 2021).
Fig. 2. Atlas morphology: sizes and shapes.
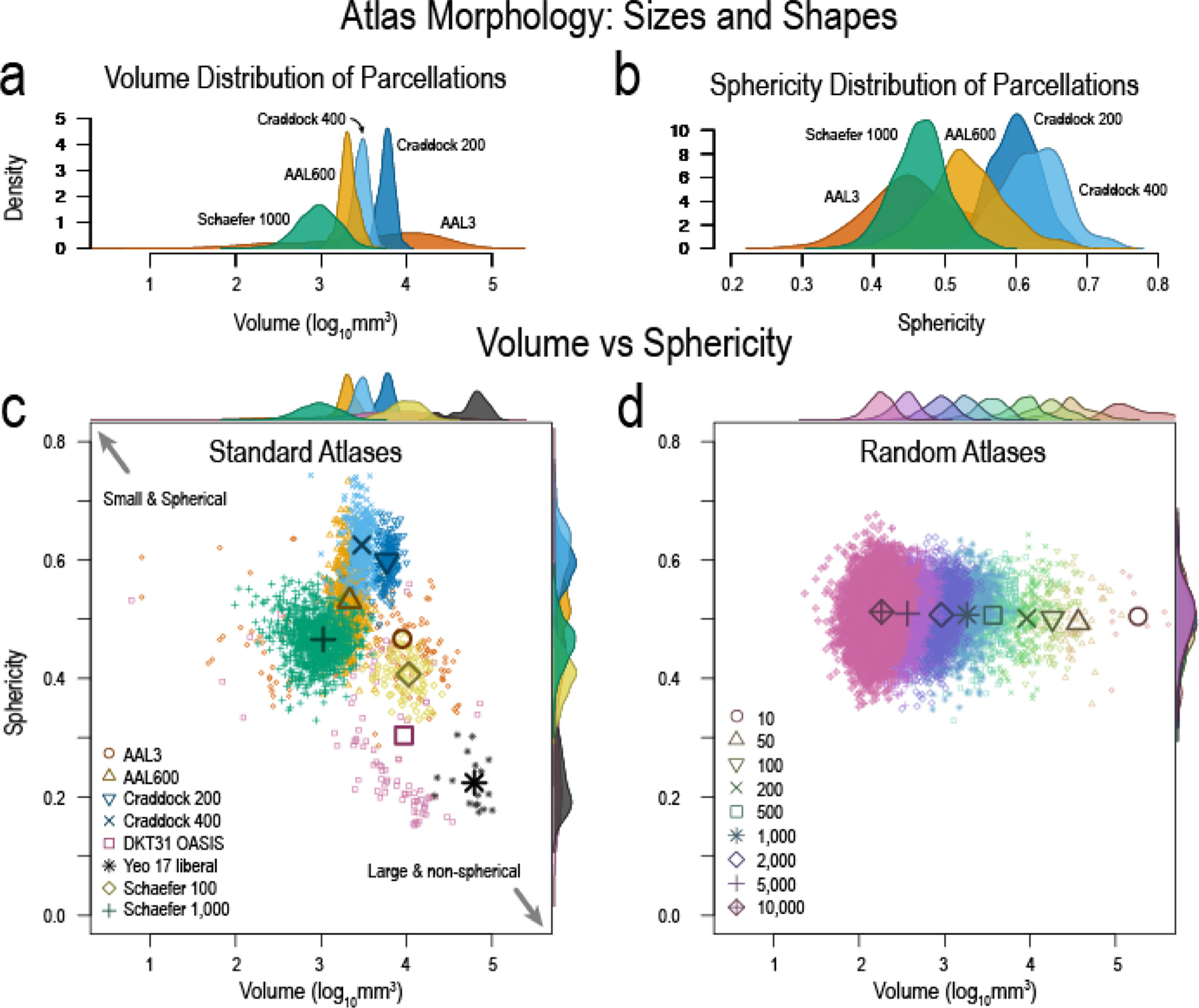
| a, Volume distribution of atlas parcellations demonstrating the diversity of parcellation sizes. b, Parcellation sphericity distributions illustrating how the shapes of different parcellations may not be uniform. c, Volumes versus sphericity showing how some atlas parcellations may be small and spherical, while others may be large and non-spherical. This illustrates the non-uniformity in atlas parcellations. d, Volumes and sphericity of random atlases showing the uniformity of sphericity with changing volumes. Random atlases allow us to study (1) the effect of parcellation scale without the confound of shape effects and (2) the need for accurate anatomical boundaries to test a hypothesis about the structure–function relationship in the brain at seizure onset. Numbers in legend represent the number of parcellations for each random atlas. Remaining atlases are in Fig. S2.
2.3. Varying atlases affect structural network topology
Although the morphology of atlas parcellations is diverse, we aimed to investigate how these morphological characteristics (particularly parcellation scale) affect structural network topology (Fig 3). Networks are the basis upon which we compute SFC, and not necessarily morphological characteristics, therefore, we measured how network density, mean degree, characteristic path length, mean clustering coefficient, and small worldness change as a function of parcellation scale (Fig. 3 a). We found that the change in these network measures are congruent between standard and random atlases and previous studies (Zalesky et al., 2010). We also show that mean density, a global network measure, is similar between our control (N=13) and patient (N=28) cohorts (Fig. 3 b).
Fig. 3. Structural network differences between atlases.
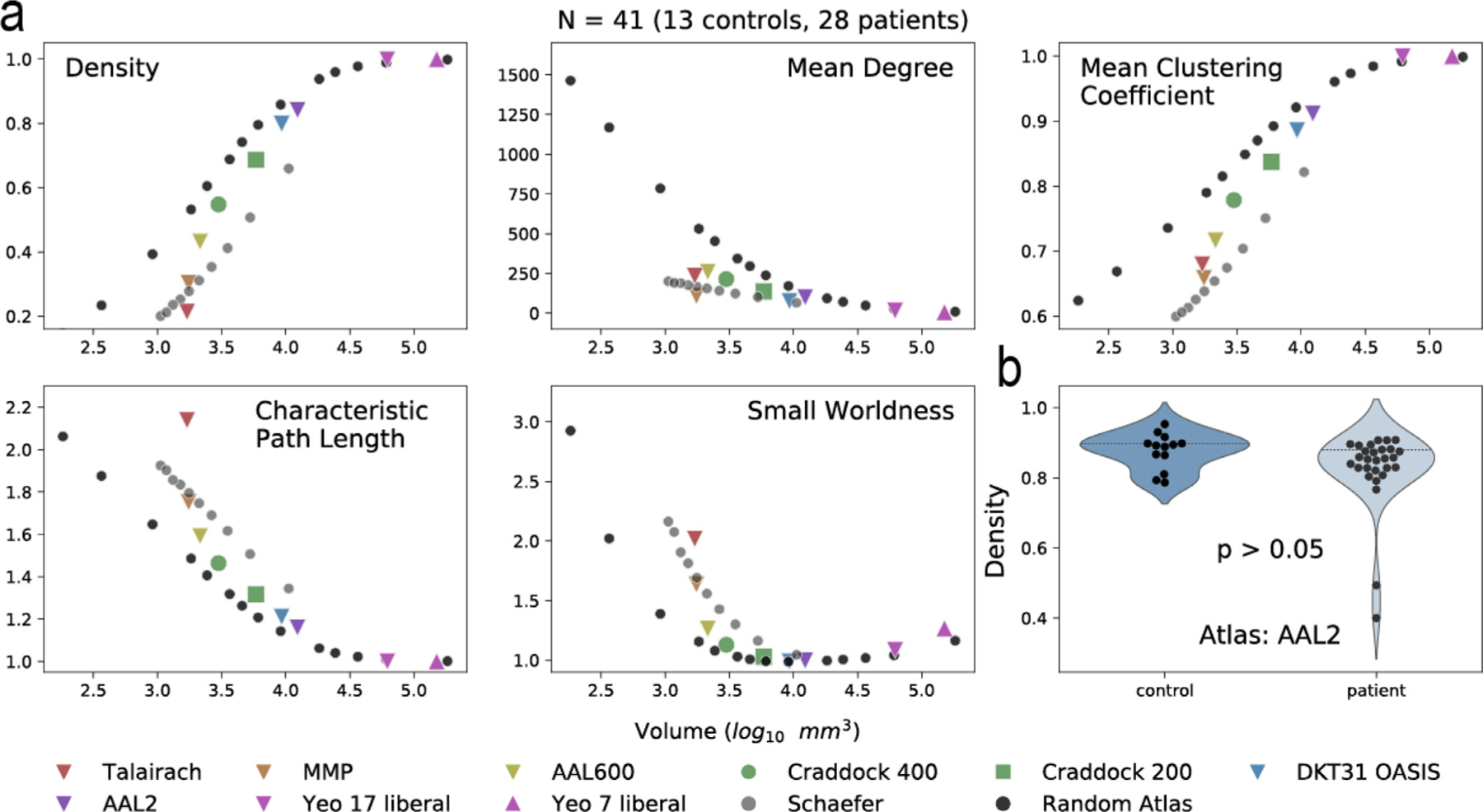
| a, Density, mean degree, mean clustering coefficient, characteristic path length, and small worldness were calculated for structural connectivity networks. A subset of atlases is shown. Remaining atlases studied are shown in Fig. S3. The average parcellation volume was calculated for each atlas and the corresponding network measure was graphed as the mean of all subjects (N=41; 13 controls, 28 patients). b, Controls and patients were not significantly different in density for the AAL2 atlas (Mann-Whitney U test), illustrating that global structural network measures are similar between cohorts. However, specific edge-level connections between cohorts may be different, and characterizing these differences is out of the scope of this manuscript. Controls and patients were separated and shown in Fig. S4. Network measures using different threshold are shown in Fig. S5.
2.4. Varying atlases affect SFC: Single subject
Fig 4 illustrates an overview of how SFC is calculated. Structure is measured with high angular resolution diffusion imaging (HARDI) and function is measured with SEEG electrode contacts. Structural connectivity matrices are generated based on the atlas chosen (Fig. 4 a) and functional connectivity matrices are generated based on broadband (1 – 127 Hz) cross-correlation of neural activity between the electrode contacts in widows of time (Fig. 4 b, see Methods section on ”Functional Connectivity Network Generation”). Thus, the structural network is static while the functional network is computed across time. The connectivity matrices shown are example data from a single patient, sub-patient07. Functional connectivity matrices are shown for 6 hours before seizure onset, 90 s before seizure onset (t = −90), 40 s after seizure onset (t = 40), 88 seconds after seizure onset (seizure duration = 89 s), and 180 seconds after seizure onset (91 s after seizure termination). Each functional connectivity matrix time window was correlated to each structural connectivity matrix, yielding a SFC at each time window (Fig. 4 c). Each point represents the structural edge weight between two brain regions and their corresponding functional connectivity edge weight in broadband cross-correlation. A line of best fit is shown for visualization, and r values represent Spearman rank correlation for that time point. SFC was graphed for all time points during the interictal, preictal, ictal, and postictal periods for this patient in Fig. 4 d.
Fig. 4. Structure–Function correlation in a single patient using different atlases.
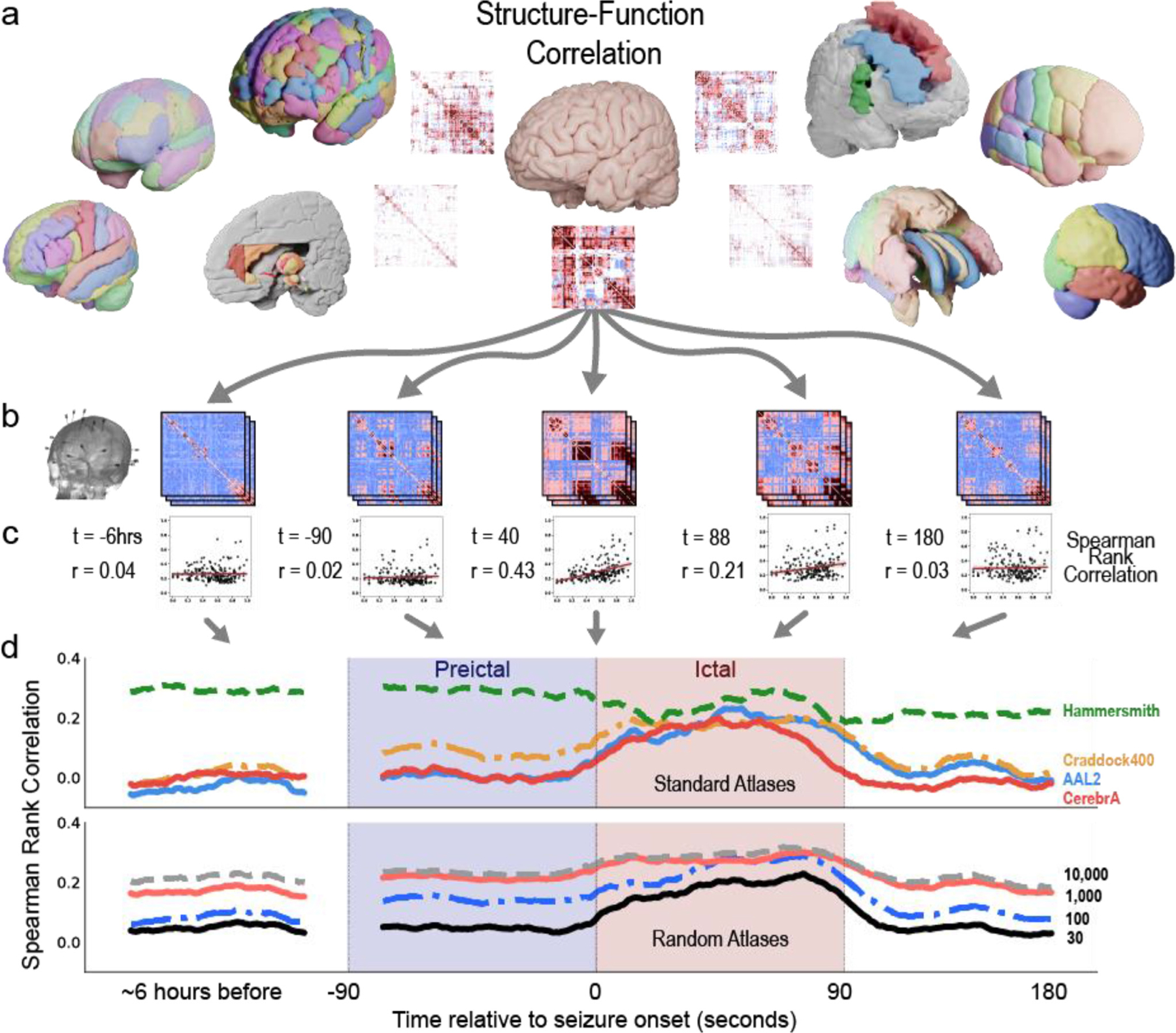
| a, Example atlases and structural connectivity matrices. b, Functional connectivity matrices are computed from SEEG recordings during the interictal, preictal, ictal, and postictal periods. During each period, the SEEG data is binned into non-overlapping windows (the vertically stacked matrices) to create time varying representations of functional connectivity. Broadband cross correlation matrices are shown for sub-patient07 at 6 h before seizure onset, 90 s before seizure onset, 40 s after seizure onset (t = 40), 88 s after seizure onset (seizure duration = 89 s), and 180 seconds after seizure onset (or 91 s after seizure termination). c, Each functional connectivity matrix is correlated to a structural connectivity matrix of a given atlas. Spearman Rank Correlation is measured between all time points and all atlases for each patient. Lines of best fit are for visualization purposes only. d, SFC is graphed at each time point for four example standard atlases (Hammersmith, Craddock400, AAL2, and CerebrA), and four example random atlases (30, 100, 1k, and 10k parcellations). SFC increases during seizure state for some standard atlases (Craddock 400, AAL2, and CerebrA atlases). This result follows previous SFC publications with ECoG (Ashourvan et al., 2021; Shah et al., 2019). However, SFC does not increase for the Hammersmith atlas. These findings highlight that the power to detect a change in the structure–function correlation at seizure onset, and thus the ability to probe the hypothesis that seizure activity is correlated to brain structure, may be reduced using some atlases. The use of different atlases may contradict previous studies.
Four example standard and random atlases are graphed. We show that SFC increases during the ictal state for many atlases (CerebrA, AAL2, Craddock 400), but not all atlases (Hammersmith). The increase in SFC during seizures follows previous SFC studies using ECoG (Ashourvan et al., 2021; Shah et al., 2019). Similarly, SFC increases for a subset of random whole-brain atlases. While parcellation scale may affect SFC, it is not the only feature affecting SFC - the Hammersmith and AAL2 atlases have similar parcellation scales yet diverging neuroanatomical properties and SFC dynamics. These findings highlight inference from one type of atlas may suggest that seizure activity is not correlated to brain structure, contradicting previous studies (Shah et al., 2019).
2.5. Varying atlases affect SFC: Multiple subjects
Fig 5 shows SFC for ten standard atlases and five random atlases using SEEG broadband cross-correlation metrics averaged across all patients with clinically annotated seizures (N = 10). The AAL2 atlas shows a statistically significant increase in SFC from preictal to ictal periods (p < 0.05 by Wilcoxon signed rank test after Bonferroni correction for 55 tests). This change from preictal to ictal SFC is denoted Delta SFC. Using the AAL2 atlas, this finding supports the hypothesis that seizure activity propagates and spreads via axon tracts making up the underlying structural connectivity of the brain (Ashourvan et al., 2021; Shah et al., 2019). SFC was similarly calculated for random whole-brain atlases. A notable finding is that during the interictal period, resting state SFC (rsSFC) increases at larger number of parcellations (i.e. smaller parcellation volumes). We show that rsSFC is observably affected by parcellation scale when plotting the random atlases in Fig 5 (bottom row). These findings may be concerning given that the inherent structure-function relationship in the brain is not necessarily changing at resting state, but its measurement is greatly affected by atlas choice alone.
Fig. 5. Structure–Function Correlation in multiple patients using different atlases.
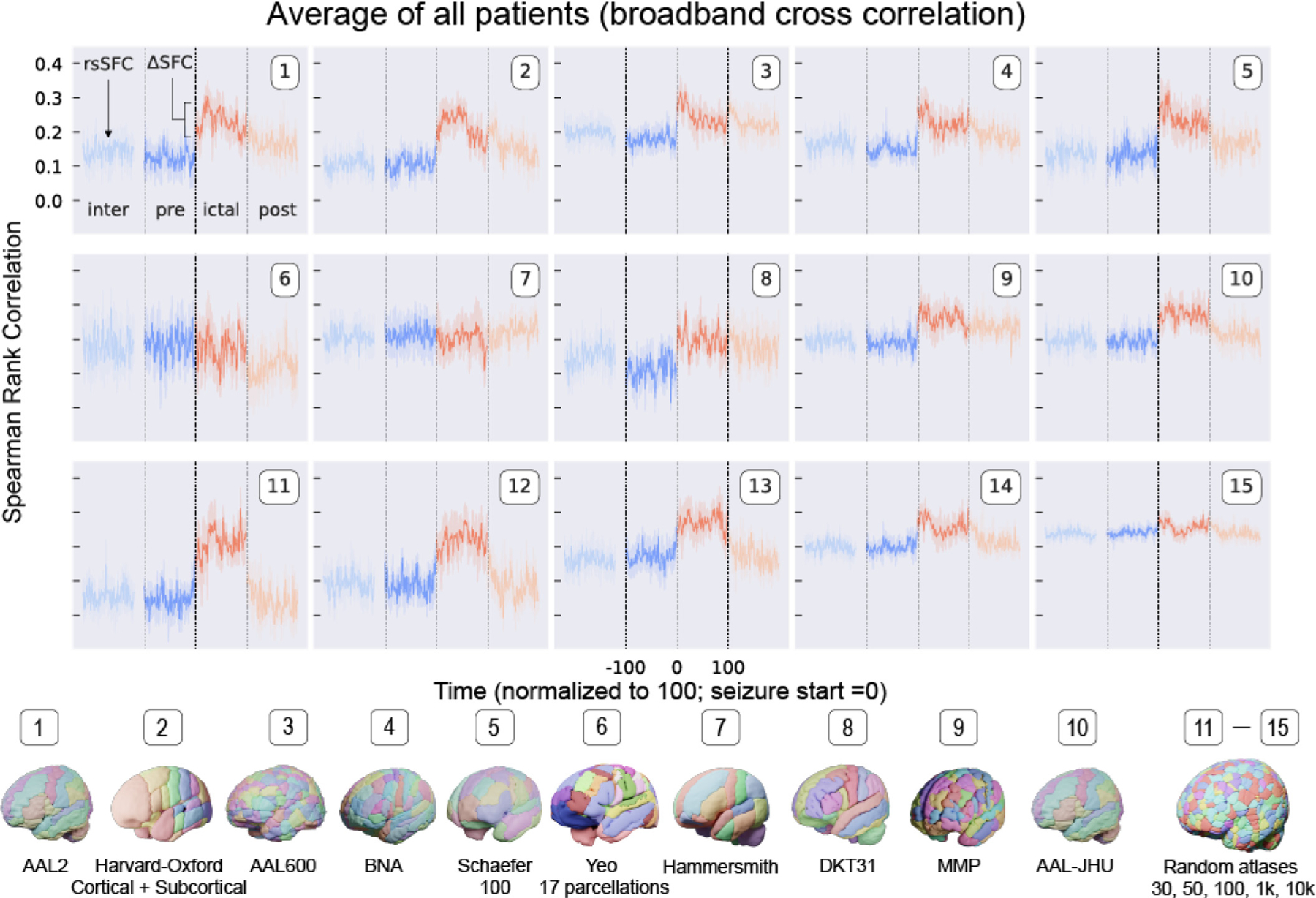
| SFC for ten standard atlases and five random atlases using SEEG broadband cross-correlation matrices averaged across all patients with clinically annotated seizures (N = 10). Resting state SFC (rsSFC) is the SFC during the interictal period. The change from preictal to ictal SFC is ΔSFC. SFC was similarly calculated for random atlases and shows that rsSFC and ΔSFC may change with parcellation scale. These findings may be concerning given that the inherent structure—function relationship in the brain is not necessarily changing at resting state, but its measurement is greatly affected by atlas choice alone.
2.6. Varying atlases affect resting state SFC and ΔSFC
Resting state SFC (rsSFC) and the change in SFC (ΔSFC) from preictal to ictal periods are affected by parcellation scale (Fig 6). Fig. 6 a shows how rsSFC decreases with larger average parcellation volumes (moving left to right). A large average parcellation volume for a given atlas generally means there is a fewer number of total parcellations (e.g. the MNI structural atlas has a large average parcellation volume given only nine parcellations). In contrast, Fig. 6 b shows ΔSFC increases with larger parcellation volumes (moving left to right). Broadly, ΔSFC may be interpreted as the change in SFC with respect to a disease (e.g. a seizure) and non-disease states. This change metric has been used to characterize and make inferences in many neurological disorders (Cocchi et al., 2014; Sathian and Crosson, 2015). Only a subset of atlases show a change in SFC at seizure onset (Fig. 6 c). These results exemplify that either overly coarse or fine parcellations may not adequately capture the underlying SFC of the brain or its dynamics with relation to a neurological disease.
Fig. 6. The power to test a hypothesis about epilepsy pathophysiology changes depending on atlas choice.
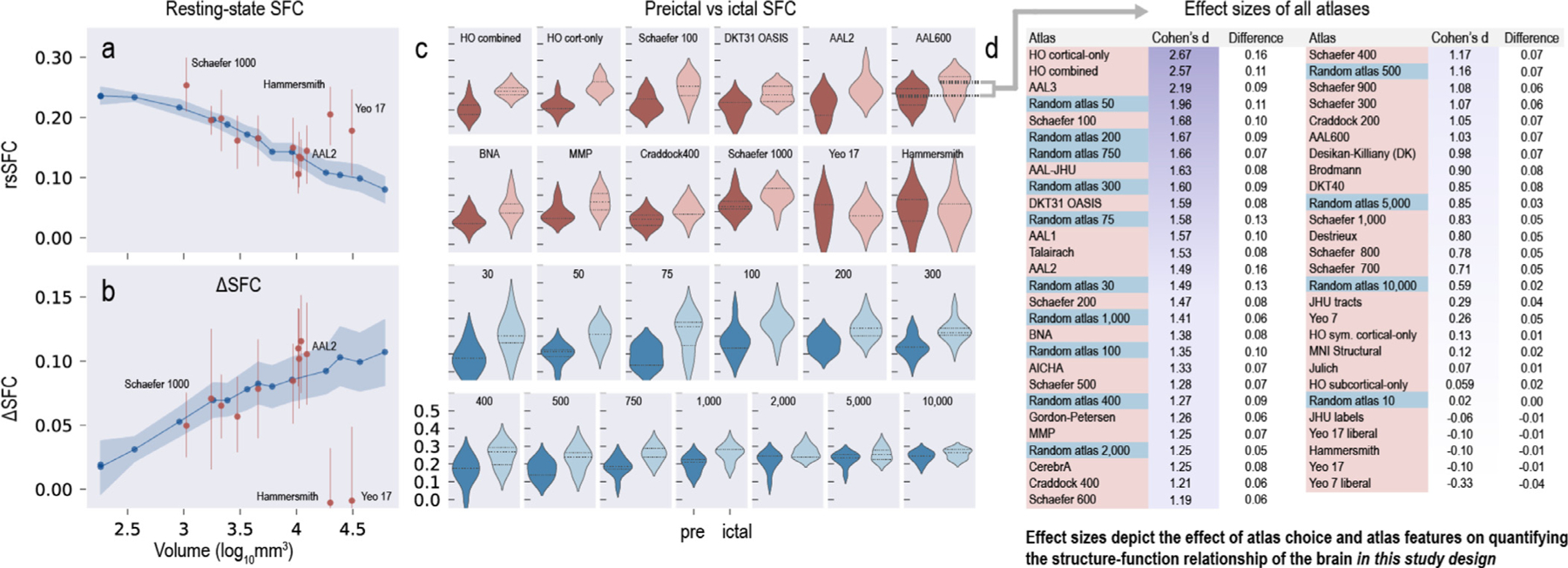
| a, Resting state SFC (rsSFC) decreases with larger parcellation volumes (moving left to right). Random atlases are shown in blue, and select standard atlases are shown in red. Points represent the average across all patients, and bands represent 95% confidence intervals. b, ΔSFC increases with larger parcellation volume (moving left to right). Broadly, DeltaSFC may be interpreted as the change in SFC with respect to disease (e.g. a seizure) and non-disease states, and this change has been used to characterize and make inferences on many neurological diseases. These results exemplify that parcellations that are either too coarse (large volumes) or too fine (small volumes) may not adequately capture the underlying SFC of the brain or its dynamics with relation to a neurological disease. c, A subset of atlases show a difference in preictal and ictal SFC. d, The effect size between preictal and ictal SFC is calculated for all 55 atlases used in this study. Many atlases commonly used in the neuroscience literature have comparable effect sizes to random atlases. The standard atlases with the greatest effect size (and thus power) are the Harvard-Oxford and AAL3 atlases. These atlases outperform many random atlases (where anatomical boundaries are not followed) and may indicate that their parcellation scheme captures the structure–function relationship in the brain at seizure onset with DTI and iEEG.
2.7. Atlas choice affects the power to test a hypothesis
The effect size between preictal and ictal SFC is calculated for all 55 atlases used in this study (Fig. 6 d). Cohen’s d and the difference between the mean ictal and mean preictal SFC are shown. Atlases are ordered by Cohen’s d.
We found that different atlases may alter the power to test the hypothesis about epilepsy pathophysiology that seizures propagate through the underlying structural tracts of the brain, measured with diffusion MRI. This hypothesis has been previously supported in prior studies(Ashourvan et al., 2021; Proix et al., 2017; Shah et al., 2019; Wirsich et al., 2016)
Many atlases commonly used in the neuroscience literature have comparable effect sizes to random atlases (where anatomical boundaries are not followed). The standard atlases with the greatest effect size (and thus power, given equal significance levels and sample sizes) are the Harvard-Oxford and AAL3 atlases. These atlases outperform many random atlases and may indicate that their parcellations may adequately capture the structure–function relationship in the brain. These atlases may capture the “true” structural network architecture (see Fig. 1 c) because these network architectures better differentiate and are more correlated to functional changes seen at seizure onset.
Despite the effect sizes of the Harvard-Oxford and AAL3 atlases, however, there may not be a “true gold standard” atlas or parcellation scheme given that resolution is more critical than the exact border location of parcels(Albers et al., 2021), there may be no single functional atlas for an individual across all brain states(Salehi et al., 2020), and many standard atlases yield similar effect sizes to randomly generated atlases (this study).
3. Discussion
In this study, we performed an extensive evaluation of the available structural, functional, random, and multi-modal atlases in the neuroscience literature (Table 1). We detailed morphological (Fig 2) and network (Fig 3) differences between these atlases. We showed the effect of atlas choice on the measurement of structure-function correlation (SFC) in epilepsy patients (Fig 4 and Fig 5). We also showed how various atlases may affect the power to test a hypothesis about seizure propagation (Fig 6). This work has implications for investigators because the ability to test hypotheses and make predictions about the brain’s function may depend on atlas choice. In light of our study using an extensive list of available brain atlases, we propose a general framework below for evaluating and selecting an atlas (Fig 7).
Fig. 7. A Framework for brain atlases.

| a, Which atlas should be chosen for a study? We propose a framework that helps select an atlas in the context of its descriptive, explanatory, and predictive validity. Descriptive validity means the features of an atlas appropriately resembles the experimental system. An atlas is also a tool to solve a variety of problems in neuroscience. It may be used as part of a methodology to explain causality (explanatory validity), or it may be used to make predictions (predictive validity). These two goals are distinct, and the differences between explanation and prediction ”must be understood for progressing scientific knowledge”Shmueli (2010). These aspects (to explain or to predict) should be considered when selecting an atlas. b, Non-mutually exclusive atlas features related to descriptive validity. c, A list of questions to consider when choosing an atlas. Gray lines connect related questions. d, An algorithm for atlases selection a priori and post hoc. Please see the main text for further details.
3.1. A framework For brain atlases
Various publications have highlighted the Atlas Concordance Problem (Bohland et al., 2009; Dickie et al., 2017; Evans et al., 2012; Mandal et al., 2012), curated several atlases in freely accessible databases (Gorgolewski et al., 2015; Lawrence et al., 2021), and made arguments for why specific atlas features (Fig. 7 b) may be superior in certain situations (Alexander et al., 2017; Brennan et al., 2019; Cabezas et al., 2011; Caspers et al., 2013; Coalson et al., 2018; Diedrichsen et al., 2009; Salehi et al., 2020). There have been great efforts to publish accurate and precise parcellations as seen with an exponential rise in atlas-related publications over the last three decades (Fig. S8). However, none have found a general solution to the underlying problem: Does atlas choice matter?
We provide a framework that allows us to determine if the choice of an atlas is appropriate in the context of its (1) descriptive, (2) explanatory, and (3) predictive validity(Bassett et al., 2018). This framework is borrowed from the logic for assessing network models(Bassett et al., 2018), animal models, (Belzung and Lemoine, 2011; Willner, 1984), and psychometric tests (Association, 2014; 1954), where assessment of these models with standard statistical model-selection methods is particularly challenging. Thus, theoretical constructs already formulated in other fields may provide guidance.
Descriptive validity of an atlas refers to an atlas that appropriately resembles the system in which we work. In other words, it has “face value”Willner (1984). An atlas should include features (Fig. 7 b) relevant to the study (e.g., parcellations containing subcortical structures relevant to epilepsy). Importantly, the descriptive validity of an atlas also relates to the modality scale we use to measure the brain - for example, DWI and fMRI at the macroscale (Sporns et al., 2005), iEEG and tracers at the meso scale (Fornito et al., 2016), and microscopy at the microscale (Sporns, 2011). It is important to select a parcellation scale that resembles the measurement modality resolution (Fig. 6 a). When correlating DWI with iEEG in our study at larger parcellation sizes, we lose our ability to discern precise anatomical locations that are structurally and functionally related (Fig 6 b. Similarly at smaller parcellation sizes (tending to voxel resolution), we may not capture the “true” structural network architecture (Fig. 1 c), and thus we lose our ability to capture structure–function relationship changes at seizure onset.
An atlas is a tool to tackle a wide variety of problems in neuroscience. It may be part of a methodology to explain causality (explanatory validity) or it may be part of a methodology to make predictions (predictive validity). These two goals are distinct, and the differences between explanation and prediction “must be understood for progressing scientific knowledge” as described in “To Explain or Predict?” by Shmueli, 2010(Shmueli, 2010). In the context of building scientific models, a model with a high explanatory ability may not have a high predictive ability.
Similar to models, atlases are also part of a scientific methodology to (1) explain how the brain functions or (2) predict new observations (i.e., they are one part of the overall methodological pipeline to test hypotheses or make predictions about the brain - for studies using atlases). Thus, atlases are tools. An atlas may be suitable for hypothesis testing, for example, because it includes subcortical structures like the hippocampus (also high descriptive validity) to support a hypothesis about seizure propagation through subcortical structures. Intuitively, without subcortical structures, it would be impossible to test hypotheses about subcortical structures. Less intuitively, explanatory validity of an atlas may also relate to the power to test hypotheses, which we show in our study. Some atlases may not be suitable for scientific inquiry because they provide little statistical power to detect differences in disease states, for example, to detect changes in SFC at seizure onset (Fig. 6 b). It may be impossible to accurately predict power using an atlas before conducting a study, however, other studies asking similar questions using similar atlases may provide reasonable estimates of effect sizes (our study has similar effect sizes to a previous study(Shah et al., 2019)). Power may also depend on the accuracy of anatomical boundaries, or in our study, other atlas features such as parcellation scale and configuration (Fig. 6 d). For example, the Harvard-Oxford and AAL3 atlases have similar parcellation configurations and similar power.
Some atlases may or may not be not be suitable for making predictions about new or future observations about the brain. For example, many network properties change with atlas choice (Fig. 3), and thus it is reasonable to suspect model prediction outputs may change with respect to the atlas used to build and train such models. Importantly, the exclusion of some anatomical structures, like white matter or the cerebellum in some atlases, may affect the training data used to build predictive models. In our study, a translational goal is to predict functional seizure activity from structural data. SEEG records activity from both gray matter and white matter; however, recent studies have shown that white matter functional recordings may provide different information than gray matter(Greene et al., 2021; Mercier et al., 2017; Revell et al., 2021; Young et al., 2019). Thus, excluding some anatomical labels may affect model predictions. Another example is the use of network models to predict spread, such as α-synuclein across the brain connectome (Henderson et al., 2019). Without the incorporation of all brain structures related to α-synuclein spread, models to predict and monitor spread may be inaccurate.
3.2. Are accurate anatomical or functional parcellations needed?
During the course of conducting this study, and while undergoing peer review, other atlases with more accurate or relevant parcellations to the study’s population were published in different areas of neuroscience(Callaway et al., 2021; Doucet et al., 2021; Huang et al., 2021; Joglekar et al., 2021; Lewis et al., 2021; Muñoz-Castañeda et al., 2021; 2021; Syversen et al., 2021; Wang et al., 2021; Zhu et al., 2021). Here, we cautiously propose a question: Are efforts to publish more atlases created with different algorithms or slightly modified parcellations from existing atlases providing any advantages over already existing atlases? Naturally, accurate and precise parcellations are needed when probing specific hypotheses about exact structures that depend on accurate segmentation of such structures (particularly at the sub-field or cellular level); however, few studies compare an atlas to a null atlas (one with randomly generated parcellations). Studies that do are Gordon et al. 2016Gordon et al. (2016)and Lewis et al. 2021Lewis et al. (2021).
In this study, we show that random atlases provide similar power to detect differences in SFC between preictal and ictal states (Fig. 6 d). Indeed, it is difficult or nearly impossible to evaluate a newly proposed atlas, given that the performance metrics to evaluate an atlas may be infinite (given infinite experimental designs). Only one such metric, SFC, was used in this study. But given new deep learning methods and other computationally expensive methods using trained classifiers for segmentation, existing atlases may be adequate for labs with limited funding resources, trained personnel, and access to GPUs. These labs may still be capable of answering important questions in neuroscience.
3.3. Which atlas should be used for my study?
One of the most difficult challenges as scientific investigators is to make optimal methodological decisions to discover useful findings for the scientific community. Selecting an atlas is one such decision we may make in some of our studies. We realize the framework provided above may be abstract to some readers; we also provide a concrete list of questions to consider when choosing an atlas (Fig. 7 c) for a neuroimaging study. However, in conducting this study, we also found that researchers may face three problems when choosing an atlas (Fig. 7 d) and these problems are worth further discussion. The first two problems are in selecting an atlas a priori, or before conducting a study. They deal with selecting one or a few atlases to preserve power, or in selecting a standard set of atlas to publish public data for other researchers to use. The third problem is the issue of conflicting results between two atlases and what to do after a study is conducted (post hoc). We provide a further discussion on these problems below.
3.4. Considerations in selecting one or a few atlases
Selecting one atlas may preserve power and avoid a multiple comparisons problem by testing every atlas. Selecting an additional atlas may also be chosen to confirm the robustness of results. In these cases, a balance of time, availability of tools, and atlas features logical for your study as outlined in Fig. 7 a-c need to be considered. For example, if a custom atlas is used, how will that affect replicability and meta analysis in the long-run for the field? What are the atlas features needed (such as scale and coverage of regions)? What are the computational costs and personnel training needed to use particular atlases? (See questions in Fig. 7 c).
3.5. Considerations in selecting a standard set of atlases
When publishing results and/or making data publicly available for other investigators to use, another approach is to select a set of atlases based on the perceived needs of other investigators, atlas features covered, prevalence of atlases used in the literature (Fig. S9a), and dummyTXdummy-(the prevalence of “turn-key” neuroimaging software that incorporate these atlases (Fig. S9b). Studies are emerging with data publicly available for use based on one or a few select atlases(Royer et al., 2021; Sinha et al., 2021). Many turn-key neuroimaging software also inevitably have to make the decision to employ a set of atlases to meet the needs of many researchers. A problem may arise, however, when other researchers need the published data at other atlas resolutions or with other structures. And unfortunately, the value of the data may be lessened and the effort put in by the publishing researchers may be in waste if this happens. What may help with the atlas concordance problem is perhaps a “standard set” of atlases – a set to benchmark studies across the neuroimaging field. Furthermore, turn-key tools like FreeSurfer, QSIprep, DSI-studio, FSL, and many others may benefit from a standard set of incorporated atlases that captures enough features useful to the majority of the neuroscience community, even if not every available atlas is included. Based on our exhaustive search of atlases in the neuroimaging literature, the ability to collect them for use in a single study, the prevalence of certain atlases already in-use (Fig. S9a), and the prevalence of neuroimaging software (Fig. S9b) we propose an initial set of atlases (Fig. 7 d).
The AAL atlas is one of the most commonly used volumetric atlases (Fig. S9a), and along with the Harvard-Oxford atlas, may provide complimentary results when published together. The Brainnetome atlas(Fan et al., 2016) is another structural atlas at a finer resolution, having gained popularity since its introduction in 2016. The Destrieux and DKT atlases are also structural atlases, and already incorporated into one of the most commonly used neuroimaging software, FreeSurfer (https://surfer.nmr.mgh.harvard.edu). FreeSurfer provides surface-based registration, which may more accurately label cortical structures than volumetric registration (Fig. S6). Accurate segmentation of sub-cortical structures may also be acquired from FSLPerlaki et al. (2017) (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki). In addition, the MMP, or “Glasser” atlas was created from multi-modal imaging data. A commonly used atlas provided at different scales is Schaefer atlases provide, however, it does not include subcortical structures.
Random atlases may also provide robust conclusions by allowing researchers to manipulate the resolution, size, and shape of parcellations and iterate over many atlases. Although random parcellations may forgo accuracy because they do not follow true anatomical boundaries, these atlases may still provide similar conclusions to other standard atlases with the added benefit of permuting results over many atlases (Fig 6). An alternative to random atlases is to divide or combine the parcellations of another standard atlas (a “derived” atlas in Fig. 7 d. For example, the AAL 600 is derived from the AAL atlas in which its parcellations are further sub-divided using a specified algorithm. Parcellations may also be sub-divided randomly.
3.6. Considerations in conflicting results between atlases
When more than one atlas is used, results may conflict. We define conflicting results as two different atlases giving alternating predictions (e.g., good vs poor outcomes, increase in SFC rather than decrease in SFC) or support alternating working hypotheses (e.g., the temporal lobe is involved in one atlas, but another atlas highlights the involvement of the frontal lobe in the pathophysiology of a disease). We do not mean that conflicting results arise due to lack of statistical power (e.g., one atlas gives a p-value of 0.06 and another atlas 0.04).
One way to understand if the observed effect is not an artifact of the atlas choice is to select a few atlases with varying features and figure out what is causing the conflict. Unfortunately, there may be no other way given that every study will have different parameters and measurements to know what gives rise to conflicting results. In the matter where conflicting results arise due to atlas selection, then it may troubleshooting may be needed to understand what gives rise to the conflict (surface vs volumetric registration, parcellation scale, missing relevant structures, etc.). Fortunately, however, most atlases in this study affect power rather than conflicting results (Fig. 6 d. We hope this discussion, our study, and our figures provide insight to others.
3.7. Limitations
Our study is not without limitations. A major limitation is that we did not evaluate atlases in a diverse set of experimental systems, but rather limited our analysis to a contemporary topic in epilepsy using SEEG implantations and to a study of the structure–function of the brain, potentially appealing to a wider audience. The question we were trying to answer (“Which atlas should we use?”) is a difficult problem to solve, given that it would be impossible to evaluate all atlases in all experimental designs. We attempted to generalize a framework given our findings after an extensive search for, and curation of, available neuroimaging atlases.
We also did not perform a feature selection analysis post-hoc to maximize Delta SFC at seizure onset; rather, we performed a comprehensive evaluation of many atlases to set a general framework and describe the nuances between the different atlases and their features. Ideally in our study, we required a whole-brain, volumetric atlas that covered the implanted SEEG electrode contacts. No such atlas existed. We opted for combining different atlases or developing randomly parcellated atlases used in previous publications (Mišić et al., 2015; Zalesky et al., 2010). However, no general framework existed to determine which atlas should be used or clearly outlined the feature space of these atlases. We had no formal basis for how changing an atlas could change our results and eventual goal for translating network models to better treat epilepsy patients.
Another limitation, we assume a change in SFC supports the hypothesis that seizures harness the underlying structural connectome of the brain (along with support from prior literature (Ashourvan et al., 2021; Betzel et al., 2019; Shah et al., 2019)). We may be biasing our results to select an atlas that maximizes Delta SFC. However, we wish to select a methodology that allows us to measure any change in brain state that accompanies seizure onset (explanatory validity), permitting us to probe epilepsy biology and understand the processes that govern seizure spread.
An additional limitation concerns the effect of parcellation volume on SFC. In probing this effect across our random atlases and atlases used in the literature, we did not perform controlled experiments to separate the effects of parcellation size from parcellation N (number of parcellations). A future experiment could fix the number of parcellations while changing parcellation volume (or vice versa). This would allow us to test whether parcellation volume or N drives changes in SFC. However, this was outside the scope of our study.
Our goal was to highlight the importance of selecting an appropriate atlas from an array of possibilities, using a data-driven, validated experimental paradigm(Shah et al., 2019). We acknowledge new studies that show that streamline counts may not completely reflect the underlying diffusion data(Smith et al., 2015); however, comparing such techniques were outside the scope and goal of our focused study. We also note that few patients had lesions on imaging. Misalignment due to non-linear distortion may add noise to our data; however, few patients had lesions. Our study was not conducted to necessarily make the claim that SFC changes exist in the brain at seizure onset, but rather to show how varying atlases may change SFC.
Finally, our analysis relies on the assumption that an atlas approach must be used to quantify SFC and does not consider an atlas-agnostic approach nor if such an approach is appropriate. To study SFC using networks, both structural and functional networks must have nodes representing the same entity - neuroanatomical structures. The atlases defining anatomical structures (whether they are functionally, histologically, genetically, procedurally, multi-modally, or randomly defined) are the link between structural connectivity and functional connectivity measurements of the brain. To study SFC, we must rely on the neuroanatomical structures defined by an atlas, then localize electrodes to these regions and correlate the structural measurements (e.g., streamlines, fractional anisotropy, mean diffusivity) with functional measurements (e.g., cross-correlation, coherence, mutual information). Fundamentally, we are defining the nodes of the brain in advance, which can alter our results; a more comprehensive discussion on defining the nodes of the brain are in Fornito et al., 2016 and Bijsterbosh et al., 2017 Bijsterbosch et al. (2017); Fornito et al. (2016).
3.8. Conclusion
The publication of atlases and their distribution across neuroimaging software platforms has risen exponentially over the last three decades. Our study illustrates the critical need to evaluate the reproducibility of neuroscience research using atlases published alongside tools and analysis pipelines already established in the neuroscience community (e.g., FreeSurfer, DSI studio, FSL, SPM, QSIprep, fMRIprep, MRIcron, ANTs, and others).
4. Materials and methods
4.1. Human dataset
MRI data was collected from forty-one individuals, including thirteen healthy controls and twenty-eight drug-resistant epilepsy patients at the Hospital of the University of Pennsylvania. Twenty-four patients underwent stereoelectroencephalography (SEEG) implantation and four underwent electrocorticography (ECoG) implantation. Ten of the SEEG patients had clinically annotated seizures and were used for SFC analyses. Inclusion criteria consisted of all individuals who agreed to participate in our research scanning protocol, and (if they had implantations) allowed their de-identified intracranial EEG (iEEG) data to be publicly available for research purposes on the International Epilepsy Electrophysiology Portal (https://www.ieeg.org) Kini et al. (2016); Wagenaar et al. (2013). Seizure evaluation was determined via comprehensive clinical assessment, which included multimodal imaging, scalp and intracranial video-EEG monitoring, and neuropsychological testing. This study was approved by the Institutional Review Board of the University of Pennsylvania, and all subjects provided written informed consent prior to participating. See Table S2 for subject demographics.
4.2. Structure
Methods and pipelines for structural connectivity generation and analysis are described in the following sections. Specific GitHub files and code are included where applicable.
4.3. Imaging protocol
Prior to electrode implantation, MRI data were collected on a 3T Siemens Magnetom Trio scanner using a 32-channel phased-array head coil. High-resolution anatomical images were acquired using a magnetization prepared rapid gradient echo (MPRAGE) T1-weighted sequence (repetition time = 1810 ms, echo time = 3.51 m, flip angle = 9, field of view = 240 mm, resolution = 0.94×0.94×1.0 mm3 ). High Angular Resolution Diffusion Imaging (HARDI) was acquired with a single-shot EPI multi-shell diffusion-weighted imaging (DWI) sequence (116 diffusion sampling directions, b-values of 0, 300, 700, and 2000 s/mm2, resolution = 2.5×2.5×2.5 mm3, field of view = 240 mm). Following electrode implantation, spiral CT images (Siemens) were obtained clinically for the purposes of electrode localization. Both bone and tissue windows were obtained (120 kV, 300 mA, axial slice thickness = 1.0 mm)
4.4. Diffusion weighted imaging (DWI) preprocessing
HARDI images were subject to the preprocessing pipeline, QSIPrep, to ensure reproducibility and implementation of the best practices for processing of diffusion images (Cieslak et al., 2021). Briefly, QSIPrep performs advanced reconstruction and tractography methods in curated workflows using tools from leading software packages, including FSL, ANTs, and DSI Studio with input data specified in the Brain Imaging Data Structure (BIDS) layout.
4.5. Structural network generation
DSI-Studio (http://dsi-studio.labsolver.org, version: December 2020) was used to reconstruct the orientation density functions within each voxel using generalized q-sample imaging with a diffusion sampling length ratio of 1.25 (Fang-Cheng et al., 2010). Deterministic whole-brain fiber tracking was performed using an angular threshold of 35 degrees, step size of 1 mm, and quantitative anisotropy threshold based on Otsu’s threshold (Otsu, 1979). Tracks with length shorter than 10 mm or longer than 800 mm were discarded, and a total of 1,000,000 tracts were generated per brain. Deterministic tractography was chosen based upon prior work indicating that deterministic tractography generates fewer false positive connections than probabilistic approaches, and that network-based estimations are substantially less accurate when false positives are introduced into the network compared with false negatives (Zalesky et al., 2010). To calculate structural connectivity, atlases listed in Table 1 were used. Structural networks were generated by computing the number of streamlines passing through each pair of structural regions in each specific atlas. Streamline counts were log-transformed and normalized to the maximum streamline count, as is common in prior studies (Bonilha et al., 2015; Park et al., 2017; Taylor et al., 2018; Wirsich et al., 2016). GitHub: packages/imaging/tractography/tractography.py
4.6. Atlases
Atlas descriptions and sources used in this study are found in Table S1. The 55 atlases used are listed explicitly in the reporting of effect sizes in Fig. 7 d. All atlases were sourced in MNI space and if not already, resliced to dimensions 182×218×182. Atlases were linear and non-linear registered to T1w subject space using the ICBM 2009c Nonlinear Asymmetric template (Fonov et al., 2011) and FSL flirt and fnirt(Jenkinson et al., 2012).
We also included three atlases registered using surface-based approaches. These atlases (the DKT, DK, and Destrieux atlases) are output from FreeSurfer’s recon-all pipeline(Dale et al., 1999). Many neuroimaging studies and software use volumetric approaches for registration(Coalson et al., 2018), yet surface-based approaches may yield more accurate labeling of the cortical surface (Fig. S6). The DKT40 atlas referred in this study is the surface version, while the DKT31 OASIS is the publicly available volumetric version (see Table S1).
In addition to published standard atlases above, we used whole-brain random atlases. A limitation of standard atlases is that they may not have anatomical definitions for all regions of the brain, and therefore, implanted electrodes may not be assigned properly to a region. This limitation was the impetus of our study (i.e., selecting an appropriate atlas for SEEG electrode localization and quantifying SFC). Whole-brain random atlases, in contrast, provide coverage to all implanted electrodes. They allow for the ability to change some morphological properties (i.e. parcellation size), while keeping other morphologies the same (e.g., parcellation shape; Fig. 2 d). However, a limitation of random atlases is that their regions may not represent true anatomical or functional boundaries. Random atlases were built in the ICBM 2009c Nonlinear Asymmetric template space and covered all voxels, excluding those labeled as CSF or outside the brain. To fill these points, a pseudo grassfire algorithm was applied (Zalesky et al., 2010). Briefly, N points representing the number of parcels of the atlas were randomly chosen as seed points. These seed points were iteratively expanded in all six Cartesian directions until all points were covered by one of the initial N seeds. After each iterative step, the smallest volume region expanded first. Random atlases created were of N equal to 10, 30, 50, 75, 100, 200, 300, 400, 500, 750, 1000, 2000, 5000, and 10,000 parcels. Five permutations for each N were created. GitHub code to generate random atlases: packages/imaging/randomAtlas/randomAtlasGeneration.py
4.7. Atlas morphology: volume and sphericity
Atlas morphological measurements included parcellation size (volume) and shape (sphericity) (Fig 2). Parcellation volume was calculated as the number of voxels in an parcel and log10 transformed. Parcellation sphericity was calculated as the ratio of the surface area of a sphere with an equal volume of the parcellation to the actual surface area of the atlas parcellation. Under this definition, sphericity is bounded from 0 to 1 where 1 is a perfect sphere. For reference, a perfect cube and a hemi-sphere have a sphericity of 0.8 and 0.7 respectively. GitHub: packages/imaging/regionMorphology/regionMorphology.py
4.8. Structural network measures
We characterized the structural network topology of 52 atlases (Fig 3 and Fig. S3). The three surface-based atlases (DKT40, DK, and Destrieux atlases output from the FreeSurfer recon-all pipeline(Dale et al., 1999)) were excluded from analyses of Fig 3 and Fig 3 because they were individually registered to each subjects’ T1w image. To quantify network topology, we examined density, mean degree, mean clustering coefficient, characteristic path length, and small worldness. Connectivity matrices were first binarized, using a threshold of 0, and a distance matrix was computed. The same binarization process and threshold was used across all atlases. The distance of any nodes that were disconnected from the main graph was set to the maximum distance between any pair of nodes in the main graph. Density, mean degree, clustering coefficient, and characteristic path length were then calculated on the binary, undirected graphs. Small worldness was calculated as the s-ratio where s = g/I and is the ratio of the average, normalized clustering coefficient, C, to the normalized characteristic path length, I. g = CG/CR and l = 1G/lR where G is the graph of interest and R represents a “random’ graph that is equivalent to G. To approximate the equivalent random graph R due to intractable computational costs (Maslov, 2002), a well-known analytical equivalent CR = d/N and IR = log N/log d were used, where d denotes average nodal degree. All network measures were calculated using the Brain Connectivity Toolbox for Python. GitHub: papers/brainAtlas/Script_05_structure_02_network_measures.py
4.9. Function
Methods and pipelines for functional connectivity generation and analysis are described in the following sections. Specific GitHub files and code are included where applicable.
4.10. Intracranial EEG acquisition
Stereotactic Depth Electrodes were implanted in patients based on clinical necessity. Continuous SEEG signals were obtained for the duration of each patient’s stay in the epilepsy monitoring unit. Intracranial data was recorded at either 512 or 1024 Hz for each patient. Seizure onset times were defined by the unequivocal onset (Litt et al., 2001). All annotations were verified and consistent with detailed clinical documentation. If a patient had more than one seizure annotated, the first seizure longer than 30 seconds without artifacts was used.
4.11. Electrode localization
In-house software (Azarion et al., 2014) was used to assist in localizing electrodes after registration of pre-implant and post-implant neuroimaging data. All electrode coordinates and labels were saved and matched with the electrode names on IEEG.org. All electrode localizations were verified by a board-certified neuroradiologist (J.S.). Electrode contact assignment to atlas region assignment was performed by rounding electrode coordinates (x,y,z) to the nearest voxel and indexing the given atlas at that voxel in the same space as the patient’s T1w image. Electrodes that fell outside the atlas of interest were excluded from subsequent analysis. Please see Fig. S10 for visualization. We also show the percentage of contacts assigned a region given an atlas (Fig. S7) GitHub: packages/atlasLocalization/atlasLocalization.py
4.12. Functional connectivity network generation
Functional connectivity networks were generated from four periods: interictal, preictal, ictal, and postictal. (1) The interictal period consisted of the time approximately 6 hours before the ictal period. (2) The preictal period consisted of the time immediately before the ictal period. (3) The ictal period consisted of the time between the seizure unequivocal onset and seizure termination. (4) The postictal period consisted of the time immediately after the ictal period. Interictal, preictal, and postictal periods were 180 seconds in duration. Following removal of artifact-ridden electrodes, SEEG signals inside either GM or WM for each period were common-average referenced to reduce potential sources of correlated noise (Ludwig et al., 2009). Next, each period was divided into 2 s time windows with 1 s overlap (Khambhati et al., 2017; 2016; 2015; Kramer et al., 2010). To generate a functional network representing broadband functional interactions between SEEG signals (Fig. 4 b), we carried out a method described in detail previously (Khambhati et al., 2016; Shah et al., 2019). Namely, signals were notch-filtered at 60 Hz to remove power line noise, low-pass and high-pass filtered at 127 Hz and 1 Hz to account for noise and drift, and pre-whitened using a first-order autoregressive model to account for slow dynamics. Functional networks were then generated by applying a normalized cross correlation function ρ between the signals of each pair of electrodes within each time window, using the formula:
where x and y are signals from two electrodes, k is the 2 s time window, t is one of the T samples during the time window, and τ is the time lag between signals, with a maximum lag of 0.5 s. Here, σ represents the standard deviation of the signal. Note that functional connectivity measurements were also calculated for coherence and zero time-lag Pearson and Spearman rank correlations with associated p-values in defined frequency bands reviewed in Newson and Thiagarajan 2019 (Newson and Thiagarajan, 2019), but were not analyzed or used in hypothesis testing in the study. For data, available data, please see “Data availability and Reproducibility” section below. Networks are represented as fully-weighted connectivity matrices. GitHub Code: GitHub:code/tools/echobase.py
4.13. Structure–function correlation
To quantify the relationship between structure and function in the epileptic brain, we computed the Spearman rank correlation coefficient between the edges of the structural connectivity network and the edges of the functional connectivity networks (Fig. 4 c). To avoid redundancy given the symmetric nature of the matrices, only the upper triangle was analyzed. In brief, the structural connectivity network, representing normalized streamline counts between each atlas region, was first down sampled to only include regions that contained at least one SEEG contact Fig. S10. This gave one static representation of structural connectivity. In the case where multiple electrodes fell in the same atlas region, a random electrode was selected to represent the functional activity of that neuroanatomically defined region. Next, for every time-window of the functional network, the functional network edges were correlated with the down sampled, static structural network edges. This resulted in a structure–function correlation time series. Note that atlases with very small region volumes included more electrodes for SFC calculation. Electrodes that did not localize to an atlas were excluded from analysis. To average the SFC for all patients and each atlas (Fig 5), SFC time-series was resampled to 100 s for each period and each sample was averaged together. GitHub code: packages/eeg/echobase/echobase.py
4.14. Rssfc and Delta SFC
Resting-state SFC (rsSFC) was defined as the SFC during the interictal period, approximately 6 hours before the ictal period. The mean SFC of that period was computed. Delta SFC was defined as the change in the mean SFC from the preictal to the ictal period (Fig 5 top left panel). rsSFC and Delta SFC was calculated for each atlas (Fig 6).
4.15. Statistics
Preictal and ictal SFC for each atlas were compared using effect sizes across the 55 atlases shown in Fig. 6 d. Cohen’s d and the difference between preictal and ictal SFC was calculated.
4.16. Data availability and reproducibility
All code files used in this manuscript are available at https://github.com/andyrevell/revellLab. All de-identified raw and processed data (except for patient MRI imaging) are available for download by following the links on the GitHub. Raw imaging data is available upon reasonable request from Principal Investigator K.A.D. iEEG snippets used specifically in this manuscript are also available, while full iEEG recordings are publicly available at https://www.ieeg.org. The Python environment for the exact packages and versions used in this study in contained in the environment directory within the GitHub. The QSIPrep docker container was used for DWI preprocessing.
Supplementary Material
Acknowledgments
We thank Adam Gibson, Carolyn Wilkinson, Jacqueline Boccanfuso, Magda Wernovsky, Ryan Archer, Kelly Oechsel, members of Andrew’s Thesis Committee, and all other members and staff of the Center for Neuroengineering and Therapeutics for their continued help and support in this work.
5. Funding
This work was supported by National Institutes of Health grants 5-T32-NS-091006–07, 1R01NS116504, 1R01NS099348, 1R01NS085211, and 1R01MH112847. We also acknowledge support by the Thornton Foundation, the Mirowski Family Foundation, the ISI Foundation, the John D. and Catherine T. MacArthur Foundation, the Sloan Foundation, the Pennsylvania Tobacco Fund, and the Paul Allen Foundation.
Footnotes
Data and Code Availability Statement
All code files used in this manuscript are available at https://github.com/andyrevell/revellLab. All de-identified raw and processed data (except for patient MRI imaging) are available for download by following the links on the GitHub. Raw imaging data is available upon reasonable request from Principal Investigator K.A.D. iEEG snippets used specifically in this manuscript are also available, while full iEEG recordings are publicly available at https://www.ieeg.org. The Python environment for the exact packages and versions used in this study in contained in the environment directory within the GitHub. The QSIPrep docker container was used for DWI preprocessing.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at 10.1016/j.neuroimage.2022.118986
Declaration of Competing Interest
The authors declare no competing interests.
Credit authorship contribution statement
Andrew Y. Revell: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Alexander B. Silva: Methodology, Software, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. T. Campbell Arnold: Software, Data curation, Methodology, Writing – review & editing. Joel M. Stein: Supervision, Data curation, Methodology, Writing – review & editing. Sandhitsu R. Das: Supervision, Data curation, Methodology, Writing – review & editing. Russell T. Shinohara: Methodology, Writing – review & editing. Dani S. Bassett: Methodology, Writing – review & editing. Brian Litt: Supervision, Project administration, Resources, Funding acquisition, Data curation, Writing – review & editing. Kathryn A. Davis: Supervision, Project administration, Resources, Funding acquisition, Data curation, Writing – original draft, Writing – review & editing.
References
- Albers KJ, Ambrosen KS, Liptrot MG, Dyrby TB, Schmidt MN, Morup M, 2021. Using connectomics for predictive assessment of brain parcellations. Neuroimage 238, 118170. doi: 10.1016/j.neuroimage.2021.118170. [DOI] [PubMed] [Google Scholar]
- Alexander B, Murray AL, Loh WY, Matthews LG, Adamson C, Beare R, Chen J, Kelly CE, Rees S, Warfield SK, Anderson PJ, Doyle LW, Spittle AJ, Cheong JLY, Seal ML, Thompson DK, 2017. A new neonatal cortical and subcortical brain atlas: the melbourne children’s regional infant brain (m-CRIB) atlas. Neuroimage 147, 841–851. doi: 10.1016/j.neuroimage.2016.09.068. [DOI] [PubMed] [Google Scholar]
- Ashourvan A, Shah P, Pines A, Gu S, Lynn CW, Bassett DS, Davis KA, Litt B, 2021. Pairwise maximum entropy model explains the role of white matter structure in shaping emergent co-activation states. Commun. Biol 4 (1). doi: 10.1038/s42003-021-01700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association, A.E.R. (Ed.), 2014. Standards for Educational and Psychological Testing American Educational Research Association, Lanham, MD. [Google Scholar]
- Association, A.P., 1954. Technical recommendations for psychological tests and diagnostic techniques. Psychol Bull 51 (2:2), 1–38. doi: 10.1037/h0053479. [DOI] [PubMed] [Google Scholar]
- Azarion AA, Wu J, Pearce A, Krish VT, Wagenaar J, Chen W, Zheng Y, Wang H, Lucas TH, Litt B, Gee JC, Davis KA, 2014. An open-source automated platform for three-dimensional visualization of subdural electrodes using CT-MRI coregistration. Epilepsia 55 (12), 2028–2037. doi: 10.1111/epi.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker FG, 1995. Phineas among the phrenologists: the american crowbar case and nineteenth-century theories of cerebral localization. J. Neurosurg 82 (4), 672–682. doi: 10.3171/jns.1995.82.4.0672. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Zurn P, Gold JI, 2018. On the nature and use of models in network neuroscience. Nat. Rev. Neurosci 19 (9), 566–578. doi: 10.1038/s41583-018-0038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal DS, Lerch JP, Cameron B, Henderson R, Gracco VL, De Nil LF, 2015. The trajectory of gray matter development in broca’s area is abnormal in people who stutter. Front. Hum. Neurosci 9. doi: 10.3389/fnhum.2015.00089. [DOI] [PMC free article] [PubMed]
- Belzung C, Lemoine M, 2011. Criteria of validity for animal models of psychiatric disorders: focus on anxiety disorders and depression. Biol. Mood Anxiety Disord 1, 9. doi: 10.1186/2045-5380-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel RF, Medaglia JD, Kahn AE, Soffer J, Schonhaut DR, Bassett DS, 2019. Structural, geometric and genetic factors predict interregional brain connectivity patterns probed by electrocorticography. Nat. Biomed. Eng doi: 10.1038/s41551-019-0404-5. [DOI] [PubMed]
- Bijsterbosch J, Smith SM, Beckmann CF, 2017. An Introduction to Resting State FMRI Functional Connectivity Oxford University Press. [Google Scholar]
- Bohland JW, Bokil H, Allen CB, Mitra PP, 2009. The brain atlas concordance problem: quantitative comparison of anatomical parcellations. PLoS ONE 4 (9), e7200. doi: 10.1371/journal.pone.0007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Gleichgerrcht E, Nesland T, Rorden C, Fridriksson J, 2015. Gray matter axonal connectivity maps. Front. Psychiatry 6. doi: 10.3389/fpsyt.2015.00035. [DOI] [PMC free article] [PubMed]
- Brennan BP, Wang D, Li M, Perriello C, Ren J, Elias JA, Van Kirk NP, Krompinger JW, Pope HG, Haber SN, Rauch SL, Baker JT, Liu H, 2019. Use of an individual-level approach to identify cortical connectivity biomarkers in obsessive-compulsive disorder. Biol. Psychiatry. Cognit. Neurosci. Neuroimaging 4 (1), 27–38. doi: 10.1016/j.bpsc.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas M, Oliver A, Lladó X, Freixenet J, Cuadra MB, 2011. A review of atlas-based segmentation for magnetic resonance brain images. Comput. Methods Programs Biomed 104 (3), e158–77. doi: 10.1016/j.cmpb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Callaway EM, Dong H-W, Ecker JR, Hawrylycz MJ, Huang ZJ, Lein ES, Ngai J, Osten P, Ren B, Tolias AS, White O, Zeng H, Zhuang X, Ascoli GA, Behrens MM, Chun J, Feng G, Gee JC, Ghosh SS, Halchenko YO, Hertzano R, Lim BK, Martone ME, Ng L, Pachter L, Ropelewski AJ, Tickle TL, Yang XW, Zhang K, Bakken TE, Berens P, Daigle TL, Harris JA, Jorstad NL, Kalmbach BE, Kobak D, Li YE, Liu H, Matho KS, Mukamel EA, Naeemi M, Scala F, Tan P, Ting JT, Xie F, Zhang M, Zhang Z, Zhou J, Zingg B, Armand E, Yao Z, Bertagnolli D, Casper T, Crichton K, Dee N, Diep D, Ding S-L, Dong W, Dougherty EL, Fong O, Goldman M, Goldy J, Hodge RD, Hu L, Keene CD, Krienen FM, Kroll M, Lake BB, Lathia K, Linnarsson S, Liu CS, Macosko EZ, McCarroll SA, McMillen D, Nadaf NM, Nguyen TN, Palmer CR, Pham T, Plongthongkum N, Reed NM, Regev A, Rimorin C, Romanow WJ, Savoia S, Siletti K, Smith K, Sulc J, Tasic B, Tieu M, Torkelson A, Tung H, van Velthoven CTJ, Vanderburg CR, Yanny AM, Fang R, Hou X, Lucero JD, Osteen JK, Pinto-Duarte A, Poirion O, Preissl S, Wang X, Aldridge AI, Bartlett A, Boggeman L, O’Connor C, Castanon RG, Chen H, Fitzpatrick C, Luo C, Nery JR, Nunn M, Rivkin AC, Tian W, Dominguez B, Ito-Cole T, Jacobs M, Jin X, Lee C-T, Lee K-F, Miyazaki PA, Pang Y, Rashid M, Smith JB, Vu M, Williams E, Biancalani T, Booeshaghi AS, Crow M, Dudoit S, Fischer S, Gillis J, Hu Q, Kharchenko PV, Niu S-Y, Ntranos V, Purdom E, Risso D, de Bzieux HR, Somasundaram S, Street K, Svensson V, Vaishnav ED, Van den Berge K, Welch JD, An X, Bateup HS, Bowman I, Chance RK, Foster NN, Galbavy W, Gong H, Gou L, Hatfield JT, Hintiryan H, Hirokawa KE, Kim G, Kramer DJ, Li A, Li X, Luo Q, Muñoz-Castañeda R, Stafford DA, Feng Z, Jia X, Jiang S, Jiang T, Kuang X, Larsen R, Lesnar P, Li Y, Li Y, Liu L, Peng H, Qu L, Ren M, Ruan Z, Shen E, Song Y, Wakeman W, Wang P, Wang Y, Wang Y, Yin L, Yuan J, Zhao S, Zhao X, Narasimhan A, Palaniswamy R, Banerjee S, Ding L, Huilgol D, Huo B, Kuo HC, Laturnus S, Li X, Mitra PP, Mizrachi J, Wang Q, Xie P, Xiong F, Yu Y, Eichhorn SW, Berg J, Bernabucci M, Bernaerts Y, Cadwell CR, Castro JR, Dalley R, Hartmanis L, Horwitz GD, Jiang X, Ko AL, Miranda E, Mulherkar S, Nicovich PR, Owen SF, Sandberg R, Sorensen SA, Tan ZH, Allen S, Hockemeyer D, Lee AY, Veldman MB, Adkins RS, Ament SA, Bravo HC, Carter R, Chatterjee A, Colantuoni C, Crabtree J, Creasy H, Felix V, Giglio M, Herb BR, Kancherla J, Mahurkar A, McCracken C, Nickel L, Olley D, Orvis J, Schor M, Hood G, Dichter B, Grauer M, Helba B, Bandrowski A, Barkas N, Carlin B, D’Orazi FD, Degatano K, Gillespie TH, Khajouei F, Konwar K, Thompson C, Kelly K, Mok S, Sunkin S, BRAIN NB, I.C.C., B.C.a. BRAIN, I.C.C.N., BICCN, c.p.i., Principal, m.e., Manuscript, w.a.f.g., Analysis, c., Integrated, d.a., d.g.a.p. scRNA seq, a.s.-s., ATAC-seq, d.g.a.p., Methylcytosine, d.p.a.a., Epi-retro seq, d.g.a.p., Omics, d.a., Tracing, a.c.d.g., Morphology, d.g.a.r., OLST/STPT, a.o.d.g., Morphology, c.a.i.a., Spatially, r.s.-c.t.M., Multimodal, p.P.-s., Transgenic, t., NeMO, a.a.a., Brain, I.L.B.a., DANDI, a., Brain, C.D.C.B., Project, m., 2021. A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598 (7879), 86–102. doi: 10.1038/s41586-021-03950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers S, Eickhoff SB, Zilles K, Amunts K, 2013. Microstructural grey matter parcellation and its relevance for connectome analyses. Neuroimage 80, 18–26. doi: 10.1016/j.neuroimage.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak M, Cook PA, He X, Yeh FC, Dhollander T, Adebimpe A, Aguirre GK, Bassett DS, Betzel RF, Bourque J, Cabral LM, Davatzikos C, Detre JA, Earl E, Elliott MA, Fadnavis S, Fair DA, Foran W, Fotiadis P, Garyfallidis E, Gies brecht B, Gur RC, Gur RE, Kelz MB, Keshavan A, Larsen BS, Luna B, Mackey AP, Milham MP, Oathes DJ, Perrone A, Pines AR, Roalf DR, Richie-Halford A, Rokem A, Sydnor VJ, Tapera TM, Tooley UA, Vettel JM, Yeatman JD, Grafton ST, Satterthwaite TD, 2021. QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat. Methods 18 (7), 775–778. doi: 10.1038/s41592-021-01185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coalson TS, Van Essen DC, Glasser MF, 2018. The impact of traditional neuroimaging methods on the spatial localization of cortical areas. Proc. Natl. Acad. Sci 115 (27), E6356–E6365. doi: 10.1073/pnas.1801582115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi L, Harding IH, Lord A, Pantelis C, Yucel M, Zalesky A, 2014. Disruption of structure-function coupling in the schizophrenia connectome. Neuroimage Clin 4, 779–787. doi: 10.1016/j.nicl.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI, 1999. Cortical surface-based analysis. Neuroimage 9 (2), 179–194. doi: 10.1006/nimg.1998.0395.https://linkinghub.elsevier.com/retrieve/pii/S1053811998903950 [DOI] [PubMed] [Google Scholar]
- Dickie DA, Shenkin SD, Anblagan D, Lee J, Blesa Cabez M, Rodriguez D, Boardman JP, Waldman A, Job DE, Wardlaw JM, 2017. Whole brain magnetic resonance image atlases: a systematic review of existing atlases and caveats for use in population imaging. Front. Neuroinform 11, 1. doi: 10.3389/fninf.2017.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N, 2009. A probabilistic MR atlas of the human cerebellum. Neuroimage 46 (1), 39–46. doi: 10.1016/j.neuroimage.2009.01.045. [DOI] [PubMed] [Google Scholar]
- Doucet GE, Labache L, Thompson PM, Joliot M, Frangou S, Alzheimer’s DNI, 2021. Atlas55+: brain functional atlas of resting-state networks for late adulthood. Cereb. Cortex 31 (3), 1719–1731. doi: 10.1093/cercor/bhaa321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AC, Janke AL, Collins DL, Baillet S, 2012. Brain templates and atlases. Neuroimage 62 (2), 911–922. doi: 10.1016/j.neuroimage.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, Laird AR, Fox PT, Eickhoff SB, Yu C, Jiang T, 2016. The human brainnetome atlas: a new brain atlas based on connectional architecture. Cereb. Cortex 26 (8), 3508–3526. doi: 10.1093/cercor/bhw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang-Cheng Y, Wedeen VJ, Tseng W-YI, 2010. Generalized q-sampling imaging. IEEE Trans. Med. Imaging 29 (9), 1626–1635. doi: 10.1109/TMI.2010.2045126. [DOI] [PubMed] [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL, 2011. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54 (1), 313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bullmore E, 2016. Fundamentals of Brain Network Analysis Academic Press. [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, Smith SM, Van Essen DC, 2016. A multi-modal parcellation of human cerebral cortex. Nature 536 (7615), 171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE, 2016. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cerebr. Cortex 26 (1), 288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Varoquaux G, Rivera G, Schwarz Y, Ghosh SS, Maumet C, Sochat VV, Nichols TE, Poldrack RA, Poline J-B, Yarkoni T, Margulies DS, 2015. Neurovault.org: a web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front. Neuroinform 9, 8. doi: 10.3389/fn-inf.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P, Li A, González-Martínez J, Sarma SV, 2021. Classification of stereo-EEG contacts in white matter vs. gray matter using recorded activity. Front. Neurol 11. doi: 10.3389/fneur.2020.605696. [DOI] [PMC free article] [PubMed]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS, 2003. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum. Brain Mapp 19 (4), 224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MX, Cornblath EJ, Darwich A, Zhang B, Brown H, Gathagan RJ, Sandler RM, Bassett DS, Trojanowski JQ, Lee VMY, 2019. Spread of - synuclein pathology through the brain connectome is modulated by selective vulnerability and predicted by network analysis. Nat. Neurosci 22 (8), 1248–1257. doi: 10.1038/s41593-019-0457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-C, Rolls ET, Feng J, Lin C-P, 2021. An extended human connectome project multimodal parcellation atlas of the human cortex and subcortical areas. Brain Struct. Funct doi: 10.1007/s00429-021-02421-6. [DOI] [PubMed]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. Neuroimage 62 (2), 782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Joglekar A, Prjibelski A, Mahfouz A, Collier P, Lin S, Schlusche AK, Marrocco J, Williams SR, Haase B, Hayes A, Chew JG, Weisenfeld NI, Wong MY, Stein AN, Hardwick SA, Hunt T, Wang Q, Dieterich C, Bent Z, Fedrigo O, Sloan SA, Risso D, Jarvis ED, Flicek P, Luo W, Pitt GS, Frankish A, Smit AB, Ross ME, Tilgner HU, 2021. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun 12 (1), 463. doi: 10.1038/s41467-020-20343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambhati AN, Bassett DS, Oommen BS, Chen SH, Lucas TH, Davis KA, Litt B, 2017. Recurring Functional Interactions Predict Network Architecture of Interictal and Ictal States in Neocortical Epilepsy, 4 doi: 10.1523/ENEURO.0091-16.2017. ENEURO.0091–16.2017 [DOI] [PMC free article] [PubMed]
- Khambhati AN, Davis KA, Lucas TH, Litt B, Bassett DS, 2016. Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron 91 (5), 1170–1182. doi: 10.1016/j.neuron.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambhati AN, Davis KA, Oommen BS, Chen SH, Lucas TH, Litt B, Bassett DS, 2015. Dynamic network drivers of seizure generation, propagation and termination in human neocortical epilepsy. PLoS Comput. Biol 11 (12), e1004608. doi: 10.1371/journal.pcbi.1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini LG, Davis KA, Wagenaar JB, 2016. Data integration: combined imaging and electrophysiology data in the cloud. Neuroimage 124, 1175–1181. doi: 10.1016/j.neuroimage.2015.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Tourville J, 2012. 101 Labeled brain images and a consistent human cortical labeling protocol. Front. Neurosci 6. doi: 10.3389/fnins.2012.00171. [DOI] [PMC free article] [PubMed]
- Kramer MA, Eden UT, Kolaczyk ED, Zepeda R, Eskandar EN, Cash SS, 2010. Coalescence and fragmentation of cortical networks during focal seizures. J. Neurosci 30, 10076–10085. doi: 10.1523/JNEUROSCI.6309-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RM, Bridgeford EW, Myers PE, Arvapalli GC, Ramachandran SC, Pisner DA, Frank PF, Lemmer AD, Nikolaidis A, Vogelstein JT, 2021. Standardizing human brain parcellations. Sci. Data 8 (1), 78. doi: 10.1038/s41597-021-00849-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Bezgin G, Fonov VS, Collins DL, Evans AC, 2021. A sub+cortical fmri-based surface parcellation. Hum. Brain Mapp doi: 10.1002/hbm.25675. [DOI] [PMC free article] [PubMed]
- Litt B, Esteller R, Echauz J, D’Alessandro M, Shor R, Henry T, Pennell P, Epstein C, Bakay R, Dichter M, Vachtsevanos G, 2001. Epileptic seizures may begin hours in advance of clinical onset. Neuron 30 (1), 51–64. doi: 10.1016/s0896-6273(01)00262-8. [DOI] [PubMed] [Google Scholar]
- Ludwig KA, Miriani RM, Langhals NB, Joseph MD, Anderson DJ, Kipke DR, 2009. Using a common average reference to improve cortical neuron recordings from microelectrode arrays. J. Neurophysiol 101 (3), 1679–1689. doi: 10.1152/jn.90989.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris N, Goldstein JM, Kennedy D, Hodge SM, Caviness VS, Faraone SV, Tsuang MT, Seidman LJ, 2006. Decreased volume of left and total anterior insular lobule in schizophrenia. Schizophr. Res 83 (2–3), 155–171. doi: 10.1016/j.schres.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Mandal PK, Mahajan R, Dinov ID, 2012. Structural brain atlases: design, rationale, and applications in normal and pathological cohorts. J. Alzheimers Dis 31 Suppl 3, S169–88. doi: 10.3233/JAD-2012-120412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, 2002. Specificity and stability in topology of protein networks. Science 296 (5569), 910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Goualher GL, Boomsma D, Cannon T, Kawashima R, Mazoyer B, 2001. A probabilistic atlas and reference system for the human brain: international consortium for brain mapping (ICBM). Philos. Trans. R. Soc. Lond. Ser. B: Biol. Sci 356 (1412), 1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier MR, Bickel S, Megevand P, Groppe DM, Schroeder CE, Mehta AD, Lado FA, 2017. Evaluation of cortical local field potential diffusion in stereotactic electro-encephalography recordings: a glimpse on white matter signal. Neuroimage 147, 219–232. doi: 10.1016/j.neuroimage.2016.08.037. [DOI] [PubMed] [Google Scholar]
- Mišić B, Betzel RF, Nematzadeh A, Goñi J, Griffa A, Hagmann P, Flammini A, Ahn YY, Sporns O, 2015. Cooperative and competitive spreading dynamics on the human connectome. Neuron 86 (6), 1518–1529. doi: 10.1016/j.neuron.2015.05.035. [DOI] [PubMed] [Google Scholar]
- Muñoz-Castañeda R, Zingg B, Matho KS, Chen X, Wang Q, Foster NN, Li A, Narasimhan A, Hirokawa KE, Huo B, Bannerjee S, Korobkova L, Park CS, Park Y-G, Bienkowski MS, Chon U, Wheeler DW, Li X, Wang Y, Naeemi M, Xie P, Liu L, Kelly K, An X, Attili SM, Bowman I, Bludova A, Cetin A, Ding L, Drewes R, D’Orazi F, Elowsky C, Fischer S, Galbavy W, Gao L, Gillis J, Groblewski PA, Gou L, Hahn JD, Hatfield JT, Hintiryan H, Huang JJ, Kondo H, Kuang X, Lesnar P, Li X, Li Y, Lin M, Lo D, Mizrachi J, Mok S, Nicovich PR, Palaniswamy R, Palmer J, Qi X, Shen E, Sun Y-C, Tao HW, Wakemen W, Wang Y, Yao S, Yuan J, Zhan H, Zhu M, Ng L, Zhang LI, Lim BK, Hawrylycz M, Gong H, Gee JC, Kim Y, Chung K, Yang XW, Peng H, Luo Q, Mitra PP, Zador AM, Zeng H, Ascoli GA, Josh Huang Z, Osten P, Harris JA, Dong H-W, 2021. Cellular anatomy of the mouse primary motor cortex. Nature 598 (7879), 159–166. doi: 10.1038/s41586-021-03970-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National, 2022a. Geographic Society Encyclopedic entry Atlas. https://www.nationalgeographic.org/encyclopedia/atlas/. [Google Scholar]
- National, 2022b. Geographic Society Encyclopedic entry Border. https://www.nationalgeographic.org/encyclopedia/border/. [Google Scholar]
- Newson JJ, Thiagarajan TC, 2019. EEG Frequency bands in psychiatric disorders: a review of resting state studies. Front. Hum. Neurosci 12, 521. doi: 10.3389/fn-hum.2018.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N, 1979. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern 9 (1), 62–66. doi: 10.1109/tsmc.1979.4310076. [DOI] [Google Scholar]
- Park B, Eo J, Park H-J, 2017. Structural brain connectivity constrains within-a-day variability of direct functional connectivity. Front. Hum. Neurosci 11, 408. doi: 10.3389/fnhum.2017.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlaki G, Horvath R, Nagy SA, Bogner P, Doczi T, Janszky J, Orsi G, 2017. Comparison of accuracy between FSL’s FIRST and freesurfer for caudate nucleus and putamen segmentation. Sci. Rep 7 (1), 2418. doi: 10.1038/s41598-017-02584-5. http://www.nature.com/articles/s41598-017-02584-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proix T, Bartolomei F, Guye M, Jirsa VK, 2017. Individual brain structure and modelling predict seizure propagation. Brain 140 (3), 641–654. doi: 10.1093/brain/awx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell AY, Silva AB, Mahesh D, Armstrong L, Arnold TC, Bernabei JM, Stein JM, Das SR, Shinohara RT, Bassett DS, Litt B, Davis KA, 2021. White matter signals reflect information transmission between brain regions during seizures. bioRxiv doi: 10.1101/2021.09.15.460549. [DOI]
- Royer J, Rodríguez-Cruces R, Tavakol S, Larivire S, Herholz P, Li Q, de Wael RV, Paquola C, Benkarim O, Park B. y., Lowe AJ, Margulies D, Smallwood J, Bernasconi A, Bernasconi N, Frauscher B, Bernhardt BC, 2021. An open MRI dataset for multiscale neuroscience. preprint doi: 10.1101/2021.08.04.454795. 10.1101/2021.08.04.454795 [DOI] [PMC free article] [PubMed]
- Salehi M, Greene AS, Karbasi A, Shen X, Scheinost D, Constable RT, 2020. There is no single functional atlas even for a single individual: functional parcel definitions change with task. Neuroimage 208, 116366. doi: 10.1016/j.neuroimage.2019.116366. [DOI] [PubMed] [Google Scholar]
- Sathian K, Crosson B, 2015. Structure-function correlations in stroke. Neuron 85 (5), 887–889. doi: 10.1016/j.neuron.2015.02.031. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo X-N, Holmes AJ, Eickhoff SB, Yeo BTT, 2018. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cerebr. Cortex 28 (9), 3095–3114. doi: 10.1093/cercor/bhx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ashourvan A, Mikhail F, Pines A, Kini L, Oechsel K, Das SR, Stein JM, Shinohara RT, Bassett DS, Litt B, Davis KA, 2019. Characterizing the role of the structural connectome in seizure dynamics. Brain: A J. Neurol doi: 10.1093/brain/awz125. [DOI] [PMC free article] [PubMed]
- Shmueli G, 2010. To explain or to predict. Stat. Sci 25 (3). doi: 10.1214/10-sts330. [DOI] [Google Scholar]
- Sinha N, Peternell N, Schroeder GM, Tisi J, Vos SB, Winston GP, Duncan JS, Wang Y, Taylor PN, 2021. Focal to bilateral tonic-clonic seizures are associated with widespread network abnormality in temporal lobe epilepsy. Epilepsia 62 (3), 729–741. doi: 10.1111/epi.16819. 10.1111/epi.16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Tournier JD, Calamante F, Connelly A, 2015. SIFT2: enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage 119, 338–351. doi: 10.1016/j.neuroimage.2015.06.092. [DOI] [PubMed] [Google Scholar]
- Sporns O, 2011. The human connectome: a complex network. Ann. N. Y. Acad. Sci 1224, 109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R, 2005. The human connectome: a structural description of the human brain. PLoS Comput. Biol 1 (4), e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syversen IF, Witter MP, Kobro-Flatmoen A, Goa PE, Navarro Schröder T, Doeller CF, 2021. Structural connectivity-based segmentation of the human entorhinal cortex. Neuroimage 245, 118723. doi: 10.1016/j.neuroimage.2021.118723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PN, Sinha N, Wang Y, Vos SB, de Tisi J, Miserocchi A, McEvoy AW, Winston GP, Duncan JS, 2018. The impact of epilepsy surgery on the structural connectome and its relation to outcome. Neuroimage Clin 18, 202–214. doi: 10.1016/j.nicl.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL, 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol 106 (3), 1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, WU-Minn HC, 2013. The WU-minn human connectome project: an overview. Neuroimage 80, 62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Irimia A, Torgerson CM, Chambers MC, Kikinis R, Toga AW, 2012. Mapping connectivity damage in the case of phineas gage. PLoS ONE 7 (5), e37454. doi: 10.1371/journal.pone.0037454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar JB, Brinkmann BH, Ives Z, Worrell GA, Litt B, 2013. A multimodal platform for cloud-based collaborative research. IEEE doi: 10.1109/ner.2013.6696201. [DOI]
- Wang HE, Scholly J, Triebkorn P, Sip V, Medina Villalon S, Woodman MM, Le Troter A, Guye M, Bartolomei F, Jirsa V, 2021. VEP Atlas: an anatomic and functional human brain atlas dedicated to epilepsy patients. J. Neurosci. Methods 348, 108983. doi: 10.1016/j.jneumeth.2020.108983. [DOI] [PubMed] [Google Scholar]
- Willner P, 1984. The validity of animal models of depression. Psychopharmacology (Berl.) 83 (1), 1–16. doi: 10.1007/BF00427414. [DOI] [PubMed] [Google Scholar]
- Wirsich J, Perry A, Ridley B, Proix T, Golos M, Bnar C, Ranjeva JP, Bartolomei F, Breakspear M, Jirsa V, Guye M, 2016. Whole-brain analytic measures of network communication reveal increased structure-function correlation in right temporal lobe epilepsy. Neuroimage Clin 11, 707–718. doi: 10.1016/j.nicl.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xu D, Potter T, Zhang Y, The ADNI, 2019. Effects of brain parcellation on the characterization of topological deterioration in alzheimer’s disease. Front. Aging Neurosci 11, 113. doi: 10.3389/fnagi.2019.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ, Friedman JS, Panov F, Camara D, Yoo JY, Fields MC, Marcuse LV, Jette N, Ghatan S, 2019. Quantitative signal characteristics of electrocorticography and stereoelectroencephalography: the effect of contact depth. J. Clin. Neurophysiol 36 (3), 195–203. doi: 10.1097/WNP.0000000000000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Ycel M, Pantelis C, Bullmore ET, 2010. Whole-brain anatomical networks: does the choice of nodes matter? Neuroimage 50 (3), 970–983. doi: 10.1016/j.neuroimage.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang H, Chong Y-S, Shek LP, Gluckman PD, Meaney MJ, Fortier MV, Qiu A, 2021. Integrated structural and functional atlases of asian children from infancy to childhood. Neuroimage 245, 118716. doi: 10.1016/j.neuroimage.2021.118716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code files used in this manuscript are available at https://github.com/andyrevell/revellLab. All de-identified raw and processed data (except for patient MRI imaging) are available for download by following the links on the GitHub. Raw imaging data is available upon reasonable request from Principal Investigator K.A.D. iEEG snippets used specifically in this manuscript are also available, while full iEEG recordings are publicly available at https://www.ieeg.org. The Python environment for the exact packages and versions used in this study in contained in the environment directory within the GitHub. The QSIPrep docker container was used for DWI preprocessing.


