Abstract
In oxygenic photosynthesis, NADP+ acts as the final acceptor of the photosynthetic electron transport chain and receives electrons via the thylakoid membrane complex photosystem I (PSI) to synthesize NAPDH by the enzyme ferredoxin:NADP+ oxidoreductase. The NADP+/NADPH redox couple is essential for cellular metabolism and redox homeostasis. However, how the homeostasis of these two dinucleotides is integrated into chloroplast biogenesis remains largely unknown. Here, we demonstrate the important role of NADP+ supply for the biogenesis of PSI by examining the nad kinase 2 (nadk2) mutant in Arabidopsis (Arabidopsis thaliana), which demonstrates disrupted synthesis of NADP+ from NAD+ in chloroplasts. Although the nadk2 mutant is highly sensitive to light, the reaction center of photosystem II (PSII) is only mildly and likely only secondarily affected compared to the wild-type. Our studies revealed that the primary limitation of photosynthetic electron transport, even at low light intensities, occurs at PSI rather than at PSII in the nadk2 mutant. Remarkably, this primarily impairs the de novo synthesis of the two PSI core subunits PsaA and PsaB, leading to the deficiency of the PSI complex in the nadk2 mutant. This study reveals an unexpected molecular link between NADK activity and mRNA translation of psaA/B in chloroplasts that may mediate a feedback mechanism to adjust de novo biosynthesis of the PSI complex in response to a variable NADPH demand. This adjustment may be important to protect PSI from photoinhibition under conditions that favor acceptor side limitation.
Mutation of NAD KINASE2 leads to NADP+ deficiency and impairs the synthesis of photosystem I in chloroplasts of Arabidopsis.
Introduction
The nicotinamide nucleotides (NAD+ and NADP+) and the corresponding reduced forms (NADH and NADPH) are essential cofactors in cellular redox reactions of most organisms (Goodman et al., 2018; Xiao et al., 2018). The biosynthesis and distribution of NADH and NADPH are always highly compartmentalized (Goodman et al., 2018; Xiao et al., 2018). In plant cells, NADP+ is mostly distributed in chloroplasts (Heber and Santarius, 1965; Wigge et al., 1993) and mainly functions as the final electron acceptor of the photosynthetic electron transport chain (Arnon and Chain, 1975; Hanke and Mulo, 2013; Goss and Hanke, 2014). This reducing energy is not only used for carbon and nitrogen assimilation, as well as lipid and chlorophyll metabolism, but also functions in maintaining redox homeostasis by regulating the scavenging of reactive oxygen species in plant cells (Baier and Dietz, 2005; Noctor, 2006). Accordingly, perturbations of NADP+ pools and/or NADP+/NADPH imbalances in chloroplasts usually cause pleiotropic effects on plant growth and development (Takahashi et al., 2009; Takahara et al., 2010; De Souza Chaves et al., 2019). However, how the homeostasis of these two dinucleotide pairs is integrated into chloroplast biogenesis is largely unknown.
In thylakoid membranes of chloroplasts, electrons are derived from water splitting at photosystem II (PSII), transferred to the cytochrome b6f complex (Cyt b6f) by plastoquinone (PQ), and from there through the thylakoid lumen via plastocyanin to photosystem I (PSI), which is referred to as the linear electron transfer (LET; Goss and Hanke, 2014.) At the end of the LET chain, the small soluble protein ferredoxin (Fd) transfers electrons to the Fd:NADP(H) oxidoreductase (FNR), which can then reduce NADP+ to NADPH (Hanke and Mulo, 2013). Concomitantly with LET, a proton gradient used for ATP generation is formed across the thylakoid membrane. Alternatively, photosynthetic electrons can be directed to the cyclic electron transfer (CET) routes around PSI, which also contributes to the proton gradient across the thylakoid membrane and ATP synthesis but does not produce NADPH in chloroplasts (Yamori and Shikanai, 2016; Nawrocki et al., 2019). CET around PSI is thought to be essential for balancing the ATP/NADPH production ratio and for protecting both photosystems from damage caused by stromal over-reduction (Yamori and Shikanai, 2016; Nawrocki et al., 2019). Overall, the PSI complex not only determines how reducing power is ultimately partitioned but also plays an important role in the regulation of the photosynthetic electronic transport, which is essential for chloroplasts to cope with dynamic fluctuations in light intensity and the variable demand for ATP and NADPH.
PSI is among the largest and most complex multi-subunit pigment–protein structures in nature and consists of at least 19 subunits, approximately 175 chlorophyll molecules, 2 phylloquinones, and 3 [4Fe–4S] clusters in plants (Amunts et al., 2007; Jensen et al., 2007; Busch and Hippler, 2011; Qin et al., 2015). Among the PSI subunits, two large subunits PsaA and PsaB form the heterodimer core of the reaction center and harbor the cofactors for the electron transport chain together with PsaC, which binds two [4Fe–4S] clusters (Busch and Hippler, 2011). Although the PSI structure and function are well understood at high spatial and temporal resolution, respectively, it remains largely unexplored and is of growing interest how the subunits and cofactors are incorporated into the functional complex. To date, a set of protein factors involved in PSI assembly, stability, and cofactor biosynthesis has been identified in plants, algae, and, cyanobacteria (Gross et al., 2006; Schwenkert et al., 2009; Schöttler et al., 2011; Chi et al., 2012; Yang et al., 2015). Interestingly, the PSI biogenesis is also regulated by the stromal redox state (Heinnickel et al., 2016; Zhu et al., 2016); however, the underling mechanism is still unclear.
The NAD kinase (NADK) catalyzes the de novo biosynthesis of NADP+ from NAD+ and ATP. There are three NADK isoforms (NADK1–3) in Arabidopsis (Arabidopsis thaliana), one of which is localized in chloroplasts (NADK2; Chai et al., 2005; Waller et al., 2010). The knockout of NADK2 in Arabidopsis leads to the downregulation of NADP+ and NADPH levels in chloroplasts, which accordingly influences the carbon and nitrogen metabolism (Chai et al., 2005; Takahashi et al., 2006, 2009). Here, we found an unexpected function of NADK2 in photosynthetic electronic transport via regulating de novo biogenesis of the PSI complex. We hypothesize that NADP+ supply in the chloroplast stroma affects the translation activity of psaA/B mRNA, allowing chloroplasts to modulate the accumulation of the PSI complex as the demand for NADPH production varies.
Results
Deletion of NADK2 in Arabidopsis affects photosynthetic electron transport
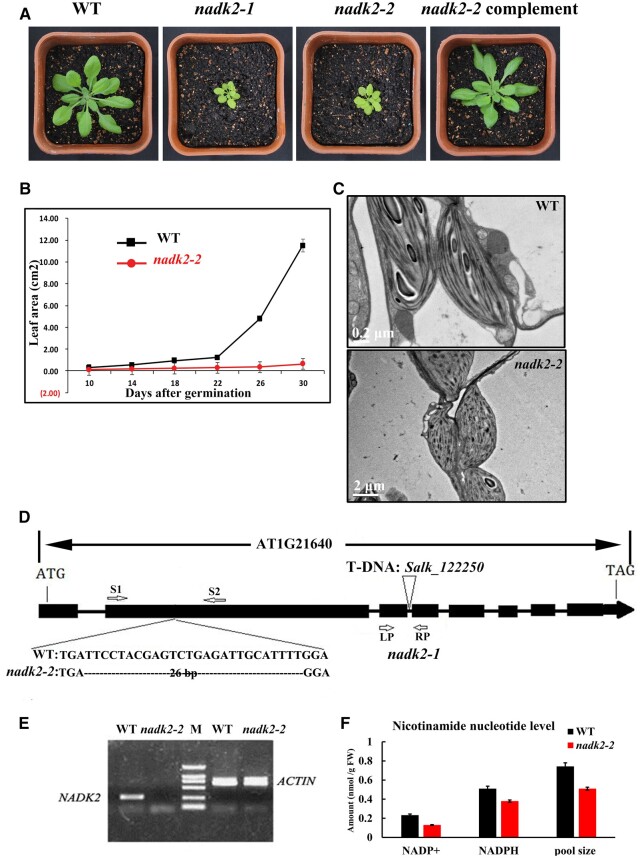
To identify critical factors involved in chloroplast biogenesis, we screened Arabidopsis T-DNA insertion lines for a pale green phenotype. A mutant assigned as nadk2-2 came to our attention. The mutant appeared pale green and exhibited severely delayed growth when in vitro grown plants were transferred to soil (Figure 1, A and B). The chlorophyll content of nadk2-2 was decreased to 50%–60% of wild-type (WT) levels, suggesting that chloroplast development was affected in the nadk2-2 mutant. WT chloroplasts displayed well-developed thylakoid membrane systems composed of grana stacks connected with stroma lamellae. In contrast, chloroplasts were smaller, the thylakoid membrane was much less organized, and starch grains were rarely visible in the mutant (Figure 1C).
Figure 1.
Phenotype, genotyping, and NADP+/NADP contents in the WT and nadk2-2 A, Phenotype of 4-week-old WT, nadk2-1, nadk2-2, and NADK2 cDNA-complemented plants grown in a growth chamber. B, Growth of the nadk2-2 mutant compared with WT plants. Values shown are means ± sd of six replicate experiments. C, Electron micrographs of chloroplasts from nadk2-2 and WT leaves. Bar = 0.2 μm for WT and 2 μm for nadk2-2 mutant. D, Genotyping of nadk2-2. The deleted nucleotides in nadk2-2 are indicated in dashed line. The localization of the T-DNA insertion site in nadk2-1 is shown (Chai et al., 2005). S1, S2, LP, and RP indicate the positions of primers used for genotyping. E, RT-PCR detection of ACTIN and NADK2 mRNA in total RNA extracted from WT and nadk2-2 mutants. The position of AT1G21640 primers S1 and S2 is shown in (D). F, The contents of NADP+, NADPH, and NADP pool (sum of NADP+ and NADPH) in WT and nadk2-2 plants. Results are presented as means ± sds (n = 3).
Map-based cloning of the mutant revealed a 26-nucleotide deletion (nucleotide positions 801- to 826-bp downstream of the ATG codon) in the AT1G21640 gene (Figure 1D), which was named NADK2 in a previous study (Chai et al., 2005). RT-PCR analysis showed that the nadk2-2 mutant failed to express AT1G21640 (Figure 1E). Indeed, the levels of NADP+ and NADPH and the size of the NADP pool (sum of NADPH and NADP+) were apparently reduced in nadk2-2 plants (Figure 1F), consistent with the putative defect in the chloroplast NADK activity in the mutant (Figure 1F). We included an additional mutant line, salk_122250, containing a T-DNA insertion in the third intron, previously named nadk2-1 (Chai et al., 2005). Both nadk lines virtually displayed the same phenotype. Expression of the NADK2 cDNA fully complemented the nadk2-2 mutant, confirming that the mutation of AT1G21640 was entirely responsible for the nadk2-2 phenotype (Supplemental Figure1A).
PSII is more sensitive to light in the nadk2-2 mutant
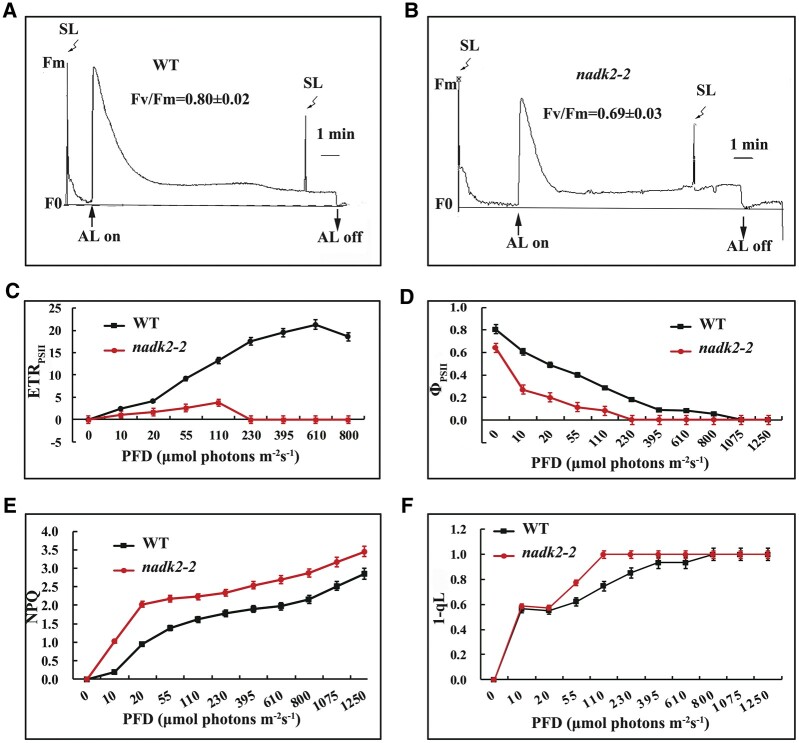
The pale green phenotype and alterations of the chloroplast ultrastructure might result from the defects of the photosynthetic machinery in nadk2-2 plants. To check this possibility, we measured the chlorophyll a fluorescence induction of intact leaves of nadk2-2 mutants and WT plants. Although the ratio Fv/Fm, reflecting the maximum potential capacity of PSII, was somewhat reduced in the mutant (0.69 ± 0.02 versus 0.80 ± 0.02 in WT) (Figure 2, A and B), it was considered not to necessarily reflect primary defects of PSII because mutants directly affected in PSII often have Fv/Fm values below 0.5 (Meurer et al., 1996; Peng et al., 2006; Armbruster et al., 2010; Wang et al., 2013).
Figure 2.
PSII activity of the WT and nadk2-2 mutants as revealed by chlorophyll fluorescence. A and B, Chlorophyll a fluorescence induction of WT and nadk2-2 mutants grown on soil for 2 weeks under 60-μmol photons m−2 s−1. AL, actinic light; SL, saturating light. Fm, maximum chlorophyll fluorescence, Fv, variable fluorescence. Fv/Fm represents the maximum photochemical yield of PSII. C–F, The ETR of PSII (ETRPSII), effective quantum yield (ΦPSII), NPQ, and (1-qL) of WT and nadk2-2 mutant plants grown under the same condition as in (A) and (B). The actinic light intensities for the PAM measurements were 55-μmol photons m−2 s−1. Results are presented as means ± sds (n = 3).
Even though the reaction center of PSII does not seem to be the main target of damage, the relative electron transport rate (ETR) and ΦPSII of nadk2-2 leaves were much lower than those of WT plants (Figure 2, C and D). In addition, the ETR was severely reduced and decreased with increasing light intensity of more than 110-μmol photons m−2 s−1 (Figure 2C), whereas in the WT it increased up to 610-μmol photons m−2 s−1, suggesting a serious inhibition and light sensitivity of the photosynthetic apparatus in the mutant. At the same time, the levels of non-photochemical quenching (NPQ), which mostly represents the thermal dissipation of excessive excitation energy to protect the PSII machinery under high light (De Bianchi et al., 2010; Ruban, 2016), was increased substantially even under low light in nadk2-2 plants (Figure 2E). It appeared that the photoprotective response of nadk2-2 plants is activated already at very low light intensities compared to the WT. Therefore, we conclude that the photosynthetic activity of nadk2-2 plants was more light sensitive compared to WT, which probably causes a slightly reduced Fv/Fm ratio already at normal growth conditions.
The qL value represents the fraction of PSII reaction centers that are open, while the value 1−qL reflects the excitation pressure of PSII (Armbruster et al., 2013). We found a slightly higher value in the nadk2-2 plants (Figure 2F), indicating a more reduced PQ pool in mutant plants. These results suggest that nadk2-2 is deficient in efficiently transferring electrons from PQ to the downstream electron transport chain in the light. Indeed, the slower post-illumination fluorescence rise, reflecting the reduction of the PQ pool in the dark, confirmed this hypothesis and also reflects lack of sufficient NADPH for efficient PQ reduction in the nadk2-2 mutant (Supplemental Figure S1). Considering that enhanced photoinhibition of PSII activity was also observed in mutants deficient in the cytochrome b6f complex and PSI (Stoppel et al., 2011; Liu et al., 2012; Fristedt et al., 2014), we speculate that the primary defect may occur in subsequent electron transport processes downstream of PSII.
The function of PSI is affected in nadk2-2
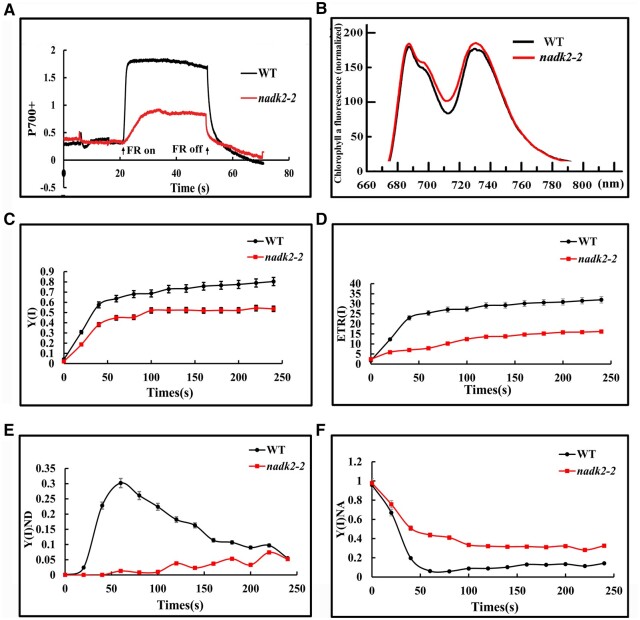
To investigate PSI activity, the PSI redox level was assessed using the pulse amplitude modulation (PAM) system. The total amplitude of changes in P700 absorbance at 820 nm, induced by far-red light and reflecting the amount of photosynthetically active PSI complexes (Asada et al., 1992), was less than 30% of WT values (Figure 3A). Considering the decrease in the chlorophyll content in nadk2-2, we also normalized P700 signals to the chlorophyll content (Supplemental Figure S2). The degree of decrease in P700 signal, normalized to chlorophyll content, was still obviously weaker in nadk2-2 plants (60% of WT level; Supplemental Figure S2). This suggests a severely decreased amount of photooxidizable PSI in the mutant. Furthermore, the far-red light-induced oxidation rate of P700 was much slower in nadk2-2 compared to WT indicating acceptor side limitation and/or an increased reduction rate of the PQ pool.
Figure 3.
Photosynthetic electron transfer of PSI in the WT and nadk2-2 mutant. A, Redox kinetics of P700 induced by far-red light at 720 nm. Absorbance at 820 nm indicates the P700 redox state. B, 77K fluorescence emission spectra of thylakoid membranes after excitation at 436 nm. The fluorescence emission signals were normalized to the PSII emission maximum at 685 nm. C, Quantum yield of PSI (ΦPSI). D, the ETR of PSI, ETR(I). E, Donor-side limitation of PSI,Φ (ND). F, Acceptor side limitation of PSI, Φ(NA). Results in (C–F) are presented as means ± sds (n = 3). Photosynthetic parameters of PSI were measured in the steady state of photosynthesis (820-μmol photons m−2 s−1) under 21 kPa O2 and 100 Pa CO2. PSI complementary quantum yields of energy conversion were measured based on the P700 redox kinetics.
The 77K fluorescence emission spectra were also analyzed in nadk2-2 and WT plants. After normalization to the PSII emission peak at 685 nm, PSI emission was slightly increased in nadk2-2 compared to WT despite reduced PSI levels (Figure 3B), suggesting an increased light harvest protein I (LHCI)/PSI ratio leading to a substantial disconnection of the LHCI antenna complex from PSI and presumably also a slight decrease in the LHCII antenna.
PSI activity was also monitored by measuring the effective quantum yield of PSI (ΦPSI) and ETR of PSI (ETRPSI). Both of them were distinctly lower in the nadk2-2 mutant compared to the WT (Figure 3, C and D). The decrease in the ΦPSI can be attributed to the donor and/or the acceptor side limitation of PSI (Pfündel et al., 2008; Suzuki et al., 2011). Our results showed that the PSI limitation level at its donor side, the Φ(I)ND value, was much lower in the nadk2-2 than that in the WT especially under lower light intensities (Figure 3E), suggesting no limitation at the PSI donor side in the mutant. In sharp contrast, the PSI limitation level at its acceptor side, Φ(I)NA), was severely increased at all light intensities (Figure 3F), pointing at a pronounced PSI acceptor side limitation most likely due to lack of the electron acceptor NADP+.
It has been demonstrated that the limitation of electron acceptors from PSI would induce the CET around PSI (Yamori and Shikanai, 2016), which mediates the NADPH-dependent electron transport from ferredoxin to plastoquinone. We took the re-reduction rate of P700+ following FR illumination in the absence or presence of 3-(3, 4-dichlorophrenyl)-1, 1-dimethylurea (DCMU) as a measure for the capacity of PSI-driven CET (Nandha et al., 2007; Lintala et al., 2007; Plöchinger et al., 2016). As shown in Supplemental Figure S3 and Supplemental Table S1, the dark reduction rate of P700+ was obviously faster in nadk2-2 than in WT, both in the presence and absence of DCMU, suggesting that the CET rate around PSI is increased in nadk2-2 mutants.
Accumulation of PSI was impaired in nadk2-2
The functional defects in photosynthesis suggested by the data presented may be due to reduced levels of the PSI complex in nadk2-2. To verify this, thylakoid membranes from Arabidopsis leaves were solubilized with n-dodecyl-β-d-maltoside (β-DM) and separated by blue native polyacrylamide gel electrophoresis (BN-PAGE) (Figure 4A). Six major bands labeled I–VI were resolved in the first dimension, apparently representing PSI and PSII supercomplexes (band I), PSI and dimeric PSII (band II), monomeric PSII (band III), CP43-less PSII (band IV), trimeric LHCII (band V), and free proteins (band VI; Stoppel et al., 2011). Our data clearly showed that the amount of band II was lower in thylakoid preparations from the nadk2-2 mutant as compared to WT plants. Analyses of the Coomassie Brilliant Blue (CBB)-stained second dimension revealed that the amounts of PSI core subunits PsaA and PsaB, and several smaller PSI proteins were clearly reduced in the mutant, whereas levels of PSII core subunits and of the LHCII were only slightly reduced in the mutant (Figure 4B), which is in agreement with the results of the first dimension. We conclude, that the decline of PsaA/B levels seems to be the major deficiency in nadk2-2. A decrease in the PSII supercomplex was also observed in nadk2-2, which could indirectly result from decreased PSI activity, although other possibilities cannot be completely ruled out. Indeed, such a decrease in PSII supercomplex levels was also observed in several other mutants with defects in PSI synthesis and/or assembly (Albus et al., 2010; Liu et al., 2012; Roose et al., 2014).
Figure 4.
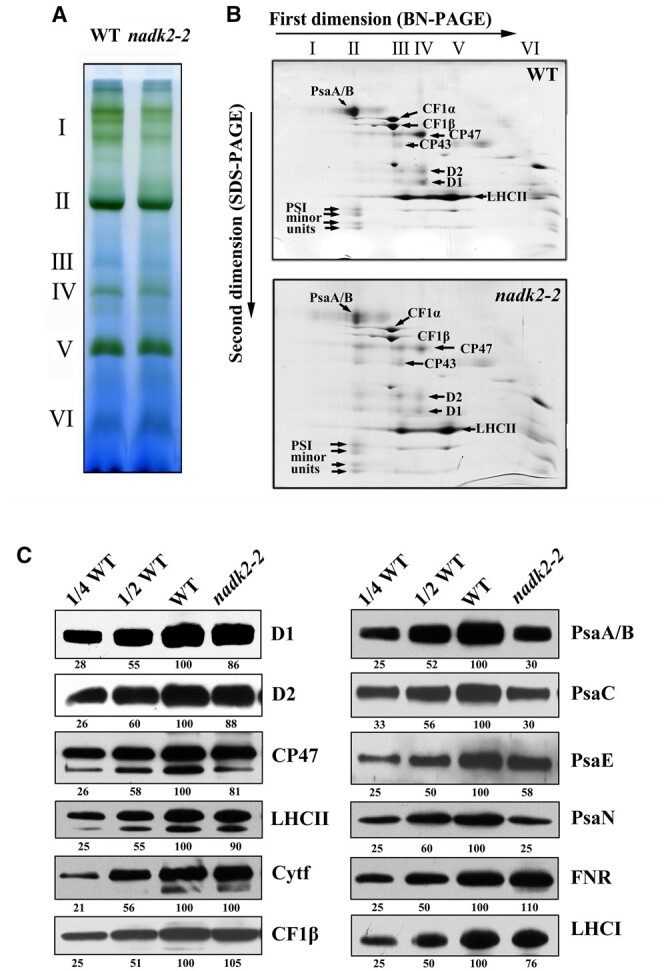
Accumulation of thylakoid proteins in WT and nadk2-2 plants. A, BN gel analysis of thylakoid membrane protein complexes. Thylakoid membranes (containing 10 µg chlorophyll) from 3-week-old WT and nadk2-2 leaves were solubilized with 1% β-DM and separated on a 6%–12% gradient BN gel. Positions of distinct protein complexes are shown on the left. I, PSII supercomplex; II, PSI and dimeric PSII; III, monomeric PSII; IV, CP43-less PSII; V, trimeric LHCII and VI, free proteins. B, 2D separation of protein complexes of thylakoid membranes. After being separated on the BN gel, the protein complexes in a single lane were further separated by 15% SDS–urea–PAGE and stained with CBB. Identities of relevant proteins are indicated by arrows. C, Immunoblotting assay of thylakoid proteins in WT and nadk2-2 plants. Ten microgram of total protein extracts from 3-week-old WT and nadk2-2 leaves were separated by SDS–urea–PAGE and probed with specific antibodies. The appearing bands were scanned and analyzed using an AlphaImager 2200 Documentation and Analysis System and quantification of their relative levels are shown below the blots.
To further verify the altered accumulation of photosynthetic membrane complexes, we performed immunoblot analyses using monospecific antibodies raised against individual thylakoid subunits. It appeared that the abundance of PSI core components is specifically and severely reduced in the nadk2-2 mutant (Figure 4C). The contents of PsaA/B, PsaC, and PsaN proteins were reduced to about 25%–35% of WT levels in nadk2-2, whereas PsaE amounts to ∼60% of WT levels. Compared with core subunits, the reduction in LHCI levels was less pronounced, indicating an increase in LHCI proteins not associated with PSI (76% of WT levels) in nadk2-2 (Figure 4C). In contrast, the levels of representative subunits of PSII (D1, D2, CP47, and LHCII) decreased only slightly in nadk2-2 compared to the WT. Subunits of the cytochrome b6f complex, FNR, and the chloroplast ATP synthase accumulated at levels similar to the WT. The immunoblot analyzes clearly showed that the nadk2-2 mutation mainly affects the accumulation of the PSI core complex, while the LHCI is less affected and levels of the other thylakoid protein complexes are hardly changed at all.
The presence of dimeric PSII complexes, detectable in band II (Figure 4B), indicates that their assembly is not affected in the mutant. To further confirm this notion, thylakoid proteins were separated by 2D BN/SDS–PAGE and subsequently subjected to immunoblot assays using the antibodies against PsaA, LHCI, and D1 (Supplemental Figure S4). We found that the distribution of PsaA and D1 in various PSII and PSI complexes was not changed markedly, suggesting that the assembly of PSI and PSII is not affected in nadk2-2. Interestingly, in contrast to WT, PsaA levels were reduced and much free LHCI proteins occurred and less LHCI was associated with PSI to form a PSI supercomplex in nadk2-2, suggesting that a large fraction of LHCI is not associated with the PSI supercomplex, consistent with data from 77K fluorescence emission spectra (Figure 3B).
Photoinhibition susceptibility is increased in nadk2-2
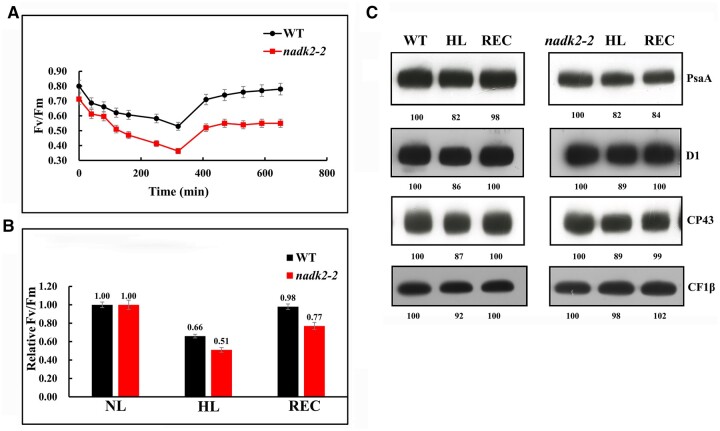
The above analysis suggests an increased sensitivity of PSII activity to high-light intensities in nadk2-2 due to PSI deficiency. To prove this experimentally, we performed photoinhibition tests. Four-week-old leaves from WT and mutant plants were illuminated with light intensities of 600-µmol photons m−2 s−1 for 5 h, after which the samples were illuminated with a low intensity (20-µmol photons m−2 s−1), and their recovery was monitored over 6 h. Changes in the Fv/Fm ratio were recorded during high-light illumination and recovery. As shown in Figure 5, A and B, during the high-light treatment, the Fv/Fm values for the WT and nadk2-2 decreased from 0.80 to 0.60 and 0.69 to 0.35, respectively. The larger decrease observed for the nadk2-2 mutant indicated that the absence of NADK increased sensitivity to high light. After 6 h recovery, the WT Fv/Fm was almost completely restored to pre-high-light illumination. In contrast, the recovery from photoinhibition was largely delayed in nadk2-2 (78% of the level before high-light illumination) (Figure 5, A and B). Therefore, susceptibility to PSII photoinhibition is actually increased in nadk2-2.
Figure 5.
Response of the WT and nadk2-2 to high-light (HL) treatment. A, Change of Fv/Fm values of the detached leaves from the WT and nadk2-2 exposed to HL illumination (600-µmol photons m−2 s−1) for 5 h, followed by 6 h of recovery at low light (20-µmol photons m−2 s−1). Prior to HL treatment, the soil-grown Arabidopsis seedlings were grown at 60 µmol photons m−2 s−1 for 3 weeks. Results are presented as means ± sds (n = 6). B, The Fv/Fm values mearsured at normal light (NL, 60 µmol photons m−2 s−1) were regarded as 1. The relative Fv/Fm values of Arabidopsis plants after 5 h of HL treatment and 6 h of recovery (REC) are shown. Results are presented as means ± sds (n = 6). C, Change of thylakoid protein levels of nadk2-2 and WT plants after HL treatment and recovery. Their relative levels are shown below the blots.
To determine whether photoinhibition affects the accumulation of the PSII or PSI complexes, we analyzed the abundance of D1, CP43, and PsaA from WT and mutants under high-light illumination and after recovery (Figure 5C). After 5 h of high-light illumination, the abundance of D1, CP43, and PsaA proteins was similarly reduced in both WT and the nadk2-2 mutant. After a 6-h recovery, accumulation of D1 and CP43 was restored to levels prior to high-light illumination in both WT and nadk2-2. However, unlike PSII subunits, the recovery of PsaA was obviously impaired in nadk2-2 compared with that of the WT (Figure 5C). The abundance of CF1β served as a control and was not alternated in all samples. This evidence indicates that the susceptibility of nadk2-2 to photoinhibition involves the activity and accumulation of PSI rather than PSII.
Biosynthesis of PSI subunits is impaired in nadk2-2
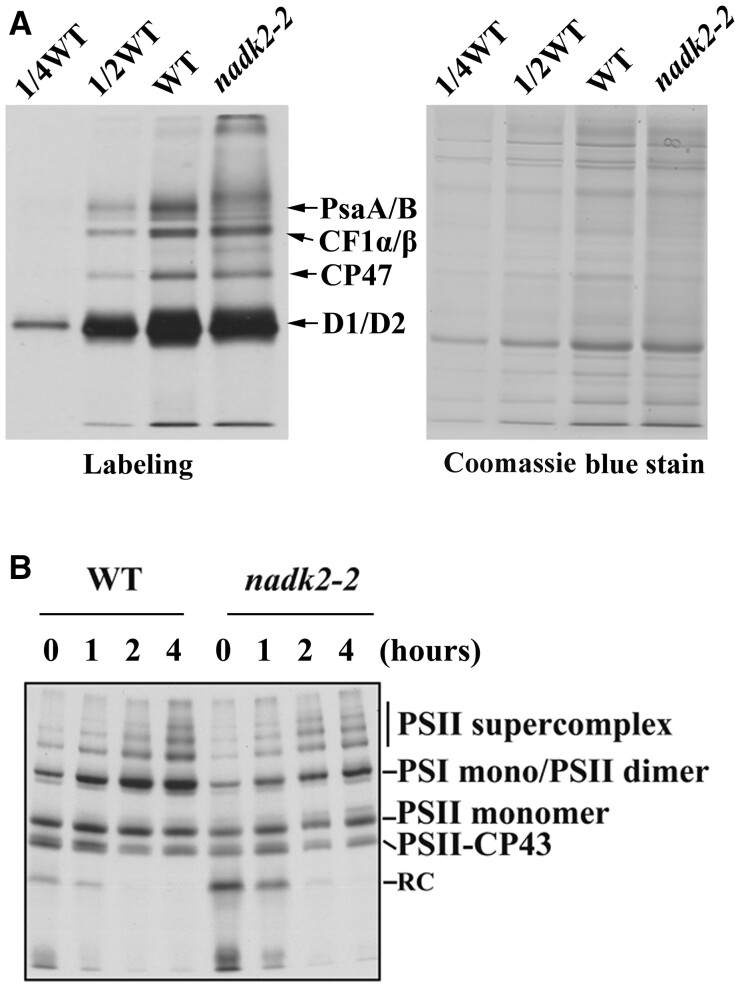
To investigate whether the diminished accumulation of PSI is due to either impaired translation or accelerated degradation of its components, the synthesis of thylakoid membrane proteins was investigated by in vivo labeling and pulse-chase experiments. For this purpose, leaf proteins were pulse-labeled with [35S] Met in the presence of cycloheximide, which blocks protein synthesis in the cytosol. As shown in Figure 6A, only plastid-encoded proteins were detected indicating efficient inhibition of cytoplasmic translation. The synthesis rate of the PSII reaction center (D1, D2, CP47, and CP43) and the α- and β-subunits of the chloroplast ATP synthase (CF1-α/β) remained almost unchanged in the mutant (Figure 6A). However, the incorporation of [35S] Met into PsaA/B proteins was considerably reduced in the nadk2-2 mutant. Pulse labeling for 20 min was followed by a chase with unlabeled Met to monitor the turnover rates of thylakoid protein complexes (Figure 6B). After 4 h of chasing, the radioactivity in the thylakoid membrane preparations was found to be associated mainly with PSII in both WT and nadk2-2 mutants (Figure 6B). Unassembled proteins were barely detectable in preparations from both WT and mutant plants, indicating that assembly and stability of PSI is not affected in the mutant.
Figure 6.
The synthesis of chloroplast-encoded thylakoid proteins in WT and nadk2-2 plants. A, Pulse labeling of thylakoid proteins of nadk2-2 and WT plants. After pulse labeling in the presence of cycloheximide for 20 min, thylakoid membranes were isolated from Arabidopsis leaves. The thylakoid proteins were separated by SDS–urea–PAGE and visualized autoradiographically. As a loading control, the same samples were separated on an additional gel and stained with CBB. The CBB-stained gel is shown on the right. B, Pulse-chase labeling of thylakoid membrane proteins from WT and nadk2-2 plants. After pulse labeling with [35S]-Met for 20 min as in (A) but without cycloheximide, leaves were incubated with an excess of cold Met for an additional 1, 2, and 4 h. Labeled proteins were then visualized by subsequent autoradiography.
Stability of PSI proteins is not affected in nadk2-2
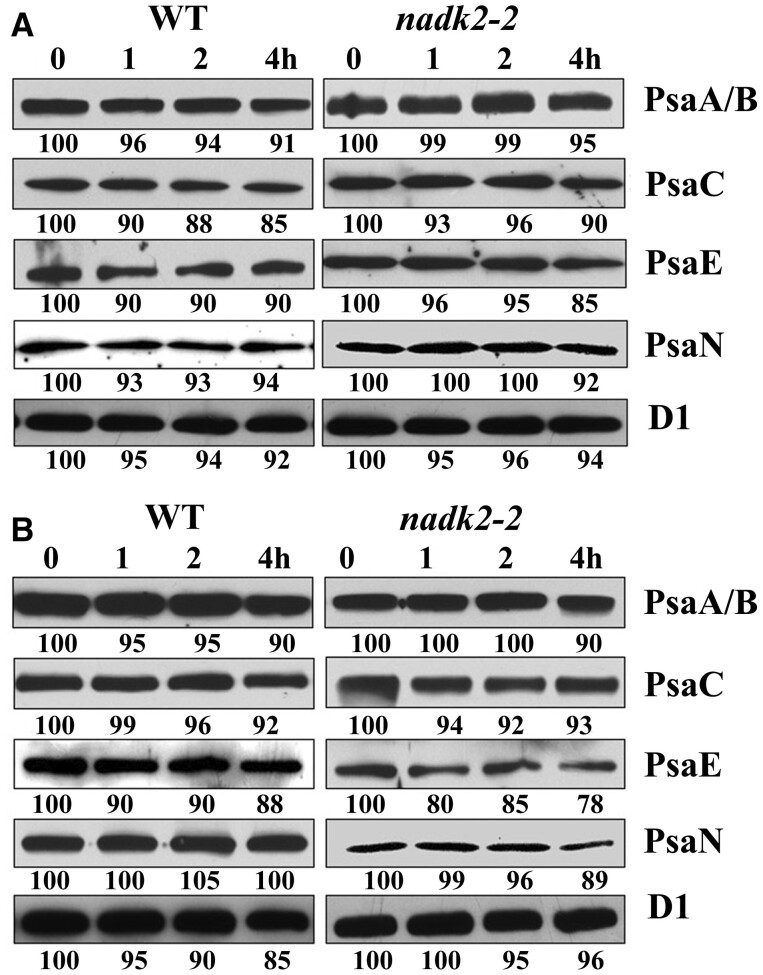
To validate the stability of assembled PSI complexes in the nadk2-2 mutant further, we treated leaves of mutant and WT plants with lincomycin to block chloroplast protein synthesis and analyzed the contents of PSI proteins by immunoblot analysis in a time course of 4 h. As shown in Figure 7A, levels of PsaA, PsaB, PsaC, PsaE, and PsaN in nadk2-2 were as stable as in the WT. Nevertheless, we cannot rule out the possibility that slight differences resulted from the reduced activity of nuclear-encoded factors which are required to protect newly synthesized PSI subunits from degradation. To exclude this possibility, a control experiment in the presence of both inhibitors, lincomycin, and cycloheximide, was performed (Figure 7B). The results showed that PSI subunits of nadk2-2 were as stable as those of the WT. Thus, we conclude that the decrease of subunits PsaA and PsaB in nadk2-2 is not due to a decreased stability but rather to their low translation efficiency.
Figure 7.
Stability of PSI proteins in WT and nadk2-2. A, Immunoblotting analysis of thylakoid membrane proteins from Arabidopsis plants treated with lincomycin. The detached Arabidopsis leaves were incubated with 100-µg/mL lincomycin for 30 min in the dark and then illuminated for 1, 2, and 4 h. After this treatment, total proteins from Arabidopsis leaves were isolated and the contents of distinct thylakoid proteins were determined by immunoblot analysis. The relative protein levels were estimated using an AlphaImager 2200 documentation and analysis system and are shown below the lanes. B, Immunoblotting analysis of thylakoid membrane proteins from Arabidopsis treated with lincomycin and cycloheximide. Arabidopsis leaves were incubated with 100-µg/mL lincomycin and 20-µg/mL cycloheximide for 30 min and illuminated for 1, 2, and 4 h. The subsequent experimental procedure was the same as in (A).
Accumulation of plastid transcripts was not affected in nadk2-2
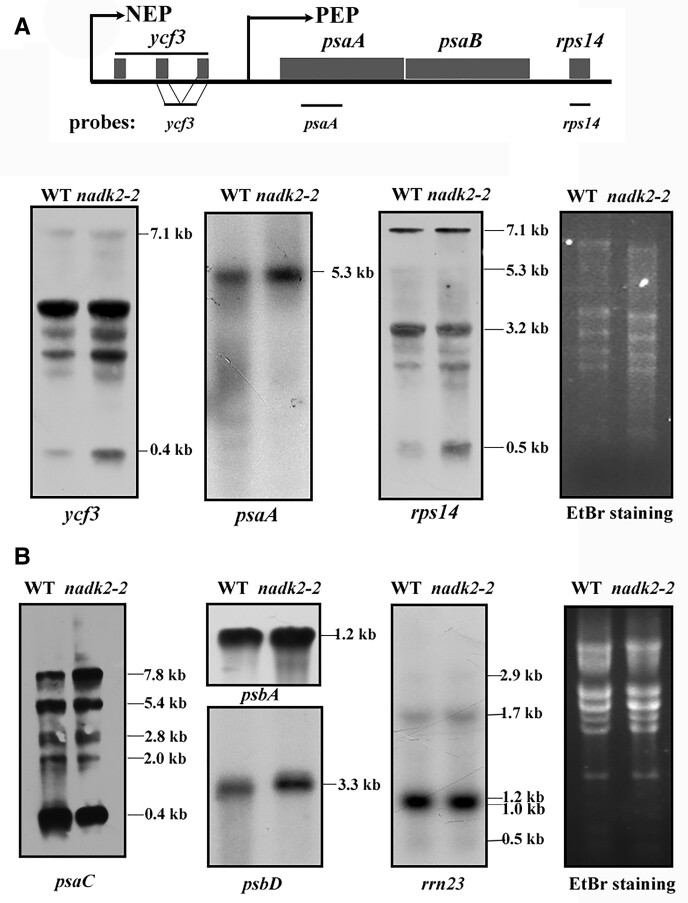
One possible explanation for the decreased PsaA/B synthesis could be inefficient transcription of psaA/B genes in nadk2-2. To address this possibility, levels of several mRNAs encoding subunits of thylakoid complexes were investigated by RNA gel blot analysis. In Arabidopsis, a pronounced tricistronic psaA-psaB-rps (small-subunit ribosomal protein)14 mRNA of 5.3 kb is generated by the plastid-encoded RNA polymerase, while a larger ycf3-psaA-psaB-rps14 transcript of 7.1 kb containing the two intron sequences of ycf3 is generated by the nuclear-encoded RNA polymerase (Figure 8A;Lezhneva and Meurer, 2004). The RNA-gel blots showed that the abundance of both primary transcripts was unchanged in nadk2-2 (Figure 8A). In addition, the levels of distinct forms of primary and processed transcripts containing psaC (encoding the PsaC subunit of PSI), psbA (encoding D1 of PSII), psbD (encoding D2 of PSII), and rrn23 (encoding 23S rRNA of chloroplasts) apparently were also not changed in nadk2-2 except an increase in the primary psaC containing transcript of 7.8 kb and a slight decrease of the monocistronic psaC transcript of 0.4 kb (Figure 8B). Thus, it is highly unlikely that decreased PsaA/B synthesis results from changes of psaA/psaB/rps14 and psaC mRNA abundance in nadk2-2.
Figure 8.
Plastid RNA accumulation in the WT and nadk2-2. A, RNA gel blot analysis of the psaA-psaB-rps14 gene cluster in WT and nadk2-2 plants. Twenty micrograms of total RNA from WT and nadk2-2 mutant plants were size-fractionated by agarose gel electrophoresis, transferred onto a nylon membrane, and probed with labeled cDNA probes. The genetic composition of this gene cluster and the locations of probes used for hybridization are shown above. Distinct transcription initiation sites of the plastid- (PEP) and nucleus-encoded (NEP) polymerases are indicated by arrows. The rRNAs stained by ethidium bromide (EtBr) are shown on the right of the blots as loading control. B, RNA gel blot analysis of other plastid genes in WT and nadk2-2 plants. The experimental procedure was the same as in (A).
The translation activity of psaA/B mRNA was reduced in nadk2-2
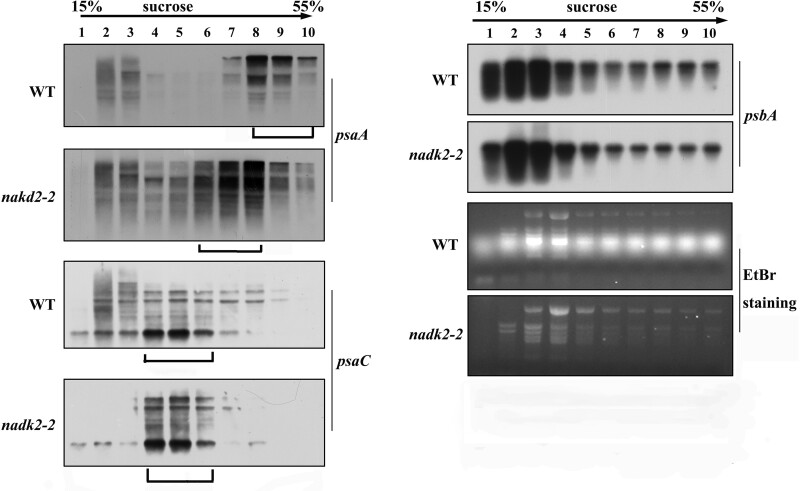
Because the levels of psaA-psaB-rps14 mRNAs were not altered in nadk2-2, but fewer radiolabeled amino acids were incorporated into PsaA and PsaB, decreased ribosome loading of the psaA-psaB-rps14 mRNA and/or other RNAs encoding components of PSI may be responsible for the decreased PsaA and PsaB synthesis. Therefore, we analyzed the polysome loading of chloroplast mRNAs by sucrose density-gradient fractionation (Figure 9), which determines the coverage of mRNAs with translating ribosomes and is thus a measure of translational activity. We found that most psaA-psaB-rps14 mRNAs were distributed in the dense-gradient fractions 8–10 in the WT, but in lower density fractions 6–8 in the mutant, indicating a shift to lighter fractions in the sucrose density gradient in the mutant. This again strongly confirms a reduced translation activity of the psaA-psaB-rps14 mRNA in nadk2-2. No obvious difference in the polysome loading of psaC and psbA mRNAs was found between the mutant and WT (Figure 9). Expression of rps14 was not affected, as indicated by an unchanged overall translation rate in the mutant. Thus, we conclude that decreased PSI levels are due to the specific reduction in translation efficiency of psaA and psaB in nadk2-2.
Figure 9.
Polysome loading assay of chloroplast mRNAs in the WT and nadk2-2. Ten fractions of equal volume were collected from the top to the bottom of 15%−55% sucrose gradients and equal proportions of the RNA purified from each fraction were analyzed by RNA gel blot analysis. The probes are shown on the right. The gradient fractions containing the bulk of the mRNAs are shown by brackets below the blots. rRNAs were detected by EtBr staining.
Discussion
The chloroplast NADP(H) pool size (sum of NADP+ and NADPH) and its redox status (the NADPH/NADP+ ratio) are crucial for the homeostasis of plant cells (Gakière et al., 2018; Hashida et al., 2018). Previous studies have shown that the size of the NADP(H) pool was reduced, whereas the NADPH/NADP+ ratio was increased in the nadk2-1 mutant (Chai et al., 2005; Takahashi et al., 2006, 2009), suggesting that the loss of NADK2 severely affects chloroplast NADP homeostasis causing a change in plant metabolism, as NADPH provides the reducing power for carbon and redox metabolism. Indeed, several metabolites associated with the Calvin cycle as well as some amino acids were changed in both nadk2-1 mutants and plants overexpressing NADK2 (Takahashi et al., 2009). In this study, we found that NADK2 is important for the formation of the PSI acceptor pool as well as translation and accumulation of PsaA/B in addition to metabolic changes.
In agreement with our findings, the study of Takahashi et al. (2006) also showed that the photosynthetic activity was affected in nadk2 mutants as revealed by chlorophyll a fluorescence analysis. However, defects in photosynthetic electron transport in the nadk2 mutant and the physiological consequences have not been determined. Our spectroscopic and immunological data showed that the primary deficiency of the photosynthetic electronic transport was indeed related to PSI and not PSII. The photoinhibition experiment showed that PSI was more affected in the nadk2-2 mutant under strong light, further strengthening this hypothesis. Interestingly, the study by Takahashi et al. (2006) reported an increased NPQ in the nadk2 mutant, which was attributed to the alteration of the xanthophyll cycle, probably due to the inhibition of zeaxanthin epoxidase activity by NADPH depletion. Our evidence showed that the CET around PSI was apparently increased, which provides an alternative or additional explanation for the increased NPQ in nadk2-2. It is possible that increased CEF leads to increased acidification of the lumen, resulting in heat dissipation in the nadk2-2 mutant. Based on the current evidence, these two possibilities are not mutually exclusive. Moreover, our results showed that the NPQ component qE rather than qI was mainly responsible for the increased NPQ in the nadk2-2 mutant (Supplemental Table S1). Given that qI is related to PSII photoinhibition (Ruban, 2016, 2017), this line of evidence also excluded the possibility that PSII activity was largely affected in nadk2-2.
In accordance with the decreased PSI activity, the level of the PSI complex was substantially reduced. The FNR, which catalyzes the conversion of NADP+ to NADPH, has been reported to be attached to the thylakoid membrane and associated with the PsaE subunit of PSI (Andersen et al., 1992; Marco et al., 2019). Considering this, one might argue that the decreased PSI abundance and/or acceptor side limitation is related to FNR. However, the accumulation of FNR was not changed in nadk2-2 excluding this possibility. In Chlamydomonas reinhardtii, perturbations of the redox state of the stroma affect the oxidative disruption of PSI during assembly (Heinnickel et al., 2016). Nevertheless, our pulse-chase labeling experiments showed that assembly and turnover of PSI subunits is very efficient once they were synthesized in nadk2-2. Instead, the pulse labeling data clearly showed that the synthesis of PsaA and PsaB was severely impaired in the nadk2-2 mutant, which likely accounts for the decreased amount of PSI complex.
It is likely that acceptor side limitation of PSI occurs when less NADP+ is available due to a deficiency in its synthesis in this mutant. PSI acceptor side limitation causes highly reduced P700 levels, which produces reactive oxygen species and initiates PSI photoinhibition (Tiwari et al., 2016). It has been demonstrated that downregulation of the cytochrome b6f complex, photorespiration, cyclic electron flow, the water–water cycle, as well as dynamic thylakoid stacking and state transitions represent mechanisms to avoid PSI photoinhibition (reviewed by Shimakawa and Miyake, 2018; Hepworth et al., 2021). Here we propose a model in which the translation of PsaA and PsaB is adjusted to the availability of the PSI acceptor pool size. Thus, a reduced translation rate of PsaA and PsaB provides another mechanism to adapt PSI levels to the available acceptor pool size and thus to avoid PSI photoinhibition.
The impaired translation capacity of PsaA and PsaB does not result from reduced mRNA abundance since the content of psaA–psaB–rps14 transcripts was not changed in nadk2-2. Instead, the polysome loading assays clearly indicated a primary defect in psaA/B translation activity. A slight decrease of the monocistronic psaC mRNA might also contribute to the loss of PSI in nadk2-2, but increased levels of psaC containing precursor transcripts should compensate for this as revealed by unchanged polysomal loading of this RNA. Nevertheless, the failure to assemble PsaC has only little, if any, effect on the synthesis of PsaA and PsaB (Shen et al., 2017; Yang et al., 2017). Considering this, we propose that psaC expression does not contribute primarily to the reduced translation of PsaA and PsaB in nadk2-2.
By which mechanism does NADK affect the translation of psaA/B mRNA? Our data showed that the decreased translation efficiency was specific for the psaA/B mRNA within all tested mRNAs, suggesting that NADK might likely affect psaA/B translation through one or more specific translation factor(s). One candidate of such factors is HCF145 which is able to specifically bind to the 5′-UTR (Untranslated Region) of the psaA/B mRNA and affect its stability and presumably translation activity (Lezhneva and Meurer, 2004; Manavski et al., 2015). The HCF145 protein contains two C-terminal tandem repeated TMR domains responsible for binding to the psaA 5′-UTR and two homologous N-terminal domains which display structural similarity to the ubiquitous SRPBCC superfamily encoding a deep ligand binding pocket. We hypothesize that nicotinamide nucleotides may affect the RNA-binding ability of HCF145 by binding to HCF145 as possible ligands. Nevertheless, the RNA-binding ability of HCF145 was not affected in vitro in the presence of NADP/NADPH or NAD/NADH, as shown by electrophoretic mobility shift assays (EMSAs; Supplemental Figure S5A), casting doubt on this hypothesis. In addition, it seems that the NADK2 mutation does not have any impacts on the accumulation of HCF145 proteins in vivo (Supplemental Figure S5B). Therefore, an additional role of HCF145 in regulating PsaA/B translation, and thus in adjusting PSI levels under conditions favoring acceptor side limitation, remains an open question. It is also possible that other previously unknown translation factors, instead of HCF145, mediate NADP/NADPH-dependent regulation of psaA/B translation and PSI levels under such condition.
Considering that alteration of the NADP(H) pool in planta would lead to a variety of pleotropic effects, their contributions to the adjustment of PSI synthesis in nadk2-2 should not be completely excluded. NADP(H) pools in chloroplasts collectively regulate the redox status of the stroma, to which initiation and elongation of plastid translation are known to be sensitive (Barnes and Mayfield, 2003). Light-mediated redox control of chloroplast translation initiation which is performed by a multicomponent protein complex including RB47 and RB60 has been well established for the psbA mRNA in Chlamydomonas (Danon and Mayfield, 1991; Kim and Mayfield, 1997). Such a redox-based regulatory mechanism for the translation of psbA mediated by LPE1 has also been proposed for Arabidopsis chloroplasts (Jin et al., 2018). Although RB60-like redox-sensitive translation activators for psaA/B have not been identified, we speculate that changes in the NADP homeostasis in nadk2-2 may affect psaA/B mRNA translation by a similar, yet unknown mechanism. Moreover, changes in stromal redox homeostasis, as well as over-reduction of the plastoquinone pool in this mutant, would lead to an alteration in ROS production in chloroplasts, which can probably directly affect plastid translation through inactivation of elongation factor G (Jimbo et al., 2018). In addition to effects on redox and ROS homeostasis, a previous study has shown that NADK activity is even relevant to Ca2+ homeostasis and CaM interacting with the PSI subunit PsaN, suggesting that NADK may affect the PSI complex via Ca2+ and/or CaM (Turner et al., 2004; Stael et al., 2012). Overall, the metabolic link between NADK activity and de novo biosynthesis of the PSI complex leaves questions unanswered and further possibilities should be explored in future studies.
Conclusion
In conclusion, our study established a molecular link between the chloroplast NADK activity and the biogenesis of the PSI complex. Loss of the NADK activity in chloroplasts disrupts the NADP(H) status in the stroma, leading to a variety of pleotropic effects including imbalanced redox homeostasis of plastids. In response to this redox shift and/or other changes in chloroplast metabolism, the amount of PSI complex was downregulated by a reduction in psaA/B translation activity, presumably to adapt to conditions favoring acceptor side limitation and thus avoid photoinhibition.
Materials and methods
Plant material and growth conditions
The nadk2-2 mutant line was in Col-0 background and obtained from the Arabidopsis Biological Resource Center. The homozygous mutant was verified by PCR using primer pairs LP/RP and S1/S2. All the primer sequences used in this study are presented in Supplemental Table S2. The PCR products were validated by DNA sequencing.
WT and mutant seeds were sterilized with 10% (v/v) sodium hypochlorite for 15 min, washed 5 times with distilled water, and placed on solid Murashige and Skoog (MS) medium supplemented with 2% (w/v) sucrose. To ensure synchronous germination, the plates were kept in the dark at 4°C for 2 d. Then plants were grown on MS medium for 15 d in a growth chamber at 22°C with a 12-h light/12-h dark photoperiod. They were then placed in soil and grown at a photon flux density of 60-μmol photons m−2 s−1.
Chloroplast ultrastructure
WT and mutant leaves from 3-week-old Arabidopsis seedlings were used for transmission electron microscopy analysis. The leaves were chopped into 1 × 2 mm2 pieces and immersed in fixative solution (2.4% (v/v) glutaraldehyde in phosphate buffer) for 4 h at 4°C. After fixation, the samples were rinsed and fixed in 1% OsO4 (v/v) overnight at 4°C and then dehydrated in an ethanol series, infiltrated with a graded series of epoxy resin in epoxy propane and embedded in Epon 812 resin (SPI Supplies 02660-AB). Thin (80–100 nm) sections were obtained using a diamond knife on a Reichert OM2 ultramicrotome, stained in 2% (v/v) uranyl acetate, pH 5.0, followed by 10 mM lead citrate, pH 12, and observed with a transmission electron microscope (Jem-1230; JEOL).
Determination of the NADPH/NADP+ contents
About 0.1 g leaves were collected from 3-week-old WT and nadk2-2 plants. Samples were then immediately frozen in liquid nitrogen and ground into powder. The amount of NADP+ and NADPH in leaves was determined using NADP+/NADPH Detection Kit (Beijing Solarbio Science & Technology, http://www. Solarbio.com/) according to the instruction manual.
Complementation of nadk2-2
To complement nadk2-2, the full-length cDNA of NADK2 was amplified with the primers (nadk coms and coma, shown in Supplemental Table S2) and cloned into the KpnI and BamH I sites of pSN1301 vector under the control of the CaMV 35S promoter. The constructed plasmids were then transformed into the Agrobacterium tumefaciens strain C58 and introduced into nadk2-2 mutants using the floral dip method as described previously (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 50-µg/mL hygromycin and were then transferred and grown on soil for 3 weeks. The complemented plants were confirmed by polymerase chain reaction (PCR) genotyping and chlorophyll fluorescence analysis.
Spectroscopic assay
The photosynthetic activity of PSII was determined based on chlorophyll fluorescence measured with the Dual-PAM-100 system (Walz, Lancaster, PA, USA). Red light (630 nm) irradiation of 55-μmol photons m−2 s−1 was used as actinic light. A saturating pulse (800 ms, 6,000-μmol photons m−2 s−1) was applied to determine the maximal fluorescence (Fm or Fm′). The minimum fluorescence (Fo), variable fluorescence (Fv), maximum photochemical efficiency of PSII (Fv/Fm), ETR , effective quantum yield of PSII (ΦPSII), and NPQ were measured and calculated according to Maxwell and Johnson (2000) and Krause and Jahns (2003). The transient increase in chlorophyll fluorescence after turning off actinic light was monitored according to Shikanai et al. (1998).
In order to resolve NPQ components, induction and relaxation of NPQ were monitored as described by Ruban (2017) with minor changes. Dark-incubated leaves (30 min) were exposed to actinic light illumination (610 μmol m−2 s−1) for two 2 h and recovery was followed for 1h after switching off actinic light using saturation pulses. Saturating light-induced Fm′ was collected at the end of a 2-h actinic light exposure and Fm″ was obtained after 45-min recovery in the dark. Measurements were performed on 4-week-old plants. qE and qI are the fast and slow reversible components of NPQ, respectively. NPQ was calculated as Fm/Fm′-1, qI was identified as (Fm–Fm″)/Fm″ and qE was identified as NPQ-qI.
The P700 oxidation state and 77K fluorescence emission spectra were measured as described previously (Meurer et al., 1996; Liu et al., 2012). CET was assessed by measuring the re-reduction of P700+ in the dark after exposure to far-red light as previously described (Nandha et al., 2007; Lintala et al., 2012; Plöchinger et al., 2016). Intact leaves from WT and nadk2-2 were vacuum infiltrated with 20-μM DCMU (Sigma, UK) in 300-mM sorbitol (Sigma, UK) in the darkness or dark adapted for 30 min. P700+ was measured with the JTS 10 equipment (Biologic, http://www.bio-logic.info) by oxidization with far-red light (720 nm, 200 µmol m−2 s−1, 20 s), and re-reduction was then monitored in the darkness. The half-life of non-photochemical P700 re-reduction in darkness after turning off far-red light was calculated as described (Plöchinger et al., 2016).
Three types of PSI-complementary quantum yield of energy conversion were measured based on the P700 induction curve as previously described (Pfündel et al., 2008). The leaves of 3-week-old WT and nadk2-2 mutant plants were dark-adapted for 20 min prior to measurement. The maximal change of P700 signal (Pm) was measured through application of a 200-ms saturation pulse (625 nm, 10,000-μmol photons m−2 s−1) at the end of 10 s FR light (720 nm, 20-μmol photons m−2 s−1) illumination. Po was determined at the end of 1-s dark period using a saturation pulse. About 40 s after FR-off, the actinic light (AL, 625 nm, 60-μmol photons m−2 s−1) was turned on. In the presence of actinic light, Pm′ was induced by a 200-ms saturating pulse (625 nm, 10,000-μmol photons m−2 s−1) applied every 20 s. The P700 signal (P) was recorded just before application of a saturation pulse. The P700 induction curve was recorded for 350 s to achieve the steady state of the photosynthetic apparatus and the actinic light was then turned off. Parameters were calculated as follows: ΦPSI = (Pm′-P)/Pm; ΦPSI (ND) = (P-Po)/Pm; ΦPSI (NA) = (Pm-Pm′)/Pm.
Thylakoid membrane isolation
Thylakoid membranes were isolated according to Zhang et al. (1999). The chlorophyll a and b contents were measured as described (Porra et al., 1989). The thylakoid membranes were used immediately or rapidly frozen in liquid nitrogen and then stored at −80°C for further analysis.
BN–PAGE, SDS–PAGE, and immunoblotting
BN–PAGE was performed as described by Schägger et al. (1994) with minor modifications. Thylakoid membranes were resuspended in suspension buffer (20% (v/v) glycerol and 25-mM Bis Tris–HCl, pH 7.0) at 1.0-mg chlorophyll/milliliter and then β-DM was added to a final concentration of 1% (w/v). Thylakoid membrane samples were incubated on ice for 10 min. After centrifugation at 15,000 g for 10 min, the supernatant was carefully transferred to a new tube and was combined with 1/10 volume of 5% (w/v) Serva Blue G in 100-mM Bis–Tris–HCl, pH 7.0, 0.5 M 6-amina-n-caproic acid, and 30% (w/v) glycerol. Subsequently, samples were loaded onto a first dimension 4.5%–12.5% (w/v) acrylamide separation gel and ran in a Hoefer Mighty Small vertical electrophoresis unit connected to a cooling circular. Lanes from the first dimension were cut and incubated in sample buffer containing SDS and 5% (v/v) 2-mercaptoethanol at room temperature for at least 30 min. The second dimension separation was performed on denaturing 15% (w/v) SDS–PAGE containing 6–M urea.
Total proteins were extracted from 3-week-old WT and mutant plants in 125-mM Tris–HCl, pH 8.8, 1% (w/v) SDS, 10% (v/v) glycerol, 50-mM Na2S2O5 as described (Martínez-García et al., 1999). About 10 µL of total protein extracts were reserved for determination of protein concentration with the BioRad Dc Protein Assay according to the manufacturer’s instructions (BioRad, Hercules, CA, USA). Subsequently total protein was separated by SDS–PAGE, transferred onto nitrocellulose membranes, incubated with specific primary antibodies, and signals from secondary conjugated antibodies were detected by the enhanced chemiluminescence method.
RT-PCR, RNA gel blot, and polysome loading analyses
For reverse transcription-PCR (RT-PCR) analysis, total RNA was isolated from 3-week-old leaves using total RNA isolation kit (RNeasy Plant Mini Kit; QIAGEN, Hilden, Germany). RT-PCR reactions were performed with the SuperScript III First-Strand Synthesis System (Invitrogen, Waltham, MA, USA) using gene-specific primers. For RNA gel blot analysis, the same amount of RNA from WT and nadk2-2 was separated on 1.5% (w/v) agarose-formaldehyde gels, blotted onto a positively charged nylon membrane, and subsequently hybridized with the probes of chloroplast genes. The probes were prepared by PCR amplification and labeled using the random primer method with 32P-dCTP or biotin (Thermo Scientific, Waltham, MA, USA). Polysomes were isolated from 3-week-old leaves according to Barkan (1988). The RNA in each fraction was extracted and subjected to RNA gel blot analysis.
Pulse-chase labeling assays in vivo
Pulse-chase labeling was performed essentially according to Meurer et al. (1998). For pulse labeling, primary leaves from 20-d-old plants were first incubated with 1-μCi/μL [35S] methionine (Met) in the presence of 20-μg/mL cycloheximide for 20 min at 25°C. The chase of the labeled protein was performed in the presence of 1-mM unlabeled Met and 20-μg/mL cycloheximide for 4 h. After labeling, thylakoid membranes were isolated and their proteins were separated by SDS–PAGE or β-DM gel. The signals were detected by autoradiography.
Analysis of PSI subunits stability
Leaves of 4-week-old WT and mutant plants were incubated in 100-µg/mL lincomycin and 20-µg/mL cycloheximide for 30 min to block the synthesis of plastid and nuclear-encoded proteins, respectively. Leaves were then subjected to illumination under normal growth conditions for various times. Thylakoid membranes were extracted from the treated leaves and the content of PSI proteins was determined by immunoblotting.
Preparation of recombinant proteins
To produce recombinant MBP-HCF145 proteins, the coding sequence of mature At-HCF145 was PCR amplified and inserted into the BamHI sites of pMAL-c5E. MBP-HCF145 was expressed in Escherichiacoli BL21(DE3) pLysS cells at 17°C overnight by adding IPTG. Cells were lysed by sonication in lysis buffer (10-mM Tris, 200-mM NaCl, pH 7.5). The cleared lysate was incubated with MBP agarose at 4°C, rotated for 2 h. Washing and elution steps were performed with the lysis buffer and 10-mM maltose, respectively. Recombinant proteins were further purified via size exclusion chromatography using Superdex 200pg, HiLoad 16/600 GL (GE Healthcare Life Sciences, Piscataway, NJ, USA). Proteins were stored at −80°C in elution buffers.
EMSA
EMSA experiments were performed as described previously (Manavski et al., 2015). 5′-biotin-labeled RNA probes were generated by Sangon Biotech (the primer sequence was shown in Supplemental Table S2). Trace amounts of labeled RNAs were incubated with HCF145 protein and different concentrations of NAD+ and NADP+ in EMSA binding buffer containing 10-mM HEPES, pH 7.3, 20-mM KCl, 1-mM MgCl2, 0.1-mg/mL BSA, 1-mM DTT, and 0.5-mg/mL heparin at 25°C for 30 min. Samples were separated on non-denaturing 5% (w/v) polyacrylamide gels containing 0.5× TBE(Tris-Borate-EDTA) buffer and then transblot to the nylon membrane for the chemiluminescence detection.
Accession numbers
Sequence data from this article can be found at Arabidopsis.org under accession number AT1G21640 (NADK2).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the transient increase in chlorophyll fluorescence after termination of AL illumination.
Supplemental Figure S2. P700 signals normalized to chlorophyll contents in WT and nadk2-2 plants.
Supplemental Figure S3. CET rate in WT and nadk2-2 plants.
Supplemental Figure S4. 2D BN/SDS–PAGE analysis of thylakoid proteins.
Supplemental Figure S5. Effect of the NADK2 mutation on HCF145 activity and accumulation.
Supplemental Table S1. Photosynthetic parameters of WT and nadk2-2.
Supplemental Table S2. PCR primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Lixin Zhang for critical reading of this manuscript. We thank Dr. Chunyan Zhang and Dr. Yan Yin from Plant Science Facility of the Institute of Botany, CAS for helpful technical assistance in the spectroscopic assay.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA26030202 to W.C.), National Key Research and Development Program (grant no. 2020YFA0907601 to W.C. and D. J.), National Science Foundation of China (grant no. 31000339 to D.J.), and the German Science Foundation (DFG, SFB TRR175, TP A03, to J.M.).
Conflict of interest statement. None declared.
Contributor Information
Daili Ji, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.
Qiuxin Li, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yinjie Guo, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wenjing An, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Nikolay Manavski, Faculty of Biology, Plant Molecular Biology, Ludwig-Maximilians University, Munich, D-82152, Germany.
Jörg Meurer, Faculty of Biology, Plant Molecular Biology, Ludwig-Maximilians University, Munich, D-82152, Germany.
Wei Chi, Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; University of Chinese Academy of Sciences, Beijing 100049, China; The Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China.
D.J. and W.C. designed the research. D.J., Y.G., W.A., Q.L., and N.M. performed the research. D.J., W.C., and J.M. analyzed the data. W.C., J.M., and D.J. wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Wei Chi (chiweimr@ibcas.ac.cn).
References
- Albus CA, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22:2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Drory O, Nelson N (2007) The structure of a plant photosystem I supercomplex at 3.4 Å resolution. Nature 447:58–63 [DOI] [PubMed] [Google Scholar]
- Andersen B, Scheller HV, Moller BL (1992) The PSI-E subunit of photosystem I binds ferredoxin: NADP+ oxidoreductase. FEBS Lett 311:169–173 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22:3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Labs M, Pribil M, Viola S, Xu W, Scharfenberg M, Hertle AP, Rojahn U, Jensen PE, Rappaport F, et al. (2013) Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25:2661–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI, Chain RK (1975) Regulation of ferredoxin-catalyzed photosynthetic phosphorylations. Proc Natl Acad Sci USA 72:4961–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K, Heber U, Schreiber U (1992) Pool size of electrons that can be donated to P700+, as determined in intact leaves: donation to P700+ from stromal components via the intersystem chain. Plant Cell Physiol 33:927–932 [Google Scholar]
- Baier M, Dietz KJ (2005) Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J Exp Bot 56:1449–1462 [DOI] [PubMed] [Google Scholar]
- Barkan A (1988) Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J 7:2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D, Mayfield SP (2003) Redox control of posttranscriptional processes in the chloroplast. Antioxid Redox Signal 5:89–94 [DOI] [PubMed] [Google Scholar]
- Busch A, Hippler M (2011) The structure and function of eukaryotic photosystem I. Biochim Biophy Acta 1807:864–877 [DOI] [PubMed] [Google Scholar]
- Chai MF, Chen QJ, An R, Chen YM, Chen J, Wang XC (2005) NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol Biol 59:553–564 [DOI] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang L (2012) Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos Trans R Soc Lond B Biol Sci 367:3420–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Danon A, Mayfield SP (1991) Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J 10:3993–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bianchi S, Ballottari M, Dall’osto L, Bassi R (2010) Regulation of plant light harvesting by thermal dissipation of excess energy. Biochem Soc Trans 38:651–660 [DOI] [PubMed] [Google Scholar]
- De Souza Chaves I, Feitosa-Araújo E, Florian A, Medeiros DB, da Fonseca-Pereira P, Charton L, Heyneke E, Apfata JAC, Pires MV, Mettler-Altmann T, et al. (2019) The mitochondrial NAD+ transporter (NDT1) plays important roles in cellular NAD+ homeostasis in Arabidopsis thaliana. Plant J 100:487–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Williams-Carrier R, Merchant SS, Barkan A (2014) A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J Biol Chem 289:30657–30667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakière B, Hao J, de Bont L, Pétriacq P, Nunes-Nesi A, Fernie AR (2018) NAD + biosynthesis and signaling in plants. Crit Rev Plant Sci 37:1–49 [Google Scholar]
- Goodman RP, Calvo SE, Mootha VK (2018) Spatiotemporal compartmentalization of hepatic NADH and NADPH metabolism. J Biol Chem 293:7508–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss T, Hanke G (2014) The end of the line: can ferredoxin and ferredoxin NADP(H) oxidoreductase determine the fate of photosynthetic electrons? Curr Protein Pept Sci 15:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Cho WK, Lezhneva L, Falk J, Krupinska K, Shinozaki K, Seki M, Herrmann RG, Meurer J (2006) A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem 281:17189–17196 [DOI] [PubMed] [Google Scholar]
- Hanke G, Mulo P (2013) Plant type ferredoxins and ferredoxin dependent metabolism. Plant Cell Environ 36:1071–1084 [DOI] [PubMed] [Google Scholar]
- Hashida SN, Miyagi A, Nishiyama M, Yoshida K, Hisabori T, Yamada MK (2018) Ferredoxin/thioredoxin system plays an important role in the chloroplastic NADP status of Arabidopsis. Plant J 95:947–960 [DOI] [PubMed] [Google Scholar]
- Heber UW, Santarius KA (1965) Compartmentation and reduction of pyridine nucleotides in relation to photosynthesis. Biochim Biophys Acta 109:390–408 [DOI] [PubMed] [Google Scholar]
- Heinnickel M, Kim RG, Wittkopp TM, Yang W, Walters KA, Herbert SK, Grossman AR (2016) Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc Natl Acad Sci USA 113:2774–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C, Wood WHJ, Emrich-Mills TZ, Proctor MS, Casson S, Johnson MP (2021) Dynamic thylakoid stacking and state transitions work synergistically to avoid acceptor-side limitation of photosystem I. Nat Plants 7:87–98 [DOI] [PubMed] [Google Scholar]
- Jimbo H, Yutthanasirikul R, Nagano T, Hisabori T, Hihara Y, Nishiyama Y (2018) Oxidation of translation factor EF-Tu inhibits the repair of photosystem II. Plant Physiol 176:2691–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Fu M, Duan Z, Duan S, Li M, Dong X, Liu B, Feng D, Wang J, Peng L, Wang HB (2018) LOW PHOTOSYNTHETIC EFFICIENCY 1 is required for light-regulated photosystem II biogenesis in Arabidopsis. Proc Natl Acad Sci USA 115:E6075–E608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Bassi R, Boekema EJ, Dekker JP, Jansson S, Leister D, Robinson C, Scheller HV (2007) Structure, function and regulation of plant photosystem I. Biochim Biophy Acta 1767:335–352 [DOI] [PubMed] [Google Scholar]
- Kim J, Mayfield SP (1997) Protein disulfide isomerase as regulator of chloroplast translational activation. Science 78:1954–1957 [DOI] [PubMed] [Google Scholar]
- Krause G H, Jahns P (2003) Pulse Amplitude Modulated Chlorophyll Fluorometry and its Application in Plant Science. Springer, Netherlands [Google Scholar]
- Lezhneva L, Meurer J (2004) The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J 38:740–753 [DOI] [PubMed] [Google Scholar]
- Lintala M, , AllahverdiyevaY, , KidronH, , PiippoM, , BattchikovaN, , SuorsaM, , RintamäkiE, , SalminenTA, , Aro EM, Mulo P (2007) Structural and functional characterization of ferredoxin-NADP+-oxidoreductase using knock-out mutants of Arabidopsis. Plant J 49: . 1041–1052. [DOI] [PubMed] [Google Scholar]
- Lintala M, Lehtimäki N, Benz JP, Jungfer A, Soll J, Aro EM, Bölter B, Mulo P (2012) Depletion of leaf-type ferredoxin-NADP(+) oxidoreductase results in the permanent induction of photoprotective mechanisms in Arabidopsis chloroplasts. Plant J 70:809–817 [DOI] [PubMed] [Google Scholar]
- Liu J, Yang H, Lu Q, Wen X, Chen F, Peng L, Zhang L, Lu C (2012) PsbP-domain protein1, a nuclear-encoded thylakoid lumenal pro-tein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24:4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N, Torabi S, Lezhneva L, Arif MA, Frank W, Meurer J (2015). HIGH CHLOROPHYLL FLUORESCENCE145 binds to and stabilizes the psaA 5′ UTR via a newly defined repeat motif in Embryophyta. Plant Cell 27:2600–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco P, Elman T, Yacoby I (2019). Binding of ferredoxin NADP+ oxidoreductase (FNR) to plant photosystem I. Biochim Biophys Acta 1860:689–698 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Monte E, Quail PH (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20:251–257 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence-a practical guide. J Exp Bot 51:659–668 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and northern hybridization. Planta 198:385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17:5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandha B, Finazzi G, Joliot P, Hald S, Johnson GN (2007) The role of PGR5 in the redox poising of photosynthetic electron transport. Biochim Biophys Acta 1767:1252–1259 [DOI] [PubMed] [Google Scholar]
- Nawrocki WJ, Bailleul B, Picot D, Cardol P, Rappaport F, Wollman FA, Joliot P (2019) The mechanism of cyclic electron flow. Biochim Biophys Acta 1860:433–438 [DOI] [PubMed] [Google Scholar]
- Noctor G (2006) Metabolic signaling in defence and stress: the central roles of soluble redox couples. Plant Cell Environ 29:409–425 [DOI] [PubMed] [Google Scholar]
- Peng LW, Ma JF, Chi W, Guo JK, Zhu S, Lu QT, Lu CM, Zhang LX (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18:955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfündel E, Klughammer C, Schreiber U (2008) Monitoring the effects of reduced PSII antenna size on quantum yields of photosystems I and II using the Dual-PAM-100 measuring system. PAM Appl Notes 1:21–24 [Google Scholar]
- Plöchinger M, Torabi S, Rantala M, Tikkanen M, Suorsa M, Jensen PE, Aro EM, Meurer J (2016) The low molecular weight protein psaI stabilizes the light-harvesting complex II docking site of photosystem I. Plant Physiol 172:450–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim Biophys Acta 975:384–394 [Google Scholar]
- Qin X, Suga M, Kuang T, Shen JR (2015. )Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348:989–995 [DOI] [PubMed] [Google Scholar]
- Roose JL, Frankel LK, Bricker TM (2014) The PsbP domain protein 1 functions in the assembly of lumenal domains in photosystem I. J Biol Chem 289:23776–23785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV (2016) Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiol 170:1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV (2017) Quantifying the efficiency of photoprotection. Philos Trans R Soc Lond B Biol Sci 372:20160393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217:220–230 [DOI] [PubMed] [Google Scholar]
- Schöttler MA, Albus CA, Bock R (2011) Photosystem I: its biogenesis and function in higher plants. J. Plant Physiol 168:1452–1461 [DOI] [PubMed] [Google Scholar]
- Schwenkert S, Netz DJ, Frazzon J, Pierik AJ, Bill E, Gross J, Lill R, Meurer J (2009) Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem J 425:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Williams‐Carrier R, Barkan A (2017) PSA3, a protein on the stromal face of the thylakoid membrane, promotes photosystem I accumulation in cooperation with the assembly factor PYG7. Plant Physiol 174:1850–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakawa G, Miyake C (2018) Oxidation of P700 ensures robust photosynthesis. Front Plant Sci 9:1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95:9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S, Wurzinger B, Mair A, Mehlmer N, Vothknecht U C, Teige M (2012) Plant organellar calcium signalling: an emerging field. J Exp Bot 63:1525–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R, Lezhneva L, Schwenkert S, Torabi S, Felder S, Meierhoff K, Westhoff P, Meurer J (2011) Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23:2680–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohmori Y, Ratel E (2011) High root temperature blocks both linear and cyclic electron transport in the dark during chilling of the leaves of rice seedlings. Plant Cell Physiol 52:1697–1707 [DOI] [PubMed] [Google Scholar]
- Takahara K, Kasajima I, Takahashi H, Hashida SN, Itami T, Onodera H, Toki S, Yanagisawa S, Kawai-Yamada M, Uchimiya H (2010) Metabolome and photochemical analysis of rice plants overexpressing Arabidopsis NAD kinase gene. Plant Physiol 152:1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Takahara K, Hashida S, Hirabayashi T, Fujimori T, Kawai-Yamada M, Yamaya T, Yanagisawa S, Uchimiya H (2009) Pleiotropic modulation of carbon and nitrogen metabolism in Arabidopsis plants overexpressing the NAD kinase 2 gene. Plant Physiol 151:100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Watanabe A, Tanaka A, Hashida SN, Kawai-Yamada M, Sonoike K, Uchimiya H (2006) Chloroplast NAD kinase is essential for energy transduction through the xanthophyll cycle in photosynthesis. Plant Cell Physiol 47:1678–1682 [DOI] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Vanderbeld B, Snedden WA (2004) Cloning and characterization of two NAD kinases from Arabidopsis. identification of a calmodulin binding isoform Plant Physiol 135:1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Mamedov F, Grieco M, Suorsa M, Jajoo A, Styring S, Tikkanen M, Aro EM (2016) Photodamage of iron-sulphur clusters in photosystem I induces non-photochemical energy dissipation. Nat Plants 2:16035. [DOI] [PubMed] [Google Scholar]
- Waller JC, Dhanoa PK, Schumann U, Mullen RT, Snedden WA (2010) Subcellular and tissue localization of NAD kinases from Arabidopsis: compartmentalization of de novo NADP biosynthesis. Planta 231:305–317 [DOI] [PubMed] [Google Scholar]
- Wang P, Liu J, Liu B, Feng D, Da Q, Wang P, Shu S, Su J, Zhang Y, Wang J, Wang HB (2013) Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol 163:1710–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge B, Krömer S, Gardeström P (1993) The redox levels and subcellular distribution of pyridine nucleotides in illuminated barley leaf protoplasts studied by rapid fractionation. Physiol Plant 88:10–18 [Google Scholar]
- Xiao W, Wang RS, Handy DE, Loscalzo J (2018) NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxid Redox Signa 28:251–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Shikanai T (2016) Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol 67:81–106 [DOI] [PubMed] [Google Scholar]
- Yang H, Liu J, Wen X, Lu C (2015) Molecular mechanism of photosystem I assembly in oxygenic organisms. Biochim Biophy Acta 1847:838–848 [DOI] [PubMed] [Google Scholar]
- Yang H, Li P, Zhang A, Wen X, Zhang L, Lu C (2017) Tetratricopeptide repeat protein Pyg7 is essential for photosystem I assembly by interacting with PsaC in Arabidopsis. Plant J 91:950–961 [DOI] [PubMed] [Google Scholar]
- Zhang LX, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274:16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Liberton M, Pakrasi HB (2016) A novel redoxin in the thylakoid membrane regulates the titer of photosystem I. J Biol Chem 291:18689–18699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.