Abstract
Aluminum (Al) toxicity is one of the key factors limiting crop production in acid soils; however, little is known about its transcriptional regulation in plants. In this study, we characterized the role of a NAM, ATAF1/2, and cup-shaped cotyledon 2 (NAC) transcription factors (TFs), ANAC017, in the regulation of Al tolerance in Arabidopsis (Arabidopsis thaliana). ANAC017 was localized in the nucleus and exhibited constitutive expression in the root, stem, leaf, flower, and silique, although its expression and protein accumulation were repressed by Al stress. Loss of function of ANAC017 enhanced Al tolerance when compared with wild-type Col-0 and was accompanied by lower root and root cell wall Al content. Furthermore, both hemicellulose and xyloglucan content decreased in the anac017 mutants, indicating the possible interaction between ANAC017 and xyloglucan endotransglucosylase/hydrolase (XTH). Interestingly, the expression of XTH31, which is responsible for xyloglucan modification, was downregulated in the anac017 mutants regardless of Al supply, supporting the possible interaction between ANAC017 and XTH31. Yeast one-hybrid, dual-luciferase reporter assay, and chromatin immunoprecipitation-quantitative PCR analysis revealed that ANAC017 positively regulated the expression of XTH31 through directly binding to the XTH31 promoter region, and overexpression of XTH31 in the anac017 mutant background rescued its Al-tolerance phenotype. In conclusion, we identified that the tTF ANAC017 acts upstream of XTH31 to regulate Al tolerance in Arabidopsis.
The membrane-bound NAC transcription factor ANAC017 acts upstream of Xyloglucan Endotransglucosylase-Hydrolase31 in regulating Al resistance via modulating the cell wall xyloglucan content.
Introduction
As one of the most abundant metal elements in the earth’s crust, Aluminum (Al) exists in the acid soil as the form of Al3+ that can cause Al toxicity. Micromolar concentrations of Al3+ rapidly inhibit the root elongation through reducing the division and extension of the cell, which in turn restricts the absorption of water and nutrients (Larsen et al., 1997; Ma, 2005; Kochian et al., 2015). Therefore, Al toxicity acts as a major limiting factor to affect the quality and the yield of crops in acid soils (Kochian, 1995), and any approaches that could increase the crops’ Al tolerance would subsequently resolve the above problems of the biofuel production and food shortage, respectively (Ding et al., 2013).
Plants employ a set of strategies to withstand Al stress (Liu et al., 2014), for example, secreting substances such as organic acids, phenolic, etc. to chelate Al (Ma et al., 2001); transporting toxic Al compounds into vacuole for sequestering Al into the vacuoles (Rauser, 1987); fixing Al in the cell wall to decrease Al entering into the cell (Yang et al., 2011); inducing the secretion of proteins such as metal thionein and phytochelatin that can bind Al (Kochian, 1995); increasing the antioxidant enzymes activity (Basu et al., 1994); and enhancing rhizosphere pH values (Pellet et al., 1995; Pineros and Kochian, 2001; Devi et al., 2003). However, accumulating evidences have focused on the role of the cell wall, especially the hemicellulose, in detoxifying Al (Huang et al., 2010; Yang et al., 2011). For instance, Al modified the content or the structure of the hemicellulose in rice (Oryza sativa; Yang et al., 2008), wheat (Triticum aestivum; Tabuchi et al., 2001), Arabidopsis (Arabidopsis thaliana; Zhu et al., 2012, 2014a), etc. Huang et al. (2010) demonstrated that the tonoplast-localized half-size ATP-binding cassette AtSTAR1 modified the cell wall to detoxify Al. Then, Zhu et al. (2012) demonstrated that hemicellulose, especially xyloglucan, both of which are not only very sensitive to Al stress (Al rapidly alters the content and the structure of the hemicellulose), but also act as the major binding site for Al, a process that Xyloglucan Endotransglucosylase-Hydrolase (XTH) participates in Zhu et al. (2012, 2014a, 2014b). Furthermore, either the T-DNA insertion mutant of xth31 or xth17 exhibited lower xyloglucan content and had lower Al retention in the cell wall (Zhu et al., 2012, 2014a), and loss of TRICHOME BIREFRINGENCE-LIKE27 (TBL27) function resulted in lower O-acetylation level of xyloglucan and also lower cell wall Al retention (Zhu et al., 2014b). However, little is known about the regulatory mechanisms of these cell wall-related genes that are responsible for detoxifying Al.
In fact, accumulating evidences have demonstrated that the transcription factors (TFs) play pivotal roles in Al toxicity. For example, Iuchi et al. (2007) identified an important Al-tolerant TF SENSITIVE TO PROTON RHIZOTOXICITY 1 (STOP1) in A.thaliana by mutant analysis. A set of genes responsible for Al stress such as MULTI-DRUG AND TOXIC COMPOUND EXTRUSION (MATE), ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 (ALMT1), and ALUMINUM SENSITIVE 3 (ALS3) are regulated by STOP1, although Al has no effect on the expression of STOP1 itself (Iuchi et al., 2007; Sawaki et al., 2009; Delhaize et al., 2012). STOP1 was reported to have a zinc finger structure of C2H2 and respond to low pH, subsequent evidences showed that this might be controlled by posttranscriptional and/or posttranslational regulation. For instance, Al stress induced the accumulation of STOP1 protein, and three genes, REGULATION OF ATALMT1 EXPRESSION 1 (RAE1), RAE3, and RAE5 participate in the posttranscriptional regulation of STOP1 (Zhang et al., 2019). RAE1, which encodes an F-box protein and negatively regulates the expression of ALMT1, directly interacts with STOP1 to promote its degradation by the ubiquitin–26S proteasome pathway (Zhang et al., 2019). RAE3, which encodes HYPERRECOMBINATION PROTEIN 1 (HPR1) that acts as a core component of the Suppressors of the Transcription Defects of hpr1Δ mutants by Overexpression–Transcription and Export complex, could regulate Al tolerance through modulating the nucleocytoplasmic export of STOP1 mRNA (Guo et al., 2020). RAE5, which encodes a small ubiquitin-like modifier (SUMO) protease Arabidopsis mutant early in short days4 (ESD4), could deSUMOylate STOP1, a process which is pivotal for the function of STOP1 in regulating Al tolerance (Fang et al., 2020). In addition, a rice homolog of STOP1, Al RESISTANCE TRANSCRIPTION FACTOR 1 (ART1), was reported as another TF to regulate the expression of genes that participate in Al detoxification in rice (Yamaji et al., 2009). Then, a homologous protein of ART1-TF ART2 was also demonstrated to respond to Al stress. Further analysis found that ART2 could not activate the expression of the gene regulated by ART1, although the expression of ART2 was induced by Al, indicating that ART1 and ART2 work independently under Al stress in rice (Che et al., 2018). In fact, STOP1-like proteins are demonstrated to be evolutionarily conserved in the detoxification of Al among land plants (Ohyama et al., 2013). However, whether other TFs such as ANAC017 participate in the regulation of Al toxicity still remains unclear.
NAM, ATAF1/2, and cup-shaped cotyladon 2 (NAC) is one of the largest TF families in plants (Riechmann et al., 2000). There are over 117 family members in Arabidopsis, 163 members in Populus euphratica, 152 in Glycine max, and 151 members in rice (Nuruzzaman et al., 2010; Le et al., 2011). The N-terminal region of the NAC family members contains a highly conserved NAC domain, while the diverse C-terminal domains constitute a variable transcriptional activation domain (Ooka et al., 2003). Typically, the NAC domains which consisted of ∼150 amino acids (aa) in length can form a helix–turn–helix structure to repress or activate transcription through directly binding to the promoters of the target genes. Among the target genes, their promoters usually harbor CGTG, CGTA, or CACG motifs that can act as the NAC Recognition Sequence (NACRS; Tran et al., 2004; Olsen et al., 2005; Nakashima et al., 2012). Many studies have shown the involvement of several NACs in plant development (Fujita et al., 2004; Tran et al., 2004; Weir et al., 2004; Li et al., 2014); biotic stress responses such as disease resistance (Ren et al., 2000; Hegedus et al., 2003), pathogen invasion (Huang et al., 2017); and abiotic stress response such as heat stress (Guo et al., 2015), drought and salt stresses in rice (Hu et al., 2008). However, the function and regulation of most NAC genes are still largely unknown. Through screening the Al-sensitive mutants of the NAC TFs family, two mutants that lacks the function of ANAC017, a typical NAC family TF that previously demonstrated to be participated in the mitochondrial proteotoxic stress (Kacprzak et al., 2020) and flood stress (Bui et al., 2020; Meng et al., 2020), were identified. Therefore, ANAC017 was selected for the study here, and through analysis, we found that ANAC017 is required for the induction of XTH31 expression in roots and a regulatory cascade of the Al stress response by ANAC017 was also proposed.
Results
Al tolerance is increased in the anac017 mutants
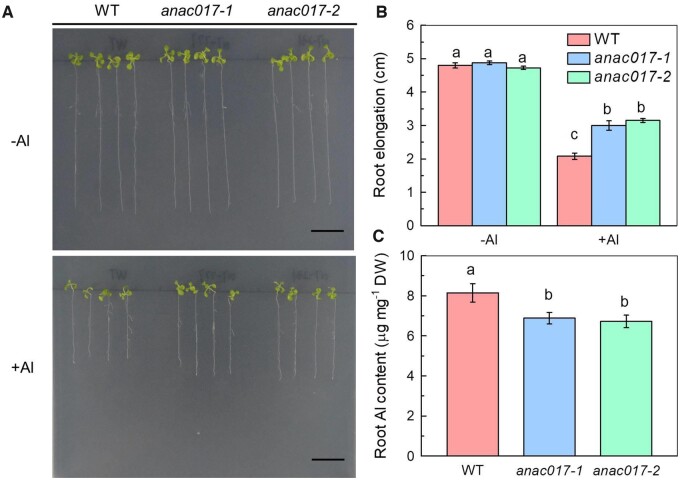
To determine whether the TF ANAC017 is in responsive to Al stress in Arabidopsis, we first characterized two T-DNA insertion mutants anac017-1 (SALK_044777) and anac017-2 (SALK_070231). Sequencing analysis suggested that the T-DNA are inserted into the fourth exon of the coding region for anac017-1 and the 3'-untranslated region (3'-UTR) for anac017-2 (Supplemental Figure S1, A and B), and both of the anac017 mutants exhibited significant reduction in the ANAC017 transcripts in the homozygous lines (Supplemental Figure S1, B and C), further suggesting the loss function of ANAC017 protein in the anac017-1 and anac017-2 mutants. It was interesting that both anac017-1 and anac017-2 mutants exhibited enhanced Al tolerance when compared with the wild-type (WT) plants (Col-0), as indicated by the inhibition of the root length (Figure 1, A and B; only 39% and 34% for anac017-1 and anac017-2 mutants, while 57% for WT), suggesting that ANAC017 plays an important role in response to Al toxicity in Arabidopsis. This conclusion was further confirmed by the decreased Al accumulation in the roots of anac017 mutants (Figure 1C), although under normal growth conditions, no evident phenotypic differences were observed between WT and anac017 mutants in both vegetative and reproductive stage, either in the growth process or development process (Supplemental Figure S2). To further validate the function of ANAC017, we made the complementary lines under its native promoter (anac017-1-com) and the expression of ANAC017 was detected by quantitative PCR (qPCR; Supplemental Figure S3C). The complementary lines exhibited similar phenotypes to those of WT in both normal and Al stress conditions (Supplemental Figure S3, A and B), verifying the hypothesis that the Al-tolerant phenotype of the anac017 mutant is the consequence of the loss function of ANAC017. Furthermore, transgenic lines that overexpressing ANAC017 showed no detectable phenotypic differences from the WT (Supplemental Figure S4), which may be attributed to the not so high expression of the ANAC017 in the ANAC017 overexpression lines (ANAC017-OX), that means, the expression of the ANAC017 in the ANAC017-OX lines was only two- to five-fold to the expression of the ANAC017 in the WT (Supplemental Figure S4), thus under Al treatment, the increased expression of the ANAC017 in the ANAC017-OX lines may not be high enough to make the phenotypic changes in the ANAC017-OX lines when compared with the WT, which needs further study.
Figure 1.
Phenotypes of WT and anac017 mutants. A, WT and anac017 mutants (anac017-1 and anac017-2) were grown on 1/2 MS medium with or without 100 μM Al for 7 days. Seedlings with roots ∼1 cm long were selected and then transferred to the Al-untreated or Al-treated medium. B, Root elongation of WT and anac017 mutants in the presence or absence of 100 µM Al. Root elongation was measured after treatment as indicated by (A). Data are means ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. C, Root Al content of Col-0 and anac017 mutants. Data are means ± sd (n = 4). DW, Dry weight. Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis.
Al Stress decreased the transcription and protein accumulation of ANAC017
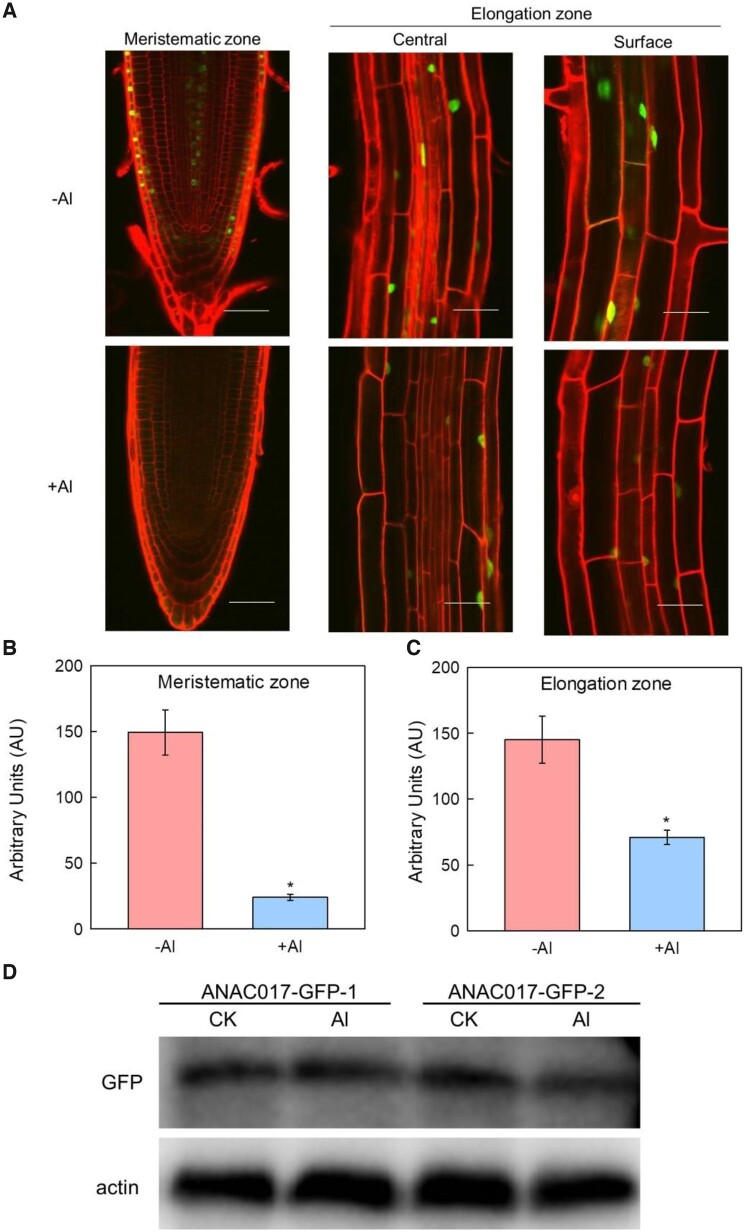
To investigate whether the Al inhibited root elongation is attributed to the transcriptional regulation of ANAC017, WT seedlings were used to conduct the RT-qPCR analysis (Figure 2). The expression of ANAC017 was substantially inhibited by Al stress (Figure 2A), which was further confirmed by the decreased β-glucuronidase (GUS) staining after Al treatment (Figure 2B), suggesting that the expression of ANAC017 is indeed in responsive to Al stress. To further determine if the accumulation of the ANAC017 protein is also in responsive to Al stress, the ANAC017-GFP transgenic lines with the GFP tag were used. It is interesting that Al stress significantly decreased the ANAC017-GFP protein accumulation (Figure 3, A–C), which was further confirmed by the western blot analysis (Figure 3D). In addition, besides Al stress, the expression of the ANAC017 was also decreased in exposure to cold or Lanthanum (La) stress (Supplemental Figure S5A), which was further confirmed by the action of pANAC017-GUS (Supplemental Figure S5, C–F), suggesting the inhibition of ANAC017 expression is not unique to Al.
Figure 2.
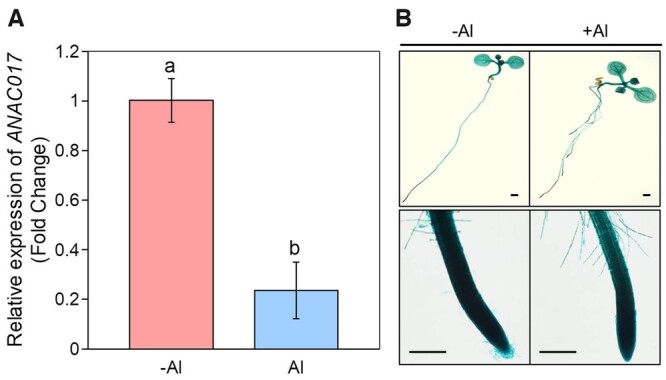
Expression of ANAC017 in response to Al stress. A, RT-qPCR analysis of the ANAC017 expression in WT. Four-week-old WT seedlings were treated with or without Al for 24 h. RNA was prepared from root tissues. TUBULIN was used as an internal control. Data are means ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. B, GUS staining. pANAC017-GUS seedlings with roots ∼1 cm long were treated with or without Al for 7 days. Roots of seedlings were subjected to GUS staining. Scale bars, 100 mm.
Figure 3.
Al stress decreases ANAC017 protein accumulation. A, Fluorescence of ANAC017 fused GFP in root tip region (A), the elongation zone with central view (B) and the elongation zone with surface view (C) in ANAC017-GFP transgenic plants under control and Al treatment. Cell wall was stained with PI dye (red). Scale bars: 50 µm. Data shown are means ± sd of three biological replicates. Asterisks above the bars indicate significant differences at P < 0.05 by Student’s t test. D, Relative protein level of ANAC017-GFP under −Al or +Al conditions. Four-week-old seedlings of two ANAC017-GFP transgenic lines (ANAC017-GFP-1 and ANAC017-GFP-2) were treated without Al or with 50 μM Al for 24 h and roots were sampled for immunoblot analysis of GFP and Actin control.
The expression of the Al stress-responsive genes were disrupted in the anac017 mutants
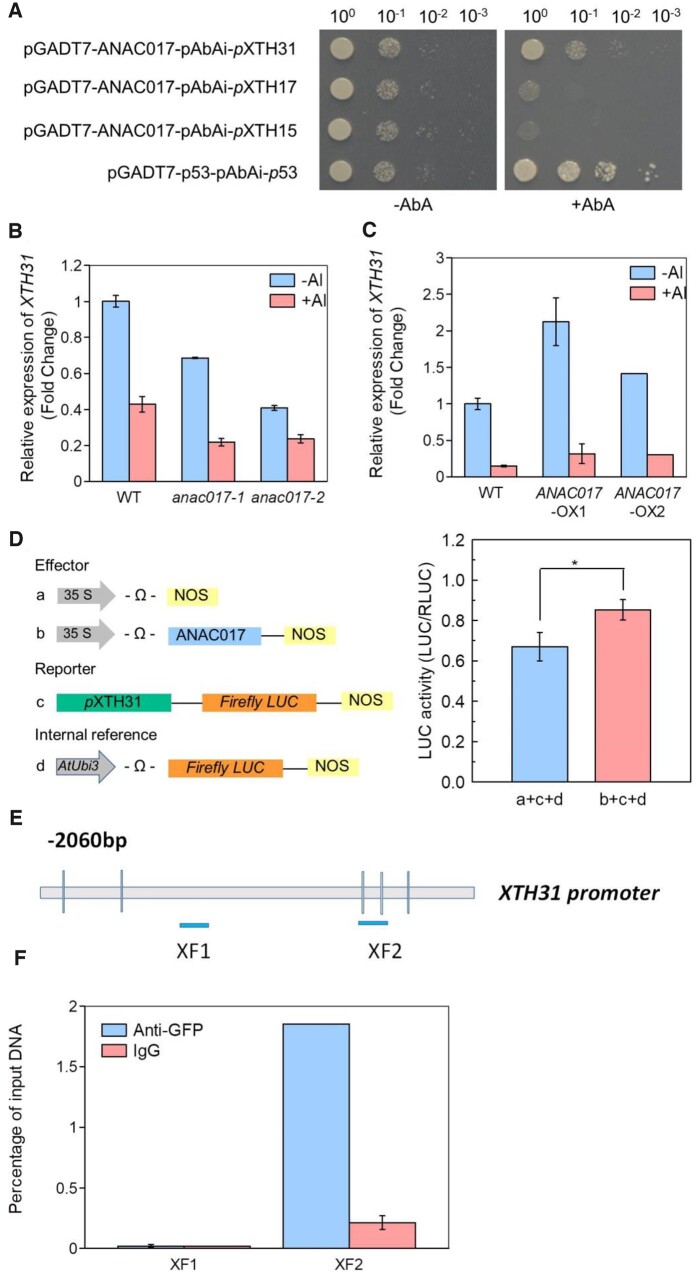
As loss of ANAC017 function resulted in Al tolerance phenotype in Arabidopsis, we speculated whether the expression of other Al stress-responsive genes in the anac017 mutants were also affected. RT-qPCR analysis was conducted to detect their expression (Table 1). Several TFs are key regulators when in response to Al stress, including STOP1, WRKY DNA-BINDING PROTEIN 46 (WRKY 46), WRKY 47, ARABIDOPSIS THALIANA HOMEOBOX 7 (HB7), and HB12 (Iuchi et al., 2007; Ding et al., 2013; Li et al., 2019; Liu et al., 2020). All of them were significantly upregulated in the anac017 mutants under Al stress except STOP1 and HB12 (Table 1). ALMT1 and MATE are two key genes related to the secretion of malate and citrate, respectively (Hoekenga et al., 2006), and their upregulation induced by Al is modulated by STOP1 (Iuchi et al., 2007; Sawaki et al., 2009). MATE expression was dramatically higher in the anac017 mutants than in the WT under control conditions, while no difference was found in the expression of ALMT1 and MATE under Al stress condition (Table 1). An analysis of eight other Al tolerance-associated genes (Larsen et al., 2005, 2007; Kumari et al., 2008; Huang et al., 2010; Zhang et al., 2019) showed that the expression of Sensitive to Al rhizotoxicity1 (STAR1), XTH17, ALS3, and RAE1 were increased, and the expression of XTH31 and ALS1 were decreased in the anac017 mutants under normal condition, while under Al stress, only the expression of STAR1 was increased and XTH31 was decreased in the anac017 mutants, although the expression of XTH15 and TBL27 were similar between the WT and the anac017 mutants both under normal condition (Table 1), further confirming the reduced Al toxicity in the anac017 mutants, that means, as loss function of ANAC017 decreased Al accumulation in roots (Figure 1C); therefore, the different expression of Al-responsive genes listed in Table 1 may be solely the results of less Al toxicity in the anac017 mutants.
Table 1.
Differential expression of the genes that responsible for Al detoxifying in WT and anac017 mutants
| Gene | WT(CK) | anac017-1(−Al) | anac017-2(−Al) | WT(+Al) | anac017-1(+Al) | anac017-2(+Al) |
|---|---|---|---|---|---|---|
| STOP1 | 1.03 ± 0.28c | 1.21 ± 0.08c | 1.23 ± 0.37c | 1.75 ± 0.15b | 2.31 ± 0.02a | 1.82 ± 0.30b |
| WRKY46 | 1.02 ± 0.22d | 2.68 ± 0.1b | 2.26 ± 0.14b | 1.48 ± 0.05c | 4.03 ± 0.37a | 3.49 ± 0.23a |
| WRKY47 | 1.01 ± 0.12d | 1.81 ± 0.1b | 1.46 ± 0.03c | 1.35 ± 0.04c | 3.39 ± 0.4a | 3.01 ± 0.22a |
| HB7 | 1 ± 0.11d | 1.63 ± 0.15c | 1.43 ± 0.09 c | 16.96 ± 0.56 b | 21.51 ± 1.85a | 19.18 ± 1.3a |
| HB12 | 1.01 ± 0.15c | 1.45 ± 0.13 b | 1.42 ± 0.18 b | 3.15 ± 0.15a | 3.33 ± 0.19a | 3.49 ± 0.19a |
| ALMT1 | 1.00 ± 0.09b | 0.93 ± 0.23b | 1.03 ± 0.16b | 20.08 ± 4.52a | 18.77 ± 4.52a | 16.90 ± 1.29a |
| MATE | 1.01 ± 0.17c | 2.06 ± 0.08b | 2.23 ± 0.01b | 76.7 ± 2.22a | 80.33 ± 9.47a | 67.49 ± 7.15a |
| RAE1 | 1 ± 0.05c | 1.72 ± 0.42b | 1.69 ± 0.18b | 7.09 ± 0.82a | 7.94 ± 1.52a | 7.86 ± 1.37a |
| ALS1 | 1 ± 0.09b | 0.54 ± 0.08c | 0.63 ± 0.05c | 1.24 ± 0.18a | 1.17 ± 0.07a | 1.16 ± 0.15a |
| ALS3 | 1.01 ± 0.09c | 1.38 ± 0.14b | 1.45 ± 0.06 b | 9.36 ± 1.33a | 8.81 ± 1.79a | 9.35 ± 0.67a |
| STAR1 | 1 ± 0.06b | 1.79 ± 0.12a | 1.65 ± 0.27a | 0.5 ± 0.02c | 0.98 ± 0.09b | 1.03 ± 0.06b |
| XTH15 | 1.00 ± 0.11a | 0.93 ± 0.36a | 0.86 ± 0.04a | 0.96 ± 0.06a | 0.82 ± 0.07a | 0.78 ± 0.02b |
| XTH17 | 1.00 ± 0.08c | 1.67 ± 0.11a | 1.36 ± 0.18b | 0.72 ± 0.06d | 0.71 ± 0.02d | 0.68 ± 0.01d |
| XTH31 | 1.00 ± 0.03a | 0.69 ± 0.00b | 0.41 ± 0.01c | 0.43 ± 0.04c | 0.22 ± 0.02d | 0.24 ± 0.02d |
| TBL27 | 1.00 ± 0.07c | 1.00 ± 0.13c | 1.06 ± 0.14c | 1.62 ± 0.03b | 1.77 ± 0.05a | 1.70 ± 0.01a |
Four-week-old WT and anac017 mutants were treated with or without Al for 24 h. RNA was prepared from root tissues. TUBULIN was used as an internal control. Data are means ± sd (n = 4). Different letters indicate significant differences at P < 0.05.
Decreased cell wall Al accumulation in the anac017 mutant roots
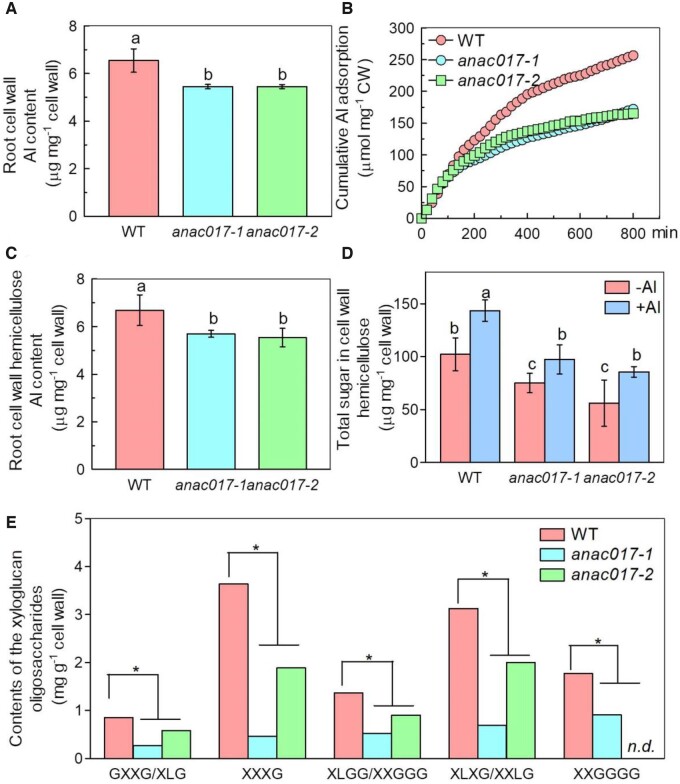
Cell wall is the main Al binding site in Arabidopsis roots (Yang et al., 2011), thus root cell wall Al accumulation was compared between WT and anac017 mutants. As shown in Figure 4, less Al was accumulated in the anac017 root cell walls (Figure 4A), which was further confirmed by the adsorption kinetics of the whole cell wall to Al (Figure 4B).
Figure 4.
Cell wall composition and its Al binding capacity analysis in WT and anac017 mutants. A, Al content in WT and anac017 mutants root cell wall. Cell walls were extracted from roots of 4-week-old WT and anac017 mutants with Al for 24 h. Data are means ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. B, Al adsorption kinetics of WT and anac017 mutants cell wall. Cell walls were extracted from 4 -week-old WT and anac017 mutants roots without Al. C, Al content in the root cell wall hemicellulose of WT and anac017 mutants. Cell walls were extracted from roots of 4-week-old WT and anac017 mutants with Al for 24 h. Data are means ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. D, Total sugar content in the root cell wall hemicellulose of WT and anac017 mutants. Seedlings were treated with or without Al for 24 h. Data are means ± sd (n = 4). Different letters above the bars represent significant differences at P < 0.05 by one-way ANOVA analysis. E, The relative abundance of xyloglucan oligosaccharides by the MALDI-TOF MS analysis. Cell walls were extracted from Col-0 and anac017 mutants roots in the absence of Al and digested with XEG. The asterisk shows a significant difference between WT and anac017 mutants at P < 0.05 by Student’s t test.
Then, a question raised why less Al was accumulated in the anac017 mutants root cell wall. As hemicellulose contributes greatly to the Al binding capacity of the cell wall, thus both Al retention in the hemicellulose and hemicellulose content were measured between WT and anac017 mutants. It was interesting that less Al was accumulated in the cell wall hemicellulose of the anac017 mutants, in company with the reduced hemicellulose content in the anac017 mutants, irrespectively under Al stress or not (Figure 4, C and D), implying that cell wall hemicellulose was responsible for the lower root Al accumulation in the anac017 mutants.
Furthermore, xyloglucan is the main component of the cell wall hemicellulose to bind Al, thus the xyloglucan content was also measured by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF) after digestion by the xyloglucanase. As expected, total xyloglucanase-accessible xyloglucan repeat units were significantly reduced in the anac017 mutants (the sum of the xyloglucan repeat units; Supplemental Figure S6), especially GXXG (and/or XLG), XXXG, XLGG (and/or XXGGG), XLXG (and/or XXLG), XXGGGG (Figure 4E). It was noteworthy that the content of xyloglucan in the two anac017 mutants showed quite big differences, for example, the difference of XXGGGG, one xyloglucan repeat unit for anac017-2 mutant to WT is larger than the anac017-1 mutant to WT (Figure 4E). However, the Al-tolerant phenotype showed no difference between these two anac017 mutants, one possible explanation is that there may be exist a tight threshold of the total xyloglucan content that leads to the observed Al effects (Supplemental Figure S6). If the xyloglucan content falls <50% of the WT, Al tolerance becomes apparent and is not increased further, even though the xyloglucan content drops even lower (down to 27%). Once the threshold of 50% of the xyloglucan content is reached, the Al binding capacity is exhausted (Figure 4, A and B), which needs to be clarified further.
Tissue-specific localization of ANAC017 expression
RT-qPCR was performed to detect the expression pattern of ANAC017. As shown in Figure 5A, the transcripts of ANAC017 was higher in flowers and siliques, but relatively lower in roots, leaves, and stems, which was further confirmed by the in vivo GUS staining. A DNA fragment consisting 2,545-bp promoter region of the ANAC017 was used to drive the expression of the GUS reporter gene to obtain the pANAC017-GUS lines. It is interesting that the GUS was constitutively stained in the roots, cotyledons, inflorescences, siliques, hypocotyls, and leaves (Figure 5B). In roots, pANAC017-GUS was not only expressed in root tips, but also expressed in the roots elongation zone, roots differentiation zone, and roots maturation zone (Figure 5B).
Figure 5.
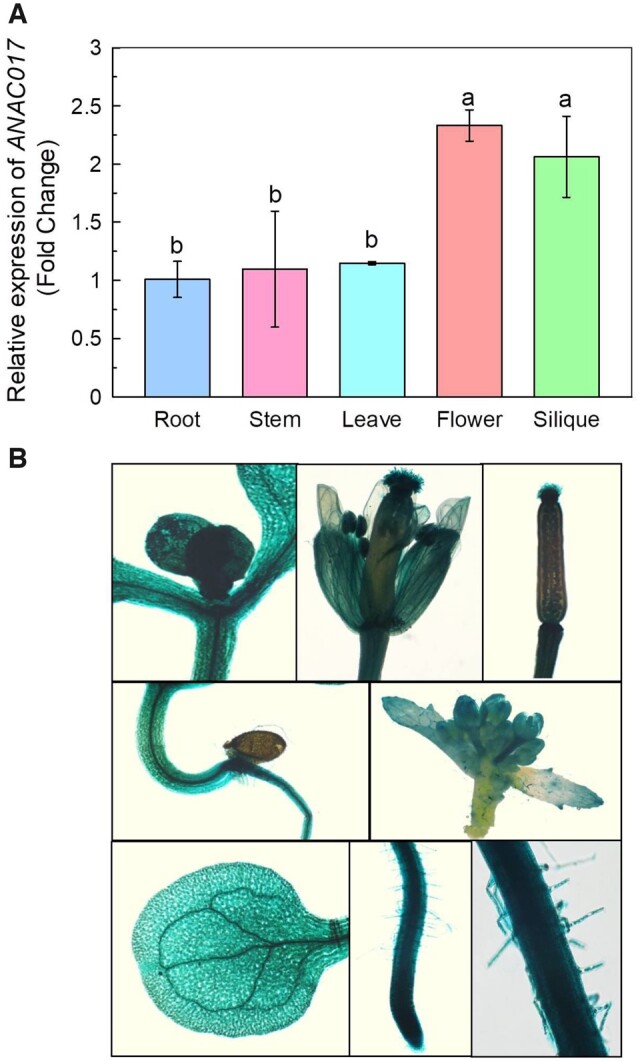
Tissue specificity of ANAC017. A, Expression of ANAC017 in various tissues, as revealed by RT-qPCR. The data were normalized to the expression level of ANAC017 in the root, and TUBULIN was used as an internal control. Data are means ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. B, Expression of ANAC017 in cotyledons, inflorescences, siliques, hypocotyls, leaf, and roots, as revealed by GUS staining. Scale bar = 400 μm.
ANAC017 is a nuclear-localized TF
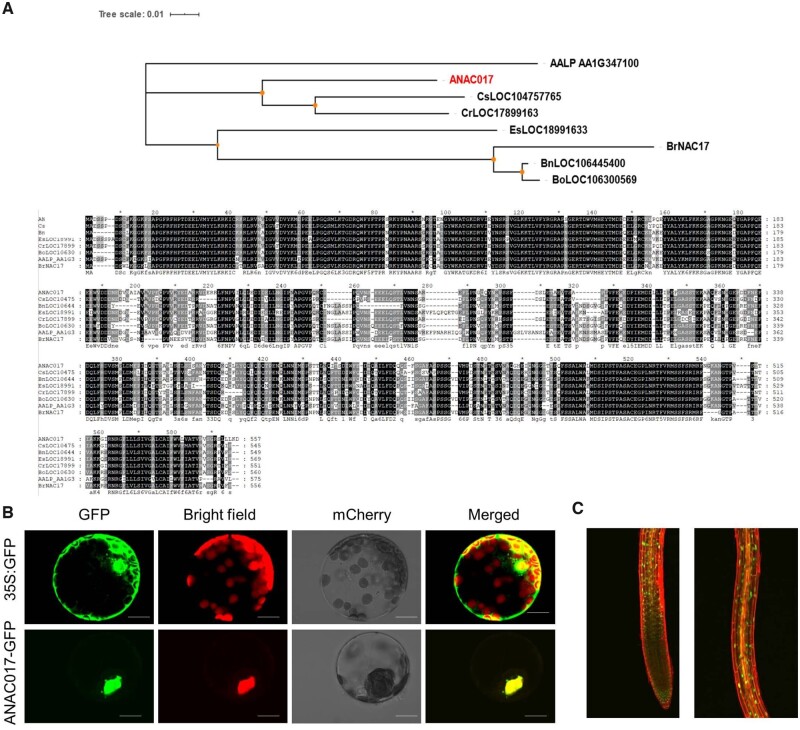
ANAC017 belongs to the Group III-a NAC TF family which contains 25 members in Arabidopsis (Jin et al., 2020). ANAC017 encodes a 557 aa protein with a typical NAC domain (Olsen et al., 2005). To further clarify the ANAC017 protein, we conducted the BLAST analysis of ANAC017 in the nonredundant protein database of NCBI and found other proteins with NAC domain sharing high sequence similarity with ANAC017, such as LOC104757765 (XP_010478849.1, Camelina sativa), LOC106445400 (XP_013742409.1, Brassica napus), LOC18991633 (XP_006396080, Eutrema salsugineum), LOC17899163 (XP_006307171.1,Capsella rubella), LOC106300569 (XP_013592194.1, Brassica oleracea var. Oleracea), LOC8054749 (XP_002462597.1, Sorghum bicolor), AALP_AA1G347100 (KFK45123.1, Arabis alpina), and BrNAC17 (XP_009114752.2, Brassica rapa). Then a phylogenetic tree was constructed to investigate the divergence of ANAC017 protein from these NAC proteins during the evolution (Figure 6A). To determine whether ANAC017 has transcriptional activation capacity, first, the subcellular localization of ANAC017 was observed. It was interesting that the ANAC017-GFP fusion protein was found to be exclusively present at nucleus, while the GFP protein alone was present at both nucleus and cytoplasm (Figure 6B;Supplemental Figure S7). This nuclear localization of ANAC017 was further confirmed by the ANAC017-GFP transgenic lines (Figure 6C), although this nuclear localization of ANAC017 was inconsistent with Ng et al. (2013), and the reason for this discrepancy may be that we used transgenic Arabidopsis, Arabidopsis protoplast and Nicotiana benthamiana leaves here, while they used onion (Allium cepa) epidermal cells, which needs further study.
Figure 6.
ANAC017 is identified as the nuclear-localized TF. A, Phylogenetic tree and sequences similarity were analyzed during different species. B, The subcellular localization of ANAC017 by transient expression in Arabidopsis protoplast. Scale bar = 10 μm. C, The subcellular localization of ANAC017 in pANAC017:ANAC017-GFP transgenic lines. Scale bar = 0.1 mm.
Next, a yeast expression system was further used to test the transactivation potential ability of the ANAC017 TF. Plasmid carrying the DNA-binding domain of the yeast GAL4 TF (DBGAL4) fused with different fragments of ANAC017 protein was transformed to the yeast strain AH109. As shown in Figure 7, ANAC017 full length was divided into three regions: 1–143 aa (plasmid b), 17–143 aa (plasmid c), and 144–557 aa (plasmid d), and only the cells carrying plasmids a and d grew well on the synthetically defined (SD) medium (in the presence of X-gal), indicating that C-terminal region of ANAC017 is critical for the transactivation potential (Figure 7).
Figure 7.
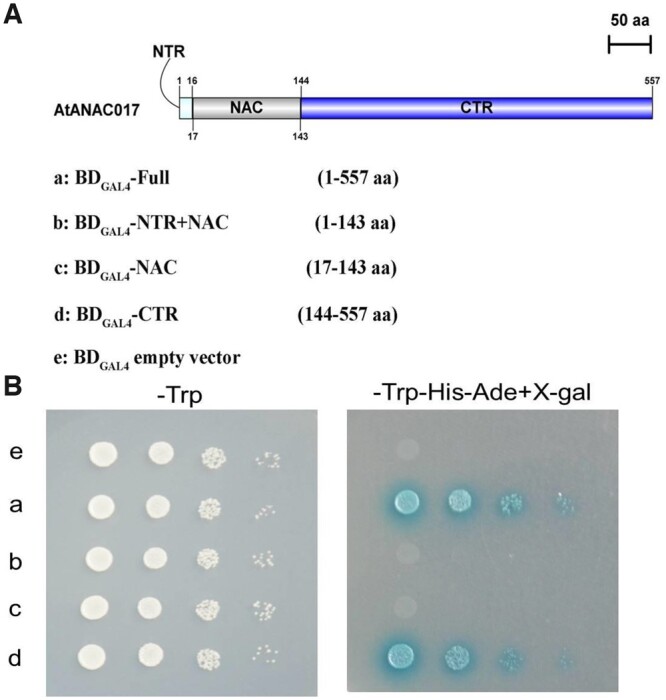
Transcriptional activation assay of ANAC017 protein in yeast. A, Schematic diagram of the constructs used for transcriptional activation assay in yeast. ANAC017 was truncated to generate four fragments including that encodes full-length cDNA. B, Transcriptional activation assay in yeast. Growth of yeast cells in medium SD/–Trp (left) and SD/–Trp–His–Ade (right) containing X-gal.
ANAC017 directly regulates the expression level of XTH31
Due to the fact that the enhanced Al tolerance in the anac017 mutants was associated with decreased Al accumulation in the root cell walls and hemicelluloses, which prompted us to determine whether a cell wall dependent exclusion mechanism is involved in this ANAC017 regulated Al tolerance. Thus, yeast one-hybrid assay was conducted to confirm the interaction between the ANAC017 and its potential downstream target, as based on our previous results that similar to anac017 mutants, xth15, xth17, and xth31 mutants are more tolerant to Al stress and also have lower Al retention in their root cell walls and hemicelluloses (Zhu et al., 2012, 2013, 2014a). It was interesting that the transcriptional activation only occurred when co-transforming the pGADT7-ANAC017 and pAbAi-pXTH31 together (Figure 8A), suggesting that XTH31 most likely acted as a target gene for the ANAC017 TF, and ANAC017 directly regulates its expression through binding to its promoter region.
Figure 8.
Regulation of XTH31 by ANAC017 TF. A, A pair of plasmids, pAbAi-pXTH31, pAbAi-pXTH17, pAbAi-pXTH15, and pGADT7-ANAC017 were introduced into Y1HGold and cultured on SD medium without Ura containing AbA (200 ng/mL; AbA200) or not (−AbA) at 30°C for 3 days. A pair of plasmids pAbAi-p53 and pGADT7-p53 were used as the positive control. B and C, The expression of XTH31 in WT, anac017 mutants, and ANAC017 overexpression lines in the WT background were detected with or without Al toxicity. D, A schematic of the XTH31-promoter-reporter construct, the effector plasmid, and the transfection control plasmid. Relative LUC activity of pXTH31-LUC reporter after co-expression of p35S:ANAC017 in Arabidopsis protoplasts. 35S-empty vector was used as the effector plasmid control. Error bars represent ± sd (n = 3). Asterisks above the bars indicate significant differences at P < 0.05 by Student’s t test. E and F, Diagram of XTH31 promoter showing five motifs. XF1-2 indicates genomic DNA fragments. The lgG was used as negative control. Error bars indicate ±sd (n = 3).
Then, the expression level of XTH31 in the WT and anac017 mutants in response to Al stress was measured. As shown in Figure 8, B and C, the Al repressed XTH31 expression in the anac017 mutants was substantially reduced but not completely eliminated, implying that the Al repressed XTH31 expression is partially dependent on ANAC017, and other unknown factors may also be involved in the XTH31 expression under Al stress. One possible explanation is that ANAC017 acts to decrease the expression of XTH31 to a similar degree (approximately two-fold), irrespective of the Al stress, a hypothesis further confirmed by the reduced Al triggered inhibition of XTH31 in ANAC017-OX transgenic lines (Figure 8C). In fact, besides Al stress, the expression of XTH31 was also decreased in exposure to cold or La stress (Supplemental Figure S5B), which showed a similar tendency to the expression of ANAC017, thus raised a question whether XTH31 is a target gene of the ANAC017. For this purpose, the pXTH31-luciferase (LUC) reporter and p35S-ANAC017 effector were transiently co-expressed in A.thaliana protoplast and the LUC activity was measured. It was interesting that co-expression of pXTH31-LUC with p35S-ANAC017 led to the activation of the LUC reporter (Figure 8D), further indicating that ANAC017 is a transcriptional activator of XTH31.
As most ANAC TFs regulate their target stress-related genes via binding to the NACRS (CGTG, CGTA, and CACG motifs) of the promoters. Thus, through analysis the sequence of the promoter, we identified five possible binding sites in XTH31 promoter region (Figure 8E). To test whether ANAC017 can directly bind to the XTH31 promoter through these potential motifs, chromatin immunoprecipitation (ChIP) assay was conducted. As shown in Figure 8F, ANAC017 was able to bind to the XTH31 promoter, further confirming that XTH31 is a target gene for the ANAC017, and ANAC017 directly regulates its expression through directly binding to its promoter.
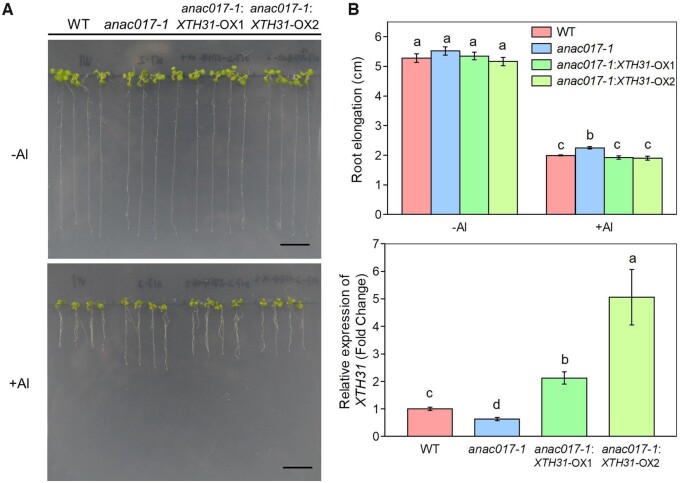
ANAC017 acts upstream of XTH31 to negatively regulate Al tolerance
As XTH31 acts as a direct target for ANAC017, therefore, in the Al response signaling, it should function downstream of ANAC017. To test this hypothesis, we observed the phenotype between WT, anac017-1, and anac017-1:XTH31-OX lines (overexpression of XTH31 in the anac017-1 mutant) in the presence or absence of Al stress. As expected, anac017-1:XTH31-OX lines exhibited more Al sensitive phenotype when compared with anac017-1 mutant, although similar to the WT seedlings, as indicated by the inhibition of the root elongation (Figure 9). Taken together, all the above results suggest that ANAC017 functions upstream of XTH31 in the Al response signaling pathway.
Figure 9.
ANAC017-regulated Al tolerance is controlled by XTH31. Phenotype (A) and root elongation (B) in WT, anac017-1 mutant and anac017-1:XTH31-OX lines in the presence or absence of Al. Data are means ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. C, RT-qPCR analysis of the expression ANAC017 in WT, anac017-1 mutants, and anac017-1:XTH31-OX lines. Error bars represent ± sd (n = 4). Different letters above the bars indicate significant differences at P < 0.05 by one-way ANOVA analysis. Scale bar = 1 cm.
ANAC017 works independently of WRKY46 and STOP1
In Arabidopsis, two different kinds of TFs, STOP1 and WRKY46 proteins have been considered as the key regulators of Al uptake or homeostasis (Iuchi et al., 2007; Ding et al., 2013), thus the stop1 and wrky46 mutants were included to connect the action between ANAC017 and known Al responsible cascades. As shown in Supplemental Figure S8A, the expression of ANAC017 was upregulated in both stop1 and wrky46 mutant under Al treatment, while no significant difference of STOP1 mRNA level in anac017 mutant was detected compared with WT, regardless of Al status, although the expression level of WRKY46 was increased in the anac017 mutant, irrespectively of the Al status (Table 1), indicating that STOP1 and WRKY46 might negatively regulate the expression ANAC017, and there might exist a feedback regulation of ANAC017 and WRKY46. However, no direct interaction was found between ANAC017 and STOP1 or WRKY46 in the yeast system (Supplemental Figure S8B), further suggesting that ANAC017 and STOP1 or WRKY46 might be work independently when Arabidopsis responses to Al stress, which needs further study.
Discussion
Here, we conclude that ANAC017 contributed substantially to the Al tolerance in A.thaliana. This conclusion is based on the following evidences. First, the inhibition of root elongation in anac017 mutants was less than that of WT under Al stress, as the obvious symptom of Al toxicity in A.thaliana is indicated by the inhibition of the root elongation (Kochian et al., 2015; Figure 1). Second, less Al was accumulated in anac017 mutants roots (Figure 1), as root Al accumulation also corresponds well with the Al tolerance (Zhu et al., 2013). Third, less Al retained in the root cell wall, especially in the hemicellulose of anac017 mutants, as retention of the Al in the cell wall contributed greatly to the tolerance of the plants to Al (Zhu et al., 2012; Figures 1 and 4). Finally, the expression of ANAC017 transcripts and the accumulation of ANAC017 protein were significantly decreased under Al stress (Figures 2 and 3), which is similar with the expression of XTH31 that when in response to Al stress (Table 1), indicating that ANAC017 play a negative role in the Al tolerance in A.thaliana. In fact, in this study, we not only reported that ANAC017 negatively regulates the Al tolerance in Arabidopsis, but also demonstrated that ANAC017 is a transcriptional activator of XTH31 and acts upstream of XTH31 to negatively regulating the Al tolerance. which is in consistent with our previous results that overexpression of XTH31 has no effect on the Al tolerance (Zhu et al., 2012). As knockout of ANAC017 results in more Al-tolerant phenotype (Figure 1, A and B) and Al decreased ANAC017 expression (Figure 2), indicating ANAC017 functions in destroy the Al tolerance of the Arabidopsis and decreased ANAC017 expression somehow protect Al-induced root growth inhibition.
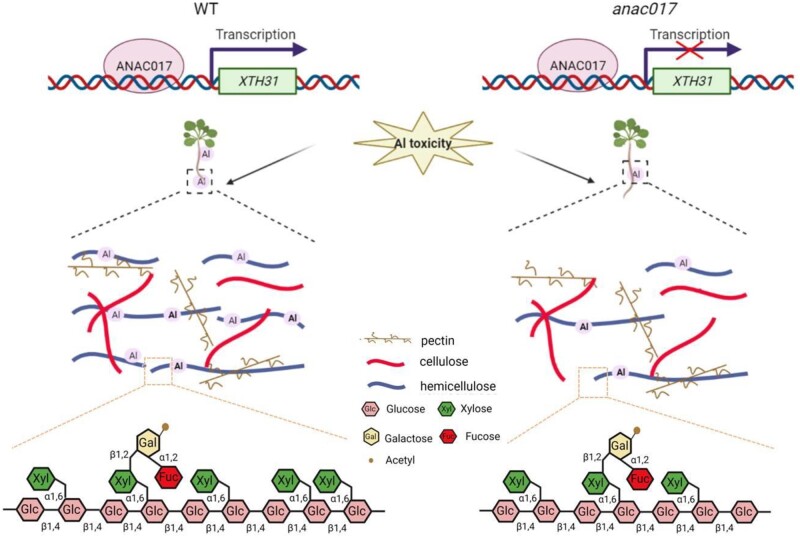
When plants suffering from Al toxicity, two strategies usually adopt by them. One is to prevent Al entering the cell through fixing Al in the root cell wall (Maron et al., 2013), while the other is to store the cell Al to the organs that are less sensitive to Al, such as vacuoles (Wu et al., 2015). After binding to the cell wall polymer, the elasticity, viscosity, and rigidity of the cell wall are impaired, thus the root growth and development are inhibited (Horst, 1995; Ma et al., 2004). Therefore, high Al tolerance in Arabidopsis is inextricably linked to low Al retention in cell wall, as demonstrated in Zea mays (Eticha et al., 2005), Secale sylvestre (Liu et al., 2008), Vigna umbellata (Zhou et al., 2012), rice (Yang et al., 2008), and Arabidopsis (Zhu et al., 2012). Interestingly, our data confirmed the low accumulation of Al in root cell wall and high Al tolerance in the anac017 mutants (Figure 4). As we known, hemicellulose and pectin, the major cell wall components, have the ability to bind Al, while another cell wall component, cellulose, which is synthesized in the plasma membrane, cannot bind to Al. Hemicellulose and pectin are synthesized in Golgi apparatus (Driouich et al., 2012), and pectin is composed of acidic polysaccharides, while hemicellulose is made up of neutral and slightly acidic polysaccharides, including xyloglucan, mannans, GX, and glucomannans (Scheller and Ulvskov, 2010). In fact, it is hemicellulose, rather than the pectin, that was demonstrated to be contributed greatly to the cell wall Al binding (Yang et al., 2011). Here, we displayed that less Al was accumulated in anac017 mutants cell wall hemicellulose (Figure 4C), in company with the lower hemicellulose content in the anac017 plants (Figure 4D), indicating a hemicellulose-dependent pathway is existed in the regulation of Al tolerance by ANAC017. In fact, xyloglucan, which accounts for 20% of cell wall content, is the main polysaccharide in hemicellulose that can bind Al (Fry, 1989; Hayashi, 1989). Thus, the xyloglucan content in WT and anac017 mutants were analyzed and less xyloglucan was existed in the mutants of anac017 (Figure 4E), suggesting ANAC017 regulated Al tolerance may be dependent on the xyloglucan content.
Then, a question raised how ANAC017 regulated the xyloglucan content. Xyloglucan, a major component of the hemicellulose, was used to hydrogen bond cellulose microfibers to form one of the three coextended frameworks (Fry, 1989; Hayashi, 1989), and XET/XEHs are abundant in plants for the modification of the xyloglucan. In Arabidopsis, three are 33 XTH genes (Yokoyama and Nishitani, 2001), while in rice and Lycopersicum esculentum, there existed 29 and 25 XTH genes, respectively (YokoyaMa et al., 2004; Saladie et al., 2006). XTHs encodes the enzymes that have the activity of XET or XEH (Tabuchi et al., 2001; Rose et al., 2002), or both (De-Silva et al., 1993; Fanutti et al., 1993), which catalyzes the molecular graft or hydrolysis the xyloglucan, respectively (Rose et al., 2002; Osato et al., 2006), to loosens the cell walls (Van-Sandt et al., 2007). Indeed, a subset of XTHs had a strong expression in A.thaliana roots, particularly XTH14, XTH15 and XTH31 (Becnel et al., 2006; Yang et al., 2011), and the expression of XTH31 was significantly down-regulated after Al treatment, in company with the reduced XET action in Arabidopsis roots when under Al stress (Zhu et al., 2012). Loss function of XTH15 resulted in the reduced accumulation of Al in the root cell wall, thus render the Arabidopsis plants more tolerant to Al stress (Zhu et al., 2013). Moreover, XTH31 was demonstrated to interact with XTH17 to play a greater role in XET, which in turn regulating the Al tolerance in Arabidopsis (Zhu et al., 2012). Thus, given that XTH31, XTH15, and XTH17 play important roles in modulating the hemicellulose content, we hypothesized that ANAC017 might also regulate them and represent the missing link between the Al stress and Al fixation in cell wall (Zhu et al., 2012, 2013, 2014a). Through yeast one-hybrid screening, it is interesting that ANAC017 only can directly bind to the promoters of XTH31 (Figure 8A), which is in company with the reduced or increased expression of the XTH31 in the anac017 mutant and ANAC017 overexpressing lines (Figure 8, B and C), and these results were further confirmed by the LUC activity detection and ChIP-qPCR (Figure 8, D–F). In addition, overexpression of XTH31 could compensate the phenotype of anac017 mutants under Al toxicity (Figure 9), further suggesting that ANAC017 act as the upstream of XTH31 in response to Al stress.
The TFs-ANAC are not only involved in biological process in plants, such as development, aging, and the formation of secondary wall (Olsen et al., 2005; Christianson et al., 2010; Zhong et al., 2010), but also play major roles when plants in response to the biotic or abiotic stress including disease resistance, pathogen invasion, and heat stress, etc. (Ren et al., 2000; Rushton et al., 2010; Guo et al., 2015; Huang et al., 2017), although no literature was found to determine whether there is a relationship between ANAC and Al stress. In general, ANAC regulate their downstream genes through recognizing the NACRS (CGTG, CGTA, and CACG) in their promoters (Tran et al., 2004; Olsen et al., 2005; Nakashima et al., 2012), that means, ANAC proteins can bind to the CGTG, CGTA, and CACG motifs, which are abounded in the XTH31 promoter region, thus ANAC017 interact with it and subsequently regulate its expression. In fact, in the promoters of many genes responsible for Al tolerance, the CGTG, CGTA, or CACG motifs are also abounded (Supplemental Figure S9), suggesting that ANAC017 protein may be enriched in these regions. Therefore, it is worthy to identify more Al responsive NACRS and help broaden the transcriptional regulation of Al-tolerant genes, as well as specific signaling pathways.
In conclusion, we identified ANAC017, a member of the ANAC macrogene family that participated in the regulation of the Al retention in the A.thaliana root cell wall. We proposed that ANAC017 functions as a component of a transcriptional response to Al stress and upregulated the XTH31 expression through binding to its promoter (Figure 10).
Figure 10.
A proposed model illustrates the ANAC017-XTH31 cascade responds to Al toxicity through modulating the xyloglucan content and its Al binding capacity in Arabidopsis.
Materials and methods
Materials and growth conditions
All the A.thaliana plants that used in this study were Col-0 ecotype. The T-DNA insertion mutants anac017-1 (SALK_044777) and anac017-2 (SALK_070231) were obtained from the Arabidopsis Biological Resource Center, while the mutants stop1 (SALK_114108) and wrky46 (SAIL_1230_H01) were obtained from Prof. Shao Jian Zheng (College of life science, Zhejiang University). All the above mutants were identified through amplifying and sequencing the flanking regions, and the primers used can be seen in Supplemental Table S1. First, the homozygous mutant seeds were washed with deionized water for 3 times after surface sterilized by 75% (v/v) ethanol, then seeds were grown in 1/2 Murashige and Skoog (MS) medium containing 1% (w/v) agar with the pH modified to 4.5 in a plant climate incubator with alternating cycles of 16 hours of light at 24°C and 8 h of darkness at 22°C (Hoekenga et al., 2006).
For hydroponic experiment, seeds were first grown in a nutrient solution with the pH modified to 4.5 (Zhu et al., 2012). Then, after 4 weeks cultivation, seedlings with similar rosette diameters were treated separately in 0.5-mM CaCl2 with or without 50-μM Al for 24 h. Finally, roots were collected not only for the extraction of the RNA and the cell wall, but also harvested for the Al content analysis.
Sequence alignment and phylogenetic analysis
Alignment of selected aa sequences was performed using the ClustalX method (http://www.clustal.org/clustal2/), selecting the “Show Low Scoring Segments” option from the quality menu to perform alignment quality control. The phylogenetic tree was reconstructed using MEGA version 11 (https://www.megasoftware.net/) based on the neighbor-joining and Poisson methods (Lin and Nei, 1991). Bootstrap values were then calculated for 1,000 trials.
Generation transgenic lines of pANAC017-GUS, ANAC017-GFP, anac017-com (complementation), ANAC017-OX (overexpression), and anac017:XTH31-OX
For pANAC017-GUS lines, DNA products of a 2,545-bp region that located upstream of the translational start site of ANAC017 was generated, and then ligated to the pQTY vector which harbors the GUS reporter gene; For ANAC017-GFP lines, the full ANAC017 CDS was cloned, and ligated to the P-super-N-GFP vector which harbors the cauliflower mosaic virus 35S promoter; for anac017-com lines, a 2545-bp length of native promoter coupled with ANAC017 gDNA sequence with terminator was cloned into a binary vector (pCAMBIA1300); for ANAC017-OX lines, the full ANAC017 coding sequence was ligated to the P-super-1300 vector which harbors the cauliflower mosaic virus 35S promoter; while for anac017:XTH31-OX transgenic lines, the full XTH31 coding sequence was also ligated to the P-super-1300 vector. All the above PCR fragments were amplified by specific primers and confirmed through sequencing (Table 1), and the constructed plasmids were transferred into Arabidopsis Col-0 (to obtain the pANAC017-GUS, ANAC017-GFP, and ANAC017-OX transgenic lines) and anac017 mutants (to obtain the anac017-com and anac017:XTH31-OX transgenic lines), Finally, these plasmids were transferred into Agrobacterium GV3101 by vacuum osmosis method (Zhu et al., 2012), and the positive lines were screened by hygromycin.
Effect of Al on root growth
Seedlings whose root length reached about 1 cm were transplanted into the following agar-solidified nutrient mediums with or without Al. After 7 days of growth, measured the roots length and then taken the photographs.
Extraction of root cell wall
After 4-week-old anac017 mutants and Col-0 were transferred to 0.5 -mM CaCl2 with or without 50-μM Al for 24 h (pH 4.5), roots were collected to grind. Subsequently, after incubated with 8 mL of ethanol that with a concentration of 75% (v/v) for 20 min, pellets were collected after centrifuged at 12,600g for 15 min. Subsequently, pellets were further washed by 8 mL of acetone, 8 mL of a 1:1 chloroform:methyl alcohol mixture and 8-mL methyl alcohol for another 20 min, respectively. Finally, the remaining pellets were regarded as cell wall material which stored in a freeze dryer.
For the extraction of cell wall hemicellulose, about 5 mg of the above crude cell walls were weighed, and then incubated with 1-mL ultrapure water at 100°C for 1 h. Then the pellets were collected after centrifugation. The above steps were repeated for another twice and subsequently, the remaining pellets were further extracted by 1 mL 24% (w/v) KOH for 12 h for 2 times. Finally, the supernatant was collected after centrifugation, and was regarded as hemicellulose fraction.
Total sugar measurement
Through using the method of phenol sulfuric acid (Dubois et al., 1956), total sugar content was detected as the indicator of hemicellulose content. In brief, 200-μL hemicellulose, 1-mL 98% (v/v) H2SO4, and 10 μL 80% (w/v) phenol were mixed evenly before boiled at 100°C for 15 min. After cooling, the absorbance at 490 nm was measured with a spectrophotometer. Glucose was used as equivalent here.
Detection of Al in root cell wall
Al retention in the root cell wall was extracted by 2-M HCl. In brief, about 5 mg of the above crude cell wall was weighed, and incubate with 1 mL 2-M HCl for 24 h, with oscillated intermittently. Then, after centrifugation, supernatants were collected and the content of Al in the supernatants was determined by inductively coupled plasma-atomic emission spectrometry (IRIS/AP optical emission spectrometry). Similarly, Al content in the hemicellulose fraction was also measured by the ICP-AES after centrifugation.
Adsorption kinetics
For evaluation, the binding capacity of the Col-0 and anac017 mutants root cell walls to Al, adsorption kinetics analysis was used. In brief, about 6-mg cell wall was weighed, and then put in a column with a filter at the bottom. After rehydrated the cell wall by 0.5-mM CaCl2 (pH 4.5) for 2 h, the adsorption solution that contained 20-μM AlCl3 in the above 0.5-mM CaCl2 was passed through the bed of cell walls by a peristaltic pump at 12 mL h−1. Eluates were collected, and their Al concentration was assayed spectrophotometrically by pyrocatechol violet according to Zheng et al. (2004). The experiments were performed twice independently, and one set of the adsorption curves is exhibited.
Xyloglucan content analysis
To analyze the xyloglucan content, cell wall materials were first incubated with 50 μL 50-mM ammonium formate with a pH of 4.5 according to Pauly et al. (1999), then 0.2 U xyloglucan-specific endoglucanase (XEG, Megazyme, E-CELTR) was added. After the above samples were incubate in a shaking incubator at 37°C for 16 h, the solvent of the digest was removed and 10-μL Bio-Rex MSZ 501(D) resin (Bio-Rad, Hercules, CA, USA) and 10-μL water were added to dissolve the XGOs. Finally, 2 μL dissolved matrix (containing 1 μL of Super-DHB, Sigma-Aldrich, St Louis, MO, USA; 10 mg mL−1) was put in a MALDI-TOF sample plate and air dried the target plate. Under the condition of 20 kV acceleration voltage, the samples were performed on Ultraflextreme instrument (Bruker, Billerica, MA, USA) in a positive reflection mode.
Gene expression analysis
The expression of the genes was measured in accordance with the manufacturer’s protocols and instructions. First, the root RNAs that extracted by the Trizol (Takara, Shiga, Japan) were transformed into cDNAs using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan). Finally, through using specific primers (Supplemental Table S1), RT-qPCR analysis was conducted by a Roche LightCycler480 system with the SYBR Premix Ex Taq II (TOYOBO). Transcription level of each mRNA was measured by the △Ct method (Ding et al., 2013).
GUS staining
To observe whether the GUS staining of pANAC017-GUS was altered under Al, cold, and La stress, pANAC017-GUS transgenic lines were first cultured on the above nutrient medium before Al, cold and La treatment or not. Then, after seedlings were immersed in a 5-mL GUS staining solution for 12 h in darkness at 37°C, samples were photographed by a charge-coupled device camera after being washed twice by 70% (v/v) ethanol. The GUS staining solution was consisted of 2-mM X–Gluc, 2-mM K3Fe(CN)6, 100-mM sodium–phosphate buffer, 10-mM EDTA, 2-mM K4Fe(CN)6 and 2% (w/v) Triton X-100 with a pH of 7.
Determination of root Al content
After 4-week-old anac017 mutants and Col-0 were transferred to 0.5-mM CaCl2 that containing 50-μM Al with a pH of 4.5 for 24 h, roots were weighed and then digested with 2-mL HNO3 until the dissolving liquid became clear and transparent to detect the Al content by ICP-AES.
Subcellular localization
The ANAC017 CDS (no termination codon) was ligated to the 35S-1300-C-GFP vector with its GFP located in the C-terminal. Four-week-old Arabidopsis leaves were selected for preparation of protoplasts (Yoo et al., 2007). In brief, through using a fresh sharp razor blade, the middle part of the leaf was cut to the 0.5–1 mm leaf strips to immerse in a prepared 20-mM MES enzymatic solution (0.4% Macerozyme R10 (w/v), 20-mM KCl, 10-mM CaCl2, 0.4-M mannitol, 1.5% cellulase R10 (w/v), and 0.1% (w/v) bovine serum albumin (BSA), pH 5.7) at 25°C in the dark for 4 h. Then, added the equal amount of W5 solution that contained 2-mM MES containing 154-mM NaCl, 125-mM CaCl2, and 5-mM KCl, pH 5.7, and after centrifuged at 100 g for 2 min to collect the pellet, 20-mL W5 solution was applied to resuspend the pellet at 0°C for 30 min. Finally, samples were centrifuged at 100 g for 2 min and the solution MMG (4-mM MES containing 15-mM MgCl2 and 0.4 M mannitol, pH 5.7) was applied to resuspend the pellet. The isolated protoplasts were further used for both subcellular localization and dual- LUC reporter analysis.
For DNA-PEG-calcium transfection,10-μL DNA (plasmid 10–20 μg) was added to 100-μL fresh protoplasts and mixed gently, then, incubated with 110-μL PEG solution (20% PEG4000 (w/v)), 100-mM CaCl2 and 0.2-M mannitol for 15 min at 25°C. After adding 500-μL W5 solution, inverted the Eppendorf tube to finish this reaction, and collected the pellet after centrifuged at 200g for 2 min. Then, 1-mL W1 solution that constitute of 4-mM MES which contained 20-mM KCl and 0.5-M mannitol, pH 5.7 was added to resuspend the pellet for 18 h at 25 °C. Finally, the confocal laser microscope (Carl zeiss LSM 710 scanning system) was used to observe GFP fluorescence with excitation and emission wavelengths set as follows: 488 nm and 485–545 nm for GFP, and 559 and 600–650 nm for propidium iodide (PI) staining.
To further confirm the above subcellular localization, the indicated constructs were also introduced into Arabidopsis WT (Col-0) and N. benthamiana leaves by Agrobacterium-mediated infiltration (strain GV3101). Homozygous T3 lines of the Arabidopsis were applied for GFP analysis, a Leica confocal microscope (Leica SP8) was used to observe the GFP fluorescence. GFP fluorescence with excitation and emission wavelengths set as follows: 488 nm and 524 nm for GFP, and the gains were four. The excitation and emission wavelengths of the chlorophyll auto-fluorescence were 559 nm and 650–710 nm, respectively.
Dual-LUC reporter assays
Through using 190fLUC and internal control plasmid AtUbi:rLUC, protoplasts were transformed by the constructed vectors. The promoter of XTH31 was ligated to the AtUbi:rLUC vector and the coding region of ANAC017 was ligated to 190fLUC vector through using specific primers (Supplemental Table S1). After incubation for 16 h, the frozen protoplasts were broken by the addition of 100-µL protoplast lysis buffer with intense shaking for 2 s. Then centrifuged at 1,000g for 2 min after incubation at 0°C for 5 min. Then 20-µL lysate and 100-mL LUC mix (Promega Madison, WI, USA) were used. Firefly and Renilla LUC were quantified using PicaGene Dual-LUC Assay kit (PG-DUAL SP; Toyo Ink). Relative LUC activity was calculated by normalizing LUC activity with rLUC activity (Yoo et al., 2007).
Yeast one-hybrid experiment
In order to evaluate the crosslink between ANAC017 and XTH31, the promoter of XTH31 was ligated to the pAbAi vector and the coding region of ANAC017 was cloned into the pGADT7 vector through using specific primers. Plasmids were mixed, and then transformed to the Y1HGold to culture on SD medium without Ura with or without 200-ng mL−1 Aureobasidin A (AbA) for 3 days.
ChIP assay analysis
Four-week-old ANAC017-OX transgenic lines were treated with 50-μM Al solution for 24 h, and roots were collected. The samples were fixed in 1.0% (v/v) formaldehyde for 10 min. After vacuumized, 0.125-M glycine was applied, and samples were cleaned and then ground. The chromatin DNA was then extracted and sheared, and immunoprecipitation was performed using anti-HA (Abmart, Shanghai, China). Finally, through using specific primers, quantitative PCR (ChIP-qPCR) was used to detect the amount of genomic DNA co-purified by specific primers with anti-HA or mouse IgG (Supplemental Table S1). The immunoprecipitated DNA was divided by the input DNA to calculate the relative enrichment value.
Observation of ANAC017-GFP subcellular localization
To observe whether the subcellular localization of ANAC017-GFP was altered under Al stress, transgenic lines of ANAC017-GFP were first cultured on the above nutrient medium. When the roots length reached about 1 cm, the seedlings were transferred to the above gel medium with or without 200-μM Al for 24 h. Finally, GFP and PI fluorescence of roots were observed by a Leica SP8 after the roots were stained with 10-μM PI solution for 10 s. The ImageJ software quantified GFP fluorescence intensity by integrating all the pixels in selected area. The GFP fluorescence with excitation and emission wavelengths was set as follows: 488 nm and 485–545 nm for GFP, and 559 and 600–650 nm for PI staining.
Extraction and gel-blot analysis of protein
To determine the protein level of ANAC017 in roots, 4-week-old transgenic lines of ANAC017-GFP were exposed to a 0.5-mM CaCl2 solution with or without 50-μM Al for 24 h. The total proteins were extracted and the concentration were determined by Bradford protein assay according to Zhu et al. (2014a). Polyclonal anti-GFP antibody was used to detect the ANAC017-GFP protein, through using ACTIN proteins as the internal reference.
Transactivation activity assay in yeast
The Clontech’s matchmaker gold system was used to test the transcriptional activation. By using specific primers, ANAC017 were divided into different regions to construct to the pGBKT7 vector. Then, transformed to the AH109 (a name of one yeast strain) with the plasmids that harbors different ANAC017 regions or not, and grown in the SD–Trp medium for 3 days at 30°C. Finally, the transformed AH109 were screened by the SD–Trp/–His/–Ade medium.
Statistical analysis
Every experiment is repeated at least 3 times. One-way ANOVA is used for data analysis, and Duncan’s multi-range test is used to compare the mean values. The different letters in the histogram indicate that the mean value is statistically different at P < 0.05 level. Asterisks in the histogram indicated significant differences at P < 0.05 based on Student’s t test.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: XTH15 (AT4G14130), XTH17 (AT1G65310), XTH31 (AT3G44990), TBL27 (AT1G70230), ALS1 (At5g39040), ALS3 (At2g37330), ALMT1 (At1g08430), MATE (At1g51340), STAR1 (At1g67940), STOP1 (At1g34370), RAE1 (At1g80670), RAE3 (At5g09860), WRKY46 (At2g46400), WRKY47 (AT4G01720), HB7 (AT2G46680), and HB12 (AT3G61890).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used in the study.
Supplemental Figure S1. Identification of the anac017 mutants.
Supplemental Figure S2. Phenotype of WT and anac017 mutants in both vegetative and reproductive stage under normal growth conditions.
Supplemental Figure S3. Phenotypes of WT, anac017 mutants, and anac017-com lines (complementary lines).
Supplemental Figure S4. Phenotypes of WT and ANAC017 overexpression (ANAC017-OX) lines.
Supplemental Figure S5. Expression of ANAN017 and XTH31 in response to Al, cold, and La stresses.
Supplemental Figure S6. Total contents of the xyloglucan oligosaccharides by the MALDI-TOF MS analysis.
Supplemental Figure S7. The subcellular localization of ANAC017 by transient expression into tobacco (N. benthamiana) leaves.
Supplemental Figure S8. Relationship between ANAC017 and known Al responsible cascades.
Supplemental Figure S9. Occurrence of NACRS in the promoter regions of Al tolerance genes.
Supplementary Material
Acknowledgments
We thank Prof Shao Jian Zheng (College of Life Science, Zhejiang University, China) for the stop1 and wrky46 mutants. Thanks are also given to the editor and the two anonymous reviewers for their valuable comments to improve the quality of our work.
Funding
This study was supported by the Foundation for Distinguished Young Scholars of Jiangsu Province, China (Grant No. BK20190050), National Natural Science Foundation of China (Grant No. 42020104004), Youth Innovation Promotion Association of Chinese Academy of Sciences (Grant No. 2015250), and National Science Foundation of China (Grant No. 31501825).
Conflict of interest statement. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Ye Tao, State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Science, Nanjing 210008, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jiang Xue Wan, State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China (in preparation), Rice Research Institute, Sichuan Agricultural University, Chengdu 611130, China.
Yu Song Liu, State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Science, Nanjing 210008, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xiao Zheng Yang, State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Science, Nanjing 210008, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Ren Fang Shen, State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Science, Nanjing 210008, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xiao Fang Zhu, State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Science, Nanjing 210008, China; University of Chinese Academy of Sciences, Beijing 100049, China.
X.F.Z. and R.F.S. conceived the project. Y.T., J.X.W., Y.S.L., and X.Z.Y. performed experiments. R.F.S. and X.F.Z. wrote the manuscript, and all authors discussed and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Xiao Fang Zhu (xiaofangzhu@issas.ac.cn).
References
- Basu A, Basu U, Taylor GJ (1994) Induction of microsomal membrane proteins in roots of an aluminum resistant cultivar of Triticum. Plant Physiol 104: 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becnel J, Natarajan M, Kipp A, Braam J (2006) Developmental expression patterns of Arabidopsis XTH genes as reported by transgenes and genevestigator. Plant Mol Biol 61: 451–467 [DOI] [PubMed] [Google Scholar]
- Bui LT, Shukla V, Giorgi FM, Trivellini A, Perata P, Licausi F, Giuntoli B (2020) Differential submergence tolerance between juvenile and adult Arabidopsis plants involves the ANAC017 transcription factor. Plant J 104: 979–994 [DOI] [PubMed] [Google Scholar]
- Che J, Tsutsui T, Yokosho K, Yamaji N, Ma JF (2018) Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol 220: 209–218 [DOI] [PubMed] [Google Scholar]
- Christianson JA, Dennis ES, Llewellyn DJ, Wilson IW (2010) ATAF NAC transcription factors: regulators of plant stress signaling. Plant Signal Behav 5: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Jian FM, Ryan PR (2012) Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci 17: 341–348 [DOI] [PubMed] [Google Scholar]
- De-Silva J, Jarman CD, Arrowsmith DA, Stronach MS, Chengappa S, Sidebottom C, Reid JSG (1993) Molecular characterization of a xyloglucan-specific endo-(1/4)-β-d-glucanase (xyloglucan endotransglycosylase) from nasturtium seeds. Plant J 3: 701–711 [PubMed] [Google Scholar]
- Devi SR, Yamamoto Y, Matsumoto H (2003) An intracellu1ar mechanism of aluminium tolerance associated with high antioxidant status in cultured tobacco cells. J Inorg Biochem 97: 59–68 [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Xu XY, Li GX, Zheng SJ (2013) WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J 76: 825–835 [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye ML, Bernard S, Kousar S, Chevalier L, Vicre-Gibouin M, Lerouxel O (2012) Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front Biogeogr 3: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colori metric method for determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Eticha D, Stass A, Horst WJ (2005) Cell-wall pectin and its degree of methylation in the maize root-apex: significance for genotypic differences in aluminium resistance. Plant Cell Environ 28: 1410–1420 [Google Scholar]
- Fang Q, Zhang J, Yang DL, Huang CF (2020) Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 32: 3921–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanutti C, Gidley MS, Reid JSG (1993) Xyloglucan endotransglycosylase (formerly called xyloglucan-specific endo-(1/4)-β-d glucanase) from the cotyledons of germinated nasturtium seeds. Plant J 3: 691–700 [DOI] [PubMed] [Google Scholar]
- Fry SC (1989) The structure and functions of xyloglucan. J Exp Bot 40: 1–11 [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LSP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Guo JL, Zhang Y, Gao HL, Li SB, Wang ZY, Huang CF (2020) Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. New Phytol 228: 179–193 [DOI] [PubMed] [Google Scholar]
- Guo WW, Zhang JX, Zhang N, Xin MM, Peng HR, Hu ZR, Ni ZF, Du JK (2015) The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and posttranslational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoS One 10: e0135667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T (1989) Xyloglucans in the primary cell wall. Ann Rev Plant Biol 40: 139–168 [Google Scholar]
- Hegedus D, Yu M, Baldwin D, Gruber, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53: 383–397 [DOI] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Pinero MA, Cancado GMA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, et al. (2006) AtALMT1, which encodes a malate transporter,is identified as one of several genes criticalfor aluminum tolerance inArabidopsis. Proc Natl Acad Sci USA 103: 9738–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst WJ (1995) The role of the apoplast in aluminium toxicity and resistance of higher plants: a review. J Plant Nutri Soil Sci 158: 419–428 [Google Scholar]
- Hu HH, You J, Fang YJ, Zhu XY, Qi ZY, Xiong LZ (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67: 169–181 [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Ma JF (2010) Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol 153: 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li T, Xu ZS, Wang F, Xiong AS (2017) Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol Biochem 120: 61–74 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M (2007) Zinc finger protein stop1 is critical for proton tolerance in arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104: 9900–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JF, Wang ZQ, He QY, Wang JY, Li PF, Xu JM, Zheng SJ, Fan W, Yang JL (2020) Genome-wide identification and expression analysis of the nac transcription factor family in tomato (solanum lycopersicum) during aluminum stress. BMC Genet 2: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzak SM, Dahlqvist A, Van-Aken O (2020) The transcription factor ANAC017 is a key regulator of mitochondrial proteotoxic stress responses in plants. Phil Trans R Soc B 375: 20190411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Biol 46: 237–260 [Google Scholar]
- Kochian LV, Pineros MA, Liu J, Magalhaes JV (2015) Plant adaptation to acid soils: the molecular basis for crop Al resistance. Ann Rev Plant Biol 66: 571–598 [DOI] [PubMed] [Google Scholar]
- Kumari M, Taylor GJ, Deyholos MK (2008) Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol Genet Genom 279: 339–357 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Cancel J, Rounds M, Ochoa V (2007) Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225: 1447–1458 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJB, Jones CA, Williams KM, Cancel JD (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41: 353–363 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Stenzler LM, Tai CY, Degenhardt J, Howell SH, Kochian KV (1997) Molecular and physiological analysis of Arabidopsis mutants exhibiting altered sensitivities to aluminum. Plant Soil 192: 3–7 [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Yan JY, Ren JY, Sun L, Xu C, Li GX, Ding ZJ, Zheng SJ (2019) A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J Integr Plant Biol 62: 1176–1192 [DOI] [PubMed] [Google Scholar]
- Li XL, Yang X, Hu YX, Yu XD, Li QL (2014) A novel NAC transcription factor from SuaedaliaotungensisK. Enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep 33: 767–778 [DOI] [PubMed] [Google Scholar]
- Lin J, Nei M (1991) Relative efficiencies of the maximum-parsimony and distance-matrix methods of phylogeny construction for restriction data. Mol Biol Evol 8: 356–365 [DOI] [PubMed] [Google Scholar]
- Liu JP, Pineros MA, Kochian LV (2014) The role of aluminum sensing and signaling in plant aluminum resistance. J Integr Plant Biol 56: 221–230 [DOI] [PubMed] [Google Scholar]
- Liu Y, Xu J, Guo S, Yuan X., Zhao S, Tian H., Dai S, Kong X, Ding Z (2020) AtHB7/12 regulate root growth in response to aluminum stress. Int J Mol Sci 21: 4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yang JL, He LS, Li YY, Zheng SJ (2008) Effect of aluminum on cell wall, plasma membrane, antioxidants and root elongation in triticale. Biol Plant 52: 87–92 [Google Scholar]
- Ma JF (2005) Plant root responses to three abundant soil minerals: silicon, aluminum and iron. CRC Crit Rev Plant Sci 24: 267–281 [Google Scholar]
- Ma JF, Ryan PR, Delhaize E (2001) Aluminum tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6: 273–278 [DOI] [PubMed] [Google Scholar]
- Ma JF, Shen RF, Nagao S, Tanimoto E (2004) Al targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol 45: 583–589 [DOI] [PubMed] [Google Scholar]
- Maron LG, Guimaraes CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, Buckler ES, Coluccio AE, Danilova TV, Kudrna D, et al. (2013) Al tolerance in maize is associated with higher MATE1 gene copy number. Pro Natl Acad Sci USA 110: 5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XX, Li L, Narsai R, De-Clercq I, Whelan J, Berkowitz O (2020) Mitochondrial signalling is critical for acclimation and adaptation to flooding in Arabidopsis thaliana. Plant J 103: 227–247 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. BBA-Gene Regu Mech 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465: 30–44 [DOI] [PubMed] [Google Scholar]
- Ohyama Y, Ito H, Kobayashi Y, Ikka T, Morita A, Kobayashi M, Imaizumi R, Aoki T, Komatsu K, Sakata Y, et al. (2013) Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol 162: 1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Osato Y, Yokoyama R, Nishitani K (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res 119: 153–162 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A (1999) A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pellet DM, Grunes DL, Kochian LV (1995) Organic acid exudation as all alumihum tolerance mechanism in maize (Zea mays L.). Planta 196:788–795 [Google Scholar]
- Pineros MA, Kochian LV (2001) A patchclamp study on the physiology of aluminum toxicity and aluminum tolerance in maize, identification and characterization of Al induced anion channels. Plant Physiol 125: 292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauser WE (1987) Compartmental efflux analysis and removal of extracellular cadmitim from roots. Plant Physiol 85: 62–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Qu F, Morris TJ (2000) HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12: 1917–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddle J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) Wrky transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Saladie M, Rose JKC, Cosgrove DJ, Catala C (2006) Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. Plant J 47: 282–295 [DOI] [PubMed] [Google Scholar]
- Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, et al. (2009) Stop1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol 150: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Ann Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Mori H, Kamisaka S, Hoson T (2001) A new type of endoxyloglucan transferase devoted to xyloglucan hydrolysis in the cell wall of azuki bean epicotyls. Plant Cell Physiol 42: 154–161 [DOI] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van-Sandt VS, Suslov D, Verbelen JP, Vissenberg K (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B (2004) CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 131: 915–922 [DOI] [PubMed] [Google Scholar]
- Wu D, Shen H, Yokawa K, Baluska F (2015) Overexpressing OsPIN2 enhances aluminium internalization by elevating vesicular trafficking in rice root apex. J Exp Bot 66: 6791–6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF (2009) Zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Li YY, Zhang YJ, Zhang SS, Wu YR, Wu P, Zheng SJ (2008) Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol 146: 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Zhu XF, Peng YX, Zheng C, Li GX, Liu Y, Shi YZ, Zheng SJ (2011) Cell wall hemicellulose contributes signifificantly to Al absorption and root growth in Arabidopsis. Plant Physiol 155: 1885–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K (2001) A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol 42: 1025–1033 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Rose JKC, Nishitani K (2004) A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol 134: 1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Guo JL, Zhou FL, Singh S, Xu X, Xie Q, Yang ZB, Huang CF (2019) F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc Natl Acad Sci USA 116: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SJ, Lin XY, Yang JL, Liu Q, Tang C (2004) The kinetics of aluminum adsorption and desorption by root cell walls of an aluminum resistance wheat (Triticum aestivum L.) cultivar. Plant Soil 261: 85–90 [Google Scholar]
- Zhong RQ, Lee CH, Ye ZH (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Xu XY, Chen LQ, Yang JL, Zheng SJ (2012) Nitric oxide exacerbates Al-induced inhibition of root elongation in rice bean by affecting cell wall and plasma membrane properties. Phytochemistry 76: 46–51 [DOI] [PubMed] [Google Scholar]
- Zhu XF, Lei GJ, Wang ZW, Shi YZ, Braam J, Li GX, Zheng SJ (2013) Coordination between apoplastic and symplastic detoxification confers plant Al resistance. Plant Physiol 162: 1947–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Shi YZ, Lei GJ, Fry SC, Zhang BC, Zhou YH, Braam J, Jiang T, Xu XY, Mao CZ, et al. (2012) XTH31, encoding an in vitro XEH/XET-active enzyme, regulates Al sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and Al binding capacity in Arabidopsis. Plant Cell 24: 4731–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Sun Y, Zhang BC, Mansoori N, Wan JX, Liu Y, Wang ZW, Shi YZ, Zhou YH, Zheng SJ (2014b) Trichome birefringence-like27 affects aluminum sensitivity by modulating the o-acetylation of xyloglucan and aluminum-binding capacity in Arabidopsis. Plant Physiol 166: 181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Wan JX, Sun Y, Shi YZ, Braam J, Li GX, Zheng SJ (2014a) Xyloglucan endotransglucosylase-hydrolase17 interacts with xyloglucan endotransglucosylase-hydrolase31 to confer xyloglucan endotransglucosylase action and affect aluminum sensitivity in Arabidopsis. Plant Physiol 165: 1566–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.