Abstract
Plant organ abscission, a process that is important for development and reproductive success, is inhibited by the phytohormone auxin and promoted by another phytohormone, jasmonic acid (JA). However, the molecular mechanisms underlying the antagonistic effects of auxin and JA in organ abscission are unknown. We identified a tomato (Solanum lycopersicum) class III homeodomain-leucine zipper transcription factor, HOMEOBOX15A (SlHB15A), which was highly expressed in the flower pedicel abscission zone and induced by auxin. Knocking out SlHB15A using clustered regularly interspaced short palindromic repeats-associated protein 9 technology significantly accelerated abscission. In contrast, overexpression of microRNA166-resistant SlHB15A (mSlHB15A) delayed abscission. RNA sequencing and reverse transcription-quantitative PCR analyses showed that knocking out SlHB15A altered the expression of genes related to JA biosynthesis and signaling. Furthermore, functional analysis indicated that SlHB15A regulates abscission by depressing JA-isoleucine (JA-Ile) levels through inhabiting the expression of JASMONATE-RESISTANT1 (SlJAR1), a gene involved in JA-Ile biosynthesis, which could induce abscission-dependent and abscission-independent ethylene signaling. SlHB15A bound directly to the SlJAR1 promoter to silence SlJAR1, thus delaying abscission. We also found that flower removal enhanced JA-Ile content and that application of JA-Ile severely impaired the inhibitory effects of auxin on abscission. These results indicated that SlHB15A mediates the antagonistic effect of auxin and JA-Ile during tomato pedicel abscission, while auxin inhibits abscission through the SlHB15A–SlJAR1 module.
The antagonistic effect of auxin and jasmonic acid during tomato pedicel abscission.
Introduction
The process of abscission in plants, which involves the detachment of multicellular organs from the main body at specialized regions called abscission zones (AZs), has been studied in various organs, including flowers, fruits, leaves, and roots, using physiological, molecular, and genetic approaches (Butenko et al., 2003; Meir et al., 2010; Ma et al., 2015; Shi et al., 2018; Li et al., 2019). Abscission is triggered by various developmental cues and environmental stimuli (Patterson, 2001) and is precisely regulated by phytohormones, with auxin acting as a major inhibitor (Abeles and Rubinstein, 1964; Meir et al., 2006). Polar auxin transport across the AZ forms an auxin gradient from the distal to the proximal side, which is vital for inhibiting abscission; impairing this auxin polar flow results in abscission initiation. This is accompanied by changes in transcript abundance of thousands of genes; however, the mechanism by which auxin inhibits abscission remains largely unknown (Roberts et al., 2002; Meir et al., 2006, 2010).
Jasmonic acid (JA) is a phytohormone involved in various aspects of development, including root growth, stamen development, flowering, leaf senescence, and abscission (Hartmond et al., 2000; Howe and Jander, 2008; Wasternack and Hause, 2013; Chini et al., 2016). Application of JA and its methyl ester (JA-Me) has been shown to promote the abscission of bean (Phaseolus vulgaris) petiole explants in the dark and light, without induced ethylene production, but by changing sugar metabolism (Ueda et al., 1996). Both a knock-out (KO) mutant defective in the JA biosynthetic gene, CYTOCHROME P450 74A (CYP74A), and the JA precursor 12-oxophytodienoic acid-mutant, opr3, have a delayed flower abscission phenotype, which can be compensated by exogenous JA application (Park et al., 2002). Moreover, a mutation in the JA receptor gene CORONATINE INSENSITIVE 1 (COI1) of eggplant (Solanum melongena) results in loss of anther dehiscence and delayed flower abscission, with a greater force required to remove petals from the early positions of flowers, when compared with that in wild-type (WT) plants (Zhang et al., 2020). These studies are consistent with the role of JA in floral organ abscission. Notably, jasmonoyl-isoleucine (JA-Ile) is required for COI1 binding to, and subsequent ubiquitin-dependent degradation of, jasmonate ZIM domain (JAZ) proteins, which trigger transcription of JA-responsive genes (Katsir et al., 2008). JAR1, which encodes a Group I Gretchen Hagen (GH3) family of proteins, catalyzes the conjugation of isoleucine to JA, the final step in the formation of JA-Ile (Staswick and Tiryaki, 2004; Svyatyna et al., 2014). In rice (Oryza sativa), two OsJAR1 recessive lines, osjar1-2 and osjar1-3, exhibit anther dehiscence defects accompanied by low fertility (Xiao et al., 2014), while in tomato (Solanum lycopersicum), SlJAR1 has been shown to synthesize the bioactive JA-Ile conjugate (Suza et al., 2010).
Interestingly, JA has been widely reported to counteract the effects of auxin on plant developmental programs. For example, JA inhibits auxin-induced elongation of oat (Avena sativa) coleoptile segments in a sugar metabolism-dependent manner (Ueda et al., 1995), while in Arabidopsis (Arabidopsis thaliana), treatment with JA has been shown to suppress auxin-inducible genes during lateral root formation, and cause decreased accumulation of auxin, as indicated by the expression of the reporter DR5::GUS (Ishimaru et al., 2018). JA signaling has also been reported to impair gravistimulation-induced lateral auxin redistribution, by altering the subcellular distribution of the PIN FORMED 2 (PIN2) auxin transporter, in a COI1-dependent manner. Auxin, on the other hand, upregulates the expression of the GH3 genes GH3.3, GH3.10, and GH3.6, to inactivate JA, which in turn promotes adventitious root formation (Huang et al., 2017). In Arabidopsis flower buds, the JA biosynthetic genes, OPR3 and DAD1, are upregulated in the auxin-perception mutants afb1-3 and tir1 afb2 afb3, and downregulated after treatment with the synthetic auxin, naphthalene acetic acid, suggesting that auxin suppresses JA biosynthesis (Cecchetti et al., 2013). Finally, the JA-Me-induced formation of secondary AZs and senescence in stems of Bryophyllum calycinum was reported to be substantially inhibited by the natural auxin, indole-3-acetic acid (IAA; Saniewski et al., 2000). However, to date, the molecular details of how auxin interacts with JA in the AZ are not well resolved.
Auxin is also known to regulate a class of transcription factors (TFs) that are important for plant development, the class III homeodomain-leucine zipper (HD-ZIP III) TFs. These feature a Leu zipper domain, sterol-binding domain, DNA-interacting PAS-related MEKHLA domain, and a homeodomain (Izhaki and Bowman, 2007; Du and Wang, 2015). There are five HD-ZIP III TFs in Arabidopsis: REVOLUTA(REV)/INTERFASCICULAR FIBERLESS1, PHABULOSA (PHB), PHAVOLUTA (PHV), AtHB8, and CORONA (AtHB15) (Izhaki and Bowman, 2007; Robischon et al., 2011; Baima et al., 2014; Ursache et al., 2014). Lowering auxin levels in the Arabidopsis protoxylem leads to downregulation of AtHB8, while ectopic expression of ATHB8 in Arabidopsis increases the production of the xylem tissue (Baima et al., 2014). The Arabidopsis mutants rp2-12 and wei8 tar2 or quintuple yucca auxin biosynthesis mutants show downregulated expression of PHB, AtHB8, and AtHB15 in the metaxylem, which in turn leads to reduced xylem formation (Ursache et al., 2014). In rice, OsHB3 regulates the leaf initiation process in the shoot apical meristem, in an auxin-dependent manner. Moreover, a growing body of evidence suggests a link between members of the HD-Zip III family and auxin as well as other hormone signaling pathways (Itoh et al., 2008). The function of REV upstream of auxin biosynthesis enhances the expression of the PYL6 gene, which encodes an abscisic acid receptor (Huang et al., 2014). PHB, PHV, and REV have been shown to interact with four type-B response regulator (ARR) TFs and these complexes in turn activate the expression of WUSCHEL (WUS), to maintain shoot apical meristem activity (Reinhart et al., 2013; Zhang et al., 2017). In tomato, SlHB15A has been found to be involved in parthenocarpy, with modified auxin and ethylene signaling, by binding to an ATGAT DNA motif, which matches the consensus binding site previously described for AtHB15 and REV. In addition to transcriptional regulation, SlHB15A is also posttranscriptionally regulated by the microRNA, miR166 (Clepet et al., 2021), which shows abscission-related expression patterns during auxin-depleted abscission (Xu et al., 2015). The timing of different steps in the abscission of tomato pedicels has been determined in previous studies, such as the loss of sensitivity of tomato pedicel AZ cells to auxin, at 4 h after flower removal (Meir et al., 2010). However, the question of whether HD-ZIP III proteins are involved in auxin-regulated abscission is yet to be answered.
In this study, we report the identification and characterization of a tomato HD-ZIP III TF, SlHB15A, as a potential key component in the crosstalk between auxin and JA during tomato pedicel abscission. Our results, and the resulting regulation model, suggest a molecular mechanism by which auxin antagonizes JA action during flower pedicel abscission.
Results
SlHB15A, an HD-ZIP III TF, is specifically expressed in AZs and positively regulated by auxin
A previous research reported that SlHB15A and SlHB15B (AGI gene ID: AT1G52150) expression was sharply downregulated during tomato pedicel abscission (Meir et al., 2010), and our reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis indicated that it is specifically expressed in the AZ. Further, in situ hybridization analysis also showed a high SlHB15A and SlHB15B signal intensity in the AZ (Figure 1, B–F). SlHB15A and SlHB15B share high amino acid sequence identity with other HD-ZIP III proteins from tomato (Supplemental Figure S1) and Arabidopsis, but lower amino acid sequence identity with the other HD-ZIP groups (Figure 1A). Since it has been reported that HD-ZIP III TFs display intensive expression induced by auxin, we further analyzed the transcript abundance of SlHB15A and SlHB15B in the pedicel, after auxin treatment, and found them to be expressed at a high level. Manual flower removal induces a wounding response, as well as auxin depletion. We used the wound response inhibitor, aspirin, to treat the pedicel explants and found higher expression of SlHB15A and SlHB15B in aspirin-treated samples, than in those not treated with aspirin; however, this increase in expression was smaller than that seen in auxin treatment (Figure 1, G–H). The ethylene perception inhibitor, 1-methylcyclopropene (1-MCP), could upregulate SlHB15A and SlHB15B expression (Figure 1I).
Figure 1.
RT-qPCR and in situ hybridization of abscission-related SlHB15A and SlHB15B expression. A, Relationship between Arabidopsis and tomato HD-ZIP III proteins. The phylogenetic analysis was performed using MEGA version 6 software. Detailed information regarding the tomato sequences is listed in Supplemental Table S3. B and C, RT-qPCR analysis of SlHB15A (B) and SlHB15B (C) expression in the AZs, as well as, proximal and distal tissues of tomato pedicels. The results represent mean of three biological replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05). D–F, In situ hybridization of SlHB15A and SlHB15B in tomato pedicel explants. The 3′-labeled SlHB15A sense probes were used as negative controls (D). WT pedicel sections were hybridized with 3′-labeled antisense (E) SlHB15A and (F) SlHB15B probes. Dis, distal; Pro, proximal. Scale bar: 100 µm. G and H, RT-qPCR analysis of SlHB15A (G) and SlHB15B (H) expression in tomato pedicel AZs from the control (tomato pedicel with flower), flower removal (FR), flower removal with auxin treatment, and flower removal with aspirin treatment groups. Values represent mean of three biological replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05). I, RT-qPCR analysis of SlHB15A and SlHB15B expression in the tomato pedicel AZs under 1-MCP treatment. Values represent mean of three biological replicates. Asterisks indicate significantly different values (Student’s t test, **P < 0.01 and *P < 0.05).
SlHB15A is involved in tomato pedicel abscission
To understand the function of SlHB15A in pedicel abscission, we first generated SlHB15A-KO lines using clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9) gene-editing technology. Two target sites were designed for SlHB15A and three independent homozygous transgenic lines (CR-slhb15a #1, 5, and 7) were selected for further studies. The CR-slhb15a #1 line has a 1-bp deletion of one allele, while CR-slhb15a #5 has a 2-bp deletion in both alleles and CR-slhb15a #7 has either a 1-bp insertion or a 5-bp deletion in the two alleles (Supplemental Figure S2). Accelerated abscission was observed in all three KO lines; for example, while WT plants did not show 100% abscission until 32 h after flower removal, all pedicels from CR-slhb15a had abscised by 24 h (Figure 2A).
Figure 2.

Pedicel abscission assay in WT plants as well as the 35S:mSlHB15A, CR-slhb15a, CR-slhb15b, and CR-slhb15aslhb15b lines. A, The effect of flower removal on WT, 35S:mSlHB15A (35S #3, 35S #5, and 35S #6), and CR-slhb15a (CR-slhb15a #1, CR-slhb15a #5, and CR-slhb15a #7) explant pedicel abscission. The results represent mean of three replicates ± sd, with at least 15 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). B, The effect of flower removal on WT, CR-slhb15a, CR-slhb15b, and CR-slhb15aslhb15b explant pedicel abscission. The results represent mean of three replicates ± sd, with at least 15 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05).
Since SlHB15A and SlHB15B both belong to the HD-ZIP III subgroup and have 88% sequence identity at the amino acid level, we investigated whether SlHB15B is also involved in flower pedicel abscission. To this end, SlHB15B single mutants and SlHB15A-SlHB15B double mutants were generated using CRISPR-Cas9 technology (Supplemental Figure S2). The results indicated that compared with WT, single SlHB15B-KO plants showed no significant difference in abscission from WT plants. Moreover, the CR-slhb15aslhb15b double mutant exhibited a similar abscission rate as the CR-slhb15a single mutant, indicating that SlHB15B does not affect abscission (Figure 2B).mRNA accumulation of SlHB15A/B TF is known to be controlled by miR166 in tomato ovule (Clepet et al., 2021) and pedicel AZs (Xu et al., 2015). It has been indicated using RT-qPCR that it is specifically expressed in the AZ, and that this expression increases after flower removal (Xu et al., 2015), and reduces upon auxin treatment (Supplemental Figure S3, A–C). In situ hybridization further revealed a special miR166 signal intensity in the AZs (Supplemental Figure S3, D and E). To further understand the function of SlHB15A in pedicel abscission, while excluding the interference of miR166 degradation on SlHB15A expression, we generated eight independent mSlHB15A (miR166-resistant) overexpression lines through Agrobacterium tumefaciens-mediated transformation. mSlHB15A carries five mutations on the third position of the five codons corresponding to the target of the miR166, and these point mutations code for the same amino acid, and are thus, synonymous mutations (Supplemental Figure S4A). The expression levels of the three lines of SlHB15A (35S:mSlHB15A #3, 35S:mSlHB15A #5, and 35S:mSlHB15A #6) were 6.2-, 5.2-, and 4.1-fold higher, respectively, in mSlHB15A lines than that in the WT plants, and thus we chose them for further studies (Supplemental Figure S4B). In addition, we found that the expression of SlHB15B in the mSlHB15A plants also decreased, but a previous study has shown that SlHB15B does not affect abscission (Supplemental Figure S4C and Figure 2B). As shown in Figure 2A, pedicels had abscised from 49% of the WT plants, at 16 h after flower removal, and from 100% of the WT plants at 32 h. However, the corresponding numbers in the overexpression lines were 6%, 6%, and 8% at 16 h, and 39%, 42%, and 51% at 32 h, for 35S:mSlHB15A #3, 35S:mSlHB15A #5, and 35S:mSlHB15A #6, respectively.
SlHB15A regulates downstream gene targets related to JA pathway
In order to identify the molecular pathways regulated by SlHB15A during abscission, we compared gene expression in the AZs of CR-slhb15a and WT plants using RNA sequencing (RNA-seq), at 2 h after flower removal. The raw sequence reads were deposited into the National Center for Biotechnology Information SRA database, with the accession number SUB10565038 (BioProject ID: PRJNA774186). A total of 1,347 significantly differentially expressed genes (DEGs; fold-change > 2 and false discovery rate [FDR] < 0.01) were identified, of which 556 genes were expressed at higher levels in CR-slhb15a plants than in WT plants, while 791 genes were expressed at lower levels (Supplemental Table S1). This was followed by a Kyoto Encyclopedia of Genes and Genomes (KEGG) classification, which assigned the 1,347 DEGs into 20 functional classes (Figure 3A). The most represented pathways were hormone signal transduction (15 DEGs) and plant–pathogen interaction (12 DEGs). Given that auxin and ethylene are known to affect abscission, we first focused on their associated pathways. Five DEGs were associated with the auxin pathway (two SMALL AUXIN-UP RNA [SAUR] genes, SlIAA8, SlLAX4, and IAA-amido synthetase GH3.6), while only two DEGs, ETHYLENE RESPONSE FACTOR 1A (SlERF1A) and SlERF1B, were associated with the ethylene pathway. In contrast, we found six DEGs that were predicted to be involved in JA biosynthesis or signal transduction of the JA pathway: SlJAR1, SlGH3.10, two cytochrome P450 genes (CYP94B3 and CYP94C1), the ninja family gene SlAFP3, and the putative TF SlMYC2-like (Table 1). In plant–pathogen interaction pathway, we found that two respiratory burst oxidase proteins, three pathogenesis-related proteins, and six calcium-binding proteins were significantly downregulated in CR-slhb15a plants (Table 1). To elucidate the positive function of JA on abscission and plant immune response, we focused on the genes related to JA pathways that displayed changed expression in CR-slhb15a plants relative to WT plants.
Figure 3.
The KEGG classification of assembled DEGs and variation in expression of JA-related genes in the AZs of CR-slhb15a and 35S:mSlHB15A lines, when compared with those of WT plants. A, KEGG classification of assembled DEGs. The most represented pathway was hormone signal transduction (15 DEGs). Two biological replicates were used in this RNA-seq experiment. B, RT-qPCR analysis of SlAFP3, SlMYC2-like, SlJAR1, SlGH3.10, cytochrome P450 (CYP)94B3, and CYP 94C1 in the AZs from WT plants, CR-slhb15a lines, and 35S:mSlHB15A lines. The results represent mean of three replicates ± sd, with at least 20 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05).
Table 1.
DEGs in tomato pedicel AZs of the CR-slhb15a line, when compared with those of WT plants
| Functional category | Gene locus | Annotation | Log2FC |
|---|---|---|---|
| Hormone signal transduction | Solyc04g005380 | Ninja-family protein (SlAFP3) | 1.93 |
| Solyc10g009270 | BHLH TF MYC2-LIKE (SlJA3) | 1.72 | |
| Solyc10g011660 | JA-amido synthetase (SlJAR1) | 1.23 | |
| Solyc10g008520 | JA-amido synthetase JAR1-like (SlGH3.10) | 1.19 | |
| Solyc06g074420 | Cytochrome P450 94C1 | 1.44 | |
| Solyc02g094110 | Cytochrome P450 94B3 | −2.13 | |
| Solyc12g007230 | Auxin-regulated IAA8 (SlIAA8) | 1.14 | |
| Solyc10g076790 | Auxin transporter-like protein 1 (SlLAX4) | 1.01 | |
| Solyc08g079150 | Small auxin upregulated RNA36-like | 2.42 | |
| Solyc12g005310 | IAA-amido synthetase GH3.6 | −1.59 | |
| Solyc03g082520 | Small auxin upregulated RNA32-like | −1.21 | |
| Solyc04g014530 | Ethylene-responsive TF 1B-like (SlERF1B) | 2.34 | |
| Solyc08g078170 | Ethylene-responsive TF 1A-like (SlERF1A) | −1.15 | |
| Solyc10g079600 | Two-component response regulator ARR9 | 1.57 | |
| Solyc05g006420 | Two-component response regulator ARR5 | 1.28 | |
| Pathogen interaction | Solyc03g117980 | Respiratory burst oxidase-like protein | −1.63 |
| Solyc01g099620 | Respiratory burst oxidase homolog protein B | −1.80 | |
| Solyc09g007020 | Pathogenesis-related protein | −1.73 | |
| Solyc02g077370 | Pathogenesis-related genes transcriptional activator PTI5 | −2.06 | |
| Solyc01g106600 | Pathogenesis-related protein | −2.79 | |
| Solyc02g088090 | Calcium-binding protein CML30 | −1.38 | |
| Solyc02g094000 | Calcium-binding protein CML19 | −1.34 | |
| Solyc04g058170 | Calcium-binding protein CML44 | −2.57 | |
| Solyc01g010020 | Calcium-binding protein CML15 | −1.07 | |
| Solyc06g083000 | Calcium-binding protein CML3 | −1.92 | |
| Solyc06g053930 | Calcium-binding protein CML8 | −1.57 |
Two biological replicates were used in this RNA-seq experiment.
To further validate the RNA-seq data, we measured the transcript levels of select genes in the AZs of CR-slhb15a and WT plants using RT-qPCR (Figure 3B). The results were consistent with the RNA-seq data, showing that SlAFP3, SlMYC2-like, SlJAR1, SlGH3.10, and Cytochrome P450 94C1 transcripts were all more abundant in the AZs of the CR-slhb15a lines, when compared with those of the WT plants, while the CYP94B3 transcripts were less abundant. We also measured expression of the same genes in the AZs of the 35S:mSlHB15A line and found that SlAFP3, SlMYC2-like, SlJAR1, SlGH3.10, and CP450 94C1 were expressed at higher levels in the 35S:mSlHB15A line, when compared with those in the WT plants, while CYP94B3 was expressed at lower levels (Figure 3B).
SlHB15A regulates JA-Ile levels in the AZ
Since JA and JA-Ile biosynthesis genes showed significantly altered expression in CR-slhb15a and 35S:mSlHB15A plants, to investigate whether SlHB15A affects the JA pathway during abscission, we quantified the levels of endogenous JA and JA-Ile in the AZs of CRISPR-edited plants, overexpressing transgenic lines, and WT plants during pedicel abscission, using liquid chromatography-tandem mass spectrometry. In WT AZs, JA concentration increased after flower removal, peaked after 4 h, and then decreased, while JA-Ile concentration was highest at 12 h. Generally, the JA concentrations in CR-slhb15a and SlHB15A-overexpressing lines were similar to those in the WT (Figure 4A). However, the JA-Ile concentrations were nearly two-fold higher in the AZs of the SlHB15A-KO plaints during abscission, than in those of the WT plants, while they were 39%, 44%, and 42% lower in the AZs of the SlHB15A-overexpressing line than in those of the WT plants, at 4, 12, and 24 h, respectively (Figure 4B).
Figure 4.

JA and JA-Ile concentrations in pedicel AZs. A, Quantification of JA levels in the AZs of WT, CR-slhb15a, and 35S:mSlHB15A lines during abscission, using LC-MS/MS. The results represent mean of three replicates ± sd, with at least 20 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). B, Quantification of JA-Ile in AZs of WT, CR-slhb15a, and 35S:mSlHB15A lines during abscission, using LC-MS/MS. The results represent mean of three replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05).
SlJAR1 is involved in abscission by means of JA-Ile biosynthesis
The observation that both SlJAR1 and SlGH3.10 were expressed at lower levels in the AZs of 35S:SlHB15A lines and at higher levels in the AZs of the CR-slhb15a lines, when compared with those of the WT plants is consistent with the hypothesis that they contribute to JA-Ile biosynthesis during abscission. Moreover, we compared the expression of other group I GH3 genes known to be related to JA-Ile biosynthesis (SlGH3.1, SlGH3.6, SlGH3.8, SlGH3.11, SlGH3.12, and SlGH3.1) in the AZs of WT as well as CR-slhb15a and 35S:mSlHB15A lines, and observed no differences (Supplemental Figures S5 and S6).
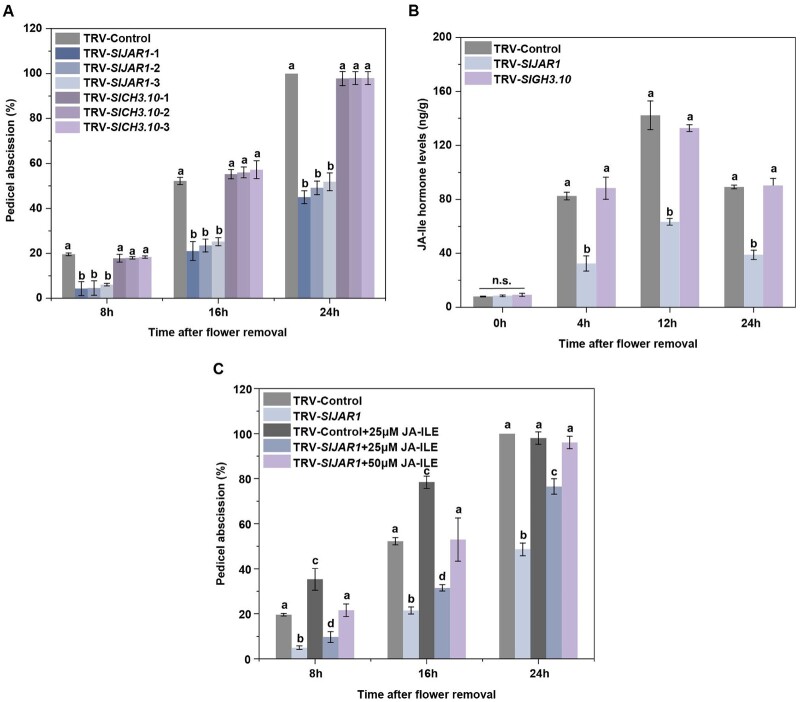
We next examined the effect of silencing SlJAR1 and SlGH3.10 on abscission using virus-induced gene silencing (VIGS). Compared with 100% abscission of pedicels at 24 h after flower removal in WT plants, 49% and 98% had abscised in the SlJAR1- and SlGH3.10-silenced lines, respectively (Figure 5A and Supplemental Figures S7A and S8). Silencing SlJAR1 expression resulted in significantly lower JA-Ile concentrations, while silencing SlGH3.10 had little effect on JA-Ile concentrations (Figure 5B). Application of 25 µM JA-Ile significantly accelerated abscission, while that of 50 µM JA-Ile complemented the abscission defect in the SlJAR1-silenced lines (Figure 5C). These results indicated that SlJAR1 contributes to pedicel abscission by increasing the levels of JA-Ile in the AZ.
Figure 5.
Effects of silencing SlJAR1 and SlGH3.10 on the kinetics of tomato pedicel abscission and JA-Ile content. A, Kinetics of pedicel abscission in TRV, TRV-SlJAR1, and TRV-SlGH3.10 lines. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of three replicates ± sd, with at least 15 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). B, Quantification of JA-Ile in the AZs of TRV, TRV-SlJAR1, and TRV-SlGH3.10 lines during abscission, using LC-MS/MS. The results represent mean of three replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05). C, The effect of JA-Ile on abscission of TRV and TRV-SlJAR1 lines. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of three replicates ± sd, with at least 15 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05).
Next, we analyzed the relationship between ethylene and SlJAR1-dependent JA-Ile function during abscission. Treatment with 1-MCP could depress SlJAR1 expression during abscission (Figure 6A). However, under 1-MCP condition, JA-Ile treatment could significantly accelerate abscission, with abscission reaching 50% at 38.6 h, when compared with 42.6 h under the condition of no JA-Ile treatment (Figure 6B). Meanwhile, we also noticed a significantly sharp decrease in JA-Ile-induced abscission upon 1-MCP treatment, which implied that the ethylene pathway might change after JA-Ile treatment. Since 1-MCP could block ethylene perception, we hypothesized that the ethylene production changed in JA-Ile-treated plants. The results indicated that application of JA-Ile could significantly promote ethylene production (Figure 6C). Moreover, silencing SlJAR1 expression significantly delayed abscission, compared with that observed in TRV lines, during exposure to 1-MCP (Figure 6D). Our previous work showed that SlEIN2 is a key element for ethylene-induced abscission (Wang et al., 2018), since JA-Ile treatment could significantly accelerate abscission in TRV-SlEIN2 plants (Figure 6E and Supplemental Figure S9). The above results indicated the SlJAR1-dependent JA-Ile-induced abscission is both dependent and independent of ethylene action.
Figure 6.
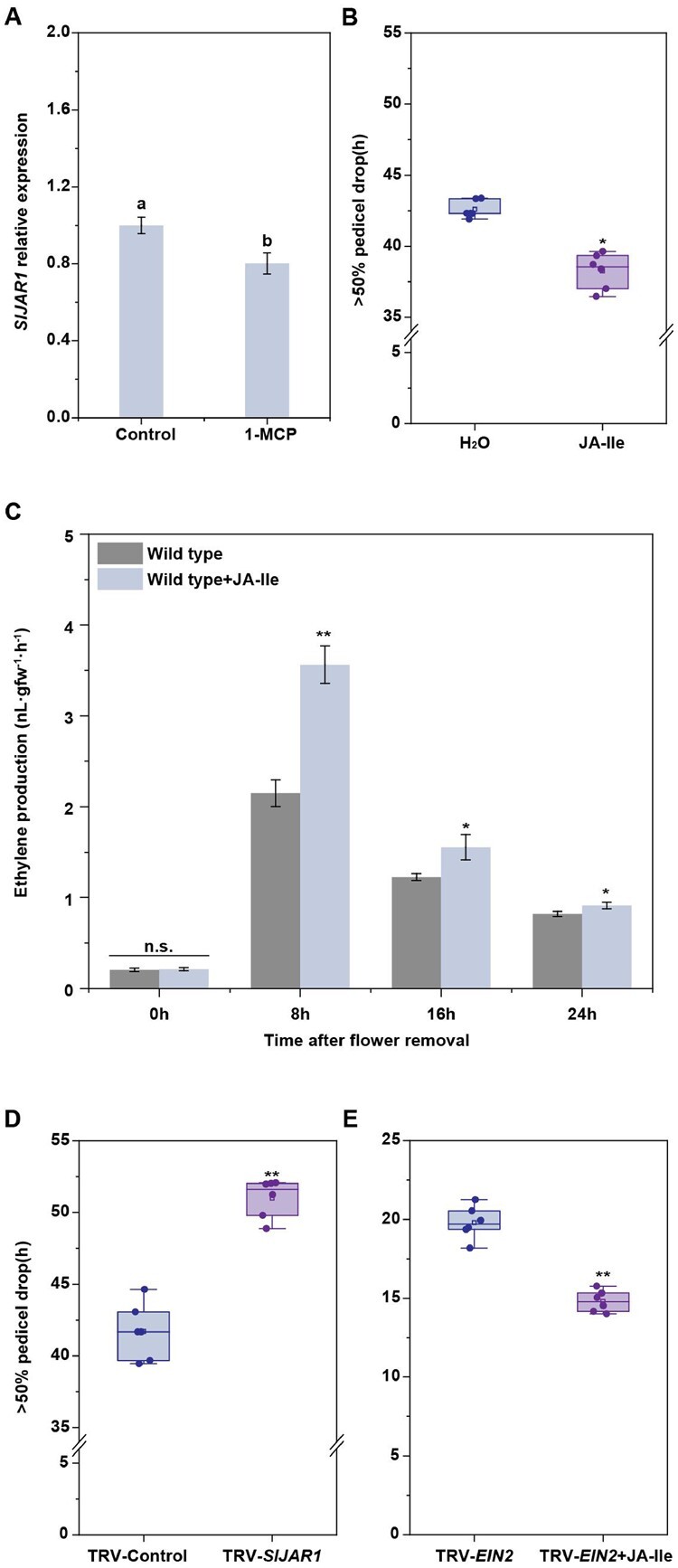
SlJAR1-dependent JA-Ile-induced abscission is ethylene dependent as well as independent. A, RT-qPCR analysis of SlJAR1 expression in the AZs of WT lines treated with or without the ethylene inhibitor, 1-MCP. The results represent mean of three replicates ± sd, with at least 20 samples per replicate (Student’s t test, P < 0.05). B, The effect of JA-Ile on abscission in 1-MCP-treated WT plants. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent means of six biological replicates, with at least 15 samples per replicate. Upmost/lowest lines indicate the maximum/minimum values; Box limits indicate the upper and lower quartiles; Lines in boxes indicate the median values of these data. Open squares represent the mean value and filled circles represent each data points. Asterisks indicate significantly different values (Student’s t test, *P < 0.05). C, The effect of JA-Ile on ethylene production in WT AZ explants. Three independent experiments were performed. Data have been expressed as mean ± sd (Student’s t test, *P < 0.05 and **P < 0.01). D, Kinetics of pedicel abscission in TRV and TRV-SlJAR1 lines, after 1-MCP treatment. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of six replicates, with at least 15 samples per replicate. Upmost/lowest lines indicate the maximum/minimum values; Box limits indicate the upper and lower quartiles; Lines in boxes indicate the median values of these data. Open squares represent the Figure 6 (continued) mean value and filled circles represent each data points. Asterisks indicate significantly different values (Student’s t test, **P < 0.01). E, Kinetics of pedicel abscission in TRV-SlEIN2 lines, after JA-Ile treatment. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of six replicates, with at least 15 samples per replicate. Upmost/lowest lines indicate the maximum/minimum values; Box limits indicate the upper and lower quartiles; Lines in boxes indicate the median values of these data. Open squares represent the mean value and filled circles represent each data points. Asterisks indicate significantly different values (Student’s t test, **P < 0.01).
SlHB15A directly binds to the SlJAR1 promoter
To investigate the possible regulation of SlJAR1 by SlHB15A, we used a yeast one-hybrid (Y1H) assay and tested whether the ATGAT DNA motif serves as a putative cis-element in the SlJAR1 promoter, for binding of SlHB15A (Figure 7A). The binding activity of SlHB15A to the ATGAT sequence in the SlJAR1 promoter was verified using electrophoretic mobility shift assay (EMSA), and mutating this sequence eliminated the binding activity (Figure 7B). To test whether SlHB15A could directly regulate the expression of SlJAR1 in vitro, we performed chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR)-qPCR to analyze the binding of SlHB15A to the promoters of SlJAR1. The promoter sequences of SlJAR1 were only precipitated (using green fluorescent protein [GFP] antibody) from the AZs of SlHB15A-overexpressing lines, with auxin treatment significantly enhanced the binding of SlHB15A to the promoters of SlJAR1, while this precipitation was not observed in the WT plants (Figure 7C).
Figure 7.
SlHB15A represses SlJAR1 expression by binding to its promoter. A, The interaction between SlHB15A and the SlJAR1 promoter ATGAT sequence was assessed using Y1H assays. Co-transformed AD-Rec-P53 and P53-promoter fragments in Y1HGold were used as positive controls. Empty vector and SlJAR1 promoter fragments were used as negative controls. B, The interaction between SlHB15A and the SlJAR1 promoter ATGAT sequence was assessed using EMSA. The hot probe was a biotin-labeled SlJAR1 promoter fragment containing the ATGAT element, while the cold probe was a nonlabeled competitive probe (at a 200-fold concentration of the hot probe). The mutant cold probe was the unlabeled hot probe sequence with two mutated nucleotides. The bands and free probes have been annotated using arrowheads. C, ChIP-qPCR analysis of direct SlHB15A binding to the SlJAR1 promoter. Cross-linked chromatin samples were extracted from mSlHB15A-GFP overexpressing AZs and precipitated using an anti-GFP antibody. Eluted DNA was used to amplify the sequences neighboring the ATGAT element, using qPCR. The 35S:GFP line was used as negative control. Values represent mean of three replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05).
The SlHB15A–SlJAR1 module is involved in auxin-repressed JA-Ile-induced abscission
As the above results showed, knocking-out SlHB15A increased the JA-Ile levels in the AZ, while over-expressing SlHB15A decreased JA-Ile accumulation. Moreover, the 35S:SlHB15A abscission defect could be restored upon JA-Ile application (Figure 8A), and silencing SlJAR1 in the CR-slhb15a lines significantly delayed abscission, when compared with that in CR-slhb15a plants (Figure 8B and Supplemental Figure S7B). Treatment with the JA-Ile biosynthesis inhibitor, jarin-1, caused significantly delayed abscission in SlHB15A-KO plants (Figure 8C), further indicating that SlHB15A mediates abscission, at least in part, via an SlJAR1-dependent increase in JA-Ile concentration.
Figure 8.
The SlHB15A-SlJAR1 module is involved in tomato pedicel abscission. A, The effect of JA-Ile on abscission in the WT and 35S:mSlHB15A lines. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of three replicates ± sd, with at least 12 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). B, Abscission assay in CR-slhb15a-TRV and CR-slhb15a-TRV-SlJAR1 lines. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of three replicates ± sd, with at least 12 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). C, Abscission assay in CR-slhb15a plants treated with or without the JA-Ile inhibitor, jarin-1. The percentages of pedicel abscission were determined at intervals after flower removal. The results represent mean of three replicates ± sd, with at least 12 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). D, Quantification of JA-Ile in the AZs of WT plants treated with or without auxin during abscission, using LC-MS/MS. The results represent mean of three replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05). E, RT-qPCR analysis of SlJAR1 expression in WT AZs treated with or without auxin. The results represent mean of three replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05). F, Auxin inhibition abscission assay in the WT, 35S:mSlHB15A, and CR-slhb15a lines. The percentages of pedicel abscission were determined at intervals after flower removal and the pedicels were incubated with 30 µg·L−1 auxin. Values represent mean of three replicates ± sd, with 15 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05). G, RT-qPCR analysis of SlJAR1 expression in AZs of WT, 35S: mSlHB15A, and CR-slhb15a plants treated with auxin. The results represent mean of three replicates ± sd. Different letters indicate significant differences (Student’s t test, P < 0.05). H, Abscission assay in auxin-treated explants treated with or without JA-Ile. The percentages of pedicel abscission were determined at intervals after flower removal. Values represent mean of three replicates ± sd, with 15 samples per replicate. Different letters indicate significant differences (Student’s t test, P < 0.05).
Auxin treatment significantly reduced JA-Ile levels, concomitant with increased SlHB15A expression and decreased SlJAR1 expression during abscission (Figures 1G and 8, D and E). Next, we investigated the role of SlHB15A in auxin-delayed abscission, by observing abscission in WT, 35S:mSlHB15A, and CR-slhb15a lines following auxin treatment. WT plants reached 50% abscission at 39 h, but this was not observed in CR-slhb15a lines until 33.0 h, and in 35S:mSlHB15A lines until 61.5 h (Figure 8F). Accordingly, knocking-out SlHB15A significantly impaired the auxin-repressed SlJAR1 expression, while overexpression of SlHB15A had the opposite effect (Figure 8G). The above results indicated that SlHB15A is involved in auxin-mediated tomato flower pedicel abscission. We assayed the effect of JA-Ile on auxin-delayed abscission and found that adding JA-Ile significantly impaired the effect of auxin (Figure 8H). The above results indicated that auxin acts through the SlHB15A–SlJAR1 module, to reduce JA-Ile biosynthesis and suppress abscission.
Discussion
Abscission is a part of plant growth and development that involves the integration of many environmental and endogenous signals, together with the intrinsic genetic program. Several plant hormones are fundamental to this process. In this study, we showed that auxin and JA-Ile antagonistically regulated tomato flower pedicel abscission, in part by inducing SlHB15A. SlHB15A could bind to the promoter of a JA-Ile biosynthetic gene, SlJAR1, to repress its expression. This regulatory module reveals the interaction between plant hormone networks during abscission.
SlJAR1-dependent JA-Ile plays an important role in abscission
The active JA conjugate, JA-Ile, which is synthesized by JAR1/4, interacts with the COI1/JAZ receptor complex to ubiquitinate the negative JAZ regulator and release JA-dependent MYC2/3/4 TFs (Thines et al., 2007; Bozorov et al., 2017; Major et al., 2017). Although JA is well known as an abscission accelerator, to our knowledge, a specific role for JA-Ile in abscission is yet to be described. Previous reports have indicated that the coi1-mutant has impaired JA signaling and delayed organ abscission (Kim et al., 2013). It has also been suggested that Nicotiana attenuata NaJAZd affects JA and JA-Ile levels and/or JA-Ile-mediated signaling during flower abscission (Oh et al., 2013). We observed that the JA-Ile content increased significantly in the AZ, at 4 h after flower removal, during tomato pedicel abscission, which is the period when the AZ auxin levels decrease and obtain abscission capacity (Meir et al., 2010). We found that application of JA-Ile accelerated abscission and that SlJAR1 is a key factor in JA-Ile-mediated flower pedicel abscission. This hypothesis is supported by the observation that SlJAR1 expression was substantially upregulated during flower removal-induced abscission and that silencing SlJAR1 caused a reduction in JA-Ile levels and delayed abscission.
JA and ethylene play roles in floral organ abscission, both independently and inter-dependently (Kim, 2014). Our results indicated that JA-Ile could significantly induce ethylene production, while 1-MCP could significantly depress the JA-Ile-induced abscission, indicating that JA-Ile-induced abscission is dependent on ethylene production. Application of JA-Ile could significantly accelerate abscission under the 1-MCP treatment, while silencing SlJAR1 expression could delay abscission under the 1-MCP treatment. Interestingly, JA-Ile treatment could accelerate abscission of SlEIN2-silenced plants. Although these results indicated that SlJAR1-dependent JA-Ile-induced tomato pedicel abscission is at least partially independent of ethylene, we assume that even a JA-dependent and ethylene-dependent pathway might exist in tomato pedicel abscission. Interesting bean, citrus, litchi, and olive are nonclimacteric fruits that produce little or no ethylene, and JA can induce bean abscission without enhanced ethylene production, while it can induce citrus abscission by stimulating levels of ethylene, thus suggesting the JA-dependent pathway plays an important role in nonclimacteric fruit abscission. This is further supported by the findings that apart from that in tomato pedicel abscission, upregulation of other group I GH3-encoding genes has also been observed during melon fruit (Cucumis melo) abscission, citrus (Citrus) fruitlet drop, litchi (Litchi chinensis) fruit abscission, and olive (Olea europaea) fruit abscission (Corbacho et al., 2013; Gil-Amado and Gomez-Jimenez, 2013; Li et al., 2013; Kim et al., 2016; Ma et al., 2019), suggesting that the acceleration of abscission by GH3 group I proteins in response to an increase in JA-Ile levels is a conserved phenomenon.
The contribution of SlHB15A to tomato pedicel abscission involves repression of SlJAR1 expression
HD-ZIP TFs are known to be involved in shedding of organs, with examples including tomato flower pedicels, soybean leaves, and litchi fruit (Lü et al., 2014; Xu et al., 2015; Kim et al., 2016; Ma et al., 2019). In this study, we showed that SlHB15A, which encodes an HD-ZIP III TF, is specifically expressed in tomato pedicel AZs, and is downregulated during abscission and induced by auxin (Hu et al., 2014). SlHB15A has also been reported to be posttranscriptionally regulated by miR166 in the ovule (Clepet et al., 2021). We previously reported that miR165/166 target SlHB15A in the AZ, with its expression increasing at 2 h after flower removal (Xu et al., 2015), which is opposite to the expression patterns of SlHB15A (Supplemental Figure S10A). To overcome this experimental limitation owing to miR166, we generated miR166-resistant mSlHB15A lines. Overexpression of an miRNA166-resistant version of SlREV in tomato resulted in continuous flower development at the pedicel AZ (Hu et al., 2014). We also carefully checked the AZ development in 35S:mSlHB15A and CR-slhb15a plants and found no difference in AZ phenotypes among these plants (Supplemental Figure S11), thus verifying that the difference in abscission was not caused by developmental defects in the AZ. Knocking-out SlHB15A significantly accelerated abscission, while overexpression of miR166-resistant mSlHB15A delayed abscission after flower removal, indicating that SlHB15A negatively regulates pedicel abscission.
Interestingly, the decrease in SlHB15A expression was accompanied by an increase in SlJAR1 expression during abscission, with knocking-out SlHB15A causing significantly enhanced SlJAR1 expression. Y1H and EMSA analyses indicated that SlHB15A can directly bind to an ATGAT motif in the SlJAR1 promotor. In addition, in contrast to the accelerated abscission phenotype of CR-slhb15a plants, silencing SlJAR1 significantly delayed abscission, while silencing SlJAR1 in CR-slhb15a plants caused a lessening of the defective abscission phenotype. These results showed that SlHB15A directly binds to the SlJAR1 promoter, thereby causing reduced expression, which is central to repressing abscission. It is also notable that CYP94B3, which encodes an enzyme that oxidizes JA-Ile to hydroxy (12OH-JA-Ile) derivatives, has a negative feedback control over JA-Ile levels and attenuates JA responses (Koo et al., 2011). SlHB15A overexpression caused an upregulation of CYP94B3 expression, while knocking out SlHB15A had the opposite effect. Future studies will investigate whether SlHB15A also upregulates CYP94B3 expression to suppress JA-Ile accumulation.
SlHB15A regulates the antagonist effect between auxin and JA-Ile during pedicel abscission
An antagonistic relationship has been established between auxin and JA in many aspects of plant growth and development, including coleoptile segment elongation, lateral root formation, and abscission; however, the underlying molecular mechanism has been unclear (Izhaki and Bowman, 2007; Sun et al., 2011; Huang et al., 2017). Auxin signaling is essential for mediating development and responses to environmental stimuli, and numerous studies have shown that auxin HD-ZIP III are involved in the auxin response (Izhaki and Bowman, 2007; Robischon et al., 2011; Baima et al., 2014). A continuous polar auxin flow in the AZ is vital for inhibiting abscission, while flower removal or other activities that disrupt the auxin flow lead to abscission (Dong et al., 2021). We showed in this study that maintaining the auxin polar flow or application of exogenous auxin in flower explants significantly delays abscission and represses JA-Ile accumulation.
Wounding induces a high auxin-to-cytokinin accumulation in the basal region of the hypocotyl explants, while decreasing active auxin levels, by draining auxin via its basipetal transport and internalization, to maintain a minimum auxin-to-cytokinin ratio in the apical region (Larriba et al., 2021). Flower removal-mediated decrease in SlHB15A expression can largely be attributed to auxin depletion, as treatment with wounding response inhibitor, aspirin, only slightly promoted SlHB15A expression, suggesting that wounding impaired AZ auxin flow, resulting in decreased expression of SlHB15A. Knocking-out SlHB15A severely impaired auxin-mediated delayed abscission and suppression of SlJAR1 expression, while overexpression of mSlHB15A had the opposite effect. Accordingly, compared with WT plants, the SlHB15A-KO lines accumulated higher JA-Ile levels, and application of the JA-Ile biosynthesis inhibitor, jarin-1, significantly delayed abscission in these lines. Application of JA-Ile reversed the 35S:mSlHB15A abscission defect and we hypothesize that SlHB15A negatively regulates abscission by decreasing JA-Ile levels in the AZ. Similarly, JA-Ile severely impaired the effects of auxin on abscission. These results suggested that SlHB15A mediates an antagonistic relationship between auxin and JA-Ile during pedicel abscission.
Tissue-specific activity of SlHB15A is posttranscriptionally regulated
Recently, SlHB15A has been reported to be a sentinel that prevents fruit set in the absence of fertilization (Clepet et al., 2021). In our work, we found that SlHB15A inhibits tomato pedicel abscission by mediating an antagonistic effect between auxin and JA-Ile. It seems contrary, since parthenocarpy inhibited abscission and the SlHB15A mutant set parthenocarpic fruits. It is reasonable to deduce that SlHB15A mutant-set parthenocarpic fruits would produce a large amount of auxin in the ovule, which would generate an auxin polar flow across the AZ, which in turn, would inhibit the effect of JA-Ile, as the AZ has lost the SlHB15A function of inhibiting abscission. However, biotic or abiotic stress could induce fertilized or parthenocarpic fruit abscission by means of impaired auxin synthesis, transport, and signaling (Else et al., 2004; Kühn et al., 2016). We found that compared with that in the WT lines, the slhb15a mutant lines had higher fruit drop rate under drought (Supplemental Figure S10B). Hence, we proposed that SlHB15A plays different roles in the ovule and AZ. This is also supported by the fact that the SlHB15A-regulated downstream genes are substantially different in the ovule and AZ. In ovule development and fruit set, SlHB15A represses auxin biosynthesis, transport, and signaling genes, such as TOMATO FLOOZY 2 (ToFYZ2), ToFYZ3, SlPIN4, and AUXIN RESPONSE FACTOR 7, while it induces ethylene biosynthesis and signaling genes, such as 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID OXIDASE 4, ETHYLENE INSENSITIVE 3-LIKE 2, and SlERF.1. However, we observed in this study that the most varied are JA-related genes and few auxin and ethylene pathway genes that showed altered expression in the AZs of CR-slhb15a plants, both in terms of upregulation and downregulation.
It is not rare that a TF performs different functions in different growth and developmental processes. For example, the homeodomain TFs WUS can physically interact with HAIRY MERISTEM TFs and then repress CLV3 expression in the organizing center, while the physical interaction of WUS with SHOOTMERISTEM LESS induces CLV3 expression in the central zone (Zeng et al., 2017; Weits et al., 2019). The activities of the HD-ZIP III TFs are regulated by KANADIs, YABBYs, PLT and HD-ZIP IIIs, and LITTLE ZIPPER proteins, through the formation of different heterodimers with them (Wenkel et al., 2007; Kim et al., 2008; Smith and Long, 2010). Different SlHB15A heterodimers in the ovule and AZ might contribute to the different functions of SlHB15A in parthenocarpy and abscission.
miR166, in general, contributes to spatially modulating the expression of HD-ZIPIII, as in the case of leaf polarity (Merelo et al., 2016). Moreover, SlHB15A is posttranscriptionally regulated by miR166 and the expression of miRNAs is a complex mechanism that depends on plant internal and external cues, including tissue specific and environmental factors (Candar-Cakir et al., 2016; Cedillo-Jimenez et al., 2020). In ovule development, miR166 is cold-induced and inhibits SlHB15A expression, leading to parthenocarpic fruit set. Removal of flowers accelerates the abscission process by reducing the auxin flow (Dermastia et al., 2012), leading to upregulation of miR166 expression (Xu et al., 2015). In this study, we found that application of auxin significantly depressed the flower removal-induced expression of miR166 and there was no significant change after cold in AZ (Supplemental Figure S3F). This different miRNA regulatory mechanism also affects the function of SlHB15A.
In summary, our work identified that biotic or abiotic stresses disrupt AZ auxin polar flow, leading to flower and fruit abscission. After the AZ auxin polar flow has been impaired, the expression of miR166 is induced, which downregulates the expression of SlHB15A, while the expression of SlJAR1 increases meanwhile, leading to JA-Ile accumulation. This, in turn, impairs auxin function and further accelerates abscission by ethylene-dependent and independent pathways. We conclude that auxin antagonizes the effects of JA-Ile in mediating abscission, at least partially via the SlHB15A/SlJAR1 regulatory module (Figure 9).
Figure 9.
Model showing SlHB15A-mediated repression of SlJAR1 expression by auxin, which prevents JA-Ile accumulation, resulting in inhibition of abscission. Under a normal environment (left panel), auxin promotes SlHB15A expression to inhibit SlJAR1 expression and prevent abscission. After auxin depletion (right panel), miR166 expression increases and SlHB15A expression decreases, following which SlJAR1 is induced to synthesize JA-Ile, which accelerates abscission. The arrows represent positive regulation and the bars represent inhibition. miR166, micro-RNA 166; HB15A, homeobox 15A; JAR1, jasmonate-resistant 1; ETR, ethylene receptor; CTR1, constitutive triple response 1; EIN2, ethylene insensitive 2.
Materials and methods
Plant materials and growth conditions
The WT tomato (S.lycopersicum) cultivar Ailsa Craig and transgenic lines were grown as previously described (Wang et al., 2018).
Pedicel abscission assay
Tomato pedicel explants were harvested from newly opened flowers and incubated in water, in a high-humidity chamber. For auxin treatment, the explants were incubated in water containing 20 ng·L−1 IAA. For JA-Ile treatment, the explants were in submerged in water containing 50 μM JA-Ile. Pedicels from the peduncle were monitored at intervals, as previously described (Wang et al., 2005).
In situ hybridization
WT pedicel explants, treated with or without auxin, were sampled and fixed in 4% (w/v) paraformaldehyde in 0.1 M sodium phosphate (pH 7.4). An ethanol gradient was used for dehydration, following which the samples were embedded in Paraplast Plus (Leica, Wetzlar, Germany) at room temperature. A microtome (Leica) was used to cut 10 μm sections from the samples, which were then used for in situ hybridization analysis of SlHB15A, SlHB15B, and miR166 expression, as previously described (Wang et al., 2018). The DIG Oligonucleotide 3ʹ-End Labeling Kit (Roche, Basel, Switzerland) was used to generate antisense and sense RNA probes. Oligonucleotides are listed in Supplemental Table S2.
Plasmid construction and plant transformation
To generate a Sly-miR166-resistant SlHB15A mutant (mSlHB15A), mutations were inserted into the Sly-miR166 mutant-binding site of SlHB15A using the MutanBEST Kit (Takara, Tokyo, Japan) and assembly PCR primers. Full-length mSlHB15A was cloned into the pENTR/D-TOPO vector (Invitrogen) using BP Clonase (Invitrogen, Waltham, MA, USA) and then into the binary vector pCAMBIA1300 using LR Clonase (Invitrogen). All primers used are listed in Supplemental Table S2.
Generation and genotyping of CRISPR-edited plants
For knocking-out SlHB15A, SlHB15B, or both, each corresponding construct was designed to harbor two single guide RNAs targeting two gene-specific sequences. Specifically, two 20-bp fragments from the SlHB15A coding sequence (CDS) (57–76 and 373–392 bp) and SlHB15B CDS (1,836–1,855 and 2,069–2,088 bp) were amplified using PCR, with the pHSE401 vector used as a template, in a reaction with primers containing the gRNA sequences. The U6-26-SlHB15A-gRNA, U6-26-SlHB15B-gRNA, and U6-26-SlHB15ASlHB15B-gRNA cassettes were cloned into the CRISPR-Cas9 binary vector, pCBC-DT1T2_tomatoU6, to generate the pCBC-DT1T2_tomatoU6-SlHB15A, pCBC-DT1T2_tomatoU6-SlHB15B, and pCBC-DT1T2_tomatoU6-SlHB15ASlHB15B-gRNA vectors, respectively. All constructs were introduced into A. tumefaciens strain LB4404, followed by stable tomato transformation, as described previously (Wang et al., 2021).
EMSA
The full-length SlHB15A sequence was cloned into the pET30 vector and expressed in Escherichia coli Rosetta cells. The bacterial cultures were induced with 1 mM Isopropyl β-D-Thiogalactoside (IPTG, TaKaRa) for 6 h, following which Ni-NTP Spin Columns (Qiagen, Hilden, Germany) were used to purify SlHB15A. The biotin-labeled SlJAR1 promoter sequences were as shown in Figure 7A. DNA fragments were labeled at their respective 5′-ends with biotin by the Life Company (Shanghai, China). EMSA was performed as previously described (Wang et al., 2021).
Y1H assays
The SlHB15A CDS was ligated into the pGADT7 vector (Clontech, Mountain View, CA, USA), while the SlJAR1 promoter fragment was ligated into the pAbAi vector (Clontech). Y1H assays were conducted as previously described (Wang et al., 2021). All primers used are listed in Supplemental Table S2. The growth assays were performed at least three times independently, with consistent results.
Assessment of JA and JA-Ile concentrations
For quantitative analysis of JA and JA-Ile levels, 0.3 g AZ tissue was homogenized in liquid nitrogen and then extracted with 500 μL ethanol. After centrifugation (4°C, 12,000 r/min, 5 min), the supernatant was collected and diluted with nine volumes of water, and then subjected to solid phase extraction on a HR-XC column (Macherey-Nagel, Düren, Germany), with 900 μL acetonitrile used as the eluent. Five microliter of the eluate was subjected to ultra-performance liquid chromatography-mass spectrometry (LC-MS/MS), as described previously by Sato et al. (2009) and Yamamoto et al. (2015).
Determination of transcript abundance
Twenty-five segments containing AZs (0.5 mm in length) and similar size distal and proximal regions of the pedicel were excised and collected at different time-points (0, 4, 8, 16, 20, 24, and 32 h) from WT and CR-slhb15a tomato lines. Total RNA extraction was performed, as described previously (Xu et al., 2015). First-strand complementary DNA (cDNA) was synthesized using M-MLV reverse transcriptase and oligo(dT) primers (Takara, Shiga, Japan). RT-qPCR was performed on an 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the SYBR Green PCR Master Mix kit (Takara). The reaction volumes and cycling conditions were used as described previously (Li et al., 2021).
The 2−ΔΔCT method was employed for relative quantification of the transcript abundance of each gene (Livak and Schmittgen, 2001). All primers used are listed in Supplemental Table S2.
Measurement of ethylene production
Ethylene production was carried out in three biological replicates, as previously described (Roberts et al., 1984). Approximately, 15 pedicel AZ samples were assessed for each biological replicate; they were placed in a sealed 5-mL glass jar, and after 1 h, 1 mL of gas was removed from the headspace with a syringe. The gas was analyzed by means of gas chromatography using a Varian GC-3800 system equipped with a GDX-102 column (Dalian Institute of Chemical Physics, China), with nitrogen (20 mL min−1) as the carrier gas.
VIGS
The 300-bp SlJAR1, 300-bp SlGH3.10, and 300-bp SlEIN2 fragments were amplified using PCR with tomato cDNA generated from WT AZ and appropriate primers (Supplemental Table S2). The resulting respective products were cloned into the tobacco rattle virus construct, pTRV2, to generate pTRV2-SlJAR1, pTRV2-SlGH3.10, and pTRV2-SlEIN2 constructs. WT and transgenic seedlings were infected with a mixed A. tumefaciens GV3101 culture containing the pTRV1, pTRV2-SlJAR1, pTRV2-SlGH3.10, and pTRV2-SlEIN2 vectors. The infection was carried out as described previously (Wang et al., 2018).
ChIP-qPCR
Approximately 1 g AZ tissue from WT and 35S:mSlHB15A:GFP plants were collected and cross-linked in 1% (v/v) formaldehyde for 15 min under vacuum, before being quenched in 2 M glycine for 5 min. The AZ samples were frozen and ground in liquid nitrogen, following which the nuclei were isolated. The nuclei were then sheared for 16 cycles of 30 s on and 90 s off, at 4°C, using a sonicator on high setting, to reduce the average DNA fragment size to approximately 500 bp. The chromatin complexes were precleared with protein-A agarose and precipitated with anti-GFP (Transgen, Beijing, China), at 4°C overnight. Protein–DNA cross-links were digested with proteinase K and reverse-crosslinked. The immunoprecipitated DNA was obtained using a PCR purification column (Qiagen) and analyzed using RT-qPCR with SYBR Green dye and a 7500 Real-Time PCR System. The reaction volumes and cycling conditions used were as previously described (Li et al., 2021). All primers used are listed in Supplemental Table S2.
RNA-Seq analysis
TRIzol reagent was used for extraction of total RNA from three biological replicates of WT and SlHB15A-KO plant AZs. The concentration and quality of the RNA were evaluated using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and 3 μg of RNA was used for RNA-Seq library construction. Sequencing was performed using an HiSeq 2500 platform (Illumina, San Diego, CA, USA), and library construction as well as RNA-seq analysis were performed by Biomarker Biotechnology Corporation (Beijing, China). In order to select 150–200 bp cDNA fragments, the library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, MA, USA). The DEGs were identified using the DESeq tool (http://www.bioconductor.org/) and filtered based on the criteria of fold-change > 2.0 and FDR < 0.01. The expression of randomly selected genes from the RNA-seq data was confirmed using RT-qPCR. The information about the primers used is listed in Supplemental Table S2.
Phylogenetic analysis
All Arabidopsis and tomato HD-Zip III proteins and GH3 I proteins were aligned under ClustalW (http://www.ebi.ac.uk/Tools/clustalw). Phylogenetic trees were constructed in MEGA version 6 (http://www.megasoftware.net/index.html). The statistical significance of clades was evaluated with 1,000 bootstrap replicates using the same search criteria. Detailed information regarding the sequences is listed in Supplemental Table S3.
Statistical analysis
All experiments were carried out in at least three replicates. Student’s t test was used to assess the statistical significance of the results, as described in the above corresponding sections of the methods and the figure legends.
Accession numbers
Sequence data from this article can be found in the Genome Database for Tomato (https://www.solgenomics.net/) or MIRbase (https://mirbase.org) under accession numbers SlHB15A (Solyc03g120910); SlHB15B (Solyc12g044410); SlGH3.10 (Solyc10g008520); SlJAR1 (Solyc10g011660); SlEIN2 (Solyc09g007870); SlAFP3 (Solyc04g005380); SlMYC2 like (Solyc10g009270); Cytochrome P450 94C1 (Solyc06g074420); Cytochrome P450 94B3 (Solyc02g094110); SlGH3.13 (Solyc10g009620); SlGH3.1 (Solyc01g095580); SlGH3.5 (Solyc05g050280); SlGH3.6 (Solyc06g048710); SlGH3.8 (Solyc07g054580); SlGH3.11 (Solyc10g009600); SlGH3.12 (Solyc10g009610); and miR166 (MIMAT0035444).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of full-length tomato HD-ZIP III protein sequences, with the START and MEKHLA motifs highlighted.
Supplemental Figure S2. CRISPR/Cas9 mutations.
Supplemental Figure S3. RT-qPCR and in situ hybridization of miR166.
Supplemental Figure S4. Information on mSlHB15A lines.
Supplemental Figure S5. Phylogenetic relationships between Arabidopsis and tomato group I GH3 proteins.
Supplemental Figure S6. Expression profiles for group I GH3 proteins in the AZs of WT, 35S:mSlHB15A, and CR-slhb15a lines.
Supplemental Figure S7. Expression of SlJAR1 in SlJAR1-silenced and TRV control plants.
Supplemental Figure S8. Expression of SlGH3.10 in SlGH3.10-silenced and TRV control plants.
Supplemental Figure S9. Expression of SlEIN2 in SlEIN2-silenced and TRV control plants.
Supplemental Figure S10. Expression analysis of SlHB15A and fruit drop rate under drought in SlHB15A mutants.
Supplemental Figure S11. Flower pedicel morphology of WT, CR-slhb15a, and mSlHB15A plants.
Supplemental Table S1. List of genes that are upregulated and downregulated in CR-SlHB15A plants.
Supplemental Table S2. Primer sequences.
Supplemental Table S3. HD-Zip III and GH3 I family protein sequences.
Funding
This work was supported by the National Key Research and Development Program of China (grant number: 2018YFD1000800), National Natural Science Foundation of China (grant number: 31991184, 31861143045, U1708232, and 31672197), and Liaoning Revitalization Talent Program (grant number: 2018050).
Conflict of interest statement. None declared.
Supplementary Material
Contributor Information
Xianfeng Liu, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Lina Cheng, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Ruizhen Li, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Yue Cai, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Xiaoyang Wang, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Xin Fu, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Xiufen Dong, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Mingfang Qi, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Cai-Zhong Jiang, Department of Plant Sciences, University of California at Davis, Davis, California 95616, USA; Crops Pathology and Genetic Research Unit, USDA-ARS, Davis, California 95616, USA.
Tao Xu, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
Tianlai Li, College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture of Ministry of Education, Shenyang 110866, China.
T.X. and T.L. conceived the original screening and research plans, supervised the experiments, and designed the research. X.L., L.C., R.L., Y.C., X.W., M.Q., X.F., and X.D. performed the experiments. T.X., T.L., and C.-Z.J. conceived the project and wrote the manuscript, with contributions from all authors. T.X. agrees to serve as the author responsible for contact and ensuring communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Tianlai Li (tianlaili@126.com) and Tao Xu (syauxutao@syau.edu.cn).
References
- Abeles F, Rubinstein B (1964) Regulation of ethylene evolution and leaf abscission by auxin. Plant Physiol 39:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Forte V, Possenti M, Peñalosa A, Leoni G, Salvi S, Felici B, Ruberti I, Morelli G (2014) Negative feedback regulation of auxin signaling by ATHB8/ACL5–BUD2 transcription module. Mol Plant 7:1006–1025 [DOI] [PubMed] [Google Scholar]
- Bozorov TA, Dinh ST, Baldwin IT (2017) JA but not JA‐Ile is the cell‐nonautonomous signal activating JA mediated systemic defenses to herbivory in Nicotiana attenuata. J Integr Plant Biol 59:552–571 [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik GE, Amundsen SS, Mandal A, Aalen RB (2003) Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15:2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candar-Cakir B, Arican E, Zhang B (2016) Small RNA and degradome deep sequencing reveals drought-and tissue-specific microRNAs and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J 14:1727–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Brunetti P, Petrocelli V, Falasca G, Ljung K, Costantino P, Cardarelli M (2013) Auxin controls A rabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J 74:411–422 [DOI] [PubMed] [Google Scholar]
- Cedillo-Jimenez CA, Feregrino-Perez AA, Guevara-González RG, Cruz-Hernández A (2020) MicroRNA regulation during the tomato fruit development and ripening: a review. Sci Hortic 270:109435 [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R (2016) Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol 33:147–156 [DOI] [PubMed] [Google Scholar]
- Clepet C, Devani RS, Boumlik R, Hao Y, Morin H, Marcel F, Verdenaud M, Mania B, Brisou G, Citerne S, et al. (2021) The miR166-SlHB15A regulatory module controls ovule development and parthenocarpic fruit set under adverse temperatures in tomato. Mol Plant 14:1185–1198 [DOI] [PubMed] [Google Scholar]
- Corbacho J, Romojaro F, Pech JC, Latché A, Gomez-Jimenez MC (2013) Transcriptomic events involved in melon mature-fruit abscission comprise the sequential induction of cell-wall degrading genes coupled to a stimulation of endo and exocytosis. PLoS ONE 8:e58363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermastia M, Kladnik A, Bar-Dror T, Lers A (2012) Endoreduplication preferentially occurs at the proximal side of the abscission zone during abscission of tomato leaf. Plant Signal Behav 7:1106–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Ma C, Xu T, Reid MS, Jiang CZ, Li T (2021) Auxin response and transport during induction of pedicel abscission in tomato. Hortic Res 8:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Wang H (2015) The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal Behavior 10: e1078955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else MA, Stankiewicz-Davies AP, Crisp CM, Atkinson CJ (2004) The role of polar auxin transport through pedicels of Prunus avium L. in relation to fruit development and retention. J Exp Bot 55:2099–2109 [DOI] [PubMed] [Google Scholar]
- Gil-Amado JA, Gomez-Jimenez MC (2013) Transcriptome analysis of mature fruit abscission control in olive. Plant Cell Physiol 54:244–269 [DOI] [PubMed] [Google Scholar]
- Hartmond U, Yuan R, Burns JK, Grant A, Kender WJ (2000) Citrus fruit abscission induced by methyl-jasmonate. J Am Soc Hortic Sci 125:547–552 [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66 [DOI] [PubMed] [Google Scholar]
- Hu G, Fan J, Xian Z, Huang W, Lin D, Li Z (2014) Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci 229:86–95 [DOI] [PubMed] [Google Scholar]
- Huang H, Liu B, Liu L, Song S (2017) Jasmonate action in plant growth and development. J Exp Bot 68:1349–1359 [DOI] [PubMed] [Google Scholar]
- Huang T, Harrar Y, Lin C, Reinhart B, Newell NR, Talavera-Rauh F, Hokin SA, Barton MK, Kerstetter RA (2014) Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26:246–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Hayashi K, Suzuki T, Fukaki H, Prusinska J, Meester C, Quareshy M, Egoshi S, Matsuura H, Takahashi K, et al. (2018) Jasmonic acid inhibits auxin-induced lateral rooting independently of the CORONATINE INSENSITIVE1 receptor. Plant Physiol 177:1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh JI, Hibara KI, Sato Y, Nagato Y (2008) Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147:1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki A, Bowman JL (2007) KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19:495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105:7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J (2014) Four shades of detachment: regulation of floral organ abscission. Plant Signal Behav 9: e976154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Dotson B, Rey C, Lindsey J, Bleecker AB, Binder BM, Patterson SE (2013) New clothes for the jasmonic acid receptor COI1: delayed abscission, meristem arrest and apical dominance. PLoS ONE 8:e60505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yang J, Yang R, Sicher RC, Chang C, Tucker ML (2016) Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Front Plant Sci 7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH. , et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20:920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ, Cooke TF, Howe GA (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-l-isoleucine. Proc Natl Acad Sci USA 108:9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn N, Serrano A, Abello C, Arce A, Espinoza C, Gouthu S, Deluc LG, Arce-johnson P (2016) Regulation of polar auxin transport in grapevine fruitlets (Vitis vinifera L.) and the proposed role of auxin homeostasis during fruit abscission. BMC Plant Biol 16:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larriba E,, Sánchez-García AB,, Justamante MS,, Martínez-Andújar C,, Albacete A,, Pérez-Pérez JM (2021) Dynamic Hormone Gradients Regulate Wound-Induced de novo Organ Formation in Tomato Hypocotyl Explants. Int J Mol Sci 22: 11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R,, Shi C-L,, Wang X,, Meng Y,, Cheng L,, Jiang C-Z,, Qi M,, Xu T,, Li T (2021) Inflorescence abscission protein SlIDL6 promotes low light intensity-induced tomato flower abscission. Plant Physiol 186: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang Y, Huang X, Li J, Wang H, Li J (2013) De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading. BMC Genomics 14:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhao M, Ma X, Wen Z, Ying P, Peng M, Ning X, Xia R, Wu H, Li J (2019) The HD-Zip transcription factor LcHB2 regulates litchi fruit abscission through the activation of two cellulase genes. J Exp Bot 70:5189–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lü P, Zhang C, Liu J, Liu X, Jiang G, Jiang X, Khan MA, Wang L, Hong B, Gao J (2014) Rh HB 1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J 78:578–590 [DOI] [PubMed] [Google Scholar]
- Ma C, Meir S, Xiao L, Tong J, Liu Q, Reid MS, Jiang CZ (2015) A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol 167:844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Li C, Huang X, Wang H, Wu H, Zhao M, Li J (2019) Involvement of HD-ZIP I transcription factors LcHB2 and LcHB3 in fruitlet abscission by promoting transcription of genes related to the biosynthesis of ethylene and ABA in litchi. Tree Physiol 39:1600–1613 [DOI] [PubMed] [Google Scholar]
- Major IT, Yoshida Y, Campos ML, Kapali G, Xin XF, Sugimoto K, de Oliveira Ferreira D, He SY, Howe GA (2017) Regulation of growth-defense balance by the JASMONATE ZIM‐DOMAIN (JAZ)‐MYC transcriptional module. New Phytol 215:1533–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S, Hunter DA, Chen JC, Halaly V, Reid MS (2006) Molecular changes occurring during acquisition of abscission competence following auxin depletion in Mirabilis jalapa. Plant Physiol 141:1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KSV, Burd S, Ophir R, Kochanek B, Reid MS, Jiang CZ, Lers A (2010) Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol 154:1929–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merelo P, Ram H, Pia Caggiano M, Ohno C, Ott F, Straub D, Graeff M, Cho SK, Yang SW, Wenkel S, et al. (2016) Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc Natl Acad Sci USA 113:11973–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Baldwin IT, Galis I (2013) A jasmonate ZIM-domain protein NaJAZd regulates floral jasmonic acid levels and counteracts flower abscission in Nicotiana attenuata plants. PLoS ONE 8:e57868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock‐out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31:1–12 [DOI] [PubMed] [Google Scholar]
- Patterson SE (2001) Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol 126:494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK (2013) Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell Physiol 25:3228–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Schindler CB, Tucker GA (1984) Ethylene-promoted tomato flower abscission and the possible involvement of an inhibitor. Planta 160:159–163 [DOI] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH (2002) Abscission, dehiscence, and other cell separation processes. Ann Rev Plant Biol 53:131–158 [DOI] [PubMed] [Google Scholar]
- Robischon M, Du J, Miura E, Groover A (2011) The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol 155:1214–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saniewski M, Ueda J, Miyamoto K (2000) Methyl jasmonate induces the formation of secondary abscission zone in stem of Bryophyllum calycinum Salisb. Acta Physiol Plant 22:17–23 [Google Scholar]
- Sato C, Seto Y, Nabeta K, Matsuura H (2009) Kinetics of the accumulation of jasmonic acid and its derivatives in systemic leaves of tobacco (Nicotiana tabacum cv. Xanthi nc) and translocation of deuterium-labeled jasmonic acid from the wounding site to the systemic site. Biosci Biotechnol Biochem 73:1962–1970 [DOI] [PubMed] [Google Scholar]
- Shi CL, Von Wangenheim D, Herrmann U, Wildhagen M, Kulik I, Kopf A, Ishida T, Olsson V, Anker MK, Albert M, et al. (2018) The dynamics of root cap sloughing in Arabidopsis is regulated by peptide signalling. Nat Plants 4:596–604 [DOI] [PubMed] [Google Scholar]
- Smith ZR, Long JA (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464:423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16:2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen Q, Qi L, Jiang H, Li S, Xu Y, Liu F, Zhou W, Pan J, Li X, et al. (2011)Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol 191:360–375 [DOI] [PubMed] [Google Scholar]
- Suza WP, Rowe ML, Hamberg M, Staswick PE (2010)A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-l-isoleucine in wounded leaves. Planta 231:717–728 [DOI] [PubMed] [Google Scholar]
- Svyatyna K, Jikumaru Y, Brendel R, Reichelt M, Mithöfer A, Takano M, Kamiya Y, Nick P, Riemann M (2014) Light induces jasmonate‐isoleucine conjugation via OsJAR1‐dependent and‐independent pathways in rice. Plant Cell Environ 37:827–839 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signalling. Nature 448:661–665 [DOI] [PubMed] [Google Scholar]
- Ueda J, Miyamoto K, Hashimoto M (1996) Jasmonates promote abscission in bean petiole explants: its relationship to the metabolism of cell wall polysaccharides and cellulase activity. J Plant Growth Regul 15:189–195 [Google Scholar]
- Ueda J, Miyamoto K, Kamisaka S (1995) Inhibition of the synthesis of cell wall polysaccharides in oat coleoptile segments by jasmonic acid: relevance to its growth inhibition. J Plant Growth Regul 14:69–76 [Google Scholar]
- Ursache R, Miyashima S, Chen Q, Vatén A, Nakajima K, Carlsbecker A, Zhao Y, Helariutta Y, Dettmer J (2014) Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141:1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Li R, Cheng L, Wang X, Fu X, Dong X, Qi M, Jiang C, Xu T, Li T (2021) SlERF52 regulates SlTIP1; 1 expression to accelerate tomato pedicel abscission. Plant Physiol 185:1829–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li T, Meng H, Sun X (2005) Optimal and spatial analysis of hormones, degrading enzymes and isozyme profiles in tomato pedicel explants during ethylene-induced abscission. Plant Growth Regul 46:97–107 [Google Scholar]
- Wang Y, Zou W, Xiao Y, Cheng L, Liu Y, Gao S, Shi Z, Jiang Y, Qi M, Xu T. , et al. (2018) MicroRNA1917 targets CTR4 splice variants to regulate ethylene responses in tomato. J Exp Bot 69:1011–1025 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann Bot 111:1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits DA, Kunkowska AB, Kamps NCW, Portz KMS, Packbier NK, Venza ZN, Gaillochet C, Lohmann JU, Pedersen O, van Dongen JT, et al. (2019) An apical hypoxic niche sets the pace of shoot meristem activity. Nature 569:714–717 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MM, Barton MK (2007) A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19:3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Chen Y, Charnikhova T, Mulder PPJ, Heijmans J, Hoogenboom A, Agalou A, Michel C, Morel JB, Dreni L, et al. (2014) OsJAR1 is required for JA-regulated floret opening and anther dehiscence in rice. Plant Mol Biol 86:19–33 [DOI] [PubMed] [Google Scholar]
- Xu T, Wang Y, Liu X, Lv S, Feng C, Qi M, Li T (2015) Small RNA and degradome sequencing reveals microRNAs and their targets involved in tomato pedicel abscission. Planta 242:963–984 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Ohshika J, Takahashi T, Ishizaki K, Kohchi T, Matusuura H, Takahashi K (2015) Functional analysis of allene oxide cyclase, MpAOC, in the liverwort Marchantia polymorpha. Phytochemistry 116:48–56 [DOI] [PubMed] [Google Scholar]
- Zeng J, Dong Z, Wu H, Tian Z, Zhao Z (2017) Redox regulation of plant stem cell fate. EMBO J 36:2844–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SW, Yuan C, An LY, Niu Y, Song M, Tang QL, Wei DY, Tian SB, Wang YQ, Yang Y. , et al. (2020) SmCOI1 affects anther dehiscence in a male-sterile Solanum melongena line. Plant Biotechnol 37:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29:1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.