Abstract
Chinese jujube (Ziziphus jujuba) is an important fruit tree in China, and soil salinity is the main constraint affecting jujube production. It is unclear how arbuscular mycorrhizal (AM) symbiosis supports jujube adaptation to salt stress. Herein, we performed comparative physiological, ion flux, fatty acid (FA) metabolomic, and transcriptomic analyses to examine the mechanism of AM jujube responding to salt stress. AM seedlings showed better performance during salt stress. AM symbiosis altered phytohormonal levels: indole-3-acetic acid and abscisic acid contents were significantly increased in AM roots and reduced by salt stress. Mycorrhizal colonization enhanced root H+ efflux and K+ influx, while inducing expression of plasma membrane-type ATPase 7 (ZjAHA7) and high-affinity K+ transporter 2 (ZjHAK2) in roots. High K+/Na+ homeostasis was maintained throughout salt exposure. FA content was elevated in AM leaves as well as roots, especially for palmitic acid, oleic acid, trans oleic acid, and linoleic acid, and similar effects were also observed in AM poplar (Populus. alba × Populus. glandulosa cv. 84K) and Medicago truncatula, indicating AM symbiosis elevating FA levels could be a conserved physiological effect. Gene co-expression network analyses uncovered a core gene set including 267 genes in roots associated with AM symbiosis and conserved transcriptional responses, for example, FA metabolism, phytohormone signal transduction, SNARE interaction in vesicular transport, and biotin metabolism. In contrast to widely up-regulated genes related to FA metabolism in AM roots, limited genes were affected in leaves. We propose a model of AM symbiosis-linked reprogramming of FA metabolism and provide a comprehensive insight into AM symbiosis with a woody species adaptation to salt stress.
Arbuscular mycorrhizal symbiosis improves salt tolerance of jujube plants by maintaining K+/Na+ homeostasis and reprogramming plant fatty acid metabolism.
Introduction
Chinese jujube (Ziziphus jujuba Mill.) is an important fruit tree species in China. Total jujube cultivation area was estimated to be around 2 million ha and delivering four million tons annually in China, and contributed greatly to rural development (Huang et al., 2016; Liu et al., 2020a). Drought and soil salinity is the major constraints affecting jujube production in the main cultivation region of China (Liu et al., 2014). Plants often recruit microorganism in the rhizosphere to improve their fitness while encountering abiotic stress (Compant et al., 2019). Among the beneficial soil microbes, arbuscular mycorrhizal (AM) fungi, belonging to phylum Glomeromycota, develop mutualistic symbioses with the majority of land plants and improve host plants’ resilience to unfavorable environment (Smith and Read, 2008). Therefore, AM fungi inoculants may serve as promising biostimulants in sustainable agriculture and especially under harsh conditions (Berruti et al., 2015). Jujube is a typical AM forming woody species, whose AM roots are known to develop extensively under field condition. However, their adaptation toward salt stress is still not known.
Mycorrhizal colonization establishes “mycorrhizal pathway” in host roots, enhancing the ability of host foraging nutrients and water (Smith and Read, 2008). In the peri-arbuscular membrane (PAM) of root cells accommodating the AM fungal arbuscules hosts a range of specific proteins responsible for nutrient uptake, with inorganic phosphate (Pi) transporters and ammonium transporters is well studied, generating a new way of host obtaining nutrients (Ezawa and Saito, 2018; Banasiak et al., 2021). Regulating the activity of ion channels, transporters, and other molecular aspects enhances selective absorption and vacuole sequestration of salt ions, leading to favorable K+/Na+ homeostasis in plant (Zelm et al., 2020). High affinity K+ transporters (OsHKT2;1, OsHKT1;5) and Na+/H+ antiporters (OsNHX3, OsSOS1) were up-regulated in AM rice (Oryza sativa L.) and delivering higher K+/Na+ under salt stress (Porcel et al., 2016). Maize (Zea mays L.) inoculated with native AM fungi led to elevated K+ and lower Na+ accumulation when compared with non-mycorrhizal (NM) plant; with concomitantly up-regulated expression of ZmAKT2, ZmSOS1, and ZmSKOR in roots during salt stress (Estrada et al., 2013). These studies were however limited to several known transporters, and other potential ion channels/transporters may be involved in maintaining K+/Na+ homeostasis within AM roots during salt stress.
Phytohormones are instrumental in orchestrating plant responses to the adverse conditions, and also the mediators of mycorrhizal establishment and function (Pozo et al., 2015). Strigolactones (SLs) exuded from plant roots could induce AM fungal spore germination and hyphal branching, and then initiate the pre-symbiotic stage (Akiyama et al., 2005). The regulation of abscisic acid (ABA) in response to salt stresses is altered in mycorrhizal roots; conversely, an increase in SL production was observed under salinity (Aroca et al., 2013). ABA could promote AM fungal colonization in Medicago truncatula at low concentrations but not at high concentrations (Charpentier et al., 2014). In addition, plants colonized by AM fungi were known to contain lower levels of ABA, but with higher levels of auxins, jasmonates, and cytokinins (CKs). Conversely, gibberellic acid (GA3) treatment or constitutive GA3 signaling in DELLA-deficient mutants suppresses arbuscule formation, and a reduction of GA3 signaling by stabilized DELLA proteins promotes arbuscule development (Floss et al., 2013). Auxin has a role in AM symbiosis and higher auxin levels in mycorrhizal roots were reported (Etemadi et al., 2014). Considering phytohormones are integrators of environmental signals in the regulation of mycorrhizal symbioses, holistic phytohormonal profiling to describe the AM symbiosis processes undergoing transitional salinity changes is therefore important.
Recent discoveries have revealed that lipids as the major carbon source are transferred from the plant host to AM fungi to sustain mycorrhizal colonization (Jiang et al., 2017). During mycorrhizal development, fatty acid (FA) metabolism in host is extensively regulated and this process is mediated by reduced arbuscular mycorrhization 1 (RAM1), which later up-regulates the genes involved in lipid biosynthesis and export, such as RAM2, stunted arbuscule (STR/STR2), etc. (Jiang et al., 2017; Luginbuehl et al., 2017). AP2-domain transcription factor 5 (WRI5) regulates RAM1 and together they function as the master regulators of AM symbiosis controlling the lipid transfer and periarbuscular membrane formation (Jiang et al., 2018). Lately, phosphate starvation response (PHR) transcription factors were identified as the center of the gene regulatory network for mycorrhizal symbiosis and regulate symbiosis-related genes (Shi et al. 2021). On the other hand, the components of phospholipids, polyunsaturated FAs in cell membranes determine the structural integrity and functionality and contribute to plant resilience (Zhang et al., 2005). The level of unsaturation in FAs substantially increased by salt stress, which is closely associated with salt tolerance (Sui et al., 2018; Zaman et al., 2019). Wu and coworkers (2019) found that AM symbiosis could elevate the unsaturation level of FAs in roots of trifoliate orange associated with the upregulation of PtFAD2, PtFAD6, and Δ15 FA desaturase (PtΔ15) genes during drought stress, which could alleviate oxidative damage. However, the lipid metabolism in salt-stressed AM plants is still unknown.
Transcriptional profiling depicting the progress of AM symbiosis has been studied widely, but most studies focused on roots (Guether et al., 2009; Hogekamp et al., 2011; Sugimura and Saito, 2017). Generally, the aboveground organs such as leaves have not received much attention and particularly the transcriptional responses linked to physiological responses to salt stress. Wild jujube (Z.jujuba var. spinosa) is usually used as root stock material for commercial jujube cultivars, which has naturally strong adaptation to salt tolerance (Shen et al., 2021). Considering the potential role of AM symbiosis in supporting jujube adaptation to saline soils, we performed a comprehensive study encompassing physiological, biochemical, and transcriptomic aspects of wild jujube plants with and without AM symbiosis during NaCl exposure. Our study aims to understand the following: (1) how mycorrhizal symbiosis reprograms the FA metabolism in jujube during salt stress; (2) how AM symbiosis maintains K+/Na+ homeostasis during salt stress by reprogramming ion flux profile in roots and expression of ion transporters; and (3) how the transcriptomic signals of jujube seedlings associate with mycorrhizal symbiosis when exposed to salt stress.
Results
Seedling growth and physiological responses to salt treatments
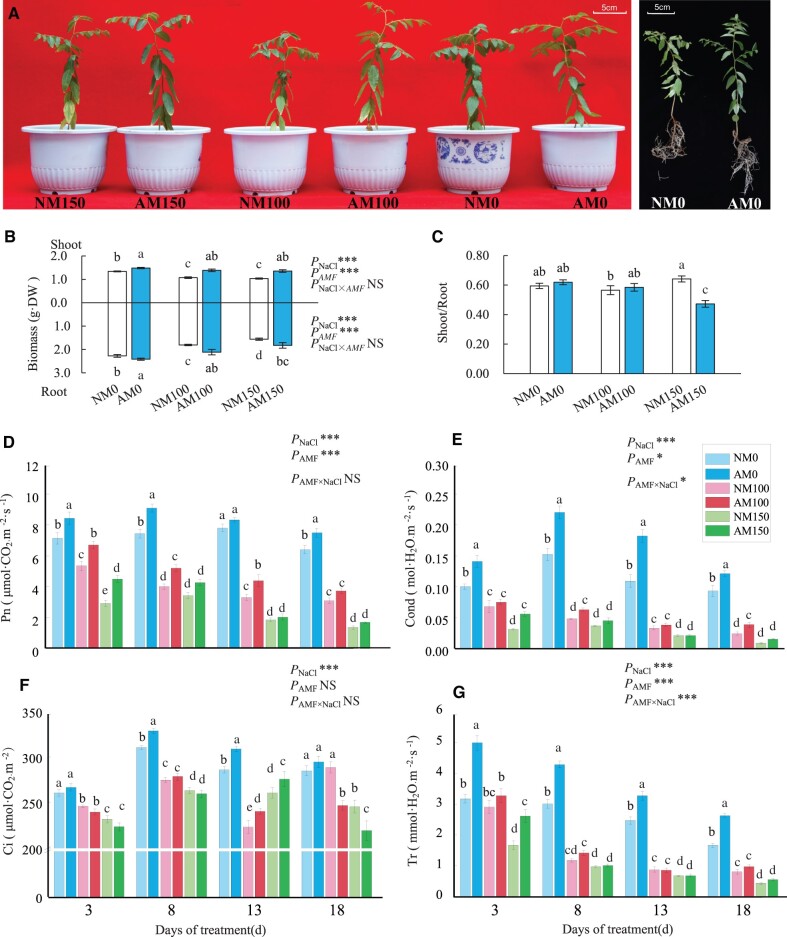
Salt stress reduced seedling growing and necrosis appeared along the foliar tips of seedlings treated with 150 mM NaCl at 18 days after treatment (DAT) and stress effect was more pronounced in NM plants than in AM plants (Figure 1A). The biomass of AM plants was always larger than that of NM plants and the largest difference in biomass between AM plants and NM plants was observed under 100-mM NaCl treatment (Figure 1B). There was no difference in shoot/root ratio between NM and AM plants under 0 and 100 mM NaCl, and contrast with this, it was much lower in AM plants when treated with 150 mM NaCl compared with the corresponding NM plants (Figure 1C). The gas exchange parameters, including net photosynthetic rate (Pn), stomatal conductance (Cond), intercellular CO2 concentration (Ci), and transpiration rate (Tr), of both AM and NM plants showed a decreasing trend with the increasing of salt concentration and exposure time (Figure 1, D–G). Under 100-mM NaCl treatment, AM plants displayed higher Pn than NM plants until 18 DAT and higher Cond until 8 DAT. The beneficial effects of symbiosis were marginal in AM plants under 150-mM NaCl treatment. Both salt treatment and AM symbiosis had significant effects on chlorophyll fluorescence. At 18 DAT, AM plants showed higher maximum quantum yield in dark-adapted state (Fv/Fm), the maximum quantum yield in light-adapted state (Fv′/Fm′), the actual quantum yield in light-adapted steady state (ϕPSII), and the photochemical quenching values (qP) compared with NM plants under 0 and 100 mM NaCl treatments, and lower non-photochemical quenching (NPQ) parameter of AM plants than NM plants under all levels of salt treatments (Supplemental Figure S1, C–G). The interaction effect between AM symbiosis and salt treatment was detected on Cond, Tr, Fv′/Fm′, ϕPSII, and qP. Therefore, AM symbiosis enhanced the gas exchange ability and the efficiency of photosystem II in jujube, but this effect was reduced when exposed to 150 mM NaCl.
Figure 1.
Wild jujube seedlings growth and physiological responses to salt stress and AM symbiosis. A, AM and NM seedlings growth condition under 18 days of 0-, 100-, and 150-mM NaCl treatment. B, Changes of AM and NM seeding biomass during salt stress. C, Changes of shoot/root ratio of AM and NM seedlings during salt stress. D, Changes of net photosynthetic rate (Pn). E, Stomatal conductance (Cond). F, Intercellular CO2 concentration (Ci). G, Transpiration rate (Tr). NM, NM inoculation; AM, mycorrhizal inoculation; DW, dry weight; 0, 100, and 150 represent the addition of 0, 100, and 150 mM NaCl, respectively. Significance of difference among treatments was assessed by two-way ANOVA; *** indicate p < 0.001, * indicate p< 0.05, NS indicate no significant difference. Means followed by the different letter indicate significant difference among treatments at p < 0.05. n = 3 in biomass, n = 7 in Pn, Cond, Ci, and Tr. Error bars represent standard error.
In addition, salt treatments elevated malondialdehyde (MDA) content significantly in leaves and roots of both NM plants and AM plants at 18 DAT, especially in leaves. Comparably, AM symbiosis mainly enhanced catalase (CAT) and peroxidase (POD) activities of protective enzyme systems while reducing MDA accumulation (Supplemental Figure S1, A and B). Therefore, AM symbiosis enhanced the scavenging enzymes of reactive oxygen species (ROS) to increase salt tolerance in wild jujube.
Effects of AM symbiosis on plant nutrient content and ion homeostasis under salt stress
With increasing salt concentration, Na+ accumulated more in both AM and NM plants, in particular, AM plants accumulated more Na+ in roots than NM plants under same level of salinity conditions. In contrast, AM leaves did not accumulate more Na+ than that in NM leaves under 150-mM NaCl treatment (Supplemental Table S1-1). On the other hand, K+ concentration in the leaves of both AM and NM plants was significantly elevated under 100-mM NaCl treatments compared with those without salt treatments. In addition, K+ content in both leaves and roots was elevated by AM symbiosis compared with NM plants under each level of salinity condition. K+ content in AM leaves was increased by 30.99% and 13.48% than NM leaves under 100- and 150-mM NaCl treatment, respectively. K+/Na+ ratio in the leaves of AM plants was 25.5% higher than that in NM plants under 100-mM NaCl treatment (p < 0.05), in contrast with this, it was 42.10% lower than that in NM leaves under 150-mM NaCl stress. It indicated that maintenance of higher K+/Na+ ratio in AM leaves under salt stress depends on more K+ accumulation by AM symbiosis, not by restricting Na+ accumulation.
P levels were significantly elevated in AM plants under salinity condition or not. It was increased by 55.9% and 69.5% in AM leaves and roots compared with NM plants under 100-mM NaCl treatment, respectively. In addition, AM symbiosis reduced N content in both leaves and roots under none salinity condition. After salt treatment, N content improved in both leaves and roots of AM and NM plants, but there was no significant difference between NM and AM plants under salinity condition. The effects of salt stress and AM symbiosis were weaker on Mg2+ and Ca2+ accumulation in all parts of seedlings. Thus, AM symbiosis increasing jujube adaptation to salt stress was mainly contributed by enhancing P and K acquisition.
Effects of AM symbiosis and NaCl stress on ion flux at root tip
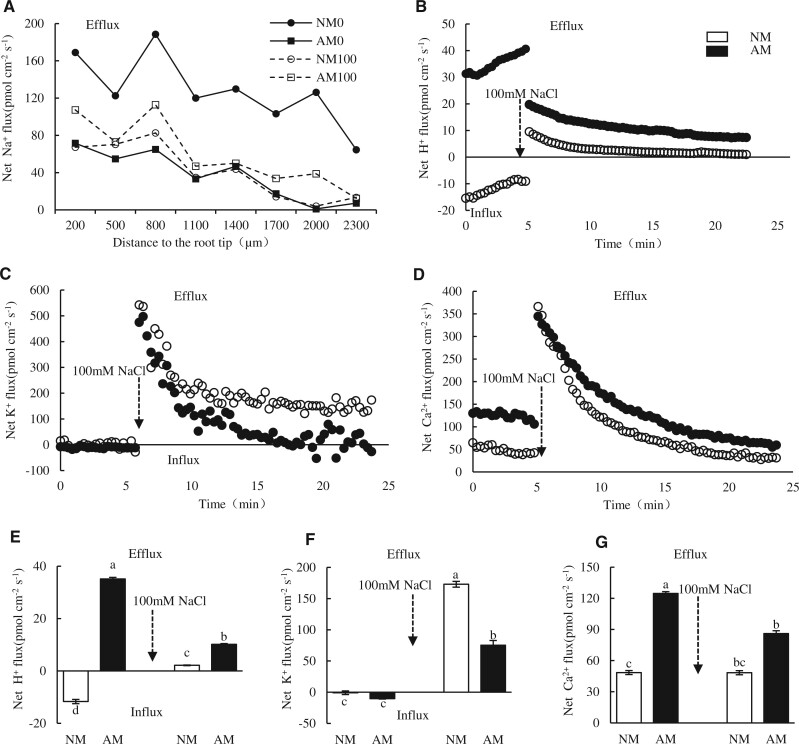
In general, Na+ efflux decreased with distance from the tip and the maximal Na+ efflux occurred at the range from 500 to 1,000 μm far from the apex of AM and NM plants under salinity condition or not (Figure 2A). At 8 DAT, AM colonization reduced Na+ efflux under non-salinity condition and Na+ efflux was weaker in NM roots than AM roots at the position of 500–1000 μm far from the apex under salinity condition. We selected the position of 600 μm far from apex to detect the temporal dynamics of K+, H+, and Ca2+ fluxes. Before salt addition, H+ net influx (11.68 pmol cm−2·s−1) was observed in NM roots, and contrast with this, it tended to efflux (35.16 pmol cm−2·s−1) in AM roots. After salt treatment, H+ net efflux occurred in both types of root tips, but was more active in AM roots (Figure 2, B and E). These observations were indicative that the AM symbiosis was regulating the H+ flux direction and enhancing H+ efflux. Prior to salt treatment, K+ influx (10.39 pmol cm−2·s−1) was detected on AM roots while its influx (0.87 pmol cm−2·s−1) in NM roots. After salt stress, K+ efflux was stronger in NM roots (172.97 pmol cm−2·s−1) than AM roots (75.30 pmol cm−2·s−1), additionally, K+ flux in AM tips tended to be influx when the flux was stabilized (Figure 2, C and F). Ca2+ efflux was detected in both types of roots and was more intense in AM roots under salt treatment and the control (Figure 2, D and G). Taken together, AM symbiosis altered the H+ influx into efflux in roots and promoted H+, Ca2+ efflux while reduced the K+ efflux during exposure to salt treatment. These results provide evidence of AM symbiosis maintaining K+/Na+.
Figure 2.
Ion fluxes profile of the fine root tips of AM and NM wild jujube seedlings under salt treatments detected by Non-invasive Micro-test Technology. A, Net Na+ flux of root tips of NM and AM seedlings from the apex to 2300 μm away when exposed to 0 mM or 100 mM NaCl. B–D, Net H+, K+, and Ca2+ fluxes at the position of 600 μm from the apex during 5 min before and 20 min after treatment with 100 mM NaCl in measuring chamber, each point is the mean of four individual plants. E–G, The mean net flux of H+, K+, and Ca2+ at 0–5 min and 15–25 min after detected start, respectively. The arrow indicates the addition of 100 mM NaCl to the determination solution. Significance of difference among treatments was assessed by one-way ANOVA; * indicate p < 0.05, n = 3.
Phytohormonal levels were regulated by AM symbiosis and NaCl stress
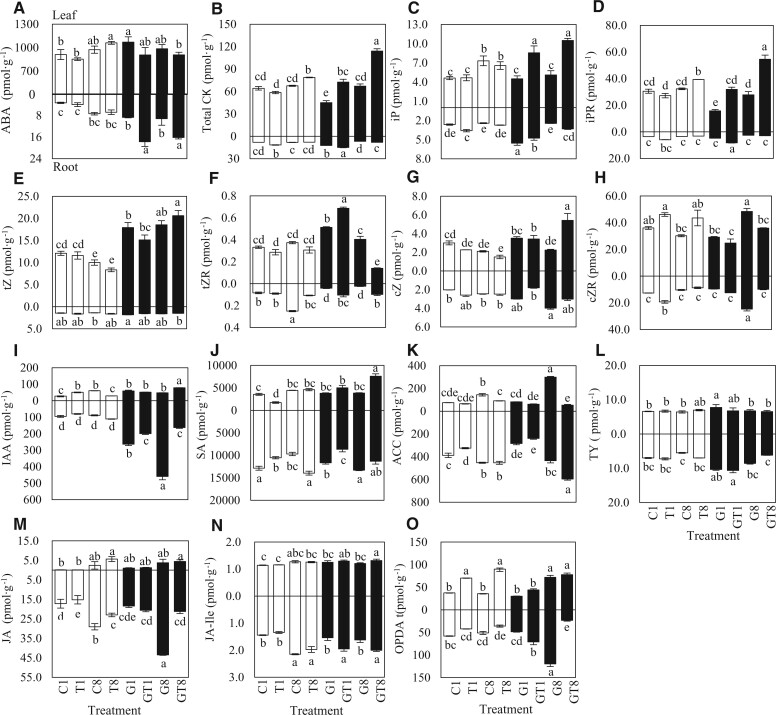
Fourteen phytohormones were quantified in plants at 1 and 8 DAT while undergoing 100-mM NaCl treatments (with control) (Figure 3). ABA content was higher in leaves than roots, it was generally higher in AM roots than NM roots, and further elevated by salt treatments (Figure 3A). CKs, including isopentenyladenine (iP), iP riboside (iPR), trans-zeatin (tZ), tZ riboside (tZR), cis-zeatin (cZ), and cZ riboside (cZR), were mainly distributed in leaves. After salt stress, the content of CK in AM leaves increased significantly compared with NM plants (Figure 3B). iP and iPR were significantly elevated in leaves of AM plant during salt stress while iPR was only elevated in leaves of NM plant at 8 DAT (Figure 3, C and D). The level of tZ was significantly elevated in leaves of AM plants than that of NM plants and was further improved in AM leaves at 8 DAT (Figure 3E). tZR was slightly affected by NaCl in NM leaves; however, tZR in AM leaves increased significantly under salt stress at 1 DAT, on the contrary was significantly decreased in AM leaves at 8 DAT (Figure 3F). The cZ content in leaves of AM plants was significantly elevated to higher level compared with NM plants under salt stress (Figure 3G), and cZR was generally higher in leaves than roots (Figure 3H). On the other hand, AM symbiosis significantly enhanced indole-3-acetic acid (IAA) content in leaves and roots, as shown in Figure 3I, IAA content in leaves increased at G1 and GT8, and the level was lowered in roots by salt stress.
Figure 3.
Phytohormone content in leaves and roots of AM and NM jujube seedlings during salt treatments. A, ABA; B, total CKs content; C, iP; D, iPR; E, tZ; F, tZR; G, cZ; H, cZR; I, IAA; J, SA; K, ACC; L, TY; M, JA; N, JA-Ile; and O, OPDA. 1, NM plant under 1 day of 0-mM NaCl treatment; T1, NM plant under 1 day of 100-mM NaCl treatment; G1, AM plant under 1 day of 0-mM NaCl treatment; GT1, AM plant under 1 day of 100-mM NaCl treatment; C8, NM plant under 8 day of 0-mM NaCl treatment; T8, NM plant under 8 day of 100-mM NaCl treatment; G8, AM plant under 8 day of 0-mM NaCl treatment; GT8, AM plant under 8 day of 100-mM NaCl treatment. Significance of difference among treatments was assessed by one-way ANOVA: * indicate p < 0.05, n = 3.
Salicylic acid (SA) content was much higher in roots than in leaves. It was elevated to higher level in leaves of AM plants compared with NM leaves under salt stress at 8 DAT (Figure 3J). 1-aminocyclopropanecarboxylic acid (ACC) decreased in both NM and AM leaves at 8 DAT, whereas it increased significantly in AM roots at 8 DAT (Figure 3K). Typhasterol (TY), as the precursor of brassinosteroids, was only elevated in AM roots compared with NM roots at 1 DAT. (Figure 3L). Jasmonic acid (JA) showed increased in AM roots at 1 DAT under salt treatment (Figure 3M). In addition, jasmone I-isoleucine (JA-Ile) levels increased significantly in both leaves and roots by AM symbiosis under salt treatment at 1 DAT (Figure 3N). 12-oxo-phytodienoic acid (OPDA), as a primary precursor of (−)-JA, was significantly increased by salt treatment in AM and NM leaves, and was significantly increased in AM roots under salt stress at 1DAT (Figure 3O). From the phytohormonal perspectives, AM symbiosis induced the following salient effects: IAA increased in all parts of AM plants while the levels decreased in AM roots during salt stress. ABA were largely elevated in AM roots by salt treatments, SA was elevated in AM leaves under long duration of salt. The level of tZ was elevated significantly in the leaves of AM plants while iP and iPR were higher in AM leaves by salt stress.
Effects of AM symbiosis and salt stress on triacylglycerol and FA content
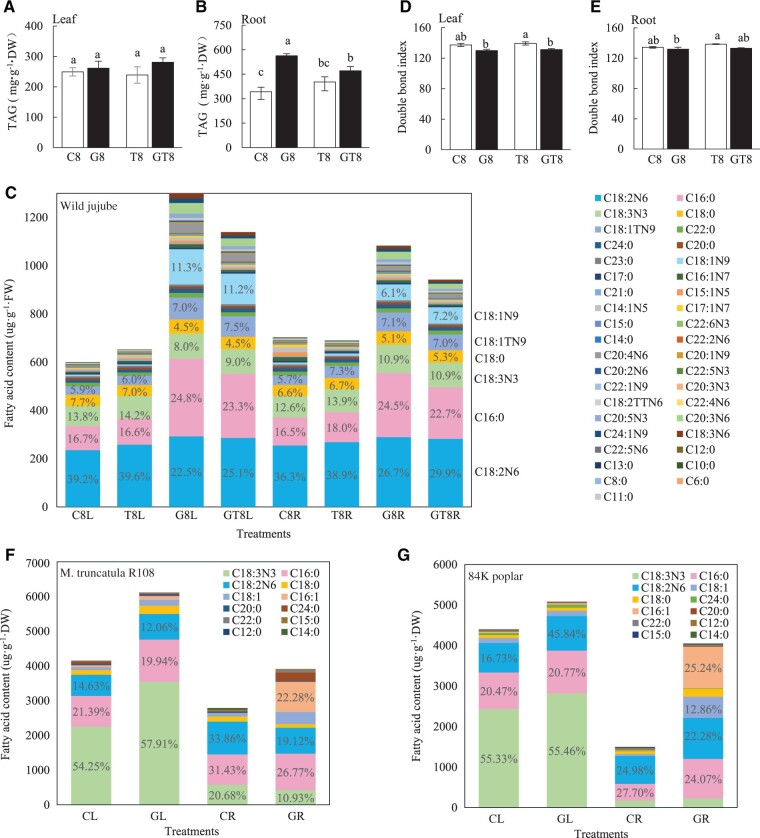
Triacylglycerol (TAG) was significantly improved in roots and marginally elevated in leaves by AM symbiosis, salt stress reduced TAG in AM roots while unaltered it in NM plants (Figure 4, A and B). The medium and long chain FAs determination quantified 16 saturated FAs (SFAs) and 23 unsaturated FAs (UFAs) (Supplemental Table S1-2). In AM plants, total amount of FAs was 1,299.43 μg/g in leaves and 1,083.46 μg/g in roots under none salinity condition, which were 116.4% and 50.4% higher than those in NM plants, respectively. Salt stress reduced the total FAs in AM roots while remaining unaltered in NM seedlings. In NM plants, ca. 40% linoleic acid (C18:2N6), ca. 17% palmitic acid (C16:0), and ca.14% linolenic acid (C18:3N3) formed most of the FAs (>80 μg/g). Most of FAs were elevated by AM colonization, among which, palmitic acid (C16:0), oleic acid (C18:1N9), trans oleic acid (C18:1TN9), and linoleic acid (C18:2N6) increased greatly (Figure 4C and Supplemental Table S1-2). The C16:0 (321.94 μg/g) increased to be the top FA in leaves and the second highest FA (265.85 μg/g) in roots of AM plants, which was comparable to C18:2N6 in leaves or roots. Overall, the ratio of C16:0/total FAs increased ca. 7% after AM colonization; C18:1N9 increased from ca. 1% in NM tissues to ca. 11% in leaves; and ca. 7% in roots of AM plants; the C18:2N6 decreased by ca. 15%. Most of those low abundant FAs contents (levels < 2%) were increased by AM colonization (Figure 4C). Total contents of UFAs in leaves and roots were not substantially changed in NM plants after salt treatment while it was increased by 114.0% and 44.1% after AM colonization (Supplemental Table S1-2). On the other hand, the double bond index (DBI) of leaves was not affected by salt treatment in both NM plants and AM plants, but it was significantly decreased by AM symbiosis at 8 DAT. Moreover, it remained unchanged in roots with salt treatment and AM symbiosis (Figure 4, D and E).
Figure 4.
TAG and FAs components in leaves and roots of AM and NM plants. A and B, TAG levels in leaves and roots, respectively, of AM and NM jujube plants after 18 days of 0- and 100-mM NaCl treatment. C, FAs component in leaves and roots of AM and NM jujube plants under salt treatments. D and E, The DBI in leaves and roots, respectively, of AM or NM jujube seedlings under NaCl treatments. L, leaf; R, root; DW, dry weight; FW, fresh weight. DBI as the index of unsaturation of FAs was calculated as follows: 1× mol% (C14:1 + C15:1 + C16:1 + C17:1 + C18:1 + C20:1 + C22:1 + C24:1) + 2× mol% (C18:2 + C20:2) + 3× mol% (C18:3 + C20:3) + 4× mol% (C20:4 + C22:4) + 5× mol% (C22:5) + 6× mol% (C22:6) to estimate the unsaturation level of all FAs (means ± se, n = 3). Column labeled different letters indicate significant difference among treatments at P < 0.05. “8,” 8 days of NaCl treatments. F and G, FAs component in leaves and roots of M. truncatula and 84K poplar, respectively. CL, Leaves of seedling without mycorrhizal symbiosis. GL, Leaves of seedling with mycorrhizal symbiosis. CR, Roots of seedling without mycorrhizal symbiosis. GL, Roots of seedling with mycorrhizal symbiosis.
Besides, we quantified long-chain FAs in poplar (P. alba × P. glandulosa cv. 84K) and barrel medic (M.truncatula). Four types of SFAs and eight types of UFAs were detected in both leaves and roots of poplar and barrel Medic, among which, C18:3N3, C16:0, C18:2N6, and C18:0 were the major FAs in the NM roots and in the leaves of the two plant species with AM colonization or not (Figure 4, F and G and Supplemental Table S1-3 and 1-4). The total of detected FAs were higher in leaves than roots in both species. AM symbiosis elevated the total concentration of FA substantially in leaves as well as roots of both plants, interestingly, methyl palmitoleate (C16:1) was hugely elevated in AM roots of both plants. Taken together, AM symbiosis elevating FAs in leaves as well as roots should be a conserve physiological effect in AM plants.
Transcriptome regulation of jujube by AM symbiosis and salt stress
These obtained RNA-seq data after quality control showed good reproducibility and high mapping rates (Supplemental Table S2). Comparative transcriptome analysis revealed that 606 and 632 genes in leaves and roots were regulated in NM seedlings at 1 DAT (T1 versus C1), respectively, and differential expression genes (DEGs) increased to 1,656 in leaves and 2,121 in roots at 8 DAT (T8 versus C1), respectively. It demonstrated a clear durational effect of salt treatment on transcriptome regulation. Under none salinity condition, 565 genes in leaves and 1,261 genes in root were regulated between NM and AM (G1 versus C1), respectively. Following 1 DAT on AM seedlings, the treatment induced 551 and 39 DEGs in leaves and roots (GT1 versus G1), respectively, which were lesser than T1 versus C1. However, when salt treatments were extended longer to 8 days, 2,628 and 2,818 genes (GT8 versus G1) were regulated in leaves and roots, respectively (Supplemental Figure S2A). Using the Venn diagram analysis (Supplemental Figure S2, B and C), there were fewer DEGs especially in GT1L versus G1L (194) and GT1R versus G1R (14), and many DEGs were duly detected in GT8L versus G1L (1,164) and GT8R versus G1R (1,520). The number of observed DEGs was a good indicator of the AM effects and the durational effect of salt stress.
Gene co-expression networks with AM symbiosis and NaCl exposure
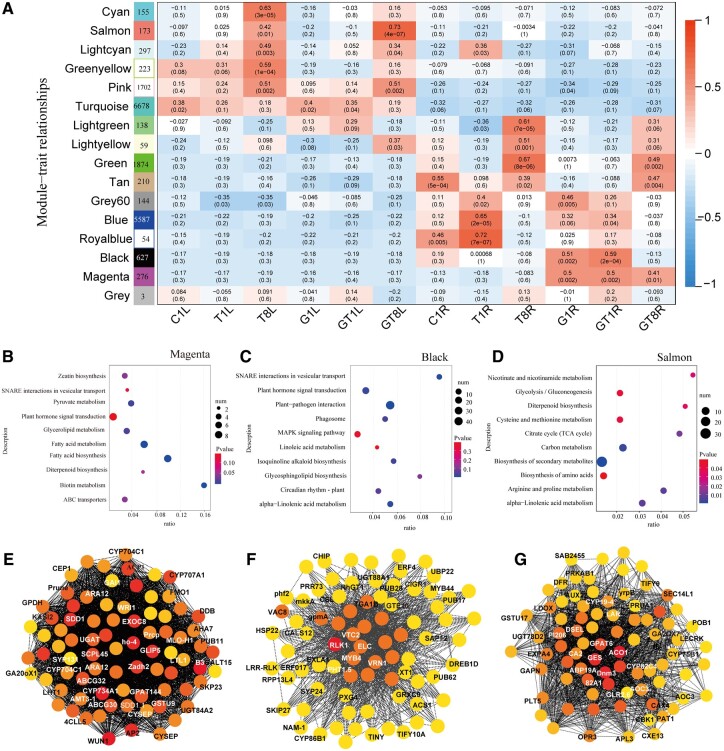
Weighted gene co-expression network analysis (WGCNA) generated 15 distinct modules and the number of genes clustered within a single module and ranging from 54 to 6,678 (Figure 5A). Interestingly, clear tissue effect and AM effect were demonstrated through the observed module–trait relationships. We found that no modules were specifically associated with leaves of AM plants receiving salt treatments or not. Among those modules related with leaves, Cyan, Green-yellow modules were specially correlated with leaves of NM plants at 8 DAT, while Salmon and Pink modules were correlated with both NM and AM plants. The correlation of Salmon module was more pronounced for AM leaves’ responses (GT8L, r = 0.73) to salt stress than NM leaves (T8L, r = 0.42). In roots, two modules (Black and Magenta) were specifically correlated with AM symbiosis under salt stress or not. These observations highlighted the specific effects of AM symbiosis on host roots but were not observable in leaves. These co-expression gene modules correlated with the different treatments and highlighted the effects of salt stress, AM symbiosis, and tissue specificity.
Figure 5.
WGCNA of NM plant and AM jujube seedling during salt stress. A, Heatmap of module–trait associations. Each row corresponds to one module (labeled by color) and each column corresponds to one treatment. The squares of red represent high adjacency (positive correlation) and blue color represents low adjacency (negative correlation). B–D, KEGG enrichment of co-expression genes in Magenta, Black, and Salmon module, respectively. E–G, Gene co-expression network marked with top 80 hub genes in Magenta, Black, and Salmon module, respectively. Hub genes within each network are highlighted with circles filled yellow to red, and red represents with higher connectivity in the module. ‘1’, 1 days of NaCl treatments.‘8’, 8 days of NaCl treatments. CL, Leaves of seedling without mycorrhizal symbiosis. GL, Leaves of seedling with mycorrhizal symbiosis. CR: Roots of seedling without mycorrhizal symbiosis. GL, Roots of seedling with mycorrhizal symbiosis.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on each module were performed to assess the effects of AM colonization and salt stress (Supplemental Table S3). Some top enriched pathways in those modules associated with leaves under salinity conditions (Figure 5D), such as betalain biosynthesis and anthocyanin biosynthesis in Cyan module, biosynthesis of secondary metabolites, arginine and proline metabolism, and alpha-linolenic acid metabolism in Salmon module; glycerophospholipid metabolism, glycerolipid metabolism, biosynthesis of UFAs and ether lipid metabolism in greenyellow module; autophagy in lightcyan module that highlighted the common plant molecular responses during exposure to high salinity. The analyses also highlighted the importance of FA metabolism in plant responses to salt stress, which were more pronounced in leaves of AM plants.
In Magenta module (267 genes) associated with AM roots, most of the high connective hub genes in this module have been identified as the AM symbiosis markers or inducers (Supplemental Table S4; Shi et al., 2021), such as ammonium transporter 3 member 1 (AMT3-1), inorganic phosphate transporter 1-3 (PHT1.3), ATPase 7, plasma membrane-type (AHA7), were involved in nutrient transportation, and the AP2-like/ethylene-responsive transcription factor (AP2/ERF), WRI1/ERF, Acyl carrier protein 1 (ACP1), limonoid UDP-glucosyltransferase, ABC transporter G family member (ABCG30, 32), were involved in lipid metabolism, which were defined as the AM marker genes previously (Figure 5E and Supplemental Table S4). These analyses indicated that the 267 genes with known or unknown function in Magenta module may represent the essential responses of host plant during AM colonization. KEGG pathways including FA metabolism and biosynthesis, biotin metabolism, zeatin biosynthesis, SNARE interaction in vesicular transport, and plant hormone signal transduction were highly enriched (Figure 5B), these were described as the conserved transcriptomic response to AM symbiosis (Rich et al., 2021).
In Black module, the top enriched pathways, such as SNARE interactions in vesicular transport, alpha-linolenic acid metabolism, plant–pathogen interaction, phytohormone signal transduction, and glycerophospholipid metabolism, were also highly relative to AM symbiosis (Figure 5C). There are 36 transcription regulators in the black module, including 16 AP2/ERFs, 11 WRKYs, 2 MYBs, etc. Top hub genes in this module, such as heat shock protein (HSP90-1), zinc finger protein (ZAT10), dehydration responsive element binding proteins (DREB1D), and wall-associated receptor kinase (WAK2), have potential roles in enhancing plant tolerance to environmental stresses. Myb-related protein (MYB4), inorganic phosphate transporter (PHT1.5), and auxin-induced protein were potentially related to AM colonization. Conversely, Green and Tan modules were associated with both AM roots (GT8R) and NM roots (T8R) at 8 DAT, containing information about environmental processes (Supplemental Table S4 and Figure 5F). Thus, these gene co-expression modules were useful in displaying distinct signatures associated with the effects of AM symbiosis and salt stress.
Responses of ion/cation channels and transporters are related to salt stress and AM symbiosis
We analyzed the transcriptional profile of 50 genes associated with ion/cation channel/transporters (Supplemental Table S5-1). PHT1 family is the most important P transporters and regulated by AM symbiosis. Seven ZjPHT1s were phylogenetically clustered into three clusters, ZjPHT1.11 and ZjPHT1.3 in subfamily I (monocot, dicot, and AMF exclusively inducible); ZjPHT1.9 in subfamily II (monocot and dicot); and ZjPHT1.1, ZjPHT1.4, ZjPHT1.5, and ZjPHT1.7 in subfamily III (dicot) (Supplemental Figure S2D). Transcriptional analysis showed that ZjPHT1.3, ZjPHT1.7, and ZjPHT1.11 were specifically induced in AM roots, and weakly expressed in NM roots. In addition, ZjPHT1.5 was strongly up-regulated in AM roots at 8 DAT. In contrast, no ZjPHT1 genes were specifically induced in leaves by AM symbiosis (Supplemental Figure S3).
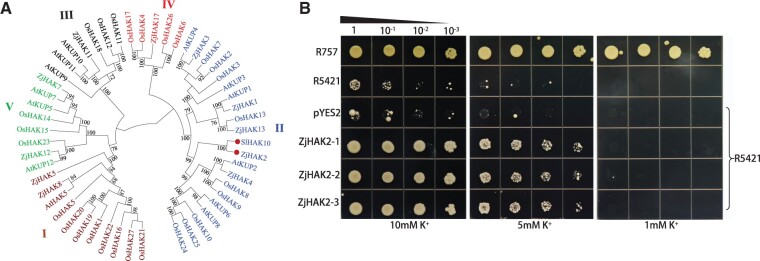
K+ channels (KAT), cation/H+ antiporters (CHXs), and Na+/H+ exchangers (NHXs) function as K+/Na+ transporters. Among these genes involved in Na+/K+ uptake or sequestration, ZjCHX4 and ZjCHX14 were substantially up-regulated in leaves of AM plants under all treatments compared with those of NM plants and other ZjCHXs that expressed lowly in roots, which might contribute to enhanced Na+ accumulation. ZjNHX1 and ZjNHX7 function as sequestrating Na+ in vacuole and these were up-regulated in both leaves and roots after salt treatments (Supplemental Figure S3). However, two SOS1 homologs, ZjNHX3 and ZjNHX5, remained unaltered under both salt treatment and AM symbiosis. These observations indicated that Na+ efflux or sequestration in vacuole was not substantially influenced by AM symbiosis when exposure to salt stress. High-affinity K+ transporters (HAKs), stelar K+ outward rectifier (SKORs) and gated outwardly K+ channels (GORKs) function as the K+ transportation. ZjAKT1 and ZjAKT2 were up-regulated in leaves by salt stress, but not influenced by AM symbiosis. We found that the ZjHAK2 expression was specifically induced in AM root, interestingly, it will be induced in NM roots at 8 DAT. The ZjHAK8 expression was concurrently down-regulated in AM roots compared with that in NM roots under non-salinity condition, but it will be up-regulated in roots of both AM plants and AM plants after salt treatments. In addition, ZjHAK4 and ZjHAK5 expression were also up-regulated in both NM and AM roots after salt stress. Phylogenetic analysis displayed that five ZjHAKs were clustered into group II with the AM-induced SlHAK10 in Solanum lycopersicum belonging to KT/KUP/HAK family and ZjHAK2 was the putative ortholog of SlHAK10 (Liu et al., 2019; Figure 6A). The ZjGORK1 and ZjGORK2, exporting K+, were down-regulated in AM roots compared with NM roots at 1 DAT. SKOR could promote K+ translocation from root to leaves and ZjSKOR1 was up-regulated in root by AM symbiosis. The expression profile of K+ transporters suggested that AM symbiosis promoted K+ uptake and alleviated K+ leakage under salinity condition. In addition, we found a plasma membrane-type ATPase (ZjAHA7) expression that was specially induced in roots by AM symbiosis and was also induced in NM roots at 8 DAT (Supplemental Figure S3). The induction of ZjAHA7 coincided with ZjHAK2 up-regulated expression, which was also associated with the profile of H+ efflux in AM root tip. Another plasma membrane-type ATPase (ZjPMA5) was down-regulated in AM roots compared with NM roots, but was up-regulated after salt stress. We proposed that AM symbiosis activated a special plasma membrane-type ATPase to enhance H+ efflux, while coordinating with ZjHAK2 expression to promote K+ influx. These K+ transporters and channels jointly regulated ion homeostasis in AM jujube plants while undergoing salt stress.
Figure 6.
Phylogenetic analysis and functional characterization of ZjHAK2 in Yeast. A, Phylogenetic tree of plant HAK proteins constructed using the neighbor-joining method. HAK transporters and corresponding plant species are: tomato, SlHAK10; Z. jujuba (11), Arabidopsis thaliana, 13; Rice, 27. B, Functional complementation of ZjHAK2 for K+ acquisition in the K+ uptake-defective yeast strain R5421. From top to bottom, the growth status of R757 wild-type yeast strain, R5421, empty vector (pYES2), and R5421 cells expressing ZjHAK2 (three biological replicates) converters on AP supplemented medium with 10, 5, and 1 mM KCl. Each transformant was precultured in AP liquid medium containing 100 mM KCl. The yeast cells were diluted on AP medium at a gradient of 10 times and then incubated in a constant temperature incubator at 30°C for 6 days.
K+ transport capacity of ZjHAK2 was further evaluated by complementation analysis using a yeast mutant strain R5421 defective in K+ uptake, which could not grow under low-K+ culture conditions. Under high-K+ conditions (10 mM), no difference was observed between the mutant expressing ZjHAK2 and wild-type yeast strain (R757) (Figure 6B). Along with the decline of K+ concentration from 10 to 5 mM, the growth of R5421 harboring empty vector was almost entirely suppressed, the R5421 cells transformed with ZjHAK2 could indeed rescue its growth defect at 5 mM K+, but were not able to grow under 1 mM K+, suggesting that ZjHAK2 has K+ transport activity.
The responses of FA synthesis and transport genes to salt stress and AM symbiosis
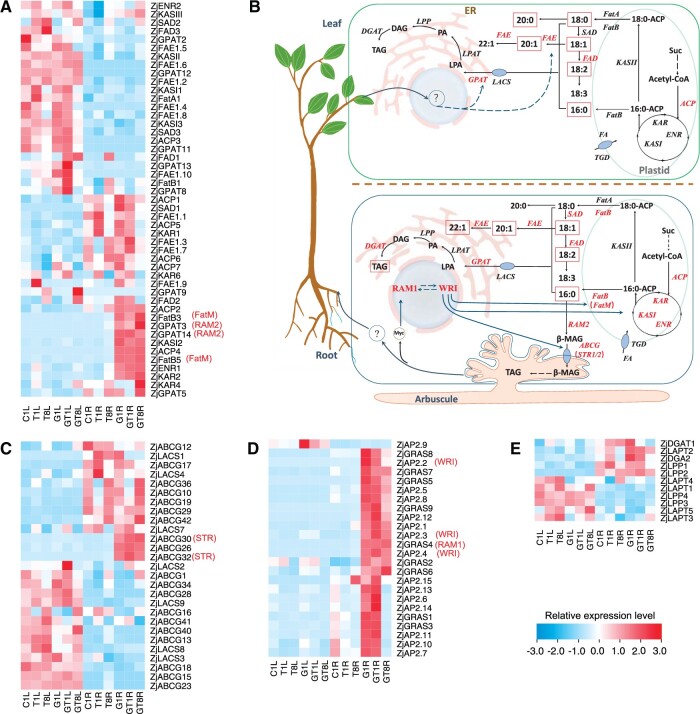
We identified all potential genes involved in FA biosynthesis and transportation in wild jujube colonized by AM fungi or not (Figure 7 and Supplemental Table S5-2). The hierarchical cluster analysis of all those genes displayed the contrasting expression profile between leaves and roots. In the FAS cycle, ACP (ZjACP2), ketoacyl-ACP synthase I (ZjKASI2), enoyl-ACP reductase (ZjENR1), and β-ketoacyl-ACP reductase (ZjKAR2) were strongly up-regulated in AM roots and maintained higher expression level during salt stress than those in NM roots, and ZjACP4 was specifically induced in AM roots (Figure 7A). On the other hand, ZjACP4 and ZjENR1 were up-regulated and ZjACP3 was down-regulated in NM leaves by salt stress while no genes were regulated in NM roots. No genes involved in FA biosynthesis were specifically induced in leaves by AM symbiosis. Those genes, ZjACP1, ZjACP3, ZjACP5, ZjACP7, and ZjKASI2, showed higher FPKM values in leaves of AM plants than those in NM leaves, but it is less than the standard of up regulated expression (log2(fold change) ≥1). These results revealed that the AM symbiosis enhanced the activity of the FAS cycle greatly in roots but only slightly in leaves at transcript level.
Figure 7.
Expression profile of genes involved in FA biosynthesis, processing, and transportation in AM and NM wild jujube seedlings during salt treatments. A, Expression profile of genes involved in FAS cycle, FA elongation, and processing. B, The proposed model of FA biosynthesis and transportation in mycorrhizal jujube. C, Expression profile of genes related with FA transportation. D, Transcriptional expression of AP2/EREBP transcription factor (ZjAP2) and GRAS transcription factor (ZjGRAS). E, Expression profile of genes related with TAG biosynthesis. Grids represent the expression patterns of genes shown as FPKM values. The genes highlighted in red indicate regulated by AM symbiosis. ZjABCG30/32, ZjAP2.2/2.3/2.4, and ZjGRAS4 are hypothetical stunted arbuscule (STR), AP2-domain transcription factor (WRI), and RAM1, respectively. KAR, β-ketoacyl-ACP reductase; KASII, ketoacyl-ACP synthase II; SAD, stearoyl-ACP desaturase; ENR, enoyl-ACP reductase; FatA, acyl-ACP thioesterase A; FatB, acyl-ACP thioesterase B; FAD, omega FA desaturase; FAE, acid elongation; FatM, acyl-ACP thioesterase M; GPAT, glycerol-3-phosphate acyltransferase; ABCG, ABC transporter; LACS, long chain acyl-CoA synthetase; LPAT, lysophosphatidyl acyltransferase; ER, endoplasmic reticulum; Suc, sucrose; 16:0-ACP, palmitoyl-ACP; 18:0-ACP, stearoyl-ACP; 16:0, palmitic acid; 18:0, stearic acid; 18:1, oleic acid; CoA, coenzyme A; Acetyl-CoA, acetyl-coenzyme A; MAG, monoacylglycerol; LPA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol. ‘1’, 1 days of NaCl treatments.‘8’, 8 days of NaCl treatments. CL, Leaves of seedling without mycorrhizal symbiosis. GL, Leaves of seedling with mycorrhizal symbiosis. CR: Roots of seedling without mycorrhizal symbiosis. GL, Roots of seedling with mycorrhizal symbiosis.
In the following pathway of FA elongation and processing, four genes, including acyl-ACP thioesterase (ZjFatB3, ZjFatB5) and glycerol-3-phosphate acyltransferase (ZjGPAT3, ZjGPAT14), were exclusively induced in AM roots, and five genes, such as stearoyl-ACP desaturase (ZjSAD1) and acid reductase (ZjFAE1.3, ZjFAE1.5, ZjFAE1.7, ZjFAE1.9), were largely up-regulated in AM roots than those in NM roots. On the other hand, ZjFAE1.10 was specifically induced in leaves by AM symbiosis, and ZjGPAT13, ZjGPAT8, Omega FA desaturase (ZjFAD1), and ZjFAE1.10 showed higher expression level in AM leaves compared with NM leaves. In addition, ZjGPAT5, ZjGPAT9, ZjFAE1.9, ZjFAD1, and ZjFAD2 were up-regulated in leaves while ZjFAD1 and ZjFAE1.8 were up-regulated in roots of NM plants under salt stress. Among those genes, ZjFatB3 and ZjFatB5 have a closer phylogenetic relationship with MtFatM, which was specifically induced in AM roots and has a role to enhance FA biosynthesis, ZjGPAT3 and ZjGPAT14 were phylogenetically closer to MtGPAT7 (MtRAM2), which is known to convert C16:0 into β-monoacylglycerol (β-MAG) (Wang et al., 2017; Supplemental Figure S4). Thus, FA elongation and unsaturation were extensively reprogrammed in roots and leaves during AM symbiosis.
Potentially genes involved in FA transportation were also annotated, including 20 ABC transporter (ABCG) and 7 long chain acyl-CoA synthetase (LACS). Among ZjABCGs, ZjABCG26, ZjABCG30, and ZjABCG32 were specifically induced in AM roots, whereas ZjABCG30 and ZjABCG32 were also slightly induced in NM roots at 8 DAT. ZjBACG30 and ZjBACG32 were phylogenetically closer to MtSTR1 and MtSTR2 (Hwang et al., 2016), respectively, which export β-MAG from plant cells into AM fungal cells (Figure 7C and Supplemental Figure S4). ZjLACSs, ZjLACS1 and ZjLACS2, were up-regulated in AM roots compared with them in NM roots under salt treatment or not. ZjLACS4 expression was up-regulated in both NM and AM leaves by salt stress. Among AP2/ERFBP transcription factor, ZjAP2.2, ZjAP2.3, and ZjAP2.4 were specifically induced in roots by AM colonization, and ZjAP2.10, ZjAP2.7, ZjAP2.11, and ZjAP2.9 were also up-regulated in leaves (Figure 7D). Among GRAS transcription factor, ZjGRAS4, ZjGRAS5, and ZjGRAS9 were specifically induced in AM roots, but their prominence remained unchanged in leaves by either AM symbiosis or salt stress. ZjAP2.2, ZjAP2.3, and ZjAP2.4 were phylogenetically closer to MtWRI5s, and ZjGRAS4 was phylogenetically closer to MtRAM1, both of which are the master regulators of FA metabolism during AM colonization (Luginbuehl et al., 2017). During the TAG synthesis pathway, except for the up-regulated expression of diacylglycerol O-acyltransferase 2 (DGAT2) in AM roots, 1-acyl-sn-glycerol-3-phosphate acyltransferase (LPAT), lipid phosphate phosphatase (LPP), and DGAT were not substantially regulated between AM and NM plants. This indicated that AM symbiosis only had slight regulation on host plant TAG synthesis. Taken together, the FA metabolism was reprogrammed greatly in roots and regulated in leaves by AM symbiosis and salt stress.
To validate reproducibility and accuracy of the transcriptomic data, 22 genes were selected for RT-qPCR analysis. Their gene expression levels were highly consistent for the two different methods (R2 = 0.782, P < 0.01) (Supplemental Figure S3), confirming that our transcriptomic results were valid (Supplemental Figure S5).
Discussion
Mycorrhizal symbioses generally improve resilience of host plants undergoing abiotic stress. Past research has generally focused along the physiological and biochemical aspects in AM roots, such as photosynthetic characteristics, nutrient absorption, and antioxidant enzymes, or limited to several well-known genes involved in ion homeostasis (Estrada et al., 2013; Dastogeer et al., 2020). To gain deeper insights, we designed and planned a series of physiological, biochemical, and molecular analyses to better understand the mechanism underlying host plant and AM symbiosis, and how some of these salient interactions change while undergoing salt stress.
Phytohormones are regulated by AM symbiosis during salt stress
Mycorrhizal colonization altered phytohormone homeostasis in host plants and these changes are associated with their tolerance to stresses. Root architecture is regulated by the concerted action of CKs, auxins, and SLs (Pozo et al., 2015). In jujube, several phytohormones were altered by AM symbiosis, especially ABA, CK, and IAA. It was known that ABA and IAA are required for arbuscular development and functionality (Martin-Rodriguez et al., 2011) and they were positively regulated by AM symbiosis in present study. As the key signaling phytohormone for abiotic stress, ABA regulates water uptake and maintains adequate plant water status under unfavorable conditions (Nakashima and Yamaguchi-Shinozaki, 2013). Previous studies have showed that ABA have a direct effect on arbuscule formation and functionality in M. truncatula (Charpentier et al., 2014). ABA levels may be unaltered or even lower in mycorrhizal plants under non-stress conditions, but it is always higher in these plants under osmotic stress (Aroca et al., 2013). In this study, ABA were elevated in AM roots compared with NM roots and these increased further at 3 DAT to 8 DAT. Conversely, ABA contents in leaves remained unaltered during AM symbiosis and salt treatments. These observations are indicative that ABA has an important role in jujube mycorrhizal development and salt adaptation.
Auxin has a role for arbuscule development and the levels were increased in mycorrhizal roots of M. truncatula, tomato, and trifoliate orange. The treatment of tomato, M. truncatula, and rice roots with low concentration of synthetic auxin analogs (2, 4-D) increased AM fungal infection rates, especially the abundance of AM formation, suggesting that auxin can also participate in the later process of AM symbiosis (Calvo-Polanco et al., 2014; Etemadi et al., 2014). Consistently, there were higher IAA contents in AM roots of jujube compared with NM roots, but it was lowered significantly by salt stress in AM roots (Figure 3I). In addition, several important genes involved in auxin response, auxin-responsive protein (ARF7), auxin binding protein (ABP19A), and auxin-induced protein, were clustered in the core gene network of the Magenta module associated with AM roots (Supplemental Table S4). These observations indicated that IAA was involved in AM symbiosis. Since auxins and gibberellins were found in AM fungi and their spores, these observations highlighted the possibility for AM fungi to use phytohormones to interact with their host plants, or to regulate their own development (Pons et al., 2020). We postulate that elevated IAA levels in AM jujube roots were contributed in part by AM fungi. The rate of mycorrhizal formation was decreased by salt stress and this observation was associated with lower IAA levels.
AM symbiosis conferred salt tolerance to jujube by maintaining ion homeostasis
Maintenance of K+/Na+ homeostasis is essential for plant cells to cope with high salinity condition, where cytosolic K+ ions are the common denominator of plant adaptive responses to salt stress (Zelm et al., 2020). Past researches have shown that AM colonization decreased the accumulation of Na+ in host leaves while the K+ absorption was enhanced substantially (Porcel et al., 2016; Zhang et al., 2017). SOS signaling pathway is one major pathway to help plants to attain greater salt tolerance (Zhu, 2016). However, two candidate SOS1 genes in jujube, ZjNHX3 and ZjNHX5, showed similar expression pattern between AM and NM roots under salt stress (Supplemental Figure S2E). The transcript profile is generally consisted with those observation by ion flux and nutrient detection. It indicated that AM symbiosis might not be necessary to increase Na+ exclusion to regulate ion homeostasis in jujube plants.
A few K+ channels and transporters have been found to be upregulated in AM roots, which might contribute to the higher K+/Na+ under salt stress (Estrada et al., 2013; Porcel et al., 2016). Among those, HAK has strong capacity for K+ uptake and would be up-regulated rapidly in response to any K+ deficiency (Yang et al., 2014). Liu et al. (2019) identified an AM-induced SlHAK10 in tomato, which mediate mycorrhizal K+ uptake under low-K+ condition thereby promoting carbohydrate accumulation in roots and facilitating the process of AM fungal colonization. We found that ZjHAK2, putative ortholog of SlHAK10 in jujube, was specifically induced in AM roots. Interestingly, ZjHAK2 was also activated in NM roots when exposed to long duration of salt treatment. It suggested that K+ uptake via ZjHAK2 would be activated by AM symbiosis and also in response to salt stress.
AM symbiosis could induce specific H+-ATPase genes in PAM (Xue et al., 2018). Mutations of the plasma membrane ATPase gene, such as OsHA1, MtHA1, and SiHA8, led to not only a severe defect in arbuscular formation, but also reduction in mycorrhizal-facilitated Pi uptake (Wang et al., 2014; Liu et al., 2020b). In addition, plasma membrane H+-ATPase could drive Na+/H+ antiporter to exclude Na+ while also reducing K+ loss through K+ outward rectifying conductance (KORCs) or nonselective cation channels (NSCCs) (Chen et al., 2019). In this study, AM colonization switched the mode of root H+ influx into efflux, and promoting greater H+ efflux in AM roots during an exposure to 100 mM NaCl and the concomitant Na+ and Ca2+ effluxes were enhanced. A plasma membrane ATPase gene (ZjAHA7) that was specifically induced by AM symbiosis and its expression pattern was consistent with the H+ flux profile. Li and coworkers (2012) found that the ectomycorrhizal (ECM) symbiosis of Populus × canescens activated Na+ extrusion and H+ uptake while reducing K+ efflux in salinized ECM roots. Taken together, we inferred that mycorrhizal colonization activated ZjHAH7 to initiate H+ efflux. These salient observations provided good evidence that AM colonization might stimulate the activity of plasma membrane H+-ATPase in AM roots and subsequently promoting other transporters by pumping more protons. Following which, ZjHAK2 was concomitantly up-regulated to enhance K+ accumulation in AM plants; high K+/Na+ homeostasis was maintained throughout salt exposure.
AM symbiosis reprogrammed FA synthesis and transportation in jujube
Lipids are major carbon source transferred to AM fungal partners (Rich et al., 2017; Wang et al., 2017). However, little is known about FA metabolism during the processes of AM symbiosis in woody species, during salt stress. In this study, AM symbiosis improved the FA contents substantially, particularly for C16:0, in both roots and leaves of jujube at a similar ratio (Figure 4 and Supplemental Table S1-2). The elevation of FAs in leaves as well as roots induced by AM symbiosis was not reported previously. We further confirmed this observation in poplar and M. truncatula. Thus, we can define that AM symbiosis elevating FAs in leaves as well as roots should be a conserved physiological effect. At molecular aspects, AM fungi initiated the signal transduction process and a series of transcriptional regulation on FA metabolism will commence (He et al., 2019). This process is mediated by MtRAM1 and MtWRI5a, which later regulate the expression of MtFatM, MtRAM2, and MtSTR1/2 (Wang et al., 2017). We also observed conserved transcriptional responses of FA metabolisms in AM roots and identified some AM-induced genes in the roots of AM jujube, including ZjAP2, ZjGRASs, ZjGPATs, ZjABCGs, and ZjFatBs. In contrast, genes involved in FA biosynthesis were only marginally up-regulated in leaves of AM plants. Therefore, further research is needed to assess whether the up-regulation of these genes support the elevation of FA metabolism in leaves as well as in roots and in plants undergoing symbiosis.
TAGs are the most abundant energy-dense storage compounds in eukaryotes, as well as the principal storage form of lipids in AM fungi allocated from host plants via the form of β-MAG (Xu and Shanklin, 2016; Jiang et al., 2018). In contrast to the synchronized changes in medium and long chain FAs across leaves and roots induced by AM symbiosis, TAG was only increased in AM roots while not affected in leaves. At transcriptional level, TAG synthesis pathway of jujube plant was not substantially regulated by AM symbiosis in leaves, while FA transformation and transportation for AM fungal partner was widely up-regulated in AM root. Therefore, we believe that the increase of TAG in roots was mainly generated for the sustainable AM symbiosis (Rosli et al., 2018). It also indicated that the elevated FAs in leaves of AM plants were not stored in TAGs and further processed to phospholipids, galactolipid, cutin, and other lipids, or they could be directly used as a source of energy through b-oxidation (Rich et al., 2017). Genetic manipulation of the involved players and further metabolomic analysis are needed to address these possibilities.
Unsaturation FA level in plants is positively related to salt tolerance while the fluidity of the membrane is altered by changes in the DBI index (Kerkeb et al., 2001). Mycorrhization induced significant changes in root FA compositions of trifoliate orange, such as oleate (C18:1N9), linoleate (C18:2), and linolenate (C18:3N3). These changes translated into higher unsaturation index of root FAs thereby improving salt tolerance by reducing the oxidative damage (Wu et al., 2019). In this study, UFAs in leaves and roots were elevated by AM symbiosis, root’s DBI of poplar and M. truncatula was increased by AM symbiosis. Conversely, DBI of jujube remained unchanged by AM symbiosis and salt stress, which might be due to the increased proportion of C16:0 was and decreased C18:2N6. Recent research shown that α-linolenic acid (C18:2N6), linoleic acid (C18:3N3), and hexadecanoate (C16:0) were required to maintain membrane system, ion transport, and K+/Na+ homeostasis in CK signaling deficient Arabidopsis plants undergoing salt stress (Abdelrahman et al., 2021). It is plausible that higher levels of FAs are needed to develop salt tolerance in AM jujube plants. UFA level increase was likely to be contributed by PtFAD2, PtFAD6, and Δ15 FA desaturase (PtΔ15) upregulation in root, SAD catalyzes the formation of C18:1 from the substrate C18:0, which is a key regulatory step to modulate the levels of UFAs in cells (Rodríguez et al., 2015; Wu et al., 2019). Some FA desaturase genes, ZjSAD1, ZjFAD1, ZjFAD2, and ZjFAEs, were up-regulated in AM roots of jujube, in particular, ZjSAD1 was specifically induced by AM symbiosis, which might lead to the increase of C18:1N9 and C:181TN9. In addition, the upregulation of ZjFADs catalyze C18:1 to generate more UFAs. On the other hand, ZjFAD1 was up-regulated by AM symbiosis in leaves, promoted the increase of C18:2 (Saini and Kumar, 2019), however, ZjSAD was unchanged by AM symbiosis. More research is needed to clarify the apparent gaps.
In conclusion, AM symbiosis helped jujube to attain better growth and physiological performance. During salt stress, AM symbiosis facilitated root K+ absorption capacity and translocation to maintain K+/Na+ homeostasis. Our metabolomics profiling revealed that AM symbiosis enhanced FA levels in roots as well as leaves of AM plants, and the levels of UFAs. There is some evidence that AM symbiosis offered various avenues of salt adaptations in this species. Transcriptomic analyses highlighted a conserved core gene sets associated with the jujube-AM symbiosis. A series of genes involved in ion homeostasis, phosphorus transportation, FA biosynthesis, and transportation were identified during AM symbiosis and under salt stress. Based on our findings, there are two broad areas for future research: are there differences between the synthesis and transport of FAs in leaves and roots after AM symbiosis? Is there any other plausible pathway to regulate the FA metabolism? Secondly, it will be interesting to clarify the mechanisms underlying AM symbiosis regulating ion transport channels and transporters while undergoing different biotic and abiotic challenges.
Materials and methods
Mycorrhizal seedling preparation and salt treatments
Rhizophagus irregularis (RI) inoculants were donated from Chinese Academy of Sciences. Wild jujube (Z.jujuba var. spinosa) seeds (Germplasm Resource Nursery, Xingtai, China) were grown up to 8th-leaf stage and they were later transplanted to pots (12 × 10 cm; 1,000 g clean sands; 100 g mycorrhizal fungal inoculants). For control, autoclaved inoculants were used for the NM seedlings. Seedlings were irrigated with 50 mL 0.5 Hoagland weekly. After 45 days, mycorrhizal colonization was assessed using trypan blue stain method (Phillips and Hayman, 1970). When mycorrhizal colonization rate was over 70%, the inoculated and non-inoculated seedlings with similar growth status were selected and subjected to salt treatments. Pot experiment was conducted in a greenhouse under a temperature of 22°C–30°C. This experiment was factorial combination of two factors: (1) NM inoculation and mycorrhizal inoculation (AM); (2) three NaCl level of 0 (no additional NaCl), 100, and 150 mM, and resulted into six treatments, each of which was composed of 50 seedlings. After 1, 8, and 18 days of salt treatment (DAT), seedlings were harvested for each experiment, respectively. We also prepared AM and NM seedlings of 84K poplar (P. alba × P. glandulosa cv. 84K) propagated by tissue culture and M.truncatula Gaertn. ecotypes R108 for FA analysis.
Photosynthesis parameters and chlorophyll fluorescence measurement
Gas exchange parameters were determined at 3, 8, and 13 DAT, using a portable photosynthesis system (model LI-6400XT, Li-COR Inc., Lincoln, NE, USA) (Yong et al., 2000); detailed chlorophyll fluorescence measured at 18 DAT with reference to Song et al. (2019). For each test, three leaves were monitored from single seedling and at least seven seedlings were replicated for each treatment.
Determination of MDA content and antioxidant enzyme activities
For enzyme extractions, leaves and roots samples (0.2 g) were homogenized with 0.05 M sodium phosphate buffer (pH =7.0) containing 1% (w/v) polyvinylpolypyrrolidone. The centrifugated supernatants were used for determining MDA and activities of SOD, POD, and CAT according to Seckin et al. (2008).
Determination of nitrogen, phosphorus, and mineral elements in leaves and roots
Total K+, Ca2+, Mg2+, and Na+ contents were detected by Z-2000 atomic absorption spectrophotometer (Shimadzu, Kyoto, Japan) after wet digestion with HNO3 and HClO4 mixture (4:1, v/v) according to the protocols of Huang et al. (2012). P concentration was quantified using the molybdenum antimony anti-colorimetric method (Kitson and Mellon, 1944) and N contents were determined on continuous flow analytical system (AutoAnalyzer3, Hamburg, Germany).
Measurements of the net flux of Na+, H+, K+, and Ca2+
Net ion flux (Na+, H+, K+, and Ca2+) of fine roots was monitored using a non-invasive Micro-test Technology (NMT100 Series, Younger, USA) (Sun et al., 2009). At first, roots of NM and AM plants at 8 DAT were excised and transferred to a petri-dish containing 10 mL Na+ buffer solution (0.1 mM NaCl, 0.5 mM KCl, 0.2 mM CaCl2, and 0.1 mM MgCl2, pH 5.7) for equilibrating (30 min), and then to another dish containing the Na+ buffer solution to measure the net Na+ flux by moving Na+ selective microelectrode along root tips. The range of 200–2300 μm from apex was scanned with 300 μm graduation steps and six fine roots were detected to determine the positions of Na+ flux occurred actively. Along the root tip where the maximal Na+ net fluxes occurred, net fluxes of Ca2+, H+, and K+ were further detected on AM roots and NM roots (without NaCl treatments) by adding 10 mL of 200 mM NaCl into the measuring solution using the selective electrodes, and were recorded for 20 min.
Quantification of phytohormones
Six treatments were selected for phytohormone determination, including C1 (NM plant at 1 DAT, under 0 mM NaCl treatment), T1 (NM plant at 1 DAT, under 100 mM NaCl treatment), G1 (AM plant at 1 DAT, under 0 mM NaCl treatment), GT1 (AM plant at 1 DAT, under 100 mM NaCl treatment), C8 (NM plant at 8 DAT, under 0 mM NaCl treatment), T8 (NM plant at 8 DAT, under 100 mM NaCl treatment), G8 (AM plant at 8 DAT, under 0 mM NaCl treatment), and GT8 (AM plant at 8 DAT, under 100-mM NaCl treatment). Eighteen phytohormones belonging to eight categories, including ABA, SA, IAA, CKs, GA, JA, and ACC. In short, 0.1 g of sample was ground into fine powder in liquid nitrogen and then transferred into a tube added with 50 μL of internal standard (50 ng/mL) of each phytohormones (Olchemim, Olomouc, Czech Republic). Following which, LC-MS steps were performed according to Tureckova et al. (2009). The experiments were performed with three biological replicates.
Quantification of FAs
Triglyceride content in leaves and roots was determined in accordance to Li et al. (2017). Using 500 μg 17:0 TAG as internal standard and the final FA methyl ester (FAME) were analyzed on a GC (GC-2010 Plus GC system, SHIMAZDZU, Japan; with a DB23 capillary column (60 m × 0.25 mm, 0.25 µm). The composition of medium and long chain FAs in leaves and roots was determined with reference to Wu et al. (2019). Briefly, a gas chromatograph FAME (GC-FAME) method, using Agilent Model 7890A/5975C GC-MS system with DB-WAX column (30m × 0.25 mm ID × 0.25 μm). The 39-component FAME mix (NU-CHEK-PREP) was used to construct a calibration curve for the concentration range of 0.5–1,000 mg/L. The chromatographic peak area and retention time were extracted by MSD Chem Station. In addition, the content of FAs in leaves and roots of M. truncatula, and poplar hybrid under different treatments were also determined. The extraction and determination of the total FAs were performed using a TRACE1310-ISQLT system with HP—INNOWAX column (30 m × 0.25 mm × 0.25 μm), for details refer to Fei et al. (2020).
Transcriptomic analysis
Roots and leaves of six treatment groups (including C1, G1, T1, GT1, T8, and GT8) were selected for RNA sequencing. Sequencing libraries generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, USA), in which 250–300 bp cDNA fragments were screened by AMPure XP beads (Beckman Coulter, Beverly, USA) and enriched by PCR. Then the cDNA libraries were subjected for paired-end sequencing on Illumina Novaseq6000 platform in Novogene Bioinformatics Technology Co. Ltd (Tianjin, China). The generated raw reads were quality control and filtration, and the obtained clean reads were mapped to the Z. jujuba “Junzao” reference genomes (LPXJ00000000) (Huang et al., 2016) using HISAT2 (v2.0.5) with default parameters (Mortazavi et al., 2008). DEG analysis between two treatments was performed using the DEseq2 R package1.16.1 (Anders and Huber, 2010).
Weighted gene co-expression network analysis
WGCNA were performed using a wgcna package (v1.6.6) (Langfelder and Horvath, 2008) in R program. Genes were filtrated based on the threshold setting of FPKM ≥1 in one or more samples (18,200 genes). Adjacency matrix was constructed with these settings: soft-thresholding power (β) were set to 6 based on the scale-free topology criterion, then adjacency matrix was converted to a topological overlap matrix via TOM similarity algorithm, and the genes were hierarchically clustered and generated dendrogram was cut using dynamic tree-cutting algorithm and modules were defined after decomposing (minModuleSize = 20, minimum height = 0.2). Genes with identical expression patterns were grouped into one module. KEGG pathway enrichment was performed on each module by setting P-value < 0.05 as a cut-off criterion with the genome of Z.jujuba “Junzao” using TBtools (Chen et al., 2020). Hub genes were visualized using the software, Cytoscape 3.5.1 (http://cytoscape.org).
Phylogenetic analysis
To discover all potential PHT1 genes in jujube, the known PHT1 genes in Arabidopsis, M.truncatula, and S.lycopersicum (Nagy et al., 2006; Liu et al., 2016) were employed as queries for TBLASTN against jujube genome. Multiple alignment of amino acid sequences of ZjPHT1s and reported PHT1s (Supplemental Table S6-1) (gap opening penalty, 10; gap extension penalty, 0.2; and delay divergent sequences set to 30%) and Neighbor-Joining phylogenetic tree was constructed in MEGA7 (Bootstrap = 1,000) (Tamura et al., 2013). Phylogenetic analysis of ZjHAKs, ZjNHX1s, ZjGPATs, ZjABCGs, ZjFatB, ZjFatA, ZjGRAS, and ZjAP2s was also performed using the same strategy, respectively.
Yeast complementation assays
Yeast complementation was performed as described in Liu et al. (2019). Briefly, a yeast mutant strain R5421 defective in K+ uptake (MATα ura3-52 leu2 trk1Δ his3Δ200 his4-15 trk2Δ1:: pCK64) was transformed with the coding sequence of ZjHAK2 cloned in the yeast expression plasmid pYES2 or with the empty plasmid. Yeast strain R5421 and R5421 transformed with empty plasmid were used as negative control. Yeast strain R757 was used as a positive control. The yeast solution with OD600 = 1.0 was spotted on AP solid medium containing different potassium ion concentrations, and cultured at 30°C for 6 days. The yeast growth assays were carried out on arginine phosphate (AP) medium containing different concentrations of K+.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Those primer pairs were designed for selected gene using Primer-BLAST online in NCBI, and separated by at least one intron on the corresponding genomic DNA (Supplemental Table S6-2). RT-qPCR was performed using a TB Green Premix Ex Taq II kit (Takara, Dalian, China) on the Light Cycler 96 assay system (Roche, Switzerland). The PCR procedure and expression level analyses were performed following our previous method (Zhang et al., 2015).
Statistical analyses
Statistical analysis was performed using an ANOVA (SPSS 20.0.0, IBM, Armonk, NY, USA). Duncan’s multiple range test was performed employed for mean comparisons and statistical significance tests at significance levels of p < 0.05. All data are expressed as mean ± se.
Accession numbers
All RNA-seq data used in this article can be found in Sequence Read Archive at NCBI. The accession number is PRJNA767821.
Supplemental data
The following supplement materials are available.
Supplemental Figure S1. AM symbiosis and salt stress effects on plant antioxidant enzyme activity and fluorescence parameters at 18 DAT.
Supplemental Figure S2. Transcriptome analysis and phylogenetic analysis of jujube PHT1 and NHX1 family members.
Supplemental Figure S3. Expression profile of genes involved in phosphorus and ion transportation in leaves and root.
Supplemental Figure S4 . Neighbor-joining phylogenetic analysis of plant ZjFatA, B, ZjABCG, ZjGRAS1, ZjAP2, and ZjGPAT genes with other known genes.
Supplemental Figure S5. RT-qPCR analysis of selected genes at 12 treatments.
Supplemental Table S1. Mineral element absorption and fatty acid content.
Supplemental Table S2. Quality control of transcriptome data and gene expression.
Supplemental Table S3. KEGG enrichment in each module.
Supplemental Table S4. Hub genes in magenta and black modules.
Supplemental Table S5. Gene expression level of ion transport and fatty acid synthesis genes.
Supplemental Table S6. Gene details for phylogenetic tree and gene primers for RT-qPCR.
Supplementary Material
Acknowledgments
We thank Ms. Yu Liu, Ms. Ju Wang, from College of Forestry, Northwest A&F University, for helping us to perform the pot experiments and enzymes’ measurements. We also thank Prof. Yi Wang (College of Biological Sciences, China Agricultural University) for kindly providing the yeast mutant strain R5421 defective in K+ uptake (MATα ura3-52 leu2 trk1Δ his3Δ200 his4-15 trk2Δ 1:: pCK64).
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31870584), Xinjiang Production and Construction Corps (XPCC) (Grant No. 2017DB006-1), and Cyrus Tang Foundation to J.H.
Conflict of interest statement. The authors declare no potential competing interests.
Contributor Information
Zhibo Ma, Key Laboratory of National Forestry and Grassland Administration on Silviculture in Loess Plateau, College of Forestry, Northwest A&F University, Yangling 712100, China.
Xinchi Zhao, Key Laboratory of National Forestry and Grassland Administration on Silviculture in Loess Plateau, College of Forestry, Northwest A&F University, Yangling 712100, China.
Aobing He, Key Laboratory of National Forestry and Grassland Administration on Silviculture in Loess Plateau, College of Forestry, Northwest A&F University, Yangling 712100, China.
Yan Cao, Key Laboratory of National Forestry and Grassland Administration on Silviculture in Loess Plateau, College of Forestry, Northwest A&F University, Yangling 712100, China.
Qisheng Han, Farmland Irrigation Research Institute, Chinese Academy of Agricultural Sciences, Xinxiang 453002, China.
Yanjun Lu, Key Laboratory of National Forestry and Grassland Administration on Silviculture in Loess Plateau, College of Forestry, Northwest A&F University, Yangling 712100, China.
Jean Wan Hong Yong, Department of Biosystems and Technology, Swedish University of Agricultural Sciences, Alnarp 75007, Sweden.
Jian Huang, Key Laboratory of National Forestry and Grassland Administration on Silviculture in Loess Plateau, College of Forestry, Northwest A&F University, Yangling 712100, China.
J.H. and J.W.H.Y. conceived and designed the experiments. Z.M., X.Z., A.H., Y.C., and Y.L. performed the experiments. Z.M., J.H., J.W.H.Y., and Q.H. analyzed the data. J.H., Z.M., and J.W.H.Y. wrote the manuscript. All authors have read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Jian Huang (huangj@nwsuaf.edu.cn).
References
- Abdelrahman M, Nishiyama R, Tran CD, Kusano M, Nakabayashi R, Okazaki Y, Matsuda F, Chavez Montes RA, Mostofa MG, Li W, et al. (2021) Defective cytokinin signaling reprograms lipid and flavonoid gene-to-metabolite networks to mitigate high salinity in Arabidopsis. Proc Natl Acad Sci USA 118: 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber WJGB (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca R, Ruiz-Lozano JM, Zamarreno AM, Paz JA, Garcia-Mina JM, Pozo MJ, Lopez-Raez JA (2013) Arbuscular mycorrhizal symbiosis influences strigolactone production under salinity and alleviates salt stress in lettuce plants. J Plant Physiol 170: 47–55 [DOI] [PubMed] [Google Scholar]
- Banasiak J, Jamruszka T, Murray JD, Jasinski M (2021) A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume–rhizobium symbioses. Plant Physiol 187: 2071–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti A, Lumini E, Balestrini R, Bianciotto V (2015) Arbuscular mycorrhizal fungi as natural biofertilizers: Let’s benefit from past successes. Front Microbiol 6: 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Polanco M, Molina S, Zamarreno AM, Garcia-Mina JM, Aroca R (2014) The symbiosis with the arbuscular mycorrhizal fungus Rhizophagus irregularis drives root water transport in flooded tomato plants. Plant Cell Physiol 55: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Charpentier M, Sun J, Wen J, Mysore KS, Oldroyd GE (2014) Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol 166: 2077–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13: 1194–1202 [DOI] [PubMed] [Google Scholar]
- Chen T, Wang W, Xu K, Xu Y, Ji D, Chen C, Xie C (2019) K+ and Na+ transport contribute to K+/Na+ homeostasis in Pyropia haitanensis under hypersaline stress. Algal Res 40: 101526. [DOI] [PubMed] [Google Scholar]
- Compant S, Samad A, Faist H, Sessitsch A (2019) A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J Adv Res 19: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastogeer KMG, Zahan MI, Tahjib-Ul-Arif M, Akter MA, Okazaki S (2020) Plant salinity tolerance conferred by arbuscular mycorrhizal fungi and associated mechanisms: A meta-analysis. Front Plant Sci 11: 588550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Aroca R, Maathuis FJ, Barea JM, Ruiz-Lozano JM (2013) Arbuscular mycorrhizal fungi native from a Mediterranean saline area enhance maize tolerance to salinity through improved ion homeostasis. Plant Cell Environ 36: 1771–1782 [DOI] [PubMed] [Google Scholar]
- Etemadi M, Gutjahr C, Couzigou JM, Zouine M, Lauressergues D, Timmers A, Audran C, Bouzayen M, Becard G, Combier JP (2014) Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol 166: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezawa T, Saito K (2018) How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol 220: 1116–1121 [DOI] [PubMed] [Google Scholar]
- Fei X, Ma Y, Hu H, Wei A (2020) Transcriptome analysis and GC-MS profiling of key genes in fatty acid synthesis of Zanthoxylum bungeanum seeds. Industrial Crops and Products 156: 1–2 [Google Scholar]
- Floss DS, Levy JG, Levesque-Tremblay V, Pumplin N, Harrison MJ (2013) DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 110: E5025–E5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- He J, Zhang C, Dai H, Liu H, Zhang X, Yang J, Chen X, Zhu Y, Wang D, Qi X, et al. (2019) A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol Plant 12: 1561–1576 [DOI] [PubMed] [Google Scholar]
- Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Kuster H (2011) Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol 157: 2023–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Nara K, Lian C, Zong K, Peng K, Xue S, Shen Z (2012) Ectomycorrhizal fungal communities associated with Masson pine (Pinus massoniana Lamb.) in Pb-Zn mine sites of central south China. Mycorrhiza 22: 589–602 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang C, Zhao X, Fei Z, Wan K, Zhang Z, Pang X, Yin X, Bai Y, Sun X, et al. (2016) The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet 12: e1006433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, Yim S, Lee E, Khare D, Kim K, et al. (2016) Plant ABC transporters enable many unique aspects of a terrestrial plant's lifestyle. Mol Plant 9: 338–355 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang W, Xie Q, Liu N, Liu L, Wang D, Zhang X, Yang C, Chen X, Tang D, et al. (2017) Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xie Q, Wang W, Yang J, Zhang X, Yu N, Zhou Y, Wang E (2018) Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol Plant 11: 1344–1359 [DOI] [PubMed] [Google Scholar]
- Kerkeb L, Donaire JP, Venema K, Rodriguez-Rosales MP (2001) Tolerance to NaCl induces changes in plasma membrane lipid composition, fluidity and H+-ATPase activity of tomato calli. Physiol Plant 113: 217–224 [DOI] [PubMed] [Google Scholar]
- Kitson RE, Mellon MG (1944) Colorimetric determination of phosphorus as molybdivanadophosphoric acid. Ind Eng Chem (Anal) 16: 379–383 [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Cheng X, Jia Q, Song H, Liu X, Wang K, Zhao C, Zhang Y, Ohlrogge J, Zhang M (2017) Investigation of plant species with identified seed oil fatty acids in Chinese literature and analysis of five unsurveyed Chinese endemic species. Front Plant Sci 8: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bao S, Zhang Y, Ma X, Mishra-Knyrim M, Sun J, Sa G, Shen X, Polle A, Chen S (2012) Paxillus involutus strains MAJ and NAU mediate K+/Na+ homeostasis in ectomycorrhizal Populus x canescens under sodium chloride stress. Plant Physiol 159: 1771–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xu Y, Jiang H, Jiang C, Du Y, Gong C, Wang W, Zhu S, Han G, Cheng B (2016) Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int J Mol Sci 17: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen J, Xie K, Tian Y, Yan A, Liu J, Huang Y, Wang S, Zhu Y, Chen A, et al. (2020a) A mycorrhiza-specific H+-ATPase is essential for arbuscule development and symbiotic phosphate and nitrogen uptake. Plant Cell Environ 43: 1069–1083 [DOI] [PubMed] [Google Scholar]
- Liu J, Liu J, Liu J, Cui M, Huang Y, Tian Y, Chen A, Xu G (2019) The potassium transporter SlHAK10 is involved in mycorrhizal potassium uptake. Plant Physiol 180: 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang J, Wang L, Liu P, Zhao J, Zhao Z, Yao S, Stănică F, Liu Z, Wang L, et al. (2020b) The historical and current research progress on jujube—a superfruit for the future. Hortic Res 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Zhao J, Cai QL, Liu GC, Wang JR, Zhao ZH, Liu P, Dai L, Yan G, Wang WJ, et al. (2014) The complex jujube genome provides insights into fruit tree biology. Nat Commun 5: 5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ (2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178 [DOI] [PubMed] [Google Scholar]
- Martin-Rodriguez JA, Leon-Morcillo R, Vierheilig H, Ocampo JA, Ludwig-Muller J, Garcia-Garrido JM (2011) Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol 190: 193–205 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold BJNM (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagy R, Vasconcelos MJ, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol 8: 186–197 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32: 959–970 [DOI] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS (1970) Improved procedures for clearing roots and staining parasitic and vesicular—arbuscular mycorrhizal fungi for rapid assessment of infection. Trans Br Mycol Soc 55: 158–161 [Google Scholar]
- Pons S, Fournier S, Chervin C, Becard G, Rochange S, Frei Dit Frey N, Puech Pages V (2020) Phytohormone production by the arbuscular mycorrhizal fungus Rhizophagus irregularis. PLoS ONE 15: e0240886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcel R, Aroca R, Azcon R, Ruiz-Lozano JM (2016) Regulation of cation transporter genes by the arbuscular mycorrhizal symbiosis in rice plants subjected to salinity suggests improved salt tolerance due to reduced Na+ root-to-shoot distribution. Mycorrhiza 26: 673–684 [DOI] [PubMed] [Google Scholar]
- Pozo MJ, López‐Ráez JA, Azcón‐Aguilar C, García‐Garrido JM (2015) Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol 205: 1431–1436 [DOI] [PubMed] [Google Scholar]
- Rich MK, Nouri E, Courty PE, Reinhardt D (2017) Diet of arbuscular mycorrhizal fungi: Bread and butter? Trends Plant Sci 22: 652–660 [DOI] [PubMed] [Google Scholar]
- Rich MK, Vigneron N, Libourel C, Keller J, Xue L, Hajheidari M, Radhakrishnan GV, Le Ru A, Diop SI, Potente G, et al. (2021) Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science 372: 864–868 [DOI] [PubMed] [Google Scholar]
- Rodríguez M, Sánchez-García A, Salas J, Garcés R, Martínez Force E (2015) Characterization of soluble acyl-ACP desaturases from Camelina sativa, Macadamia tetraphylla and Dolichandra unguis-cati. J Plant Physiol 178: 35–42 [DOI] [PubMed] [Google Scholar]
- Rosli R, Chan PL, Chan KL, Amiruddin N, Low EL, Singh R, Harwood JL, Murphy DJ (2018) In silico characterization and expression profiling of the diacylglycerol acyltransferase gene family (DGAT1, DGAT2, DGAT3 and WS/DGAT) from oil palm, Elaeis guineensis. Plant Sci 275: 84–96 [DOI] [PubMed] [Google Scholar]
- Saini R, Kumar SJPG (2019) Genome-wide identification, characterization and in-silico profiling of genes encoding FAD (fatty acid desaturase) proteins in chickpea (Cicer arietinum L.). Plant Genetic 18: 100180 [Google Scholar]
- Seckin B, Sekmen AH, Türkan İ (2008) An enhancing effect of exogenous mannitol on the antioxidant enzyme activities in roots of wheat under salt stress. J Plant Growth Regul 28: 12–20 [Google Scholar]
- Shen L-Y, Luo H, Wang X-L, Wang X-M, Qiu X-J, Liu H, Zhou S-S, Jia K-H, Nie S, Bao Y-T, et al. (2021) Chromosome-scale genome assembly for Chinese sour jujube and insights into its genome evolution and domestication signature. Front in Plant Sci 12: 773090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao B, Zheng S, Zhang X, Wang X, Dong W, Xie Q, Wang G, Xiao Y, Chen F, et al. (2021) A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell 184: 5527–5540 e5518 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read FD (2008) Mycorrhizal symbiosis, Ed3. Academic Press, New York. [Google Scholar]
- Song Q, Liu Y, Pang J, Yong JWH, Chen Y, Bai C, Gille C, Shi Q, Wu D, Han X, et al. (2019) Supplementary calcium restores Peanut (Arachis hypogaea) growth and photosynthetic capacityunder low nocturnal temperature. Front Plant Sci 10: 1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura Y, Saito K (2017) Comparative transcriptome analysis between Solanum lycopersicum L. and Lotus japonicus L. during arbuscular mycorrhizal development. Soil Sci Plant Nutr 63: 127–136 [Google Scholar]
- Sui N, Wang Y, Liu S, Yang Z, Wang F, Wan S (2018) Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front Plant Sci 9: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Dai S, Wang R, Chen S, Li N, Zhou X, Lu C, Shen X, Zheng X, Hu Z, et al. (2009) Calcium mediates root K+/Na+ homeostasis in poplar species differing in salt tolerance. Tree Physiol 29: 1175–1186 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tureckova V, Novak O, Strnad M (2009) Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80: 390–399 [DOI] [PubMed] [Google Scholar]
- Wang W, Shi J, Xie Q, Jiang Y, Yu N, Wang E (2017) Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol Plant 10: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Piñeros MA, Wang Z, Wang W, Li C, Wu Z, Kochian LV, Wu PJPC (2014) Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ 37: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Wu QS, He JD, Srivastava AK, Zou YN, Kuca K (2019) Mycorrhizas enhance drought tolerance of citrus by altering root fatty acid compositions and their saturation levels. Tree Physiol 39: 1149–1158 [DOI] [PubMed] [Google Scholar]
- Xu C, Shanklin J (2016) Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu Rev Plant Biol 67: 179–206 [DOI] [PubMed] [Google Scholar]
- Xue L, Klinnawee L, Zhou Y, Saridis G, Vijayakumar V, Brands M, Dormann P, Gigolashvili T, Turck F, Bucher M (2018) AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc Natl Acad Sci USA 115: E9239–E9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L, et al. (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong JW, Wong SC, Letham DS, Hocart CH, Farquhar GD (2000) Effects of elevated CO2 and nitrogen nutrition on cytokinins in the xylem sap and leaves of cotton. Plant Physiol 124: 767–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Hu S, Alam MA, Du H, Che S (2019) The accumulation of fatty acids in different organs of purslane under salt stress. Sci Hortic 250: 236–242 [Google Scholar]
- Zelm EV, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annu Rev Plant Biol 71: 403–433 [DOI] [PubMed] [Google Scholar]
- Zhang C, Huang J, Li X (2015) Identification of appropriate reference genes for RT-qPCR analysis in Ziziphus jujuba Mill. Sci. Hortic 197: 166–169 [Google Scholar]
- Zhang H, Wei S, Hu W, Xiao L, Tang M (2017) Arbuscular mycorrhizal fungus Rhizophagus irregularis increased potassium content and expression of genes encoding potassium channels in Lycium barbarum. Front Plant Sci 8: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Barg R, Yin M, Gueta-Dahan Y, Leikin-Frenkel A, Salts Y, Shabtai S, Ben-Hayyim G (2005) Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J 44: 361–371 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.