Abstract
High temperature (HT) causes male sterility and decreases crop yields. Our previous works have demonstrated that sugar and auxin signaling pathways, Gossypium hirsutum Casein kinase I (GhCKI), and DNA methylation are all involved in HT-induced male sterility in cotton. However, the signaling mechanisms leading to distinct GhCKI expression patterns induced by HT between HT-tolerant and HT-sensitive cotton anthers remain largely unknown. Here, we identified a GhCKI promoter (ProGhCKI) region that functions in response to HT in anthers and found the transcription factor GhMYB4 binds to this region to act as an upstream positive regulator of GhCKI. In the tapetum of early-stage cotton anthers, upregulated expression of GhMYB4 under HT and overexpressed GhMYB4 under normal temperature both led to severe male sterility phenotypes, coupled with enhanced expression of GhCKI. We also found that GhMYB4 interacts with GhMYB66 to form a heterodimer to enhance its binding to ProGhCKI. However, GhMYB66 showed an expression pattern similar to GhMYB4 under HT but did not directly bind to ProGhCKI. Furthermore, HT reduced siRNA-mediated CHH DNA methylations in the GhMYB4 promoter, which enhanced the expression of GhMYB4 in tetrad stage anthers and promoted the formation of the GhMYB4/GhMYB66 heterodimer, which in turn elevated the transcription of GhCKI in the tapetum, leading to male sterility. Overall, we shed light on the GhMYB66–GhMYB4–GhCKI regulatory pathway in response to HT in cotton anthers.
GhMYB4 and GhMYB66 are crucial factors for cotton anther response to high temperature, and the GhMYB66/GhMYB4 heterodimer enhances the high temperature-induced male sterility through Casein kinase I.
Introduction
With the latest global weather changes, extreme high temperature (HT) occurs more frequently around the world than before (Xu et al., 2018). HT often has a substantial negative impact on the growth and development of various crops, resulting in much lower yields (Wilczek et al., 2014; Lesk et al., 2016). Statistics showed that every 3–4°C rise in temperature could lead to a drop in crop yields up to 15%–25% in Africa and Asia, and 25%–35% in the Middle East (Bita and Gerats, 2013). The male organs of plants are more sensitive to HT than the female organs under HT stress (Sage et al., 2015; Djanaguiraman et al., 2018). As a result, the HT-stressed crops usually show phenotypes such as microspore abortion, abnormal degeneration of the tapetum layer cell, reduced pollen activity, and indehiscent anthers (Abiko et al., 2005; Hedhly et al., 2009), leading to male sterility. Cotton (Gossypium hirsutum) is the most important fiber crop in the world, whose yield is also greatly impacted by HT stress. When the temperature is higher than 35°C, cotton anthers show male sterile phenotypes such as anther indehiscence and reduced pollen activity (Min et al., 2014), resulting in great yield loss. However, molecular studies on how cotton male organs respond to HT stress are still lacking.
Transcription factors play very important roles in plant growth, development, and response to stresses (Han et al., 2014). In recent years, there are increasing reports showing many transcription factors are involved in plant responses to the heat stress. For example, heat stress response factors (HSFs) can regulate the expression of heat shock proteins (HSPs) in response to heat stress (Guo et al., 2016). Overexpression of the Lilium longiflorum heat-stress transcription factor A3A gene in Arabidopsis thaliana enhanced both the basal and acquired heat tolerances in Arabidopsis plants (Wu et al., 2018a). The dehydration response element binding protein 2B, a AP2/CBF transcription factor from lily, participated in the HsfA3 heat response pathway (Wu et al., 2018b). Moreover, overexpression of AtWRKY30 in wheat (Triticum aestivum) or TaWRKY33 in Arabidopsis, greatly improved the heat tolerance of wheat and Arabidopsis plants, respectively (El-Esawi et al., 2019). NAM/ATAF/CUC (NAC)-like transcription factors are also involved in the plant heat tolerance responses. In Arabidopsis, NAC019 responds to HT by binding to the promoters of HSF genes such as HSFA1b and HSFA6b. NAC019 overexpression plants showed increased heat tolerance, while NAC019 silencing plants were more sensitive to HT stress (Guan et al., 2014).
MYB transcription factors in plants contain 1–4 conserved incompletely repeated MYB structural domains, named R1, R2, R3, and R4 (Rosinski and Atchley, 1998). According to the number of MYB structural domains contained, MYB transcription factors are divided into four categories, namely 1R-MYB, 2R-MYB (R2R3-MYB), 3R-MYB (R1R2R3-MYB), and 4R-MYB, with R2R3-MYB being the most abundant one in plants (Dubos et al., 2010). MYB transcription factors play important roles in response to abiotic stress. For example, AtMYB30 responds to heat stress by binding to the promoters of ANNEXIN 1 and ANNEXIN 4 to repress their expressions, which in turn leads to the modification of cytosolic calcium concentrations (Liao et al., 2017). AtMYB96 promotes drought tolerance by activating cuticular wax biosynthesis (Seo et al., 2011). AtMYB30 modulates salt tolerance by repressing the toxic reactive oxygen species production via AOX1a-mediated alternative respiration (Gong et al., 2020). Phosphorylation of AtMYB15 by mitogen-activated protein kinase 6 is critical for cold tolerance in Arabidopsis (Kim et al., 2017). Overexpression of OsMYB55 in rice (Oryza sativa) and maize (Zea mays) increased the total amino acid content and enhanced heat tolerance during vegetative growth stage (El-Kereamy et al., 2012; Casaretto et al., 2016). Furthermore, MYB4, an R2R3-MYB transcription factor, is involved in plant UV defense (Hemm et al., 2001), flavonoid synthesis (Wang et al., 2020), drought and cold tolerance (Pasquali et al., 2008). MYB transcription factors are also broadly involved in plant reproductive development. The overexpression of GhMYB24 in Arabidopsis caused malformation of flowers, shorter filaments, nondehiscent anthers, and reduced viable pollen grains (Li et al., 2013). MYB98 plays a very important role in pollen tube guidance and synergid cell differentiation in Arabidopsis. Its mutant myb98 plants exhibited dramatically reduced seed set (Kasahara et al., 2005). However, MYB transcription factors have not been studied well in cotton during heat stress response compared to other plants, especially their roles in heat stress-induced male sterility.
DNA methylation is a common epigenetic modification that regulates gene expression (He et al., 2011; Matzke and Mosher, 2014). DNA methylation is usually classified into three types, CHH, CHG, and CG (Law and Jacobsen, 2010). In plants, DNA methylation can be established through the RNA-directed DNA methylation pathway (Matzke and Mosher, 2014). Our previous study found that HT significantly reduced the CHH methylation in anthers of a HT-sensitive cotton line, which distinctly affected the sugar metabolism and auxin signaling, leading to male sterility (Min et al., 2014; Ma et al., 2018). GhCKI, which is involved in the sugar metabolism as well as the auxin signaling, was induced by HT to be expressed earlier during anther development (Min et al., 2013, 2014). GhCKI interacts with starch synthase and inhibits its activity, leading to more glucose accumulated during early anther development (Min et al., 2013). However, in late developmental stage anthers, GhCK1 inhibits glucose uptake and induces starch synthesis, which leads to glucose deficiency, resulting in delayed tapetum programmed cell death (PCD) and anther abortion (Min et al., 2013). GhCKI plays an important role in the HT response, but the mechanism of upstream regulation of GhCKI is unclear.
Here, we identified a GhCKI promoter region that responds to HT, and found that GhMYB4 can bind to two MYB transcription factor-binding sites within this region and positively regulate the expression of GhCKI. GhMYB4 is normally expressed in anthers at the tapetum degradation stage (TDS), but is expressed earlier at the tetrad stage (TS) under HT. This process might be directly regulated by siRNA-mediated CHH DNA methylation. Overexpression lines of GhMYB4 appeared to be male sterile, mimicking plants under HT stress. In addition, we found that GhMYB66 can form a heterodimer with GhMYB4 and enhance the binding activity of GhMYB4 to the GhCKI promoter. Our study illustrated an important molecular mechanism of male sterility caused by HT through GhMYB66–GhMYB4–GhCKI signaling in cotton anthers.
Results
Analysis of GhCKI promoter region in response to HT
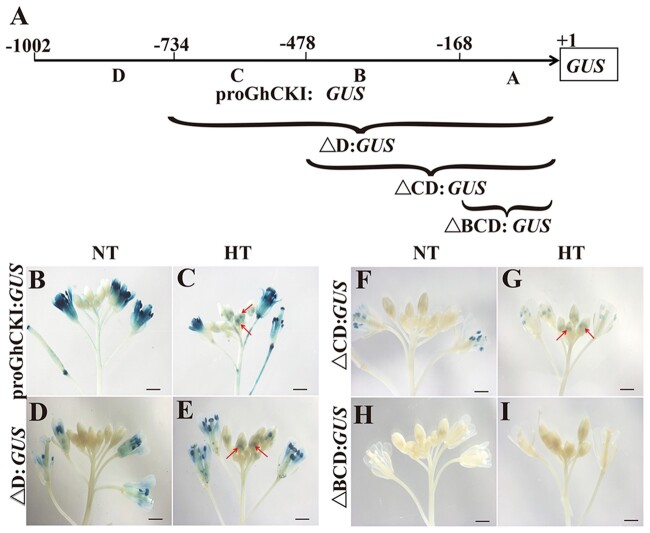
Our previous study showed that GhCKI is expressed in late-stage anthers (Stages 12–14) and was induced to express in anthers from Stages 7 to 14 by HT, specifically in tapetal cells and microspores between Stages 7 and 10 (Min et al., 2013). To investigate the function of GhCKI core promoter region in response to HT, we divided the 1-kb GhCKI promoter region immediately upstream of the start codon (1,002 bp) into four parts (A, B, C, and D) and generated different constructs containing truncated promoters on GUS expression vectors, which were transformed into A.thaliana plants (Min et al., 2015; Figure 1A). Homozygous Arabidopsis plants containing these constructs were then subjected to HT treatment at blooming stage, with normal temperature (NT) condition as controls. Under both NT and HT conditions, the GUS staining results showed that the full-length GhCKI promoter (ProGhCKI), the ProGhCKI with D region removed (△D), and the ProGhCKI with CD region removed (△CD) were all able to drive the expression of GUS reporter gene in anthers after anthesis, but the GUS activity gradually decreased with the progressive 5′-truncation of ProGhCKI (Figure 1, B–G). To the contrary, promoter containing only the A region (lacking B, C, and D regions, △BCD) could not drive the GUS expression under both NT and HT conditions (Figure 1, H and I), suggesting that the region B of ProGhCKI (−169 to −478 bp) may contain cis-elements that are responsive to HT during anther development, and this response may be reinforced by other elements in regions C (−479 to −734 bp) and D (−733 to −1,002 bp).
Figure 1.
The region B of ProGhCKI is key for anther development in response to HT. A, The 1-kb region of GhCKI promoter upstream of the start codon is divided into four parts as A, B, C, and D, ranging from −1 to −168 bp (region A), −169 to −478 bp (region B), −479 to −733 bp (region C), and −734 to −1,002 bp (region D). Δ represents truncation. (B–I) Qualitative analysis of GUS genes driven by ProGhCKI of different lengths under NT (B, D, F, and H) and HT (C, E, G, and I). Bars = 1 mm. Arrows indicate the expressions of GUS reporter gene driven by ProGhCKI and its derivatives in anthers.
GhMYB4 binds to the region B of ProGhCKI to activate the expression of GhCKI
GhCKI was substantially induced in the anthers in the HT-sensitive line H05 under HT condition but not in the HT-tolerant cotton line 84021 (Min et al., 2013). However, no sequence difference was detected between 84021 and H05 cotton lines in their promoter regions (1,002 bp) as characterized in Supplemental Figure S1. Thus, we speculated that the different responses of these two cotton lines to HT might be due to the transcription factors involved in the transcriptional regulation of GhCKI under HT. Based on our previous RNAseq data (Min et al., 2014), we performed a clustering analysis of the differentially expressed genes in anthers between NT and HT at the TS, TDS, and anther dehiscence stage (ADS) in 84021 and H05. The results showed that genes in clusters 3, 4, 11, 15, 16 were likely co-regulated with GhCKI, as they had similar expression patterns as GhCKI in both 84021 and H05 plants under both NT and HT conditions (Supplemental Figure S2A). Interestingly, the transcription factors in these clusters were mainly the MYB, ERF, and bHLH types, with the largest number of transcription factors belonging to the MYB family (Supplemental Figure S2B and Supplemental Table S1). Further analysis of the expression patterns of these MYB-like transcription factors found they were induced by HT in H05 but not in 84021 (Supplemental Figure S3A). For example, Ghir_D03G016220 and Ghir_A12G003840, Ghir_A11G034860 and Ghir_D13G017770, and Ghir_A05G037220 and Ghir_D06G001350 were all induced by HT at TS, TDS, and ADS stages in H05, but no expression changes were found in 84021 (Supplemental Figure S3A). We also found many cis-acting elements related to MYB transcription factors in region B (−169 to −478 bp) of the ProGhCKI, such as MYB26PS, MYBPLANT, and MYB1AT (Supplemental Table S2). Therefore, we speculate that MYB transcription factors may play a key role in the regulation of GhCKI expression.
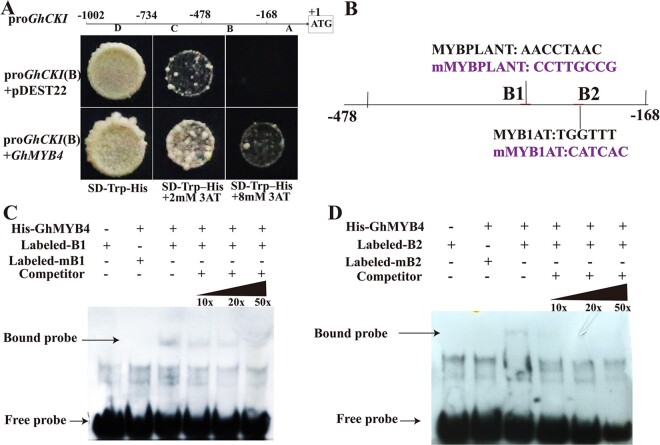
To search for the potential upstream activators of GhCKI under HT, the B region of ProGhCKI was used in a yeast one-hybrid screen against a cotton anther cDNA library. 73 transcription factors were screened, of which 18 were MYB-related transcription factors, among which Ghir_A12G003840 was included (Supplemental Figure S3A and Supplemental Table S1). Further, Ghir_A12G003840 was indeed found to bind to the region B (Figure 1A) of ProGhCKI (Figure 2A). Ghir_A12G003840 is homologous to the MYB4 protein from many other plant species, among which it has 85% identity with TcMYB4 from Theobroma cacao. (Supplemental Figure S3, B and C). Therefore, Ghir_A12G003840 was designated as GhMYB4. The full-length open reading frame of GhMYB4 consists of 774 nucleotides (nt) encoding a peptide of 257 amino acids with a predicted molecular mass of 28.86 kDa (Supplemental Figure S4).
Figure 2.
GhMYB4 activates the expression of GhCKI in vitro. A, Yeast one-hybrid analysis of GhMYB4 binding to the B region of ProGhCKI (−168 to −478 bp). Interaction was determined on SD medium lacking tryptophan and histidine in the presence of 3-AT. ProGhCKI(B)+pDEST22 was used as the negative control. B, Schematic diagram of the GhCKI promoter B region (−168 to −478 bp). B1 and B2 represent the predicted MYB binding site 1 (AACCTAAC) and 2 (TGGTTT) respectively, mB1 and mB2 represent modified sequences of B1 and B2 that changed AACCTAAC to CCTTGCCG and TGGTTT to CATCAC, respectively. C and D, EMSA assays of GhMYB4’s binding to ProGhCKI (B). B1, B2, mB1, mB2 probes were labeled with biotin and incubated with recombinant GhMYB4-His protein. Unlabeled probes were added to compete with biotin-labeled probes. The sequences of WT and mutant probes are shown in Supplemental Table S5.
To identify the specific binding site of GhMYB4 within the region B of ProGhCKI (Figure 1A), we modified the two predicted MYB binding sites AACCTAAC (B1, −275 to −334 bp) and TGGTTT (B2, −215 to −274 bp) to CCTTGCCG (mB1) and CATCAC (mB2), respectively (Figure 2B). A shifted band indicative of GhMYB4 binding to the probe containing the B1 or B2 region was detected (Figure 2, C and D). When excess amounts of unlabeled competitor probes (10X, 20X, 50X) were present, the intensity of the shifted band was substantially reduced (Figure 2, C and D). Similar binding analysis using oligonucleotides with mutated B1 site (AACCTAAC to CCTTGCCG) and B2 site (TGGTTT to CAACCC) did not yield the shifted binding bands with His-GhMYB4 (Figure 2, C and D). These results indicated that GhMYB4 can bind to the GhCKI promoter through B1 and B2 sites.
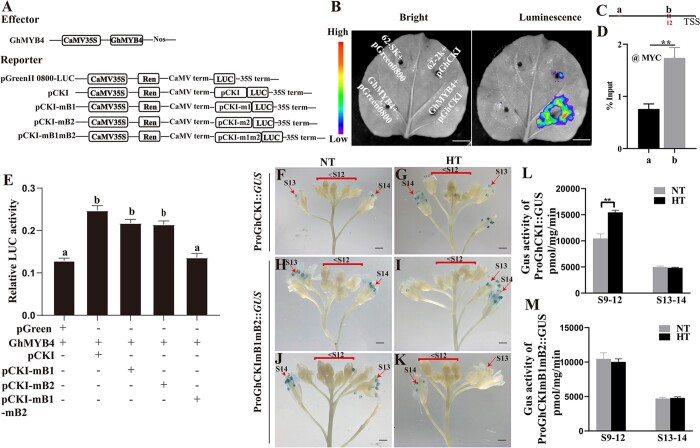
Alternatively, a dual-luciferase reporter system-based luminescence assay in N. benthamiana was used to show GhMYB4’s binding to ProGhCKI in vivo (Figure 3, A and B). GhMYB4 was cloned into the pGreenII 62-SK vector to serve as the effector and various forms of ProGhCKI were cloned into the pGreen II 0800-LUC vector individually to serve as reporters (Figure 3A). We found that co-expression of ProGhCKI:LUC with GhMYB4 in N. benthamiana leaves led to much stronger LUC activity than expressing ProGhCKI:LUC alone (Figure 3B), indicating that GhMYB4 activated the expression of LUC when driven by the GhCKI promoter. Similarly, ChIP-qPCR showed GhMYB4 binding to the B regions (contains the two predicted MYB binding sites) in cotton protoplasts (Figure 3, C and D). Furthermore, co-expression of intact GhCKI promoter with GhMYB4 in cotton protoplasts also resulted in higher LUC activity than the negative control (Figure 3E). Compared with the endogenous GhCKI promoter (pCKI) co-expressed with GhMYB4, the co-expression of GhCKI promoter containing mB1 or mB2 site with GhMYB4 (pCK1-mB1, pCK1-mB2) did not significantly reduce the LUC activity (Figure 3E). However, when the GhCKI promoter containing both mB1 and mB2 sites (pCK1-mB1mB2) was co-expressed with GhMYB4, the LUC activity became much lower, similar to the negative control when the pCK1 was expressed alone without effector (Figure 3E). These results suggested that GhMYB4 binds to both B1 and B2 sites in the promoter of GhCKI and positively regulates its expression, and the deletion of either binding site does not affect the regulation of GhCKI by GhMYB4 significantly.
Figure 3.
Expression of GhCKI is directly activated by GhMYB4. A, Diagrams of the effectors and reporters used in the dual-luciferase reporter assays. Nos, nopaline synthase; CaMV, cauliflower mosaic virus; 35S, promoter of cauliflower mosaic virus; mB1, mutation of B1 from AACCTAAC to CCTTCGGC. mB2, mutation of B2 from TGGTTT to CATCAC. B, Visualization of luminescence signals on N. benthamiana leaves using a cryogenically cooled CCD camera. Construct pGreenII 0800-LUC containing the GhCKI promoter (−1 to −478 bp) and construct pGreenII 62-SK containing GhMYB4 coding region were transiently expressed in N. benthamiana leaves. Bars = 1 cm. C, Schematic of GhCKI promoter. Red lines represent regions amplified by PCR. Black box represent the MYB-binding sites in B region. D, ChIP-qPCR analyses using anti-MYC antibody at chromatin regions (see C) of GhCKI in cotton protoplast. Error bars represent se from three independent biological replicates. Asterisks indicate statistically significant differences (*P <0.05, **P <0.01) by Student’s t test. E, The LUC/REN activity ratios of GhMYB4 binding to ProGhCKI (−1 to −478 bp) in cotton protoplasts. The values are means ± standard deviations (sd), n = 4. Different letters indicate significant differences, as determined by ANOVA followed by Tukey’s multiple comparison test (P <0.05). F–K, GUS expressions in Arabidopsis driven by the endogenous GhCK1 promoter (ProGhCKI) or the mutant GhCKI promoters containing mB1 and mB2 sites (ProGhCKImB1mB2) during anther development under NT and HT conditions. Bars = 1 mm. mB1, mutation of B1. mB2, mutation of B2. Red arrows represent GUS staining in anthers. L and M, GUS enzymatic activity quantifications at different anther developmental stages (see labels in F–K) of ProGhCKI::GUS and ProGhCKImB1mB2::GUS transgenic Arabidopsis under NT and HT conditions, respectively. Error bars represent se from three independent biological replicates. Asterisks indicate statistically significant differences (*P <0.05, **P <0.01) by Student’s t test.
GhCKI responds to HT through the two GhMYB4 binding sites within region B of ProGhCKI
To further confirm that GhCKI responds to HT through the two GhMYB4 binding sites within the B region of its promoter, and the functions of B1 and B2 binding sites may be redundant (Figure 3E), we cloned the GhCKI promoter (−1 to −478 bp) containing mB1 and mB2 mutant sites into a GUS expression vector (ProGhCKImB1mB2::GUS) and transformed the construct into A.thaliana plants for in vivo testing. Ten independent transgenic plants were selected and subjected to HT treatment at flowering time. Under the NT condition, GUS constructs driven by the native promoter ProGhCKI and the altered promoter ProGhCKImB1mB2 showed similar expression patterns, active GUS staining presented in stage 13 and stage 14 (S13 and S14) anthers but not the earlier stages (<S12) (Figure 3, F, H, and J). However, under HT, the native promoter ProGhCKI was able to drive GUS expression at the early stages of anther development (<S12) in addition to S13 and S14 (Figure 3G). On the other hand, ProGhCKImB1mB2::GUS under HT showed expression patterns similar to that under NT, only in S13 and S14 anthers (Figure 3, I and K). In addition, we also detected GUS protein activity at different developmental stages of buds under NT and HT conditions. HT significantly reduced the GUS activity in the anther of ProGhCKI:GUS from S9 to S12, but did not change the GUS activity in S13 and S14 anthers of ProGhCKI:GUS (Figure 3L). However, HT did not change the GUS activity at the early stages (S9–S12) and the late stage (S13 and S14) in the anther of ProGhCKImB1mB2::GUS (Figure 3M). These observations suggest that the mutations in the two GhMYB4 binding sites (B1 and B2) within ProGhCKI have no significant impact on the GhCKI expression under NT, but played an essential role under HT for the induction of GhCKI expression in early flowering stages (<S12).
Overexpression of GhMYB4 leads to male sterility
To better understand the role GhMYB4 plays in HT-induced male sterility, we investigated its biological and physiological properties. By green fluorescent protein (GFP)-fusion localization, GhMYB4 protein was found to reside in the nucleus and plasma membrane (Supplemental Figure S5). Reverse transcription–quantitative PCR (RT-qPCR) analysis of various tissues showed that GhMYB4 was mainly expressed in anthers (Supplemental Figure S6A). The expression level of GhMYB4 was higher at the later stage than the early stage of anther development (Supplemental Figure S6B). More specifically, the expression pattern of GhMYB4 was examined in the HT-tolerant line 84021 and the HT-sensitive line H05 at three different anther stages (TS, TDS, ADS) under both NT and HT conditions (Supplemental Figure S6C). We found that the HT-tolerant line 84021 did not show any significant difference of GhMYB4 expression at all the three stages comparing HT and NT. However, in the HT-sensitive line H05, GhMYB4 was highly induced by HT at the TS stage but not at the TDS and ADS (Supplemental Figure S6C).
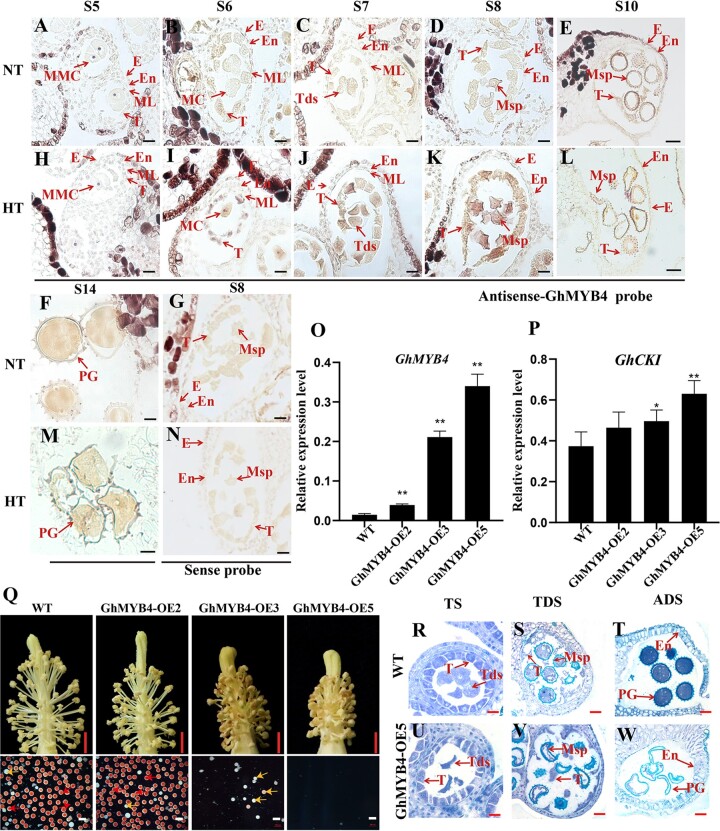
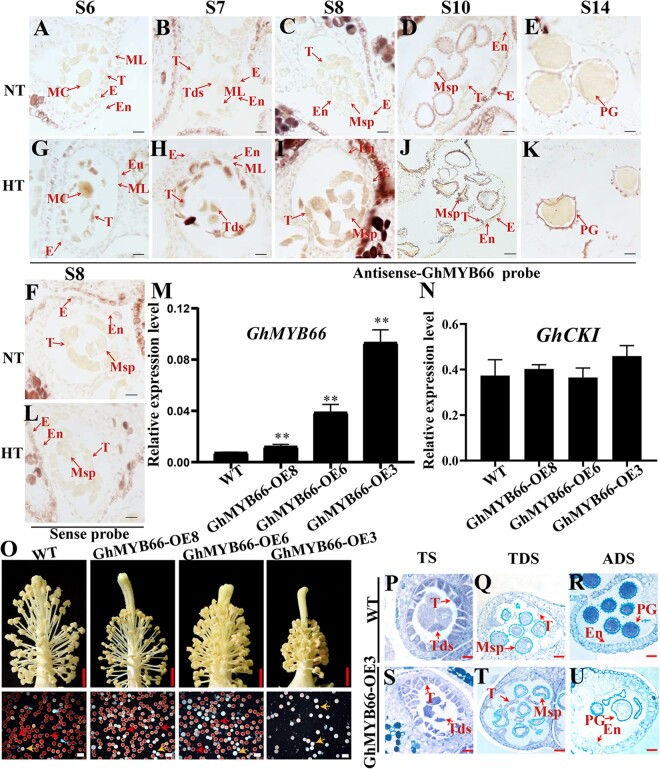
More specific expression pattern of GhMYB4 was investigated in the HT-sensitive line H05 by in situ RNA hybridization (Figure 4, A–N). At Stage 5 (S5), the expression of GhMYB4 was relatively low under NT (Figure 4A). With the development of anthers, the expression of GhMYB4 gradually increased from Stages 6 to 14 (Figure 4, B–F), similar to the trend showed by the RT–qPCR results (Supplemental Figure S6B). GhMYB4 was mainly expressed in tapetum and microspores at different stages (Figure 4, B–F), and induced by HT in the HT-sensitive line H05 during Stages 6–10 (Figure 4, I–L). However, no difference was detected among the expressions of GhMYB4 between NT and HT at Stages 5 (pre-meiosis) and 14 (ADS) (Figure 4, A, H, F, and M). As a negative control, no hybridization signal was detected with a GhMYB4 sense probe (Figure 4, G and N). These results showed that GhMYB4 expression can be induced early during the anther development under HT, which was similar to the expression pattern of GhCKI (Min et al., 2013).
Figure 4.
HT-induced expression of GhMYB4 in early-stage anthers and male sterility caused by overexpression of GhMYB4. A–N, RNA in situ hybridization analysis of GhMYB4 expression in anther of HT-sensitive cotton line H05 under NT (A–F) and HT (H–M) conditions. A and H, B and I, C and J, D, and K, E, and L, F, and M correspond to the anther development stage of S5, S6, S7, S8, S10, and S14, respectively. G and N, GhMYB4 sense probe negative controls at S8. Bars, 50 μm. O and P, Relative expression of GhMYB4 (O) and GhCKI (P) in the T1 generation of GhMYB4 overexpression lines compared with the WT. GhUBQ7 (Ghir_A11G011460) was used as the internal reference. The values are means ± sd, n = 4. Statistical analyses were performed using a Student’s t test compared to WT: *P <0.05; **P <0.01. Q, Flowers and anthers of GhMYB4 over-expressing plants and WT plants under NT. Scale bars above = 5 mm. Red pollen grains are fertile and white pollen grains are sterile. Scale bars below = 200 μm. R–W, Semi-thin sections of GhMYB4 overexpression line GhMYB4-OE5 and WT. Scale bars = 50 μm. TS, tetrad stage; E, epidermis; ML, middle layer; MMC, microspore mother cell; Msp, microspore; MC, meiotic cell; T, tapetum; PG, pollen grain. Tds, tetrad; En, endothecium.
Next, cotton plants overexpressing GhMYB4 were created by Agrobacterium tumefaciens mediated transformation. Three independent plants (GhMYB4-OE2, GhMYB4-OE3, GhMYB4-OE5) with increased expressions of GhMYB4 (2.7, 14.3, 23 folds higher respectively, compared to the wild-type [WT] plants) were selected (Figure 4O, Supplemental Figure S7). Since GhMYB4 can bind to the promoter of GhCKI to regulate its expression, the expression of GhCKI in the GhMYB4-overexpression plants was examined. Results showed that the expression of GhCKI was upregulated significantly in GhMYB4-OE3 and GhMYB4-OE5 plants (Figure 4P) where the expression of GhMYB4 were much higher than the WT control (Figure 4O), which further supported our conclusion that GhMYB4 can activate the expression of GhCKI.
Our previous study showed that overexpressed GhCKI or pre-expression of GhCKI in anther caused cotton anther abortion (Min et al., 2013). The GhMYB4-OE2 plant with relatively lower GhMYB4 expression showed normal anther dehiscence and active pollen grains (Figure 4Q;Supplemental Figure S7). However, GhMYB4-OE3 and GhMYB4-OE5 with much higher GhMYB4 expressions showed male sterile phenotypes such as smaller and indehiscent anthers, less and nonviable pollen grains (Figure 4Q;Supplemental Figure S7). These sterile phenotypes were more severe in GhMYB4-OE5 than that in GhMYB4-OE3 plants (Figure 4Q;Supplemental Figure S7), similar to those of H05 plants under HT conditions (Ma et al., 2018). These results indicated that GhMYB4 was a negative regulator for cotton anther development under HT, with a dosage effect on cotton male sterility.
To explore more on the male sterility in GhMYB4 over-expressing plants, cross sections of anthers at TS, TDS, ADS from GhMYB4-OE5 and WT plants were compared for morphological differences. Although tetrads can be formed in GhMYB4-OE5 anthers (Figure 4, R and U), they were shrunken, while the WT tetrads were circular (Figure 4, R and U). In addition, the microspores of GhMYB4-OE5 at TDS and ADS were severely shrunken too (Figure 4, V and W) when compared to the WT controls (Figure 4, S and T). Thus, the arrest of anther development in GhMYB4 overexpression lines might be due to the microspore abortion. Combined with the gene expression data (Figure 4, O and P), the anther abortion caused by the overexpression of GhMYB4 might be achieved through GhMYB4’s binding to ProGhCKI and the induction of increased expression of GhCKI.
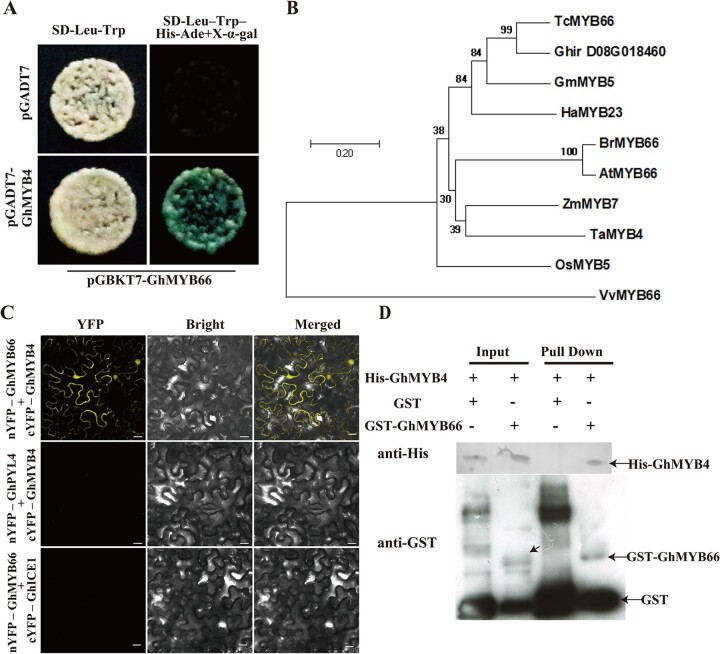
GhMYB66 interacts with GhMYB4 and promotes its binding to ProGhCKI
To explore more how GhMYB4 functions, GhMYB4 was fused with the coding region of GAL4 DNA-binding domain to generate the bait vector pGADT7-GhMYB4, which was used to screen a cotton anther yeast-two-hybrid library. Another R2–R3 type MYB transcription factor, Ghir_D08G018460, was found to have strong interaction with GhMYB4 (Figure 5A). Ghir_D08G018460 is homologous to many MYB genes from other plants. Among them, the highest identity (72%) was with TcMYB66 from T.cacao (Figure 5B;Supplemental Figure S8). Thus, this gene was designated as GhMYB66. The full-length open reading frame of GhMYB66 consists of 693 nt, encoding a 230 amino acid peptide with a predicted molecular mass of 26.74 kDa (Supplemental Figure S9, A and B). Similar to the localization pattern of GhMYB4 (Supplemental Figure S5), the transient expression of 35S::GhMYB66::GFP in N. benthamiana leaves showed GhMYB66 protein in the nucleus and the plasma membrane (Supplemental Figure S10). To verify the interaction between GhMYB4 and GhMYB66, biomolecular fluorescence complementation (BiFC) and GST pull-down assays were performed. In the BiFC assay, strong yellow fluorescent signals appeared in the nucleus and plasma membrane only when both nYFP-GhMYB66 and cYFP-GhMYB4 were expressed together (Figure 5C). In this assay, expressions of nYFP-GhPYL4 plus cYFP-GhMYB4 and nYFP-GhMYB66 plus cYFP-GhICE1 were used as negative controls, from which no yellow fluorescent signal was detected (Figure 5C). In the GST pull-down assay, we observed that GST-GhMYB66, but not GST protein alone, successfully pulled down His-GhMYB4 (Figure 5D). These results revealed that GhMYB4 and GhMYB66 can interact with each other, and the interaction occurred in the nucleus and plasma membrane.
Figure 5.
GhMYB4 interacts with GhMYB66. A, Yeast two-hybrid assays showing the interaction between GhMYB4 and GhMYB66. Blue colonies on SD–Trp–Leu–His–Ade (with X-α-Gal) medium indicates positive interactions. B, Phylogenetic analysis of Ghir_D08G018460 and its homologous genes from other plants: Theobroma cacao (TC), Vitis vinifera (Vv), Helianthus annuus (Ha), Glycine max (Gm), Arabidopsis thaliana (At), O. sativa (Os), Z. mays (Zm), Brassica napus (Br), Triticum aestivum (Ta). The neighbor-joining tree was constructed using the MEGA7 program. The scale bar indicates the number of amino acid substitutions per site. C, BiFC assays between GhMYB66-nYFP and GhMYB4-cYFP in N. benthamiana leaf epidermal cells. YFP florescence indicates a positive interaction. GhPYL4 (REGULATORY COMPONENTS OF ABA RECEPTOR 10)-nYFP and GhMYB4-cYFP, and GhMYB66-nYFP and GhICE1 (INDUCER OF CBF EXPRESSION 1)-cYFP as negative controls. Scale bars = 10 μm. D, GST pull-down assays between GST-GhMYB66 and His-GhMYB4. The GST protein was used as the negative control. Immunoprecipitated proteins were detected using an anti-GST or an anti-His antibody.
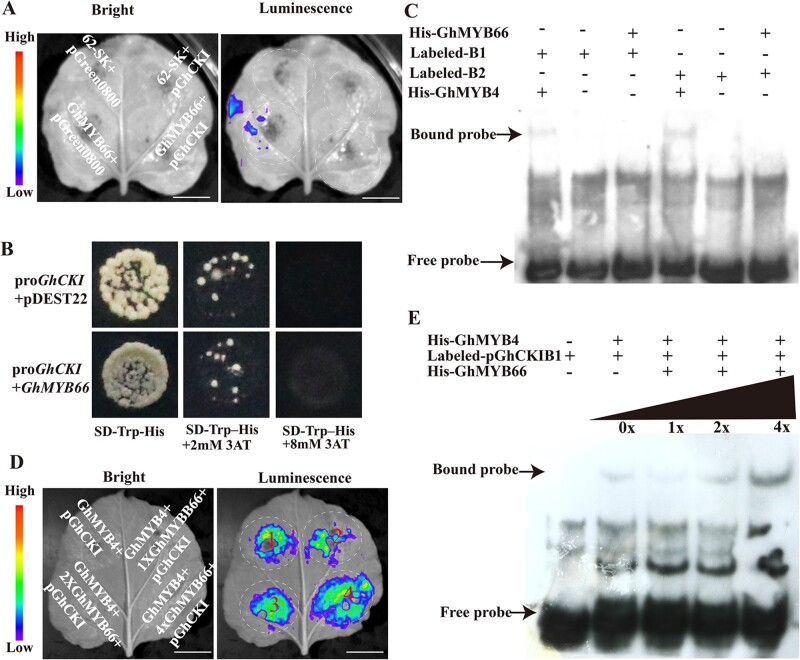
To determine whether GhMYB66 can bind to the ProGhCKI, the luciferase expression vector with ProGhCKI (pGreen0800-ProGhCKI) and GhMYB66 (62-SK-GhMYB66) were generated. When equal amounts of different constructs were co-infiltrated into N. benthamiana leaves, the LUC luminescence intensity, which indicates the transcriptional ability of ProGhCKI, did not show any increase in the presence of GhMYB66 (Figure 6A). Next, GhMYB66 was used as prey and ProGhCKI was used to generate baits in Y1H assay to further examine the interaction between GhMYB66 and ProGhCKI. When co-transformed with ProGhCKI, no difference was detected between GhMYB66 and the negative control pDEST22. In both cases, yeast cells did not grow on SD medium lacking tryptophan and histidine in the presence of 8 mM 3-Amino-1, 2, 4-triazole (3-AT) (Figure 6B). Furthermore, GhMYB66 was cloned into the PET28a vector to generate an N-terminal His-tagged fusion protein His-GhMYB66 (Supplemental Figure S9A). The purified recombinant protein was incubated with oligonucleotide probes containing the B1 site or B2 site in EMSAs, but no band shifting was observed (Figure 6C). These results suggest that GhMYB66 cannot bind to the GhCKI promoter through the B1 or B2 regions.
Figure 6.
GhMYB66 promotes the binding activity of GhMYB4 to GhCKI promoter.A, Luminescence imaging of transient dual-luciferase reporter assay between GhMYB66 and GhCKI promoter (−1 to −1,002 bp). Luminescence signals on N. benthamiana leaves were visualized using a cryogenically cooled CCD camera. Bars = 1 cm. B, Yeast one-hybrid analysis of GhMYB66 binding to the GhCKI promoter (−1 to −1,002 bp). Interaction was determined on SD medium lacking tryptophan and histidine in the presence of 3-AT. ProGhCKI + pDEST22 was used as the negative control. C, EMSA assay of the DNA binding activity of GhMYB66 for the GhCKI promoter. B1, B2 probes were labeled with biotin and incubated with recombinant His-GhMYB66 protein. The His-GhMYB4 was used as the positive control. D, Luminescence imaging of transient dual-luciferase reporter assay. GhMYB4, the GhCKI promoter, and different concentrations of GhMYB66 were co-injected into tobacco (N. tabacum) leaves. 1X: OD (A. tumefaciens strain GV3101 with 62-SK-GhMYB66) = 0.2, 2X: OD = 0.4, 4X: OD = 0.8. Bars = 1 cm. E, EMSA assay of GhMYB66’s impact on GhMYB4 binding to the GhCKI promoter. B1 probes were labeled with biotin and incubated with purified recombinant His-GhMYB4 protein and different concentrations of His-GhMYB66 proteins. 0X indicates no His-GhMYB66 protein, 1X, 2X, 4X represents the GhMYB66 protein amount is the same as the GhMYB4 protein amount, the GhMYB66 protein amount is twice of the GhMYB4 protein amount, and the GhMYB66 protein amount is four times of the GhMYB4 protein amount, respectively.
To explore the effect of GhMYB66–GhMYB4 interaction on the binding of GhMYB4 to the GhCKI promoter, we co-transformed N. benthamiana leaves with GhMYB4 protein, ProGhCKI, and different concentrations of GhMYB66 proteins. With increasing concentrations of GhMYB66, the LUC luminescence intensity also increased significantly (Figure 6D). Under the same conditions, GhCKI promoter and different concentrations of GhMYB66 proteins were subjected to EMSA experiments. The binding signal between ProGhCKI and GhMYB4 protein gradually increased along with the increasing of GhMYB66 protein concentration (Figure 6E). These results indicated that the interaction of GhMYB66 and GhMYB4 could enhance the binding of GhMYB4 protein to the GhCKI promoter.
GhMYB66 expression is induced by HT, and overexpression of GhMYB66 leads to male sterility
To better understand the biological function of GhMYB66, the expression pattern of GhMYB66 was analyzed in the root, stem, leaf, and anthers at different developmental stages. GhMYB66 was found highly expressed in Stage 8 anthers (uninucleate microspore stage, Wu et al., 2015) (Supplemental Figure S11A). Comparing the patterns of expression in anthers from the HT-sensitive line H05 and the HT-tolerant line 84021, GhMYB66 expression was significantly higher in H05 during TS and ADS under HT than that under NT, but not during TDS (Supplemental Figure S11B). On the other hand, GhMYB66 expression in 84021 was only slightly induced by HT compared to NT during ADS but not TS and TDS (Supplemental Figure S11B).
In situ RNA hybridization signals of GhMYB66 were generally faint at Stages 6–14 under NT (Figure 7, A–E), but relatively strong in microspores and pollen grains at Stages 10 and 14 (Figure 7, D and E). However, the in situ RNA hybridization signals were significantly stronger under HT at all stages tested, especially in the tetrad and tapetum of stage 7 (Figure 7, G–K). As a negative control, no hybridization signal was detected with a GhMYB66 sense probe under both NT and HT (Figure 7, F and L). These results showed GhMYB66 was induced to express earlier during the anther development under HT, which was similar to the patterns of GhMYB4 and GhCKI.
Figure 7.
HT-induced expression of GhMYB66 in early-stage anthers and male sterility caused by overexpression of GhMYB66. A–L, Expression analysis of GhMYB66 in anthers of cotton line H05 under NT and HT conditions by RNA in situ hybridization. The antisense-GhMYB66 probe was used for A and G, B and H, C and I, D and J, E and K at anther development stages of S6, S7, S8, S10, S14 respectively, under NT (A–E) and HT (G–K). The sense-GhMYB66 probe was used for F and L at stage 8 anthers under NT (F) and HT (L), respectively. Bars, 50 μm. M and N, Relative expression of GhMYB66 (M) and GhCKI (N) in the T1 generation of GhMYB66 overexpression lines compared with the WT. GhUBQ7 (Ghir_A11G011460) was used as the internal reference. The values are means ± sd, n = 4. Statistical analyses were performed using a Student’s t test: *P <0.05; **P <0.01. O, The phenotype of GhMYB66 over-expressing plants and WT under NT. Scale bars above = 5 mm. Scale bars below = 200 μm. The red pollen grains represent fertile pollen grains and the white pollen grains represent sterile pollen grains. The red arrow represents fertile pollen and the yellow arrow represents sterile pollen. P–U, Semi-thin section results of GhMYB66 overexpression line GhMYB66-OE3 and WT. Scale bars = 50 μm. TS, tetrad stage; Msp, microspore; E, epidermis; ML, middle layer; MC, meiotic cell; T, tapetum; PG, pollen grain; Tds, tetrads; En, endothecium.
Next, GhMYB66 overexpression plants were developed (Supplemental Figure S12). In GhMYB66-OE8, GhMYB66-OE6, and GhMYB66-OE3 plants, the expression of GhMYB66 was increased by 2-, 5-, 12-fold, respectively, compared to the WT control (Figure 7M, Supplemental Figure S12). The anthers of GhMYB66-OE8 plant with relatively lower GhMYB66 expression were able to dehiscence normally, and the pollen grains of the GhMYB66-OE8 plant were viable (Figure 7O). In GhMYB66-OE6 plant, more anther indehiscence and more pollen grains were inactive, compared with those in GhMYB66-OE8 (Figure 7O). However, GhMYB66-OE3 with even higher GhMYB66 expression showed very clear sterile phenotypes such as smaller and indehiscent anthers, and much less viable pollen grains (Figure 7O). These sterile phenotypes are the same as those of H05 plant under HT and the GhMYB4 overexpression lines under NT (Figure 4Q). The cross-sections of anthers showed that the tetrad of GhMYB66-OE3 was shrunken during TS, while WT had the normal tetrad (Figure 7, P and S). At TDS and ADS, the microspores and pollen grains from GhMYB66-OE3 were shrunken and lacked contents (Figure 7, Q–R and T–U). The above results indicated that GhMYB66 had a dosage effect on inducing cotton male sterility and acted as a negative regulator for cotton anther development under HT, similar to GhMYB4.
As GhMYB66 protein can promote the binding of GhMYB4 to the GhCKI promoter (Figure 6E), and GhMYB4 can activate the transcription of GhCKI (Figure 3E), the expression of GhCKI in the anthers of GhMYB66 overexpression plants was examined. Surprisingly, the GhCKI expressions from the three OE plants showed no significant difference compared to the WT (Figure 7N). Thus, we speculate that although GhMYB66 can interact with GhMYB4, thereby increasing the binding of GhMYB4 to the GhCKI promoter, other mechanisms may exist to account for the abortion caused by GhMYB66 overexpression.
DNA methylation is involved in the regulation of GhMYB4 expression in response to HT
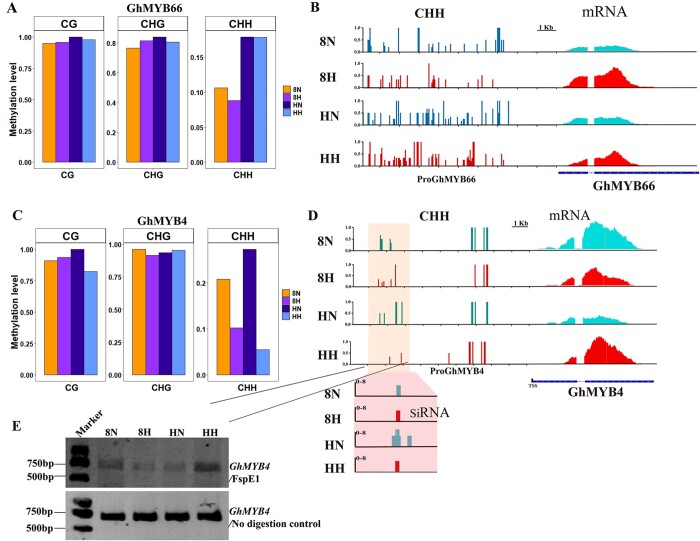
Both GhMYB4 and GhMYB66 were upregulated by HT at TS (Supplemental Figures S6, C and S11, B). To explore how GhMYB4 and GhMYB66 responds to HT at TS, the sequence compositions of GhMYB4 and GhMYB66 promoters were compared between HT-sensitive lines and HT-tolerant lines within a native population of 510 upland cotton lines. No difference within the 2-kb promoter regions of GhMYB4 or GhMYB66 was found between HT-sensitive lines and HT-tolerant lines in the population (Supplemental Figures S13 and S14). Therefore, epigenetic modification may play a role in regulating the expression of GhMYB4 and GhMYB66. DNA methylation is often associated with gene expression regulation (Jullien et al., 2012) and our previous study showed that inhibiting DNA methylation in H05 plants led to shrunken microspores under both NT and HT conditions (Ma et al., 2018). Thus, the methylation patterns of GhMYB4 and GhMYB66 promoters were examined during TS. Compared to NT, no obvious difference was detected in GhMYB66 promoter methylation pattern or level in both H05 and 84021 plants under HT (Figure 8, A and B). However, the CHH methylation level of GhMYB4 promoter differed remarkably between HT and NT for both H05 and 84021 plants, while CG and CHG methylation patterns remained similar (Figure 8C). The distributions of CHH methylation on the length of GhMYB4 promoter under NT and HT also differed significantly at TS (Figure 8D). Comparing the two different cotton lines, much less CHH DNA methylation was found in H05 anthers than in 84021 anthers at the TS under HT in the ProGhMYB4 region (−1,350 to −1,800 bp) (Figure 8D). The methylation-sensitive Chop-PCR testing verified the substantial differences in methylation patterns on the GhMYB4 promoter (Figure 8E). To the contrary, the mRNA level of GhMYB4 was higher in H05 and lower in 84021 at TS anthers under HT (Figure 8B). This result indicated that the expression of GhMYB4 might be specifically regulated by CHH methylation changes under HT. Consistent with this hypothesis, the abundance of siRNA also showed the same trend as the methylation level of CHH in these two cotton lines under NT and HT conditions (Figure 8D). Therefore, we hypothesize that siRNA-mediated CHH methylation might regulate the expression of GhMYB4 in response to HT.
Figure 8.
DNA methylation distribution on GhMYB66 and GhMYB4 promoter. A, Levels of CG, CHG, CHH methylations on the GhMYB66 promoter in cotton anther during TS under NT and HT. The degree of methylation is calculated as mCs/totalCs, where mCs = number of mCs; totalCs = mCs + unmethylated Cs. B, A genome browser snapshot showing the CHH DNA methylation level on GhMYB66 promoter at tetrad stage in anther of cotton line H05 and 84021 under NT and HT. C, Levels of CG, CHG, CHH methylations on the GhMYB4 promoter in cotton anther during TS under NT and HT. The degree of methylation is calculated as mCs/totalCs, where mCs = number of mCs; totalCs = mCs + unmethylated Cs. D, A genome browser snapshot showing the CHH DNA methylation level and the siRNA abundance in GhMYB4 promoter, and the mRNA level of GhMYB4 at tetrad stage in anther of H05 (HT-sensitive line) and 84021 (HT-tolerant line) under NT and HT. The marked areas indicate substantial differences. Red represents high temperature, blue represents NT. E, Analysis of DNA methylation status on the GhMYB4 promoter by methylation-sensitive PCR (Chop-PCR) at tetrad stage in anther of cotton line H05 and 84021 under NT and HT. Undigested genomic DNA was used as a control. The 8N and 8H refer to 84021 (HT-tolerant line) under NT and HT conditions, respectively; HN and HH refer to H05 (the HT-sensitive line) under NT and HT conditions, respectively.
Discussion
Cotton is the most important economic crop in the world, and it is the foundation of the textile industry (Wang et al., 2017, 2021). However, with the increasing frequency of global HT stress, cotton has also been greatly affected (Snider et al., 2011). Under HT stress, the male organs are most susceptible to infertility (Djanaguiraman et al., 2018). As the male organs of cotton are sterile, the subsequent fertilization process cannot be completed normally, resulting in a significant reduction in cotton fiber production (Sage et al., 2015). Therefore, it is very important to analyze the molecular mechanism of how cotton anther responds to HT stress.
Transcription factors regulate downstream genes through binding to specific promoter regions. The common binding sites of MYB transcription factors include CNGTT(A/G), ACC(A/T)A(A/C), TTAGGG, and others (Prouse and Campbell, 2012). In this report, we demonstrated that the B region of GhCKI promoter has a core function in responding to HT (Figure 1). The core promoter region of GhCKI involved in responding to HT contains two MYB binding sites, B1 (AACCTAAC) and B2 (TGGTTT) (Figure 2B); both sequences were reported before to be recognized by MYB transcription factors (Abe et al., 2003; Chen et al., 2006; Zhou et al., 2009). When these two MYB binding sites in ProGhCKI were altered (ProGhCKImB1mB2), ProGhCKI could not respond to HT any more to induce the accumulation of GUS protein in the early anthers in Arabidopsis (Figure 3, F–M). Therefore, these MYB binding sites are essential for GhCKI’s response to HT. A previous study showed that both AtCKL2 and AtCKL7 are GhCKI homologs in Arabidopsis, which were induced by HT in the early stage of anther development (Li et al., 2018). Both AtCKL2 and AtCKL7 have two MYB binding sites in their promoters, including the B2 (TGGTTT) region on which AtMYB4 was predicted to bind (Li et al., 2018). Therefore, it is possible that Arabidopsis has a similar HT-response mechanism as cotton, with these two MYB binding sites being essential to respond to HT. In that direction, editing the B region or the binding site of the two cis-elements on the CKI promoter may prevent CKI from being expressed in advance in the early anthers under HT, and thus to prevent male sterility that is normally induced by HT. This may provide an approach for the creation of temperature-tolerant plants in the future.
We observed that the expressions of GhMYB4 and GhMYB66 showed increases of 33 and 4.3 fold, respectively, at the TS stage in the HT-sensitive line H05 but not in the HT-tolerant line 84021 under HT (Supplemental Figures S6, C and S11, B). However, HT-mimic male sterility phenotypes appeared when the expression of GhMYB4 was increased to about 11 times in the GhMYB4 overexpression line (GhMYB4-OE8) and when the expression of GhMYB66 is increased about five times in the GhMYB66 overexpression line (GhMYB66-OE6) (Supplemental Figures S7 and S12). This may be due to the fact that the gene expressions we measured using molecular biological techniques may not fully represent the true cellular expression levels of these genes in specific cell types or tissues. We speculate that the engineered expressions of GhMYB4 (increased 11 times) and GhMYB66 (increased five times) may be able to mimic the in-planta expressions of GhMYB4 and GhMYB66 in H05 under HT. Besides, GhMYB4 protein can recognize the GhCKI promoter to activate the expression of GhCKI (Figure 3E) and the expression of GhCKI was upregulated significantly in GhMYB4 overexpression plants (Figure 4P). Therefore, the induced expression of GhMYB4 under HT may have caused male sterility through the signaling pathway of GhCKI. However, the GhMYB66 protein could not bind to the GhCKI promoter (Figure 6, A–C) and the expression of GhCKI showed no significant difference in the anthers of GhMYB66 overexpression plants compared to controls (Figure 7N). Therefore, it seems that GhMYB66 overexpression did not lead to higher expression of GhCKI. This may be due to GhMYB4 becoming the limiting factor in this regulation signaling cascade under HT, so even when GhMYB66 is overexpressed, there is not enough GhMYB4 protein around to induce a higher expression of GhCKI. Since overexpression of GhMYB66 did not induce GhCKI expression (Figure 7N) but still led to male sterility (Figure 7O), there must be other genes/pathways downstream of GhMYB66 that function under HT to lead to male sterility.
The regulation of gene expression seems to play a very important role in response to HT in our study. DNA methylation of promoter is typically associated with gene repression, disruption of which usually leads to altered gene expression (Hu et al., 2014; Harris et al., 2018). In plants, the RNA-directed DNA methylation (RdDM) pathway is responsible for the initial establishment of CG, CHG, and CHH methylations (He et al., 2011). In this study, we observed that under HT, both GhMYB4 and GhMYB66 were upregulated in the HT-sensitive line H05 (Supplemental Figures S6, C and S11, B), the CHH methylation on the GhMYB4 promoter was significantly reduced (Figure 8, C–E), and the corresponding siRNA was also significantly reduced (Figure 8D). We believe that HT may have affected the deposition of siRNA-mediated DNA methylation on the promoter of GhMYB4 to regulate its expression. This is consistent with our previous observation that disrupted CHH DNA methylation caused by HT enhanced the expression of amylase genes, which resulted in excessive consumption of starch, leading to male sterility in H05 (Ma et al., 2018). In addition, we also found many MYB binding sites on the promoter of GhMYB4 (Supplemental Table S3), so GhMYB4 may be regulated by other MYB transcription factors. However, the DNA methylation on the GhMYB66 promoter did not show significant change under HT (Figure 8, A and B). At the same time, the GhMYB66 promoters showed no sequence difference between the HT-sensitive lines and the HT-tolerant lines from a population of native cotton collection (Supplemental Figures S13 and S14). We speculate that there may be other ways to regulate the expression of GhMYB66 under HT, such as histone modification or regulation by other transcription factors. In Arabidopsis, H3K27me3 binds to the promoter of flowering repressor gene FLOWERING LOCUS C (FLC) (MADS-box transcription factor) and the florigen gene FLOWERING LOCUS T (FT) to play essential roles in flowering time control (Zhang et al., 2020). In the process of cotton fiber development, GhWRKY16 binds to the W-box (WRKY binding site) on the GhMYB25 promoter to regulate the expression of GhMYB25, thereby regulating fiber initiation (Wang et al., 2021). At the same time, GhWRKY16 can bind to the W-box (WRKY binding site) on the GhMYB109 promoter to modulate the expression of GhMYB109, thus regulating the fiber elongation process (Wang et al., 2021). When the GhMYB66 promoter region (2 kb) was examined for cis-regulatory elements, we found some transcription factor binding sites, including the MYB binding site and the W-box (Supplemental Table S4). Therefore, other transcription factors may have been involved in regulating the expression of GhMYB66 under HT.
MYB transcription factors have been shown to have versatile biological functions in plants (Dubos et al., 2010), especially during the plant reproduction development. For example, MYB103, MYB33, MYB65, MYB32, and MYB26 are all essential for the development of Arabidopsis anthers (Higginson et al., 2003; Steiner-Lange et al., 2003; Preston et al., 2004; Millar and Gubler, 2005). AtMYB103 was highly expressed in the tapetum. When the expression of ATMYB103 decreased, the development of pollen and tapetum was affected (Higginson et al., 2003). The myb33 myb65 double mutant has defects in the development of anthers, the tapetum was hypertrophy at the pollen mother cell stage, leading to abortion before meiosis during pollen development (Millar and Gubler, 2005). AtMYB32 was strongly expressed in the tapetum, and the pollen grains of atmyb32 mutant plant was distorted and lacked cytoplasm (Preston et al., 2004). With a transposon inserted in AtMYB26, the Arabidopsis mutant plants showed nondehiscent anthers even though the pollen grain maturation was not affected (Steiner-Lange et al., 2003). Oryzasativa Carbon Starved Anther (OsCSA), an R2R3 MYB transcription factor, is a key transcriptional regulator for sugar partitioning in rice during male reproductive development (Zhang et al., 2010). In our study, the overexpression lines of GhMYB4 and GhMYB66 showed a sterile phenotype similar to H05 under HT (Figures 4Q and 7O). So both GhMYB4 and GhMYB66 are negative regulators of cotton anther development in response to HT. We also found many other MYB-related transcription factors that share the same expression trend as GhCKI in response to HT (Supplemental Figures S2 and S3A). Therefore, we speculate that GhMYB4 and GhMYB66 are not the only regulators of GhCKI expression, and many other MYB transcription factors may be involved in the process of cotton anther response to HT. Interestingly, MYB66 is implied to play a role during ovule development as it is expressed almost 200× higher in aposporous samples compared to sexual ones in Hypericum perforatum (Table S7 from Galla et al., 2019). In this study, MYB66 was further shown to have an expression pattern similar to a few genes whose homologs in Arabidopsis are involved in RNA splicing/processing, including AtCDC5 (A.thaliana Cell Division Cycle 5), RDM16 (RNA-directed DNA methylation 16), At3g47120 (RNA-binding motif protein, X-linked 2), and MAC5A (MOS4-associated complex subunit 5A) (Table S10 from Galla et al., 2019). It will be interesting to explore the molecular mechanisms that how GhMYB66 regulates male sterility in HT-sensitive cotton plants under HT.
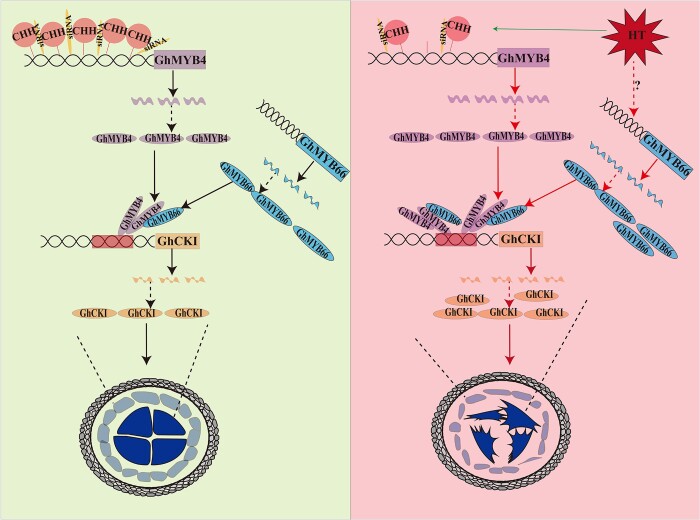
In this study, we explored the upstream regulatory mechanism of GhCKI, a HT induced key kinase, and found that MYB transcription factor regulates the expression of GhCKI (Figure 3). We found that GhMYB4 can bind to the promoter of GhCKI in response to HT and GhMYB66 can interact with GhMYB4 to promote the binding of GhMYB4 to the GhCKI promoter, which ultimately leads to an increase in the expression of GhCKI. The increased expression of GhCKI in the early anther development stage led to abnormal development of the anthers, ultimately leading to the male sterile phenotype of the plants. We also found that both the abundance of siRNA and the CHH methylation modification on the GhMYB4 promoter decreased under HT, which might be a factor causing the increase of GhMYB4 expression in anthers at TS. Based on our results, we propose a working model for the GhMYB66–GhMYB4–GhCKI regulatory pathway (Figure 9).
Figure 9.
A model for GhMYB4 together with GhMYB66 respond to HT stress to regulate GhCKI in cotton anther. This model shows that the abundance of sRNA and the CHH methylation modification on the GhMYB4 promoter decreased under HT, which increased the expression of GhMYB4 in anther at TS. GhMYB4 can bind to the promoter of GhCKI in response to HT and GhMYB66 can interact with GhMYB4 to promote the binding of GhMYB4 to the GhCKI promoter, which ultimately leads to an increase in the expression of GhCKI. The increased expression of GhCKI in the early anther development stage will lead to abnormal development of the anthers, ultimately leading to the male sterile phenotype of the plants. The red arrow represents promotion, the green arrow represents suppression, the dotted line represents possibility, the solid line represents supported by evidence. HT, high temperature.
Materials and methods
Plant materials and growth conditions
G. hirsutum cv. YZ1 was used for transformation. Transgenic lines derived from YZ1 were planted in the field under standard farming conditions during the normal season, or grown in the greenhouse during the winter at Huazhong Agricultural University, Wuhan, China. The greenhouses were kept at NT (28–35°C/20–27°C day/night) or HT (35–39°C/29–31°C day/night) with 12-h light/12-h dark conditions. Arabidopsis thaliana ecotype Columbia (Col-0) was used to generate transgenic plants, which were grown under long day conditions (16-h light/8-h dark) with white fluorescent light at 20°C and a relative humidity of 60%. For HT treatment, the A.thaliana plants at blooming stage were transferred to growth chambers at 33°C for 1 day under long-day conditions (16-h light/8-h dark) before the primary inflorescence were collected. Seedlings of N.benthamiana were grown in a greenhouse under a 16-h/8-h light/dark photoperiod at 25°C.
Vector construction and transformation
To generate the overexpression constructs, the full-length CDS of GhMYB4 and GhMYB66 without their stop codons were amplified and inserted into the pGWB409 and pGWB417 vectors via Gateway BP and LR recombination reactions (Invitrogen) separately. These vectors were then introduced into G. hirsutum cv. YZ1 using A.tumefaciens-mediated transformation (Jin et al., 2006). For expression pattern analysis, the promoter regions of GhCKI of different lengths were fused with the GUS reporter gene in pGWB433 (Li et al., 2018). All constructs were introduced into A.tumefaciens GV3101. The proGhCKI::GUS constructs were then transformed into A.thaliana Col-0 plants using the floral dipping method (Zhang et al., 2006). The primers used in this study are listed in Supplemental Table S5.
Histochemical assay of GUS activity
GUS activity assay was performed as described previously (Li et al., 2018). In brief, fresh tissues were collected from A.thaliana plants and incubated in staining solution immediately at 37°C overnight and then washed with 75% (v/v) ethanol. The staining solution was composed of 0.9 g L−1 5-bromo- 4-chloro-3-indolylglucuronide, 50 mmol L−1 sodium phosphate buffer (pH 7.0), 20% (v/v) methanol and 100 mg L−1 Chloromycetin. The samples were examined and photographed with a stereomicroscope (Leica Microsystems, Germany). The quantitative analysis of GUS activity in ProGhCKI::GUS and ProGhCKImB1mB2::GUS transformants was performed as described previously (Li et al., 2018). Briefly, the quantitative analyses of GUS activity were expressed as pmol 4-methylumbelliferone mg−1 protein min−1. Fluorescence was measured at an excitation wavelength of 365 nm and an emission wavelength of 455 nm using the Infinite 200 PRO multimode reader (Tecan).
Yeast one-hybrid assay
The full-length CDS sequence of GhMYB4 was inserted into the entry vector pDONR221 by the BP reaction. Then the LR reaction was carried out with the target vector pDEST22 to obtain the recombinant vector, which was transformed into the yeast strain Y187. The 310-bp (−169 to −478 bp) region of the GhCKI promoter was integrated into the yeast bait vector pHisi-1 by the infusion method (Thermo Fisher). The vector was linearized by XhoI and transferred to the yeast strain YM4271. The recombinant bait and prey-positive plasmid yeast Y187 and Y4271 were co-cultured in YPDA medium at 30°C for 24 h. About 5 μL of fusion yeast was spotted onto SD–Trp–His solid medium containing 2 or 8 mM 3-AT (Sigma), and incubated at 30°C for 3 days, to analyze the relationship between GhMYB4 and the promoter regions of GhCKI. The primer sequences used in this study are shown in Supplemental Table S5.
RNA in situ hybridization
A fragment of 170 bp and another one at 188 bp were amplified from the coding sequences of GhMYB4 and GhMYB66, respectively, using the corresponding specific primer pairs (Supplemental Table S5). These two fragments were inserted into the pGEM-T vector (Promega, http://www.promega.com/) separately for sequencing to confirm their identities, and used for RNA in situ hybridization according to Min et al., 2013.
Yeast two-hybrid assay
To characterize the interaction between GhMYB4 and GhMYB66 protein, a bait vector carrying the full-length GhMYB4 ORF fused to the GAL4 DNA-binding domain in a pDEST32 (Invitrogen) vector and a prey vector carrying full-length GhMYB66 ORF fused to the GAL4 DNA activation domain in a pDEST22 (Invitrogen) vector were generated according to the manufacturer’s instructions (ProQuest Two-Hybrid System; Invitrogen). pDEST32 and pDEST22 were introduced into AH109 and Y189 strains, respectively. The primers used in the study are listed in Supplemental Table S5.
Subcellular localization and BiFC assays
The CDS of GhMYB4 and GhMYB66 were amplified and inserted into pMDC83 (BioVector NTCC) to obtain vectors in which GFP was fused with GhMYB4 and GhMYB66, respectively, at their C-termini. The 35S::GFP vector was used as a control. The vector was then transformed into N. benthamiana leaves by agroinfiltration. After 48 h, the green fluorescence in leaves was detected using a confocal microscope (488-nm excitation wavelength, 44% transmissivity, 100-nm collection bandwidth and gain was 1). For BiFC assays, the CDSs of GhMYB4 and GhMYB66 were cloned into pS1301nYFP or pS1301cYFP vectors. Each pair of the two-gene combination constructs were transformed into A. tumefaciens strain GV3101, then transiently expressed in N. benthamiana leaves by injection with needleless syringes. The fluorescence in the epidermal cells was observed ∼60 h later using a confocal microscope (559-nm excitation wavelength, 35% transmissivity, 100-nm collection bandwidth, and gain was 1). The primers used in this study are listed in Supplemental Table S5.
Dual-luciferase reporter assays
The CDS of GhMYB4 and GhMYB66 was cloned into the pGreen II 62-SK vector as the effectors, the promoter of GhCKI was cloned into the pGreen II 0800-LUC vector as the reporter. The effector and reporter were co-transformed into protoplasts of G. hirsutum cv. YZ1. The transformed protoplasts were cultured at 23°C in the dark for 24 h. The LUC and REN activity assays were conducted using a Dual-Luciferase Reporter Assay System (DLR, Promega) according to the manufacturer’s instructions and quantified using a Multimode Plate Reader (PerkinElmer). The DLR system assays were employed in N. benthamiana leaves. Different combination pairs were injected into the N. benthamiana leaves, the infiltrated plants were grown at 25°C for 48 h, and measurements of LUC luminescence were performed as described previously (Hu et al., 2018).
Reverse transcription–quantitative PCR
Various plant samples were collected and immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from the collected cotton tissues using previously published methods (Deng et al., 2012). First-strand cDNA was generated from 3-μg total RNA using the M-MLV reverse transcriptase (Invitrogen). The cDNA was used as a template for RT-qPCR. The RT-qPCR reactions were performed using the 7500 Real-Time PCR System (Applied Biosystems). The primers used in this study were listed in Supplemental Table S5.
ChIP-qPCR
Protoplasts were isolated from G.hirsutum cv. YZ1 embryogenic calli (described by Yang et al., 2008) and transformed with 35S::GhMYB4:MYC constructs. ChIP-based transient expression testing was performed as previously described with minor modification (Lee et al., 2017). Briefly, protoplasts were fixed with 1% (v/v) formaldehyde for 10 min at room temperature first, and the remaining formaldehyde was neutralized with 0.125-M glycine for 5 min. Fixed protoplasts were collected by centrifuging at 1,000g at 4°C for 5 min. Bioruptor was used at high power with 30-s-on/30-s-off cycles for 15 times until the average chromatin size was approximately 300 bp. Anti-MYC (Abcam, ab16898) antibodies were used to perform immunoprecipitations. Immune complexes were eluted from the protein A or G agarose/salmon sperm DNA beads (Millipore) and reverse cross-linked by incubation for 6 h at 65°C. Then qPCR reactions were performed. The primers are listed in Supplemental Table S5.
Protein expression and pull-down assays
The CDSs of GhMYB4 and GhMYB66 were cloned into the pET-28-a vector to produce His-tagged proteins, and the CDSs of GhMYB66 were cloned into the pGEX-4T-1 vector to produce GST-tagged proteins using Gateway BP and LR recombination reactions. The plasmid constructs were transferred into Escherichia coli strain BL21 and induced at 16°C with 0.5 mM isopropyl-β-d-1-thio-galactopyranoside. Pull-down assays between the GhMYB4-His and GhMY66-GST proteins were conducted using MangeGST Protein Purification System (Promega) according to the manufacturer’s instructions, and immunoblotted with an anti-His or an anti-GST antibody.
Electrophoretic mobility shift assay
EMSAs were performed using a Chemiluminescent EMSA kit (Beyotime Biotechnology) according to the manufacturer’s protocol. The oligonucleotide probes contained B1, B2, mB1, and mB2 were synthesized individually and labeled with biotin by Sangon Biotech (Shanghai). His-tagged recombinant GhMYB4 was generated by cloning GhMYB4 into the pET-28a vector with a His-tag fusion at the N-terminus. The His-tagged recombinant GhMYB4 (His-GhMYB4) was purified with MangeHis Protein Purification System (Promega), then incubated with the oligonucleotide probes containing B1, B2, mB1, and mB2 regions, respectively. The competition experiment was performed with different amounts of non-labeled oligonucleotides. Double-stranded oligonucleotides used in this study are listed in Supplemental Table S5.
Tissue sectioning, staining, and imaging
Tissue dissection, staining, and imaging was performed as described previously (Ma et al., 2018). In brief, anthers were fixed in FAA (50-mL absolute ethanol, 10-mL 37% (v/v) formaldehyde solution, 5-mL acetic acid and diluted with water to 100 mL) after removal of bracts and petals. Dehydration used a graded ethanol series (30%, 50%, 70%, 95%, and 100%). Tissue was embedded in epoxy and sectioned at 10-µm thickness. Toluidine blue solution (1%) and aniline blue solution (1%) were used to stain the anther sections. A Zeiss Axio Scope A1 microscope was used to image the samples under bright field for toluidine blue staining and at 395-nm excitation for aniline blue staining.
Bioinformatics analysis
The promoter sequence (2-kb upstream of transcription start site) of GhMYB4 and GhMYB66 in the 20 HT-sensitive lines and 20 HT-tolerance lines were extracted from our cotton resequencing database (Ma Y, Min L, Zhang X, et al., unpublished data), with TM-1 as the reference genome (Wang et al., 2019). Sequence alignment was performed using CLUSTALX (Thompson et al., 2002). The modified abundance of DNA methylation and small RNA on the GhMYB4 and GhMYB66 promoters were analyzed (2-kb upstream of transcription start site) based on our pervious DNA methylation and small RNA data (Ma et al., 2018) (NCBI Accession Number: PRJNA393079). The expression pattern of GhMYB4 gene was obtained from our previous RNA-seq data (Ma et al., 2018) (Accession Numbers of NCBI database: PRJNA393079). Different omics data were visualized using the Integrative Genomics Viewer (https://software.broadinstitute.org/software/igv/).
Chop-PCR
For Chop-PCR assays, briefly, 1 μg of genomic DNA of each sample was completely digested with the methylation-sensitive enzyme FspEl (NEB company, R0662S), and then used as template for PCR amplification. Undigested genomic DNA was used as a control (Dasgupta and Chaudhuri, 2019; Kong et al., 2020). The band intensity after electrophoresis serves as a reflection of methylation levels at the tested loci. The primers used in this study were listed in Supplemental Table S5.
Availability of data and materials
The data that support the results are included within the article and its additional files. Other relevant materials are available from the corresponding authors on reasonable request.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: GhCKI (XP_016719690), GhMYB4 (NP_001314278), GhMYB66 (XP_016716195), GhUB7 (DQ116441). The whole-genome bisulfite sequencing reads, small RNA sequencing reads, and RNA-sequencing reads have been deposited with the National Center for Biotechnology Information under Sequence Read Archive under accession number PRJNA393079.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The sequence of GhCKI promoter.
Supplemental Figure S2. Cluster analysis and statistics of transcription factor types.
Supplemental Figure S3. The expression patterns of MYB transcription factors and sequence alignment of GhMYB4.
Supplemental Figure S4. Purification of GhMYB4 protein.
Supplemental Figure S5. Subcellular localization of GhMYB4.
Supplemental Figure S6. Expression profiles of GhMYB4.
Supplemental Figure S7. The fertility of T1 generation of GhMYB4 overexpression lines.
Supplemental Figure S8. Multiple sequence alignments between GhMYB66 and TcMYB66 of Theobroma cacao.
Supplemental Figure S9. Purification of GhMYB66 protein.
Supplemental Figure S10. Subcellular localization of GhMYB66.
Supplemental Figure S11. Expression profiles of GhMYB66.
Supplemental Figure S12. The fertility of T1 generation of GhMYB66 overexpression lines.
Supplemental Figure S13. The sequence alignment of the GhMYB4 promoter in the resequencing population.
Supplemental Figure S14. The sequence alignment of the GhMYB66 promoter in the resequencing population.
Supplemental Table S1. Transcription factors with the same expression pattern as GhCKI.
Supplemental Table S2. The predicted cis elements on the GhCKI promoter (−168 bp to −478 bp).
Supplemental Table S3. The predicted cis elements on the GhMYB4 promoter (2 kb).
Supplemental Table S4. The predicted cis elements on the GhMYB66 promoter (2 kb).
Supplemental Table S5. All primers used in this study.
Supplementary Material
Acknowledgments
We thank Prof. Qing Li (Huazhong Agricultural University, China) for assistance with Chop-PCR and Prof. Yu Zhao (Huazhong Agricultural University, China) for technical support with ChIP-qPCR.
Funding
Project supported by the National Natural Science Foundation of China (32072024), the Fundamental Research Funds for the Central Universities (2021ZKPY019), the Open Funds of the National Key Laboratory of Crop Genetic Improvement.
Conflict of interest statement. None declared.
Contributor Information
Yanlong Li, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Yaoyao Li, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China; College of Life Sciences, State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou 510642, Guangdong, China.
Qian Su, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Yuanlong Wu, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Rui Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Yawei Li, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Yizan Ma, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Huanhuan Ma, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Xiaoping Guo, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Longfu Zhu, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Ling Min, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
Xianlong Zhang, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China.
YL.L. and YY.L. carried out the experiments and wrote the main manuscript text, Q.S., YL.W., R.Z., YW.L. performed anther sampling and RNA extraction. YZ.M. and HH.M. performed pollen staining and pollen counts. L.M., XL.Z., LF.Z., and XP.G. designed and supervised the research, and L.M. and XL.Z. revised the manuscript. All authors reviewed the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Ling Min (lingmin@mail.hzau.edu.cn).
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15(1): 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abiko M, Akibayashi K, Sakata T, Kimura M, Kihara M, Itoh K, Asamizu E, Sato S, Takahashi H, Higashitani A (2005) High-temperature induction of male sterility during barley (Hordeum vulgare L.) anther development is mediated by transcriptional inhibition. Sex Plant Reprod 18(2): 91–100 [Google Scholar]
- Bita CE, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaretto JA, El-Kereamy A, Zeng B, Stiegelmeyer SM, Chen X, Bi YM, Rothstein SJ (2016) Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PW, Chiang CM, Tseng TH, Yu SM (2006) Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell 18(9): 2326–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta P, Chaudhuri S (2019) Analysis of DNA methylation profile in plants by Chop-PCR. Methods Mol Biol 1991:79–90 [DOI] [PubMed] [Google Scholar]
- Deng F, Tu L, Tan J, Li Y, Nie Y, Zhang X (2012) GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158(2): 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djanaguiraman M, Perumal R, Jagadish SVK, Ciampitti IA, Welti R, Prasad PVV (2018) Sensitivity of sorghum pollen and pistil to high-temperature stress. Plant Cell Environ 41(5): 1065–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10): 573–581 [DOI] [PubMed] [Google Scholar]
- El-Esawi MA, Al-Ghamdi AA, Ali HM, Ahmad M (2019) Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.). Genes 10(2): 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kereamy A, Bi YM, Ranathunge K, Beatty PH, Good AG, Rothstein SJ (2012) The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One 7(12): e52030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galla G, Basso A, Grisan S, Bellucci M, Pupilli F, Barcaccia G (2019) Ovule gene expression analysis in sexual and aposporous apomictic Hypericum perforatum L. (Hypericaceae) accessions. Front Plant Sci 10: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Li S, Zheng Y, Duan H, Xiao F, Zhuang Y, He J, Wu G, Zhao S, Zhou H, et al. (2020) SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J 102(6): 1157–1171 [DOI] [PubMed] [Google Scholar]
- Guan Q, Yue X, Zeng H, Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26(1): 438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Liu J-H, Ma X, Luo D-X, Gong Z-H, Lu M-H (2016) The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front Plant Sci 7: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Kumar D, Chen H, Wu S, Kim JY (2014) Transcription factor-mediated cell-to-cell signalling in plants. J Exp Bot 65(7): 1737–1749 [DOI] [PubMed] [Google Scholar]
- Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z et al. (2018) A DNA methylation reader complex that enhances gene transcription. Science 362(6419): 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK (2011) Regulation and function of DNA methylation in plants and animals. Cell Res 21(3): 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedhly A, Hormaza JI, Herrero M (2009) Global warming and sexual plant reproduction. Trends Plant Sci 14(1): 30–36 [DOI] [PubMed] [Google Scholar]
- Hemm MR, Herrmann KM, Chapple C (2001) AtMYB4: a transcription factor general in the battle against UV. Trends Plant Sci 6(4): 135–136 [DOI] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35(2): 177–192 [DOI] [PubMed] [Google Scholar]
- Hu L, Li N, Xu C, Zhong S, Lin X, Yang J, Zhou T, Yuliang A, Wu Y, Chen YR, et al. (2014) Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc Natl Acad Sci U S A 111(29): 10642–10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Zhu L, Zhang X, Guan Q, Xiao S, Min L, Zhang X (2018) GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol 178(2): 876–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Liang S, Zhang X, Nie Y, Guo X (2006) An efficient grafting system for transgenic plant recovery in cotton (Gossypium hirsutum L.). Plant Cell Tiss Org 85(2): 181–185 [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22(19): 1825–1830 [DOI] [PubMed] [Google Scholar]
- Kasahara RD, Portereiko MF, Sandaklie-Nikolova L, Rabiger DS, Drews GN (2005) MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell. 17(11): 2981–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS (2017) Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res 45(11): 6613–6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Hong Y, Hsu YF, Huang H, Liu X, Song Z, Zhu JK (2020) SIZ1-mediated sumoylation of ros1 enhances its stability and positively regulates active DNA demethylation in Arabidopsis. Mol Plant 13(12): 1816–1824 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3): 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Jin S, Kim SY, Kim W, Ahn JH (2017) A fast, efficient chromatin immunoprecipitation method for studying protein-DNA binding in Arabidopsis mesophyll protoplasts. Plant Methods 13: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk C, Rowhani P, Ramankutty N (2016) Influence of extreme weather disasters on global crop production. Nature 529(7584): 84–87 [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang J, Du ML, Li L, Wang XL, Li XB (2013) A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol 54(6): 893–906 [DOI] [PubMed] [Google Scholar]
- Li Y, Min L, Zhang L, Hu Q, Wu Y, Li J, Xie S, Ma Y, Zhang X, Zhu L (2018) Promoters of Arabidopsis Casein kinase I-like 2 and 7 confer specific high-temperature response in anther. Plant Mol Biol 98(1–2): 33–49 [DOI] [PubMed] [Google Scholar]
- Liao C, Zheng Y, Guo Y (2017) MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol 216(1): 163–177 [DOI] [PubMed] [Google Scholar]
- Ma Y, Min L, Wang M, Wang C, Zhao Y, Li Y, Fang Q, Wu Y, Xie S, Ding Y, et al. (2018) disrupted genome methylation in response to high temperature has distinct affects on microspore abortion and anther indehiscence. Plant Cell 30(7): 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15(6): 394–408 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17(3):705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Hu Q, Li Y, Xu J, Ma Y, Zhu L, Yang X, Zhang X (2015) LEAFY COTYLEDON1-CASEIN KINASE I-TCP15-PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin homeostasis. Plant Physiol 169(4): 2805–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Li Y, Hu Q, Zhu L, Gao W, Wu Y, Ding Y, Liu S, Yang X, Zhang X (2014) Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol 164(3): 1293–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Zhu L, Tu L, Deng F, Yuan D, Zhang X (2013) Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J 75(5): 823–835 [DOI] [PubMed] [Google Scholar]
- Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M (2008) Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep 27(10): 1677–1686 [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40(6): 979–995 [DOI] [PubMed] [Google Scholar]
- Prouse MB, Campbell MM (2012) The interaction between MYB proteins and their target DNA binding sites. Biochim Biophys Acta 1819(1): 67–77 [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46(1): 74–83 [DOI] [PubMed] [Google Scholar]
- Sage TL, Bagha S, Lundsgaard-Nielsen V, Branch HA, Sultmanis S, Sage RF (2015) The effect of high temperature stress on male and female reproduction in plants. Filed Crop Res 182: 30–42 [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23(3): 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider JL, Oosterhuis DM, Kawakami EM (2011) Diurnal pollen tube growth rate is slowed by high temperature in field-grown Gossypium hirsutum pistils. J Plant Physiol 168(5): 441–448 [DOI] [PubMed] [Google Scholar]
- Steiner-Lange S, Unte US, Eckstein L, Yang C, Wilson ZA, Schmelzer E, Dekker K, Saedler H (2003) Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J 34(4): 519–528 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Wang NN, Li Y, Chen YH, Lu R, Zhou L, Wang Y, Zheng Y, Li XB (2021) Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum). Plant Cell 33(8): 2736–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tu L, Lin M, Lin Z, Wang P, Yang Q, Ye Z, Shen C, Li J, Zhang L, et al. (2017) Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat Genet 49(4): 579–587 [DOI] [PubMed] [Google Scholar]
- Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G et al. (2019) Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet 51(2): 224. [DOI] [PubMed] [Google Scholar]
- Wang XC, Wu J, Guan ML, Zhao CH, Geng P, Zhao Q (2020) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J 101(3): 637–652 [DOI] [PubMed] [Google Scholar]
- Wilczek AM, Cooper MD, Korves TM, Schmitt J (2014) Lagging adaptation to warming climate in Arabidopsis thaliana. Proc Natl Acad Sci U S A 111(22): 7906–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liang J, Wang C, Zhao X, Zhong X, Cao X, Li G, He J, Yi M (2018a) Overexpression of lily HsfA3s in Arabidopsis confers increased thermotolerance and salt sensitivity via alterations in proline catabolism. J Exp Bot 69(8): 2005–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liang J, Zhang S, Zhang B, Zhao Q, Li G, Yang X, Wang C, He J, Yi M (2018b) A Canonical DREB2-Type Transcription Factor in Lily Is Post-translationally Regulated and Mediates Heat Stress Response. Front Plant Sci 9: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Min L, Wu Z, Yang L, Zhu L, Yang X, Yuan D, Guo X, Zhang X (2015) Defective pollen wall contributes to male sterility in the male sterile line 1355A of cotton. Sci Rep 5: 9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ramanathan V, Victor DG (2018) Global warming will happen faster than we think. Nature 564(7734): 30–32 [DOI] [PubMed] [Google Scholar]
- Yang X, Tu L, Zhu L, Fu L, Min L, Zhang X (2008) Expression profile analysis of genes involved in cell wall regeneration during protoplast culture in cotton by suppression subtractive hybridization and macroarray. J Exp Bot 59(13): 3661–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1(2): 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liang W, Yang X, Luo X, Jiang N, Ma H, Zhang D (2010) Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22(3): 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, Yuan J, Zhang L, Chen C, Wang Y, Zhang G, Peng L, Xie SS, Jiang J, Zhu JK, et al. (2020) Coupling of H3K27me3 recognition with transcriptional repression through the BAH-PHD-CPL2 complex in Arabidopsis. Nat Commun 11(1): 6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21(1): 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the results are included within the article and its additional files. Other relevant materials are available from the corresponding authors on reasonable request.